Abstract
Xenbase is the Xenopus model organism database (www.xenbase.org), a web-accessible resource that integrates the diverse genomic and biological data for Xenopus research. It hosts a variety of content including current and archived genomes for both X. laevis and X. tropicalis, bioinformatic tools for comparative genetic analyses including BLAST and GBrowse, annotated Xenopus literature, and catalogs of reagents including antibodies, ORFeome clones, morpholinos, and transgenic lines. Xenbase compiles gene-specific pages which include manually curated gene expression images, functional information including gene ontology (GO), disease associations, and links to other major data sources such as NCBI:Entrez, UniProtKB, and Ensembl. We also maintain the Xenopus Anatomy Ontology (XAO) which describes anatomy throughout embryonic development. This chapter provides a full description of the many features of Xenbase, and offers a guide on how to use various tools to perform a variety of common tasks such as identifying nucleic acid or protein sequences, finding gene expression patterns for specific genes, stages or tissues, identifying literature on a specific gene or tissue, locating useful reagents and downloading our extensive content, including Xenopus gene-Human gene disease mapping files.
Keywords: Xenopus, Genome database, Polyploid genome, Gene expression analysis, Anatomy ontology, BLAST, GBrowse, Textpresso
1. Introduction
Modern cell and developmental biologists have relied on the large externally developing embryos of amphibians, particularly in the African clawed frogs of the genus Xenopus, since the late 1950s. Early cloning experiments in Xenopus demonstrated that differentiated cells contained the full complement of nuclear material, the principle of genomic equivalence [1, 2], and this finding revolutionized the understanding of cell differentiation, and thus paved the way, decades later, to induce pluripotent stem cells which in turn has revolutionized regenerative biomedical research. While Xenopus has been an outstanding system to make fundamental discoveries such as these, it has also played a major role in understanding pathological processes and elucidating the function of an increasing number of human disease genes (reviewed in [3]). Importantly, as the major nonmammalian tetrapod model in biomedical research, Xenopus research bridges the gap between the mammalian models and the more evolutionarily distant vertebrates such as teleosts [3].
Today, genomic data is at the core of all modern experimental design and interpretation. Xenbase is the Xenopus Model Organism Database (MOD), launched in 2005 (see [4]), and now running in a virtual environment [5], whose mission is to integrate and widely disseminate key molecular, cell, developmental, and bioinformatic data about Xenopus. We aim to accelerate discovery and to support the use of Xenopus for modeling human disease. To this end, Xenbase content is integrated with other MODs (MGI, Zfin, Geisha, WormBase; see Table 1 for a full list of abbreviations and website links used) and human disease databases (OMIM, Decipher, MalaCards, Gene Cards, HGNC). Our system associates Xenopus genes through “Gene Pages” to the orthologous human genes, and reciprocal data exchanges with numerous external databases and knowledgebases (e.g., NCBI, Entrez Gene, UniProtKB, and Ensembl). Thus, Xenbase not only supports Xenopus researchers but also makes Xenopus data broadly available to researchers in diverse fields, from cell and developmental biology, to environmental toxicology and human disease research.
Table 1.
Glossary of abbreviations for online resources, databases, and tools referred to in text, and/or linked to from Xenbase Gene Pages, with website address
| Resource | Description | Website address |
|---|---|---|
| Allen Brain Atlas | A comprehensive database with a suite of tools to view neurobiology in humans, mouse and nonhuman primates. | www.brain-map.org |
| CRB | Center for Xenopus Biological Resources, based in France. | xenopus.univ-rennes1.fr |
| Decipher | Mapping database to comparison Human clinical phenotypic and genomic data. | decipher.sanger.ac.uk |
| DRYAD | A curated data repository for scientific and medical literature. | datadryad.org |
| Ensembl | A genome browser for comparative vertebrate genomics. | www.ensembl.org/index.html |
| Eurexpress | A Transcriptome Atlas Database for the Mouse Embryo. | www.eurexpress.org/ |
| EXRC | European Xenopus Resource Center based in UK. | xenopusresource.org |
| FlyBase | A Database for Drosophila Genes and Genomes. | flybase.org |
| GBrowse | An interactive tool used by most MODs to manipulate and display genomes. | |
| Geisha | A Chicken Embryo Gene Expression Database. | geisha.arizona.edu/geisha/index.jsp |
| GeneCards | The Human Gene Database with integrated genomic, transcriptomic, proteomic, genetic, clinical and functional information. | www.genecards.org |
| Genomicus | Genomes in Evolution. A genome browser to display genes/genomes across taxa, through time and in predicted ancestral species. | www.genomicus.biologie.ens.fr/genomicus-88.01/cgi-bin/search.pl |
| GitHub | Online version control depository, where open source software and code, like the Xenopus Anatomy Ontology, is available. | github.com/ |
| GO | The Gene Ontology, from the GO Consortium. | www.geneontology.org |
| HGNC | Human Gene Nomenclature Committee. | www.genenames.org |
| iHOP | Information Hyperlinked Over Protein. | www.ihop-net.org/UniPub/iHOP/ |
| IMPC | International Mouse Phenotyping Consortium. | www.mousephenotype.org |
| JBrowse | JBrowse is a new genome browser which will replace GBrowse on Xenbase, (over ~2 years phase-out period) because GBrowse is no longer supported or being developed. | jbrowse.org |
| JGI-Xenopus | Joint Genome Institute, Xenopus genome project. | jgi.doe.gov/xenopus-frog-genome-project-on-cbc/ |
| JGI-Metazome | Genome database that organizes the proteomes of metazoans into gene families in evolutionary context. | metazome.jgi.doe.gov/pz/portal.html |
| JGI/KOG | Functional protein annotations from fungal genomics resource at Joint Genome Institute. | genome.jgi.doe.gov/help/kogbrowser.jsf |
| KEGG | The Kyoto Encyclopedia of Genes and Genomes. | www.kegg.jp/kegg |
| MalaCards | Human Disease Database with clinical and genetic annotations. | www.malacards.org |
| MGI | Mouse Genomic Informatics. | www.informatics.jax.org |
| miRBase | A searchable database of published miRNA sequences and annotations. | www.mirbase.org/index.shtml |
| NBRP | National BioResource project, based in Japan. | www.nbrp.jp/report/reportProject.jsp?project=xenopus |
| NCBI | National Center for Biotechnology Information. Hosts a extensive range of biomedical and genomic databases and analysis tools to support advances in science and human health. | www.ncbi.nlm.nih.gov |
| NCBI/BLAST | The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. | blast.ncbi.nlm.nih.gov/Blast.cgi |
| NCBI/EntrezGene | A portal to gene-specific content based on NCBI’s RefSeq project, model organism databases and others. | www.ncbi.nlm.nih.gov/gene |
| NCBI/GEO | Gene Expression Omnibus, functional genomics data repository at NCBI. | www.ncbi.nlm.nih.gov/geo |
| NCBI/HomoloGene | A tool to construct putative homology groups from gene sequences. | www.ncbi.nlm.nih.gov/homologene |
| NCBI/SRA | Sequence Read Archive, stores raw sequence data from next-generation sequencing projects. | trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi |
| NXR | National Xenopus Resource based in USA. | www.mbl.edu/xenopus |
| OBO Foundry | Open Biomedical Ontologies. | www.obofoundry.org |
| OMIM | Online Mendelian Inheritance in Man, An Online Catalog of Human Genes and Genetic Disorders. | omim.org |
| Panther | Protein Annotation Through Evolutionary Relationship, a large-scale gene function analysis tool. | pantherdb.org |
| RRID | Research Resource Identifiers, which are persistent and unique identifiers we use to reference research resources, such as antibodies and transgenic Xenopus lines. | scicrunch.org/resources |
| The Human Protein Atlas | Database of protein coding genes, their expression and localization at tissue and cellular levels. | www.proteinatlas.org |
| TrEMBL | A computer-annotated supplement of SwissProt that contains all the translations of EMBL nucleotide sequence entries not yet integrated in SwissProt. | www.uniprot.org/uniprot |
| Uberon | Integrated multispecies anatomy ontology, available on GitHub. | uberon.github.io |
| UniProtKB/Swiss-Prot | A protein sequence and function database. | www.uniprot.org |
| WormBase | A database for genetics, genomics and biology of C. elegans and related nematodes. | www.wormbase.org |
| XenMARK | Heatmap-based Xenopus gene expression image annotation tool. | genomics.crick.ac.uk/apps/XenMARK |
| XenMine | Multitool analysis resource for published Xenopus genomic data. | www.xenmine.org |
| XGNC | Xenopus Gene Nomenclature Committee, the scientific group charged with gene nomenclature review and approval, coordinated by Xenbase. | |
| Zfin | The Zebrafish Information Network. | zfin.org |
The DNA sequencing revolution of the 2000s quickly focused on model organisms, and the first amphibian species to be sequenced was the diploid Western clawed frog Xenopus tropicalis [6]. The larger Xenopus species, the African clawed frog, X. laevis, which is widely used as the nonmammalian tetrapod model in biomedical research, posed a more intractable problem to sequence because it is an allotetraploid (2n = 36). X. laevis likely arose via the interspecific hybridization of two diploid progenitors with 2n = 18, followed by subsequent genome doubling which restored meiotic pairing and disomic inheritance [7]. The sequencing, genome assembly, and annotation of X. laevis was, not surprisingly, very complicated [8] and took several years to complete [7]. Simultaneous integration of the two X. laevis homologs (referred to as “L” and “S” for long and short chromosomes, respectively [9]) into the Xenbase genome module was finalized in 2016. As a result, Xenbase currently provides cell and developmental biologists the most up-to-date genomic information based on both frog species, and this data is displayed on our genome browser and on Gene Pages, with both the X. tropicalis and corresponding X laevis L and S genes. Combined with an extensive catalog of curated literature, that covers over 48,000 published Xenopus articles, and a vast catalog of manually curated, tissue-specific gene expression images (66,000+), Xenbase is the go-to site for the most-up-to-date genomic Xenopus data. In addition, Xenbase hosts a vast amount of technical and reference material on Xenopus development, anatomy (including the extensive Xenopus Anatomy Ontology (XAO) [10]), and husbandry. Xenbase also provides an online hub for researchers, as we host personal profiles and laboratory descriptions, list conferences, workshops, a jobs board, discussion forum, and an array of links to other resources.
This chapter aims to give a practical guide on how to access the major features of Xenbase in a step-by-step manner, first covering how to navigate the home page, the extensive data on Gene Pages, then how to use the Quick Search Menu. We continue with a discussion of how to utilize Xenbase to its full potential-the remaining topics are presented in the order as they appear of the drop-down menus (except Gene Pages), going from left to right. We discuss how to find markers for a specific organ system, download large NextGen Sequence (NGS) data, use genomic tools (like BLAST and GBrowse), find guidelines on gene and transgenic nomenclature, and locate Xenopus specific protocols or reagents.
2. Navigating the Xenbase Home Page
The home page (http://www.xenbase.org) combines the horizontal navigation bars that are common to all Xenbase pages with additional information in subject based “tiles” and an additional vertical navigation bar. The tiled lay-out covers the same areas that are accessible via drop-down menus in the header. Many search functions are also available in a quick search bar (aka the mini-bar), in the top right corner of the home page. Centrally placed on the Xenbase home page is a rotating image carousel, where we spotlight the latest high impact Xenopus research publications, and which serves as a community notice board covering, for example, conferences and workshops, awards and journal special issues. These are reiterated in the “Announcements” column on the right-hand side of the home page. This side column also gives links to static content on the website, including an introduction to Xenopus as a model organism, links to various features and data on Xenbase, the Xenopus Stock Centers, and other databases and external resources useful to Xenopus researchers.
3. Genes and Gene Pages
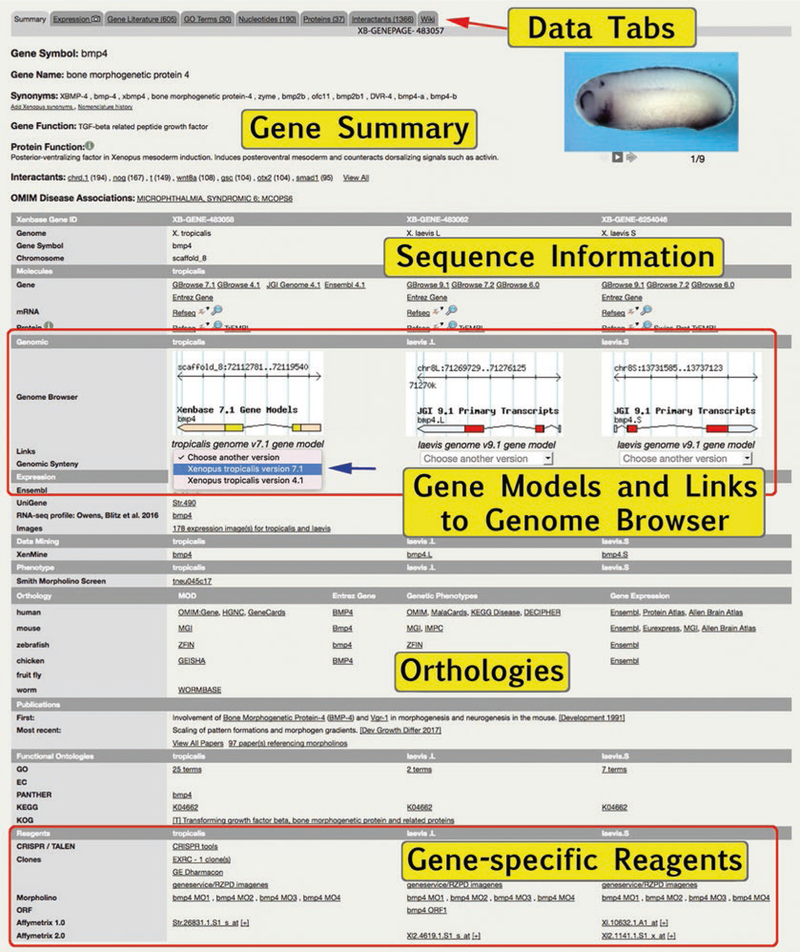
Xenbase is fundamentally a “gene-centric” database. The Genes module is a catalog of genes in the diploid X. tropicalis and polyploid X. laevis—all three genes (one X. tropicalis gene, and two X. laevis genes) are represented on a single “umbrella” Gene Page, which details all information about the Xenopus gene and its products. Each Gene Page carries a stable Xenbase Gene Page ID (e.g., XB-GENEPAGE-483057 is the bmp4 Gene Page), and each gene has its own stable Xenbase gene identification number. Here we describe the information on a Gene Page, and the how to find a specific Gene Page.
3.1. How to Find a Specific Gene Page
Select “Gene Search” under the “Genes” menu to find specific Gene Pages or gene families. The default is to “search all,” but to scale down or speed up results, choose one of the more specific search options which include a partial or full gene name (e.g., “bone” or “bone morphogenetic protein 4”), gene symbol (e.g., bmp4) or synonym (e.g., bmp-4), orthologs (if any with different symbols/names), or gene function (e.g., “morphogenetic protein” which will return all bmp genes as well as related gene families). The menu will autofill with the matched text highlighted in yellow.
Alternatively, enter an NCBI accession number, Entrez gene ID, Unigene ID, OMIM ID, GO ID, or GO term. Also, a Xenbase accession number such as a “Gene Page ID” can be entered (e.g., XB-GENEPAGE-483057) to find Gene Page(s).
Checkboxes permit you to filter results to include only “manually curated Gene Pages” or “Gene Pages with expression images.”
Gene Pages can also be browsed alphabetically.
The “Advanced search” offers additional filters: to text-match specific letter combination (e.g., “rsp”) or parts of names (e.g., “receptor”).
3.2. Gene Pages
The most utilized, useful data and salient features for each gene are presented on the “Summary” tab on the Gene Page, under the following headings (as an example, enter “bmp4” into the quick search bar in the top right corner of the Xenbase hompage):
Summary: Official gene symbol and full name, synonyms, gene function, protein function, a list of cocited interactants (and a thumbnail of an interactive graphical display of interactants), and associated OMIM diseases are all detailed on the top of the Gene Page. Images that summarize the gene expression throughout a range of embryonic stages are shown to the right. Click the + link (the Xenbase symbol that additional text is available) to see all OMIM associations (if present), and click the link to “Nomenclature history” to open the Wiki tab, where changes to gene names and gene symbols are recorded. Xenopus gene names and symbols are identical to human gene names, whenever possible, and orthology to human genes is usually assigned by synteny. Gene names for X. laevis homeologs are appended with “L homeolog” or “Shomeolog” to distinguish the sub-genome with which they are associated.
Xenbase Gene ID: Xenbase IDs are allocated to each species/sub-genome specific gene. The chromosome location is indicated when known, and scaffold positions are given in cases where the location has not been fully determined (e.g., due to incomplete or in-progress genome annotations).
Molecules section lists and links-out to NCBI/Entrez Gene IDs, nucleotide, and protein data at Swiss-Prot and/or TrEMBL. mRNA RefSeq data has BLAST functionality (click on the rocket icon) and sequence files in FASTA format (click magnifying glass icon to pop up sequence file), can be viewed for any listed sequence. Complete data for Nucleotides and Proteins associated with the gene are listed on relevant “tabs” at the top of the Gene Page.
Genomic data is illustrated by gene model snapshots from the genome browser JBrowse. Clicking on these options will open the full view of the gene in JBrowse. The default display is the most current genome with an annotated model for the gene displayed, and earlier versions and GBrowse view can be selected from drop-down menus under each gene model snapshot.
Expression section links out to Ensembl and UniGene entries, and RNA-Seq profiles illustrating temporal and tissue expression patterns.
Data Mining section allows researchers to access a specific gene’s entry on XenMine, a comprehensive toolbox for NGS data analysis that is part of the Intermine project, and is hosted by Stanford University.
Phenotype section currently links to the morpholino screen data produced by the Smith Lab at the Gurdon Institute at the University of Cambridge. Full phenotype curation is a major priority for Xenbase in the coming year, and phenotype annotations will be posted in this section on Gene Pages.
Orthology section provides direct links to the orthologous genes recorded in human (OMIM:gene, HGNC, and GeneCards) and the relevant other model organism databases: mouse (MGI), zebrafish (Zfin), chicken (GEISHA), fruit fly (FlyBase) and worm (WormBase).
Publications lists the first article to mention the gene, and the most recent article. Click on the journal reference in parenthesis to see the Article Page in Xenbase, or click the “View All Papers” link to go to the complete list (which can also be access via the Gene Literature tab). A camera icon indicates the paper has images displayed.
Functional Ontologies section provides gene-specific links to GO terms (sourced from UniProt), gene information at PANTHER [11] (a Gene Ontology consortium project), KEGG orthology entry for this gene, and KOG classification of the gene (sourced from JGI).
Reagents section provides links to reagents and resources tailored to the relevant gene. A link is provided to our list of design tools for CRISPR/Cas constructs that includes information on which Xenopus genome builds are compatible with the various tools. Links are provided to several sources for sequence clones including the EXRC and GE Dharmacon, and to our own catalog of antibodies, morpholinos and ORF clones used in Xenopus research involving the specific gene (see Subheading 9 for more reagent details). We also provide links to the details of Affymetrix array probe-sets for Xenopus (note these require an Affymetrix log-in to access).
3.3. Expression Tab: Viewing Gene Expression Data and Images
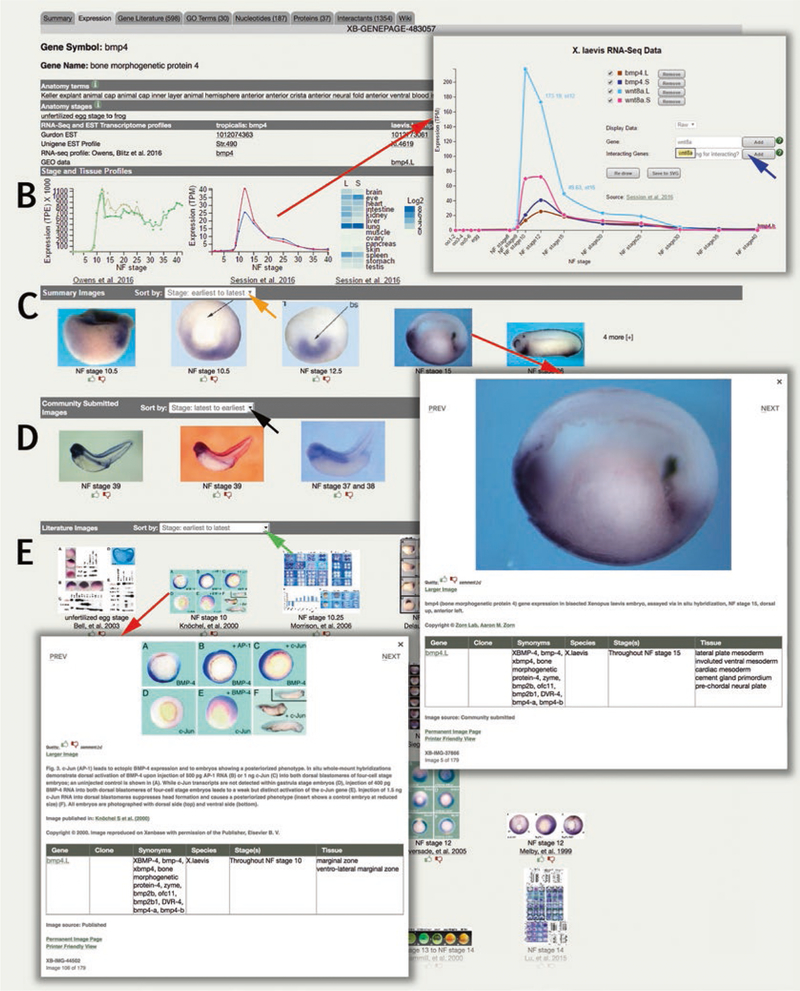
Xenbase displays 66,000+ in situ hybridization and immunohisto-chemistry images that are posted on the Gene Page “Expression” tab, under two main headings: “Community Submitted” (mostly unpublished images from large scale screens) and “Literature Images” (from journal articles). Curators manually annotate the observed gene expression in these images using terms from the Xenopus Anatomy Ontology, the XAO, to generate a gene expression annotation table for each curated image. Out of the 15,878 Gene Pages currently in Xenbase (v4.7, January 2018), c. 24% (3775 genes) have gene expression images, mostly from in situ hybridization (a camera icon indicates that images are posted for that gene). Additionally, about 95% of genes have expression data from RNA-Seq and EST Transcriptome profiles and/or developmental stage profiles determined by microarray analyses (see Fig. 1B, C).
Fig. 1.
Xenbase is built around the “Gene Page,” where a file-like tab system provides comprehensive coverage of data about each gene. This example is the “Summary” tab for ‘bone morphogenetic protein 4’ (bmp4) with the Xenbase gene page ID ‘XB-GENEPAGE-483057’. The salient features of gene (official name, synonyms, gene and protein function, cocited interactants, and human disease associations) are all shown in the upper summary panel, along with a developmental expression series (where available). Sequence information and JBrowse snapshots of the gene models are shown for X. tropicalis and X. laevis L and X. laevis S homeologs (upper red box). To view a different gene model, select from “choose another version” (blue arrow). The rest of the Gene Page provides links to more data covering orthology, first and most recent publications, and functional ontology, with curated gene-specific reagents (e.g., MOs, primary antibodies, and ORFeome clones) in the lower panels (lower red box). Frequently accessed tabs include the “Expression” tab (details in B–D below with a camera icon that indicates presence of images, “Gene Literature,” and “GO terms” from UniProtKB, where number in parenthesis indicates number of citations or terms respectively. (B) The Expression tab of a Gene Page displays gene expression data in several useful formats. Interactive graphs plot X. laevis L and S homeolog expression from RNA-Seq data [7] with ability to add more genes (blue arrow) to the graph via dialog boxes (red arrow, click to pop-up). Heat-maps from adult tissues compare X. laevis L and S homeologs, data from [7]. (C) Summary images are selected to represent gene expression over a range of embryonic stages and can be sorted by stage (orange arrow). (D) Community submitted images from large scale screens, which generally use ISH and IHC, can also be sorted by stage (black arrow). (E) Literature images from published articles can be sorted by stage or publication date (green arrow), and include link to the Xenbase Article Page. Click on the image to pop up a larger image (red arrow), along with caption and annotation table
Gene expression data is organized on the “Expression” tab, under the following headings:
Anatomy terms: XAO terms compiled from manual curation by Xenbase, and NBCI cDNA libraries. Use the [+/−] toggle to expand or hide terms.
Anatomy stages in which gene expression has been recorded, often unfertilized egg to adult frog stage.
- RNA-Seq and EST Transcriptome profiles. We link out to:
- Gurdon Institute EST database (e.g., X. tropicalis bmp4)
- Unigene EST Profiles, with heat map of tissue-specific expression (e.g., X. tropicalis bmp4)
- GEO data: links to this NCBI resource and runs an automatic search for the gene symbol and “Xenopus.” Currently there are about 185k GEO entries for Xenopus, but not all genes are represented.
- Developmental Stage Profiles:
- X. laevis RNA-Seq data is displayed in dynamic graphs generated from the X. laevis genome sequencing project data [7] (see pop-out in Fig. 1B). These graphs plot transcripts per million (TPM) values against developmental stage (oocyte stage 1–2 to NF stage 40) for the X. laevis L and X. laevis S homeologs.
- Click the graph thumbnail to open a larger interactive graph.
- Use dialog boxes to add additional gene symbols to plot: type ahead suggests gene symbols from gene catalog, and there is no limit (Fig. 1B, blue arrow). After selecting click “Add.”
- Interacting genes is limited to cocited genes.
- Use “Display data” box to choose either “Raw” or “log2” transformation.
- Mouse over a data point to display the underlying value and the stage.
- Click “save to svg” button to download graph.
- X. laevis L versus X. laevis S homeolog expression in various tissues illustrated via a heatmap. Click to open a larger view.
Summary Images. A curated selection of gene expression images from in situ hybridization (ISH) or immunohistochemistry (IHC) across embryonic developmental stages. These are the same images that appear in Summary section of Gene Page, and they are selected from either “Community Submitted images” or “Literature Images” by Xenbase curators (Fig. 1C). Click image to enlarge and view annotation table.
Community Submitted Images come mostly from large scale screens, and are generally ISH. Laboratory of origin holds the copyright to these images. Double click the image to enlarge it and view the annotation table.
-
Literature images display the curated figures from research papers where we have redisplay permission or which are open access. Figures are often multipaneled, and gene expression annotation table is viewed by double clicking on the figure. These images may be protected by copyright; if so, this is indicated.
Notes/Troubleshooting on viewing Expression on Gene Page:- Some genes are very well studied with hundreds of images posted. Click the [+] to toggle between more and less data [−].
- Use “Sort By” to organize by developmental stage: “earliest to latest” or “latest to earliest.”
- Literature images are also sortable by earliest or latest publication data.
- Use thumbs up or thumbs down tool to vote for high quality images
- Xenbase welcomes high quality images via community submission to populate poorly studied genes! Submit new gene expression images via the “Contact Us” (email: xenbase@ucal-gary.ca) in the footer of every Xenbase page.
- As there is strong conservation in gene expression in the vast majority of the expressed orthologs and in situ probes designed for one species generally work equally well in the alternate Xenopus species [14], gene expression tables are largely accepted as applicable to both species, although there are exceptions.
- Species (X. tropicalis or X. laevis) is indicated in the image caption for community submitted and large scale screen data.
3.4. Other Gene Page Tabs
At the top of each Gene Page, a series of file-like “tabs” collate additional gene-specific data as follows:
-
1
Gene Literature lists all articles that refer to the gene in its data or text.
-
2
GO Terms provide a quick overview of the cellular role of a gene and can also be used for analysis of high-throughput proteomics data. GO terms are presented under the three categories—Molecular Function, Biological Process, Cellular Component (sourced from UniProt). Click on the GO term for a full definition or the information button for evidence metadata.
-
3
Nucleotides tab provides links to all gene models and mRNA data from JGI, Ensembl, NCBI, Unigene clusters, mRNA and ESTs for the gene. The rocket icon will autofill a BLAST request, and the magnifying glass icon will provide a pop-up of the sequence in FASTA format. Click on Clone name or Accession number for more details.
-
4
Proteins tab links to all protein model data from JGI, NCBI, Ensembl and protein sequence from specific accessions in NCBI Protein, RefSeq and Swiss-Prot/UniProKB. The rocket icon will auto fill a BLAST request and the magnifying glass icon will provide a pop-up of the sequence in FASTA format.
-
5Interactants: An interactive graph illustrates the genes cocited with the gene of interest, which is placed in the center of the graph.
- Drag the nodes to move them, and set them in place.
- Double click to release node position.
- Number of cocitations are marked on the edges of the graph.
- Click on the gene symbol to go to the corresponding Gene Page.
- Graph is downloadable in two formats: use buttons “save to svg” or “save to png.”
Cocited genes are then listed in ranked descending order in two columns, with links to Gene Pages and to literature (e.g., 1358 genes have been cocited with bmp4, the top hit being chrd.1 (chor-din, gene 1) in 190 articles; status June 2017). Finally, links are also provided to IHOP (Information Hyperlinked over Proteins) for both X. tropicalis and X. laevis. In the near future, interactants will include data on physically interacting proteins from human networks, and also genes in coexpression or coregulated networks.
-
6
Wiki: Nomenclature changes are recorded on the Wiki tab, which can be also accessed by clicking the “Nomenclature History” link. In addition, the Wiki is used to record any information about a gene that is not recorded elsewhere on Xenbase, such as synteny analysis methods, reagent or protocol notes. Registered users can add to Wiki content
3.5. Notes/Troubleshooting Genes Module
Genes can also be searched using the Quick Search Menu (see Subheading 4 below).
Can’t find a gene? If you cannot find a Gene Page for a gene of interest, try our Search Help page for hints. Xenopus genes are following human gene nomenclature, so searching by an old name may not work. We store old or “legacy” gene names as synonyms. If your search fails, it may mean Xenbase does not have that gene name or symbol in the database. Try the human, mouse, chicken, or zebrafish gene symbol. If this fails also, the ultimate gene finder requires you to BLAST the Xenbase genome database as detailed in Subheading 5.
Gene nomenclature issues? Xenbase is the clearing house for Xenopus gene nomenclature. Gene Nomenclature Guidelines are posted under the Genes menu. As gene nomenclature is updated constantly by the HGNC, many gene names and symbols completely change over time. Although gene symbol synonyms are a powerful tool to track down the new name for a gene, they also can be misleading, especially when the same gene symbol has been used/reused in different species/model organisms. NCBI databases record a more comprehensive list of legacy synonyms and symbols than Xenbase, as we try to concentrate on just those symbols used/referred to in Xenopus literature. Note that our gene search does not search Wiki entries, which is where gene nomenclature changes are recorded, however the “Search with Google” in the Quick Search menu does search the Wiki (and everywhere else). Suggest adding a gene name, missing synonyms, or report errors or omissions by contacting Xenbase (xenbase@ucalgary.ca).
Why is not there an L or S model for this gene? Not all X. laevis genes have both X. laevis homeologs. After the hybridization event that created X. laevis, there was a genome reduction that resulted in loss of some homeologous genes with a higher proportion of S genes being removed than L genes [7]. It is also possible that the homeologous locus is still being assembled fully, or both gene models exist, but only one has been properly annotated.
Where did the A and B genes go? With the discovery that X. laevis contains two independently interacting legacy genomes that can be distinguished from each other, “A” and “B” genes were migrated to the more informative L and S nomenclature.
4. Quick Search Menu
The quickest way to get to the most popular and well-used content on Xenbase is to use the Quick Search Menu (aka the mini-bar) in the top right hand corner of the home page and every Xenbase page (in red box, Fig. 2). Select the search topic from the drop-down options, and enter a term to search as follows:
Genes: Enter a partial or full gene symbol (e.g., “fgf”) or partial gene name (e.g., “fibroblast”) to return all “fgf” family genes as well as “fgfr” genes, genes with “FGF” in the name, function or synonyms. Get precise, single gene return by entering an exact gene name or symbol (e.g., “fgf3,” or ‘fibro-blast growth factor 3’) (see Fig. 3A).
Xenbase with Google: Search Xenbase for any text, e.g., a partial article title or phrase, gene symbol, clone ID, or author with the Google search option to pull every match in the Xenbase database, including Wiki entries. Searching for “fgf3,” for example, returns the Gene Page record, in situ data, expression profiles, literature for that gene, the anatomy term expression page (for which it has gene expression curations), a list of potential gene regulatory network interactants and cocited genes, as well as all ORFeome clones and plasmids mapped to this gene. We control which pages google indexes, so if you note something missing from these search results please let us know and staff will ensure that the missing content is included in future crawls.
Anatomy Items: Enter an anatomical term (e.g., “heart,” see Fig. 3B) to find all “Xenopus Anatomy Ontology” (XAO) terms [10] used in gene expression annotations. Select a term as it autofills from the XAO, text matches are high-lighted in yellow, in addition to showing all elements that are “part of” the term (e.g., “cardiac mesoderm”). Selecting any option from drop down will take you to the specific XAO term page.
People: Find any of the 1900+ researchers with Xenbase profiles. Enter any part of a person’s first or last name and it auto-fills a list, highlighting in yellow the text match. Hit search to display all results.
Labs: Find any of the over 270 Xenopus research labs with Xenbase profiles. Laboratories are generally named with group leader’s last name (e.g., Smith Lab).
Organizations: Find contact information for stock centers and other organizations that supply reagents, frogs and husbandry equipment, as well as publishers of key life science journals and scientific societies (e.g., NXR, see Fig. 3C).
Paper Authors: Enter a surname to search all authors of all 48,000+ Xenopus research papers in the literature module. Enter any part of an author’s last name, and autofill options will highlight matched text in yellow (see Fig. 3D). Select a specific author or hit search to display all results. This search will also find letter combinations, e.g., “vg” will find all instances in both the first and last names and as an author’s initials.
Paper Title: Enter the entire paper title to find a specific paper, or a partial title or any word or phrase from the title of a published article to run a quick literature search for Xenopus specific articles on a topic (e.g., “left–right” to return all papers on “left–right patterning,” “left–right asymmetry,” and “left–right axis determination”; Fig. 3E).
Clones: Search for data from over one million clone entries in the Xenbase database. Enter either the gene symbol to which the clone/plasmid specifies (e.g., “fgf3”) or an existing clone ID number (e.g., IMAGE:7029804 or xl301j22).
- Xenbase Accession: This search finds specific data using the unique Xenbase identifiers with our numbering and cataloguing systems. After working with Xenbase data, researchers may record a specific Xenbase accession number to easily return to this specific database page. The following are examples of valid Xenbase Accession numbers:
- XB-GENE-484294 (Gene Page)
- XB-ART-53013 (Article Page)
- XB-PERS-3515 (Person/Researcher Page)
- XB-LAB-702 (Lab Page)
- XB-ANTIBODY-14574796 (Antibody Page)
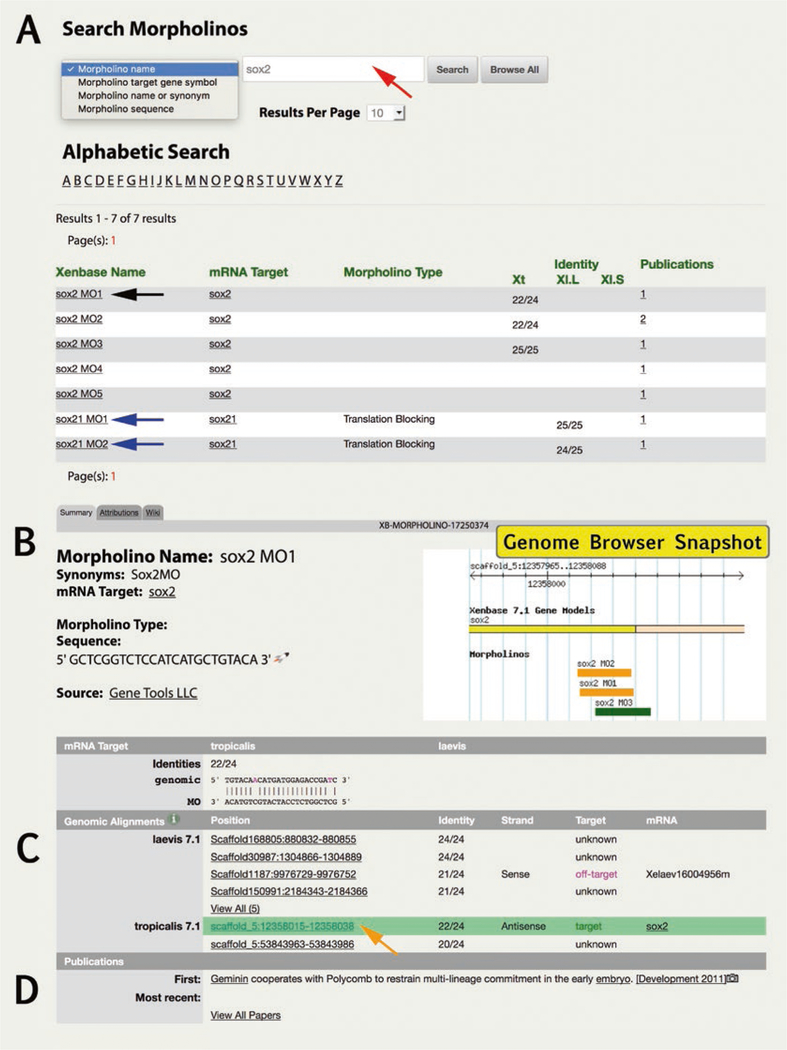
- XB-MORPHOLINO-17249870 (Morpholino Page)
OMIM ID: Enter an OMIM ID number for any disease from the Online Mendelian Inheritance in Man (OMIM) database to find associated Xenopus-Human disease model data. For example, enter “219700,” the OMIM ID for “Cystic Fibrosis,” to return two Gene Pages associated with this disease, cftr and tgfb1. This is a quick way to find the Xenopus literature from cell biology to phenotypic models that are applicable to, or associated with, a specific human disease.
OMIM Description: Enter a term from the OMIM disease name or description (e.g., “diabetes”) to return all homologous Xenopus Gene Pages to discover all Xenopus literature and associated genomic data associated with that specific human disease or family of diseases. This is a fast way to find the known Xenopus gene expression data and associated literature from cell biology to phenotypic models, that is applicable to, or associated with, a range of related or similar human diseases and syndromes.
GO Terms: Gene Ontology (GO) terms cover three areas: molecular function, biological process and cellular component. Enter a full or partial GO term (e.g., “axial”) and the drop-down menu autofills and text matches highlight in yellow. Mouse down to select the specific term of interest and hit search. Single returns will direct to the gene page, and multiple returns will be shown in a table. Click the gene symbol to go to that Gene Page, where this GO term, and all others annotated for the gene, are listed under the GO Terms tab. Approximately 8400+ GO terms are currently associated with X. laevis genes (both L and S) and 7400+ GO terms are currently associated with X. tropicalis genes. As Xenbase further develops this feature, reciprocal data exchange with the GO Consortium will update and add more GO terms to Xenopus genes. Xenbase curators will also manually add GO annotations extracted from the published literature to Gene Pages and Articles Pages.
Fig. 2.
The Xenbase home page (http://www.xenbase.org) features a rotating image carousel to spotlight new articles and announce relevant news to the Xenopus community. Log-in and the Quick Search minibar are in the upper right corner. The drop-down menu bar spans the top of the web page, and reiterates the links in the subject tiles below. Additional links to Xenopus resources are in the side column, and social media and contact Xenbase links are in the footer. This layout visually describes the database architecture and is designed to accommodate different workflows and preferences
Fig. 3.
Quick Search Menu is located at the upper right corner on every Xenbase page. Options available from drop downs include: (A) Select “Genes” to search for partial of full gene name or symbol (e.g., “fgf”); (B) “Anatomy Items” link an XAO term page search (e.g., “heart”); (C) “Organization” quickly finds contact details for stock centers of suppliers (e.g., NXR); (D) “Paper Authors” will text-match partial and full names to Xenopus literature (e.g., “Blum”); (E) “Paper Titles” searches full or partial article titles, and effectively searches for keywords
Notes/Troubleshooting the Quick Search Menu
Note that there is no wildcard (*) search in the quick search menu.
If you get no results, check for typing errors (remove all spaces before or after the text, check for erroneous spelling, symbols, or Greek letters that did not copy correctly, or extra punctuation marks), as this is an “exact” text match algorithm, so only perfect matches will be returned, then search again.
For GO term searches, users must select a specific GO term from the drop-down menu to return results.
For a comprehensive text match search of the entire paper, not just the title, use Textpresso; see Subheading 6 below.
Google is continually adding more content from Xenbase to their search engine, however, the Google search may not include all content from Xenbase.
If you cannot find what you are looking for, try choosing one of the specific search areas from the menu, and ensure that you are searching the right item from the appropriate menu option.
4.1. Accessing Xenbase Features from the Main Menus and Home Page Tiles
The following sections cover how to access and use the database features of Xenbase via the Main Navigation Menu, remembering that these options are reiterated on the Home Page Tiles. All topics discussed can be accessed via both options, and we discuss them here in order of the main menu, from left to right, excluding the Gene Search, which is covered above in Subheading 3.
5. BLAST Menu
BLAST (Basic Local Alignment Search Tool) is a tool that finds regions of similarity between two nucleotide or protein sequences [15].
Use BLAST to:
Identify sequence fragments.
Calculate sequence conservation across taxa.
Identify orthologs across taxa.
Check for target versus off-target sites for a PCR primer or morpholino (MO).
The main BLAST menu offers options to align the query sequence against Xenopus mRNA, Xenopus proteins, various genome versions and the mitochondrial genomes for three Xenopus species (X. laevis, X. borealis, and X. victorianus).
5.1. How to Use Xenbase BLAST
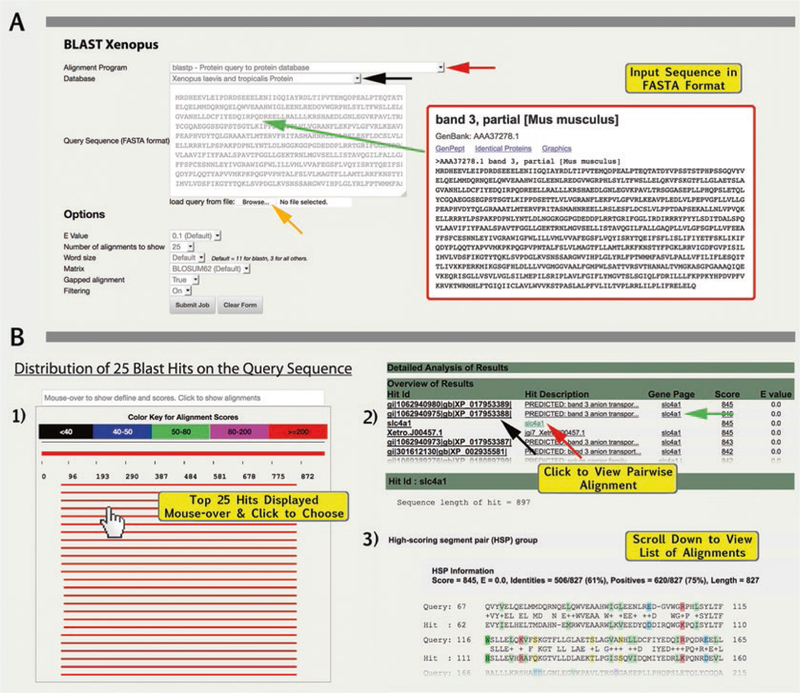
Choose from the alignment program query options (e.g., blastn: DNA query to DNA database or blastp: Protein query to protein database) (red arrow, Fig. 4A).
Choose the target database or genome build to which you want to compare/align your sequence (e.g., Xenopus laevis and tropicalis mRNA or X. laevis J-strain 9.1) (black arrow, Fig. 4B). Xenbase BLAST allows users to compare nucleotide or protein sequences to the latest (and legacy) X. laevis and X. tropicalis genome builds, mRNA and protein sequences, with these options available in the second drop-down menu labeled “Database.”
Enter (i.e., type or copy and paste) a single query sequence into the data box in GenBank/FASTA format (green arrow, Fig. 4A). Alternatively, upload a query sequence file using the “choose file” dialog box (orange arrow, Fig. 4A).
For almost all Xenopus-to-Xenopus comparisons, the default “Options” settings will result in a high scoring, statistically significant alignment, although more advanced users can choose a custom set of options.
Click the “Submit Job” button to compute the sequence alignment.
- BLAST results are displayed graphically in three sections:
- An “Overview of Results” table has five columns: “Hit ID” (i.e., accession ID number of hit or scaffold number), “Hit Description” (name of the sequence hit), Gene Page (i.e., link via gene symbol), HSP “Score” and E-value. Click “Hit ID” (black arrow, Fig. 4B.2) to open the match on GBrowse. Click “Hit Description” (Fig. 4B.2, white arrow) to skip to the High-scoring Segment Pair (HSP) alignments, with computed percent identity, and links to chromosome locations and GBrowse.
- Click the gene symbol (green arrow, Fig. 4B.2) to go to the Xenbase “Gene Page.”
Fig. 4.
Using BLAST on Xenbase. (1) Access BLAST from drop-down menu or tile; (2) Choose Alignment program and (3) the database to which your search will be aligned; (4). Paste the query sequence into the box, in FASTA format; (5) Set and adjust options and (6) click “Submit Job” button. (A) In this example to assess evolutionary conservation of the protein Slc4a1 between mouse and frog, we used “blastp” (protein query-toprotein database) and entered the amino acid sequence for mouse Slc4a1 (Gene ID: 20533; protein_id=AAA37278.1) in FASTA format. We selected “X. laevis and X. tropicalis proteins” from database options. (B) Results of BLAST for mouse Slc4a1 vs. Xenopus are displayed sequentially in three formats: (1) Distribution of the top 25 hits on the query sequence with red indicating alignment scores >200; (2) Table with “Overview of Results” showing high scoring segment pair alignments (with alignment score). (3) Click hit ID or scroll down page to view pairwise alignments and identity calculated as a percentage
5.2. How to Use BLAST to Inform Design of Xenopus-Specific Primer or Morpholino
Select “blastn - DNA-to DNA query.”
Select the database “Xenopus laevis and tropicalis mRNA.”
Enter the sequence into the query sequence box (e.g., CTCACTGGACATCCAGGTCTGAG, a potential scl4a1 PCR primer sequence).
Click “Submit Job.” Results are displayed in the same formats as shown in Fig. 4B in the above example, indicating that 24/24 bases match X. laevis scl4a1.L homeolog, and 23/24 bases match X. laevis scl4a1.S homeolog.
5.3. Troubleshooting BLAST
BLAST searches are usually almost instant, but occasionally can take some time to complete. Very long sequences (e.g., a scaffold), sequences with repeats, or sequences with low complexity increase the chance of a BLAST run being slow, or even timing out. In these cases, try entering a smaller sequence, or change the E-value to get a more sensitive alignment.
If a BLAST query results in no alignments, check that the correct database and BLAST program has been selected, increase the E-value, or rerun the same BLAST. A warning message will be displayed if an incorrect database is selected for the selected alignment program.
Mitochondrial genomes form a distinct data unit in BLAST and therefore must be selected from the option in the main menu. No mitochondrial annotations are currently available.
If BLAST times out after ~30 s, it can be due to heavy use of the service. Try again during an “off peak” time slot and if problems persist, please contact Xenbase. This typically only occurs with very large jobs or complex tblastn or tblastx runs. Once again, feel free to email us if this occurs.
There is no fully annotated genome available for X. victorianus, only mtDNA.
6. Genomes Menu
6.1. Download Xenopus Genomes
The Xenbase Data Downloads page provides access to genome assemblies, gene models, sequences, and database reports. Most files are in a tab-delimited format. Use the toggle [+] to see all files. Click the [readme] link to view information on the files, including the header row for these files. To download a file, click on the corresponding FASTA link. More files are located at our FTP File Browser.
6.2. GBrowse
GBrowse is an open source, browser based, interactive genome visualization software that allows gene models to be viewed within the genome next to RNA-seq and ChIP-Seq data. Xenbase GBrowse can be accessed from the menu bar, via BLAST against a genome, or clicking a snapshot on a Gene Page, or a snapshot morpholino page. Xenbase hosts the most recent and several legacy genome assemblies for both X. tropicalis and X. laevis. The main view of GBrowse on Xenbase shows all selected tracks for the chosen genome. Tracks can include gene model annotations, RNA-Seq alignments, ChIP-Seq alignments, and morpholino alignments. Tracks are binned into categories, such as gene models, tissue RNA-Seq, stage RNA-Seq, and methylation ChIP-Seqs. To customize which tracks are displayed, click the “Select Tracks” tab, and use checkboxes.
6.2.1. How to Use GBrowse
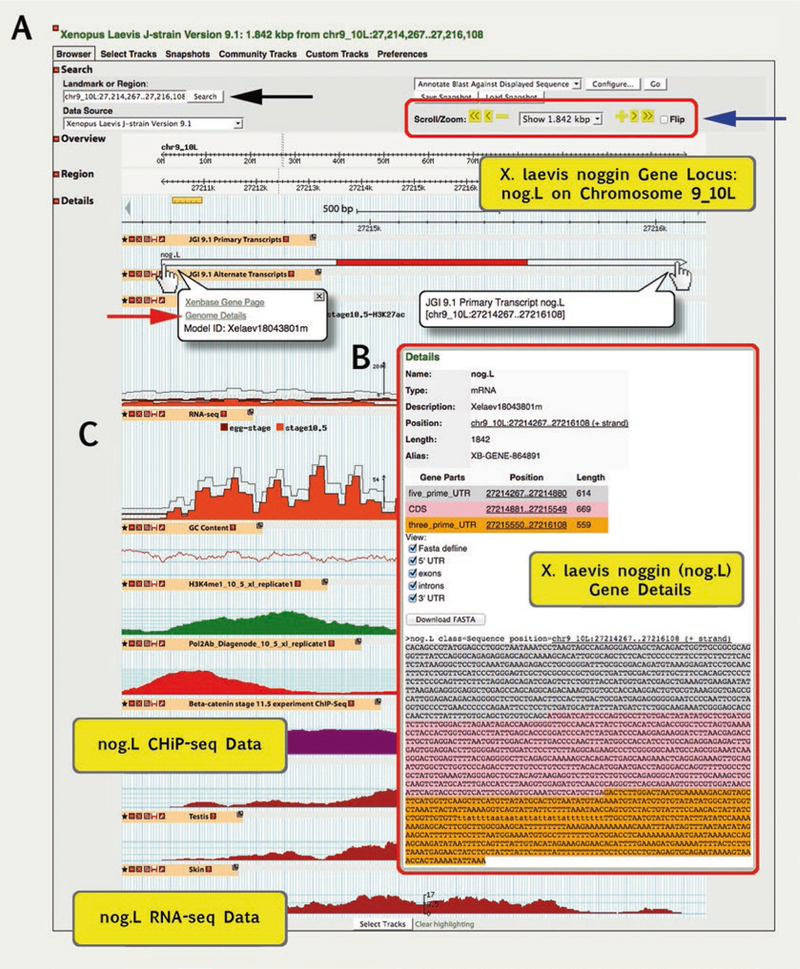
Open GBrowse via the Genomes menu, or home page tile, by selecting a genome model version (e.g., X. laevis 9.1 (J-Strain) on GBrowse).
Use the “Landmark or Region” dialog box (black arrow, Fig. 5A) to search for a scaffold position (e.g., chr9_ 0S:3,571,719..3,581,718).
If the scaffold position is unknown, enter a gene symbol (e.g., pax3), to identify which chromosome the gene is on (e.g., chr5L or chr5S) then click to choose a region to view (e.g., pax3.L, chr5L:123,000,255..123,046,707) from the results table.
Scroll/Zoom tools allow you to move left and right along a GBrowse view (blue arrow, Fig. 5A). Alternatively, use the drop-down menu to select options from 100 bp to 2 Mbp to zoom in and out. This is very helpful when identifying sur-rounding gene models.
Click on specific track to access additional information.
Hover/mouse over a track to popup its precise scaffold position.
Click on a gene model to give a pop-up box that provides a link to the Xenbase Gene Page, as well as gene model details for the given transcript. Click the gene model “Details” (red arrow, Fig. 5A) to show metadata for the model (e.g., nog.L in box, Fig. 5B), including the type, position, and length of each exon, and an interactive FASTA display, which allows the sequence to be copied for further use.
Click and hold/drag a track to rearrange track position.
On each track, a series of buttons on the far left side allow users to save a track as a favorite [star], show or hide a track [−], turn off at rack [×], share [radio], save [disc icon], or configure [tool icon] tracks. The [?] button gives more information including an option to download the data for the track.
Use check boxes on the “Select Tracks” tab to customize the data displayed (e.g., include or exclude BAC and Fosmid end data or Methylation ChIP-Seq data) and whether to show the RNA-Seq and ChIP-Seq data stacked in Topoview.
Additional tabs s “Snapshots,” view “Community Tracks” and upload “Custom Tracks.”
Changes to the color scheme and grid width can be set in “Preferences” tab.
Fig. 5.
Using GBrowse, in conjunction with BLAST, to visualize alignments against gene models and Next-Gen sequence data. (A) In this example, we BLAST X. tropicalis nog mRNA against X. laevis 9.1 genome. GBrowse gene model details are shown for X. laevis nog.L. Moving the cursor over the gene model (hand cursor) generates a pop-up with links to “Xenbase Gene Page” and “Genome Details.” Click on Genome Details (red arrow) to access metadata (pop up in B) about the given gene model, including the specific Xenopus gene (L or S) to which the model is associated. (B) The Gene Details provides a quick overview of the structure of the model, and size of exons, introns, and 5′ and 3′ UTRs. The interactive sequence section allows the UTRs and introns to be toggled on and off for easy copying and can be edited to include/exclude exons. (C) Next-Gen RNA-Seq and ChIP-Seq datasets are shown below the gene models
6.2.2. Troubleshooting GBrowse
Some gene models (e.g., pax1.S) in GBrowse may give “Xelaevis” model IDs and not link to Gene Pages. The frequency of these legacy mappings will decrease with ongoing improvements to the gene model annotations.
Occasionally tracks within GBrowse will not display, and will show a rendering error. Changing the zoom level will usually fix this problem.
If the gene search does not work, a gene symbol synonym may be being searched, rather than the official gene symbol. Refer to Xenbase Gene Pages for the official gene symbol.
If a gene model is still not being found, it is best to BLAST the sequence against the genome (as shown in Fig. 5), and then follow the BLAST links to try to identify the correct model.
JBrowse [16], a newer genome browser with increased functionality, has just been launched on Xenbase (under “Genomes” menu, X. laevis v9.2 on JBrowse is now at the top of the list). We will continue to support both genome browsers for ~2 years, as GBrowse is phased out, and new genomic data will only be added to JBrowse.
6.3. Xenbase UCSC Track Hub
Track hubs are web-accessible directories of genomic data that can be viewed on an external genome browser, and are helpful tools for quickly visualizing large genome-wide data sets, including numerous custom tracks [17]. Xenbase hosts a University of California, Santa Cruz (UCSC) Track Hub that can be loaded into a UCSC instance. A link to the track hub is accessible from the Xenbase home page under the Genomes menu. The UCSC Track Hub includes gene models for X. tropicalisv7.1, v8.0, and v9.0, and X. laevis v9.1 genome builds, plus a large number of RNA-Seq and ChIP-Seq tracks. Xenbase is currently processing additional NGS datasets including them in the track hub (sese RNA-Seq (red) and ChIP-Seq (orange) tracks in Fig. 5C).
6.4. Other Genome Assemblies
Xenbase also links out to other genome resources from the Genomes menu, including the Japanese National Institute of Genetics X. laevis genome project.
7. Expression Menu
Gene expression patterns can be searched via two routes on Xenbase, both under the Expression Menu. The first method is the “Expression Search” and the second method is the “Anatomy Search.” These two options take you to two different types of gene expression search, and can help answer different questions—a more detailed explanation follows.
7.1. Method 1: Expression Search
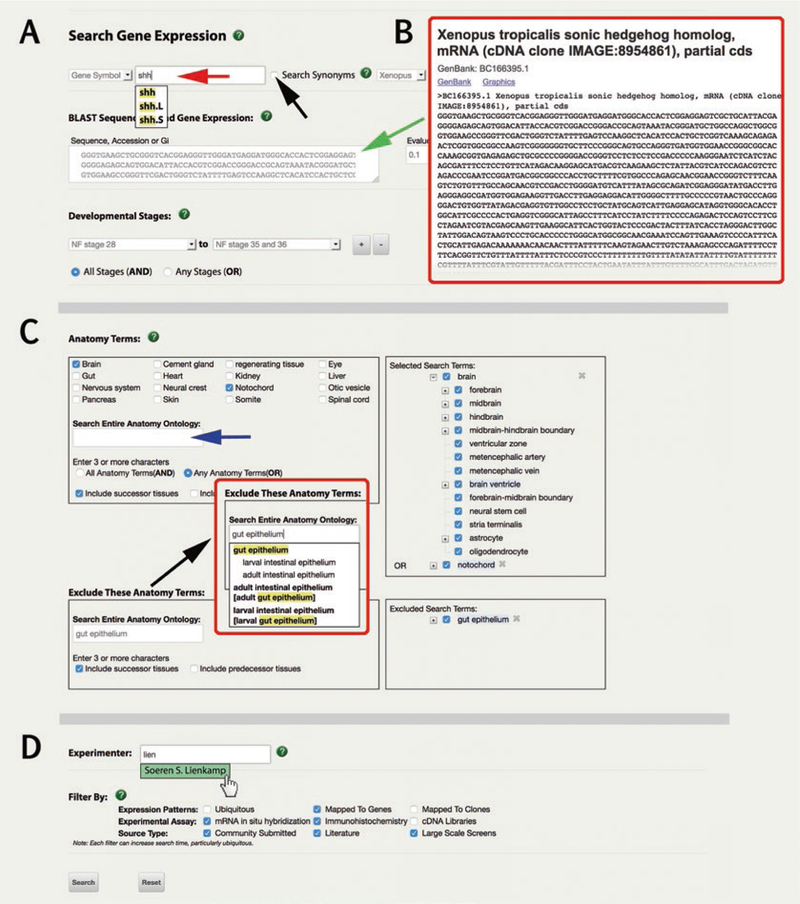
The “Search Gene Expression” interface offers numerous user-defined criteria to include/exclude data types, the goal of which is to filter a large catalog of images to answer specific or general questions. As such it can be used to find all examples of gene expression in a specific gene, tissue, or combinations of these, as well as a range of additional criteria. Put simply, there are three tiers of filters that can be selected. First, choose from variables such as species or embryonic stage (details in Subheading 7.1.1). Secondly, choose the anatomy terms to search (details in Subheading 7.1.2), and third, add optional filters based on experimenter (i.e., laboratory or researcher) or database associations (details in Subheading 7.1.3). An example of a three-tiered query might be for “genes expressed in the ‘pronephric duct’ in tadpole stages (NF stage 28 to NF stage 35 and 36) from the Lienkamp laboratory screen,” or “all genes expressed in the foregut progenitor tissues, but not the heart, in gastrula stage embryos.” Equally a query might focus on the unknown tissues, “where is shh expressed besides the notochord”?
7.1.1. Gene Expression Search Options
The top section of the “Search Gene Expression” interface (Fig. 6A, B) presents a set of options to effectively “filter” the annotated expression database. Fields include gene symbol, clone name or sequence, species, and/or developmental stage. If a gene symbol is entered, the search effectively “filters” all gene expression image data for that gene (which are also shown in the Expression tab of the Gene Page), using a user-defined set of criteria (e.g., all shh expression in X. tropicalis at NF stage 28), thus omitting nonapplicable and/or redundant and/or semiautomated curations, which are common to large scale screens. Note that not all fields need to be entered, and combinations are acceptable. The options include:
Enter a gene symbol in the top entry box (e.g., shh, Fig. 6A red arrow), a clone or Affymetrix ID (Optional).
Select the “Search Synonyms” box (black arrow Fig. 6A) so that legacy names will also be searched (i.e., xshh and vhh-1 are legacy gene symbols for the gene now called “sonic hedgehog” with the gene symbol “shh”) (Optional).
Specify either X. tropicalis or X. laevis from the menu box, or the default “Xenopus” returns all data (Optional).
In the expandable box, paste your sequence in FASTA format or simply provide a GenBank accession identifier (e.g., mRNA accession BC166395 for X. tropicalis shh; Fig. 6B). Set the E-value in the box to the right (the default is 0.1) (Optional).
Limit by developmental stage via drop-down menus to select a start and end embryonic stage range. Options include specific stages (e.g., NF stage 10.5) or general terms (e.g., blastula). Click the + and − buttons to add and remove stage(s). Use the “All Stages” and “Any Stages” radio buttons to select the Boolean operator for your search criteria (AND or OR, respectively) (Optional).
Continue to next section to choose additional filters, or scroll to the bottom of the page and hit “Search.”
Fig. 6.
Search Gene Expression via “Expression” menu. (A) Gene expression can be searched by entering a gene symbol (red arrow), or by entering a sequence in FASTA format into the dialog box (green arrow, see (B). Additional filters can include search synonyms (checkbox, black arrow) and choosing which species (X. laevis or X. tropicalis, the default is for both). (C) Anatomical terms (organs, tissues, or cell types) can be included (top box) and/or excluded (black arrow). Select XAO terms either by marking check boxes, or manually entering terms. Selected anatomy terms move right to the “Selected Search Terms” box. Toggle between child terms using + and − buttons, mark or unmark checkboxes to select/deselect terms (selected terms have tick in a small blue box). Choose “All Anatomy terms” (AND) or “Any Anatomy terms” (OR) functionality via radio buttons. Choose to “Include predecessor tissues” and/or “Include successor tissues” via checkboxes as needed. (D) The bottom fields include additional filter options including “Experimenter,” which autosuggests (highlighted in green) from the list of paper authors and Xenopus community members
7.1.2. Specifying Anatomy (XAO) Terms to Include and/or Exclude Organs/Tissues
The central section of the Search Gene Expression interface will set up a query of the database for expression patterns in specific organ(s), tissue(s), or cell types by using terms from the XAO. Queries can be submitted as follows:
A “free standing” XAO query (e.g., all records of expression in the “brain”).
A “combined” XAO query (e.g., all records of expression in “brain” AND/OR “notochord”).
An “include-exclude” XAO query (e.g., all records that include “brain” but exclude “notochord” (e.g., “brain” NOT “notochord”).
Any XAO query (option 1, or 2, or 3 above) in conjunction with 1 or more options chosen in the top section as described above (e.g., all shh expression in X. tropicalis at NF stages 28–34, expressed in “brain” AND/OR “notochord,” but NOT in “liver diverticulum”).
Select XAO terms as follows:
Choose from a set of 16 common anatomy terms, available as checkboxes. In the example in Fig. 6C, “brain” and “notochord” have been checked, and they automatically move to the “Selected Search Terms” box to the right.
Enter anatomy term(s) (using three or more characters) using the “Search Entire Anatomy Ontology” suggestion box (Fig. 6C, blue arrow). All terms that match your text will autofill in bold below, with matched text highlighted in yellow. Synonyms of an XAO term appear in square brackets. Mouse down to select a term from this menu to add it to the list of search terms (Fig. 6C).
The XAO sub-parts of a term can be expanded by clicking the [ + ] icon (e.g., “brain” has parts including “hindbrain,” “mid-brain,” and “forebrain”). By default, all subparts are checked, but users can exclude any from the search by unchecking them.
To exclude an anatomy term(s) enter your “excluded” term(s) in the section marked “Exclude These Anatomy Terms” in the same manner as those in the included terms box above (e.g., “gut epithelium” is excluded in Fig. 6C).
Choose to “Include predecessor tissues” and/or “Include successor tissues” via checkboxes as needed. This option applies to embryonic anlage terms, such as “anterior neural tube,” which develops_into successor tissues, “brain” that has_parts, “hindbrain,” “midbrain,” and “forebrain”.
Continue to next section to choose additional filters, or scroll to the bottom of the page and hit “Search.”
7.1.3. Specifying “Experimenter” and Using “Filter By” Options
Specifying “Experimenter” and using “Filter By” options are the third tier of filters for a gene expression query (Fig. 6D). This is an excellent way to find all, or a subset of, the images from large data sets that Xenbase hosts. These large data sets include an angiogen-esis screen (Patient Lab, see [18]), retinal marker screen (Perron Lab and Pollet Lab, see [19]), pronephric marker screens (Brandli Lab, see [20]; Lienkamp lab, see [21]); MO-synphenotype screen (Smith Lab, see [22]), XenMARKimages [23]; or ISH images for clones supplied by the European Xenopus Resource Centre (EXRC), among others. To find images from published literature and community submissions:
Enter a full, or partial, first or last name of a researcher or author in the “Experimenter” field. Use cursor to select a name from the autofilled options.
- Additional advanced filtering options to either reduce or increase the number of results can be selected via checkboxes as follows:
- Expression patterns: Ubiquitous (i.e., annotated with five or more tissues); Mapped to Genes, or Mapped to Clones.
- Experimental Assay type: ISH, IHC or cDNA libraries.
- Source types: Community submitted, Literature, or “Large Scale Screens” (e.g., defined from cDNA libraries).
Scroll to the bottom of the page and hit “Search.”
7.1.4. Navigating the Gene Expression Search Results
Here we show two examples of gene expression queries and guide the user through features of the results tables. The first example, shown in Fig. 7A, is gene expression for the gene “sonic hedgehog” (shh), which is an early marker of notochord, but is later expressed in the foregut. Here we combine a gene symbol and an XAO term in a query. We checked the upper level XAO term “gut,” which is a synonym for “alimentary system,” to include all parts of the gut from embryo to adult frog stages without stage restriction. We choose AND, but deselected predecessor and successor tissue. Figure 7A shows a subset of the returns, with the source (e.g., citations from literature or community submitted data) to left, then species, a thumbnail of the data image, NF stages, and the XAO terms annotated (and thus matched) to the image to the right. In a second example Gene expression query, the Experimenter “Lienkamp” returns all annotated images from 52 genes in a screen for pronephric markers submitted and published from this researcher (Fig. 7B; [21]). We then used “modify search” (black arrow, Fig. 7B) to further filter returned images from this screen, by choosing more specific terms that are part_of the pronephric kidney, such as the “pronephric duct” and/or “early distal tubule” (Fig. 7C).
Fig. 7.
Gene expression search results. (A) Gene expression query output for the gene “sonic hedgehog” (shh) and the XAO term “alimentary system.” A subset of the returns, with the experimental source (e.g., citations from literature or community submitted data) to left, then species, a thumbnail of the data image, NF stages and the XAO terms annotated (and thus matched) to that image to the right. Click on the image to enlarge it and view annotation table. Click on the source to open the Article or Lab page. (B) Gene expression query output for XAO term “pronephric kidney” plus the Experimenter “Lienkamp,” returns all annotated images from a screen for pronephric markers, plus any images from publications with this author (not shown). Images can be filtered for more specific terms using the “Modify Search” button (black arrow). (C) Adding the anatomy term “pronephric duct” filters the results (shown in B) to a smaller, annotated set of images from the “Lienkamp” laboratory
7.1.5. Notes/Troubleshooting the Gene Expression Search
Hitting the return key on your keyboard will not execute this search—always click the “Search” button—bottom left-hand corner of the screen.
Select either a gene symbol or enter a sequence: entering both will give an error.
If you get no results, try again, with or without changing a few parameters, as sometimes the search times out.
Use the “Modify Search” button to return to the search interface, to expand or reduce returned results.
Follow the “Too many results?” or “Too few results?” links for more advice on how to refine your gene expression search.
The gene expression search is a complex set of algorithms with numerous variables: as such it is particularly temperamental, and can take several iterations of options to find the data you are looking for.
Contact Xenbase (xenbase@ucalgary.ca) to report bugs if you think the search is broken.
7.2. Method 2: Gene Expression via Anatomy Search
What are the best markers for cardiac mesoderm?
Which genes are known to be expressed on the migrating neural crest cells?
Are there any clones/plasmids available for this gene?
Searching for gene expression in a specific anatomical feature is an especially useful query to find both standard and novel molecular markers for a tissue/organ via the image catalog; a comprehensive list of all genes observed to be expressed in a tissue; available clones for that marker; or the body of literature that contains gene expression data for a specific cell type, tissue or organ. This is a simple two-step process. Firstly, find the XAO page for the tissue or organ (see Subheading 7.2.1 below), then secondly, click the “Expression” tab for that term. Details of how to assess the results table from this page are given in Subheading 7.2.2 below.
7.2.1. Finding the XAO Term Page
Under the “Expression” menu, choose the “Anatomy Search” option to arrive at the term search function in the XAO module.
Enter for the specific anatomy term (e.g., “heart,” “migrating neural crest cell,” or “intermediate mesoderm”) in the dialog box. The matched text will autofill: matches are highlighted in yellow, synonyms are in square brackets. Menu options alsoinclude XAO ID numbers (e.g., heart, XAO ID: 0000064) or anatomy page number (e.g., heart, XB-ANAT-63).
Hit Search (or Browse All).
Use the + and − buttons in the navigable view of the entire XAO, located to the right of the page (Optional).
Use Nieuwkoop & Faber (NF) stage restrictions from dropdown menus, using either broad categories (e.g., “early tailbud stage” to “tadpole stage”) or precise stages (e.g., NF stage 20 to NF stage 28) to focus results (Optional).
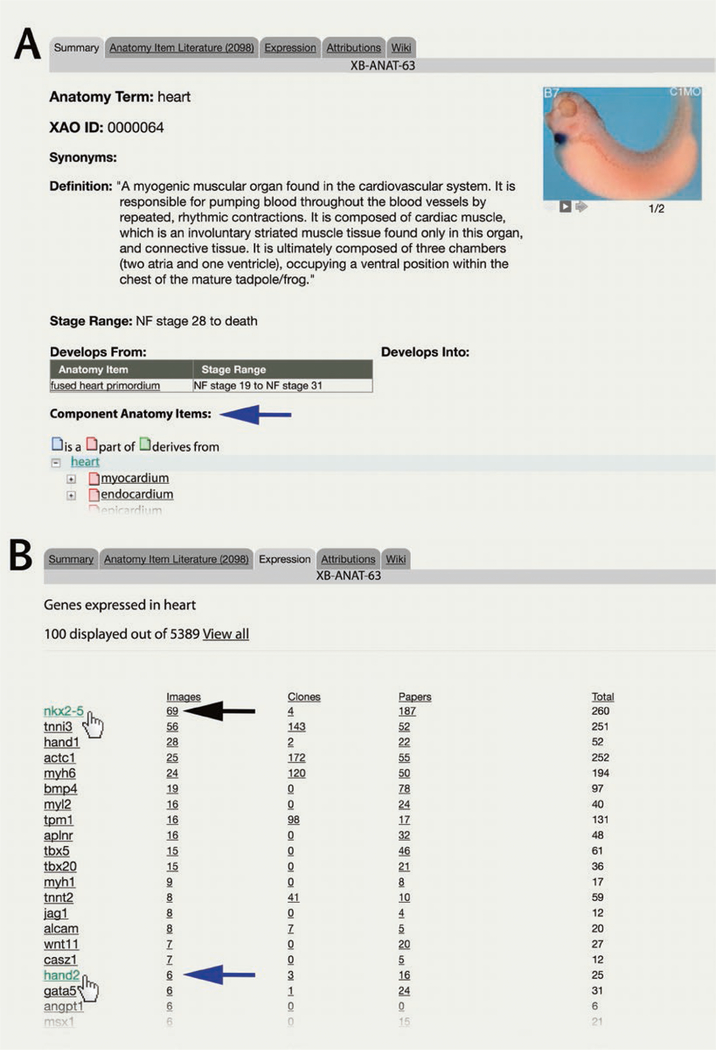
Multiple matches (e.g., “heart,” “primary heart field,” “left lymph heart”) are displayed in a table. Click term name to go to the XAO term page (Fig. 8A). Single results go directly to the XAO term which has an “Expression” tab, just like a Gene Page.
Click “Expression” tab to display gene expression for this specific XAO term.
Fig. 8.
Using the Anatomy Search to explore gene expression, and the XAO. Here we use the XAO module to ask “Which genes are expressed in the heart?” (A) Each XAO term page gives the term definition, NF stage restrictions for its use and relationships to other terms (see “Component Anatomy Items,” blue arrow). (B) From the XAO term page for heart click the “Expression” tab. The first few results (of top 100) are shown, resorted by clicking “images” to be ranked in descending order by number of images (e.g., nkx2–5 69 images, tnni3, 56 images, and hand1, 28 images etc.) with data from clones, papers and total columns on the left. Lower down the column lesser known genes with heart expression are shown (e.g., hand2, 6 images). Mouse over (hand cursor) to select
7.2.2. Assessing Results Table on “Expression” Tab of an XAO Term
The genes annotated as having expression in the XAO term appear in a table, with gene symbols to the left, and associated data types for that gene organized in four columns: Images, Clones, Papers (i.e., articles/literature), then a combined Total count of records (see Fig. 8B). Each column can be sorted in descending order by clicking the column tile. All table entries are underlined indicating they are live links to further data.
Click the View All link to open the entire list of genes matched to the XAO term.
Click “Images” column header to find the top marker genes.
Click the gene symbol (e.g., nkx2–5 or hand2) to open that Gene Page (hand cursors, Fig. 8B).
Click the number of images available for a gene to see annotated gene expression images matching the XAO term.
Click the number of clones to execute a query for clones for that gene.
Click the number of papers to see a full literature list associated with the XAO term and the gene of interest.
Here we use the XAO module to explore gene expression in “heart” (Fig. 8A, XAO page XB-ANAT-63, for “heart,” XAO I:0000064). After selecting the “Expression” tab, the top 100 results for “genes expressed in heart” are shown from 5300+ records on the first page of the results. Genes expressed in “heart” are ranked in descending order by total count of data records), and here have been reordered by “Images” (black arrow, Fig. 8B): nkx2–5 has 69 images, tnni3 has 56 images, hand1 has 28 images, etc. Further down the column, more, but less well-studied, genes with heart expression can be assessed (e.g., hand2, 6 images) (blue arrow, Fig. 8B).
7.2.3. Notes/Troubleshooting the Anatomy Search for Gene Expression
Additional routes get to this feature: from the home page “Gene expression” tile/“Anatomy Search” link, or from the “Anatomy and Development” tile, choose “XAO,” then the “Search Anatomy” tab.
Adult tissue terms (e.g., “bladder”) and many cell types (e.g., “cementoblast”) have few gene expression annotations.
“Attributions” on this page is an attribution to the definition of the XAO term.
A Wiki is provided to record notes not recorded elsewhere on Xenbase.
If no data is available for a particular class of data (i.e., no clones) clicking on the zero will execute a query for clones, but will show no results.
For higher level ontology terms, such as “heart,” data returned using this search includes matches for predecessor and successor tissues (e.g., “cardiac mesoderm” and “endocardial tube”). Use the Expression search (described above, Subheading 7.1) to exclude these results.
There are three additional expression data sets under the “Expression” Menu. These are:
7.3. miRNA Catalog
MicroRNAs (miRNAs) are small, noncoding RNAs that play a role in regulating gene expression [24, 25]. The data in the miRNA Catalog contains miRNA in situ expression in Xenopus embryos that was submitted by courtesy of the Wheeler Laboratory [24] and XenMARK [25]. The miRNAs have been correlated with Xenopus records in miRBase to provide more information. Click the miRNA links (e.g., xtr-miR-133a) to view more information about the miRNA, including in situ images.
7.4. Expression Data at GEO
Select this menu option to run a preset search for Xenopus NGS data sets through the NCBI Gene Expression Omnibus (GEO) database.
7.5. RNA-Seq Data at the NCBISRA
Select this menu option to run a preset search for Xenopus sequence data through the NCBI Sequence Read Archive (SRA) database.
8. Anatomy and Development Menu
The Anatomy and Development section of Xenbase covers a wide range of reference material used by researchers and students. The following headings can be selected via either drop-down menu or home page tile. A brief description of the content available under each subject follows.
8.1. Organ Atlas
The organ systems in Xenopus are illustrated here with a variety of imaging methodologies, including confocal microscopy. Currently, the organ atlas covers only heart and pronephric kidney development, and is undergoing a significant expansion to cover more organ systems in the future (e.g., cranial cartilages from the XenHead project [26], the nervous system and musclular skeletal system).
8.2. NF Developmental Stages
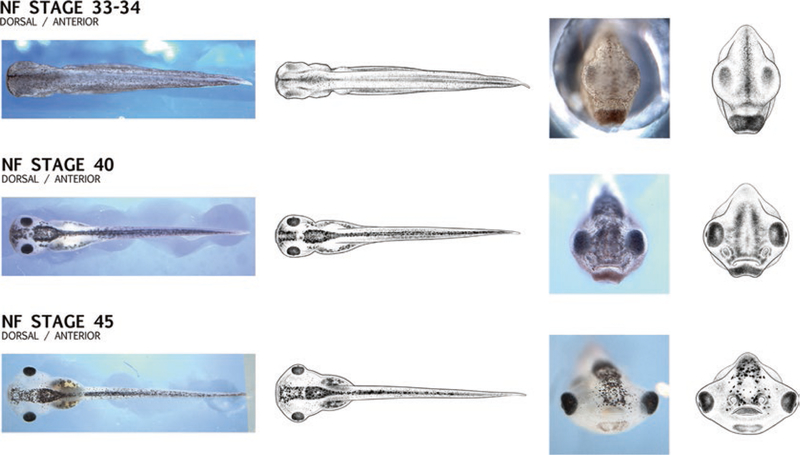
A complete Xenopus laevis stage series (NF stage 1–NF stage 66) [http://www.xenbase.org/anatomy/alldev.do] based on Nieuwkoop and Faber [27] illustrations are shown. A new developmental stage series, the Zahn drawings, and complementary bright field photographs, all of which are open access, posted here on Xenbase [26]. The Zahn drawings can be downloaded and used in the laboratory setting to illustrate gene expression domains, pheno-types, and other changing patterns during normal and abnormal development, and can be reused under the creative commons license under which they will be published. Examples of the new images, which include anterior, dorsal, and ventral views, perspectives not included in Nieuwkoop and Faber [27], are shown in Fig. 9.
Fig. 9.
New developmental series illustrations: Zahn series. The newly published, open access, Zahn drawings will be posted on Xenbase under the Anatomy and Development menu. This developmental stage series for Xenopus, based on multiple individuals, includes views that have not been previously published (e.g., dorsal and anterior as show here) as well as ventral views (not shown). The drawings call attention to morphological changes during critical stages of organogenesis, with a focus on changes in the shape and size of the head as seen in the images here demonstrating changes through NF stages 33 and 34, NF stage 40 and NF stage 45. The image series also includes bright field photographs (left) to compare with drawings (right). Images reproduced here are open access, and appear in Zahn, Levin and Spencer Adams, (2017) Development.
8.3. Images of Xenopus Embryos
These image files are in the Wiki, and are generally whole-mount microscopy, illustrating each developmental stage as a researcher would see the live embryo.
8.4. Development Stage/Temperature Charts
The rate of Xenopus development is influenced by temperature, and although X. laevis and X. tropicalis embryos develop at similar rates, X. tropicalis tolerate a narrower range of temperatures [14]. The charts provide a standard reference with which to plan experiments and were supplied by the Khokha Laboratory, Yale University.
8.5. Movies of Xenopus Development
High quality movies of the developing Xenopus embryos are pro vided as educational resources, covering key developmental pro cesses including cleavage, gastrulation and neurulation, and the synchronous development of Xenopus laevis embryos during early embryogenesis.
8.6. Cell Fate Maps
Cell fate is illustrated with mouse-over animations in forward direction (blastomere-to-tissue) from NF stage 5 (16-cell) to NFstage 10.5 (beginning of gastrulation) (Fig. 10A), and reverse direction (tissue-to-blastomere) (Fig. 10B), based on the classic studies by Moody [28, 29], and Bauer et al. [30]. To use these dynamic fate maps, simply move the cursor over the blastomere to highlight which cells in later stage embryos are derived from the 16-cell and 32-cell stage blastomeres. The 16-cell blastomeres and their descendants appear in orange (upper panels), while 32-cell descendants will appear in blue (lower panels). Due to the two-dimensional nature of the illustrations, and that NF stage 8 and NF stage 10.5 embryos are shown as sections, only some derivatives of the blastomeres of the NF stage 8 (32-cell) embryo show up in blue on later stage figures. In the reverse fate maps (Fig. 10B), move the cursor over an anatomy term to highlight blastomeres that make major contribution (in red), a minor contribution (in green) or rarely contribute (in orange) cells to the adult tissue.
Fig. 10.
Dynamic cell fate maps by Xenbase, based on classic studies by Moody [28, 29] and Bauer et al. [30]. (A) Cell fate in a forward direction, from blastomere to tissue. To use these animations, move the cursor over a blastomere, and the cells in later developmental stages are highlighted for NF stage 5 (16-cell) in orange, and NF stage 6 (32-cell) embryos in blue. Click on any blastomere to see its derivatives. (B) Cell fate in the reverse direction (i.e., tissue from blastomere). To use this tool, mouse over an anatomy term (e.g., “cement gland”), from a primary germ layer category (e.g., ectoderm, neurectoderm, mesoderm, or endoderm) to highlight the blastomeres that contribute to these tissues. Color coding indicates the degree of contribution from the NF stage 6 (32-cell) embryo: major (red), minor (green), or rarely incorporates cells (orange)
8.7. The Xenopus Anatomy Ontology
The Xenopus Anatomy Ontology, aka the XAO, is a comprehensive set of anatomical terms that describe the entire course of development and organogenesis in Xenopus from unfertilized egg to the adult frog [31]. The XAO forms the backbone of our gene expression curation and is updated frequently in response to the latest research and community input. The goal of the XAO is to describe all anatomical structures in a formal language hierarchy, with each term being defined and related to other terms. XAO terms have “is_a”, “part_of”, “develops_from”, and “develops_ into” relationships, as well as specific developmental timing boundaries, using NF stages [27], such that each term has “starts_during” and “ends_during” stage relationships (see [31]). Cross referencing the XAO to mouse and human phenotype ontologies will ensure the interoperability of Xenbase phenotype annotation (new feature to be launched on Xenbase), with human disease phenotype.
8.7.1. Downloading the XAO
The latest XAO (v5 released January 2017) is available for download from Xenbase, in either OWL or OBO formats, and from the Open Biomedical Ontologies site (OBO Foundry).
8.7.2. Requesting New XAO Terms
Request new XAO terms via GitHub (a log-in is required). New term requests require a definition and additional supporting information about relationships to other terms, developmental timing, cross-references to other ontologies (e.g., Uberon or ZFA), and literature reference (s). Submissions suggesting many new XAO terms, can be made as a file attachment through the GitHub portal.
8.7.3. Illustating the XAO
Xenbase is currently working to illustrate XAO terms and developmental stages with exemplary figures from anatomical dissections, histology, whole mount microscopy, textbook figures and/or with marker gene expression. Images will appear on the XAO term page. Figure 8A illustrates the XAO page for “heart,” showing its definition, relationships, and place in the hierarchy of other ontology terms and a key marker gene as an in situ hybridization. We will post a variety of images including gene expression, dissections and histology to illustrate XAO terms. Note that ontology phrases can be read in both forward and reverse order in the hierarchy, for example “endocardium” is part_of “heart,” while “heart” has_parts “endocardium” is also true. Contact Xenbase (xenbase@ucal-gary.ca) to suggest or submit images to illustrate the XAO term pages and/or for the anatomy atlas.
8.8. Notes/Troubleshooting
Anomalies in the time temperature charts have been reported by some researchers and this table is currently under revision. Contact Xenbase (xenbase@ucalgary.ca) with any questions.
9. Reagents and Protocols Menu
Access the following modules and catalogs under the “Reagents & Protocols” menu of the main website banner or the tile on the home page.
9.1. CRISPr and TALEN Support
New genome editing technologies work well in Xenopus [32, 33]. CRISPr/Cas and TALEN/ZFN editing technologies function by inducing site-specific DNA strand breaks anywhere in the Xenopus genome. Mutations are induced by inefficient, error-prone nonhomologous end joining (NHEJ). In addition, site-specific DNA breaks promote precise knockin, homologous recombination (HR) gene editing. This module provides a review of the techniques with links to Xenopus literature, protocol guides, and other resources for CRISPr and TALENS.
9.2. Antibodies
Antibodies used in Xenopus research are curated from published articles, and the Xenbase antibody catalog has over 1200 entries (at time of press). Xenbase antibodies are named from either the antigen/gene symbol or tissue (where antigen is unknown), in the order they exit our curation pipeline, not the order in which they are published. Antibodies may be searched by common name (e.g., Xlim-1), synonym (e.g., WGA), catalog number (e.g., 3G8), Xenbase name (e.g., Kidney Ab2), antigen gene symbol (e.g., lhx1), or anatomy/XAO terms (e.g., kidney or visual system). Antibody pages contain relevant experimental information including host source, antigen, posttranslational modifications, cross-species interactivity, RRID (Research Resource Identifiers), and a list of experimental applications with confirmed utility in Xenopus research (see Fig. 11). Additional tabs give information on “Attributions,” a “Wiki” for additional notes, images in Xenopus (when available) and links to commercial sources for the antibody.
Fig. 11.
Xenbase Antibody catalog is accessed under the Reagents & Protocols menu/tile. (A) To find antibody entries, use the “Search All” (default) by entering antigen gene symbol, catalog number, or common name (red arrow). Select an antibody from the results table (e.g., Sox2 Ab1, black arrow) to open the antibody entry. (B) Each Antibody page includes Xenbase name, common name, source and catalog number, an image illustrating reactivity plus details such as tissue-specific expression (XAO terms). (C) Properties including validated activity and citation (including RRID numbers) are recorded when available (blue arrow). (D) Immunogen details and, (E) “Reported Usage” (e.g., western blot, immunofluorescence; orange arrow) are recorded. (F) Publications using the antibody are listed as “First” and “Most recent,” with a “View All Papers” option, which is reiterated on the “Attributions” tab
Notes/Troubleshooting Antibodies
Start with “Search All” (default setting) to cover all options.
Use browser back button to return from the Wiki pages.
Contact Xenbase (xenbase@ucalgary.ca) to submit images and additional usage data for validated antibodies.
Text box does not autofill from XAO terms or gene symbols.
GO terms may also be matched to Antibody entries.
9.3. Morpholinos
Morpholinos (MOs) are chemically modified oligonucleotides used to reduce the expression of a gene of interest [34, 35]. MOs knockdown gene expression by inhibiting mRNA translation, blocking RNA splicing, or inhibiting miRNA activity and maturation [34]. MOs have been shown to be effective in both X. laevis and X. tropicalis [36] and are widely used in experimental Xenopus embryology. Xenbase has manually curated 2400+ published Xenopus-specific MOs. How to use the MO search interface, and an example MO entry (e.g., sox2 MO1) is illustrated in Fig. 12. Search for published MOs through the interface, via the MO name or target gene symbol (e.g., sox2), a MO sequence, or use the “Alphabetic Search” (Fig. 12A). Note that the search for “sox2” also returns MOs for “sox21” (blue arrows, Fig. 12A). Each MO is assigned a unique name based on the targeted mRNA/gene (Fig. 12B), and we record any synonyms, the 5′ to 3′ sequence of the oligonucleotide, and whether it is designed to be splice-blocking or translation-blocking. We BLAST the MO sequence to identify on-target and off-target hits (Fig. 12C), and display the MO’s scaffold position in a GBrowse snapshot of each MO page (Fig. 12B), as well as give scaffold positions orange arrow, Fig. 12C). All curated MOs are mapped to the genome and are displayed in the full genome view on GBrowse. Note that MOs are listed on Gene Pages under “Reagents,” and are also displayed on associated Article Page(s), the latter being accessible under “publications” section (Fig. 12D).
Fig. 12.
Xenbase morpholino catalog is accessed via the “Search Morpholino” interface, under the Reagents & Protocols menu/tile. (A) Enter a gene symbol or MO sequence (red arrow) and click the Search button. Select the MO (black arrow), from the results table to open the MO page. (B) Each MO page includes all recorded details and a GBrowse snapshot showing the aligned position on the MO to the target mRNA. (C) Genomic alignments illustrate on-target (green) and off-target (pink) hits for X. tropicalis, X. laevis L and X. laevis S. Select a scaffold (highlighted in green) to view in GBrowse (orange arrow). (D) Publications using the same MO are listed, with a “View All Papers” option, which is reiterated on the ‘Attributions’ tab
Our catalog of MOs can be searched under the Reagents and Protocols menu via the Search Morpholinos using the steps:
- Choose search options from drop-down menu and enter text in the search field (red arrow, Fig. 12A). Options include:
- Target gene/mRNA symbol (e.g., sox2).
- MO name (e.g., sox2 MO2).
- MO synonym, as used in a publication(s) (e.g., MO-sox2).
- MO sequence (e.g., AGCTCGGTCTCCATCATGCT GTAC).
Hit Search button.
A single search hit will go straight to that MO page (e.g., sox2 MO2: XB-MORPHOLINO-17250375). Multiple search hits (such as resulting from a search for “sox2”) will be displayed in a table (Fig. 12A).
Click the Xenbase MO name from left hand column to examine details on the MO page (black arrow, Fig. 12A).
Click the GBrowse snapshot (Fig. 12B) or the specific scaffold position (orange arrow, Fig. 12C) to view MOs in the full genome browser tool,
Use browser back button to return to the MO page or search results table.
Notes/Troubleshooting MOs
An “Alphabetic Search” is also enabled on the MO search interface.
MOs can also be found using the Quick Search Menu, using the Xenbase accession number option (e.g., XB-MOR PHOLINO-17249151).
Nucleotide searches must be exact matches to find a specific MO, and exclude the 5′ prefix and 3′ suffix.
“Browse All” will provide an alphanumeric list of the entire MO catalog of 2400+ entries, so using at least a gene symbol or synonym will help finding a specific record.
Click on the GBrowse snapshot to view positional information or search for off-target interactions.
Xenbase curates MOs (and numbers them in serial sequence) in the order in which they exit our curation pipeline, not in order of publication.
Gene symbol search uses text-matching, so that a search for MOs to the gene symbol “apln” will return MOs for apln, aplnr, and hapln3.
Phenotypes generated using a specific MO will be posted on each MO Page as well as on the Article Page, when Phenotypes are launched on Xenbase.
9.4. ORFeome
The Xenopus ORFeome project generated a comprehensive set of 8600+ full-length, end-sequence validated, high quality open reading frame clones in the Gateway cloning system, suitable for recombinant protein expression [37]. ORF sequences represent 7800+ unique genes, including 2724 genes with human ortholog disease association (e.g., optn ORF1, associated with glaucoma (OMIM #137760)). In total the ORFeome clones represent approximately 40% of the nonredundant X. laevis genome [37]. ORFeome reagents allow high-throughput in vivo functional-genomic screening of frog genes in a manner previously not feasible. Details of each ORF clone are given on individual “ORF Page” including gene symbol, gene name, Entrez ID, 5′ and 3′ sequence, predicted translation, confidence estimates, and links to suppliers.
Notes/Troubleshooting ORFeome clones
Xenbase ORF Page IDs in the form “XB-ORF-#” (e.g., XB-ORF-17287509), can be used on the ORF search page or from the quick search menu using Xenbase Accession option.
Xenbase does not supply ORFeome clones. Researchers must contact the supplier(s) directly.
9.5. Small Molecules Wiki
Useful small molecules are manually catalogued from published literature in a Wiki format. Entries are listed under the following headings:
Alphabetic list with literature references.
Small Molecules affecting Pathways: e.g., Retinoic Acid: Citral; or Hedgehog: cyclopamine.
Small Molecules affecting Biological Functions and Processes: e.g., angiogenesis: suramin, apoptosis: cyclohexaminde, or neurotransmission: diazepam.
Drugs by class: e.g., kinases: KT5720, a specific, cell-permeable inhibitor of protein kinase A (PKA).
Notes/Troubleshooting Small Molecules Wiki
Registered users can add to this Wiki.
Minimal information required includes a description, genes/pathways/functions affected, source/supplier, reference(s), as well as a structural diagram (such as those available in PubChem).
Click the Textpresso link at the bottom of a Wiki entry, to run a search for the term in Xenopus articles.
9.6. Protocols Wiki
Entries are listed under the following headings:
Books for Xenopus Research and Protocols
- Online Resources
- Journal of Visualized Experiments (JOVE) video demon-strations, showing protocols and techniques (e.g., host transfer methods of oocyte fertilization; dissections of retinal tissue; electroporation; live-cell imaging for quantitative analysis; and patch clamp and perfusion techniques).
- Cold Spring Harbor (CSH) Xenopus Protocols.
General Research Protocols covering Animal Husbandry, Lab solution recipes and reagents, Generating Embryos, Transgenesis, in situ Hybridization, Immunohistochemistry, ChIP-Seq protocols, Histology, Embryo Staining Protocols, Immunohistochemistry and Protein Protocols, Nucleic Acid Protocols, Oocyte Transfer Technique, Xenopus Oocyte and Egg Extracts, and Xenopus Tissue Culture.
Notes/Troubleshooting Protocols Wiki
Reference books, text-books and chapters without PubMed IDs cannot be added to the literature module in Xenbase.
Both JOVE and CSH Xenopus Protocols require institutional licensing to access.
Registered Xenbase users can add to the Protocols Wiki.
Protocols are submitted to Xenbase by specific laboratories, as indicated, and researchers should contact the lab directly (via Xenbase Laboratory profile) to troubleshoot the specific protocol. Contact Xenbase if there are errors or updates in the protocol as published.
9.7. Search Clones
The clone catalog in Xenbase houses data on over one million plasmids developed from both X. laevis and X. tropicalis. Access the clone search interface via Reagents and Protocols menu, and choose from search options: search all, gene symbol (e.g., aldh1a2.L), NCBI/GenBank accession number, clone name (e.g., IMAGE:3421129), source tissue (e.g., cornea, gonad, or oocyte), clone page ID (e.g., XB-CLONE-242125). Each Clone Page gives the clone name, the gene to which it is mapped, and source species, as well as sequence data, Unigene accession number, source/external databases, a description of tissue or embryonic stage from which it was generated, the vector details, and a vector map.
Notes/Troubleshooting Clones
Wildcard * can be used to search clones.
Use the checkbox to filter clones only available from the EXRC.
Alphabetic search is for clone name, not gene symbol (these often do not match).
9.8. Clone Libraries
Several cDNA libraries were used to generate the Xenopus ESTs, many of which are from the Xenopus Gene Collection Library. The Library name and tissue used are listed here for X. tropicalis and X. laevis.
9.9. Vectors
Select this menu option to see a list of standard vectors used in Xenopus research. Click the vector name (e.g., pCS107) to view the plasmid and phagemid Vector Page, with map and supplier details. Use the + toggle to see the sequence if available.
9.10. Obtain Frogs
This table gives contact details of Stock Centers, commercial sup pliers of frogs, oocytes, wet lab and aquarium equipment.
10. Literature Menu
The Xenbase literature module houses an extensive catalog of 49,000+ Xenopus research papers that are available from NCBI’s PubMed service, 90% of which are searchable by Textpresso, and a books catalog. Articles are automatically uploaded each week by text-matching “Xenopus” or “Silurana” in the title, abstract or keywords of newly released papers. Latest Xenopus research articles added to Xenbase are listed at the top of the Announcements column on the home page. Each article is represented on an “Article Page” (details below in Subheading 10.1), a recent example ofwhich is shown in Fig. 13. Articles can be searched via the quick search menu (discussed above, Subheading 4), and via the “Literature” menu/tile link, by entering an author name, partial or full paper title, or using a Xenbase accession number (e.g., XB-ART-45000).
Fig. 13.
Publications in the Xenbase Literature module are represented on an “Article Page.” We assign a database accession number (e.g., XB-ART-51882) and pull the full abstract and associated data from PubMed. Authors with Xenbase profiles have their name underlined (red arrow), indicating a link to their personal profile page. The abstract is automatically text-matched for gene names, gene symbols, and XAO terms (underlined). Direct links are provided for PubMed, PMC, and the Journal entries for the article (upper red box), enabling quick access to the full article and PDFs. Genes referenced in the article are either mentioned in abstract or added manually by curators, as are antibodies and morpholinos (lower red box). Matched and curated terms are underlined and linked to Gene Page(s), XAO, AB, and MO pages, respectively. Figures from the article (if available) are shown as thumbprints, and references cited in the paper follow. Use the [+] to expand to show captions (black arrow) or the full reference list. Double click the image to open the larger figure and the annotation table. Additionally, links to OMIM diseases and GO terms reverenced in the research, and to raw or supplementary data (e.g., NCBI/GEO and DRYAD) are shown when available. Images with new gene expression can be selected from figures and be placed as ‘summary images’ on a Gene Page (green arrow)
10.1. Article Pages
An Article Page header includes the Xenbase accession number (sequentially numbered as they are added to the database), and each article page carries the following information:
Reference: Journal, Title and Authors appear at the top of each entry, followed by a full Abstract, PubMed ID and PMC ID. PubMed/PMC links redirect to the article record at those resources.
Article link goes directly to journal website. If the article link is missing, click the PubMed Link to access the journal website. This may require a subscription.
Grant support gives details of sources of funding (when available), with a link to the funding website.
Genes referenced in the article come from both text matching and manual curation.
Antibodies referenced lists the curated primary antibodies used in the research.
Morpholinos referenced lists the curated morpholinos used in the research.
References cited in the article are listed here, use the [+] to expand. Links to the PubMed record and/or the Xenbase Article Page for the references are provided.
Resources URL (when shown) provides links to external databases where raw or supplementary data files are deposited, such as NCBI/GEO or DRYAD (e.g., XB-ART-42630 with micro-array data at NCBI/GEO). Not all article have this link.
Article Images and their captions are posted for open access articles and for those journals with whom Xenbase has negotiated permission to redisplay figures. Use the [+] toggle to see full captions. These images may be subject to copyright, and if so, this is indicated.
10.2. Notes/Troubleshooting Article Pages
Xenbase only curates gene expression data from images that we can display (i.e., when open access or redisplay permission has been obtained from the copyright holder).
Two new data links will soon be added to the Article Page: ORFeome clones and curated Phenotypes described in the article.
Use the “thumbs up” or “thumbs down” icons to “vote” on image/data quality.
Articles can only be uploaded via PubMed ID.
Articles that do not cite “Xenopus,” and use alternate terms such as “vertebrate” or “amphibian” instead, may be missed by the loader, yet can be manually uploaded via PubMed ID by Xenbase staff and registered users. To do this, log in, click the “Literature/papers” menu heading, then the “Click here to manually add an article by PubMed ID” link.
Books and book chapters are entered separately under the Literature/Books menu and can be searched via Title, Author, ISBN number, publisher, or Xenbase accession (e.g., XB-BOOK-202).
10.3. Textpresso
Advanced querying of the full text of most Xenopus papers can be achieved by using a Xenbase-specific instance of Textpresso, an information extraction and processing software package for scientific literature developed by the California Institute of Technology, and used under license on Xenbase. Simple keyword searches can be entered in a free text field, and/or up to five category terms can be chosen from a preset list which includes higher level GO terms, experimental techniques and relational terms. The most advanced and flexible option for a Textpresso search is the “Query Language” which is available toward the top of the page, in the menu under the Textpresso logo. This tool allows the user to assemble sets of commands to build complex queries, and allows users not only to specify a variety of query fields, categories, and keywords, but also to set specific thresholds for the number of instances of those key-words and combine previously defined searches using Boolean operators (i.e., AND, OR, NOT).
Notes/Troubleshooting Textpresso
The user guide for Textpresso can be found here on Xenbase: http://www.xenbase.org/cgi-bin/textpresso/xenopus/user_guide
The “Advanced Search” allows users to specify which sections of the paper should be searched. Select “on” next to the “Advanced search options” text.
“Body” option is used for papers that could not be properly sectioned into the other elements. “Scope” option allows a choice between “sentence,” “document,” and “field.” “Sort by” option offers a variety of criteria including year of publication, number of citations, title, author, or score.
Option available to exclude supplementary data and/or abstracts.
Filter by “Author,” “Journal,” “Year,” or “Doc ID” (i.e., PubMed ID).
Too many results? Exclude terms using filter option by entering “+” or “−” signs in front of terms. Enter the field type (e.g., author or title) in square brackets and phrases in double quotes (e.g., “+Patel-Zheng[author]”). Click on the “Filter!” button to activate.
Too few results? Broaden the scope by allowing the search to include synonyms for your keywords or by increasing the sections of the literature searched, including “unsectioned” manuscripts.
11. Community Support
Xenbase has several features designed to facilitate and encourage communication within the Xenopus research community. This includes an extensive catalog of individual Xenopus researchers, research labs, institutes such as Universities, governmental and nongovernmental organizations, funding bodies, publishers, and companies. Xenbase also provides announcements for upcoming meetings relevant to Xenopus research, job postings, the “Xine” community newsletter, Xenbase forums and links to a variety of relevant professional societies.
11.1. Find Researcher, Lab, and Organization Profiles
Register with Xenbase to make a personal and/or lab profile page, with contact details and a description of your research and link papers, which are shown on the “Publications” tab. Click the “Register” link in the top right corner of the site and follow prompts to record your name and contact information, and log-in details. Lab Pages are associated with individual profiles of the Principle Investigator as well as Lab members (e.g., postdocs, graduate students). Find a People, Lab, or Organization profile (e.g., a Xenopus supplier or stock center), by clicking the Community Menu/Tile to the search interface. Search via researcher name, institution or research interests.
11.2. Job Postings
A jobs board is available for registered users to post open positions in a laboratory or institution, and covers all levels of research from entry-level lab technician and student scholarships to professorships and division directors.
11.3. Xine
Xine is a Xenopus community email group hosted at the National Xenopus Resource (NXR). Xine’s goal is to inform researchers about important developments and to disseminate information of wide interest. Xenbase archived Xine releases and supplements from 2001–2011. Xine depends on contributions from members of the Xenopus community for its content. Not a member? Join here: https://lists.mbl.edu/mailman/listinfo/xenopus. If you encounter problems, contact the Xine editor (xenopus@mbl.edu).
11.4. Xenbase Forums
Xenbase forums, an avenue for researchers to take part in Xenopus-related discussion, was relaunched in May 2017, and users must register to post items.
11.5. Meetings and Resources
We provide details to upcoming conferences, workshops, and technical courses relevant to the Xenopus research community. These are updated biannually.
11.6. Xenopus White Papers
The Xenopus Community White Papers, which provide a succinct, authoritative report and literature review of recent Xenopus research, are posted on Xenbase to maximize their distribution. White Papers provide recommendations on how continued, focused funding from the National Institutes of Health (NIH) can maximize the impact of biomedical research using the Xenopus system (see [3]), and therefore they serve as a very useful resource. Researchers are encouraged to reference the most recent Xenopus Community White Papers in their NIH grant proposals.
11.7. International Xenopus Board
We host information and history about the International Xenopus Board (IXB). The IXB was incorporated in 2015 as a tax-exempt, nonprofit organization with a remit to organize a biennial International Xenopus Conference, organize an annual Resources and Emerging Technologies meeting, represent and promote communication among Xenopus researchers, and promote the development and use of Xenopus resources.
Xenbase provides a list of the current members of the IXB and posts documents containing a report on the creation of the IXB, minutes from previous Resources and Emerging Technologies meetings, the by-laws and certificate of incorporation of the IXB.
11.8. Notes/Troubleshooting Community Support Pages
Contact Xenbase (xenbase@ucalgary.ca) to post a meeting or work-shop, or if you find omissions, errors, or bugs in these pages.
12. Stock Center Support
The international Xenopus community has established several Xenopus stock centers for obtaining reagents and frogs for biomedical and immunological research. A major goal of Xenbase is to support the Stock Centers by curating and cataloguing the frogs (lines and strains) and other reagents they supply. Xenopus lines and strains are given Research Resource Identifiers (RRID) numbers, a measure which helps disambiguate which lines were used in the research, promoting reproducibility, rigor, and transparency. The catalog is searchable through the “Lines and Strains” option on the “Stock Center” menu, or the “Transgenic lines” link in the “Reagents & Protocols” tile of the Xenbase home page. There are currently five Stock Centers worldwide:
NXR: The National Xenopus Resource is the US frog stock center and training center for advanced technologies (Contact: xenopus@mbl.edu).
EXRC: The European Xenopus Resource Centre is the stock center in the UK (Contact: EXRC@xenopusresource.org).
CRB: Centre de Ressources Biologiques Xenopes (Biological Resource Center Xenopus) is the stock center in France (Contact: crb-xenopes@univ-rennes1.fr).
NBRP: The National BioResource Project, is the stock center in Japan (Contact: oakkashi@hiroshima-u.ac.jp).
URMC: The X. laevis Research Resource for Immunology is based at the University Rochester, NY (Contact: jacques_rob-ert@urmc.rochester.edu).
12.1. Xenopus Lines
Standardized nomenclature is critical to make research accessible to the broader scientific community and to ensure consistency and provenance, and researchers are encouraged to follow the working Transgenic Nomenclature Guidelines which are posted on Xenbase under the Genes tile on the home page. Xenbase names transgenic (Tg) and mutant lines according to these guidelines, which were developed in consultation with the Xenopus stock centers, following best practices used by all other model organisms (see review in [38]). We only curate published, stable lines, and those available at stock centers using these criteria. Common names or Stock Center shorthand names are recorded as synonyms. For example the Tg line called line “511” or line “275,” has been named officially as Xla.Tg(actc1:GFP)Amaya. It is a X. laevis (Xla) line with a transgene (Tg) which has the promoter for the X. laevis cardiac actin gene (actc1), driving expression of a green fluorescent protein (GFP); the line comes from the Amaya lab (Amaya). Each Tg line page (e.g., XB-LINE-935) has an “Attributions” tab which details associated papers, and the “Transgene” tab gives details of the transgenic constructs used to produce the line.
12.2. Xenopus Strains
Wild type strains of Xenopus are named with a species code (Xla or Xtr), geographic origin or common names, and supplier/source (e.g., Xla.J-strainEXRC or Xtr.NigerianNBRP).
12.3. Transgenes
We curate transgenic constructs from published research articles. “Transgenes” are searchable via the “Stock Center” menu. While some of the transgenes are the basis for Tg lines, many have been used for transient transgenesis or are expression constructs. Transgenes are named following the mutant and Tg line nomenclature guidelines, with two exceptions: we omit species prefix and the originating lab code.
12.4. Notes/Troubleshooting
- Consult nomenclature guidelines for Genes, Chromosomes and Tg/Mutant lines when naming your transgenic constructs and frogs.
-
–Gene nomenclature: http://www.xenbase.org/gene/static/geneNomenclature.jsp
-
–Chromosome nomenclature: http://www.xenbase.org/gene/static/chromosomeNomenclature.jsp
-
–Transgenic and Mutant Line nomenclature: http://www.xenbase.org/gene/static/tgNomenclature.jsp
-
–
Contact Xenbase (xenbase@ucalgary.ca) if you experience omissions, errors or bugs in these pages, or if you need advice with nomenclature.
13. Downloading and Submit Data Menu
Xenbase hosts an assortment of files of general utility on our FTP server. Users can find regularly updated reports from Xenbase database content relating to gene expression, sequence information, accession numbers and genomic location, and other specific mappings files (e.g., Xenopus gene-human disease gene mappings), by navigating through the expandable folders on the FTP page. Additionally, users are encouraged to submit their own data to be shared with the broader research community. We can accept gene expression images (see template page for minimum metadata required), protocols, development movies and cell-fate maps, and welcome discussions on hosting other types of data. Click “Submit your data” option from the menu to open a data submission form for uploading the files.
14. New Features and Future Developments on Xenbase
Phenotypes including anatomical, gene function, and gene expression as phenotype.
JBrowse will replace GBrowse as genome viewer tool, with a 2-year phase-out period.
RNA-Seq and ChIP-Seq data will be shown in stacked views.
Illustrated XAO terms and expanded Organ Atlas, including images from XenHead [26] and EctoMap projects.
Many NGS and genome feature tracks added to genome browser, older NGS content will be mapped to latest genome builds.
Expanded curation of mutant and transgenic lines available from Xenopus Stock Centers.
“How to use Xenbase” videos will be expanded to cover more topics.
Educational pages on Xenopus in biomedical research will be expanded.
OMIM disease, GO terms and References cited will be posted on Article Pages.
Protein–protein interaction data and enhanced gene network support.
Revamped gene expression interface.
RRID numbers for all antibodies.
Acknowledgements
Funding for Xenbase is provided by the Eunice and Kennedy Shriver National Institute of Child Health and Human Development, grant P41 HD064556 (Zorn and Vize, Joint-PIs). We thank James Coulombe (NIH/NICHD), the Xenbase EAB members, and Xenopus researchers around the world for continued support and feedback that help us set priorities for development and new features on Xenbase.
References
- 1.Gurdon JB (1960) The developmental capacity of nuclei taken from differentiating endoderm cells of Xenopus laevis. J Embryol Exp Morphol 8:505–526 [PubMed] [Google Scholar]
- 2.Gurdon JB, Elsdale TR, Fischberg M (1958) Sexually mature individuals of Xenopus lae-vis from the transplantation of single somatic nuclei. Nature 182(4627):64–65 [DOI] [PubMed] [Google Scholar]
- 3.Sater AK, Moody SA (2017) Using Xenopus to understand human disease and developmental disorders. Genesis 55(1–2). 10.1002/dvg.22997 [DOI] [PubMed] [Google Scholar]
- 4.Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, Noumen E, Pollet N, Vize PD (2008) Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res 36(Database issue):D761–D767. 10.1093/nar/gkm826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karimi K, Vize PD (2014) The Virtual Xenbase: transitioning an online bioinformatics resource to a private cloud. Database (Oxford) 2014:bau108 10.1093/database/bau108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS (2010) The genome of the Western clawed frog Xenopus tropicalis. Science 328(5978):633–636. 10.1126/science.1183670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, van Heeringen SJ, Quigley I, Heinz S, Ogino H, Ochi H, Hellsten U, Lyons JB, Simakov O, Putnam N, Stites J, Kuroki Y, Tanaka T, Michiue T, Watanabe M, Bogdanovic O, Lister R, Georgiou G, Paranjpe SS, van Kruijsbergen I, Shu S, Carlson J, Kinoshita T, Ohta Y, Mawaribuchi S, Jenkins J, Grimwood J, Schmutz J, Mitros T, Mozaffari SV, Suzuki Y, Haramoto Y, Yamamoto TS, Takagi C, Heald R, Miller K, Haudenschild C, Kitzman J, Nakayama T, Izutsu Y, Robert J, Fortriede J, Burns K, Lotay V, Karimi K, Yasuoka Y, Dichmann DS, Flajnik MF, Houston DW, Shendure J, DuPasquier L, Vize PD, Zorn AM, Ito M, Marcotte EM, Wallingford JB, Ito Y, Asashima M, Ueno N, Matsuda Y, Veenstra GJ, Fujiyama A, Harland RM, Taira M, Rokhsar DS (2016) Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538(7625):336–343. 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vize PD, Liu Y, Karimi K (2015) Database and informatic challenges in representing both diploid and tetraploid Xenopus species in Xenbase. Cytogenet Genome Res 145(3–4):278–282. 10.1159/000430427 [DOI] [PubMed] [Google Scholar]
- 9.Matsuda Y, Uno Y, Kondo M, Gilchrist MJ, Zorn AM, Rokhsar DS, Schmid M, Taira M (2015) A new nomenclature of Xenopus lae-vis chromosomes based on the phylogenetic relationship to Silurana/Xenopus tropicalis. Cytogenet Genome Res 145(3–4):187–191. 10.1159/000381292 [DOI] [PubMed] [Google Scholar]
- 10.Segerdell E, Bowes JB, Pollet N, Vize PD (2008) An ontology for Xenopus anatomy and development. BMC Dev Biol 8:92 10.1186/1471-213X-8-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mi H, Muruganujan A, Casagrande JT, Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8(8):1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Owens ND, Blitz IL, Lane MA, Patrushev I, Overton JD, Gilchrist MJ, Cho KW, Khokha MK (2016) Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell Rep 14(3):632–647. 10.1016/j.celrep.2015.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanai I, Peshkin L, Jorgensen P, Kirschner MW (2011) Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell 20(4): 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, Papalopulu N, Smith JC, Zorn Am , Harland RM, Grammer TC (2002) Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn 225(4):499–510. 10.1002/dvdy.10184 [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 16.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH (2009) JBrowse: a next-generation genome browser. Genome Res 19(9):1630–1638. 10.1101/gr.094607.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, Wang T, Nguyen N, Paten B, Zweig AS, Karolchik D, Kent WJ (2014) Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 30(7):1003–1005. 10.1093/bioinformatics/btt637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciau-Uitz A, Pinheiro P, Kirmizitas A, Zuo J, Patient R (2013) VEGFA-dependent and -independent pathways synergise to drive Scl expression and initiate programming of the blood stem cell lineage in Xenopus. Development 140(12):2632–2642. 10.1242/dev.090829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parain K, Mazurier N, Bronchain O, Borday C, Cabochette P, Chesneau A, Colozza G, El Yakoubi W, Hamdache J, Locker M, Gilchrist MJ, Pollet N, Perron M (2012) A large scale screen for neural stem cell markers in Xenopus retina. Dev Neurobiol 72(4):491–506. 10.1002/dneu.20973 [DOI] [PubMed] [Google Scholar]
- 20.Raciti D, Reggiani L, Geffers L, Jiang Q, Bacchion F, Subrizi AE, Clements D, Tindal C, Davidson DR, Kaissling B, Brandli AW (2008) Organization of the pronephric kidney revealed by large-scale gene expression mapping. Genome Biol 9(5):R84 10.1186/gb-2008-9-5-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminski MM, Tosic J, Kresbach C, Engel H, Klockenbusch J, Muller AL, Pichler R, Grahammer F, Kretz O, Huber TB, Walz G, Arnold SJ, Lienkamp SS (2016) Direct reprogramming of fibroblasts into renal tubular epithelial cells by defined transcription factors. Nat Cell Biol 18(12):1269–1280. 10.1038/ncb3437 [DOI] [PubMed] [Google Scholar]
- 22.Rana AA, Collart C, Gilchrist MJ, Smith JC (2006) Defining synphenotype groups in Xenopus tropicalis by use of antisense morpholino oligonucleotides. PLoS Genet 2(11):e193 10.1371/journal.pgen.0020193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilchrist MJ, Pollet N (2012) Databases of gene expression in Xenopus development. Methods Mol Biol 917:319–345. 10.1007/978-1-61779-992-1_19 [DOI] [PubMed] [Google Scholar]
- 24.Ahmed A, Ward NJ, Moxon S, Lopez-Gomollon S, Viaut C, Tomlinson ML, Patrushev I, Gilchrist MJ, Dalmay T, Dotlic D, Munsterberg AE, Wheeler GN (2015) A database of microRNA expression patterns in Xenopus lae-vis. PLoS One 10(10):e0138313 10.1371/journal.pone.0138313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armisen J, Gilchrist MJ, Wilczynska A, Standart N, Miska EA (2009) Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res 19(10):1766–1775. 10.1101/gr.093054.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahn N, Levin M, Adams DS (2017) The Zahn drawings: new illustrations of Xenopus embryo and tadpole stages for studies of craniofacial development. Development 144(15):2708–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieuwkoop PD, Faber J (1994) Normal table of Xenopus laevis (Daudin): a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Garland Pub, New York, NY [Google Scholar]
- 28.Moody SA (1987) Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev Biol 119(2):560–578 [DOI] [PubMed] [Google Scholar]
- 29.Moody SA (1987) Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Dev Biol 122(2):300–319 [DOI] [PubMed] [Google Scholar]
- 30.Bauer DV, Huang S, Moody SA (1994) The cleavage stage origin of Spemann’s Organizer: analysis of the movements of blastomere clones before and during gastrulation in Xenopus. Development 120(5):1179–1189 [DOI] [PubMed] [Google Scholar]
- 31.Segerdell E, Ponferrada VG, James-Zorn C, Burns KA, Fortriede JD, Dahdul WM, Vize PD, Zorn AM (2013) Enhanced XAO: the ontology of Xenopus anatomy and development underpins more accurate annotation of gene expression and queries on Xenbase. J Biomed Semantics 4(1):31 10.1186/2041-1480-4-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blitz IL, Biesinger J, Xie X, Cho KW (2013) Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis 51(12):827–834. 10.1002/dvg.22719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharya D, Marfo CA, Li D, Lane M, Khokha MK (2015) CRISPR/Cas9: an inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Dev Biol 408(2):196–204. 10.1016/j.ydbio.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasevicius A, Ekker SC (2000) Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet 26(2):216–220. 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- 35.Heasman J, Kofron M, Wylie C (2000) Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol 222(1):124–134. 10.1006/dbio.2000.9720 [DOI] [PubMed] [Google Scholar]
- 36.Nutt SL, Bronchain OJ, Hartley KO, Amaya E (2001) Comparison of morpholino based translational inhibition during the development of Xenopus laevis and Xenopus tropicalis. Genesis 30(3):110–113 [DOI] [PubMed] [Google Scholar]
- 37.Grant IM, Balcha D, Hao T, Shen Y, Trivedi P, Patrushev I, Fortriede JD, Karpinka JB, Liu L, Zorn AM, Stukenberg PT, Hill DE, Gilchrist MJ (2015) The Xenopus ORFeome: a resource that enables functional genomics. Dev Biol 408(2):345–357. 10.1016/j.ydbio.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knowlton MN, Smith CL (2017) Naming CRISPR alleles: endonuclease-mediated mutation nomenclature across species. Mamm Genome 28:367 10.1007/s00335-017-9698-3 [DOI] [PMC free article] [PubMed] [Google Scholar]