Abstract
Despite the cytopathic nature of influenza A virus (IAV) replication, we recently reported that a subset of lung epithelial club cells is able to intrinsically clear virus and survive infection. However, the mechanisms that drive cell survival during a normally lytic infection remained unclear. Using a loss-of-function screening approach, we discovered that the DNA mismatch repair (MMR) pathway is essential for club cell survival of IAV infection. Repair of virally-induced oxidative damage by the DNA MMR pathway not only allowed cell survival of infection but also facilitated host gene transcription, including the expression of antiviral and stress response genes. Enhanced viral suppression of the DNA MMR pathway prevented club cell survival and increased the severity of viral disease in vivo. Altogether, these results identify previously unappreciated roles for DNA MMR as a central modulator of cellular fate and a contributor to the innate antiviral response, which together, control influenza viral disease severity.
Keywords: Influenza A virus, DNA mismatch repair, cellular survival, club cell, oxidative damage, siRNA screening, non-lytic clearance
INTRODUCTION
Influenza A virus (IAV) causes an acute respiratory infection, and neither viral replication nor transmission requires long-term persistence of an infected cell. As a result, IAV infection typically proceeds with a characteristic cytopathic phenotype culminating in the death of infected cells through a variety of cell death pathways including necrosis, necroptosis, and pyroptosis1. Additionally, part of the innate immune response to viruses includes the induction of apoptosis, a pathway of controlled cell death2,3. While cellular viability must be maintained for a long enough time to facilitate sufficient production of viral progeny (and indeed, IAV has evolved mechanisms to block early apoptosis of the host cell4,5), the long-term fate of the infected cells is thought to be irrelevant for viral replication/transmission. In addition to virally-induced death of infected cells, professional immune cells can effectively recognize and clear infected cells from the host6. Thus, until recently, it had been assumed that viral clearance from the host is dependent on the death of all infected cells through one of the processes described above.
It was therefore surprising when we and others reported that a subset of respiratory epithelial cells, named club cells, can survive direct infection with IAV and persist in the host long-term7,8. We have shown that these cells are not persistently infected, but rather cells that have been infected and have cleared the virus through a non-lytic process7. Non-lytic clearance of Sindbis virus from neurons9 and hepatitis B virus and lymphocytic choriomeningitis virus (LCMV) from hepatocytes10,11 have also been reported with the underlying hypothesis that in some tissues, extensive cell death would compromise organ architecture and function12,13. While autophagy plays a role in cell-intrinsic control of some viral infections14, the mechanisms that promote non-lytic viral clearance remain incompletely understood.
To identify genes and pathways that promote non-lytic viral clearance after IAV infection, we performed an siRNA screen to identify modulators of cellular survival. This screen revealed a number of genes that promote club cell survival following IAV infection, including a gene involved in the DNA mismatch repair (MMR) pathway. Since IAV is not generally thought to lead to cellular death via lethal genomic DNA damage, it was unclear how this pathway would promote survival from IAV infection. We found that the DNA MMR pathway was required to prevent the accumulation of oxidative DNA lesions in antiviral gene loci, which likely contributes to the non-lytic viral clearance phenotype in club cells. Additionally, we engineered viruses that artificially enhance suppression of the DNA MMR pathway and showed that under these conditions, cells were unable to survive IAV infection and the overall severity of viral disease was increased in vivo. Together, these data have revealed an unappreciated role for a DNA repair mechanism that is required for the innate cellular response and controls cellular fate during IAV infection.
RESULTS
Differential cellular fate after influenza A virus infection can be modeled in vitro.
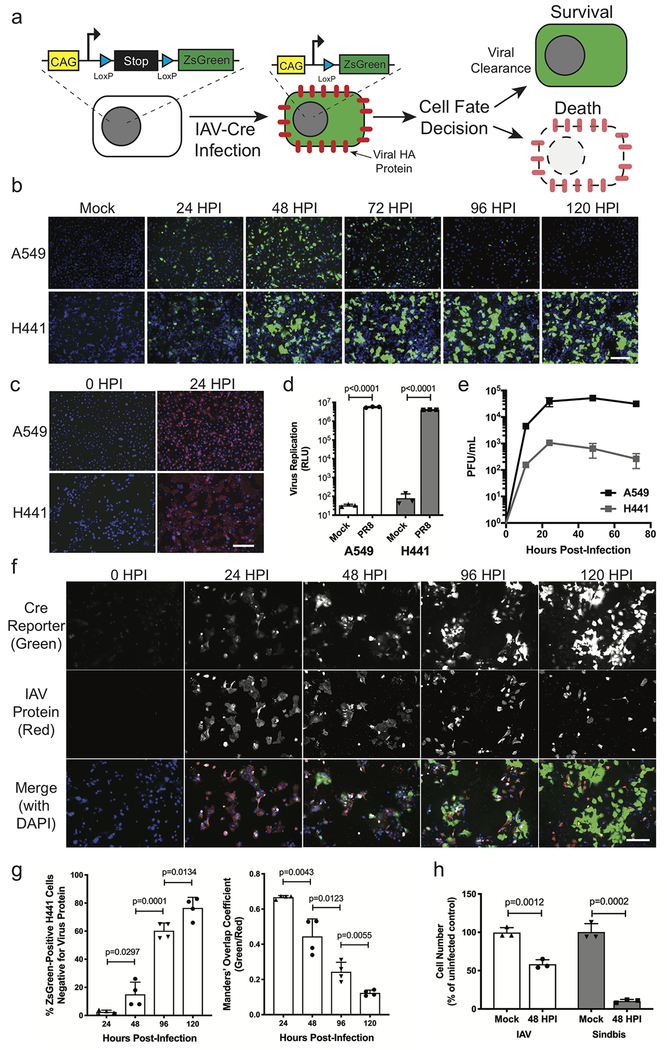
To observe cellular fate after IAV infection in vitro, we previously utilized LoxP-Stop-LoxP-reporter-containing NCI-H441 cells which activate a fluorescent reporter after infection with a Cre-expressing influenza virus (IAV-Cre)15. For this study, we generated an analogous reporter in an alveolar epithelial cell line (A549) to compare rates of survival after infection (Fig. 1a). Upon IAV-Cre infection, we found that while both cell types initially activate the ZsGreen reporter, the infected A549 cells ultimately succumbed while the H441-CR cells survived (Fig. 1b). A549-CR cells divide much more rapidly than H441-CR cells (Supplementary Fig. 1), discounting the possibility that higher numbers of survivors at later time points is simply due to rapid growth of surviving H441-CR cells.
Figure 1. NCI-H441 cells survive direct infection with influenza A virus.
(a) Diagram of the ZsGreen-Cre-reporter system used to label cells that survive direct IAV infection. (b) Visualization of ZsGreen-positive survivor cells (green) and nuclei (blue) at indicated time points following infection of A549-CR and H441-CR cells. Representative of two independent experiments. (c) Staining for extracellular viral HA glycoprotein (PY102 antibody, red) and nuclei (DAPI, blue) at 24 hpi. Representative of two independent experiments. (d) Viral RdRP replication assay comparing A549 and H441 cells. Data shown as mean ± SD, n=3 independent samples. Representative of two independent experiments. (e) Titer of infectious virus collected in supernatants of A549 or H441 cells at the indicated time points following WT PR8 infection. Data shown as mean ± SD, n=4 independent samples. Representative of two independent experiments. (f) Time course of WT PR8 infection in H441 cells with viral HA glycoprotein (PY102 antibody, red), ZsGreen-reporter (green), and nuclei (DAPI, blue) over 120 hours. Representative of two independent experiments. (g) Analysis of f showing the number of ZsGreen-positive cells that have cleared viral protein (green+ red-blue+), and the co-localization of green and red signal over time. Data shown as mean ± SD, n=4 independent samples. Representative of two independent experiments. (h) Viability of H441 cells following infection with IAV (WT PR8) or Sindbis virus at 48 hpi, normalized to Mock controls. Data shown as mean ± SD, n=3 independent samples. Representative of two independent experiments. For all panels: p-values calculated using unpaired two-tailed t tests; scale bars = 200 μm.
To better understand the nature of infection in both A549 and H441 cells, we monitored infection and hemagglutinin (HA) surface trafficking via microscopy as well as the activity of the viral RNA-dependent RNA polymerase (RdRP); however, no significant differences were observed (Fig. 1c–d). We also conducted a multicycle growth curve and found that infectious progeny viruses are released from H441 cells, albeit at lower levels than A549 cells (Fig. 1e). These data suggested that H441 cells have the ability to clear actively replicating virus rather than being incompletely permissive. To monitor the kinetics of viral infection and clearance in H441 cells, we conducted an infection time course and visualized viral protein production (red) and induction of the ZsGreen reporter (green) over time on a per cell basis. Despite early viral protein expression, we observed rapid clearance of replicating virus from infected cells, which subsequently survive (Fig. 1f–g). H441 cells however, were killed after infection with an alphavirus (Sindbis virus), indicating that these cells are not just generally resistant to virus-dependent cytotoxicity (Fig 1h).
siRNA screening identifies a DNA mismatch repair protein as required for H441 cell survival after IAV infection.
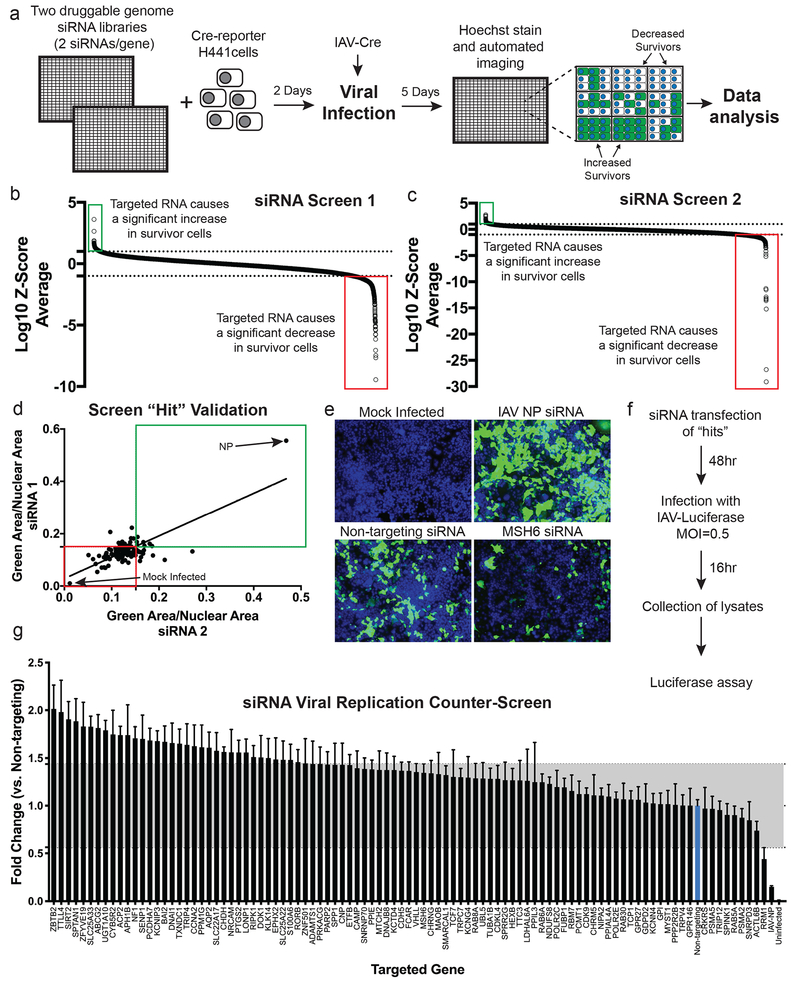
In order to identify the genes that allow H441 cells to clear and survive IAV infection, we conducted an RNAi screen using a library targeting the druggable genome (23,349 siRNAs targeting 7,783 genes). We transfected H441 cells containing our ZsGreen reporter with siRNAs arrayed in 384-well plates, infected with IAV-Cre virus for 120 hours, and used automated imaging and analysis to determine genes that, when targeted by siRNA, altered H441 survival (Fig. 2a). We treated cells with type I interferon (IFN) as a negative control (which blocks infection and prevents ZsGreen signal) and transfected cells with an siRNA targeting the viral nucleoprotein (NP) as a positive control (which increases cell survival by suppressing viral replication). We performed the screen in duplicate, with two unique siRNAs per gene per well in two wells (for a total of four unique siRNAs per gene). A Z-score was calculated based on the gene’s ability to either significantly increase or decrease H441 cell survival (% ZsGreen+ cells) when knocked down, and a “hit” was defined as having a log10 Z-score of greater than 1.0 or less than −1.0 in both replicates of the screen (Fig. 2b–c, Supplementary Table 1).
Figure 2. An siRNA screen of the druggable genome reveals host factors that control H441 cell survival during IAV infection.
(a) Experimental diagram of the siRNA screen. (b,c) Results of the two independent siRNA screens. The log10 transformation of the Z-scores are plotted for each gene targeted by siRNA and their effect on survival (two siRNA per gene per independent screen). Highlighted are those genes that significantly increase H441 survival (green boxes) when targeted and those that significantly decrease H441 survival (red boxes) when targeted (n=4 independent samples per gene per screen). Average log10 Z-scores for all targeted genes are reported in Supplementary Table 1. (d) Validation of the hits (96 genes) from the high-throughput screen with two additional siRNAs (n=4 independent samples per siRNA). Mock Infected = no Cre present, negative control. NP = siRNA targeting the IAV NP gene, positive control. Boxes label targeted genes that reproducibly increased survival (green) or decreased survival (red). P-values listed in Supplementary Table 2. (e) Representative images of the screen validation controls and MSH6, one of the screen hits. Survivor cells (green) were visualized in H441 monolayers following transfection with the specified siRNA and then infection with IAV-Cre for 120 h. Scale bar = 200 μm. Representative of two independent experiments. (f) Experimental protocol used for the viral replication counter-screen. (g) Results of the viral replication counter-screen. The average luciferase values of both individual siRNAs are plotted. The gray shaded box indicates two standard deviations above and below the control siRNA. All replication values were normalized to the non-targeting siRNA control (blue bar). Data shown as mean ± SEM, n=6 independent samples (3 for each siRNA). A detailed version of this graph is available in Supplementary Fig. 2.
Using these criteria, we identified 87 candidate genes. In addition to those 87 genes, we also selected 9 additional genes that had large phenotypes (log10 Z-score of greater than 1.3) in only one of the two replicate screens, for a total of 96 candidate genes for further testing. We obtained two additional siRNAs per gene and performed an RNAi validation screen (Fig. 2d–e, Supplementary Table 2). Because genes that directly affect viral infection could also indirectly impact cellular survival, we infected siRNA-transfected H441 cells with a luciferase-expressing influenza virus (IAV-Luc) and eliminated genes that modulated luciferase levels at 16 hpi (Fig. 2f–g, Supplementary Fig. 2). Finally, because some siRNAs can induce cellular toxicity irrespective of viral treatment, we focused on genes that did not decrease total cell numbers by more than 50% relative to control. Together, these analyses revealed 15 genes that significantly affected H441 survival in both the primary and validation screens, with only minor effects on virus replication levels and cell viability (Table 1).
Table 1. Genes required for H441 cell survival after influenza virus infection.
The 15 genes that significantly decreased H441 cell survival in both the primary siRNA screen and the validation screen, with only minor effects on viral replication levels and cell viability. aAverage log10 z-scores for each gene in the primary siRNA screen (n=4 samples per gene per screen replicate). bP-values (unpaired two-tailed t tests) for two new siRNAs targeting each gene in the validation screen (n=4 samples per siRNA). cAverage fold change in IAV-Luc replication in the viral replication counter-screen when the indicated gene was targeted by two separate siRNAs (n=3 samples per siRNA). dTotal cell number for each gene when targeted by siRNA compared to the overall median cell number for all genes on the same 384-well plate in the primary siRNA screen (n=4 samples per gene).
| NCBI Gene ID | Gene Symbol | Official Full Gene Name | Gene Function | aAvg. Log10 Z-Score Screen 1 | aAvg. Log10 Z-Score Screen 2 | bValidation Screen siRNA 1 p-value | bValidation Screen siRNA 2 p-value | cEffect on Virus Replication | dTotal Cell Number (% of Overall Median) |
|---|---|---|---|---|---|---|---|---|---|
| 1133 | CHRM5 | Cholinergic receptor, muscarinic 5 | GPCR signaling | −1.72 | −1.20 | 0.0038 | 0.0016 | 1.11-fold change | 55.6% |
| 51755 | CDK12 | Cyclin-dependent Kinase 12 | Transcription regulation | −1.15 | −1.10 | 0.0002 | <0.0001 | 0.97-fold change | 67.3% |
| 8880 | FUBP1 | Far upstream element (FUSE) binding protein 1 | Nucleic acid binding | −1.01 | −1.32 | <0.0001 | 0.0033 | 1.19-fold change | 95.6% |
| 2821 | GPI | Glucose phosphate isomerase | Protein metabolism | −1.11 | −1.38 | 0.0002 | <0.0001 | 1.02-fold change | 80.4% |
| 115330 | GPR146 | G protein-coupled receptor 146 | Plasma Membrane Signaling | −1.30 | −1.07 | <0.0001 | 0.0002 | 1.00-fold change | 75.1% |
| 3783 | KCNN4 | Potassium calcium-activated channel subfamily N member 4 | Ion channel | −1.21 | −1.38 | 0.0076 | 0.0008 | 1.02-fold change | 132.6% |
| 2956 | MSH6 | mutS homolog 6 (E. coli) | DNA repair | −1.30 | −1.02 | 0.0078 | 0.0345 | 1.34-fold change | 86.6% |
| 5432 | POLR2C | Polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | Transcription | −1.07 | −2.00 | <0.0001 | <0.0001 | 1.20-fold change | 88.2% |
| 5434 | POLR2E | Polymerase (RNA) II (DNA directed) polypeptide E, 25kDa | Transcription | −1.49 | −2.33 | <0.0001 | <0.0001 | 1.07-fold change | 76.1% |
| 653505 | PPIAL4A | Peptidylprolyl isomerase A like 4A | Protein folding | −1.39 | −1.04 | <0.0001 | 0.0155 | 1.09-fold change | 96.1% |
| 5521 | PPP2R2B | Protein Phosphatase 2 Regulatory Subunit beta | Cell growth/division | −1.41 | −1.57 | <0.0001 | <0.0001 | 1.01-fold change | 64.8% |
| 5683 | PSMA2 | Proteasome subunit alpha 2 | Protease activity | −2.56 | −1.11 | <0.0001 | <0.0001 | 0.88-fold change | 57.5% |
| 5686 | PSMA5 | Proteasome subunit alpha 5 | Protease activity | −5.06 | −1.80 | 0.0005 | <0.0001 | 0.97-fold change | 59.0% |
| 5870 | RAB6A | RAB6A, member RAS oncogene family | Intracellular trafficking | −2.44 | −1.20 | 0.0034 | <0.0001 | 1.24-fold change | 79.8% |
| 6950 | TCP1 | T-complex 1 | Protein folding | −2.75 | −1.04 | <0.0001 | <0.0001 | 1.06-fold change | 94.2% |
Many of the validated genes play roles in cellular stress response pathways. Examples include genes involved in protein degradation (PSMA2, PSMA5), host transcription (POLR2C, POLR2E, CDK12), and protein folding (PPIAL4A, TCP1). One of the more unexpected hits was MSH6, a core member of the DNA mismatch repair (MMR) pathway. This pathway is responsible for excising and repairing mismatched nucleotides that arise during DNA replication or some types of DNA damage16. As an RNA virus, IAV infection is not generally thought to affect host DNA metabolic processes, and thus it was unclear why this gene would be required for cellular survival from IAV infection; we therefore selected this gene for further study.
DNA MMR is differentially regulated in A549 and H441 cells during IAV infection.
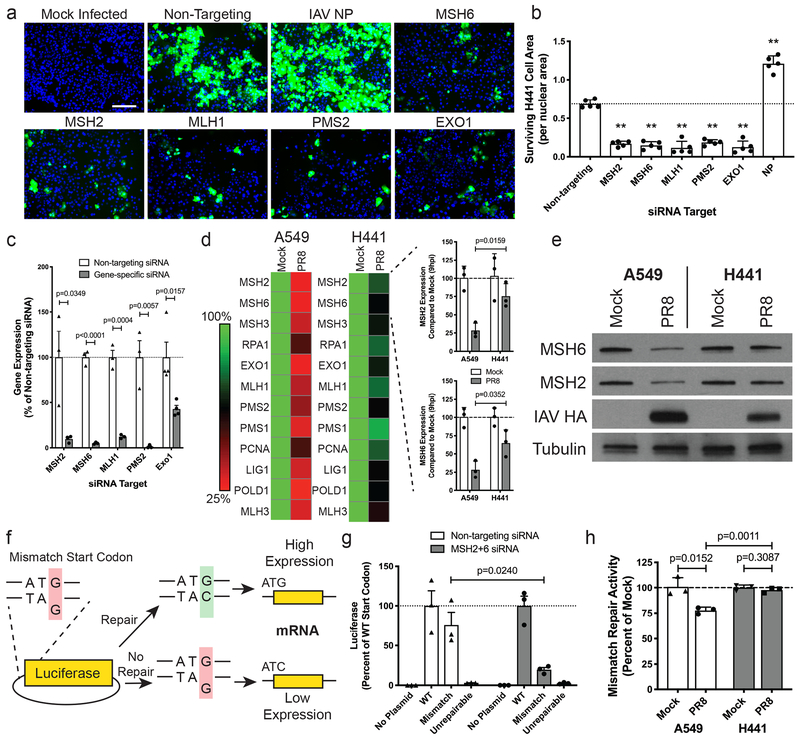
First, we determined whether MSH6 was required for H441 cell survival in the context of DNA MMR, or whether it had an alternative “moonlighting” activity. Knockdown of other key proteins involved in the canonical DNA MMR pathway (MSH2, MLH1, PMS2, EXO1) revealed that each member of the pathway was required for H441 cell survival (Fig. 3a–c, Supplementary Fig. 3). We next explored whether differences in the regulation of the DNA MMR pathway could explain the differential survival phenotypes of H441 and A549 cells. We therefore monitored expression of DNA MMR genes in cells mock-infected or infected with wild-type A/Puerto Rico/8/1934 (WT PR8), a laboratory-adapted H1N1 strain of IAV. At 9 hpi, PR8-infected A549 cells display a significant reduction in the gene expression of all core DNA MMR genes compared to uninfected cells, whereas PR8-infected H441 cells maintain relatively high levels of expression of each DNA MMR gene comparable to uninfected cells (Fig. 3d, Supplementary Table 3). Western blot analysis of two main DNA MMR proteins (MSH2 and MSH6) also displayed a reduction at 24 hpi in A549 cells, but not in H441 cells (Fig. 3e).
Figure 3. DNA MMR genes are important for survival and their expression is maintained in H441 cells during infection.
(a) Visualization of survivor cells (green) and nuclei (blue) in H441-CR cell monolayers following siRNA transfection and infection with IAV-Cre for 120 h. Scale bar represents 200 μm. Representative of two independent experiments. (b) Quantification of survivor cells (normalized to nuclei) in the samples described in a. Data shown as mean ± SD, n=5 independent samples; **p<0.0001. Representative of two independent experiments. (c) Validation of siRNA knockdown of each of the DNA MMR genes from a. Data shown as mean ± SD, n=3 independent samples. Representative of two independent experiments. (d) Heat map displaying the level of RNA expression for each of the DNA MMR genes 9 hpi with WT PR8 in A549 and H441 cells, each normalized to mock-infected cells (averaged across replicates). Inset bar graphs highlight the difference in MSH2 and MSH6 RNA levels between cell types at 9 hpi. Data shown as mean ± SD, n=3 independent samples. Representative of two independent experiments. (e) Western blot comparing MSH2 and MSH6 protein levels at 24 hpi with WT PR8 in A549 and H441 cells. IAV HA = infection control; tubulin = loading control. Representative of two independent experiments. (f) Schematic showing the engineered “mismatch-reporter” NanoLuc plasmid used to determine the level of DNA MMR activity. (g) Validation of the MMR activity assay using A549 cells with or without depletion of MSH2+MSH6. Luciferase was measured following transfection with the WT, mismatch (G-G), or an unrepairable NanoLuc plasmids. Data shown as mean ± SD, n=3 independent samples. Representative of two independent experiments. (h) Comparison of DNA MMR activity in A549 and H441 cells at 24 hpi with WT PR8. Each bar shown is the ratio of mismatch plasmid luciferase/WT plasmid luciferase, normalized to the mock-infected control for each cell type. Data shown as mean ± SD, n=3 independent samples. Representative of two independent experiments. For all panels: p-values calculated using unpaired two-tailed t tests.
To determine whether differences in MMR protein levels impacted DNA MMR activity, we adapted a previously described17 DNA MMR assay by generating a NanoLuc plasmid with a single nucleotide mismatch (G-G) in the start codon (Fig. 3f). We compared NanoLuc expression from the ‘mismatch plasmid’ to the ‘WT plasmid’ to measure the level of DNA MMR activity. Cells transfected with non-targeting control siRNAs repaired and expressed the ‘mismatch plasmid’ in an MSH2/6 dependent manner (Fig. 3g). After infection of A549 and H441 cells, we observed that A549 cells lost >25% of their MMR activity while H441 cells maintained levels equal to mock-infected cells (Fig. 3h).
DNA MMR is required to repair ROS-induced DNA damage during IAV infection.
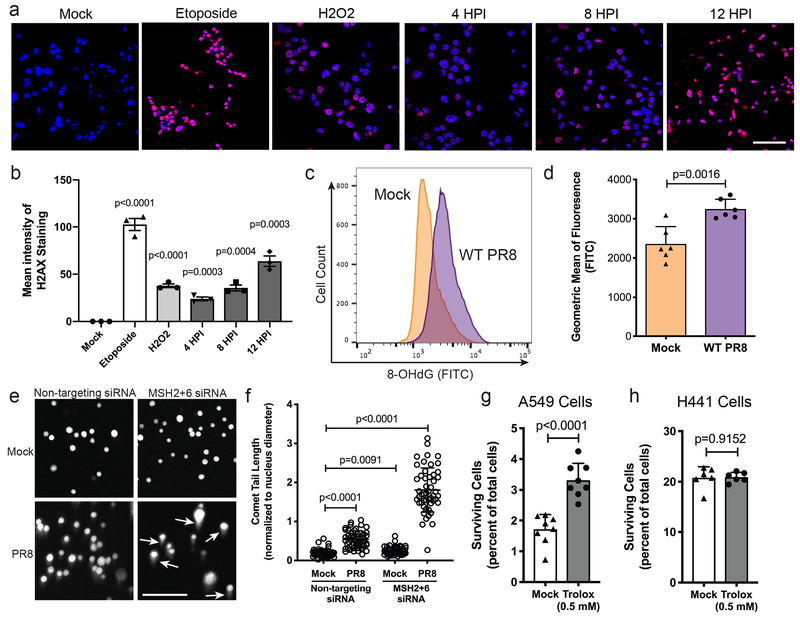
Since we identified a DNA repair pathway as critical for surviving viral infection, we next verified that DNA damage was induced during IAV infection. We infected H441 cells with WT PR8 and monitored phospho-H2AX levels, a histone modification associated with DNA repair (Fig. 4a–b). We observed strong induction of phospho-H2AX 8–12 hours after PR8 infection, on par with treatment with the control DNA damaging agents Etoposide and hydrogen peroxide (H2O2).
Figure 4. DNA MMR activity remains high in H441 cells allowing repair of virally-induced ROS-mediated DNA damage.
(a) Representative phospho-H2AX (red) and nuclei (blue) staining of H441 cells to measure the level of DNA damage present during WT PR8 infection. Etoposide and H2O2 are used as positive controls. (b) Quantification of the mean intensity of the phospho-H2AX staining from samples in a. Data shown as mean ± SD, n=3 independent samples. Representative of two independent experiments. (c) Histogram of 8-Oxo-2’-deoxyguanosine (8-OHdG) in H441 cells comparing mock-infected to PR8-infected at 48 hpi. Representative of two independent experiments. (d) Quantification of the geometric mean of fluorescence of the 8-OHdG staining from c. Data shown as mean ± SD, n=6 independent samples. Representative of two independent experiments. (e) Representative images of the modified Comet assay used to compare the level of oxidative DNA damage following siRNA knockdown of control or DNA MMR genes MSH2+MSH6 and mock or WT PR8 infection. Longer tails (indicated by white arrows) correspond to greater DNA damage. (f) Quantification of comet tail lengths (normalized to nuclear diameter) in the samples displayed in e. Data shown as mean ± SD, n=50 nuclei. Representative of two independent experiments. (g) Percent of surviving A549-CR cells after treatment with 0.5 mM Trolox or mock control and infection with IAV-Cre for 120 h. Data shown as mean ± SD, n=8 independent samples. Representative of five experiments. (h) Percent of surviving H441-CR cells after treatment with 0.5 mM Trolox or mock control and infection with IAV-Cre for 120 h. Data shown as mean ± SD, n=6 independent samples. Representative of four experiments. For all panels: p-values calculated using unpaired two-tailed t tests; scale bars = 100 μm.
Apart from its major role in repairing spontaneous nucleotide mismatches following DNA replication, the DNA MMR pathway also plays a role in repairing reactive oxygen species (ROS)-induced DNA damage18. IAV infection is known to alter the redox state of infected cells19,20 and increase intracellular ROS levels and oxidative damage in a variety of cell lines including A549 and MDCK cells21–26. These data suggested a possible link between IAV infection and the DNA MMR pathway. Although total ROS levels have been reported to increase during IAV infection, it was unknown whether this leads to ROS-induced DNA damage. We therefore employed an assay to measure 8-hydroxy-2’-deoxyguanosine (8-OHdG), a common biomarker of ROS-induced DNA damage27. We found increased levels of 8-OHdG upon PR8 infection, suggesting that IAV infection increases ROS-induced DNA damage (Fig. 4c–d).
To determine whether DNA MMR is required to repair the oxidative DNA damage induced by IAV infection, we employed a modified comet assay, wherein cells are treated with Fpg, a restriction enzyme that cleaves DNA only at sites of oxidative damage28,29. H441 cells were transfected with non-targeting control siRNAs or depleted of DNA MMR genes, and subsequently infected with WT PR8. Comet tail lengths increased only 2.8-fold following PR8 infection in cells competent for DNA MMR, but tail lengths increased 9.4-fold in infected cells lacking MSH2+6 (Fig. 4e–f). Our data suggested that maintaining DNA MMR during infection controls ROS-induced DNA damage. Therefore, artificially decreasing ROS levels could potentially increase the rate of cellular survival. When we infected A549-CR cells (which normally succumb to IAV infection) with IAV-Cre in the presence of the ROS scavenger Trolox30, cell survival roughly doubled (Fig. 4g). However, the same treatment of H441-CR cells had no effect on the percentage of survivors (Fig. 4h), likely due to their maintenance of DNA MMR activity.
DNA MMR impacts expression of innate antiviral genes.
We hypothesized that the avoidance of lethal oxidative genomic damage was part of a larger program required for epithelial cells to survive viral infection. ROS-induced DNA damage increases stalling of RNA polymerase II during transcription31,32 and DNA MMR is associated with transcription-coupled repair33–35. Although DNA MMR and antiviral gene expression have not been previously linked, we tested whether the reduction in DNA MMR activity and the subsequent accumulation of DNA damage in IAV-infected cells also attenuated transcription of host antiviral genes.
We first tested whether ROS-induced DNA damage affected cellular transcription in club cells. H2O2 treatment reduced expression of a cellular NanoLuc reporter by ~80% compared to PBS-treated control cells (Fig. 5a). The decrease in NanoLuc expression, however, was not due to cell death, as cell viability in both samples was comparable (Fig. 5b). We also monitored the effects of oxidative damage on expression of endogenous antiviral genes. Expression of the interferon-stimulated gene (ISG) Mx1 was significantly reduced upon treatment with H2O2 at both the RNA (Fig. 5c) and the protein level (Fig. 5d).
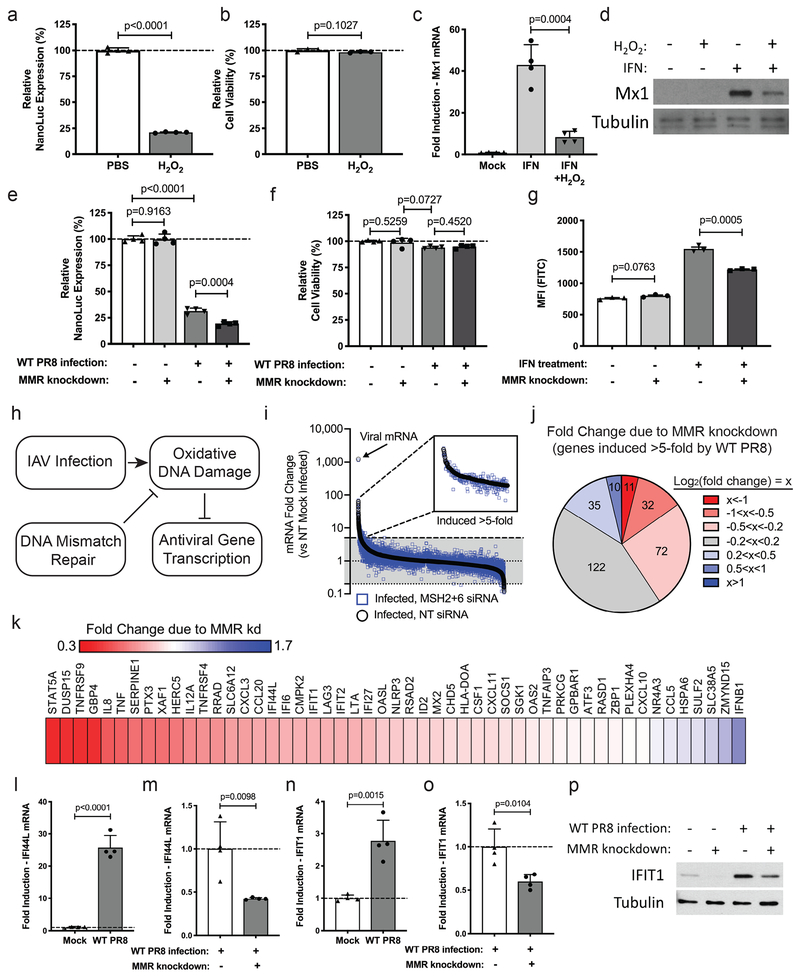
Figure 5. Loss of DNA MMR activity reduces the innate antiviral transcriptional response against influenza A virus.
(a) NanoLuc reporter expression and (b) relative cell viability in H441 cells that have been treated with PBS or H2O2 (for 30 min). Data shown as mean ± SD, n=4 independent samples. (c) Fold change of Mx1 RNA levels in H441 cells following treatment with PBS or IFN-alpha +/− H2O2 treatment (for 30 min). Data shown as mean ± SD, n=4 independent samples. (d) Western blot for Mx1 in H441 cells following the specified treatments. Tubulin = loading control. (e) NanoLuc reporter expression and (f) relative cell viability in H441 cells following the specified treatments. Data shown as mean ± SD, n=4 independent samples. (g) Median fluorescent intensity of the ISRE-GFP reporter in 293T cells following the specified treatments. Data shown as mean ± SD, n=3 independent samples. (h) Model depicting the role of DNA MMR in preserving antiviral gene expression. (i) RNAseq data showing fold change of mRNA levels in H441 cells comparing PR8-infected cells transfected with non-targeting siRNA (black) or MSH2+MSH6 siRNA (blue) to mock-infected cells. Inset is a magnified view of all genes induced >5-fold in PR8-infected cells treated with non-targeting siRNA. (j) Chart grouping all of the genes induced >5-fold in PR8-infected cells based on the effect MMR knockdown has on their mRNA levels. (k) Heat map displaying the effect of MMR knockdown on ISG and antiviral genes from the group of genes displayed in j. (l-o) Fold induction of (l) IFI44L and (n) IFIT1 RNA levels after viral infection as well as the difference in infection-induced (m) IFI44L and (o) IFIT1 RNA levels (48 hpi) after knockdown of control or MMR genes. Data shown as mean ± SD, n=4 independent samples. Data are representative of at least three independent experiments. (p) Western blot of IFIT1 in H441 cells following the specified treatments. Tubulin = loading control. For all panels: p-values calculated using unpaired two-tailed t tests; representative of two independent experiments, unless otherwise indicated.
Previous reports have shown that DNA repair pathways in general are frequently downregulated during IAV infection36–38. To our knowledge, the contribution of virally-induced oxidative DNA damage to host shutoff, however, is unexplored. Infection of our NanoLuc reporter club cells led to the expected reduction in luciferase levels (Fig. 5e–f). In order to test if the maintenance of DNA MMR activity was helping to limit host shutoff, we repeated this experiment after knockdown of DNA MMR genes and observed a further reduction of NanoLuc expression (Fig. 5e–f). To determine whether the DNA MMR mediated effects on host translation could extend to antiviral gene expression, we made use of an interferon reporter construct (ISRE-GFP). Reporter expression was significantly reduced upon knockdown of DNA MMR genes (Fig. 5g). Together, these data are consistent with a model in which DNA MMR activity repairs oxidative DNA damage to allow for transcriptional induction of antiviral genes (Fig. 5h).
In order to more broadly investigate the impact of DNA MMR on antiviral gene expression, we performed RNAseq on H441 cells transfected with control or DNA MMR siRNA and then infected with WT PR8 for 24 h (Fig. 5i, Supplementary Table 4, NCBI GEO Series GSE130189). We confirmed depletion of the MMR genes, as well as viral infection (Supplementary Fig. 4a–c). Out of the 282 genes induced >5-fold by PR8 infection, the expression of 115 genes was reduced (defined as a log2(fold change)<−0.2) in cells transfected with MSH2+6 siRNA (Fig. 5i–j, Supplementary Table 5). More specifically, out of the 51 ISG and stress response genes induced >5-fold, the expression of 28 of those genes was reduced >10% by MSH2+6 depletion (Fig. 5k, Supplementary Table 5). We confirmed that induction of two of the ISGs identified in the RNA sequencing (IFI44L and IFIT1) was significantly attenuated in cells depleted of DNA MMR genes (Fig. 5l–o). We also verified that the reduction of IFIT1 mRNA led to reduced IFIT1 protein in infected H441 cells depleted of DNA MMR genes (Fig. 5p).
Enhanced viral suppression of the DNA MMR pathway decreases cellular survival and increases the severity of viral disease.
We wanted to generate a recombinant IAV strain that would suppress the DNA MMR pathway in all infected cells. In order to accomplish this goal, we inserted artificial microRNAs (amiRNAs) into an extension of the 3’ UTR of segment 6 (Fig. 6a). Processing of amiRNAs leads to the generation of small RNAs which are functionally equivalent to siRNAs39. We generated two separate viruses: one with an amiRNA designed to target GFP as a control (GFP-amiRNA virus), and a second targeting mouse MSH6 in order to suppress DNA MMR activity (MSH6-amiRNA virus). We also inserted Cre recombinase into segment 8 of both viruses (Fig. 6a) to facilitate measurement of cellular survival. We verified functional Cre recombinase expression and monitored processing of the cleaved, mature amiRNAs via northern blot (Fig. 6b–c). We also confirmed that the mature amiRNAs reduced MSH6 RNA levels in mouse epithelial cells (Fig. 6d). In chicken cells where the amiRNA against mouse MSH6 is non-functional (Fig. 6e), we performed growth curves and observed that the growth of the GFP-amiRNA and MSH6-amiRNA viruses was identical (Fig. 6f).
Figure 6. DNA MMR is required for cellular survival and protection from virulence in mice.
(a) Diagram of the engineered IAV PR8 Seg8-Cre + Seg6-amiRNA viruses. (b) PR8-Cre+amiRNA viruses activate Cre-reporter (ZsGreen) expression in H441-CR cells (nuclei, blue). Scale bars = 200 μm. Representative of two independent experiments. (c) Northern blot of total RNA from MLE15 cells infected with the indicated PR8-Cre+amiRNA virus. Probes are specific for the GFP amiRNA, MSH6 amiRNA, or U6 snRNA loading control. Red arrows = precursor-amiRNA; black arrows = mature amiRNA. Representative of two independent experiments. (d) Relative MSH6 RNA levels in MLE15 cells co-transfected with a pLEX plasmid (for puromycin selection) and the indicated Seg6-amiRNA plasmid. Data shown as mean ± SD, n=3 samples. Representative of three independent experiments. (e) Alignment of the antisense MSH6 amiRNA sequence and the MSH6 gene target sequence (both mouse and chicken). Bases are colored green (complementary) or red (non-complementary). (f) Growth curve in fertilized chicken eggs comparing WT PR8 and PR8-Cre+amiRNA viruses. Data shown as mean ± SEM, n=3 samples; *p<0.05, ns=not significant. Representative of two independent experiments. (g) Experimental timeline for differentiation and infection of air-liquid interface (ALI) cultures and validation of differentiation by staining for mature ciliated cells (ACTUB+, green), basal cells (Krt5+, red), and nuclei (blue). Scale bar = 25 μm. Representative of two independent experiments. (h) ALI cultures derived from tdTomato-Cre-reporter transgenic mice were infected with the indicated viruses. Quantification of the number of HA+ (PY102+) cells per field of view at 1 dpi (n=5), and the number of tdTomato+ cells per field of view at 8 dpi (n=10). Data shown as mean ± SD. Representative of two independent experiments. (i) Nuclei staining (DAPI, blue) and survivor cells (tdTomato, red) in ALI cultures at 8 dpi. Scale bars = 50 μm. Representative of two independent experiments. (j-l) Weight loss and survival curves for mice infected with (j) 4000, (k) 1200, or (l) 400 PFU of the indicated virus. Data shown as mean ± SD, n=4 mice per group. Representative of two independent experiments. For all panels: p-values calculated using unpaired two-tailed t tests.
In order to evaluate the effect of MSH6 targeting in primary cells, we generated air-liquid interface (ALI) epithelial cultures (Fig. 6g) from transgenic mice harboring a LoxP-Stop-LoxP-tdTomato cassette and infected them with our amiRNA viruses. An equal number of cells were initially infected with the GFP-amiRNA and MSH6-amiRNA viruses, but at later timepoints, we observed a significant reduction in cellar survival when MSH6 was targeted by the virally-encoded amiRNA (Fig. 6h–i). Next, we set out to determine the roles of DNA MMR and club cell survival in viral pathogenesis. We infected 8-week-old female C57BL/6J mice intranasally with a range of doses of the parental WT PR8, GFP-amiRNA, or MSH6-amiRNA viruses. Mice that were infected with the MSH6-amiRNA virus experienced increased morbidity and mortality relative to GFP-amiRNA-infected mice (Fig. 6j–l), although both modified viruses were attenuated relative to parental PR8. The MSH6-amiRNA virus also replicated to higher titers in the mouse lungs at 4 dpi (Supplementary Fig. 5), which is consistent with dampened antiviral gene expression.
DISCUSSION
Until recently, it was assumed that IAV clearance from the host was the result of the elimination of all infected cells. However, it is now clear that some cells can clear the virus and survive infection. In this report, we demonstrate that DNA MMR is critical for club cell survival of IAV infection. We found that IAV infection generally leads to a reduction in DNA MMR, while club cells are uniquely able to maintain normal levels of DNA MMR activity. The higher level of DNA MMR activity allows club cells to repair virus-induced oxidative DNA damage, facilitating the transcriptional expression of antiviral genes that together likely contribute to viral clearance and cell survival. This has important consequences in vivo, as loss of DNA MMR led to decreased cell survival and increased severity of viral disease.
Our observation of IAV-induced oxidative DNA damage is consistent with previous reports of DNA damage after IAV infection22,40,41. In addition to IAV, many other RNA viruses are known to cause significant DNA damage during infection42. This includes RNA viruses that exclusively replicate in the cytoplasm, such as HCV and alphaviruses, which have been found to induce DNA damage and activate DNA damage response elements in infected cells43–46. Altogether, these data suggest a broader role for DNA MMR and the innate cellular response against many RNA viruses. Although the sites of oxidative DNA damage were not mapped in our study, it is known that DNA damage preferentially accumulates at sites of actively transcribed DNA47,48. It is therefore likely that during viral infection, DNA damage accumulates at genetic loci involved in the antiviral and stress response pathways that are highly upregulated after viral infection.
In addition to inducing oxidative DNA damage, infection with a number of RNA viruses is linked to the general downregulation of DNA MMR pathways in infected cells36–38,49,50. While it is currently unclear whether DNA repair pathway suppression is the result of a host response, or subversion by viruses, we propose that DNA MMR is a key host-pathogen interface that impacts infection by a range of pathogens. In support of this, we mined publicly available transcriptomic studies and found many DNA viruses and bacterial pathogens whose infection is also associated with the downregulation of DNA MMR including Bombyx mori nucleopolyhedrovirus51, Vaccinia virus52, Escherichia coli53, Yersinia enterocolitica54, and Helicobacter pylori55. Given that this response to infection appears to be more common than previously appreciated, determining how the DNA MMR pathway and other DNA repair pathways are controlled during infection is an important area of future study.
One question raised by this study is why the DNA MMR pathway is essential for survival when other oxidative DNA damage repair pathways exist in the cell. Nucleotide excision repair (NER), base excision repair (BER), and mismatch repair (MMR) are all used by the cell to repair damage restricted to a single strand of the DNA, but show different specificity based on the type of damage they recognize and repair56–58. While we did not identify these genes in our screen, future studies will dissect the role of these pathways in cellular survival. Another question is how IAV inhibits DNA MMR activity in most cells. Possible mechanisms likely relate to host shutoff which is largely implemented by the viral polymerase, NS1, and PA-X proteins59–61. We hypothesize that club cells are able to partially evade the IAV shutoff mechanisms, and the pathways used to accomplish this will be explored in the future. Finally, it will also be important to determine the relative DNA MMR activity and magnitude of oxidative DNA damage after infection with a number of different influenza virus subtypes to define any strain specific effects on this process.
In this study, we found that IAV-induced oxidative DNA damage impacts both cellular survival and antiviral gene expression. Therefore, we propose that in addition to promoting the maintenance of cellular genomic integrity, an important aspect of DNA MMR is to also promote strong induction of antiviral genes; perhaps including those genes responsible for non-lytic viral clearance from infected cells. Determining the pathways required for non-lytic clearance, and their mechanisms of action, will be important not only to understand viral infection but also as a novel strategy to boost viral clearance from cells that cannot be replaced. Altogether, our study has identified a previously unappreciated interface between the virus and host. DNA MMR, and DNA repair pathways in general, are commonly attenuated during infection with diverse pathogens, and this may be the result of common regulatory mechanisms. Preventing DNA MMR pathway suppression in host cells, or inducing repair pathways, may represent a general antimicrobial strategy for the future.
METHODS
Cells.
All cells were obtained from ATCC and grown at 37°C in 5% CO2. H441 cells were grown in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal bovine serum, HEPES, glucose, sodium pyruvate, and penicillin-streptomycin. 293T and A549 cells (ATCC) were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 5% fetal bovine serum, GlutaMAX, and penicillin-streptomycin. Madin-Darby canine kidney (MDCK) cells were grown in minimal essential medium (MEM) supplemented with 5% fetal bovine serum, HEPES, NaHCO3, GlutaMAX, and penicillin-streptomycin. MLE15 cells were grown in DMEM supplemented with 10% fetal bovine serum, GlutaMAX, and penicillin-streptomycin. The Cre-reporter cassette was constructed and transduced into H441 and A549 cells as previously described15. The growth curve comparing division rates of A549-CR and H441-CR was conducted in 6-wells. Cells were seeded at 150,000 cells per well and were collected and counted at 24, 48, 72, and 96 h.
Viruses.
Recombinant viruses were generated using reverse genetics methods as previously described62,63, using the bicistronic pDZ rescue plasmid system. All viruses were based off of the A/Puerto Rico/8/1934 (PR8) H1N1 influenza virus background. The WT PR8 virus contains all WT PR8 segments. The NEBuilder HiFi DNA assembly kit (NEB) was used to assemble DNA fragments to create the desired recombinant DNA gene segments. The IAV-Cre recombinant virus was engineered to express Cre recombinase within the PR8 PB2 segment (segment 1), as previously described7. The time-course measuring ZsGreen expression during IAV-Cre infection in A549-CR and H441-CR cells was conducted by infecting cells at MOI=1. Every 24 h, cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) and nuclei were stained with Hoescht dye before pictures were taken with the ZOE fluorescent cell imager (Bio-Rad). The IAV-Luc recombinant virus was also engineered to express luciferase in the PR8 PB2 segment (segment 1), as previously described64. For the PR8-Cre+amiRNA viruses, the Cre recombinase was inserted into the PR8 NS segment (segment 8) following separation of the NS1 and NEP coding regions, and the amiRNA expression sequence was inserted into the PR8 NA segment (segment 6). After assembly and transformation of the constructs into high-efficiency competent cells (NEB), the plasmids were collected and used in separate rescue transfections in 293T/MDCK co-cultures. Rescued virus was expanded in day 10 fertilized chicken eggs and then plaque-purified on MDCK cells. Individual plaques were picked and expanded in day 10 fertilized chicken eggs before being validated via RT-PCR and sequencing of the recombinant segments. For the viruses containing Cre recombinase, Cre activity was confirmed by infecting H441-CR at MOI=2 and observing the expression of the Cre-controlled ZsGreen reporter in cell monolayers at 24–48 hpi.
siRNA screen.
Single siRNAs (2 sites/target, 25 nM final concentration, Qiagen) were spotted in collagen-coated 384-well plates complexed with HiPerFect (0.6 μL/well in 9.4 μL OptiMEM). Following complexing, H441-CR cells (1,625 cells/well) were added to each well and infected 48 h later with IAV-Cre at an MOI=5. After 120 h, cells were fixed with 4% PFA, washed, and stained with Hoechst stain. All solutions were added using automated liquid handling (Well-Mate, Thermo Fisher) to limit well-to-well variability. Images were captured (3 sites/well) at 10X using an ImageXpressMicro Microscope (Molecular Devices). Automated image analysis (MetaXpress) was used to calculate the total number of cells (Hoechst+) and the number of survivor cells (ZsGreen+). These values were used to calculate the median and interquartile range (on log transformed data), which were then used to calculate robust Z-scores. The median of each plate in the screen was used to calculate baseline. Hits were identified as those wells that exhibited a change in survivor cell percentage of total cells by the indicated standard deviations from calculated Z-scores within each plate.
siRNA survivor cell assays.
H441-CR cells were transfected with 0.2 μM siRNA using HiPerFect reagent (Qiagen) in OptiMEM. After 48 h, cells were infected with IAV-Cre at MOI=1 for 1 h before virus was removed and replaced with complete media. Cells were incubated at 37°C for 5 days before being fixed with 4% PFA and stained with Hoescht dye for nucleus labeling. Images were then collected using the ZOE fluorescent cell imager (Bio-Rad) to identify the level of ZsGreen-positive cells indicating survival of direct infection. Image analysis was done using ImageJ (NIH).
Viral replication counter-screen.
H441-CR cells were transfected with the siRNA mini-library and then infected after 48 h with IAV-Luc at MOI=0.5. At 16 hpi, cells were lysed and luciferase activity was measured using a luciferase assay system (Promega) as a readout of viral replication. All siRNAs were compared relative to the non-targeting control siRNA and those which altered viral replication greater than two standard deviations were eliminated from the final list of screen hits.
Viral growth comparisons.
The RdRP replication assay was conducted as previously described64. For the multi-cycle growth assay comparing A549 and H441 cells, WT PR8 was added to cells at MOI=1 for 1 h before being maintained in OptiMEM with 0.3 μg/mL TPCK-treated trypsin protease at 37°C. For the experiment comparing IAV and Sindbis survival, H441-CR cells were infected with MOI=1 of WT PR8 influenza virus or Sindbis virus (gift from Benjamin tenOever). After 48 h, the total number of remaining cells was quantified compared to mock-infected controls using flow cytometry and counting beads. For the amiRNA virus growth curves, eggs were injected with 1000 PFU virus and incubated at 37°C for 24–72 h, or MLE15 cells were infected at MOI=0.1 for one hour before being maintained in OptiMEM with 0.4 μg/mL TPCK-treated trypsin protease for 24–72 h at 37°C. All samples were then titered using plaque assays.
Plaque assays.
Viral titers were determined using standard plaque assay procedures on MDCK cells. Serial virus dilutions were incubated on cells for 1 h before removing the virus and adding the agar overlay. Cells were then incubated at 37°C for 48 h before being fixed in 4% PFA for at least 12 h. The 4% PFA was then aspirated, and the agar layer was removed before washing cells with PBS. Serum from WT PR8-infected mice was diluted 1:2,000 in antibody dilution buffer (5% (w/v) nonfat dried milk and 0.05% Tween-20 in PBS) and incubated on cells at 4°C for 2 h. Cells were then washed three times with PBS and then incubated for 2 h in anti-mouse IgG horseradish peroxidase (HRP)-conjugated sheep antibody (GE Healthcare) diluted 1:4,000 in antibody dilution buffer. Assays were then washed three times with PBS and exposed to 0.5 mL of True Blue peroxidase substrate (KPL) for 15 min. Plates were then washed with water and dried before plaques were counted.
Cell infections.
For experiments measuring RNA and protein levels of MMR genes and ISGs, H441-CR cells were infected with WT PR8 at MOI=5 for one hour before virus was removed and complete media was added. Cells were incubated at 37°C for 3–24 h before total RNA was collected, or for 24–48 h before total protein was collected. For experiments using siRNA-treated cells, infection was done at 48 h after siRNA transfection to ensure maximum knockdown of target genes.
qRT-PCR assays.
For cell culture-based qRT-PCR experiments, total RNA was collected using Monarch Total RNA Miniprep Kits (NEB). For the mouse lung IAV NP qRT-PCR, lungs were collected at 4 dpi (with 1200 PFU virus) and homogenized directly in Trizol, before using chloroform and isopropyl alcohol to complete the RNA extraction. For the confirmation of siRNA knockdown, measurement of ISG RNA levels, the comparison of relative MSH6 RNA levels in amiRNA-transfected cells, and the quantification of mouse lung IAV NP levels, samples were analyzed using the EXPRESS One-Step Superscript Universal qRT-PCR Kit (Thermo) and gene-specific TaqMan Expression Assay Probes (Thermo) (Supplementary Table 6). For the MMR gene panel, the qRT-PCR was performed in two separate steps. First, single-stranded cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Thermo). Second, cDNA was loaded into Custom TaqMan Array 96-well Plates (Thermo) we designed to include all major genes directly involved in the DNA MMR pathway as well as two housekeeping gene controls for normalization.
Western blotting.
Equal amounts of protein were loaded into 4–20% acrylamide gels (Bio-Rad) and transferred to nitrocellulose membrane. PBS with 5% (w/v) nonfat dried milk and 0.1% Tween-20 was used to block for 2 h at 4°C. Primary antibodies were then incubated with the membrane overnight at 4°C. Antibodies used were rabbit anti-MSH2 (Cell-Signaling Technology D24B5), rabbit anti-MSH6 (Abcam EPR3945), mouse anti-HA (PY102, gift from Thomas Moran at Mt. Sinai), rabbit anti-Mx1 (Abcam 95926), and mouse anti-α-Tubulin (Sigma T5168). Membranes were washed five times in PBS with 0.1% Tween-20 and then anti-mouse-HRP or anti-rabbit-HRP secondary antibody was added for 1 h. The membrane was then washed five times and Clarity or Clarity Max ECL substrate (Bio-Rad) was added before being exposed to film and developed. Uncropped scans of all Western blots are displayed in Supplementary Figure 6.
DNA MMR activity assay.
The assay was designed as previously described17, with some adjustments. In short, we digested our synthesized pLCMV-NanoLuc plasmid with NheI and PflFI to remove the NanoLuc start codon region. We then ligated in annealed oligos containing a G-G mismatch in the third position of the NanoLuc start codon; this was referred to as our “Mismatch plasmid.” A set of annealed oligos with a WT start codon was also ligated in to be used as a normalization control. The annealed oligos also contained a disrupted HpaI restriction site, allowing us to specifically eliminate any uncut parental pLCMV-NanoLuc plasmid (which contains an intact HpaI site directly upstream of start codon) following plasmid ligation. Validation of the MMR activity assay was done using A549 cells transfected with MSH2 and MSH6 siRNA. For the comparison of A549 and H441 cells, the cells were infected at MOI=10, transfected with 0.5 μg of the WT or Mismatch plasmid at 24 hpi, and then analyzed at 16 h post-transfection. To measure NanoLuc expression, cells were lysed with Luciferase Cell Lysis Buffer (NEB) and then evaluated using the Nano-Glo Luciferase Assay Reagent Kit (Promega).
DNA damage assays.
For phospho-H2AX staining, H441 cells were plated on collagen-coated cover slides and infected with WT PR8 (MOI=5) for indicated lengths of time before fixation with 4% PFA. Positive controls were treated with 1 μM Etoposide (Abcam) or 5 mM H2O2 (EMD Millipore) for 30 min before fixation. Cells were stained using the Histone H2A.XS139ph antibody (Active Motif, #39117). DAPI was added in the mounting solution for nuclei staining. Images were collected using a Leica SP5 inverted confocal microscope. Images within the same panel were collected at the same time using the same settings and analyzed using ImageJ (NIH). Quantification of 8-OHdG was done using the OxyDNA Assay Kit (Calbiochem) following the manufacturer’s instructions with H441 cells infected with WT PR8 (MOI=10) for 48 h. The gating strategy used for the 8-OHdG flow cytometry analysis is shown in Supplementary Figure 7. The modified comet assay was performed as previously described28,29, with some alterations. H441 cells were transfected with MSH2+6 or non-targeting control siRNA, and then infected with WT PR8 (MOI=10) 48 h after siRNA transfection. At 24 hpi, cells were collected and mixed with 1% TopVision Low Melting Point Agarose (ThermoScientific) before plating on agarose coated microscope slides. Agar-embedded cells were then lysed and treated with Fpg restriction enzyme. After alkaline treatment to neutralize the restriction enzyme, the cells were exposed to gel electrophoresis to induce migration of digested nuclei. Cells were washed with neutralization buffer and then stained with DAPI. Images were collected using a Leica SP5 inverted confocal microscope. Images within the same panel were collected at the same time using the same settings and analyzed using ImageJ (NIH). For the ROS scavenger assays, A549-CR and H441-CR cells were treated with 0.5 mM Trolox ((±)-6-Hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid, Sigma-Aldrich) for 1 h before infection with IAV-Cre (MOI=5). Fresh Trolox (0.5 mM) was then maintained in the cell media for 3 dpi before being removed and replaced with media containing no Trolox. At 5 dpi, cells were fixed with 4% PFA and stained with Hoescht dye before analyzing the percentage of ZsGreen+ cells using a Cellomics Arrayscan VTI HCS.
Expression of Nanoluc and ISRE-GFP reporters.
All experiments measuring expression of the pLCMV-NanoLuc plasmid were conducted in H441 cells and done using the uncut reporter plasmid we originally synthesized for the MMR activity assay described above. For the ROS experiments, 5 mM H2O2 was added to the media on the cells and incubated at 37°C for 30 min before being washed and replaced with fresh complete media. Immediately following the addition of fresh media, the cells were transfected with 0.5 μg of pLCMV-NanoLuc plasmid using Lipofectamine2000 (Thermo). After 6 h, the cells were lysed with Luciferase Cell Lysis Buffer (NEB) and luminescence was measured using the Nano-Glo Luciferase Assay Reagent Kit (Promega). For the IAV infection experiments, cells were transfected with siRNA and incubated for 48 h before being infected with WT PR8 at MOI=10. At 18 hpi, the infected cells were transfected with 0.5 μg of the pLCMV-NanoLuc plasmid. After 6 h, cells were lysed with Luciferase Cell Lysis Buffer (NEB) and luminescence was measured using the Nano-Glo Luciferase Assay Reagent Kit (Promega). Cell viability was confirmed in all experiments using cells treated/infected in parallel and the CellTiter-Glo Luminescent Cell Viability Assay (Promega). We designed the ISRE-GFP reporter in the pTRIP vector by inserting seven consensus ISRE sites directly upstream of a minimal CMV promoter. This promoter drives super-folder GFP fused to a mouse ornithine decarboxylase domain to decrease the protein half-life. For the ISRE-GFP reporter experiments, 293T cells were transfected with siRNA and incubated for 48 h before transfecting in the reporter. After 24 h, 1000 U/mL IFN-alpha was added to the cells and flow analysis was done to look at the level of GFP reporter expression.
RNAseq.
H441 cells were transfected with siRNA and incubated for 48 h before being infected with WT PR8 (MOI=10). Total RNA was collected at 24 hpi using Monarch Total RNA Miniprep Kits (NEB). RNA was then prepped for RNAseq submission using the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB), NEBNext Ultra II RNA Library Prep Kit for Illumina (NEB), and NEBNext Multiplex Oligos for Illumina (NEB). Mapping of raw reads to the human hg19 reference genome was accomplished using a custom app on the Illumina BaseSpace Sequence Hub7. After data normalization, average read values were compared across samples. For comparisons in which some samples had zero reads detected for a specific gene, 1 read was added to all values in the sample to complete analyses that required non-zero values. Raw data is available at NCBI GEO (Series GSE130189).
ISG expression assays.
All experiments measuring ISG expression were conducted in H441 cells. For experiments including ROS treatment, cells were transfected with siRNA and after 48 h, 5 mM H2O2 was added to the media for 30 min. Media was then aspirated and replaced with media containing 500 U/mL IFN-alpha for activation of ISG expression. Cells were then incubated at 37°C for 3 h before total RNA was collected and for 24 h before total protein samples were collected. For IFI44L and IFIT1 experiments, cells were transfected with siRNA and incubated for 48 h before infecting with WT PR8 at MOI=10 and collecting total RNA at 48 hpi.
Northern blot.
Total RNA samples were collected from either MLE15 cells infected in a 10 cm dish with the PR8-Cre+amiRNA viruses for 24 h, or from MLE15 cells transfected in a 10 cm dish with 15 μg of the Seg6-amiRNA plasmids for 24 h. Equal amounts of RNA was loaded into a 15% TBE-urea gel (Bio-Rad) and transferred to a nitrocellulose membrane. The dried membrane was UV-crosslinked and then exposed to pre-hybridization solution for 1 h. Probes specific for the GFP amiRNA, MSH6 amiRNA, and U6 control snRNA were labeled with 32P-ATP and then added to the membrane for 1 h. Membrane was washed two times with saline-sodium citrate (SSC) buffer plus 0.05% SDS and then exposed to film for 96 h before developing. Uncropped scans are displayed in Supplementary Figure 6.
amiRNA activity confirmation.
MLE15 cells were co-transfected in 24-well plates with 500 ng of the Seg6-amiRNA plasmids and 500 ng of a pLEX helper plasmid with puromycin resistance marker using Lipofectamine2000 reagent (Thermo). After 24h, co-transfected cells were selected for with 2.5 μg/mL puromycin. After 48 h of selection (72 h post-transfection), total RNA was collected using TRIzol reagent. MSH6 RNA levels were then quantified from each of the samples using one-step qRT-PCR.
ALI culture infections.
Tracheal epithelial cells were isolated from transgenic Cre-tdTomato-reporter mice and cultured at air-liquid interface for 14 days to allow formation of a pseudostratified epithelium. This was monitored using markers for mature differentiated cell types (KRT5 – basal stem cells, ACTUB – mature ciliated cells). These cultures were then infected with 500,000 PFU of WT PR8, GFP-amiRNA, or MSH6-amiRNA virus. At 1 and 8 dpi, cultures were collected, rinsed two times with PBS, and fixed with 2% PFA at room temperature for 15 min. Cultures at 1 dpi were stained with PY102 to quantify the number of infected cells and cultures at 8 dpi were imaged using tdTomato as a marker of survivor cells. All cultures were imaged on a Leica SP5 inverted confocal microscope using a 40X oil objective. Images within the same panel were collected at the same time using the same settings. Images were analyzed using Fiji (NIH) using macros that were written to automatically quantify the number of PY102+ and tdTomato+ cells per field of view.
Mouse infections.
Eight-week-old female C57BL/6J mice were used for all experiments, with a sample size of 4 mice per dose of virus. Prior to infection, mice were anesthetized with a 100-μL injection of a ketamine-xylazine mixture. After being tail-marked and weighed, mice were intranasally infected with 40 μL of virus diluted in pharmaceutical-grade PBS. Mice were weighed daily and euthanized once their body weight reached <80% of the starting weight as a humane endpoint. Euthanasia was performed via CO2 as the primary method and a bilateral thoracotomy was performed as the secondary method. All procedures were approved by the Duke University IACUC.
Statistical analysis.
Comparison of data sets was performed using an unpaired, two-tailed Student t tests unless otherwise stated. Analysis was performed using Prism 7 and Prism 8 (GraphPad) software.
DATA AVAILABILITY
The raw RNAseq data files from Fig. 5i–k are available at NCBI GEO (Series GSE130189). The raw data for Fig. 2b–d, 3d, and 5i-k are available in Supplementary Tables 1–5. Raw data from all other figures and unique materials, including viruses and plasmids, are available from the corresponding authors upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Hal Bogerd and Brian Cullen (Duke University) for their help with the amiRNA Northern blots. We would like to thank Peter Palese (Mt. Sinai) for support and reagents during preliminary optimization experiments. We would also like to thank Benjamin tenOever (Mt. Sinai) for his help in designing the amiRNA-expressing viruses. We are also grateful for contributions made by Heather Froggatt (Duke University) in researching the literature on other pathogens that downregulate DNA MMR. The RNAseq mapping pipeline was developed by David Sachs. N.S.H. is partially supported by NIH K22-AI116509–01, R21-AI133444–01, R01-HL142985, R01-AI137031 and the Duke School of Medicine Whitehead Scholarship. B.S.C. is supported by NIH training grant T32-CA009111. R.E.D. is supported by NIH Training grant T32-GM007184–41. S.C. is supported by NIH grants R01AI074951, R01AI140539, and R01AI122749, and is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award.
Footnotes
REQUESTS FOR MATERIALS
Please address any requests to Nicholas S. Heaton (nicholas.heaton@duke.edu).
COMPETING INTERESTS
Duke University has filed a provisional patent for targeting DNA MMR as a method to enhance the growth of influenza vaccine strains.
REFERENCES
- 1.Downey J, Pernet E, Coulombe F & Divangahi M Dissecting host cell death programs in the pathogenesis of influenza. Microbes Infect, doi: 10.1016/j.micinf.2018.03.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turpin E et al. Influenza virus infection increases p53 activity: role of p53 in cell death and viral replication. J Virol 79, 8802–8811, doi: 10.1128/JVI.79.14.8802-8811.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orzalli MH & Kagan JC Apoptosis and Necroptosis as Host Defense Strategies to Prevent Viral Infection. Trends Cell Biol 27, 800–809, doi: 10.1016/j.tcb.2017.05.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrhardt C et al. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J Virol 81, 3058–3067, doi: 10.1128/JVI.02082-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhirnov OP & Klenk HD Control of apoptosis in influenza virus-infected cells by up-regulation of Akt and p53 signaling. Apoptosis 12, 1419–1432, doi: 10.1007/s10495-007-0071-y (2007). [DOI] [PubMed] [Google Scholar]
- 6.van de Sandt CE, Kreijtz JH & Rimmelzwaan GF Evasion of influenza A viruses from innate and adaptive immune responses. Viruses 4, 1438–1476, doi: 10.3390/v4091438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heaton NS et al. Long-term survival of influenza virus infected club cells drives immunopathology. J Exp Med 211, 1707–1714, doi: 10.1084/jem.20140488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reuther P et al. Generation of a variety of stable Influenza A reporter viruses by genetic engineering of the NS gene segment. Scientific Reports 5, doi: 10.1038/srep11346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burdeinick-Kerr R & Griffin DE Gamma interferon-dependent, noncytolytic clearance of sindbis virus infection from neurons in vitro. J Virol 79, 5374–5385, doi: 10.1128/JVI.79.9.5374-5385.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidotti LG et al. Intracellular Inactivation of the Hepatitis B Virus by Cytotoxic T Lymphocytes. Immunity 4, 25–36, doi: 10.1016/s1074-7613(00)80295-2 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Guidotti LG et al. Noncytopathic Clearance of Lymphocytic Choriomeningitis Virus from the Hepatocyte. The Journal of Experimental Medicine 189, 1555–1564, doi: 10.1084/jem.189.10.1555 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chisari FV Viruses, Immunity, and Cancer: Lessons from Hepatitis B. The American Journal of Pathology 156, 1117–1132, doi: 10.1016/s0002-9440(10)64980-2 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin DE Recovery from viral encephalomyelitis: immune-mediated noncytolytic virus clearance from neurons. Immunol Res 47, 123–133, doi: 10.1007/s12026-009-8143-4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudchodkar SB & Levine B Viruses and autophagy. Rev Med Virol 19, 359–378, doi: 10.1002/rmv.630 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton JR et al. Club cells surviving influenza A virus infection induce temporary nonspecific antiviral immunity. Proc Natl Acad Sci U S A 113, 3861–3866, doi: 10.1073/pnas.1522376113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bridge G, Rashid S & Martin SA DNA mismatch repair and oxidative DNA damage: implications for cancer biology and treatment. Cancers (Basel) 6, 1597–1614, doi: 10.3390/cancers6031597 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei X, Zhu Y, Tomkinson A & Sun L Measurement of DNA mismatch repair activity in live cells. Nucleic Acids Res 32, e100, doi: 10.1093/nar/gnh098 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macpherson P et al. 8-oxoguanine incorporation into DNA repeats in vitro and mismatch recognition by MutSalpha. Nucleic Acids Res 33, 5094–5105, doi: 10.1093/nar/gki813 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nencioni L et al. Influenza A virus replication is dependent on an antioxidant pathway that involves GSH and Bcl-2. FASEB J 17, 758–760, doi: 10.1096/fj.02-0508fje (2003). [DOI] [PubMed] [Google Scholar]
- 20.Sgarbanti R et al. Redox regulation of the influenza hemagglutinin maturation process: a new cell-mediated strategy for anti-influenza therapy. Antioxid Redox Signal 15, 593–606, doi: 10.1089/ars.2010.3512 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Amatore D et al. Influenza virus replication in lung epithelial cells depends on redox-sensitive pathways activated by NOX4-derived ROS. Cell Microbiol 17, 131–145, doi: 10.1111/cmi.12343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li N et al. Influenza infection induces host DNA damage and dynamic DNA damage responses during tissue regeneration. Cell Mol Life Sci 72, 2973–2988, doi: 10.1007/s00018-015-1879-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin X et al. The Influenza Virus H5N1 Infection Can Induce ROS Production for Viral Replication and Host Cell Death in A549 Cells Modulated by Human Cu/Zn Superoxide Dismutase (SOD1) Overexpression. Viruses 8, doi: 10.3390/v8010013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell AB, Trapnell C & Bloom JD Extreme heterogeneity of influenza virus infection in single cells. Elife 7, doi: 10.7554/eLife.32303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin N, Pyo CW, Jung KI & Choi SY Influenza A virus PB1-F2 is involved in regulation of cellular redox state in alveolar epithelial cells. Biochem Biophys Res Commun 459, 699–705, doi: 10.1016/j.bbrc.2015.03.010 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Wang QW et al. Anti-influenza A virus activity of rhein through regulating oxidative stress, TLR4, Akt, MAPK, and NF-kappaB signal pathways. PLoS One 13, e0191793, doi: 10.1371/journal.pone.0191793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valavanidis A, Vlachogianni T & Fiotakis C 8-hydroxy-2’ -deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 27, 120–139, doi: 10.1080/10590500902885684 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Singh NP, McCoy MT, Tice RR & Schneider EL A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 175, 184–191 (1988). [DOI] [PubMed] [Google Scholar]
- 29.Collins AR, Dusinska M & Horska A Detection of alkylation damage in human lymphocyte DNA with the comet assay. Acta Biochima Polonica 48, 611–614 (2001). [PubMed] [Google Scholar]
- 30.Hamad I, Arda N, Pekmez M, Karaer S & Temizkan G Intracellular scavenging activity of Trolox (6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid) in the fission yeast, Schizosaccharomyces pombe. J Nat Sci Biol Med 1, 16–21, doi: 10.4103/0976-9668.71667 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuraoka I et al. Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J Biol Chem 278, 7294–7299, doi: 10.1074/jbc.M208102200 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Charlet-Berguerand N et al. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J 25, 5481–5491, doi: 10.1038/sj.emboj.7601403 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellon I & Champe GN Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc Natl Acad Sci U S A 93, 1292–1297 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellon I, Rajpal DK, Koi M, Boland CR & Champe GN Transcription-Coupled Repair Deficiency and Mutations in Human Mismatch Repair Genes. Science 272, 557–560 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Ni TT, Marsischky GT & Kolodner RD MSH2 and MSH6 Are Required for Removal of Adenine Misincorporated Opposite 8-Oxo-Guanine in S. cerevisiae. Molecular Cell 4, 439–444, doi: 10.1016/s1097-2765(00)80346-9 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Bercovich-Kinori A et al. A systematic view on influenza induced host shutoff. Elife 5, doi: 10.7554/eLife.18311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domingues P et al. Global Reprogramming of Host SUMOylation during Influenza Virus Infection. Cell Rep 13, 1467–1480, doi: 10.1016/j.celrep.2015.10.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noh H, Shoemaker JE & Gunawan R Network perturbation analysis of gene transcriptional profiles reveals protein targets and mechanism of action of drugs and influenza A viral infection. Nucleic Acids Res 46, e34, doi: 10.1093/nar/gkx1314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varble A et al. An in vivo RNAi screening approach to identify host determinants of virus replication. Cell Host Microbe 14, 346–356, doi: 10.1016/j.chom.2013.08.007 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Khanna M et al. Detection of influenza virus induced ultrastructural changes and DNA damage. Indian J Virol 21, 50–55, doi: 10.1007/s13337-010-0004-1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vijaya Lakshmi AN, Ramana MV, Vijayashree B, Ahuja YR & Sharma G Detection of influenza virus induced DNA damage by Comet assay. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 442, 53–58, doi: 10.1016/s1383-5718(99)00058-3 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Ryan EL, Hollingworth R & Grand RJ Activation of the DNA Damage Response by RNA Viruses. Biomolecules 6, 2, doi: 10.3390/biom6010002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machida K et al. Hepatitis C virus infection activates the immunologic (type II) isoform of nitric oxide synthase and thereby enhances DNA damage and mutations of cellular genes. J Virol 78, 8835–8843, doi: 10.1128/JVI.78.16.8835-8843.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machida K et al. Hepatitis C virus inhibits DNA damage repair through reactive oxygen and nitrogen species and by interfering with the ATM-NBS1/Mre11/Rad50 DNA repair pathway in monocytes and hepatocytes. J Immunol 185, 6985–6998, doi: 10.4049/jimmunol.1000618 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clavarino G et al. Induction of GADD34 is necessary for dsRNA-dependent interferon-beta production and participates in the control of Chikungunya virus infection. PLoS Pathog 8, e1002708, doi: 10.1371/journal.ppat.1002708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nargi-Aizenman JL, Simbulan-Rosenthal CM, Kelly TA, Smulson ME & Griffin DE Rapid activation of poly(ADP-ribose) polymerase contributes to Sindbis virus and staurosporine-induced apoptotic cell death. Virology 293, 164–171, doi: 10.1006/viro.2001.1253 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Datta A & Jinks-Robertson S Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science 268, 1616–1619, doi: 10.1126/science.7777859 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Morey NJ, Greene CN & Jinks-Robertson S Genetic analysis of transcription-associated mutation in Saccharomyces cerevisiae. Genetics 154, 109–120 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naganuma A, Dansako H, Nakamura T, Nozaki A & Kato N Promotion of microsatellite instability by hepatitis C virus core protein in human non-neoplastic hepatocyte cells. Cancer Res 64, 1307–1314 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Olejnik J et al. Ebolaviruses Associated with Differential Pathogenicity Induce Distinct Host Responses in Human Macrophages. J Virol 91, doi: 10.1128/JVI.00179-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue J et al. Dynamic interactions between Bombyx mori nucleopolyhedrovirus and its host cells revealed by transcriptome analysis. J Virol 86, 7345–7359, doi: 10.1128/JVI.07217-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beard PM et al. A loss of function analysis of host factors influencing Vaccinia virus replication by RNA interference. PLoS One 9, e98431, doi: 10.1371/journal.pone.0098431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maddocks OD, Scanlon KM & Donnenberg MS An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. MBio 4, e00152–00113, doi: 10.1128/mBio.00152-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauvonnet N, Pradet-Balade B, Garcia-Sanz JA & Cornelis GR Regulation of mRNA expression in macrophages after Yersinia enterocolitica infection. Role of different Yop effectors. J Biol Chem 277, 25133–25142, doi: 10.1074/jbc.M203239200 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Kim JJ et al. Helicobacter pylori impairs DNA mismatch repair in gastric epithelial cells. Gastroenterology 123, 542–553, doi: 10.1053/gast.2002.34751 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Iyer RR, Pluciennik A, Burdett V & Modrich PL DNA mismatch repair: functions and mechanisms. Chem Rev 106, 302–323, doi: 10.1021/cr0404794 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Marteijn JA, Lans H, Vermeulen W & Hoeijmakers JH Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 15, 465–481, doi: 10.1038/nrm3822 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Robertson AB, Klungland A, Rognes T & Leiros I DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci 66, 981–993, doi: 10.1007/s00018-009-8736-z (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jagger BW et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337, 199–204, doi: 10.1126/science.1222213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nemeroff ME, Barabino SM, Li Y, Keller W & Krug RM Influenza Virus NS1 Protein Interacts with the Cellular 30 kDa Subunit of CPSF and Inhibits 3’ End Formation of Cellular Pre-mRNAs. Molecular Cell 1, 991–1000 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Vreede FT, Chan AY, Sharps J & Fodor E Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 396, 125–134, doi: 10.1016/j.virol.2009.10.003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlivan M et al. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol 79, 8431–8439, doi: 10.1128/JVI.79.13.8431-8439.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heaton NS et al. In vivo bioluminescent imaging of influenza a virus infection and characterization of novel cross-protective monoclonal antibodies. J Virol 87, 8272–8281, doi: 10.1128/JVI.00969-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heaton BE et al. A CRISPR Activation Screen Identifies a Pan-avian Influenza Virus Inhibitory Host Factor. Cell Rep 20, 1503–1512, doi: 10.1016/j.celrep.2017.07.060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNAseq data files from Fig. 5i–k are available at NCBI GEO (Series GSE130189). The raw data for Fig. 2b–d, 3d, and 5i-k are available in Supplementary Tables 1–5. Raw data from all other figures and unique materials, including viruses and plasmids, are available from the corresponding authors upon request.