Abstract
Brain somatic mutations confer genomic diversity in the human brain and cause neurodevelopmental disorders. Recently, brain somatic activating mutations in MTOR have been identified as a major etiology of intractable epilepsy in patients with cortical malformations. However, the molecular genetic mechanism of how brain somatic mutations in MTOR cause intractable epilepsy has remained elusive. In this study, translational profiling of intractable epilepsy mouse models with brain somatic mutations and genome-edited cells revealed a novel translational dysregulation mechanism and mTOR activation–sensitive targets mediated by human MTOR mutations that lead to intractable epilepsy with cortical malformation. These mTOR targets were found to be regulated by novel mTOR-responsive 5′-UTR motifs, distinct from known mTOR inhibition–sensitive targets regulated by 5′ terminal oligopyrimidine motifs. Novel mTOR target genes were validated in patient brain tissues, and the mTOR downstream effector eIF4E was identified as a new therapeutic target in intractable epilepsy via pharmacological or genetic inhibition. We show that metformin, an FDA-approved eIF4E inhibitor, suppresses intractable epilepsy. Altogether, the present study describes translational dysregulation resulting from brain somatic mutations in MTOR, as well as the pathogenesis and potential therapeutic targets of intractable epilepsy.
Keywords: Neuroscience, Therapeutics
Keywords: Epilepsy, Genetic variation, Translation
Introduction
Brain somatic mutations are genetic variations arising in somatic cells of the brain during development and aging, and confer genomic diversity in the human brain (1, 2). However, their existence or pathogenic roles in human brain disorders remain poorly understood. Recently, we and other groups have reported that brain somatic activating mutations in MTOR are the major genetic cause of focal malformations of cortical development (FMCD), encompassing focal cortical dysplasia (FCD) and hemimegalencephaly (HME); FMCD is the most common cause of intractable epilepsy in children and is strongly associated with developmental delay and autism spectrum disorder (3–7). MTOR plays a critical role in growth and metabolism (8). Thus, single activating mutations in MTOR have been rarely reported to cause human inherited disease, perhaps due to their strong mutational effect or embryonic lethality (9). Nevertheless, brain somatic activating mutations in MTOR have been found to account for up to 40% of patients with sporadic FMCD (7, 10). Interestingly, somatic activating mutations in MTOR with low mutational burden (e.g., mutated allele frequency of ~1% in the affected tissue) have been shown to be sufficient to lead to intractable focal epilepsy (4). The molecular mechanism of how somatic activating mutations in MTOR cause intractable epilepsy, however, has remained elusive.
mTOR kinase is a major regulator of translation, which is a fundamental molecular mechanism regulating gene expression (11). Translational dysregulation is thought to underlie the molecular pathogenesis of various human diseases, particularly cancers and neurodevelopmental disorders, as they are associated with the aberrant activation of mTOR (11, 12). Accordingly, we hypothesized that translational dysregulation induced by human MTOR mutations may underlie the molecular pathogenesis of intractable epilepsy and other pathologies found in FMCD. However, owing to a lack of disease models of single activating mutations in MTOR, we found understanding of the molecular basis of translational dysregulation directly induced by human MTOR mutations to be lacking: previous studies of mTOR-driven translational control have relied on pharmacological inhibition of mTOR (13, 14).
To date, translational profiles in disease models with single MTOR mutations are not available, and questions of how human MTOR mutations lead to disease, which genes are controlled, and how they are regulated remain largely unanswered. In this study using FMCD mouse models, genome-edited cells, and patient brain tissues, we describe, for the first time to our knowledge, the translational landscape induced by human MTOR mutations, as well as the molecular pathogenesis and novel therapeutics of FMCD.
Results
Ribosome profiling in mouse models with brain somatic mutations reveals mTOR target genes contributing to FMCD.
We aimed to document whether the translational landscape of FMCD mouse models with brain somatic activating mutations in MTOR contributes to the molecular pathogenesis of FMCD. Firstly, we used 2 previously established FMCD mouse models with brain somatic mutations in MTOR (mTOR p.Cys1483Tyr or p.Leu2427Pro), as well as an mTOR wild-type (WT) mouse model (4, 15). Mice underwent in utero electroporation of mTOR mutant– and WT–expressing constructs with an IRES-GFP reporter at embryonic day 14 (E14). The mice faithfully recapitulated the major phenotypes of FMCD, including intractable epilepsy, cytomegalic dysmorphic neurons, and cortical dyslamination (4, 15) (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI127032DS1). We further performed immunofluorescence analysis of brain slices, and noted that mTOR p.Cys1483Tyr and p.Leu2427Pro elicited the activation of mTOR downstream translational machinery upon measurement of the phosphorylation of S6 and 4E-BPs in mutation-carrying neurons expressing GFP reporter at E18 (Supplemental Figure 2A) and adult mouse brains (Supplemental Figure 2B). We conducted ribosome profiling at E18 in order to focus on the direct translational outputs of mTOR mutants and to avoid potential secondary effects via any developmental processes and epileptic seizures occurring around P21 (Supplemental Figure 1B). The observed mTOR mutant–related activity was reversed upon treatment with rapamycin, an mTOR inhibitor (Supplemental Figure 1A and Supplemental Figure 2, A and B).
In analyzing translational profiles in MTOR mutation–carrying neurons isolated from electroporated mouse brains, we encountered a technical hurdle in that MTOR mutation–carrying neurons were present in only about 4%–8% of affected brain regions at E18 (Figure 1A). This low percentage of mutation-carrying neurons hampered our ability to acquire enough cells for translational profiling at an in vivo level (Figure 1A). To overcome this, we dissected mouse brain (Supplemental Figure 3A) and enriched mutant or WT mTOR–expressing GFP-positive neurons using FACS (Figure 1A). Then, we performed genome-wide ribosome profiling in mutation-carrying neurons from mTOR p.Cys1483Tyr, mTOR p.Leu2427Pro, and mTOR WT mice to monitor the impact of MTOR activating mutations (Figure 1A). We purified sufficient ribosome-protected fragments (RPFs) and mRNA for Ribo-Seq and RNA-Seq analysis, respectively (Supplemental Figure 3A). The qualities of our Ribo-Seq and RNA-Seq were validated by proper size selection of RPFs and mRNAs during library preparation (Supplemental Figure 3B); by the enrichment of RPF and mRNA reads in coding sequences (Supplemental Figure 3C and Supplemental Table 1); by the occupancy of RPF and mRNA reads around start and stop codons (Supplemental Figure 3D); by 3-nt periodicity of the Ribo-Seq reads, which is a representative signature of the movement of ribosomes in translation (Supplemental Figure 3E); and by reproducibility in Pearson’s correlation analysis (Supplemental Figure 3F).
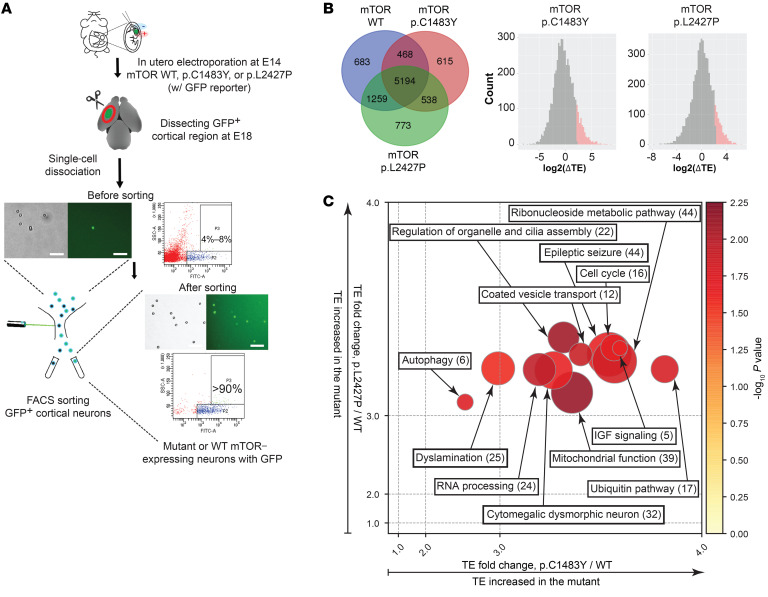
Figure 1. Ribosome profiling in the intractable epilepsy mouse models with brain somatic mutations in MTOR reveals mTOR target genes contributing to FMCD.
(A) Schematic diagram depicts the enrichment of low-level mutation-carrying neurons by FACS from FMCD mice using GFP reporter. Scale bars: 200 μm. (B) Venn diagrams show the number of genes identified from Ribo-Seq and RNA-Seq analysis of mTOR WT (WT), mTOR p.Cys1483Tyr (p.C1483Y), and mTOR p.Leu2427Pro (p.L2427P) mice. Distribution of TE changes in p.C1483Y or p.L2427P mice relative to WT mice. Upregulated mRNAs (Z score ≥ 1.2) are labeled as red in p.C1483Y (left) and p.L2427P (right) mice. See also Supplemental Tables 2 and 3. (C) Functional enrichment analysis of mTOR activation–sensitive genes in FMCD. Each circle represents a functional cluster. Color intensity indicates the statistical significance of functional enrichment as determined by average P value. The size of the circle corresponds to the number of genes, whereas the coordinates indicate average fold change values of p.C1483Y versus WT (x axis) and p.L2427P versus WT (y axis). Numbers of genes are given in parentheses. See also Supplemental Figure 3H.
To identify mRNAs sensitive to mTOR activity at the translational level, we calculated the translational efficiency (TE; TE = RPF-normalized read counts/mRNA-normalized read counts) of each mRNA in mTOR p.Cys1483Tyr and mTOR p.Leu2427Pro mice relative to mTOR WT mice (Figure 1B and Supplemental Table 2). In our Ribo-Seq and RNA-Seq analysis, we were able to detect 5194 genes that were expressed in all groups and 538 genes that were expressed in both mTOR p.Cys1483Tyr and p.Leu2427Pro mice but not in mTOR WT mice (Figure 1B). We noted that various neuronal markers were highly expressed in GFP-positive cells isolated from mTOR WT mouse brains, suggesting that our protocol successfully enriched GFP-positive cortical neurons (Supplemental Figure 3G). We identified 256 mTOR target genes that were translationally activated in cortical neurons from FMCD mice carrying brain somatic mutations (hereafter referred to as mTOR activation–sensitive genes in FMCD mice) (Figure 1B and Supplemental Tables 2 and 3). Subsequently, we conducted functional cluster analysis of these 256 mTOR activation–sensitive genes using ClueGO and Ingenuity Pathway Analysis (IPA) (16, 17). They were found to be highly enriched in the clusters of epileptic seizures, cytomegalic dysmorphic neurons, and dyslamination, all of which are reflected in the phenotype of FMCD (Figure 1C and Supplemental Figure 3H).
Increased expression of mTOR activation–sensitive genes in FMCD patient and mouse brain tissue.
Among the novel mTOR activation–sensitive genes found to be translationally activated in our FMCD mice, we were intrigued by the activation of adenosine kinase (ADK), insulin receptor substrate of 53 kDa (IRSp53), and cAMP responsive element–binding protein 1 (CREB1), which are implicated in epileptic seizures, dysmorphic neurons, and defective neuronal migration, respectively (Supplemental Figure 4). ADK is an adenosine kinase that converts adenosine to AMP, and thus is important in energy production (18, 19). Overexpression of ADK is sufficient to trigger epileptic seizures, and inhibition of ADK has been found to suppress epileptic seizures (20–24). IRSp53, also known as brain-specific angiogenesis inhibitor 1–associated protein 2 (BAIAP2), is an important regulator of membrane and actin dynamics (25–28). Dysregulation of IRSp53 expression in neurons affects dendritic and neuronal branching (29–31). CREB1 is a transcription factor involved in the expression of genes regulating synaptic function and neuronal survival (32–36). Dysregulation of CREB1 has been shown to elicit epileptic seizures and dyslamination (37, 38). To validate the increased expression of ADK, IRSp53, and CREB1 in the FMCD mice, we performed immunofluorescence, and found that the expressions of ADK, IRSp53, and CREB1 were robustly increased in adult mice (Supplemental Figure 5, A and B) and E18 mouse brains (Supplemental Figure 6) expressing mTOR p.Cys1483Tyr and p.Leu2427Pro, compared with mTOR WT mice. The aberrant increases in the expressions of ADK, IRSp53, and CREB1 were rescued by treatment with rapamycin in FMCD mice (Supplemental Figure 5, A and B, and Supplemental Figure 6).
We then examined whether the expression of proteins encoded by these 3 genes is significantly increased in FMCD patient brain tissue carrying brain somatic mutations or germline mutations activating mTOR kinase. To do this, we collected brain tissue from various FMCD patients, including those with FCD, HME, and tuberous sclerosis complex (TSC) (Supplemental Table 4). We particularly sought to analyze brain tissue from an HME patient with an MTOR p.Cys1483Tyr somatic mutation, an FCD patient with an MTOR p.Ala1459Gly somatic mutation, and a TSC patient with a TSC2 p.Asp1119* germline mutation leading to the activation of mTOR kinase (Supplemental Table 5). We further performed coimmunofluorescence staining of ADK, IRSp53, and CREB1 with a neuronal marker (NeuN), and found that the expression of ADK, IRSp53, and CREB1 and the phosphorylation of S6 and 4E-BPs were significantly increased in neuronal cells compared with postmortem, non-FMCD, or unaffected brain tissue (Figure 2, A and B, and Supplemental Figures 7 and 8). Using Western blot analysis, we found that the expression levels of ADK (including ADK short isoforms and ADK long isoforms in humans), IRSp53, CREB1, and phosphorylated S6 (p-S6) and p–4E-BPs were significantly increased in patient brain lysates (Figure 2, C and D). Interestingly, we also found increased expression of ADK, IRSp53, and CREB1 in surrounding cells (Figure 2, A and B, and Supplemental Figure 5, A and B), implying that brain somatic activating mutations in MTOR might have non–cell-autonomous effects on surrounding cells. In addition, we found that the intensity of p-S6 fluorescence was significantly greater in GFP-positive and GFP-negative cells from FMCD mice than in those from WT mice (Supplemental Figure 5, C and D). Consistent with the results from the mouse models, the expression of mTOR activation–sensitive genes was significantly increased in FMCD patient brain tissues.
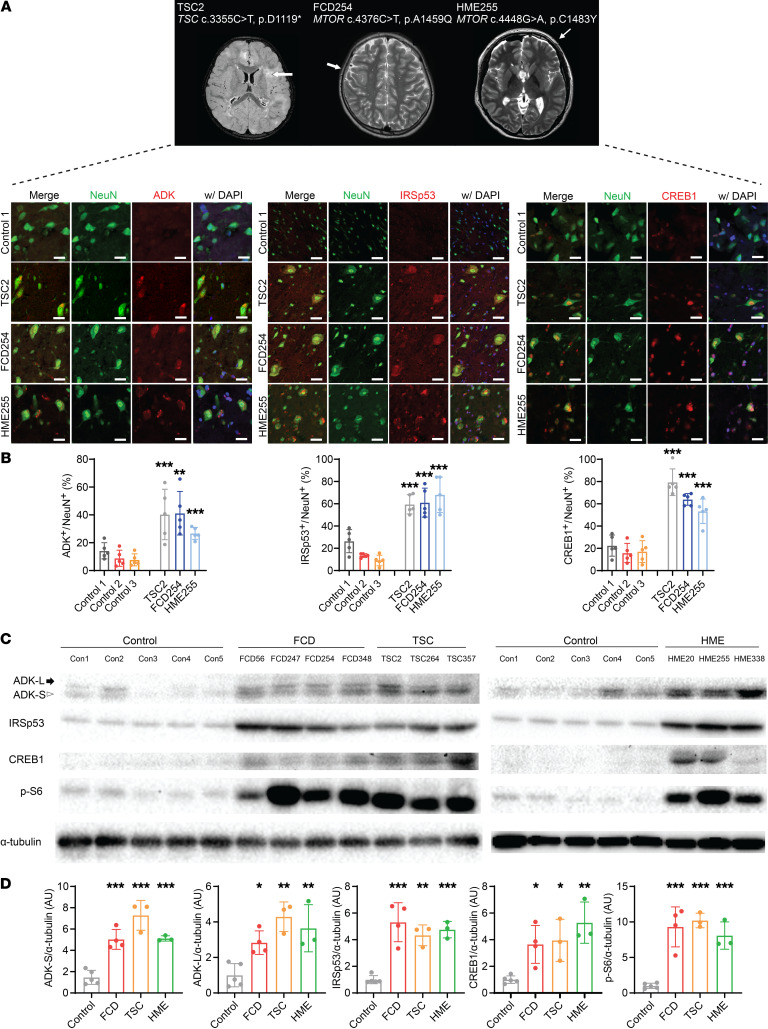
Figure 2. mTOR activation–sensitive genes in patients’ brain tissues with somatic mutations in MTOR.
(A) Representative brain MRI from a TSC, an FCD, and an HME patient with MTOR activating mutation. Arrows highlight the affected cortical region. Representative immunofluorescence staining of translationally upregulated mTOR targets (ADK, IRSp53, and CREB1 [red]) in NeuN+ (green) cells from the patient’s brain tissues stained with DAPI (blue). Scale bars: 25 μm for ADK and CREB1 and 60 μm for IRSp53. Control 1 refers to the postmortem brain tissues of UMB5309. (B) Quantification of samples in A. ADK, IRSp53, or CREB1 positivity in NeuN+ cells from TSC, FCD, and patients with HME. Five images were quantified in each section. Mean ± SD. (C) Western blots of ADK, IRSp53, CREB1, and p-S6 in lysates from patients with FMCD and control brain specimens. Arrow indicates long isoform of ADK (ADK-L), and arrowhead indicates short isoform of ADK (ADK-S). Blotting of α-tubulin in lysates was used as a loading control. Control 1 (Con1) refers to the postmortem brain region of UMB5309, Con2 to the postmortem brain region of UMB5408, Con3 to the unaffected brain region of FCD247, Con4 to the postmortem brain region of UMB1712, and Con5 to the postmortem brain region of UMB4917. (D) Quantification of C. The level of each target protein is presented as a percentage change relative to the average of 5 control samples. n = 5 in control (Con1, Con2, Con3, Con4, and Con5), n = 4 in FCD (FCD56, FCD247, FCD254, and FCD348), n = 3 in TSC (TSC2, TSC264, and TSC357), n = 3 in HME (HME20, HME255, and HME338). Mean ± SD. *P <0.05, **P < 0.01, ***P < 0.001. One-way ANOVA with Bonferroni’s post hoc test.
Translational landscape of mTOR activation–sensitive genes is distinct from that of mTOR inhibition–sensitive genes.
Next, we sought to determine how mTOR activation–sensitive genes, including ADK, IRSp53, and CREB1, are selectively regulated by MTOR activating mutations. Previously, in ribosome profiling studies applying pharmacological inhibition of mTOR, researchers demonstrated that mTOR selectively regulates the translation of mRNAs that primarily encode translational machinery (i.e., ribosomal proteins) and that contain 5′ terminal oligopyrimidine (5′ TOP) motifs (13, 14). Interestingly, we discovered no significant effects on the translation of 5′ TOP mRNAs in either of the mTOR activation conditions (mTOR p.Cys1483Tyr or p.Leu2427Pro mice) compared with mTOR WT mice (Figure 3A and Supplemental Table 6). Accordingly, we hypothesized that the translational control mediated by MTOR activating mutations would be distinct from that mediated by mTOR pharmacological inhibition.
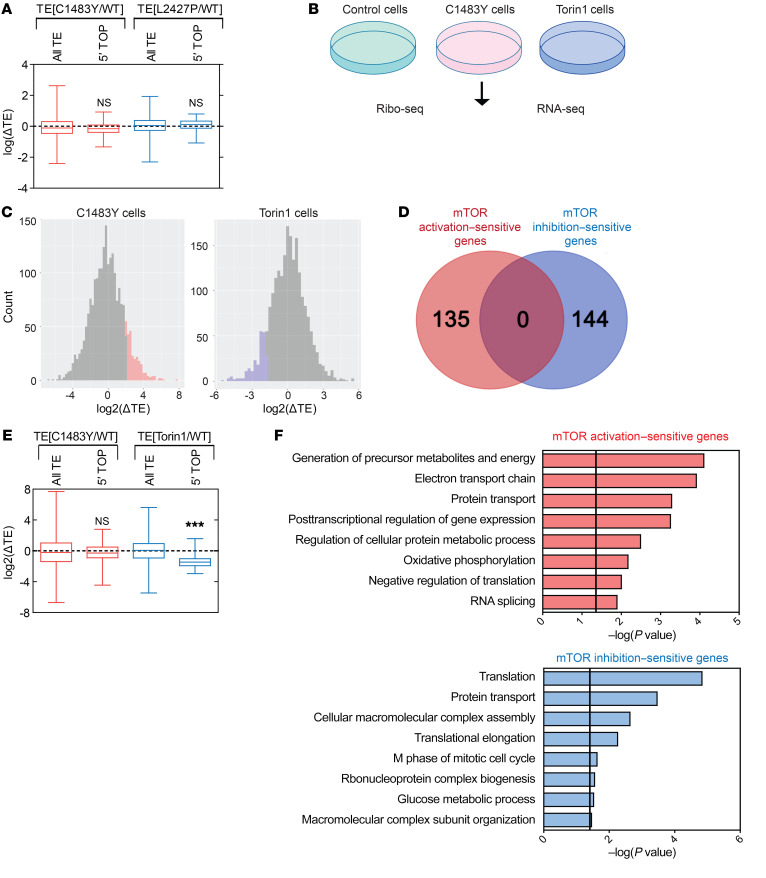
Figure 3. Translational landscape of mTOR activation–sensitive genes is distinct from that of mTOR inhibition–sensitive genes.
(A) Box plots show the log ratio of fold changes [log(ΔTE)] in the TEs of mRNAs encoding all genes and 5′ TOP mRNAs in p.C1483Y mice (TE[C1483Y/WT]) and p.L2427P mice (TE[L2427P/WT]) relative to WT mice. Mean ± SEM. See also Supplemental Table 6. (B) Schematic diagram of Ribo-Seq and RNA-Seq in control, C1483Y, and Torin1 cells. (C) Distribution of changes in the TEs of mRNAs from C1483Y and Torin1 cells relative to control cells. Upregulated mRNAs (Z score ≥ 1.5, mTOR activation–sensitive genes) in C1483Y cells (left) are indicated in red, and downregulated mRNAs (Z score ≤ –1.5, mTOR inhibition–sensitive genes) in Torin1 cells (right) are indicated in blue. See also Supplemental Table 8. (D) Venn diagram of mTOR activation–sensitive genes in C1483Y cells (red) and mTOR inhibition–sensitive genes in Torin1 cells (blue). (E) Box plots showing the log2 ratios of fold changes [log2(ΔTE)] in the TEs of mRNAs encoding all genes and 5′ TOP in C1483Y cells [TE(C1483Y/WT)] and Torin1 cells [TE(Torin1/WT)] relative to control cells. Mean ± SEM. See also Supplemental Table 9. (F) The top 8 clusters in DAVID functional annotation were associated with mTOR activation–sensitive genes in C1483Y cells (top) and mTOR inhibition–sensitive genes in Torin1 (bottom). Black lines reflect a P value of 0.05. ***P < 0.001. One-way ANOVA with Bonferroni’s post hoc test.
Unfortunately, translational profiles of MTOR activating mutations have not been compared with that of mTOR pharmacological inhibition at the genome scale. Thus, to compare the translational landscape of human activating mutations in MTOR with that of pharmacological inhibition of mTOR, we used CRISPR-mediated genome-edited NIH 3T3 cells carrying the mutation MTOR c.4448G>A (p.Cys1483Tyr) (hereafter referred to as C1483Y cells) (15), cells treated with the mTOR inhibitor Torin1 (200 nM, 3 hours) (hereafter referred to as Torin1 cells), and vehicle-treated WT cells (hereafter referred to as control cells) (Figure 3B). MTOR c.4448G>A (p.Cys1483Tyr) is recurrently found in human FMCD, as well as various cancers, and reportedly causes aberrant activation of mTOR kinase (3, 15, 39, 40). We confirmed the hyperactivation of mTOR kinase in C1483Y cells and its inhibition in Torin1 cells by performing Western blotting of p-S6 ribosomal protein, a major readout of mTORC1 activity (Supplemental Figure 9).
Then, we performed ribosome profiling in C1483Y, Torin1, and control cells to compare the impact of mTOR activating mutation with that of pharmacological inhibition of mTOR on translational profiles. The qualities of the Ribo-Seq and RNA-Seq libraries in C1483Y, Torin1, and control cells were validated in a manner similar to that for FMCD mice (Supplemental Figure 10, A–E, and Supplemental Table 7). We calculated the TEs (RPF-normalized read counts/mRNA-normalized read counts) of each mRNA in control, C1483Y, and Torin1 cells (Figure 3C and Supplemental Table 8). Using Z score cutoffs of 1.5 for an upregulated TE and –1.5 for a downregulated TE, we were able to select 135 mRNAs exhibiting significantly increased TEs in C1483Y cells and 144 mRNAs experiencing a significant decrease in TE in Torin1 cells, compared with control cells (Figure 3C). While it has been suggested that upregulated genes upon mTOR activation would overlap with downregulated genes upon mTOR inhibition (41), surprisingly, we found no overlap between mRNAs with upregulated TEs in C1483Y cells (hereafter referred to as mTOR activation–sensitive genes in C1483Y cells) and those with downregulated TEs in Torin1 cells (hereafter referred to as mTOR inhibition–sensitive genes in Torin1 cells) (Figure 3D). Also, mTOR inhibition–sensitive genes in Torin1 cells and mTOR activation–sensitive genes in C1483Y cells were not affected in C1483Y cells and Torin1 cells, respectively (Supplemental Figure 11).
In line with previous studies, the TEs of 5′ TOP mRNAs in Torin1 cells were significantly inhibited (Figure 3E and Supplemental Table 9). However, as in FMCD mice, the TEs of 5′ TOP mRNAs were not affected in C1483Y cells (Figure 3E and Supplemental Table 9). The translation of 5′ TOP mRNAs, which, as stated above, primarily encode translational machinery, positively regulates protein synthesis (13, 14). Here, we noted that bulk protein synthesis was not significantly changed in C1483Y cells, as determined by puromycin incorporation assays, while prolonged Torin1 treatment (14 hours) resulted in significant decreases in global protein synthesis (13) (Supplemental Figure 12A). This suggested that, in the conditions used for translational profiling, mTOR activation and inhibition elicited gene-specific translational control. In line with the bulk protein synthesis results in C1483Y cells, brain tissue from mTOR mutant mouse embryos, as well as mTOR mutant–expressing primary cortical neurons and heterologous cell lines, exhibited no significant alterations in protein synthesis rate as a result of carrying the mTOR p.Cys1483Tyr or p.Leu2427Pro mutation (Supplemental Figure 12, B–E).
To further analyze the biological functions of mTOR activation– and inhibition–sensitive genes, we conducted functional cluster analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) and ClueGO (16, 42). We found that mTOR inhibition–sensitive genes in Torin1 cells were significantly enriched in translation function (Figure 3F and Supplemental Figure 13), which is consistent with previous studies (13, 14). Meanwhile, however, mTOR activation–sensitive genes in C1483Y cells were enriched in mitochondrial activity and energy production (Figure 3F and Supplemental Figure 13). Consistent with our functional cluster analysis in the mTOR activation–sensitive genes in C1483Y cells, mTOR activation–sensitive genes in FMCD were also enriched in mitochondrial function and energy production (Figure 1C and Supplemental Figure 3H). Consistent with our findings, other studies have provided evidence suggesting that activation of the mTOR pathway leads to increased translation of a subset of mitochondrial and energy production–related mRNAs (43, 44) and that 5′ TOP genes are insensitive to the activation of mTOR (45, 46). Taken together, our data indicated that translational control and functions mediated by somatic activating mutations in MTOR are distinct from those mediated by pharmacological inhibition of mTOR.
mTOR activation–sensitive genes confer mTOR-responsive 5′-UTR motifs.
In light of the above, we wondered how mTOR activation–sensitive genes are differently regulated from mTOR inhibition–sensitive genes regulated by 5′ TOP motif. Studies have reported that the translation of target mRNAs is regulated by recognition of their 5′-UTR regions (13, 14, 47). Thus, we hypothesized that the 5′-UTR regions of mTOR activation–sensitive genes, rather than 5′ TOP motifs, would be responsible for the selective increases in translation upon activation of mTOR kinase. To test this idea, we first performed 5′-UTR luciferase reporter assays of the 5′-UTR regions of Adk-S (ADK short isoform), Adk-L (ADK long isoform), IRSp53, and Creb1 in heterologous cell lines expressing mTOR WT, p.Cys1483Tyr, or p.Leu2427Pro (Figure 4A). The 5′-UTR regions of Adk-S, Adk-L, IRSp53, and Creb1 did not contain a 5′ TOP motif. As a result, we found that the 5′-UTRs of these mTOR activation–sensitive genes were sufficient to selectively increase their expression in response to somatic activating mutations in MTOR, whereas the 5′-UTRs of control genes, such as Actb (β-actin) and Gapdh, were not (Figure 4A).
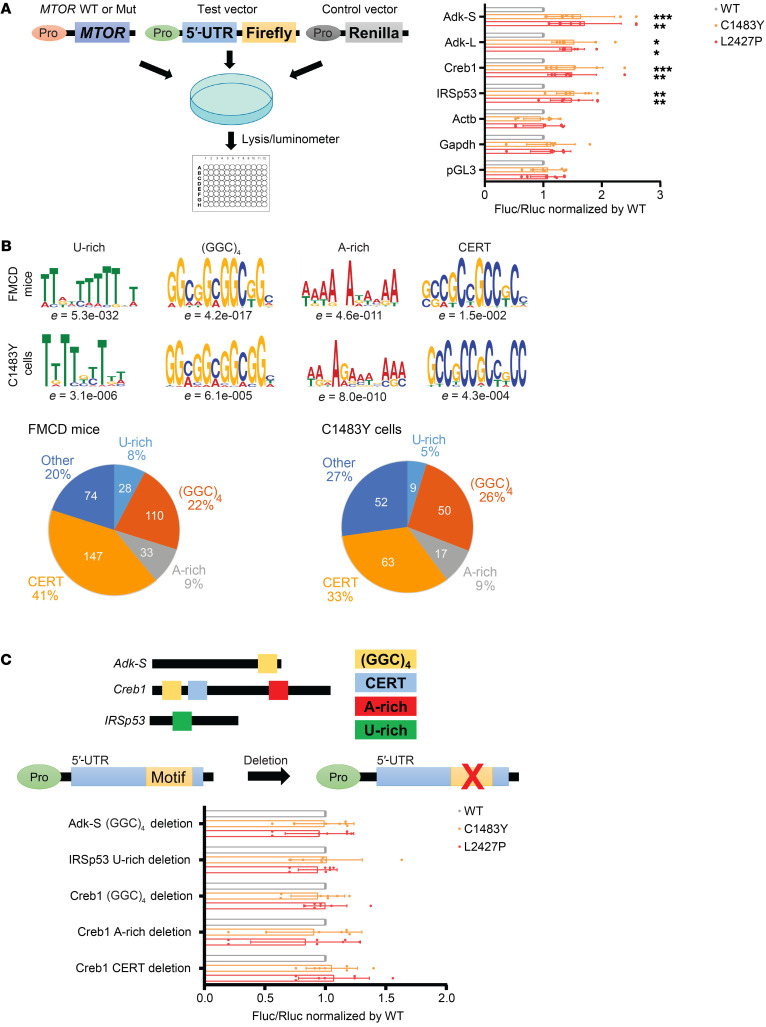
Figure 4. mTOR activation–sensitive genes confer mTOR-responsive 5′-UTR motifs.
(A) 5′-UTR–mediated translation of target mRNAs (Adk-S, Adk-L, Creb1, and IRSp53) and control mRNAs (Actb and Gapdh) by 5′-UTR luciferase reporter assay in HEK293T cells transfected with mTOR WT (WT) and mTOR p.C1483Y or p.L2427P. Pro denotes promoter. Actb denotes β-actin gene. pGL3 denotes the test vector lacking a 5′-UTR. Results are normalized to 5′-UTR reporter activity in transfected mTOR WT cells. n = 7 in each case. Mean ± SD. (B) Consensus sequence and enrichment values (e value) of the U-rich, guanine quartet (GGC)4, A-rich, and CERT motifs identified in mTOR activation–sensitive genes in FMCD mice and C1483Y cells by MEME analysis. Diagram illustrating the frequencies of mTOR activation–sensitive genes in FMCD mice and C1483Y cells containing U-rich, guanine quartet (GGC)4, A-rich, and CERT motifs. Several motifs in the same genes were counted independently. See also Supplemental Tables 11 and 13. (C) Location of 5′-UTR motifs in Adk-S, IRSp53, and Creb1 is indicated. Effect of deletion mutations in 5′-UTR motif domains in Adk-S, IRSp53, and Creb1 on 5′-UTR luciferase reporter activity in mTOR-activated p.C1483Y or p.L2427P cells relative to WT-transfected HEK293T cells. Pro denotes promoter. n = 7 in each case. Mean ± SD. See also Supplemental Table 14. *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA with Bonferroni’s post hoc test.
Then, we wondered whether the 5′-UTRs of the mTOR activation–sensitive genes reflected common features responsive to MTOR activating mutation. We examined canonical 5′-UTR features (length, GC content, and Gibbs free energy) in mTOR activation–sensitive genes in FMCD mice and C1483Y cells, as well as in mTOR inhibition–sensitive genes in Torin1 cells. However, these canonical 5′-UTR features in mTOR activation– and inhibition–sensitive genes in the FMCD mice, C1483Y cells, and Torin1 cells were not statistically distinguishable from those of background genes (Supplemental Figure 14 and Supplemental Table 10). To identify consensus 5′-UTR motifs responsive to the activity of mTOR kinase, we performed an unbiased Multiple EM for Motif Elicitation (MEME) analysis for 5′-UTRs of mTOR activation– or inhibition–sensitive genes. Interestingly, we found that U-rich, guanine quartet (GGC)4, and A-rich motifs and cytosine-enriched regulator of translation (CERT) domains were significantly enriched in both mTOR activation–sensitive genes in FMCD mice and C1483Y cells and accounted for approximately 80% of mTOR activation–sensitive genes in FMCD mice and approximately 70% of mTOR activation–sensitive genes in C1483Y cells (Figure 4B and Supplemental Table 11). However, 5′ pyrimidine-rich translational element (PRTE), A-rich, and GGAGG motifs were identified in the 5′-UTRs of mTOR inhibition–sensitive genes in Torin1 cells (Supplemental Figure 15 and Supplemental Table 11). PRTE motif is a broad motif that encompasses 5′ TOP or 5′ TOP–like motifs (14). The finding of PRTE motifs is consistent with previous studies on acute mTOR inhibition–sensitive genes (13, 14) (Supplemental Figure 15).
Interestingly, guanine quartet (GGC)4, CERT domains, and U-rich motifs, which we noted in mTOR-responsive motifs, have been described as being dependent on mTOR downstream effector eIF4F activity for their translation (47–50). In addition, mTOR is known to activate eIF4F through phosphorylation of 4E-BPs and S6Ks (8). Although its relevance to eIF4F activity remains unknown, A-rich was found in 9% of mTOR activation– and inhibition–sensitive genes. Upon upstream regulator analysis with IPA, we found that eIF4E, a core component of eIF4F, was a significant upstream regulator of mTOR activation–sensitive genes in C1483Y cells (Supplemental Table 12). Among the identified mTOR activation–sensitive genes, we noted that ATOX1, BIRC6, CDC34, CKS2, and NUDT3, which are known as eIF4E target genes, indeed contained guanine quartet (GGC)4 and CERT motifs, similarly to Adk-S, IRSp53, and Creb1 (Supplemental Table 13). These results suggested that mTOR activation–sensitive genes contain 5′-UTR motifs of U-rich, guanine quartet (GGC)4, A-rich, or CERT domains, thereby making them dependent on eIF4F activity for their translation. Accordingly, we sought to determine whether guanine quartet (GGC)4 motifs in ADK-S, U-rich motifs in IRSp53, and guanine quartet (GGC)4, A-rich, and CERT motifs in CREB1 are necessary for their increased expression in response to somatic activating mutations in MTOR. To do this, we performed mutagenesis of individual motifs followed by 5′-UTR luciferase reporter assays, and found that deletions introduced in each motif sufficiently reversed the increased expression of luciferase in response to somatic activating mutations in MTOR (Figure 4C and Supplemental Table 14). Taken altogether, these results demonstrated that mTOR activation–sensitive genes, including Adk, IRSp53, and Creb1, show specific 5′-UTR motifs that confer dependence on eIF4F activity for their translation. These results implied that the translational control mechanism of mTOR activation–sensitive genes is mediated by 5′-UTR motifs that confer dependence on eIF4F activity, differing from the translational control mechanism of mTOR inhibition–sensitive genes.
Pharmacological or genetic inhibition of eIF4E rescues intractable epilepsy and other pathologies observed in FMCD mice.
We found that most of the mTOR activation–sensitive genes in FMCD and C1483Y cells contained eIF4F-sensitive 5′-UTR motifs that regulated their expression. However, the pathogenic role of eIF4F in intractable epilepsy and cortical malformations is unknown. Thus, we wondered whether the translational dysregulation mediated by increased activity of eIF4F in response to the activation of mTOR kinase would lead to the major phenotypes of FMCD, such as epilepsy, cytomegalic dysmorphic neurons, and defective cortical lamination. To investigate this hypothesis, we performed in vivo knockdown of eIF4E, a key component of the eIF4F complex, to reduce eIF4F activity in FMCD mice via in utero electroporation of mTOR mutant or WT constructs coexpressing scrambled shRNAs (shScramble) or shRNAs against eIF4E (sheIF4E) (Supplemental Figure 16A). Efficient knockdown of eIF4E by sheIF4E was validated via Western blot analysis (Supplemental Figure 16A). We also confirmed that eIF4E expression was significantly reduced in sheIF4E-expressing neurons upon immunostaining (Supplemental Figure 16B). Accordingly, we performed further immunostaining to examine the effect of in vivo knockdown of eIF4E on the expression of ADK, IRSp53, and CREB1 in FMCD mice, and found that their expressions were significantly reduced (Supplemental Figure 17).
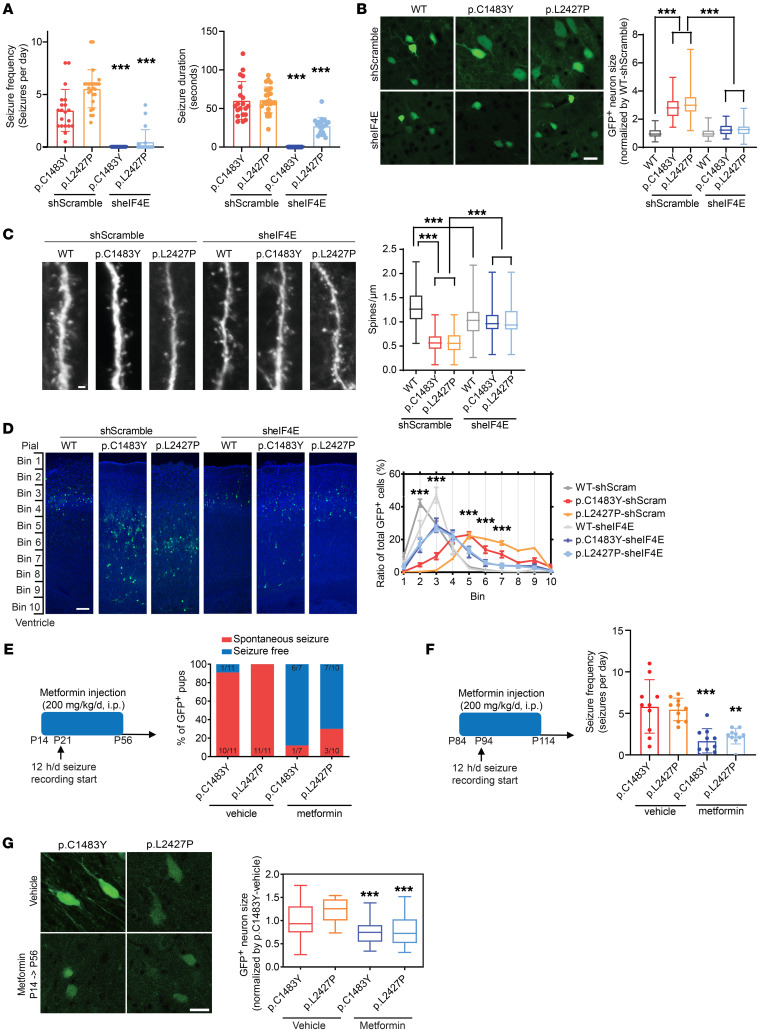
We further examined whether in vivo knockdown of eIF4E could alleviate the major phenotypes of FMCD. Surprisingly, we found that eIF4E knockdown was able to successfully rescue all of the pathological phenotypes of FMCD, including epileptic seizure (Figure 5A), hypertrophic soma size (Figure 5B), spine density (Figure 5C), and migration defect (Figure 5D). In detail, we found that eIF4E knockdown almost completely rescued spontaneous seizures observed in mTOR p.Cys1483Tyr and p.Leu2427Pro mice (Figure 5A). Additionally, we noted that eIF4E knockdown rescued hypertrophic soma and decreased spine density, which are representative morphologies of dysmorphic neurons found in FMCD (51, 52) (Figure 5, B and C); consistent with this finding, it was recently reported that normalizing translation through constitutive active 4E-BP1 prevents mTOR-driven cortical dyslamination and hypertrophy of soma (52). Also, eIF4E knockdown significantly alleviated defective neuronal migration in mTOR mutant mice (Figure 5D). We pharmacologically inhibited eIF4E activity via the FDA-approved drug metformin to cure epilepsy in FMCD mice. Metformin, a first-line drug for treating type 2 diabetes, has been found to inhibit eIF4E activity via decreased eIF4E phosphorylation in the brain and to show effectiveness in treating fragile X syndrome via eIF4E inhibition (53). We injected metformin i.p. at a dose of 200 mg/kg from P14 to P56 (early treatment) and from P84 to P114 (late treatment) (Figure 5, E and F). Twelve-hour recording and video-EEG analysis demonstrated that early treatment with metformin prevented seizure onset and late treatment suppressed seizure frequency in FMCD mice (Figure 5, E and F). In addition, early treatment with metformin rescued hypertrophic soma (Figure 5G). Accordingly, these results showed that the increased activity of eIF4F in response to somatic activating mutations in MTOR leads to the phenotypes of FMCD via the increased translation of mTOR activation–sensitive genes and that epileptic seizures could be prevented through eIF4E inhibition by metformin.
Figure 5. Pharmacological or genetic inhibition of eIF4E rescues intractable epilepsy and other pathologies observed in FMCD mice.
(A) Quantification of seizure frequency (left) and duration (right) in FMCD adult mice treated with sheIF4E (from P21 to P120). p.C1483Y-shScramble: n = 20; p.L2427P-shScramble: n = 25; p.C1483Y-sheIF4E: n = 15; p.L2427P-sheIF4E: n =20. Mean ± SD. (B) Images of layers 2/3 of GFP+ cells from FMCD mice treated with shScramble and sheIF4E at P21 and soma area quantification. n = 5 in each case. Scale bar: 20 μm. Mean ± SEM. (C) Spine density quantification and images of basal dendrites in neurons from FMCD mice treated with shScramble or sheIF4E at P21. Five mice per condition. Scale bar: 2 μm. Mean ± SEM. (D) Representative images of GFP+ cell migration in the cortices of FMCD mice treated with shScramble or sheIF4E at P7. Quantification of the distributions of GFP+ neurons in the cortex. n = 5 in each case. Scale bar: 100 μm. Mean ± SEM. (E) Metformin was injected i.p. once from P14 to P56 (200 mg/kg/d). Quantification of seizure frequency in adult FMCD mice (P21–P56) treated with metformin. p.C1483Y-vehicle: n = 11; p.L2427P-vehicle: n = 11; p.C1483Y-metformin: n = 7; p.L2427P-metformin: n = 10. (F) Metformin was injected i.p. once for 30 days (200 mg/kg/d). Quantification of seizure frequency in adult FMCD mice (P94–P114) treated with metformin. Ten mice per condition. Mean ± SD. (G) Images of layers 2/3 of GFP+ cells from FMCD mice treated with vehicle and metformin (P14–P56) and soma area quantification. n = 5 in each case. Scale bar: 25 μm. Mean ± SEM. **P < 0.01, ***P < 0.001. One-way ANOVA with Bonferroni’s post hoc test.
Pharmacological or genetic inhibition of ADK, a novel downstream target of the mTOR/eIF4F axis, alleviates intractable epilepsies in FMCD mice.
Next, we examined whether a downstream target of the mTOR/eIF4F axis directly contributes to intractable epilepsy, the clinically important phenotype in FMCD. To do this, we focused on the epileptogenic function of ADK, whose expression was upregulated in patients with FMCD and in mouse models (Figure 2 and Supplemental Figures 5 and 6). ADK is known to lose its expression in cortical neurons after P21 in mice, upon which it is shifted to astrocytes (54). In line with this, previous studies have shown that the overexpression of ADK in astrocytes contributes to epileptic seizures (55). However, in the case of FMCD caused by mTOR somatic mutations, we discovered that the expression of ADK was aberrantly increased in cortical neurons expressing mutant mTOR ahead of ADK expression in astrocytes (Supplemental Figures 6 and 18). Thus, we first tested whether the selective inhibition of ADK in mutant mTOR–expressing cortical neurons could reduce epileptic seizures. To do so, we generated mTOR mutant constructs coexpressing shADK, and validated the efficient knockdown of ADK in vitro and in vivo (Supplemental Figure 19).
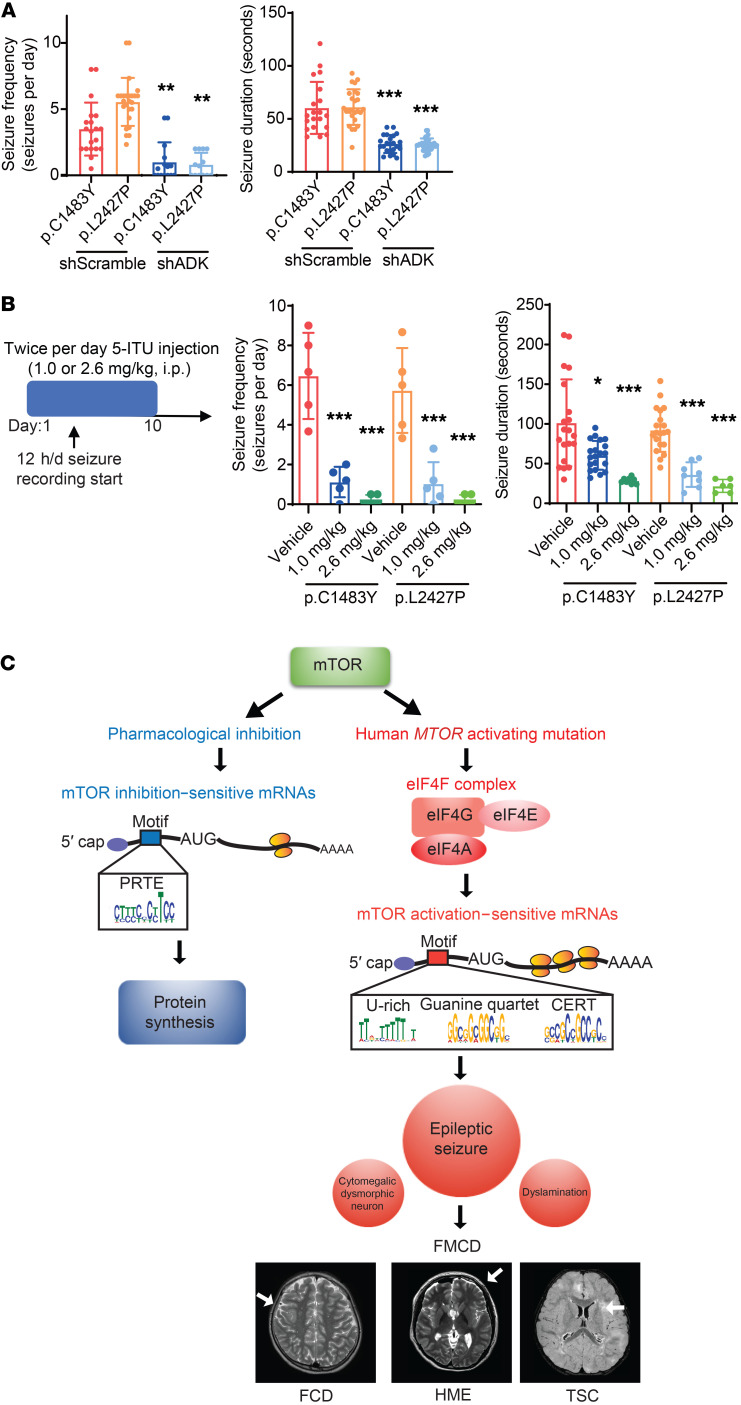
Then, we performed in utero electroporation of these constructs, and found that the frequency and duration of spontaneous behavioral seizures were significantly decreased in shADK-expressing mTOR mutant mice compared with control mice (Figure 6A). Furthermore, we pharmacologically inhibited the function of ADK in our FMCD mutant mice, in which we intraperitoneally injected 5-iodotubercidin (5-ITU), a well-known ADK inhibitor, at a dose of 1.0 mg/kg or 2.6 mg/kg twice per day for 10 days (56). Video-EEG analysis demonstrated that treatment with 5-ITU robustly suppresses seizure frequency and seizure duration in FMCD mice in a dose-dependent manner (Figure 6B). Altogether, these findings showed that the translational dysregulation of ADK in mTOR mutation–carrying cortical neurons contributes to epileptogenesis and that ADK could be a therapeutic target in FMCD.
Figure 6. Pharmacological or genetic inhibition of ADK alleviates intractable epilepsies in FMCD mice.
(A) Quantification of seizure frequency (left) and duration (right) in adult FMCD mice treated with shADK (from P21 to P112, 3 times per 1 week). p.C1483Y-shScramble: n = 20; p.L2427P-shScramble: n = 25; p.C1483Y-shADK: n = 15; p.L2427P-shADK: n =13. Mean ± SD. (B) 5-ITU or vehicle was injected i.p. twice for 10 days (1.0 or 2.6 mg per kg of body weight per day), with 12 h/d seizure recording. Quantification of seizure frequency (left) and duration (right) in adult FMCD mice (P112–P140) treated with 5-ITU. n = 5 in each case. Mean ± SD. (C) A model of translation dysregulation mediated by human MTOR activating mutation, compared with pharmacological inhibition of mTOR. Acute pharmacological inhibition of mTOR kinase mainly downregulates the translation of PRTE mRNAs mainly encoding ribosomal proteins and translation factors. However, human MTOR–activating mutations upregulate the translation of mRNAs with 5′-UTR motifs responsive to eIF4F activity, which underlie major pathological phenotypes in FMCD. *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA with Bonferroni’s post hoc test.
In this study, our comprehensive translation profiling in FMCD mouse models and genome-edited cells with brain somatic mosaicisms revealed novel mTOR activation–sensitive genes contributing to intractable epilepsy and other phenotypes of FMCD. These mTOR target genes show specific 5′-UTR motifs conferring dependence on eIF4F for their translation (Figure 6C). Finally, we demonstrated that pharmacological or genetic inhibition of eIF4E, a core component of the eIF4F complex, rescued all of the major pathological phenotypes of FMCD and that pharmacological or genetic inhibition of ADK alleviated intractable epilepsy.
Discussion
Via ribosome profiling in FMCD mouse models, we were able to discover novel mTOR target genes accounting for intractable epilepsy and other pathological phenotypes of FMCD arising from somatic activating mutations in MTOR in the brain. Notably, we demonstrated that normalizing translation through in vivo knockdown of eIF4E corrects intractable epilepsy of FMCD stemming from hyperactivation of mTOR kinase, thereby suggesting that eIF4E is a novel therapeutic target of intractable epilepsy associated with hyperactivation of the mTOR pathway. Inhibiting eIF4E, metformin is an FDA-approved drug widely used to treat diabetic individuals, including children. We found that metformin ameliorated epileptic seizures in FMCD mice. The dose of metformin administered to mice (200 mg/kg/d) in this study was calculated according to a clinically relevant human dose based on body surface area (57). The maximum prescribed dose of this drug for a human of average weight is 2000 mg/d. According to the surface area rule, the ratio of mouse (20 g) to human (60 kg) is 0.0026, and hence the highest dose of metformin for mice was calculated to be 260 mg/kg (57). With proven long-term safety and tolerability, metformin could be a promising drug of use in treating FMCD. Meanwhile, we also showed that increased expression of ADK in mutation-carrying neurons, as a specific downstream target gene of eIF4F activity, was responsible for epileptic seizures in FMCD mice. Previously, the role of ADK in epileptogenesis has been studied exclusively in astrocytes (58). While ADK overexpression in astrocytes has been shown to be sufficient to induce epileptogenesis (20, 21, 24), our findings further suggest that the increased expression of neuronal ADK also contributes to epileptic seizures (Figure 6, A and B). Altogether, this study provides new insights into translational dysregulation in and the molecular pathogenesis of FMCD arising from brain somatic mutations in MTOR, as well as novel therapeutic targets for the potential treatment thereof.
To date, the translational regulation mechanisms mediated by mTOR activity have been primarily studied in the context of acute mTOR inhibition using mTOR inhibitors, hypoxia, and serum or amino acid starvation (13, 14, 59). However, the condition of somatic mutations in MTOR, which is actually implicated in human diseases, seems to be more physiologically relevant to understanding the biological consequences and mechanisms of translational control mediated by mTOR activation, compared with the condition of acute mTOR inhibition. In line with this, our translation profiling in genome-edited cells and FMCD mouse models with human MTOR mutations demonstrated that, while mTOR inhibition–sensitive mRNAs, such as those encoding ribosomal proteins, exhibit pyrimidine-enriched sequence (PES) motifs (e.g., 5′ TOP, 5′ TOP–like, and PRTE motifs), consistent with previous studies (13, 14, 60), mTOR activation–sensitive mRNAs contain specific 5′-UTR motifs [guanine quartet (GGC)4, CERT, and U-rich motifs] known to be dependent on eIF4F activity for their translation (48–50). Regarding the mechanistic difference in translation control between mTOR activation– and inhibition–sensitive genes, we suspect that there might be different translational switches for mTOR activation and inhibition, respectively. For example, LARP1 has recently been identified as a trans-acting factor for the cis-regulatory element of PES motifs (61, 62). LARP1 inhibits translation of PES motif–containing mRNAs by hampering binding to eIF4E (63). The phosphorylation of LARP1, induced by the activation of mTOR kinase, relieves its inhibitory activity on the translation of PES mRNA (60). Interestingly, LARP1 has been shown to bind to the 5′-UTR of ribosomal mRNAs upon mTOR inactivation for its inhibitory function on translation, whereas the binding of LARP1 to the 5′-UTR of ribosomal mRNAs was almost 0% in normal growth conditions associated with mTOR activation (60). Collectively, our findings and previous reports imply that LARP1 and eIF4F may play key roles in translational control upon mTOR inhibition and activation, respectively.
In addition to the above, we found non–cell-autonomous effects of increased expression of p-S6, ADK, IRSp53, and CREB1 in cells surrounding dysmorphic neurons from our patient tissues and GFP-positive cells in FMCD mice (Figure 2, A and B, and Supplemental Figure 5). One possible mechanism of non–cell-autonomous activation of mTOR has been proposed in Tsc1–/– cell lines: Tsc1–/– cells secrete exosomes that contain Rheb RNA, which results in increased mTOR pathway activity in surrounding cells (64).
Studies have shown that a mutational burden of about 1% for somatic mutations in mTOR pathway genes consistently leads to the same phenotypes observed in FMCD mice generated by in utero electroporation (65–67). According to a previous study (67), 1%–2% of mutation-carrying neurons were present in adult mice after in utero electroporation at E14. Thus, such a low percentage of mutation-carrying neurons is sufficient to lead to the observed phenotypes of FMCD in mice. Nevertheless, more research is needed to determine the minimum mutational burden of somatic mutations that would cause abnormal excessive or synchronous neuronal activity throughout the brain.
Methods
Subject ascertainment
Individuals diagnosed with HME, FCD, or TSC who had undergone epilepsy surgery at Severance Children’s Hospital since 2004 were identified. Enrolled individuals met the study entry criteria for FCDII and underwent extensive presurgical evaluations, including video-EEG monitoring, high-resolution MRI, fluorodeoxyglucose PET, and subtraction ictal single-photon emission computed tomography coregistered to MRI, to localize anatomic lesions. Complete resection was defined as resection of all areas of seizure-onset and irritative zones on intracranial EEG. The pathological diagnoses of studied individuals with HME, FCD, or TSC were reconfirmed for this study according to the recent consensus classification by the International League Against Epilepsy Diagnostic Methods Commission. The study was performed and all human tissues were obtained in accordance with protocols approved by Severance Children’s Hospital and the Korea Advanced Institute of Science and Technology (KAIST) Institutional Review Board and Committee on Human Research. Informed consent was obtained from the parents of individuals with HME, FCD, and TSC. Healthy adult, control brain samples were obtained from the University of Maryland Brain and Tissue Bank: brain tissues of individuals without neurological disease were obtained.
Transfection
HEK293T, Neuro2A, and NIH 3T3 cells were transfected with plasmid using iNfect transfection reagent (Intron Biotechnology, 15081).
In utero electroporation
Timed pregnant mice (E14) were anesthetized with isoflurane (maintained at 0.4 L/min of oxygen and isoflurane via a vaporizer during surgery). The uterine horns were exposed, and a lateral ventricle of each embryo was injected using pulled glass capillaries with 2 μg/mL Fast Green (MilliporeSigma, F7252) combined with 2 to 3 mg MTOR WT, MTOR p.Cys1483Tyr, or MTOR p.Leu2427Pro with or without mU6-shRNA (eIF4E, ADK, scramble) plasmids, as indicated. Plasmids were electroporated on the head of the embryo by discharging of 35 V via an ECM830 electroporator (BTX-Harvard apparatus) in 5 electric pulses of 100 milliseconds at 950-millisecond intervals. Embryonic mice were electroporated at E14.
Single-cell dissociation of cortical neurons for FACS and primary cortical neuron culture
From in utero–electroporated mice, cortical neurons expressing MTOR WT, MTOR p.Cys1483Tyr, or MTOR p.Leu2427Pro, along with GFP reporter, were dissected at E18.5 in HBSS (Life Technologies) supplemented with 10 mM HEPES (Gibco, 15630-080) and Pen/Strep (Life Technologies, 15140-122). For samples to be sorted by FACS and frozen for further analysis, all procedures were performed with 100 μg/mL cycloheximide (MilliporeSigma, 239763). For Ribo-Seq and RNA-Seq, we dissected about 100 electroporated mouse brains per group at E18, and enriched mutant or WT mTOR–expressing GFP-positive neurons up to 90% of the total cells using FACS. In utero–electroporated regions were excised in Hibernate-E medium (Gibco, A12476-01) supplemented with GlutaMAX-I (Gibco, 35050-061) and B27 (Gibco, 17504-044) visualizing GFP reporter. GFP-positive cortices were digested in dissection medium containing 0.05% trypsin (Life Technologies, 15090-046) for 30 minutes at 4°C. After digestion, pellets were centrifuged and resuspended in Hibernate-E medium supplemented with GlutaMAX-I and B27 three times. To dissociate cells, fire-polished Pasteur pipettes (Corning, CO-7095B-9) were used. Sequentially, 100-μm and 40-μm strainers were used to remove large debris. Cell sorting was performed with the BD FACSAria II Flow Cytometer (BD Biosciences) with FITC gating. Sorted cortical neurons were stored at −80°C for up to 6 months.
For primary cortical neuron culture, all procedures were performed in the same manner as that for FACS sorting, except the use of cycloheximide was omitted. FACS-sorted or unsorted neurons were dissociated in DMEM containing 10% FBS and 0.5% Pen/Strep, and plated on plates coated with poly-d-lysine (MilliporeSigma, P6407) at 2 × 104 to 3 × 104 cells per coverslip for low-density culture. After 3 hours, medium was replaced with Neurobasal medium (GIBCO, 21103049) supplemented with GlutaMAX-I and B27. Cells were grown at 37°C and 5% CO2. Half of the culture medium was replaced with fresh medium every 3–4 days. Neurons from multiple mouse embryos from a single pregnant female mouse (including both male and female embryos) were combined for experiments. Neurons were typically used at 18 days in vitro.
Western blotting
Brain tissue and primary cortical neuron cultures were lysed in RIPA buffer; other cells were lysed with PBS containing 1% Triton X-100 with Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, 78440). Proteins were resolved by SDS-PAGE and transferred to PVDF membranes (MilliporeSigma, M-SLGV033RS). The membranes were blocked with 3% BSA in TBS containing 0.1% Tween-20 (TBST) for 1 hour at room temperature, and then washed 4 times with TBST. The membranes were incubated with primary antibodies against p-S6 (Ser240/244) (Cell Signaling Technology, 5364; 1:1000), puromycin (Kerafast, EQ0001; 1:1000), ADK (Human Protein Atlas, HPA038409; 1:500), CREB1 (Cell Signaling Technology, 9197; 1:1000), eIF4E (Cell Signaling Technology, 9742L; 1:1000), IRSp53 (Novus, NBP1-88711; 1:1000), S6 (Cell Signaling Technology, 2217; 1:1000), and α-tubulin (MilliporeSigma, T6047; 1:5000) in TBST overnight at 4°C. After incubation, the membranes were washed 4 times with TBST. They were incubated with HRP-linked anti-rabbit (Cell Signaling Technology, 7074; 1:5000) or anti-mouse secondary antibodies (Cell Signaling Technology, 7076; 1:2500) for 2 hours at room temperature. After washing with TBST, immunodetection was performed using ECL reaction reagents. Western blots were quantified by normalization of each blot to α-tubulin or EZBlue (MilliporeSigma, G1041) to control for loading differences.
Immunofluorescence and microscopy
For surgical tissue blocks, brain tissue was fixed in freshly prepared phosphate-buffered 4% paraformaldehyde overnight, cryoprotected overnight in 30% buffered sucrose, frozen in OCT on dry ice, and stored at −80°C. Cryostat-cut sections (20 μm thick) were collected and placed on glass slides.
For mouse brain blocks, brain tissue was harvested at the time of investigation and fixed in freshly prepared phosphate-buffered 4% paraformaldehyde for 2 hours, cryoprotected overnight in 30% buffered sucrose, frozen in OCT on dry ice, and stored at −80°C. Cryostat-cut sections (20-μm thick) were collected and placed on glass slides.
Tissue sections were blocked in PBS-GT (0.2% gelatin and 0.2% Triton X-100 in PBS) for 1 hour at room temperature, incubated in primary antibodies diluted into blocking buffer at 4°C overnight, and washed in PBS (3 × 5 minutes). Secondary antibodies were applied in blocking buffer for 1 hour at room temperature. Following another round of washing, the coverslips were mounted onto mounting solution containing DAPI (Life Technologies, P36931).
Cells were fixed in 4% paraformaldehyde in PBS (pH 7.4) for 10 minutes at room temperature, washed in PBS (3 × 5 minutes), permeabilized in 0.1% Triton X-100 in PBS for 5 minutes, and washed in PBS (3 × 5 minutes). Coverslips were blocked for 1 hour in 5% goat serum, incubated in primary antibodies diluted in blocking buffer at 4°C overnight, and washed 3 times for 5 minutes again in PBS. Secondary antibodies were applied in blocking buffer for 1 hour at room temperature. Following another round of washing, the coverslips were mounted onto mounting solution containing DAPI (Life Technologies, P36931).
Antibodies used included those against p-S6K (Thr389) (Cell Signaling Technology, 9205; 1:200), p-S6 (Ser240/244) (Cell Signaling Technology, 5364; 1:800), p–4E-BP (Thr37/46) (Cell Signaling Technology, 2855; 1:200), puromycin (Kerafast, EQ0001; 1:100), GFP (Abcam, ab290; 1:100), NeuN (MilliporeSigma, MAB377; 1:100), ADK (Human Protein Atlas, HPA038409; 1:200), CREB1 (Cell Signaling Technology, 9197; 1:800), GFAP (Thermo Fisher Scientific, MA5-15086; 1:300), and IRSp53 (Novus, NBP1-88711; 1:100).
Confocal images were obtained with a Zeiss LSM780 or LSM800 (Carl Zeiss) confocal microscope with sequential acquisition set at a resolution of 2048 × 2048 pixels. The number of cells positive for puromycin, NeuN, p-S6 (Ser240/244), p–4E-BP (Thr37/46), ADK, CREB1, and IRSp53 was determined using the ×10 or ×20 objective lens; 4 or 5 fields were acquired per subject. The number of DAPI-positive cells revealed total cell counts. We determined fluorescence-positive signals as those with an intensity 2.5 times stronger than background signals. Neuronal cell size was measured in ImageJ software (NIH; http://rsbweb.nih.gov/ij/). Dendritic spines were determined using the ×63 objective lens. Ten basal dendrites were acquired per subject, and measured in electroporated cells expressing GFP reporter using manual counting.
Analysis of global protein synthesis
For in vivo puromycin labeling, in utero–electroporated pregnant mice were injected i.p. with 0.04 μmol puromycin per gram of body weight using a 10-mg/mL solution of puromycin dihydrochloride (Thermo Fisher Scientific, A11138-03) in saline. After 30 minutes, mice were perfused, and immunofluorescence was analyzed.
For SUnSET–Western blotting, NIH 3T3 cells incubated with 200 nM Torin1 (MilliporeSigma, 47591) for 3 hours or with 200 nM Torin1 for 14 hours and CRISPR-edited MTOR(G4448A)-p.C1483Y cells (15) and NIH 3T3–WT cells incubated with 10 mg/mL cycloheximide (Calbiochem, 239763) for 2 hours were incubated with 5 μg/mL puromycin dihydrochloride for 30 minutes. HEK293T cells transfected with FLAG-tagged MTOR WT or with FLAG-tagged MTOR p.Cys1483Tyr or p.Leu2427Pro were incubated with 200 nM rapamycin (Calbiochem, M-553210) or vehicle for 14 hours, followed by 5 μg/mL puromycin or vehicle for 30 minutes. Cells were then lysed and blotted as described in the “Western blotting” section. For whole protein detection, EZBlue staining solution (MilliporeSigma, G1041) was used as recommended by the manufacturer.
For SUnSET-immunocytochemistry, cortical neurons were cultured. Neurons at 18 days in vitro were incubated with 5 μg/mL puromycin dihydrochloride for 30 minutes. Immunocytochemical analysis was performed as described in the “Immunofluorescence and microscopy” section.
Ribosome profiling (Ribo-Seq) and RNA sequencing
Footprint libraries were prepared as described previously with minor modifications (68). Cells were seeded in 10-cm dishes at 50% confluence. We ensured that cells had not reached confluence by the following day. NIH 3T3 cells were treated with 200 nM Torin1 or vehicle, and CRISPR-edited MTOR(G4448A)-p.C1483Y NIH 3T3 cells were treated with vehicle for 3 hours. Cells were washed once with ice-cold PBS supplemented with 100 μg/mL cycloheximide and lysed in mammalian polysome buffer (10 mM Tris-HCl at pH 7.4, 5 mM MgCl2, 100 mM KCl, 2 mM DTT, 1% Triton X-100, 100 μg/mL cycloheximide supplemented with 1 μL protease inhibitor cocktail, 2 μL of RNasin, and 2 μL SUPERase) to maintain binding between the ribosomes and mRNA. The samples were incubated for 20 minutes on a rotator to induce cell lysis. For FACS-sorted cortical neurons, the same treatments were applied. The cell lysates were separated into 2 tubes, 75% of the sample for Ribo-Seq and the rest for RNA-Seq, which were used to prepare Ribo-Seq and RNA-Seq libraries, respectively, as follows.
Ribo-Seq.
Cell lysates were treated with RNase I (Ambion, AM2294) to remove the ribosome-unbound region of mRNA from the ribosome-mRNA complexes. Subsequently, the ribosome-mRNA complexes were purified using Sephacryl S-400 columns (GE Healthcare, 27-5140-01). RNA footprints were extracted from the sample using TRIzol LS (Ambion, 10296-010). rRNA was removed from the sample using a Ribo-zero rRNA removal kit (Epicentre, RZH110424).
RNA-Seq.
Total RNA was purified from the cell lysates using TRIzol LS. Subsequently, the RNA sample was enriched using RNA Clean & Concentrator-5 or -25 (Zymo Research, R1015 or R1017). rRNA was removed using a Ribo-zero rRNA removal kit (Epicentre, RZH110424). After rRNA removal, the sample RNAs were fragmented via alkaline hydrolysis using the NEBNext Magnesium RNA Fragmentation Module (New England BioLabs, E6150S).
Ribosome footprints (~30 nt in size) and RNA fragments (40–60 nt in size) were size-fractionated by Urea-PAGE and purified from the gel elutes.
Preparation of Ribo-Seq and RNA-Seq libraries: Antarctic Phosphatase and PNK treatment
To do 3′ end repair, the samples were incubated in 20 μL of 1× reaction mixture containing 1 μL of Antarctic Phosphatase (New England BioLabs, M0289S) for 1 hour at 37°C, and subsequently incubated for 5 minutes at 65°C to inactivate the enzyme. Following the reaction, the RNAs were incubated in 45 μL of 1× reaction mixture containing 2 μL of PNK (Takara, 2021A) and 1 μL of [γ-32P]ATP (PerkinElmer, NEG502Z) for 5 minutes at 37°C to label the RNA. Following the labeling, 5 μL of 1 mM ATP (New England BioLabs, P0756L) was added, and the samples were incubated for 10 minutes at 37°C to phosphorylate the 5′ end of the RNAs for subsequent 5′ linker ligation. The ribosome footprints and RNA fragments were subsequently size-fractionated and purified using Urea-PAGE to remove free ATP.
Preparation of Ribo-Seq and RNA-Seq libraries: linker ligation, reverse transcription–PCR, and sequencing
TruSeq Small RNA kits (Illumina, RS-200-0012) were used for 3′ linker, 5′ linker, reverse transcription primer, 5′ primer, and 3′ primer ligation. To ligate the 3′ linker, 1.5 μL T4 RNA ligase 2, truncated (Epicentre, LR2D1132K), 1 μL 10× buffer, 0.5 μL of 3′ linker (RA3, TruSeq Small RNA kit), and 1 μL SUPERase-In were added to each RNA sample and incubated for 4 hours at 22°C. The linker-ligated RNAs were size-fractionated by Urea-PAGE and then purified from gel elute. Subsequently, to ligate the 5′ linker, 1 μL T4 RNA ligase, 1 μL 10× buffer, 1 μL 10 mM ATP, 0.5 μL 5′ linker (RA5, TruSeq Small RNA kit), and 1 μL SUPERase-In were added to each sample and incubated for 16 hours at 22°C. To construct cDNA libraries, the linker-ligated RNAs were reverse-transcribed in 20 μL 1× reaction mixture containing 4 μL 2.5 mM deoxynucleoside triphosphate (dNTP), 1 μL Superscript II RT enzyme, 4 μL 5× buffer, 2 μL 0.1 M DTT, 0.5 μL RT primer (RTP, TruSeq Small RNA kit), and 1 μL RNasin. To amplify the cDNA for Illumina DNA sequencing, 1 μL from a total of 20 μL RT sample was mixed with 1 μL Phusion HF polymerase (Thermo Fisher Scientific, F-530L), 5 μL 5× buffer, 4 μL 2.5 mM dNTP, 0.2 μL 5′ primer (TruSeq Small RNA kit), 0.2 μL 3′ primer (TruSeq Small RNA kit), and 33.6 μL of distilled water. This PCR mixture was run for 22–25 cycles of 98°C for 10 seconds, 60°C for 30 seconds, and 72°C for 20 seconds. Amplified cDNA libraries were purified by Native-PAGE and from gel elute, and then sequenced using an Illumina HiSeq 2000 or HiSeq 2500 sequencing system. All original sequencing data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE129816 and GSE129818).
Sequence processing and alignment of Ribo-Seq and RNA-Seq libraries
The first steps in the sequence analysis were done using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The reads were trimmed from the 3′ end so that the remaining reads were 26 nt in length. The reads were aligned with Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) version 1.1.2 to UCSC mm10 assembly to remove rRNA and tRNA. Then, the reads were aligned to UCSC mm10 genome sequences using TopHat (http://ccb.jhu.edu/software/tophat/index.shtml) version 2.1.1 with default options, except for no novel junctions.
mRNA transcript quantification and normalization of Ribo-Seq and RNA-Seq libraries
To quantify mRNA abundance of individual genes, we counted the number of Ribo-Seq and RNA-Seq tags per individual transcript after aligning with HTSeq (https://htseq.readthedocs.io/en/release_0.9.1/). Transcripts with low read counts in the Ribo-Seq and RNA-Seq libraries (i.e., FMCD mouse model <100 raw reads and NIH 3T3 control, C1483Y, and Torin1 cells <200 in all of the libraries) were excluded to reduce noise. Each read count per individual mRNA transcript was normalized to median values of the read counts in each library.
Quantification of translation efficiencies of individual mRNA transcripts
TE of mRNA transcripts was calculated as the normalized read counts of an individual transcript in RPFs divided by the normalized RNA-Seq tag counts mapped to the coding sequence (CDS). The CDS-mapped RNA-Seq tag counts were also normalized to their medians in the RNA-Seq libraries.
Trinucleotide periodicity
For trinucleotide periodicity analysis, reads were trimmed to 25 nt in length, underwent rRNA and tRNA removal, and were aligned using Bowtie with default options selected. The analysis was performed using riboSeqR (http://bioconductor.org/packages/release/bioc/html/riboSeqR.html).
Cluster analysis
To assess enrichment of gene ontology categories, mTOR activation–sensitive genes in FMCD and C1483Y cells and mTOR inhibition–sensitive genes in Torin1 cells were analyzed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov), version 6.7 (42); ClueGO (http://apps.cytoscape.org/apps/cluego), version 2.3.4 with Cluepedia, version 1.3.4 (16); and Ingenuity Pathway Analysis (IPA) (https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) (17). For DAVID, functional categories were clustered using the Functional Annotation clustering tool. For ClueGO, enrichment analysis was conducted with P values less than 0.05. For IPA, core analysis was performed. Epileptic seizure, cytomegalic dysmorphic neuron, and dyslamination terms were recategorized based on Gene Ontology biological process subterm and IPA disease function with P values less than 0.05.
Analysis of general 5′-UTR features
5′-UTRs were obtained from the UCSC Genome Browser (GRCm38/mm10; https://genome.ucsc.edu/) for all genes present on the ribosome profiling. The longest 5′-UTR sequence of individual mRNAs annotated in RefSeq was selected for analysis. The mTOR activation–sensitive genes in FMCD were compared with all detected genes for 5′-UTR percentage G+C content, length, and Gibbs free energy. The same procedures were applied for mTOR activation–sensitive genes in C1483Y cells and mTOR inhibition–sensitive genes in Torin1 cells. Minimum folding ΔG° was predicted for each sequence using QuikFold2 version 3.0 (http://mfold.rna.albany.edu/?q=DINAMelt/Quickfold) set with default parameters of RNA folding energy rules.
Motif analysis
MEME (http://meme-suite.org/tools/meme) was performed using version 4.12.0 (69). mTOR activation–sensitive genes in FMCD and C1483Y cells and mTOR inhibition–sensitive genes in Torin1 cells were tested. 5′-UTRs were called from the UCSC Genome Browser (GRCm38/mm10), and RefSeq-annotated mRNAs with known 5′-UTRs were selected for further analysis. The longest 5′-UTR for each gene was compiled for input into MEME as a training set. We used motif search parameters for a 12-nt sequence with any number of repeats using only a given strand.
For the FMCD mice and C1483Y cells, mTOR activation–sensitive genes in FMCD and C1483Y cells with RefSeq-annotated mRNAs containing known 5′-UTRs were evaluated for the presence of guanine quartet (GGC)4, CERT, A-rich, and U-rich motifs using Find Individual Motif Occurrences (FIMO; http://meme-suite.org/tools/fimo).
5′-UTR luciferase reporter assays
5′-UTRs were obtained from the UCSC Genome Browser (GRCm38/mm10). 5′-UTRs with WT, transversion, and deletion sequences (see also Supplemental Table 14) were synthesized by Macrogen and subcloned into the pGL3-promoter vector (Promega, E1761) between the SV40 promoter and the firefly luciferase open reading frame using HindIII (New England BioLabs, R3104) and NcoI (New England BioLabs, R3193). Renilla pGL4.74 was used as a control reporter. HEK293T cells were cotransfected with pGL3-SV40 5′-UTR reporter; FLAG-tagged mTOR WT, FLAG-tagged mTOR p.Cys1483Tyr, or FLAG-tagged mTOR p.Leu2427Pro; and pGL4.74 control reporter, at a ratio of 30:30:1. Cells were harvested for lysates 24 hours later, and luciferase activity was measured using a Dual Luciferase Assay kit (Promega, E1960). Firefly luciferase activity was normalized to Renilla activity.
Plasmid constructs for knockdown experiments
shRNA sequences were obtained from the Broad Institute Genetic Perturbation Platform (GPP) Web Portal (https://portals.broadinstitute.org/gpp/public/) and RNAi central (http://katahdin.cshl.org/siRNA/RNAi.cgi?type=shRNA). shRNA oligos were synthesized by Macrogen and subcloned into the pSicoR vector (Addgene, 11579) after the mU6 promoter using HpaI (New England BioLabs, R0105S) and XhoI (New England BioLabs, R0146S).
shRNA sequences
shRNA sequences were as follows: shADK #1, TTGATATCCTCTTTGGAAA; shADK #2, GCTGTAGTAGACAAAGATT; shADK #3, TAGAGAAAGCCAGAGTTTA; shADK #4, CTGTCTGCACCGTTCATTA; sheIF4E #1 (TRCN0000077475), CCGAAGATAGTGATTGGTTAT; sheIF4E #2 (TRCN0000077477), CGATTGATCTCTAAGTTTGAT; sheIF4E #3, TGTGTGTGGAGCTGTTGTTAA; sheIF4E #4, GAGATGCAGTCACACACATAG.
shRNA knockdown validation by Western blot in Neuro2A cell lines
Neuro2A cells were transfected with pSicoR-empty, pSicoR-shScramble, pSicoR-shADKs, and pSicoR-sheIF4Es as indicated in the “Transfection” section. Cells were lysed and Western-blotted.
shRNA knockdown validation in in utero–electroporated FMCD mouse brain tissue
Timed pregnant mice (E14) were electroporated in utero as described in the “In utero electroporation” section with mTOR WT, p.Cys1483Tyr, or p.Leu2427Pro, with or without mU6-shScramble, mU6-shADK, or mU6-sheIF4E. Immunofluorescence analysis was then performed.
In vivo rapamycin treatment
Rapamycin (LC Labs, R5000200MG) was dissolved initially in 20 mg/mL 100% ethanol stock solution and stored at –20°C. Immediately before injection, stock solution was diluted in 5% polyethylene glycol 400 and 5% Tween-80 to final concentrations of 1 mg/mL rapamycin and 4% ethanol. Adult (>P56) mice were injected daily through an i.p. route with 10 mg/kg rapamycin or vehicle alone for 2 weeks, and embryos were injected daily through an i.p. route with 5 mg/kg rapamycin or vehicle alone for 4 days for in utero electroporated pregnant mice, from E14.
In vivo metformin treatment
Metformin (MilliporeSigma, PHR1084-500MG) was dissolved initially in 20 mg/mL saline stock solution and stored at –20°C. Mice were injected daily through an i.p. route with 200 mg/kg of metformin or vehicle alone for 30 days.
In vivo 5-ITU treatment
In brief, 5-ITU (Tocris 1745) was dissolved initially in 100% DMSO, diluted to 0.775 mg/mL (20% DMSO in saline) of stock solution, and stored at –20°C. Immediately before injection, stock solution was diluted to 5% DMSO in saline. Mice were injected twice per day through an i.p. route with 1.0 or 2.6 mg/kg of 5-ITU or vehicle alone for 10 days.
Video-EEG monitoring
EEG signals from epidural electrodes positioned on the frontal lobes (anterior to posterior + 2.8 mm, medial to lateral ± 1.5 mm) and temporal lobes (anterior to posterior – 2.4 mm, medial to lateral ± 2.4 mm) were recorded using the cerebellum as a reference. After more than 10 days of recovery from the surgery, EEG signals were recorded for more than 2 days (12 hours per day). Signals were amplified with an RHD2000 amplifier chip (Intan Technologies), recorded with an RHD2000 USB interface board (Intan Technologies), and analyzed with the open-source MATLAB toolbox EEGLAB (http://sccn.ucsd.edu/eeglab).
Statistics
Data are presented as the mean ± SEM or SD. Results were analyzed using 1-way ANOVA where appropriate using GraphPad Prism version 8 (GraphPad Software Inc.). A Bonferroni’s post hoc test was used to analyze significant differences in the 1-way ANOVA test. A P value less than 0.05 was considered statistically significant. All experiments requiring the use of animals, directly or as a source of cells, were subjected to randomization. Sample size was predetermined based on the variability observed in preliminary and similar experiments. The researchers were not blinded to allocation during experiments. Detailed methods and P values for statistical significance are described in the figure legends and Methods. In box plots, the top of the box represents the third quartile, the bottom of the box represents the first quartile, the central line of the box represents the median value, and the whisker extends up to the highest value of the upper limit and down to the lowest value of the lower limit.
Study approval
The study was performed and all human tissues were obtained in accordance with protocols approved by Severance Children’s Hospital and KAIST Institutional Review Board and Committee on Human Research. Informed consent from the parents of individuals with FMCD was obtained.
Author contributions
JKK and JHL conceived and designed the study. JKK performed most of the experiments and subsequent analyses, including the in vitro and in vivo studies. JC and VNK helped with the ribosome profiling and analysis. DSK, HCK, and SHK collected patient samples and managed patient information and tissue samples. JKK and JHL wrote the manuscript with input from all of the authors.
Supplementary Material
Acknowledgments
This work was supported by grants from the Suh Kyungbae Foundation (to JHL); the Korean Health Technology R&D Project; the National Research Foundation of Korea (NRF), funded by the Korean government, Ministry of Science and ICT (2019R1A3B2066619, to JHL), and the Ministry of Health and Welfare, Republic of Korea (H15C3143 and H16C0415 to JHL). JKK is a Cheong-Am Science Fellow supported by POSCO Cheong-Am Foundation. Some tissue, biological specimens, and data used in this research were obtained from Autism BrainNet, which is sponsored by the Simons Foundation and Autism Speaks. The authors also acknowledge the Autism Tissue Program that was the predecessor to Autism BrainNet.
Version 1. 09/04/2019
Electronic publication
Version 2. 10/01/2019
Print issue publication
Version 3. 02/16/2024
added 8 missing supplemental tables
Footnotes
Conflict of interest: JHL is a cofounder and chief technology officer of SoVarGen Inc., which develops new diagnostics and therapeutics for brain disorders. SoVarGen Inc. plans to apply for patents on the basis of the results of the current study.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(10):4207–4223.https://doi.org/10.1172/JCI127032.
Contributor Information
Jun Cho, Email: jun_cho@hms.harvard.edu.
Se Hoon Kim, Email: paxco@yuhs.ac.kr.
Hoon-Chul Kang, Email: hipo0207@yuhs.ac.
Dong-Seok Kim, Email: dskim33@yuhs.ac.
Jeong Ho Lee, Email: jhlee4246@kaist.ac.kr.
References
- 1.Lee JH. Somatic mutations in disorders with disrupted brain connectivity. Exp Mol Med. 2016;48:e239. doi: 10.1038/emm.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae T, et al. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science. 2018;359(6375):550–555. doi: 10.1126/science.aan8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JH, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44(8):941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim JS, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med. 2015;21(4):395–400. doi: 10.1038/nm.3824. [DOI] [PubMed] [Google Scholar]
- 5.Leventer RJ, et al. Hemispheric cortical dysplasia secondary to a mosaic somatic mutation in MTOR. Neurology. 2015;84(20):2029–2032. doi: 10.1212/WNL.0000000000001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakashima M, et al. Somatic mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann Neurol. 2015;78(3):375–386. doi: 10.1002/ana.24444. [DOI] [PubMed] [Google Scholar]
- 7.Kim JK, Lee JH. Mechanistic target of rapamycin pathway in epileptic disorders. J Korean Neurosurg Soc. 2019;62(3):272–287. doi: 10.3340/jkns.2019.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shor B, Cavender D, Harris C. A kinase-dead knock-in mutation in mTOR leads to early embryonic lethality and is dispensable for the immune system in heterozygous mice. BMC Immunol. 2009;10:28. doi: 10.1186/1471-2172-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsan E, Baulac S. Review: Mechanistic target of rapamycin (mTOR) pathway, focal cortical dysplasia and epilepsy. Neuropathol Appl Neurobiol. 2018;44(1):6–17. doi: 10.1111/nan.12463. [DOI] [PubMed] [Google Scholar]
- 11.Tahmasebi S, Khoutorsky A, Mathews MB, Sonenberg N. Translation deregulation in human disease. Nat Rev Mol Cell Biol. 2018;19(12):791–807. doi: 10.1038/s41580-018-0034-x. [DOI] [PubMed] [Google Scholar]
- 12.Crino PB. The mTOR signalling cascade: paving new roads to cure neurological disease. Nat Rev Neurol. 2016;12(7):379–392. doi: 10.1038/nrneurol.2016.81. [DOI] [PubMed] [Google Scholar]
- 13.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SM, et al. Brain somatic mutations in MTOR disrupt neuronal ciliogenesis, leading to focal cortical dyslamination. Neuron. 2018;99(1):83–97.e7. doi: 10.1016/j.neuron.2018.05.039. [DOI] [PubMed] [Google Scholar]
- 16.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bontemps F, Van den Berghe G, Hers HG. Evidence for a substrate cycle between AMP and adenosine in isolated hepatocytes. Proc Natl Acad Sci U S A. 1983;80(10):2829–2833. doi: 10.1073/pnas.80.10.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bontemps F, Vincent MF, Van den Berghe G. Mechanisms of elevation of adenosine levels in anoxic hepatocytes. Biochem J. 1993;290(pt 3):671–677. doi: 10.1042/bj2900671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouder N, Scheurer L, Fritschy JM, Boison D. Overexpression of adenosine kinase in epileptic hippocampus contributes to epileptogenesis. J Neurosci. 2004;24(3):692–701. doi: 10.1523/JNEUROSCI.4781-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedele DE, et al. Astrogliosis in epilepsy leads to overexpression of adenosine kinase, resulting in seizure aggravation. Brain. 2005;128(pt 10):2383–2395. doi: 10.1093/brain/awh555. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Lan JQ, Boison D. Uncoupling of astrogliosis from epileptogenesis in adenosine kinase (ADK) transgenic mice. Neuron Glia Biol. 2008;4(2):91–99. doi: 10.1017/S1740925X09990135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masino SA, et al. A ketogenic diet suppresses seizures in mice through adenosine A1 receptors. J Clin Invest. 2011;121(7):2679–2683. doi: 10.1172/JCI57813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen HY, et al. Overexpression of adenosine kinase in cortical astrocytes and focal neocortical epilepsy in mice. J Neurosurg. 2014;120(3):628–638. doi: 10.3171/2013.10.JNS13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18(2):52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed S, Goh WI, Bu W. I-BAR domains, IRSp53 and filopodium formation. Semin Cell Dev Biol. 2010;21(4):350–356. doi: 10.1016/j.semcdb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Oikawa T, Okamura H, Dietrich F, Senju Y, Takenawa T, Suetsugu S. IRSp53 mediates podosome formation via VASP in NIH-Src cells. PLoS One. 2013;8(3):e60528. doi: 10.1371/journal.pone.0060528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh TC, Ogawa W, Danielsen AG, Roth RA. Characterization and cloning of a 58/53-kDa substrate of the insulin receptor tyrosine kinase. J Biol Chem. 1996;271(6):2921–2928. doi: 10.1074/jbc.271.6.2921. [DOI] [PubMed] [Google Scholar]
- 29.Chen CJ, et al. SH2B1 and IRSp53 proteins promote the formation of dendrites and dendritic branches. J Biol Chem. 2015;290(10):6010–6021. doi: 10.1074/jbc.M114.603795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung W, et al. Social deficits in IRSp53 mutant mice improved by NMDAR and mGluR5 suppression. Nat Neurosci. 2015;18(3):435–443. doi: 10.1038/nn.3927. [DOI] [PubMed] [Google Scholar]
- 31.Crespi A, et al. LIN7 regulates the filopodium- and neurite-promoting activity of IRSp53. J Cell Sci. 2012;125(pt 19):4543–4554. doi: 10.1242/jcs.106484. [DOI] [PubMed] [Google Scholar]
- 32.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35(4):605–623. doi: 10.1016/S0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 33.Josselyn SA, Nguyen PV. CREB, synapses and memory disorders: past progress and future challenges. Curr Drug Targets CNS Neurol Disord. 2005;4(5):481–497. doi: 10.2174/156800705774322058. [DOI] [PubMed] [Google Scholar]
- 34.Kaindl AM, Asimiadou S, Manthey D, Hagen MV, Turski L, Ikonomidou C. Antiepileptic drugs and the developing brain. Cell Mol Life Sci. 2006;63(4):399–413. doi: 10.1007/s00018-005-5348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papadia S, Stevenson P, Hardingham NR, Bading H, Hardingham GE. Nuclear Ca2+ and the cAMP response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J Neurosci. 2005;25(17):4279–4287. doi: 10.1523/JNEUROSCI.5019-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riccio A, Ahn S, Davenport CM, Blendy JA, Ginty DD. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science. 1999;286(5448):2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 37.Díaz-Ruiz C, et al. Regulation of neural migration by the CREB/CREM transcription factors and altered Dab1 levels in CREB/CREM mutants. Mol Cell Neurosci. 2008;39(4):519–528. doi: 10.1016/j.mcn.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Jancic D, Lopez de Armentia M, Valor LM, Olivares R, Barco A. Inhibition of cAMP response element-binding protein reduces neuronal excitability and plasticity, and triggers neurodegeneration. Cereb Cortex. 2009;19(11):2535–2547. doi: 10.1093/cercor/bhp004. [DOI] [PubMed] [Google Scholar]
- 39.Grabiner BC, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4(5):554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh AP, et al. Point mutations of the mTOR-RHEB pathway in renal cell carcinoma. Oncotarget. 2015;6(20):17895–17910. doi: 10.18632/oncotarget.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fonseca BD, Smith EM, Yelle N, Alain T, Bushell M, Pause A. The ever-evolving role of mTOR in translation. Semin Cell Dev Biol. 2014;36:102–112. doi: 10.1016/j.semcdb.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Dennis G, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 43.Morita M, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18(5):698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Schieke SM, et al. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281(37):27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 45.Qin L, et al. Differentially expressed proteins underlying childhood cortical dysplasia with epilepsy identified by iTRAQ proteomic profiling. PLoS One. 2017;12(2):e0172214. doi: 10.1371/journal.pone.0172214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wesseling H, Elgersma Y, Bahn S. A brain proteomic investigation of rapamycin effects in the Tsc1+/- mouse model. Mol Autism. 2017;8:41. doi: 10.1186/s13229-017-0151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truitt ML, et al. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162(1):59–71. doi: 10.1016/j.cell.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modelska A, et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015;6:e1603. doi: 10.1038/cddis.2014.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubio CA, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15(10):476. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolfe AL, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513(7516):65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci. 2005;25(49):11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin TV, Hsieh L, Kimura T, Malone TJ, Bordey A. Normalizing translation through 4E-BP prevents mTOR-driven cortical mislamination and ameliorates aberrant neuron integration. Proc Natl Acad Sci U S A. 2016;113(40):11330–11335. doi: 10.1073/pnas.1605740113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gantois I, et al. Metformin ameliorates core deficits in a mouse model of fragile X syndrome. Nat Med. 2017;23(6):674–677. doi: 10.1038/nm.4335. [DOI] [PubMed] [Google Scholar]
- 54.Kiese K, Jablonski J, Boison D, Kobow K. Dynamic Regulation of the adenosine kinase gene during early postnatal brain development and maturation. Front Mol Neurosci. 2016;9:99. doi: 10.3389/fnmol.2016.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aronica E, et al. Upregulation of adenosine kinase in astrocytes in experimental and human temporal lobe epilepsy. Epilepsia. 2011;52(9):1645–1655. doi: 10.1111/j.1528-1167.2011.03115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiesner JB, et al. Adenosine kinase inhibitors as a novel approach to anticonvulsant therapy. J Pharmacol Exp Ther. 1999;289(3):1669–1677. [PubMed] [Google Scholar]
- 57.Qu Z, Zhang Y, Liao M, Chen Y, Zhao J, Pan Y. In vitro and in vivo antitumoral action of metformin on hepatocellular carcinoma. Hepatol Res. 2012;42(9):922–933. doi: 10.1111/j.1872-034X.2012.01007.x. [DOI] [PubMed] [Google Scholar]
- 58.Boison D. The adenosine kinase hypothesis of epileptogenesis. Prog Neurobiol. 2008;84(3):249–262. doi: 10.1016/j.pneurobio.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2015;1849(7):801–811. doi: 10.1016/j.bbagrm.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 60.Hong S, et al. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. Elife. 2017;6:e25237. doi: 10.7554/eLife.25237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fonseca BD, et al. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1) J Biol Chem. 2015;290(26):15996–16020. doi: 10.1074/jbc.M114.621730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tcherkezian J, et al. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5’TOP mRNA translation. Genes Dev. 2014;28(4):357–371. doi: 10.1101/gad.231407.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lahr RM, et al. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife. 2017;6:e24146. doi: 10.7554/eLife.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel B, et al. Exosomes mediate the acquisition of the disease phenotypes by cells with normal genome in tuberous sclerosis complex. Oncogene. 2016;35(23):3027–3036. doi: 10.1038/onc.2015.358. [DOI] [PubMed] [Google Scholar]
- 65.Baek ST, et al. An AKT3-FOXG1-reelin network underlies defective migration in human focal malformations of cortical development. Nat Med. 2015;21(12):1445–1454. doi: 10.1038/nm.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsieh LS, et al. Convulsive seizures from experimental focal cortical dysplasia occur independently of cell misplacement. Nat Commun. 2016;7:11753. doi: 10.1038/ncomms11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lim JS, et al. Somatic mutations in TSC1 and TSC2 cause focal cortical dysplasia. Am J Hum Genet. 2017;100(3):454–472. doi: 10.1016/j.ajhg.2017.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cho J, et al. Multiple repressive mechanisms in the hippocampus during memory formation. Science. 2015;350(6256):82–87. doi: 10.1126/science.aac7368. [DOI] [PubMed] [Google Scholar]
- 69.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.