In this review, Cosby et al. argue that the evolutionary success of TEs cannot be explained solely by evasion from host control mechanisms; rather, some TEs have evolved commensal and even mutualistic strategies that mitigate the cost associated with their propagation. These coevolutionary processes promote the emergence of complex cellular activities, which in turn pave the way for cooption of TE sequences for organismal function.
Keywords: gene regulation, genomics, KRAB zinc finger, fetrotransposons, transposable elements, piRNA
Abstract
Transposable elements (TEs) are mobile DNA sequences that colonize genomes and threaten genome integrity. As a result, several mechanisms appear to have emerged during eukaryotic evolution to suppress TE activity. However, TEs are ubiquitous and account for a prominent fraction of most eukaryotic genomes. We argue that the evolutionary success of TEs cannot be explained solely by evasion from host control mechanisms. Rather, some TEs have evolved commensal and even mutualistic strategies that mitigate the cost of their propagation. These coevolutionary processes promote the emergence of complex cellular activities, which in turn pave the way for cooption of TE sequences for organismal function.
Transposable elements (TEs) are mobile repetitive DNA sequences that comprise a substantial fraction of eukaryotic genomes (Bourque et al. 2018). For instance, TEs and their remnants account for more than half the nuclear DNA content of maize, zebrafish, and humans and approximately a third of the nuclear DNA content of Drosophila melanogaster and Caenorhabditis elegans (The C. elegans Sequencing Consortium 1998; Schnable et al. 2009; de Koning et al. 2011; Howe et al. 2013; Hoskins et al. 2015). The evolutionary success of TEs lies in their ability to replicate independently of and faster than the host genome (Doolittle and Sapienza 1980; Orgel and Crick 1980). TEs are classified into two broad groups based on their molecular transposition intermediate: Class I elements (retrotransposons) mobilize via an RNA intermediate, while class II elements (DNA transposons) mobilize via a DNA intermediate. Retrotransposons include endogenous retroviruses (ERVs) and related long terminal repeat (LTR) retrotransposons as well as non-LTR retrotransposons (Wicker et al. 2007; Bourque et al. 2018). DNA transposons include “cut and paste” DNA transposons as well as non-“cut and paste” elements (Wicker et al. 2007; Bourque et al. 2018). If transposition occurs in cells that contribute to the next generation (i.e., the germline), the newly integrated TE copy becomes inheritable and may spread vertically within the population. TEs also spread horizontally between species at an appreciable frequency, which is crucial to their evolutionary persistence by allowing the colonization of new genomes (Gilbert and Feschotte 2018).
As for any heritable mutational event, natural selection and genetic drift dictate the fate of a new TE insertion in the population. Because of their sheer length (∼100–10,000 bp) and propensity to carry regulatory elements (i.e., promoters, splice sites, and poly-A signals), TE insertions are particularly prone to disrupt gene function. For instance, ∼10% and ∼50% of spontaneous mutant phenotypes isolated in laboratory strains of mice and flies, respectively, are caused by de novo TE insertions within the coding or noncoding portion of genes (Eickbush and Furano 2002; Gagnier et al. 2019). An equally sizeable fraction of maize mutants arose from transposition events (Neuffer et al. 1997). In humans, at least 120 Mendelian diseases have been attributed to de novo TE insertions (Hancks and Kazazian 2016). These observations, together with more direct measurement of fitness effects in Drosophila caged populations (e.g., Pasyukova 2004), indicate that TE mobilization is highly mutagenic and represents a significant source of genome instability in a wide range of organisms.
Even TEs that are no longer mobile still pose a threat to organismal fitness. The repetitive nature of TE families provides a substrate for ectopic recombination events that can lead to chromosomal rearrangements, often with deleterious consequences (Montgomery et al. 1991; Ade et al. 2013; Bennetzen and Wang 2014; Hancks and Kazazian 2016). TE-encoded products (RNA, cDNA, and proteins), even with compromised functionalities, can be toxic, and their accumulation to aberrant amounts are associated with and increasingly recognized as directly contributing to various disease states, including cancer, senescence, and chronic inflammation (Lee et al. 2012a; Tubio et al. 2014; Burns 2017; Tang et al. 2017; Bourque et al. 2018; Dubnau 2018; Schauer et al. 2018; De Cecco et al. 2019).
The manifold capacity of TEs to compromise genome integrity and interfere with normal cellular function suggests that uncontrolled TE amplification can have catastrophic consequences for both organisms and TEs, as their fitness is inextricably linked. Thus, like any parasitic system, natural selection will eliminate TEs with excessive activity and overly deleterious effects on their host. It will also select for the emergence of host-encoded mechanisms that suppress or dampen TE activity. If a defense mechanism completely blocks a given TE family, it will in turn place selective pressure on these elements to evolve a counterdefense or escape mechanism to avoid extinction, pressuring the host to evolve further mechanisms to compensate. Such genetic conflict is often framed in terms of an arms race (Hurst and Werren 2001; Burt and Trivers 2006; Werren 2011; McLaughlin and Malik 2017; Ozata et al. 2019), which builds on the Red Queen model for host–pathogen interactions (Van Valen 1973). Alternatively, the arms race could be avoided if TEs mitigate the conflict with their host either through self-control or by providing a benefit offsetting their cost. Such strategies would reduce pressure on the host to evolve systems to counteract TEs, promoting equilibrium rather than precipitating an arms race.
In the first part of this review, we examine the evidence for and against the arms race model of host–TE interactions. We posit that the extent to which TEs outcompete their host is constrained by their dependence on organismal fitness, which favors TEs that evolve strategies that circumvent or attenuate, rather than block, host defenses. We then explore alternative models, including self-regulatory mechanisms and mutualistic interactions, that reduce the cost of TE activity. We speculate that cooperative strategies may be more widespread than currently appreciated and pave the way for cooption of TE sequences for host function.
Arms races
Consistent with a need to prevent the deleterious effects of rampant transposition, a variety of host-encoded mechanisms are known to repress eukaryotic TEs at both the transcriptional (chromatin modification and DNA methylation) and posttranscriptional (modifying and degrading TE transcripts) level (Yoder et al. 1997; Borges and Martienssen 2015; Goodier 2016; Czech et al. 2018; Ozata et al. 2019). This leads to suppression of TE expression, mobility, and/or ability to promote recombination. However, all of these mechanisms do not act exclusively on TE sequences but also regulate host gene expression or protect against exogenous pathogens (Klose and Bird 2006; Fedoroff 2012; Klemm et al. 2019). In fact, it has been proposed that epigenetic control systems, which suppress recombination, might facilitate the accumulation of TEs (Fedoroff 2012). These observations beg the following question: Are TEs the raison d’être and primary target for these mechanisms or are they caught in the cross fire? One way to address this question is to examine whether some of these mechanisms have evolved the ability to distinguish TEs from host sequences. Another predicted hallmark of a TE defense system is that it should be able to adapt to control new or variant TEs that evade repression, triggering an arms race between TEs and components of the pathway. Below, we discuss two pathways that appear to possess these attributes: piwi-interacting small RNAs (piRNAs) and Kruppel-associated box-containing zinc finger proteins (KRAB-ZFPs).
piRNA-mediated TE silencing
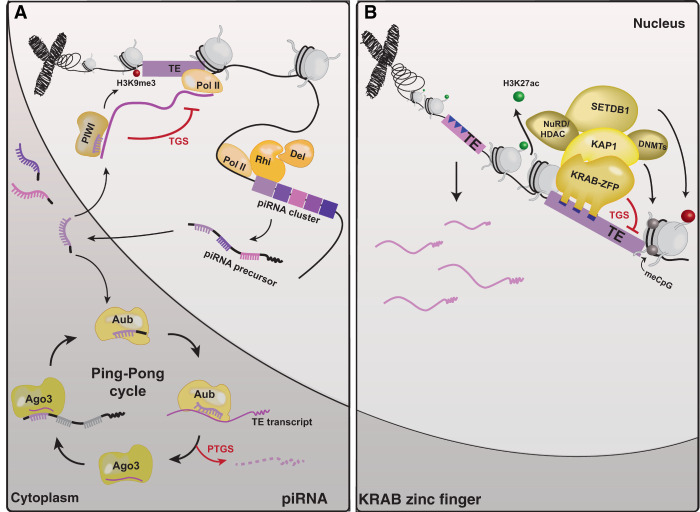
piRNAs are small RNAs found in most metazoans that are typically produced from long precursor transcripts derived from specialized loci called piRNA clusters (for review, see Ernst et al. 2017; Ozata et al. 2019). Once expressed, primarily in the gonads but also in the soma of some organisms, piRNA precursors are processed into mature piRNAs, which then complex with PIWI clade Argonaute proteins. These ribonucleoprotein (RNP) complexes recognize complementary target mRNAs in the cytoplasm or nascent RNAs in the nucleus, triggering a cascade of biochemical processes that ultimately reduces target RNA expression via posttranscriptional or transcriptional mechanisms, respectively (Fig. 1A; Ozata et al. 2019). piRNAs produced from TE loci act as potent trans-repressors of related TEs located throughout the genome (Ozata et al. 2019). In some organisms, piRNA-mediated TE repression appears necessary to preserve the integrity of the germline. This is best documented in D. melanogaster, where the loss of piRNAs normally repressing certain TEs (P element and I element) leads to rampant transposition and hybrid dysgenesis, which is characterized by extensive DNA damage, gonadal atrophy, and sterility (Kidwell et al. 1977; Bucheton et al. 1984; Brennecke et al. 2008; Wang et al. 2018). Increased piRNA production against P elements in aging flies rescues fertility, further underlining the importance of the piRNA pathway in maintaining germline integrity (Khurana et al. 2011). In mice, elimination of PIWI proteins also results in massive accumulation of retrotransposon transcripts in sperm and oocytes (Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008; De Fazio et al. 2011; Kabayama et al. 2017). This activation precedes—and may cause—defects in spermatogenesis due to meiotic arrest and apoptosis (Deng and Lin 2002; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007; Frost et al. 2010), although whether defects are due to transposition remains unclear (Newkirk et al. 2017). Similar phenotypes are also seen in C. elegans upon loss of PIWI protein PRG-1, which leads to transposon activation and impaired fertility (Batista et al. 2008; Wang and Reinke 2008; Bagijn et al. 2012; Lee et al. 2012b). Thus, it is indisputable that one function of the piRNA pathway is to recognize and silence TEs.
Figure 1.
piRNA and KRAB-ZFPs: two host systems that recognize and silence TEs. (A) The piRNA pathway in D. melanogaster. Mature piRNAs re-enter the nucleus to perform transcriptional gene silencing (TGS) or participate in the ping-pong cycle to perform posttranscriptional gene silencing (PTGS) of TEs. (Rhi) Rhino; (Del) Deadlock; (Aub) Aubergine; (Ago3) Argonaute 3. (B) The KRAB-ZFP pathway in tetrapods (see the text). (NURD) Nucleosome remodeling deacetylase; (HDAC) histone deacetylase; (DNMTs) DNA methyltransferases.
The piRNA pathway evolves rapidly, but why?
Is the piRNA pathway engaged in an arms race with TEs? A common hallmark of an arms race between host and pathogen is the rapid diversification of host and pathogen proteins that are engaged in conflicting interactions. This may be a direct physical interaction (e.g., host protein blocks pathogen protein), but indirect or secondary interactions may also drive rapid evolution (McLaughlin and Malik 2017; Parhad and Theurkauf 2018). Diversification and adaptation of protein sequences often manifest as so-called positive selection at the gene level, which is characterized by an excess of nonsynonymous changes in codons relative to synonymous mutations over time. Signatures of positive selection are pervasive in invertebrate piRNA pathway proteins, operating at virtually every step of piRNA biogenesis both within and between species (Begun et al. 2007; Larracuente et al. 2008; Obbard et al. 2009; Mackay et al. 2012; Simkin et al. 2013; Blumenstiel et al. 2016). Signatures of positive selection are also apparent in the evolution of several piRNA genes across fish species, including Piwil1, but less evident in their mammalian homologs (Yi et al. 2014).
Another prediction of a rapidly evolving host defense system is that it can lead to functional incompatibilities, introducing a breach in the defense of the progeny of individuals carrying divergent components. Support for this scenario came from an elegant study of the rapidly evolving piRNA pathway components Rhino and Deadlock in hybrids of D. melanogaster and Drosophila simulans (Parhad et al. 2017). In drosophilids, Rhino and Deadlock interact directly as part of a complex that binds to and promotes the transcription of piRNA clusters (Fig. 1A; Ozata et al. 2019). Genetic loss of Rhino (Klattenhoff et al. 2009) or depletion of Deadlock (Mohn et al. 2014) in D. melanogaster leads to loss of cluster-derived piRNAs and results in massive transcriptional activation of TEs in the germline. While introduction of a D. melanogaster rhino transgene rescues this mutant phenotype, the D. simulans rhino transgene does not (Fig 2A; Parhad et al. 2017). Domain-swapping experiments and coimmunoprecipitation and colocalization assays indicate that this is due to an inability of the D. simulans Rhino protein to interact with D. melanogaster Deadlock due to species-specific amino acid changes at the interaction interface of the two orthologous proteins (Parhad et al. 2017). However, D. simulans Rhino does interact with D. simulans Deadlock, indicating that coevolution has occurred between these proteins in the D. simulans lineage as well. The investigators speculate that the rapid coevolution of Rhino and Deadlock proteins was precipitated by a yet-unknown TE-encoded antisilencing factor present in one or both species’ lineages that may directly compete or interfere with their interaction (Parhad et al. 2017).
Figure 2.
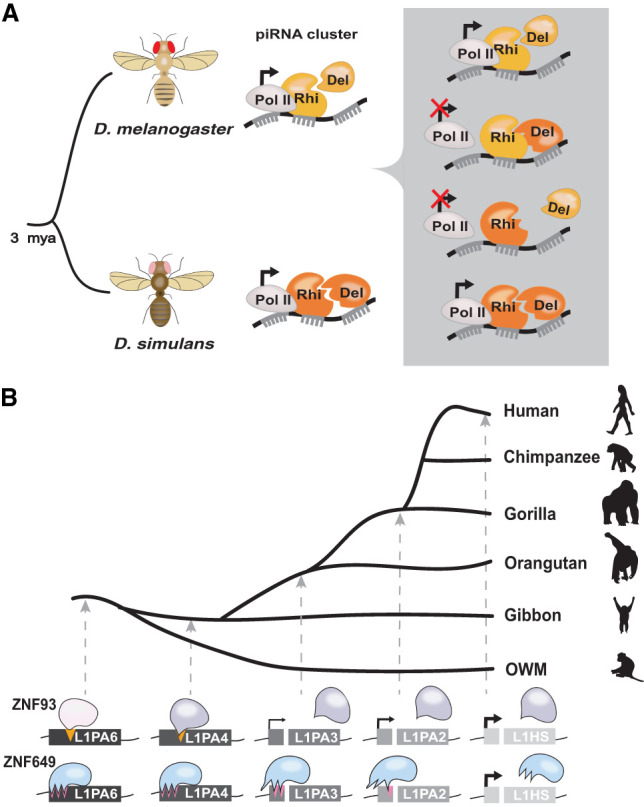
Evidence of host–TE arms races. (A) D. melanogaster–D. simulans Rhino–Deadlock incompatibility. (Rhi) Rhino; (Del) Deadlock; (Pol II) RNA polymerase II. D. melanogaster–D. simulans Rhino/Deadlock proteins cannot complement to transcribe piRNA clusters (see the text). (B) ZNF93- and ZNF649-mediated silencing of primate L1 elements. Older L1 primate families are silenced by ZNF93 and ZNF649, but younger L1 families lack ZNF93- and ZNF649-binding sites and are not silenced.
Another indicator of adaptive evolution of the piRNA pathway lies in the recurrent duplication and turnover of genes involved in the pathway. Again, this is most striking in invertebrates. For example, a phylogenomic survey of Piwi clade Argonaute genes across 84 dipteran species revealed that these genes have duplicated 27 times independently (Lewis et al. 2016). Interestingly, this study revealed a disproportionate number of duplication events in mosquitoes, which carry a large and diverse TE load (Arensburger et al. 2010; Matthews et al. 2018). More recently, a broader survey of arthropods identified an additional 17 gene duplication events of Piwi, 14 of which occurred in the lineage of the pea aphid alone (Lewis et al. 2018), perhaps reflecting their abundant and diverse TE content (∼38% of the genome) (The International Aphid Genomics Consortium 2010).
The rapid diversification of the piRNA pathway machinery in insects via positive selection and recurrent gene duplication is consistent with the idea that the system is engaged in an arms race. What remains unclear, however, is whether adaptation in the pathway is driven primarily by a need to adjust to TE activity or evasion or by another conflict. If TE activity is the primary driver, one would predict that the timing and intensity of positive selection should track with TE load and diversity across species’ lineages. However, in Drosophila, where this idea has been modeled and examined, this appears to not be the case. TE abundance across the Drosophila genus was found to be correlated with the level of purifying selection (constraint) on piRNA pathway components but not with the rate at which these proteins have diversified (Castillo et al. 2011). Another prediction of an arms race model is that variation in TE activity would be accompanied by commensurate changes in expression of piRNA pathway components or changes in abundance or composition of the piRNA pool. The former possibility was examined across wild-type strains of D. simulans with variable TE content (Fablet et al. 2014). While piRNA pathway genes exhibited wide variation in transcript levels across strains, there was no positive correlation with TE copy number. In fact, RNA sequencing of 16 inbred lines from the Drosophila Genetic Reference Panel identified only minor variation in piRNA expression, and piRNA cluster expression did not correlate with the presence of strain-specific TE insertions (Song et al. 2014). Another study investigated the genomic factors that contribute to the piRNA pool by integrating genomic, mRNA, and small RNA data for two laboratory strains of D. melanogaster (Kelleher and Barbash 2013). While variation of piRNA abundance between strains appears to be positively correlated with total TE content and expression, the most recently active TEs did not produce the most abundant piRNAs in ovaries (Kelleher and Barbash 2013). Collectively, the evidence so far in Drosophila does not support the notion that TE activity is a primary driver of rapid evolution in the piRNA pathway. It should be noted, however, that correlation between TE content and piRNA pathway expression variants may not be detectable due to the low levels of linkage disequilibrium in Drosophila (Mackay et al. 2012). Further characterization of the relationship between piRNA pathway evolution and TE composition across species with more drastic variation in TE content might reveal a different picture.
piRNAs function beyond TE silencing and outside the germline
What other forces could drive adaptive evolution of the piRNA pathway? In addition to TEs, it is well established that piRNAs also target host genes and viruses (Ozata et al. 2019), and both targets could have considerable influence on piRNA pathway evolution. The fact that host genes are not immune to, but actually are frequent targets of piRNAs in a wide range of organisms implies a need to minimize off-target effects. Indeed, if piRNA targeting is not sufficiently specific to TEs, it could interfere with host gene expression. The autoimmunity hypothesis (Blumenstiel et al. 2016) posits that positive selection in piRNA genes reflects alternating periods of high and low TE activity, which impose opposite constraints on the piRNA response: High TE activity requires high piRNA specificity, while low activity calls for greater sensitivity. In this model, TE activity indirectly influences piRNA evolution. Measurable fitness defects caused by off-targeting effects of TE-derived small RNAs (Hollister and Gaut 2009; Lee 2015) bring empirical support to the model. Further evidence comes from a recent study that compared off-target effects of three piRNA pathway components—Aubergine (aub), Armitage (armi), and Spindle E (spnE)—in D. melanogaster mutant backgrounds trans-complemented with the D. simulans protein (Wang et al. 2019). When mutant flies were trans-complemented, more D. melanogaster protein-coding genes were repressed than when complemented with the D. melanogaster version, suggesting that the D. melanogaster proteins have adapted to avoid genic off-targeting in the D. melanogaster background, whereas the D. simulans proteins have not (Wang et al. 2019). These results support the idea that the avoidance of off-target effects on “self” sequences is a plausible driver of adaptation in the piRNA pathway.
There is also mounting evidence that the piRNA pathway functions in antiviral defense, which may trigger another conflict driving rapid evolution. The strongest evidence for antiviral activity of piRNAs thus far comes from studies in mosquitoes. These insects have a dramatically expanded repertoire of Piwi genes (Lewis et al. 2016) and deploy somatic piRNAs to combat arbovirus infection (Morazzani et al. 2012; Vodovar et al. 2012; Léger et al. 2013; Schnettler et al. 2013; Miesen et al. 2016). Small RNA profiling also revealed abundant viral-derived piRNAs in Drosophila cell culture—thought to reflect naturally occurring infections (Wu et al. 2010)—but not in actual flies subjected to experimental viral infection (Petit et al. 2016). Chickens that harbor endogenized avian leukosis virus (ALV) sequences also produce copious amount of piRNAs from these loci in their testes (Sun et al. 2017). Since one of these ALV loci, ALVE6, has been historically associated with ALV resistance (Robinson et al. 1981), it is tempting to speculate that piRNAs derived from this locus offer antiviral protection. Likewise, koalas infected by the KoRV retrovirus produce abundant piRNAs that map to endogenized KoRV insertions, although it remains unclear whether these piRNAs protect against KoRV infection (Yu et al. 2019). A similar mechanism has been proposed to operate in primates and rodents, where endogenous bornavirus-like elements generate piRNAs in the testes (Parrish et al. 2015). Together, these data suggest that endogenous viral sequences are a common source of piRNAs in diverse animals. Thus, both gene targeting and viral targeting by piRNAs add a layer of complexity that must be considered as potential drivers of piRNA pathway rapid evolution.
KRAB-ZFPs as an adaptive TE silencing system
The expansion of KRAB-ZFPs in mammalian genomes is increasingly recognized as an adaptive response to TE invasion. KRAB-ZFPs minimally contain an N-terminal KRAB domain followed by a variable array of C2H2-type zinc fingers (Imbeault et al. 2017; Yang et al. 2017b). Most KRAB-ZFPs so far characterized act as transcriptional repressors (for review, see Ecco et al. 2016; Yang et al. 2017b). Typically, KRAB-ZFPs bind DNA through their zinc fingers and recruit the corepressor KAP1 (TRIM28) via their KRAB domain. In turn, KAP1 recruits a variety of epigenetic modifiers such as histone and DNA methyltransferases that nucleate the formation of repressive chromatin at the target locus (Fig. 1B). Several KRAB-ZFPs have been implicated in the silencing of particular TE families recognized through sequence-specific DNA-binding interactions (Najafabadi et al. 2015; Schmitges et al. 2016; Imbeault et al. 2017). Perhaps the best-characterized example is mouse Zfp809, which was initially identified as a transcriptional repressor of proviral insertions of murine leukemia virus (MLV) in embryonic stem cells (ESCs) (Wolf and Goff 2009) but is also able to establish stable repression of ERV-like VL30 retrotransposons in early mouse development (Wolf et al. 2015). Thus, there is growing evidence that KRAB-ZFPs frequently target and silence TEs.
Several observations suggest that the rapid evolution of KRAB-ZFP genes is driven in part by an arms race with TEs. First, KRAB-ZFPs are massively expanded in all tetrapod lineages examined (with the notable exception of birds), ranging from 200 to 400 genes in most mammals, but very few are deeply conserved, implying pervasive evolutionary turnover (Imbeault et al. 2017). Second, the copy number of ZFPs in a given genome, including KRAB-ZFPs, correlates strikingly with the amount of LTR retroelements, and this correlation is observed across a wide range of vertebrate species and timescales (Thomas and Schneider 2011), which suggests a persistent coevolutionary relationship between TEs and ZFPs. Furthermore, the vast majority of mammalian KRAB-ZFPs profiled thus far bind only a single or a few TE families, and often the evolutionary emergence of a KRAB-ZFP closely follows the expansion of the TE family that it targets (Najafabadi et al. 2015; Schmitges et al. 2016; Imbeault et al. 2017). Finally, the DNA-contacting residues of many mammalian ZFPs exhibit telltale signatures of positive selection (Schmidt and Durrett 2004; Emerson and Thomas 2009; Nowick et al. 2010), suggesting that some KRAB-ZFPs adapt their targeting capacity in order to repress new DNA sequences, which may be introduced by newly expanded TE families.
Studies of two KRAB-ZFPs—ZNF93 and ZNF649—offer a compelling example of an arms race with the LINE1 (L1) family of non-LTR retrotransposons in the primate lineage (Fig. 2B; Jacobs et al. 2014; Fernandes et al. 2018). ZNF93 is a primate-specific gene whose zinc fingers underwent a series of adaptive changes in the hominoid lineage that enabled its binding to the L1PA6 and L1PA5 subfamilies (L1PA6-PA5) shortly after these elements started to expand in the hominoid ancestor (Jacobs et al. 2014). ZNF93 binding to L1PA6-PA5 elements had seemingly moderate repressive effects on these subfamilies, since they continued to amplify, but ZNF93 gained increased binding affinity for their descendants (L1PA4-PA3), which must have led to tighter repression. Indeed, it took a derivative of L1PA3 (L1PA3-6030) acquiring a 129-bp deletion—removing the ZNF93-binding site within its 5′ untranslated region (UTR)—to evade repression and enable further waves of L1 amplification, yielding the most recent L1PA2/L1HS subfamilies (Fig. 2B). Accordingly, reinserting the 129-bp segment deleted during evolution back to its original position within the modern active L1HS element restores ZNF93 binding and transcriptional repression and significantly dampens transposition of L1HS in cell culture (Jacobs et al. 2014).
A recent study revealed yet another layer of coevolution between primate L1 and KRAB-ZFPs, implicating an older gene, ZNF649, not directly related to ZNF93, in the silencing of L1PA6 elements (Fernandes et al. 2018). Like ZNF93, the zinc fingers of ZNF649 seem to have adapted to bind a motif within the 5′ UTR of the ancestral L1PA6 element but upstream of the 129-bp region bound by ZNF93. However, descendants of L1PA6 progressively evaded ZNF649 binding through a series of point mutations within their 5′ UTR, in parallel to their evasion from ZNF93 binding (Fig. 2B; Fernandes et al. 2018). This intricate game of cat and mouse between KRAB-ZFPs and L1 elements provides a vivid illustration of an arms race spanning ∼30 million years of primate evolution. Similar interactions may explain the extensive 5′ UTR diversification of L1 subfamilies throughout mammalian evolution (Khan et al. 2006). The wide diversity of TEs and KRAB-ZFPs across tetrapods implies that many other arms races must have been at play to fuel their coevolutionary relationship (Thomas and Schneider 2011).
KRAB-ZFPs also regulate host genes and viral activity
Although evidence for a host–TE arms race is currently stronger for KRAB-ZFPs than for piRNAs, it can only partially explain KRAB-ZFP evolution. It is well established that several KRAB-ZFPs bind non-TE sequences and play important roles in host physiology and development that now appear independent of TE repression (Imbeault et al. 2017; Yang et al. 2017a; for review, see Yang et al. 2017b). Additionally, KRAB-ZFPs can persist in the genome long after their identified TE targets have lost transposition activity, and new KRAB-ZFPs can evolve to target TEs that have long ceased to be active (Imbeault et al. 2017). These and other observations have led to the hypothesis that the recurrent interaction of KRAB-ZFPs with TEs is not a defensive strategy but rather a “massive and sophisticated enterprise of TE domestication for the evolutionary benefit of the host” (Friedli and Trono 2015). Under this model, KRAB-ZFPs are selected to exploit a vast reservoir of previously dispersed TE families and their various cis-regulatory activities (Chuong et al. 2017) to modulate gene expression networks in a species- and cell type-specific fashion (Trono 2015; Ecco et al. 2016; Yang et al. 2017b). While this scenario does not preclude an occasional arms race with TEs, it offers a host-centric alternative worth considering as an additional driver of KRAB-ZFP evolution.
Some KRAB-ZFPs are also known to repress the activity of exogenous retroviruses. ZFP809, for example, protects mouse ESCs from MLV replication, and its expression in differentiated cell lines is sufficient to render cells resistant to MLV infection (Wolf and Goff 2009). ZFP809 restricts MLV by binding to the proline tRNA primer-binding site of proviral DNA, which represses its transcription. Interestingly, the same primer sequence is used by various retroviruses, which suggests that a single KRAB-ZFP could potentially restrict a wide range of retroviruses. Recently, a pair of KRAB-ZFPs, known as Suppressor of nonecotropic ERV1 (Snerv1) and Snerv2 were shown via genetic analysis to be required for silencing of nonecotropic ERV (NEERV) expression (Treger et al. 2019). In immunodeficient mice, NEERV loci can recombine to generate infectious retroviruses, and expression of NEERV glycoprotein gp70 contributes to lupus nephritis susceptibility (Ito et al. 2013; Ottina et al. 2018). Similar to ZFP809, SNERV1 recruits KAP1 to silence NEERV elements by binding sequences overlapping their LTR, including a glutamine tRNA primer-binding site (Treger et al. 2019). Together, these findings suggest that antiviral activity may be a recurrent theme promoting the selection of novel KRAB-ZFPs. Furthermore, the fact that multiple KRAB-ZFPs have repeatedly evolved the ability to target tRNA primer-binding sites, which are some of the most evolutionarily constrained sequences in retroviral genomes, attests to the ability of retroviruses to frequently evade KRAB-ZFP binding to other parts of their genome. These observations point to retroviruses as common targets and important drivers of KRAB-ZFP evolution.
Counterdefense mechanisms
Invoking an arms race between TEs and host control systems implies that TEs commonly evade silencing. However, there are very few explicit cases of TEs having evolved active escape mechanisms. To our knowledge, only three examples of TE-encoded antisilencing mechanisms have been reported so far—all from plants (Nosaka et al. 2012, 2013; Fu et al. 2013; McCue et al. 2013; Hosaka et al. 2017). In cultivated rice (Oryza sativa), a family of CACTA DNA transposons carries a microRNA (miRNA) gene, mir820, which down-regulates the expression of the de novo methyltransferase gene OsDRM2 (Fig. 3B; Nosaka et al. 2012). mir820 binding to the 3′ UTR of OsDRM2 mRNA modestly reduces OsDRM2 expression, and independent RNAi-mediated knockdown of OsDRM2 resulted in reduced DNA methylation of a variety of TEs and concomitantly elevated TE expression (Nosaka et al. 2012, 2013). These results indicate that inhibition of OsDRM2 by miR820 would enable several TEs, including CACTA elements, to evade silencing. Interestingly, compensatory mutations appear to have been selected during rice evolution to maintain interactions between the TE-encoded miRNA and its binding site within the OsDRM2 mRNA, which may be the signature of an ongoing arms race (Nosaka et al. 2012).
Figure 3.
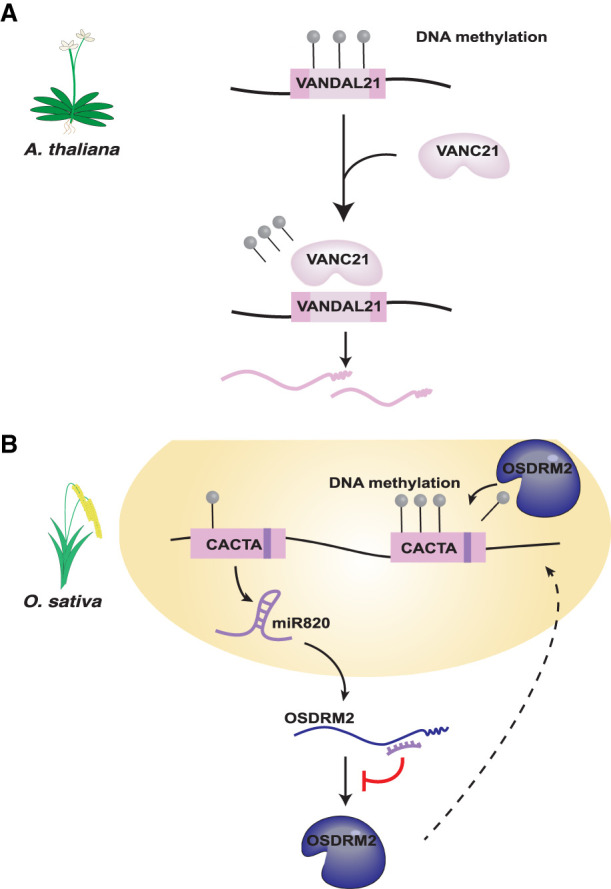
Evidence of TE counterdefense (A) VANDAL21 elements in Arabidopsis thaliana encode VANC21, which inhibits host DNA methylation (gray circles) of VANDAL21 elements. (B) Some CACTA DNA transposons in O. sativa encode a miRNA, mir820, which basepairs with OsDRM2 mRNA and reduces translation of OsDRM2, a DNA methyltransferase.
In Arabidopsis, some TEs produce siRNAs that can affect host gene expression in trans (trans-acting siRNAs [tasiRNAs]). One of these tasiRNAs, derived from Athila6 retrotransposons, was shown to target the 3′ UTR of the UPB1b mRNA, which encodes a host protein involved in global translational repression under stress conditions (McCue et al. 2013). tasiRNA-mediated repression of UPB1b results in elevated transcript and protein levels of Athila6 elements, supporting a countersilencing role of this tasiRNA. It is ironic that small RNA-based regulation, which is usually perceived as a prominent mechanism to silence TEs, would be deployed by a TE to promote its propagation.
VANDAL DNA elements in Arabidopsis provide perhaps the most convincing case reported thus far of transposons encoding a suppressor of silencing. At least two distantly related families, VANDAL21 and VANDAL6, were shown to encode an accessory protein, VANC21 and VANC6, respectively, that, when transiently expressed from a plasmid, induces demethylation of cognate VANDAL family members without affecting methylation of any other TE families in the Arabidopsis genome, including each other or closely related VANDAL families (Fig 3A; Fu et al. 2013; Hosaka et al. 2017). Mechanistically, it remains unclear how VANC promotes hypomethylation of VANDAL elements, but the process is dependent on a short tandem sequence motif that is found in high copy number within VANDAL21/6 elements but at low copy number elsewhere in the genome (Hosaka et al. 2017). By achieving sequence-specific antisilencing, VANDAL elements have evolved a powerful selfish strategy that promotes their own mobility without affecting that of other transposons, thereby limiting the deleterious impact of their antisilencing system on host fitness.
To our knowledge, no TE-encoded antisilencing factors have been described against either the piRNA or KRAB-ZFP pathways despite the proposed arms race long engaged between TEs and these control systems (Jacobs et al. 2014; Parhad et al. 2017; Wang et al. 2018). The dearth of antisilencing strategies described in eukaryotic TEs is all the more surprising given the plethora of strategies described for viruses and other pathogens to counteract host defense mechanisms. These include many examples of virus-encoded proteins that directly antagonize or degrade host defense systems, such as RNAi, CRISPR, and nucleic acid sensors to name just a few (for review, see Crow et al. 2016; Hynes et al. 2018; Landsberger et al. 2018).
Escape and self-control strategies
Why are there seemingly so few antisilencing mechanisms encoded by TEs? One fundamental difference between TEs and viruses is that TEs must replicate in the germline in order to propagate within a population, whereas viruses generally do not (Haig 2016). Thus, TE fitness is intimately intertwined with the reproductive fitness of their host organisms. This dependency places an important limitation on the ability of TEs to evolve broadly effective antisilencing mechanisms. For instance, a mechanism blocking the entire piRNA pathway would lead to massive mobilization of diverse TEs, simultaneously compromising host fertility and dooming TE propagation (Blumenstiel et al. 2016; Haig 2016), as documented in piRNA mutant backgrounds (e.g., see Wang et al. 2018). Consistent with this quandary, all TE-encoded antisilencing mechanisms described thus far have narrow effects or modes of action either resulting in modest decreases of host regulatory proteins (mir820 and Athila6 tasiRNA) or selectively targeting individual families (VANC). This is in stark contrast to viruses, which evolve mechanisms that achieve broad and/or highly effective blocks of the targeted pathways (Crow et al. 2016; Hynes et al. 2018; Landsberger et al. 2018).
Bypassing host surveillance
Alternatively, TEs may evade but in contexts in which their activity does not impact host fitness. For example, the I elements of D. melanogaster hijack ovarian nurse cells, which are permissive to their transcription but apparently refractory to transposition, as factories to assemble RNP complexes that serve as transposition intermediates (Van De Bor et al. 2005; Hamilton et al. 2009; Wang et al. 2018). These products are then trafficked via microtubules and delivered to the oocyte, where transposition takes place (Wang et al. 2018). I-element mRNA trafficking to the oocyte further requires an RNA stem–loop secondary structure present in its ORF2p sequence (Van De Bor et al. 2005). Although these retrotransposons may be susceptible to piRNA silencing upon entry into the oocyte, assembling the transposon products in permissive cells reduces the number of transposition steps exposed to host silencing. Given that other transposons, including LTR elements HMS-Beagle and 3S18, also preferentially localize to the oocyte (Wang et al. 2018) and that the RNA stem–loop structure required for I-element trafficking is present in G2 and jockey retrotransposons (Hamilton et al. 2009), it seems likely that such a transport mechanism may be a recurrent evasion strategy. Similarly, the virus-like particles produced by EVADÉ retrotransposons in Arabidopsis partially protect their mRNAs against small RNA-mediated degradation (Marí-Ordóñez et al. 2013). These bypass strategies suggest that, in eukaryotes, selection favors TEs that circumvent rather than antagonize or block host silencing pathways.
Targeting preferences
TEs have also repeatedly evolved mechanisms to direct their insertion to “safe havens” or regions of the genome where insertion will cause minimal harm. Studies that map de novo transposition events have shown that TEs, especially those colonizing compact genomes, have repeatedly evolved mechanisms to target benign or highly redundant regions of the genome. For example, Ty1 and Ty3 LTR retrotransposons in yeast, Skipper retroelements in Dictyostelium discoideum, and Dada DNA transposons in fish independently evolved preferences for integration in the immediate vicinity of tRNA genes, with apparently little to no impact on tRNA expression (for review, see Sultana et al. 2017; Cheung et al. 2018). Another safe harbor has been adopted by R1 and R2 non-LTR retroelements in arthropods (Pérez-González and Eickbush 2002) as well as Pokey DNA transposons in Daphnia (Penton and Crease 2004), which independently evolved targeting to ribosomal DNA arrays. Ty5 in yeast (Zou et al. 1996), Het-A/TAHRE/TART in Drosophila (Pardue and DeBaryshe 2011), and TRAS/SART (Fujiwara et al. 2005) in silkworms all independently evolved the ability to target telomeric regions. Several TE families also show preference for insertion upstream of protein-coding genes, including Tf retrotransposons in fission yeast (Levin and Boeke 1992), Drosophila P elements (Liao et al. 2000), maize Mutator elements (Dietrich et al. 2002), and rice mPing transposons (Naito et al. 2009). While insertion of these elements in this compartment must occasionally perturb host gene expression, it is still less likely to be detrimental than in coding regions. It also provides these elements with the added benefit of an “open” chromatin environment that will facilitate further mobilization and might shield them against host silencing. Remarkably, TEs can also evolve preference for insertion into other TEs, such as Tx1 non-LTR retrotransposons that target Tx1d DNA transposons in Xenopus laevis (Christensen et al. 2000) and Tourist elements that preferentially insert into other Tourist elements in rice and maize (Jiang and Wessler 2001), a strategy shared by various TE families in maize (Stitzer et al. 2019). Collectively, these data indicate that TEs have repeatedly adapted to occupy genomic niches that minimize the cost of transposition on host fitness.
Self-regulatory mechanisms
TEs can also minimize their deleterious impacts on their hosts by evolving self-regulatory mechanisms. A prominent theme is the spatiotemporal restriction of transposition activity. TEs are present in the genome of all cells and could, in principle, mobilize in both germline and somatic tissues. However, mobilization in somatic tissue has a greater potential of harming organismal fitness, whereas mobilization in the germline has lesser immediate effects on host function as long as it does not affect fertility (Charlesworth and Langley 1986; Haig 2016). Thus, suppression of TE activity in the soma is advantageous for both hosts and TEs and therefore predicts that TEs should evolve mechanisms to restrict expression to the germline. A classic example is the Drosophila P element, whose transposase open reading frame (ORF) is interrupted by an intron that is spliced only in the germline, preventing P-element mobility in the soma, where only prematurely truncated inhibitory transposase is produced (Laski et al. 1986). It appears that this regulatory switch was evolved through the gain of sequence elements within the transposon that recruit somatically expressed splicing inhibiting factors (for review, see Majumdar and Rio 2015). A recent study adds another layer of intricacy by implicating the piRNA pathway in repressing the splicing of this intron in the germ cells through piRNA-mediated chromatin changes within the P element (Teixeira et al. 2017). Thus, spatiotemporal regulation of the P element in flies involves an interplay of mechanisms evolved by the transposon and by the host.
In mammals, several retroelements are known to have evolved exquisite stage-specific expression during early embryonic development (for review, see Rodriguez-Terrones and Torres-Padilla 2018). For instance, in mice, MaLR/MT elements are transcribed exclusively in oocytes (Peaston et al. 2004; Brind'Amour et al. 2018), while transcription of mouse ERV type L (MERVL) (Macfarlan et al. 2012) and young L1 subfamilies (Jachowicz et al. 2017; Percharde et al. 2018) peaks at the two-cell (2C) stage and coincides with zygotic genome activation. Human ERV type K (HERV-K) expression peaks at the eight-cell stage of human embryonic development (Grow et al. 2015), while HERVH/LTR7 are expressed in the pluripotent stem cells (PSCs) of the blastocyst (Wang et al. 2014; Göke et al. 2015). The mechanisms enabling such developmental precision of expression are becoming increasingly clear: Each TE family recruits a unique cocktail of host transcription factors that precisely specify these developmental stages. For example, HERVH/LTR7 recruits NANOG, OCT4, SOX2, and KLF4 (Kunarso et al. 2010; Ohnuki et al. 2014; Göke et al. 2015; Ito et al. 2017; Pontis et al. 2019), while MERVL recruits Dux (De Iaco et al. 2017; Hendrickson et al. 2017). Because these stages precede the differentiation of germ cells, they allow each of these different elements to generate inheritable insertions while occupying distinct expression niches, which might reduce their competition for cellular resources.
Another mitigating strategy is the evolution of suboptimal transposition and self-restraining copy number control mechanisms. In order to persist, TEs must be active enough to generate new insertions but not so active as to impair host fitness. To this effect, some TEs have evolved mechanisms to reduce their own activity. This is most studied in Tc1/mariner transposons, which self-regulate their mobility in at least three ways: evolution of suboptimal transposases (supported by the isolation of hyperactive mutants) (Lampe et al. 1999; Mátés et al. 2009; Liu and Chalmers 2014), inhibition of transposase function by aggregation when transposase expression reaches a certain threshold (also known as overproduction inhibition) (Lohe and Hartl 1996), and selection for imperfect terminal inverted repeats (TIRs) that reduce transposase affinity and excision efficiency (Augé-Gouillou et al. 2001). These strategies are likely to be widespread among DNA transposons. Indeed, hyperactive transposases have been readily obtained by experimental mutagenesis for various elements (Yusa et al. 2011; Lazarow et al. 2012; Voigt et al. 2016), diverse transposases are known to form aggregates when overexpressed (Heinlem et al. 1994; Liu and Chalmers 2014; Woodard et al. 2017), and many consensus transposon sequences have imperfect TIRs. A subset of Ty1 elements in yeast also encodes a truncated Gag protein with a dominant-negative effect on Ty1 copy number (Saha et al. 2015). These examples illustrate that TEs commonly evolve self-control mechanisms that mitigate their deleterious impact.
Host–transposon mutualism
TE self-regulation and targeting may reduce the detrimental impact that TEs can have on host fitness but do not directly provide a selective advantage to the host. Is it conceivable that TEs and their hosts could achieve such a mutualistic relationship? Mutualism can be attained if maintenance of TE activity immediately benefits the host. This form of host–TE cooperation is commonplace in bacteria, where transposons and conjugative plasmids frequently shuttle antibiotic resistance genes and other factors allowing bacterial hosts to adapt to environmental challenges (Wintersdorff et al. 2016). Thus far, very few examples akin to host–TE mutualism have been documented in eukaryotes. Here we highlight three possible cases: telomeric retroelements in Drosophila, telomere-bearing element (TBE)-mediated genome rearrangement in the ciliate Oxytricha trifallax, and an emerging role for mammalian retrotransposons in early embryonic development.
Retrotransposons maintain Drosophila telomeres (and centromeres?)
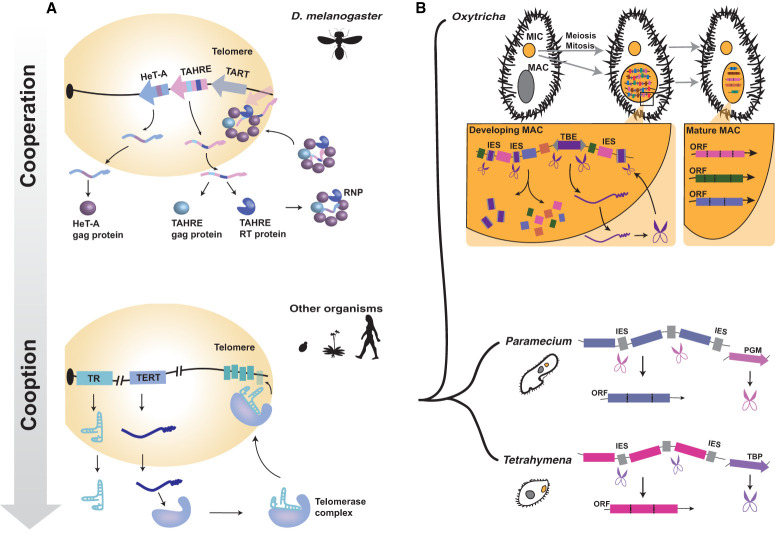
Eukaryotes have adopted several mechanisms to ensure replication of the termini of linear chromosomes. In most, chromosomes are protected by telomeric repeats maintained by a specialized enzyme called telomerase (Fig 4A; Kordyukova et al. 2018). Drosophilid species, however, have lost the gene encoding telomerase, and, in all Drosophila species examined thus far (with one exception, which is discussed below), their telomeric repeats have been replaced by arrays of non-LTR retrotransposons, collectively referred to as telomeric retroelements (Fig 4A; Pardue and DeBaryshe 2008; Casacuberta 2017). In D. melanogaster, telomeric elements belong to three related families of jockey-like non-LTR retrotransposons (HeT-A, TAHRE, and TART) that are continuously inserted head to tail at chromosome ends via target-primed reverse transcription supported by proteins produced by autonomous TAHRE and TART copies (Fig. 4A). mRNA transcribed from all three retroelement families are imported into the nucleus by virtue of their association with their respective gag-like proteins, but targeting of TAHRE RNPs to telomeres requires association with the Het-A gag (Rashkova et al. 2002; Pardue and DeBaryshe 2008). Het-A in turn requires pol proteins from TAHRE/TART to be reverse-transcribed (Pardue and DeBaryshe 2008). Thus, telomeric retroelements cooperate with each other for their own amplification and for telomere maintenance (Capkova Frydrychova et al. 2008). As such, telomeric retroelements have long been regarded as the prototypical example of host–TE mutualism (Kidwell and Lisch 2001), although recent data have challenged this view (see below). Intriguingly, evidence is mounting that a distantly related family of non-LTR retrotransposons (G2/jockey3) contributes directly to the organization and function of centromeres of D. melanogaster and its sister species, D. simulans (Chang et al. 2019). Thus, several Drosophila retroelements appear directly involved in the faithful replication and segregation of chromosomes.
Figure 4.
Cooperation paves the way for cooption. (A) Drosophila telomeric transposons as a potential model for telomerase evolution. In D. melanogaster, three non-LTR transposons families maintain telomeres. Telomeric elements form a head-to-tail array in the telomere. Gag and reverse transcriptase (RT) proteins from HeT-A, TAHRE, and TART complex with their cognate mRNAs, forming a RNP complex capable of telomere elongation. In other organisms, telomerase reverse transcriptase (TERT) and telomerase RNA (TR) form the telomerase complex, which maintains telomeres. (B) Transposase proteins necessary for germline internal eliminated sequence (IES) elimination in Tetrahymena and Paramecium may have evolved from a mechanism analogous to TBE excision in Oxytricha (see the text). In the developing Oxytricha somatic macronucleus (MAC), TBE transposases excise IESs. Subsequent steps stitch together exons into single-gene chromosomes, which are amplified to thousands of copies in the mature MAC. In Paramecium and Tetrahymena, PiggyBac-derived proteins are domesticated for IES recognition and excision. (Pgm) PiggyMac and interactors; (TBP) Tetrahymena piggyBac-like. Scissors represent transposase proteins, and arrows represent genes.
Active transposons are required for Oxytricha development
The role of TBEs in O. trifallax genome rearrangement offers another tantalizing case of host–TE mutualism. Ciliates, such as O. trifallax, are single-celled eukaryotes that possess two types of nuclei: a transcriptionally silent diploid germline micronucleus (MIC) and a somatic macronucleus (MAC) that maintains gene expression in vegetative cells and is derived from the MIC through extensive genome rearrangements that occur shortly after sexual conjugation (Fig. 4B; Chen et al. 2014). Genes in the MIC genome are interrupted by noncoding sequences referred to as internal eliminated sequences (IESs) that must be excised for proper MAC function. Furthermore, in O. trifallax, exons are arranged out of order and often inverted in the MIC, so they must also be unscrambled to assemble a functional MAC genome (Chen et al. 2014). TEs comprise a large fraction of IESs and contribute substantially to the size of the MIC genome (Chen et al. 2014; Hamilton et al. 2016). The process of IES excision and unscrambling removes essentially all TEs from the MAC genome and reduces the 1-Gb MIC genome to an ∼50-Mb MAC genome consisting of thousands of single-gene chromosomes (∼2 kb long), each subsequently amplified to thousands of copies (Fig. 4B; Swart et al. 2013; Chen et al. 2014). In O. trifallax, this complex remodeling of the genome requires the cooperation of a family of DNA transposons, called TBEs, actively mobilized during meiosis (Nowacki et al. 2009). Experimental silencing of all three families of TBE transposases via RNAi impairs cell growth and causes cell death due to defects in germline elimination of both TBEs and IESs (Nowacki et al. 2009). However, silencing of a single TBE family is insufficient to cause this phenotype, indicating that all three TBE families cooperate to promote MAC development (Nowacki et al. 2009). Therefore, TBEs must remain active in the Oxytricha genome to provide an indispensable role for the development of its host organism, suggesting a mutualistic arrangement established during Oxytricha evolution (Vogt et al. 2013). Additional support for this model comes from evidence that TBE-encoded transposases have evolved under purifying selection, suggesting evolutionary constraint to serve both transposon and host functions (Chen and Landweber 2016).
Is mammalian embryogenesis addicted to TEs?
There is growing evidence that retrotransposons and ERVs are intimately intertwined with mammalian embryonic development. MERVL and HERVL appear to contribute to zygotic genome activation in mice and humans, respectively (Kigami et al. 2003; Svoboda et al. 2004; Macfarlan et al. 2012; De Iaco et al. 2017; Hendrickson et al. 2017; Whiddon et al. 2017). Furthermore, MERVL transcription in the 2C embryo triggers the formation of hundreds of chromatin loops that fold the genome in a 3D organization that is unique and possibly critical to totipotency (Kruse et al. 2019). In human ESCs, a distinct family, HERVH, is highly expressed and marks pluripotent cell populations (Santoni et al. 2012; Ohnuki et al. 2014; Wang et al. 2014; Göke et al. 2015). The high level of transcription of several HERVH loci also promotes the formation of chromatin loops and topological domains that are unique to ESCs (Zhang et al. 2019b) as well as the expression of chimeric long noncoding RNAs (lncRNAs) (Kelley and Rinn 2012; Kapusta et al. 2013; Lu et al. 2013, 2014; Wang et al. 2014). RNAi knockdown of some of these individual HERVH-derived lncRNAs in induced PSCs (iPSCs) results in loss of pluripotency (for review, see Izsvák et al. 2016), and CRISPR knockout of a single HERVH locus increases the capacity of iPSCs to differentiate into cardiomyocytes (Zhang et al. 2019b). A recent study also implicated HERVK and SVA elements as enhancers in naïve human ESCs, a model for preimplantation development (Pontis et al. 2019). The regulatory activities emanating from retrotransposons during mammalian development are intriguing, but it remains unclear whether they are merely a relic of selfish or cooperative strategies that these TEs deployed to occupy a niche, facilitating their transmission, or whether they have been coopted for lineage-specific developmental innovations (Haig 2016; Izsvák et al. 2016; Chuong et al. 2017).
Recently, evidence has surfaced for another potential partnership between TEs and mammalian embryonic development, implicating the L1 retrotransposon. There is a striking nuclear accumulation of the RNA produced by the youngest transpositionally active L1 subfamilies in the mouse 2C embryo. Experimental depletion of endogenous L1 RNAs at that stage elicited a wide range of chromatin and regulatory defects that block embryonic development (Jachowicz et al. 2017; Percharde et al. 2018). Interestingly, a similar phenotype was observed when L1 transcription was experimentally prolonged beyond the 2C stage by targeted chromatin manipulation (TALE-VP64) but not when full-length L1 mRNA was injected, and the phenotype could not be rescued by inhibiting L1 reverse transcriptase (Jachowicz et al. 2017). These results suggest that the precise transcriptional activation of L1 in the nuclei of preimplantation embryos is required for the establishment of a chromatin state that promotes developmental progression. Mechanistically, nuclear L1 RNAs appear to exert these activities by forming a RNP complex with at least two host proteins, Nucleolin and KAP1, that activates ribosomal RNA transcription and represses the Dux locus, which in turn stimulates exit from the 2C stage (Percharde et al. 2018). It is unknown whether these findings extend beyond the mouse embryo, but it is worth noting that Nucleolin was reported previously to bind human L1HS RNA (Peddigari et al. 2013). Also, these results should be interpreted with caution because it is difficult to rule out nonspecific effects with the experimental approaches used to manipulate L1 expression. Undoubtedly, these provocative observations will stimulate further investigation.
Conflicts in disguise?
Although the examples described above suggest that mutualistic interactions between TEs and their hosts may be more widespread than currently appreciated, they remain open to alternative interpretations. Rather than true mutualisms, they may be viewed as commensalisms benefiting the TEs but of no real benefit to the host or even as “addictions,” whereby the TEs have supplanted ancestral functions essential to the host without providing an adaptive innovation but nevertheless creating a dependency on active transposition (Jangam et al. 2017). Such addiction may precipitate into a conflict if transposition occurs in excess or otherwise incurs a net cost on host fitness, which may set an arms race with the host to evolve mechanisms to control transposition (Fig. 4A). There is growing evidence that such instability may be at play at Drosophila telomeres. First, intraspecific and interspecific evolutionary analyses of 29 genes encoding proteins associated with telomere maintenance and function revealed that they have experienced repeated bouts of positive selection, indicating the existence of recurrent conflicts necessitating rapid adaptation of telomeric proteins (Lee et al. 2017). Furthermore, a recent study tracing the evolution of D. melanogaster telomeric retroelements in closely related species shows that this lineage of retrotransposons has experienced rapid turnover with drastic changes in their abundance across species (Saint-Leandre et al. 2019). These results uncover a paradoxical level of evolutionary instability for seemingly essential components of the genome. Surprisingly, it appears that one species, Drosophila biarmipes, has even lost altogether the telomeric retroelements, which are now replaced by a mixture of unrelated TEs at the tip of its chromosomes (Saint-Leandre et al. 2019). It remains to be seen how D. biarmipes copes with the loss of telomeric retroelements, as the TEs found at their telomeres appear to be no longer active (Saint-Leandre et al. 2019). These findings suggest that replacing vital host functions with TE activity may be an evolutionarily contentious and ultimately untenable situation.
En route to cooption
An advantage of adopting a cooperation-centric model of host–TE interaction is that cooperation provides a facile path for TE cooption. Cooption is the process by which natural selection taps into TE sequences to evolve new host function. Once considered rare, numerous examples of TE cooption have now been described, although the evolutionary forces and molecular path that lead to cooption remain poorly understood (Feschotte and Pritham 2007; Sinzelle et al. 2009; Chuong et al. 2017; Frank and Feschotte 2017; Jangam et al. 2017; Zhang et al. 2019a). We propose that the type of cooperative activities that TEs and host engage in directly influence the function that TEs get coopted for.
The case of Drosophila telomeric retroelements provides a good example to illustrate this model. There are striking similarities between this system and telomerase, the mechanism used by most eukaryotes to maintain telomere length (Fig 4A; Pardue and DeBaryshe 2008; Kordyukova et al. 2018). Telomerase is a reverse transcriptase that appears most closely related to that currently encoded by Penelope-like retrotransposons (Gladyshev and Arkhipova 2007). Moreover, in organisms lacking telomerase (Drosophila) or with low telomerase expression (e.g., the silkworm Bombyx mori), telomeric retrotransposons can supplant telomerase function (Fujiwara 2015; Servant and Deininger 2016). These observations support a long-standing model that telomere maintenance via telomerase originated from an ancient retrotransposon-based mechanism (Eickbush 1997; Nakamura and Cech 1998; Pardue and DeBaryshe 2008). If so, then the cooperation between Drosophila telomeric retrotransposons and their hosts may be an evolutionary replay of how an ancient group of retroelements (perhaps Penelope-like) maintained their activity prior to being fully coopted for telomere maintenance in the last eukaryotic common ancestor (Fig. 4A).
O. trifallax and its TBEs appear to be engaged in a mutualistic or perhaps addicted relationship. However, other ciliates, such as Paramecium and Tetrahymena, appear to have advanced that relationship to full domestication: These species use immobilized transposase proteins to mediate their MIC-to-MAC transition (Fig. 4B). In Paramecium, PiggyMac (Pgm) is a piggyBac-derived transposase required for DNA elimination that catalyzes and interacts with five additional related Pgm-like proteins to ensure complete IES targeting (Bétermier and Duharcourt 2014; Bischerour et al. 2018). Tetrahymena also uses a suite of related transposase-derived genes (TBP, TBP2, TBP6, and LIA5) to ensure proper genome rearrangement and IES deletion (Vogt and Mochizuki 2013; Cheng et al. 2016; Feng et al. 2017). Although these proteins are all clearly related to piggyBac transposases, they are extremely diverged from each other and share no close similarity to any extant TEs in these species, indicating that they have been fully and possibly independently coopted (Cheng et al. 2010; Vogt et al. 2013; Bétermier and Duharcourt 2014). Whether these transposases were once derived from elements with an activity similar to TBEs in Oxytricha is unknown, but there are clear sequence similarities between ciliate IESs and the TIRs of transposons (Fass et al. 2011; Cheng et al. 2016; Hamilton et al. 2016). Furthermore, the extent to which each domesticated piggyBac transposase is fully domesticated varies, with some (Pgm and TBP6) retaining features of piggyBac transposition, including precise excision and TIR dependence, while others (TBP2) are entirely dependent on host factors for IES targeting (Feng et al. 2017). These observations suggest that the Pgm and TBP transposases may have been coopted as a mean to resolve an “addiction” to their cognate elements similar to that seen in Oxytricha, which would link their cooption directly to their initial cooperation with their hosts (Fig 4B).
Outlook
In this review, we examined two host-encoded TE silencing systems, piRNAs and KRAB-ZFPs, that carry strong signatures of adaptive evolution in some lineages, suggesting that they are engaged in an arms race with TEs (McLaughlin and Malik 2017). However, there remains no evidence that TEs have evolved mechanisms directly neutralizing either of these two pathways, and a variety of other factors may explain their rapid evolution. Clearly, more work is needed to better understand the forces driving the diversification of these systems and gain a fuller picture of their interactions with TEs across a broad range of species.
Adaptive evolution of piRNAs has been studied extensively in insects but not in mammals, and the few studies that have addressed this gap suggest that mammalian piRNA proteins have not diversified extensively (Yi et al. 2014). One explanation for this contrast could be that mammals have replaced the antiviral arm of the piRNA pathway with other systems. Indeed, mammalian cells possess a variety of antiviral responses, including nucleic acid sensors and interferon responses, which might have supplanted or relieved the piRNA pathway to carry out this immune function (for review, see tenOever 2016). Such a scenario would support the hypothesis that piRNA pathway evolution outside of mammals is driven primarily by its antiviral function.
Detailed studies of KRAB-ZFPs so far have been carried out only in mice and humans despite dramatic expansion of these genes in diverse tetrapods (Imbeault et al. 2017). While evidence has built for intricate coevolution of KRAB-ZFP and L1 retrotransposons in primates (Fig. 2B; Jacobs et al. 2014; Fernandes et al. 2018), it is still unclear how common such a tug of war is. It may be illuminating to investigate these questions in tetrapods with more aggressive TE activity, such as frogs, axolotls, opossums, or bats (Mikkelsen et al. 2007; Pritham and Feschotte 2007; Ray et al. 2008; Hellsten et al. 2010; Sotero-Caio et al. 2017; Nowoshilow et al. 2018; Rogers et al. 2018).
Another take-home message is that the arms race is only one, but perhaps not the most prevalent, form of host–TE interaction in eukaryotes. TEs have also evolved subtle evasive strategies as well as self-control and targeting mechanisms that attenuate the cost of transposition on host fitness. Some TEs even appear to have engaged in cooperative strategies with their host organism in a way that resembles a mutualistic or symbiotic relationship. While few cases of mutualism have surfaced thus far in eukaryotes, this strategy is commonplace in prokaryotes (Wintersdorff et al. 2016). It is possible that symbiotic interactions are widespread in eukaryotes but more difficult to capture because they are more challenging to identify and test experimentally, in part because of the large amounts of TEs that need to be manipulated simultaneously (e.g., retrotransposons in mammalian embryogenesis). The advent of genome editing and other large-scale perturbations offers new powerful tools to overcome these challenges (Bourque et al. 2018; Fuentes et al. 2018; Smith et al. 2019; Todd et al. 2019). It is also possible that many mutualistic interactions are evolutionarily unstable and volatile because they are prone to tilt back and forth between disproportionately benefiting the TE (e.g., Drosophila telomeres) or the host to turn into full cooption events (Fig. 5).
Figure 5.
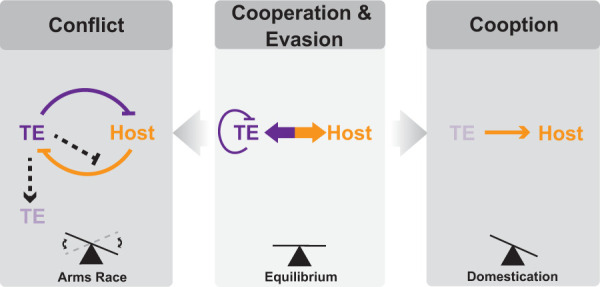
Model for host–TE interactions. Conflict: TEs (purple) harm the host (orange), leading to host silencing of TEs. TEs occasionally evolve direct antisilencing mechanisms (dashed line). Most host–TE conflict leads to TE death. Cooperation and evasion: TEs evolve self-regulatory mechanisms to mitigate impacts on host fitness. Hosts and TEs can also evolve a mutualistic relationship. Cooperation can lead to both conflict and cooption. Cooption: Host repurposes all or part of a TE for novel host function at the expense of the TE.
We therefore envision a model in which the host and TE cooperate for a period of time, which resolves in one of three ways: (1) The TE no longer cooperates, leading to reactivation and possible loss of the family in the population if too active (arms race); (2) the TE fades into obscurity due to relaxed selection pressure on its sequence; or (3) maintenance of TE features for cellular function rather than the TE family as a whole leads to eventual loss of the TE family (cooption) (Fig. 5). Validating the model will require the study of transitional systems such as those described in this review and others that are bound to surface.
Acknowledgments
We thank Jason Fernandez (University of California at Santa Cruz) and Yifan Liu (University of Michigan) for feedback on the KRAB-ZFP and Tetrahymena sections, respectively. We also thank three anonymous reviewers for their comments and suggestions. We thank members of the Feschotte laboratory for helpful discussion. We apologize to those whose research we could not cite due to space and scope limitations. This review was supported by R35GM122550, U01HG009391, and R01GM112972 from the National Institutes of Health to C.F.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.327312.119.
Freely available online through the Genes & Development Open Access option.
References
- Ade C, Roy-Engel AM, Deininger PL. 2013. Alu elements: an intrinsic source of human genome instability. Curr Opin Virol 3: 639–645. 10.1016/j.coviro.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, et al. 2010. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 330: 86–88. 10.1126/science.1191864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augé-Gouillou C, Hamelin MH, Demattei MV, Periquet M, Bigot Y. 2001. The wild-type conformation of the Mos-1 Inverted terminal repeats is suboptimal for transposition in bacteria. Mol Gen Genomics 265: 51–57. 10.1007/s004380000385 [DOI] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick E-M, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. 2012. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578. 10.1126/science.1220952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78. 10.1016/j.molcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Kern AD, Jones CD. 2007. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176: 1131–1137. 10.1534/genetics.106.069245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Wang H. 2014. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65: 505–530. 10.1146/annurev-arplant-050213-035811 [DOI] [PubMed] [Google Scholar]
- Bétermier M, Duharcourt S. 2014. Programmed rearrangement in ciliates: paramecium. Microbiol Spectr 2. 10.1128/microbiolspec.MDNA3-0035-2014 [DOI] [PubMed] [Google Scholar]
- Bischerour J, Bhullar S, Denby Wilkes C, Régnier V, Mathy N, Dubois E, Singh A, Swart E, Arnaiz O, Sperling L, et al. 2018. Six domesticated PiggyBac transposases together carry out programmed DNA elimination in Paramecium. Elife 7: e37927. 10.7554/eLife.37927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel JP, Erwin AA, Hemmer LW. 2016. What drives positive selection in the Drosophila piRNA machinery? the genomic autoimmunity hypothesis. Yale J Biol Med 89: 499–512. [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA. 2015. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16: 727–741. 10.1038/nrm4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS, et al. 2018. Ten things you should know about transposable elements. Genome Biol 19: 199. 10.1186/s13059-018-1577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392. 10.1126/science.1165171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brind'Amour J, Kobayashi H, Albert JR, Shirane K, Sakashita A, Kamio A, Bogutz A, Koike T, Karimi MM, Lefebvre L, et al. 2018. LTR retrotransposons transcribed in oocytes drive species-specific and heritable changes in DNA methylation. Nat Commun 9: 3331. 10.1038/s41467-018-05841-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ. 1984. The molecular basis of I-R hybrid dysgenesis in Drosophila Melanogaster: identification, cloning, and properties of the I factor. Cell 38: 153–163. 10.1016/0092-8674(84)90536-1 [DOI] [PubMed] [Google Scholar]
- Burns KH. 2017. Transposable elements in cancer. Nat Rev Cancer 17: 415–424. 10.1038/nrc.2017.35 [DOI] [PubMed] [Google Scholar]
- Burt A, Trivers R. 2006. Genes in conflict. Belknap Press of Harvard University Press, Cambridge, MA. [Google Scholar]
- Capkova Frydrychova R, Biessmann H, Mason JM. 2008. Regulation of telomere length in Drosophila. Cytogenet Genome Res 122: 356–364. 10.1159/000167823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 Is Essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514. 10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Casacuberta E. 2017. Drosophila: retrotransposons making up telomeres. Viruses 9: E192. 10.3390/v9070192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo DM, Mell JC, Box KS, Blumenstiel JP. 2011. Molecular evolution under increasing transposable element burden in Drosophila: a speed limit on the evolutionary arms race. BMC Evol Biol 11: 258. 10.1186/1471-2148-11-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. 10.1126/science.282.5396.2012 [DOI] [PubMed] [Google Scholar]
- Chang C-H, Chavan A, Palladino J, Wei X, Martins NMC, Santinello B, Chen C-C, Erceg J, Beliveau BJ, Wu C-T, et al. 2019. Islands of retroelements are major components of Drosophila centromeres. PLoS Biol 17: e3000241. 10.1371/journal.pbio.3000241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH. 1986. The evolution of self-regulated transposition of transposable elements. Genetics 112: 359–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Landweber LF. 2016. Phylogenomic analysis reveals genome-wide purifying selection on TBE transposons in the ciliate Oxytricha. Mob DNA 7: 2. 10.1186/s13100-016-0057-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Bracht JR, Goldman AD, Dolzhenko E, Clay DM, Swart EC, Perlman DH, Doak TG, Stuart A, Amemiya CT, et al. 2014. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell 158: 1187–1198. 10.1016/j.cell.2014.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-Y, Vogt A, Mochizuki K, Yao M-C. 2010. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. MBoC 21: 1753–1762. 10.1091/mbc.e09-12-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-Y, Young JM, Lin C-YG, Chao J-L, Malik HS, Yao M-C. 2016. The piggyBac transposon-derived genes TPB1 and TPB6 mediate essential transposon-like excision during the developmental rearrangement of key genes in Tetrahymena thermophila. Genes Dev 30: 2724–2736. 10.1101/gad.290460.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung S, Manhas S, Measday V. 2018. Retrotransposon targeting to RNA polymerase III-transcribed genes. Mob DNA 9: 14. 10.1186/s13100-018-0119-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S, Pont-Kingdon G, Carroll D. 2000. Comparative studies of the endonucleases from two related Xenopus laevis retrotransposons, Tx1L and Tx2L: target site specificity and evolutionary implications. Genetica 110: 245–256. 10.1023/A:1012704812424 [DOI] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71–86. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MS, Lum KK, Sheng X, Song B, Cristea IM. 2016. Diverse mechanisms evolved by DNA viruses to inhibit early host defenses. Crit Rev Biochem Mol Biol 51: 452–481. 10.1080/10409238.2016.1226250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Munafò M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, Hannon GJ. 2018. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet 52: 131–157. 10.1146/annurev-genet-120417-031441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, Caligiana A, Brocculi G, Adney EM, Boeke JD, et al. 2019. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566: 73–78. 10.1038/s41586-018-0784-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263. 10.1038/nature10547 [DOI] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. 2017. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49: 941–945. 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384. 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830. 10.1016/S1534-5807(02)00165-X [DOI] [PubMed] [Google Scholar]
- Dietrich CR, Cui F, Packila ML, Li J, Ashlock DA, Nikolau BJ, Schnable PS. 2002. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160: 697–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284: 601–603. 10.1038/284601a0 [DOI] [PubMed] [Google Scholar]
- Dubnau J. 2018. The Retrotransposon storm and the dangers of a Collyer's genome. Curr Opin Genet Dev 49: 95–105. 10.1016/j.gde.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. 2016. Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev Cell 36: 611–623. 10.1016/j.devcel.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH. 1997. Telomerase and retrotransposons: which came first? Science 277: 911–912. 10.1126/science.277.5328.911 [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Furano AV. 2002. Fruit flies and humans respond differently to retrotransposons. Curr Opin Genet Dev 12: 669–674. 10.1016/S0959-437X(02)00359-3 [DOI] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet 5: e1000325. 10.1371/journal.pgen.1000325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C, Odom DT, Kutter C. 2017. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat Commun 8: 1411. 10.1038/s41467-017-01049-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fablet M, Akkouche A, Braman V, Vieira C. 2014. Variable expression levels detected in the Drosophila effectors of piRNA biogenesis. Gene 537: 149–153. 10.1016/j.gene.2013.11.095 [DOI] [PubMed] [Google Scholar]
- Fass JN, Joshi NA, Couvillion MT, Bowen J, Gorovsky MA, Hamilton EP, Orias E, Hong K, Coyne RS, Eisen JA, et al. 2011. Genome-scale analysis of programmed DNA elimination sites in Tetrahymena thermophila. G3 (Bethesda) 1: 515–522. 10.1534/g3.111.000927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV. 2012. Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338: 758–767. 10.1126/science.338.6108.758 [DOI] [PubMed] [Google Scholar]
- Feng L, Wang G, Hamilton EP, Xiong J, Yan G, Chen K, Chen X, Dui W, Plemens A, Khadr L, et al. 2017. A germline-limited piggyBac transposase gene is required for precise excision in Tetrahymena genome rearrangement. Nucleic Acids Res 45: 9481–9502. 10.1093/nar/gkx652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes JD, Haeussler M, Armstrong J, Tigyi K, Gu J, Filippi N, Pierce J, Thisner T, Angulo P, Katzman S, et al. 2018. KRAB zinc finger proteins coordinate across evolutionary time scales to battle retroelements. bioRxiv. 10.1101/429563 [DOI] [Google Scholar]
- Feschotte C, Pritham E. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41: 331–368. 10.1146/annurev.genet.40.110405.090448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank JA, Feschotte C. 2017. Co-option of endogenous viral sequences for host cell function. Curr Opin Virol 25: 81–89. 10.1016/j.coviro.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli M, Trono D. 2015. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol 31: 429–51. 10.1146/annurev-cellbio-100814-125514 [DOI] [PubMed] [Google Scholar]
- Frost RJA, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. 2010. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci 107: 11847–11852. 10.1073/pnas.1007158107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Kawabe A, Etcheverry M, Ito T, Toyoda A, Fujiyama A, Colot V, Tarutani Y, Kakutani T. 2013. Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor. EMBO J 32: 2407–2417. 10.1038/emboj.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes DR, Swigut T, Wysocka J. 2018. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. Elife 7: e35989. 10.7554/eLife.35989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H. 2015. Site-specific non-LTR retrotransposons. Microbiol Spectr 3: 1147–1163. [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Osanai M, Matsumoto T, Kojima KK. 2005. Telomere-specific non-LTR retrotransposons and telomere maintenance in the silkworm, Bombyx mori. Chromosome Res 13: 455–467. 10.1007/s10577-005-0990-9 [DOI] [PubMed] [Google Scholar]
- Gagnier L, Belancio VP, Mager DL. 2019. Mouse germ line mutations due to retrotransposon insertions. Mob DNA 10: 15. 10.1186/s13100-019-0157-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Feschotte C. 2018. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr Opin Genet Dev 49: 15–24. 10.1016/j.gde.2018.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev EA, Arkhipova IR. 2007. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci 104: 9352–9357. 10.1073/pnas.0702741104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, Lu X, Chan Y-S, Ng HH, Ly L-H, Sachs F, Szczerbinska I. 2015. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16: 135–141. 10.1016/j.stem.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Goodier JL. 2016. Restricting retrotransposons: a review. Mob DNA 7: 16. 10.1186/s13100-016-0070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. 2015. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522: 221–225. 10.1038/nature14308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. 2016. Transposable elements: self-seekers of the germline, team-players of the soma. Bioessays 38: 1158–1166. 10.1002/bies.201600125 [DOI] [PubMed] [Google Scholar]
- Hamilton RS, Hartswood E, Vendra G, Jones C, Van De Bor V, Finnegan D, Davis I. 2009. A bioinformatics search pipeline, RNA2DSearch, identifies RNA localization elements in Drosophila retrotransposons. RNA 15: 200–207. 10.1261/rna.1264109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, et al. 2016. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. Elife 5: e19090. 10.7554/eLife.19090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH. 2016. Roles for retrotransposon insertions in human disease. Mob DNA 7: 9. 10.1186/s13100-016-0065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlem M, Brattig T, Kunze R. 1994. In vivo aggregation of maize Activator (Ac) transposase in nuclei of maize endosperm and petunia protoplasts. Plant J 5: 705–714. 10.1111/j.1365-313X.1994.00705.x [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. 2010. The genome of the western clawed frog Xenopus tropicalis. Science 328: 633–636. 10.1126/science.1183670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim J-W, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. 2017. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49: 925–934. 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res 19: 1419–1428. 10.1101/gr.091678.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka A, Saito R, Takashima K, Sasaki T, Fu Y, Kawabe A, Ito T, Toyoda A, Fujiyama A, Tarutani Y, et al. 2017. Evolution of sequence-specific anti-silencing systems in Arabidopsis. Nat Commun 8: 2161. 10.1038/s41467-017-02150-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, Booth BW, Pfeiffer BD, George RA, Svirskas R, et al. 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res 25: 445–458. 10.1101/gr.185579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst GDD, Werren JH. 2001. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet 2: 597–606. 10.1038/35084545 [DOI] [PubMed] [Google Scholar]
- Hynes AP, Rousseau GM, Agudelo D, Goulet A, Amigues B, Loehr J, Romero DA, Fremaux C, Horvath P, Doyon Y, et al. 2018. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat Commun 9: 2919. 10.1038/s41467-018-05092-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M, Helleboid P-Y, Trono D. 2017. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543: 550–554. 10.1038/nature21683 [DOI] [PubMed] [Google Scholar]
- The International Aphid Genomics Consortium. 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol 8: e1000313. 10.1371/journal.pbio.1000313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Baudino L, Kihara M, Leroy V, Vyse TJ, Evans LH, Izui S. 2013. Three Sgp loci act independently as well as synergistically to elevate the expression of specific endogenous retroviruses implicated in murine lupus. J Autoimmun 43: 10–17. 10.1016/j.jaut.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Sugimoto R, Nakaoka H, Yamada S, Kimura T, Hayano T, Inoue I. 2017. Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet 13: e1006883. 10.1371/journal.pgen.1006883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvák Z, Wang J, Singh M, Mager DL, Hurst LD. 2016. Pluripotency and the endogenous retrovirus HERVH: conflict or serendipity? Bioessays 38: 109–117. 10.1002/bies.201500096 [DOI] [PubMed] [Google Scholar]
- Jachowicz JW, Bing X, Pontabry J, Bošković A, Rando OJ, Torres-Padilla M-E. 2017. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet 49: 1502–1510. 10.1038/ng.3945 [DOI] [PubMed] [Google Scholar]
- Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245. 10.1038/nature13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangam D, Feschotte C, Betrán E. 2017. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet 33: 817–831. 10.1016/j.tig.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Wessler SR. 2001. Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13: 2553–2564. 10.1105/tpc.010235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabayama Y, Toh H, Katanaya A, Sakurai T, Chuma S, Kuramochi-Miyagawa S, Saga Y, Nakano T, Sasaki H. 2017. Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes. Nucleic Acids Res 12: 5387–5398. 10.1093/nar/gkx027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470. 10.1371/journal.pgen.1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Barbash DA. 2013. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol Biol Evol 30: 1816–1829. 10.1093/molbev/mst081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D, Rinn J. 2012. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13: R107. 10.1186/gb-2012-13-11-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. 2006. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 16: 78–87. 10.1101/gr.4001406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE. 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147: 1551–1563. 10.1016/j.cell.2011.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55: 1–24. 10.1111/j.0014-3820.2001.tb01268.x [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA. 1977. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86: 813–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigami D, Minami N, Takayama H, Imai H. 2003. MuERV-L Is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod 68: 651–654. 10.1095/biolreprod.102.007906 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149. 10.1016/j.cell.2009.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, Greenleaf WJ. 2019. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20: 207–220. 10.1038/s41576-018-0089-8 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31: 89–97. 10.1016/j.tibs.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Kordyukova M, Olovnikov I, Kalmykova A. 2018. Transposon control mechanisms in telomere biology. Curr Opin Genet Dev 49: 56–62. 10.1016/j.gde.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Kruse K, Diaz N, Enriquez-Gasca R, Gaume X, Torres-Padilla M-E, Vaquerizas JM. 2019. Transposable elements drive reorganisation of 3D chromatin during early embryogenesis. bioRxiv. 10.1101/523712 [DOI] [Google Scholar]
- Kunarso G, Chia N-Y, Jeyakani J, Hwang C, Lu X, Chan Y-S, Ng HH, Bourque G. 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634. 10.1038/ng.600 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849. 10.1242/dev.00973 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917. 10.1101/gad.1640708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci 96: 11428–11433. 10.1073/pnas.96.20.11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberger M, Gandon S, Meaden S, Rollie C, Chevallereau A, Chabas H, Buckling A, Westra ER, van Houte S. 2018. Anti-CRISPR phages cooperate to overcome CRISPR–Cas immunity. Cell 174: 908–916.e12. 10.1016/j.cell.2018.05.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, Sturgill D, Zhang Y, Oliver B, Clark AG. 2008. Evolution of protein-coding genes in Drosophila. Trends Genet 24: 114–123. 10.1016/j.tig.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Laski FA, Rio DC, Rubin GM. 1986. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell 44: 7–19. 10.1016/0092-8674(86)90480-0 [DOI] [PubMed] [Google Scholar]
- Lazarow K, Du M-L, Weimer R, Kunze R. 2012. A hyperactive transposase of the maize transposable element activator (Ac). Genetics 191: 747–756. 10.1534/genetics.112.139642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG. 2015. The role of piRNA-mediated epigenetic silencing in the population dynamics of transposable elements in Drosophila melanogaster. PLoS Genet 11: e1005269. 10.137/journal.pgen.1005269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, Lohr JG, Harris CC, Ding L, Wilson RK, et al. 2012a. Landscape of somatic retrotransposition in human cancers. Science 337: 967–971. 10.1126/science.1222077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-C, Gu W, Shirayama M, Youngman E, Conte D Jr, Mello CC. 2012b. C. elegans piRNAs Mediate the Genome-wide surveillance of germline transcripts. Cell 150: 78–87. 10.1016/j.cell.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Leek C, Levine MT. 2017. Recurrent innovation at genes required for telomere integrity in Drosophila. Mol Biol Evol 34: 467–482. 10.1093/molbev/msw248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léger P, Lara E, Jagla B, Sismeiro O, Mansuroglu Z, Coppée JY, Bonnefoy E, Bouloy M. 2013. Dicer-2- and Piwi-mediated RNA interference in rift valley fever virus-infected mosquito cells. J Virol 87: 1631–1648. 10.1128/JVI.02795-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL, Boeke JD. 1992. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J 11: 1145–1153. 10.1002/j.1460-2075.1992.tb05155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SH, Salmela H, Obbard DJ. 2016. Duplication and diversification of dipteran argonaute genes, and the evolutionary divergence of piwi and aubergine. Genome Biol Evol 8: 507–518. 10.1093/gbe/evw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, Smith SA, Sharma PP, Cordaux R, Gilbert C, Giraud I, et al. 2018. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol 2: 174–181. 10.1038/s41559-017-0403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G-C, Rehm EJ, Rubin GM. 2000. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci 97: 3347–3351. 10.1073/pnas.97.7.3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Chalmers R. 2014. Hyperactive mariner transposons are created by mutations that disrupt allosterism and increase the rate of transposon end synapsis. Nucleic Acids Res 42: 2637–2645. 10.1093/nar/gkt1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Hartl DL. 1996. Autoregulation of mariner transposase activity by overproduction and dominant-negative complementation. Mol Biol Evol 13: 549–555. 10.1093/oxfordjournals.molbev.a025615 [DOI] [PubMed] [Google Scholar]
- Lu Q, Ren S, Lu M, Zhang Y, Zhu D, Zhang X, Li T. 2013. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genomics 14: 651. 10.1186/1471-2164-14-651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Sachs F, Ramsay L, Jacques P-É, Göke J, Bourque G, Ng HH. 2014. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol 21: 423–425. 10.1038/nsmb.2799 [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Rio DC. 2015. P transposable elements in Drosophila and other eukaryotic organisms. Microbiol Spectr 3: 727–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A, Marchais A, Etcheverry M, Nature AM. 2013. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet 45: 1029–1039. 10.1038/ng.2703 [DOI] [PubMed] [Google Scholar]
- Mátés L, Chuah MKL, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. 2009. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41: 753–761. 10.1038/ng.343 [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, Glassford WJ, Herre M, Redmond SN, Rose NH, et al. 2018. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563: 501–507. 10.1038/s41586-018-0692-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue AD, Nuthikattu S, Slotkin RK. 2013. Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol 10: 1379–1395. 10.4161/rna.25555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RN Jr, Malik HS. 2017. Genetic conflicts: the usual suspects and beyond. J Exp Biol 220: 6–17. 10.1242/jeb.148148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesen P, Ivens A, Buck AH, van Rij RP. 2016. Small RNA profiling in dengue virus 2-infected Aedes mosquito cells reveals viral piRNAs and novel host miRNAs. PLoS Negl Trop Dis 10: e0004452. 10.1371/ournal.pntd.0004452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. 2007. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447: 167–177. 10.1038/nature05805 [DOI] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. 2014. The rhino–deadlock–cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157: 1364–1379. 10.1016/j.cell.2014.04.031 [DOI] [PubMed] [Google Scholar]
- Montgomery EA, Huang SM, Langley CH, Judd BH. 1991. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics 129: 1085–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. 2012. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito. PLoS Pathog 8: e1002470. 10.1371/journal.ppat.1002470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR. 2009. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134. 10.1038/nature08479 [DOI] [PubMed] [Google Scholar]
- Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, et al. 2015. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotech 33: 555–562. 10.1038/nbt.3128 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Cech TR. 1998. Reversing time: origin of telomerase. Cell 92: 587–590. 10.1016/S0092-8674(00)81123-X [DOI] [PubMed] [Google Scholar]
- Neuffer MG, Coe EH, Wessler SR. 1997. Mutants of Maize Revised. Cold Spring Harbor Laboratory Press. [Google Scholar]
- Newkirk SJ, Lee S, Grandi FC, Gaysinskaya V, Rosser JM, Vanden Berg N, Hogarth CA, Marchetto MCN, Muotri AR, Griswold MD, et al. 2017. Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc Natl Acad Sci 114: E5635–E5644. 10.1073/pnas.1701069114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka M, Itoh J-I, Nagato Y, Ono A, Ishiwata A, Sato Y. 2012. Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice. PLoS Genet 8: e1002953. 10.1371/journal.pgen.1002953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka M, Ishiwata A, Shimizu-Sato S, Ono A, Ishimoto K, Noda Y, Sato Y. 2013. The copy number of rice CACTA DNA transposons carrying MIR820 does not correlate with MIR820 expression. Plant Signal Behav 8: e25169. 10.4161/psb.25169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. 2009. A functional role for transposases in a large eukaryotic genome. Science 324: 935–938. 10.1126/science.1170023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Hamilton AT, Zhang H, Stubbs L. 2010. Rapid sequence and expression divergence suggest selection for novel function in primate-specific KRAB-ZNF genes. Mol Biol Evol 27: 2606–2617. 10.1093/molbev/msq157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowoshilow S, Schloissnig S, Fei J-F, Dahl A, Pang AWC, Pippel M, Winkler S, Hastie AR, Young G, Roscito JG, et al. 2018. The axolotl genome and the evolution of key tissue formation regulators. Nature 554: 50–55. 10.1038/nature25458 [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM. 2009. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci 364: 99–115. 10.1098/rstb.2008.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, Nakamura M, Tokunaga Y, Nakamura M, Watanabe A, et al. 2014. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc Natl Acad Sci 111: 12426–12431. 10.1073/pnas.1413299111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE, Crick FHC. 1980. Selfish DNA: the ultimate parasite. Nature 284: 604–607. 10.1038/284604a0 [DOI] [PubMed] [Google Scholar]
- Ottina E, Levy P, Eksmond U, Merkenschlager J, Young GR, Roels J, Stoye JP, Tüting T, Calado DP, Kassiotis G. 2018. Restoration of endogenous retrovirus infectivity impacts mouse cancer models. Cancer Immunol Res 6: 1292–1300. 10.1158/2326-6066.CIR-18-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O'Carroll D, Zamore PD. 2019. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 20: 89–108. 10.1038/s41576-018-0073-3 [DOI] [PubMed] [Google Scholar]
- Pardue M-L, DeBaryshe PG. 2008. Drosophila telomeres: a variation on the telomerase theme. Fly 2: 101–110. 10.4161/fly.6393 [DOI] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG. 2011. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci 108: 20317–20324. 10.1073/pnas.1100278108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parhad SS, Theurkauf WE. 2018. Rapid evolution and conserved function of the piRNA pathway. Open Biol 9: 180181. 10.1098/rsob.180181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parhad SS, Tu S, Weng Z, Theurkauf WE. 2017. Adaptive evolution leads to cross-species incompatibility in the piRNA transposon silencing machinery. Dev Cell 43: 60–70.e5. 10.1016/j.devcel.2017.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish NF, Fujino K, Shiromoto Y, Iwasaki YW, Ha H, Xing J, Makino A, Kuramochi-Miyagawa S, Nakano T, Siomi H, et al. 2015. piRNAs derived from ancient viral processed pseudogenes as transgenerational sequence-specific immune memory in mammals. RNA 21: 1691–1703. 10.1261/rna.052092.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova EG. 2004. Accumulation of transposable elements in the genome of Drosophila melanogaster is associated with a decrease in fitness. J Hered 95: 284–290. 10.1093/jhered/esh050 [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606. 10.1016/j.devcel.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Peddigari S, Li PW-L, Rabe JL MARTINSL. 2013. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res 41: 575–585. 10.1093/nar/gks1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton EH, Crease TJ. 2004. Evolution of the transposable element pokey in the ribosomal DNA of species in the subgenus daphnia (crustacea: cladocera). Mol Biol Evol 21: 1727–1739. 10.1093/molbev/msh189 [DOI] [PubMed] [Google Scholar]
- Percharde M, Lin C-J, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, Ramalho-Santos M. 2018. A LINE1-nucleolin partnership regulates early development and esc identity. Cell 174: 391–405.e19. 10.1016/j.cell.2018.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-González CE, Eickbush TH. 2002. Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics 162: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M, Mongelli V, Frangeul L, Blanc H, Jiggins F, Saleh M-C. 2016. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc Natl Acad Sci 113: E4218–E4227. 10.1073/pnas.1607952113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontis J, Planet E, Offner S, Turelli P, Duc J, Coudray A, Theunissen TW, Jaenisch R, Trono D. 2019. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell 24: 724–735.e5. 10.1016/j.stem.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham EJ, Feschotte C. 2007. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci 104: 1895–1900. 10.1073/pnas.0609601104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova S, Karam SE, Kellum R, Pardue M-L. 2002. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J Cell Biol 159: 397–402. 10.1083/jcb.200205039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Feschotte C, Pagan HJT, Smith JD, Pritham EJ, Arensburger P, Atkinson PW, Craig NL. 2008. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res 18: 717–728. 10.1101/gr.071886.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson HL, Astrin SM, Senior AM, Salazar FH. 1981. Host susceptibility to endogenous viruses: defective, glycoprotein-expressing proviruses interfere with infections. J Virol 40: 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Terrones D, Torres-Padilla M-E. 2018. Nimble and ready to mingle: transposon outbursts of early development. Trends Genet 34: 806–820. 10.1016/j.tig.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Rogers RL, Summers K, Wu Y, Guo C, Zheng J, Xun X, Xiong Z, Yang H, Zhou L, Zhang G, et al. 2018. Genomic takeover by transposable elements in the strawberry poison frog. Mol Biol Evol 35: 2913–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Mitchell JA, Nishida Y, Hildreth JE, Ariberre JA, Gilbert WV, Garfinkel DJ. 2015. A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J Virol 89: 3922–3938. 10.1128/JVI.03060-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Leandre B, Nguyen SC, Levine MT. 2019. Diversification and collapse of a telomere elongation mechanism. Genome Res 29: 920–931. 10.1101/gr.245001.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni FA, Guerra J, Luban J. 2012. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 9: 111–111. 10.1186/1742-4690-9-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SN, Carreira PE, Shukla R, Gerhardt DJ, Gerdes P, Sánchez-Luque FJ, Nicoli P, Kindlova M, Ghisletti S, Santos Dos A, et al. 2018. L1 retrotransposition is a common feature of mammalian hepatocarcinogenesis. Genome Res 28: 639–653. 10.1101/gr.226993.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Durrett R. 2004. Adaptive evolution drives the diversification of zinc-finger binding domains. Mol Biol Evol 21: 2326–2339. 10.1093/molbev/msh246 [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Radovani E, Najafabadi HS, Barazandeh M, Campitelli LF, Yin Y, Jolma A, Zhong G, Guo H, Kanagalingam T, et al. 2016. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res 26: 1742–1752. 10.1101/gr.209643.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Schnettler E, Donald CL, Human S, Watson M, Siu RWC, McFarlane M, Fazakerley JK, Kohl A, Fragkoudis R. 2013. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J Gen Virol 94: 1680–1689. 10.1099/vir.0.053850-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G, Deininger PL. 2016. Insertion of retrotransposons at chromosome ends: adaptive response to chromosome maintenance. Front Genet 6: 358. 10.3389/fgene.2015.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A, Wong A, Poh Y-P, Theurkauf WE, Jensen JD. 2013. Recurrent and recent selective sweeps in the piRNA pathway. Evolution (N Y) 67: 1081–1090. 10.1111/evo.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinzelle L, Izsvák Z, Ivics Z. 2009. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol Life Sci 66: 1073–1093. 10.1007/s00018-009-8376-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Castanon O, Said K, Volf V, Khoshakhlagh P, Hornick A, Ferreira R, Wu C-T, Güell M, Garg S, et al. 2019. Enabling large-scale genome editing by reducing DNA nicking. bioRxiv. 10.1101/574020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Liu J, Schnakenberg SL, Ha H, Xing J, Chen KC. 2014. Variation in piRNA and transposable element content in strains of Drosophila melanogaster. Genome Biol Evol 6: 2786–2798. 10.1093/gbe/evu217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotero-Caio CG, Platt RN II, Suh A, Ray DA. 2017. Evolution and diversity of transposable elements in vertebrate genomes. Genome Biol Evol 9: 161–177. 10.1093/gbe/evw264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzer MC, Anderson SN, Springer NM, Ross-Ibarra J. 2019. The genomic ecosystem of transposable elements in maize. bioRxiv. 10.1101/559922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana T, Zamborlini A, Cristofari G, Lesage P. 2017. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat Rev Genet 18: 292–308. 10.1038/nrg.2017.7 [DOI] [PubMed] [Google Scholar]
- Sun YH, Xie LH, Zhuo X, Chen Q, Ghoneim D, Zhang B, Jagne J, Yang C, Li XZ. 2017. Domestic chickens activate a piRNA defense against avian leukosis virus. Elife 6: 8634. 10.7554/eLife.24695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. 2004. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol 269: 276–285. 10.1016/j.ydbio.2004.01.028 [DOI] [PubMed] [Google Scholar]
- Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, et al. 2013. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol 11: e1001473. 10.1371/journal.pbio.1001473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Steranka JP, Ma S, Grivainis M, Rodić N, Huang CRL, Shih I-M, Wang T-L, Boeke JD, Fenyo D, et al. 2017. Human transposon insertion profiling: analysis, visualization and identification of somatic LINE-1 insertions in ovarian cancer. Proc Natl Acad Sci 114: E733–E740. 10.1073/pnas.1619797114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Okuniewska M, Malone CD, Coux R-X, Rio DC, Lehmann R. 2017. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 8: 272–272. 10.1038/nature25018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever BR. 2016. The evolution of antiviral defense systems. Cell Host Microbe 19: 142–149. 10.1016/j.chom.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. 2011. Coevolution of retroelements and tandem zinc finger genes. Genome Res 21: 1800–1812. 10.1101/gr.121749.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd CD, Deniz Ö, Taylor D, Branco MR. 2019. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. Elife 8: 1473. 10.7554/eLife.44344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger RS, Pope SD, Kong Y, Tokuyama M, Taura M, Iwasaki A. 2019. The lupus susceptibility locus Sgp3 encodes the suppressor of endogenous retrovirus expression SNERV. Immunity 50: 334–347.e9. 10.1016/j.immuni.2018.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D. 2015. Transposable elements, polydactyl proteins, and the genesis of human-specific transcription networks. Cold Spring Harb Symp Quant Biol 80: 281–288. 10.1101/sqb.2015.80.027573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubio JMC, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K, et al. 2014. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345: 1251343. 10.1126/science.1251343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Bor V, Hartswood E, Jones C, Finnegan D, Davis I. 2005. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev Cell 9: 51–62. 10.1016/j.devcel.2005.04.012 [DOI] [PubMed] [Google Scholar]
- Van Valen L. 1973. A new evolutionary law. Evol Theory 1–30. [Google Scholar]
- Vodovar N, Bronkhorst AW, van Cleef KWR, Miesen P, Blanc H, van Rij RP, Saleh M-C. 2012. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One 7: e30861. 10.1371/journal.pone.0030861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A, Mochizuki K. 2013. A domesticated PiggyBac transposase interacts with heterochromatin and catalyzes reproducible DNA elimination in Tetrahymena. PLoS Genet 9: e1004032. 10.1371/journal.pgen.1004032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A, Goldman AD, Mochizuki K, Landweber LF. 2013. Transposon domestication versus mutualism in ciliate genome rearrangements. PLoS Genet 9: e1003659. 10.1371/journal.pgen.1003659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt F, Wiedemann L, Zuliani C, Querques I, Sebe A, Mátés L, Izsvák Z, Ivics Z, Barabas O. 2016. Sleeping Beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat Commun 7: 11126. 10.1038/ncomms11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Reinke V. 2008. A C. elegans Piwi, PRG-1, Regulates 21U-RNAs during spermatogenesis. Curr Biol 18: 861–867. 10.1016/j.cub.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xie G, Singh M, Ghanbarian AT, Raskó T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. 2014. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 516: 405–409. 10.1038/nature13804 [DOI] [PubMed] [Google Scholar]
- Wang L, Dou K, Moon S, Tan FJ, Zhang ZZ. 2018. Hijacking oogenesis enables massive propagation of LINE and retroviral transposons. Cell 174: 1082–1094.e12. 10.1016/j.cell.2018.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Barbash D, Kelleher E. 2019. Divergence of piRNA pathway proteins affects piRNA biogenesis but not TE transcript level. bioRxiv. 10.1101/521773 [DOI] [Google Scholar]
- Werren JH. 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci 108: 10863–10870. 10.1073/pnas.1102343108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong C-J, Zhong JW, Tapscott SJ. 2017. Conservation and innovation in the DUX4-family gene network. Nat Genet 49: 935–940. 10.1038/ng.3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8: 973–982. 10.1038/nrg2165 [DOI] [PubMed] [Google Scholar]
- Wintersdorff von CJH, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, Savelkoul PHM, Wolffs PFG. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol 7: 305–173. 10.3389/fmicb.2016.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204. 10.1038/nature07844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Yang P, Füchtbauer AC, Füchtbauer E-M, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS. 2015. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev 29: 538–554. 10.1101/gad.252767.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard LE, Downes LM, Lee Y-C, Kaja A, Terefe ES, Wilson MH. 2017. Temporal self-regulation of transposition through host-independent transposase rodlet formation. Nucleic Acids Res 45: 353–366. 10.1093/nar/gkw1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Schweizer C, Rudinskiy N, Taylor DM, Kazantsev A, Luthi-Carter R, Fraering PC. 2010. Novel γ-secretase inhibitors uncover a common nucleotide-binding site in JAK3, SIRT2, and PS1. FASEB J 24: 2464–2474. 10.1096/fj.09-148031 [DOI] [PubMed] [Google Scholar]
- Yang P, Wang Y, Hoang D, Tinkham M, Patel A, Sun M-A, Wolf G, Baker M, Chien H-C, Lai K-YN, et al. 2017a. A placental growth factor is silenced in mouse embryos by the zinc finger protein ZFP568. Science 356: 757–759. 10.1126/science.aah6895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang Y, Macfarlan TS. 2017b. The role of KRAB-ZFPs in transposable element repression and mammalian evolution. Trends Genet 33: 871–881. 10.1016/j.tig.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Chen F, Luo M, Cheng Y, Zhao H, Cheng H, Zhou R. 2014. Rapid evolution of piRNA pathway in the teleost fish: implication for an adaptation to transposon diversity. Genome Biol Evol 6: 1393–1407. 10.1093/gbe/evu105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13: 335–340. 10.1016/S0168-9525(97)01181-5 [DOI] [PubMed] [Google Scholar]
- Yu T, Koppetsch BS, Chappell K, Pagliarani S, Johnston S, Weng Z, Theurkauf WE. 2019. The piRNA response to retroviral invasion of the koala genome. bioRxiv. 10.1101/599852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Zhou L, Li MA, Bradley A, Craig NL. 2011. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci 108: 1531–1536. 10.1073/pnas.1008322108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng TC, Huang G, Lu Q, Surleac MD, Mandell JD, Pontarotti P, Petrescu AJ, Xu A, Xiong Y, et al. 2019a. Transposon molecular domestication and the evolution of the RAG recombinase. Nature 569: 79–84. 10.1038/s41586-019-1093-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li T, Preissl S, Grinstein J, Farah E, Destici E, Lee AY, Chee S, Qiu Y, Ma K, et al. 2019b. 3D Chromatin architecture remodeling during human cardiomyocyte differentiation reveals a role of HERV-H in demarcating chromatin domains. bioRxiv. 10.1101/485961 [DOI] [Google Scholar]
- Zou S, Ke N, Kim JM, Voytas DF. 1996. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev 10: 634–645. 10.1101/gad.10.5.634 [DOI] [PubMed] [Google Scholar]