Abstract
Ability to promote completion of mitotic cycling of adult mammalian cardiomyocytes remains an intractable and vexing challenge despite being one of the most sought after ‘holy grails’ of cardiovascular research. While some of the struggle is attributable to adult cardiomyocytes themselves that are notoriously post-mitotic, another contributory factor rests with difficulty in definitive tracking of adult cardiomyocyte cell cycle and lack of rigorous measures to track proliferation in situ. This review summarizes past, present, and future directions to promote adult mammalian cardiomyocyte cell cycle progression, proliferation, and renewal. Establishing relationship(s) between cardiomyocyte cell cycle progression and cellular biological properties is sorely needed to understand the mechanistic basis for cardiomyocyte cell cycle withdrawal to enhance cardiomyocyte cell cycle progression and mitosis.
Keywords: cardiomyocyte, cell cycle, proliferation, ploidy, senescence
I. Cardiomyocyte proliferation: everything old is new again
Retrospective views on cardiomyocyte mitosis.
Considering the universally accepted conclusion that loss of cardiomyocytes is a major underlying cause of heart failure from acute pathologic injury or chronic stress, the answer of generating additional cardiomyocytes to restore structural and functional integrity of the heart seems a simple, clever, and achievable solution. However, inherent biological properties of the adult mammalian myocardium have rendered this overtly straightforward approach frustratingly difficult. Indeed, decades of research and thousands of publications have been dedicated to the singular goal of prompting adult mammalian cardiomyocytes to re-enter cell cycle and complete mitosis. The inescapable conclusion from collective efforts put forth is that adult mammalian cardiomyocytes are remarkably refractory to mitotic activity, unlike those found in either early postnatal mice or zebrafish. Nevertheless, new publications appear every year touting major advances in understanding and augmenting cardiomyocyte proliferation.[1-4] Therefore, it seems reasonable to briefly reflect upon where we are in this process, what factors are obstructing forward progress, and how the field could re-center with renewed focus and purpose to empower the ultimate goal of developing interventional approaches for therapeutic cardiomyogenesis.
Evolution of thinking on cardiomyocyte renewal.
Current literature is replete with masterful reviews on the topic of cardiomyogenesis that summarize the sophisticated and elegant studies carried out by hundreds of laboratories around the world.[5-10] The consensus opinion for many years remains that mammalian cardiomyocyte proliferation is readily observed in prenatal and early postnatal development.[11, 12] Furthering evidence for immature mammalian cardiomyocyte proliferative capacity, similar conclusions were reached observing cultured neonatal cardiomyocytes.[13-15] However, scant evidence exists to support adult mammalian cardiomyocyte division in vitro, but rather a “de-differentiation” process characterized by loss of myofibril organization, return to immature phenotypic properties, and expression of stem cell marker c-kit.[16-19] Even less encouraging, adult mammalian cardiomyocyte division in situ remained elusive, with reports of occasional mitotic figures without definitive proof of completed cytokinesis [20, 21], since labeling with proliferating cell nuclear antigen and bromodeoxyuridine (BrdU) are not definitive evidence of completed cell division.[22, 23] The new millennium witnessed a number of controversial turns in the search for evidence of adult mammalian cardiomyogenesis with the advent of cardiac stem cells[24, 25], carbon-14 estimates of turnover from nuclear bomb blasts[26, 27], as well as the rise and fall of related studies from the Anversa lab.[28] Retrospectively considering the arc of thinking on adult mammalian cardiomyocyte replacement, there is no disputing that researchers have failed to unlock regenerative potential of the adult mammalian myocardium sufficient to restore structure or function lost from pathologic damage, chronic stress or aging.[12] Acceptance of this humbling defeat stands in stark contrast to myocardial repair in lower vertebrates or neonatal mice where acute injury promotes cardiomyocyte replacement.[29, 30] Profound differences in reparative potential between mammalian neonatal versus adult hearts are intriguing, but the underlying explanation might simply be chalked up to neonatal mammalian cardiomyocytes having more in common with zebrafish than adult mammals.[31] So without a clear path forward, many have returned to re-examination of the adult cardiomyocyte armed with novel approaches and unflagging optimism, intent upon succeeding where so many others have failed before them.
Current renewed interest in cardiomyocyte proliferation.
Several excellent reviews have previously covered many aspects of current knowledge and obstacles in the pursuit of cardiomyogenesis for adult mammalian hearts.[23, 29, 32, 33] Highlighting the distinction of adult mammalian hearts is important, as substantial time and effort has been expended defining the indisputable cardiomyogenic activity inherent to postnatal mouse myocardium as well as zebrafish hearts. Yet there is abundant evidence that the inherent biological milieu of hearts from postnatal mice or zebrafish is profoundly distinct from adult mammalian myocardium, leaving translatability of such research unresolved. Clearly, it stands to reason that cardiomyogenic testing for adult mammalian hearts is best tested in the setting of an in vivo adult mammal model to achieve the most dependable and reliable results. And yet, even in the setting of adult murine models there has been lack of consensus on cardiomyogenic cell sources, proliferative activity, and quantitation of mitotic activity. For example, a rigorous study of cardiomyogenesis in mice during postnatal development concluded that a very brief period of cardiomyogenic potential exists after birth that disappears in the adult heart,[34] consistent with more recent re-visitation of this topic using the apical resection model[35, 36] as well as neonatal pigs.[37, 38] None of these studies address potential induction on cardiomyogenesis following pathologic damage in an adult setting, but a recent consensus statement from the American Heart Association focused upon endogenous cardiomyogenesis (rather than cell-based therapeutic approaches) concluded “1) Cardiomyocyte renewal rates may be higher after injury than under normal conditions, and 2) The experimental determination of cardiomyocyte turnover after cardiac injury can be challenging owing to inflammation, proliferation of stromal and vascular cells, and scar formation.”[39] After decades of unrelenting investigation, the consensus is that answers related to cardiomyocyte turnover in the pathological setting remain unresolved. Clearly, new approaches and additional knowledge are required. Selected primary considerations that have hampered the field are presented in the next few paragraphs, highlighting longstanding limitations as well as the way forward proposed in this review.
To avoid detours and stay on the main path toward mitosis, we need to understand where the offramps are and bypass them.
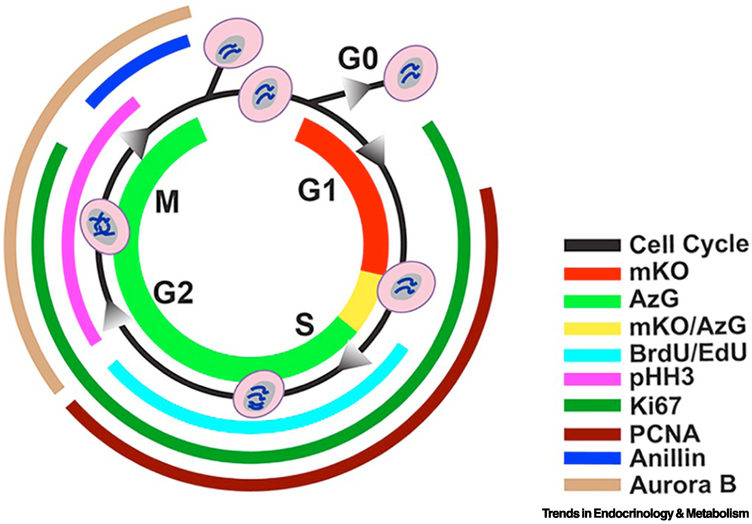
Paradoxically, a primary issue hampering studies of adult mammalian cardiomyogenesis has been the difficulty of determining cardiomyocyte proliferation using markers of cell cycling. While such demonstrations are readily reproduced in neonatal mice or zebrafish, the biological responses of adult cardiomyocytes to mitotic stimuli render typical measures of cell division irrelevant. For example, multiple markers of cell cycle have been developed for investigations of non- myocardial cell biology and co-opted to assess cardiomyocyte proliferation (Fig. 1). Each of these markers has been used to infer mitotic activity, yet none of them alone are truly definitive indicators of authentic cell division when working with cardiomyocytes. Specifically, these markers indicate progression through cell cycle including mitosis. However, in the context of cardiomyocytes, many of these markers are present at multiple stages of cell cycle and it is impossible to distinguish cells that are progressing through mitosis from those that arrest at various mitotic checkpoints.
Figure. 1. Markers of division and cell-cycle status.
FUCCI fluorescence mKO (red) presents in G1 phase, and AzG (green) presents during S/G2/M phases, where during the G1/S transition both fluorescence (mKO/AzG) present simultaneously and merge into a yellow colour. BrdU or Edu, both thymidine analogs incorporate into DNA during synthesis (cyan). Phosphorylated Histone 3 (pHH3) is responsible for chromatin condensation and is thus present during G2 through M phase (magenta). Nuclear antigen Ki67 is present from G1 to M phase (emerald). PCNA is presents between G1 and G2 phase in response to DNA synthesis (burgundy). Anillin plays a role in creating the cleavage furrow formation and begin to accumulate in late G2 through late M phase (blue). Aurora B plays a role in mitosis, present from G2 through M phase (sand). Reproduced from Alvarez et al.[22]
II. The Janus-faced cardiomyocyte: deceptively progressive
Numerous approaches for assessing mitosis – most inauthentic.
All sorts of results have been reported in the adult mammalian context with widely varying observations of cardiomyocyte “proliferation” using a plethora of markers and metrics to assess de novo cardiomyogenesis.[34, 40, 41] Lack of standardization, varied experimental approaches, and underappreciation for distinctive cell cycle regulation of cardiomyocytes has led to substantial confusion and, in some cases, hyperbolic claims of translational potential that have not as yet been borne out through the passage of time and practical experience.
Warnings from published articles on flawed methodology.
Limitations of using these markers to document cardiomyocyte proliferation have been highlighted in previous publications,[40, 42] but despite these admonitions the presentation of these labels as evidence of cardiomyocyte mitotic activity continues. This serious problem for the field is indicative of disconnects in recognizing the atypical mitotic resistance of cardiomyocytes relative to other cell types where such labels could be accurate and appropriate. A recent study pointed out these limitations and offered a way forward using two novel proteins (RhoA and IQGAP3)[43] or midbody positioning[44] as definitive markers of cardiomyocyte division, but unfortunately use of these markers also rests upon a tour-de-force confocal analysis of intracellular localization at a critical transient moment in the penultimate steps of mitosis. Demonstration of cardiomyocyte mitosis using individual proteins or structures will require further development of tools to monitor these proteins in real-time to follow intracellular localization that is beyond the capabilities of current typical investigations of cardiomyocyte analyses.
Complexity of demonstrating mitosis in vivo.
Cumulative background information presented thus far in this section certainly is sufficiently disconcerting to prompt skepticism and reservations related to recent publications of enhanced cardiomyocyte proliferation. Of note, one recent publication from 2017 asserts that frequency of mononuclear diploid cardiomyocytes correlates with increased cardiac regenerative potential[45] with an associated editorial,[46] yet this study did not rigorously discriminate between ploidy levels resulting from endomitosis, endoreplication, or cellular division. In a different publication from 2017, administration of miRNA mimics was touted to induce cardiomyocyte passage through mitosis, yet these conclusions were based upon Aurora B and phospho-histone 3 immunolocalization[3] spawning an editorial comment in the same issue.[47] A third high profile study based upon a defined set of four cyclin-related factors (4F) concludes adult cardiomyocyte proliferation was evident based upon EdU and phospho-histone 3 as well as histologic assessment of Mosaic Analysis with Double Markers (MADM) transgenic mice.[2] Cardiac-specific mouse models for clonal analysis including MADM have been comprehensively covered in an excellent review by Leone et al.[23] MADM has been used to assess mitosis in postnatal cardiomyocytes or adult cardiomyocytes with relatively low labeling efficiency of 0.78% or 0.9%, respectively.[48] Low efficiency cardiomyocyte labeling in MADM is likely due to the requirement for cell division coupled with Cre-mediated recombination in G2 phase to allow for recombinant alleles to segregate into separate daughter cells representative of mitosis.[49] [50] Catching mitosis and Cre-recombination simultaneously given the rarity of cardiomyocyte division in an adult heart is clearly challenging, and the requirement for three separate alleles (Cre as well as two MADM) into a single mouse for MADM presents a daunting prospect for mouse breeding schemes to introduce additional genetic modifications.[51] MADM is much more amenable to use with delivery of inductive agents to adult mice, as in a recent study touting unprecedentedly high adult cardiomyocyte mitotic activity following combined adenoviral delivery of four cell-cycle regulators.[2] Lastly, yet another publication in 2018 shows “birth of new cardiomyocytes in adult mice” following 8 weeks of running exercise identified based on incorporation of 15N-thymidine by multi-isotope imaging mass spectrometry (MIMS) and on being mononucleate/diploid.[1] MIMS also relies upon quantitation from a very limited number of 15N labeled diploid mononuclear cardiomyocytes (0.14% - 0.09% in non-injured hearts in one study; 0.25% in sedentary mice in a second study) leaving the technique susceptible to substantial influence from finding small numbers of additional labeled cells.[1, 52] MIMS technology, while impressive, is certainly not a widely adopted technique for assessing cardiomyocyte proliferation and validation using more broadly available techniques is warranted to substantiate the conclusion that exercise prompts a ~4.6 fold increase in new cardiomyocytes.[1] Whether such profound elevation of adult mammalian cardiomyocyte mitosis in these studies can be authenticated by other laboratories remains to be seen, as previous controversial claims for a postnatal burst of cardiomyocyte proliferation[53] were subsequently challenged by multiple laboratories unable to replicate these results.[54-56]
Considerable resources, time, and effort have been poured into studies of cardiomyogenesis in experimental models characterized by repair after acute injury, most notably in neonatal mice and zebrafish. The excitement and enthusiasm with which these models have been pursued is indisputable, but translating findings from these models to promote productive adult mammalian cardiomyocyte cell cycle progression and mitosis remains unfulfilled.[29] Aside from controversies of cardiomyogenesis versus ‘regeneration’ in the neonatal mouse apical resection model[30, 31, 36, 57-59] and the role of stem cells[60-62], shared reparative capabilities of neonatal mice and zebrafish appear to rest with the immature phenotype of the tissues relative to the adult mammalian myocardium.[31] The proteomic analysis concluded “the profound differences in structural gene expression place the (regenerative) zebrafish heart rather in the vicinity of the (proliferative) neonatal, but not the adult mouse hearts… It is therefore questionable if promitotic stimuli that drive cardiac regeneration in zebrafish may be capable of inducing cardiac regeneration in adult mammalian cardiomyocytes.” [29] Additional significant differences includes presence of intact centrosomes in cardiomyocytes of adult zebrafish or neonatal mammalian rodents versus absence in adult mammalian hearts.[63] Narrowing to the nexus of this challenge leads to defining causes and consequences for mammalian adult cardiomyocytes to withdraw from and their intractability to re-enter cell cycle.
III. Many causes, one consequence – the withdrawn adult cardiomyocyte
Decades of studying the cardiomyocyte cell cycle and current barriers to proliferation induction has produced far too much information than can be adequately summarized in this review, but fortunately has been covered in recent overviews.[5, 64-66] One inescapable conclusion from digesting the avalanche of prior studies on this topic is that, when pressured by manipulation of cell cycle to progress toward mitosis, adult mammalian cardiomyocytes respond uncooperatively with abortive mitosis from checkpoint arrest, polyploidy with DNA synthesis without cytokinesis, hypertrophic growth, or even death. Perhaps the failed forced entry when pushing adult mammalian cardiomyocytes to advance to mitosis is inextricably linked to their biological contractile function, which is inevitably compromised as a consequence of structural remodeling linked to acquiring immature status that (as noted in the preceding paragraph) is likely part and parcel of authentic mitosis.[16] Since cardiomyocyte cell cycle is replete with various offramps from the mitotic highway, defining specific stage(s) of progression and exit points will be crucial to developing interventional approaches to keep these reluctant travelers on the road to productive cytokinesis.
IV. Where do reluctant cardiomyocytes get off?
Fluorescent Ubiquitin Cell Cycle Indicator (FUCCI) to study cardiomyocyte cell cycle.
Adult mammalian cardiomyocytes are notoriously indifferent to stimuli well known to drive mitosis in other cell types such as serum stimulation, oncogenic stimuli, or forced cell cycle reentry, yet it is clear that they do respond in alternative ways. These longstanding ambiguities have rendered claims of induced cardiomyocyte cell cycling open to debate and skepticism, although pervasive doubts are sharply contrasted against the abundance of publications in support of induced cardiomyocyte mitotic activity. Our group assessed cardiomyocyte regulation from a new perspective using FUCCI reporters [67] as a tool to dissect cell cycle progression (Fig. 1). Implementation of FUCCI labeling has yielded advances in biological systems ranging from cell culture to zebrafish, flies, mice, and embryonic stem cells.[67] The novel transgenic mouse is based upon well documented and proven FUCCI technology adapted to in vivo cell cycle monitoring via cardiomyocyte-specific transgenesis (FUCCI-Tg).[22] Although the FUCCI system has previously been studied in the cardiovascular context,[68-71] none of these prior studies used cardiomyocyte-specific expression and none were concerned with demonstration of enhanced adult cardiomyogenesis. Cardiomyocyte division is not the derived readout of the FUCCI-Tg, but rather the study of cell cycle progression. Indeed, to address the eventual outcome of cell cycle progression to discriminate between endoreplication, endomitosis, or mitosis, additional readouts are required for incorporation with imaging to determine ploidy state of nuclei as well as nuclearity of cardiomyocytes. The FUCCI-Tg is particularly valuable as a novel tool to assess cardiomyocyte proliferation because: 1) every cardiomyocyte in the heart is visualized for cell cycle status, not just “cycling myocytes”, 2) four distinct stages of cell cycle progression are revealed with inherent implications for cardiomyocyte mitosis, 3) quantitation of cell cycle status for collective myocyte populations is possible, 4) the system can be used in combination with DNA labeling to correlate cell cycle progression with DNA synthesis versus DNA damage, and most importantly 5) in vivo labeling is assessed in the adult mammalian heart – the ultimate testing milieu for authentic proliferative activity (to validate observations from in vitro or postnatal environments). The oscillation of FUCCI signal occurs in real time unlike long term tracking or genetic tracing approaches, so several time points need to be analyzed to find optimal timing for detection, for example after treatment, when cardiomyocyte proliferation occurs. Functionality and utility of the FUCCI-Tg documented in our publication[22] not only reinforced many prior studies of cardiomyocyte biological properties, but also revealed a previously unappreciated aspect of withdrawal from cell cycle: the canonical restriction point (R-point) first described by Pardee in 1974.[72]
R-point.
R-point cell cycle withdrawal could be mistaken for another cell cycle checkpoint, but the processes involved differ substantially. Whereas a checkpoint involves primarily intracellular sensors of metabolic state, genome integrity, and sequential execution of prior cell cycle steps, the R-point transition rests upon cellular integration of signals received from the environment over an extended period of time to determine whether growth is warranted.[73] Without permission to proceed past R-point, the cell withdraws from cycling and enters an arrested state. The R-point has been narrowed to a mid-to-late G1 stage known as the G1/S boundary when cellular resources are focused upon maintenance and preservation of ongoing processes rather than proliferative growth. Cardiomyocyte arrest at the R-point is intuitively attractive given the high metabolic demands of contractile function and the need to maintain structural integrity for normal function. The R-point integrates a multifaceted “knot of mitogen and inhibitory signaling” intrinsically dedicated to preventing cell cycle progression[74], which likely accounts for both the extended lifespan of cardiomyocytes as well as their notorious reluctance to undergo mitotic activity.
Polyploidy.
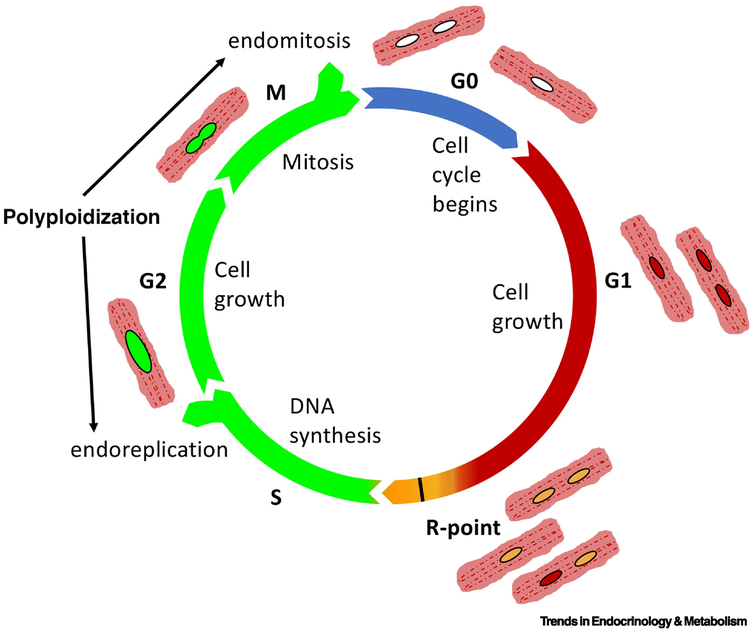
Contributing to the confusion, biological phenomena of endomitosis, endoreplication, and DNA damage are often underappreciated or unaccounted for in assessments of cardiomyocyte proliferation.[75] Consistent with their atypical nature and resistance to cell division, cardiomyocytes enter mitosis and exit without generating daughter cells but rather by mere duplication of DNA without new nucleation or by adding additional nuclei.[45, 76-81] Although the terms ‘endoreplication’, ‘endomitosis’, and ‘endoreduplication’ are often used interchangeably,[82] for this proposal endoreplication refers to DNA duplication without karyokinesis, whereas endomitosis refers to karyokinesis without cytokinesis. These two processes involve progression through cell cycle that presents as DNA synthesis often misrepresented as mitotic activity (Fig. 2). Cardiomyocytes exhibit increased levels of ploidy within single nuclei well as by accumulating multiple nuclei, even as a normal process of aging.[34, 80, 83] So too, cardiomyocytes incorporate DNA labeling agents consequential to attempting DNA repair response following environmental challenge such as oxidative stress.[84, 85] Failure to appreciate these normal aspects of cardiomyocyte cell biology leads to controversial claims of proliferation rates and potentially erroneous claims of regeneration.[86-88]
Figure 2.
Adult mammalian cardiomyocytes withdraw from cell cycle progression at two primary points of R-point restriction (R-point) and acquisition of higher level ploidy (Polyloidy) through genomic duplication (endoreplication) multinucleation (endomitosis). Nuclei shown in varying coloration corresponding to the FUCCI reporter system.
V. Redirecting the driver rather than hijacking the vehicle
Cardiomyocytes have good reasons for bailing out with R-point or polyploidy, as these represent biologically sensible choices in the face of proliferative stimuli. The structural and functional demands placed upon the adult mammalian heart are incompatible with widespread coordinated adult mammalian cardiomyocyte mitosis that would compromise tissue integrity and hemodynamic output. And yet even now, pieces continue to emerge in the puzzle of the recalcitrant cardiomyocyte. Among the candidate directions to follow, prevention of stresses that prompt cardiomyocytes to bail out of cell cycle such as metabolic shifts, endothermy, phenotypic maturation, and reactive oxygen species have all received recent attention.[89-91] Lest we forget, promoting a youthful lifestyle for cardiomyocytes on an environmental and molecular level helps to stave off cellular senescence and decrepitude.[8, 92] Perhaps the answer lies not simply with brute force bludgeoning adult mammalian cardiomyocytes into submission to cell cycle, but gentle persuasion by offering a conducive environment and involving cellular crosstalk. Recent examples from studying liver biology demonstrate ploidy state plays an essential role in regulation of cellular proliferation and tissue regeneration.[93, 94] Regulation of cellular proliferation is a recurring theme in studies of polyploidy with particular emphasis in liver aging and repair. Acquisition of stable higher ploidy state prompts cell cycle withdrawal and potential emergence of senescence-associated characteristics.[95-97] However, hepatocytes undergo ploidy reversal during liver repopulation, senescent human hepatocytes are “rejuvenated” after cell transplantation, and polyploidy in hepatocytes does not necessarily equate with senescence.[98] A regulatory role for inhibition of proliferation in highly regenerative liver tissue and cultured cells appears to be exerted by tetraploid cells upon diploid brethren.[99] In support of an anti-proliferative action, the polyploid state plays a tumor-suppressive role in the liver.[100] Concurrently, tetraploid hepatocytes also give rise to aneuploid progeny and can facilitate adaptation to chronic liver disease.[93] As evident from these few selected examples, incontrovertible evidence that regulation of ploidy in liver is fundamentally important for determination of proliferative activity, even as mechanisms of ploidy determination and ensuing biological actions remain frustratingly elusive. Similar observations of ploidy-based regulation of cardiac repair occur in zebrafish heart regeneration.[101] If such concepts could be adapted to adult mammalian myocardium, then cardiomyocytes therein might be more amenable to stating on the road to mitosis rather than taking the offramps to quiescence or, alternatively, running out of gas and ending in cytokinesis failure.
VI. Where does the road lead (Concluding remarks / future perspectives)?
We are only as strong as our weakest links – factoring in the entire organism.
Given everything written up to this point certainly is sufficient to give one pause regarding prospects for restoration of myocardial function through promotion of adult cardiomyocyte cell cycle. In keeping with allusions to the cell cycle highway, staying on track for adult mammalian cardiomyocyte may be facilitated by shifting gears rather than hitting the accelerator. Namely, focusing upon cell biology rather than narrow heavy-handed molecular interventions, recognizing that changing fundamental phenotype of cardiomyocyte to a more pliable and accommodating condition is inextricably linked to changing the potential for cell cycle progression, and taking cues from other adult organs and cells such as the liver.[93, 94] Lastly, although this review has centered upon adult mammalian cardiomyocytes, the involvement of the cardiac interstitial cell population should not be discounted or overlooked since those support cells regulate the surrounding environment.[102-106] And in the final analysis, a complex web of intrinsic and extrinsic factors all provide signposts in a medley that influences receptivity of the tissue to reparative action.[107, 108]
Realistic expectations, believable outcomes, and achievable destinations.
The quest for cardiomyogenic approaches in the adult mammalian heart remains a top priority for cardiovascular research and therapeutic interventional strategies to treat heart failure, even after decades of frustration and what can be characterized, at best, as modest outcomes. As previously observed, some of the impasse is attributable to the plethora of approaches and interpretations used in prior published studies. Even today, new tools allowing for increased understanding and improved accuracy for assessments are desperately needed. Application of rigorous and consistent measures to determine induction of cardiomyocyte cell cycle progression in the adult mammalian myocardium is essential to validate and compare the ever-expanding series of methods and practices developed throughout the world. Inconsistent measures, inappropriately applied measures, and overinterpretation of findings have been and continue to be problematic for achieving resolution in advancing mechanistic understanding of cardiomyocyte cell cycle regulation. Paradoxically, while substantial information has been gathered on the unique characteristics of the cardiomyocyte cell cycle relative to other cell types, assessments of outcomes often fail to fully and faithfully encompass the spectrum of possibilities with high rigor and reproducibility. The research community should coalesce around a commonly shared set of principles used to guide measurement of cardiomyocyte cell cycle and allow all researchers to benefit by comparative measures with standardized references such as the FUCCI-Tg serving as a platform for adult mammalian myocardial cell-cycle analysis.[22] Accomplishing the goal of unraveling cardiomyocyte cell cycle control will provide a path forward to reconcile disparate observations, thereby improving the accuracy and reproducibility of interventions intended to enhance adult mammalian cardiomyocyte cell cycle progression.
Outstanding questions.
Why have decades of concerted efforts to promote completion of adult mammalian cardiomyocyte cell cycle resulted in so little tangible progress?
How relevant are studies of cardiomyocytes with proven proliferative capabilities in neonatal animals or lower vertebrates such as zebrafish to furthering understanding of adult mammalian cardiomyocyte cell cycle progression?
What measures can provide definitive, readily demonstrable, and unambiguous evidence of adult mammalian cardiomyocyte cell cycle completion?
Can canonical points of adult cardiomyocyte cell cycle withdrawal such as R-point or multinucleation be overcome to promote completion of cytokinesis?
What is the role of the myocardial environment and interstitial cell populations in limiting cell cycle progression and mitotic activity of adult mammalian cardiomyocytes?
Trends.
Adult mammalian cardiomyocytes are remarkably refractory to completion of cell cycle progression through mitosis.
Despite ongoing study for decades, progress to promote adult mammalian cardiomyocyte cell cycle completion has been frustratingly ineffective.
Fundamental biological differences exist between adult mammalian cardiomyocytes versus those derived from neonatal mice or lower vertebrates such as zebrafish that both possess relatively immature phonotypes.
Studies reporting cardiomyocyte proliferation often lack definitive proof of authentic cardiomyocyte mitotic activity due to methodologies misrepresented as completion of cell cycle progression.
Two major points of cell cycle withdrawal for adult mammalian cardiomyocytes are the Restriction point (R-point) and acquisition of higher level ploidy through multinucleation (Ploidy).
Glossary box
- R-point
The restriction point (R) is a point in G1 of the animal cell cycle at which the cell becomes “committed” to the cell cycle and after which extracellular proliferation stimulants are no longer required. A cell’s decision to enter, or reenter, the cell cycle is determined by collective progressive and inhibitory extracellular signals that are received and processed.
- Diploid
A cell or an organism possessing paired sets of chromosomes.
- Polyploid
A cell or organism having more than typical diploid paired chromosomes
- Ploidy
The number of paired chromosome sets in a cell or organism.
- Karyokinesis
nuclear division resulting in doubling of the nuclear number
- Cytokinesis
physical separation that completes cell division resulting in two comparable daughter cell progeny.
- Endomitosis
karyokinesis without cytokinesis leading to multinucleation
- Endoreplication
genomic duplication without karyokinesis leading to polyploidization
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vujic A et al. (2018) Exercise induces new cardiomyocyte generation in the adult mammalian heart. Nat Commun 9 (1), 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohamed TMA et al. (2018) Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell 173 (1), 104–116 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lesizza P et al. (2017) Single-Dose Intracardiac Injection of Pro-Regenerative MicroRNAs Improves Cardiac Function After Myocardial Infarction. Circ Res 120 (8), 1298–1304. [DOI] [PubMed] [Google Scholar]
- 4.Diez-Cunado M et al. (2018) miRNAs that Induce Human Cardiomyocyte Proliferation Converge on the Hippo Pathway. Cell Rep 23 (7), 2168–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan X and Braun T (2017) Multimodal Regulation of Cardiac Myocyte Proliferation. Circ Res 121 (3), 293–309. [DOI] [PubMed] [Google Scholar]
- 6.He L and Zhou B (2017) Cardiomyocyte proliferation: remove brakes and push accelerators. Cell Res 27 (8), 959–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazar E et al. (2017) Cardiomyocyte renewal in the human heart: insights from the fall-out. Eur Heart J 38 (30), 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siddiqi S and Sussman MA (2014) The heart: mostly postmitotic or mostly premitotic? Myocyte cell cycle, senescence, and quiescence. Can J Cardiol 30 (11), 1270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yutzey KE (2017) Cardiomyocyte Proliferation: Teaching an Old Dogma New Tricks. Circ Res 120 (4), 627–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasumarthi KB and Field LJ (2002) Cardiomyocyte cell cycle regulation. Circ Res 90 (10), 1044–54. [DOI] [PubMed] [Google Scholar]
- 11.de Carvalho A et al. (2017) Early Postnatal Cardiomyocyte Proliferation Requires High Oxidative Energy Metabolism. Sci Rep 7 (1), 15434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kretzschmar K et al. (2018) Profiling proliferative cells and their progeny in damaged murine hearts. Proc Natl Acad Sci U S A 115 (52), E12245–E12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prosdocimo G and Giacca M (2017) Manipulating the Proliferative Potential of Cardiomyocytes by Gene Transfer. Methods Mol Biol 1553, 41–53. [DOI] [PubMed] [Google Scholar]
- 14.Belostotskaya GB and Golovanova TA (2014) Characterization of contracting cardiomyocyte colonies in the primary culture of neonatal rat myocardial cells: a model of in vitro cardiomyogenesis. Cell Cycle 13 (6), 910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bon-Mathier AC et al. (2019) Oxygen as a key regulator of cardiomyocyte proliferation: New results about cell culture conditions! Biochim Biophys Acta Mol Cell Res. [DOI] [PubMed] [Google Scholar]
- 16.Jopling C et al. (2010) Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464 (7288), 606–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y et al. (2010) Dedifferentiation and proliferation of mammalian cardiomyocytes. PLoS One 5 (9), e12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubin T et al. (2011) Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell 9 (5), 420–32. [DOI] [PubMed] [Google Scholar]
- 19.Wang WE et al. (2017) Dedifferentiation, Proliferation, and Redifferentiation of Adult Mammalian Cardiomyocytes After Ischemic Injury. Circulation 136 (9), 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beltrami CA et al. (1997) Proliferating cell nuclear antigen (PCNA), DNA synthesis and mitosis in myocytes following cardiac transplantation in man. J Mol Cell Cardiol 29 (10), 2789-802. [DOI] [PubMed] [Google Scholar]
- 21.Quaini F et al. (1994) End-stage cardiac failure in humans is coupled with the induction of proliferating cell nuclear antigen and nuclear mitotic division in ventricular myocytes. Circ Res 75 (6), 1050–63. [DOI] [PubMed] [Google Scholar]
- 22.Alvarez R Jr. et al. (2019) Cardiomyocyte cell cycle dynamics and proliferation revealed through cardiac-specific transgenesis of fluorescent ubiquitinated cell cycle indicator (FUCCI). J Mol Cell Cardiol 127, 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leone M et al. (2015) Cardiomyocyte proliferation in cardiac development and regeneration: a guide to methodologies and interpretations. Am J Physiol Heart Circ Physiol 309 (8), H1237–50. [DOI] [PubMed] [Google Scholar]
- 24.Torella D et al. (2015) Generation of new cardiomyocytes after injury: de novo formation from resident progenitors vs. replication of pre-existing cardiomyocytes. Ann Transl Med 3 (Suppl 1), S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vicinanza C et al. (2017) Adult cardiac stem cells are multipotent and robustly myogenic: c-kit expression is necessary but not sufficient for their identification. Cell Death Differ 24 (12), 2101–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergmann O et al. (2009) Evidence for cardiomyocyte renewal in humans. Science 324 (5923), 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergmann O et al. (2012) Cardiomyocyte renewal in humans. Circ Res 110 (1), e17–8; author reply e19-21. [DOI] [PubMed] [Google Scholar]
- 28.(2019) Expression of Concern. Circulation 139 (3), e5–e6. [DOI] [PubMed] [Google Scholar]
- 29.Foglia MJ and Poss KD (2016) Building and re-building the heart by cardiomyocyte proliferation. Development 143 (5), 729–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadek HA et al. (2014) Multi-investigator letter on reproducibility of neonatal heart regeneration following apical resection. Stem Cell Reports 3 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes RS et al. (2016) “Young at heart”: Regenerative potential linked to immature cardiac phenotypes. J Mol Cell Cardiol 92, 105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yester JW and Kuhn B (2017) Mechanisms of Cardiomyocyte Proliferation and Differentiation in Development and Regeneration. Curr Cardiol Rep 19 (2), 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leach JP and Martin JF (2018) Cardiomyocyte Proliferation for Therapeutic Regeneration. Curr Cardiol Rep 20 (8), 63. [DOI] [PubMed] [Google Scholar]
- 34.Walsh S et al. (2010) Cardiomyocyte cell cycle control and growth estimation in vivo--an analysis based on cardiomyocyte nuclei. Cardiovasc Res 86 (3), 365–73. [DOI] [PubMed] [Google Scholar]
- 35.Zebrowski DC et al. (2017) Cardiac injury of the newborn mammalian heart accelerates cardiomyocyte terminal differentiation. Sci Rep 7 (1), 8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen DC et al. (2016) Persistent scarring and dilated cardiomyopathy suggest incomplete regeneration of the apex resected neonatal mouse myocardium--A 180 days follow up study. J Mol Cell Cardiol 90, 47–52. [DOI] [PubMed] [Google Scholar]
- 37.Zhu W et al. (2018) Regenerative Potential of Neonatal Porcine Hearts. Circulation 138 (24), 2809–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye L et al. (2018) Early Regenerative Capacity in the Porcine Heart. Circulation 138 (24), 2798–2808. [DOI] [PubMed] [Google Scholar]
- 39.Eschenhagen T et al. (2017) Cardiomyocyte Regeneration: A Consensus Statement. Circulation 136 (7), 680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zebrowski DC et al. (2016) Towards regenerating the mammalian heart: challenges in evaluating experimentally induced adult mammalian cardiomyocyte proliferation. Am J Physiol Heart Circ Physiol 310 (9), H1045–54. [DOI] [PubMed] [Google Scholar]
- 41.Zebrowski DC and Engel FB (2013) The cardiomyocyte cell cycle in hypertrophy, tissue homeostasis, and regeneration. Rev Physiol Biochem Pharmacol 165, 67–96. [DOI] [PubMed] [Google Scholar]
- 42.Raulf A et al. (2015) Transgenic systems for unequivocal identification of cardiac myocyte nuclei and analysis of cardiomyocyte cell cycle status. Basic Res Cardiol 110 (3), 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leone M et al. (2018) Cardiomyocyte binucleation is associated with aberrant mitotic microtubule distribution, mislocalization of RhoA and IQGAP3, as well as defective actomyosin ring anchorage and cleavage furrow ingression. Cardiovasc Res. [DOI] [PubMed] [Google Scholar]
- 44.Hesse M et al. (2018) Midbody Positioning and Distance Between Daughter Nuclei Enable Unequivocal Identification of Cardiomyocyte Cell Division in Mice. Circ Res 123 (9), 1039–1052. [DOI] [PubMed] [Google Scholar]
- 45.Patterson M et al. (2017) Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat Genet 49 (9), 1346–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.da Costa Martins PA (2017) Mononuclear Diploidy at the Heart of Cardiomyocyte Proliferation. Cell Stem Cell 21 (4), 421–422. [DOI] [PubMed] [Google Scholar]
- 47.Zangi L and Hajjar RJ (2017) Synthetic MicroRNAs Stimulate Cardiac Repair. Circ Res 120 (8), 1222–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali SR et al. (2014) Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc Natl Acad Sci U S A 111 (24), 8850–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gitig D (2010) Transcriptomics: individuality in the cellular world. Biotechniques 48 (6), 439–43. [DOI] [PubMed] [Google Scholar]
- 50.Zong H et al. (2005) Mosaic analysis with double markers in mice. Cell 121 (3), 479–92. [DOI] [PubMed] [Google Scholar]
- 51.Kadow ZA and Martin JF (2018) Distinguishing Cardiomyocyte Division From Binucleation. Circ Res 123 (9), 1012–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Senyo SE et al. (2013) Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493 (7432), 433–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naqvi N et al. (2015) Cardiomyocytes Replicate and their Numbers Increase in Young Hearts. Cell 163 (4), 783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alkass K et al. (2015) No Evidence for Cardiomyocyte Number Expansion in Preadolescent Mice. Cell 163 (4), 1026–36. [DOI] [PubMed] [Google Scholar]
- 55.Hirai M et al. (2016) Revisiting Preadolescent Cardiomyocyte Proliferation in Mice. Circ Res 118 (6), 916–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soonpaa MH et al. (2015) Cardiomyocyte Cell-Cycle Activity during Preadolescence. Cell 163 (4), 781–2. [DOI] [PubMed] [Google Scholar]
- 57.Andersen DC et al. (2014) Do neonatal mouse hearts regenerate following heart apex resection? Stem Cell Reports 2 (4), 406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bryant DM et al. (2015) A systematic analysis of neonatal mouse heart regeneration after apical resection. J Mol Cell Cardiol 79, 315–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sampaio-Pinto V et al. (2018) Neonatal Apex Resection Triggers Cardiomyocyte Proliferation, Neovascularization and Functional Recovery Despite Local Fibrosis. Stem Cell Reports 10 (3), 860–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jesty SA et al. (2012) c-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc Natl Acad Sci U S A 109 (33), 13380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tallini YN et al. (2009) c-kit expression identifies cardiovascular precursors in the neonatal heart. Proc Natl Acad Sci U S A 106 (6), 1808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elhelaly WM et al. (2019) C-Kit Cells Do Not Significantly Contribute to Cardiomyogenesis During Neonatal Heart Regeneration. Circulation 139 (4), 559–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zebrowski DC et al. (2015) Developmental alterations in centrosome integrity contribute to the post-mitotic state of mammalian cardiomyocytes. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hesse M et al. (2018) Heart regeneration and the cardiomyocyte cell cycle. Pflugers Arch 470 (2), 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirai M et al. (2016) Tissue-Specific Cell Cycle Indicator Reveals Unexpected Findings for Cardiac Myocyte Proliferation. Circ Res 118 (1), 20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ponnusamy M et al. (2017) Understanding cardiomyocyte proliferation: an insight into cell cycle activity. Cell Mol Life Sci 74 (6), 1019–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zielke N and Edgar BA (2015) FUCCI sensors: powerful new tools for analysis of cell proliferation. Wiley Interdiscip Rev Dev Biol 4 (5), 469–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abe T et al. (2013) Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development 140 (1), 237–46. [DOI] [PubMed] [Google Scholar]
- 69.Hashimoto H et al. (2014) Time-lapse imaging of cell cycle dynamics during development in living cardiomyocyte. J Mol Cell Cardiol 72, 241–9. [DOI] [PubMed] [Google Scholar]
- 70.Hashimoto H et al. (2015) Analysis of cardiomyocyte movement in the developing murine heart. Biochem Biophys Res Commun 464 (4), 1000–7. [DOI] [PubMed] [Google Scholar]
- 71.Sakaue-Sawano A and Miyawaki A (2014) Visualizing spatiotemporal dynamics of multicellular cell-cycle progressions with fucci technology. Cold Spring Harb Protoc 2014 (5). [DOI] [PubMed] [Google Scholar]
- 72.Pardee AB (1974) A restriction point for control of normal animal cell proliferation. Proc Natl Acad Sci U S A 71 (4), 1286–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Planas-Silva MD and Weinberg RA (1997) The restriction point and control of cell proliferation. Curr Opin Cell Biol 9 (6), 768–72. [DOI] [PubMed] [Google Scholar]
- 74.Blagosklonny MV and Pardee AB (2002) The restriction point of the cell cycle. Cell Cycle 1 (2), 103–10. [PubMed] [Google Scholar]
- 75.Bergmann O et al. (2011) Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp Cell Res 317 (2), 188–94. [DOI] [PubMed] [Google Scholar]
- 76.Gonzalez-Rosa JM et al. (2018) Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev Cell 44 (4), 433–446 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kadow ZA and Martin JF (2018) A Role for Ploidy in Heart Regeneration. Dev Cell 44 (4), 403–404. [DOI] [PubMed] [Google Scholar]
- 78.Lee Y (2010) To proliferate or not to proliferate. Cardiovasc Res 86 (3), 347–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richardson GD (2016) Simultaneous Assessment of Cardiomyocyte DNA Synthesis and Ploidy: A Method to Assist Quantification of Cardiomyocyte Regeneration and Turnover. J Vis Exp (111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sukhacheva TV et al. (2015) Age-Related Features of Cardiomyocyte Ploidy in Patients with Hypertrophic Obstructive Cardiomyopathy. Bull Exp Biol Med 159 (1), 95–9. [DOI] [PubMed] [Google Scholar]
- 81.Liu Z et al. (2010) Regulation of cardiomyocyte polyploidy and multinucleation by CyclinG1. Circ Res 106 (9), 1498–506. [DOI] [PubMed] [Google Scholar]
- 82.Lee HO et al. (2009) Endoreplication: polyploidy with purpose. Genes Dev 23 (21), 2461-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Silva IS et al. (2018) Polyploidy and nuclear phenotype characteristics of cardiomyocytes from diabetic adult and normoglycemic aged mice. Acta Histochem 120 (2), 84–94. [DOI] [PubMed] [Google Scholar]
- 84.Zhao H et al. (2011) Induction of DNA damage signaling by oxidative stress in relation to DNA replication as detected using “click chemistry”. Cytometry A 79 (11), 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao H et al. (2017) ATM Activation and H2AX Phosphorylation Induced by Genotoxic Agents Assessed by Flow- and Laser Scanning Cytometry. Methods Mol Biol 1599, 183–196. [DOI] [PubMed] [Google Scholar]
- 86.Beltrami AP et al. (2001) Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med 344 (23), 1750–7. [DOI] [PubMed] [Google Scholar]
- 87.Hsieh PC et al. (2007) Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med 13 (8), 970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Soonpaa MH and Field LJ (1998) Survey of studies examining mammalian cardiomyocyte DNA synthesis. Circ Res 83 (1), 15–26. [DOI] [PubMed] [Google Scholar]
- 89.Hirose K et al. (2019) Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science 364 (6436), 184–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kimura W et al. (2015) Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature 523 (7559), 226–30. [DOI] [PubMed] [Google Scholar]
- 91.Nakada Y et al. (2017) Hypoxia induces heart regeneration in adult mice. Nature 541 (7636), 222–227. [DOI] [PubMed] [Google Scholar]
- 92.Siddiqi S and Sussman MA (2013) Cardiac Hegemony of Senescence. Curr Transl Geriatr Exp Gerontol Rep 2 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wilkinson PD et al. (2019) Polyploid Hepatocytes Facilitate Adaptation and Regeneration to Chronic Liver Injury. Am J Pathol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilkinson PD et al. (2019) The Polyploid State Restricts Hepatocyte Proliferation and Liver Regeneration in Mice. Hepatology 69 (3), 1242–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang MJ et al. (2017) Hepatocyte polyploidization and its association with pathophysiological processes. Cell Death Dis 8 (5), e2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gorla GR et al. (2001) Polyploidy associated with oxidative injury attenuates proliferative potential of cells. J Cell Sci 114 (Pt 16), 2943–51. [DOI] [PubMed] [Google Scholar]
- 97.Duncan AW (2013) Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol 24 (4), 347–56. [DOI] [PubMed] [Google Scholar]
- 98.Wang MJ et al. (2014) Reversal of hepatocyte senescence after continuous in vivo cell proliferation. Hepatology 60 (1), 349–61. [DOI] [PubMed] [Google Scholar]
- 99.Wilkinson PD et al. (2018) The polyploid state restricts hepatocyte proliferation and liver regeneration. Hepatology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang S et al. (2018) The Polyploid State Plays a Tumor-Suppressive Role in the Liver. Dev Cell 47 (3), 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cao J et al. (2017) Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue. Dev Cell 42 (6), 600–615 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chistiakov DA et al. (2016) The role of cardiac fibroblasts in post-myocardial heart tissue repair. Exp Mol Pathol 101 (2), 231–240. [DOI] [PubMed] [Google Scholar]
- 103.Furtado MB et al. (2014) Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res 114 (9), 1422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shinde AV and Frangogiannis NG (2014) Fibroblasts in myocardial infarction: a role in inflammation and repair. J Mol Cell Cardiol 70, 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Talman V and Ruskoaho H (2016) Cardiac fibrosis in myocardial infarction-from repair and remodeling to regeneration. Cell Tissue Res 365 (3), 563–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sussman MA (2019) Cardiac nonmyocyte subpopulations: a secular congregation. Regen Med. [DOI] [PubMed] [Google Scholar]
- 107.Gude NA et al. (2018) Cardiac ageing: extrinsic and intrinsic factors in cellular renewal and senescence. Nat Rev Cardiol 15 (9), 523–542. [DOI] [PubMed] [Google Scholar]
- 108.Broughton KM et al. (2018) Mechanisms of Cardiac Repair and Regeneration. Circ Res 122 (8), 1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]