Abstract
Single-cell RNA-seq (scRNA-seq) has emerged as a powerful technique for characterizing cellular heterogeneity, but it is currently impractical on large sample cohorts and cannot be applied to fixed specimens collected as part of routine clinical care. We previously developed an approach for digital cytometry, called CIBERSORT, that enables estimation of cell type abundances from bulk tissue transcriptomes. We now introduce CIBERSORTx, a machine learning method that extends this framework to infer cell-type-specific gene expression profiles without physical cell isolation. By minimizing platform-specific variation, CIBERSORTx also allows the use of scRNA-seq data for large-scale tissue dissection. We evaluated the utility of CIBERSORTx in multiple tumor types, including melanoma, where single-cell reference profiles were used to dissect bulk clinical specimens, revealing cell type-specific phenotypic states linked to distinct driver mutations and response to immune checkpoint blockade. We anticipate that digital cytometry will augment single-cell profiling efforts, enabling cost-effective, high-throughput tissue characterization without the need for antibodies, disaggregation, or viable cells.
Introduction
Tissues are complex ecosystems comprised of diverse cell types that are distinguished by their developmental origins and functional states. While strategies for studying tissue composition have generated profound insights into basic biology and medicine, comprehensive assessment of cellular heterogeneity remains challenging. Traditional immunophenotyping approaches, such as flow cytometry and immunohistochemistry (IHC), generally rely on small combinations of preselected marker genes, limiting the number of cell types that can be simultaneously interrogated. In contrast, single-cell mRNA sequencing (scRNA-seq) enables unbiased transcriptional profiling of thousands of individual cells from a single-cell suspension. Despite the power of this technology1, analyses of large sample cohorts are not yet practical, and most fixed clinical specimens (e.g., formalin-fixed, paraffin embedded (FFPE) samples) cannot be dissociated into intact single-cell suspensions. Furthermore, the impact of tissue disaggregation on cell type representation is poorly understood.
Over the last decade, a number of computational techniques have been described for dissecting cellular content directly from genomic profiles of mixture samples2–8. The majority of these methods rely on a specialized knowledgebase of cell type-specific “barcode” genes, often called a “signature matrix,” which is generally derived from FACS-purified or in vitro differentiated/stimulated cell subsets2,3. Although useful when cell types of interest are well defined, such gene signatures are suboptimal for the discovery of novel cellular states and cell type-specific gene expression profiles (GEPs), and for capturing the full spectrum of major cell phenotypes in complex tissues. To overcome these limitations, previous studies have explored the utility of deconvolution methods for inferring cell type GEPs2,3 and the potential of single-cell reference profiles for in silico tissue dissection5,9–14. However, the accuracy of these strategies on real bulk tissues remains unclear.
Here we introduce CIBERSORTx, a computational framework to accurately infer cell type abundance and cell type-specific gene expression from RNA profiles of intact tissues (Fig. 1). To accomplish this, we extended CIBERSORT, a method that we previously developed for enumerating cell composition from tissue GEPs15, with new functionalities for cross-platform data normalization and in silico cell purification. The latter allows the transcriptomes of individual cell types to be digitally “purified” from bulk RNA admixtures without physical isolation. As a result, changes in cell type-specific gene expression can be inferred without cell separation or prior knowledge. By leveraging cell type expression signatures from single-cell experiments or sorted cell subsets, CIBERSORTx can provide detailed portraits of tissue composition without physical dissociation, antibodies, or living material.
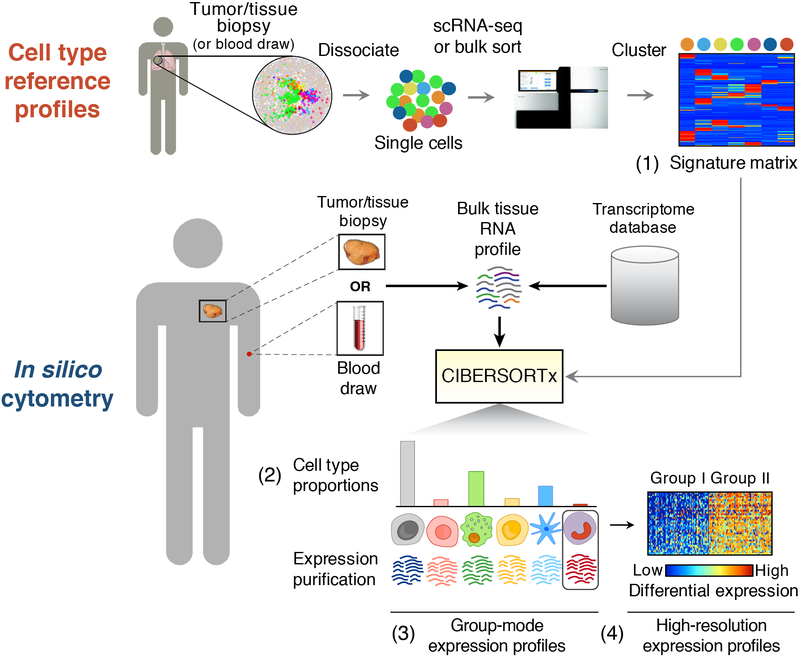
Figure 1.
Framework for in silico cell enumeration and purification. A typical CIBERSORTx workflow involves a serial approach, in which molecular profiles of cell subsets are first obtained from a small collection of tissue samples and then repeatedly used to perform systematic analyses of cellular abundance and gene expression signatures from bulk tissue transcriptomes. This process involves: (1) transcriptome profiling of single cells or sorted cell subpopulations to define a “signature matrix” consisting of barcode genes that can discriminate each cell subset of interest in a given tissue type; (2) applying the signature matrix to bulk tissue RNA profiles in order to infer cell type proportions and (3) representative cell type expression signatures; and (4) purifying multiple transcriptomes for each cell type from a cohort of related tissue samples. Using metastatic melanomas as an example, Figure 6 illustrates the application of each step.
Results
Tissue dissection with scRNA-seq
CIBERSORTx was designed to enable large-scale tissue characterization using cell signatures derived from diverse sources, including single-cell reference profiles (Fig. 1). To achieve this goal, we developed analytical tools for deriving a signature matrix from single-cell or bulk sorted transcriptional data while minimizing batch effects as a source of confounding technical variation (Supplementary Figs. 1,2; Supplementary Note 1). We then investigated the utility of commonly used scRNA-seq technologies16 for enumerating cell proportions in RNA admixtures derived from bulk tissues.
We started by generating a single-cell RNA-seq library from peripheral blood mononuclear cells (PBMCs) obtained from a patient with non-small cell lung cancer (NSCLC) using 10x Genomics Chromium v2 (3’ assay). Unsupervised clustering and canonical marker gene assessment revealed six major leukocyte subsets (B cells, CD4 T cells, CD8 T cells, NKT cells, NK cells, and monocytes; Fig. 2a). To assess deconvolution performance, we built a signature matrix to distinguish these cell subsets and tested it on a validation cohort of bulk RNA-seq profiles of blood obtained from 12 healthy adults (Supplementary Tables 1 and 2).
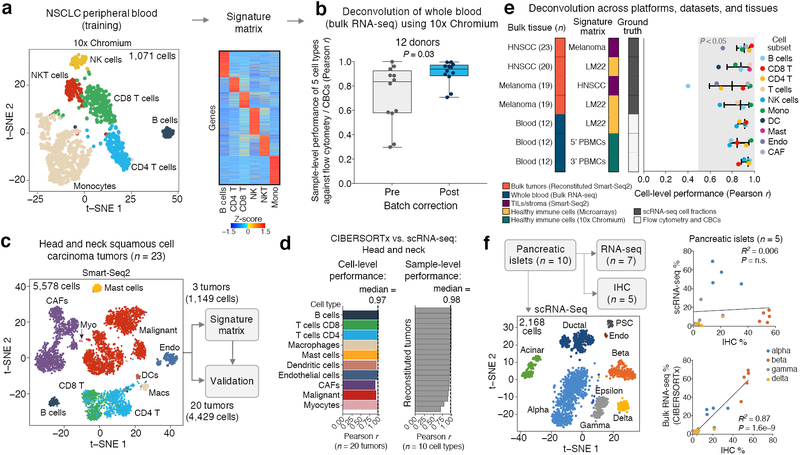
Figure 2.
Bulk tissue deconvolution with single-cell reference profiles. (a) Left: t-SNE projection of scRNA-seq data from the peripheral blood of a patient with NSCLC, with six major leukocyte populations indicated. Right: Heat map of signature matrix genes distinguishing these six subsets. (b) Enumeration of leukocyte frequencies in RNA-seq profiles of whole blood (n = 12 healthy adults) using the PBMC signature matrix from panel a, shown before and after cross-platform normalization. Performance gains are shown as Pearson correlations and assessed for each sample across five cell types quantified by flow cytometry and automated complete blood counts (CBCs): B cells, NK cells, CD8 T cells, CD4 T cells, and monocytes. Statistical significance was determined using a two-sided Wilcoxon signed-rank test. Data are presented as boxplots (n = 12 per group; center line, median; box limits, upper and lower quartiles; whiskers, maximum and minimum values). (c) t-SNE projection of scRNA-seq data from 23 head and neck squamous cell carcinoma (HNSCC) tumors9 (left) and training/validation approach for assessing single-cell deconvolution performance (right). (d) Concordance between cell type proportions measured by scRNA-seq and CIBERSORTx deconvolution for 20 held-out HNSCC tumors for validation from panel c. All tumor GEPs were reconstructed from single-cell data. (e) Analysis of cell subset enumeration across diverse signature matrices, tissues, and platforms. Deconvolution was run with batch correction (Methods). Ground truth cell proportions were determined by scRNA-seq (HNSCC, melanoma) or by flow cytometry and automated hematology leukocyte differential counter (blood). Cell subsets within the gray band are significantly concordant with ground truth by Pearson correlation (P < 0.05). Signature matrices are provided in Supplementary Table 2, with the exception of LM2215. Data are presented as medians ± interquartile range. Additional details are provided in Supplementary Note 1. (f) Left: t-SNE plot of pancreatic islet subsets from ten human subjects, five of which were profiled by scRNA-seq, bulk RNA-seq, and IHC21 (also see Supplementary Fig. 3f-i). Right: Scatterplots depicting concordance between the frequencies of four major islet subsets quantitated by IHC versus scRNA-seq (top) and CIBERSORTx deconvolution of bulk RNA-seq (bottom), as determined by linear regression. The significance of the result was assessed by a two-sided t test. Cell subset abbreviations: Mono, monocytes; Macs, macrophages; DCs, dendritic cells; Mast, mast cells; CAFs, cancer-associated fibroblasts; Endo, endothelial cells; Myo, myocytes.
Compared to ground truth cell proportions as determined by direct cytometry and fluorescence immunophenotyping, uncorrected deconvolution results showed clear estimation biases for some cell types in bulk admixtures (Fig. 2b, Supplementary Fig. 1d). We hypothesized that these biases could be driven by platform-specific variation between the signature matrix and bulk RNA-seq data, as might be introduced, for example, by the variable use of unique molecular identifiers during library preparation. Indeed, following application of a batch correction scheme that we developed (Supplementary Fig. 1a, Supplementary Note 1, Methods), deconvolution results substantially improved and compared favorably to ground truth cell proportions (Fig. 2b, Supplementary Fig. 1d). We observed similar gains in performance through batch correction when analyzing other datasets and signature matrices, including publicly available Chromium v2 PBMC data (3’ and 5’ kits; Supplementary Fig. 2a-c) and purified leukocyte subsets profiled using microarrays (Supplementary Fig. 2d). Given these systematic improvements, we therefore applied batch correction in all subsequent cross-platform analyses, unless stated otherwise (Supplementary Table 1).
We next extended our analysis to solid tumor biopsies where single cells were profiled by SMART-Seq2. Focusing on head and neck squamous cell carcinomas9 (HNSCC) and melanomas10 (n = 18 and 19 patients, respectively), we initially tested deconvolution performance on simulated tumors reconstructed from single cells. This allowed us to evaluate the utility of single-cell reference profiles in a manner that controlled for dissociation-related artifacts and heterogeneity in phenotypic definitions. In a dataset of 18 primary tumors and 5 lymph node metastases from patients with HNSCC9, we created a signature matrix from a training cohort consisting of 2 primary tumor specimens and 1 lymph node biopsy (Supplementary Table 2). This matrix distinguished malignant cells, CD4 and CD8 T cells, B cells, macrophages, dendritic cells, mast cells, endothelial cells, myocytes, and cancer-associated fibroblasts (Fig. 2c). When evaluated using reconstructed tumor samples, deconvolution results were highly concordant with ground truth cell proportions (Fig. 2d, Supplementary Fig. 3a). Strong performance was also maintained when considering deconvolution results across distinct tumor types and cell types, including within rare or difficult to isolate cell subpopulations, such as distinct CD8 T cell effector subsets infiltrating melanomas (Supplementary Figs. 3b-d, 4, Supplementary Table 2).
To explore the impact of key signature matrix-related parameters on single cell-guided deconvolution, we next examined CIBERSORTx performance as a function of the number of cells per phenotype, the number of donor samples, and the number of genes considered. Across a range of values for these factors, we observed a surprisingly modest effect on cell proportion estimates (Supplementary Fig. 5a-c). Moreover, regardless of their primary biological source, CIBERSORTx signature matrices exhibited strong generalizability across diverse expression profiling platforms, datasets, and tissues after batch correction was applied (Fig. 2e).
Leveraging the single cell-derived signature matrix from melanoma biopsies described above, we then applied CIBERSORTx to dissect melanoma RNA-seq profiles from The Cancer Genome Atlas (TCGA)17. We observed substantial differences in the fractional representation of B/T lymphocytes and macrophages when comparing predicted cell type proportions in bulk tumors and the original scRNA-seq results (Supplementary Fig. 3e). Since these cell subsets were unselected relative to one another in the scRNA-seq dataset10, such compositional distortions may have arisen either from technical artifacts owing to single-cell isolation and sequencing18,19, or from the deconvolution approach itself. In support of the former, IHC estimates of TIL subsets in an independent melanoma cohort20 were far more similar to TIL fractions estimated by CIBERSORTx than those determined by scRNA-seq (Supplementary Fig. 3e). Moreover, we observed the same distortion phenomenon in a dataset of human pancreatic islets profiled by scRNA-seq (SMART-Seq2), bulk RNA-seq, and IHC21 (Fig. 2f, Supplementary Fig. 3f–i). In a direct comparison of paired islet specimens, cell fractions determined by IHC in bulk tissues were significantly correlated with bulk islet deconvolution results, but not scRNA-seq (Fig. 2f, Supplementary Fig. 3i). These data further validate CIBERSORTx and highlight its value for mitigating dissociation-related distortions resulting from the physical isolation of intact single cells (for additional discussion, see Supplementary Note 2).
Cell type-specific gene expression without physical cell isolation
Cell-type-specific transcriptome profiles can provide valuable insights into cell identity and function. However, such profiles are generally derived from single cells or bulk sorted populations, which can be difficult to obtain for large cohorts and fixed clinical samples. Even when purified cell types are available, tissue dissociation and preservation conditions can cause non-biological alterations in gene expression that obscure downstream analyses18,19. While mathematical separation of bulk tissue RNA profiles into cell type-specific transcriptomes can potentially overcome these problems22–28, the accuracy of this technique on real tissue samples remains unclear. We therefore set out to evaluate whether a signature matrix, consisting of highly optimized marker genes, can be used to faithfully reconstruct cell type-specific transcriptome profiles from non-disaggregated tissue samples, including fresh/frozen and fixed tumors (Fig. 3a).
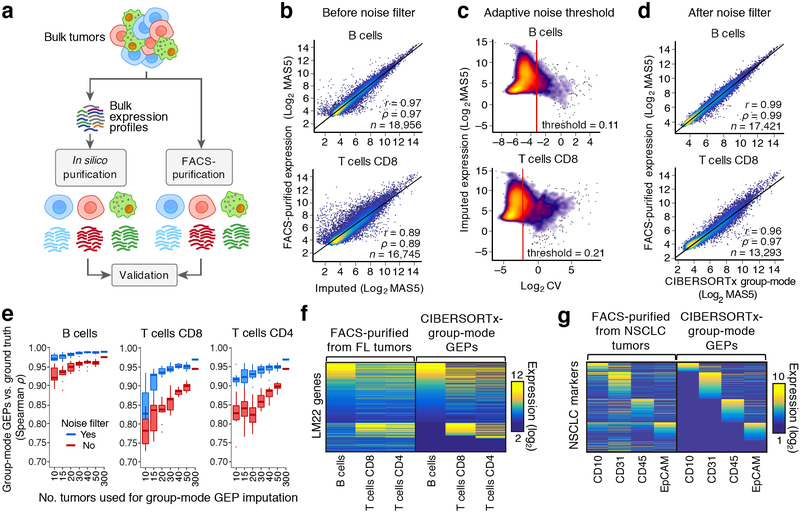
Figure 3.
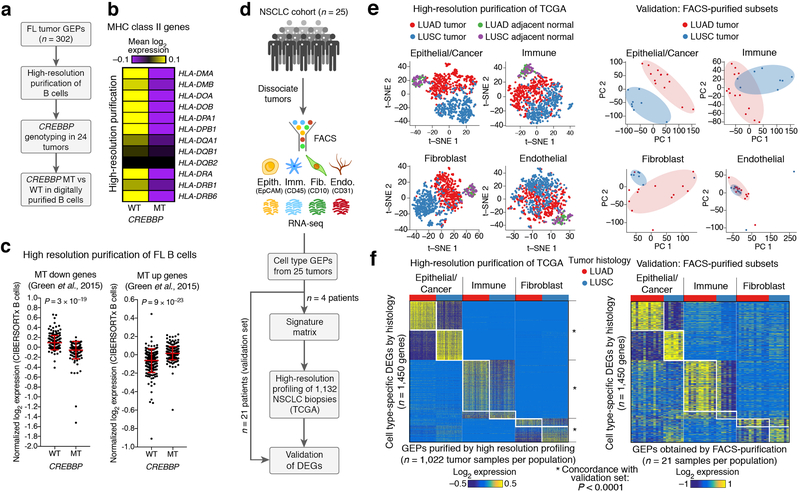
Purification of representative cell type-specific transcriptome profiles from a group of specimens. (a) Approach for in silico purification and validation of group-mode cell type-specific GEPs. (b) Scatterplots comparing genome-wide expression profiles of mathematically purified (x-axis) and FACS-purified (y-axis) B cells and CD8 T cells from FL lymph nodes. (c) Scatterplots showing the predicted expression level of each gene in panel b (y-axis) as a function of its uncertainty, as captured by the geometric coefficient of variation (c.v.) (x-axis). (d) Same as b, but after applying an adaptive noise filter based on the transcriptome-wide distribution of c.v. values for each cell type. Concordance in b,d was determined by applying Pearson correlation (r), Spearman correlation (ρ), and linear regression (diagonal line) to genes with detectable expression on both platforms. (e) Analysis of the impact of sample size on group-mode gene expression purification for three FL tumor cell subsets, as assessed by Spearman correlation against corresponding FACS-purified GEPs (“ground truth”) in log2 space. For each sample size, tumors were subsampled without replacement from a larger cohort (n = 302) 10 times, and the results are shown with and without adaptive noise filtration. Data are presented as boxplots (center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers). (f) Heat map comparing imputed and ground truth expression profiles for three FL immune subsets, with LM22 genes as rows and immune cell types as columns. Genes that were not predicted to be expressed or that were removed by adaptive noise filtration are colored navy blue. (g) Same as f, but for immune (CD45+), epithelial/cancer (EpCAM+), and stromal (CD10+ or CD31+) subpopulations imputed from bulk RNA-seq profiles of 26 NSCLC tumors. Ground truth GEPs (y-axis) were obtained from FACS-purified populations.
We began by profiling 302 fresh/frozen primary tumor biopsies from patients with untreated follicular lymphoma (FL) and tested a common approach25 in which cell type proportions are used to infer a single representative GEP for each cell type from a group of mixture samples (Methods). Since B cells, CD8 T cells, and CD4 T cells comprise the vast majority of FL tumor cellularity and can be readily purified by FACS29, we focused on these three subsets to assess the accuracy of the approach. To enumerate FL immune proportions, we applied LM2215, a microarray-derived signature matrix for distinguishing 22 human hematopoietic cell subsets in bulk tissues, including tumors30,31. We started by examining B cells and CD8 T cells as examples of highly abundant and less abundant cell types in FL lymph nodes (>50% versus ~5–10%, respectively15). Although imputed and FACS-purified cell type transcriptomes were reasonably well correlated, considerable noise distorted expression estimates of many genes (Fig. 3b). We therefore developed an adaptive noise filter to eliminate unreliably estimated genes for each cell type (Fig. 3c). After implementing this novel strategy within CIBERSORTx, we observed consistently improved correlations between in silico purified transcriptomes and those from FACS-purified cells (Fig. 3d, Supplementary Fig. 6a).
We next investigated key factors that influence the accuracy of transcriptome purification (Fig. 3e, Supplementary Fig. 6). Using the set of 302 FL GEPs, we observed a predictable relationship between the number of tumors profiled and the accuracy of transcriptome imputation across B cells, CD4 T cells, and CD8 T cells (Fig. 3e, Supplementary Fig. 6b). The largest gains were achieved when analyzing at least 4–5 fold more mixture samples than cell types (Fig. 3e). Nevertheless, our filtration scheme uniformly improved performance over a previous approach25 irrespective of cohort size, the number of cell types, and overall cell type abundance (Fig. 3e, Supplementary Fig. 6b-e,g). We also observed favorable performance for resolving previously identified markers of cell identity and additional cell type transcriptomes (Fig. 3f,g, Supplementary Fig. 6d-h). As expected, the fraction of recovered genes after adaptive filtration was proportional to both the number of evaluated samples and the proportion of each cell subset (Supplementary Fig. 6h). Moreover, the use of different single cell-derived signature matrices did not significantly impact results, provided that identical cell types were interrogated (Supplementary Fig. 5c).
Cell type-specific expression purification at high-resolution
Having shown that cell type-specific GEPs can be reliably estimated, we next turned to the problem of inferring cell type-specific differential expression from bulk specimens. Despite the utility of the above technique, it is limited to learning a single representative expression profile for each cell type given a group of mixture samples (i.e., group-mode GEPs, Fig. 1). While useful for comparing two groups of specimens, such profiles are not sample-specific and must be generated for each group of interest in order to study differentially expressed genes (DEGs) between them. Although approaches for sample-level deconvolution have been previously described, they only consider mixtures with two or three cellular components32,33. We therefore developed a framework that generalizes to multiple (>3) components by modeling gene expression deconvolution as a unique non-negative matrix factorization problem with partial observations (Supplementary Fig. 7, Methods). Briefly, our approach attempts to separate a single matrix of mixture GEPs into a set of underlying cell type-specific expression matrices using imputed cell proportions (Fig. 4a,b). Once these expression profiles are obtained, they can be analyzed post hoc to gain insights into sample-level variation and patterns of gene expression for individual cell types of interest. To solve the matrix factorization problem, we implemented a novel divide and conquer algorithm that produces biologically realistic solutions (Supplementary Figs. 7 and 8, Methods).
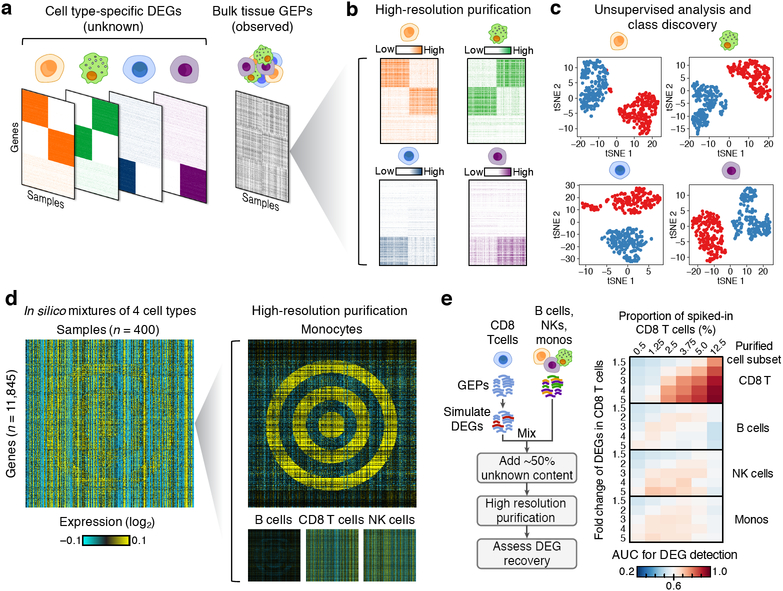
Figure 4.
High-resolution purification of cell type-specific expression from synthetic mixtures. (a) Schematic illustrating a hypothetical example of unobserved (left) and observed (right) expression data for a series of four randomly admixed immune subsets, each with one or two sets of ground truth DEGs (indicated by colored block-like patterns). (b) Results of in silico purification applied to the bulk tissue expression matrix in panel a. (c) t-SNE representation of purified expression matrices in b (n = 400 samples), performed separately for each cell type and color-coded by the class corresponding to each block pattern in b (red = leftmost block, blue = rightmost block). (d) Left: Synthetic GEP matrix of four randomly admixed immune subsets, one of which contains DEGs in the shape of a bullseye (monocytes). Right: Results of in silico purification. (e) Analysis of the sensitivity and specificity of DEG recovery in synthetic mixtures (n = 50) across 30 conditions (six CD8 T cell proportions and five DEG fold changes) when sample classes are known. Left: DEGs with a given fold change were added into the reference profile of CD8 T cells, which were spiked at a predetermined proportion into random mixtures of three other immune subsets. A colon cancer cell line GEP was added into each mixture to simulate ~50% unknown content. After in silico purification, known CD8 T cell DEGs were assessed in each purified cell type by ranking genes by fold change with respect to known DEG classes. The area under the curve (AUC) for DEG recovery was calculated for each combination of fold change and cell type spike-in fraction, and shown as a heat map. Data in panels a-b,d were log2 adjusted and median-centered for each gene prior to rendering the heat maps.
To test the method’s capability for “high-resolution” cell purification (Fig. 1), we created a series of synthetic mixtures, each containing DEGs in one or more cell types. These DEGs were simulated to include overlapping block-like patterns, reminiscent of those seen in real tissues34–36, and non-linear geometries, all of which would be difficult to ascertain by previous computational techniques. Remarkably, the method recovered expected DEG patterns in all tested cases, including an obscured target (“bullseye”) (Fig. 4b-d, Supplementary Fig. 9). Moreover, unlike group mode (Fig. 1), the resulting high-resolution profiles were amenable to standard methods for unsupervised analysis (e.g., Fig. 4c).
Using modeled tumor admixtures, we next evaluated the analytical performance of the method across several parameters, including cell type abundance and the magnitude of differential expression. First, simulated DEGs were ‘spiked’ into CD8 T cell transcriptomes to create two phenotypic classes. These CD8 GEPs were then randomly admixed in silico with three other immune subsets in modeled tumors, and a colon cancer cell line was included to simulate 50% unknown content (Fig. 4e, left). Following high-resolution purification, cell type-specific transcriptomes were grouped into defined DEG classes. Across a broad range of cell spike-in levels and expression fold changes, previously defined DEGs were recovered in CD8 T cells with high sensitivity and specificity (Fig. 4e, right). We observed similar performance in a second experiment, where we simulated melanoma tumors using pooled scRNA-seq data10 and assessed the above parameters in relation to cohort size (Supplementary Fig. 10).
High-resolution profiling of diverse tumor subpopulations
Diffuse large B cell lymphoma (DLBCL) can be classified into two major molecular subtypes based on differences in B cell differentiation states: germinal center-like (GCB) and activated B cell-like (ABC) DLBCL34. Using GEPs of 150 DLBCL lymph node biopsies with previously annotated cell-of-origin subtypes37, we asked whether high-resolution profiling of 10 major leukocyte subsets could correctly attribute known cell-of-origin differences to B cells. Although our approach was blinded to class labels, we identified DLBCL subtype-specific expression differences in malignant B cells that were (1) highly consistent with those of normal GC and activated B cells and (2) ~9-fold more significant, on average, than in bulk DLBCL tumors (Supplementary Fig. 11a-c). Notably, we obtained similar results when repeating this analysis using 10x Chromium-derived signature matrices derived from either peripheral blood or FL tumors, demonstrating the generalizability of our approach (Supplementary Fig. 11d,e).
We then compared our results with two alternative methods: (1) a common strategy for assigning bulk tissue expression patterns to individual cell types based on correlations with cell abundance38, and (2) a previously described technique for imputing cell type-specific DEGs when phenotypic classes and cell type frequencies are known25. In both cases, CIBERSORTx exhibited superior performance, both in relation to cell type specificity and the number of detectable DEGs at a given significance threshold (Supplementary Figs. 12, 13a-b).
FL is the most common indolent Non-Hodgkin lymphoma, and CREBBP mutations in FL tumors are associated with reduced antigen presentation in B cells29,39. Since we originally discovered this association using FACS-purified FL B cells29, we asked whether high-resolution purification could recapitulate this result starting from bulk tumor GEPs and paired tumor genotypes (Fig. 5a). Indeed, after stratifying tumors by CREBBP mutation status (Supplementary Table 3), previously described signatures were detectable in digitally sorted B cell GEPs, including loss of MHC II expression in CREBBP-mutant tumors (Fig. 5b-c). Notably, this result was reproducible across microarray and 10x Chromium-derived signature matrices covering leukocyte subsets derived from distinct biological sources (Fig. 5b-c, Supplementary Fig. 14a,b). Moreover, as observed for DLBCL (Supplementary Fig. 11), the majority of CREBBP mutation-associated genes did not correlate with B cell abundance, hindering their discovery in bulk tissues without deconvolution (Supplementary Fig. 14c).
Figure 5.
High-resolution expression profiling of bulk tumor biopsies. (a–c) Analysis of CREBBP mutation-associated expression changes in B cells from follicular lymphoma (FL) tumors. (a) Schema outlining the application of high-resolution purification to identify CREBBP mutation-associated DEGs in follicular lymphoma B cells. (b) Heat map confirming loss of MHC class II expression in CREBBP-mutant FL B cell GEPs inferred by CIBERSORTx. Expression values were median-centered prior to plotting. (c) Analysis of published gene sets29 associated with lower (left) or higher (right) B cell expression in CREBBP-mutant FL tumors. Scatter plots show the corresponding log2 expression of each gene in digitally sorted B cell GEPs, after median centering and averaging by CREBBP mutation status. Group comparisons in b and c were assessed by a two-sided Wilcoxon signed-rank test. Data are presented as means ± s.d. WT, wildtype (n = 10); MT, mutant (n = 14). (d–f) High-resolution expression profiling of tumor cell subpopulations from non-small cell lung cancer (NSCLC) tumors. (d) Schema for profiling and validating expression signatures of epithelial and cancer cells (Epith., EpCAM+), immune cells (Imm., CD45+), fibroblasts (Fib., CD10+), and endothelial cells (Endo., CD31+) in 1,022 NSCLC tumor and 110 adjacent normal GEPs from TCGA. (e) Left: tSNE plots showing population-specific transcriptional diversity imputed from 1,132 bulk NSCLC GEPs, color-coded to denote tumor histological subtype (LUAD vs. LUSC) and adjacent normal tissues. Plots were created with perplexity set to 10. Right: PCA plots of 21 GEPs from corresponding FACS-purified populations, color-coded by tumor histological subtype. Histological differences are highlighted by ovals. (f) Heat maps showing DEGs identified in epithelial/cancer, immune, and fibroblast populations identified in NSCLC tumors from TCGA (left, n = 1,022 tumor samples) and RNA-seq profiles of corresponding FACS-purified populations from 21 NSCLC patients with LUAD or LUSC (right; Supplementary Table 1). The same genes are shown in both heat maps and are ordered identically. For clarity, a maximum of 300 over- and under-expressed genes are shown for each cell type (all DEGs are provided in Supplementary Table 4). Median centering was applied to each cell population separately. To quantify DEG concordance between CIBERSORTx and the bulk-sorted validation profiles in panel f, we used a Monte Carlo strategy described in Supplementary Note 1. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
To extend our assessment to solid tumors, we obtained surgically-resected primary NSCLC tumor biopsies (n = 26 patients), and for each tumor, generated RNA-seq libraries of four major subpopulations purified by FACS: epithelial/cancer, hematopoietic, endothelial, and fibroblast subsets (Fig. 5d, Supplementary Table 1). After deriving a signature matrix from a subset of four patients, we applied high-resolution profiling to digitally dissect these four populations in bulk tumors from the remaining 22 patients, all of which had RNA-seq profiles available. In a direct comparison against FACS-purified cell populations on matching patient samples, in silico profiles showed strong evidence of expression purification (Supplementary Fig. 13c-f). In addition, CIBERSORTx outperformed other methods6,33 for purifying GEPs of epithelial cells from bulk tumors while also enabling the digital purification of more cell types (Supplementary Fig. 13c-e).
We applied the same signature matrix to resolve epithelial/cancer, hematopoietic, endothelial, and fibroblast GEPs from bulk RNA-seq profiles of 518 lung adenocarcinoma (LUAD) tumors, 504 lung squamous cell carcinoma (LUSC) tumors, and 110 adjacent normal tissues from TCGA40,41 (Fig. 5d). Using t-SNE to visualize the resulting GEPs, we identified striking patterns of histology-specific gene expression for most cell types, including distinct phenotypic shifts in cancer-associated fibroblasts (CAFs) (Fig 5e). Histological differences were far less pronounced in tumor-associated endothelial cells, and adjacent normal tissues clustered together regardless of histology. We compared these results with bulk RNA-seq profiles of FACS-purified NSCLC cell subpopulations from 21 patients with LUAD or LUSC. We observed similar clustering tendencies at the whole transcriptome level and strong concordance in relation to patterns of cell type-specific differential expression between histological subtypes (Fig. 5e,f, Supplementary Table 4, Supplementary Note 1). Our results were further corroborated by histology-specific DEGs and tumor-specific DEGs identified from a recently published scRNA-seq atlas of NSCLC tumors and adjacent normal tissues42 (Supplementary Fig. 15, Supplementary Table 4).
Applications of CIBERSORTx to melanoma
We implemented the set of CIBERSORTx techniques into a comprehensive toolkit (http://cibersortx.stanford.edu). We then explored three potential applications of CIBERSORTx for characterizing cellular heterogeneity in resected tumor biopsies from patients with melanoma. The following techniques were applied in turn: high-resolution expression purification (Fig. 6a-b), group-mode expression purification (Fig. 6c), and enumeration of cell composition across diverse platforms using single-cell reference profiles (Fig. 6d-f).
Figure 6.
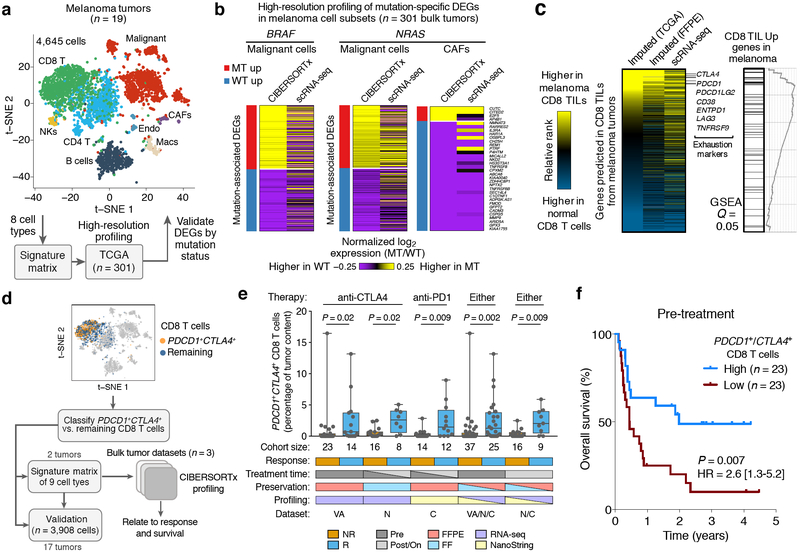
Cellular signatures of melanoma driver mutation status and immunotherapy response. (a,b) High-resolution expression profiling of mutation-associated phenotypic states in distinct tumor cell types. (a) Top: tSNE plot showing 8 major tumor subpopulations profiled from 19 melanomas by scRNA-seq10. Bottom: schema for characterizing and validating context-dependent expression variation in 342 bulk melanoma tumors from TCGA. (b) Heat maps depicting cell type-specific DEGs identified by CIBERSORTx that are associated with BRAF or NRAS driver mutations in melanoma. Expression values were averaged across TCGA tumor samples (CIBERSORTx, left) and single-cell GEPs (scRNA-seq, right) for clarity. Underlying data are provided in Supplementary Fig. 16 and Supplementary Table 4. Only cell types with DEGs detected by CIBERSORTx and with available single-cell validation data are shown. For clarity, a maximum of 300 over- and under-expressed genes are shown for each cell type. CAFs, cancer associated fibroblasts; MT, mutant; WT, wildtype. (c) Left: Heat map showing the absolute difference in gene expression, expressed as ranks, between a normal CD8 T cell reference profile61 and the following three melanoma CD8 TIL GEPs: ‘Imputed (FF)’, a group-mode CD8 TIL GEP imputed from 473 bulk RNA-seq profiles of fresh/frozen (FF) tumors generated by TCGA44; ‘Imputed (FFPE)’, a group-mode CD8 TIL profile predicted from 42 FFPE tumors50; ‘scRNA-seq’, a representative CD8 TIL profile derived from aggregated single-cell data of 19 melanoma patients10. The heat map is ordered by the difference in ranks between the “Imputed (FF)” vector and the normal CD8 T cell GEP. All imputed profiles were processed by adaptive noise filtration. Genes predicted in the FF but not FFPE cohorts (owing to noise filtration) are colored gray in the latter. Selected T cell exhaustion genes are indicated. Right: Gene set enrichment analysis (GSEA)62 of a previously published melanoma CD8 TIL-associated gene set49, assessed relative to the ordering of genes in the heat map. (d) Framework for characterizing the clinical relevance of PDCD1+CTLA4+ CD8 T cells in bulk melanoma tumors using single-cell reference profiles. (e) Estimated levels of PDCD1+CTLA4+ CD8 T cells in bulk tumor expression profiles from melanoma patients who received immunotherapy, stratified by clinical response. VA, Van Allen and colleagues50 (n = 37); N, Nathanson and colleagues55 (n = 24); C, Chen and colleagues20 (n = 26). Statistical comparisons were performed using a two-sided Mann-Whitney test. Data are expressed as boxplots (center line, median; box limits, upper and lower quartiles; whiskers, maximum and minimum values). (f) Kaplan-Meier plot showing differences in overall survival between melanoma patients with high and low estimated levels of PDCD1+CTLA4+ CD8 T cells in biopsies preceding immune checkpoint blockade (pre-treatment). Patients were split by the median estimated fractional abundance of PDCD1+CTLA4+ CD8 T cells into high and low groups, and subsequently pooled across two studies with available overall survival data50,55. Statistical significance was calculated by a two-sided log-rank test. HR, hazard ratio. 95% HR confidence intervals are shown in brackets.
Oncogenic BRAF mutations occur in over half of melanomas and can be inhibited by approved targeted therapies43,44 whereas NRAS mutations occur in approximately half of non-BRAF mutant melanoma tumors but lack such therapies45. Understanding how key mutations influence cellular states could potentially lead to novel treatment strategies. Using single-cell reference profiles from melanomas to build a signature matrix, we applied high-resolution expression purification to dissect 8 major cell types from the transcriptomes of 342 bulk melanoma tumors profiled by TCGA44 (Fig. 6a). Within digitally purified cell subsets, we discovered many significant DEGs within malignant cells and CAFs that distinguish melanomas according to BRAF or NRAS mutation status (Fig. 6b, Supplementary Table 4). We verified these findings using scRNA-seq data from primary melanomas where mutation data were available, allowing us to confirm GEPs associated with BRAF and NRAS genotypes within individual malignant cells and/or CAFs (Fig. 6b, Supplementary Fig. 16).
Tumor-infiltrating CD8 T cells are driven to a state of “exhaustion” by chronic antigen stimulation or by overexposure to inflammatory signals46. Given the importance of these cells for current and emerging cancer immunotherapies47,48, we next used CIBERSORTx to examine expression changes that characterize the exhaustion phenotype. Using LM22, which is derived from healthy peripheral blood leukocytes, we enumerated immune composition in fresh/frozen melanomas profiled by TCGA44. We then performed group-mode expression purification to impute a representative CD8 TIL GEP. By rank-ordering the estimated CD8 TIL GEP against a baseline reference profile of normal peripheral blood CD8 T cells, we confirmed the expression of key exhaustion markers in the inferred CD8 TIL GEP (Fig. 6c). In addition, CD8 TIL-specific genes were consistent with those observed for CD8 TILs isolated from melanomas by single-cell RNA-seq10 and by FACS49 (Fig. 6c). Similar results were obtained when repeating the analysis on FFPE tumors50 (Fig. 6c, Supplementary Fig. 17).
The most effective regimens for metastatic melanoma currently employ checkpoint blockade targeting PD-1 and/or CTLA4 expression on exhausted T cells47. Although a subset of patients achieve durable anti-tumor T cell responses, clinical outcomes remain heterogeneous and effective predictive biomarkers are lacking51. CD8 TILs expressing high levels of PDCD1 (encoding PD-1) or CTLA4 are key targets of these therapies52,53, suggesting that CD8 TILs expressing both markers might correlate with response, as was recently shown in melanoma patients receiving PD1 blockade54. To test this hypothesis, we used single-cell reference profiles of melanoma tumors to build a signature matrix containing PDCD1+/CTLA4+ CD8 T cells along with eight other major tumor cell types (Fig. 6d, Supplementary Figs. 3b, 18a,b). We then applied the signature matrix to interrogate three publicly available melanoma expression datasets (Fig. 6d). These included bulk expression data of FFPE and fresh/frozen melanoma tumors that were profiled by RNA-seq or NanoString50,55. In support of our hypothesis, imputed levels of PDCD1+/CTLA4+ CD8 T cells were significantly associated with response in all three studies (P < 0.05; Fig. 6e). Moreover, the detection of these cells stratified overall survival in this meta-analysis, separated survival curves in individual datasets, and more robustly associated with survival and response than key marker genes and other cell types (Fig. 6f, Supplementary Fig. 18c–f).
Discussion
In this study, we present CIBERSORTx as a new platform for in silico tissue dissection. Key features that distinguish it from previous work include dedicated normalization schemes to suppress cross-platform variation and improved approaches for separating RNA admixtures into cell-type specific expression profiles. In our analysis of peripheral blood, pancreatic islet specimens, and nearly 2,300 malignant tumors, 444 of which were profiled in this work, we found that CIBERSORTx delivers accurate portraits of human tissue heterogeneity using expression profiles derived from disparate sources.
Efforts to define comprehensive cell atlases are now underway1,56. Given the rapid pace of data generation coupled with emerging techniques to combine scRNA-seq datasets57,58, methods to broadly apply single-cell reference maps will become increasingly important, especially in settings where tissue is limited, fixed, or challenging to disaggregate into intact single cells. In our analysis of neoplastic and healthy tissues, we demonstrated that single-cell reference profiles can enable detailed interrogation of tissue composition and that inter-subject heterogeneity is not a major factor influencing results. While significant differences in performance between reference signatures derived from bulk populations and those derived from scRNA-seq data were not observed, the latter has several advantages for CIBERSORTx (Supplementary Note 2). These include (1) the ability to customize signature matrices for nearly any tissue type without the need for complicated antibody panels or cell sorting schemes, and (2) the ability to study poorly understood or unknown transcriptional states at scale.
CIBERSORTx also enables robust molecular profiling of cell subset GEPs from complex tissues, independent of expression profiling platform or tissue preservation state. By incorporating two techniques for expression analysis, we showed that CIBERSORTx outperforms previous methods to facilitate rapid assessment of cell type GEPs when phenotypic categories are known, and high-resolution profiling of expression variation when additional detail is desired. With these features, we anticipate that CIBERSORTx will improve our understanding of heterotypic interactions within complex tissues, including tumor microenvironments, with implications for informing diagnostic and therapeutic approaches that rely on targeting specific cell types.
CIBERSORTx currently requires multiple bulk tissue samples for expression purification. Although further developments are needed to better accommodate smaller sample sizes (e.g., <15), we expect expression purification to be feasible in many situations, particularly given the abundance of publicly available tissue GEPs and the affordability of bulk RNA sequencing. Second, while the fidelity of cell reference profiles remains an important consideration for deconvolution applications3,15, single-cell RNA-seq can mitigate this issue, as shown in this work. Finally, like its predecessor59,60, we hypothesize that the algorithmic principles underlying CIBERSORTx are likely to generalize to other species and genomic data types. Future studies will be needed to formally demonstrate these possibilities.
In summary, CIBERSORTx represents a broadly applicable framework for decoding cellular heterogeneity in complex tissues. This strategy can be used to “digitally gate” cell subsets of interest from single-cell transcriptomes, profile the identities and expression patterns of these cells in cohorts of bulk tissue GEPs (e.g., fixed specimens from clinical trials), and systematically determine their associations with diverse metadata, including genomic features and clinical outcomes. CIBERSORTx should therefore have utility for increasing the statistical power for biological discovery. Given the versatility of the approach and its potential for seamless integration with other techniques, we envision that in silico cytometry will enhance the analysis of multicellular systems in humans, mice, and other metazoans.
Online Methods
Additional details are described in Supplementary Note 1.
Human subjects.
All patient samples in this study were collected with informed consent for research use and were approved by the Stanford Institutional Review Board in accordance with the Declaration of Helsinki. For a patient with metastatic NSCLC treated with an immune checkpoint inhibitor (Pembrolizumab, Merck), peripheral blood was obtained on the first day of treatment prior to infusion (Fig. 2a; NCT00349830 and NCT02955758). Fresh tumor biopsies from patients with early stage NSCLC were obtained during routine primary surgical resection (Figs. 3g and 5, Supplementary Fig. 13c-f). Fresh or frozen surgical biopsies of follicular lymphoma tumors were obtained from previously untreated FL patients enrolled in a phase III clinical trial (NCT0001729063), as well as from patients seen as part of the Stanford University Lymphoma Program Project (NCT00398177; Figs. 3b-f, 5a-c, Supplementary Figs. 6a-c, 14). Whole blood samples from 12 healthy adult donors were obtained from the Stanford Blood Center (Fig. 2b,e and Supplementary Figs. 1d,k,l, 2a,d).
External datasets.
Full details of each dataset9,10,17,20,21,37,42,50,55,64–69, including data type, sample type, source, and normalization approach, are available in Supplementary Table 1. Briefly, next generation sequencing datasets were downloaded and analyzed using the authors’ normalization settings unless otherwise specified; these consisted of transcripts per million (TPM), reads per kilobase of transcript per million (RPKM), or fragments per kilobase of transcript per million (FPKM) space. For analyses in log2 space, we added 1 to expression values prior to log2 adjustment. Affymetrix microarray datasets were summarized and normalized as described in ‘Gene expression profiling – Microarrays’ (Supplementary Note 1), using RMA in cases where bulk tissues and ground truth cell subsets were profiled on the same Affymetrix platform, and otherwise using MAS5 normalization. NanoString nCounter data were downloaded from the supplement of Chen et al.20 and analyzed with batch correction in non-log linear space, but without any additional preprocessing.
Two publicly available PBMC datasets from healthy donors profiled by Chromium v2 (5’ and 3’ kits) were downloaded (Supplementary Table 1) and preprocessed as described in ‘Gene expression profiling – Single-cell RNA-seq’ (Supplementary Note 1), with the following minor modifications. During quality control, we excluded cells with >5000 expressed genes for 5’ PBMCs, >4000 expressed genes for 3’ PBMCs, and <200 expressed genes for both datasets. Seurat “FindClusters” was applied on the first 20 principal components, with the resolution parameter set to 0.6. Cell labels were assigned as described above. In addition, myeloid cells were defined by high CD68 expression, megakaryocytes by high PPBP expression, and dendritic cells by high FCER1A expression.
For the 3’ FL signature matrix in Supplementary Figs. 11d, and 14a-b, publicly available 10x Chromium v2 scRNA-seq data (3’ kit)70 were downloaded (Supplementary Table 1) and preprocessed as described for the 10x PBMC signature matrices above, but with the following differences. Seurat “FindClusters” was applied on the first 10 principal components, with the resolution parameter set to 0.6. Cell labels were assigned based on the following canonical marker genes (MS4A1 = B cells; CD3E, CD8A and CD8B = CD8 T cells; CD3E and CD4 = CD4 T cells).
Single-cell signature matrix construction.
Expression data from input datasets (Supplementary Table 1) were summarized as described above. Given the variability in cell type representation and the inherent noise in scRNA-seq data, several techniques have been developed to address stochastic dropout, impute cell-level scaling factors, and model technical and biological noise components in single-cell differential expression analysis71–73. Although useful for defining single cell phenotypes, we did not observe any significant gains in deconvolution performance when applying such techniques after cell labels were assigned (data not shown). Since the discovery of cell subpopulations in scRNA-seq data was not the focus of this work, we limited our pre-processing steps to the following approach, which performed well in our experiments. Single-cell expression values were first normalized to TPM and divided by 10 to better approximate the number of transcripts per cell10. For each cell phenotype, genes with low average expression (<0.75 transcript per cell) in log2 space were then set to 0 as a quality control filter. Although an expression threshold of <0.75 was used in this work, downstream results remained comparable when using modestly different thresholds (data not shown). Owing to sparser gene coverage relative to SMART-Seq2, this filter was not applied to data generated by 10x Chromium. For each cell type represented by at least 3 single cells, we selected 50% of all available single-cell GEPs using random sampling without replacement (fractional sample sizes were rounded up such that 2 cells were sampled if only 3 were available). We then aggregated the profiles by summation in non-log linear space and normalized each population-level GEP into TPM. This process was repeated in order to generate 5 aggregated transcriptome replicates per cell type. Unless stated otherwise, scRNA-seq and bulk RNA-seq signature matrices were generated as described previously15 using the following parameters: minimum number of genes per cell type = 300, maximum number of genes per cell type = 500, q-value <0.01 for differential gene expression, and no quantile normalization.
All single cell-derived signature matrices are available in Supplementary Table 2.
Overview of CIBERSORTx analytical framework.
Introduction.
A number of computational methods have been proposed to infer cell type abundance, cell type-specific GEPs, or both from bulk tissue expression profiles2–8. These methods generally assume that biological mixture samples can be modeled as a system of linear equations, where a single mixture transcriptome m with n genes is represented as the product of H and f, where H represents an n × c cell type expression matrix consisting of expression profiles for the same n genes across c distinct cell types, and f represents a vector of size c, consisting of cell type mixing proportions.
To infer cell type abundance using this linear model within CIBERSORTx, let M be an n × k matrix with n genes and k mixture GEPs, let matrix B be a subset of H containing discriminatory marker genes for each of the c cell subsets (i.e., signature or basis matrix15,74,75), and let M’ be the subset of M that contains the same marker genes as B. Given M’ and B, the following equation can then be used to impute F, a c × k fractional abundance matrix with columns [f1,f2,…,fk]:
| (1) |
where Fi,j ≥ 0 for all i, j, the system is overdetermined (i.e., n > c), and expression data in M’ and B are represented in non-log linear space76. (Note that Mi,• and M•,j denote row i and column j of matrix M, respectively). Many methods either normalize F or impose an additional constraint on F such that for each mixture sample, the inferred mixing coefficients sum to one, allowing F to be directly interpreted as cell type proportions (with respect to the cell subsets in B)3. We previously introduced CIBERSORT as a method to estimate F using an implementation of ν-support vector regression, a machine learning technique that is robust to noise, unknown mixture content, and collinearity among cell type reference profiles15. CIBERSORT was used to impute F in this work, and within this imputation workflow, the batch correction scheme described below was used for all cross-platform analyses, unless stated otherwise (Supplementary Table 1).
Group-mode expression purification.
The fractional abundance matrix F can be determined for the bulk GEP matrix M, either through expression deconvolution as described above2,3 or with prior empiric knowledge of the compositional representation of cell types within the bulk specimen (e.g., by an automated hematology analyzer, or by flow cytometry)25. Once F is determined for a given M, a representative imputed GEP for each cell type in F can be estimated by solving the following system of linear equations:
| (2) |
where H is a n × c expression matrix of n genes and c cell types, Hi,j ≥ 0 for all i, j, and F is defined as above with the constraint that relative cell fractions sum to one for each mixture sample. Like Equation 1 above, the system should be overdetermined (k > c), with a greater difference between k and c generally leading to improved GEP estimation (Fig. 3e, Supplementary Fig. 6). To ensure biologically realistic estimates of gene expression, we employ non-negative least squares regression (NNLS), an optimization framework to solve the least squares problem with non-negativity constraints. Although NNLS is robust on simple mixtures and toy examples, its performance on more complex mixtures inherent within real tissue samples can be affected by noise, imprecision, and missing data in the linear system15. We therefore developed a series of novel data normalization and filtering techniques to help mitigate these issues (Fig. 3b-d, see ‘Imputation of group-mode expression profiles’ in Supplementary Note 1).
Rationale for high-resolution expression purification.
Despite the utility of Equation 2 for imputing cell type-specific gene expression from bulk tissues (e.g., in silico purifications in Fig. 3), it can only estimate a single representative GEP for each cell type. Therefore, to explore cell type-specific differentially expressed genes (DEGs) between more than one condition of interest, cell type transcriptomes must be re-generated for each condition (e.g., responders versus non-responders to a given therapy, early versus advanced stage disease, etc.). To more broadly address this issue, we extended the model in Equation 2 within a novel method for “high-resolution” in silico cell purification.
Our approach decomposes a matrix of bulk tissue GEPs (i.e., M) into c gene expression matrices of equal size, one for each cell subset in the cell fraction matrix F. Unlike previous approaches22,23,25–28, this method is entirely agnostic to phenotypic class structure and can be formulated as a unique non-negative matrix factorization (NMF) problem with partial observations. Let M and F be defined as above (n × k and c × k matrices, respectively), and assume the latter is estimated by CIBERSORT15. Then, for each gene i, we seek to determine an expression matrix Gi, defined here as a k × c expression matrix of k mixture samples by c cell types. Fixing M and F to solve for G yields the following constrained matrix decomposition problem:
| (3) |
where Gi,j,k ≥ 0 for all i, j, k. Unlike the linear systems in Equations 1 and 2, there are no closed-form solutions for G, which is a 3D n × k × c matrix, and existing numerical techniques are unlikely to yield biologically plausible estimates without additional constraints (e.g., regularization). Since conventional approaches for non-negative matrix factorization77 are not directly applicable to Equation 3, we therefore developed a novel heuristic algorithm to estimate G, depicted schematically in Supplementary Fig. 7 and described below.
Our approach for inferring G makes two distinct assumptions that improve the tractability of the problem while generating biologically plausible solutions. First, we assume each gene can be analyzed independently. Although ignoring gene-gene covariance relationships will likely impact the resolving power for some genes, we found this assumption to be effective in practice (e.g., Figs. 4, 5, 6a-b, Supplementary Fig. 9). Second, for a given gene, we assume that at least some evidence of cell type-specific differential expression is detectable in bulk tissue samples, even if statistically insignificant.
We evaluated this hypothesis using a previously published dataset of cell type-specific DEGs in the human pancreas21. In this dataset, the authors analyzed 5 pancreatic endocrine islet cell types (ranging from ~5% to 44% median fractional abundance within whole islets) for DEGs exhibiting >1.2 fold-change between non-diabetic normal subjects (n = 6) and patients with type 2 diabetes mellitus (T2D; n = 4)21. Importantly, these DEGs were identified by scRNA-seq profiling21, allowing us to test for evidence of differential expression in bulk islets reconstructed in silico.
Consistent with our hypothesis, when grouped by known phenotypic classes (non-diabetic versus T2D), 98% of DEGs associated with T2D showed at least some concordant change in the expected orientation in reconstructed islets (Supplementary Fig. 8a). This striking concordance suggested that cell type-specific DEGs might be discernible in bulk tissues without prior knowledge of phenotypic class labels. Indeed, when we independently ordered each gene in reconstructed islets by expression levels, and split each vector evenly into high and low expression groups (i.e., 50th percentile split), diabetic patients were skewed to low or high expression for 80% of previously defined cell type-specific DEGs. Among these genes, the enrichment direction matched the orientation of the known fold change in 99% of cases (Supplementary Fig. 8b). We therefore conclude that variation in bulk gene expression data can be leveraged to infer latent phenotypic class structure in the underlying cell subpopulations.
Pseudocode for high-resolution expression purification.
Given these foundational assumptions along with a mixture GEP matrix M and cell type fractional abundance matrix F, we developed a heuristic algorithm for high-resolution purification. An overview of this heuristic is outlined in the text below, with a corresponding graphical summary in Supplementary Figure 7 and additional algorithmic details provided in Supplementary Note 1:
For a given gene i, estimate whether the gene is significantly expressed by at least one cell type (Supplementary Fig. 7c, step 1). If so, proceed to step 2. If not, proceed to the next gene.
Sort the gene’s corresponding bulk mixture vector Mi,• into ascending order (Supplementary Fig. 7c, step 2). Any cell type-specific DEGs with detectable signal in Mi,• will influence this ordering, and the most prominent DEGs are likely to skew to one side of the distribution (e.g., Supplementary Fig. 8b).
Split the vector into two groups using a sliding window strategy (Supplementary Figure 7c, step 3). For each group, impute cell type-specific gene expression coefficients for c cell types (denoted g1 for group 1 and g2 for group 2) (Supplementary Figure 7c, step 3, left). Find the g1, g2 pair that best explains Mi,• (Supplementary Figure 7c, step 3, right).
Perform significance testing to determine whether the gene is statistically different between g1 and g2 for each cell type (Supplementary Figure 7c, step 4).
Refine estimates of g1 and g2 based on the significance of their differential expression (Supplementary Figure 7c, step 5).
Impute smooth expression values for each cell type using a sliding window strategy, and store as matrix Ĝ (Supplementary Figure 7c, step 6).
Adjust smooth expression vectors in Ĝ to maximize their agreement with g1 and g2 (Supplementary Figure 7c, step 7).
Restore the original sample ordering of Ĝ (based on sample ordering in step 2; Supplementary Figure 7c, step 8).
Repeat steps 1 through 8 for all genes.
Within this framework, cell type-specific gene expression vectors are imputed using NNLS (detailed in Supplementary Note 1). In order to capture variation in expression across Mi,•, Equation 2 is iteratively solved on subsets of mixture samples grouped by similar expression values. The size of each subset is governed by a sliding window of length w. In order to satisfy NNLS constraints and to avoid overlapping phenotypic classes, w is bounded by the interval c < w ≤ (k/2), where c denotes the number of cell types and k denotes the number of samples. We observed favorable performance of this approach across a broad range of w values. Nevertheless, given the marginal gains observed with increasingly large w values within our saturation analysis of group-mode expression purification (Fig. 3e, Supplementary Fig. 6), we set w to 4–5 fold greater than c, which balances performance with practical considerations based on our empirical observations. To address potential instability in the linear system and to infer expression coefficients with robust standard errors and confidence intervals, NNLS is run using bootstrapping. Additional details are provided in Supplementary Note 1.
Cross-platform deconvolution.
While several prior studies have applied CIBERSORT to RNA-seq, including to reference phenotypes derived from single-cell transcriptome profiling11,14, our original description of CIBERSORT did not explicitly handle technical variation between the signature matrix and bulk mixture profiles. To the best of our knowledge, there is no previously described approach that can be applied to eliminate technical variation between mixture sample expression profiles (denoted M) and a signature matrix comprised of cell type reference profiles (denoted B) while preserving biological signal. This is because, unlike technical batches of the same sample type, both biological differences and technical batches are inherently conflated in the setting of deconvolution. Since previous techniques for addressing technical dropout and cross-platform normalization in bulk GEP and scRNA-seq data are not directly applicable to this problem57,58,78,79, we developed a novel approach for cross-platform deconvolution (Supplementary Fig. 1). By exploiting the fact that M can be modeled as a linear combination of B, and by estimating M from B (denoted M*), batch correction can be directly applied to M and M*. Our approach comprises two distinct strategies (B-mode and S-mode) to apply gene expression deconvolution across distinct platforms and tissue storage types (e.g., fresh/frozen versus FFPE). A decision tree to guide users in selecting the most appropriate strategy is provided in Supplementary Figure 1a. A comprehensive description of our approach is provided in Supplementary Note 1.
B-mode.
Bulk-mode batch correction (i.e., B-mode) removes technical differences between a signature matrix derived from bulk sorted reference profiles (e.g., bulk RNA-seq or microarrays) and an input set of mixture samples (Supplementary Fig. 1c). The technique can also be applied to signature matrices derived from scRNA-seq platforms, provided that transcripts are measured analogously to bulk mixture GEPs (e.g., full-length transcripts without UMIs profiled by SMART-Seq2). Given a set of mixture samples M, the approach creates a series of estimated mixture samples M*, where the latter consists of a linear combination of imputed cell type proportions in M along with the corresponding signature matrix profiles (in non-log linear space). Although the strategy is general and can flexibly accommodate different deconvolution and batch correction methods, we used ComBat79 (an empirical Bayesian method) to eliminate technical variation between M and M* after log2-adjustment. Once cross-platform variation has been minimized, cell proportions are re-estimated using the adjusted mixture samples in non-log linear space. In addition to cell type enumeration, this approach can also be applied to cell type-specific gene expression purification, allowing for down-weighting of genes whose expression levels are low or absent from the collection of cell types in the signature matrix. An absolute minimum of 3 mixture samples is required to perform the batch correction procedure, though at least 10 is recommended. We found that application of this strategy to LM22, a microarray-derived signature matrix, results in improved deconvolution performance across multiple datasets and platforms (Supplementary Fig. 2d).
S-mode.
Despite the benefits of the above technique, deconvolution may fail when excessive technical variation is present (e.g., when cell types that are expected to be present are observed to “drop out” in bulk GEP samples after deconvolution). In this study, we observed this phenomenon when using signature matrices derived from 10x Chromium without batch correction (Fig. 2b, Supplementary Fig. 1d,k). This discrepancy was not surprising given the major differences in transcriptome representation between UMI-based and 3’/5’-biased methods, such as 10x Chromium, and those that capture full transcripts without UMIs, such as SMART-Seq2 (Figure S3E in Ziegenhain et al.16). Because B-mode requires an initial round of fractional abundance estimates prior to normalization, it inherently cannot address cellular dropout and is insufficient to overcome excessive variation (Supplementary Fig. 1d). We therefore developed a second strategy for single-cell batch correction (i.e., S-mode), tailored for signature matrices derived from droplet-based or UMI-based scRNA-seq techniques, including 10x Chromium, but also applicable to other challenging datasets (Supplementary Fig. 1b).
Like B-mode, the primary objective of S-mode is to obtain cell frequencies from a set of mixture GEPs, while minimizing technical variation. However, unlike B-mode, S-mode directly adjusts the signature matrix, rather than the mixture matrix. We found this strategy to deliver superior performance on datasets with considerable technical variation (e.g., Supplementary Fig. 1d). Details of S-mode are provided in Supplementary Note 1 and validation data are shown in Supplementary Fig. 2a-c.
Software implementation and website.
Similar to its predecessor, CIBERSORTx was developed within a web framework with its back-end based on R and PHP and hosted at http://cibersortx.stanford.edu. This web framework minimizes inherent dependencies on specific hardware, software packages and libraries, and file-system attributes. Users are presented with a detailed guide employing several step-by-step Tutorials, and allowing the recreation of key figures in this work, including for each step depicted in Figure 1. Through this interface, CIBERSORTx allows users to process gene expression data representing a bulk admixture of different cell types, along with (1) a signature gene file that enumerates the genes defining the expression profile for each cell type of interest. For the latter, users can either use existing/curated signature matrices for reference cell types, or can create custom signature gene files by providing the reference gene expression profiles of pure cell populations. Specifically, to create a custom signature gene matrix, users can provide single-cell RNA sequencing data or data from bulk sorted samples, along with the phenotypic identities of single cell types or cell populations of interest.
Given these input files, CIBERSORTx allows (2) imputation of the fractional representations of each cell type present in the mixture, similar to its predecessor. However, unlike CIBERSORT, CIBERSORTx now supports deconvolution from bulk RNA-seq data by implementing the critical batch correction methods described above. CIBERSORTx also allows imputation of GEPs for individual in silico purified cell-types in two distinct modes as described above (i.e., (3) group-mode and (4) high-resolution). The resulting imputed cell fractions and imputed cell type-specific GEPs are then rendered as heat maps, tables, stacked bar plots for visualization and downloading. In addition, customizable t-SNE plots are automatically generated for high-resolution purification results.
The interactive CIBERSORTx user interface is powered by the jQuery JavaScript library and various open source libraries (including phpMailer, idiorm, blueimp jQuery-File-Upload, DataTables, phpExcel and mPDF), with the graphical user interface of the website powered by Twitter Bootstrap 2.3.2 and R Shiny. The site runs on an Apache server on a virtual machine and stores user and job data in a MySQL database. However, users have complete control over their data and can delete them at will. Each user’s environment includes example datasets used for benchmarking, tutorials for the use of CIBERSORTx and preparation of input data, and other example files.
Statistical analysis.
Linear concordance between known and predicted cell-type features (i.e., proportions or GEPs) was determined by Pearson correlation (r), Spearman correlation (rho), or linear regression (R2), as indicated, and a two-sided t test was used to assess whether the result was significantly nonzero. Lin’s concordance correlation coefficient (CCC) was determined by comparing predicted and expected log2-adjusted expression profiles using the CCC function from the R package, DescTools. When data were normally distributed, group comparisons were determined using a two-sided t test with unequal variance or a paired t test, as appropriate; otherwise, a two-sided Wilcoxon test was applied. Multiple hypothesis testing was performed using the Benjamini and Hochberg method unless stated otherwise. Results with P < 0.05 were considered significant. Statistical analyses were performed with R and Prism v7 (GraphPad Software, Inc.). The investigators were not blinded to allocation during experiments and outcome assessment. No sample-size estimates were performed to ensure adequate power to detect a pre-specified effect size.
Code availability.
CIBERSORTx v1.0 was used to generate the results in this work and is freely available for academic research use at http://cibersortx.stanford.edu.
Data availability.
All expression datasets analyzed in this work, including accession codes, file names, and web links (if available), are listed in Supplementary Table 1. Expression data generated in this study are available at http://cibersortx.stanford.edu and through the Gene Expression Omnibus with accession code GSE127472.
Supplementary Material
Acknowledgements
We are grateful to R. Levy, S.K. Plevritis, B. Chen, B. Nabet, and M. Matusiak for assistance with this study. This work was supported by grants from the National Cancer Institute (A.M.N., R00CA187192; A.A.A., U01CA194389; A.A.A./.M.D., R01CA188298; S.K.P., U01CA154969), the Stinehart-Reed foundation (A.M.N., A.A.A.), the Stanford Bio-X Interdisciplinary Initiatives Seed Grants Program (IIP) (A.M.N.), the Virginia and D.K. Ludwig Fund for Cancer Research (A.M.N.), the US Department of Defense (A.M.N., W81XWH-12–1-0498), the V Foundation for Cancer Research (A.A.A.), the Leukemia and Lymphoma Society (A.A.A.), the Damon Runyon Cancer Research Foundation (A.A.A.), and the American Society of Hematology (A.A.A.).
Footnotes
Competing Interests
A.M.N. has patent filings related to expression deconvolution and cancer biomarkers and has served as a consultant for Roche, Merck, and CiberMed. A.A.A. has patent filings related to expression deconvolution and cancer biomarkers and has served as a consultant or advisor for Roche, Genentech, Janssen, CiberMed, Pharmacyclics, Gilead, and Celgene. M.D. has patent filings related to cancer biomarkers and has served as a consultant for Roche, Novartis, CiberMed, and Quanticel Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Wagner A, Regev A & Yosef N Revealing the vectors of cellular identity with single-cell genomics. Nat Biotech 34, 1145–1160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen-Orr SS & Gaujoux R Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol 25, 571–578 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman AM & Alizadeh AA High-throughput genomic profiling of tumor-infiltrating leukocytes. Curr Opin Immunol 41, 77–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aran D, Hu Z & Butte AJ xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biology 18, 220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Racle J, de Jonge K, Baumgaertner P, Speiser DE & Gfeller D Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife 6, e26476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quon G, et al. Computational purification of individual tumor gene expression profiles leads to significant improvements in prognostic prediction. Genome Medicine 5, 29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelova M, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biology 16, 64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becht E, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biology 17, 218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puram SV, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 171, 1611–1624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baron M, et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lappalainen T & Greally JM Associating cellular epigenetic models with human phenotypes. Nat Rev Genet 18, 441–451 (2017). [DOI] [PubMed] [Google Scholar]
- 13.He Z, et al. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat Neurosci 20, 886–895 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Schelker M, et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nature Communications 8, 2032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziegenhain C, et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Molecular Cell 65, 631–643.e634 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas, N. Genomic Classification of Cutaneous Melanoma. Cell 161, 1681–1696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvinge H, et al. Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proceedings of the National Academy of Sciences 111, 16802–16807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadić E, Moniz RJ, Huo Y, Chi A & Kariv I Effect of cryopreservation on delineation of immune cell subpopulations in tumor specimens as determined by multiparametric single cell mass cytometry analysis. BMC Immunology 18, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen P-L, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discovery (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segerstolpe A, et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab 24, 593–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaujoux R & Seoighe C CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics 29, 2211–2212 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Liebner DA, Huang K & Parvin JD MMAD: microarray microdissection with analysis of differences is a computational tool for deconvoluting cell type-specific contributions from tissue samples. Bioinformatics 30, 682–689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffitt RA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 47, 1168–1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen-Orr SS, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods 7, 287–289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong Y, Wan YW, Pang K, Chow LM & Liu Z Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC Bioinformatics 14, 89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuckerman NS, Noam Y, Goldsmith AJ & Lee PP A self-directed method for cell-type identification and separation of gene expression microarrays. PLoS Comput Biol 9, e1003189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onuchic V, et al. Epigenomic Deconvolution of Breast Tumors Reveals Metabolic Coupling between Constituent Cell Types. Cell Reports 17, 2075–2086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green MR, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proceedings of the National Academy of Sciences 112, E1116–E1125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentles AJ, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 21, 938–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorsson V, et al. The Immune Landscape of Cancer. Immunity 48, 812–830.e814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn J, et al. DeMix: deconvolution for mixed cancer transcriptomes using raw measured data. Bioinformatics 29, 1865–1871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, et al. Transcriptome Deconvolution of Heterogeneous Tumor Samples with Immune Infiltration. iScience 9, 451–460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Golub TR, et al. Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring. Science 286, 531–537 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439, 353–357 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Lenz G, et al. Stromal Gene Signatures in Large-B-Cell Lymphomas. New England Journal of Medicine 359, 2313–2323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitney AR, et al. Individuality and variation in gene expression patterns in human blood. Proceedings of the National Academy of Sciences 100, 1896–1901 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Y, et al. CREBBP Inactivation Promotes the Development of HDAC3-Dependent Lymphomas. Cancer Discovery 7, 38–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med (2018). [DOI] [PubMed] [Google Scholar]
- 43.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Akbani R, et al. Genomic Classification of Cutaneous Melanoma. Cell 161, 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtin JA, et al. Distinct Sets of Genetic Alterations in Melanoma. New England Journal of Medicine 353, 2135–2147 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Wherry EJ & Kurachi M Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Postow MA, Callahan MK & Wolchok JD Immune Checkpoint Blockade in Cancer Therapy. Journal of Clinical Oncology 33, 1974–1982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson Ana C., Joller N & Kuchroo Vijay K. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 44, 989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baitsch L, et al. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. The Journal of Clinical Investigation 121, 2350–2360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redman JM, Gibney GT & Atkins MB Advances in immunotherapy for melanoma. BMC Medicine 14, 20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kvistborg P, et al. Anti–CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Science Translational Medicine 6, 254ra128–254ra128 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Daud AI, et al. Tumor immune profiling predicts response to anti–PD-1 therapy in human melanoma. The Journal of Clinical Investigation 126, 3447–3452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nathanson T, et al. Somatic Mutations and Neoepitope Homology in Melanomas Treated with CTLA-4 Blockade. Cancer Immunology Research (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao J, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology 36, 411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haghverdi L, Lun ATL, Morgan MD & Marioni JC Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nature Biotechnology 36, 421 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakravarthy A, et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nature Communications 9, 3220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abbas AR, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun 6, 319–331 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levy R, et al. Active idiotypic vaccination versus control immunotherapy for follicular lymphoma. J Clin Oncol 32, 1797–1803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allantaz F, et al. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS One 7, e29979 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 459, 717–721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jourdan M, et al. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood 114, 5173–5181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kiaii S, et al. Follicular lymphoma cells induce changes in T-cell gene expression and function: potential impact on survival and risk of transformation. J Clin Oncol 31, 2654–2661 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 12, 786–795 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tatlow PJ & Piccolo SR A cloud-based workflow to quantify transcript-expression levels in public cancer compendia. Sci Rep 6, 39259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milpied P, et al. Germinal Center Program De-Synchronization and Intra-Patient Heterogeneity in Follicular Lymphoma B-Cells Revealed By Integrative Single-Cell Analysis. Blood 130, 41–41 (2017). [Google Scholar]
- 71.Vallejos CA, Risso D, Scialdone A, Dudoit S & Marioni JC Normalizing single-cell RNA sequencing data: challenges and opportunities. Nat Meth 14, 565–571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stegle O, Teichmann SA & Marioni JC Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet 16, 133–145 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Hicks SC, Townes FW, Teng M & Irizarry RA Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics, kxx053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venet D, Pecasse F, Maenhaut C & Bersini H Separation of samples into their constituents using gene expression data. Bioinformatics 17 Suppl 1, S279–287 (2001). [DOI] [PubMed] [Google Scholar]
- 75.Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z & Clark HF Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One 4, e6098 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong Y & Liu Z Gene expression deconvolution in linear space. Nat Methods 9, 8–9; author reply 9 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Lee DD & Seung HS Algorithms for non-negative matrix factorization. in Proceedings of the 13th International Conference on Neural Information Processing Systems 535–541 (MIT Press, Denver, CO, 2000). [Google Scholar]
- 78.Bacher R, et al. SCnorm: robust normalization of single-cell RNA-seq data. Nature Methods 14, 584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson WE, Li C & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All expression datasets analyzed in this work, including accession codes, file names, and web links (if available), are listed in Supplementary Table 1. Expression data generated in this study are available at http://cibersortx.stanford.edu and through the Gene Expression Omnibus with accession code GSE127472.