Abstract
mRNA has broad potential as a therapeutic. Current clinical efforts are focused on vaccination, protein replacement therapies, and treatment of genetic diseases. The clinical translation of mRNA therapeutics has been made possible through advances in the design of mRNA manufacturing and intracellular delivery methods. However, broad application of mRNA is still limited by the need for improved delivery systems. In this review, we discuss the challenges for clinical translation of mRNA-based therapeutics, with an emphasis on recent advances in biomaterials and delivery strategies, and we present an overview of the applications of mRNA-based delivery for protein therapy, gene editing, and vaccination.
Keywords: mRNA delivery, mRNA vaccine, CRISPR, gene editing, gene therapy, protein replacement, biomaterials, mRNA nanoparticles, lipid nanoparticles, clinical trial
This review addresses the challenges for clinical translation of mRNA-based therapeutics, with an emphasis on recent advances in biomaterials and delivery strategies, and the authors present an overview of the applications of mRNA-based delivery for protein therapy, gene editing, and vaccination.
Main Text
mRNA holds the potential to revolutionize vaccination, protein replacement therapies, and the treatment of genetic diseases. Since the first pre-clinical studies in the 1990s,1 significant progress in the clinical translation of mRNA therapeutics has been made through advances in the design of mRNA manufacturing and intracellular delivery methods.2 The translatability and stability of mRNA as well as its immunostimulatory activity are the key factors to be optimized for specific therapeutic application.3 Increased translation and stability can be affected by many regions of the RNA. mRNA 5′ and 3′ UTRs are responsible for recruiting RNA-binding proteins and microRNAs, and they can profoundly affect translational activity.2, 4 The modification of rare codons in protein-coding sequences with synonymous frequently occurring codons, so-called codon optimization, can result in order-of-magnitude changes in expression levels.5, 6 Modification of the 5′ mRNA cap can also enhance mRNA translation by inhibiting RNA decapping and improving resistance to enzymatic degradation.7 Chemical modification of RNA bases can be used to modify mRNA immunostimulatory activity.8, 9 The importance of immunostimulation can depend on the application,10 and, in some cases, it may actually improve performance, as in the case of vaccines.11
Finally, methods and vehicles for intracellular delivery remain the major barrier to the broad application of mRNA therapeutics.12 With some exceptions, the intracellular delivery of mRNA is generally more challenging than that of small oligonucleotides, and it requires encapsulation into a delivery nanoparticle, in part due to the significantly larger size of mRNA molecules (300–5,000 kDa, ∼1–15 kb) as compared to other types of RNAs (small interfering RNAs [siRNAs], ∼14 kDa; antisense oligonucleotides [ASOs], 4–10 kDa).10, 13 In this review, we discuss the challenges for clinical translation of mRNA-based therapeutics, with an emphasis on recent advances in biomaterials and delivery strategies, and we present an overview of the applications of mRNA-based delivery for protein therapy, gene editing, and vaccination.
Materials for mRNA Delivery
Structural Aspects of Material Design
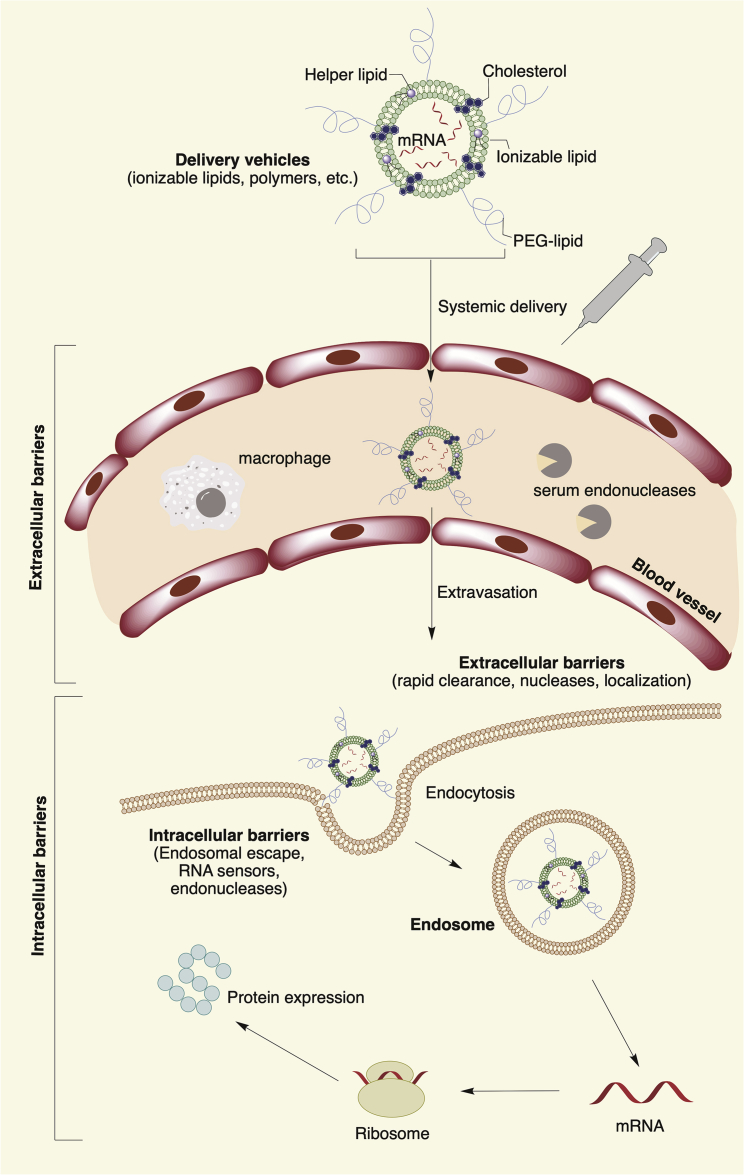
Among the many barriers to function, mRNA must cross the cell membrane in order to reach the cytoplasm (Figure 1). The cell membrane is a dynamic and formidable barrier to intracellular delivery. It is made up primarily of a lipid bilayer of zwitterionic and negatively charged phospholipids, where the polar heads of the phospholipids point toward the aqueous environment and the hydrophobic tails form a hydrophobic core.14 Various ion pumps and ion channels help maintain a negative potential (–40 to –80 mV) across the cell membrane, and they keep the cytoplasmic space negatively charged by controlling the balance of most of the essential metal ions (e.g., K+, Na+, Ca2+, and Mg2+).15 The negative potential across the cell membrane creates a formidable barrier for highly negatively charged mRNA molecules. Unsaturated lipids, especially the cis-double-bonded ones, increase cell membrane fluidity by introducing defects in the membrane structure. The other major components of the bilayer include sterols (∼30% of total lipids). Cholesterol, the major sterol of the lipid bilayer, helps maintain a balance between fluidization and condensation of the lipid bilayer by either creating or filling up bilayer defects.16
Figure 1.
Schematic Representation of Extra- and Intracellular Barriers for mRNA Delivery
Apart from the barrier of cell membrane, mRNA faces degradation by extracellular ribonucleases abundantly present in skin and blood.17, 18 To protect mRNA against degradation by nucleases and shield its negative charge, amine-containing materials are commonly used as non-viral vectors. One of the most developed methods for mRNA delivery is co-formulation into lipid nanoparticles (LNPs).10 LNP formulations are typically composed of (1) an ionizable or cationic lipid or polymeric material (see Materials Used for Non-viral mRNA Delivery), bearing tertiary or quaternary amines to encapsulate the polyanionic mRNA; (2) a zwitterionic lipid (e.g., 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine [DOPE]) that resembles the lipids in the cell membrane; (3) cholesterol to stabilize the lipid bilayer of the LNP; and (4) a polyethylene glycol (PEG)-lipid to lend the nanoparticle a hydrating layer, improve colloidal stability, and reduce protein absorption.19, 20
Although the mechanism of mRNA delivery by the LNPs is not fully understood, it is generally accepted that these multicomponent LNPs are taken up by endocytosis and can electrostatically attach and fuse with the cell membrane using inverted non-bilayer lipid phases.21 Interestingly, LNPs can also be exocytosed, posing a challenge to cellular delivery.22 Initial clathrin-dependent endocytosis, followed by macropinocytosis, has been identified as a common mechanism for LNP delivery inside cells.22, 23 Once inside the cell, LNPs are routed into early endosomes, followed by late endosomes, and finally the lysosomes where the mRNA contents are enzymatically degraded.24 A hypothesis termed the proton sponge effect proposes that a few percentage (1%–2%) of LNPs evade degradation as the ATP-driven gradual acidification of the compartments from pH 6.5 to 5–6 promotes protonation of the residual amines of the LNPs and disrupts the endosomal membrane that leads to the endosomal escape of the mRNA content.25 Other studies indicate that the actual mechanism could be much more complex than the classical proton sponge effect and may depend on fusion with the endosomal membrane and additional factors, such as the endosome size, membrane leakiness, late endosome formation, Rab7A localization on the surface of endosomes, and activation of mTORC1 for downstream signaling for protein synthesis.26, 27
Materials Used for Non-viral mRNA Delivery
Ionizable Lipids
One well-studied class of non-viral mRNA delivery agents includes the cationic or ionizable lipids and lipid-like materials. Cationic lipids bear alkylated quarternary ammonium groups and retain their cationic nature in a pH-independent fashion, while ionizable lipids acquire positive charges by protonation of free amines as pH is lowered. Lipid-like materials bear more hydrophobic side chains than natural lipids. Early studies with cationic lipids, such as N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium chloride (DOTMA), have been reviewed elsewhere28 and are not discussed here.
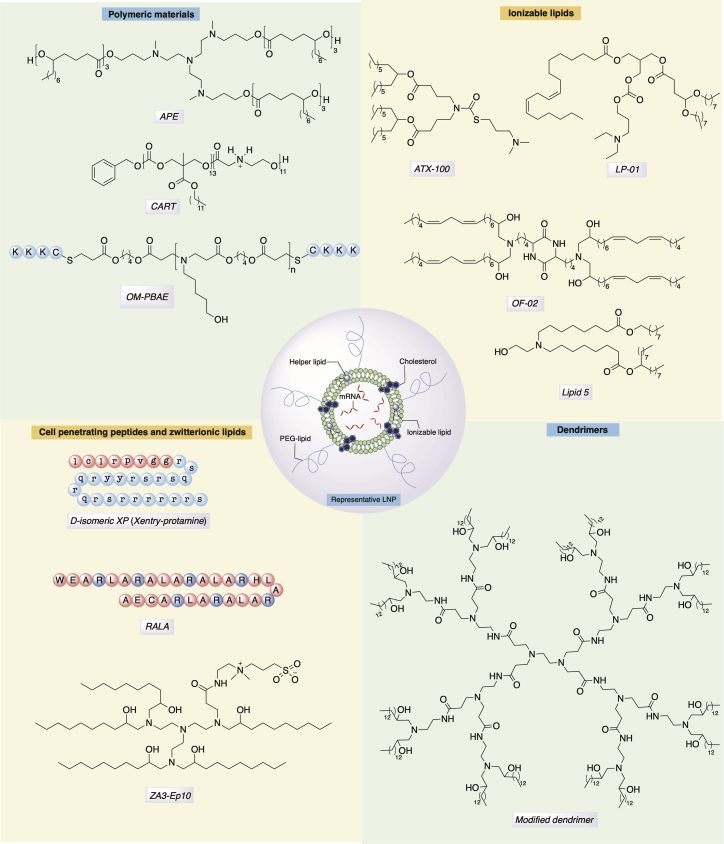
Early work focused on the use of cationic lipids, but more recent work has focused on pH-dependent ionizable materials.29, 30 Recently, the FDA approved the first siRNA drug (patisiran [Onpattro]), which contains an ionizable lipid named Dlin-MC3-DMA (MC3).31 Inspired by the success of LNPs, various groups showed that MC3 can also be used to safely transfect mRNA in order to express therapeutic proteins.32, 33 Researchers at Intellia Therapeutics have reported an ionizable lipid named LP-01 (Figure 2) as part of the LNP formulation for the co-delivery of Cas9 mRNA and single guide RNA (sgRNA) for transthyretin gene, enabling successful editing of the mouse transthyretin gene in the liver.34 In another example, Ramaswamy et al.35 reported successful protein replacement with human recombinant factor IX mRNA in a mouse model of hemophilia B, using an LNP formulation that utilized a proprietary ionizable lipid (representative example shown as ATX-100 in Figure 2) developed by Arcturus Therapeutics for their lipid-enabled and unlocked nucleic acid-modified RNA (LUNAR) delivery platform.35, 36, 37
Figure 2.
Representative Structures of Various Classes of Materials Developed for mRNA Delivery
Researchers from Moderna Therapeutics have reported a biodegradable ionizable lipid (lipid 5, Figure 2), which can deliver mRNA to non-human primates (NHPs).38, 39, 40 The presence of a primary ester on one of the hydrophobic tails of lipid 5 enhanced the liver clearance, while having the terminal alcohol on the head group showed a superior mRNA expression profile. Another diketopiperazine-based ionizable lipid, cKK-E12 (also known as MD1), has been developed for siRNA and mRNA delivery.10, 41 This compound, which has also shown activity in primates, has been used for cancer immunotherapy and genome editing.42, 43, 44 The introduction of unsaturated fatty chains to cKK-E12 structure yielded the lipid named OF-02 (Figure 2), which further increased mRNA expression compared to cKK-E12.13 It has been hypothesized that unsaturated lipid tails similar to linoleic acid introduce fluidity and structural defects that facilitate fusion of the LNP with the cell membrane as well as later endosomal escape.45 The degree of unsaturation is an important aspect, as either more or less than two cis-alkenyl groups led to inferior expression of the target protein.13 Further, a biodegradable ester version of OF-02, named OF-Deg-Lin, was shown to promote protein expression selectively in the spleen, whereas the non-biodegradable OF-02 promoted expression in mouse liver.46
Polymers
Polymeric materials are not as clinically advanced for nucleic acid delivery as ionizable lipids, with few formulations used for the delivery of therapeutic siRNA.47, 48 Relative to lipids, polymeric materials face additional challenges related to polydispersity and clearance or biodegradation for large molecular weight polymers. Low-molecular-weight polyethyleneimine (PEI) modified with fatty chains has been used for siRNA and mRNA delivery to reduce toxicity of high-molecular weight PEI.49, 50, 51, 52 Poly(glycoamidoamine) polymers modified with fatty chains, such as TarN3C10 that contains a tartrate backbone, were shown to be potent in delivering erythropoietin (EPO) mRNA in mice.53 Poly(β-amino)esters (PBAEs) are biodegradable polymers that have been investigated for nucleic acid delivery and were originally developed for DNA transfection.54, 55, 56 Early studies from Zugates et al.57 showed that PBAE-mediated in vitro mRNA transfection was ∼6-fold higher in the absence of serum proteins than in their presence. This led to the development of new PBAEs formulated with PEG-lipid to increase their serum stability, and it allowed for the use of these polymers for in vivo mRNA delivery.58, 59, 60 Recently, hyperbranched PBAEs were utilized to stabilize the nanoformulations designed to enable mRNA delivery to lung epithelium via inhalation.61
Polymethacrylates with amine-bearing side chains,62, 63 polyaspartamides with oligoaminoethylene side chains,64 and polyacrylic acids amidated with tetramine with alternating ethyl-propyl-ethyl spacers65 have also been reported to transfect mRNA, potentially by combining efficient endosomal escape at pH < 5.5 as well as optimal polyplex stability. Recent work by McKinlay and colleagues66, 67, 68 reported self-immolative polycarbonate-block-poly(α-amino)esters (Figure 2), which the authors named charge-altering releasable transporters (CARTs). These polymers were able to release mRNA upon rearrangement followed by degradation at pH 7.4. This was hypothesized to facilitate endosomal escape, producing a diketopiperazine derivative as a by-product. Kowalski et al.69 reported biodegradable amino polyesters (APEs), which could be synthesized with low dispersity from tertiary amino alcohols as initiators in ring-opening polymerization of various lactones, capable of tissue-selective mRNA delivery (Figure 2). Following a similar trend as seen with siRNA carriers, new biodegradable polymers with biocompatible degradation products and enhanced endosomal escape capabilities are expected to emerge for mRNA delivery, which may facilitate clinical translation of these materials.70
Dendrimers
Polyamidoamine (PAMAM) or polypropylenimine-based dendrimers have been extensively studied for gene delivery.71 Khan et al.72 synthesized fatty chain-modified PAMAM dendrimers for siRNA delivery, which were subsequently used by Chahal et al.73, 74 to develop a single-dose, adjuvant-free, intramuscularly delivered, self-replicating mRNA vaccine platform to express antigens for Ebola, H1N1 influenza, Toxoplasma gondii, and Zika. In another report, Islam et al.75 utilized a modified PAMAM (generation 0) dendrimer co-formulated with poly(lactic-co-glycolic acid) (PLGA) and ceramide-PEG in a polymer-lipid hybrid nanoparticle to transfect phosphatase and tensin homolog (PTEN) mRNA in vivo. As dendrimer-repeating units branch out in a tree-like shape, their enzymatic biodegradation may be hindered due to steric factors, leading to toxicity stemming from the accumulation of these materials in tissue. We expect advances in biodegradable dendrimers may allow for increased usage of these materials.76
Cell-Penetrating Peptides
Cell-penetrating peptides (CPPs) have been studied for their potential as vectors for intracellular delivery of nucleic acid delivery.77 Although their internalization mechanisms are not fully understood, it is hypothesized that CPPs may promote clustering of the negatively charged glycosaminoglycans on the cell surface, which in turn triggers macropinocytosis and lateral diffusion or directly disrupts the lipid bilayer.78, 79 A CPP with arginine-rich amphipathic RALA sequence repeats (Figure 2) was reported to function as an mRNA vector to dendritic cells and evoke T cell immunity in vivo.80 Bell et al.81 recently reported a D-amino acid-based truncated protamine fused to the CPP Xentry, named Xentry-protamine (Figure 2), which enabled transfection of cystic fibrosis transmembrane regulator (CFTR) mRNA into epithelial cells in the presence of a transfection enhancer, Toll-like receptor (TLR) antagonist E6446. However, CFTR protein expression attained via the use of this CPP was less than that obtained by transfection reagent (MessengerMax).81
Other Materials
Recently, Miller et al.82 reported a combination of cationic and zwitterionic lipids, reminiscent of cationic and helper lipids in usual LNP formulations, which the authors termed zwitterionic amino lipids (ZALs). Among the ZALs, ZA3-EP10 (Figure 2) was the most potent in co-delivering Cas9 mRNA and sgRNA.82 Moreover, virus-like particles (VLPs) coated with CPPs83 and Zr-based metal-organic framework (Zr-MOF) functionalized with polycationic ethanolamine-conjugated poly(glycidyl methacrylate) were also reported to be able to transfect mRNA.84 Kim et al.85 demonstrated that self-assembled mRNA nanoparticles (mRNA-NPs) produced via rolling circle transcription can be coated with ionizable lipid or polymer that better protects mRNA from degradation and prolongs protein expression.
Biodegradability and Targeting Issues with Non-viral Vectors
Systemically delivered LNPs carrying mRNA face multiple barriers to delivery (Figure 1). The mononuclear phagocytic system (MPS), especially in the liver and spleen, is a frequent destination for injected nanoparticles, owing to its native role in policing the body for nano-sized infectious agents.86 The kidney filters off naked mRNA or any nanoparticle with hydrodynamic diameter less than 5.5 nm.87 Most of the LNPs are about 100 nm in diameter and large enough to prevent them from escaping the MPS and reaching other organs of interest.88 The liver, which forms a major part of the MPS, has a fenestrated vasculature and contains phagocytic cells such as Kupffer cells, which retain cationic LNPs. Furthermore, large cationic LNPs fail to extravasate from the capillaries found in the lungs and cannot be filtered from the bloodstream by the kidney.86 This can lead to the accumulation of delivery materials in the liver, lungs, or other organs.89 Therefore, the clearance and biodegradability of the delivery system components is one consideration when developing mRNA delivery materials.
Ester groups are the most commonly used functional group for enhancing the biodegradability of biomaterials, but the in vivo degradation of different ester bonds can depend on the overall chemistry of the molecules and formulations.90 For example, both LP-01 and lipid 5 (discussed earlier) are reported to clear from the liver rapidly (half-life [t1/2] ∼6 h), compared to DLin-MC3-DMA (t1/2 > 50 h), with comparable if not more protein expression. In certain tissues, the presence of ester functionality may accelerate the degradation of LNPs, potentially even limiting protein expression. For example, OF-Deg-Lin induced protein expression selectively in the spleen, even though it was able to reach the liver cells.46 It was hypothesized that this may be due to the rapid degradation of the LNPs by liver enzymes. Rational design of degradable lipids or polymers based on polyesters or polycarbonates91 could offer better control over the degradation of the delivery systems.
Targeting specific organs or tissues beyond the liver with LNPs has proven more challenging. Some LNPs accumulate or are taken up by the liver via an apolipoprotein E-dependent fashion.92 For APEs, it was found that the core structure of the tertiary amino alcohol of the polymers could play an important role in targeting specific organs.69 Recently, end-capping the acrylate groups on PBAEs with CKKK oligopeptide via Michael addition afforded oligopeptide-modified PBAEs (OM-PBAEs) (Figure 2), which targeted antigen-presenting cells (APCs).93 Such targeting strategies have their limitations and generally rely on high-throughput screening to find successful targeting solutions for specific tissues. Rationally designed targeting approaches, such as including ligands to tissue-specific receptors, are an alternative strategy.94 Recently, Lou et al.95 reported an in vitro active targeting of sialic acid-ended glycoproteins on the surface of dendritic cells, mediated by a 30-amino-acid synthetic peptide with glutamic acid-alanine-leucine-alanine (GALA) repeats conjugated to polyplex carrying EGFP-mRNA. However, any opsonization on the LNPs forming a protein corona can be detrimental to active-targeting strategies, due to masking of the targeting ligands.96 Therefore, new strategies toward targeting specific tissues need to be explored to address clinical challenges.
Therapeutic mRNA Delivery for Protein Therapy and Gene Editing
Protein Therapy
The use of mRNA for the expression of therapeutic proteins holds the potential to treat a wide range of diseases. Therapeutic applications include (1) protein replacement to restore the function of a single protein for rare monogenic diseases; (2) cell reprogramming where mRNA can be used to modulate cell behavior by expressing transcription or growth factors; and (3) immunotherapies where mRNA-encoded transcripts evoke specific immune responses against target cells, e.g., therapeutic antibodies.12 In general, mRNAs designed to express therapeutic proteins are engineered to display low immunogenicity, prolonged stability, and potent translation. Intracellular delivery of mRNA enables the expression of virtually any desired protein inside the host cells and tissues, and it allows the preservation of post-translational modifications of the encoded proteins innate to the host cells.1, 97 This approach can also address formulation and delivery challenges encountered with protein-based drugs, especially those aimed at restoring intracellular and transmembrane proteins.98 Other important features of mRNA-based therapeutics include low risk of insertional mutagenesis, transient production of encoded protein, and cytoplasmic activity of mRNA lowering the cellular barriers for functional delivery.2
Repeated administrations of mRNA therapeutics are required to sustain therapeutic levels of protein. The dosing frequency may depend on the half-life of the protein, its activity, as well as the turnover rate of the target cell. Average protein production half-life from transfected modified mRNA ranges from 50 h in vitro to 7–30 h in vivo, depending on the administration route.99 The Anderson group100 has recently shown that synthetically designed exogenous circular RNAs provide up to a 3-fold increase in the half-life of protein production and potent expression in vitro, as compared to linearly modified mRNA. The majority of mRNA-based protein replacement therapies are currently directed toward the liver, lungs, and heart, owing to relatively efficient methods for mRNA delivery into these tissues. Accessing other organs and cell types may require the continued development of new delivery strategies, including novel biomaterials, active targeting, or different administration methods. In this section, we discuss different delivery strategies and types of carriers used for mRNA-based protein therapy.
Local Transfection
Direct injection of mRNA or mRNA complexes has been investigated for delivery into cardiac tissue, where modified mRNA has been utilized for the expression of growth factors (e.g., vascular endothelial growth factor-A [VEGF-A], insulin-like growth factor 1 [IGF1], epidermal growth factor [EGF], hepatocyte growth factor [HGF], transforming growth factor [TGF]b1, TGFb2, stromal cell-derived factor 1 [SDF-1], fibroblast growth factor 1 [FGF-1], growth hormone [GH], and stem cell factor [SCF]).95, 96 Intramyocardial injection of modified RNA encoding human VEGF-A, complexed with RNAiMAX, has been reported to improve heart function and long-term survival of mice with myocardial infarction.101 Turnbull et al.102 compared intracoronary administration and direct myocardial injection of mRNA formulated into the LNPs (C14-113) in rodents and pigs, showing that the latter led to higher cardiac and lower off-target mRNA expression. In contrast, recent studies have reported that the delivery of naked modified mRNA in a sucrose-citrate buffer or saline could yield higher local protein expression in the heart as compared to mRNA complexed with transfection reagents, including Lipofectamine 2000, jetPEI, RNAiMAX, and Invivofectamine.103, 104 This study suggests that complexation of the mRNA may potentially hinder translation in cardiac tissue and increase the apoptosis of cardiac cells in vivo.
Therapeutic effects of direct transfection of mRNA have also been demonstrated in other tissues. De novo synthesis of elastin in porcine skin after intradermal microinjection of modified mRNA encoding tropoelastin and complexed with Lipofectamine 2000 was recently reported.105 The remarkably long elastin half-life (74 years) presents the potential of this approach to restore the loss of tissue elasticity in congenital or acquired elastin deficiencies.105 Intratumoral delivery of naked mRNA via electroporation, encoding mixed-lineage kinase domain-like protein, was used to boost T cell responses, and it showed marked tumor growth inhibition in several subcutaneous tumor models in mice.106 mRNA-mediated lung delivery of surfactant protein B (SP-B) via intratracheal high-pressure spraying protected mice from respiratory failure.107 Moreover, the delivery of LNP-formulated mRNA directly into the colon was significantly augmented by ultrasound resulting in localized translation of firefly luciferase protein, demonstrating the potential utility of ultrasound-mediated mRNA delivery for gastrointestinal diseases.108
LNPs
As discussed above, LNPs have been widely investigated for their potential as mRNA delivery reagents. Most mRNA protein therapies use liver as a biological factory for the production and secretion of therapeutic proteins. The capacity of producing human EPO (hEPO) in NHPs has been demonstrated using mRNA LNPs formulated with cationic lipid C12-200, ionizable lipid MC3, and Moderna’s degradable amino lipids (lipid 5) at doses as low as 0.25, 0.03, and 0.01 mg/kg, respectively.33, 40, 44 Expression of hEPO could be sustained for over a month when dosed weekly, with a peak expression between 6 and 12 h after LNP administration.40 In all of the above cases, mRNA-containing LNPs were tolerated, and they did not cause liver injury or show signs of inflammation or complement activation in NHPs at the doses tested. In vivo efficacy of mRNA-based protein replacement therapy utilizing systemic delivery of liver-targeting LNPs was demonstrated in pre-clinical models of hemophilia B, liver fibrosis, and metabolic diseases such as ornithine transcarbamylase deficiency (OTCD) and methylmalonic acidemia/aciduria (MMA).39, 44, 109, 110 LNPs containing porphobilinogen deaminase (PBGD) mRNA were used to restore liver metabolic function in large animals.111 Sustained therapeutic efficacy was reported after repeat dosing in mice and rabbits with acute intermittent porphyria, and the safety of this approach was demonstrated in NHPs.111
mRNA formulations that induce liver expression have also been reported to produce therapeutic antibodies. Systemic delivery of mRNA complexed with TransIT reagent was reported to produce bispecific antibodies in the liver tissue, facilitating the recruitment of cytotoxic T cells to tumors and, ultimately, tumor cell lysis.112 Interestingly, the mRNA-based approach achieved sustained serum protein expression leading to greater cytotoxic activity, as compared to injections of recombinant bispecific antibodies, which have a half-life of less than 2 h. In another study, broadly neutralizing antibodies against HIV-1 were produced in the mouse liver transfected with systemically administered mRNA LNPs. High antibody serum concentrations, up to 170 μg/mL, could be measured for 9 days and maintained by repeated weekly injection.113 Recently, nasal application of MC3 LNPs with mRNA encoding cystic fibrosis transmembrane conductance regulator (CFTR) protein was reported to restore chloride response in airway epithelium for 2 weeks post-administration in CFTR-knockout mice.114 Other studies have also reported LNP-mediated delivery of mRNA to various cell types in the spleen, which may have potential therapeutic applications for vaccination.46, 115
Polymeric Nanoparticles
To date, several polymeric delivery systems have been developed or adopted for mRNA-based delivery of therapeutic proteins, predominantly into the lungs. Chitosan-coated PLGA nanoparticles were reported to provide expression of human CFTR (hCFTR) mRNA in the lungs after intravenous and intratracheal administrations in CFTR-knockout mice.116 Cationic polyaspartamides, composed from repeat aspartamide units with both primary and secondary amides, were used for the delivery of neutrophilic factor (brain-derived neurotrophic factor [BDNF])-expressing mRNA into the brain.117 Intranasal administration of PEG-b-polyaspartamide nanomicelles loaded with BDNF mRNA enhanced the neurological recovery of olfactory function and the repair of the olfactory epithelium in a mouse model of olfactory dysfunction. In addition, a number of polymers, including PEI, poly(2-dimethylamino-ethylmethacrylate) (PDMAEMA), chitosan, and PBAEs, previously developed for the delivery of DNA and siRNA have been adopted for use with mRNA (reviewed by Kauffman et al.118). Recently, hybrid LNPs based on amino polyesters and PBAEs were shown to transfect mRNA into lung endothelium and APCs in the spleen.59, 69
Gene Editing
Gene editing is a promising therapeutic area for the application of mRNA technology expressing programmable nucleases, including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), or CRISPR-Cas.119 These genome-engineering tools enable the replacement or alteration of gene expression by introducing site-specific modifications into the genome of cells, including correction of deleterious or introduction of protective mutations.120 With ZFNs, TALENs, and CRISPR-Cas, editing occurs after cellular DNA repair pathways resolve the DNA double-stranded break (DSB) by non-homologous end joining (NHEJ), which can introduce insertions or deletions, or by homology-directed repair (HDR), with a donor sequence present at the site of the break.121 ZFNs and TALENs facilitate sequence recognition by protein-DNA interactions; however, complicated protein engineering required to create specific DNA-targeting protein domains restricts their broad application.112, 113 The discovery of RNA-guided DNA endonucleases, such as CRISPR-Cas9, Cpf1 (Cas12a), and Cas12b, equipped scientists with a relatively easy-to-use platform to alter genomic information.122 In addition, catalytically impaired Cas9 variants such as dead Cas9 (dCas9) have been fused to diverse functional domains to achieve targeted genetic and epigenetic modifications of DNA sequences, including base editing.123, 124, 125 Recently, novel nuclease Cas13a has been shown to preferentially bind and cleave RNA rather than DNA substrates, further expanding the CRISPR toolbox.126, 127
The barriers to broad clinical translation of CRISPR-Cas systems include (1) imperfect DNA-targeting specificity leading to off-target effects,128 (2) low efficiency of genome editing using HDR,129 and (3) challenging delivery of CRISPR-Cas components.130 More recently, concerns were raised about the immune response toward Cas9 and the presence of neutralizing antibodies;131 the potential for large deletions and complex rearrangement induced by Cas9 cleavage;132 and activation of the p53 pathway, which can antagonize the efficiency of Cas9-mediated gene editing.133 The therapeutic protein itself can also be a source of immune response, in particular when treating genetic diseases.134
Despite these challenges, mRNA formulations offer significant potential as vehicles for in vivo gene editing. To date, the most widely used and well-characterized gene-editing technology is the CRISPR-Cas9 system, with an effector domain originating from Streptococcus pyogenes (SpCas9). Non-viral delivery of the CRISPR-Cas9 components can be achieved using plasmid DNA, mRNA, or the Cas9/sgRNA ribonucleoprotein (Cas9-RNP) complex.135 In contrast to plasmid DNA, mRNA-based delivery carries no risks associated with the integration of the nuclease into the host genome. In addition, the transient nature of the mRNA expression limits the presence of the nuclease inside the cells, reducing the risks of off-target cutting and immune response toward Cas9 protein; however, the intracellular presence of Cas9 protein has been more persistent after mRNA expression as compared to the delivery of Cas9-RNPs.131 Thus far several studies have demonstrated the therapeutic potential of local in vivo Cas9-RNP delivery,136, 137 while systemic RNP delivery still needs to be evaluated in contrast to more established mRNA-based approaches.130 The use of mRNA could also mitigate obstacles associated with Cas9-RNPs, such as manufacturing and preserving the activity of Cas9 protein as well as challenging in vivo protein delivery.130, 138 Moreover, the ability to modify mRNA sequence to encode regulatory elements (e.g., K-turn motifs and microRNA [miRNA]-binding sites) could provide the means to control expression of gene-editing tools in a cell-specific manner.139 However, mRNA-based approaches require effective co-delivery of Cas9 mRNA and sgRNA, in addition to a donor DNA template for gene replacement, that poses a significant challenge.140 Base modifications of the sgRNA, such as altering the RNA 2ʹOH group to 2ʹOMe and 2ʹF or partial replacement of RNA with DNA nucleotides, have been shown to extend intracellular stability of sgRNA and maximize the efficacy of co-delivery by compensating for the delayed translation of Cas9 mRNA.141, 142
mRNA-Based Approaches for Gene Editing
Yin et al.142 demonstrated C12-200 LNP-mediated co-delivery of Cas9 mRNA and modified sgRNA to knock out proprotein convertase subtilisin/kexin type 9 (PCSK9) in mouse hepatocytes, showing high efficacy (>80% insertions or deletions [indels]) and low off-target effects associated with this approach. Similarly, co-delivery of CRISPR-Cas9 components targeting mouse transthyretin (Ttr) gene in the liver with LNP composed of degradable lipid LP-01 (Figure 1) led to >97% reduction in serum transthyretin levels that persisted for at least 12 months after a single administration of the nanoparticles.34 Lipid-like nanoparticles (N1,N3,N5-tris(2-aminoethyl)benzene-1,3,5-tricarboxamide [TT]-derived lipid-like nanoparticles [TT-LLNs]) were also able to achieve effective co-delivery and in vivo targeting of hepatitis B virus (HBV) DNA and PCSK9 in the liver.143 Moreover, ZAL nanoparticles were capable of inducing the expression of floxed tdTomato in the liver, kidneys, and lungs of TdTomato, Lox-Stop-Lox TdTomato (LSL-TdTomato) mice after co-delivery of Cas9 mRNA and sgRNA targeting LoxP site (sgLoxP).82 The effective correction of the fumarylacetoacetate hydrolase (Fah) gene in more than 6% of liver hepatocytes was demonstrated after nanoparticle-mediated delivery of Cas9 mRNA in combination with adeno-associated viruses (AAVs) encoding an sgRNA and a repair template.43 This treatment rescued the symptoms of hereditary tyrosinemia in a mouse model, highlighting both potential and challenges for gene correction via systemic non-viral delivery. Mahiny et al.144 used PLGA-coated chitosan nanoparticle mRNA encoding ZFNs in combination with an AAV6-expressing donor template to realize site-specific genome editing in lungs. This approach resulted in correction of the gene encoding SP-B in mice with SP-B deficiency, and it extended their survival.144
Ex Vivo versus In Vivo Delivery
Co-delivery of Cas9 mRNA and sgRNA or ZFN mRNA for ex vivo gene editing can be achieved via physical methods that bypass the extracellular and cytoplasmic barriers to deposit the molecular cargoes, such as electroporation145, 146 or microinjection into embryos.147, 148, 149 These approaches reduce the risk of off-target genetic perturbations, as only the pre-selected and correctly edited cells can be implanted into patients.119 Utilizing mRNA for in vivo delivery of gene-editing tools holds the promise for lasting correction of monogenic disease or knockout of disease-related genes. Thus far, mRNA-based gene editing has been achieved primarily in the liver and lungs via co-delivery with nanoparticle-based systems or in combination with viral delivery.12, 119 The potential risk of off-target editing in unsolicited cells and tissues highlights the importance of tissue-specific delivery and transient expression of gene-editing tools in vivo.
Clinical Development of mRNA for Protein Therapy and Gene Editing
Most mRNA-based protein therapies have demonstrated translational potential for both secreted and intracellular protein targets (Table 1). Currently, Moderna and its partner AstraZeneca are investigating local delivery of VEGF mRNA for heart regeneration after myocardial infarction in a phase II clinical trial. Moderna also launched a phase I trial for intratumoral delivery of mRNA targeting OX40-binding partner (OX40L), a member of the tumor necrosis factor receptor (TNFR) and TNF superfamily expressed on activated CD4 and CD8 T cells as well as a number of other lymphoid and non-lymphoid cells. Translate Bio is investigating the inhalation of LNPs for lung delivery of mRNA encoding CFTR protein in phase I/II clinical trials for the treatment of cystic fibrosis, and their mRNA-based treatment for Ornithine transcarbamylase (OTC) deficiency is expected to enter clinical testing in the first half of 2019.150 Germany-based companies Ethris151 and BioNTech152 are also developing their own mRNA-based drugs for cystic fibrosis and tumor immunotherapy that are nearing clinical evaluation.
Table 1.
Clinical Trials Involving Non-viral Delivery for RNA-Based Protein Therapy and Gene Editing
| Name | Therapeutic Modality | Protein Target | Delivery Vehicle | Administration Method | Disease | ClinicalTrials.gov Identifier | Phase |
|---|---|---|---|---|---|---|---|
| mRNA-2416 | mRNA | OX40L | lipid nanoparticle | intratumoral | solid tumor or lymphoma | NCT03323398 | I |
| MRT5005 | mRNA | CFTR | lipid nanoparticle | inhalation | cystic fibrosis | NCT03375047 | I/II |
| AZD-8601 | mRNA | VEGF-A | naked mRNA (modified) | intracardiac injection | heart failure | NCT03370887 | II |
| NY-ESO-1 | CRISPR-Cas9 | PD-1 and TCR | autologous T cells | ex vivo (AAV + sgRNA) | multiple myeloma, synovial sarcoma, melanoma | NCT03399448 | I |
| – | CRISPR-Cas9 | PD-1 and TCR | CAR-T cells | ex vivo | mesothelin positive multiple solid tumors | NCT03545815 | I |
| CRISPR/TALEN-HPV E6/E7 | CRISPR/Cas9, TALEN | E6 and E7 | plasmid DNA in gel | ND | cervical intraepithelial neoplasia | NCT03057912 | I |
| CTX001 | CRISPR-Cas9 | BCL11A | modified CD34+ hHSPCs | ex vivo | β-thalassemia | NCT03655678 | I/II |
| – | CRISPR-Cas9 | CD19 and CD20 | dual specificity CAR-T cells | ex vivo | B cell leukemia and lymphoma | NCT03398967 | I/II |
| – | CRISPR-Cas9 | CD19 | CAR-T cells | ex vivo | B cell leukemia and lymphoma | NCT03166878 | I/II |
| – | CRISPR-Cas9 | PD-1 | cytotoxic T lymphocytes | ex vivo | Epstein-Barr virus-associated malignancies | NCT03044743 | I/II |
| SB-728mR-HSPC | ZFN mRNA | CCR5 | CD34+ hHSPCs | ex vivo (mRNA) | HIV | NCT02500849 | I |
| SB-728mR-T | ZFN mRNA | CCR5 | T cells | ex vivo (mRNA) | HIV | NCT02225665 | I/II |
Gene-editing technology has entered the early stages of clinical development. Current clinical efforts by academia and industry are primarily focused on ex vivo applications of gene-editing tools (Table 1). The University of Pennsylvania is evaluating ex vivo CRISPR-Cas9 delivery to disrupt the expression of endogenous T cell receptors (TCRs) and programmed cell death protein 1 (PD-1) in T cells isolated from cancer patients for autologous T cell therapy.153 Similar approaches utilizing CRISPR-Cas9 are being pursued by Chinese institutions (Table 1). CRISPR Therapeutics, together with its partner Vertex, has obtained approvals of clinical trial applications to conduct phase I/II testing of ex vivo-edited cells harvested from patients for the treatment of β-thalassemia and sickle cell disease. Other leading CRISPR biotech companies Editas Medicine154 and Intellia Therapeutics155 have programs in advanced pre-clinical stages of development focusing on disorders affecting liver, lungs, and hematopoiesis, and they will likely soon follow the clinical route. Sangamo Therapeutics is leading clinical efforts for mRNA-based delivery of ZFNs for gene editing in T cells and hematopoietic stem cells (HSCs) to treat β-thalassemia and HIV.
mRNA Vaccines
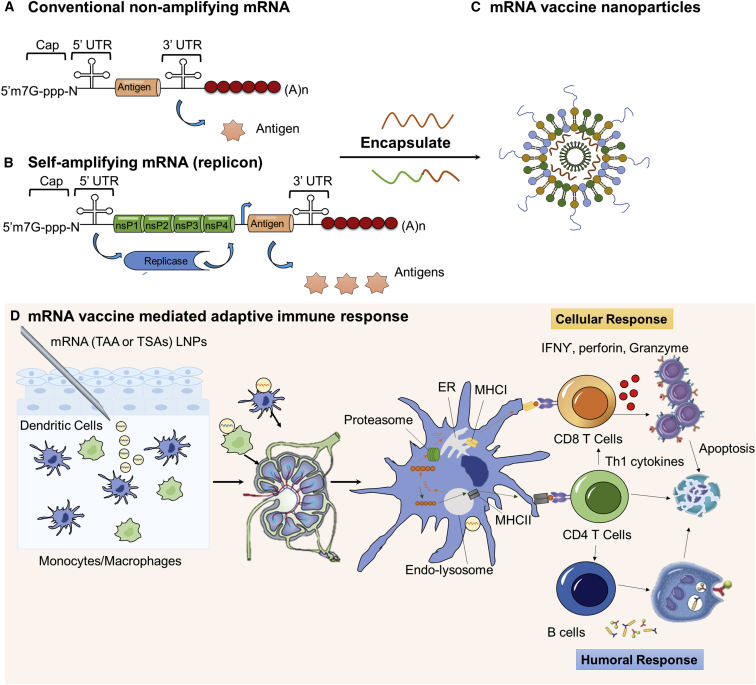
Vaccines have been utilized to provide specific immune responses against infectious diseases or cancer. Conventional live attenuated vaccines that contain the majority of antigens from virus or bacteria have demonstrated durable protection against a variety of infectious pathogens.11, 156, 157 Despite their broad use, the clinical application of conventional vaccines is largely limited to infectious disease, and it faces challenges associated with rapid deployment.11 Viral proteins or peptides are being developed as alternatives, but generally they require combination with adjuvant therapy to boost immunogenicity.158 DNAs have also been applied to encode antigens, but with genome integration potential.11 DNAs have also been investigated as vaccines and adjuvants (CpG) but have been limited in their usage.159 More recently, mRNA-based vaccines are being investigated due to their ability to encode a wide range of antigens, self-adjuvanting effects, and the ease of scalable manufacturing.11, 160, 161 The exploration of mRNA to induce adaptive immune responses in cancer started in 1995, when Conry et al.162 found that protective antitumor immunity could be obtained by intramuscular injection of carcinoembryonic antigen (CEA) mRNA. Since then, RNA vaccines have been classified into two subtypes: non-amplifying mRNA-based vaccines and self-amplifying mRNA (SAM) vaccines (Figures 3A and 3B), both of which utilize the host cell translational machinery to produce the targeted antigens and elicit specific adaptive immune responses.163
Figure 3.
Non-amplifying and Self-Amplifying mRNA Vaccine and Adaptive Immune Response
(A) Schematic structure of conventional non-amplifying mRNA vaccine. (B) Schematic structure of self-amplifying mRNA vaccine (replicon), which contains the sequence-encoding antigens and the non-structural proteins that facilitate RNA capping and replication. (C) An illustration of mRNA vaccine or replicon encapsulated into nanoparticles for improved in vivo performance. (D) The process of antigen presentation and adaptive immune activation after subcutaneous injection of mRNA vaccine LNPs. Briefly, the mRNA vaccine can be captured by antigen-presenting cells (APCs; macrophages or dendritic cells) at the injection site and transported to a draining lymph node, where mRNA is translated into protein and processed by proteasome in the APCs. Then it is presented by major histocompatibility complex (MHC) class I or MHC class II molecules to CD8+ T cells or CD4+ T cells, thus activating both cellular and humoral responses.
Non-amplifying mRNA Vaccines
Non-amplifying mRNA vaccines contain the basic structure of mRNA, with an open reading frame (ORF) encoding the desired antigens (Figure 3A). Major attributes of non-replicating mRNA vaccines include (1) the relatively small size of mRNA as compared to a self-amplifying vaccine (∼2–3 versus ∼10 kb);11 (2) the absence of additional proteins (relative to viral systems), minimizing the possibility of eliciting undesired immunogenic interactions with the host; (3) relatively easy to scale up and manufacture, enabling rapid deployment in case of an outbreak; and (4) facile sequence engineering to improve vaccine performance and minimize off-target effects.164 The biggest barrier to mRNA vaccine utility is the need for intracellular delivery. Chemical modifications and sequence engineering have improved both translation and shelf life of synthetic mRNA vaccines.11 To achieve an enhanced adaptive immune response, mRNA has been electroporated ex vivo into dendritic cells used for adoptive transfer.165 This strategy has been applied in several clinical trials against melanoma, myeloma, and leukemia (Table 1). However, the adoptive transfer is laborious, difficult to scale up, and can induce unfavorable immunogenicity. Therefore, the field has moved on to developing more efficient and potentially less toxic mRNA carriers using lipids or polymer-based materials.
Protamine Complexes
CureVac developed the RNActive vaccine platform based on protamine-formulated RNA complexes, which serve as a TLR 7/8 agonist to induce Th1 T cell response, whereas the nucleotide-modified mRNA functions as an antigen producer.166, 167, 168 Antigen expression strongly depends on the ratio between protamine and mRNA.169 This vaccine platform has raised favorable immune activations against cancer and infectious diseases in both pre-clinical animal models and in patients.169, 170, 171
Lipids and Polymers
Both cationic lipids and polymers have been investigated as vehicles for mRNA vaccines in the past decades. Two recent studies demonstrated that the cationic polymer PEI could be condensed with the mRNA encoding HIV-1 gag or gp120 to raise specific antibodies against HIV infections through subcutaneous and intranasal injection, respectively.42, 172 Despite this success, clinical translation of PEI-based mRNA complexes is restricted by safety concerns. Lipid materials with good biocompatibility have become the most appealing mRNA vaccine delivery system.11, 161, 173 Cationic liposomes composed of the cationic lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) or DOTMA, together with the helper lipid DOPE in combination with mRNA, have been developed as mRNA vaccines.173, 174, 175
BioNTech has reported preferential expression of mRNA in dendritic cells via systemic delivery using DOTMA- and DOPE-containing lipoplexes.174 The first melanoma patients treated with this formulation showed a positive immune response. Several other clinical trials using this formulation are under investigation.174, 176, 177, 178 LNPs containing ionizable lipid MC3 together with other helper lipids, originally developed for siRNA delivery, have also shown promise as mRNA vaccines.179 This early success of MC3 LNPs in mRNA delivery propelled the development of proprietary ionizable lipids for mRNA vaccine delivery by a number of pharmaceutical companies (e.g., Acuitas, Moderna, and Precision). Recently, both Acuitas and Moderna have reported LNPs for the delivery of Zika virus mRNA vaccines. These LNPs produced virus-neutralizing antibody after single or two low-dose vaccinations in mice, rabbit, or NHPs.164, 180 It is noteworthy that antigen expression is not the ultimate criteria for assessing the effectiveness of a vaccine, and T cell activation may better reflect whether an immune response will be protective.42 Based on this rationale, Oberli et al.42 have investigated multiple LNP formulations based on their capability to induce antigen-specific T cell activation using ovalbumin (OVA) mRNA as a model antigen, and they identified formulations with potential as cancer vaccines.
SAM Vaccines
SAM encodes an engineered RNA virus genome from a positive single-stranded RNA virus, such as alphaviruses and flaviviruses.173, 181 The resultant RNA, termed replicon, usually contains two different ORFs, with one encoding nonstructural proteins that help RNA capping and replication and the other expressing the antigen that replaces the viral structural protein (Figure 3B).163 SAM vaccines have several attractive features, such as extending the duration (approximately 2 months) and magnitude of expression compared to their non-replicating counterparts. However, in order to maintain self-amplifying activity, these RNA replicons are not able to tolerate many of the synthetic nucleotide modifications and sequence alterations. Other limitations of SAM include (1) the inclusion of unrelated proteins, which may induce a potential host response; and (2) large replicon size (∼10 kb), which could limit cell internalization efficiency.181, 182
Non-viral Delivery of Self-Replicating mRNA Vaccines
Despite the large replicon size, early work with SAM focused on non-viral delivery using cationic lipids. Two generations of five component particles have been developed. The first lipid particles comprised 1,2-dilinoleyloxy-3-dimethylaminopropane (DLin-DMA) and helper lipids 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG)-PEG2000, yielding uniform small particles encapsulating mRNA encoding F-specific immunoglobulin G (IgG) for the treatment of respiratory syncytial virus (RSV).181 The same team recently developed another lipid formulation, utilizing the first FDA-approved nanoemulsion system MF59,183 composed of squalene, DOTAP, and sorbitan trioleate, to condense the replicon.184, 185 Protective RSV vaccination was obtained after two doses of mRNA vaccine. Besides LNPs, lipid-complexed PRINT (particle replication in nonwetting template) protein particle systems (LPP particle) have also been applied to deliver SAMs.186 Although these methods have been used for SAM delivery in vivo and in vitro, they have not yet shown protection against lethal pathogens.73 Chahal et al.73 developed dendrimer formulations with SAMs to deliver antigenic RNA payloads. They found that the dendrimer system could provide protective immunity and survival against multiple lethal pathogens, including Ebola, H1N1 influenza, and Toxoplasma gondii. They further hypothesized that this polyamine dendrimer could provide optimal protection from nucleases while allowing for functional release in the cytoplasm as compared to lipid delivery, due to the high charge density.73
Immunogenicity and mRNA Vaccines
One characteristic of mRNA is its potential to act as self-adjuvant.11 Exogenously delivered mRNA can resemble an RNA virus and stimulate TLRs 3, 7, and 8, a class of pattern recognition receptors (PRRs) expressed on the cell surface or in the endosomal compartment of major APCs. mRNA can also bind to cytosolic RNA sensors retinoic acid-inducible protein 1 (RIG-1) and MDA5.187 Activation of these molecules further facilitates APC maturation and boosts the secretion of inflammatory cytokines, and it can improve the function of adaptive lymphocytes (Figure 3C). Stimulation of innate immunity may improve a vaccine’s efficacy, however, indiscriminate immune activation can induce mRNA degradation and reduce antigen expression.11
The effects of type I interferons (IFNs) on mRNA vaccines are a topic of debate.187 De Beuckelaer et al. have demonstrated that intrinsic activation of type 1 IFNs by mRNA inhibited subsequent protein translation,175 also confirming that the inhibition can subsequently lead to diminished CD8+ T cell activation.187 Nucleotide modification or purification has been utilized to decrease type I IFN activation.11 Conversely, Kranz et al.174 reported that intravenous (i.v.) injection of unmodified mRNA-lipid complexes raised T cell stimulation through TLR 7-mediated activation of type I IFN. Similarly, CureVac’s protamine-containing RNActive vaccine platform also utilized TLR 7 and 8 to stimulate type I IFNs and improve anti-cancer immunity.168 One possible explanation of this paradoxical effect proposed by De Koker et al.187 is the kinetics of type I IFN signaling relative to TCR activation. If TCR activation coincides or shortly precedes type I IFN signaling, mRNA-mediated activation of type I IFN would facilitate an immune response; otherwise, it would be a concern limiting T cells’ activity.187, 188
Optimizing Injection Routes
The kinetics between TCR activation and IFN signaling are related in part to the distance between the APCs and T cells. Since APCs are one of the major sources of type I IFN molecules, a greater distance between the APCs and CD8+ T cells (e.g., in the case of local vaccination) increases IFN expression within APCs, potentially leading to type I IFN signaling preceding TCR activation. Furthermore, activation of type I IFN in local APCs facilitated mRNA degradation. These may explain the inhibitory T cell activation efficacy caused by subcutaneous or intramuscular injection of mRNA LNPs.187 However, when the lipid complexes were dosed intravenously, mRNA-lipid complexes were directly delivered to APCs within the lymphatic organs, where type I IFN activation and TCR activation occurred simultaneously.174
Aside from affecting the kinetics of IFN signaling, vaccination routes also have drastic impacts on several other aspects, such as but not limited to type of cells that encounter the mRNA vaccine and the rate of antigen presentation.187 Until now, subcutaneous and intramuscular injections have been the two most frequently used injection routes for mRNA vaccination, due to their less invasive nature; however, companies such as BioNTech are investigating intravenous injection of mRNA-lipid complexes in several phase I clinical trials, and they have reported several promising first-in-patient results.177 Other researchers have also started to develop alternative approaches to tune the immune activation kinetics. For example, Jarzębińska et al.65 have designed an intranasal mRNA vaccination system (HIV gp120) for the efficient generation of mucosal HIV antibodies. Intratumoral mRNA vaccination is also being investigated, since it may offer the advantage of rapid and specific activation of tumor-resident T cells.1, 189
Another approach to tune the kinetics is to utilize different delivery components to activate type I IFN response. For example, the protamine complex in the RNActive platform was utilized to activate type I IFN, while free stabilized mRNA was used for antigen expression. Recent work suggests that lipid or polymeric carriers could activate type I IFN while chemically modified mRNA does not induce type I IFN activation.190 Rapid release of mRNA into the cytoplasm together with activation of a type I IFN using delivery materials may result in a kinetic that improves the overall vaccine response.
Co-encapsulation of Other Stimulatory Molecules
Besides type I IFNs, other cytokines, chemokines, and co-factors also participate in the activation of immune systems. mRNA technology has, therefore, been extended to encode immunomodulatory molecules such as CD40L, CD70, OX40L, or granulocyte-macrophage colony-stimulating factor (GM-CSF) (e.g., TriMix).191, 192 In addition to the above-mentioned immunomodulatory mRNAs, small-molecule or lipid-like adjuvants can also be incorporated into RNA delivery vehicles to adjust immunogenicity through non-IFN-related pathways.42, 186
Clinical Development of Non-viral mRNA Vaccine Delivery System
Transfection of mRNA into dendritic cells (DCs) for adoptive transfer against cancer was the first mRNA-based vaccine to enter clinical testing.193 Although DC-based therapies still account for the majority of mRNA vaccines in clinical trials, in vivo mRNA vaccination using non-viral vectors is being explored. Three companies, CureVac, BioNTech, and Moderna, have reported on the development of multiple mRNA formulations with several ongoing pre-clinical and clinical investigations.152, 194, 195 Recently, other large pharmaceutical companies, such as Genentech, Amgen, and Merck, have entered this field, and the number of mRNA vaccine clinical trials is growing.196 For example, Moderna’s cytomegalovirus (CMV) vaccine, mRNA-1647, combines six mRNAs encoding different viral proteins, including five proteins that comprise the CMV gH Pentamer complex as well as another CMV antigen, the herpesvirus glycoprotein (Table 2). Recently, both BioNTech and Moderna have been investigating the delivery of mRNA against Zika virus. Single low-dose vaccinations have shown to be protective against virus infection.180, 197
Table 2.
Clinical Trial of mRNA Vaccine
| Sponsor Institution | Brand | API | Delivery Vehicle | Injection Route | Antigen | Disease | ClinicalTrials.gov Identifier/Trial Number |
|---|---|---|---|---|---|---|---|
| CureVac | CV7201 | mRNA | RNActive, protamine | i.d. or i.m. | rabies virus glycoprotein (RABV-G) | rabies | NCT02241135 |
| CV9201 | mRNA | RNActive, protamine | i.d. | TAAs: MAGEC1, MAGEC2, NY-ESO-1, survivin, 5 T4 | NSCLC | NCT00923312 | |
| CV9202 | mRNA | RNActive, protamine | i.d. | TAAs: NY-ESO-1, MAGEC1, MAGEC2, 5 T4, survivin, and MUC1 | NSCLC | NCT03164772 | |
| CV9103 | mRNA | RNActive, protamine | i.d. | TAAs: PSA, PSCA, PSMA, and STEAP1. | prostate carcinoma | NCT00831467/EudraCT 2008-003967-37 | |
| CV9104 | mRNA | RNActive, protamine | i.d. | TAAs: PSA, PSCA, PSMA, STEAP1, PAP and MUC1 | prostate carcinoma | NCT02140138 | |
| BioNTech | NA | mRNA | naked RNA | ultrasound-guided i.n. | HPV antigen CD40 | HPV-driven squamous cell carcinoma | NCT03418480 |
| Lipo-MERIT | mRNA | Lipo-MERIT, DOTMA(DOTAP)/DOPE lipoplex | i.v. | TAAs: NYESO-1, MAGE-A3, tyrosinase, and TPTE | advanced melanoma | NCT02410733 | |
| IVAC | mRNA | Lipo-MERIT, DOTMA(DOTAP)/DOPE lipoplex | i.v. | (1) 3 TAAs selected from a warehouse and p53 RNA; (2) Neo-Ag based on NGS screening | TNBC | NCT02316457 | |
| NA | mRNA | naked mRNA | ultrasound-guided i.n. | TAAs (RBL001/RBL002) | melanoma | NCT01684241 | |
| IVAC MUTANOME | mRNA | naked mRNA | ultrasound-guided i.n. | Neo-Ag | melanoma | NCT02035956 | |
| RO7198457 | mRNA | naked mRNA | i.v. | Neo-Ag | melanoma | NCT03289962 | |
| NSCLC | |||||||
| bladder cancer | |||||||
| Moderna | mRNA-1325 | mRNA | lipid nanoparticle-encapsulated mRNA | i.d. | Zika virus antigen | Zika virus | NCT03014089 |
| mRNA-1653 | mRNA | lipid nanoparticle-encapsulated mRNA | i.d. | human metapneumovirus and human parainfluenza virus type 3 vaccine | human metapneumovirus and parainfluenza infection | NCT03392389 | |
| VAL-506440 | mRNA | lipid nanoparticle-encapsulated mRNA | i.d. | H10N8 antigen | influenza | NCT03076385 | |
| VAL-339851 | mRNA | lipid nanoparticle-encapsulated mRNA | i.d. | H7 influenza antigen | influenza | NCT03345043 | |
| mRNA-1647/1443 | mRNA | lipid nanoparticle Encapsulated mRNA | i.d. | CMV glycoprotein H (gH) pentamer complex | cytomegalovirus infection | NCT03382405 | |
| mRNA-2416 | mRNA | lipid nanoparticle-encapsulated mRNA | intratumoral | human OX40L | solid tumor maliganancies or lymphoma | NCT03323398 | |
| mRNA-4157 | mRNA | lipid nanoparticle-encapsulated mRNA | i.d. | Neo-Ag | solid tumor | NCT03313778 | |
| mRNA 4650 | mRNA | naked mRNA | i.m. | Neo-Ag | melanoma | NCT03480152 | |
| colon cancer | |||||||
| gastrointestinal cancer | |||||||
| genitourinary cancer | |||||||
| hepatocellular cancer | |||||||
| Hospital Clínic de Bacelona | NA | mRNA | naked Trimix (CD40. CD70 and IL2) | i.d. | HIV mRNA + 300 μg TriMix mRNA | HIV infections | NCT02413645 |
| Memorial Sloan Kettering Cancer Center | NA | mRNA | dendritic cell (DC)-loaded mRNA | i.d. | CT7, MAGE-A3, and WT1 mRNA-electroporated Langerhans cells (LCs) | malignant melanoma | NCT01995708 |
| Massachusetts General Hospital | NA | mRNA | dendritic cell (DC)-loaded mRNA | i.d. | HIV-1 Gag- and Nef-transfected DCs | HIV infections | NCT00833781 |
| Changhai Hospital Stemirna Therapeutics | NA | mRNA | naked mRNA | s.c. | Neo-Ag | solid tumor maliganancies or lymphoma | NCT03468244 |
| University Hospital Tuebingen | NA | mRNA | naked mRNA | i.d | TAA for melanoma (Melan-A, Mage-A1, Mage-A3, survivin, GP100, and tyrosinase) | melanoma | NCT00204516 |
| The Norwegian Radium Hospital | NA | mRNA | dendritic cell (DC)-loaded mRNA | i.d. or i.n. | TAA-transfected DC | malignant melanoma | NCT01278940 |
| Oslo University Hospital | NA | mRNA | dendritic cells (DC)-loaded mRNA | i.d. | TAA-transfected DCs | prostate cancer | NCT01278914 |
| AlphaVax | AVX601 | replicon | – | i.m. or s.c. | alphavirus replicon vaccine expressing cytomegalovirus genes | cytomegalovirus | NCT00439803 |
| AVX502 | replicon | – | i.m. or s.c. | alphavirus replicon vaccine expressing an influenza HA protein | influenza | NCT00440362; NCT00706732 | |
| AVX101 | replicon | i.m. or s.c. | alphavirus replicon, HIV-1 subtype C gag vaccine | HIV infections | NCT00097838; NCT00063778 | ||
| AVX701 | replicon | – | i.m. or s.c. | an alphavirus replicon (VRP) encoding the protein (CEA) | colon cancer | NCT01890213; NCT00529984 |
NA, not applicable; i.m., intramuscular; s.c., subcutaneous; i.d., intradermal; i.n., intranodal; TAA, tumor-associated antigens; HA, hemagglutinin; Neo-Ag, neo-antigen; TPTE, putative tyrosine-protein phosphatase.
mRNA Anti-cancer Vaccine Using Tumor-Associated Antigens
Most cancer vaccines seek to stimulate cell-mediated responses, such as those from activated CD8+T cells.198 Anti-cancer vaccines can be designed to target tumor-associated antigens that are preferentially expressed in cancerous cells, for example, growth-associated factors or antigens that are unique to malignant cells, owing to somatic mutation.199, 200, 201 The success of pre-clinical studies has led to the initiation of clinical trials using intranodally injected naked mRNA encoding tumor-associated antigens into patients with advanced melanoma and with hepatocellular carcinoma (Table 2). In patients with castration-resistant prostate cancer, an RNActive vaccine expressing multiple prostate cancer-associated proteins elicited antigen-specific T cell responses in the majority of recipients (Table 2).
Neoantigen and Personalized Vaccines
Neoepitopes formed by cancer mutations can be presented by human leukocyte antigen (HLA) molecules and recognized by T cells.202 As many cancer mutations are unique to individual patients, the concept of developing individualized mutanome vaccines has been proposed by Sahin and colleagues.202 Mutanome is a comprehensive map of somatic mutations in individual tumors. Once the mutanome has been identified using next-generation sequencing (NGS), somatic mutations with immunogenicity can be further evaluated and create neoantigens that are not subject to central immune tolerance, conferring antitumor vaccine activity. RNA-based vaccinations have the flexibility to incorporate multiple neoantigen epitopes within the same backbone (poly-neoepitope), and the ease of manufacturing has led to their use for personalized vaccines. Proof-of-concept work has been reported by Kreiter et al.,203 who demonstrated that a substantial portion of non-synonymous cancer mutations was immunogenic when delivered via mRNA and that these peptides were mainly recognized by CD4+ T cells. First-in-human application of this concept has been tested in 13 patients with metastatic melanoma by Sahin et al.204 Patients were immunized against ten neoepitopes per individual by injecting naked mRNA intranodally. Since then, non-viral vector-based delivery of mRNA encoding a personalized vaccine has drawn significant attention, with several ongoing clinical trials. Moderna and its partner Merck tested LNP-delivered personalized vaccine against KRAS (mRNA-5671) in combination with Merck’s PD-1-specific antibody (Keytruda) to augment immune response.205 The vaccine is designed to target most of the KRAS mutations that occur in non-small cell lung cancer (NSCLC), colorectal cancer, and pancreatic cancer. Germany-based BioNTech has launched phase 1/2 trials, for its lipid-delivered individualized cancer vaccine in patients with multiple tumors, with its partner Genentech (summarized in Table 2).
Conclusions
Since the first report detailing proof-of-concept delivery of exogenous mRNA encapsulated in liposomes to mouse lymphocytes for functional protein expression was published in 1978,206 mRNA therapeutics have gained significant momentum. Various therapeutic applications of mRNA, including protein replacement, gene editing, and vaccination, are currently being investigated, both by academia and commercial entities. Thus far, clinical efforts are focused largely on vaccination, where mRNA therapeutics have a number of advantages over conventional strategies. However, mRNA also has strong potential as a vehicle for local and systemic protein replacement therapy. Finally, mRNA is being investigated for its potential for ex vivo and in vivo delivery of genome-editing tools, such as ZFNs and CRISPR-Cas nucleases, paving the way for non-viral genome-editing therapies. Broad application of mRNA is still limited by the need for improved delivery systems. However, we believe continued advances in mRNA nanoformulation, using a range of different materials, will ultimately lead to the use of mRNA for the treatment of a wide range of diseases.
Authors Contributions
P.S.K., A.R., L.M., and D.G.A. conceived and co-wrote the manuscript. P.S.K., A.R., and L.M. provided equal contributions in manuscript preparation. All authors read and approved the final manuscript.
Conflicts of Interest
D.G.A. is a consultant with Translate Bio and CRISPR Tx.
Acknowledgments
P.S.K. acknowledges funding from the Juvenile Diabetes Research Foundation (JDRF) postdoctoral fellowship grant 3-PDF-2017-383-A-N. L.M. is supported by Misrock Fellowship. We thank Dr. Joshua Doloff for proofreading and correcting the manuscript.
References
- 1.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:17056. [Google Scholar]
- 2.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 3.Granot Y., Peer D. Delivering the right message: Challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics-An innate immune system standpoint. Semin. Immunol. 2017;34:68–77. doi: 10.1016/j.smim.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Türeci O., Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 5.Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R., Coller J. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wojtczak B.A., Sikorski P.J., Fac-Dabrowska K., Nowicka A., Warminski M., Kubacka D., Nowak E., Nowotny M., Kowalska J., Jemielity J. 5′-Phosphorothiolate Dinucleotide Cap Analogues: Reagents for Messenger RNA Modification and Potent Small-Molecular Inhibitors of Decapping Enzymes. J. Am. Chem. Soc. 2018;140:5987–5999. doi: 10.1021/jacs.8b02597. [DOI] [PubMed] [Google Scholar]
- 8.Li B., Luo X., Dong Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjug. Chem. 2016;27:849–853. doi: 10.1021/acs.bioconjchem.6b00090. [DOI] [PubMed] [Google Scholar]
- 9.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 11.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenton O.S., Kauffman K.J., McClellan R.L., Appel E.A., Dorkin J.R., Tibbitt M.W., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Bioinspired Alkenyl Amino Alcohol Ionizable Lipid Materials for Highly Potent In Vivo mRNA Delivery. Adv. Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harayama T., Riezman H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018;19:281–296. doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 15.Honig B.H., Hubbell W.L., Flewelling R.F. Electrostatic interactions in membranes and proteins. Annu. Rev. Biophys. Biophys. Chem. 1986;15:163–193. doi: 10.1146/annurev.bb.15.060186.001115. [DOI] [PubMed] [Google Scholar]
- 16.Krause M.R., Regen S.L. The structural role of cholesterol in cell membranes: from condensed bilayers to lipid rafts. Acc. Chem. Res. 2014;47:3512–3521. doi: 10.1021/ar500260t. [DOI] [PubMed] [Google Scholar]
- 17.Houseley J., Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 18.Tsui N.B., Ng E.K., Lo Y.M. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin. Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 19.Blanco E., Shen H., Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 21.Yanez Arteta M., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Székely N., Radulescu A., Bergenholtz J., Lindfors L. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. USA. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 24.Ur Rehman Z., Hoekstra D., Zuhorn I.S. Mechanism of polyplex- and lipoplex-mediated delivery of nucleic acids: real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano. 2013;7:3767–3777. doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- 25.Martens T.F., Remaut K., Demeester J., De Smedt S.C., Braeckmans K. Intracellular delivery of nanomaterials: How to catch endosomal escape in the act. Nano Today. 2014;9:344–364. [Google Scholar]
- 26.Vermeulen L.M.P., Brans T., Samal S.K., Dubruel P., Demeester J., De Smedt S.C., Remaut K., Braeckmans K. Endosomal Size and Membrane Leakiness Influence Proton Sponge-Based Rupture of Endosomal Vesicles. ACS Nano. 2018;12:2332–2345. doi: 10.1021/acsnano.7b07583. [DOI] [PubMed] [Google Scholar]
- 27.Patel S., Ashwanikumar N., Robinson E., DuRoss A., Sun C., Murphy-Benenato K.E., Mihai C., Almarsson Ö., Sahay G. Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA. Nano Lett. 2017;17:5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B., Zhang X., Dong Y. Nanoscale platforms for messenger RNA delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019;11:e1530. doi: 10.1002/wnan.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landesman-Milo D., Peer D. Toxicity profiling of several common RNAi-based nanomedicines: a comparative study. Drug Deliv. Transl. Res. 2014;4:96–103. doi: 10.1007/s13346-013-0158-7. [DOI] [PubMed] [Google Scholar]
- 30.Ma Z., Li J., He F., Wilson A., Pitt B., Li S. Cationic lipids enhance siRNA-mediated interferon response in mice. Biochem. Biophys. Res. Commun. 2005;330:755–759. doi: 10.1016/j.bbrc.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 31.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 32.Nabhan J.F., Wood K.M., Rao V.P., Morin J., Bhamidipaty S., LaBranche T.P., Gooch R.L., Bozal F., Bulawa C.E., Guild B.C. Intrathecal delivery of frataxin mRNA encapsulated in lipid nanoparticles to dorsal root ganglia as a potential therapeutic for Friedreich’s ataxia. Sci. Rep. 2016;6:20019. doi: 10.1038/srep20019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sedic M., Senn J.J., Lynn A., Laska M., Smith M., Platz S.J., Bolen J., Hoge S., Bulychev A., Jacquinet E. Safety Evaluation of Lipid Nanoparticle-Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet. Pathol. 2018;55:341–354. doi: 10.1177/0300985817738095. [DOI] [PubMed] [Google Scholar]
- 34.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., van Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 35.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. USA. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payne J.E., Chivukula P., Karmali P., Tanis S.P. US patent US9670152B2; 2018. Ionizable cationic lipid for RNA delivery. filed November 9, 2016, and granted June 6, 2017. [Google Scholar]
- 37.Li B., Luo X., Deng B., Wang J., McComb D.W., Shi Y., Gaensler K.M., Tan X., Dunn A.L., Kerlin B.A., Dong Y. An Orthogonal Array Optimization of Lipid-like Nanoparticles for mRNA Delivery in Vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benenato K.E., Kumarasinghe E.S., Cornebise M. US patent US20170210697A1; 2018. Compounds and compositions for intracellular delivery of therapeutic agents. filed March 31, 2017, and granted January 16, 2018. [Google Scholar]
- 39.An D., Schneller J.L., Frassetto A., Liang S., Zhu X., Park J.S., Theisen M., Hong S.J., Zhou J., Rajendran R. Systemic Messenger RNA Therapy as a Treatment for Methylmalonic Acidemia. Cell Rep. 2017;21:3548–3558. doi: 10.1016/j.celrep.2017.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Y., Love K.T., Dorkin J.R., Sirirungruang S., Zhang Y., Chen D., Bogorad R.L., Yin H., Chen Y., Vegas A.J. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. USA. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid Nanoparticle Assisted mRNA Delivery for Potent Cancer Immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeRosa F., Guild B., Karve S., Smith L., Love K., Dorkin J.R., Kauffman K.J., Zhang J., Yahalom B., Anderson D.G., Heartlein M.W. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016;23:699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Fenton O.S., Kauffman K.J., Kaczmarek J.C., McClellan R.L., Jhunjhunwala S., Tibbitt M.W., Zeng M.D., Appel E.A., Dorkin J.R., Mir F.F. Synthesis and Biological Evaluation of Ionizable Lipid Materials for the In Vivo Delivery of Messenger RNA to B Lymphocytes. Adv. Mater. 2017;29:1606944. doi: 10.1002/adma.201606944. [DOI] [PubMed] [Google Scholar]
- 47.Wong S.C., Cheng W., Hamilton H., Nicholas A.L., Wakefield D.H., Almeida A., Blokhin A.V., Carlson J., Neal Z.C., Subbotin V. HIF2α-Targeted RNAi Therapeutic Inhibits Clear Cell Renal Cell Carcinoma. Mol. Cancer Ther. 2018;17:140–149. doi: 10.1158/1535-7163.MCT-17-0471. [DOI] [PubMed] [Google Scholar]
- 48.Ramot Y., Rotkopf S., Gabai R.M., Zorde Khvalevsky E., Muravnik S., Marzoli G.A., Domb A.J., Shemi A., Nyska A. Preclinical Safety Evaluation in Rats of a Polymeric Matrix Containing an siRNA Drug Used as a Local and Prolonged Delivery System for Pancreatic Cancer Therapy. Toxicol. Pathol. 2016;44:856–865. doi: 10.1177/0192623316645860. [DOI] [PubMed] [Google Scholar]
- 49.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Khan O.F., Kowalski P.S., Doloff J.C., Tsosie J.K., Bakthavatchalu V., Winn C.B., Haupt J., Jamiel M., Langer R., Anderson D.G. Endothelial siRNA delivery in nonhuman primates using ionizable low-molecular weight polymeric nanoparticles. Sci. Adv. 2018;4:eaar8409. doi: 10.1126/sciadv.aar8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dahlman J.E., Barnes C., Khan O., Thiriot A., Jhunjunwala S., Shaw T.E., Xing Y., Sager H.B., Sahay G., Speciner L. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 2014;9:648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao M., Li M., Zhang Z., Gong T., Sun X. Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 2016;23:2596–2607. doi: 10.3109/10717544.2015.1038856. [DOI] [PubMed] [Google Scholar]
- 53.Dong Y., Dorkin J.R., Wang W., Chang P.H., Webber M.J., Tang B.C., Yang J., Abutbul-Ionita I., Danino D., DeRosa F. Poly(glycoamidoamine) Brushes Formulated Nanomaterials for Systemic siRNA and mRNA Delivery in Vivo. Nano Lett. 2016;16:842–848. doi: 10.1021/acs.nanolett.5b02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lynn D.M., Langer R. Degradable Poly(β-amino esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA. J. Am. Chem. Soc. 2000;122:10761–10768. [Google Scholar]
- 55.Akinc A., Lynn D.M., Anderson D.G., Langer R. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. J. Am. Chem. Soc. 2003;125:5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 56.Green J.J., Shi J., Chiu E., Leshchiner E.S., Langer R., Anderson D.G. Biodegradable polymeric vectors for gene delivery to human endothelial cells. Bioconjug. Chem. 2006;17:1162–1169. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- 57.Zugates G.T., Peng W., Zumbuehl A., Jhunjhunwala S., Huang Y.-H., Langer R., Sawicki J.A., Anderson D.G. Rapid optimization of gene delivery by parallel end-modification of poly(beta-amino ester)s. Mol. Ther. 2007;15:1306–1312. doi: 10.1038/mt.sj.6300132. [DOI] [PubMed] [Google Scholar]
- 58.Capasso Palmiero U., Kaczmarek J.C., Fenton O.S., Anderson D.G. Poly(β-amino ester)-co-poly(caprolactone) Terpolymers as Nonviral Vectors for mRNA Delivery In Vitro and In Vivo. Adv. Healthc. Mater. 2018;7:e1800249. doi: 10.1002/adhm.201800249. [DOI] [PubMed] [Google Scholar]
- 59.Kaczmarek J.C., Kauffman K.J., Fenton O.S., Sadtler K., Patel A.K., Heartlein M.W., DeRosa F., Anderson D.G. Optimization of a Degradable Polymer-Lipid Nanoparticle for Potent Systemic Delivery of mRNA to the Lung Endothelium and Immune Cells. Nano Lett. 2018;18:6449–6454. doi: 10.1021/acs.nanolett.8b02917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaczmarek J.C., Patel A.K., Kauffman K.J., Fenton O.S., Webber M.J., Heartlein M.W., DeRosa F., Anderson D.G. Polymer-Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chem. Int. Ed. Engl. 2016;55:13808–13812. doi: 10.1002/anie.201608450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel A.K., Kaczmarek J.C., Bose S., Kauffman K.J., Mir F., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Inhaled Nanoformulated mRNA Polyplexes for Protein Production in Lung Epithelium. Adv. Mater. 2019;31:1805116. doi: 10.1002/adma.201805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nuhn L., Kaps L., Diken M., Schuppan D., Zentel R. Reductive Decationizable Block Copolymers for Stimuli-Responsive mRNA Delivery. Macromol. Rapid Commun. 2016;37:924–933. doi: 10.1002/marc.201600046. [DOI] [PubMed] [Google Scholar]
- 63.Cheng C., Convertine A.J., Stayton P.S., Bryers J.D. Multifunctional triblock copolymers for intracellular messenger RNA delivery. Biomaterials. 2012;33:6868–6876. doi: 10.1016/j.biomaterials.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uchida H., Itaka K., Nomoto T., Ishii T., Suma T., Ikegami M., Miyata K., Oba M., Nishiyama N., Kataoka K. Modulated protonation of side chain aminoethylene repeats in N-substituted polyaspartamides promotes mRNA transfection. J. Am. Chem. Soc. 2014;136:12396–12405. doi: 10.1021/ja506194z. [DOI] [PubMed] [Google Scholar]
- 65.Jarzębińska A., Pasewald T., Lambrecht J., Mykhaylyk O., Kümmerling L., Beck P., Hasenpusch G., Rudolph C., Plank C., Dohmen C. A Single Methylene Group in Oligoalkylamine-Based Cationic Polymers and Lipids Promotes Enhanced mRNA Delivery. Angew. Chem. Int. Ed. Engl. 2016;55:9591–9595. doi: 10.1002/anie.201603648. [DOI] [PubMed] [Google Scholar]
- 66.McKinlay C.J., Vargas J.R., Blake T.R., Hardy J.W., Kanada M., Contag C.H., Wender P.A., Waymouth R.M. Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals. Proc. Natl. Acad. Sci. USA. 2017;114:E448–E456. doi: 10.1073/pnas.1614193114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKinlay C.J., Benner N.L., Haabeth O.A., Waymouth R.M., Wender P.A. Enhanced mRNA delivery into lymphocytes enabled by lipid-varied libraries of charge-altering releasable transporters. Proc. Natl. Acad. Sci. USA. 2018;115:E5859–E5866. doi: 10.1073/pnas.1805358115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haabeth O.A.W., Blake T.R., McKinlay C.J., Waymouth R.M., Wender P.A., Levy R. mRNA vaccination with charge-altering releasable transporters elicits human T cell responses and cures established tumors in mice. Proc. Natl. Acad. Sci. USA. 2018;115:E9153–E9161. doi: 10.1073/pnas.1810002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kowalski P.S., Capasso Palmiero U., Huang Y., Rudra A., Langer R., Anderson D.G. Ionizable Amino-Polyesters Synthesized via Ring Opening Polymerization of Tertiary Amino-Alcohols for Tissue Selective mRNA Delivery. Adv. Mater. 2018;30:1801151. doi: 10.1002/adma.201801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kowalski P.S., Bhattacharya C., Afewerki S., Langer R. Smart Biomaterials: Recent Advances and Future Directions. ACS Biomater. Sci. Eng. 2018;4:3809–3817. doi: 10.1021/acsbiomaterials.8b00889. [DOI] [PubMed] [Google Scholar]
- 71.Abedi-Gaballu F., Dehghan G., Ghaffari M., Yekta R., Abbaspour-Ravasjani S., Baradaran B., Dolatabadi J.E.N., Hamblin M.R. PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy. Appl. Mater. Today. 2018;12:177–190. doi: 10.1016/j.apmt.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khan O.F., Zaia E.W., Yin H., Bogorad R.L., Pelet J.M., Webber M.J., Zhuang I., Dahlman J.E., Langer R., Anderson D.G. Ionizable amphiphilic dendrimer-based nanomaterials with alkyl-chain-substituted amines for tunable siRNA delivery to the liver endothelium in vivo. Angew. Chem. Int. Ed. Engl. 2014;53:14397–14401. doi: 10.1002/anie.201408221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chahal J.S., Khan O.F., Cooper C.L., McPartlan J.S., Tsosie J.K., Tilley L.D., Sidik S.M., Lourido S., Langer R., Bavari S. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. USA. 2016;113:E4133–E4142. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chahal J.S., Fang T., Woodham A.W., Khan O.F., Ling J., Anderson D.G., Ploegh H.L. An RNA nanoparticle vaccine against Zika virus elicits antibody and CD8+ T cell responses in a mouse model. Sci. Rep. 2017;7:252. doi: 10.1038/s41598-017-00193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Islam M.A., Xu Y., Tao W., Ubellacker J.M., Lim M., Aum D., Lee G.Y., Zhou K., Zope H., Yu M. Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng. 2018;2:850–864. doi: 10.1038/s41551-018-0284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ping Y., Wu D., Kumar J.N., Cheng W., Lay C.L., Liu Y. Redox-responsive hyperbranched poly(amido amine)s with tertiary amino cores for gene delivery. Biomacromolecules. 2013;14:2083–2094. doi: 10.1021/bm400460r. [DOI] [PubMed] [Google Scholar]
- 77.Guidotti G., Brambilla L., Rossi D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017;38:406–424. doi: 10.1016/j.tips.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 78.Ziegler A., Seelig J. Binding and clustering of glycosaminoglycans: a common property of mono- and multivalent cell-penetrating compounds. Biophys. J. 2008;94:2142–2149. doi: 10.1529/biophysj.107.113472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Verdurmen W.P.R., Brock R. Biological responses towards cationic peptides and drug carriers. Trends Pharmacol. Sci. 2011;32:116–124. doi: 10.1016/j.tips.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 80.Udhayakumar V.K., De Beuckelaer A., McCaffrey J., McCrudden C.M., Kirschman J.L., Vanover D., Van Hoecke L., Roose K., Deswarte K., De Geest B.G. Arginine-Rich Peptide-Based mRNA Nanocomplexes Efficiently Instigate Cytotoxic T Cell Immunity Dependent on the Amphipathic Organization of the Peptide. Adv. Healthc. Mater. 2017;6:1601412. doi: 10.1002/adhm.201601412. [DOI] [PubMed] [Google Scholar]
- 81.Bell G.D., Yang Y., Leung E., Krissansen G.W. mRNA transfection by a Xentry-protamine cell-penetrating peptide is enhanced by TLR antagonist E6446. PLoS ONE. 2018;13:e0201464. doi: 10.1371/journal.pone.0201464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miller J.B., Zhang S., Kos P., Xiong H., Zhou K., Perelman S.S., Zhu H., Siegwart D.J. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. Engl. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun Y., Sun Y., Zhao R., Gao K. Intracellular delivery of messenger RNA by recombinant PP7 virus-like particles carrying low molecular weight protamine. BMC Biotechnol. 2016;16:46. doi: 10.1186/s12896-016-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun P., Li Z., Wang J., Gao H., Yang X., Wu S., Liu D., Chen Q. Transcellular delivery of messenger RNA payloads by a cationic supramolecular MOF platform. Chem. Commun. (Camb.) 2018;54:11304–11307. doi: 10.1039/c8cc07047d. [DOI] [PubMed] [Google Scholar]
- 85.Kim H., Park Y., Lee J.B. Self-assembled Messenger RNA Nanoparticles (mRNA-NPs) for Efficient Gene Expression. Sci. Rep. 2015;5:12737. doi: 10.1038/srep12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilhelm S., Tavares A.J., Dai Q., Ohta S., Audet J., Dvorak H.F., Chan W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016;1:16014. [Google Scholar]
- 87.Albanese A., Tang P.S., Chan W.C. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 88.De Jong W.H., Hagens W.I., Krystek P., Burger M.C., Sips A.J., Geertsma R.E. Particle size-dependent organ distribution of gold nanoparticles after intravenous administration. Biomaterials. 2008;29:1912–1919. doi: 10.1016/j.biomaterials.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 89.Barz M., Luxenhofer R., Zentel R., Vicent M.J. Overcoming the PEG-addiction: well-defined alternatives to PEG, from structure–property relationships to better defined therapeutics. Polym. Chem. 2011;2:1900–1918. [Google Scholar]
- 90.Hajj K.A., Ball R.L., Deluty S.B., Singh S.R., Strelkova D., Knapp C.M., Whitehead K.A. Branched-Tail Lipid Nanoparticles Potently Deliver mRNA In Vivo due to Enhanced Ionization at Endosomal pH. Small. 2019;15:e1805097. doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- 91.Watanabe J., Kotera H., Akashi M. Reflexive Interfaces of Poly(trimethylene carbonate)-Based Polymers: Enzymatic Degradation and Selective Adsorption. Macromolecules. 2007;40:8731–8736. [Google Scholar]
- 92.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fornaguera C., Guerra-Rebollo M., Ángel Lázaro M., Castells-Sala C., Meca-Cortés O., Ramos-Pérez V., Cascante A., Rubio N., Blanco J., Borrós S. mRNA Delivery System for Targeting Antigen-Presenting Cells In Vivo. Adv. Healthc. Mater. 2018;7:e1800335. doi: 10.1002/adhm.201800335. [DOI] [PubMed] [Google Scholar]
- 94.Field L.D., Delehanty J.B., Chen Y., Medintz I.L. Peptides for specifically targeting nanoparticles to cellular organelles: quo vadis? Acc. Chem. Res. 2015;48:1380–1390. doi: 10.1021/ar500449v. [DOI] [PubMed] [Google Scholar]
- 95.Lou B., De Koker S., Lau C.Y.J., Hennink W.E., Mastrobattista E. mRNA Polyplexes with Post-Conjugated GALA Peptides Efficiently Target, Transfect, and Activate Antigen Presenting Cells. Bioconjug. Chem. 2018 doi: 10.1021/acs.bioconjchem.8b00524. Published online September 6, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Salvati A., Pitek A.S., Monopoli M.P., Prapainop K., Bombelli F.B., Hristov D.R., Kelly P.M., Åberg C., Mahon E., Dawson K.A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 97.Wong C.H. Protein glycosylation: new challenges and opportunities. J. Org. Chem. 2005;70:4219–4225. doi: 10.1021/jo050278f. [DOI] [PubMed] [Google Scholar]
- 98.Mitragotri S., Burke P.A., Langer R. Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat. Rev. Drug Discov. 2014;13:655–672. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turnbull I.C., Eltoukhy A.A., Fish K.M., Nonnenmacher M., Ishikawa K., Chen J., Hajjar R.J., Anderson D.G., Costa K.D. Myocardial Delivery of Lipidoid Nanoparticle Carrying modRNA Induces Rapid and Transient Expression. Mol. Ther. 2016;24:66–75. doi: 10.1038/mt.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carlsson L., Clarke J.C., Yen C., Gregoire F., Albery T., Billger M., Egnell A.C., Gan L.M., Jennbacken K., Johansson E. Biocompatible, Purified VEGF-A mRNA Improves Cardiac Function after Intracardiac Injection 1 Week Post-myocardial Infarction in Swine. Mol. Ther. Methods Clin. Dev. 2018;9:330–346. doi: 10.1016/j.omtm.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sultana N., Magadum A., Hadas Y., Kondrat J., Singh N., Youssef E., Calderon D., Chepurko E., Dubois N., Hajjar R.J., Zangi L. Optimizing Cardiac Delivery of Modified mRNA. Mol. Ther. 2017;25:1306–1315. doi: 10.1016/j.ymthe.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lescan M., Perl R.M., Golombek S., Pilz M., Hann L., Yasmin M., Behring A., Keller T., Nolte A., Gruhn F. De Novo Synthesis of Elastin by Exogenous Delivery of Synthetic Modified mRNA into Skin and Elastin-Deficient Cells. Mol. Ther. Nucleic Acids. 2018;11:475–484. doi: 10.1016/j.omtn.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Van Hoecke L., Van Lint S., Roose K., Van Parys A., Vandenabeele P., Grooten J., Tavernier J., De Koker S., Saelens X. Treatment with mRNA coding for the necroptosis mediator MLKL induces antitumor immunity directed against neo-epitopes. Nat. Commun. 2018;9:3417. doi: 10.1038/s41467-018-05979-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 108.Schoellhammer C.M., Lauwers G.Y., Goettel J.A., Oberli M.A., Cleveland C., Park J.Y., Minahan D., Chen Y., Anderson D.G., Jaklenec A. Ultrasound-Mediated Delivery of RNA to Colonic Mucosa of Live Mice. Gastroenterology. 2017;152:1151–1160. doi: 10.1053/j.gastro.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prieve M.G., Harvie P., Monahan S.D., Roy D., Li A.G., Blevins T.L., Paschal A.E., Waldheim M., Bell E.C., Galperin A. Targeted mRNA Therapy for Ornithine Transcarbamylase Deficiency. Mol. Ther. 2018;26:801–813. doi: 10.1016/j.ymthe.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schrom E., Huber M., Aneja M., Dohmen C., Emrich D., Geiger J., Hasenpusch G., Herrmann-Janson A., Kretzschmann V., Mykhailyk O. Translation of Angiotensin-Converting Enzyme 2 upon Liver- and Lung-Targeted Delivery of Optimized Chemically Modified mRNA. Mol. Ther. Nucleic Acids. 2017;7:350–365. doi: 10.1016/j.omtn.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jiang L., Berraondo P., Jericó D., Guey L.T., Sampedro A., Frassetto A., Benenato K.E., Burke K., Santamaría E., Alegre M. Systemic messenger RNA as an etiological treatment for acute intermittent porphyria. Nat. Med. 2018;24:1899–1909. doi: 10.1038/s41591-018-0199-z. [DOI] [PubMed] [Google Scholar]
- 112.Stadler C.R., Bähr-Mahmud H., Celik L., Hebich B., Roth A.S., Roth R.P., Karikó K., Türeci Ö., Sahin U. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 113.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Robinson E., MacDonald K.D., Slaughter K., McKinney M., Patel S., Sun C., Sahay G. Lipid Nanoparticle-Delivered Chemically Modified mRNA Restores Chloride Secretion in Cystic Fibrosis. Mol. Ther. 2018;26:2034–2046. doi: 10.1016/j.ymthe.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fenton O.S., Kauffman K.J., McClellan R.L., Kaczmarek J.C., Zeng M.D., Andresen J.L., Rhym L.H., Heartlein M.W., DeRosa F., Anderson D.G. Customizable Lipid Nanoparticle Materials for the Delivery of siRNAs and mRNAs. Angew. Chem. Int. Ed. Engl. 2018;57:13582–13586. doi: 10.1002/anie.201809056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Haque A.K.M.A., Dewerth A., Antony J.S., Riethmüller J., Latifi N., Yasar H., Weinmann P., Pedemonte N., Sondo E., Laval J. Modified hCFTR mRNA restores normal lung function in a mouse model of cystic fibrosis. bioRxiv. 2017 doi: 10.1038/s41598-018-34960-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baba M., Itaka K., Kondo K., Yamasoba T., Kataoka K. Treatment of neurological disorders by introducing mRNA in vivo using polyplex nanomicelles. J. Control. Release. 2015;201:41–48. doi: 10.1016/j.jconrel.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 118.Kauffman K.J., Webber M.J., Anderson D.G. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Release. 2016;240:227–234. doi: 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 119.Yin H., Kauffman K.J., Anderson D.G. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 2017;16:387–399. doi: 10.1038/nrd.2016.280. [DOI] [PubMed] [Google Scholar]
- 120.Cox D.B., Platt R.J., Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pawelczak K.S., Gavande N.S., VanderVere-Carozza P.S., Turchi J.J. Modulating DNA Repair Pathways to Improve Precision Genome Engineering. ACS Chem. Biol. 2018;13:389–396. doi: 10.1021/acschembio.7b00777. [DOI] [PubMed] [Google Scholar]
- 122.Strecker J., Jones S., Koopal B., Schmid-Burgk J., Zetsche B., Gao L., Makarova K.S., Koonin E.V., Zhang F. Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 2019;10:212. doi: 10.1038/s41467-018-08224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Knott G.J., Doudna J.A. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Eid A., Alshareef S., Mahfouz M.M. CRISPR base editors: genome editing without double-stranded breaks. Biochem. J. 2018;475:1955–1964. doi: 10.1042/BCJ20170793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tycko J., Myer V.E., Hsu P.D. Methods for Optimizing CRISPR-Cas9 Genome Editing Specificity. Mol. Cell. 2016;63:355–370. doi: 10.1016/j.molcel.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Komor A.C., Badran A.H., Liu D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Luther D.C., Lee Y.W., Nagaraj H., Scaletti F., Rotello V.M. Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opin. Drug Deliv. 2018;15:905–913. doi: 10.1080/17425247.2018.1517746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Crudele J.M., Chamberlain J.S. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat. Commun. 2018;9:3497. doi: 10.1038/s41467-018-05843-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 134.Salazar-Fontana L.I., Desai D.D., Khan T.A., Pillutla R.C., Prior S., Ramakrishnan R., Schneider J., Joseph A. Approaches to Mitigate the Unwanted Immunogenicity of Therapeutic Proteins during Drug Development. AAPS J. 2017;19:377–385. doi: 10.1208/s12248-016-0030-z. [DOI] [PubMed] [Google Scholar]
- 135.Wang H.X., Li M., Lee C.M., Chakraborty S., Kim H.W., Bao G., Leong K.W. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem. Rev. 2017;117:9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 136.Gao X., Tao Y., Lamas V., Huang M., Yeh W.H., Pan B., Hu Y.J., Hu J.H., Thompson D.B., Shu Y. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee K., Conboy M., Park H.M., Jiang F., Kim H.J., Dewitt M.A., Mackley V.A., Chang K., Rao A., Skinner C. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017;1:889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015;33:839–841. doi: 10.1038/nbt.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Glass Z., Lee M., Li Y., Xu Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018;36:173–185. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yin H., Song C.Q., Suresh S., Kwan S.Y., Wu Q., Walsh S., Ding J., Bogorad R.L., Zhu L.J., Wolfe S.A. Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat. Chem. Biol. 2018;14:311–316. doi: 10.1038/nchembio.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yin H., Song C.Q., Suresh S., Wu Q., Walsh S., Rhym L.H., Mintzer E., Bolukbasi M.F., Zhu L.J., Kauffman K. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017;35:1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jiang C., Mei M., Li B., Zhu X., Zu W., Tian Y., Wang Q., Guo Y., Dong Y., Tan X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017;27:440–443. doi: 10.1038/cr.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mahiny A.J., Dewerth A., Mays L.E., Alkhaled M., Mothes B., Malaeksefat E., Loretz B., Rottenberger J., Brosch D.M., Reautschnig P. In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat. Biotechnol. 2015;33:584–586. doi: 10.1038/nbt.3241. [DOI] [PubMed] [Google Scholar]
- 145.Wang J., DeClercq J.J., Hayward S.B., Li P.W., Shivak D.A., Gregory P.D., Lee G., Holmes M.C. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res. 2016;44:e30. doi: 10.1093/nar/gkv1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 148.Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 149.Wu Y., Zhou H., Fan X., Zhang Y., Zhang M., Wang Y., Xie Z., Bai M., Yin Q., Liang D. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25:67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Translate Bio (2018). https://translate.bio/pipeline/.
- 151.Ethris (2018). https://ethris.com/about-2/.
- 152.BioNTech (2018). https://biontech.de/pipeline-patients/.
- 153.Baylis F., McLeod M. First-in-human Phase 1 CRISPR Gene Editing Cancer Trials: Are We Ready? Curr. Gene Ther. 2017;17:309–319. doi: 10.2174/1566523217666171121165935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Editas Medicine (2018). https://www.editasmedicine.com/pipeline/.
- 155.Intellia Therapeutics (2018). https://www.intelliatx.com/pipeline-2/.
- 156.Medina E., Guzmán C.A. Use of live bacterial vaccine vectors for antigen delivery: potential and limitations. Vaccine. 2001;19:1573–1580. doi: 10.1016/s0264-410x(00)00354-6. [DOI] [PubMed] [Google Scholar]
- 157.Cox R.J., Brokstad K.A., Ogra P. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand. J. Immunol. 2004;59:1–15. doi: 10.1111/j.0300-9475.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- 158.Disis M.L., Bernhard H., Shiota F.M., Hand S.L., Gralow J.R., Huseby E.S., Gillis S., Cheever M.A. Granulocyte-macrophage colony-stimulating factor: an effective adjuvant for protein and peptide-based vaccines. Blood. 1996;88:202–210. [PubMed] [Google Scholar]
- 159.Kutzler M.A., Weiner D.B. DNA vaccines: ready for prime time? Nat. Rev. Genet. 2008;9:776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mullard A. mRNA vaccines get another booster. Nat. Rev. Drug Discov. 2018;17:460. doi: 10.1038/nrd.2018.113. [DOI] [PubMed] [Google Scholar]
- 161.Guan S., Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 162.Conry R.M., LoBuglio A.F., Wright M., Sumerel L., Pike M.J., Johanning F., Benjamin R., Lu D., Curiel D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–1400. [PubMed] [Google Scholar]
- 163.Iavarone C., O’hagan D.T., Yu D., Delahaye N.F., Ulmer J.B. Mechanism of action of mRNA-based vaccines. Expert Rev. Vaccines. 2017;16:871–881. doi: 10.1080/14760584.2017.1355245. [DOI] [PubMed] [Google Scholar]
- 164.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wilgenhof S., Van Nuffel A.M., Corthals J., Heirman C., Tuyaerts S., Benteyn D., De Coninck A., Van Riet I., Verfaillie G., Vandeloo J. Therapeutic vaccination with an autologous mRNA electroporated dendritic cell vaccine in patients with advanced melanoma. J. Immunother. 2011;34:448–456. doi: 10.1097/CJI.0b013e31821dcb31. [DOI] [PubMed] [Google Scholar]
- 166.Scheel B., Teufel R., Probst J., Carralot J.P., Geginat J., Radsak M., Jarrossay D., Wagner H., Jung G., Rammensee H.G. Toll-like receptor-dependent activation of several human blood cell types by protamine-condensed mRNA. Eur. J. Immunol. 2005;35:1557–1566. doi: 10.1002/eji.200425656. [DOI] [PubMed] [Google Scholar]
- 167.Kallen K.J., Heidenreich R., Schnee M., Petsch B., Schlake T., Thess A., Baumhof P., Scheel B., Koch S.D., Fotin-Mleczek M. A novel, disruptive vaccination technology: self-adjuvanted RNActive(®) vaccines. Hum. Vaccin. Immunother. 2013;9:2263–2276. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kowalczyk A., Doener F., Zanzinger K., Noth J., Baumhof P., Fotin-Mleczek M., Heidenreich R. Self-adjuvanted mRNA vaccines induce local innate immune responses that lead to a potent and boostable adaptive immunity. Vaccine. 2016;34:3882–3893. doi: 10.1016/j.vaccine.2016.05.046. [DOI] [PubMed] [Google Scholar]
- 169.Rausch S., Schwentner C., Stenzl A., Bedke J. mRNA vaccine CV9103 and CV9104 for the treatment of prostate cancer. Hum. Vaccin. Immunother. 2014;10:3146–3152. doi: 10.4161/hv.29553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sebastian M., von Boehmer L., Zippelius A., Mayer F., Reck M., Atanackovic D., Thomas M., Schneller F., Stoehlmacher J., Goekkurt E. Messenger RNA vaccination in NSCLC: Findings from a phase I/IIa clinical trial. J. Clin. Oncol. 2011;29(Suppl 15):2584. [Google Scholar]
- 171.Sebastian M., Papachristofilou A., Weiss C., Früh M., Cathomas R., Hilbe W., Wehler T., Rippin G., Koch S.D., Scheel B. Phase Ib study evaluating a self-adjuvanted mRNA cancer vaccine (RNActive®) combined with local radiation as consolidation and maintenance treatment for patients with stage IV non-small cell lung cancer. BMC Cancer. 2014;14:748. doi: 10.1186/1471-2407-14-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li M., Zhao M., Fu Y., Li Y., Gong T., Zhang Z., Sun X. Enhanced intranasal delivery of mRNA vaccine by overcoming the nasal epithelial barrier via intra- and paracellular pathways. J. Control. Release. 2016;228:9–19. doi: 10.1016/j.jconrel.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 173.Midoux P., Pichon C. Lipid-based mRNA vaccine delivery systems. Expert Rev. Vaccines. 2015;14:221–234. doi: 10.1586/14760584.2015.986104. [DOI] [PubMed] [Google Scholar]
- 174.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 175.De Beuckelaer A., Pollard C., Van Lint S., Roose K., Van Hoecke L., Naessens T., Udhayakumar V.K., Smet M., Sanders N., Lienenklaus S. Type I Interferons Interfere with the Capacity of mRNA Lipoplex Vaccines to Elicit Cytolytic T Cell Responses. Mol. Ther. 2016;24:2012–2020. doi: 10.1038/mt.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kloke B.P., Kreiter S., Vormehr M., Diken M., Kuhn A.N., Sahin U. Actively personalized cancer vaccines--the step into clinical application. Pharmazie. 2016;71:43–47. [PubMed] [Google Scholar]
- 177.Heesen L., Jabulowsky R., Loquai C., Utikal J., Gebhardt C., Hassel J., Kaufmann R., Pinter A., Derhovanessian E., Diken M. A first-in-human phase I/II clinical trial assessing novel mRNA-lipoplex nanoparticles encoding shared tumor antigens for potent melanoma immunotherapy. Ann. Oncol. 2017;28(Suppl 11) mdx711.030. [Google Scholar]
- 178.Jabulowsky R.A., Loquai C., Diken M., Kranz L.M., Haas H., Attig S., Bidmon N., Buck J., Derhovanessian E., Diekmann J. A first-in-human phase I/II clinical trial assessing novel mRNA-lipoplex nanoparticles for potent cancer immunotherapy in patients with malignant melanoma. Ann Oncol. 2018;29:viii400–viii441. [Google Scholar]
- 179.Pasi K.J., Rangarajan S., Georgiev P., Mant T., Creagh M.D., Lissitchkov T., Bevan D., Austin S., Hay C.R., Hegemann I. Targeting of Antithrombin in Hemophilia A or B with RNAi Therapy. N. Engl. J. Med. 2017;377:819–828. doi: 10.1056/NEJMoa1616569. [DOI] [PubMed] [Google Scholar]
- 180.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schwendener R.A. Liposomes as vaccine delivery systems: a review of the recent advances. Ther. Adv. Vaccines. 2014;2:159–182. doi: 10.1177/2051013614541440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.O’Hagan D.T. MF59 is a safe and potent vaccine adjuvant that enhances protection against influenza virus infection. Expert Rev. Vaccines. 2007;6:699–710. doi: 10.1586/14760584.6.5.699. [DOI] [PubMed] [Google Scholar]
- 184.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Maruggi G., Chiarot E., Giovani C., Buccato S., Bonacci S., Frigimelica E., Margarit I., Geall A., Bensi G., Maione D. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine. 2017;35:361–368. doi: 10.1016/j.vaccine.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 186.Xu J., Luft J.C., Yi X., Tian S., Owens G., Wang J., Johnson A., Berglund P., Smith J., Napier M.E., DeSimone J.M. RNA replicon delivery via lipid-complexed PRINT protein particles. Mol. Pharm. 2013;10:3366–3374. doi: 10.1021/mp400190z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.De Beuckelaer A., Grooten J., De Koker S. Type I Interferons Modulate CD8+ T Cell Immunity to mRNA Vaccines. Trends Mol. Med. 2017;23:216–226. doi: 10.1016/j.molmed.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 188.Crouse J., Kalinke U., Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 2015;15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 189.Van der Jeught K., Van Lint S., Thielemans K., Breckpot K. Intratumoral delivery of mRNA: Overcoming obstacles for effective immunotherapy. OncoImmunology. 2015;4:e1005504. doi: 10.1080/2162402X.2015.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Luo M., Wang H., Wang Z., Cai H., Lu Z., Li Y., Du M., Huang G., Wang C., Chen X. A STING-activating nanovaccine for cancer immunotherapy. Nat. Nanotechnol. 2017;12:648–654. doi: 10.1038/nnano.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Van Lint S., Goyvaerts C., Maenhout S., Goethals L., Disy A., Benteyn D., Pen J., Bonehill A., Heirman C., Breckpot K., Thielemans K. Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res. 2012;72:1661–1671. doi: 10.1158/0008-5472.CAN-11-2957. [DOI] [PubMed] [Google Scholar]
- 192.Hess P.R., Boczkowski D., Nair S.K., Snyder D., Gilboa E. Vaccination with mRNAs encoding tumor-associated antigens and granulocyte-macrophage colony-stimulating factor efficiently primes CTL responses, but is insufficient to overcome tolerance to a model tumor/self antigen. Cancer Immunol. Immunother. 2006;55:672–683. doi: 10.1007/s00262-005-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vormehr M., Schrörs B., Boegel S., Löwer M., Türeci Ö., Sahin U. Mutanome Engineered RNA Immunotherapy: Towards Patient-Centered Tumor Vaccination. J. Immunol. Res. 2015;2015:595363. doi: 10.1155/2015/595363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.CureVac (2018). https://www.curevac.com/.
- 195.Moderna Therapeutics. (2018). https://www.modernatx.com/pipeline.
- 196.Mullard A. The cancer vaccine resurgence. Nat. Rev. Drug Discov. 2016;15:663–665. doi: 10.1038/nrd.2016.201. [DOI] [PubMed] [Google Scholar]
- 197.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;169:176. doi: 10.1016/j.cell.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 198.Durgeau A., Virk Y., Corgnac S., Mami-Chouaib F. Recent Advances in Targeting CD8 T-Cell Immunity for More Effective Cancer Immunotherapy. Front. Immunol. 2018;9:14. doi: 10.3389/fimmu.2018.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kuan C.T., Wikstrand C.J., Bigner D.D. EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr. Relat. Cancer. 2001;8:83–96. doi: 10.1677/erc.0.0080083. [DOI] [PubMed] [Google Scholar]
- 200.Reynolds S.R., Celis E., Sette A., Oratz R., Shapiro R.L., Johnston D., Fotino M., Bystryn J.C. HLA-independent heterogeneity of CD8+ T cell responses to MAGE-3, Melan-A/MART-1, gp100, tyrosinase, MC1R, and TRP-2 in vaccine-treated melanoma patients. J. Immunol. 1998;161:6970–6976. [PubMed] [Google Scholar]
- 201.Walsh P.C. Combination of prostate-specific antigen, clinical stage, and Gleason score to predict pathological stage of localized prostate cancer: a multi-institutional update. J. Urol. 1997;158:1618–1619. [PubMed] [Google Scholar]
- 202.Castle J.C., Kreiter S., Diekmann J., Löwer M., van de Roemer N., de Graaf J., Selmi A., Diken M., Boegel S., Paret C. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 203.Kreiter S., Vormehr M., van de Roemer N., Diken M., Löwer M., Diekmann J., Boegel S., Schrörs B., Vascotto F., Castle J.C. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 205.Business Wire (2018). Moderna and Merck Expand mRNA Cancer Vaccines Collaboration. https://www.businesswire.com/news/home/20180503006100/en/.
- 206.Dimitriadis G.J. Translation of rabbit globin mRNA introduced by liposomes into mouse lymphocytes. Nature. 1978;274:923–924. doi: 10.1038/274923a0. [DOI] [PubMed] [Google Scholar]