Abstract
Ovarian cancer remains the most lethal type of cancer among all gynecological malignancies. The majority of patients are diagnosed with ovarian cancer at the late stages of the disease. Therefore, there exists an imperative need for the development of early ovarian cancer diagnostic techniques. Exosomes, secreted by various cell types, play pivotal roles in intercellular communication, which emerge as promising diagnostic and prognostic biomarkers for ovarian cancer. In this study, we present for the first time, at least to the best of our knowledge, the proteomics profiling of exosomes derived from the plasma of patients with ovarian cancer via liquid chromatography tandem mass spectrometry (LC-MS/MS) with tandem mass tagging (TMT). The exosomes enriched from patient plasma samples were characterized by nanoparticle tracking analysis (NTA), dynamic light scattering (DLS), transmission electron microscopy (TEM) and western blot analysis. The size of the plasma exosomes fell into the range of 30 to 100 nm in diameter. The exosomal marker proteins, CD81 and TSG101, were clearly stained in the exosome samples; however, there was no staining for the endoplasmic reticulum protein, calnexin. A total of 294 proteins were identified with all exosome samples. Among these, 225 proteins were detected in both the cancerous and non-cancerous samples. Apart from universal exosomal proteins, exosomes derived from ovarian cancer patient plasma also contained tumor-specific proteins relevant to tumorigenesis and metastasis, particularly in epithelial ovarian carcinoma (EOC). Patients with EOC often suffer from coagulation dysfunction. The function of exosomes in coagulation was also examined. Several genes relevant to the coagulation cascade were screened out as promising diagnostic and prognostic factors that may play important roles in ovarian cancer progression and metastasis. On the whole, in this study, we successfully isolated and purified exosomes from plasma of patients with EOC, and identified a potential role of these exosomes in the coagulation cascade, as well as in the diagnosis and prognosis of patients. differentially expressed genes, functional enrichment analysis, protein-protein interaction, diagnostic and prognostic biomarkers
Introduction
Ovarian cancer is the most lethal type of cancer among all frequent gynecological malignancies, not only in China but globally as well (1). The majority of patients are diagnosed with epithelial ovarian cancer (EOC) in the advanced stages of the disease and subsequent platinum/taxane chemoresistance occurs following prolonged treatment, which is responsible for the high mortality associated with EOC (2). Despite the fact that serum CA125 examination and ultrasonography are clinically accepted methods for the diagnosis of ovarian cancer, these are not satisfactory and do not fulfill the requirements for the early detection of ovarian cancer due to poor sensitivity and specificity (3). Cytoreductive surgery and subsequent platinum/taxane-based post-operative adjuvant chemotherapy have made some progress in improving the survival rate of patients with ovarian cancer; however, eventual recurrence and acquired chemoresistance are still inevitable. Thus, the identification of more sensitive diagnostic biomarkers and the establishment of novel therapeutic strategies is of utmost importance.
Exosomes are spherical and bilayered proteolipids with a diameter of 30–100 nm. They are abundant in various cellular components with diverse bioactivities, including proteins, lipids and nuclear acids (4,5). A variety of cells can release exosomes, such as epithelial cells (6,7), immune cells (8–10), reticulocytes (11), mast cells (12) and tumor cells (13,14). In addition, exosomes have also been found in various bodily fluids, including amniotic fluid (15), urine (16), malignant ascites (17), plasma (18), bile (19) and saliva (20). They have been proposed as a potential source of diagnostic and prognostic biomarkers, and have been proven to play important roles in intercellular communication (21). Moreover, certain common exosomal proteins, such as TSG101, Alix, CD9, CD81, CD63, GTPase active proteins and cytoskeletal proteins, including actin, as well as tubulin proteins have been found to be crucial for the biogenesis, secretion and translocation of exosomes (22). Exosomes may also transport some tumor-associated proteins, particularly those that originate from cancer cells (23). Therefore, exosomes in urine, saliva, plasma and other bodily fluids may be utilized for the identification of diagnostic and prognostic biomarkers in a non-invasive manner (24).
As regards ovarian cancer, exosomes have been isolated from plasma/serum or ascites of patients with ovarian cancer in some studies. In one study, the expression levels of 8 microRNAs (miRNAs or miRs; miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205 and miR-214) were found to be elevated in exosomes isolated from the serum of women with various stages of ovarian cancer as compared to those from the serum of women with benign diseases, suggesting that miRNAs of circulating tumor exosomes can potentially be used as alternative diagnostic markers (25). Peng et al proved the origin of ascites-derived exosomes from patients with ovarian cancer; two genes, namely FasL and TRAIL, were identified in the exosome and were found to be responsible for the apoptosis of different types of immune cells (26). Exosomes containing claudin 4 have also been isolated from patients with EOC and found to be a promising diagnostic marker as compared to CA125 (27). Therefore, the analysis of the biological characteristics of ovarian cancer-derived exosomes may be of great value to the diagnosis of ovarian cancer, monitoring the therapeutic efficacy, and exploring their roles in tumor progression and metastasis.
Regardless of the advances achieved in the above-mentioned studies, the results, however, have only highlighted the expression of limited individual molecules in exosomes. The systemic proteomic analysis of ovarian cancer-derived exosomes from patient plasma has seldom been reported, at least to the best of our knowledge. Additionally, exosomes isolated from clinical samples ought to possess greater significance and closer association with the real-life situations of patients. Therefore, in this study, we utilized exosomes from EOC patient plasma samples and performed the first comprehensive proteomic analysis (to the best of our knowledge) by liquid chromatography tandem mass spectrometry (LC-MS/MS) with tandem mass tagging (TMT) to identify potential biomarkers and investigate their clinical values. It should be noted that, to the best of our knowledge, this is the first research using the TMT technique to analyze the exosomal proteins. TMT is a type of chemical label for the quantification and identification of proteins, peptides, nucleic acids and other biological macromolecules (28). The chemical structures of TMTs are identical, but each contains isotopes replaced at diverse regions. The biomolecule sequence information is obtained during the fragmentation process and the amount of the biomolecules is collected in the meantime, which renders TMT an ideal tool for the high-throughput characterization of the exosomal proteins (29).
Materials and methods
Subjects and materials
The study protocol was approved by the Ethics Committee of Fudan University Shanghai Cancer Center (Shanghai, China). All patients who provided signed informed consent forms in this study were of Chinese origin and from the Department of Gynecological Oncology at the Fudan University Shanghai Cancer Center. For proteomics analysis, plasma samples from 3 patients with ovarian cancer were obtained as the experimental group and 6 non-cancerous samples served as the controls between July, 2017 to August, 2017. Another cohort of 80 female candidates (40 patients and 40 healthy volunteers) was recruited for the verification of specific biomarkers screened from the proteomics analysis from June, 2016 to May, 2017. Detailed information of the 9 patients (3 with EOC and 6 with benign disease) for proteomics and 40 patients with EOC is summarized in Tables I and II, respectively. Blood was collected into EDTA tubes and spun at 2,000 × g for 10 min at 4°C to obtain plasma. The plasma was stored at -80°C for further analysis. The exosome isolation kit (exoEasy kit) was purchased from Qiagen GmbH (Hilden, Germany). TSG101 (sc-7964) and CD81 (sc-7637) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, TX, USA). Calnexin (cat. no. 2433) was purchased from Cell Signaling (Cell Signaling Technology, Danvers, MA, USA). Enzyme-linked immunosorbent assay (ELISA) kits for gelsolin (GSN; EH0875), lipopolysaccharide binding protein (LBP; EH1560), fibrinogen alpha chain (FGA; EH3065) and fibrinogen gamma chain (FGG; EH0693) were obtained from Finetest (Fine Biotech, Wuhan, China). Other materials, if not specifically stated, were all purchased from Sigma-Aldrich (St. Louis, MO, USA).
Table I.
Clinical characteristics of the patients recruited for proteomics analysis.
| No. | Age (years) | FIGO stage | Grading | Lymph node metastasis | Histological subtype | CA125 | Post-operative residual tumor | Recurrence at time of analysis | Survival status at time of analysis |
|---|---|---|---|---|---|---|---|---|---|
| EOC1 | 46 | IIIC | G3 | NA | High-grade serous carcinoma | 533 | None | NA | NA |
| EOC2 | 67 | III | G3 | N1 | High-grade serous carcinoma | 7378 | <0.5 cm | Yes | Yes |
| EOC3 | 52 | IV | G2 | N1 | Moderately differentiated squamous cell carcinoma | 420.4 | NA | Yes | Yes |
| B1 | 44 | G1 | N1 | Cervical myoma | 14.79 | None | Yes | Yes | |
| B2 | 47 | G1 | N0 | Cervical myoma | 11.79 | None | No | Yes | |
| B3 | 45 | G1 | N0 | Hysteromyoma | 13.48 | None | No | Yes | |
| B4 | 54 | G1 | N0 | Hysteromyoma | 14.8 | None | No | Yes | |
| B5 | 44 | G1 | N0 | Uterine leiomyoma | 12.24 | None | No | Yes | |
| B6 | 53 | G1 | N0 | Uterine leiomyoma | 11.42 | None | No | Yes |
NA, not available.
Table II.
Clinical characteristics of patients with epithelial ovarian cancer recruited for ELISA.
| No. | Age (years) | FIGO stage | Grading | Histological subtype | CA125 | Post-operative residual tumor | Recurrent disease at time of analysis | Survival status at time of analysis |
|---|---|---|---|---|---|---|---|---|
| EOC1 | 51 | IIIB | G3 | HGSC | 1366 | NA | No | Yes |
| EOC2 | 58 | IIIC | G3 | HGSC | 734.7 | <0.5 cm | No | Yes |
| EOC3 | 40 | IIIC | G3 | HGSC | 89.92 | None | No | Yes |
| EOC4 | 80 | IIIC | G3 | LGSC | 600 | >2 cm | Yes | NA |
| EOC5 | 56 | IIIC | G3 | HGSC | 1201 | NA | No | Yes |
| EOC6 | 56 | IIIB | G3 | HGSC | 1327 | None | No | Yes |
| EOC7 | 63 | IIIC | G3 | HGSC | 962.4 | NA | No | Yes |
| EOC8 | 72 | IIIC | G3 | HGSC | 71.29 | <0.5 cm | No | Yes |
| EOC9 | 49 | IIIB | G3 | HGSC | 381.5 | None | No | Yes |
| EOC10 | 39 | IIIC | G3 | HGSC | 861 | NA | No | Yes |
| EOC11 | 40 | IIIC | G3 | HGSC | >5000 | None | No | Yes |
| EOC12 | 69 | IV | G3 | HGSC | >1000 | <0.2 cm | Yes | Yes |
| EOC13 | 61 | IIIC | N.A. | N.A. | 453.2 | NA | No | Yes |
| EOC14 | 63 | IIIC | G3 | HGSC | 2253 | <1 cm | No | Yes |
| EOC15 | 61 | IV | G3 | Adenocarcinoma | 154 | NA | No | Yes |
| EOC16 | 53 | IV | G3 | HGSC | 893.9 | <1 cm | No | Yes |
| EOC17 | 61 | IIIC | G3 | HGSC | 256.5 | <0.5 cm | NA | NA |
| EOC18 | 52 | IV | G3 | HGSC | N.A. | NA | Yes | Yes |
| EOC19 | 60 | IIIC | G3 | HGSC | 513.7 | NA | No | Yes |
| EOC20 | 57 | III | G3 | HGSC | 1137 | <0.2 cm | No | Yes |
| EOC21 | 47 | IIIC | G3 | HGSC | 465.7 | None | No | Yes |
| EOC22 | 64 | IVB | G3 | HGSC | >5,000 | NA | No | Yes |
| EOC23 | 50 | IVB | G3 | HGSC | 2276 | None | No | Yes |
| EOC24 | 66 | IV | G3 | HGSC | 553.9 | None | No | Yes |
| EOC25 | 58 | IIIC | G3 | HGSC | >5,000 | NA | No | Yes |
| EOC26 | 75 | IV | G3 | HGSC | 1660 | NA | NA | NA |
| EOC27 | 64 | IIIC | G3 | HGSC | 511.1 | None | NA | NA |
| EOC28 | 56 | IIIA | G3 | HGSC | 8.28 | None | No | Yes |
| EOC29 | 54 | IIIC | G3 | HGSC | 317.7 | None | No | Yes |
| EOC30 | 51 | IIIC | G3 | HGSC | 474.7 | <2 cm | NA | Yes |
| EOC31 | 68 | IIIA | G3 | HGSC | 2661 | None | Yes | Yes |
| EOC32 | 67 | III | G3 | HGSC | 295.1 | None | NA | Yes |
| EOC33 | 45 | IIIB | G3 | HGSC | 1487 | <0.3 cm | NA | NA |
| EOC34 | 47 | IIIC | G3 | HGSC | 921.2 | NA | NA | NA |
| EOC35 | 71 | IIIC | G3 | HGSC | 157.4 | 0.1–0.2 cm | No | Yes |
| EOC36 | 56 | IIIC | G3 | HGSC | 425.2 | <1 cm | NA | Yes |
| EOC37 | 48 | IIIC | G3 | HGSC | 276 | NA | No | Yes |
| EOC38 | 50 | IIIC | G3 | HGSC | 1556 | None | No | Yes |
| EOC39 | 50 | IIIC | G3 | HGSC | 1423 | NA | No | Yes |
| EOC40 | 44 | IIIC | G3 | HGSC | 17.98 | None | No | Yes |
NA, not available; HGSC, high-grade serous carcinoma; LGSC, low-grade serous carcinoma.
Exosome isolation and purification
Exosomes were isolated from plasma using the exoEasy Maxi kit as described in the manufacturer's manual. Briefly, 1 volume of buffer XBP was added into 1 volume of plasma. The mix was topped up onto the exoEasy spin column and centrifuged at 500 × g for 1 min at 4°C. The flow-through was discarded and 10 ml buffer XWP was added and centrifuged at 5,000 × g for 5 min at 4°C to remove residual buffer from the column. The spin column was then transferred to a fresh collection tube and 1 ml Buffer XE was added to the membrane and incubated for 1 min. The elute was collected by centrifugation at 500 × g for 5 min at 4°C, which contained the purified exosome fraction. After measuring the total protein concentration of the purified exosomes using BCA assays (Thermo Fisher Scientific, Waltham, MA, USA), the exosome preparations were stored at −80°C and resuspended in PBS for transmission electron microscopy (TEM), western blot analysis (WB), nanoparticle tracking analysis (NTA) and dynamic light scattering (DLS) analysis to verify the nature of the isolated particles.
NTA and DLS analysis
NTA was performed to measure the size and the concentration of the isolated exosomes using NanoSight NS300 (Malvern, Westborough, MA, USA) according to the operating instructions without any changes. In addition, DLS was conducted using Zetasizer Nano (Malvern) referring to the operating instructions without further modifications.
TEM
The isolated exosomes were processed at room temperature for TEM. The samples were diluted 1:5 in 1X PBS prior to fixation with 2.0 % glutaraldehyde (G5882, Sigma-Aldrich). Following fixation, a 75-mesh grid (G2075C; Agar Scientific, Essex, UK) was laid on a drop of sample for 10 min; the grid was then rinsed 10 times with MiliQ H2O (1 min per rinse). Subsequently, the grid was firstly laid on a drop of uranyl acetate (pH 7.0, 2624; SPI-CHEM, West Chester, PA, USA) for 10 min. After rinsing with Milli-Q H2O and methylcellulose uranyl (pH 4.0), the grid was incubated at room temperature for 10 min on a drop of methylcellulose uranyl (pH 4.0, M-6385, Sigma-Aldrich). The exosome samples were eventually analyzed with a FEI Tecnai™ T12 electron microscope (Thermo Fisher Scientific).
Western blot analysis
Exosomes were prepared as described above. RIPA buffer (#9806, Cell Signaling Technology) was used to extract the proteins from the exosomes. In brief, 500 µl of RIPA buffer were added to the exosome samples and this was maintained for 1 h at room temperature. The mixtures were then centrifuged at 12,000 × g for 5 min at 4°C. The supernatants were collected as the exosomal protein extracts. Subsequently, 5X SDS-loading buffer was added to dissolve the proteins from exosomes, diluted to 1X SDS-loading buffer, and then heated at 95°C for 5 min. The samples were centrifuged at 12,000 × g for 5 min at 4°C to remove insoluble precipitates. Supernatants were subsequently loaded onto SDS-PAGE (3% stacking gel, 12% running gel; Bio-Rad, Munich, Germany), running in a Mini Protean 2 electrophoresis system (Bio-Rad). The protein was transferred onto a polyvinylidene fluoride membrane (Bio-Rad) in transfer buffer. After being blocked with 5% non-fat milk in TBST for 1 h at room temperature, the membrane was incubated with the primary antibody, including CD81 (1:1,000), TSG101 (1:1,000), and calnexin (1:1,000) overnight at 4°C. The membrane was rinsed with 1X TBST and this was repeated 3 times (5 min per rinse). The membrane was then labeled with HRP-conjugated secondary antibody (1:2,000; cat. no. 7074; Cell Signaling Technology) for 1 h at room temperature. Again, the membrane was rinsed with 1X TBST and this was repeated 3 times (5 min per rinse). The final products were detected using an enhanced and freshly prepared chemiluminescence (ECL) system (Thermo Fisher Scientific). Cell lysates were also characterized in order to compare with exosome samples.
Proteomics of exosomes
LC-MS/MS was performed for proteomics analysis. In brief, LC-MS/MS was used to analyze the purified proteins from the exosome fractions. After extracting the protein from the exosome samples as mentioned above, the fractions were resolved in two lanes of a 10% SDS-PAGE gel respectively and subsequently stained with Coomassie blue staining solution (B6529, Sigma-Aldrich) at room temperature for 1 h. Each lane was excised and divided into 6 sections equally for in-gel trypsin digestion; following reduction and alkylation, each section was digested with trypsin overnight. Following digestion, the solution was centrifuged at 12,000 × g at room temperature for 20 min to collect the dissolved peptides. TEAB buffer was added and the solution was centrifuged at 12,000 × g at room temperature for 20 min. The filtrates were collected and lyophilized to obtain the dry powder. The peptide samples were dissolved in TEAB buffer and mixed with anhydrous acetonitrile and vortexed for 1 min. TMT reagents (Thermo Fisher Scientific) were added to the resulting solution and maintained for 1 h at room temperature. Subsequently, 5% hydroxylamine was added to terminate the reaction and the samples were ready for MS. The peptides were analyzed with a Q-Exactive LC-MS/MS (Thermo Fisher Scientific). The raw data were converted to mascot generic (mgf) files using Proteome Discoverer version 2.2 (Thermo Fisher Scientific) and the mgf files were then searched against the Uniprot human proteome database using an in-house Mascot Server version 2.4.1 (Matrix Science, London, UK).
Bioinformatics analysis
An online website, Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) is an open database which provides thorough functional annotation tools for researchers to understand versatile biological and functional meanings behind numerous genes (30). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed for the identified differentially expressed genes (DEGs) using the DAVID database (30). A P-value <0.05 was set to distinguish statistically significant enrichment results. The functional interactions between proteins can be plotted to illustrate the molecular mechanisms and signaling pathways of cellular processing. A protein-protein interaction (PPI) network of DEGs was constructed using an open-access bioinformatics tool, the Search Tool for the Retrieval of Interacting Genes (STRING, http://string.embl.de/) database (31) and subsequently was visualized using Cytoscape (32). The confidence score ≥0.7 was set as the cut-off criterion and the Molecular Complex Detection (MCODE) was performed to screen modules of PPI network with a degree cut-off of 2, a node score cut-off of 0.2, a k-core of 2, and a max. depth of 100 (33). In order to supplement the evidence of the identified genes as potential prognostic markers, progression-free survival (PFS) and overall survival (OS) were simulated on a Kaplan Meier-plotter (KM plotter, http://kmplot.com/analysis/), which is capable of assessing the effects of >50,000 genes on survival using >10,000 cancer samples, including breast, ovarian, lung and gastric cancer (GC) samples (34). The patients with ovarian cancer were split into 2 groups according to the expression of a particular gene (high vs. low). The hazard ratio (HR) with 95% confidence intervals and log-rank P-value were calculated and displayed on the webpage.
Procoagulation activity mediated by plasma exosomes in patients with EOC
Exosomes associated procoagulant activity was measured follows: Briefly, 20 µl of each sub-fraction-PBS solution were incubated for 15 min in 100 µl incubation buffer containing 10 mM HEPES, 25 µM negatively charged phospholipids which was composed of dioleoylphosphatidylserine and dioleoylphosphatidylcholine at a 1:9 ratio (840035P and 850375P; Avanti Polar Lipids, Alabaster, AL, USA), 137 mM NaCl, 4 mM KCl, 6 mM CaCl2, 5 mg/ml bovine serum albumin and 5 U/ml hirudin (pH 7.45, H0393; Sigma-Aldrich). Subsequently, 40 µl of 281 nM Factor X (HFX1010; Enzyme Research Laboratories, Ltd., Swansea, UK) was added following 40 µl 5.63 nM Factor VII (HFVII1007; Enzyme Research Laboratories, Ltd.) or blank buffer and 25 µl Factor Xa chromogenic substrate S2765 (82141339; Chromogenix, Milano, Italy) to commence the reaction. The rate of the chromophoric group p-nitroaniline formation was recorded for 90 min at 405 nm and the rate of Factor Xa generation was calculated. The procoagulant activity was expressed as the rate of Factor Xa generation. Recombinant human tissue factor (B4212, Dade® Innovin; Siemens Healthcare Diagnostics, Marburg, Germany) was used as the positive control.
ELISA for GSN, LBP, FGA, and FGG for prognostic verification
The levels of all 4 markers, namely GSN, LBP, FGA and FGG were measured according to the manufacturer's instructions (Fine Biotech). Briefly, 200 µl of Assay Diluent were added to each well. Subsequently, 50 µl of Standard, control, or sample was also added to each well. The plate was incubated at room temperature for 2 h on a horizontal orbital microplate shaker. Each well was aspirated and washed, repeating the process 2 times for a total of 3 washes. Subsequently, 200 µl of Conjugate were added to each well and the plate was incubated at room temperature for 2 h on the shaker again. Each well was aspirated and washed 3 times. A total of 200 µl Substrate Solution were then added to each well and incubated at room temperature for 30 min on the benchtop free from light. Eventually, 50 µl of Stop Solution was added to each well. UV absorption was measured at 450 nm within 30 min using a plate reader (Thermo Fisher Scientific). Wavelength correction was set to 540 or 570 nm.
Statistical analysis
Receiver Operating Characteristic curve (ROC) analysis was performed to evaluate the efficiency of GSN, LBP, FGA and FGG as diagnostic markers. The result of procoagulation assay was analyzed using a Student's t-test as each case was independent from each other. All statistical analyses were performed using GraphPad Prism® software (GraphPad Prism® Version 6).
Results
Isolation and characterization of plasma exosomes in ovarian cancer
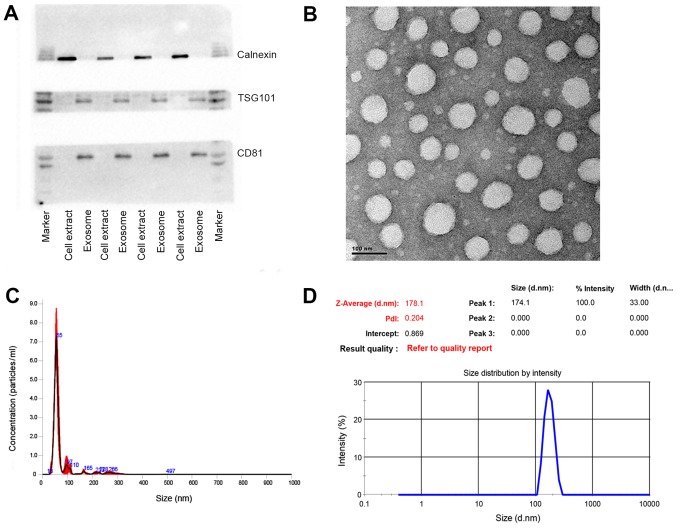
The exosomes were isolated from the patient plasma samples with a commercially available exosome isolation kit. The isolation procedure was referred to the manual without further modifications. Western blot analysis was conducted to detect the exosomal protein markers. As shown in Fig. 1A, TSG101 and CD81, as exosomal protein markers that are involved in exosome biogenesis, maturation and secretion, were detected in all the plasma exosome samples. On the contrary, calnexin, an endoplasmic reticular protein, was only found to be present in the cell extracts, proving that the isolated exosomes were highly purified and not contaminated by redundant intracellular components. TEM was utilized to further characterize the size and morphology of the exosomes. A number of spherical particles were detected on the copper meshes. The size remained within the range of 30–100 nm and approximately 70 nm in diameter (Fig. 1B). Other methodologies were applied to confirm the size as well. For instance, NTA, as a conventional method of characterizing exosomes, was exploited to measure the size and concentration based on the tracking of Brownian movement (35). The mean size was approximately 70 nm and the concentration was approximately 1.53×108 following a 1,000-fold dilution (Fig. 1C). Another technique, DLS, was also applied to measure the size that was greatly larger due to the aquated membrane covering exosomes (Fig. 1D). Another crucial parameter, the polydispersity index (PDI) was also evaluated by this technique to characterize the size distribution of the exosomes. The PDI was approximately 0.22, showing relatively even size distribution of exosomes, which could also be confirmed by the sharp single peak in NTA analysis. Taken together, these results demonstrated that the exosomes were successfully isolated from the plasma with high purity and well characterized by various methods.
Figure 1.
Various characterizations of exosomes. (A) Western blot analysis of the typical exosomal proteins, TSG 101 and CD81, and the endoplasmic reticular protein, calnexin. (B) Transmission electron microscopy (TEM) of exosomes. (C) Nanoparticle tracking analysis (NTA) of exosomes. (D) Dynamic light scattering (DLS) analysis of exosomes.
Proteomics analysis of exosomes derived from patient plasma samples
To determine the protein profile of the exosomes, total exosomal proteins were separated by SDS-PAGE on a 12% gel. After every band was cut into sections and subjected to trypsin digestion in 1.5 ml Eppendorf tubes respectively overnight, the extracted peptides were analyzed by LC-MS/MS. A total of 262 proteins in the cancerous plasma exosomes and 257 proteins in the non-cancerous plasma exosomes were identified from the protein database. In total, 225 proteins were present in both samples (Fig. 2A). Based on the protein information following crude screening, the fold change (FC) value and P-value were calculated from the t-test. An FC >1.5 or FC <2/3 and P-value <0.05 were set as the criteria for the selection of upregulated or downregulated genes. Eventually, 50 genes were screened as DEGs, which were depicted in a volcano plot (Fig. 2B). The red dots represent overexpressed genes, which contained 19 genes; while the green dots represent downregulated genes, 31 in sum.
Figure 2.
Bioinformatics analysis of proteomic results. (A) Venn diagram of the number of detected proteins in cancerous and non-cancerous patient samples and their intersection. (B) Volcano plot of differently expressed proteins. Fold change (FC) values >1.5 or <2/3 and a P-value <0.05 were set as the filter criteria. Green dots represent downregulated proteins; red dots represent upregulated proteins. (C) Gene Ontology (GO) enrichment analysis of differently expressed proteins. Top 10 items are listed in biological process, cell component and molecular function, respectively based on their statistical significance (30,31). (D) KEGG pathway enrichment analysis of differently expressed proteins. Top 10 items are listed on their statistical significance (30,31). (E) Protein-protein interaction (PPI) network of the enriched proteins and their corresponding cell signaling pathways (32,33).
Annotation of identified genes
GO analysis was performed for the classification of various molecular functions. The identified genes were categorized into several groups, including biological process, cellular components and molecular functions. In Fig. 2C, the 10 most statistically significant items in biological process, cellular components and molecular functions are listed in sequence. For biological process, the exosomal genes were mainly involved in immune response, protein activation cascade, complement activation, and so on, which may indicate that plasma exosomes contribute to the proper functioning of immune system. The aberrant regulation of body immunity could partially involve the tumorigenesis and metastasis of EOC. For instance, in one study, exosomes were revealed to be able to facilitate the intercellular communication in tumor microenvironment, thereby remodeling normal macrophages to a tumor-activated phenotype with the assistance of hypoxia-inducible factors (36). As for the GO analysis of cellular components, these DEGs were mainly associated with extracellular space, extracellular region part, blood microparticle, extracellular region, extracellular exosome, extracellular vesicle, etc., confirming that these proteins were originated from plasma exosomes. GO enrichment analysis in molecular functions was also conducted for these 50 genes. Some of the genes involved in the following molecular functions: antigen binding, immunoglobulin receptor binding, serine-type endopeptidase activity, and so on. These enriched molecular functions were closely relevant to corresponding biological process the other way around, such as immune response, indicating that the molecular function of each gene mutually interacted and participated in these meaningful biological processes. Additionally, other processes may also be related to particular functions of the exosomes, such as complement binding and receptor binding.
KEGG pathway enrichment and PPI network
As can be seen from Fig. 2D, the top 10 pathways that contain these exosomal proteins, among which complement and coagulation cascades and platelet activation were closely related to abnormal coagulation function of patients with EOC, which could be an explanation for the hypercoagulability in a number of patients with EOC. Pathways of Staphylococcus aureus infection, pertussis and systemic lupus erythematosus may result from immune dysfunction, which was in accordance with the results of GO enrichment analysis. Most other pathways ought to be involved in tumor growth, progression and apoptosis, including Fc γ Receptor-mediated phagocytosis and the NF-κB signaling pathway. Either direct or indirect interactions between DEGs were plotted in the PPI network (Fig. 2E). It is worth noting that FGG and FGA were relevant to both complement and coagulation cascades, and platelet activation, implying that exosomal FGG and FGA may be key genes regulating the coagulation cascade in patients with EOC. Moreover, GSN and LBP were linked to Fc γ Receptor-mediated phagocytosis and the NF-κB signaling pathway, respectively, which made it more explicit that GSN and LBP originated from exosomes played a role in tumor apoptosis.
Exosome-associated hypercoagulability in patients with EOC
In clinical practice, patients with EOC often suffer from venous thromboembolism (VTE) due to the aberrant activation of platelet and coagulation dysfunction (37). Most of them remain in a hypercoagulable state and the status of coagulation has emerged as an indicator for EOC. As shown in Table III, among the 50 DEGs, 10 genes participated in the complement and coagulation cascade, namely coagulation factor XIII A chain (F13A1), coagulation factor IX (F9), serpin family A member 1 (SERPINA1), FGA, FGB, FGG, complement C9 (C9), complement component 4 binding protein alpha (C4BPA), complement C8 alpha chain (C8A) and complement component 4 binding protein beta (C4BPB), the majority of which were upregulated in patients with EOC, suggesting that the overexpression of these exosomal genes mediates the coagulation cascade, as well as platelet activation and induces blood clotting. Therefore, the procoagulation assay was conducted to verify the discrepant influences of cancerous and non-cancerous plasma exosomes on the rate of clotting (Fig. 3). The EOC group exhibited a much higher degree of coagulation (P<0.0001), which proved the promotion and acceleration effects on coagulation by EOC exosomes and confirmed the estimated pathways and the subsequent regulations.
Table III.
Information of identified differentially expressed genes (DEGs).
| Accession no. | Gene ID | Fold change | P-value | Expression |
|---|---|---|---|---|
| P01706 | IGLV2-11 | 0.218133124 | 0.046131844 | Downregulated |
| P01705 | IGLV2-23 | 0.301249245 | 0.032502181 | Downregulated |
| P04211 | IGLV7-43 | 0.337616894 | 0.003289896 | Downregulated |
| A0A075B6K0 | IGLV3-16 | 0.342103003 | 0.027922644 | Downregulated |
| A0A075B6S2 | IGKV2D-29 | 0.361179729 | 0.018761308 | Downregulated |
| P01780 | IGHV3-7 | 0.413239637 | 0.000846937 | Downregulated |
| A0A0B4J1V0 | IGHV3-15 | 0.413556853 | 0.003187854 | Downregulated |
| P01701 | IGLV1-51 | 0.466774595 | 0.017830268 | Downregulated |
| P01859 | IGHG2 | 0.47139263 | 0.017141917 | Downregulated |
| P06396 | GSN | 0.47577625 | 0.000550377 | Downregulated |
| P06312 | IGKV4-1 | 0.480672648 | 0.002873312 | Downregulated |
| P00488 | F13A1 | 0.481024159 | 0.005927534 | Downregulated |
| P01860 | IGHG3 | 0.481916606 | 0.027922715 | Downregulated |
| A0A0B4J1V2 | IGHV2-26 | 0.48266588 | 0.014221922 | Downregulated |
| A0A075B7D0 | IGHV1OR15-1 | 0.486125479 | 0.032529504 | Downregulated |
| A0A0B4J1X8 | IGHV3-43 | 0.488511091 | 0.006554509 | Downregulated |
| A0A0B4J1Y9 | IGHV3-72 | 0.493082012 | 0.001689489 | Downregulated |
| A0A0C4DH35 | IGHV3-35 | 0.494979343 | 0.006757808 | Downregulated |
| P05452 | CLEC3B | 0.507434994 | 0.004828297 | Downregulated |
| A0A075B6I9 | IGLV7-46 | 0.511105442 | 0.007728489 | Downregulated |
| S4R460 | IGHV3OR16-9 | 0.520150846 | 0.010756891 | Downregulated |
| A0A075B6R9 | IGKV2D-24 | 0.52141676 | 0.035994038 | Downregulated |
| Q9NQ79 | CRTAC1 | 0.533552214 | 0.009071347 | Downregulated |
| P0DOY2 | IGLC2 | 0.540646515 | 0.025332541 | Downregulated |
| A0A0A0MS15 | IGHV3-49 | 0.567732527 | 0.014566172 | Downregulated |
| A0A0B4J1U7 | IGHV6-1 | 0.593352797 | 0.033839926 | Downregulated |
| A0A075B7B8 | IGHV3OR16-12 | 0.604561348 | 0.003882883 | Downregulated |
| A0A0B4J231 | IGLL5 | 0.608827162 | 0.049914959 | Downregulated |
| P01834 | IGKC | 0.641872888 | 0.032179959 | Downregulated |
| P01742 | IGHV1-69 | 0.646993729 | 0.042670996 | Downregulated |
| F8W1S1 | KRT74 | 0.652950133 | 0.045547508 | Downregulated |
| Q99784 | OLFM1 | 1.520266138 | 0.016352632 | Upregulated |
| P0C0L4 | C4A | 1.523579037 | 0.011662874 | Upregulated |
| Q03591 | CFHR1 | 1.528039639 | 0.000491116 | Upregulated |
| P36980 | CFHR2 | 1.539803521 | 0.02091088 | Upregulated |
| P02743 | APCS | 1.57664451 | 0.01704065 | Upregulated |
| P06702 | S100A9 | 1.644481596 | 0.01755607 | Upregulated |
| E7ETH0 | CFI | 1.732825958 | 0.031482969 | Upregulated |
| P02748 | C9 | 1.867280995 | 0.024739231 | Upregulated |
| P01009 | SERPINA1 | 1.885728943 | 0.033000428 | Upregulated |
| P02750 | LRG1 | 1.939799893 | 0.045335361 | Upregulated |
| P05546 | SERPIND1 | 2.289112647 | 0.020819823 | Upregulated |
| P02763 | ORM1 | 2.538489431 | 0.007998879 | Upregulated |
| P18428 | LBP | 2.616390498 | 0.012556488 | Upregulated |
| A0A096LPE2 | SAA2-SAA4 | 3.287876685 | 0.005920009 | Upregulated |
| P02671 | FGA | 3.31200379 | 0.02307751 | Upregulated |
| P02675 | FGB | 6.618243962 | 0.018788218 | Upregulated |
| P02679 | FGG | 8.57650775 | 0.025580522 | Upregulated |
| P00738 | HP | 10.72525182 | 0.018597109 | Upregulated |
| P02741 | CRP | 11.99122018 | 0.026727478 | Upregulated |
Figure 3.
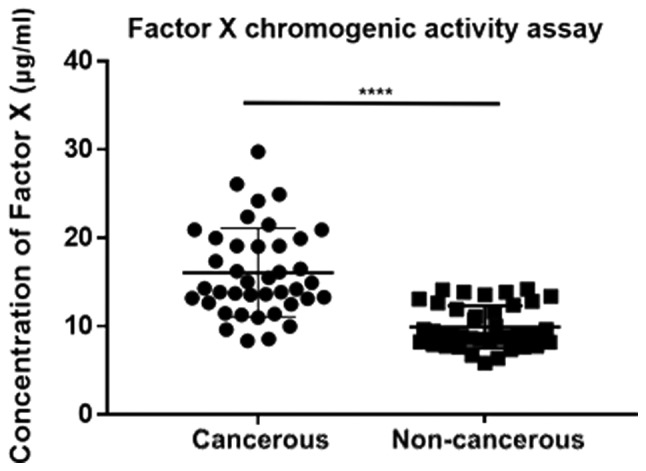
Procoagulation assay of exosomes based on the concentration of Factor X via a chromogenic methodology (****P<0.0001).
Identification of diagnostic and prognostic biomarkers based on the results of proteomics analysis
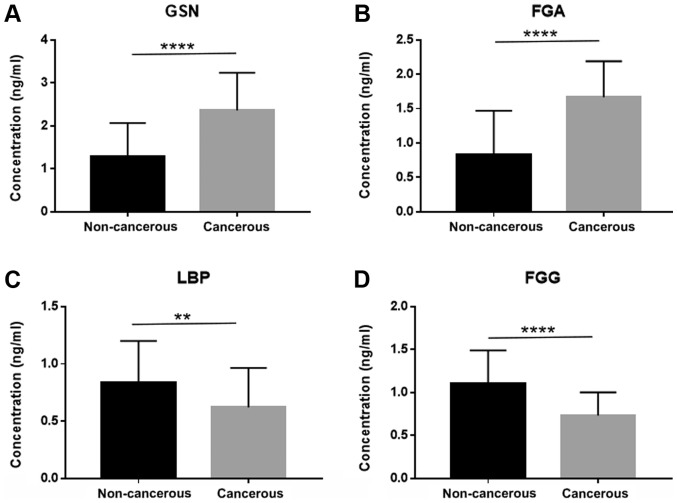
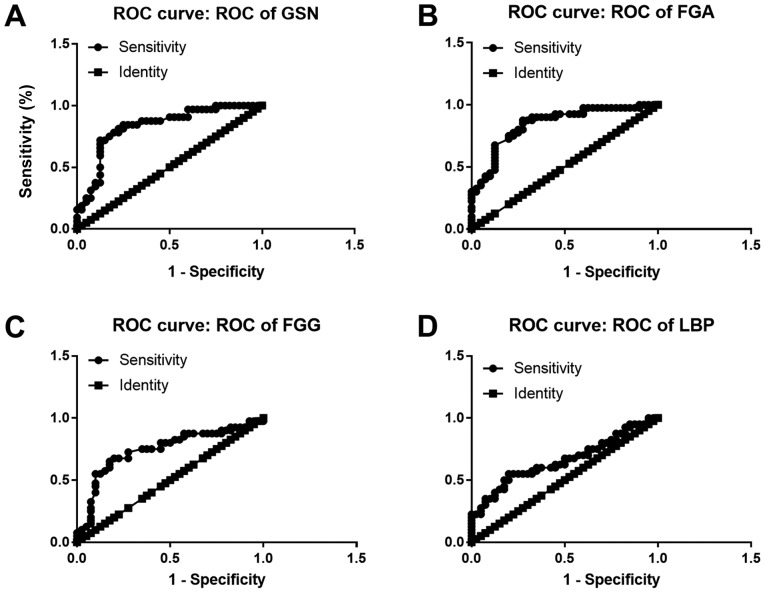
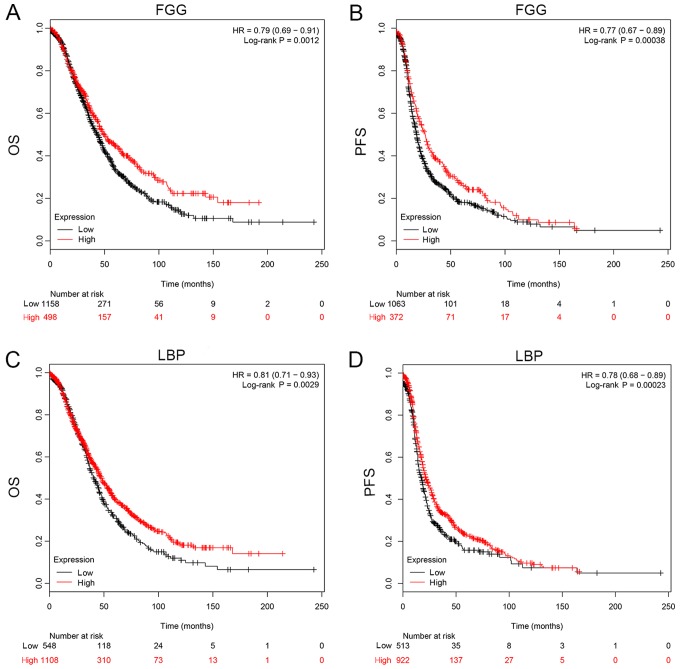
As mentioned above, 50 DEGs were identified and sorted according to FC value with statistical significance, among which 19 genes were all enriched in GO analysis, KEGG pathway analysis, as well as the PPI network. In total, 4 genes (LBP, FGG, FGA and GSN) were eventually selected as the potential diagnostic and prognostic biomarkers for the reason that they were all involved in two categories of function enrichment, namely coagulation and apoptosis related pathways. The proteomics analysis revealed that the protein levels of FGA and GSN were upregulated. FGG and LBP, on the contrary, were downregulated. Another cohort of patients (40 with EOC and 40 non-cancerous subjects) was enrolled in the validation assay of these 4 genes. ELISA was conducted and the sensitivity and specificity were accessed by ROC analysis. The expression of these 4 genes in exosomes derived from the 80 patient plasma samples is shown in Fig. 4. The FGA and GSN levels were significantly elevated in the cancer group, which was consistent with what was observed in the proteomics analysis. Similarly, the FGG and LBP levels were downregulated in the cancer patient cohort. FGA conferred the highest area under the curve (AUC) among all 4 candidates, which was approximately 0.8459 (Fig. 5B). The lowest AUC belonged to LBP, approximately 0.6588 (Fig. 5D). Thus, FGA emerged as a promising biomarker for the diagnosis of EOC. For prognostic biomarker screening, the OS and PFS of patients as regards FGG and LBP expression were estimated on the Kaplan Meier-plotter, which is an open-access online database for estimating the progression-free survival (PFS) and overall survival (OS) of over 50,000 genes on survival using >10,000 cancer samples, including breast, ovarian, lung and gastric cancer (34). The patients with ovarian cancer were divided into 2 groups according to the expression of a particular gene (high vs. low). The hazard ratio (HR) with 95% confidence intervals and log-rank P-value were calculated. As can be seen from Fig. 6, FGG (HR, 0.79 for OS and 0.77 for PFS) and LBP (HR, 0.81 for OS and 0.78 for PFS) could both be applied to predict the prognosis of patients with EOC. It was found that a high mRNA expression of FGG or LBP was associated with a shorter PFS and OS for patients with EOC, reflecting a worse prognosis. However, it should be noted that the potential for these genes as prognostic biomarkers was only investigated on a Kaplan Meier-plotter that contains thousands of cases. In fact, the survival curve was derived from the results of gene expression, but not from exosomal gene expression. Further studies are required for the validation and verification of these genes in exosomes as prognostic indicators.
Figure 4.
ELISA data of exosomal GSN, FGA, LBP and FGG expression. (A) Exosomal GSN expression in cancer and non-cancerous patients (****P<0.0001). (B) Exosomal FGA expression in cancer and non-cancerous patients (****P<0.0001). (C) Exosomal LBP expression in cancer and non-cancerous patients (**P<0.01). (D) Exosomal FGG expression in cancer and non-cancerous patients (****P<0.0001). GSN, gelsolin; FGA, fibrinogen alpha chain; LBP, lipopolysac-charide binding protein; FGG, fibrinogen gamma chain.
Figure 5.
Receiver operating characteristic (ROC) curve assay of the selected exosomal proteins as diagnostic biomarkers. (A) ROC-AUC of GSN was 0.8309 (0.7343 to 0.9274, P<0.0001). (B) ROC-AUC of FGA was 0.8459 (0.7602 to 0.9317, P<0.0001). (C) ROC-AUC of FGG was 0.7447 (0.6323 to 0.8571, P<0.0001). (D) ROC-AUC of LBP was 0.6588 (0.5381 to 0.7794, P<0.001). GSN, gelsolin; FGA, fibrinogen alpha chain; LBP, lipopolysaccharide binding protein; FGG, fibrinogen gamma chain.
Figure 6.
Prognostic significance of FGG and LBP estimated by simulated Kaplan-Meier analysis. Both overall survival (OS) and progression-free survival (PFS) were evaluated. The hazard ratio (HR) was utilized to describe the efficiency of prognostic markers and log-rank test was performed to demonstrate their statistical significance (34). (A) Estimated OS of FGG in ovarian cancer. (B) Estimated PFS of FGG in ovarian cancer. (C) Estimated OS of LBP in ovarian cancer. (D) Estimated PFS of LBP in ovarian cancer. FGG, fibrinogen gamma chain; LBP, lipopolysaccharide binding protein.
Discussion
Exosomes belong to extracellular vesicles with 30–100 nm in diameter and are secreted by various types of cells. Researchers nowadays pay more attention to the studies of exosomes to investigate the important roles in intercellular communications. Despite tremendous research being made on exosomes in a variety of types of cancer, these studies have shed light into the pivotal roles of several exosomal molecules in tumorigenesis, evasion, metastasis and recurrence (5–12). However, there were few reports on the systemic proteomic analysis of exosomes, particularly for plasma exosomes in ovarian cancer, as compared to other types of cancer (38–40). It is of great importance to obtain a more comprehensive proteomic profile of the plasma exosomes in ovarian cancer, when developing some novel diagnostic and prognostic biomarkers based on exosomes. Furthermore, investigating the protein composition of exosomes would supplement the understanding of the mechanism of their biogenesis and the functional roles in this most lethal gynecological malignancy.
In this study, exosomes were enriched from EOC patient plasma with spherical morphology and mostly 30–100 nm in diameter. TEM, NTA and DLS were also utilized to confirm the size, concentration and size distribution of the exosomes. Western blot analysis was performed to examine the expression of typical exosomal proteins, including TSG101 and CD81. The high purity of the exosomes was not only ensured during isolation procedures, but was also verified by an endoplasmic reticular marker, calnexin, which was not observed in exosome stains. Based on these multifaceted characterizations, plasma exosomes with a high purity were obtained, which laid the foundation for the subsequent proteomics analysis. In total, 262 exosomal proteins were detected from EOC and non-cancerous samples by performing an LC-MS/MS in combination with TMT, which is the first ovarian cancer exosome study based on this technique, at least to the best of our knowledge. A total of 50 DEGs were screened out with distinct annotation to cellular components, biological process, and molecular function respectively. Among these, 19 genes were plotted in the PPI network to explain their interaction and involved signaling pathways. Two particular categories of pathway should be paid more attention to, including coagulation and apoptosis-related pathways. In this study, we demonstrated the procoagulation activity of exosomes derived from EOC patient plasma. The bioinformatics analysis provided an explanation for hypercoagulation that occurs in patients with EOC at the molecular level. FGG, FGA, C9, C4BPA, C4BPB and C4A, as well as other genes were found to be relevant to coagulation in this study.
Among all DEGs, 4 genes, namely GSN, FGG, LBP and FGA were selected as diagnostic markers. FGA ought to be the most promising diagnostic biomarker based on the results of ROC analysis. FGA has been found to serve as a diagnostic marker in several studies. For instance, Davalieva et al reported the non-invasive biomarkers in urine with higher specificity than prostate-specific antigen (PSA) in patients with prostate cancer; a number of genes, including FGG and FGA were observed to be present with different abundances, which could be potential biomarker candidates (41). In another study, the serum levels of the proteins, alpha 2-HS glycoprotein (AHSG), FGA and apolipoprotein A1 (APOA-I), were identified as diagnostic biomarkers for gastric cancer (GC); ELISA data suggested that the serum levels of FGA, AHSG and APOA-I in patients with GC differed significantly as compared to the healthy volunteers; moreover, the serum levels of these three proteins were associated with TNM stages and could reflect tumor burden (42). The FGG gene mediates the formation of the fibrinogen γ chain, a subunit of the fibrinogen protein. This protein is important for coagulation, which is needed to attenuate excessive bleeding following injury (43). Besides being the diagnostic factors, FGA and FGG have also been proven to serve as novel prognostic biomarkers for GC. The elevated expression of FGA and FGG predicted poorer prognosis of GC patients according to the survival analysis (44).
In prostate cancer (PC), GSN has also been demonstrated to interact with androgen receptor (AR) in a dose-dependent manner, which can enhance the bioactivity of AR. The blockage of the interaction between GSN and AR may thus emerge as a promising therapeutic target for the treatment of PC (45). Another study focused on the prognostic value of cytoskeleton-associated proteins for ovarian cancer (46). In silico analysis was performed using the cancer genome atlas (TCGA) and 17 cytoskeletal proteins, including GSN, playing a role in tumor progression (46). GSN has also been shown to be involved in TGF-β1 induced epithelial to mesenchymal transition (EMT) in breast cancer (47). An increased GSN expression was shown to result in alterations of cell proliferation and cell cycle, due to the modification of the actin filament assembly by GSN. The expression of typical markers for EMT process, such as N-cadherin, E-cadherin, and vimentin was altered by the silencing of GSN, suggesting that TGF-β1 triggered aberrant expression of GSN could affect the EMT process in breast cancer cells (47). A high level of GSN was found to be involved in chemoresistance in gynecological cancer cells compared with their sensitive cell types. Cisplatin-induced GSN downregulation is associated with its cleavage and apoptosis. In resistant cells, GSN was highly expressed and cisplatin failed to abolish the downstream interaction, leading to the attenuated apoptosis. In addition, the overexpression of GSN was closely associated with a more aggressive behavior and a shorter OS, as well as PFS. These findings are in agreement with the notion that GSN is a crucial role in chemosensitivity in gynecological cell, which could be exploited as diagnostic and prognostic markers (48).
LBP has been studied in other types of cancer to explore its potential as the diagnostic and/or prognostic biomarkers. An increased level of LBP has been shown to be closely associated with colorectal (CRC) and gallbladder cancer (49,50). Gut microbiota dysbiosis impairs the intestinal barrier function and elevates plasma lipopolysaccharide levels, thereby promoting endotoxemia and contributing to the development of CRC. The reduction in plasma LBP levels may be a crucial parameter for patients newly diagnosed with CRC (51). Additionally, LBP may also be useful in renal cancer (52). Cox regression analysis was performed to explore the survival in association with age, sex, clinicopathological parameters and LBP expression. A high expression of LBP indicates conventional renal cell carcinoma patients at a high risk of post-operative progression that may require optimized active surveillance and prompt regime (52). Circulating exosomes containing LBP emerged as a biomarker for non-small cell lung cancer (NSCLC) (53). LBP was found to be well distinguished between patients with metastatic and patients with non-metastatic NSCLC, implying LBP as a promising and effective indicator of metastatic NSCLC (53).
Due to the heterogeneity and frequent relapse of ovarian cancer, it is of great importance to discover biomarkers for early diagnosis of ovarian cancer. The study by Liang et al demonstrated that the exosomal proteins were highly enriched in signaling pathways associated with carcinogenesis. A number of proteins were overexpressed in both tissue and exosomes, including tubulin beta-3 chain (TUBB3), epithelial cell surface antigen (EpCAM), claudin 3 (CLDN3), proliferation cell nuclear antigen (PCNA), epidermal growth factor receptor (EGFR), apolipoprotein E (APOE), fatty acid synthase (FASN), etc., which may serve as potential diagnostic markers and therapeutic targets for the treatment of ovarian cancer (38). In one study, EpCAM was demonstrated to be associated with the remote metastases in advanced endometrial cancer (54). In clinical practice, however, EpCAM was not a conventional biomarker to be evaluated, implying that the sensitivity and the specificity for ovarian cancer is not sufficient enough. A more promising way is to discover other biomarkers with high sensitivity and specificity.
Apart from ovarian cancer, proteomics analysis has been conducted in a number of other types of cancer. In one study, exosomes were enriched from the serum of patients with prostate cancer and proteomic profiling was performed using LC-MS/MS so as to reveal distinct proteins across different ethnicities (55). Large quantities of novel proteins were discovered that appeared to be ethnicity-specific in prostate cancer, including Iroquois homeobox protein 5 (IRX5), mitochondrial tumor suppressor 1 isoform 4 (MTS1) and trinucleotide repeat containing 6B isoform 3 (TNR6B). The purpose of that study was to find drug targets for the prostate cancer patients with particular ethnicity. However, those authors failed to recruit a validation cohort to verify the feasibility of these potential biomarkers which should be further improved (55).
Furthermore, proteomics analysis has also been performed for the identification of biomarkers in exosomes derived from patients with pancreatic cancer. An et al (56) reported the proteomic results of exosomes from the sera of ten patients with locally advanced pancreatic cancer. The quantitative analysis was conducted using the iTRAQ method. Each sample contained 700 to 800 exosomal proteins, several of which may be relevant to metastasis and chemoresistance of pancreatic cancer. The exosomes at various time points were also collected to compare the alterations of proteomic profiles during the treatment period. In total, 8 proteins were identified to show universal treatment-specific changes, supporting the importance of tumor-derived exosomes in pancreatic cancer progression and metastasis. OBSL1 and PLF4 displayed the highest treatment response among 8 proteins, which could be favorable candidates for biomarker development (56).
Recently, Chen et al (57) presented a quantitative proteomics analysis of exosomes purified from the colorectal cancer (CRC) patient serum samples. A total of 918 proteins were detected and 725 of these were found in the Exocarta proteins list. In comparison with normal volunteers, 36 proteins were upregulated and 22 proteins were downregulated in the serum exosomes of patients with CRC. Bioinformatics analysis revealed that upregulated proteins were involved in processes relevant to metastasis; while the downregulated proteins mainly contributed to tumorigenesis and cell survival (57). Some of these differently expressed proteins may be promising diagnostic and/or prognostic indicators; however, no further verification analysis was performed to establish the association between clinical characteristics and the proteomics results, which should be further elucidated for the development and application in clinical practice.
Notably, this study covered several important clinical needs in ovarian cancer. On the one hand, this study is the first (to the best of our knowledge) to propose exosomal GSN, FGG, FGA and LBP to be the diagnostic biomarkers of ovarian cancer, which was verified and validated by the ELISA. The diagnostic molecular signature identified in exosomes would perfectly supplement the current diagnostic procedure based on the tissue samples, reducing the number of invasive biopsies needed. This proteomics approach paves the way for the identification of proteomic signatures in exosomes that are more relevant to clinical characteristics. On the other hand, from the Kaplan Meier-plotter, we found the promising prognostic values of FGG and LBP. However, the survival results from that website were concluded based on the gene expression in tissue samples, but not in exosomes, attenuating the reliability of exosomal FGG and LBP as the potential prognostic biomarkers. Therefore, the survival data should be summarized according to the exosomal expression of FGG and LBP in future studies, which could complement the limitations of the current study. In fact, the patients have been under observations to acquaint their updated follow-up statuses. Upon completion, the survival curves will be constructed to enhance the values of exosomal FGG and LBP as the prognostic factors. Given that ovarian cancer is closely associated with the high challenge of metastatic and chemoresistant characteristics, our exosomal proteomic results may provide some new aspects and directions for future studies. Due to the limited sample size of this study, further research should be further conducted to reveal more prevalent and insightful markers. Additionally, we only explored the diagnostic and prognostic potentials of the individual exosomal protein. The combined utilization of our findings and clinical markers, such as CA125, could emerge as an encouraging perspective for a more in-depth investigation.
In conclusion, in this study, we successfully isolated and purified exosomes from plasma in patients with EOC, which were characterized by various approaches to prove the high quality of the isolated exosomes. To the best of our knowledge, this study is the first to use LC-MS/MS combined with TMT for the proteomics analysis of ovarian cancer-derived exosomes. The subsequent GO analysis, KEGG pathway and PPI network described the mutual interactions of DEGs, indicating that some of DEGs were involved in the mediation of coagulation cascade relevant to hypercoagulable state in patients with EOC. The procoagulation assay also proved that exosomes enriched from EOC patient plasma could enhance and accelerate the coagulation process. In total, 4 genes emerged as promising diagnostic and 2 as potential prognostic indicators. The common proteins associated with exosome biogenesis may provide some new information on understanding the mechanisms of exosome secretion in ovarian cancer. Further studies should be conducted to validate some candidate prognostic markers in one separate cohort of patients and confirm the functional roles of exosomes in the malignant disease. More patients should be included to reduce bias and enhance the reproducibility and reliability of the study, particularly for proteomic analysis. Limited sample size may compromise the robustness.
Acknowledgments
Special and sincere thanks were given to Polaris Biology (Shanghai, China) for their heartful and prompt assistance in this study.
Funding
This study was supported by the National Natural Science Foundation of China (no. 81602270).
Availability of data and materials
The datasets and materials used and/or analyzed in this study are available from the corresponding author on reasonable request.
Authors' contributions
WZ and XO conceived and designed, and conducted and drafted the manuscript. XW revised the initial design of the study, incorporated the ELISA for validation, and provided suggestions and solutions to the difficulties encountered in the project. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study and experimental procedures were approved by the Human Research Ethics Committee of Fudan University Shanghai Cancer Center (Shanghai, China). Written informed consent was obtained from the patients or patients' families.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. 2011;61:183–203. doi: 10.3322/caac.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seidman JD, Horkayne-Szakaly I, Haiba M, Boice CR, Kurman RJ, Ronnett BM. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol. 2004;23:41–44. doi: 10.1097/01.pgp.0000101080.35393.16. [DOI] [PubMed] [Google Scholar]
- 4.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riches A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells - a new regulatory pathway. Eur J Cancer. 2014;50:1025–1034. doi: 10.1016/j.ejca.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Skogberg G, Lundberg V, Berglund M, Gudmundsdottir J, Telemo E, Lindgren S, Ekwall O. Human thymic epithelial primary cells produce exosomes carrying tissue-restricted antigens. Immunol Cell Biol. 2015;93:727–734. doi: 10.1038/icb.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitt JM, Charrier M, Viaud S, André F, Besse B, Chaput N, Zitvogel L. Dendritic cell-derived exosomes as immunotherapies in the fight against cancer. J Immunol. 2014;193:1006–1011. doi: 10.4049/jimmunol.1400703. [DOI] [PubMed] [Google Scholar]
- 9.Lugini L, Cecchetti S, Huber V, Luciani F, Macchia G, Spadaro F, Paris L, Abalsamo L, Colone M, Molinari A, et al. Immune surveillance properties of human NK cell-derived exosomes. J Immunol. 2012;189:2833–2842. doi: 10.4049/jimmunol.1101988. [DOI] [PubMed] [Google Scholar]
- 10.Cai Z, Yang F, Yu L, Yu Z, Jiang L, Wang Q, Yang Y, Wang L, Cao X, Wang J. Activated T cell exosomes promote tumor invasion via Fas signaling pathway. J Immunol. 2012;188:5954–5961. doi: 10.4049/jimmunol.1103466. [DOI] [PubMed] [Google Scholar]
- 11.Barrès C, Blanc L, Bette-Bobillo P, André S, Mamoun R, Gabius HJ, Vidal M. Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood. 2010;115:696–705. doi: 10.1182/blood-2009-07-231449. [DOI] [PubMed] [Google Scholar]
- 12.Xiao H, Lässer C, Shelke GV, Wang J, Rådinger M, Lunavat TR, Malmhäll C, Lin LH, Li J, Li L, et al. Mast cell exosomes promote lung adenocarcinoma cell proliferation - role of KIT-stem cell factor signaling. Cell Commun Signal. 2014;12:64. doi: 10.1186/s12964-014-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, Clayton A. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi DS, Lee JM, Park GW, Lim HW, Bang JY, Kim YK, Kwon KH, Kwon HJ, Kim KP, Gho YS. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J Proteome Res. 2007;6:4646–4655. doi: 10.1021/pr070192y. [DOI] [PubMed] [Google Scholar]
- 15.Xiao GY, Cheng CC, Chiang YS, Cheng WT, Liu IH, Wu SC. Exosomal miR-10a derived from amniotic fluid stem cells preserves ovarian follicles after chemotherapy. Sci Rep. 2016;6:23120. doi: 10.1038/srep23120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKiernan J, Donovan MJ, O'Neill V, Bentink S, Noerholm M, Belzer S, Skog J, Kattan MW, Partin A, Andriole G, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2016;2:882–889. doi: 10.1001/jamaoncol.2016.0097. [DOI] [PubMed] [Google Scholar]
- 17.Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 18.Inal JM, Kosgodage U, Azam S, Stratton D, Antwi-Baffour S, Lange S. Blood/plasma secretome and microvesicles. Biochim Biophys Acta. 2013;1834:2317–2325. doi: 10.1016/j.bbapap.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Severino V, Dumonceau JM, Delhaye M, Moll S, Annessi-Ramseyer I, Robin X, Frossard JL, Farina A. Extracellular vesicles in bile as markers of malignant biliary stenoses. Gastroenterology. 2017;153:495–504e8. doi: 10.1053/j.gastro.2017.04.043. [DOI] [PubMed] [Google Scholar]
- 20.Sharma S, Gillespie BM, Palanisamy V, Gimzewski JK. Quantitative nanostructural and single-molecule force spectroscopy biomolecular analysis of human-saliva-derived exosomes. Langmuir. 2011;27:14394–14400. doi: 10.1021/la2038763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau C, Kim Y, Chia D, Spielmann N, Eibl G, Elashoff D, Wei F, Lin YL, Moro A, Grogan T, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288:26888–26897. doi: 10.1074/jbc.M113.452458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menay F, Herschlik L, De Toro J, Cocozza F, Tsacalian R, Gravisaco MJ, Di Sciullo MP, Vendrell A, Waldner CI, Mongini C. Exosomes isolated from ascites of T-cell lymphoma-bearing mice expressing surface CD24 and HSP-90 induce a tumor-specific immune response. Front Immunol. 2017;8:286. doi: 10.3389/fimmu.2017.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purushothaman A, Bandari SK, Liu J, Mobley JA, Brown EE, Sanderson RD. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J Biol Chem. 2016;291:1652–1663. doi: 10.1074/jbc.M115.686295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen SW, Sceneay J, Lima LG, Wong CS, Becker M, Krumeich S, Lobb RJ, Castillo V, Wong KN, Ellis S, et al. The biodistri-bution and immune suppressive effects of breast cancer-derived exosomes. Cancer Res. 2016;76:6816–6827. doi: 10.1158/0008-5472.CAN-16-0868. [DOI] [PubMed] [Google Scholar]
- 25.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 26.Peng P, Yan Y, Keng S. Exosomes in the ascites of ovarian cancer patients: Origin and effects on anti-tumor immunity. Oncol Rep. 2011;25:749–762. doi: 10.3892/or.2010.1119. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Sherman-Baust CA, Tsai-Turton M, Bristow RE, Roden RB, Morin PJ. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer. 2009;9:244. doi: 10.1186/1471-2407-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 29.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 30.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43D: 2015:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 35.Wright M. Nanoparticle tracking analysis for the multiparameter characterization and counting of nanoparticle suspensions. Methods Mol Biol. 2012;906:511–524. doi: 10.1007/978-1-61779-953-2_41. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Zhou J, Li X, Wang X, Lin Y, Wang X. Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor-promoted phenotype. Cancer Lett. 2018;435:80–91. doi: 10.1016/j.canlet.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Matsuo K, Hasegawa K, Yoshino K, Murakami R, Hisamatsu T, Stone RL, Previs RA, Hansen JM, Ikeda Y, Miyara A, et al. Venous thromboembolism, interleukin-6 and survival outcomes in patients with advanced ovarian clear cell carcinoma. Eur J Cancer. 2015;51:1978–1988. doi: 10.1016/j.ejca.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang B, Peng P, Chen S, Li L, Zhang M, Cao D, Yang J, Li H, Gui T, Li X, et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. 2013;80:171–182. doi: 10.1016/j.jprot.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 39.Yi H, Zheng X, Song J, Shen R, Su Y, Lin D. Exosomes mediated pentose phosphate pathway in ovarian cancer metastasis: A proteomics analysis. Int J Clin Exp Pathol. 2015;8:15719–15728. [PMC free article] [PubMed] [Google Scholar]
- 40.Sinha A, Ignatchenko V, Ignatchenko A, Mejia-Guerrero S, Kislinger T. In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochem Biophys Res Commun. 2014;445:694–701. doi: 10.1016/j.bbrc.2013.12.070. [DOI] [PubMed] [Google Scholar]
- 41.Davalieva K, Kiprijanovska S, Komina S, Petrusevska G, Zografska NC, Polenakovic M. Proteomics analysis of urine reveals acute phase response proteins as candidate diagnostic biomarkers for prostate cancer. Proteome Sci. 2015;13:2. doi: 10.1186/s12953-014-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi F, Wu H, Qu K, Sun Q, Li F, Shi C, Li Y, Xiong X, Qin Q, Yu T, et al. Identification of serum proteins AHSG, FGA and APOA-I as diagnostic biomarkers for gastric cancer. Clin Proteomics. 2018;15:18. doi: 10.1186/s12014-018-9194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tokutomi K, Tagawa T, Korenaga M, Chiba M, Asai T, Watanabe N, Takeoka S, Handa M, Ikeda Y, Oku N. Decoration of fibrinogen γ-chain peptide on adenosine diphosphate-encapsulated liposomes enhances binding of the liposomes to activated platelets. Int J Pharm. 2011;407:151–157. doi: 10.1016/j.ijpharm.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 44.Duan S, Gong B, Wang P, Huang H, Luo L, Liu F. Novel prognostic biomarkers of gastric cancer based on gene expression microarray: COL12A1, GSTA3, FGA and FGG. Mol Med Rep. 2018;18:3727–3736. doi: 10.3892/mmr.2018.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimura K, Ting HJ, Harada Y, Tokizane T, Nonomura N, Kang HY, Chang HC, Yeh S, Miyamoto H, Shin M, et al. Modulation of androgen receptor transactivation by gelsolin: A newly identified androgen receptor coregulator. Cancer Res. 2003;63:4888–4894. [PubMed] [Google Scholar]
- 46.Schiewek J, Schumacher U, Lange T, Joosse SA, Wikman H, Pantel K, Mikhaylova M, Kneussel M, Linder S, Schmalfeldt B, et al. Clinical relevance of cytoskeleton associated proteins for ovarian cancer. J Cancer Res Clin Oncol. 2018;144:2195–2205. doi: 10.1007/s00432-018-2710-9. [DOI] [PubMed] [Google Scholar]
- 47.Chen ZY, Wang PW, Shieh DB, Chiu KY, Liou YM. Involvement of gelsolin in TGF-beta 1 induced epithelial to mesenchymal transition in breast cancer cells. J Biomed Sci. 2015;22:90. doi: 10.1186/s12929-015-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abedini MR, Wang PW, Huang YF, Cao M, Chou CY, Shieh DB, Tsang BK. Cell fate regulation by gelsolin in human gynecologic cancers. Proc Natl Acad Sci USA. 2014;111:14442–14447. doi: 10.1073/pnas.1401166111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen R, Luo FK, Wang YL, Tang JL, Liu YS. LBP and CD14 polymorphisms correlate with increased colorectal carcinoma risk in Han Chinese. World J Gastroenterol. 2011;17:2326–2331. doi: 10.3748/wjg.v17.i18.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Dyke AL, Kemp TJ, Corbel AF, Zhu B, Gao YT, Wang BS, Rashid A, Shen MC, Hildesheim A, Hsing AW, et al. Lipopolysaccharide-pathway proteins are associated with gallbladder cancer among adults in Shanghai, China with mediation by systemic inflammation. Ann Epidemiol. 2016;26:704–709. doi: 10.1016/j.annepidem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.González-Sarrías A, Núñez-Sánchez MA, Ávila-Gálvez MA, Monedero-Saiz T, Rodríguez-Gil FJ, Martínez-Díaz F, Selma MV, Espín JC. Consumption of pomegranate decreases plasma lipopolysaccharide-binding protein levels, a marker of metabolic endotoxemia, in patients with newly diagnosed colorectal cancer: A randomized controlled clinical trial. Food Funct. 2018;9:2617–2622. doi: 10.1039/C8FO00264A. [DOI] [PubMed] [Google Scholar]
- 52.Kovacs G, Peterfi L, Farkas N, Javorhazy A, Pusztai C, Szanto A. Expression of inflammatory lipopolysaccharide binding protein (LBP) predicts the progression of conventional renal cell carcinoma - a short report. Cell Oncol (Dordr) 2017;40:651–656. doi: 10.1007/s13402-017-0346-4. [DOI] [PubMed] [Google Scholar]
- 53.Wang N, Song X, Liu L, Niu L, Wang X, Song X, Xie L. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 2018;109:1701–1709. doi: 10.1111/cas.13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu YT, Osmulski P, Wang Y, Huang YW, Liu L, Ruan J, Jin VX, Kirma NB, Gaczynska ME, Huang TH. EpCAM-regulated transcription exerts influences on nanomechanical properties of endometrial cancer cells that promote epithelial-to-mesenchymal transition. Cancer Res. 2016;76:6171–6182. doi: 10.1158/0008-5472.CAN-16-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turay D, Khan S, Diaz Osterman CJ, Curtis MP, Khaira B, Neidigh JW, Mirshahidi S, Casiano CA, Wall NR. Proteomic profiling of serum-derived exosomes from ethnically diverse prostate cancer patients. Cancer Invest. 2016;34:1–11. doi: 10.3109/07357907.2015.1081921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An M, Lohse I, Tan Z, Zhu J, Wu J, Kurapati H, Morgan MA, Lawrence TS, Cuneo KC, Lubman DM. Quantitative proteomic analysis of serum exosomes from patients with locally advanced pancreatic cancer undergoing chemoradiotherapy. J Proteome Res. 2017;16:1763–1772. doi: 10.1021/acs.jproteome.7b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, Xie Y, Xu L, Zhan S, Xiao Y, Gao Y, Wu B, Ge W. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int J Cancer. 2017;140:900–913. doi: 10.1002/ijc.30496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets and materials used and/or analyzed in this study are available from the corresponding author on reasonable request.