Abstract
Transposable elements (TEs) have profoundly affected the evolution of transcriptional and chromatin profiles in mammalian genomes. In a recent paper, Percharde et al. (2018) identify a lncRNA-like function for LINE1 transposable elements in regulating gene expression to facilitate embryonic stem cell self-renewal and preimplantation development.
Transposable elements (TEs) are the genetic remnants of our ancestors’ struggle against selfish nucleic acid invaders. TEs were highly successful; their descendants comprise more than 40% of our genomes. Most of these elements, however, lie dormant. They lack critical regulatory or protein-coding sequences that would permit their continued genomic expansion and horizontal transfer. Despite the parasitic relationship between TEs and their hosts, TEs have been instrumental in the evolution of mammalian genomes. For example, TEs have contributed novel protein coding genes and facilitated gene regulatory innovations by providing binding sites for transcription factors (Chuong et al., 2017). Few studies, however, have evaluated the role of TE transcripts in cellular processes. In a recent issue of Cell, Percharde et al. (2018) used antisense oligos as a tool to deplete long interspersed nuclear element 1 (LINE1) transcripts in embryonic stem cells (ESCs) and preimplantation mouse embryos as a complementary but distinct approach to the transcriptional silencing of LINE1 recently performed by Jachowicz et al. (2017). The latter paper utilized KRAB-domain fused transcription activator-like effectors (TALEs). Percharde et al. (2018) recapitulate Jachowicz’s finding that LINE1 is needed to induce global chromatin condensation. They go further, however, demonstrating lncRNA-like roles for LINE1 RNA in ESC self-renewal and preimplantation development.
LINE1 elements are the largest group of TEs in the human genome and the only family still capable of retrotransposition. Numerous studies have reported high levels of LINE1 RNA expression in embryos and germ cells despite relatively low levels of retrotransposition (Richardson et al., 2017). Percharde et al. (2018) employed an antisense oligo strategy to deplete LINE1 RNA in mouse ESCs (mESCs). LINE1 knockdown (KD) ESC cultures proliferated more slowly and had fewer cells in S phase, indicating an important role for LINE1 in self-renewal (Figure 1A). Gene expression profiling identified numerous upregulated transcripts in LINE1 KD ESCs. Many of the identified genes are normally restricted to the 2-cell (2C) stage of embryogenesis, when zygotic genome activation (ZGA) is initiated in mice (Macfarlan et al., 2012). The 2C genetic program is largely controlled by a single transcription factor, Dux (De Iaco et al., 2017; Hendrickson et al., 2017). LINE1 KD in mESCs reduced the repressive mark H3K9me2 at the Dux locus and activated Dux target gene transcription. Importantly, small interfering RNA (siRNA)-mediated reduction of Dux in LINE1 KD mESCs rescued 2C gene repression. It did not, however, alleviate the cell proliferation defects, indicating that LINE1 likely has independent roles in ESC self-renewal and 2C gene repression.
Figure 1. LINE1 Depletion Experiments and Model for LINE1 lncRNA-like Function.
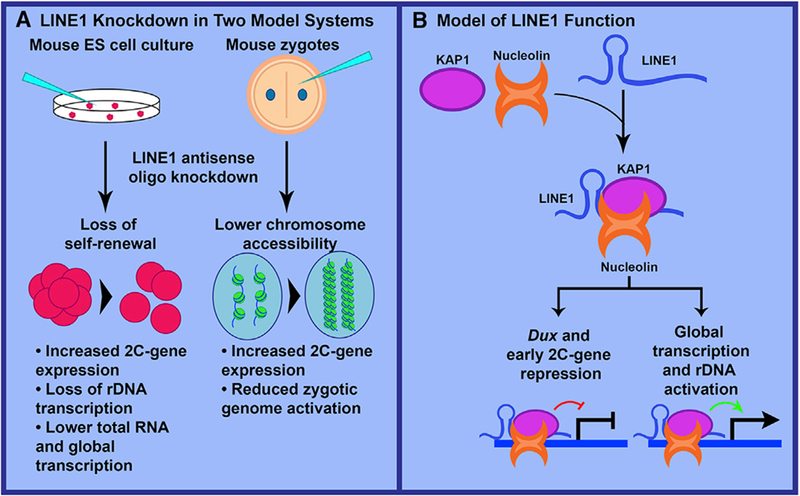
(A) Percharde et al. (2018) depleted LINE1 RNAin mouseembryonic stem cells(mESCs) and zygotes. Both ESCs and early embryos exhibited increased 2C-stage gene expression. ESCs displayed loss of selfrenewal, reduced rDNA transcription, and lower total RNA and nascent transcription. Zygotes exhibited lower chromosome accessibility and reduced zygotic genome activation.
(B) The authors propose that LINE1 RNA binds Nucleolin and KAP1 and interacts with the genome at Dux and rDNA loci. At the Dux locus, the complex represses transcription, whereas at rDNA it promotes expression of rRNAand riboproteins, facilitating the self-renewal of ESCs and thetransition through early preimplantation development.
One clue for LINE1’s role in self-renewal was the authors’ observation that LINE1 KD mESCs express low levels of genes involved in ribosomal biogenesis and translation and have lower total RNA levels than controls. These findings are consistent with those from Jachowicz et al. (2017), who found that LINE1 reduction lowers rates of nascent transcription in 2C embryos, suggesting that LINE1 may be required to activate global gene expression during early development (Jachowicz et al., 2017). These experiments led to a hypothesis for LINE1’s dual functionality: LINE1 represses Dux to prevent activation of 2C genes while activating rDNA expression to maintain mESC self-renewal (Figure 1B).
The authors proposed that LINE1’s regulation of rRNA production and Dux expression might be mediated by LINE1 RNA, an attractive hypothesis because most LINE1 elements lack coding potential and because long non-coding RNAs (lncRNAs) are increasingly implicated in gene regulatory processes (Salviano-Silva et al., 2018). Consistent with this idea, LINE1 chromatin immunoprecipitation by RNA purification (ChIRP) demonstrated LINE1 enrichment at both Dux and rDNA loci, suggesting that LINE1 RNA regulates transcription at these loci by direct chromatin interactions. Searching for potential LINE1 cofactors, the researchers turned to Nucleolin, a nucleolar protein known for promoting rRNA synthesis and repressing DUX4 in humans (Gabellini et al., 2002; Ginisty et al., 1998). They found a DNA-independent interaction between LINE1 RNA and Nucleolin. Furthermore, Nucleolin KD in mESCs recapitulated the transcriptional profile of LINE1 KD. Finally, Dux KD in a Nucleolin KD background restored 2C gene repression but not ESC selfrenewal, suggesting that LINE1 and Nucleolin cooperate to promote rRNA synthesis while repressing 2C genes via Dux silencing.
The co-repressor KAP1 was recently demonstrated to repress Dux in mESCs (De Iaco et al., 2017). Percharde et al. (2018) showed that Kap1 deletion led not only to a significant increase in Dux and 2C gene expression, but also to many of the other transcriptional changes that were observed in LINE1 KD ESCs. Furthermore, both Nucleolin and KAP1 were found to bind Dux and rDNA loci, and this binding was partially LINE1 dependent. These data suggest that LINE1 coordinates chromatin binding of a Nucleolin-KAP1 complex to repress Dux and induce rRNA expression.
Finally, the authors performed experiments to deplete LINE1 directly in 2C-stage embryos, reminiscent of previous work by Jachowicz et al. (2017), who used a TALE-KRAB strategy to reduce LINE1 transcription (and hence LINE1 transcripts) in 2C embryos. LINE1 KD in 2C embryos reduced chromatin accessibility and increased levels of heterochromatin (Figure 1A), a result that parallels that of Jachowicz et al. (2017). In late 2C-stage and 4-cell embryos, LINE1 KD caused reactivation of 2C genes, indicating the ongoing requirement of LINE1 to repress the 2C transcriptional program in early embryogenesis. LINE1 KD also led to strongly reduced zygotic genome activation, another observation consistent with Jachowicz et al. (2017). In sum, these studies support a model whereby pervasive LINE1 transcription and LINE1 transcripts themselves play an important role in early developmental gene and chromatin regulation.
Historically, LINE1 has been considered primarily as a threat to genomic integrity due to its capacity for retrotransposition and its connection to human disease. The work from Percharde et al. (2018) suggests that LINE1 elements have also evolved critical functions as lncRNAs in early development. By binding Nucleolin and KAP1, LINE1 elements facilitate both the activation of rDNA genes and the suppression of many 2C genes via silencing of Dux. Despite these intriguing results, the work leaves many unanswered questions. Does LINE1 play similar roles in other mammals including humans, despite the high degree of divergence in LINE1 promoters and coding sequence? How does LINE1 interact with KAP1/Nucleolin? Do the LINE1/KAP1/Nucleolin complexes interact specifically with rRNA genes and Dux, and if so, how? Finally, how does LINE1/KAP1/Nucleolin activate some genes and repress others? Despite these outstanding questions, the demonstration that ESC self-renewal and embryogenesis require LINE1 RNAs suggests that this highly successful mobile genetic element may be inexorably linked to the healthy development of its host.
REFERENCES
- Chuong EB, Elde NC, and Feschotte C (2017). Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet 18, 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, and Trono D (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet 49, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D, Green MR, and Tupler R (2002). Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110, 339–348. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, and Bouvet P (1998). Nucleolin functions in the first step of ribosomal RNA processing. EMBO J 17, 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet 49, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachowicz JW, Bing X, Pontabry J, Bošković A, Rando OJ, and Torres-Padilla ME (2017). LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet 49, 1502–1510. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, and Pfaff SL (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, and Ramalho-Santos M (2018). A LINE1-Nucleolin partnership regulates early development and ESC identity. Cell 174, in press. Published online June 21, 2018. 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Gerdes P, Gerhardt DJ, Sanchez-Luque FJ, Bodea GO, Muñoz-Lopez M, Jesuadian JS, Kempen MHC, Carreira PE, Jeddeloh JA, et al. (2017). Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Res 27, 1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salviano-Silva A, Lobo-Alves SC, Almeida RC, Malheiros D, and Petzl-Erler ML (2018). Besides pathology: long non-coding RNA in cell and tissue homeostasis. Noncoding RNA 4, E3. [DOI] [PMC free article] [PubMed] [Google Scholar]


