Abstract
Thirty-five years after the identification of HIV-1 as the causative agent of AIDS, we are still in search of vaccines and treatments to eradicate this devastating infectious disease. Progress has been made in understanding the molecular pathogenesis of this infection, which has been crucial for the development of the current therapy regimens. However, despite their efficacy at limiting active viral replication, these drugs are unable to purge the latent reservoir: a pool of cells that harbor transcriptionally inactive, but replication-competent HIV-1 proviruses, and that represent the main barrier to eradicate HIV-1 from affected individuals. In this review, we discuss advances in the field that have allowed a better understanding of HIV-1 latency, including the diverse cell types that constitute the latent reservoir, factors influencing latency, tools to study HIV-1 latency, as well as current and prospective therapeutic approaches to target these latently infected cells, so a functional cure for HIV/AIDS can become a reality.
Keywords: : HIV, viral reservoirs, persistent infection, latency, shock and kill, block and lock
Introduction: Classes of HIV-1 Latency and Cell Types Constituting the Latent Reservoir
According to the most recent data from UNAIDS, more than 36.7 million people are currently living with HIV-1 worldwide, with 1.8 million newly infected individuals reported in 2016. Although the introduction of combination antiretroviral therapy (cART) has allowed inhibition of HIV-1 replication by a combination of mechanisms, which effectively decrease viral loads to undetectable levels, delay disease progression, restore T cell immune function, and prevent transmission to uninfected individuals, the HIV-1 latent reservoir represents a major hurdle to completely eradicate this infection.1–3
Despite remarkable efforts to provide an accurate definition for the HIV-1 cellular reservoir, to date it still remains challenging for the scientific community. In an exercise to bring consensus, Eisele and Siliciano defined the HIV-1 reservoir as infected cells harboring full-length transcriptionally inactive, but replication-competent, HIV-1 proviruses in individuals on cART with sustained viremia suppression.4 These viral reservoirs are long-lived, self-replenishing, refractory to ART, and, due to their resting phenotype, unrecognized by the immune system.5,6 HIV-1 latency is associated with a quiescent viral state established in resting CD4+ T cells and other long-lived immune cell populations, in which the HIV-1 proviral DNA is transcriptionally silenced with no (or minimal) production of infectious viral particles.1,2,6,7 This viral latency can be achieved by two distinct mechanisms: (1) by preventing proviral integration in the cellular genome (preintegration latency), and (2) by enforcing proviral silencing at the transcriptional level (postintegration latency). Details on factors influencing postintegration latency are discussed in the next section.
Preintegration latency was first reported in CD4+ T cells,8 but there is also evidence for this type of dormant infection in monocytes. In this case, the HIV-1 complementary DNA (cDNA) is partly or completely synthesized during reverse transcription, but fails at integrating into the cellular genome, remaining as an episome.8–10 This form of latency is relieved upon T cell activation, allowing the integration of the provirus and, in consequence, leading to a productive infection.9 Unlike postintegration latency, latent infections in which the virus life cycle has been compromised at a preintegration step are rather unstable in resting CD4+ T cells, where the amounts of HIV-1 cDNA decrease rapidly.11,12 In the case of nondividing cells, such as those of the monocyte lineage, the presence of restriction factors seems to play a major role in preventing proviral integration. In these cells, SAMHD1, members of the APOBEC3 family, and MX2 strongly interfere with early steps of the HIV-1 replication cycle,13–19 and thus, one could speculate a more relevant role of preintegration latency in these cell types than in CD4+ T cells. Despite the evidence of preintegration latency, more studies are needed to assess its persistence and contribution to the size and landscape of the viral reservoir, since our current knowledge on preintegration latency is still limited.20,21
Postintegration latency (referred hereafter as latency), however, accounts for most of the HIV-1 persistent reservoir. Postintegration latency is established once the provirus is integrated into the host DNA, but fails at properly expressing its genome, having restricted viral expression—so viral antigens are inefficiently produced and, presumably, poorly presented (or not at all) to the immune system. Although latency is rather infrequent, this silent state is remarkably stable and reversible.1,2 Indeed, latently infected cells harboring replication-competent dormant proviruses maintain the potential to spontaneously activate, or reactivate, in the presence of cytokines, antigens, latency-reversing agents (LRAs), or when ART is interrupted,22–24 causing viral rebound and further replenishing the latent reservoir. It is widely recognized that latency is established within days of the initial infection. However, as mentioned above, this pool of cells remains undetected by the immune surveillance and is refractory to ART.2,25,26 Despite the fact that latently infected cells can reseed viremia upon stimulation, only few of these proviruses have the potential to re-emerge from latency. Errors during proviral integration causing deletions, insertions, or introducing mutations in the HIV-1 cDNA seem to be the main cause for their defective phenotype.27 In fact, according to a recent study by the Siliciano laboratory, only 10% of all integrated HIV-1 proviruses maintain an intact genome, and just one tenth of these replication-competent proviruses has the potential to reactivate in vitro3 (able to generate infectious virions upon cellular activation).
The latent reservoir includes a varied pool of cells, and thus, its origins are also diverse, including: (1) replication-competent proviruses in long-lived latently infected cells1,28; (2) viral sanctuary tissues where cART has poor penetration, so virus replication is maintained, generating residual viremia (i.e., central nervous system [CNS] and gut-associated lymphoid tissues)29–31; (3) cells that promote cell-to-cell virus dissemination, in which a low-level but ongoing viral replication is sustained32,33; (4) homeostatic proliferation of replication-competent proviruses in latently infected cells5,34,35; and (5) intermittent viremia from spontaneous activation of a small fraction of resting, infected CD4+ T cells during cART.5,36,37 Despite its type and origins, the latent reservoir is normally referred to three cell groups: CD4+ T cells, dendritic cells (DCs), and macrophages.
Since latency is primarily established when infected and activated CD4+ T cells transition to a resting memory phenotype, the best-characterized latently HIV-infected cells are resting memory CD4+ T cells, particularly the central memory (TCM: CD45RA− CCR7+ CD27+) and transitional memory (TTM: CD45RA− CCR7− CD27+) subsets.5,6,25,38,39 Because they can persist for long periods of time,40 resting memory CD4+ T cells carrying replication-competent proviruses represent an important long-term viral reservoir in patients on cART.1,2,6,7 In fact, latently infected CD4+ T cells are present in blood and different tissues, including lymphoid tissues and the gut.10,29,41 Besides resting memory CD4+ T cells, latency has also been reported in other T helper cell subsets such as naive,5,42 follicular,43 and memory stem cells.44,45 In addition, recent studies have demonstrated the presence of latent, replication-competent HIV-1 proviruses in the unconventional T cell subset Vδ2.46 Therefore, the contribution of other CD4+ T cell populations to the reservoir may currently be underestimated.
In addition to CD4+ T cells, HIV-1 can also remain dormant in non-T cells and thus, cause viral rebound upon cART is discontinued, as evidenced by early studies by Chun et al.47 It is now well known that HIV-1 can persist latently in cells of the myeloid lineage, including monocytes, macrophages, and DCs.48–50 Macrophages are susceptible to HIV-1 infection, and thus, they are active producers of HIV-1 virions in both ART-treated and untreated patients.51,52 Therefore, similar to CD4+ T cells, macrophages have the potential to harbor latent HIV-1 proviruses. Macrophages present longer persistence of unintegrated viral DNA and are less susceptible to the cytopathic effects caused by HIV-1 compared to CD4+ T cells.12,48,53 Moreover, these cells present an extended lifespan in the presence of infection, and greater resistance to apoptosis,54–57 since HIV-1 potently triggers the NF-κB pathway, inducing antiapoptotic genes.58–60 The fact that macrophages can engulf HIV-1-infected CD4+ T cells increases the probability of macrophage infection and latency establishment.61 Importantly, since macrophages are antigen-presenting cells, viral reactivation in these phagocytes may increase HIV-1 transmission to other immune cells through the virological synapse,62 but also through the production of chemokines, which would facilitate the recruitment and activation of other immune cells.63 Therefore, although the contribution of macrophages to the latent reservoir appears smaller than that of CD4+ T cells, they may be important contributors to viral maintenance and rebound due to their inherent cellular and immune characteristics.
Early work on HIV-1 latency already reported that proviral DNA was detected in monocytes/macrophages from infected individuals.64 More recently, Yukl et al. demonstrated that gut tissue macrophages from ART-suppressed individuals contained viral DNA, although their studies did not directly address whether macrophages constitute part of the HIV-1 latent reservoir.65 However, under ART conditions, CD4+ T cells from blood and mucosal tissues decrease dramatically, while infected macrophages undergo death at a slower rate than that of T cells, consequently giving rise to the slower second phase of decay.36 This, together with their intrinsic characteristics mentioned above, suggests that macrophages can sustain ongoing viral replication, and thereby, importantly contribute to the viral reservoir. In fact, both in vitro as well as in vivo assays revealed that latency can be established in monocyte-derived macrophages,66 and that virus reemergence from latency can be observed from macrophage-tropic HIV-infected cells, in myeloid-only humanized mice on ART67—evidencing that tissue macrophages can persist as a viral reservoir. Furthermore, macrophages can be found as latently infected cells in tissue sanctuaries, where the access of drugs is limited, serving as a source of HIV-1 viral persistence. For example, macrophages residing in the CNS, together with microglial cells and astrocytes, are anatomical viral sanctuaries representing the principal HIV-1 reservoir in the brain, and posing a major barrier for HIV-1 therapies.68 However, it remains unclear to which extent these cells can be effectively reactivated to produce infectious particles. However, in the gastrointestinal tract macrophages represent a significant cellular reservoir for HIV-1, since they are extremely abundant in this tissue,69 where HIV-1 intensively replicates and spreads.
DCs are antigen-presenting cells that act as a connection between the innate and the adaptive immune systems. Upon antigen recognition, they mature and migrate to lymph nodes for antigen presentation to T cells. DCs are classified into three major subtypes: myeloid DCs, plasmacytoid DCs, and Langerhans cells, all of them susceptible to HIV-1 infection, although inefficiently. DCs are also an important component of the HIV-1 reservoir, with follicular DCs (FDCs) being the major DC contributors.48,70,71 In fact, work from Haase et al. in ART-treated patients demonstrated the presence of a relatively stable pool of virions on the surface of FDCs, but not on other mononuclear cells.72 This was corroborated in later studies, suggesting that FDCs have the capacity to produce low-level viremia and to recruit resting CD4+ T cells, which may aid in virus spread. Hence, since the trapped virions are replication competent, FDCs may contribute to pathogenesis and also serve as a significant HIV-1 reservoir for durable viral persistence.71,73–76 Similar to macrophages, FDCs are in close contact with resting CD4+ T cells within lymphoid tissues. Actually, early studies described the capacity of DCs to promote trans HIV-1 infection to CD4+ T cells,77,78 which requires the formation of infectious synapses.79 Therefore, this DC-CD4+ T cell interaction may be critical to drive and establish latency in resting memory CD4+ T cells.33
Factors Influencing the Establishment and Maintenance of Latency
The establishment and maintenance of HIV-1 latency is a complex process modulated by many different factors6,80–82; some of which have the ability to suppress latency, such as cytokines involved in T cell activation,6,83,84 Toll-like and Rig-I-like receptor agonists,85–87 effectors of the NF-κB pathway,82,88–90 inhibitors of histone deacetylases (HDACs),91,92 or the pleiotropic effector mTOR93; whereas other factors promote proviral latency, for instance histone modifiers,82,90,94,95 inhibitors of viral transcription,96,97 cytokines that induce memory T cell differentiation,98,99 or transcriptional repressors82,90,100–102 (Fig. 1). Despite the vast number of elements that play a role in the modulation of viral gene expression and latency establishment, the mechanisms by which they ultimately achieve such regulatory effect in vivo are not fully understood. The elements presented above act at different molecular levels in the process of HIV-1 transcription, such as (1) integration site selection and epigenetic regulation, (2) transcription factor-mediated activation and repression, (3) RNA elongation, (4) transcription interference, (5) silencing mediated by microRNAs (miRNAs), (6) RNA splicing and transport, and (7) generation of antisense HIV-1 genomic transcripts, which downregulate gene expression.103–105 The influence of the environmental factors presented above, as well as their synergistic/antagonistic effects on these molecular levels, has a profound impact on latency establishment and maintenance. Therefore, investigating how these molecular networks interact with each other is crucial to increase our fundamental understanding of HIV-1 latency.
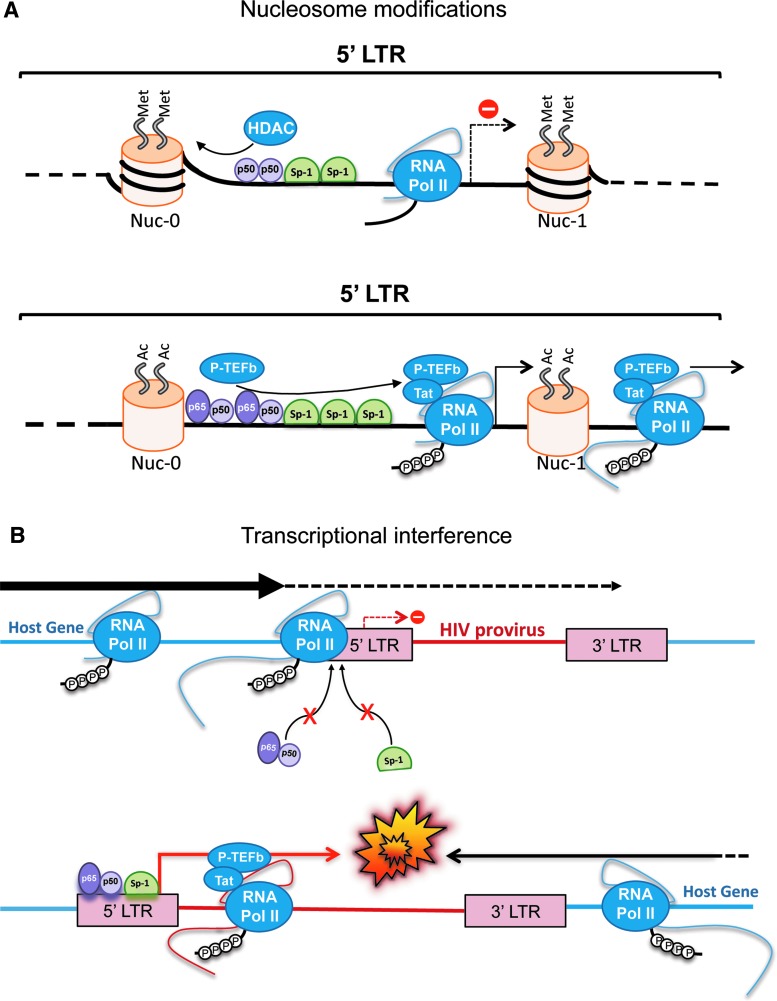
FIG. 1.
Factors influencing the establishment and maintenance of latency. (A) Effects of chromatin condensation status and epigenetic modifications in the repression (top) or activation (bottom) of HIV-1 transcription. (B) Representation of scenarios for transcriptional interference. Top: when the HIV-1 provirus is in the same orientation as a highly expressed cellular gene. Bottom: when the provirus and the cellular host gene have opposite orientations. Color images are available online at www.liebertpub.com/aid
Integration site selection and epigenetic regulation
The transcription of integrated HIV-1 proviruses, as well as the expression of host genes, is strongly regulated by variations in the condensation state of the chromatin. Genes located in high compacted DNA regions, or heterochromatin, remain silenced due to inaccessibility of transcription factors to their regulatory elements. By contrast, highly expressed genes are found in less condensed DNA areas (euchromatin).106 Contrary to what it was previously thought, the process that leads to HIV-1 proviral DNA integration does not occur in a random manner, but in a more selective way: by targeting euchromatin regions through a direct interaction between the viral integrase and the host protein LEDGF/p75, which in turn interacts with RNA-binding proteins found in areas where DNA is being actively transcribed.107,108 In addition to LEDGF/p75, other host proteins are involved in the selective integration of HIV-1 proviruses into actively transcribed chromatin areas, such as CPSF6 (through its direct interaction with capsid molecules found in the preintegration complex), and the host splicing machinery (in this case, through their association with LEDGF/p75).107,109–111 Consistent with this, a high-throughput study of the integration landscape of HIV-1, in which primary cells from HIV-1 viremic progressors, viremic controllers, and patients under cART were analyzed, revealed that HIV-1 proviruses frequently integrate within intronic regions of highly expressed genes, irrespective of the type of patient (controllers or progressors).112 However, in patients controlling viremia with cART, the majority of the proviruses analyzed corresponded to latent HIV-1, since actively infected cells are negatively selected by the antiretroviral drugs. In this context, these proviruses were rarely found within genes, and in cases where they integrated within coding regions, these genes were surrounded by heterochromatin.112 This is in agreement with findings using in vitro T cell line models for latency, where HIV-1 proviruses are frequently located in/or near heterochromatin,113 suggesting that the condensation state of the chromatin at the integration sites influences the outcome of the infection.111 However, studies in primary cells showed that HIV-1 proviruses have an analogous integration pattern in both resting memory CD4+ T cells and actively infected CD4+ T cells,114,115 reflecting that there must be factors other than chromatin condensation state and integration site selection involved in the establishment and maintenance of latency in vivo.
Although discrepancies have been found when analyzing the proviral integration sites in T cell lines and primary cells,116,117 even when using the same cell line models, common features have been defined, such as the observation that integration favors Alu repeats both in genic and intergenic regions, implying that the integration sites may harbor some consensus sequence, although unknown yet.112,115 Furthermore, the inconsistent results on the chromatin status surrounding latent proviruses may simply reflect versatile epigenetic modifications. As stated earlier, besides the HIV-1 integrase, cellular cofactors aid in the integration of the provirus, and these molecules preferentially bind to less condensed chromatin areas. Therefore, it is tempting to speculate that events occurring during the activated-to-resting transition of CD4+ T cells may influence the overall chromatin structure, causing chromatin remodeling at the locations where HIV-1 proviruses are integrated.118
The condensation state of the chromatin, and therefore, its related transcriptional levels, is regulated by a highly dynamic process, which involves posttranslational modifications (PTMs) of different DNA-associated proteins. Chromatin is wrapped around histone proteins, forming nucleosomes. However, unlike euchromatin, heterochromatin is characterized by a highly condensed status, where the chromatin–histone interactions are more rigid. Differential combinations of PTMs on these histones will determine the condensation state of surrounding chromatin, since PTMs can alter the ability of histones to recruit Mi-2/NuRD and SWI/SNF complexes, which are directly related to DNA condensation and unwinding processes, respectively. Histone acetyltransferases (HATs) promote histone acetylation and DNA relaxation, increasing transcriptional activity, whereas HDACs promote chromatin condensation, which represses gene expression.106 Similar to cellular DNA, the 5′ long-terminal repeat (LTR) of HIV-1 proviruses contains two nucleosomes: (1) nuc-0, located at the 5′ end of the LTR (between nucleotides −405 and −245, from the transcription start site), and (2) nuc-1, located after the provirus promoter (between nucleotides +20 and +165)119–121 (Fig. 1). These nucleosomes are present even when proviral DNA integration occurs within euchromatin regions.122,123 Therefore, their condensation status will directly influence proviral transcription, mainly because both nuc-0 and nuc-1 partially overlap with the transcriptional control regions in the LTR, and thus, can alter the association of transcriptional regulators to the HIV-1 promoter as well as interfere with RNA elongation123 (Fig. 1A). For instance, viral reactivation and nuc-1 remodeling was observed in different studies when HATs, such as CBP or GCN5, were recruited to the provirus integration site.124–126 Similar results were obtained when resting CD4+ T cells were exposed to HDAC inhibitors, which led to nuc-1 displacement and facilitated the recruitment of the RNA Pol II to the LTR.6,127 These results indicate that the acetylation and, in turn, condensation state of the HIV-1 nucleosomes, play an important role in the establishment and maintenance of latency.
In addition to histone acetylation status, epigenetic control can also occur by directly modifying DNA through methylation, a reversible modification that has a profound impact on gene expression. The methylation of cytosines on CpG-rich regions causes transcriptional repression. This has also been observed for HIV-1 proviruses, particularly in areas of the proviral DNA containing the transcriptional control regions. Methylation of cytosines in this location causes proviral DNA silencing and promotes latency6,82,102,128 (Fig. 1A, top). Several reports have shown a high methylation pattern on the 5′ LTRs of latent proviruses from latently infected CD4+ T cells obtained in vitro.128–131 These epigenetic modifications prevent transcription factors from binding to their cognate enhancers and thus, repress provirus transcription.132 In agreement with these results, demethylation of the provirus LTRs has been associated with transcriptional reactivation and virus reemergence from latency in vitro.128,133,134 However, DNA methylation does not seem to play a major role in HIV-1 latency in vivo, since no significant levels of CpG methylation are found within the 5′ LTR in resting CD4+ T cells from HIV-1-infected individuals on suppressive ART.135 These observations suggest that the contribution of CpG methylation in the establishment of the latent reservoir in patients is minimal compared with the other epigenetic factors mentioned above.
Transcription factors
As indicated earlier, the largest reservoir for HIV-1 is found in resting memory CD4+ T cells, after they transition from an activated status to a resting memory phenotype. Because HIV-1 RNA synthesis relies on the presence of cellular transcription factors that are only temporarily available upon antigen exposure, HIV-1 transcription declines in this activated-to-resting transition.6,118,136 Therefore, the availability of these transcription factors is a major factor governing the establishment of latency. The HIV-1 proviral DNA contains several regulatory regions on its 5′ LTR with responsive elements for different host transcription factors, which, upon binding, alter viral RNA synthesis by modulating RNA Pol II access and recruitment to the TATA box in the promoter.102,137,138 Among these factors, NF-κB, NFAT, and Sp-1 play a key role in this transactivation process.6,102,139–141 The presence of these three transcription factors in the nucleus, and their ability to interact with their enhancer elements at the LTR, is required to maintain basal HIV-1 transcriptional levels,140,142 whereas their cytoplasmic sequestration, or inability to bind to the regulatory elements due to mutations, has been linked to latency in CD4+ T cells.6,102,140,142,143 In addition to NF-κB, NFAT, and Sp-1, there are other transcription regulators, AP-1,144 C/EBPb,145 USF-1, and Ets-1,146 with the ability to cooperate in HIV-1 transcription.6,147
Above all, NF-κB seems to play a central role in the establishment and maintenance of latency. In response to pathogens and proinflammatory cytokines, NF-κB monomers, p50 and RelA (p65), dimerize and translocate into the nucleus, where they interact with the NF-κB regulatory elements in their target cellular genes, but also in the proviral LTR, strongly activating HIV-1 transcription.139,148 In resting CD4+ T cells, RelA is found sequestered in the cytoplasm by the inhibitory protein IκBα.148–150 Moreover, while IκBα retains RelA, p50/p50 homodimers are able to translocate to the nucleus and further reinforce proviral latency by recruiting HDACs to the HIV-1 provirus, promoting its heterochromatinization (Fig. 1A, top).102,127 Interestingly, a recent study uncovered that the cellular protein breast cancer associated-gene 2 (BCA2, also known as Rabring7) is an important negative effector of NF-κB, and in turn, detrimentally affects HIV-1 transcription and replication.151 Specifically, BCA2 promotes the SUMOylation of IκBα, rendering this molecule resistant to degradation and a stronger NF-κB inhibitor.151 Ongoing studies in our laboratory have revealed that BCA2 plays a pivotal role in fine-tuning NF-κB in primary CD4+ T cells, and that its depletion from naive CD4+ T cells makes HIV-1 less prone to undergo latency. In line with these observations, upregulation of BCA2 enforces and prolongs proviral silencing while the selective knockdown of this gene in latently infected cells results in spontaneous viral reactivation, highlighting the relevance of BCA2 and its NF-κB regulatory activity to proviral latency (Colomer-Lluch et al., unpublished data).
In addition to BCA2, there are other transcriptional regulators that negatively impact HIV-1 transactivation and thus, enhance the establishment of latency. Among these molecules we find CYLD and Naf1, which also inhibit NF-κB-driven viral transcription,152,153 LSF-1 and YY-1, which recruit HDAC1154; CBF-1, which reduces the amount of RNA Pol II in the LTR155; Blimp-1, which interferes with the Tat-trans-activation response (TAR) association156; or Bcl-11b, which represses HIV-1 transcription by promoting heterochromatinization.100 Therefore, besides the epigenetic elements considered above, the relative abundance of cellular transcription activators, such as NF-κB, and transcriptional repressors will dictate the fate of the HIV-1 infection in terms of latency.
Tat and RNA elongation
One of the main obstacles during HIV-1 transcription is the inefficient synthesis of genome-length transcripts due to RNA Pol II pausing, an event caused by secondary structures in the nascent RNA that halts RNA elongation.157,158 To overcome this block, HIV-1 uses the viral transactivation protein Tat, which increases the elongation efficiency during transcription by interacting with a secondary structure in the nascent HIV-1 RNA (TAR element), allowing Tat to associate with positive and negative host regulatory protein complexes. To release the RNA Pol II pause, HIV-1 requires the presence of the transcription elongation factor (P-TEFb).159,160 P-TEFb is a protein complex containing cyclin T1 and CDK9 kinase, which phosphorylates the carboxyl-terminal domain of the RNA Pol II recruited at the 5′ LTR, enhancing in turn its transcription elongation activity.159,161 However, most P-TEFb molecules are held catalytically inactive through their association with the 7SK small nuclear ribonucleoprotein (snRNP) complex. Tat promotes P-TEFb dissociation from 7SK, allowing P-TEFb to become catalytically active, hyperphosphorylate RNA Pol II, and release the transcriptional pause.162,163 By contrast, the negative elongation factor (NELF) complex plays the opposite role in the regulation of RNA Pol II activity.160,164 Specifically, NELF associates with RNA pol II, limiting its escape from the HIV-1 promoter region.165 Besides this primary role, NELF may also influence the overall structure of the chromatin. In latent proviruses, RNA pol II complexes accumulate near the transcription start site at the 5′ LTR (nucleotide +11), and at nucleotide +50165,166—the latter is analogous to the major pause observed in host genes.167–169 However, in the absence of NELF, the +50 pause site is shifted 20 nucleotides downstream, reflecting repositioning of nucleosomes.165 Therefore, NELF seems to impact HIV-1 transcription at several levels. In productive infections, however, P-TEFb counteracts this repressing effect by phosphorylating NELF and rescinding NELF's association with the nascent HIV-1 RNA.170 Consistent with this, in vitro as well as in vivo studies have associated low levels of P-TEFb with HIV-1 latency in resting CD4+ T cells.118,136
Despite the fact that Tat significantly enhances the efficiency of HIV-1 RNA synthesis, the first round(s) of viral transcription after the integration of the proviral DNA need to occur in a Tat-independent manner, since Tat is a nonstructural protein. Although these initial Tat-independent transcriptional events are very inefficient, some genome-length HIV-1 transcripts are successfully synthesized, mainly due to the action of NF-κB. Similar to Tat, RelA (a monomer of the NF-κB heterodimer) also has the ability to recruit P-TEFb to the 5′ LTR171 (Fig. 1A, bottom), increasing the efficiency of RNA elongation and generating in turn full-length transcripts that will subsequently be translated into viral proteins, among which, we find Tat. From this point on, and due to a positive feedback, a highly efficient Tat-dependent HIV-1 transcription phase begins by the mechanisms described above.82,159,161,162,172 Additionally, Tat recruits chromatin-remodeling factors, such as SWI/SNF, which facilitate transcription by promoting DNA relaxation and consequently nuc-1 remodeling.173
Tat's role in latency has been extensively documented.159,174 For instance, Tat protein alone was found to be sufficient to successfully reactivate HIV-1 transcription in latently infected CD4+ T cells isolated from patients undergoing cART.159,175,176 This was demonstrated by delivering tat either by transduction with non-HIV vectors,176 or by ectopically delivering recombinant Tat protein to these cells.175 In this particular case, a soluble Tat-GFP chimeric protein was produced from pancreatic cells of transgenic mice, and delivered to primary, latently infected CD4+ T cells of patients undergoing cART. Remarkably, ectopic delivery of Tat or T cell receptor (TCR)-mediated cell activation caused similar levels of reemergence from latency.175 Likewise, the ectopic expression of this chimeric Tat protein, or tat's stable expression through transduction, in Jurkat and THP-1 cells prevented the establishment of latency and reactivated latent proviruses.175,177–179 Consistent with these findings, HIV-1 variants encoding attenuated tat genes are more prone to undergo latency, as evidenced by studies by the Karn laboratory using primary CD4+ T cells infected either with wild-type HIV-1 NL4-3 or a NL4-3 variant carrying the attenuating H13L mutation in Tat.118,178,180 Therefore, despite the presence of cellular transcription factors that aid in HIV-1 RNA synthesis, defects in Tat will make HIV-1 more prone to latency due to the inability to relieve the RNA Pol II pause block.
Interference
Not only does the integration site of the HIV-1 DNA influence virus transcription (due to the local chromatin state), but also the presence of active host genes surrounding the provirus, and their relative orientation, have a significant impact on HIV-1 replication.181–184 As mentioned earlier, HIV-1 shows a preference of integration within transcriptionally active regions in the latently infected J-Lat cell line181 as well as in resting CD4+ T cells isolated from patients on cART.114 Despite the fact that these proviruses can easily gain access to transcription factors and RNA Pol II, the presence of actively transcribed genes nearby the proviral DNA drastically affects HIV-1 RNA synthesis. In fact, the elongation of RNAs from neighboring host genes can have both a repressive and activating effect on HIV-1 transcription through different mechanisms: (1) when the integrated provirus and the nearby host gene share the same polarity, RNA Pol II elongation may prevent transcription factors from binding to their cognate sequences on the 5′ LTR, and thus, promote proviral silencing and latency181,183 (Fig. 1B, top); (2) alternatively, the activation of host genes upstream the HIV-1 5′ LTR can cause read-through transcription of the provirus, which is an important limitation to a productive infection. This phenomenon consists of adjacent promoters initiating transcription of a host gene and continuing through the HIV-1 coding sequence, therefore impacting HIV-1 gene expression.114,181,185 Although these HIV-1 transcripts are generally nonproductive, the effect of host gene read-through transcription might prevent latency and up-regulate proviral expression if the provirus is integrated in the same orientation as the host gene. (3) On the other hand, proviral integration in the opposite orientation of an actively expressed host gene leads to (i) a premature termination of transcription due to the collision between RNA Pol II complexes moving in different directions, which inevitably reduces the amounts of HIV-1 transcripts181,184 (Fig. 1B, bottom), or (ii) the generation of antisense HIV-1 RNAs. Although this event has mainly been observed from the HIV-1 3′ LTR,103–105 one can envision a similar phenomenon occurring from host genes that are located downstream, and in the opposite direction of the HIV-1 provirus, leading to antisense HIV-1 genomic transcripts. Nevertheless, upon cellular activation, the repressive effect of these interfering mechanisms is diminished by the strong binding affinity that NF-κB exhibits for the HIV-1 5′ LTR, enhancing transcription from the virus promoter rather than from the host gene.6,186 Although these observations indicate that the proviral integration site can affect HIV-1 transcription due to (1) the chromatin condensation status, which may hinder the recruitment of the cellular transcriptional machinery, and (2) the interference of cellular genes surrounding the HIV-1 provirus, most of these studies examined only few clones of latently infected cells. Therefore, a comprehensive interrogation of more clones is needed to ascertain how the orientation of HIV-1 proviruses relative to host genes affects the outcome of the infection.
MicroRNAs
It is well established that the presence of cellular miRNAs can regulate gene expression through the RNA-induced silencing complex (RISC), either by impairing translation or by promoting the degradation of transcripts.187 This is also true for HIV-1, since it has been reported that cellular miRNAs present in memory CD4+ T cells can directly affect HIV-1 RNA synthesis.188–191 Using computational analysis, miR-29a, miR-29b, miR-149, miR-324-5p, and miR-378 were identified as miRNAs with a regulatory effect on HIV-1 transcription.189 Ahluwalia et al. confirmed that miR-29a inhibits HIV-1 transcription in Jurkat cells by targeting the nef region on HIV-1 transcripts, although Frattari et al. reported that this miRNA affects HIV-1 RNA levels by targeting the 3′ untranslated region (UTR).192,193 miR-28, miR-125b, miR-150, miR-223, and miR-382 have also been reported to reduce HIV-1 gene expression and infectivity by targeting the 3′ UTR of the HIV-1 RNA.190 In addition, miRNAs can also affect the overall expression levels of HIV-1 dependency factors, playing a role in HIV-1 latency.194 This has been observed for cyclin T1195 and Pur-α, a co-factor of Tat.196
The study of miRNAs is still an emerging field, and so is the investigation of their effects on HIV-1 infection. Since the amount and species of these small RNA molecules depend on multiple intracellular and extracellular factors, research on how miRNAs affecting HIV-1 are regulated, as well as their mechanisms of action, will not only increase our understanding on their role in HIV-1 latency, but also could open new avenues to neutralize this pool of cells.
RNA export
Even if the epigenetic signature, cellular environment, and the presence of cellular transcription factors is optimal to support HIV-1 RNA synthesis, HIV-1 might, in theory, still establish a latent infection if viral transcripts are not exported, translated, or packaged into nascent virions. An important mechanism directly involved in the regulation of HIV-1 gene expression is the transport of viral transcripts from the nucleus to the cytoplasm.197,198 The efficiency of this process relies on the presence and activity of both host and viral proteins. Rev is an auxiliary HIV-1 protein that binds to a specific nucleotide region (rev-responsive element or RRE) within viral transcripts (nonspliced and singly spliced), facilitating their cytosolic translocation.199–202 In fact, the presence of low levels of Rev is required for a productive HIV-1 infection, whereas its absence appears to play a role in proviral latency.203 In addition, Rev-independent events related to RNA transport also impact the establishment of latency. For instance, nuclear retention of HIV-1 transcripts, particularly multiply spliced HIV-1 RNAs, was observed in primary, latently infected CD4+ T cells obtained from patients undergoing cART.176 Although the mechanism by which this retention only occurs in resting, latently infected cells is unknown, the overexpression of the RNA-binding protein PTB circumvented this posttranscriptional block, allowing for the production of virions without requiring CD4+ T cell activation.176 Hence, factors that affect HIV-1 gene expression at a posttranscriptional level also influence the outcome of the infection, and can enforce latency.
Implication of these mechanisms in the establishment of latency in vivo
The relevance of each of the mechanisms presented above in the formation of latent reservoirs is widely accepted. However, how their combination affects the outcome of the HIV-1 infection is not well understood. In addition, the fact that many of the studies discussed in this review were performed in the context of cell lines or ex vivo models may not accurately reflect how latency is established in vivo, which complicates our understanding of the molecular bases influencing latency in patients. Despite these limitations, few aspects stand out for their convergence in vitro, ex vivo and in vivo, such as the roles of Tat and host transcription factors. In particular, Tat and NF-κB displace the effect of regulatory elements, such as epigenetic modulation and transcriptional interference, allowing HIV-1 RNA synthesis to proceed.138,150,159,162,163,171,173,175 Nevertheless, we need to stress that our knowledge of the roles of less investigated elements, such as miRNAs and HIV-1 RNA export in the outcome of HIV-1 infection is incomplete. Therefore, additional studies on physiologically relevant systems are needed to better comprehend their influence over the other mechanisms, and their overall contribution to HIV-1 latency.
Current Therapeutic Strategies to Eradicate the HIV-1 Latent Reservoirs
Although current cART regimens efficiently limit HIV-1 replication, reducing the viremia to undetectable levels in HIV-1+ individuals, they fail at clearing the cellular HIV-1 reservoir, posing a major challenge to achieving a sterilizing cure. As mentioned earlier, these dormant long-lived cells are resistant to ART and persist undetected by the immune system, having the capacity to emerge and support fully replicating and infectious viral particles spontaneously or if ART is discontinued.1,2,6,7 In addition, the high cost of cART, the need for lifelong adherence, and the associated toxicities to prolonged treatment are important limitations. Thus, there is an agreement among the scientific community to develop new therapies that go beyond continuous cART treatments to finally achieve the end of the HIV/AIDS pandemic. For this, numerous efforts are being devoted to explore novel strategies to either clear or permanently suppress the HIV-1 latent reservoir204–207 (Fig. 2).
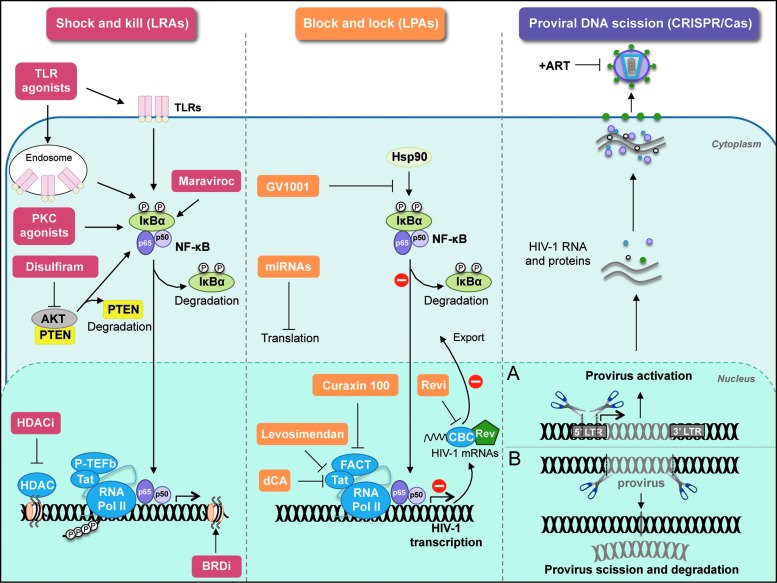
FIG. 2.
Current therapeutic strategies to eradicate the HIV-1 latent reservoirs. Schematic representation of the molecules targeted by (1) latency-reversing agents in shock and kill approaches (left panel), (2) latency-promoting agents in block and lock approaches (central panel), and (3) CRISPR-mediated DNA scission techniques (right panel). (A) Gene editing tools to reactivate proviruses. (B) Gene editing tools to excise HIV-1 proviruses. CRISPR, clustered, regularly interspaced short palindromic repeat. Color images are available online at www.liebertpub.com/aid
Shock and kill strategies
Multiple therapeutic strategies aimed at achieving a functional cure of the HIV-1 infection are focused on the “shock and kill” strategy, using LRAs which, in combination to cART, disrupt HIV-1 latently infected cells, so they can be eliminated by immune effectors (cytotoxic T cells or CTLs) or cleared by the cytopathic effect of the reactivated virus.208–211 Optimally, effective and safe LRAs should reactivate the latent provirus and promote viral transcription followed by viral protein production. This approach should enhance HIV-1-specific immune responses (CTL-mediated responses) to recognize the presentation of viral antigens and clear the reactivated cells. So far, several assays in ex vivo latency models, as well as in clinical trials, have been performed to evaluate the feasibility of small-molecule LRA candidates to effectively reverse HIV-1 latency, which have recently been reviewed elsewhere210,211 (Fig. 2, left panel). These include:
-
(1)
Protein kinase C (PKC) agonists (e.g., PMA, prostratin, bryostatin, and ingenol molecules). PKC agonists activate PKC cellular isoforms, which then initiate NF-κB signaling to induce HIV-1 transcription, thereby, potently reversing latency.88,212,213 According to recent studies, only bryostatin-1 has shown efficacy when tested in cells derived from patients on suppressive cART.214,215 However, one of the main concerns when applying PKC agonists relates to the associated side effects resulting from continuous activation and inflammation.214 To address this, alternative approaches have been proposed to reduce the PKC agonist-mediated off-target effects of T cell activation. For example, the use of PKC agonists in combination with adjuvant agents, such as the JAK inhibitor ruxolitinib, has shown a decrease in proinflammatory cytokine production while maintaining the capacity to reverse latency.210 Also, an attractive option to restrict PKC agonist-derived toxicities is the recently proposed oral therapy based on the PKC agonist ingenol, a plant-derived natural compound, which reactivates latent HIV-1 and when combined with other LRAs shows synergistic effects, facilitating a reduction in the dosing of both agents.216 Another possibility to reduce toxic effects associated with polyclonal T cell activation might be the combination of PKC agonists with Rapamycin. In a recent study by the Siliciano laboratory, the combination of LRAs with Rapamycin, a drug that triggers autophagy but also works as an immunosuppressant, showed efficient viral reactivation with no toxic effects from the systemic release of proinflammatory cytokines.217
-
(2)
Histone deacetylase inhibitors (HDACi; e.g., vorinostat/SAHA, panobinostat, and romidepsin). As previously stated, histone deacetylation (HDAC) is an epigenetic modification of chromatin that results in the repression of HIV-1 RNA synthesis.218 Small-molecule inhibitors targeting HDAC enzymes have shown to safely reverse proviral latency by promoting HIV-1 transcription both in vitro and in vivo, although with different success. For example, the HDACi vorinostat/SAHA modestly reactivates latent HIV-1 in resting CD4+ T cells.208,209,219–221 However, this proviral reactivation is not associated with an effective killing of latently infected cells in vivo,221 suggesting that CTL activation is a prerequisite for HDACi treatment success. By contrast, treatment with panobinostat not only reactivates proviruses, but also increases innate immune activity of natural killer (NK) cells, suggesting that innate immune responses can critically modulate the efficacy of LRAs.222 In the recent past, HDACi have been the pillars of latency reversal in clinical trials, evidencing that after HDACi treatment an enhancement in HIV-1 RNA and subsequent increase in viral production was observed.208,223–225 Unfortunately, to date, none of the HDACi-based treatments tested in clinical trials has succeeded at significantly reducing the size of the reservoir (reviewed in Spivak and Planelles210).
-
(3)
Disulfiram, an FDA-approved drug, reactivates latent HIV-1 in a primary CD4+ T cell model by decreasing the levels of the PTEN cytoplasmic protein with the subsequent activation of the Akt signaling pathway.90,226 Two clinical trials have recently assessed disulfiram as a LRA, with no significant reduction in the size of the latent reservoir being observed.227,228
-
(4)
Bromo and extraterminal (BET) bromodomain inhibitors, together with toll-like receptor agonists, have shown promising results as LRAs, but the efficacy of many of these compounds in vivo remains to be examined.
-
(5)
Remarkably, in a search for novel LRAs, maraviroc, a drug currently used in cART as a CCR5 antagonist, was found to reverse HIV-1 latency in infected individuals under cART in a phase II clinical trial.
-
(6)
The authors of this study suggested that the interaction of maraviroc with CCR5 induces the activation of NF-κB and the subsequent enhancement of HIV-1 transcription in resting CD4+ T cells. Therefore, maraviroc shows potential as a LRA to reduce the HIV-1 viral reservoir in ART-treated patients.235
Despite all this progress, the shock and kill strategy is beset by significant limitations before effective elimination of the reservoir can be achieved. Although LRAs may disrupt latency in CD4+ T cells, LRAs are poorly efficient in other HIV-1 target cell populations, such as macrophages and microglial cells. Thus, their potential to reactivate all the cell types that constitute the latent reservoir is partial at best. Even the most effective LRAs have been reported to fail at reactivating all replication-competent proviruses.3 An important caveat to LRAs is that they modulate gene expression in a general manner; thus, their potential for off-target or toxic effects is a major concern. For example, HDACi, such as SAHA act by opening the chromatin structure around the promoter of the HIV-1 integrated provirus, and therefore, may lead to the expression of other host genes, and potentially induce reactivation of other integrated proviruses alongside HIV-1. Additionally, some LRAs may activate uninfected HIV-1 target cells, which then may become susceptible to HIV-1 infection. In some cases, despite their combination with ART, LRA implementation might result in reinfection of target cells in anatomical viral sanctuary tissues, where ART has less penetration, including the CNS.236
Another main challenge of this approach is that LRA compounds fail at inducing sufficient viral cytopathic effects and potent HIV-1-specific CTL responses to purge HIV-1 reservoirs, showing so far a moderate impact on the reservoir size. Obstacles, such as the presence of CD8+ T cell escape mutations in the proviral genome of latently infected CD4+ T cells,237 LRAs limited success at reactivating latent proviruses to allow antigen presentation for CTL-mediated elimination,238 or the presence of HIV-1 reservoirs in lymph nodes and the CNS, where CTL infiltration is more difficult, seem to prevent the CTL-mediated clearance of latently infected cells, enhancing even further the establishment of HIV-1 reservoirs.
Additional considerations include the window opportunity of the LRAs and the fact that these compounds could alter the host immune function. In this regard, a recent study explored the effect of LRA combinations in HIV-1-specific CD8+ T cell responses and found that although the administration of romidepsin (HDACi) and bryostatin-1 (PKC agonist) effectively results in the reversal of the latent sate, this dual treatment inhibits HIV-1-specific CD8+ T cell function to recognize and eliminate HIV-1-reactivated cells.239 Interestingly, another study suggests that natural latent HIV-1 reservoirs in ex vivo CD4+ T cells from ART-treated patients present intrinsic resistance to CD8+ T cell-mediated clearance, which may complicate even further their eradication.240 The authors speculate that the observed resilience may be attributed to (i) HIV-1 Nef downmodulation of MHC-I on the surface of HIV-1-infected cells, resulting in a lack of recognition by CD8+ T cells, and/or (ii) HIV-1 latently infected cells with inherent resistance may be selected to conform a large fraction of the reservoir. These limitations underscore the need for more effective and specific LRAs for viral reactivation accompanied by potent immunotherapeutic interventions to ensure stronger HIV-1-specific immune responses, and urge the design of alternative therapeutic strategies to successfully eliminate the latent reservoir.
Block and lock strategy
The block and lock approach has recently emerged as an alternative to the shock and kill strategy to functionally cure HIV-1. This approach consists of the use of small-molecule inhibitors, HIV-1 latency-promoting agents (LPAs), to effectively silence (block) the HIV-1 promoter region in latently infected cells, and therefore, restrict (lock) viral transcription of HIV-1 integrated proviruses, impairing in turn viral rebound, and further intensifying their latent state. Several compounds are being investigated for their LPA potential, some of them with promising results (Fig. 2, central panel). These include:
-
(1)
Levosimendan, which specifically blocks the Tat-enhanced HIV-1 transcription in a PI3K pathway-dependent manner.241
-
(2)
The small-molecule Curaxin 100 (CBL0100), which impairs the access of the RNA Pol II and FACT at the HIV-1 promoter region, blocking the elongation process.242
-
(3)
The G1V001 telomerase-derived peptide, which inhibits HIV-1 transcription and viral reactivation by blocking the NF-κB pathway in an Hsp90-dependent fashion.243
-
(4)
Didehydro-Cortistatin A (dCA), a Tat inhibitor characterized by Mousseau et al.96,244 that potently silences HIV-1 transcription.
Altogether, with this study, other compunds affecting Tat biogenesis have already been reported to cause proviral silencing.245 However, to our knowledge, dCA is the only compound used in vivo that provided the first proof of concept for the potential use of a Tat inhibitor to durably suppress viral transcription.246 With the exception of dCA, the effectiveness of the other compounds needs further investigation, particularly in vivo, before their ultimate therapeutic use as anti-HIV-1 agents.
Despite some limitations, the encouraging results obtained in these studies indicate that the block and lock strategy is feasible and thus, urge to explore the potential of other molecules as LPAs. One promising locking approach is to target the HIV-1 Rev protein. Rev is critical for viral replication, since it regulates the export of nonspliced and singly spliced viral transcripts from the nucleus to the cytoplasm and, as mentioned above, defects in RNA export are thought to play an important role in HIV-1 latency. Campos and collaborators recently characterized ABX464, a Rev inhibitor that blocks viral replication in vitro as well as in humanized mice, showing long-lasting effects and no associated adverse effects in normal cellular splicing.247 Specifically, ABX464 binds to the Cap-Binding Complex (CBC) and impairs Rev-dependent export of nonspliced and singly spliced HIV-1 messenger RNAs (mRNAs), preventing their translation and neutralizing the expression of the viral genome. This molecule holds promise to affect the viral reservoir and encourages future studies to move further into clinical trials. Other attractive molecules as LPAs are miRNAs. In the past decade, more studies have revealed that miRNAs play a role in modulating latency in HIV-1-infected, quiescent cells.248 As mentioned above, miR-29a, which is one of the best-characterized miRNAs affecting HIV-1, efficiently suppresses the expression of viral transcripts. This observation led the authors to consider miR-29a as a potential tool for suppressing the HIV-1 reservoir by potentiating the endogenous levels of miR-29a, but also through miR-29a agonists. Either strategy could lead to a permanently silent state of HIV-1 proviruses as part of the block and lock strategy.193
Although still an emerging line of investigation, permanently repressing these latently infected cells appears a promising approach to keep dormant proviruses in check, given the limited success of the shock and kill strategy. A major caveat to this approach is the need to identify cellular markers that will allow to specifically target LPAs into cells harboring replication-competent proviruses and minimize off-target effects.
Proviral DNA scission
In the past few years, the use of gene-editing technologies, such as CRISPR/Cas9 (clustered, regularly interspaced short palindromic repeats—CRISPR-associated proteins) has gained momentum, including in the HIV-1 field. Genetic modulation by CRISPR/Cas9 is based on the use of a guide RNA (gRNA) that recognizes and mutates target DNA in the HIV-1 genome, offering sequence-dependent specificity and high cleavage efficiency.249,250 These properties make CRISPR/Cas9 a very attractive tool to permanently inactivate or even remove the proviral DNA from HIV-1 latently infected cells, and also to protect cells from future HIV-1 infections due to the continuous expression of the CRISPR/Cas9 system, thus, minimizing any potential harm to the host. To date, this technique has been tested in several cell types targeting critical HIV-1 genes as well as the LTR regions, since cleavage of both LTRs will result in the complete excision of the proviral DNA249 (Fig. 2, right panel, B). For example, Ebina et al. reported the successful removal of HIV-1 proviruses in infected Jurkat T cells with CRISPR/Cas9 by targeting the HIV-1 LTRs.251 In another study, by targeting the HIV-1 Rev region in the provirus, latently HIV-1-infected Jurkat cells were efficiently inactivated.252 The CRISPR/Cas9 system has also been successfully implemented in human pluripotent stem cell-derived HIV-1 reservoir cell types, where this technique was used not only to excise latent proviruses, but also to neutralize incoming infections by specifically targeting the incoming viral cDNA.253 Hu and collaborators further analyzed the capacity of the CRISPR system to eradicate the HIV-1 proviral DNA from myeloid lineage cells, the main HIV-1 cell reservoir in the brain. By targeting the U3 region in the HIV-1 LTRs, they effectively blocked viral gene expression, replication, and completely removed the proviral integrated genome that was flanked by the two LTRs, with no detectable off-target effects.254 Despite these encouraging results, some studies have reported undesired effects when using CRISPR/Cas9 to edit the human genome.255 This is mainly due to the cellular machinery that naturally repairs the cleaved DNA: nonhomologous end joining (NHEJ) or double-strand break (DSB) homologous recombination. Whereas homologous recombination has high fidelity, NHEJ is highly error prone, and the choice of one over the other is not controllable. To overcome this limitation, other cutting-edge gene-editing tools have been proposed to safely excise HIV-1 proviruses, such as a modified version of the Cre-Lox recombinase.256 Although an attractive alternative, the recent advances and improvement of the CRISPR/Cas9 system, together with the fact that errors introduced by NHEJ will mainly affect HIV-1 proviruses rather than the host genome, has allowed to reliably clear integrated HIV-1 proviral DNA. For instance, the study developed by Kaminski et al. demonstrated efficient cleavage and removal of the HIV-1 genome in latently HIV-1-infected CD4+ T cells by targeting the LTR with no associated harm to the DNA of host cells, and no undesired effects. Remarkably, in their ex vivo assays they found that their lentivirus-delivered CRISPR/Cas9 significantly reduced HIV-1 DNA and protein levels in CD4+ T cells from HIV-1-infected patients.257 The same team also provided proof of concept in an in vivo study that the CRISPR/Cas9 system can cleave HIV-1 DNA in transgenic mice and rats.258 Overall, these data support the solid potential of the CRISPR/Cas9-based tools to efficiently and safely cleave the HIV-1 integrated genome in latently infected cells, and therefore, block viral gene expression and replication as well as immunize against future HIV-1 infections.
Besides its use to inactivate proviruses, CRISPR/Cas9 can also be exploited to activate the transcriptional activity of the HIV-1 integrated provirus in latently infected cells in a site-specific manner, in line with the shock and kill strategy (Fig. 2, right panel, A). There is increasing evidence supporting the use of CRISPR-based transcriptional activation systems, alone or in combination with LRAs, to reverse HIV-1 latency and achieve the complete eradication of the latent infection. These CRISPR-based activation systems present substantial advantages over current LRA treatments, such as enhanced LTR activity compared with traditional shock and kill strategies, and minimal toxic and off-target effects,259–261 triggering fully infectious viral production.262
One of the main concerns when using CRISPR/Cas9 to edit DNA in quiescent cells, such as the HIV-1 reservoir, is the reliance on the cellular enzymatic machinery to express the gRNA and the Cas9 protein. Most CRISPR/Cas9 tools involve the use of lentiviral or adenoviral vectors that encode for these components. Therefore, it is easy to envision that the success rate for efficiently editing the HIV-1 proviral genome would significantly increase by preactivating these latently infected cells with LRAs, so the cells are metabolically active by the time they are transduced with CRISPR/Cas9. However, this does not seem to be the case. According to a recent publication by Zhu et al., latently infected cells can be efficiently edited without preactivation.252 Nevertheless, aspects such as the cell cycling status may interfere with the success of this technique, and should be investigated in detail.
There is no doubt that CRISPR/Cas9, as well as other gene-modifying techniques, has revolutionized the landscape of editing tools. Although very promising, there are some limitations and concerns that should be addressed before its general implementation as anti-HIV-1 treatments. HIV-1 viral escape is one of the main hurdles for a broad application of CRISPR/Cas9, and several studies have recently addressed whether HIV-1 could evade the CRISPR/Cas9-mediated suppression. Interestingly, Deng and collaborators found that the latent reservoir can also contain escape mutants for CRISPR/Cas9.237 However, the use of multiple gRNAs targeting different HIV-1 conserved regions could circumvent HIV-1 escape. In fact, recent reports proved that HIV-1-infected cells can be cured by a combination of two gRNAs targeting various sites in the proviral DNA, effectively blocking HIV-1 replication and preventing viral escape.263,264 Unfortunately, other studies evidenced HIV-1 evasion of the CRISPR/Cas9 suppression when mutations arose in the gRNA target sequence.265,266 In addition, due to the limited success in the delivery of these CRISPR/Cas9 systems, alternative delivery methods such as nanoparticles should be tested to enhance the applicability of this approach. Another caveat is that most of the data we have on CRISPR/Cas9 and HIV-1 is derived from in vitro studies. CRISPR/Cas9 effectiveness and safety should be analyzed in actual resting, latently infected CD4+ T cells obtained from HIV-1-infected patients to design customized CRISPR/Cas9 pairs, and this can be costly and time consuming.
Tools to Identify and Study the Latent Reservoir
One of the reasons why latency poses such a hurdle for HIV-1 eradication is the absence of effective methods to accurately detect latent reservoirs in vivo. The low frequency of infected resting CD4+ T cells present in patients undergoing cART, together with the low levels of transcribed viral RNA, greatly limit our ability to recognize, quantify, and thus assess the size of the latent reservoir in infected individuals.267–272 To date, the identification of unique cellular biomarkers that would correctly distinguish latently infected CD4+ T cells from healthy, uninfected memory CD4+ T cells still remains a challenge.267 Different studies have attempted to address this with relative success, since the proposed molecules are either not entirely exclusive for latently infected cells or are present only in a small fraction of the latent reservoir.273–276 Several molecules have been proposed as latency markers, including the membrane proteins CD2, PD-1, TIGIT, LAG-3, or HLA-DR.274,275,277,278 However, the actual proportion of latently infected cells that express these surface receptors is still undetermined. Although challenging, the investigation of biomarkers exclusively found in latently infected cells is a high priority, and several groups are investigating this through different approaches, for instance by combining transcriptomics and bioinformatics datasets.279
If the current unavailability of specific biomarkers to detect latently infected cells is a major stumbling block to purge the latent reservoir, another important limitation is the need for better techniques to accurately measure the amount of integrated HIV-1 proviruses.271,280. Current methods to detect replication-competent proviruses include polymerase chain reaction (PCR)-based assays, and quantitative virus outgrowth assays (Q-VOA).281,282
Quantitative PCR-based methods
Different quantitative PCR (qPCR)-based methods are currently used to measure the relative amount of HIV-1 cDNA or HIV-1 RNA present in a certain pool of peripheral blood mononuclear cells or plasma obtained from infected patients.283–285 For the DNA-based qPCR methods, the most widely used is the AluPCR, which measures the total integrated proviral DNA. By combining primers that target gag or the LTR together with the highly repetitive elements Alu in the human genome, this technique exclusively detects integrated proviral DNA. However, it does not discriminate between actively or latently infected cells, nor if the detected proviruses are replication competent or defective.271,286 For the RNA-based qPCR, the most common methods aim at measuring the relative levels of extracellular RNA (plasma viremia) over cell-associated RNA.271,280,283,287 However, these methods also fail at pinpointing latently infected cells. Although PCR-based assays are among the most sensitive and simple available tools, their main caveat is the unwarranted detection of defective integrated proviruses.271,280,288 In an attempt to address this limitation, Procopio et al. developed the Tat/Rev-induced limiting dilution assay (TILDA), which measures by qPCR the relative amount of cell-associated multiply-spliced HIV-1 RNA upon T cell stimulation, which in turns correlates with the presence of replication-competent proviruses.280
Quantitative virus outgrowth assays
The Q-VOA, unlike PCR-based methods, can single out resting CD4+ T cells capable of supporting a productive infection by measuring the relative release of infectious virions upon cellular stimulation. Hence, it is considered the gold standard method to quantify drug-resistant infected cells from resting CD4+ T cells isolated from patients on cART.271,281,282 However, compared with the qPCR assays, Q-VOA is more demanding, since this method is lengthy, requires large amounts of samples from donors, and seems to underestimate the actual size of the reservoir.3,27,35,281 Unfortunately, comparative experiments using both Q-VOA and PCR-based assays have shown clear discrepancies between these two techniques. In addition, they are limited to the quantitative nature of the reservoirs and they are incapable of identifying which subpopulation of the pooled cells has the potential to sustain a productive infection.288 Therefore, new improved methodologies to measure the latent reservoir in a simple, reproducible and effective manner are urgently needed. For instance, approaches based on proviral DNA sequencing are now gaining more attention, since they can accurately elucidate the composition of the proviral landscape within an individual, and discriminate between defective and replication-competent proviruses. Although they can be extremely expensive and labor intensive, the use of next-generation sequencing seems to streamline this approach significantly.35
In addition to the methods described above, several in vitro, ex vivo, as well as animal models have been developed to facilitate the study of HIV-1 latency. The in vitro systems include immortalized cell lines such as the T lymphocyte cell models, ACH2, J1.1, and Ca5, or the premonocytic U1 clone.83,289–292 These cell lines remain chronically and nonproductively infected, but retain the potential to support a productive infection upon stimulation with mitogens and cytokines such as αCD3/CD28 antibodies or the tumor necrosis factor alpha (TNFα), respectively.290,291 Jordan et al. generated the widely used J-Lat cell line, which was produced by inducing the stable and latent infection of Jurkat cells with a Δenv/Δnef HIV-1 NL4-3 carrying a GFP reporter.113 One of the weaknesses of these cell lines is that, unlike resting CD4+ T cells, they remain in a constant proliferation state and thus, they may not entirely reflect latency in vivo. To overcome this, several groups have developed ex vivo systems of latency using primary CD4+ T cells.118,293–297 Among these studies, the method developed by Bosque and Planelles is characterized by the isolation of naive CD4+ T cells to near purity, and inducing their differentiation through TCR engagement into a central memory-like phenotype. While activated, the cells are infected with a molecular clone of HIV-1, and are subsequently allowed to transition to this resting phenotype in a nonpolarized manner,296,298 which closely resembles the characteristics of TCM in vivo. Ex vivo systems have been extensively used to dissect signaling pathways that are involved in HIV-1 latency in CD4+ T cells, but fail at addressing other aspects of proviral latency, such as the role of non-T cells and the contribution of viral sanctuary tissues to the size of the reservoir. In this regard, humanized mouse models represent a more physiological scenario for the study of latency. These animal models are generated by injecting HIV-1 into immunodeficient recipient mice transplanted with either (1) fetal human thymus and liver, which procures human T cell lymphopoiesis (Thy/Liv mice model),299–301 (2) human bone marrow CD34+ hematopoietic stem cells (HSCs; huCD4 mice model),302 or (3) a combination of both, which generates the commonly used BLT mice model.303 However, many investigators are skeptical to use humanized mice as a model for the study of HIV-1 latency. Although they virtually have a human immune system, the roles of other organs and tissues, such as viral sanctuaries, may not accurately reflect the overall picture of the HIV-1 reservoir in humans. Animal models that better resemble human physiology and lentiviral latency are nonhuman primates infected with SIV (simian immunodeficiency virus) or recombinant viruses of SIV and HIV (SHIV), such as rhesus macaques or pig-tailed macaques.38,234,304–308 These models allow for (1) a more accurate study of latency and the roles of organs such as the CNS and the gut in the size and distribution of the reservoir,307–309 and (2) testing the efficiency of antiretrovirals,310 and possibly LRAs and LPAs. Despite these advantages, the use of nonhuman primates in AIDS research is expensive and requires the use of specialized facilities. Moreover, unlike the humanized mice models, this system requires long periods of experimental preparation.234,308 Therefore, in spite of the improvement of the experimental systems in the study of latency234,280,311,312 much work is still needed to develop cost and time-efficient models that would closely resemble HIV-1 latency in vivo.313
Concluding Remarks
To date, much effort has been devoted in eradicating HIV-1 from latently infected cells. However, to achieve a complete purge of the latent reservoir, a better understanding of the factors influencing the establishment and maintenance of latency, the contribution of T cells, as well as non-T cells to the latent reservoir—including HSCs314,315—as well as the identification of cellular biomarkers that will allow us to discriminate between healthy, resting cells and cells harboring replication-competent proviruses is needed. These technical advances will not only allow us to specifically target latently infected cells with the goal of clearing or silencing them, but also to assess the exact size and distribution of the reservoir in a more customized manner.
Acknowledgments
The authors would like to thank the American Foundation for AIDS Research (amfAR) and the editorial board of AIDS Research and Human Retroviruses for giving them the opportunity to contribute with this review article to this special issue to commemorate Dr. Mathilde Krim's legacy. R.S.-M. was a Krim fellow from 2012 to 2014. The work presented here has been funded in part by Texas Tech University and NIH/NIAID R21AI106400.
Author Disclosure Statement
The authors declare no competing financial interests.
References
- 1.Chun TW, Stuyver L, Mizell SB, et al. : Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997;94:13193–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finzi D, Hermankova M, Pierson T, et al. : Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997;278:1295–1300 [DOI] [PubMed] [Google Scholar]
- 3.Ho YC, Shan L, Hosmane NN, et al. : Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013;155:540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisele E, Siliciano RF: Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 2012;37:377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomont N, El-Far M, Ancuta P, et al. : HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009;15:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siliciano RF, Greene WC: HIV latency. Cold Spring Harb Perspect Med 2011;1:a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong JK, Hezareh M, Günthard HF, et al. : Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997;278:1291–1295 [DOI] [PubMed] [Google Scholar]
- 8.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS: HIV-1 entry into quiescent primary lymphocytes: Molecular analysis reveals a labile, latent viral structure. Cell 1990;61:213–222 [DOI] [PubMed] [Google Scholar]
- 9.Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M: Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 1991;254:423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun TW, Carruth L, Finzi D, et al. : Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997;387:183–188 [DOI] [PubMed] [Google Scholar]
- 11.Pierson TC, Zhou Y, Kieffer TL, Ruff CT, Buck C, Siliciano RF: Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol 2002;76:8518–8531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly J, Beddall MH, Yu D, Iyer SR, Marsh JW, Wu Y: Human macrophages support persistent transcription from unintegrated HIV-1 DNA. Virology 2008;372:300–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahouassa H, Daddacha W, Hofmann H, et al. : SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 2012;13:223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laguette N, Sobhian B, Casartelli N, et al. : SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011;474:654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger G, Durand S, Fargier G, et al. : APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog 2011;7:e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaipan C, Smith JL, Hu WS, Pathak VK: APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J Virol 2013;87:444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goujon C, Moncorgé O, Bauby H, et al. : Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 2013;502:559–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo K, Wang T, Liu B, et al. : Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol 2007;81:7238–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mbisa JL, Bu W, Pathak VK: APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J Virol 2010;84:5250–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petitjean G, Al Tabaa Y, Tuaillon E, et al. : Unintegrated HIV-1 provides an inducible and functional reservoir in untreated and highly active antiretroviral therapy-treated patients. Retrovirology 2007;4:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan CN, Trinité B, Lee CS, et al. : HIV-1 latency and virus production from unintegrated genomes following direct infection of resting CD4 T cells. Retrovirology 2016;13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douek DC, Brenchley JM, Betts MR, et al. : HIV preferentially infects HIV-specific CD4+ T cells. Nature 2002;417:95–98 [DOI] [PubMed] [Google Scholar]
- 23.Davey RT, Jr., Bhat N, Yoder C, et al. : HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A 1999;96:15109–15114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonetti FR, Sobolewski MD, Fyne E, et al. : Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 2016;113:1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS: Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A 1998;95:8869–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finzi D, Blankson J, Siliciano JD, et al. : Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999;5:512–517 [DOI] [PubMed] [Google Scholar]
- 27.Bruner KM, Murray AJ, Pollack RA, et al. : Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016;22:1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strain MC, Günthard HF, Havlir DV, et al. : Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: Intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci U S A 2003;100:4819–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chun TW, Nickle DC, Justement JS, et al. : Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis 2008;197:714–720 [DOI] [PubMed] [Google Scholar]
- 30.Gras G, Kaul M: Molecular mechanisms of neuroinvasion by monocytes-macrophages in HIV-1 infection. Retrovirology 2010;7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fletcher CV, Staskus K, Wietgrefe SW, et al. : Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 2014;111:2307–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigal A, Kim JT, Balazs AB, et al. : Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 2011;477:95–98 [DOI] [PubMed] [Google Scholar]
- 33.Evans VA, Kumar N, Filali A, et al. : Myeloid dendritic cells induce HIV-1 latency in non-proliferating CD4+ T cells. PLoS Pathog 2013;9:e1003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V: Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog 2011;7:e1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Simonetti FR, Siliciano RF, Laird GM: Measuring replication competent HIV-1: Advances and challenges in defining the latent reservoir. Retrovirology 2018;15:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu T: HIV-1 in peripheral blood monocytes: An underrated viral source. J Antimicrob Chemother 2002;50:309–311 [DOI] [PubMed] [Google Scholar]
- 37.Sahu GK, Paar D, Frost SD, Smith MM, Weaver S, Cloyd MW: Low-level plasma HIVs in patients on prolonged suppressive highly active antiretroviral therapy are produced mostly by cells other than CD4 T-cells. J Med Virol 2009;81:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitney JB, Hill AL, Sanisetty S, et al. : Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 2014;512:74–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pallikkuth S, Sharkey M, Babic DZ, et al. : Peripheral T follicular helper cells are the major HIV reservoir within central memory CD4 T cells in peripheral blood from chronically HIV-infected individuals on combination antiretroviral therapy. J Virol 2015;90:2718–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michie CA, McLean A, Alcock C, Beverley PC: Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature 1992;360:264–265 [DOI] [PubMed] [Google Scholar]
- 41.Yukl SA, Gianella S, Sinclair E, et al. : Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis 2010;202:1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wightman F, Solomon A, Khoury G, et al. : Both CD31(+) and CD31(–) naive CD4(+) T cells are persistent HIV type 1-infected reservoirs in individuals receiving antiretroviral therapy. J Infect Dis 2010;202:1738–1748 [DOI] [PubMed] [Google Scholar]
- 43.Banga R, Procopio FA, Noto A, et al. : PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 2016;22:754–761 [DOI] [PubMed] [Google Scholar]
- 44.Buzon MJ, Sun H, Li C, et al. : HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med 2014;20:139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gattinoni L, Lugli E, Ji Y, et al. : A human memory T cell subset with stem cell-like properties. Nat Med 2011;17:1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soriano-Sarabia N, Archin NM, Bateson R, et al. : Peripheral Vgamma9Vdelta2 T cells are a novel reservoir of latent HIV infection. PLoS Pathog 2015;11:e1005201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chun TW, Davey RT, Jr., Ostrowski M, et al. : Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat Med 2000;6:757–761 [DOI] [PubMed] [Google Scholar]
- 48.Coleman CM, Wu L: HIV interactions with monocytes and dendritic cells: Viral latency and reservoirs. Retrovirology 2009;6:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar A, Abbas W, Herbein G: HIV-1 latency in monocytes/macrophages. Viruses 2014;6:1837–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gama L, Abreu CM, Shirk EN, et al. : Reactivation of simian immunodeficiency virus reservoirs in the brain of virally suppressed macaques. AIDS 2017;31:5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koenig S, Gendelman HE, Orenstein JM, et al. : Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 1986;233:1089–1093 [DOI] [PubMed] [Google Scholar]
- 52.Sharova N, Swingler C, Sharkey M, Stevenson M: Macrophages archive HIV-1 virions for dissemination in trans. EMBO J 2005;24:2481–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho DD, Rota TR, Hirsch MS: Infection of monocyte/macrophages by human T lymphotropic virus type III. J Clin Invest 1986;77:1712–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM: Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science 1996;274:787–789 [DOI] [PubMed] [Google Scholar]
- 55.Wolf D, Witte V, Laffert B, et al. : HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat Med 2001;7:1217–1224 [DOI] [PubMed] [Google Scholar]
- 56.Olivetta E, Federico M: HIV-1 Nef protects human-monocyte-derived macrophages from HIV-1-induced apoptosis. Exp Cell Res 2006;312:890–900 [DOI] [PubMed] [Google Scholar]
- 57.Klase Z, Winograd R, Davis J, et al. : HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology 2009;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giri MS, Nebozyhn M, Raymond A, et al. : Circulating monocytes in HIV-1-infected viremic subjects exhibit an antiapoptosis gene signature and virus- and host-mediated apoptosis resistance. J Immunol 2009;182:4459–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choe W, Volsky DJ, Potash MJ: Activation of NF-kappaB by R5 and X4 human immunodeficiency virus type 1 induces macrophage inflammatory protein 1alpha and tumor necrosis factor alpha in macrophages. J Virol 2002;76:5274–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Busca A, Saxena M, Kumar A: Critical role for antiapoptotic Bcl-xL and Mcl-1 in human macrophage survival and cellular IAP1/2 (cIAP1/2) in resistance to HIV-Vpr-induced apoptosis. J Biol Chem 2012;287:15118–15133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baxter AE, Russell RA, Duncan CJ, et al. : Macrophage infection via selective capture of HIV-1-infected CD4+ T cells. Cell Host Microbe 2014;16:711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peressin M, Proust A, Schmidt S, et al. : Efficient transfer of HIV-1 in trans and in cis from Langerhans dendritic cells and macrophages to autologous T lymphocytes. AIDS 2014;28:667–677 [DOI] [PubMed] [Google Scholar]
- 63.Swingler S, Mann A, Jacqué J, et al. : HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat Med 1999;5:997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McElrath MJ, Steinman RM, Cohn ZA: Latent HIV-1 infection in enriched populations of blood monocytes and T cells from seropositive patients. J Clin Invest 1991;87:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yukl SA, Sinclair E, Somsouk M, et al. : A comparison of methods for measuring rectal HIV levels suggests that HIV DNA resides in cells other than CD4+ T cells, including myeloid cells. AIDS 2014;28:439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown A, Zhang H, Lopez P, Pardo CA, Gartner S: In vitro modeling of the HIV-macrophage reservoir. J Leukoc Biol 2006;80:1127–1135 [DOI] [PubMed] [Google Scholar]
- 67.Honeycutt JB, Thayer WO, Baker CE, et al. : HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med 2017;23:638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R: HIV-1 target cells in the CNS. J Neurovirol 2015;21:276–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brown D, Mattapallil JJ: Gastrointestinal tract and the mucosal macrophage reservoir in HIV infection. Clin Vaccine Immunol 2014;21:1469–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smed-Sörensen A, Loré K, Vasudevan J, et al. : Differential susceptibility to human immunodeficiency virus type 1 infection of myeloid and plasmacytoid dendritic cells. J Virol 2005;79:8861–8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith BA, Gartner S, Liu Y, et al. : Persistence of infectious HIV on follicular dendritic cells. J Immunol 2001;166:690–696 [DOI] [PubMed] [Google Scholar]
- 72.Haase AT, Henry K, Zupancic M, et al. : Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 1996;274:985–989 [DOI] [PubMed] [Google Scholar]
- 73.Spiegel H, Herbst H, Niedobitek G, Foss HD, Stein H: Follicular dendritic cells are a major reservoir for human immunodeficiency virus type 1 in lymphoid tissues facilitating infection of CD4+ T-helper cells. Am J Pathol 1992;140:15–22 [PMC free article] [PubMed] [Google Scholar]
- 74.Heath SL, Tew JG, Tew JG, Szakal AK, Burton GF: Follicular dendritic cells and human immunodeficiency virus infectivity. Nature 1995;377:740–744 [DOI] [PubMed] [Google Scholar]
- 75.Keele BF, Tazi L, Gartner S, et al. : Characterization of the follicular dendritic cell reservoir of human immunodeficiency virus type 1. J Virol 2008;82:5548–5561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J, Perelson AS: Contribution of follicular dendritic cells to persistent HIV viremia. J Virol 2013;87:7893–7901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loré K, Smed-Sörensen A, Vasudevan J, Mascola JR, Koup RA: Myeloid and plasmacytoid dendritic cells transfer HIV-1 preferentially to antigen-specific CD4+ T cells. J Exp Med 2005;201:2023–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cameron PU, Freudenthal PS, Barker JM, Gezelter S, Inaba K, Steinman RM: Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 1992;257:383–387 [DOI] [PubMed] [Google Scholar]
- 79.McDonald D, Wu L, Bohks SM, KewalRamani VN, Unutmaz D, Hope TJ: Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 2003;300:1295–1297 [DOI] [PubMed] [Google Scholar]
- 80.Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF: The multifactorial nature of HIV-1 latency. Trends Mol Med 2004;10:525–531 [DOI] [PubMed] [Google Scholar]
- 81.Mousseau G, Valente ST: Role of host factors on the regulation of Tat-mediated HIV-1 transcription. Curr Pharm Des 2017;23:4079–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Romani B, Allahbakhshi E: Underlying mechanisms of HIV-1 latency. Virus Genes 2017;53:329–339 [DOI] [PubMed] [Google Scholar]
- 83.Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS: Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 1987;238:800–802 [DOI] [PubMed] [Google Scholar]
- 84.Prins JM, Jurriaans S, van Praag RM, et al. : Immuno-activation with anti-CD3 and recombinant human IL-2 in HIV-1-infected patients on potent antiretroviral therapy. AIDS 1999;13:2405–2410 [DOI] [PubMed] [Google Scholar]
- 85.Alvarez-Carbonell D, Garcia-Mesa Y, Milne S, et al. : Toll-like receptor 3 activation selectively reverses HIV latency in microglial cells. Retrovirology 2017;14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rochat MA, Schlaepfer E, Speck RF: Promising role of Toll-like receptor 8 agonist in concert with prostratin for activation of silent HIV. J Virol 2017;91:e02084-16. doi: 10.1128/JVI.02084-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li P, Kaiser P, Lampiris HW, et al. : Stimulating the RIG-I pathway to kill cells in the latent HIV reservoir following viral reactivation. Nat Med 2016;22:807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang G, Dandekar S: Targeting NF-kappaB signaling with protein kinase C agonists as an emerging strategy for combating HIV latency. AIDS Res Hum Retroviruses 2015;31:4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang G, Mendes EA, Kaiser P, et al. : Synergistic reactivation of latent HIV expression by ingenol-3-angelate, PEP005, targeted NF-κB signaling in combination with JQ1 induced p-TEFb activation. PLoS Pathog 2015;11:e1005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xing S, Bullen CK, Shroff NS, et al. : Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. J Virol 2011;85:6060–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barton KM, Archin NM, Keedy KS, et al. : Selective HDAC inhibition for the disruption of latent HIV-1 infection. PLoS One 2014;9:e102684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Archin NM, Kirchherr JL, Sung JA, et al. : Interval dosing with the HDAC inhibitor vorinostat effectively reverses HIV latency. J Clin Invest 2017;127:3126–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Besnard E, Hakre S, Kampmann M, et al. : The mTOR complex controls HIV latency. Cell Host Microbe 2016;20:785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim KC, Lee S, Son J, et al. : Identification of novel genes associated with HIV-1 latency by analysis of histone modifications. Hum Genomics 2017;11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boehm D, Jeng M, Camus G, et al. : SMYD2-mediated histone methylation contributes to HIV-1 latency. Cell Host Microbe 2017;21:569.e6–579.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mousseau G, Kessing CF, Fromentin R, Trautmann L, Chomont N, Valente ST: The Tat inhibitor didehydro-cortistatin A prevents HIV-1 reactivation from latency. MBio 2015;6:e00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang J, Yang J, Yang Z, et al. : RbAp48, a novel inhibitory factor that regulates the transcription of human immunodeficiency virus type 1. Int J Mol Med 2016;38:267–274 [DOI] [PubMed] [Google Scholar]
- 98.Wang X, Mbondji-Wonje C, Zhao J, Hewlett I: IL-1beta and IL-18 inhibition of HIV-1 replication in Jurkat cells and PBMCs. Biochem Biophys Res Commun 2016;473:926–930 [DOI] [PubMed] [Google Scholar]
- 99.Cameron PU, Saleh S, Sallmann G, et al. : Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc Natl Acad Sci U S A 2010;107:16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cismasiu VB, Paskaleva E, Suman Daya S, Canki M, Duus K, Avram D: BCL11B is a general transcriptional repressor of the HIV-1 long terminal repeat in T lymphocytes through recruitment of the NuRD complex. Virology 2008;380:173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Panagoulias I, Karagiannis F, Aggeletopoulou I, et al. : Ets-2 acts as a transcriptional repressor of the human immunodeficiency virus type 1 through binding to a repressor-activator target sequence of 5′-LTR. Front Immunol 2017;8:1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ne E, Palstra RJ, Mahmoudi T: Transcription: Insights from the HIV-1 promoter. Int Rev Cell Mol Biol 2018;335:191–243 [DOI] [PubMed] [Google Scholar]
- 103.Saayman S, Ackley A, Turner AW, et al. : An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther 2014;22:1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kobayashi-Ishihara M, Yamagishi M, Hara T, et al. : HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology 2012;9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kobayashi-Ishihara M, Terahara K, Martinez JP, et al. : HIV LTR-driven antisense RNA by itself has regulatory function and may curtail virus reactivation from latency. Front Microbiol 2018;9:1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Allshire RC, Madhani HD: Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol 2018;19:229–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Singh PK, Plumb MR, Ferris AL, et al. : LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev 2015;29:2287–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shun MC, Raghavendra NK, Vandegraaff N, et al. : LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 2007;21:1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sowd GA, Serrao E, Wang H, et al. : A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc Natl Acad Sci U S A 2016;113:E1054–E1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rasheedi S, Shun MC, Serrao E, et al. : The cleavage and polyadenylation specificity factor 6 (CPSF6) subunit of the capsid-recruited pre-messenger RNA cleavage factor I (CFIm) complex mediates HIV-1 integration into genes. J Biol Chem 2016;291:11809–11819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lusic M, Siliciano RF: Nuclear landscape of HIV-1 infection and integration. Nat Rev Microbiol 2017;15:69–82 [DOI] [PubMed] [Google Scholar]
- 112.Cohn LB, Silva IT, Oliveira TY, et al. : HIV-1 integration landscape during latent and active infection. Cell 2015;160:420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jordan A, Bisgrove D, Verdin E: HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 2003;22:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han Y, Lassen K, Monie D, et al. : Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol 2004;78:6122–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F: HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002;110:521–529 [DOI] [PubMed] [Google Scholar]
- 116.Brady T, Agosto LM, Malani N, Berry CC, O'Doherty U, Bushman F: HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS 2009;23:1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sherrill-Mix S, Lewinski MK, Famiglietti M, et al. : HIV latency and integration site placement in five cell-based models. Retrovirology 2013;10:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tyagi M, Pearson RJ, Karn J: Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol 2010;84:6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li L, Dahiya S, Kortagere S, et al. : Impact of Tat genetic variation on HIV-1 disease. Adv Virol 2012;2012:123605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Verdin E, Paras P, Jr., Van Lint C: Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J 1993;12:3249–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Verdin E: DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J Virol 1991;65:6790–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Demarchi F, D'Agaro P, Falaschi A, Giacca M: In vivo footprinting analysis of constitutive and inducible protein-DNA interactions at the long terminal repeat of human immunodeficiency virus type 1. J Virol 1993;67:7450–7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Easley R, Carpio L, Dannenberg L, et al. : Transcription through the HIV-1 nucleosomes: Effects of the PBAF complex in Tat activated transcription. Virology 2010;405:322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lusic M, Marcello A, Cereseto A, Giacca M: Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 2003;22:6550–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Col E, Caron C, Seigneurin-Berny D, Gracia J, Favier A, Khochbin S: The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J Biol Chem 2001;276:28179–28184 [DOI] [PubMed] [Google Scholar]
- 126.Vendel AC, Lumb KJ: Molecular recognition of the human coactivator CBP by the HIV-1 transcriptional activator Tat. Biochemistry 2003;42:910–916 [DOI] [PubMed] [Google Scholar]
- 127.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC: NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 2006;25:139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Blazkova J, Trejbalova K, Gondois-Rey F, et al. : CpG methylation controls reactivation of HIV from latency. PLoS Pathog 2009;5:e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E: Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog 2009;5:e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marban C, Suzanne S, Dequiedt F, et al. : Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J 2007;26:412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.du Chéné I, Basyuk E, Lin YL, et al. : Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 2007;26:424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bednarik DP, Cook JA, Pitha PM: Inactivation of the HIV LTR by DNA CpG methylation: Evidence for a role in latency. EMBO J 1990;9:1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ishida T, Hamano A, Koiwa T, Watanabe T: 5′ Long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology 2006;3:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li F, Li L, Zhong Y, et al. : Relationship between LTR methylation and gag expression of HIV-1 in human spermatozoa and sperm-derived embryos. PLoS One 2013;8:e54801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Blazkova J, Murray D, Justement JS, et al. : Paucity of HIV DNA methylation in latently infected, resting CD4+ T cells from infected individuals receiving antiretroviral therapy. J Virol 2012;86:5390–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Budhiraja S, Famiglietti M, Bosque A, Planelles V, Rice AP: Cyclin T1 and CDK9 T-loop phosphorylation are downregulated during establishment of HIV-1 latency in primary resting memory CD4+ T cells. J Virol 2013;87:1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Patikoglou G, Burley SK: Eukaryotic transcription factor-DNA complexes. Annu Rev Biophys Biomol Struct 1997;26:289–325 [DOI] [PubMed] [Google Scholar]
- 138.Ott M, Geyer M, Zhou Q: The control of HIV transcription: Keeping RNA polymerase II on track. Cell Host Microbe 2011;10:426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.DeLuca C, Petropoulos L, Zmeureanu D, Hiscott J: Nuclear IkappaBbeta maintains persistent NF-kappaB activation in HIV-1-infected myeloid cells. J Biol Chem 1999;274:13010–13016 [DOI] [PubMed] [Google Scholar]
- 140.Romanchikova N, Ivanova V, Scheller C, Jankevics E, Jassoy C, Serfling E: NFAT transcription factors control HIV-1 expression through a binding site downstream of TAR region. Immunobiology 2003;208:361–365 [DOI] [PubMed] [Google Scholar]
- 141.Jones KA, Kadonaga JT, Luciw PA, Tjian R: Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science 1986;232:755–759 [DOI] [PubMed] [Google Scholar]
- 142.Ross EK, Buckler-White AJ, Rabson AB, Englund G, Martin MA: Contribution of NF-kappa B and Sp1 binding motifs to the replicative capacity of human immunodeficiency virus type 1: Distinct patterns of viral growth are determined by T-cell types. J Virol 1991;65:4350–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Suñé C, García-Blanco MA: Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J Virol 1995;69:6572–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Duverger A, Wolschendorf F, Zhang M, et al. : An AP-1 binding site in the enhancer/core element of the HIV-1 promoter controls the ability of HIV-1 to establish latent infection. J Virol 2013;87:2264–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mukerjee R, Sawaya BE, Khalili K, Amini S: Association of p65 and C/EBPbeta with HIV-1 LTR modulates transcription of the viral promoter. J Cell Biochem 2007;100:1210–1216 [DOI] [PubMed] [Google Scholar]
- 146.Sieweke MH, Tekotte H, Jarosch U, Graf T: Cooperative interaction of ets-1 with USF-1 required for HIV-1 enhancer activity in T cells. EMBO J 1998;17:1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rohr O, Marban C, Aunis D, Schaeffer E: Regulation of HIV-1 gene transcription: From lymphocytes to microglial cells. J Leukoc Biol 2003;74:736–749 [DOI] [PubMed] [Google Scholar]
- 148.Hay RT, Vuillard L, Desterro JM, Rodriguez MS: Control of NF-kappa B transcriptional activation by signal induced proteolysis of I kappa B alpha. Philos Trans R Soc Lond B Biol Sci 1999;354:1601–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Simeonidis S, Stauber D, Chen G, Hendrickson WA, Thanos D: Mechanisms by which IkappaB proteins control NF-kappaB activity. Proc Natl Acad Sci U S A 1999;96:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Coiras M, López-Huertas MR, Rullas J, Mittelbrunn M, Alcamí J: Basal shuttle of NF-kappaB/I kappaB alpha in resting T lymphocytes regulates HIV-1 LTR dependent expression. Retrovirology 2007;4:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Colomer-Lluch M, Serra-Moreno R: BCA2/Rabring7 interferes with HIV-1 proviral transcription by enhancing the SUMOylation of IkappaBalpha. J Virol 2017;91:pii: . doi: 10.1128/JVI.02098-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Manganaro L, Pache L, Herrmann T, et al. : Tumor suppressor cylindromatosis (CYLD) controls HIV transcription in an NF-kappaB-dependent manner. J Virol 2014;88:7528–7540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li C, Wang HB, Kuang WD, et al. : Naf1 regulates HIV-1 latency by suppressing viral promoter-driven gene expression in primary CD4+ T cells. J Virol 2017;91:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Coull JJ, Romerio F, Sun JM, et al. : The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol 2000;74:6790–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tyagi M, Karn J: CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J 2007;26:4985–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kaczmarek Michaels K, Natarajan M, Euler Z, Alter G, Viglianti G, Henderson AJ: Blimp-1, an intrinsic factor that represses HIV-1 proviral transcription in memory CD4+ T cells. J Immunol 2015;194:3267–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Palangat M, Hittinger CT, Landick R: Downstream DNA selectively affects a paused conformation of human RNA polymerase II. J Mol Biol 2004;341:429–442 [DOI] [PubMed] [Google Scholar]
- 158.Palangat M, Meier TI, Keene RG, Landick R: Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol Cell 1998;1:1033–1042 [DOI] [PubMed] [Google Scholar]
- 159.Kamori D, Ueno T: HIV-1 Tat and viral latency: What we can learn from naturally occurring sequence variations. Front Microbiol 2017;8:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Garber ME, Jones KA: HIV-1 Tat: Coping with negative elongation factors. Curr Opin Immunol 1999;11:460–465 [DOI] [PubMed] [Google Scholar]
- 161.Parada CA, Roeder RG: Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 1996;384:375–378 [DOI] [PubMed] [Google Scholar]
- 162.Muniz L, Egloff S, Ughy B, Jády BE, Kiss T: Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog 2010;6:e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McNamara RP, Bacon CW, D'Orso I: Transcription elongation control by the 7SK snRNP complex: Releasing the pause. Cell Cycle 2016;15:2115–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yamaguchi Y, Takagi T, Wada T, et al. : NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 1999;97:41–51 [DOI] [PubMed] [Google Scholar]
- 165.Jadlowsky JK, Wong JY, Graham AC, et al. : Negative elongation factor is required for the maintenance of proviral latency but does not induce promoter-proximal pausing of RNA polymerase II on the HIV long terminal repeat. Mol Cell Biol 2014;34:1911–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang Z, Klatt A, Gilmour DS, Henderson AJ: Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J Biol Chem 2007;282:16981–16988 [DOI] [PubMed] [Google Scholar]
- 167.Brannan K, Kim H, Erickson B, et al. : mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell 2012;46:311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Adelman K, Lis JT: Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet 2012;13:720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cheng B, Li T, Rahl PB, et al. : Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell 2012;45:38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM: Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 2004;24:787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM: NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 2001;8:327–337 [DOI] [PubMed] [Google Scholar]
- 172.Kao SY, Calman AF, Luciw PA, Peterlin BM: Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 1987;330:489–493 [DOI] [PubMed] [Google Scholar]
- 173.Tréand C, du Chéné I, Brès V, et al. : Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J 2006;25:1690–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Karn J: The molecular biology of HIV latency: Breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS 2011;6:4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lin X, Irwin D, Kanazawa S, et al. : Transcriptional profiles of latent human immunodeficiency virus in infected individuals: Effects of Tat on the host and reservoir. J Virol 2003;77:8227–8236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF: Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog 2006;2:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mbonye U, Karn J: The molecular basis for human immunodeficiency virus latency. Annu Rev Virol 2017;4:261–285 [DOI] [PubMed] [Google Scholar]
- 178.Pearson R, Kim YK, Hokello J, et al. : Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol 2008;82:12291–12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Donahue DA, Kuhl BD, Sloan RD, Wainberg MA: The viral protein Tat can inhibit the establishment of HIV-1 latency. J Virol 2012;86:3253–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kim YK, Mbonye U, Hokello J, Karn J: T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol 2011;410:896–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lenasi T, Contreras X, Peterlin BM: Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 2008;4:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Greger IH, Demarchi F, Giacca M, Proudfoot NJ: Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res 1998;26:1294–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Callen BP, Shearwin KE, Egan JB: Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell 2004;14:647–656 [DOI] [PubMed] [Google Scholar]
- 184.Han Y, Lin YB, An W, et al. : Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 2008;4:134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shan L, Rabi SA, Laird GM, et al. : A novel PCR assay for quantification of HIV-1 RNA. J Virol 2013;87:6521–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.De Marco A, Biancotto C, Knezevich A, Maiuri P, Vardabasso C, Marcello A: Intragenic transcriptional cis-activation of the human immunodeficiency virus 1 does not result in allele-specific inhibition of the endogenous gene. Retrovirology 2008;5:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fabian MR, Sonenberg N: The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat Struct Mol Biol 2012;19:586–593 [DOI] [PubMed] [Google Scholar]
- 188.Klase Z, Houzet L, Jeang KT: MicroRNAs and HIV-1: Complex interactions. J Biol Chem 2012;287:40884–40890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hariharan M, Scaria V, Pillai B, Brahmachari SK: Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun 2005;337:1214–1218 [DOI] [PubMed] [Google Scholar]
- 190.Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM: Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 2009;34:696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chable-Bessia C, Meziane O, Latreille D, et al. : Suppression of HIV-1 replication by microRNA effectors. Retrovirology 2009;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ahluwalia JK, Khan SZ, Soni K, et al. : Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology 2008;5:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Frattari G, Aagaard L, Denton PW: The role of miR-29a in HIV-1 replication and latency. J Virus Erad 2017;3:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chiang K, Rice AP: MicroRNA-mediated restriction of HIV-1 in resting CD4+ T cells and monocytes. Viruses 2012;4:1390–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sung TL, Rice AP: miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog 2009;5:e1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shen CJ, Jia YH, Tian RR, Ding M, Zhang C, Wang JH: Translation of Pur-α is targeted by cellular miRNAs to modulate the differentiation-dependent susceptibility of monocytes to HIV-1 infection. FASEB J 2012;26:4755–4764 [DOI] [PubMed] [Google Scholar]
- 197.Shida H: Role of nucleocytoplasmic RNA transport during the life cycle of retroviruses. Front Microbiol 2012;3:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nekhai S, Jeang KT: Transcriptional and post-transcriptional regulation of HIV-1 gene expression: Role of cellular factors for Tat and Rev. Future Microbiol 2006;1:417–426 [DOI] [PubMed] [Google Scholar]
- 199.Perales C, Carrasco L, González ME: Regulation of HIV-1 env mRNA translation by Rev protein. Biochim Biophys Acta 2005;1743:169–175 [DOI] [PubMed] [Google Scholar]
- 200.Meyer BE, Malim MH: The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev 1994;8:1538–1547 [DOI] [PubMed] [Google Scholar]
- 201.Fischer U, Meyer S, Teufel M, Heckel C, Lührmann R, Rautmann G: Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J 1994;13:4105–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cochrane A: HIV-1 Rev function and RNA nuclear-cytoplasmic export. Methods Mol Biol 2014;1087:103–114 [DOI] [PubMed] [Google Scholar]
- 203.Pomerantz RJ, Seshamma T, Trono D: Efficient replication of human immunodeficiency virus type 1 requires a threshold level of Rev: Potential implications for latency. J Virol 1992;66:1809–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.International AIDS Society Scientific Working Group on HIV Cure, Deeks SG, Autran B, et al. : Towards an HIV cure: A global scientific strategy. Nat Rev Immunol 2012;12:607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ: The challenge of finding a cure for HIV infection. Science 2009;323:1304–1307 [DOI] [PubMed] [Google Scholar]
- 206.Ruelas DS, Greene WC: An integrated overview of HIV-1 latency. Cell 2013;155:519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Martin AR, Siliciano RF: Progress toward HIV eradication: Case reports, current efforts, and the challenges associated with cure. Annu Rev Med 2016;67:215–228 [DOI] [PubMed] [Google Scholar]
- 208.Archin NM, Liberty AL, Kashuba AD, et al. : Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012;487:482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Shan L, Deng K, Shroff NS, et al. : Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 2012;36:491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Spivak AM, Planelles V: HIV-1 eradication: Early trials (and tribulations). Trends Mol Med 2016;22:10–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Spivak AM, Planelles V: Novel latency reversal agents for HIV-1 cure. Annu Rev Med 2018;69:421–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Rullas J, Bermejo M, García-Pérez J, et al. : Prostratin induces HIV activation and downregulates HIV receptors in peripheral blood lymphocytes. Antivir Ther 2004;9:545–554 [PubMed] [Google Scholar]
- 213.Spina CA, Anderson J, Archin NM, et al. : An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog 2013;9:e1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF: New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 2014;20:425–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Laird GM, Bullen CK, Rosenbloom DI, et al. : Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J Clin Invest 2015;125:1901–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Cary DC, Fujinaga K, Peterlin BM: Euphorbia kansui reactivates latent HIV. PLoS One 2016;11:e0168027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Martin AR, Pollack RA, Capoferri A, Ambinder RF, Durand CM, Siliciano RF: Rapamycin-mediated mTOR inhibition uncouples HIV-1 latency reversal from cytokine-associated toxicity. J Clin Invest 2017;127:651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Van Lint C, Emiliani S, Ott M, Verdin E: Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J 1996;15:1112–1120 [PMC free article] [PubMed] [Google Scholar]
- 219.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM: Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 2009;25:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Contreras X, Schweneker M, Chen CS, et al. : Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem 2009;284:6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ke R, Lewin SR, Elliott JH, Perelson AS: Modeling the effects of vorinostat in vivo reveals both transient and delayed HIV transcriptional activation and minimal killing of latently infected cells. PLoS Pathog 2015;11:e1005237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Olesen R, Vigano S, Rasmussen TA, et al. : Innate immune activity correlates with CD4 T cell-associated HIV-1 DNA decline during latency-reversing treatment with panobinostat. J Virol 2015;89:10176–10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Elliott JH, Wightman F, Solomon A, et al. : Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 2014;10:e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rasmussen TA, Tolstrup M, Brinkmann CR, et al. : Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: A phase 1/2, single group, clinical trial. Lancet HIV 2014;1:e13–e21 [DOI] [PubMed] [Google Scholar]
- 225.Søgaard OS, Graversen ME, Leth S, et al. : The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog 2015;11:e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Doyon G, Zerbato J, Mellors JW, Sluis-Cremer N: Disulfiram reactivates latent HIV-1 expression through depletion of the phosphatase and tensin homolog. AIDS 2013;27:F7–F11 [DOI] [PubMed] [Google Scholar]
- 227.Elliott JH, McMahon JH, Chang CC, et al. : Short-term administration of disulfiram for reversal of latent HIV infection: A phase 2 dose-escalation study. Lancet HIV 2015;2:e520–e529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Spivak AM, Andrade A, Eisele E, et al. : A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1-infected adults on antiretroviral therapy. Clin Infect Dis 2014;58:883–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Banerjee C, Archin N, Michaels D, et al. : BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol 2012;92:1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Gohda J, Suzuki K, Liu K, et al. : BI-2536 and BI-6727, dual Polo-like kinase/bromodomain inhibitors, effectively reactivate latent HIV-1. Sci Rep 2018;8:3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Thibault S, Imbeault M, Tardif MR, Tremblay MJ: TLR5 stimulation is sufficient to trigger reactivation of latent HIV-1 provirus in T lymphoid cells and activate virus gene expression in central memory CD4+ T cells. Virology 2009;389:20–25 [DOI] [PubMed] [Google Scholar]
- 232.Novis CL, Archin NM, Buzon MJ, et al. : Reactivation of latent HIV-1 in central memory CD4(+) T cells through TLR-1/2 stimulation. Retrovirology 2013;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Vibholm L, Schleimann MH, Højen JF, et al. : Short-course Toll-like receptor 9 agonist treatment impacts innate immunity and plasma viremia in individuals with human immunodeficiency virus infection. Clin Infect Dis 2017;64:1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lim SY, Osuna CE, Hraber PT, et al. : TLR7 agonists induce transient viremia and reduce the viral reservoir in SIV-infected rhesus macaques on antiretroviral therapy. Sci Transl Med 2018;10:pii: . doi: 10.1126/scitranslmed.aao4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Madrid-Elena N, García-Bermejo ML, Serrano-Villar S, et al. : Maraviroc is associated with latent HIV-1 reactivation through NF-kappaB activation in resting CD4(+) T cells from HIV-infected individuals on suppressive antiretroviral therapy. J Virol 2018;pii: . doi: 10.1128/JVI.01931-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ellis R, Langford D, Masliah E: HIV and antiretroviral therapy in the brain: Neuronal injury and repair. Nat Rev Neurosci 2007;8:33–44 [DOI] [PubMed] [Google Scholar]
- 237.Deng K, Pertea M, Rongvaux A, et al. : Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 2015;517:381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF: Latency reversal and viral clearance to cure HIV-1. Science 2016;353:aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Walker-Sperling VE, Pohlmeyer CW, Tarwater PM, Blankson JN: The effect of latency reversal agents on primary CD8+ T cells: Implications for shock and kill strategies for human immunodeficiency virus eradication. EBioMedicine 2016;8:217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Huang SH, Ren Y, Thomas AS, et al. : Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest 2018;128:876–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hayashi T, Jean M, Huang H, Simpson S, Santoso NG, Zhu J: Screening of an FDA-approved compound library identifies levosimendan as a novel anti-HIV-1 agent that inhibits viral transcription. Antiviral Res 2017;146:76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Jean MJ, Hayashi T, Huang H, et al. : Curaxin CBL0100 blocks HIV-1 replication and reactivation through inhibition of viral transcriptional elongation. Front Microbiol 2017;8:2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kim H, Choi MS, Inn KS, Kim BJ: Inhibition of HIV-1 reactivation by a telomerase-derived peptide in a HSP90-dependent manner. Sci Rep 2016;6:28896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Mousseau G, Clementz MA, Bakeman WN, et al. : An analog of the natural steroidal alkaloid cortistatin A potently suppresses Tat-dependent HIV transcription. Cell Host Microbe 2012;12:97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wan Z, Chen X: Triptolide inhibits human immunodeficiency virus type 1 replication by promoting proteasomal degradation of Tat protein. Retrovirology 2014;11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kessing CF, Nixon CC, Li C, et al. : In vivo suppression of HIV rebound by didehydro-cortistatin A, a “block-and-lock” strategy for HIV-1 treatment. Cell Rep 2017;21:600–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Campos N, Myburgh R, Garcel A, et al. : Long lasting control of viral rebound with a new drug ABX464 targeting Rev-mediated viral RNA biogenesis. Retrovirology 2015;12:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Huang J, Wang F, Argyris E, et al. : Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 2007;13:1241–1247 [DOI] [PubMed] [Google Scholar]
- 249.Wang G, Zhao N, Berkhout B, Das AT: CRISPR-Cas based antiviral strategies against HIV-1. Virus Res 2018;244:321–332 [DOI] [PubMed] [Google Scholar]
- 250.Doudna JA, Charpentier E: Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346:1258096. [DOI] [PubMed] [Google Scholar]
- 251.Ebina H, Misawa N, Kanemura Y, Koyanagi Y: Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep 2013;3:2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zhu W, Lei R, Le Duff Y, et al. : The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 2015;12:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Liao HK, Gu Y, Diaz A, et al. : Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 2015;6:6413. [DOI] [PubMed] [Google Scholar]
- 254.Hu W, Kaminski R, Yang F, et al. : RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A 2014;111:11461–11466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Tsai SQ, Zheng Z, Nguyen NT, et al. : GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015;33:187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Karpinski J, Hauber I, Chemnitz J, et al. : Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity. Nat Biotechnol 2016;34:401–409 [DOI] [PubMed] [Google Scholar]
- 257.Kaminski R, Chen Y, Fischer T, et al. : Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep 2016;6:22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Kaminski R, Bella R, Yin C, et al. : Excision of HIV-1 DNA by gene editing: A proof-of-concept in vivo study. Gene Ther 2016;23:696. [DOI] [PubMed] [Google Scholar]
- 259.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK: CRISPR RNA-guided activation of endogenous human genes. Nat Methods 2013;10:977–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ji H, Jiang Z, Lu P, et al. : Specific reactivation of latent HIV-1 by dCas9-SunTag-VP64-mediated guide RNA targeting the HIV-1 promoter. Mol Ther 2016;24:508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Saayman SM, Lazar DC, Scott TA, et al. : Potent and targeted activation of latent HIV-1 using the CRISPR/dCas9 activator complex. Mol Ther 2016;24:488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Bialek JK, Dunay GA, Voges M, et al. : Targeted HIV-1 latency reversal using CRISPR/Cas9-derived transcriptional activator systems. PLoS One 2016;11:e0158294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wang G, Zhao N, Berkhout B, Das AT: CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol Ther 2016;24:522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lebbink RJ, de Jong DC, Wolters F, et al. : A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci Rep 2017;7:41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wang G, Zhao N, Berkhout B, Das AT: A combinatorial CRISPR-Cas9 attack on HIV-1 DNA extinguishes all infectious provirus in infected T cell cultures. Cell Rep 2016;17:2819–2826 [DOI] [PubMed] [Google Scholar]
- 266.Liang C, Wainberg MA, Das AT, Berkhout BB: CRISPR/Cas9: A double-edged sword when used to combat HIV infection. Retrovirology 2016;13:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Yang W, Jackson B, Zhang H: Identification of glycoproteins associated with HIV latently infected cells using quantitative glycoproteomics. Proteomics 2016;16:1872–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Pierson T, McArthur J, Siliciano RF: Reservoirs for HIV-1: Mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu Rev Immunol 2000;18:665–708 [DOI] [PubMed] [Google Scholar]
- 269.Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF: Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol 2007;5:95–106 [DOI] [PubMed] [Google Scholar]
- 270.Zhang ZQ, Notermans DW, Sedgewick G, et al. : Kinetics of CD4+ T cell repopulation of lymphoid tissues after treatment of HIV-1 infection. Proc Natl Acad Sci U S A 1998;95:1154–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Massanella M, Richman DD: Measuring the latent reservoir in vivo. J Clin Invest 2016;126:464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Fischer M, Joos B, Wong JK, et al. : Attenuated and nonproductive viral transcription in the lymphatic tissue of HIV-1-infected patients receiving potent antiretroviral therapy. J Infect Dis 2004;189:273–285 [DOI] [PubMed] [Google Scholar]
- 273.Kok YL, Ciuffi A, Metzner KJ: Unravelling HIV-1 latency, one cell at a time. Trends Microbiol 2017;25:932–941 [DOI] [PubMed] [Google Scholar]
- 274.Iglesias-Ussel M, Vandergeeten C, Marchionni L, Chomont N, Romerio F: High levels of CD2 expression identify HIV-1 latently infected resting memory CD4+ T cells in virally suppressed subjects. J Virol 2013;87:9148–9158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Fromentin R, Bakeman W, Lawani MB, et al. : CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog 2016;12:e1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Descours B, Petitjean G, López-Zaragoza JL, et al. : CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature 2017;543:564–567 [DOI] [PubMed] [Google Scholar]
- 277.Hatano H, Jain V, Hunt PW, et al. : Cell-based measures of viral persistence are associated with immune activation and programmed cell death protein 1 (PD-1)-expressing CD4+ T cells. J Infect Dis 2013;208:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Cockerham LR, Siliciano JD, Sinclair E, et al. : CD4+ and CD8+ T cell activation are associated with HIV DNA in resting CD4+ T cells. PLoS One 2014;9:e110731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lee SY, Choi BS, Yoon CH, Kang C, Kim K, Kim KC: Selection of biomarkers for HIV-1 latency by integrated analysis. Genomics 2018;pii: . doi: 10.1016/j.ygeno.2018.02.007 [DOI] [PubMed] [Google Scholar]
- 280.Procopio FA, Fromentin R, Kulpa DA, et al. : A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2015;2:874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Prasad VR, Kalpana GV: FISHing out the hidden enemy: advances in detecting and measuring latent HIV-infected cells. MBio 2017;8:pii: . doi: 10.1128/mBio.01433-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Laird GM, Eisele EE, Rabi SA, et al. : Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog 2013;9:e1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Pasternak AO, Adema KW, Bakker M, et al. : Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol 2008;46:2206–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Fischer M, Huber W, Kallivroussis A, et al. : Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues. J Clin Microbiol 1999;37:1260–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Rouzioux C, Mélard A, Avéttand-Fénoël V: Quantification of total HIV1-DNA in peripheral blood mononuclear cells. Methods Mol Biol 2014;1087:261–270 [DOI] [PubMed] [Google Scholar]
- 286.O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH: A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol 2002;76:10942–10950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Dornadula G, Zhang H, VanUitert B, et al. : Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 1999;282:1627–1632 [DOI] [PubMed] [Google Scholar]
- 288.Eriksson S, Graf EH, Dahl V, et al. : Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 2013;9:e1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Morén C, González-Casacuberta I, Álvarez-Fernández C, et al. : HIV-1 promonocytic and lymphoid cell lines: An in vitro model of in vivo mitochondrial and apoptotic lesion. J Cell Mol Med 2017;21:402–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Folks TM, Clouse KA, Justement J, et al. : Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci U S A 1989;86:2365–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Perez VL, Rowe T, Justement JS, Butera ST, June CH, Folks TM: An HIV-1-infected T cell clone defective in IL-2 production and Ca2+ mobilization after CD3 stimulation. J Immunol 1991;147:3145–3148 [PubMed] [Google Scholar]
- 292.Seu L, Sabbaj S, Duverger A, et al. : Stable phenotypic changes of the host T cells are essential to the long-term stability of latent HIV-1 infection. J Virol 2015;89:6656–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Yang HC, Xing S, Shan L, et al. : Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest 2009;119:3473–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Saleh S, Solomon A, Wightman F, Xhilaga M, Cameron PU, Lewin SR: CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: A novel model of HIV-1 latency. Blood 2007;110:4161–4164 [DOI] [PubMed] [Google Scholar]
- 295.Marini A, Harper JM, Romerio F: An in vitro system to model the establishment and reactivation of HIV-1 latency. J Immunol 2008;181:7713–7720 [DOI] [PubMed] [Google Scholar]
- 296.Bosque A, Planelles V: Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 2009;113:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sahu GK, Lee K, Ji J, Braciale V, Baron S, Cloyd MW: A novel in vitro system to generate and study latently HIV-infected long-lived normal CD4+ T-lymphocytes. Virology 2006;355:127–137 [DOI] [PubMed] [Google Scholar]
- 298.Bosque A, Planelles V: Studies of HIV-1 latency in an ex vivo model that uses primary central memory T cells. Methods 2011;53:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Brooks DG, Kitchen SG, Kitchen CM, Scripture-Adams DD, Zack JA: Generation of HIV latency during thymopoiesis. Nat Med 2001;7:459–464 [DOI] [PubMed] [Google Scholar]
- 300.McCune JM, Namikawa R, Kaneshima H, Shultz LD, Lieberman M, Weissman IL: The SCID-hu mouse: Murine model for the analysis of human hematolymphoid differentiation and function. Science 1988;241:1632–1639 [DOI] [PubMed] [Google Scholar]
- 301.Namikawa R, Kaneshima H, Lieberman M, Weissman IL, McCune JM: Infection of the SCID-hu mouse by HIV-1. Science 1988;242:1684–1686 [DOI] [PubMed] [Google Scholar]
- 302.Herzyk DJ, Gore ER, Polsky R, et al. : Immunomodulatory effects of anti-CD4 antibody in host resistance against infections and tumors in human CD4 transgenic mice. Infect Immun 2001;69:1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Karpel ME, Boutwell CL, Allen TM: BLT humanized mice as a small animal model of HIV infection. Curr Opin Virol 2015;13:75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Deere JD, Kauffman RC, Cannavo E, et al. : Analysis of multiply spliced transcripts in lymphoid tissue reservoirs of rhesus macaques infected with RT-SHIV during HAART. PLoS One 2014;9:e87914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Igarashi T, Brown CR, Endo Y, et al. : Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A 2001;98:658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Mavigner M, Watkins B, Lawson B, et al. : Persistence of virus reservoirs in ART-treated SHIV-infected rhesus macaques after autologous hematopoietic stem cell transplant. PLoS Pathog 2014;10:e1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Dinoso JB, Rabi SA, Blankson JN, et al. : A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J Virol 2009;83:9247–9257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Pace MJ, Agosto L, Graf EH, O'Doherty U: HIV reservoirs and latency models. Virology 2011;411:344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Shen A, Zink MC, Mankowski JL, et al. : Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J Virol 2003;77:4938–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Van Rompay KK: Evaluation of antiretrovirals in animal models of HIV infection. Antiviral Res 2010;85:159–175 [DOI] [PubMed] [Google Scholar]
- 311.Charlins P, Schmitt K, Remling-Mulder L, et al. : A humanized mouse-based HIV-1 viral outgrowth assay with higher sensitivity than in vitro qVOA in detecting latently infected cells from individuals on ART with undetectable viral loads. Virology 2017;507:135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Sanyal A, Mailliard RB, Rinaldo CR, et al. : Novel assay reveals a large, inducible, replication-competent HIV-1 reservoir in resting CD4(+) T cells. Nat Med 2017;23:885–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Policicchio BB, Pandrea I, Apetrei C: Animal models for HIV cure research. Front Immunol 2016;7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Alexaki A, Wigdahl B: HIV-1 infection of bone marrow hematopoietic progenitor cells and their role in trafficking and viral dissemination. PLoS Pathog 2008;4:e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Carter CC, Onafuwa-Nuga A, McNamara LA, et al. : HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat Med 2010;16:446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]