Abstract
We use all-atom molecular simulation with explicit solvent to study the properties of selected intrinsically disordered proteins and unfolded states of foldable proteins, which include chain dimensions and shape, secondary structure propensity, solvent accessible surface area and contact formation. We find that the qualitative scaling behavior of the chains matches expectations from theory under ambient conditions. In particular, unfolded globular proteins tend to be more collapsed under the same conditions than charged disordered sequences of the same length. However, inclusion of explicit solvent in addition naturally captures temperature-dependent solvation effects, which result in an initial collapse of the chains as temperature is increased, in qualitative agreement with experiment. There is a universal origin to the collapse, revealed in the change of hydration of individual residues as a function of temperature: namely that the initial collapse is driven by unfavorable solvation free energy of individual residues, which in turn has a strong temperature dependence. We also observe that in unfolded globular proteins, increased temperature also initially favors formation of native-like (rather than non-native-like) structure. Our results help to establish how sequence encodes the degree of intrinsic disorder or order as well as its response to changes in environmental conditions.
Graphical TOC Entry
Introduction
A large class of so-called intrinsically disordered proteins (IDPs) do not fold to a single three-dimensional structure under physiological conditions. Yet, these proteins of low sequence complexity carry out important biological functions, including some traditionally associated with well-folded proteins.1 Many IDPs and intrinsically disordered regions (IDRs) of large proteins are associated with human diseases, often due to their role as signalling molecules.2–8 The unfolded states of globular proteins, while also disordered, have significantly different properties from intrinsically disordered proteins. They have been studied largely to improve our understanding of protein folding, 9–26 and because of the potential relevance of unfolded species to protein aggregation or misfolding.27–29
At the most coarse-grained level, unfolded and disordered proteins can be characterized in terms of the chain size, shape and polymer scaling, which may all influence their function, for example their binding30 or aggregation8 characteristics, or properties for tethering other proteins.31
Previous work on disordered proteins has revealed that the degree of compactness of the chain depends on its sequence composition via the hydrophobicity and number and distribution of charged residues. 24,25,32–34 Intrinsically disordered proteins are in general low in hydrophobic amino acids and have a larger mean net charge per residue than globular proteins. Moreover, the fraction of positively and negatively charged amino acids and distribution of the charged amino acids over the sequence have been shown to have a direct effect on the polymeric properties and chain dimensions of IDPs and IDRs via polymer scaling laws.34,35 Therefore, sequence composition is a major contribution to the properties of unfolded and disordered proteins. Of course, environment effects such as ionic strength and temperature also play a role. In contrast to the expected behavior for a polymer chain with a temperature-independent monomer-monomer interaction energy, which will expand as temperature increases, several studies have shown that unfolded and disordered proteins become more compact with increasing temperature.13,22,36 Our previous work has shown that the temperature-dependent solvation free energies of the constituent amino acids play a key role in this temperature-dependent collapse.37 Thus, both sequence composition and temperature are important determinants of many properties of unfolded and disordered proteins. Beyond the coarse properties of the polypeptide chain, it is clear that most unfolded and disordered proteins are far from being ideal random coils,38–40 with the existence of residual or nascent structure being a prominent aspect of unfolded and disordered protein research.41–52 Other properties of interest for unfolded and disordered protein states include the degree of long-range contact formation51,53 and solvent accessibility as a function of protein sequence and temperature. 54–57
Atomistic simulations serve as an invaluable tool for the investigation of unfolded and disordered proteins, because the unfolded and disordered proteins exist as ensembles of diverse conformations rather than having a narrowly defined native state. Using replica-exchange molecular dynamics simulations with an explicit solvent and all-atom model, which is especially designed for better representation of unfolded and disordered proteins, 58 we generate equilibrium ensembles of selected IDPs and unfolded states of globular proteins in aqueous solution to study the sequence and temperature dependence of their properties: chain dimensions and polymer scalings, secondary structure and solvent accessibility. We find that the qualitative scaling behavior of the chains matches with the expectations from theory34,59 under ambient conditions. That is, the more disordered sequences tend to have a scaling approaching that expected for a good solvent or for a theta-solvent, while the unfolded states of proteins with a native structure are closer to collapsed globules. In addition, inclusion of explicit solvent naturally captures the temperature dependent collapse without the need for temperature-dependent force field parameters.37 Remarkably, we find that as a result of the chain collapse with temperature, some of the proteins switch from the good solvent limit of polymer scaling towards a more compact theta solvent limit as temperature increases. Largely independent of the specific sequence in which they are located, the solvent accessible surface areas of different classes of amino acids exhibit common trends as a function of temperature. Solvent accessible surface areas (SASA) of hydrophobic amino acids share a U-shaped trend with a minimum in SASA at intermediate temperatures. On the other hand, negatively charged amino acids exhibit a monotonic increase in burial as temperature increases. These observations explain the observed trends in collapse behavior of the various proteins studied here. Taken together, our results help to rationalize at an atomistic level how the sequence encodes the intrinsic disorder or order at ambient temperatures, as well as its temperature dependence.
Results and Discussion
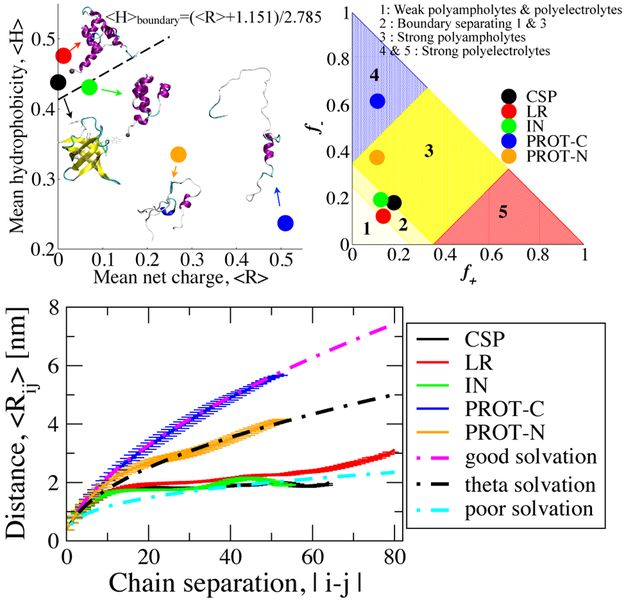
To investigate properties of unfolded and disordered proteins, we employ five sequences, namely, the cold shock protein (CSP) from Thermotoga maritima, the DNA-binding domain of λ-repressor (LR), the N-terminal domain of HIV integrase (IN), C- (PROT-C) and N-(PROT-N) terminal segments of human prothymosin-α (PROTα), which have all been experimentally characterized using single-molecule Förster resonance energy transfer (FRET). 22,24,60 CSP and LR are globular proteins, forming all-β and all-α folds respectively in their folded state while IN is an IDP which folds only in the presence of Zn2+ ions and has an all-α fold. PROT-C and PROT-N are the segments of a highly charged protein which are disordered under any condition. The proteins studied here cover a large range of charge and hydropathy encoded by their sequences which are given in supporting information (SI) Table S1. To summarize this variance and compare the sequence characteristics of the proteins, we locate all of the sequence on an Uversky diagram32 (Figure 1, top-left), and a Das and Pappu diagram34 (Figure 1, top-right). The Uversky diagram characterizes proteins as globular or intrinsically disordered based on their hydropathy and mean net charge. Hydropathy indices in this diagram are the average Kyte-Dolittle61 scores with a window size of 5-residues and scaled to fit between 0 and 1 (Figure 1, top-left). Mean net charge is ∣ f+ – f− ∣ where f+ and f− are the fraction of positively and negatively charged residues, respectively. While the proteins falling above the dashed line, which have low net charge and high hydrophobicity, are classified as globular proteins, the ones below that line are generally classified as IDPs. The Das and Pappu diagram34 predicts the compactness of IDPs under ambient conditions, in terms of intra-chain distance dependence on the chain separation, in aqueous solution based on their fractions of positively and negatively charged residues, f+ and f−.
Figure 1:
Top left: Uversky diagram32 of IDPs and globular proteins, Top right: Das and Pappu diagram of IDPs;34 symbols indicate peptides studied in this work. Bottom: Ensemble averaged intra-chain distance (Rij) profiles of the polypeptides with respect to chain separation. Theoretical polymer scaling limits are shown with dashed lines.
As conformational sampling of all but the smallest proteins (even in the disordered states) is limited if using brute force molecular dynamics simulations, we use a well-established enhanced sampling technique, replica exchange molecular dynamics, 62 to obtain their equilibrium properties. In this technique, multiple simulations are performed in parallel at different temperatures (ranging from the temperature of interest to a very high temperature) which are weakly coupled to each other by exchange attempts at periodic intervals. As the simulations at higher temperatures can overcome energy barriers easier than at the lower temperatures, this approach provides a convenient way to obtain converged equilibrium data at low temperatures. The data from these simulations is used in the following sections to provide insights into various questions related to the sequence- and temperature-dependent properties of disordered proteins. As an evidence of convergence, distributions of radius of gyration for each protein for eleven selected temperatures are provided in SI (Figures S1-S5) for two equal non-overlapping blocks of the production data. Also, statistical uncertainties are provided as error bars in radius of gyration, secondary structure and solvent accessible surface area, estimated as blocked standard errors by dividing each data set into a number of equal non-overlapping blocks. The number of blocks used are given in the caption of each figure.
Chain dimensions
Polymer scaling laws characterize the conformational preferences of polymers such as swollen coils or compact globules, which are governed by the balance of protein-solvent, protein-protein, and solvent-solvent interactions. The scaling exponent characterizing the conformational state can be determined from ensemble-averaged internal distance, ⟨Rij⟩, with respect to chain separation, ∣ i – j ∣, with the equation in the form of ⟨Rij⟩ = A ∣ i – j ∣ν. We globally use a prefactor of A = 0.55 nm to plot all limits with different exponents, ν. Here, Rij is averaged at every chain separation within the sequence and for every conformation found in the equilibrium ensemble. Figure 1 (bottom) shows ⟨Rij⟩ for each protein at 300 K temperature. We find that at this temperature, PROT-C (strong polyelectrolyte), conforms closely to the good solvent scaling limit where ⟨Rij⟩ is predicted to be proportional to ∣ i – j ∣0.6. The chain explores an expanded set of conformations with effectively repulsive interactions between the monomers.63 On the other extreme, in a poor solvent, the chain samples compact conformations for which we plot a reference limit ⟨Rij⟩ α∣ i – j ∣0.33. The globular protein sequences CSP, LR and IN are found to approximately follow this regime and the interactions between monomers are effectively attractive. At 300 K, PROT-N (strong polyampholyte) fits best to the intermediate theta solvent limit, for which ⟨Rij⟩ α∣ i – j ∣0.5 and monomer interactions are balanced, i.e., neither repulsive nor attractive.63 Here, the trends in calculated polymer scalings of all proteins match the expectations from recent literature, based on their hydropathy and electrostatic characteristics (Figure 1B).34,35,64 Experimental results on the scaling properties of unfolded and disordered proteins have yielded a range of scaling behaviors, largely as a result of the different sequences and experimental conditions considered in each case. The radii of gyration of unfolded proteins in high denaturant concentration (4 M guanidinium chloride), measured by small angle X-ray scattering experiments (SAXS) conform closely to a power law dependence on chain length with an exponent of 0.598,14 indicating good solvent. Förster resonance energy transfer also indicates a similar scaling for a number of unfolded and intrinsically disordered proteins in high denaturant concentration.25 However, at lower denaturant concentrations, the unfolded states of several proteins and some IDPs are found to become more collapsed, and consequently the scaling exponent moves closer to the value for an ideal chain (theta solvent), ~ 1/2.25 A similar result was obtained from another study in which hydrodynamic radius measurements of a number of IDPs from two different experimental methods are compiled: the scaling relationship of hydrodynamic radius of the IDPs yielded an exponent of 0.509.65 We note that there is some controversy regarding the collapse of unfolded chains in low denaturant concentration, because it has been argued that this collapse is not observed in SAXS experiments.66 However, addressing the origin of this discrepancy is beyond the scope of the present work, and we restrict ourselves to comparisons with the available data. Our finding that more hydrophobic sequences exhibit smaller power law exponents at low denaturant concentration is qualitatively consistent with the results from single-molecule FRET,25 although the fitted exponents in that case were always slightly greater than 1/3. Part of the reason for this may be that protein force fields are overall slightly too hydrophobic so that our radii of gyration are always slightly smaller than in experiment. This deficiency has only very recently been addressed67,68 using two different approaches. We note that the future work should evaluate the impact of these protein force field changes to the results presented here.
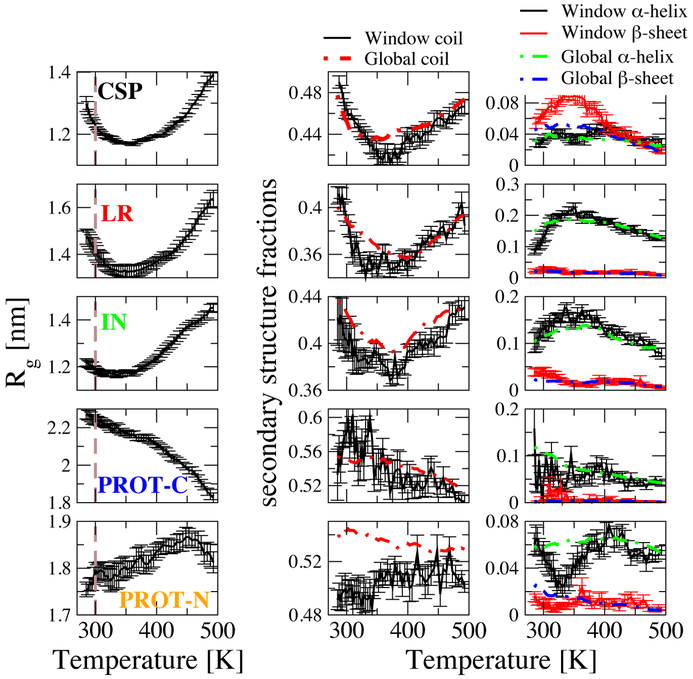
We further investigate the chain dimensions by calculating the average radius of gyration, ⟨Rg⟩, as a function of temperature (Figure 2A). At 300 K, we obtain ⟨Rg⟩ (Figure 2A, dashed lines) values for the different proteins which are qualitatively consistent with their scaling. Moreover, the temperature dependence of ⟨Rg⟩ clearly reproduces the experimentally observed temperature induced collapse for all globular protein sequences and for PROT-C. The globular proteins (CSP, LR and IN) share a very similar trend of collapse followed by re-expansion with slightly different temperatures of maximum compaction. Similar to globular proteins PROT-C also shows an initial collapse, but as opposed to the globular proteins, it continues to collapse as temperature is increased further. On the other hand, PROT-N shows a slight expansion followed by a collapse only at very high temperatures.
Figure 2:
Radius of gyration (Rg) of peptides (left) and secondary structures defined by dssp algorithm,69 Middle column: Coil fraction assigned by dssp, Right column: α -helix and β-sheet fractions. Dashed thick lines: Total data averages, solid lines with error bars: narrow Rg based-window averages, Rg-windows: 1.2-1.3 nm for CSP and IN, 1.3-1.4 nm for LR, 2.0-2.1 nm for PROT-C, 1.6-1.7 nm for PROT-N. Rg interval represented here is selected differently for different proteins to consider commonly sampled Rg in all temperatures for different proteins. Error bars are blocked standard errors using ten equal non-overlapping blocks.
In a recent single molecule FRET study, all five of these proteins were shown to collapse initially as temperature was increased, in qualitative agreement with our results.37 Due to the limited temperature range in experiment, expansion at even higher temperatures was only observed for λ-repressor. The strongest collapse was observed in experiment for the most hydrophilic (with high fraction of charged residues) chains (PROT-C and PROT-N).37 In our simulations we also observe a strong collapse in PROT-C, but very little temperature dependence of Rg for PROT-N. The mismatch in PROT-N may arise partly because we simulate a truncated N-terminal segment of PROTα, while in the study of Wuttke et al., the N-terminal segment is labelled in the context of the complete PROTα sequence. Calculated Rg values are tabulated in SI (Table S2) together with native Rg values (for globular proteins), experimental (FRET)37 Rg values and fully denatured Rg values as a reference for good solvent conditions.14 The denatured Rg is the radius of gyration predicted from the length of the peptide, following the empirical relationship described by Kohn et al.14 for proteins in high concentrations of chemical denaturant. Quantitatively, although we capture the approximate rank order of the dimensions of these proteins, the Rg of each protein was smaller than that measured in the FRET experiments. This discrepancy is most likely due to the tendency of current force fields to favor structures which are too collapsed, as mentioned earlier.67,68 We note that while the the simulated Rg values for PROT-C and PROT-N appear to be in good agreement with the Kohn predictions, this is probably due to the fortuitous cancellation of two effects: the fact that these charged sequences tend to adopt even more expanded states than typical proteins in denaturant,25 and the compensating effect of the force field favoring structures which are too collapsed.
Additionally, the Rg distributions for eleven selected temperatures with 20 K temperature difference between adjacent temperatures, are also provided in SI (Figures S6-S10). All proteins show a smooth overall shift in their Rg distributions as the overall Rg changes, suggesting that the simulations are well converged.
Secondary structures
To investigate the formation of secondary structure, we calculate the secondary structures populations for each sequence (based on the DSSP criteria), as a function of temperature. For all proteins, the average fraction of residues in coil conformation is given by dashed lines in Figure 2B-left column whereas α-helix and β-sheet fractions are shown by dashed lines in Figure 2B-right column. For all three globular proteins, the fraction of residues in coil conformation decreases with increasing temperature up to the turnover temperature, then it increases again. Populations of secondary structure of the type found in the native state (β-sheet for CSP and α-helix for LR and IN) show the opposite, increasing with temperature to a maximum and then decreasing. The limited degree of native-like secondary structure formation in unfolded CSP is consistent with the ~ 20% β-structure observed for CSP by Hoffmann et al.19 Notably, non-native secondary structures (α-helix for CSP and β-sheet for LR and IN) do not show this trend, in general decreasing with temperature. For PROT-C and PROT-N, which have no native structure, there is essentially no secondary structure formed as expected. Overall, we observe for all globular proteins that increasing temperature initially favors the formation of the type of secondary structure found in the native state in parallel with protein collapse. An obvious question which arises is whether the initial increase in structure formation is an intrinsic effect due to the change in temperature, or instead a secondary effect, driven by protein collapse. This question received conflicting answers in the literature. Work by Yang et al.70 indicated β-like structures form prior to aggregation. Similarly, the study of Kimura et al.71 suggested that collapse can be initiated by β-sheet structure. However, Doniach et al72 found that collapse should not be driven by secondary structure according to their models. Moreover, the work by Sadqi et al.13 suggested that when time scales are compared, chain collapse appears faster than the formation of secondary structure. This rapid chain dynamics is supported by the ~ 50 ns reconfiguration times determined for unfolded proteins by Nettels et al.73 To address this issue, we consider, in addition to the global secondary structure average (dashed lines) discussed above, an average over configurations lying within a 0.1 nm thick slice of Rg (solid lines). We see an overall similar trend for both, suggesting that increased formation of the type of secondary structure found in the native state is not merely a consequence of collapse, but appears to be driven by similar temperature-dependent effects. We also calculate intramolecular protein and scaled intermolecular protein-water hydrogen bond formation with respect to temperature (Figure S11), which shows that more compact chains form more intramolecular hydrogen bonds. To account for the loss in water’s hydrogen bond propensity with increasing temperature, we normalize protein-water hydrogen bonds with water-water hydrogen bonds at a given temperature. Again, there is clearly a non-monotonic trend (Figure S11) with the number of protein-water hydrogen bonds decreasing first as a function of temperature (up to the collapse temperature) and then increasing again for all the globular proteins. This clearly highlights the critical role of temperature-dependent water-mediated interactions in the observed behavior.
In addition to the temperature-dependence of secondary structures, we also calculate the per-residue secondary structure fractions at room temperature which are shown in SI along with their native structures for globular proteins (Figure S12-S16). All the unfolded populations of globular proteins sample a significant amount of secondary structure of the same type found in the native state, supporting the idea that unfolded globular proteins tend to form native-like local secondary structures in aqueous solution. Moreover, we calculate residue-residue contact propensities of these peptides at room temperature (Figure S17). For globular proteins we find that the structure formed in the unfolded state essentially reflects native-like contacts with small sequence separation.74
Solvent accessibility
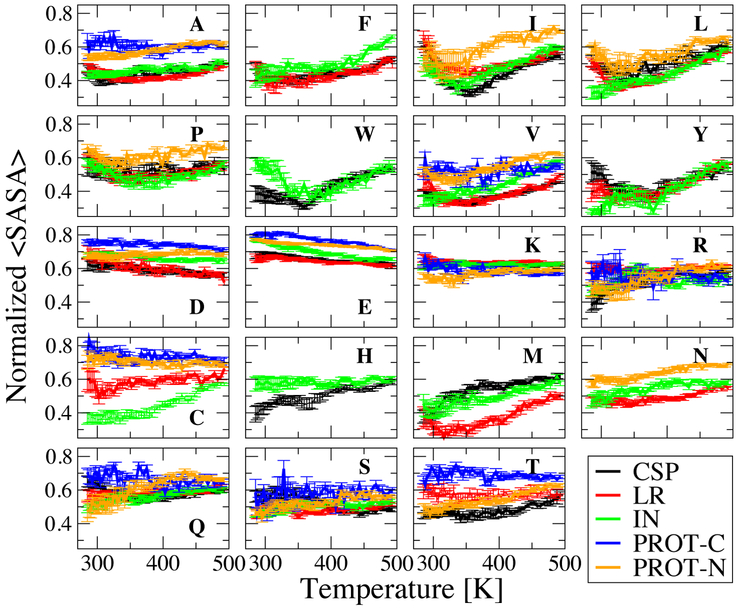
What are the factors influencing the change of protein dimensions with temperature? To gain some insight into this question, we have calculated the solvent-accessible surface area (SASA) for each type amino acid side-chain, in each sequence and determined an ensemble-averaged SASA, ⟨SASA⟩. To account for the different sizes of the side-chains and their abundance in the sequence, we normalize the ⟨SASA⟩ based on the fully exposed surface area of the side-chain of each residue X in Gly-X-Gly tripeptides,75 and by the total number of the particular amino acid in the sequence. The normalized ⟨SASA⟩ for each amino acid is shown in Figure 3 for a narrow window of Rg (0.1 nm). Using a narrow Rg window again ensures that samples at each temperature are of similar compactness so that ⟨SASA⟩ trends are not biased by overall compactness level, but reflect true differences between amino acids. The normalized ⟨SASA⟩ is lower than or about 0.5 on average for all hydrophobic amino acids (Ala, Phe, Ile, Leu, Pro, Trp, Val, Tyr) in globular sequences in general. In IDP sequences not only hydrophobic amino acids but all in general are more exposed naturally as an effect of solvation characteristics of the full length chain. Amino acids Cys, His, Met, Asn, Ser, Thr have normalized ⟨SASA⟩ of around 0.5 for globular proteins whereas all charged amino acids (Asp, Glu, Lys, Arg) and Gln have that of greater than 0.5 on average as consistent with their hydrophobicity based on Kyte and Doolittle scale.61 A scatter plot is given in SI (Figure S18) to show the observed correlation between ⟨SASA⟩ at 300 K and the Kyte and Doolittle hydrophobicity scale.61
Figure 3:
Rg-window averages normalized solvent accessible surface area (⟨SASA⟩) of each amino acid in each peptide. Rg windows are the same as in Figure 2 (secondary structures). Error bars are blocked standard errors using four equal non-overlapping blocks.
The temperature dependence of ⟨SASA⟩ shows that some amino acid groups share common features within the group, across all sequences. Hydrophobic amino acids (Figure 3-top two row) tend to have a U-shaped trend common in all five sequences. This trend is consistent with the well-known minimum in solvation free energy for idealized hydrophobic solutes, which occurs at a similar temperature. 76 On the other hand, for most sequences the negatively charged amino acids, Asp and Glu have an overall monotonic increase in their burial with respect to temperature. This effect can be understood as arising from the less favorable solvation free energy of charged groups at higher temperature, due to their large negative entropy of solvation. The situation for the cationic amino acids is different: Arg shows a tendency to get more exposed to water on average as temperature increases, whereas the ⟨SASA⟩ of Lys does not seem to be much affected by temperature This more complex behaviour probably arises from the fact that both Arg and Lys have a significant hydrophobic portion of their side-chains in addition to the charged group, which may lead to competing effects at high temperature. Other hydrophilic amino acids His, Asn, Gln, Ser and Thr do not have a clear trend in common, either. Since high-hydrophobicity amino acids are more frequent in the primary structure of globular proteins in general, and are more likely to be in contact, this helps to explain the collapse followed by re-expansion for globular sequences. Since more than 50% of constituent amino acids of PROT-C are negatively charged amino acids, the abundance of these residues in PROT-C may explain its monotonic collapse. On the other hand, the sequence of PROT-N is more balanced in hydrophobic, charged and other hydrophilic amino acids, which may explain its unclear trend in ⟨Rg⟩.
As a complement to this picture, we have also computed hydrophobic-hydrophobic, hydrophilic-hydrophilic, hydrophobic-hydrophilic (cross) and total contact formations for both backbone and side-chains of all proteins (Figure S19 and S20), Hydrophobic contacts clearly support observations from ⟨SASA⟩, as they are increasing at collapse temperatures then decreasing. This is not only true for globular sequences but also true for highly charged sequnces PROT-N and PROT-C. The trends are less clear for hydrophilic contacts, largely because all non-hydrophobic amino acids have been lumped with the hydrophilic ones.
Temperature dependence of polymer scaling
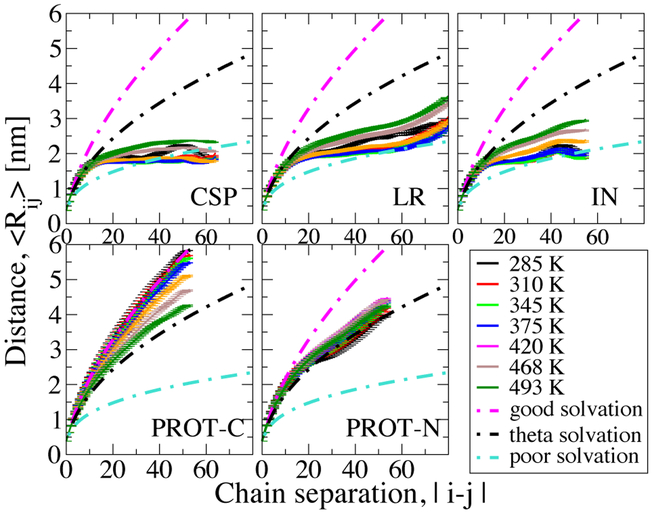
We have further investigated the polymer scaling with respect to temperature, as shown in Figure 4. The scaling did not show a clear temperature dependence in the only previous theoretical work – to the best of our knowledge – in which this was studied.34 Here we find that globular proteins initially move to a slightly more compact regime, then they move back to more expanded regimes with a further increase in temperature, consistent with their temperature dependent collapse trend. Overall, however, they remain firmly within the poor solvent limit. On the other hand, PROT-C continuously moves to more compact regimes such that it switches from good solvent limit to theta solvent limit, as can be anticipated from its strong collapse. The change in PROT-N is not as marked as other sequences. Scaling indications can further be supported with ensemble averaged asphericities which are given in Figure S10. As expected for the globular limit, the asphericity parameter, ⟨φ⟩, of all globular proteins indicate sphere-like shapes whereas PROT-N and PROT-C have a more prolate shape, on average. As temperature increases, globular proteins become even more sphere-like (decreasing ⟨φ⟩) whereas PROT-N hardly shows a change.
Figure 4:
Intra-chain distance (Rij) profiles of the peptides with respect to chain separation at different temperatures.
Characterization of unfolded ensembles at different temperatures with same average ⟨Rg⟩
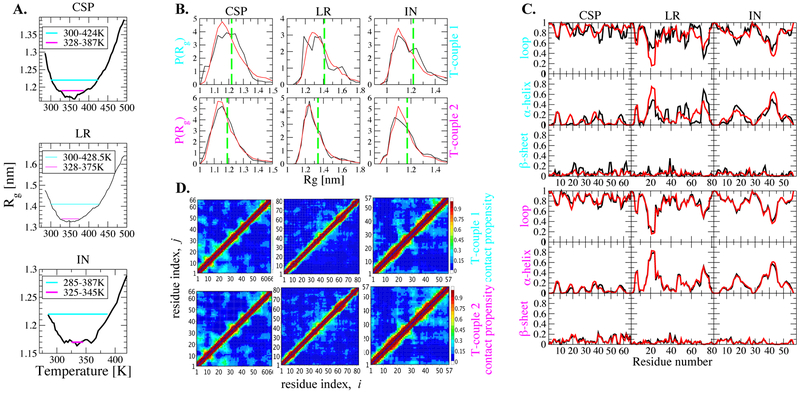
Lastly, we investigate how similar are the sampled conformations at pairs of temperatures at which the protein has the same Rg. This question is limited to globular proteins, which show an initial collapse followed by re-expansion. To answer this question at a residue level, we select two temperature couples from Rg curves of these proteins such that each member of a couple has the same average Rg (Figure 5A). We employ a significant difference in temperature of 100-125 K in T-couple 1 and a smaller one of 20-60 K in T-couple 2. Distributions of Rg at selected temperatures are shown in Figure 5B. They, in fact, show only little difference for both couples. For T-couple 1, the difference looks more pronounced; distributions are smoother for the higher temperature (red curves). However, this diffence is most likely an effect of limited sampling. Because of the much faster kinetics at high temperature, smoother distributions are obtained. At a residue level, secondary structures do not indicate a significant difference between low temperature and high temperature ensembles either (Figure 5C). Additionally, backbone contacts show that both ensembles are almost equivalent both in the context of short- and long-range contacts (Figure 5D).
Figure 5:
Comparison of properties at different temperatures at same Rg. A. Selected temperature couples are indicated on Rg change plot, B. Rg distribution of couples, black: low-T, red: high-T, green: average Rg C. Secondary structure propensities per residue black: low-T, red: high T, D. Backbone contacts of couples, upper triangle: low-T, lower triangle: high-T.
Conclusions
We have employed long replica exchange molecular dynamics simulations at constant pressure with an all-atom protein force field and explicit solvent to obtain equilibrium ensembles for selected IDPs and unfolded states of globular proteins with variety of different primary structures in aqueous solution. The inclusion of explicit solvent naturally captures the temperature dependence of hydrophobic interactions that are expected to govern the observed behavior. We find that the difference in polymeric properties of unfolded and disordered proteins arises from the differences in their sequence composition, which already encodes their intrinsic disorder or order. Specifically, we observe that the degree of burial of hydrophobic and negatively charged amino acids exhibits temperature dependent characteristics (presumably due to variations in solvation free energy) which are consistent with the continuous collapse of PROT-C, and with the collapse followed by re-expansion of unfolded states of globular proteins, CSP, LR and IN. Thus our results explain how the temperature dependent properties of each chain are modulated by those of its constituent amino acids. A;though the dimensions of the proteins in the simulations may quantitatively be too collapsed compared to the experimental values, our simulations are able to successfully capture size relevant features of these proteins qualitatively, including temperature dependent collapse and the relative compactness of these proteins with respect to each other as shown by radius of gyration and polymer scaling based on internal distances.
One particularly important observation is that while collapse can be expected for the proteins which are mostly dominated by hydrophobic amino acids because of the temperature dependence of the hydrophobic effect,76 the largest amplitude of collapse was found for the most hydrophilic negatively charged case, as also determined experimentally. 37 Thus those IDPs which have a large fraction of negatively charged amino acids may in general be expected to exhibit a sharp collapse with increasing temperature. Additionally, the temperature dependence of the intrachain distance scaling analysis reveals that solvation characteristics can also change with temperature such that scaling regime can switch from the limit of good solvent to effective theta solvent conditions. We also find evidence for residual structure in all cases, which are mostly dominated by secondary structures populated in the native state for the globular proteins as well as long-range contacts. Lastly, for globular proteins which show collapse followed by re-expansion, we find that equilibrium ensemble at high temperature appears to be quite similar to the low temperature ensemble at the same average Rg.
Our work has principally focused on the gross properties of unfolded and disordered chains in solution. In future work, it will be very interesting to perform a more detailed examination of structure formation in disordered polypeptides. In this regard, the recent availability of high-resolution experimental studies of IDPs by NMR spectroscopy, providing information on local and cooperative structure formation 48,77,78 as well as long-range interactions 51,53 will be an important benchmark. In addition, while we have focused on variation of temperature as an environmental parameter, an equally physiologically important parameter is the salt concentration, whose effect would also bear investigation in future work.
Methods
We use the Amber ff03w force field58 for the protein model and TIP4P/2005 as the water model79 and perform constant pressure simulations using Gromacs 4.5.3.80 Amber03w has been shown to provide an accurate representation of disordered and unfolded proteins,7,81,82 which is also verified by calculated chemical shift deviations from the simulations using this force field is compared with experimentally obtained ones.83 For an enhanced sampling of phase space we perform replica exchange molecular dynamics (REMD)62 simulations by using 56 different parallel temperatures between 285 K to 493 K. Further details of the simulation conditions and analysis tools are given in supporting information.
Supplementary Material
Acknowledgments
We thank Prof. Ben Schuler for helpful discussions. JM acknowledges support from the U.S. Department of Energy, Office of Basic Energy Science, Division of Material Sciences and Engineering under Award (DE-SC0013979). RB is supported by the Intramural Research Program of the National Insitute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health. Use of the high-performance computing capabilities of the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation (NSF) grant no. TG-MCB-120014, is gratefully acknowledged. This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov)
Footnotes
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- (1).Wright PE; Dyson HJ Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol 1999, 293, 321–331. [DOI] [PubMed] [Google Scholar]
- (2).Eliezer D; Kutluay E; Bussell R; Browne G Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol 2001, 307, 1061–1073. [DOI] [PubMed] [Google Scholar]
- (3).Linding R; Schymkowitz J; Rousseau F; Diella F; Serrano L A comparative study of the relationship between protein structure and β-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol 2004, 342, 345–353. [DOI] [PubMed] [Google Scholar]
- (4).Dedmon MM; Lindorff-Larsen K; Christodoulou J; Vendruscolo M; Dobson CM Mapping long-range interactions in α-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc 2005, 127, 476–477. [DOI] [PubMed] [Google Scholar]
- (5).Von Bergen M; Barghorn S; Biernat J; Mandelkow E-M; Mandelkow E Tau aggregation is driven by a transition from random coil to beta sheet structure. Biochim. Biophys. Acta, Mol. Basis Dis 2005, 1739, 158–166. [DOI] [PubMed] [Google Scholar]
- (6).Halfmann R; Alberti S; Krishnan R; Lyle N; O’Donnell CW; King OD; Berger B; Pappu RV; Lindquist S Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol. Cell 2011, 43, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Miller C; Zerze GH; Mittal J Molecular simulations indicate marked differences in the structure of amylin mutants, correlated with known aggregation propensity. J. Phys. Chem. B 2013, 117, 16066–16075. [DOI] [PubMed] [Google Scholar]
- (8).Crick SL; Ruff KM; Garai K; Frieden C; Pappu RV Unmasking the roles of N-and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc. Nat. Acad. Sci. U.S.A 2013, 110, 20075–20080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hagen SJ; Hofrichter J; Szabo A; Eaton WA Diffusion-limited contact formation in unfolded cytochrome c: estimating the maximum rate of protein folding. Proc. Nat. Acad. Sci. U.S.A 1996, 93, 11615–11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schuler B; Lipman EA; Eaton WA Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature 2002, 419, 743–747. [DOI] [PubMed] [Google Scholar]
- (11).Uzawa T; Akiyama S; Kimura T; Takahashi S; Ishimori K; Morishima I; Fujisawa T Collapse and search dynamics of apomyoglobin folding revealed by submillisecond observations of α-helical content and compactness. Proc. Nat. Acad. Sci. U.S.A 2004, 101, 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Krieger F; Fierz B; Bieri O; Drewello M; Kiefhaber T Dynamics of unfolded polypeptide chains as model for the earliest steps in protein folding. J. Mol. Biol 2003, 332, 265–274. [DOI] [PubMed] [Google Scholar]
- (13).Sadqi M; Lapidus LJ; Muñoz V How fast is protein hydrophobic collapse? Proc. Nat. Acad. Sci. U.S.A 2003, 100, 12117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kohn JE; Millett IS; Jacob J; Zagrovic B; Dillon TM; Cingel N; Dothager RS; Seifert S; Thiyagarajan P; Sosnick TR; et al. , Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Nat. Acad. Sci. U.S.A 2004, 101, 12491–12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Möglich A; Joder K; Kiefhaber T End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc. Nat. Acad. Sci. U.S.A 2006, 103, 12394–12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sherman E; Haran G Coil–globule transition in the denatured state of a small protein. Proc. Nat. Acad. Sci. U.S.A 2006, 103, 11539–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Merchant KA; Best RB; Louis JM; Gopich IV; Eaton WA Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc. Nat. Acad. Sci. U.S.A 2007, 104, 1528–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Jacob J; Dothager RS; Thiyagarajan P; Sosnick TR Fully reduced ribonuclease A does not expand at high denaturant concentration or temperature. J. Mol. Biol 2007, 367, 609–615. [DOI] [PubMed] [Google Scholar]
- (19).Hoffmann A; Kane A; Nettels D; Hertzog DE; Baumgärtel P; Lengefeld J; Reichardt G; Horsley DA; Seckler R; Bakajin O; et al. , Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy. Proc. Nat. Acad. Sci. U.S.A 2007, 104, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Arai M; Kondrashkina E; Kayatekin C; Matthews CR; Iwakura M; Bilsel O Microsecond hydrophobic collapse in the folding of Escherichia coli dihydrofolate reductase, an α/β-type protein. J. Mol. Biol 2007, 368, 219–229. [DOI] [PubMed] [Google Scholar]
- (21).Wang Y; Trewhella J; Goldenberg DP Small-angle x-ray scattering of reduced ribonuclease A: effects of solution conditions and comparisons with a computational model of unfolded proteins. J. Mol. Biol 2008, 377, 1576–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Nettels D; Müller-Späth S; Küster F; Hofmann H; Haenni D; Rüegger S; Reymond L; Hoffmann A; Kubelka J; Heinz B; Gast K; Best RB; Schuler B Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins. Proc. Nat. Acad. Sci. U.S.A 2009, 106, 20740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ziv G; Haran G Protein folding, protein collapse, and tanford’s transfer model: lessons from single-molecule FRET. J. Am. Chem. Soc 2009, 131, 2942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Müller-Späth S; Soranno A; Hirschfeld V; Hofmann H; Rüegger S; Reymond L; Nettels D; Schuler B Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Nat. Acad. Sci. U.S.A 2010, 107, 14609–14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hofmann H; Soranno A; Borgia A; Gast K; Nettels D; Schuler B Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Nat. Acad. Sci. U.S.A 2012, 109, 16155–16160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lindorff-Larsen K; Trbovic N; Maragakis P; Piana S; Shaw DE Structure and dynamics of an unfolded protein examined by molecular dynamics simulation. J. Am. Chem. Soc 2012, 134, 3787–3791. [DOI] [PubMed] [Google Scholar]
- (27).McParland VJ; Kad NM; Kalverda AP; Brown A; Kirwin-Jones P; Hunter MG; Sunde M; Radford SE Partially unfolded states of β2-microglobulin and amyloid formation in vitro. Biochemistry 2000, 39, 8735–8746. [DOI] [PubMed] [Google Scholar]
- (28).Dobson CM Protein folding and misfolding. Nature 2003, 426, 884–890. [DOI] [PubMed] [Google Scholar]
- (29).Zheng W; Schafer NP; Wolynes PG Free energy landscapes for initiation and branching of protein aggregation. Proc. Nat. Acad. Sci. U.S.A 2013, 110, 20515–20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kozlov AG; Weiland E; Mittal A; Waldman V; Antony E; Fazio N; Pappu RV; Lohman TM Intrinsically disordered C-terminal tails of E. coli single-stranded DNA binding protein regulate cooperative binding to single-stranded DNA. J. Mol. Biol 2015, 427, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Buske PJ; Mittal A; Pappu RV; Levin PA An intrinsically disordered linker plays a critical role in bacterial cell division. Sem. Cell Dev. Biol 2015, 37, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Uversky VN Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ashbaugh HA; Hatch H Natively unfolded protein stability as a coil-to-globule transition in charge/hydropathy space. J. Am. Chem. Soc 2008, 130, 9536–9542. [DOI] [PubMed] [Google Scholar]
- (34).Das RK; Pappu RV Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Nat. Acad. Sci. U.S.A 2013, 110, 13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mao AH; Crick SL; Vitalis A; Chicoine CL; Pappu RV Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Nat. Acad. Sci. U.S.A 2010, 107, 8183–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Langridge TD; Tarver MJ; Whitten ST Temperature effects on the hydrodynamic radius of the intrinsically disordered N-terminal region of the p53 protein. Proteins: Struct., Funct., Bioinf 2014, 82, 668–678. [DOI] [PubMed] [Google Scholar]
- (37).Wuttke R; Hofmann H; Nettels D; Borgia MB; Mittal J; Best RB; Schuler B Temperature-dependent solvation modulates the dimensions of disordered proteins. Proc. Nat. Acad. Sci. U.S.A 2014, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Baldwin RL; Zimm BH Are denatured proteins ever random coils? Proc. Nat. Acad. Sci. U.S.A 2000, 97, 12391–12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Pappu RV; Srinivasan R; Rose GD The Flory isolated-pair hypothesis is not valid for polypeptide chains: implications for protein folding. Proc. Nat. Acad. Sci. U.S.A 2000, 97, 12565–12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Plaxco KW; Gross M Unfolded, yes, but random? Never! Nat. Struct. Mol. Biol 2001, 8, 659–660. [DOI] [PubMed] [Google Scholar]
- (41).Chan HS; Dill KA Origins of structure in globular proteins. Proc. Nat. Acad. Sci. U.S.A 1990, 87, 6388–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Dobson CM Unfolded proteins, compact states and molten globules: Current Opinion in Structural Biology 1992, 2: 6–12. Curr. Opin. Struct. Biol 1992, 2, 6–12. [Google Scholar]
- (43).Wong K-B; Clarke J; Bond CJ; Neira JL; Freund SM; Fersht AR; Daggett V Towards a complete description of the structural and dynamic properties of the denatured state of barnase and the role of residual structure in folding. J. Mol. Biol 2000, 296, 1257–1282. [DOI] [PubMed] [Google Scholar]
- (44).Shortle D; Ackerman MS Persistence of native-like topology in a denatured protein in 8 M urea. Science 2001, 293, 487–489. [DOI] [PubMed] [Google Scholar]
- (45).Jane Dyson H; Ewright P Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv. Protein Chem 2002, 62, 311–340. [DOI] [PubMed] [Google Scholar]
- (46).Cho J-H; Sato S; Raleigh DP Thermodynamics and kinetics of non-native interactions in protein folding: a single point mutant significantly stabilizes the N-terminal domain of L9 by modulating non-native interactions in the denatured state. J. Mol. Biol 2004, 338, 827–837. [DOI] [PubMed] [Google Scholar]
- (47).Saxena AM; Udgaonkar JB; Krishnamoorthy G Characterization of intramolecular distances and site-specific dynamics in chemically unfolded barstar: evidence for denaturant-dependent non-random structure. J. Mol. Biol 2006, 359, 174–89. [DOI] [PubMed] [Google Scholar]
- (48).Jensen MR; Markwick PR; Meier S; Griesinger C; Zweckstetter M; Grzesiek S; Bernado P; Blackledge M Quantitative determination of the conformational properties of partially folded and intrinsically disordered proteins using NMR dipolar couplings. Structure 2009, 17, 1169–1185. [DOI] [PubMed] [Google Scholar]
- (49).Eliezer D Biophysical characterization of intrinsically disordered proteins. Curr. Opin. Struc. Biol 2009, 19, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zhang W; Ganguly D; Chen J Residual structures, conformational fluctuations, and electrostatic interactions in the synergistic folding of two intrinsically disordered proteins. PLoS Comput. Biol 2012, 8, e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Meng W; Lyle N; Luan B; Raleigh DP; Pappu RV Experiments and simulations show how long-range contacts can form in expanded unfolded proteins with negligible secondary structure. Proc. Nat. Acad. Sci. U.S.A 2013, 110, 2123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Keppel TR; Weis DD Mapping residual structure in intrinsically disordered proteins at residue resolution using millisecond hydrogen/deuterium exchange and residue averaging. J. Am. Soc. Mass Spectrom. 2014, 1–8. [DOI] [PubMed] [Google Scholar]
- (53).Salmon L; Nodet G; Ozenne V; Yin G; Jensen MR; Zweckstetter M; Blackledge M NMR characterization of long-range order in intrinsically disordered proteins. J. Am. Chem. Soc 2010, 132, 8407–8418. [DOI] [PubMed] [Google Scholar]
- (54).Creamer TP; Srinivasan R; Rose GD Modeling unfolded states of peptides and proteins. Biochemistry 1995, 34, 16245–16250. [DOI] [PubMed] [Google Scholar]
- (55).Creamer TP; Srinivasan R; Rose GD Modeling unfolded states of proteins and peptides. II. Backbone solvent accessibility. Biochemistry 1997, 36, 2832–2835. [DOI] [PubMed] [Google Scholar]
- (56).Bernadó P; Blackledge M; Sancho J Sequence-specific solvent accessibilities of protein residues in unfolded protein ensembles. Biophys. J 2006, 91, 4536–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Gong H; Rose GD Assessing the solvent-dependent surface area of unfolded proteins using an ensemble model. Proc. Nat. Acad. Sci. U.S.A 2008, 105, 3321–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Best RB; Mittal J Protein simulations with an optimized water model: cooperative helix formation and temperature-induced unfolded state collapse. J. Phys. Chem. B 2010, 114, 14916–14923. [DOI] [PubMed] [Google Scholar]
- (59).Rubinstein M; Colby RH Polymer physics; OUP Oxford, 2003. [Google Scholar]
- (60).Nettels D; Hoffmann A; Schuler B Unfolded protein and peptide dynamics investigated with single-molecule FRET and correlation spectroscopy from picoseconds to seconds. J. Phys. Chem. B 2008, 112, 6137–6146. [DOI] [PubMed] [Google Scholar]
- (61).Kyte J; Doolittle RF A simple method for displaying the hydropathic character of a protein. J. Mol. Biol 1982, 157, 105–132. [DOI] [PubMed] [Google Scholar]
- (62).Sugita Y; Okamoto Y Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett 1999, 314, 141–151. [Google Scholar]
- (63).Flory PJ The configuration of real polymer chains. J. Chem. Phys 1949, 17, 303. [Google Scholar]
- (64).Tran HT; Mao A; Pappu RV Role of Backbone- Solvent Interactions in Determining Conformational Equilibria of Intrinsically Disordered Proteins. J. Am. Chem. Soc 2008, 130, 7380–7392. [DOI] [PubMed] [Google Scholar]
- (65).Marsh JA; Forman-Kay JD Sequence determinants of compaction in intrinsically disordered proteins. Biophys. J 2010, 98, 2383–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Yoo TY; Meisburger SP; Hinshaw J; Pollack L; Haran G; Sosnick TR; Plaxco K Small-angle X-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state. J. Mol. Biol 2012, 418, 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Best RB; Zheng W; Mittal J Balanced protein-water interactions improve properties of disordered proteins and non-specific protein association. J. Chem. Theor. Comput 2014, 10, 5113–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Piana S; Donchev AG; Robustelli P; Shaw DE Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J. Phys. Chem. B 2015, 115, dx.doi.org://10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- (69).Kabsch W; Sander C Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- (70).Yang WY; Larios E; Gruebele M On the extended β-conformation propensity of polypeptides at high temperature. J. Am. Chem. Soc 2003, 125, 16220–16227. [DOI] [PubMed] [Google Scholar]
- (71).Kimura T; Uzawa T; Ishimori K; Morishima I; Takahashi S; Konno T; Akiyama S; Fujisawa T Specific collapse followed by slow hydrogen-bond formation of β-sheet in the folding of single-chain monellin. Proc. Nat. Acad. Sci. U.S.A 2005, 102, 2748–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Doniach S; Garel T; Orland H Phase diagram of a semiflexible polymer chain in a θ solvent: application to protein folding. J. Chem. Phys 1995, [Google Scholar]
- (73).Nettels D; Gopich IV; Hoffmann A; Schuler B Ultrafast dynamics of protein collapse from single-molecule photon statistics. Proc. Nat. Acad. Sci. U.S.A 2007, 104, 2655–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Zerze GH; Best RB; Mittal J Modest influence of FRET chromophores on the properties of unfolded proteins. Biophys. J 2014, 107, 1654–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Miller S; Janin J; Lesk AM; Chothia C Interior and surface of monomeric proteins. J. Mol. Biol 1987, 196, 641–656. [DOI] [PubMed] [Google Scholar]
- (76).Chandler D Interfaces and the driving force of hydrophobic assembly. Nature 2005, 437, 640–647. [DOI] [PubMed] [Google Scholar]
- (77).Ozenne V; Schneider R; Yao M; Huang J.-r.; Salmon L; Zweckstetter ,M; Jensen MR; Blackledge M Mapping the potential energy landscape of intrinsically disordered proteins at amino acid resolution. J. Am. Chem. Soc 2012, 134, 15138–15148. [DOI] [PubMed] [Google Scholar]
- (78).Iešmantavičius V; Dogan J; Jemth P; Teilum K; Kjaergaard M Helical propensity in an intrinsically disordered protein accelerates ligand binding. Angew. Chem., Int. Ed 2014, 53, 1548–1551. [DOI] [PubMed] [Google Scholar]
- (79).Abascal JL; Vega C A general purpose model for the condensed phases of water: TIP4P/2005. J. Chem. Phys 2005, 123, 234505. [DOI] [PubMed] [Google Scholar]
- (80).Hess B; Kutzner C; Van Der Spoel D; Lindahl E GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theor. Comput 2008, 4, 435–447. [DOI] [PubMed] [Google Scholar]
- (81).Krieger JM; Fusco G; Lewitzky M; Simister PC; Marchant J; Camilloni C; Feller SM; De Simone A Conformational recognition of an intrinsically disordered protein. Biophys. J 2014, 106, 1771–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Knott M; Best RB A preformed binding interface in the unbound ensemble of an intrinsically disordered protein: evidence from molecular simulations. PLoS Comput. Biol 2012, 8, e1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Mittal J; Yoo TH; Georgiou G; Truskett TM Structural ensemble of an intrinsically disordered polypeptide. J. Phys. Chem. B 2012, 117, 118–124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.