Abstract
Molecular targeted therapy heralded a new era for the treatment of patients with oncogene-driven advanced-stage non–small-cell lung cancer (NSCLC). Molecular testing at the time of diagnosis guides therapy selection, and targeted therapies in patients with activating mutations in EGFR, BRAF, and rearrangements in anaplastic lymphoma kinase (ALK) and ROS1 have become part of routine care. These therapies have extended the median survival from a mere few months to greater than 3 years for patients with stage 4 disease. However, despite the initial success, these treatments are eventually met with molecular resistance. Selective pressure leads to cellular adaption to maintain cancer growth, making resistance complex and the treatment challenging. This review focuses on recent advances in targeted therapy, mechanisms of resistance, and therapeutic strategies to overcome resistance in patients with lung cancer.
Introduction
Lung cancer is the most lethal cancer globally; nearly 1.6 million deaths are reported each year. Despite a reduction in lung cancer mortality in the US in the past two decades, lung cancer incidence is increasing in the developing world due to a rise in tobacco smoking in many parts of the world. Until recently, there were no proven methods to detect lung cancer early. Hence, the vast majority of patients are diagnosed at an advanced stage of the disease, which is not amenable to curative therapies. The use of low-dose CT scan in subjects at high risk for lung cancer for early detection results in a 20% reduction in lung cancer mortality (1). This approach is yet to be adopted universally, and particularly so in resource-constrained societies. Approximately 85% of patients diagnosed with lung cancer have non–small-cell lung cancer (NSCLC), which includes the histological subtypes adenocarcinoma, squamous cell carcinoma, and large-cell carcinoma. Adenocarcinoma has emerged as the most common histological subtype diagnosed in the US (50%) followed by squamous cell histology (30%).
Palliative chemotherapy was the only available systemic therapy for patients with advanced NSCLC until the year 2003. The use of platinum-based combination regimens was beneficial in improving survival and quality of life in comparison to supportive care alone, but the median survival was a modest 8–10 months (2–4). In addition, all subtypes of NSCLC were treated alike, because of limited knowledge on the biological differences between adenocarcinoma and squamous cell carcinoma. In 2018, the treatment of lung cancer is dramatically different, particularly for patients with lung adenocarcinoma (Figure 1). Molecular testing, often by next-generation sequencing (NGS), is routinely used at the time of diagnosis to determine the best treatment approach for a given patient. A third of the patients harbor an oncogenic driver event that is druggable, another third of the patients have an inflamed tumor micro-environment that can be targeted with an immune checkpoint inhibitor, and the remainder are treated with combination chemotherapy. These developments have positioned NSCLC as a prime example for personalized cancer therapy. This article reviews the development of targeted agents for lung cancer and the clinical challenges ahead.
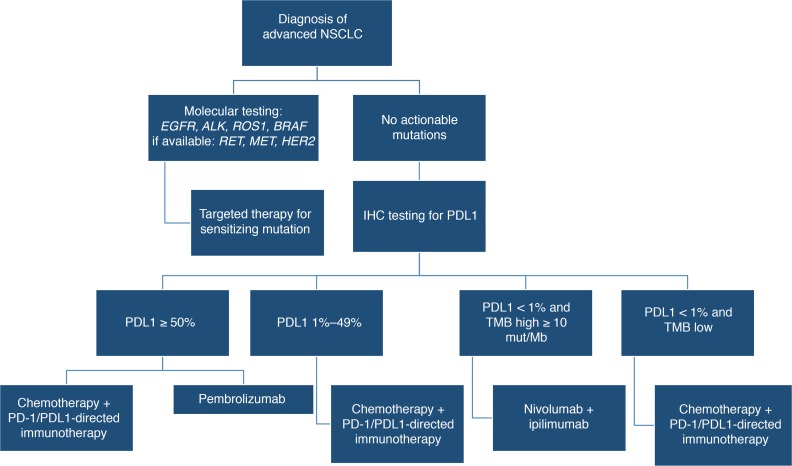
Figure 1. Treatment algorithm for stage 4 non–small-cell lung cancer.
Molecular testing is used to identify patients that are candidates for targeted therapies based on mutations. Applicable therapies for EGFR mutation include osimertinib, erlotinib, gefitinib, and afatinib; for anaplastic lymphoma kinase gene (ALK) mutation include alectinib, crizotinib, and ceritinib; second-line brigatinib; for c-ros oncogene 1 (ROS1) mutation include crizotinib and ceritinib; for v-raf murine sarcoma viral oncogene homolog B1 (BRAF) V600E include dabrafenib plus trametinib. Targeted therapy with activity against other genetic alterations have been identified for rearranged during transfection (RET) mutation (cabozantinib, vandetanib), for mesenchymal-to-epithelial transition (MET), amplification or exon 14 mutation (crizotinib) or human epidermal growth factor receptor 2 (HER2) mutation (ado-trastuzumab emtansine). Patients without actionable mutations will be tested for PDL1 expression, and treatment is determined by the overall level of expression and tumor mutational burden (TMB).
Era of targeted therapy for NSCLC
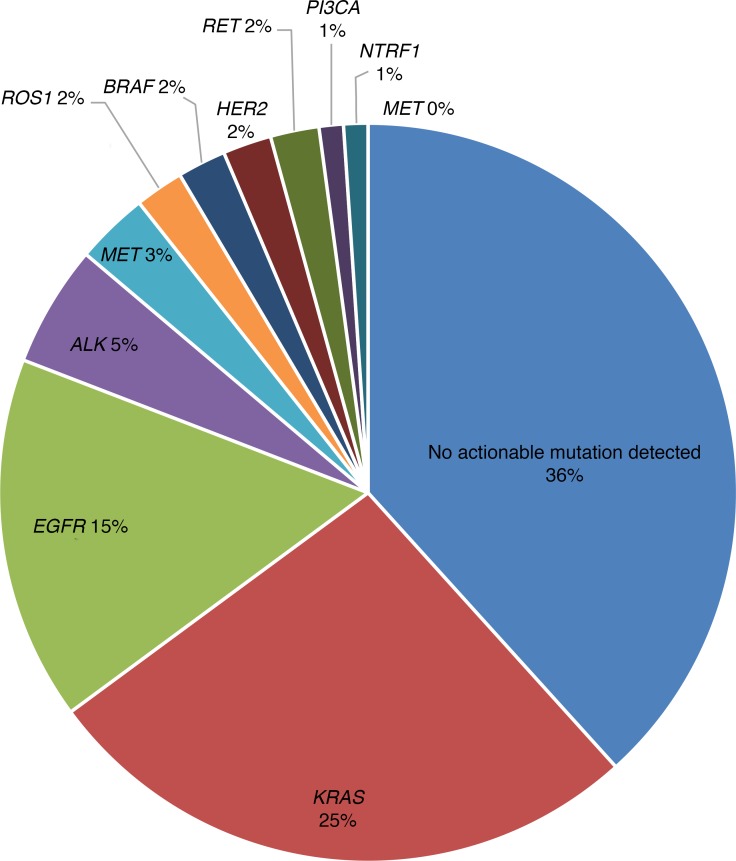
In 2003, gefitinib, an oral epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI), received FDA approval for the treatment of advanced-stage NSCLC. Gefitinib was associated with an objective tumor response rate of 10%–19% in a subset of patients, thus starting the era of personalized medicine in NSCLC. The initial clinical trial with gefitinib and other agents of its class noted that its highest efficacy was in certain subsets of mainly Asian, female, and never-smokers with adenocarcinoma histology (5, 6). Understanding the biology behind these differential responses led to the discovery of the two sensitizing EGFR mutations (exon 19 deletion [del 19] and exon 21 L858R point mutation) that were present in patients with a favorable response to EGFR inhibitor therapy. Subsequent studies noted that EGFR mutations were present in approximately 15% of patients of European descent, 35% of Asian patients, and accounted for 80%–90% of responses to EFGR TKIs (7, 8). Further molecular characterization of non–squamous cell NSCLC has revealed several oncogenic driver mutations: rearrangement of the anaplastic lymphoma kinase gene (ALK), c-ros oncogene 1 (ROS1), v-raf murine sarcoma viral oncogene homolog B1 (BRAF), rearranged during transfection (RET), human epidermal growth factor receptor 2 (HER2), and mesenchymal-to-epithelial transition (MET), which have been implicated in the development and propagation of NSCLC by signaling uncontrolled proliferation. Multicenter genomic testing in France and in the US has revealed that up to 50% of NSCLC and 64% of adenocarcinomas have a driver mutation, respectively (9, 10). KRAS is the most prevalent alteration, representing 25%–30% of adenocarcinomas, followed by EGFR (15%–30% in the US, up to 63% in Asia), ALK (5%–7%), MET (3%), BRAF (1%–4%), ROS1 (2%), HER2 (2%), RET (1%–2%), tropomyosin receptor kinase (NTRK1; 1%–2%), phosphoinositide 3-kinase (PIK3CA; 2%), and MEK1 (<1%) (Figure 2 and refs. 9–13). These mutations are generally mutually exclusive such that the presence of one driver event often excludes another in the same patient.
Figure 2. Genomic classification of lung adenocarcinoma.
The incidence of specific genetic mutations in NSCLC is reported, with mutations in KRAS, EGFR, and ALK having the highest prevalence.
EGFR mutations
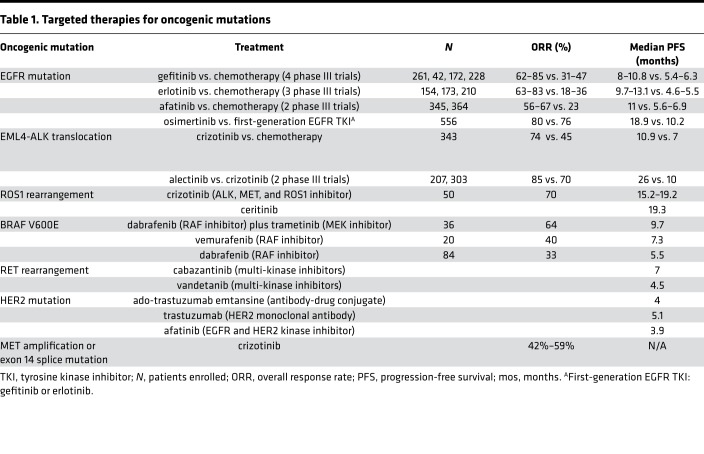
Nearly 90% of the EGFR mutations are localized to exon 19 or 21; cancer cells bearing these events are highly susceptible to EGFR TKIs (14). Erlotinib and gefitinib were the first generation of reversible EGFR inhibitors that were approved for use in advanced NSCLC. These agents result in objective response rates of 50%–70% and median progression-free survival (PFS) of 9 to 10 months for patients harboring EGFR mutations (14–20). Multiple randomized clinical trials have documented superior response rate and PFS with erlotinib or gefitinib over standard platinum-based chemotherapy for patients with EGFR-activating mutations. Afatinib and dacomitinib are referred to as second-generation irreversible EGFR inhibitors. Afatinib was associated with modest improvement in efficacy relative to gefitinib (PFS: 11 vs. 10.9 months); similarly, dacomitinib demonstrated superior PFS compared with gefitinib (PFS: 14.7 vs. 9.2 months); however, this comes at the expense of additional toxicity related to the skin and the gastrointestinal tract (grade 2 or higher adverse event: 33% vs. 9%; 37% vs. 10%, respectively) (21, 22). More recently, osimertinib, a third-generation EGFR inhibitor, has gained approval by the US FDA based on superiority over gefitinib and erlotinib. In a phase 3 study (FLAURA), patients treated with osimertinib had superior PFS (18.9 vs. 10.2 months) and duration of response (23). Activity against brain metastasis was also more favorable for patients treated with osimertinib. Taken together with a favorable toxicity profile, the results show that osimertinib has emerged as a standard first-line therapy option for patients with EGFR-activating mutations. In light of this, the role of second-generation EGFR TKIs in first-line therapy is questionable. Osimertinib was initially developed based on its ability to inhibit the T790M resistance mutation and its selectivity for the mutant receptor. Since it also inhibits the common EGFR-activating mutations, its role as first-line therapy represents a new milestone for patients with EGFR mutations. For patients with uncommon EGFR mutations (L861Q, G719X, and S768I), afatinib is the preferred agent based on modest efficacy results in nonrandomized trials (24). Exon 20 insertion mutations are refractory to the class of EGFR inhibitors currently in clinical use. Recently, poziotinib, a pan-HER TKI, has demonstrated promising preliminary results in patients with exon 20 insertions and is being investigated in ongoing studies (25).
ALK gene rearrangement
ALK fusion is observed more commonly in younger patients and never-smokers with adenocarcinoma histology. The disease phenotype is characterized by higher predilection for pleural, pericardial, and brain metastases. ALK fusion can be detected by immunohistochemistry, fluorescent in situ hybridization (FISH), or RT-PCR. Testing for ALK is recommended as part of initial diagnostic workup for patients with advanced non–squamous cell NSCLC histology. Crizotinib was the first ALK inhibitor that demonstrated efficacy in this patient subset. This agent was originally developed as a MET inhibitor; upon recognition of ALK fusions in NSCLC, crizotinib was studied in this biological subset of patients in nonrandomized studies. Based on promising response rates of approximately 60%–65% and median PFS of 10 months, this agent was approved for use in advanced NSCLC patients with ALK fusion (26). These results were confirmed in randomized studies that demonstrated superiority for crizotinib over standard chemotherapy (27). This paved the way for the development of more potent ALK inhibitors such as alectinib, ceritinib, and brigatinib. Initially, these agents were studied for patients that developed acquired resistance to crizotinib; however, two randomized trials in recent years demonstrated robust superiority for alectinib over crizotinib, with median PFS exceeding two years (28, 29). Ceritinib was also proven to be effective as first-line therapy based on superiority over standard chemotherapy in patients with ALK fusion (30). These newer agents exert greater activity against brain metastasis relative to crizotinib.
ROS1 fusion
Approximately 1% of patients with advanced NSCLC harbor the ROS1 fusion gene. Crizotinib, which also inhibits ROS1, is the only approved agent for this patient population. It demonstrated a response rate of approximately 70% and median PFS of 19 months in a nonrandomized study (31). For patients that develop acquired resistance to crizotinib, a recent study documented modest activity with a response rate of 56% for lorlatinib, a multi-kinase inhibitor (32). Entrectinib, a ROS1 and TRK inhibitor, has also demonstrated a 29-month median PFS in a recent study for patients with ROS1 fusion abnormality including those with brain metastases (33). These data demonstrate the rapid evolution of targeted therapies for this relatively small subset of NSCLC patients.
BRAF mutation
Approximately 2%–3% of patients with advanced NSCLC harbor mutations in the BRAF gene; approximately half of these are the V600E mutation, which is sensitive to BRAF inhibitors. Dabrafenib was initially evaluated as monotherapy and resulted in a response rate of approximately 30% for patients with the V600E mutation (34). Recently, the combination of dabrafenib and trametinib (MEK inhibitor) was approved by the FDA based on a response rate of 65% and a median PFS of nearly 10 months with this combination approach (35). This regimen is now the standard treatment for patients with the BRAF V600E mutation. Vemurafenib, another BRAF inhibitor, has also demonstrated anticancer activity in this patient subset with a modest response rate (36).
Other treatable driver mutations
MET exon 14 mutation is present in approximately 3%–4% of patients with NSCLC; it is more common in patients with sarcomatoid histology (37). Several MET inhibitors are being studied for this patient subset and the initial results have been promising, with a response rate of approximately 40% (38). RET fusion abnormality is observed in approximately 1% of patients with advanced NSCLC. Initial studies with RET inhibitors resulted in underwhelming results; however, more recently a new class of RET inhibitors that have higher potency and selectivity have entered clinical testing and are hoped to result in more favorable outcomes. NTRK fusions, observed in less than 1% of NSCLC, is sensitive to the new class of NTRK inhibitors. These developments underline the rapid pace at which the pursuit of personalized therapies have advanced in the treatment of lung cancer. Several other driver events are currently being targeted in ongoing clinical trials.
The pursuit of targeted therapies for patients with KRAS mutations has yet to prove successful. The combination of chemotherapy with MEK inhibition was associated with promising results in KRAS-mutated NSCLC, but could not be confirmed in a follow-up phase 3 study (39). The complexity of KRAS mutations has been further magnified by the specific mutation and the presence of co-mutations. For instance, patients with KRAS and LKB1 co-mutation have a poor overall prognosis and are insensitive to immune checkpoint inhibition (40). A class of direct KRAS inhibitors have recently entered clinical investigations.
Despite substantial improvements in efficacy outcomes and meaningful prolongation in duration of response to therapy, the effect on extending overall survival has been debated. Since the majority of patients enrolled in clinical trials ultimately receive the specific TKI being evaluated regardless of the treatment group they are assigned to, the survival results in the control group are diluted. However, population-based studies very clearly show that targeted therapies are making a major contribution to patient outcomes. In France, 51% of patients with a driver mutation who received targeted therapy had an improved survival compared with patients without such an abnormality (16.5 vs. 11.8 months; P < 0.00001) (9). While this difference could imply that presence of the mutation itself confers a survival advantage, a retrospective analysis has shown that the prognosis of treatment-naive ALK-positive NSCLC patients did not differ from a general cohort of NSCLC patients, although treatment with crizotinib improved survival (41). Additionally, in the Lung Cancer Mutation Consortium (LCMC), patients from the US treated with genome-directed therapy had a median survival of 3.5 years versus 2.4 years and 2.1 years for those with or without a mutation not treated with targeted therapy, respectively (10). A Japanese study also noted that the overall survival for NSCLC has increased with the routine use of EGFR inhibitors compared with the era before, which substantiated reports from the US and France (42).
Acquired resistance to targeted therapies
Irrespective of the advances that come with targeted therapies, NSCLC patients with oncogenic driver mutations develop resistance under selective pressures and ultimately develop disease progression. Resistance often results from acquired mutations that render the TKI ineffective, activation of alternative cell-signaling pathways, or histologic transformation. These resistance mechanisms appear to be common among various classes of targeted agents discussed earlier. In the following section we describe the major mechanisms underlying escape from targeted therapy in NSCLC.
Modifications of the oncogenic driver
Secondary mutations.
The commonly used targeted drugs in lung cancer inhibit tyrosine kinase function by either disrupting adenosine triphosphate (ATP) binding or inducing suppressive conformational changes. However, secondary mutations in the oncogenic tyrosine kinase can counter this inhibition usually with a higher affinity for ATP or steric hindrance to TKI binding. In the case of EGFR-mutated NSCLC, 50%–60% of patients treated with first-generation TKIs (gefitinib, erlotinib) develop a new mutation in exon 20 T790M that increases the affinity for ATP binding (43–46). Other secondary EGFR mutations account for only approximately 1%–2% of patients and include D761Y, T854A, and L747S (43, 47–49). Osimertinib was studied specifically for T790M-positive NSCLC in the setting of acquired resistance to EGFR TKI therapy, and demonstrated robust superiority over chemotherapy. Interestingly, none of the patients treated with osimertinib as first-line therapy developed T790M-mediated acquired resistance. Escape mechanisms of third-generation TKIs such as osimertinib include the loss of T790M, the acquisition of tertiary mutations, or the development of mutations in alternative pathways such as HER-2 and BRAF. The most common tertiary mutation appears to be C797S, which affects the TKI covalent bond residue (50). Loss of T790M, but maintenance of the original driver mutation, can be therapeutically targeted with reintroduction of the first-generation TKIs even in the setting of the tertiary C797S mutation (51, 52). While C797S appears to represent about 20% of secondary mutations of third-generation TKIs, other less frequent mutations include L718Q, L792F, L844V, and G796D (53–56). In preclinical experiments, gefitinib and afatinib demonstrate anticancer effects against C797S, L718Q, and L88V when each was added to EGFR-sensitizing mutations in the absence of T790M due to the fact that the original binding pocket is unaffected in many of these situations (54). Clinically, osimertinib resulted in a response rate of 14% after prior nazartinib failure, with a median duration of treatment of 9 months, which suggests that sequencing therapies can be effective in some cases, depending on the resistance mechanism induced (57). However, the presence of all three mutations (L858R/T790M/C797S) appears to have resulted in resistance to the available EGFR TKIs, but has shown some response to EGFR antibodies alone or in combination with brigatinib (54, 58). Additionally, the combination of first- and third-generation TKIs appears to be active against the three mutations combined, but only when the C797S and T790M mutation are on trans alleles (59). Therefore, combination therapies against the EGFR-dependent mechanisms remain effective in some cases. There are also varying mechanisms of resistance to osimertinib, depending on whether it is used as first-line therapy or second-line therapy. C797S mutations are seen in a small subset of patients and activation of other pathways driven by mutations have been reported with first-line osimertinib therapy. Effective treatment for this group of patients is continually being redefined based on the genomic shift that occurs in response to specific TKIs, taking personalized medicine to its ultimate limits.
The development of secondary ALK mutations after ALK TKI therapy occurs in approximately 20%–30% of resistant cases (60). In contrast to EGFR, resistance mutations appear quite varied, with at least 15 identified mutations, of which L1196 and G1269A mutations are the most common (Table 1) (61–64). Since crizotinib is a relatively weak inhibitor, resistance mechanisms tend to be different from those treated with the more potent second-generation ALK TKIs. The emergence of gatekeeper mutations appears to be slightly lower with crizotinib, as a subset of these patients develop resistance due to lower potency of the drug against ALK. The difference in potency likely explains the improved efficacy of the second-generation ALK inhibitors against brain metastasis relative to crizotinib, a common problem in ALK-driven NSCLC. Resistance mutations develop in 50% of patients treated with second-generation ALK TKIs and also increase when treated with more than one ALK TKI, suggesting that more potent inhibition not only leads to an increase in mutations, but also more specific alterations (60, 65). Among the acquired mutations, the G1202R mutation appears to be the most difficult type to treat. In addition, recent evidence indicates that mechanisms of resistance vary based on the specific ALK fusion variant. Fusion variant 3 is associated with a higher likelihood of development of the G1202R mutation, when compared with variant 1 (66). Lorlatinib, a promising new ALK inhibitor, appears to inhibit ALK signaling in patients with the G1202R mutation, with objective responses in greater than 50% of the patients (67–71). As with other TKIs, lorlatinib induces resistance mutations such as L1198F, which affects drug binding (72). This L1198F modification, however, results in resensitization to crizotinib by enhancing its binding to the ALK translocation, despite the persistence of a crizotinib-induced secondary resistance mutation. Similarly, alectinib has been able to overcome ceritinib’s G1123S-mediated resistance, while ceritinib has been used successfully in patients with alectinib-induced I1171T resistance (73, 74). Therefore, identifying posttreatment resistance mutations can effectively guide sequential therapies.
Table 1. Targeted therapies for oncogenic mutations.
A number of secondary ROS1 resistance mutations have been identified, with the most common being G2032R in approximately 40% of crizotinib-treated patients (75). More recently, entrectinib, a novel multi-kinase inhibitor, has demonstrated robust PFS of 30 months in a small cohort of patients with ROS1-positive NSCLC (76). However, entrectinib does not appear to be effective against G2032R in preclinical models and might be ineffective in patients that have received prior therapy with crizotinib; to the contrary, lorlatinib has shown promising activity in the acquired resistance setting for patients with ROS1-positive NSCLC. Several other promising agents are also under investigation (77–83). As TKIs are being employed in the treatment of other oncogenic drivers such as cabozantinib or vandetanib for RET and entrectinib for NTRK, the emergent resistance mutations are beginning to be reported in the literature. Similar to the ALK situation, the vandetanib-mediated KIF5B-RET810A resistance mutation paradoxically enhances its sensitivity to other TKIs, ponatinib and lenvatinib, in vitro (84). Therefore, studying the sensitivity of each gatekeeper mutation to the specific therapeutic agent will guide the management of acquired resistance.
Alterations of target expression.
Amplification of the oncogenic driver is another mechanism that can promote therapy resistance by outpacing drug effects, which is dose-limited by side effects. Often, levels of the original alteration are increased in addition to other resistance mechanisms, specifically secondary mutations that can be preferentially amplified as well. An analysis of patients who developed resistance to first-generation TKIs revealed that those with an EGFR amplification also had a T790M mutation, and in two of three cases, the T790M expression was selectively amplified (44). As noted in ALK-positive patients, amplifications can also be an independent cause of resistance and appear to occur regardless of TKI potency (60, 63). Less commonly, loss of activating mutations has been noted for EGFR- and ALK-positive patients (61, 85). This loss of an EGFR mutation has been accompanied by a compensatory MET tyrosine kinase amplification in resistant patient tumor samples and has demonstrated MET TKI sensitivity in resistant cell lines (86). Identified as an oncogenic swap, in which growth dependence switches from the original mutation to another driver, this highlights the importance of knowing the dominant resistance mechanism to not only improve selection of a therapy, but also to minimize the toxicities of unnecessary combination therapy.
Bypass pathways
Parallel pathways.
Cellular signaling primarily occurs through the JAK/STAT, PI3K/AKT/MTOR, and Ras/Raf/MEK/ERK1/2-MAPK pathways, which serve as downstream effectors of many oncogenic driver mutations. While TKI therapy against an activating mutation can effectively disrupt cellular signaling, emergence of alternative or downstream pathways can circumvent such inhibition beyond on-target alterations alone. MET amplification is a common bypass mechanism of EGFR TKIs, representing approximately 5%–10% of first-generation TKI-resistant cases and up to 30% for osimertinib-resistant cases, while upregulation of its ligand, hepatocyte growth factor (HGF), seems to occur less frequently (44, 45, 56, 87, 88). MET heterodimerizes with ERBB3 to activate the PI3K/AKT pathway independently of EGFR. In the clinic, this knowledge has prompted the investigation of combining EGFR TKIs with MET inhibitors for patients with MET amplification as the major resistance mechanism. Initial results of combination therapy with gefitinib and the MET inhibitor campmatinib (INC280) in EGFR-mutated NSCLC patients who had progressed on prior TKI therapy demonstrated a disease control rate of 80%. Patients with a MET copy number of at least 6 had a higher response rate of 30% versus 19% in those with a copy number of at least 5 (89). Similarly, the MET inhibitor tepotinib in combination with gefinitib has shown promising results (90). The effectiveness of this approach supports further evaluation of novel combination strategies to overcome acquired resistance.
Other major tyrosine kinase parallel pathways that are being targeted for inhibition in EGFR-mutated NSCLC include HER2, IGF-1R, AXL, and FGFR3 (91–95). HER2 amplification, although rare in treatment-naive EGFR-mutant patients, represents up to 13% of EGFR TKI resistance and appears to be mutually exclusive from T790M (45, 91). Unfortunately, HER2-directed therapy has not proven effective against HER2-amplified NSCLCs (96). However, afatinib, which has HER2 TKI activity, combined with cetuximab, a monoclonal EGFR antibody, demonstrated a 29% overall response rate with similar responses in both T790M-positive and -negative patients in a phase 2 trial of EGFR-mutant TKI-resistant NSCLC patients (97). Notably, the T790M-negative patients had a longer duration of response (9.4 vs. 5.6 months). Additionally, HER3 inhibitors are currently being evaluated, as the HER3 receptor heterodimerizes with EGFR, HER2, and MET, but has limited kinase activity itself.
Similarly, resistance to ALK TKIs can occur via several alternative pathways that include EGFR, IGF-1R, AXL, KIT, HER3, and SRC tyrosine kinases (63, 95, 98, 99). EGFR activation is commonly seen in ALK resistance (74), including development of EGFR-activating mutations (61, 100). EGFR activation has also been found to circumvent inhibition in ROS1-driven NSCLCs (101). Selective inhibition of the dominant pathway promotes resistance through parallel pathways, suggesting that blocking one target is not sufficient for long-term control of NSCLC.
Activation of downstream signaling.
While multi-kinase inhibitors may inhibit the initial activation of a pathway, modifications in the downstream effector proteins can maintain cellular signaling independently. Generally, KRAS, NRAS, BRAF, and MEK1 mutations do not occur concurrently with EGFR- or ALK-activating mutations, but such mutations have been reported in acquired resistance. KRAS mutations have been detected in EGFR TKI-resistant patient samples using more sensitive methods and in ALK- or ROS1-resistant patients (61, 102, 103).
More commonly, amplification of the MAPK pathway or downregulation of negative regulators is implicated in activation of downstream signaling. This includes MAPK amplification or downregulation of neurofibromin 1 (NF1), which inactivates a Ras-activating GTPase in EGFR resistance; downregulation of the enzyme dual-specificity phosphatase 6 (DUSP6), which inactivates the pathway’s tyrosine kinases in ALK-resistant cells; and KRAS amplification in ROS1 (103–106). In BRAFV600-mutant NSCLC, a truncated variant induced resistance to inhibition, but was able to maintain pathway signaling (107). MEK inhibition has been effective in not only resensitizing the NSCLC to TKIs, but also in delaying resistance. These combinations are being further investigated.
Activation of the PI3K pathway has also been linked to acquired resistance to targeted therapies. PI3K mutations have been detected in approximately 4% of resistant EGFR or ALK TKI–treated specimens, while loss of phosphatase and tensin homolog (PTEN), which inactivates phosphatidylinositol-3,4,5-triphosphate (PIP3) signaling via dephosphorylation, has also been identified as a mechanism of EGFR resistance (65, 108, 109). However, the clinical role of the PI3K pathway in mediating resistance is not clear, as pathway alterations do not appear to affect TKI response in EGFR-mutated NSCLC patients nor cell survival in ALK-positive NSCLC cells (105, 110).
Phenotypic transformations
Epithelial-to-mesenchymal transition (EMT) of tumors has been detected in EGFR TKI– and ALK TKI–resistant NSCLCs facilitating migration and invasion, but the exact role of EMT in resistance remains unclear (44, 65). While the mechanism of this transition has not been fully elucidated, AXL tyrosine kinase expression has been associated with it and IGF-1R has been noted to induce EMT in EGFR TKI–resistant cell lines (93, 111). However, once EMT occurs, IGF-1R inhibition appears to become ineffective, though a dependence on the Src/focal adhesion kinase (FAK) pathway develops such that these cells are sensitive to the Src TKI dasatinib in combination with EGFR inhibition (112). In fact, dasatinib or MEK inhibition combined with EGFR TKI has also been noted to prevent EMT, suggesting a role for both pathways in preventing this phenotype (113, 114).
Histologic transformation into small-cell lung cancer accounts for 3%–10% of EGFR TKI resistance and has also been reported in ALK TKI resistance (44, 45, 115–117). Despite this transition, the original activating mutation is retained, although molecularly these cancers appear more similar to de novo small-cell cancers (118). Tissue analysis has linked RB loss to these transformed small-cell cancers in EGFR-mutated patients, compared with only 11% in the NSCLC samples. These patients are treated with chemotherapy similar to the approach against classical small-cell cancers.
Circulating tumor DNA in understanding resistance mechanisms
It is increasingly evident that resistance to targeted therapies is a dynamic event and can not only be used to select treatment approaches, but also to define prognosis. Until recently, a tumor biopsy was necessary to determine the molecular mechanisms of resistance to targeted therapies. Recently, molecular testing in cell-free DNA has emerged as a potent tool to study resistance.
Blood-based sampling of tumor cell fragments or circulating tumor DNA (ctDNA) has recently been FDA approved for detection of activating EGFR mutations in NSCLC as a companion diagnostic test, and these assays are being further investigated in clinical trials to help guide therapeutic decisions. The commonly used ctDNA testing methods include digital PCR, amplification (via beads, emulsion, and magnetics [BEAMing]), or nondigital platforms (amplification-refractory mutation system [ARMS], ligand-targeted PCR [LT-PCR]), and next-generation sequencing (NGS) (119). PCR offers limited mutational testing with an often quick turnaround time, while NGS offers more extensive testing, but requires more time for testing, better standardization, and the need to further differentiate between detection of background noise versus clinically relevant results. ctDNA can be obtained from plasma or serum; while serum generally has a higher concentration, detection of mutations is often better in plasma samples.
ctDNA assays were found to have a high specificity (0.96; CI 0.93–0.98) and a moderate sensitivity (0.62; CI 0.51–0.72) for detecting EGFR mutations compared with tissue in a meta-analysis of 3,110 patients (120). Accuracy improves with advanced-stage disease, but is decreased after chemotherapy. Higher ctDNA concentrations predict for poorer outcomes, but are also associated with predictive potential for outcome with targeted therapies (121). In studies of third-generation TKIs, response rate and PFS were similar irrespective of the method (tissue vs. plasma) used for T790M detection; however, 20%–30% of mutations were missed by ctDNA alone, suggesting that multiple methods may need to be employed to confirm a negative result (122–124). Additionally, ctDNA can be used to predict resistance and progression by detection of the T790M mutation even before radiologic progression (125, 126). Evidence suggests that ctDNA analysis can detect multiple resistance mechanisms within a patient and can also be used to monitor responses in ALK-positive patients (102, 127). Recent data also indicate that serial monitoring of ctDNA can help determine prognosis within weeks of initiation of EGFR TKI therapy (128). As the clinical role of ctDNA is being better defined, this monitoring approach is increasingly being used as an adjunctive study to help guide therapeutic decisions.
Role of immunotherapy in NSCLC
Engaging the patient’s immune system to treat cancer has recently emerged as an effective strategy. Several immune checkpoint inhibitors that reverse T cell exhaustion by inhibiting the programmed death protein 1 (PD-1) pathway have proven to be effective for the treatment of advanced-stage NSCLC. In patients with tumors harboring high PD-L1 expression (>50%), pembrolizumab is associated with superior PFS and overall survival (129). In addition, the combination of chemotherapy with pembrolizumab results in superior overall survival when compared with platinum-based chemotherapy alone for patients with advanced non–squamous cell NSCLC, regardless of the PD-L1 expression status (130). These developments have completely altered the treatment landscape for patients with NSCLC. Other inhibitors of the PD-1 pathway such as nivolumab and atezolizumab have also demonstrated improved outcomes for patients with advanced-stage NSCLC (131, 132). Another emerging novel approach for treatment of cancer includes combinations of immune checkpoint inhibitors. A phase 3 clinical trial recently demonstrated improved PFS for the combination of nivolumab and ipilimumab (CTLA4 inhibitor) when compared with chemotherapy for patients with high tumor mutation burden (133).
These exciting developments, however, have not made a substantial impact for patients with driver mutations such as EGFR. Nearly all the studies with immune checkpoint inhibitors excluded patients with known EGFR and ALK aberrations. A meta-analysis of second-line randomized immunotherapy trials failed to show an overall survival advantage in EGFR-mutated patients treated with checkpoint inhibitors compared with docetaxel (134). Tumors of patients with EGFR mutation are associated with lower PD-L1 expression and low tumor mutation burden, which could explain the relative lack of efficacy of immune checkpoint inhibitors in this patient population (135). Additionally, the combination of TKIs and immunotherapies have proven toxic with significant grade 3 or 4 adverse events, resulting in the closing of several of these trials early (136). For these reasons, it is important that targeted therapy remain as the standard of care for patients with EGFR mutation and ALK translocation. New efforts are looking to modulate the immune system to treat oncogene-addicted tumors through novel combinations, vaccines, and other evolving strategies that will likely lead to improved patient outcomes.
Conclusions
With increasing insights into tumor biology, we have moved from a shotgun approach with chemotherapy to a more targeted treatment paradigm. Genomic profiling of The Cancer Genome Atlas (TCGA) has demonstrated that up to 90% of 3,277 tumors tested have at least 1 potentially targetable alteration, with the majority having multiple (137). Overall, nearly 75% of these alterations are estimated to be clinically relevant, with more than 80% potentially impacting treatment options for NSCLC patients (138). As our detection methods improve, the challenge becomes identifying the therapeutic significance of such alterations, especially if multiple exist. Currently, several basket trials such as the NCI MATCH (NCT02465060) and TAPUR (NCT02693535) trials are ongoing to further evaluate molecularly targeted therapies based on genomic testing.
In NSCLC, TKI therapy has already changed the therapeutic landscape and the lives of patients by reducing toxicities, improving quality of life, and outcomes. Effective targeted therapy revolves around the idea of a single oncogenic driver that sustains cancer growth; however, selective inhibition gives rise to escape mechanisms in the form of on-target alterations, bypass pathways, and to a lesser degree, histologic transformation to small-cell cancer. As such, novel drugs to target secondary mutations, multi-kinase inhibitors, or combination therapy is being used to combat resistance. Increasing evidence suggests that liquid biopsies will offer better treatment monitoring and a more comprehensive assessment of the resistance mechanisms involved, which will allow for appropriate treatment selection. With better temporal monitoring of resistance, sequential therapy may offer effective options with less toxicity than combinations. However, about one-third of EGFR TKI and ALK TKI resistance remains unknown, indicating the need for better techniques and additional research to identify, and ultimately prevent such mechanisms (139, 140). The recent success of immunotherapy in NSCLC offers hope that a person’s immune system can be harnessed to more comprehensively control cancer in even those with oncogenic drivers. Hopefully, between the crossroads of targeted therapy and immunotherapy lies the solution for better and longer lives for all cancer patients.
Address correspondences to: Suresh S. Ramalingam, Professor of Hematology and Medical Oncology, 1365 Clifton Road NE, C4014E, Atlanta, Georgia 30322, USA. Phone: 404.778.1916; Email: ssramal@emory.edu.
Version 1. 08/09/2018
Electronic publication
Footnotes
Conflict of interest: SSR has served on scientific advisory board meetings and received honoraria from Amgen, Abbvie, AstraZeneca, Bristol Myers Squibb, Genentech, Roche, Merck, Nektar, and Loxo Oncology.
Published: August 9, 2018
Reference information: JCI Insight. 2018;3(15):e120858. https://doi.org/10.1172/jci.insight.120858.
References
- 1.National Lung Screening Trial Research Team, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiller JH, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Scagliotti GV, et al. Phase III randomized trial comparing three platinum-based doublets in advanced non-small-cell lung cancer. J Clin Oncol. 2002;20(21):4285–4291. doi: 10.1200/JCO.2002.02.068. [DOI] [PubMed] [Google Scholar]
- 4.Sweeney CJ, et al. Outcome of patients with a performance status of 2 in Eastern Cooperative Oncology Group Study E1594: a Phase II trial in patients with metastatic nonsmall cell lung carcinoma. Cancer. 2001;92(10):2639–2647. doi: 10.1002/1097-0142(20011115)92:10<2639::AID-CNCR1617>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Fukuoka M, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] J Clin Oncol. 2003;21(12):2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 6.Miller VA, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22(6):1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 7.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 8.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 9.Barlesi F, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT) Lancet. 2016;387(10026):1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 10.Kris MG, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sholl LM, et al. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol. 2015;10(5):768–777. doi: 10.1097/JTO.0000000000000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9(2):154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sequist LV, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011;22(12):2616–2624. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 15.Han JY, et al. First-SIGNAL: first-line single-agent Iressa versus gemcitabine and cisplatin trial in never-smokers with adenocarcinoma of the lung. J Clin Oncol. 2012;30(10):1122–1128. doi: 10.1200/JCO.2011.36.8456. [DOI] [PubMed] [Google Scholar]
- 16.Mitsudomi T, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 17.Maemondo M, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 18.Zhou C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 19.Rosell R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 20.Wu YL, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol. 2015;26(9):1883–1889. doi: 10.1093/annonc/mdv270. [DOI] [PubMed] [Google Scholar]
- 21.Park K, et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016;17(5):577–589. doi: 10.1016/S1470-2045(16)30033-X. [DOI] [PubMed] [Google Scholar]
- 22.Wu YL, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol. 2017;18(11):1454–1466. doi: 10.1016/S1470-2045(17)30608-3. [DOI] [PubMed] [Google Scholar]
- 23.Soria JC, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378(2):113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 24.Yang JC, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16(7):830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 25. Elamin, JR, J Heymach. OA 12.01 - The preclinical and clinical activity of poziotinib, a potent, selective inhibitor of EGFR exon 20 mutant NSCLC. IASLC 18th World Conference on Lung Cancer 2017. Yokohama, Japan. http://wclc2017.iaslc.org.
- 26.Camidge DR, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012;13(10):1011–1019. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solomon BJ, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 28.Hida T, et al. Alectinib versus crizotinib in patients with ALK-positive non-small-cell lung cancer (J-ALEX): an open-label, randomised phase 3 trial. Lancet. 2017;390(10089):29–39. doi: 10.1016/S0140-6736(17)30565-2. [DOI] [PubMed] [Google Scholar]
- 29.Peters S, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med. 2017;377(9):829–838. doi: 10.1056/NEJMoa1704795. [DOI] [PubMed] [Google Scholar]
- 30.Soria JC, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet. 2017;389(10072):917–929. doi: 10.1016/S0140-6736(17)30123-X. [DOI] [PubMed] [Google Scholar]
- 31.Shaw AT, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371(21):1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Solomon, et al. OA 05.06 Phase 2 study of lorlatinib in patients with advanced ALK+/ROS1+ non-small-cell lung cancer. IASLC 18th World Conference on Lung Cancer 2017: Yokohama, Japan. http://wclc2017.iaslc.org.
- 33. Ahn MJ, CB, Siena S, et al. Entrectinib in patients with locally advanced or metastatic ROS1 fusion-positive non-small cell lung cancer (NSCLC). IASLC 18th World Conference on Lung Cancer 2017. Yokohama, Japan. http://wclc2017.iaslc.org.
- 34.Planchard D, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(5):642–650. doi: 10.1016/S1470-2045(16)00077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Planchard D, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol. 2016;17(7):984–993. doi: 10.1016/S1470-2045(16)30146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyman DM, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373(8):726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, et al. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol. 2016;34(8):794–802. doi: 10.1200/JCO.2015.62.0674. [DOI] [PubMed] [Google Scholar]
- 38.Drilon AE, et al. Efficacy and safety of crizotinib in patients (pts) with advanced MET exon 14-altered non-small cell lung cancer (NSCLC) J Clin Oncol. 2016;34(15_suppl):108 [Google Scholar]
- 39.Jänne PA, et al. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: The SELECT-1 Randomized Clinical Trial. JAMA. 2017;317(18):1844–1853. doi: 10.1001/jama.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calles A, et al. Expression of PD-1 and its ligands, PD-L1 and PD-L2, in smokers and never smokers with KRAS-mutant lung cancer. J Thorac Oncol. 2015;10(12):1726–1735. doi: 10.1097/JTO.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 41.Shaw AT, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011;12(11):1004–1012. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takano T, et al. EGFR mutations predict survival benefit from gefitinib in patients with advanced lung adenocarcinoma: a historical comparison of patients treated before and after gefitinib approval in Japan. J Clin Oncol. 2008;26(34):5589–5595. doi: 10.1200/JCO.2008.16.7254. [DOI] [PubMed] [Google Scholar]
- 43.Balak MN, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12(21):6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 44.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu HA, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105(6):2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bean J, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res. 2008;14(22):7519–7525. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa DB, Schumer ST, Tenen DG, Kobayashi S. Differential responses to erlotinib in epidermal growth factor receptor (EGFR)-mutated lung cancers with acquired resistance to gefitinib carrying the L747S or T790M secondary mutations. J Clin Oncol. 2008;26(7):1182–4;author reply1184. doi: 10.1200/JCO.2007.14.9039. [DOI] [PubMed] [Google Scholar]
- 49.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11(8):473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 50.Thress KS, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21(6):560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asahina H, et al. Phase II study of gefitinib readministration in patients with advanced non-small cell lung cancer and previous response to gefitinib. Oncology. 2010;79(5-6):423–429. doi: 10.1159/000326488. [DOI] [PubMed] [Google Scholar]
- 52.Oh IJ, Ban HJ, Kim KS, Kim YC. Retreatment of gefitinib in patients with non-small-cell lung cancer who previously controlled to gefitinib: a single-arm, open-label, phase II study. Lung Cancer. 2012;77(1):121–127. doi: 10.1016/j.lungcan.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 53.Ou Q, et al. Investigating novel resistance mechanisms to third generation EGFR TKI osimertinib in non-small cell lung cancer patients using next generation sequencing. J Clin Oncol. 2017;35(15_suppl):2572 [Google Scholar]
- 54.Ercan D, et al. EGFR mutations and resistance to irreversible pyrimidine-based EGFR inhibitors. Clin Cancer Res. 2015;21(17):3913–3923. doi: 10.1158/1078-0432.CCR-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng D, et al. EGFR G796D mutation mediates resistance to osimertinib. Oncotarget. 2017;8(30):49671–49679. doi: 10.18632/oncotarget.17913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piotrowska Z, et al. MET amplification (amp) as a resistance mechanism to osimertinib. J Clin Oncol. 2017;35(15_suppl):9020 [Google Scholar]
- 57.Arbour KC, et al. Response to osimertinib following treatment with EGF816 in patients with T790M EGFR mutant NSLCLC. J Clin Oncol. 2017;35(15_suppl):e20673 [Google Scholar]
- 58.Uchibori K, et al. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nat Commun. 2017;8:14768. doi: 10.1038/ncomms14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niederst MJ, et al. The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin Cancer Res. 2015;21(17):3924–3933. doi: 10.1158/1078-0432.CCR-15-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCoach CE, et al. Resistance mechanisms to targeted therapies in ROS1+ and ALK+ non-small cell lung cancer. J Clin Oncol. 2016;34(15_suppl):9065. doi: 10.1158/1078-0432.CCR-17-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doebele RC, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18(5):1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gainor JF, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res. 2013;19(15):4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katayama R, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4(120):120ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dagogo-Jack I, Engelman JA, Shaw AT. Overcoming on-target resistance to tyrosine kinase inhibitors in lung cancer. Annu Rev Cancer Biol. 2017;1(1):257–274. doi: 10.1146/annurev-cancerbio-050216-122044. [DOI] [Google Scholar]
- 65.Gainor JF, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov. 2016;6(10):1118–1133. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin JJ, et al. Impact of EML4-ALK Variant on resistance mechanisms and clinical outcomes in ALK-positive lung cancer. J Clin Oncol. 2018;36(12):1199–1206. doi: 10.1200/JCO.2017.76.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ou SH, et al. Alectinib in crizotinib-refractory ALK-rearranged non-small-cell lung cancer: A phase II global study. J Clin Oncol. 2016;34(7):661–668. doi: 10.1200/JCO.2015.63.9443. [DOI] [PubMed] [Google Scholar]
- 68.Shaw AT, et al. Ceritinib versus chemotherapy in patients with ALK-rearranged non-small-cell lung cancer previously given chemotherapy and crizotinib (ASCEND-5): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2017;18(7):874–886. doi: 10.1016/S1470-2045(17)30339-X. [DOI] [PubMed] [Google Scholar]
- 69.Kim DW, et al. Brigatinib in patients with crizotinib-refractory anaplastic lymphoma kinase-positive non-small-cell lung cancer: A randomized, multicenter phase II trial. J Clin Oncol. 2017;35(22):2490–2498. doi: 10.1200/JCO.2016.71.5904. [DOI] [PubMed] [Google Scholar]
- 70.Zou HY, et al. PF-06463922, an ALK/ROS1 inhibitor, overcomes resistance to first and second generation ALK inhibitors in preclinical models. Cancer Cell. 2015;28(1):70–81. doi: 10.1016/j.ccell.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Solomon B, et al. OA 05.06 phase 2 study of lorlatinib in patients with advanced ALK+/ROS1+ non-small-cell lung cancer. J Thorac Oncol. 2017;12(11):S1756. doi: 10.1016/j.jtho.2017.09.351. [DOI] [Google Scholar]
- 72.Shaw AT, et al. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med. 2016;374(1):54–61. doi: 10.1056/NEJMoa1508887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toyokawa G, et al. Identification of a novel ALK G1123S mutation in a patient with ALK-rearranged non-small-cell lung cancer exhibiting resistance to ceritinib. J Thorac Oncol. 2015;10(7):e55–e57. doi: 10.1097/JTO.0000000000000509. [DOI] [PubMed] [Google Scholar]
- 74.Katayama R, et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res. 2014;20(22):5686–5696. doi: 10.1158/1078-0432.CCR-14-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gainor JF, et al. Patterns of Metastatic Spread and Mechanisms of Resistance to Crizotinib in ROS1-Positive Non-Small-Cell Lung Cancer. JCO Precis Oncol. 2017;2017:10.1200/PO.17.00063. doi: 10.1200/PO.17.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Drilon A, et al. Abstract CT007: Entrectinib, an oral pan-Trk, ROS1, and ALK inhibitor in TKI-naïve patients with advanced solid tumors harboring gene rearrangements: Updated phase I results. Cancer Res. 2016;76(14 Supplement):CT007. doi: 10.1158/1538-7445.AM2016-CT007. [DOI] [Google Scholar]
- 77.Lim SM, et al. Open-label, multicenter, phase II study of ceritinib in patients with non-small-cell lung cancer harboring ROS1 rearrangement. J Clin Oncol. 2017;35(23):2613–2618. doi: 10.1200/JCO.2016.71.3701. [DOI] [PubMed] [Google Scholar]
- 78.Drilon A, et al. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7(4):400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chong CR, et al. Identification of existing drugs that effectively target NTRK1 and ROS1 rearrangements in lung cancer. Clin Cancer Res. 2017;23(1):204–213. doi: 10.1158/1078-0432.CCR-15-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Katayama R, et al. Cabozantinib overcomes crizotinib resistance in ROS1 fusion-positive cancer. Clin Cancer Res. 2015;21(1):166–174. doi: 10.1158/1078-0432.CCR-14-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zou HY, et al. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc Natl Acad Sci USA. 2015;112(11):3493–3498. doi: 10.1073/pnas.1420785112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cui JJ, et al. TPX-0005, a novel ALK/ROS1/TRK inhibitor, effectively inhibited a broad spectrum of mutations including solvent front ALK G1202R, ROS1 G2032R and TRKA G595R mutants. Eur J Cancer. 2016;69(suppl_1):S32 [Google Scholar]
- 83.Kiga M, et al. Preclinical characterization and antitumor efficacy of DS-6051b, a novel, orally available small molecule tyrosine kinase inhibitor of ROS1 and NTRKs. Eur J Cancer. 2016;69(suppl_1):S35–S36. [Google Scholar]
- 84.Huang Q, et al. Preclinical modeling of KIF5B-RET fusion lung adenocarcinoma. Mol Cancer Ther. 2016;15(10):2521–2529. doi: 10.1158/1535-7163.MCT-16-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tabara K, et al. Loss of activating EGFR mutant gene contributes to acquired resistance to EGFR tyrosine kinase inhibitors in lung cancer cells. PLoS One. 2012;7(7):e41017. doi: 10.1371/journal.pone.0041017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mizuuchi H, et al. Oncogene swap as a novel mechanism of acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor in lung cancer. Cancer Sci. 2016;107(4):461–468. doi: 10.1111/cas.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arcila ME, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17(5):1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamada T, et al. Hepatocyte growth factor reduces susceptibility to an irreversible epidermal growth factor receptor inhibitor in EGFR-T790M mutant lung cancer. Clin Cancer Res. 2010;16(1):174–183. doi: 10.1158/1078-0432.CCR-09-1204. [DOI] [PubMed] [Google Scholar]
- 89.Wu YL, et al. Phase (Ph) II safety and efficacy results of a single-arm ph ib/II study of capmatinib (INC280) + gefitinib in patients (pts) with EGFR-mutated (mut), cMET-positive (cMET+) non-small cell lung cancer (NSCLC) J Clin Oncol. 2016;34(15_suppl):9020 [Google Scholar]
- 90.Wu YL, et al. Tolerability, efficacy and recommended phase II dose (RP2D) of tepotinib plus gefitinib in Asian patients with c-Met-positive/EGFR-mutant NSCLC: Phase Ib data. J Clin Oncol. 2016;34(15_suppl):e20501–e20501. [Google Scholar]
- 91.Takezawa K, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2(10):922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee Y, Wang Y, James M, Jeong JH, You M. Inhibition of IGF1R signaling abrogates resistance to afatinib (BIBW2992) in EGFR T790M mutant lung cancer cells. Mol Carcinog. 2016;55(5):991–1001. doi: 10.1002/mc.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Z, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44(8):852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohashi K, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci USA. 2012;109(31):E2127–E2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crystal AS, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346(6216):1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zinner RG, et al. Trastuzumab in combination with cisplatin and gemcitabine in patients with Her2-overexpressing, untreated, advanced non-small cell lung cancer: report of a phase II trial and findings regarding optimal identification of patients with Her2-overexpressing disease. Lung Cancer. 2004;44(1):99–110. doi: 10.1016/j.lungcan.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 97.Janjigian YY, et al. Dual inhibition of EGFR with afatinib and cetuximab in kinase inhibitor-resistant EGFR-mutant lung cancer with and without T790M mutations. Cancer Discov. 2014;4(9):1036–1045. doi: 10.1158/2159-8290.CD-14-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lovly CM, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med. 2014;20(9):1027–1034. doi: 10.1038/nm.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilson FH, et al. A functional landscape of resistance to ALK inhibition in lung cancer. Cancer Cell. 2015;27(3):397–408. doi: 10.1016/j.ccell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim S, et al. Heterogeneity of genetic changes associated with acquired crizotinib resistance in ALK-rearranged lung cancer. J Thorac Oncol. 2013;8(4):415–422. doi: 10.1097/JTO.0b013e318283dcc0. [DOI] [PubMed] [Google Scholar]
- 101.Davies KD, et al. Resistance to ROS1 inhibition mediated by EGFR pathway activation in non-small cell lung cancer. PLoS One. 2013;8(12):e82236. doi: 10.1371/journal.pone.0082236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chabon JJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cargnelutti M, et al. Activation of RAS family members confers resistance to ROS1 targeting drugs. Oncotarget. 2015;6(7):5182–5194. doi: 10.18632/oncotarget.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ercan D, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov. 2012;2(10):934–947. doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hrustanovic G, Bivona TG. RAS-MAPK in ALK targeted therapy resistance. Cell Cycle. 2015;14(23):3661–3662. doi: 10.1080/15384101.2015.1096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.de Bruin EC, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov. 2014;4(5):606–619. doi: 10.1158/2159-8290.CD-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin L, et al. Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proc Natl Acad Sci USA. 2014;111(7):E748–E757. doi: 10.1073/pnas.1320956111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sos ML, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69(8):3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ludovini V, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2011;6(4):707–715. doi: 10.1097/JTO.0b013e31820a3a6b. [DOI] [PubMed] [Google Scholar]
- 110.Eng J, et al. Impact of concurrent PIK3CA mutations on response to EGFR tyrosine kinase inhibition in EGFR-mutant lung cancers and on prognosis in oncogene-driven lung adenocarcinomas. J Thorac Oncol. 2015;10(12):1713–1719. doi: 10.1097/JTO.0000000000000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou J, et al. Implication of epithelial-mesenchymal transition in IGF1R-induced resistance to EGFR-TKIs in advanced non-small cell lung cancer. Oncotarget. 2015;6(42):44332–44345. doi: 10.18632/oncotarget.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wilson C, et al. Overcoming EMT-associated resistance to anti-cancer drugs via Src/FAK pathway inhibition. Oncotarget. 2014;5(17):7328–7341. doi: 10.18632/oncotarget.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sesumi Y, et al. Effect of dasatinib on EMT-mediated-mechanism of resistance against EGFR inhibitors in lung cancer cells. Lung Cancer. 2017;104:85–90. doi: 10.1016/j.lungcan.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 114.Buonato JM, Lazzara MJ. ERK1/2 blockade prevents epithelial-mesenchymal transition in lung cancer cells and promotes their sensitivity to EGFR inhibition. Cancer Res. 2014;74(1):309–319. doi: 10.1158/0008-5472.CAN-12-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Takegawa N, et al. Transformation of ALK rearrangement-positive adenocarcinoma to small-cell lung cancer in association with acquired resistance to alectinib. Ann Oncol. 2016;27(5):953–955. doi: 10.1093/annonc/mdw032. [DOI] [PubMed] [Google Scholar]
- 116.Miyamoto S, et al. Transformation to small-cell lung cancer as a mechanism of acquired resistance to crizotinib and alectinib. Jpn J Clin Oncol. 2016;46(2):170–173. doi: 10.1093/jjco/hyv173. [DOI] [PubMed] [Google Scholar]
- 117.Fujita S, Masago K, Katakami N, Yatabe Y. Transformation to SCLC after treatment with the ALK inhibitor alectinib. J Thorac Oncol. 2016;11(6):e67–e72. doi: 10.1016/j.jtho.2015.12.105. [DOI] [PubMed] [Google Scholar]
- 118.Niederst MJ, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. doi: 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mayo-de-Las-Casas C, et al. An update on liquid biopsy analysis for diagnostic and monitoring applications in non-small cell lung cancer. Expert Rev Mol Diagn. 2018;18(1):35–45. doi: 10.1080/14737159.2018.1407243. [DOI] [PubMed] [Google Scholar]
- 120.Kuiper JL, et al. Incidence of T790M mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients. Lung Cancer. 2014;85(1):19–24. doi: 10.1016/j.lungcan.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 121.Ai B, Liu H, Huang Y, Peng P. Circulating cell-free DNA as a prognostic and predictive biomarker in non-small cell lung cancer. Oncotarget. 2016;7(28):44583–44595. doi: 10.18632/oncotarget.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wakelee HA, et al. Epidermal growth factor receptor (EGFR) genotyping of matched urine, plasma and tumor tissue from non-small cell lung cancer (NSCLC) patients (pts) treated with rociletinib. J Clin Oncol. 2016;34(15_suppl):9001 [Google Scholar]
- 123.Oxnard GR, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3375–3382. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sundaresan TK, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res. 2016;22(5):1103–1110. doi: 10.1158/1078-0432.CCR-15-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zheng D, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913. doi: 10.1038/srep20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Oxnard GR, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20(6):1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Puig O, et al. Pooled mutation analysis for the NP28673 and NP28761 studies of alectinib in ALK+ non-small-cell lung cancer (NSCLC) J Clin Oncol. 2016;34(15_suppl):9061 [Google Scholar]
- 128.Bigot F, et al. Early TKI-pharmokinetics and circulating tumor DNA (ctDNA) to predict outcome in patients with EGFR-mutated non-small cell lung cancer (NSCLC) J Clin Oncol. 2017;35(15_suppl):11544 [Google Scholar]
- 129.Reck M, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 130.Gandhi L, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 131.Brahmer J, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373(2):123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Horn L, et al. Nivolumab versus docetaxel in previously treated patients with advanced non-small-cell lung cancer: Two-year outcomes from two randomized, open-label, phase III trials (CheckMate 017 and CheckMate 057) J Clin Oncol. 2017;35(35):3924–3933. doi: 10.1200/JCO.2017.74.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hellmann MD, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee CK, et al. Checkpoint inhibitors in metastatic EGFR-mutated non-small cell lung cancer-A meta-analysis. J Thorac Oncol. 2017;12(2):403–407. doi: 10.1016/j.jtho.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 135.Wang Y, Ellis P. EGFR mutation positive non-small cell lung cancer: can we identify predictors of benefit from immune checkpoint inhibitors. Ann Transl Med. 2017;5(21):424. doi: 10.21037/atm.2017.08.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ahn MJ, Sun JM, Lee SH, Ahn JS, Park K. EGFR TKI combination with immunotherapy in non-small cell lung cancer. Expert Opin Drug Saf. 2017;16(4):465–469. doi: 10.1080/14740338.2017.1300656. [DOI] [PubMed] [Google Scholar]
- 137.Yuan Y, et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat Biotechnol. 2014;32(7):644–652. doi: 10.1038/nbt.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dienstmann R, Jang IS, Bot B, Friend S, Guinney J. Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov. 2015;5(2):118–123. doi: 10.1158/2159-8290.CD-14-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Majem M, Remon J. Tumor heterogeneity: evolution through space and time in EGFR mutant non small cell lung cancer patients. Transl Lung Cancer Res. 2013;2(3):226–237. doi: 10.3978/j.issn.2218-6751.2013.03.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Friboulet L, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4(6):662–673. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]