Abstract
Stable transmission of genetic information during cell division requires faithful mitotic spindle assembly and chromosome segregation. The Ran GTPase plays a key role in mitotic spindle assembly. However, how the generation of a chemical gradient of Ran-GTP at the spindle is coupled to mitotic post-translational modifications has never been characterized. Here, we solved the complex structure of Ran with the nucleotide release factor Mog1 and delineated a novel mitosis-specific acetylation-regulated Ran–Mog1 interaction during chromosome segregation. Our structure-guided functional analyses revealed that Mog1 competes with RCC1 for Ran binding in a GTP/GDP-dependent manner. Biochemical characterization demonstrated that Mog1-bound Ran prevents RCC1 binding and subsequent GTP loading. Surprisingly, Ran is a bona fide substrate of TIP60, and the acetylation of Lys134 by TIP60 liberates Mog1 from Ran binding during mitosis. Importantly, this acetylation-elicited switch of Ran binding to RCC1 promotes high level of Ran-GTP, which is essential for chromosome alignment. These results establish a previously uncharacterized regulatory mechanism in which TIP60 provides a homeostatic control of Ran-GTP level by tuning Ran effector binding for chromosome segregation in mitosis.
Keywords: Ran-GTP, Lys134 acetylation, TIP60, mitosis, chromosome segregation, NMR
Introduction
The process of stable transmission of genetic information through generations of living organisms requires accurate bipolar spindle assembly to segregate chromosomes into daughter cells. Aberrant assembly and/or regulation of mitotic spindle in human cells lead to aneuploidy and tumorigenesis (London and Biggins, 2014). The small GTPase Ran is essential for chromatin-driven spindle assembly (Clarke and Zhang, 2008). Ran cycling between inactive GDP-bound and active GTP-bound forms is regulated by the Ran guanine nucleotide exchange factor (RanGEF) RCC1 and GTPase-activating protein (RanGAP) (Bischoff and Ponstingl, 1991; Bischoff et al., 1995). RCC1 dynamically mediates Ran GDP/GTP exchange during the cell cycle on the chromatin surface. The association of RCC1 with chromatin generates a high level of Ran-GTP in the vicinity of chromosomes (Nemergut et al., 2001; Kalab et al., 2002; Li et al., 2003), which is necessary for cell division control (Carazo-Salas et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999; Zhang and Clarke, 2000). Ran-GTP is a substrate of PAK1 and the phosphorylation abrogates the binding of Ran-GTP to RanGAP1 and thus protects Ran from GTP hydrolysis, suggesting a context-dependent hierarchical interaction underlying spatiotemporal regulation of Ran-GTP gradient (Bompard et al., 2010).
Mog1 was initially discovered as a suppressor of conditional growth defect alleles of the Ran gene in Saccharomyces cerevisiae, which rescued the temperature-sensitive defect phenotype (Oki and Nishimoto, 1998). Mog1 stimulates GDP/GTP release from Ran and prevents nucleotide re-binding to Ran by forming a tight complex with the nucleotide-free Ran (Baker et al., 2001). However, the mechanism of action underlying the Ran–Mog1 interaction and its regulation in mitosis are largely unknown. In addition, it was unclear how Mog1 and RCC1 cooperate to generate spatiotemporal dynamics of Ran GTPase activity in mitosis.
In this study, we describe the essential and novel contribution of Ran acetylation in accurate chromosome segregation in mitosis. We combined nuclear magnetic resonance (NMR)-based structural analyses with biochemical characterization to pinpoint the binding interface of Ran to Mog1 and RCC1, respectively. Based on delineated structure information, we carried out molecular analyses of Ran interaction with Mog1 and revealed that Mog1 and RCC1 compete for binding to Ran. Surprisingly, Ran is a bona fide substrate of acetyltransferase TIP60 and the acetylation of Lys134 of Ran released its association from Mog1 and enables its binding to RCC1. This acetylation-elicited switch provides a temporal and prompt control of GTP loading to Ran and sustains a robust Ran-GTPase activity for accurate chromosome-kinetochore attachment. Interestingly, the CDK1-elicited phosphorylation of TIP60 exhibits a similar temporal profile to that of Ran-GTPase during cell cycle. Competition for Ran binding between its GEF and guanine nucleotide release factor Mog1 provides a direct and perhaps evolutionarily conserved strategy to sense and organize dynamic kinetochore microtubules for faithful mitosis.
Results
Molecular delineation of structural determinants underlying Ran–Mog1 complex formation
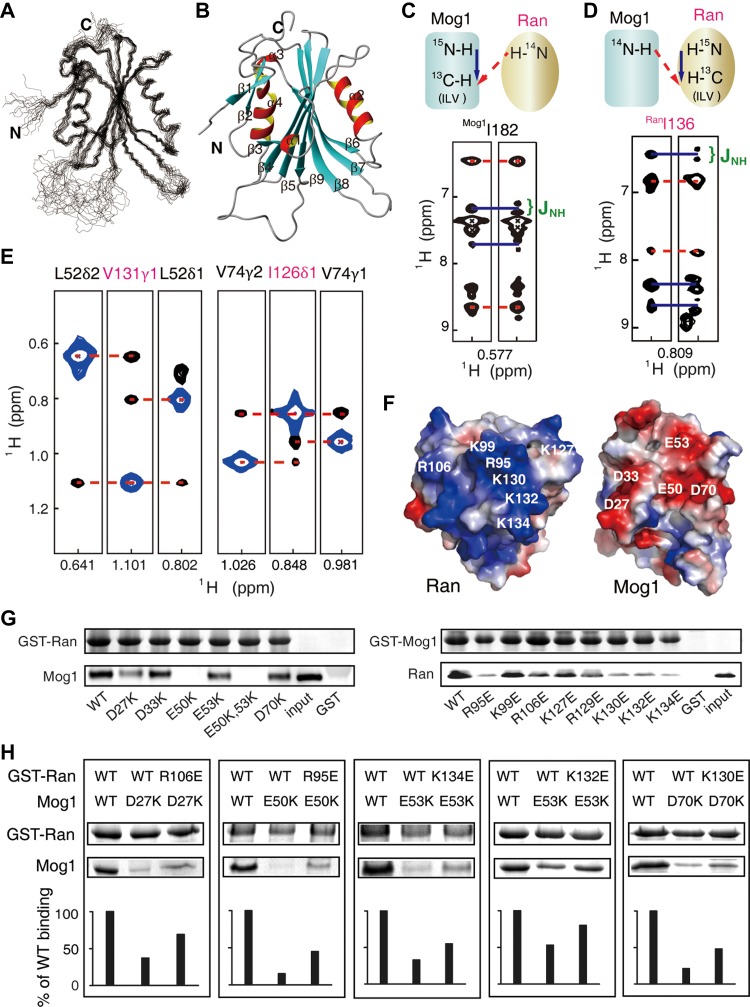
To gain structural insight into a better understanding of Ran–Mog1 interaction, we first determined the solution structure of a human Mog1 monomer by conventional NMR spectroscopy as shown in Figure 1A (PDBID: 5YFG, sequential assignment in Supplementary Figure S1, structure statistics listed in Supplementary Table S1). Mog1 is comprised of nine β-strands and four helices, where β3 to β9 assemble into the central antiparallel β-sheets packed by helices α2 and α4 on either side to form a sandwich-like structure (Figure 1B).
Figure 1.
Interaction between Mog1 and Ran mapped by NMR and mutagenesis methods. (A) The 20 representative conformers of Mog1. (B) A ribbon representation of Mog1. (C and D) The scheme of 1HN-methyl NOESY-HSQC spectra to identify the ILV residues in the interface, with or without 15N-decoupling in the indirect proton dimension for Mog1 or Ran (700 MHz, 27°C). (E) 13C-edited methyl NOESY-HMQC experiments show the intermolecular NOEs (700 MHz, 40°C). The methyl-methyl NOEs are indicated by red dashed lines. Diagonal peaks are shown in blue. (F) Electrostatic surface of Ran–Mog1 complex. Red and blue colors denote negative and positive surface charges, respectively. (G) Validation of the complex model using a GST pull-down assay by disrupting the electrostatic interaction through mutations. (H) Reverse mutagenesis recovers the affinity detected by GST pull-down.
Our initial attempt to solve the complex structure was compromised by the difficulty to obtain the corresponding crystal and to solve the structure by conventional NMR due to line broadening caused by conformational exchanges inside the complex (Supplementary Figure S2A and B). The slow conformational exchange in the NMR time scale (Supplementary Figure S2C and D) precludes the application of chemical shift perturbation or the ‘divide and conquer’ strategy to transfer the assignment of apo-form NMR peaks to the corresponding holo-form NMR peaks. To overcome these difficulties, we used methyl-TROSY-based NMR analysis coupled with Ile, Leu, and Val methyl-specific isotope-labeling method to delineate the complex structure of Ran and Mog (Gelis et al., 2007; Rosenzweig et al., 2013). We used site-directed mutagenesis in concert with the intra-molecular methyl-methyl NOE pattern recognition for Mog1 in the apo and holo forms, respectively (Supplementary Figure S3A–C). Although the chemical shifts changed upon complex formation, the ILV NOE pattern remained largely similar. NOE pattern in the apo form of Mog1 was compared with that in the holo form, which allowed the assignment of methyl groups in the complex. The holo form of Ran was assigned similarly by the intra-molecular methyl-methyl NOE patterns retrieved from the experiment and those back-calculated from the crystal structure of Ran (Supplementary Figure S3D–F). The L and V methyl group stereo-selectively labeled Ran–Mog1 allowed us to distinguish the two methyl peaks in Leu and Val at the HMQC spectrum. Using the above interleaving assignment strategy, 89% and 70% of the ILV methyl peaks of Mog1 and Ran were assigned, respectively (Supplementary Figure S3G).
To pinpoint the interface in the complex, two sets of 13C-edited NOESY spectra were acquired with and without 15N-decoupling in the indirect proton dimension, using 15N and ILV methyl group-13C-labeled Mog1 and deuterated Ran, or vice versa. The amide (bonded to 14N) to methyl NOE signals without splitting pattern changes represent the interfacial methyl groups (Figure 1C and D). Furthermore, seven pairs of intermolecular methyl-methyl NOE was identified by 13C-edited NOESY spectra with both proteins ‘ILV’-labeled (Figure 1E). Ran and Mog1 are highly positively and negatively charged around the binding surfaces, respectively (Figure 1F). Site-directed mutagenesis combined with GST pull-down assays demonstrated that most charged residues are responsible for the complex formation (Figure 1G). Then, we utilized reverse mutagenesis to pinpoint pair-wise residues on binding surface (Figure 1H). The C-terminus of Ran was not involved in the interaction with Mog1, as RanΔ181–216 truncated mutant exhibit similar spectra as the full-length one (Supplementary Figure S4).
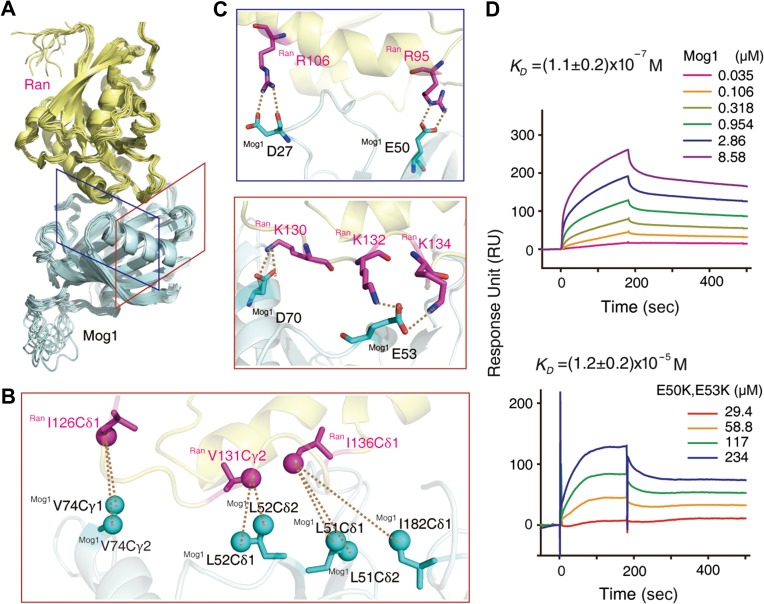
With the aforementioned information (Supplementary Table S2), a RanΔ181–216–Mog1 complex model was generated by using HADDOCK (High Ambiguity Driven protein-protein DOCKing) (de Vries et al., 2010; Wassenaar et al., 2012), resulting in 112 converged structures with an RMSD of backbone atoms 0.76 Å for ordered regions (Supplementary Table S3). The overall structure of the complex model is shown in Figure 2A. The two proteins have a large buried surface area of 1900 Å2. Twenty-one residues of Mog1 and twenty residues of Ran are involved in the contact interface (using a cut-off of 3.5 Å). According to this model, besides intermolecular methyl-methyl distances obtained from 13C-edited NOESY spectra (Figure 2B), electrostatic interactions are the major contributors to complex formation, and several salt bridges (R95Ran–E50Mog1, R106Ran–D27Mog1, K130Ran–D70Mog1, K132Ran–E53Mog1, and K134Ran–E53Mog1) are involved (Figure 2C). These bridges were confirmed by surface plasma resonance (SPR) experiments, in which the binding affinity of mutant Mog1E50K,E53K for Ran was reduced by two orders of magnitude compared with the wild-type [KD = (1.1 ± 0.2) × 10−7 M] (Figure 2D).
Figure 2.
The complex structure of Ran–Mog1. (A) Ensemble of 10 lowest energy HADDOCK structures for the Ran–Mog1 complex. (B and C) The methyl-methyl NOEs and salt bridges at the binding surface (red and blue boxes corresponding to the view perspective indicated in A). (D) SPR-binding assay between immobilized Ran and wild-type Mog1 or Mog1E50K,E53K mutant (n = 3, technical replicates).
Mog1-binding-induced conformational changes of Ran promote its nucleotide release
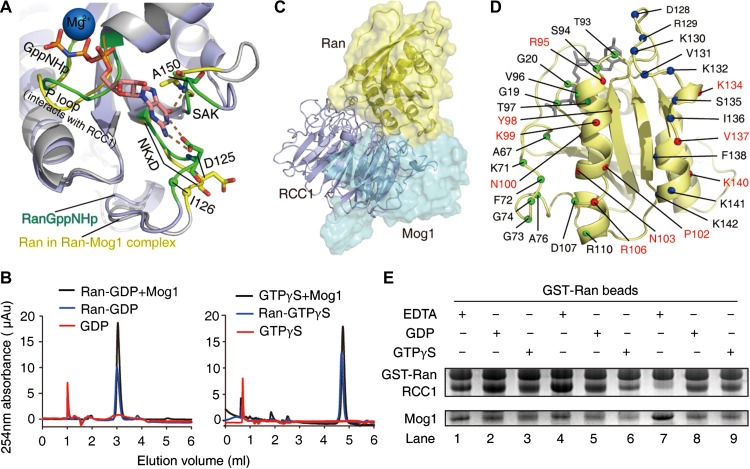
In the complex, there are large conformational flexibilities in switch I, switch II, the end of α3, the β5-α4 loop (residues, 121–133), and the β6-α5 loop (residues, 149–157) of Ran shown by 300 ns molecular dynamic simulations (Supplementary Figure S5). In Ran-GTP and Ran-GDP, the guanine base of GTP/GDP interacts with K123, D125 in the NKxD motif (NKVD, residues 122–125) and A150 in the SAK motif. The distance between methyl groups V124 and V92 is less than 5 Å shown from crystal structures of the Ran-GDP (PDB: 3GJ0) or Ran-GTP (PDB: 1K5D), but we did not find any NOE signal for V124 in the NOESY spectra of the complex. We speculate that a local conformational change of the NKxD motif leads to the movement of V124 from V92 in the complex. I126Ran, one residue downstream of the NKxD motif, shows NOE with Mog1 (Figure 1E). This suggests that the interaction between I126Ran and Mog1 displaces the nucleotide base-binding motif (NKxD), which results in nucleotide release (Figure 3A). A ultra-performance liquid chromatography (UPLC)-based guanine nucleotide release assay validated that Mog1 promotes release of the nucleotide from both Ran-GDP and Ran-GTP (Figure 3B), which demonstrates that Mog1 is a bona fide nucleotide release factor.
Figure 3.
Mog1 and RCC1 compete to bind to Ran. (A) The binding of Mog1 induces conformational changes in D125 (NKxD motif), A150 (SAK motif) (base-binding sites) as indicated by Ran in the Ran–Mog1 complex (yellow) superimposed with Ran-GTP (green, PDB: 1K5D). (B) The GDP/GTP releasing activities of Mog1 analyzed by UPLC. The blue and red lines indicate the nucleotide bound to Ran and Ran–Mog1, respectively. (C) Structural superposition of Ran–RCC1 (PDB: 1I2M) and Ran–Mog1 reveals a steric conflict between RCC1 and Mog1 as noted by a dashed circle. (D) The Ran residues contacted with Mog1 (blue), RCC1 (green), and both (red). (E) Mog1 and RCC1 compete for Ran binding in the presence of GDP/GTPγS as demonstrated by GST pull-down experiment. Lanes 1–3: Mog1 and RCC1 were added into the GST-fused Ran simultaneously. Lanes 4–6: RCC1 was added first. Lanes 7–9: Mog1 was added first.
Competition for Ran binding between RCC1 and Mog1
Our structural analyses show that Ran–Mog1 binding surface is partially overlapped with Ran–RCC1 binding surface in two regions (Figure 3C), from R95Ran to R106Ran and K134Ran to R140Ran (10 residues involved) (Figure 3D), covering an area of approximately 880 Å2, suggesting that Mog1 and RCC1 cannot bind Ran simultaneously. To confirm whether Mog1 competes RCC1 for Ran binding, we carried out pull-down assay in which GTP-Ran was used as an affinity matrix. As shown in Figure 3E, our pull-down assays demonstrate that Mog1 and RCC1 compete for Ran binding in the presence of GTP or GDP. Significantly, prior incubation of Mog1 with Ran, in the absence of GTP or GDP, forms a stable complex that prevents Ran from RCC1 binding (Figure 3E, lane 7). Collectively, these results show that Mog1 competes with RCC1 for Ran binding and hence prevents RCC1-mediated Ran-GTP cycle once Mog1 binds to Ran.
Balanced equilibrium between Mog1 and Ran ensures accurate chromosome segregation
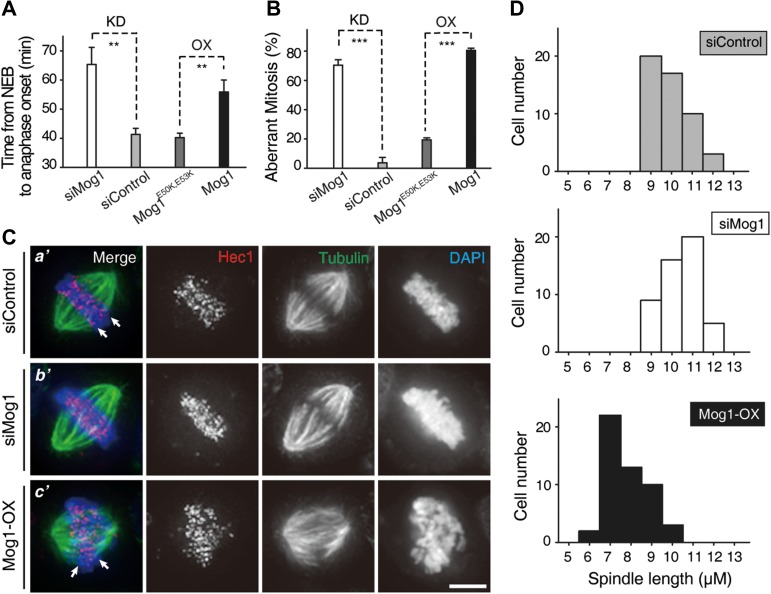
To test whether competition between Mog1 and RCC1 for Ran binding functions in cell division, we examined the impact of Mog1 levels on accurate chromosome segregation. Aliquots of HeLa cells were transfected with either Mog1 siRNA to suppress Mog1 expression or mCerulean-Mog1 construct to achieve an overexpression of Mog1 protein. We next examined whether there are any abnormality seen in Mog1-overexpressing cells. Suppression of Mog1 delays mitotic progression into anaphase (Supplementary Figure S6A) as average Mog1-depleted cells took ~65.4 min to enter anaphase while anaphase onset arrived after an average of ~41.3 min in control cells (Figure 4A). As shown in Figure 4A, chromosome segregation in Mog1-overexpressing cells exhibited a typical delay (~55.9 min versus ~40.2 min). Although most sister chromatids eventually segregated, the quality of mitosis was compromised as multipolar spindle and micronuclei phenotypes were seen in Mog1-overexpressing cells (Supplementary Figure S6B). Statistical analyses showed high ratios of abnormal mitosis when accurate level of Mog1 was perturbed (Figure 4B; ~70.4% for knockdown and ~80.5% for overexpression group, respectively). The perturbation was attenuated in cells expressing Ran-binding-deficient Mog1E50K,E53K mutant, suggesting the importance of fine balance of Ran–Mog1 levels in accurate mitosis. Western blotting analyses showed that the siRNA oligonucleotide caused remarkable suppression of Mog1 protein levels while exogenously expressed Mog1 protein was ~3–4 times of the level of endogenous Mog1 protein (Supplementary Figure S6C).
Figure 4.
Accurate level of Mog1 relative to Ran is essential for faithful chromosome segregation. (A and B) chromosome segregation in live HeLa cells transfected with Mog1 siRNA (scramble siRNA as control) or overexpressing Mog1E50K,E53K and wild-type Mog1 was scored. Statistics of the time from nuclear envelope breakdown (NEB) to anaphase onset (A) and chromosome segregation abnormality (B) in cells with Mog1 depletion (KD) or overexpression (OX) were presented as mean ± SEM. In A: n = 14, siMog1; n = 15, siControl; n = 11, Mog1E50K,E53K; n = 22, Mog1. In B: n = 36, siMog1; n = 24, siControl; n = 37, Mog1E50K,E53K; n = 40, Mog1. n, the cells per condition from three independent experiments. Statistical significance was tested by two-sided t-test; **P < 0.01, ***P < 0.001. (C) Representative phenotypes of HeLa cells transfected with Mog1 siRNA or Mog1 (OX). Scale bar, 5 μm. (D) Histograms of spindle length in control siRNA-treated cells (n = 50), Mog1 siRNA-treated cells (n = 50), and Mog1-overexpressing cells (n = 50).
It has been proposed that the Ran-GTPase controls the spatial distribution of microtubule nucleation and spindle plasticity (Ohba et al., 1999; Wilde and Zheng, 1999). To probe whether alteration of Mog1 protein level affects chromosome movements and spindle plasticity, we carried out immunofluorescent microscopic analyses of the transfected HeLa cells stained for Hec1, tubulin and DNA. As shown in Figure 4C, suppression of Mog1 resulted in hyperstablized spindle microtubule as microtubule bundling was evident (Figure 4C-b’). On the other hand, overexpression of Mog1 resulted in an aberrant kinetochore-microtubule connection and subsequent chromosome misalignment (Figure 4C-c’, arrows). Since Ran-regulated and Tpx2-dependent microtubule nucleation contributes to setting spindle leng (Petry et al., 2013), we next examined the effect of perturbing Mog1 level on spindle length control. Statistical analyses of spindle length show that suppression of Mog1 increased spindle length, while overexpression of Mog1 shortened the length (Figure 4D), consistent with previous results arguing the role of Ran-GTPase activity for spindle assembly and spindle length control (Petry et al., 2013). Thus, we conclude that homeostatic control of Ran–Mog1 level is precisely regulated for accurate mitosis.
Ran is a bona fide substrate of TIP60
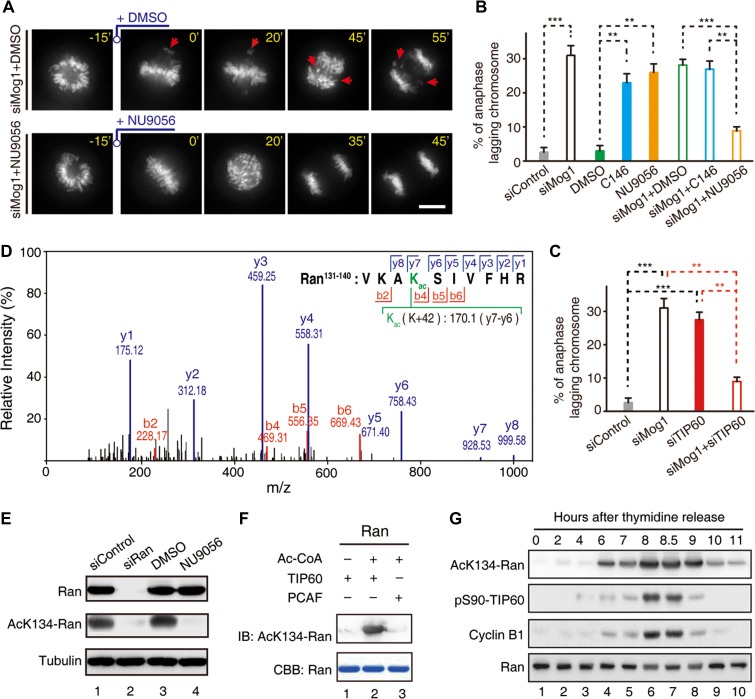
Ran is a substrate for PAK4 and the phosphorylation of Ran at S135 regulates Ran activity and mitosis (Bompard et al., 2010). The critical role of K134Ran in binding to Mog1 prompted us to conduct computational analyses for lysine acetylation (Li et al., 2006), which suggested K134Ran as a potential substrate of TIP60 (Mo et al., 2016). Given our recent demonstration that TIP60-dependent acetylation is critical for accurate chromosome segregation in mitosis (Mo et al., 2016), we postulated that TIP60 may acetylate K134Ran to retain a dynamic Ran–Mog1 interaction for accurate kinetochore-microtubule interactions in mitosis. Specifically, our structural analyses suggest that persistent acetylation of K134Ran would disrupt the Ran–Mog1 interaction. To test this hypothesis, we employed TIP60 inhibitor NU9056 to rescue the aberrant mitosis elicited by Mog1 knockdown. As shown in Figure 5A, NU9056 attenuated chromosome segregation errors seen in Mog1-suppressed cells (upper panel, arrows). Statistical analyses demonstrated that inhibition of TIP60 by NU9056 rescued the anaphase lagging chromosome phenotype resulted from Mog1 suppression (Figure 5B). As a control, PCAF inhibitor C146 did not rescue the above phenotype, suggesting that the rescue is a specific effect of TIP60. Importantly, suppression of TIP60 using siRNA also rescued the phenotype seen in Mog1-suppressed cells (Figure 5C), suggesting that TIP60 regulates homeostatic Ran–Mog1 interaction in mitosis.
Figure 5.
Ran is a novel substrate of TIP60 in mitotic cells. (A) Real-time mitosis in Mog1-suppressed cells was scored as a function of TIP60 inhibition by NU9056. Scale bar, 10 μm. (B and C) Statistics of the chromosome segregation abnormality in the presence of TIP60 inhibitor (NU9056) or PCAF inhibitor (C146) (B) and in cells transfected with TIP60 siRNA (C) (mean ± SEM, n = 62, siControl; n = 36, siMog1; n = 55, DMSO; n = 39, C146; n = 38, NU9056; n = 28, siMog1 + DMSO; n = 26, siMog1 + C146; n = 35, siMog1 + NU9056; n = 33, siTIP60; n = 35, siMog1 + siTIP60). Statistical significance was tested by two-sided t-test; **P < 0.01, ***P < 0.001. (D) Representative mass spectra of acK134-Ran from Ran immunoprecipitates isolated from mitotic HeLa cells. (E) Characterization of affinity-purified acK134-Ran antibody. Aliquots of HeLa cells were transfected with Ran siRNA followed by nocodazole synchronization or treated with TIP60 inhibitor NU9056. Cells were harvested and lysed in RIPA buffer and the acetylation level of Ran was analyzed by western blot. Note that the reactivity of acK134-Ran antibody was abolished either by treatment with TIP60 inhibitor NU9056 or siRNA-mediated Ran suppression which demonstrates the specificity of acK134-Ran antibody. (F) TIP60 or PCAF was incubated with Ran in the presence of Ac-CoA for in vitro acetylation assay. The acetylation level of Ran was analyzed by anti-acK134-Ran antibody. (G) Temporal profile of cyclin B1 accumulation, Ran acetylation (acK134-Ran), and TIP60 phosphorylation (pS90-TIP60).
To ascertain whether Ran is a bona fide substrate of TIP60, we isolated Ran from mitotic HeLa cells treated with or without NU9056 (Supplementary Figure S7A). Mass spectrometric analyses showed that K134Ran is acetylated in mitosis and the acetylation is responsive to NU9056 (Figure 5D). To assess whether Ran is a cognate substrate of TIP60 in mitosis, we generated an antibody to acK134-Ran and analyzed K134Ran acetylation in aliquots of nocodazole-synchronized HeLa cells (Figure 5E). Anti-acK134-Ran specifically recognized endogenously acetylated Ran (Figure 5E, lanes 1 and 3) and K134 acetylation was dramatically reduced after Ran suppression by siRNA (Figure 5E, lane 2) or TIP60 inhibition by NU9056 (Figure 5E, lane 4), indicating that K134Ran is a cognate substrate of TIP60 in mitotic cells. Next, we sought to determine whether TIP60 acetylates Ran in vitro. Anti-acK134-Ran blot established that Ran is a substrate of TIP60 (Figure 5F, lane 2) but not PCAF (Figure 5F, lane 3). Since phosphorylation of TIP60 by Cdk1 promotes accurate chromosome segregation in mitosis, we sought to determine the temporal dynamics of acK134-Ran and pS90-TIP60 levels during cell cycle by collecting synchronized HeLa cells at indicated time intervals after release from the G1/S phase for western blotting analyses (Figure 5G). The temporal dynamics of acK134-Ran were similar to those of pS90-TIP60, demonstrating that acK134-Ran was temporally regulated by TIP60 acetyltransferase in a cell cycle-dependent manner. These studies suggest that Cdk1-elicited robust TIP60 activity is likely coupled to Ran GTPase activity in mitotic chromosome segregation.
K134 acetylation switches Ran from Mog1-binding to RCC1-binding for accurate mitosis
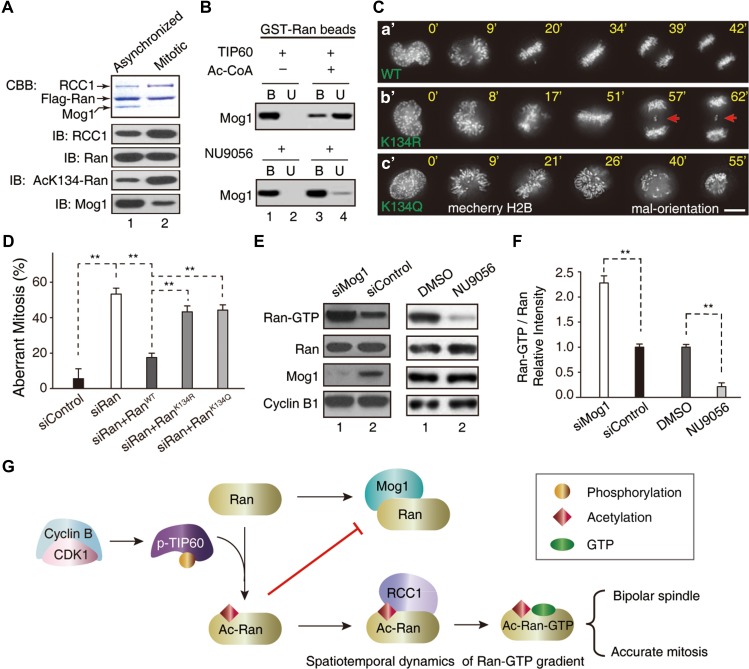
To examine whether TIP60-elicited acetylation of K134 switches Ran from binding to Mog1 to RCC1, we carried out FLAG-Ran immunoprecipitation from mitotic and asynchronized HeLa cells. As shown in Figure 6A, western blotting analyses confirmed that acK134 level of FLAG-Ran from mitotic cells is ~4-fold higher than that of asynchronized cells (fourth panel) under equal protein level of Ran (third panel). As predicted, RCC1 level is higher in FLAG-Ran immunoprecipitates from mitotic cells (second panel). Conversely, the level of Mog1 is higher in FLAG-Ran immunoprecipitates from asynchronized cells (bottom panel). To directly assess the impact of acetylation of K134Ran on Mog1 binding, we conducted a pull-down assay in which aliquots of GST-Ran affinity beads were first incubated with TIP60 in the presence or absence of Ac-CoA followed by addition of equal molar of recombinant Mog1 protein (Figure 6B). After an incubation of 2 h, GST-Ran beads-bound materials (B) were separated from unbound solution (U) by a brief centrifugation. As shown in Figure 6B (upper panel), almost all Mog1 was retained on GST-Ran-beads (lane 1). However, addition of Ac-CoA shifted Mog1 from GST-Ran beads-bound to unbound (upper panel, lanes 3 and 4). Importantly, TIP60 inhibitor NU9056 retained Mog1 bound to GST-Ran (lower panel). Thus, we conclude that acetylation of K134Ran attenuates Ran binding to Mog1 and promotes Ran binding to RCC1. The acetylation of K134Ran in mitosis prompted us to examine its functional relevance in chromosome segregation. To this end, aliquots of siRNA-resistant GFP-Ran constructs (wild-type, acetylation-mimicking K134Q, and non-acetylatable K134R) were transiently transfected to express in HeLa cells depleted of endogenous Ran. Western blotting analysis showed that exogenously expressed Ran proteins were about twice the level of endogenous Ran (Supplementary Figure S7B). As expected, expression of exogenous GFP-RanWT enabled an accurate chromosome segregation in endogenous Ran-suppressed cells (Figure 6C-a’). However, expression of RanK134R failed to restore an accurate mitosis but resulted in chromosome bridges phenotype (Figure 6C-b’; arrows). Surprisingly, persistent expression of RanK134Q failed to rescue the mitotic defects seen in Ran-suppressed cells (Figure 6C-c’). The RanK134Q-expressing cells exhibited perturbed spindle plasticity as the mal-orientation of mitotic spindle was observed. It is possible that RanK134Q did not fully mimic the dynamic properties of acetylation of Ran. Statistical analyses of transfected cells from three independent experiments show that RanWT but not RanK134Q or RanK134R restored accurate mitosis in Ran-suppressed cells (Figure 6D). This phenomenon suggests that dynamic acetylation of Ran is coupled to the fine-tuning of Ran-GTP cycle which is essential for accurate chromosome segregation and mitotic spindle length control.
Figure 6.
Acetylation of Ran by TIP60 switches Ran from Mog1-binding to RCC1-binding. (A) Flag-Ran immunoprecipitates from mitotic and asynchronized HeLa cells were analyzed by western blot with indicated antibodies. (B) Purified GST-Ran was incubated with TIP60 in the presence or absence of Ac-CoA or TIP60 inhibitor (NU9056) followed by addition of Mog1. The binding between Ran and Mog1 was analyzed by western blot. (C) Real-time imaging shows that expression of non-acetylatable RanK134R resulted in chromosome segregation error (b’, arrows). Scale bar, 10 μm. (D) Statistical analyses of efficiency of exogenously expressed siRNA-resistant RanWT, RanK134Q, and RanK134R in rescuing the mitotic defects in Ran-suppressed cells (mean ± SEM, n = 16, siControl; n = 13, siRan; n = 18, siRan + RanWT; n = 18, siRan + RanK134R; n = 16, siRan + RanK134Q, **P < 0.01). (E and F) Suppression of Mog1 promoted the level of Ran-GTP and inhibition of TIP60 reduced the level of Ran-GTP in mitosis. Western blotting analyses of Ran-GTP levels in Mog1 suppressed and TIP60-inhibited HeLa cells in mitosis, respectively (**P < 0.01). (G) Working model accounting for acetylation-elicited tuning Ran effector binding for chromosome segregation.
TIP60-elicited K134 acetylation forms a link between Ran effector switch and dynamic Ran-GTP cycle
To probe whether Ran-GTP level is an accurate reporter of TIP60-perturbed Ran–Mog1 interaction in mitosis, aliquots of HeLa cells depleted Mog1 and synchronized in mitosis were assessed for Ran-GTP level using western blotting analyses. In addition, another aliquots of mitotic HeLa cells were treated with TIP60 inhibitor NU9056 followed by western blotting analyses. As shown in Figure 6E, western blots with antibodies to Ran, Ran-GTP, and Mog1 established that suppression of Mog1 increased Ran-GTP levels without alteration of Ran protein level (left panels, lane 1). On the other hand, chemical inhibition of TIP60 by NU9056 promoted a stable Ran–Mog1 association which subsequently suppressed RCC1-mediated nucleotide loading and reduced the level of Ran-GTP (right panels, lane 2). Quantitative analyses, shown in Figure 6F, demonstrate that homeostatic level of Ran-GTP is governed by TIP60 as chemical inhibition of TIP60 acetyltransferase activity minimizes Ran-GTP level in mitosis. These findings suggest that the dynamics of Ran-GTP level senses the status of Ran–Mog1 interaction in human cells and TIP60 is an essential homeostat for fine-tuning of local Ran-GTPase at the spindle in mitosis.
Discussion
Mitotic spindle is a specialized apparatus governing genome stability during mitosis by orchestrating accurate chromosome-microtubule attachments and spindle assembly checkpoint execution. Acetylation of Ran by TIP60 therefore provided hierarchical regulation of Ran GTPase gradient for spindle plasticity control and sensing/correcting errors in kinetochore attachment before sister chromatid separation. Our identification of the TIP60–Ran–Mog1 signaling axis uncovered a new regulatory mechanism by which acetylation of Ran prevented Mog1-sequestrated nucleotide loading to enhance the Ran-GTPase activity for linking spindle microtubule length control to accurate cell division (Figure 6G). It would be of great interest, in follow-up studies, to characterize additional substrates of TIP60 in mitotic cells and delineate their precise molecular function in mitosis. Together with our recent identification of CDK1-TIP60-Aurora B axis underlying kinetochore sensing of aberrant microtubule attachment, the present study highlighted how an acetylation-regulated, reversible GTPase cycle in the sensing and correcting chromosome attachment errors during mitotic progression maintains genomic stability.
Our results suggest that there is a dynamic and complex interaction network that controls spatiotemporal dynamics of Ran-GTP in mitosis. In contrast to the situation for Xenopus egg extracts, once a bipolar spindle is established in cells, the Ran-GTP gradient and the Ran-importin-β cargo regulation appears to be largely dispensable for spindle plasticity control. This indicates that in cells, mitotic spindles are built and maintained by multiple, parallel pathways, and demonstrates that kinetochore- and chromatin-driven systems differentially use Ran-GTP to promote mitotic spindle assembly and plasticity control. On the basis of our results, we propose that the mitotic cytoplasm operates near a physiological threshold in which positive and negative regulators are at equilibrium. Such a system would be poised to break the threshold in response to fine local changes in Ran-GTP concentration and, for example, influence microtubule stability around chromatin in prophase and prometaphase cells. This behavior may allow the Ran-importin-β pathway to locally regulate its targets and to signal both chromatin- and centrosome-driven events in mitosis. We propose that acetylation of Ran could confer protection to GTP-bound Ran against Ran-GAP-elicited hydrolysis. It has been proposed that the Ran gradient controls the spatial distribution of microtubule nucleation. However, recent study argued that both Ran gradient-dependent and independent mechanism contribute to efficient spindle length control in mitosis (Oh et al., 2016). Our analyses delineate the structural determinants underlying the competitive binding of Mog1 and RCC1 to Ran. It is worth noting that the acetylation of K134 perturbs the salt bridge (K134Ran and E53Mog1), critical for Ran–Mog1 interaction (Figure 2C), which provides a homeostatic switch to enable Ran–RCC1 complex formation. Although our attempt to delineate the mechanism of action underlying phenotypic changes seen in Figure 6C using a co-immunoprecipitation assay with endogenous Mog1 and exogenously expressed RanK134R or RanK134Q did not prevail, the in vitro pull-down assay shown in Figure 6B provided direct and conclusive evidence that the physical binding between Mog1 and Ran is inhibited by TIP60 acetylation. Interestingly, acetylation of K134 did not affect RCC1–Ran interaction as K134 is located to the periphery of the physical contact of Ran–RCC1, which is consistent with literature (Renault et al., 2001; de Boor et al., 2015). To this end, acetylation-elicited switch of Ran effectors provides a conceptual novel model for context-dependent regulation of Ran-GTP level and spatiotemporal dynamics in mitosis. Thus, our structure-functional favors a working model in which a diffusible gradient of Ran-GTP emanating from the chromosomes, together with the centromere localization of TIP60, is critical to execute Ran function at the kinetochore–microtubule and spindle geometry in mitosis. It would be of great interest, in follow-up studies, to quantify the spatiotemporal dynamics of TIP60-elicited acetylation relative to Ran GTPase gradient in dividing cells using fluorescence resonance energy transfer-based sensors and delineate their precise molecular function in mitosis (Chu et al., 2012; Laviv et al., 2016).
In sum, our results establish a novel mechanism of how Mog1 competes with RCC1 for Ran binding and Ran-GTP loading is dynamically regulated by acetylation in mitosis (Figure 6G). Mog1 directly binds to Ran to create steric hindrance for RCC1 binding and further prevents persistently high level of Ran-GTP around chromosomes during mitosis. TIP60-elicited acetylation of Lys134 switches Ran from Mog1-binding to RCC1-binding, which is critical for spatial control of Ran-GTP gradients that guide the spindle geometry during mitosis. Our findings provide structural delineation of molecular mechanism by which post-translational modifications could yield sharp transitions between functionally distinct Ran-GTPase states, generating nonlinearity in Ran signaling cascades, and contributing to the spindle plasticity control during cell division cycle. It is worth noting that TIP60 localization depends on Ndc80 (Mo et al., 2016), and TIP60-elicited acetylation of Ran would provide centromere-based spatial control of Ran-GTP gradient centered on kinetochore (Figure 6G). Thus, our study points to an intricate cross-talk among Cdk1, TIP60, Ndc80, and Ran-GTPase in sensing kinetochore-microtubule attachment. On one hand, kinetochore-based Ran-GTP gradient promotes Tpx2-elicited local microtubule branching and perhaps kinetochore capture (Petry et al., 2013). On the other hand, kinetochore-based end-on microtubules compete with Mps1 for Ndc80 (Hiruma et al., 2015; Ji et al., 2015). Thus, the evolutionarily conserved TIP60–Ran–Mog1 axis may serve as a novel sensor of kinetochore-spindle attachment for accurate mitosis from yeast to human.
Materials and methods
Plasmid construction
The DNA fragments encoding full-length Ran and Mog1 were amplified by PCR from a human brain cDNA library and cloned into pET22b (+) (Novagen) vectors (for NMR and SPR) or pGEX-4T1 vectors (for GST pull-down).
To obtain a stable complex of Ran–Mog1 for NMR experiments, we co-expressed the proteins in a pRSFDuet-1 plasmid, which contained Mog1 without a tag and Ran with a His6 tag at the N-terminus. To investigate the role of Mog1 interacting with the Ran C-terminus (residue 181–216), which is disordered in complex with other cofactors, the Ran C-terminus was deleted by mutation in the pRSFDuet-1 and pGEX-4T1 plasmids.
For immunofluorescence and living cell imaging, the cDNA encoding fluorescent proteins (mCerulean and GFP) and Mog1 were cloned into pcDNA3.1(+)/myc/his B (Novagen) vector at the N- and C-terminus, respectively. All of Mog1 mutants were generated by the MutanBESTkit (TaKaRa). EGFP-tagged siRNA-resistant Ran wild-type and site-specific mutants were generated by PCR-based, site-directed mutagenesis kit from Vazyme (C212) according to the manufacturer’s instructions.
All plasmids were verified by DNA sequencing (Invitrogen).
Expression, purification, and uniform isotope-labeling of Mog1 and Ran
The Mog1 and Ran constructs were transformed into E.coli BL21 (DE3) Gold cells (Novagen) and induced with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 37°C for 6 h and 27°C for 12 h, respectively. The uniformly 15N-,13C- and 2H-labeled recombinant protein was produced by growing the bacteria in LR medium prepared with 99.9% D2O, containing 0.5 g/L 15NH4Cl and 2.5 g/L 13C6-glucose. The expressed proteins with His6-tag or GST-tag were purified by nickel-chelating column (Qiagen) or glutathione-Sepharose (GE Healthcare), followed by size-exclusion chromatography on a Hiload16/60 Superdex 75 or 200 column (GE Healthcare), respectively. The purity of the protein was confirmed by SDS-PAGE, and the concentration was determined photometrically.
To obtain Mog1 or the Ran–Mog1 complex with specific isoleucine, valine, and leucine residues that were 13CH3-labeled at only one methyl position arbitrarily, with the other methyl group 12CD3-labeled,α-ketoisovaleric acid [3-methyl-13C, 3,4,4,4-d4, sodium salt] and α-ketobutyric acid [methyl-13C,3,3-d2, sodium salt] precursors (Cambridge Isotope Laboratories) were added to a uniformly 15N-labeled LR medium into D2O with D-Glucose-d7. α-ketobutyric acid [methyl-13C, 3,3-d2, sodium salt] precursors can be obtained from α-ketobutyric acid [methyl-13C, sodium salt] by incubation at high pH in D2O before use, following the procedure of Tugarinov and Kay (2004).
Because Ran expression is unstable in D2O at 37°C, it must be co-expressed with Mog1.To obtain NOESY-HSQC spectra with a 15N-coupled or 15N-decoupled sample, a refolding procedure was utilized. Ran (fused with a His6 tag) and Mog1 (without a His6 tag) were co-expressed using the methyl labeling procedure. The complex was denatured in 8 M urea and flowed through a nickel-chelating column to separate the two proteins. Then, denatured U-[15N, 2H], Ileδ1-[13CH3], Leu, Val-[13CH3, 12CD3]-labeled Ran and U-[2H]-labeled Mog1 were mixed for refolding.
Solution structure of Mog1 and structure calculation
The 13C, 15N-labeled Mog1 was dissolved to a final concentration of 0.9 mM in buffer I (20 mM NaH2PO4, 100 mM NaCl, 5 mM EDTA and 1 mM DTT, 10% D2O, pH6.8). The NMR spectra were acquired at 30°C using a Bruker DMX600 spectrometer equipped with the cryoprobe. To obtain backbone and side chain resonance assignments, the following spectra were recorded: 2D 15N-1H HSQC, 2D 13C-1H HSQC, 2D NOESY, 2D TOCSY, 3D triple-resonance spectra HNCO, HN(CA)CO, CBCA(CO)NH, CBCANH, HNCA, HN(CO)CA, C(CO)NH-TOCSY, H(CCO)NH-TOCSY, HCCH-TOCSY, HCCH-COSY, HBHA(CBCACO)NH, and 3D 15N-separated and 13C-separated NOESY. The Mog1 backbone RDCs were determined in 29 mg/ml Pf1 phage (Hansen et al., 1998). All NMR data were processed by NMRPipe and NMRDraw software (Delaglio et al., 1995) and assigned with Sparky 3 (T.D. Goddard and D.G. Kneller, University of California, San Francisco). HNCO and HN(CO)CACB spectra of the deuterated Ran–Mog1 complex was recorded at 27°C using an Agilent 700 MHz NMR spectrometer with a cryogenic probe.
NOE distance restraints were obtained from 3D 15N-edited NOESY and 3D 13C-edited NOESY spectra. Backbone dihedral angles (φ and ψ) in secondary structures were derived from the analysis of 13Cα, 13Cβ, 13CO, 1Hα, and 15N chemical shifts by TALOS (Cornilescu et al., 1999). The structures were calculated with a simulated annealing protocol using the CNS v1.1 program (Brunger et al., 1998). In the final calculations, 200 structures were generated, from which 20 models with the lowest energies were selected to form the representative ensemble. Ramachandran plot statistics were evaluated with the program PROCHECK (Laskowski et al., 1996). Molecular illustrations are prepared with Pymol (http://pymol.org/) and MOLMOL (Koradi et al., 1996).
Chemical shift assignments for ILV methyl groups in Ran–Mog1 complex
The 2D spectra of protein samples (wide type, mutants and stereo-selective labeled samples) with a concentration of 0.1 mM were recorded in buffer II (20 mM NaH2PO4, 150 mM NaCl, 2 mM EDTA, 2 mM DTT, 10% D2O, pH6.25) at 40°C. The stereo-selective labeling for δ2/γ2 methyl groups was used to distinguish the δ1/γ1 and δ2/γ2 methyl groups in Leu and Val (www.nmr-bio.com) (Gans et al., 2010).
The 13C-edited 1H-1H nuclear overhauser effect (NOE) spectra of the complex was recorded with 0.5 mM perdeuterated ILV-labeled Ran–Mog1 complex solved in buffer III (20 mM NaH2PO4, 150 mM NaCl, 2 mM EDTA, 2 mM DTT, 100% D2O, pH6.25) at 40°C. NMR spectra were collected using an Agilent 700 MHz NMR spectrometer with a cryogenic probe.
Ran–Mog1 binding interface determination and intermolecular NOE measurement
To delineate the Ran–Mog1 binding interface, two sets of 13C-edited NOESY-HSQC spectra with or without 15N-decoupling in the t1 dimension were recorded using the same sample (the complex of U-[15N, 2H], Ileδ1-[13CH3], Leu, Val-[13CH3, 12CD3]-labeled Mog1 and U-[2H]-labeled Ran or the complex of U-[15N, 2H], Ileδ1-[13CH3], Leu, Val-[13CH3, 12CD3]-labeled Ran and U-[2H]-labeled Mog1).
The intermolecular methyl-methyl NOEs were retrieved from methyl NOESY-HMQC experiments with both proteins ‘ILV’-labeled.
Generation of the Ran–Mog1 complex model
Since the C-terminus of Ran is not involved in the interaction with Mog1, the RanΔ181–216 truncated mutant (PDB: 1K5D with GTP removed) and Mog1 were used in model building. Two rounds of docking were performed for the complex using the HADDOCK web server. The initial structures for docking were Mog1 from the lowest energy structure of the NMR ensemble and RanΔ181–216 truncated from the crystal structures (PDB: 1K5D). The first run only used seven intermolecular methyl-methyl NOEs and four intra-molecular methyl-methyl NOEs in Mog1. In the second docking run, five interacted residue pairs were added as unambiguous restraints which had been verified by reverse mutagenesis. The nine active residues corresponding to those identified at the interface were selected based on site-directed mutagenesis and GST pull-down according to the first docking run. The residues that show intermolecular methyl-methyl NOE in the complex were defined as the passive residues. The semi-flexible residues were defined automatically by analyzing intermolecular contacts (<5.0 Å). The intrinsically flexible segments were set to be fully flexible.
Starting from 5 trials of rigid body minimization, 8000 solutions were calculated and ranked according to their HADDOCK scores. The best 200 of 8000 solutions were selected and subjected to a semi-flexible simulated annealing refinement protocol in torsion angle space and further refined in explicit solvent. A cluster analysis was performed on the final ensemble of 200 solutions with a 2 Å cut-off and a minimum of 20 cluster members. 94.5% of the structures were grouped into 1 cluster.
Molecular dynamics simulation of the Ran–Mog1 complex model
A 300 ns molecular dynamics (MD) simulation was performed for the RanΔ181–216–Mog1 complex from the haddock model, during which the interface was constrained. Simulations were performed with a parallel implementation of the GROMACS package (Hess et al., 2008), version 4.5.5, using the Amber03 force field (MacKerell et al., 1998). The complex was solvated in a rhombic dodecahedron box of TIP3P water molecules (Jorgensen et al., 1983) with periodic boundary condition. The minimum distance between the solute and the box boundary was 12 Å. The energy of the system was minimized by the steepest descent method, until the maximum force was smaller than 1000 kJ·mol−1nm−1. One Na+ ion was added to neutralize the system. The solvated complex was subjected to energy minimization using the steepest descent and conjugate gradient algorithms, respectively, until the maximum force on any atom was smaller than 200 kJ·mol−1nm−1. A 100 ps equilibration simulation with positional restraints was performed using a force constant of 1000 kJ·mol−1nm−1. Initial atomic velocities were generated according to a Maxwell distribution at 310 K. The MD simulation was run using the Verlet integration scheme (Hockney et al., 1974) with a time step of 2 fs and the NPT ensemble (Berendsen et al., 1984). The pressure was maintained at 1 bar with a relaxation time of 0.5 ps, and the compressibility was 4.5 × 10−5 bar−1. The protein, solvent and ions were coupled separately to a temperature bath of 310 K using a velocity rescaling thermostat (Bussi et al., 2007) with a relaxation time of 0.1 ps. Covalent bonds were constrained using the LINCS algorithm (Hess, 2008), while the twin-range cut-off distances for van der Waals interactions were set to 0.9 and 1.4 nm, respectively. The long-range electrostatic interactions were treated by the PME algorithm (Essmann et al., 1995) with a tolerance of 1 × 10−5 and an interpolation order of 4.
GST pull-down assay
Aliquots of 15 μg of GST-tagged Mog1 or Ran (wild-type or mutants) and GST (control) bound glutathione-Sepharose 4B beads (GE Healthcare) in PBS were incubated at 4°C for 2 h followed by washing four times with 800 μl of PBS. Then, 15 μg Ran or Mog1 (wild-type or mutants) was added into the resulting beads, and the binding reactions were incubated for 2 h at 4°C. After five washes with washing buffer (PBS with 0.1% Triton X-100), 10 μl of each sample was subjected to SDS-PAGE and stained with Coomassie blue. ImageJ (NIH) was used to quantify the intensity of the bands (normalized to wild-type).
In Mog1 and RCC1 competition assays, GST-fused Ran was treated with 10 mM EDTA to obtain nucleotide-free Ran, then 10 MgCl2 and 10 mM GTPγS or GDP was added to obtain GTP- or GDP-bound form Ran. The sample was washed and maintained in EDTA (PBS and 2 mM EDTA), GTPγS (PBS, 2 mM GTPγS, and 2 mM MgCl2), GDP (PBS, 2 mM GDP, and 2 mM MgCl2) buffer during the experiments. Then, Mog1, RCC1 or both was added to GST-fused Ran. The samples were incubated for 2 h at 4°C. After five times washing, the second protein was added to the reaction tubes and incubated for another 2 h at 4°C. The samples were washed for another five times, and then analyzed by SDS-PAGE.
Surface plasmon resonance
Ran protein was immobilized on a CM5 sensor chip (Biacore) using the Amine Coupling Kit (Biacore). The surface of the sensor chip was activated with 70 μl EDC/NHS containing 100 mM N-ethyl-N0-(dimethyl-aminopropyl)-carbodimide-hydrochloride and 400 mM (N-hydroxysuccinimide) using a flow rate of 10 μl/min. Ran protein was diluted in 10 mM sodium acetate pH4.0 at a concentration of 240 μg/ml. Subsequently, the sensor chip was deactivated with 70 μl of 1 M ethanolamine hydrochloride at pH8.5 (flow rate: 10 μl/min), and PBS flowed for 5 min. Binding analysis were performed with multiple injections of solutions of different protein concentration over the immobilized surfaces at 25°C for 3 min at a flow rate of 30 μl/min. The equilibrium association constant KD was calculated using the BIAevaluation 3.0 software.
Ultra-performance liquid chromatography
Purified Ran was incubated with a 10-fold excess of GDP or GTPγS and 10 mM MgCl2 at 4°C overnight with subsequent purification from the unbound nucleotide by gel filtration (buffer composed of 50 mM NaH2PO4, 200 mM NaCl, 5 mM MgCl2, pH8.0). To validate the nucleotide bound to Ran, the nucleotide-bound Ran-GTP and Ran-GDP collected from gel filtration were then heated at 100°C for 10 min, subsequently cooled on ice, and centrifuged at 14000× g for 10 min to denature protein and to release the Ran-bound nucleotide to supernatants. Then the analysis was performed on a C18 reversed phase column using 93% buffer A [100 mM KH2PO4, 10 mM tetra-n-butylammonium tribromide (TBAB) at pH6.5] and 7% buffer B (10% methanol) with GDP/GTPγS as a control.
To study the Ran-bound nucleotide release role of Mog1, Ran-GTP or Ran-GDP was mixed with Mog1 at a 1:1 molar ratio followed by gel filtration to obtain the complex. To validate the nucleotide bound to the complex, the complex sample was analyzed as described above using UPLC.
Cell culture, synchronization, and transfection
HeLa cells, from American Tissue Culture Collection, were maintained as subconfluent monolayers in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) with 10% (v/v) fetal bovine serum (FBS; Hyclone) and 100 IU/ml penicillin plus 100 mg/ml streptomycin (Gibco) at 37°C with 5% CO2. For cell synchronization, aliquots of HeLa cells were synchronized at G1/S with 2.5 mM thymidine (Sigma-Aldrich) for 16 h, washed with PBS three times, and then cultured in thymidine-free medium for appropriate time intervals.
All the siRNAs or constructs were transfected into HeLa cells with Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol.
Antibodies
In western blotting analyses to detect proteins expression level, the following antibodies were used: rabbit anti-Mog1 antibody (sc-292432, Santa Cruz), mouse anti-Ran antibody (sc-58467, Santa Cruz), mouse anti-RCC1 antibody (sc-374325, Santa Cruz), mouse anti-cyclin B1 antibody (554177, BD Pharmingen), mouse anti-α-Tubulin antibody (T9026, clone DM1A, Sigma-Aldrich), mouse anti-active Ran (Ran-GTP) antibody (26915, NewEast Biosciences). Rabbit anti-pS90-TIP60 was used as described previously (Mo et al., 2016). Rabbit anti-acK134-Ran antibody was generated by YenZym LLC. To generate anti-acK134-Ran antibody, peptide containing acetylated K134 (C-KDRKVKA-acK-SIVFHR; synthesized by YenZym, LLC) was conjugated to rabbit albumin (Sigma-Aldrich) and immunized into rabbits as previously described (Yao et al., 1997). The serum was collected and pre-absorbed by unacetylated Ran peptide (C-KDRKVKA-K-SIVFHR) followed by affinity-purification using (C-KDRKVKA-acK-SIVFHR)-conjugated sulftone sepharose beads (Sigma-Aldrich).
In immunofluorescence assay, the following antibodies were used: mouse anti-Hec1 (ab3613, Abcam) and rabbit anti-tubulin (2871-1, Epitomics). Secondary antibodies were purchased from Jackson ImmunoResearch.
RNA interference and inhibitor treatment
Gene silencing by small interfering RNA (siRNA) was conducted with control (scrambled siRNA), Ran siRNA (5′-GAAAUUCGGUGGACUGAGAUU-3′), or the Mog1-targeted siRNA: mixture of Mog1-1 (5′-CCCUUCACGAUCCUAACAUTT-3′) and Mog1-2 (5′-AUGUUAGGAUCGUGAAGGGTT-3′). All the siRNAs were synthesized from GenePharma. Previously described siRNA duplex was used to repress TIP60 (Cheng et al., 2008).
NU9056 was from Tocris Bioscience and used at 20 μM. C146 was from Sigma and used at 1 μM. Nocodazole (100 ng/ml), NAM (10 mM) and TSA (10 μM) were from Sigma. The protease inhibitors cocktail was from Sigma-Aldrich.
Immunofluorescence and live cell imaging
HeLa cells transfected with Mog1 siRNA or mCerulean-Mog1 were fixed using PTEM buffer (60 mM PIPES, pH6.8, 10 mM EGTA, 2 mM MgCl2, 0.2% Triton X-100) supplemented with 3.7% paraformaldehyde. After blocking with 1% bovine serum albumin (Sigma-Aldrich) in PBST (PBS with 0.05% Tween-20) buffer for 45 min at room temperature, the fixed cells were incubated with primary antibodies in a humidified chamber for 1 h followed by secondary antibodies for 1 h at room temperature. The DNA was stained with DAPI (Sigma-Aldrich). Images were acquired by DeltaVision softWoRx software (Applied Precision) and processed by deconvolution and z-stack projection.
For live cell imaging, HeLa cells were cultured in glass-bottom culture dishes (MatTek) and maintained in CO2-independent media (Gibco) supplemented with 10% (v/v) FBS and 2 mM glutamine (Ding et al., 2010). During imaging, the dishes were placed in a sealed chamber at 37°C. Images of living cells were taken with a DeltaVision microscopy system (Applied Precision Inc.). Image processing was performed with SoftWoRx (Applied Precision Inc.). To trace chromosomes in mitosis, frames were collected at 3–5 min intervals. Images were prepared for publication using Adobe Photoshop software.
Immunoprecipitation and western blot
Cells transfected with FLAG-Ran were trypsinized and lysed in lysis buffer (50 mM Tris-HCl, pH8.0, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitor cocktail (Sigma-Aldrich). The FLAG-M2 resin was added to the lysates and incubated for 4 h before washing. Four hours later, the binding fraction was washed with lysis buffer five times and analyzed by western blot. Samples were subjected to SDS-PAGE and transferred onto nitrocellulose membrane. Proteins were probed by appropriate primary and secondary antibodies and detected using ECL. The band intensity was then quantified using ImageJ (NIH).
In vitro acetylation assay
The acetylation reaction was performed essentially as previously described (Xia et al., 2012; Mo et al., 2016). Basically, purified TIP60 (Mo et al., 2016) or PCAF (Xia et al., 2012) was incubated with GST-Ran in HAT buffer (20 mM Tris-HCl, pH8.0, 10% glycerol, 100 mM NaCl, 1 mM DTT, 1 mM EDTA; 10 μM TSA, 10 mM NAM) containing 100 μM acetyl-CoA for 2 h at 30°C. The reaction was stopped by addition of 5× sample buffer and heated at 95°C for 5 min before being resolved by SDS-PAGE and immunoblotted with indicated antibodies.
Characterization of Ran–Mog1 interaction regulated by TIP60
GST-Ran affinity beads were incubated with purified TIP60 in HAT buffer (20 mM Tris-HCl, pH8.0, 10% glycerol, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, 10 μM TSA, 10 mM NAM) containing 20 μM NU9056 in the presence or absence of 100 μM acetyl-CoA for 1.5 h at 30°C as previously described (Mo et al., 2016). Equal molar of recombinant Mog1 protein was added into the reaction. After another incubation of 30 min, GST-Ran beads-bound materials (B) were separated from unbound solution (U) by a brief centrifugation. The reaction was stopped by addition of 5× sample buffer and heated at 95°C for 5 min before being resolved by SDS-PAGE and immunoblotted with Mog1.
Immunoprecipitation and mass spectrometry
For immunoprecipitation, mitotic HeLa cells were treated with indicated inhibitors before being harvested by mitotic shake-off and lysed in lysis buffer (50 mM Tris-HCl, pH8.0, 120 mM NaCl, 0.2% NP-40) supplemented with protease inhibitor cocktail (Sigma-Aldrich). After pre-clearing with Protein A/G resin (Pierce Chemicals, Inc.), aliquots of lysate were incubated with Ran peptide antibody at 4°C for 4 h with gentle rotation. Aliquots of Protein A/G resin were then added to the lysates followed by additional incubation of 1 h. The Ran-bound Protein A/G resin was then washed three times with lysis buffer plus twice with lysis buffer without NP-40. The Protein A/G matrix-bound Ran protein was then eluted with Ran peptide (150 μg/ml; five volume of Protein A/G matrix) in lysis buffer. The pooled elution fractions were subjected to SDS-PAGE analyses and proteins were visualized by Coomassie blue staining.
Protein samples from immunoprecipitations were digested by trypsin (Promega) and then using StageTip C18 desalting. An Easy-nLC 1000 system (Thermo Fisher Scientific) was connected to the Orbitrap Q Executive mass spectrometer (Thermo Fisher Scientific) to analyze the samples. The peptide samples were directly loaded onto the analytical column (100 m i.d. × 20 cm) with integrated spray tip packed with 1.9 m and 120 Å ReproSil-Pur C18 resins (Dr. Maisch GmbH). The tryptic peptides were separated by a binary buffer system of 0.1% (v/v) FA in water (buffer A) and 0.1% (v/v) FA in ACN (buffer B) at a flow rate of 250 nl/min. The gradient was set as follows: from 3% to 7% (v/v) buffer B in 2 min, from 7% to 22% (v/v) buffer B in 50 min, from 22% to 35% (v/v) buffer B in 10 min, from 35% to 90% (v/v) buffer B in 2 min, holding at 90% (v/v) buffer B for 6 min, declining to 3% (v/v) buffer B in 2 min, and holding at 3% (v/v) buffer B for 8 min. The spray voltage was set at +1.9 kV, and collision energy was set at 30%. The data-dependent acquisition method in the top speed mode with cycle time of 3 sec was used. The normalized collision energy of HCD fragmentation was set to 30 and the dynamic exclusion time was set to 60 sec. The MS scans were acquired at a resolution of 70000. The Human NCBI Reference Sequence database (downloaded on Sep 29, 2016) was searched by Sequest HT node (Version 1.4, Thermo Fisher Scientific) with the following parameters: mass tolerance: 5 ppm on MS and 0.6 Daltons on MS/MS; maximum missed cleavages: 2; fixed modification: carbamidomethyl; variable modification: oxidation and acetylation.
Characterization of acK134-Ran antibody
To test the specificity of the acK134-Ran antibody, aliquots of mitotic cells were harvested by shake-off followed by sedimentation and treated with TIP60 inhibitor NU9056 (20 μM) for 30 min before being lysed in RIPA buffer (25 mM Tris-HCl, pH7.5, 5 mM EDTA, 0.5% SDS, and 1% deoxycholate). In a separated experiment, two aliquots of HeLa cells were synchronized by mitotic shake-off followed by transfection of Ran siRNA in early G1 as previously described (Yao et al., 2000). Twenty-four hours after the transfection, HeLa cells were then synchronized with 100 ng/ml nocodazole for an additional 18 h before the cells are harvested and lysed in RIPA buffer. The extracts were then sonicated and centrifuged to remove the residual insoluble materials. Before electrophoresis, an appropriate amount of extract was diluted with 5× sample buffer and boiled at 95°C for 2 min. After separation in SDS-PAGE, the proteins were transferred onto a nitrocellulose membrane and incubated with anti-acK134-Ran antibody followed by HRP-conjugated goat anti-rabbit antibody as described above.
Statistical analyses
All statistics were described in the figure legends. Two-tailed unpaired Student’s t-test was applied for experimental comparisons using GraphPad Prism. All data were taken from three separated experiments.
Supplementary Material
Acknowledgements
We thank Drs Chuanmao Zhang (Peking University), Hongtao Yu (UT Southwestern Medical Center) and Lingluo Chu (Harvard University) for helpful discussions; Dr Zhijun Liu (National Center for Protein Science Shanghai) and High Magnetic Field Laboratory, Chinese Academy of Sciences for NMR data collection. The WeNMR project (European FP7 e-Infrastructure grant, contract no. 261572, www.wenmr.eu), supported by the European Grid Initiative (EGI) through the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, South Africa, Malaysia, Taiwan, the Latin America GRID infrastructure via the Gisela project, and the US Open Science Grid (OSG), is acknowledged for the use of web portals, computing, and storage facilities.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was financially supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08030302, XDB08010101, XDB19040000), Chinese Academy of Sciences Center of Excellence (2015HSC-UE010), the Ministry of Science and Technology of China (2016YFA0500700, 2017YFA0503600, 2016YFA0100500), the National Basic Research Program of China (973 Program 2012CB917201), the National Natural Science Foundation of China (31330018, 31270760, 31430054, 31320103904, 31621002, 31671405), Chinese Ministry of Education (IRT_17R102), and the US National Institutes of Health Grants (CA164133, DK56292, and DK115812).
Conflict of interest: none declared.
References
- Baker R.P., Harreman M.T., Eccleston J.F., et al. (2001). Interaction between Ran and Mog1 is required for efficient nuclear protein import. J. Biol. Chem. 276, 41255–41262. [DOI] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., Vangunsteren W.F., et al. (1984). Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690. [Google Scholar]
- Bischoff F.R., Krebber H., Kempf T., et al. (1995). Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc. Natl Acad. Sci. USA 92, 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., and Ponstingl H. (1991). Catalysis of guanine-nucleotide exchange on Ran by the Mitotic Regulator Rcc1. Nature 354, 80–82. [DOI] [PubMed] [Google Scholar]
- Bompard G., Rabeharivelo G., Frank M., et al. (2010). Subgroup II PAK-mediated phosphorylation regulates Ran activity during mitosis. J. Cell Biol. 190, 807–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., Adams P.D., Clore G.M., et al. (1998). Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bussi G., Donadio D., and Parrinello M. (2007). Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Guarguaglini G., Gruss O.J., et al. (1999). Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Ke Y., Ding X., et al. (2008). Functional characterization of TIP60 sumoylation in UV-irradiated DNA damage response. Oncogene 27, 931–941. [DOI] [PubMed] [Google Scholar]
- Chu L., Zhu T., Liu X., et al. (2012). SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J. Mol. Cell Biol. 4, 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P.R., and Zhang C.M. (2008). Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 9, 464–477. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio F., and Bax A. (1999). Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302. [DOI] [PubMed] [Google Scholar]
- de Boor S., Knyphausen P., Kuhlmann N., et al. (2015). Small GTP-binding protein Ran is regulated by posttranslational lysine acetylation. Proc. Natl Acad. Sci. USA 112, E3679–E3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S.J., van Dijk M., and Bonvin A.M. (2010). The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., et al. (1995). NMRPipe—a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Ding X., Yan F., Yao P., et al. (2010). Probing CENP-E function in chromosome dynamics using small molecule inhibitor syntelin. Cell Res. 20, 1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., et al. (1995). A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593. [Google Scholar]
- Gans P., Hamelin O., Sounier R., et al. (2010). Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew. Chem. Int. Ed. Engl. 49, 1958–1962. [DOI] [PubMed] [Google Scholar]
- Gelis I., Bonvin A.M., Keramisanou D., et al. (2007). Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M.R., Mueller L., and Pardi A. (1998). Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 5, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Hess B. (2008). P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122. [DOI] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., et al. (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447. [DOI] [PubMed] [Google Scholar]
- Hiruma Y., Sacristan C., Pachis S.T., et al. (2015). CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science 348, 1264–1267. [DOI] [PubMed] [Google Scholar]
- Hockney R.W., Goel S.P., and Eastwood J.W. (1974). Quiet High-Resolution Computer Models of a Plasma. J. Comput. Phys. 14, 148–158. [Google Scholar]
- Ji Z., Gao H., and Yu H. (2015). CELL DIVISION CYCLE. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science 348, 1260–1264. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., et al. (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. [Google Scholar]
- Kalab P., Weis K., and Heald R. (2002). Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295, 2452–2456. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., and Wuthrich K. (1996). MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics 14, 51. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann J.A.C., MacArthur M.W., et al. (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Laviv T., Kim B.B., Chu J., et al. (2016). Simultaneous dual-color fluorescence lifetime imaging with novel red-shifted fluorescent proteins. Nat. Methods 13, 989–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Xue Y., Jin C., et al. (2006). Prediction of Nepsilon-acetylation on internal lysines implemented in Bayesian Discriminant Method. Biochem. Biophys. Res. Commun. 350, 818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.Y., Wirtz D., and Zheng Y. (2003). A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 160, 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., and Biggins S. (2014). Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A.D., Bashford D., Bellott M., et al. (1998). All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys.Chem. B 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- Mo F., Zhuang X., Liu X., et al. (2016). Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat. Chem. Biol. 12, 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut M.E., Mizzen C.A., Stukenberg T., et al. (2001). Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292, 1540–1543. [DOI] [PubMed] [Google Scholar]
- Oh D., Yu C.H., and Needleman D.J. (2016). Spatial organization of the Ran pathway by microtubules in mitosis. Proc. Natl Acad. Sci. USA 113, 8729–8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., et al. (1999). Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284, 1356–1358. [DOI] [PubMed] [Google Scholar]
- Oki M., and Nishimoto T. (1998). A protein required for nuclear-protein import, Mog1p, directly interacts with GTP-Gsp1p, the Saccharomyces cerevisiae ran homologue. Proc. Natl Acad. Sci. USA 95, 15388–15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S., Groen A.C., Ishihara K., et al. (2013). Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152, 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L., Kuhlmann J., Henkel A., et al. (2001). Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell 105, 245–255. [DOI] [PubMed] [Google Scholar]
- Rosenzweig R., Moradi S., Zarrine-Afsar A., et al. (2013). Unraveling the mechanism of protein disaggregation through a ClpB-DnaK Interaction. Science 339, 1080–1083. [DOI] [PubMed] [Google Scholar]
- Tugarinov V., and Kay L.E. (2004). An isotope labeling strategy for methyl TROSY spectroscopy. J. Biomol. NMR 28, 165–172. [DOI] [PubMed] [Google Scholar]
- Wassenaar T.A., van Dijk M., Loureiro-Ferreira N., et al. (2012). WeNMR: Structural Biology on the Grid. J. Grid Comput. 10, 743–767. [Google Scholar]
- Wilde A., and Zheng Y. (1999). Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284, 1359–1362. [DOI] [PubMed] [Google Scholar]
- Xia P., Wang Z., Liu X., et al. (2012). EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl Acad. Sci. USA 109, 16564–16569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Abrieu A., Zheng Y., et al. (2000). CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491. [DOI] [PubMed] [Google Scholar]
- Yao X., Anderson K.L., and Cleveland D.W. (1997). The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.M., and Clarke P.R. (2000). Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 288, 1429–1432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.