Abstract
Motivation
Large pharmacogenomic screenings integrate heterogeneous cancer genomic datasets as well as anti-cancer drug responses on thousand human cancer cell lines. Mining this data to identify new therapies for cancer sub-populations would benefit from common data structures, modular computational biology tools and user-friendly interfaces.
Results
We have developed GDSCTools: a software aimed at the identification of clinically relevant genomic markers of drug response. The Genomics of Drug Sensitivity in Cancer (GDSC) database (www.cancerRxgene.org) integrates heterogeneous cancer genomic datasets as well as anti-cancer drug responses on a thousand cancer cell lines. Including statistical tools (analysis of variance) and predictive methods (Elastic Net), as well as common data structures, GDSCTools allows users to reproduce published results from GDSC and to implement new analytical methods. In addition, non-GDSC data resources can also be analysed since drug responses and genomic features can be encoded as CSV files.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Cancers occur due to genetic alterations in cells accumulated through the lifespan of an individual. Cancers are genetically heterogeneous and as a consequence patients with similar diagnoses may vary in their response to the same therapy. The path towards precision cancer medicine requires the identification of specific biomarkers, such as genetic alterations, allowing effective patient selection strategies for therapy. Large-scale pharmacological screens such as the Genomics of Drug Sensitivity in Cancer (GDSC) (Garnett et al., 2012) and Cancer Cell Line Encyclopaedia projects (Barretina et al., 2012) have been used to identify potential new treatments and to explore biomarkers of drug sensitivity in cancer cells. In particular, the GDSC project releases database resources periodically (www.cancerRxgene.org) (Yang et al., 2013). A recent installment of this resource (version 17) includes cancer-driven alterations identified in 11 289 tumors from 29 tissues across 1001 molecularly annotated human cancer cell lines, and cell line sensitivity data for 265 anti-cancer compounds. A systematic identification of clinically-relevant markers of drug response uncovered numerous alterations that sensitize to anti-cancer drugs (Iorio et al., 2016).
Here, we present GDSCTools, a Python library that allows users to perform pharmacogenomic analyses as those presented in (Iorio et al., 2016). Our software complements an existing tool (Smirnov et al., 2016) by giving access to the full GDSC dataset and providing a powerful platform for statistical analyses and data mining through visualization tools.
2 Data formats and data wrangling tools
The GDSC database provides large-scale genomics and drug sensitivity datasets. The drug sensitivity dataset contains dose-response curves (e.g. cell viability for 5–9 drug concentrations) which can be used to derive drug sensitivity indicators (Garnett et al., 2012; Vis et al., 2016), such as the half-maximal inhibitory concentration () or the area under the curve (AUC) (Fig. 1A). In GDSCTools, logged indicators are encoded as a Nc × Nd matrix, where Nc is the number of cell lines labeled with their COSMIC identifier (http://cancer.sanger.ac.uk/cosmic) and Nd is the number of drugs. For a given drug, we denote with Yd the vector of logged s across the Nc cell lines. The genomic feature dataset is also encoded as a Nc × Nf matrix, where Nf is the number of genomic features. In addition to a subset of the data files available in GDSCTools (version 17 only), users can also retrieve additional datasets online (e.g. methylation data, copy number variants, etc.) Database-like queries can be used to extract and use specific features (e.g. only gene amplifications or deletions). These database-like functionalities are part of the OmniBEM builder (Supplementary Material).
Fig. 1.
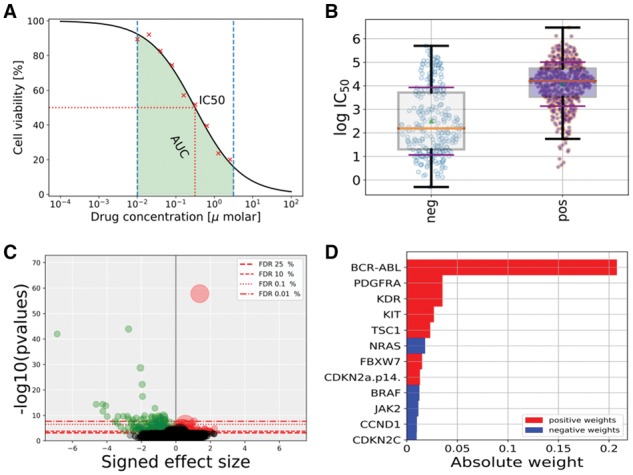
(A) Drug response (cell viability versus drug concentrations) and derived drug response metrics (AUC and s). (B) Distribution of s in response to a given drug across a dichotomy of cell lines induced by the status of a genomic feature. (C) P-values from an ANOVA analysis versus signed effect sizes (all drug-genomic feature interactions). (D) Weight distributions resulting from training a sparse linear regression model of a given drug response using all the genomic features
3 Data analysis tools
Using GDSCTools, genomic features can be investigated as possible predictors of differential drug sensitivity across screened cell lines. The statistical interaction Yd ∼ X between drug response and genomic features can be tested within a sample population of cell lines from the same cancer type with a t-test. However, to account for possible confounding factors (including the tissue of origin, when performing pan-cancer analyses) a more versatile analysis of variance (ANOVA) is implemented. In this model, the variability observed in Yd is first explained using the tissue covariate, subsequently using additional factors (e.g. microsatellite instability denoted by MSI), and finally by each of the genomic features in X (one model per feature). This can be mathematically expressed as Yd ∼ C(tissue) + C(MSI) + … + feature, where the operator indicates a categorical variable. An ANOVA test is performed for each combination of drug and genomic feature (Fig. 1B). Outcomes of this large number of tests (Nd × Nf) are corrected for multiple hypothesis testing using Bonferroni or Benjamini-Hochberg corrections. To account for P-value inflations due to differences in sample sizes, the effect sizes of the tested statistical interactions (computed with the Cohen and Glass models) are also included (Fig. 1C).
Unlike the ANOVA analysis that is performed on a one drug/one feature basis, linear regression models assume that drug response can be expressed as a linear combination of the status of a set of genomic features. GDSCTools includes three linear regression methods: (i) Ridge, based on an L2 penalty term, which limits the size of the coefficient vector; (ii) Lasso, based on an L1 penalty term, which imposes sparsity among the coefficients (i.e. makes the fitted model more interpretable) and (iii) Elastic Net, a compromise between Ridge and Lasso techniques with a mix penalty between L1 and L2 norms (see Supplementary Material for details). These three methods require the optimization of an α parameter (importance of L1 and L2 penalties) and a ρ parameter (mix ratio between L1 and L2 penalties; ElasticNet case only). This is performed via a cross validation to avoid over-fitting. The best model is determined using as objective function the Pearson correlation between predicted and actual drug responses on the training set. The final regressor weights are outputted as shown in Figure 1D. Significance of the final selected models is computed against.
4 Implementation and future directions
GDSCTools is available on http://github.com/CancerRxGene/gdsctools. It is fully documented on http://gdsctools.readthedocs.io. Pre-compiled versions of the library are available on https://bioconda.github.io/. GDSCTools can be used via standalone applications to analyse a user defined set of drugs (and genomic features) and assemble the results in an HTML report. We also provide solutions based on the Snakemake framework (Köster and Rahmann, 2012) to parallelize the analysis on distributed cluster farm architectures such as LSF or SLURM (Supplementary Material). Besides analysis of pharmacogenomic datasets, GDSCTools can provide the framework for discovering new biomarkers through integration/mining of novel and heterogeneous datasets, including pharmacological, RNA interference or increasingly available genetic screens (e.g. CRISPR), alternative drug response metrics (e.g. AUC) or implementing new analytical tools. The augmentation of genomic features with information obtained from online web services (Cokelaer et al., 2013) like pathway enrichment [e.g. via OmniPath (Turei et al., 2016)] will further extend functionality and usefulness of GDSCTools.
Conflict of Interest: none declared.
Supplementary Material
References
- Barretina J. et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 492, 290.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokelaer T. et al. (2013) BioServices: a common Python package to access biological Web Services programmatically. Bioinformatics, 29, 3241–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M.J. et al. (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature, 483, 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F. et al. (2016) A landscape of pharmacogenomic interactions in cancer. Cell, 166, 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J., Rahmann S. (2012) Snakemake–a scalable bioinformatics workflow engine. Bioinformatics, 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Smirnov P. et al. (2016) PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics, 32, 1244–1246. [DOI] [PubMed] [Google Scholar]
- Turei D. et al. (2016) OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods, 13, 966–967. [DOI] [PubMed] [Google Scholar]
- Vis D.J. et al. (2016) Multilevel models improve precision and speed of IC50 estimates. Pharmacogenomics, 17, 691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. et al. (2013) Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res., 41, D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


