Abstract
KRAS is the most commonly mutated oncogene in human cancer. Most KRAS-mutant cancers depend on sustained expression and signaling of KRAS, thus making it a high-priority therapeutic target. Unfortunately, development of direct small molecule inhibitors of KRAS function has been challenging. An alternative therapeutic strategy for KRAS-mutant malignancies involves targeting codependent vulnerabilities or synthetic lethal partners that are preferentially essential in the setting of oncogenic KRAS. KRAS activates numerous effector pathways that mediate proliferation and survival signals. Moreover, cancer cells must cope with substantial oncogenic stress conferred by mutant KRAS. These oncogenic signaling pathways and compensatory coping mechanisms of KRAS-mutant cancer cells form the basis for synthetic lethal interactions. Here, we review the compendium of previously identified codependencies in KRAS-mutant cancers, including the results of numerous functional genetic screens aimed at identifying KRAS synthetic lethal targets. Importantly, many of these vulnerabilities may represent tractable therapeutic opportunities.
RAS mutations occur in approximately 30% of human cancers, including the majority of pancreatic ductal adenocarcinoma (PDAC), half of colorectal cancers, and one-third of all lung cancers (Pylayeva-Gupta et al. 2011; Cox et al. 2014). The three RAS genes (KRAS, NRAS, and HRAS) have high sequence homology except for a carboxy-terminal hypervariable region (Cox et al. 2014), and RAS gene mutations are typically mutually exclusive in human cancer. Despite their similarity, KRAS mutations are far more common in human cancer than NRAS or HRAS mutations. Whereas much of the early studies of RAS signaling and biology focused on HRAS, KRAS has now become the dominant focus for cancer modeling and therapeutic development efforts (Stephen et al. 2014; Papke and Der 2017). The similarities and differences between RAS isoforms have been reviewed elsewhere (Pylayeva-Gupta et al. 2011; Cox et al. 2014; Stephen et al. 2014), and the remainder of this review will primarily focus on studies related to the KRAS oncogene.
Experiments in cell culture and animal models have shown that the majority of tumors that harbor KRAS mutations depend on sustained expression of the oncogene for cell proliferation and viability (Podsypanina et al. 2008; Singh et al. 2009; Collins et al. 2012; Ying et al. 2012; Kapoor et al. 2014; Shao et al. 2014; Hayes et al. 2016), making oncogenic KRAS a high-priority therapeutic target. However, KRAS is a small, structurally dynamic protein that has not been particularly amenable to direct therapeutic targeting. Unfortunately, attempts to develop drugs that target mutant RAS proteins have thus far been unsuccessful (Stephen et al. 2014; Papke and Der 2017), leading investigators to explore alternative opportunities for targeting KRAS-driven cancers.
Oncogenic KRAS activates more than 10 different effector signaling pathways, and the therapeutic potential of inhibiting these effectors has been the focus of intensive investigation. The most well-studied and critical KRAS effector pathways include the mitogen-activated protein kinase (MAPK) signaling cascade (Moodie et al. 1993; Vojtek et al. 1993; Warne et al. 1993; Zhang et al. 1993), the phosphatidyl inositol 3-kinase (PI3K)-AKT-MTOR pathway (Sjolander et al. 1991; Rodriguez-Viciana et al. 1994), and the Ras-like (RAL)-guanine nucleotide exchange factor (GEF) family of GEFs for the RAL GTPases (Hofer et al. 1994; Kikuchi et al. 1994; Spaargaren and Bischoff 1994). Most genetic and functional studies have suggested that the MAPK pathway is the dominant oncogenic RAS signaling pathway, and KRAS mutations generally occur in a mutually exclusive manner with mutations affecting other MAPK pathway components (e.g., BRAF, EGFR mutation). RAS-MAPK signaling has been shown to mediate tumor cell proliferation and survival in a variety of in vitro and in vivo experiments (Blasco et al. 2011; Karreth et al. 2011; Collisson et al. 2012; Yuan et al. 2014), and numerous small molecule inhibitors of this pathway are in clinical development (Ryan et al. 2015). Although some KRAS-driven cancers such as pancreatic cancers depend on the PI3K pathway, other KRAS-mutant tumors such as some lung or colorectal cancers do not (Ebi et al. 2011; Eser et al. 2013). Thus, PI3K dependence in KRAS-mutant cancers is context-specific. Therapeutic targeting of PI3K isoforms in KRAS-mutant cancers remains an active area of study. RAL-GEFs activate multiple processes, including nuclear factor (NF)-κB signaling and cell motility and have also been shown to be important for transformation of human cells (Hamad et al. 2002; Lim et al. 2005). Therapeutic inhibition of this pathway may also hold promise for KRAS-mutant cancers (Barbie et al. 2009; Yan et al. 2014; Kitajima et al. 2016). Several other RAS effectors have been identified, including TIAM1 and PLCε, but their role in RAS signaling remains less well understood and their therapeutic value has not yet been proven.
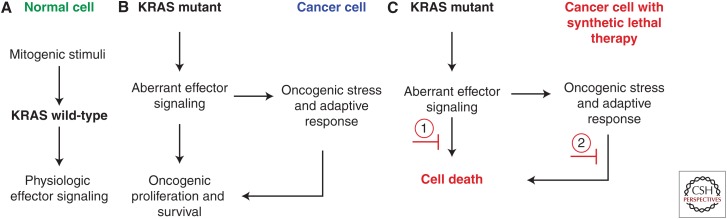
An alternative approach to direct targeting of mutant cancer genes is to exploit the concept of synthetic lethality, in which gene products are identified that, when suppressed or inhibited, result in cell death only in the presence of the cancer-causing alteration (Fig. 1) (Kaelin 2005; Ngo et al. 2006; McLornan et al. 2014). Thus, in principle, targeting synthetic lethal vulnerabilities in cancer should reduce the potential for side effects, because cells harboring the biomarker oncogenic lesion should have more differential sensitivity to the perturbation than normal cells, which do not have the oncogenic lesion. The most salient example of this concept in cancer biology and therapeutics is the discovery that homologous-recombination-deficient BRCA1 or BRCA2 mutant cancers show profound sensitivity to inhibition of poly-(ADP-ribose) polymerase (PARP) (Bryant et al. 2005; Farmer et al. 2005). PARP inhibitors have subsequently shown therapeutic efficacy in BRCA1/2 mutant breast, ovarian, and some pancreatic cancers (McLornan et al. 2014; Lord and Ashworth 2017).
Figure 1.
Synthetic lethality as a therapeutic paradigm in cancer. (A) In normal healthy cells, wild-type (WT) KRAS is activated by appropriate mitogenic stimuli such as receptor tyrosine kinase–mediated growth factor signaling. (B) In cancer cells with KRAS mutation, aberrant effector signaling mediates oncogenic proliferation and survival but also creates oncogenic stress to which cancer cells must adapt to sustain oncogenic growth. These downstream aberrant effector signaling pathways as well as parallel adaptive pathways that mitigate oncogenic stress represent unique features of KRAS-mutant cells that may include selective vulnerabilities and synthetic lethal targets. (C) Synthetic lethal targets refer to those targets whose inhibition results in cell death only in the presence of another mutation (i.e., KRAS mutation). Broadly applied, this concept may represent targets that act (1) downstream of aberrant effector signaling, or (2) in parallel adaptive pathways. In theory, synthetic lethal therapies result in cell death of the KRAS-mutant cancer cells, but not in normal cells (shown in A) because the aberrant oncogenic signals or parallel adaptive processes are not present in these KRAS WT cells.
Given the relative intractability of KRAS itself as a therapeutic target, identification of synthetic lethal partners of oncogenic KRAS has been the focus of intense investigation by many groups. The fundamental premise underlying the concept of synthetic lethality in KRAS-mutant tumors is that oncogenic KRAS signaling establishes a distinct cell state, marked by altered KRAS effector signaling, adaptation to oncogenic stress, and transcriptional and metabolic reprogramming. Disruption of this KRAS-driven cell state may impair proliferation and viability of KRAS-mutant cells. Furthermore, many of these adaptive features may have partial selectivity or even unique specificity for the KRAS-driven oncogenic cell state over a KRAS wild-type (WT) state, and thus may represent nononcogene dependencies or vulnerabilities between these cell contexts. Because most cancer cells and normal cells use RAS effector signaling in some capacity for mitogenic processes, it should be expected that many of the oncogenic KRAS codependencies will not show unique specificity to the KRAS-mutant context and thus will not strictly conform to the genetic definition of synthetic lethality (Fig. 1). However, the term “synthetic lethal” has also been applied more liberally to denote such vulnerabilities that show some quantitative selectivity for the KRAS-mutant state over the KRAS WT state. Many recent studies, including hypothesis-driven focused studies as well as large small-molecule and genetic screens performed over the past decade, have identified several putative KRAS synthetic lethal partners of varying strength and specificity. Whereas the long-sought-after universal synthetic lethal target for all KRAS-mutant cancers has not yet been identified, these studies have informed our understanding of KRAS biology and have identified numerous context-selective vulnerabilities in KRAS-mutant cancers. Whether these synthetic lethal candidates identified in experimental systems will translate to effective therapeutic targets remains to be shown. Here, we review recent work on KRAS synthetic lethal vulnerabilities and provide a conceptual framework for how to interpret these observations and prioritize synthetic lethal targets for therapeutic development.
INHIBITING KRAS EFFECTOR PATHWAYS IN KRAS-MUTANT CANCERS
Numerous studies have shown that KRAS effectors and related receptor tyrosine kinase (RTK)-mediated signaling pathways may be preferential vulnerabilities in the KRAS-mutant context. Through large-scale small-molecule screens in many different cancer cell lines with a variety of genotypes and lineages, several groups have compared compound sensitivity in RAS-mutant and RAS WT cell lines. These studies have repeatedly identified inhibitors of the MAPK pathway as the most selective compounds for KRAS-mutant cancers, including inhibitors of MAPK kinase 1 and 2 proteins (MEK1/2) and RAF kinases (Garnett et al. 2012; Basu et al. 2013; Molina-Arcas et al. 2013), although inhibition of these pathways leads primarily to cytostasis. These studies have also shown that KRAS-mutant cells harbor increased dependence on RTK signaling pathways, as small-molecule inhibitors of IGF1R and MET show some preferential selectivity for subsets of KRAS-mutant lines over KRAS WT lines (Ebi et al. 2011; Molina-Arcas et al. 2013). In addition, MAPK inhibition leads to increased dependence on RTK signaling, with combined targeting of MEK1/2 and IGF1R (Ebi et al. 2011; Molina-Arcas et al. 2013) or FGFR1 (Manchado et al. 2016), demonstrating enhanced differential impact on KRAS-mutant cells over WT cells. Moreover, KRAS-MAPK effector signaling ultimately leads to activation of AP-1 transcription factors and up-regulation of cell-cycle regulatory proteins, such as cyclin D1 (CCND1), which promotes G1/S cell-cycle progression. Inhibition of CDK4/6 has been reported as a promising combination therapeutic strategy in KRAS-mutated malignancies (Puyol et al. 2010). Unfortunately, clinical trials testing single-agent inhibition of the critical KRAS effectors (e.g., MAPK or PI3K) in KRAS-mutant cancers have been disappointing to date (Adjei et al. 2008; Infante et al. 2012, 2013; Zhao and Adjei 2014). However, inhibition of these pathways will undoubtedly form the foundation for future combination therapy strategies in KRAS-mutated malignancies (Engelman et al. 2008; Britten 2013; Ebi et al. 2014; Ryan et al. 2015).
RAS signaling through both the MAPK and PI3K-AKT-MTOR pathways has been shown to mediate antiapoptotic signaling through down-regulation of proapoptotic mediators and up-regulation of antiapoptotic proteins (Pylayeva-Gupta et al. 2011). Engelman and colleagues have shown that disruption of RAS effector signaling unveils selective vulnerabilities in the KRAS-mutant context. MEK1/2 inhibition leads to induction of the proapoptotic protein BIM, but this protein is bound by antiapoptotic BCL-XL proteins (Corcoran et al. 2013). Exploiting this observation, they showed through genetic and pharmacologic means that combined MEK inhibition with genetic ablation or small-molecule inhibition of BCL-XL led to robust apoptosis and synthetic lethality in KRAS-mutant cells. This therapeutic strategy has now advanced to early-phase clinical trials (NCT02079740). Further work by the Engelman laboratory has also shown that KRAS- or BRAF-mutant colorectal cancer cells compared with WT cells harbor increased sensitivity to combined inhibition of the antiapoptotic proteins MCL-1, BCL-XL, and BCL-2 through dual treatment with an MTOR inhibitor and the BCL2/BCL-XL inhibitor ABT-263, with selectivity stemming from disruption of BIM/MCL-1 complexes in the KRAS- or BRAF-mutant context (Faber et al. 2014).
Several studies have shown that KRAS-associated inflammatory signaling mediates key proliferative and survival programs (Kitajima et al. 2016). NF-κB signaling has been shown to play an important role in KRAS-mutant cancers. As noted above, KRAS has been shown to activate NF-κB signaling through activation of RAL-GEFs. Genetic or pharmacologic suppression of NF-κB signaling inhibits tumorigenesis in the KrasG12D;p53 (KP) mouse model of lung cancer (Meylan et al. 2009; Basseres et al. 2010). In a KrasG12D;Ink4a/Arf mutant PDAC model, oncogenic Kras signaling activates an interleukin (IL)-1a/p62 feedforward loop to induce NF-κB signaling (Ling et al. 2012). Small-molecule inhibitors of RAL GTPase proteins are under development (Yan et al. 2014), and such compounds could be used to directly abrogate KRAS-mediated activation of NF-κB signaling. Through an arrayed RNA-interference (RNAi)-based synthetic lethal screening (discussed below), the IκB kinase (IKK)–related kinase Tank-binding kinase-1 (TBK1) was identified as a synthetic lethal target with oncogenic KRAS (Barbie et al. 2009). This finding expanded upon the prior observation that the RALB GTPase activates TBK1, thus suggesting a mechanistic link to KRAS signaling through RAL-GEFs (Chien et al. 2006). Subsequent work has shown that TBK1 promotes KRAS-driven tumorigenesis by regulating an autocrine CCL5 and IL-6 cytokine signaling loop (Zhu et al. 2014). Numerous other studies have also shown an important role for Janus kinase (JAK)-signal transducers and activators of transcription (STAT) signaling in PDAC tumorigenesis (Corcoran et al. 2011; Fukuda et al. 2011; Lesina et al. 2011). MEK inhibition in KRAS-mutant cancer cells, as well as those driven by other oncogenes such as epidermal growth factor receptor (EGFR) or MET, leads to feedback activation of IL-6 signaling and STAT3 activation to drive resistance to this therapy (Lee et al. 2014). The multitargeted kinase inhibitor momelotinib inhibits both JAK1/2 and the TBK1 (and IKKe) kinases. Suppression of both JAK-STAT and TBK1/NF-κB signaling with momelotinib has shown strong preclinical efficacy in cell culture models and murine Kras-driven animal models (Zhu et al. 2014). Clinical trials of momelotinib in combination with chemotherapy or MEK inhibition in KRAS-mutant malignancies are currently underway. In addition, further development of more potent and specific inhibitors of TBK1 and JAK-STAT signaling is an area of current activity. Thus, KRAS-directed inflammatory signaling networks and compensatory cytokine signaling represent important vulnerabilities for therapeutic development.
KRAS ONCOGENIC STRESS AND ADAPTATION
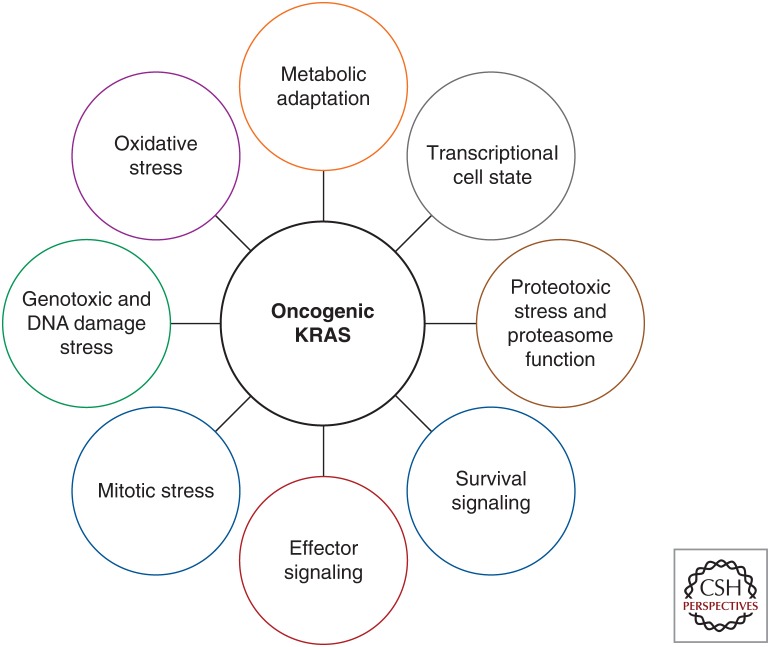
Cancer cells harbor a variety of oncogenic stresses, including DNA damage and replicative stress, proteotoxic stress, oxidative stress, and metabolic stress (Luo et al. 2009b). KRAS signaling establishes an oncogenic cell state that gives rise to adaptive changes to oncogenic stress that are necessary for cancer cell proliferation and survival. The myriad of parallel cell processes that are essential in the context of KRAS signaling have been termed nononcogene addictions (Luo et al. 2009b), and these processes form the basis for KRAS synthetic lethality (Fig. 2).
Figure 2.
KRAS oncogenic stress and adaptation.
RAS signaling has been reported to directly lead to genotoxic stress stemming from generation of reactive oxygen species (ROS) and DNA hyperreplication (Grabocka et al. 2015). Cancer cells respond to this stress through activation of DNA damage repair (DDR) checkpoints and up-regulation of the function of DDR pathways (Grabocka et al. 2015). Bar-Sagi and colleagues have shown an important role for WT H-/N-Ras in the activation of the ATR/CHK1-mediated DDR and maintenance of genomic stability in KRAS-mutant cancers (Grabocka et al. 2014). They have shown that WT HRAS and NRAS negatively regulate MAPK and AKT signaling to control inhibitory phosphorylation of CHK1 on serine 280. Loss of WT HRAS or NRAS expression in KRAS-mutant cells resulted in impaired CHK1 activation and checkpoint failure, leading to increased genomic instability and sensitivity to DNA-damaging chemotherapy agents. These studies more generally suggest that inhibition of WT HRAS/NRAS signaling or inhibition of the ATR-CHK1 pathway may have important therapeutic value in combination with DNA damaging chemotherapy in KRAS-mutant cancers. Furthermore, combination small molecule screening studies in cancer-cell lines has identified synergistic interactions of DDR checkpoint inhibitors specifically in the KRAS-mutant context, with dual inhibition of the of CHK1 and MK2 cell-cycle checkpoint inhibitors having a potent effect in KRAS-mutant contexts (Dietlein et al. 2015). Whereas these therapeutic strategies involving DDR checkpoint blockade or DNA-damaging chemotherapy are by no means specific for the KRAS-mutant context, the above evidence suggests that there may be partial selectivity for KRAS-mutant cells that may offer a sufficient therapeutic window to enable therapeutic efficacy.
Using an integrative approach combining gene expression and RNAi screening data, White and colleagues recently described a specific dependency of KRAS-mutant non-small-cell lung cancer (NSCLC) cells to receptor-dependent nuclear export (Kim et al. 2016). Specifically, they showed that chemical inhibition of nuclear export with a clinically available inhibitor of XPO1 led to a synthetic lethal interaction with oncogenic KRAS. XPO1 inhibition was mechanistically linked to inhibition of NF-κB transcription factor activity. Thus, addiction to XPO1-dependent nuclear and cytoplasmic trafficking is a druggable liability in KRAS-mutant lung cancers.
Multiple groups have shown that KRAS-mutant cancers harbor proteotoxic or endoplasmic reticulum (ER) stress, resulting in activation of the unfolded protein response (UPR) signaling pathway (Denoyelle et al. 2006; De Raedt et al. 2011). Enhancement of this proteotoxic stress can overwhelm the cell’s ability to compensate and leads to cell death. Cichowski and colleagues showed that Kras/p53 mutant lung cancers are sensitive to agents that enhance proteotoxic stress (De Raedt et al. 2011). They showed that HSP90 inhibition with IPI-504 in combination with MTOR inhibition with rapamycin results in catastrophic ER stress and tumor regressions in murine lung tumor models. They argued that this combination is clinically feasible and should be evaluated in human clinical trials in KRAS-mutant lung cancers.
Downward (2015) has highlighted a fundamental role for protein synthesis machinery and proteasome function in KRAS-mutant cells. RNAi screening was performed on approximately 7000 druggable genes and identified the GATA2 transcription factor as a synthetic lethal target with preferential dependency in KRAS-mutant cells (Kumar et al. 2012; Steckel et al. 2012). GATA2 was shown to up-regulate proteasome components through the transcription factor Nrf1; however, loss of GATA2 impaired proteasome activity in both KRAS-mutant and WT lung cancer cells. GATA2 also up-regulates other essential pathways, including IL-1 and Rho signaling cascades, and the concurrent regulation of these three pathways was proposed to be the basis for partial selectivity of GATA2 essentiality in KRAS-mutant versus WT cells. Supporting these observations, proteasome components were also identified in other synthetic lethal screening efforts (Barbie et al. 2009; Luo et al. 2009a) discussed below. However, proteasome inhibitors such as bortezomib do not have selectivity for KRAS-mutant cells in culture (Garnett et al. 2012; Basu et al. 2013) and have a relatively narrow therapeutic window in human trials; thus, whether the current clinical grade proteasome inhibitors will show efficacy in KRAS-driven cancers remains uncertain.
Several studies suggest that RAS mutant cells may have enhanced sensitivity to oxidative stress. Stockwell and colleagues have pursued synthetic lethal small molecule screening in isogenic HRAS G12V mutant or WT fibroblasts and have identified a series of small molecules that induce ferroptosis, a nonapopototic cell death characterized by iron-dependent accumulation of lethal lipid ROS in oncogenic RAS-harboring cells (Dolma et al. 2003; Yagoda et al. 2007; Yang and Stockwell 2008). Furthermore, Shaw and colleagues (2011) have performed synthetic lethal screening of over 50,000 compounds in mouse embryonic fibroblasts (MEFs) isogenic for a KrasG12D allele and identified lanperisone, a compound that induces nonapoptotic cell death selectively in KrasG12D MEFs through induction of ROS leading to oxidative stress and cell death. Notably, lanperisone showed suppression of tumor growth in a xenograft model with KrasG12D;p53–/– transformed MEFs without significant toxicity; however, human KRAS-mutant lung cancer cell lines were relatively resistant to this compound, suggesting context specificity for this vulnerability (Shaw et al. 2011). Moreover, as noted above, the proteotoxic stress and Kras-mutant tumor regressions invoked by combined HSP90 and MTOR inhibition, described by Cichowski and colleagues, is thought to be fueled by oxidative stress (De Raedt et al. 2011). Last, work by Tuveson and colleagues has shown that KRAS and other oncogenes promote detoxification of ROS through transcriptional up-regulation of an NFE2L2/NRF2-mediated antioxidant program, and genetic targeting of Nrf2 impairs KrasG12D-driven tumorigenesis (DeNicola et al. 2011). Subsequent work has shown that loss of NRF2 impairs autocrine EGFR signaling and leads to oxidation of translational regulatory proteins that inhibit messenger RNA (mRNA) translation and pancreatic cancer cell proliferation (Chio et al. 2016). Targeting of these NRF2-mediated signaling pathways with combined inhibition of AKT and glutathione synthesis was shown to be a synthetic lethal strategy resulting in impaired pancreatic cancer growth in animal models.
Oncogenic KRAS drives altered cellular metabolism to support anabolic processes that enable proliferation and survival (Ying et al. 2012; Bryant et al. 2014). Studies of KRAS-driven metabolism have unveiled numerous potential synthetic lethal vulnerabilities. KRAS-mediated metabolic reprogramming results in increased utilization of autophagy for recycling of intracellular components to fuel biosynthetic pathways and promote KRAS-mutant pancreatic cancer cell proliferation (Guo et al. 2011; Lock et al. 2011; Yang et al. 2011, 2014). Moreover, rigorous studies in KRAS-mutant PDAC mouse models and human PDAC patient-derived xenografts (PDXs) showed that autophagy inhibition impairs PDAC tumor growth, thus highlighting this process as an important dependency that may be amenable to therapeutic targeting.
KRAS-driven tumors consume extracellular protein through the process of macropinocytosis (Commisso et al. 2013; Davidson et al. 2017). Macropinocytosis results in internalization of extracellular fluid and nutrients within vesicles, where proteins undergo degradation to yield amino acids that can enter central carbon metabolism. Pharmacologic inhibition of micropinocytosis results in growth inhibition of KRAS-mutant but not KRAS-WT PDAC xenografts (Commisso et al. 2013). Thus, the dependence of KRAS-mutant cancer cells on macropinocytosis is a metabolic vulnerability with therapeutic potential.
Kimmelman and colleagues have reported that KRAS directs reprogramming of glutamine metabolism in PDAC cells through transcriptional up-regulation of metabolic enzymes in a noncanonical pathway of glutamine utilization (Son et al. 2013). Through RNAi experiments, they showed that suppression of expression of multiple enzymes in this pathway was essential for the growth of PDAC cells in vitro and in vivo, but not essential for normal cell growth. Glutaminase is the initial enzyme in this pathway and is currently being investigated as a therapeutic target in KRAS-mutated PDAC. However, it is becoming increasingly appreciated that the cellular environment is a major determinant of metabolic phenotypes and that metabolic dependencies may be highly context specific. For example, whereas Kras-mutant lung cancer cells depend on glutaminase in culture, Kras-driven lung tumors do not depend on glutaminase in vivo (Davidson et al. 2016). Moreover, metabolic adaptations are not uniform in KRAS-driven cancers. Vander Heiden and colleagues have shown distinct utilization of branched-chain amino acids (BCAAs) between Kras;Trp53 mutant mouse models of NSCLC and PDAC (Mayers et al. 2016). NSCLC incorporates BCAAs into protein and uses them as a nitrogen source; furthermore, loss of the BCAA- processing enzymes Bcat1 and Bcat2 impairs NSCLC growth. However, PDAC tumors have decreased BCAA uptake and do not depend on Bcat1 or Bcat2. This context-selectivity of metabolic dependencies highlights the importance of studying KRAS-driven metabolic adaptations using in vivo model systems that most closely represent human tumor physiology.
Cantley and colleagues have recently harnessed the metabolic properties of KRAS-mutant cells and sensitivity of these cells to oxidative stress (Yun et al. 2015). They showed that high-dose vitamin C shows selective toxicity in KRAS (or BRAF) mutant tumor cells because of uptake of oxidized vitamin C via the GLUT1 transporter, leading to buildup of ROS and inactivation of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Notably, high GLUT1 expression is driven by the KRAS or BRAF oncogene-induced glycolytic addiction; thus, oxidative stress leads to an energetic crisis and cell death specifically in the KRAS- or BRAF-driven tumor cell context.
While there is not yet a clinically effective direct inhibitor of oncogenic KRAS, multiple groups have modeled KRAS inhibition through inducible suppression of KRAS expression in cell culture or mouse tumor models (Podsypanina et al. 2008; Collins et al. 2012; Ying et al. 2012; Kapoor et al. 2014; Shao et al. 2014). These studies have revealed important KRAS-independent escape mechanisms in the context of KRAS suppression. Using an inducible KrasG12D (iKras) mouse model of pancreatic cancer, Draetta and colleagues have shown that metabolic reprogramming from a decreased reliance on glycolysis to a strong dependence on oxidative phosphorylation enables survival in the absence of KRAS signaling (Viale et al. 2014). This finding has prompted efforts to combine KRAS signaling blockade (e.g., MEK1/2) with inhibition of oxidative phosphorylation, although the efficacy and tolerability of this strategy needs to be evaluated in human clinical trials. Hahn and colleagues used a cDNA overexpression screen to identify the YAP1 transcription factor as a mediator of KRAS-independent growth (Shao et al. 2014). Using the iKras mouse model, DePinho and colleagues also showed that YAP1 could overcome Kras suppression to promote PDAC growth (Kapoor et al. 2014). YAP1 has also been shown to be essential for KRAS-induced cell transformation (Shao et al. 2014), as well as PDAC development in mice (Zhang et al. 2014). Moreover, Bivona and colleagues have described a role for YAP1 as a node of resistance to RAF and MEK inhibition, and therefore propose that YAP1 may be a synthetic lethal interactor that is an attractive candidate for combination therapy in KRAS- (or BRAF-) mutant malignancies (Lin et al. 2015).
RAS signaling is well known to lead to epithelial-mesenchymal transition (EMT) in certain epithelial cell contexts. Settleman and colleagues have previously described an important role for EMT in mediating KRAS-independent growth of KRAS-mutant cancer cells (Singh et al. 2009). Moreover, aggressive mesenchymal subpopulations have been shown to arise through Smarcb1-Myc network-driven reprogramming upon extinction of KrasG12D expression in animal models of PDAC (Genovese et al. 2017). KRAS and YAP1 have also been shown to converge to activate an EMT transcriptional program (Shao et al. 2014). Additionally, the SNAI2 gene encoding the SNAIL transcription factor, a known regulator of EMT, has been previously identified as a KRAS synthetic lethal target in an RNAi screen of an isogenic pair of colorectal cancer cell lines (Wang et al. 2010). Thus, the mesenchymal cell state itself, partially induced by KRAS signaling, may harbor synthetic lethal vulnerabilities for therapeutic targeting in a subset of KRAS-mutant cancers.
FUNCTIONAL GENETIC SCREENING FOR KRAS SYNTHETIC LETHAL TARGETS
Loss-of-function genetic screens provide a means to define the compendium of genes that are essential for cancer cell proliferation and viability in a context-selective manner (Boehm and Hahn 2011). Unbiased, genome-scale functional genetic screens hold promise to identify novel and unpredicted synthetic lethal relationships with oncogenic KRAS. Multiple different approaches have been used to screen for KRAS synthetic lethal relationships, including both arrayed/multiwell and pooled screening formats in both panels of KRAS-mutant or WT cell lines, as well as isogenic KRAS-mutant and WT cell systems. Most of these screens have been performed using RNAi, although the demonstration that CRISPR-Cas9 technology can be used in mammalian cells for genome scale screens has led several groups to begin using this technology. The strengths and disadvantages of these methodologic approaches to KRAS synthetic lethal screening have been recently reviewed elsewhere (Downward 2015). We summarize a wide range of KRAS synthetic lethal screens in Table 1, but focus on a few illustrative examples below.
Table 1.
RAS synthetic lethal functional genetic screens
| Synthetic lethal genes or pathways | Library (assay and format) | Cells in primary screen | Drug inhibition | References |
|---|---|---|---|---|
| RAN, TPX2, SCD1 | ∼3700 druggable genes, small interfering RNA (siRNA), arrayed cell death | NCIH1299 (NRASQ6K NSCLC) | Not tested | Morgan-Lappe et al. 2007 |
| BIRC5 (survivinn), CDK1, RBCK1, | ∼4000 genes, siRNA, arrayed cell death | Isogenic DLD1 (CRC, KRASG3D) | Not tested | Sarthy et al. 2007 |
| PLK1, APC/C, proteosome | 74,905 retroviral short hairpin RNAs (shRNAs) to 32,293 human transcripts (pooled proliferation screen) | Isogenic DLD1 (CRC, KRASG3D) | BI-2536 (PLK) | Luo et al. 2009a |
| STK33, AKT3, CPNE1, CAMPK1, MLKL, FLT3LG, and DGKZ | ∼1000 druggable genes, shRNA, arrayed proliferation | Pan-cancer cell-line panel (four KRAS-mutant, four KRAS-wild-type [WT]) and two immortalized cell lines | STK33 kinase inhibitor, failed to suppress proliferation in KRAS-mutant cells (Babij et al. 2011; Luo et al. 2012) | Scholl et al. 2009 |
| TBK1, PSKH2, PTCH2, CPNE1, MAP3K8, proteasome genes | ∼1000 druggable genes, shRNA, arrayed proliferation | Pan-cancer cell-line panel (seven KRAS-mutant, 10 KRAS-WT) and two immortalized cell lines | CYT387 (TBK1 and JAK inhibitor) assessed in Zhu et al. (2014) | Barbie et al. 2009 |
| WT1, RAC1, PHB2 | 162 KRAS-related genes, shRNA, in vitro and in vivo pooled proliferation screens with bead array readout | LKR10 and LKR13 (Kras;Trp53 mutant mouse lung-tumor-derived cell lines) | Not tested | Vicent et al. 2010 |
| SNAI2 (SNAIL2) | ∼2500 druggable genes, shRNA, pooled proliferation | Isogenic HCT116 (CRC, KRASG13D) | Not tested | Wang et al. 2010 |
| GATA2, CDC6, proteasome | >7000 druggable genes, siRNA pools (arrayed apoptosis and cell proliferation) | Isogenic HCT116 (KRASG13D) and pan-cancer cell-line panel (14 KRAS-mutant, 12 KRAS-WT) | Bortezomib with fasudil (GATA2) | Kumar et al. 2012; Steckel et al. 2012 |
| MAP3K7 (TAK1) | 17 kinases highly expressed in KRAS-dependent CRC, shRNA, arrayed proliferation | KRAS-dependent SW620 and KRAS-independent SW837 (CRC, KRAS-mutant) | 5Z-7-oxozeaenol | Singh et al. 2012 |
| Cttnb1 (β-catenin), Mllt6, Raf1, Akt3 | Genome scale, shRNA, pooled in vivo proliferation with NGS readout | Mouse keratinocytes (H-rasG12V) | Not tested | Beronja et al. 2013 |
| COPI coatomer subunits ARCN1, COPB1, COPA dependent in KRAS;LKB1-mutant lung cancer cells | Genome scale, siRNA, arrayed proliferation | 17 KRAS- and LKB1-mutant lung cancer cell lines, matched tumor (KRAS- mutant), and normal NSCLC cell-line pair | Saliphenylhalamide A | Kim et al. 2013 |
| ARHGEF2 (GEFH1) | Genome scale, shRNA, pooled proliferation | 72 human cancer cell lines | Not tested | Marcotte et al. 2012; Cullis et al. 2014 |
| BCL2L1 (BCLXL) | ∼1200 druggable genes in presence of MEK inhibitor (selumetinib), shRNA, pooled proliferation screen for enhancement of MEK inhibitor effect | HCT116 and SW620 (CRC, KRAS-mutant) | Selumetinib and navitoclax | Corcoran et al. 2013 |
| RAF1, BRAF | 535 kinases and related genes, shRNA, pooled proliferation screen for enhancement with MEK inhibitor selumetinib | SW480 KRAS-mutant colon cancer cell line | RAF265 or AZ628 (RAF inhibitors) with selumetinib | Lamba et al. 2014 |
| YAP1 | 5046 signaling components, shRNA, pooled proliferation screen for enhancement of BRAF-inhibitor response | HCC364 BRAF V600E human NSCLC cell line with validation in KRAS mutants with MEK inhibition | Not tested | Lin et al. 2015 |
| CDK1 | 853 genes, primarily kinases, siRNA, arrayed cell viability | LIM1215 isogenic lines with KRAS WT, G12D, G12S, or G12V | Not tested | Costa-Cabral et al. 2016 |
| FGFR1, BRAF, RAF1, ERK2 | 526 kinases, shRNA, pooled proliferation screen for enhancement with MEK inhibitor trametinib | H23 KRAS (G13C) mutant lung cancer cell line | Ponatinib (FGFR inhibitor) | Manchado et al. 2016 |
| XPO1 | Genome scale, siRNA arrayed toxicity screens | Panel of human NSCLC cell lines | KPT-330 (selinexor) | Kim et al. 2016 |
| RAF1, SHOC2, PREX1, RCE1, ICMT | Genome-scale, CRISPR-Cas9, pooled proliferation screen | 14 leukemia cell lines (six KRAS or NRAS, six RAS WT) | Not tested | Wang et al. 2017 |
Modeled after similar tables in Downward (2015) and Ebi et al. (2014).
NSCLC, Non-small-cell lung cancer; JAK, Janus kinase.
A team led by Elledge and colleagues performed a pooled primary screen of a genome-scale lentivirally delivered short hairpin RNA (shRNA) library in an isogenic KRASG13D and WT DLD1 colorectal cancer cell line (Luo et al. 2009a). Their library contained 75,905 shRNAs targeting 32,293 unique human transcripts to approximately 12,000 genes. Stringent criteria identified a subset of 379 shRNAs targeting 368 genes that were candidate KRAS synthetic lethal interactors. Notably, the DLD1 KRAS WT cell line showed decreased MAPK signaling and slower in vitro proliferation than its KRAS-mutant counterpart. Follow-up studies were performed using a multicolor competition assay in both the DLD1 isogenic pair as well as a second isogenic HCT116 KRAS-mutant and WT pair. They identified 77 candidate KRAS synthetic lethal interactors across a variety of biological processes, including protein modification, nucleic acid metabolism, and cell-cycle and signal transduction. In particular, they described a striking number of genes involved in regulation of mitosis as being candidate KRAS synthetic lethals, including subunits of the anaphase-promoting complex/cyclosome (APC/C), the proteasome and the mitotic kinase polo-like kinase (PLK1). They showed that KRAS-mutant cells harbor heightened mitotic stress compared with WT cells, and both genetic and small molecule inhibition of PLK1 revealed greater sensitivity in the KRAS-mutant isogenic cancer cells over the KRAS WT cells. Moreover, inhibition of proteasome function with bortezomib also appeared to demonstrate selectivity in the KRAS isogenic cell systems. Mitotic stress is not unique to KRAS-mutant cancers; however, these studies suggest that KRAS-mutant cells may be selectively more sensitive to targeting mitotic regulation than WT cells.
Scholl et al. (2009) performed an arrayed format RNAi screen in four KRAS-mutant and four KRAS-WT cancer cell lines, as well as normal human fibroblasts and immortalized human mammary epithelial cells (HMECs). They used a library of 5024 lentivirally delivered shRNA constructs targeting 1011 human kinases, phosphatases, and known cancer genes. Comparing KRAS-mutant to WT cells, they identified the serine-threonine protein kinase STK33 as selectively essential in KRAS-mutant cells. Through evaluation of STK33 dependency in 25 additional cell lines, they determined that STK33 is preferentially required by KRAS mutants that also depend on sustained KRAS expression for viability and proliferation. STK33 has been subsequently investigated as a KRAS synthetic lethal target by multiple other groups, some with conflicting findings and interpretations (Babij et al. 2011; Frohling and Scholl 2011). Much of the excitement regarding STK33 stemmed from its kinase function and thus potential therapeutic tractability. Potent small molecule inhibitors of STK33 have been developed, and these molecules have conclusively shown that STK33 kinase activity is not required for KRAS-mutant cancers (Babij et al. 2011; Luo et al. 2012), although other strategies that result in loss of STK33 protein may still hold therapeutic potential (Azoitei et al. 2012).
Barbie et al. (2009) performed an arrayed format primary RNAi screen in 19 cancer cell lines, using a lentivirally delivered shRNA library targeting kinases, phosphatases, and oncogenes. They compared KRAS-mutant and WT cells to identify those genes that selectively killed KRAS-mutant cells and identified 45 candidates that were screened in a larger panel of mutant and WT cells. In addition to KRAS itself, they found RAF1 and the serine-threonine kinase TBK1 as top-scoring candidate KRAS synthetic lethals. They validated TBK1 as a synthetic lethal target in KRAS-mutant and -dependent cells and confirmed a link between RALB-mediated activation of TBK1 (Chien et al. 2006) to promote NF-κB survival signaling, in part, through c-Rel and BCL-XL. As discussed above, pharmacologic inhibition of the TBK1 induces cell death in preclinical animal models of KRAS-driven lung cancers (Zhu et al. 2014), and TBK1 inhibition is being tested in clinical studies as a potential therapeutic approach.
The lack of overlap in genes scoring from KRAS synthetic lethal screens has caused much consternation. However, the KRAS synthetic lethal screens performed to date have had several important differences and limitations. Each study used different experimental systems and were limited in scale in terms of the cell lines screened and the number of genes evaluated. None of the RNAi-based screens reached genetic saturation. Moreover, RNAi studies have previously suffered from an incomplete understanding of off-target effects. Advances in RNAi-screening efforts have improved upon these limitations. Recent technical advances in RNAi screening have enabled much higher density screens across many more cell lines (Luo et al. 2008; Cheung et al. 2011; Marcotte et al. 2012). Additionally, refinements in bioinformatic methodology have also improved the signal-to-noise resolution and distinction of off-target effects (Shao et al. 2013; Marcotte et al. 2016; Tsherniak et al. 2017). Furthermore, the advent of CRISPR-Cas9 screening technology as an alternative means of genetic perturbation has enabled a new approach to loss-of-function genetic screening (Shalem et al. 2014, 2015). CRISPR-Cas9 technology enables knockout of target genes, as compared to suppression of mRNA expression conferred by RNAi. Multiple groups have now shown that CRISPR-Cas9 genome-scale screens can effectively elucidate essential genes in cancer cell lines (Koike-Yusa et al. 2014; Wang et al. 2014, 2015; Hart et al. 2015; Parnas et al. 2015; Aguirre et al. 2016).
Sabatini and colleagues recently reported genome-scale CRISPR-Cas9 screens in 14 acute myeloid leukemia (AML) cell lines and compared six KRAS or NRAS mutant lines with six RAS WT cell lines to identify synthetic lethal genes (Wang et al. 2017). These studies identified a surprisingly small number of genes in the RAS processing (RCE1 and ICMT) and MAPK signaling pathways (RAF1, SHOC2). A second pair of screens was performed in isogenic murine Ba/F3 cell lines, one of which was engineered to express oncogenic NRAS, and this screen also discovered the same set of genes as well as additional MAPK signaling components. PREX1 was identified as a novel RAS synthetic lethal partner in the human AML screen. PREX1 is a GEF for Rac GTPases, and the investigators showed that PREX1 is an AML-specific activator of MAPK signaling.
NEXT STEPS IN DISCOVERY AND UTILIZATION OF KRAS SYNTHETIC LETHAL TARGETS
While the results of Sabatini and colleagues suggest that in AML there are only a limited number of RAS synthetic lethal targets that are largely restricted to regulators of RAS itself or the MAPK signaling pathway, it is important to note that these findings reflect only one of many KRAS-mutant cancer contexts. Indeed, there is profound heterogeneity of KRAS-mutant cancers that complicates uniform identification of KRAS synthetic lethal targets. This multidimensional heterogeneity includes (1) variability in KRAS dependency across cancers; (2) differing lineage specificity across cancer types; (3) variation in effector signaling between different KRAS alleles (e.g., G12D vs. G12V or Q61H); and (4) variability in the co-occurring mutational and copy-number landscape across cancers. It may be unreasonable to expect that a universal KRAS synthetic lethal target will apply across all contexts. To adequately address this heterogeneity and to determine biomarker-driven predictive models of these synthetic lethal relationships, genome-scale screens will need to be performed in a large number of diverse cell lines. Toward this goal, the Achilles Consortium at the Broad Institute has now screened over 300 cell lines with genome-scale CRISPR-Cas9 knockout technology, including over 60 KRAS-mutant cell lines across a variety of contexts. These efforts have not found a single strong universal synthetic lethal target across all KRAS-mutant contexts. Instead, numerous strong vulnerabilities have emerged with preferential dependency in a subset of KRAS-mutant cell lines. Understanding these vulnerabilities and their context specificity across KRAS-mutant cell lines remains a top priority.
Besides standard CRISPR-Cas9 knockout screening, additional new screening approaches may prove useful for identifying KRAS synthetic lethal candidates. CRISPR-inhibitor (CRISPRi) technology targets a transcriptional repressor to specific genes, and CRISPRi approaches have been developed for genome-scale screening (Gilbert et al. 2014). Moreover, CRISPR-Cas9 libraries have been developed that enable simultaneous knockout of two different genes within the same cell, and will allow examination of pairwise genetic interactions in KRAS-mutant cancer cells (Rosenbluh et al. 2016; Han et al. 2017). Last, because most treatment strategies in KRAS-mutant cancers will require combination therapies, functional genetic screening in the context of small-molecule inhibition of key KRAS effector signaling pathways may prove most fruitful for the identification of high-priority KRAS synthetic lethal interactions. A number of screens have been performed using RNAi technologies to identify combination therapy targets with MAPK pathway inhibition, including FGFR1 (Manchado et al. 2016), BCL-XL (Corcoran et al. 2013), YAP1 (Lin et al. 2015), and PTPN11 (Prahallad et al. 2015).
In addition to performing screens in two-dimensional cell culture growth conditions for established cancer cell lines, it will also be important to evaluate for synthetic lethal candidates in early passage patient-derived cell lines as well as other potentially more physiologically relevant cancer model systems, such as three-dimensional organoid cultures (Sachs and Clevers 2014; Boj et al. 2015) or animal models of KRAS-mutant tumors (Zender et al. 2008; Zuber et al. 2011; Beronja et al. 2013; Carugo et al. 2016). Indeed, utilization of KRAS-mutant cancer models that recapitulate tumor–stroma interactions may enable elucidation of an entirely novel set of KRAS-driven non-cell-autonomous vulnerabilities that could impair tumor growth. Whether these alternative culture techniques or model systems will yield a different set of KRAS synthetic lethal interactors remains an open and important question. In collaboration with the National Cancer Institute’s RAS Initiative, the RAS Synthetic Lethal Network (RSLN) of laboratories has been organized to examine these novel approaches to identify KRAS synthetic lethal targets and to share these data with the larger RAS scientific community to promote rapid translation of clinically useful therapeutic targets.
Beyond identification in functional genetic screens, extensive validation efforts will be required to rigorously demonstrate the value of novel KRAS synthetic lethal targets. Validation will require confirmation of the potency and specificity of the synthetic lethal relationship using multiple genetic and pharmacologic approaches in both established cell lines and patient-derived models. Moreover, although cross comparison of one tumor cell with a KRAS mutation to another that does not have a KRAS mutation may reveal vulnerabilities that are stronger in the KRAS-mutant cancer context, this approach does not guarantee tolerable toxicity in KRAS WT normal cells. Investigators will also need to consider the essentiality of targets within normal cells and tissues that would be most likely to lead to dose-limiting toxicities of targeted cancer therapies. Prioritizing synthetic lethal partners through patterns of expression of the dependent target or its biomarker in normal human tissues or through functional modeling in normal cell culture systems may help address this important issue.
Ultimately, clinical translation of novel synthetic lethal vulnerabilities of oncogenic KRAS into viable therapeutic approaches will require strategies for targeted inhibition of these proteins. Some candidates may possess protein domains that are therapeutically tractable, such as kinases or other enzymes; however, they most likely will not be inherently targetable. As with the oncogenic KRAS protein itself, novel approaches to target “undruggable” proteins are urgently needed. Phthalimide conjugation strategies for targeted protein degradation have recently been described and may be one viable path forward (Winter et al. 2015). It is expected that novel small molecules that directly target mutant KRAS will be soon be developed. Whereas one may argue that the advent of such therapies may obviate the need for targeting synthetic lethal partners, it is inevitable that combination treatments will be needed with direct KRAS inhibitors to overcome compensatory signaling and resistance mechanisms. It is likely that the catalog of synthetic lethal vulnerabilities discussed above as well as novel ones identified through future screening approaches will provide a critical framework for developing multifaceted combination therapy strategies in KRAS-mutant cancers.
ACKNOWLEDGMENTS
We thank Belinda Wang, PhD, for her assistance with the generation of some figures and tables used in this review. We also acknowledge the following funding sources: R01 CA130988 (W.C.H.), U01 CA199253 (W.C.H.), U01 CA 176058 (W.C.H.), P01 CA154303 (W.C.H), and P50 CA127003 (W.C.H. and A.J.A.). A.J.A. is supported by the Hope Funds for Cancer Research Postdoctoral Fellowship, Dana Farber Cancer Institute Hale Center for Pancreatic Cancer, Perry S. Levy Endowed Fellowship, and the Harvard Catalyst and Harvard Clinical and Translational Science Center (UL1 TR001102).
Footnotes
Editors: Linda VanAelst, Julian Downward, and Frank McCormick
Additional Perspectives on Ras and Cancer in the 21st Century available at www.perspectivesinmedicine.org
REFERENCES
- Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, Hanson LJ, Gore L, Chow L, Leong S, et al. 2008. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinasekinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol 26: 2139–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. 2016. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov 6: 914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoitei N, Hoffmann CM, Ellegast JM, Ball CR, Obermayer K, Gossele U, Koch B, Faber K, Genze F, Schrader M, et al. 2012. Targeting of KRAS mutant tumors by HSP90 inhibitors involves degradation of STK33. J Exp Med 209: 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babij C, Zhang Y, Kurzeja RJ, Munzli A, Shehabeldin A, Fernando M, Quon K, Kassner PD, Ruefli-Brasse AA, Watson VJ, et al. 2011. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res 71: 5818–5826. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. 2009. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basseres DS, Ebbs A, Levantini E, Baldwin AS. 2010. Requirement of the NF-κB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res 70: 3537–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, et al. 2013. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154: 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beronja S, Janki P, Heller E, Lien WH, Keyes BE, Oshimori N, Fuchs E. 2013. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature 501: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco RB, Francoz S, Santamaria D, Canamero M, Dubus P, Charron J, Baccarini M, Barbacid M. 2011. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small-cell lung carcinoma. Cancer Cell 19: 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Hahn WC. 2011. Towards systematic functional characterization of cancer genomes. Nat Rev Genet 12: 487–498. [DOI] [PubMed] [Google Scholar]
- Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, et al. 2015. Organoid models of human and mouse ductal pancreatic cancer. Cell 160: 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten CD. 2013. PI3K and MEK inhibitor combinations: Examining the evidence in selected tumor types. Cancer Chemother Pharmacol 71: 1395–1409. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917. [DOI] [PubMed] [Google Scholar]
- Bryant KL, Mancias JD, Kimmelman AC, Der CJ. 2014. KRAS: Feeding pancreatic cancer proliferation. Trends Biochem Sci 39: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carugo A, Genovese G, Seth S, Nezi L, Rose JL, Bossi D, Cicalese A, Shah PK, Viale A, Pettazzoni PF, et al. 2016. In vivo functional platform targeting patient-derived xenografts identifies WDR5-Myc association as a critical determinant of pancreatic cancer. Cell Rep 16: 133–147. [DOI] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. 2011. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci 108: 12372–12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M Jr, et al. 2006. RalB GTPase-mediated activation of the IκB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127: 157–170. [DOI] [PubMed] [Google Scholar]
- Chio II, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Ohlund D, et al. 2016. NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell 166: 963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. 2012. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 122: 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, Charles RP, Rabinovich BA, Hann B, Dankort D, et al. 2012. A central role for RAF→MEK→ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov 2: 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. 2013. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497: 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Contino G, Deshpande V, Tzatsos A, Conrad C, Benes CH, Levy DE, Settleman J, Engelman JA, Bardeesy N. 2011. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res 71: 5020–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, Greninger P, Brown RD, Godfrey JT, Cohoon TJ, et al. 2013. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 23: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Cabral S, Brough R, Konde A, Aarts M, Campbell J, Marinari E, Riffell J, Bardelli A, Torrance C, Lord CJ, et al. 2016. CDK1 is a synthetic lethal target for KRAS mutant tumours. PLoS ONE 11: e0149099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. 2014. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 13: 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis J, Meiri D, Sandi MJ, Radulovich N, Kent OA, Medrano M, Mokady D, Normand J, Larose J, Marcotte R, et al. 2014. The RhoGEF GEF-H1 is required for oncogenic RAS signaling via KSR-1. Cancer Cell 25: 181–195. [DOI] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. 2016. Environment impacts the metabolic dependencies of Ras-driven non-small-cell lung cancer. Cell Metab 23: 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Jonas O, Keibler MA, Hou HW, Luengo A, Mayers JR, Wyckoff J, Del Rosario AM, Whitman M, Chin CR, et al. 2017. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med 23: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. 2011. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475: 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoyelle C, Abou-Rjaily G, Bezrookove V, Verhaegen M, Johnson TM, Fullen DR, Pointer JN, Gruber SB, Su LD, Nikiforov MA, et al. 2006. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol 8: 1053–1063. [DOI] [PubMed] [Google Scholar]
- De Raedt T, Walton Z, Yecies JL, Li D, Chen Y, Malone CF, Maertens O, Jeong SM, Bronson RT, Lebleu V, et al. 2011. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer Cell 20: 400–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietlein F, Kalb B, Jokic M, Noll EM, Strong A, Tharun L, Ozretic L, Kunstlinger H, Kambartel K, Randerath WJ, et al. 2015. A synergistic interaction between Chk1- and MK2 inhibitors in KRAS-mutant cancer. Cell 162: 146–159. [DOI] [PubMed] [Google Scholar]
- Dolma S, Lessnick SL, Hahn WC, Stockwell BR. 2003. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell 3: 285–296. [DOI] [PubMed] [Google Scholar]
- Downward J. 2015. RAS synthetic lethal screens revisited: Still seeking the elusive prize? Clin Cancer Res 21: 1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, Ryan DP, Meyerhardt JA, Benes C, Settleman J, et al. 2011. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest 121: 4311–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebi H, Faber AC, Engelman JA, Yano S. 2014. Not just gRASping at flaws: Finding vulnerabilities to develop novel therapies for treating KRAS mutant cancers. Cancer Sci 105: 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. 2008. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 14: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler M, Hieber M, Arbeiter A, Klein S, Kong B, et al. 2013. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer cell 23: 406–420. [DOI] [PubMed] [Google Scholar]
- Faber AC, Coffee EM, Costa C, Dastur A, Ebi H, Hata AN, Yeo AT, Edelman EJ, Song Y, Tam AT, et al. 2014. mTOR inhibition specifically sensitizes colorectal cancers with KRAS or BRAF mutations to BCL-2/BCL-XL inhibition by suppressing MCL-1. Cancer Discov 4: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921. [DOI] [PubMed] [Google Scholar]
- Frohling S, Scholl C. 2011. STK33 kinase is not essential in KRAS-dependent cells—Letter. Cancer Res 71: 7716; author reply 7717. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Wang SC, Morris JPt, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ, et al. 2011. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell 19: 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. 2012. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Carugo A, Tepper J, Robinson FS, Li L, Svelto M, Nezi L, Corti D, Minelli R, Pettazzoni P, et al. 2017. Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature 542: 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, Jeng HH, Bar-Sagi D. 2014. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer cell 25: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabocka E, Commisso C, Bar-Sagi D. 2015. Molecular pathways: Targeting the dependence of mutant RAS cancers on the DNA damage response. Clin Cancer Res 21: 1243–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. 2011. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25: 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, Der CJ, Counter CM. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev 16: 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. 2017. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol 35: 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163: 1515–1526. [DOI] [PubMed] [Google Scholar]
- Hayes TK, Neel NF, Hu C, Gautam P, Chenard M, Long B, Aziz M, Kassner M, Bryant KL, Pierobon M, et al. 2016. Long-term ERK inhibition in KRAS-mutant pancreatic cancer is associated with MYC degradation and senescence-like growth suppression. Cancer Cell 29: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer F, Fields S, Schneider C, Martin GS. 1994. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci 91: 11089–11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante JR, Fecher LA, Falchook GS, Nallapareddy S, Gordon MS, Becerra C, DeMarini DJ, Cox DS, Xu Y, Morris SR, et al. 2012. Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: A phase 1 dose-escalation trial. Lancet Oncol 13: 773–781. [DOI] [PubMed] [Google Scholar]
- Infante JR, Papadopoulos KP, Bendell JC, Patnaik A, Burris HA III, Rasco D, Jones SF, Smith L, Cox DS, Durante M, et al. 2013. A phase 1b study of trametinib, an oral Mitogen-activated protein kinase kinase (MEK) inhibitor, in combination with gemcitabine in advanced solid tumours. Eur J Cancer 49: 2077–2085. [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr. 2005. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 5: 689–698. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y, Wu CJ, Sadanandam A, Hu B, et al. 2014. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Frese KK, DeNicola GM, Baccarini M, Tuveson DA. 2011. C-Raf is required for the initiation of lung cancer by K-RasG12D. Cancer Discov 1: 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Demo SD, Ye ZH, Chen YW, Williams LT. 1994. ralGDS family members interact with the effector loop of ras p21. Mol Cell Biol 14: 7483–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Mendiratta S, Kim J, Pecot CV, Larsen JE, Zubovych I, Seo BY, Kim J, Eskiocak B, Chung H, et al. 2013. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell 155: 552–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, McMillan E, Kim HS, Venkateswaran N, Makkar G, Rodriguez-Canales J, Villalobos P, Neggers JE, Mendiratta S, Wei S, et al. 2016. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature 538: 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Thummalapalli R, Barbie DA. 2016. Inflammation as a driver and vulnerability of KRAS mediated oncogenesis. Semin Cell Dev Biol 58: 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. 2014. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32: 267–273. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, Armenteros-Monterroso E, Lassailly F, Matthews N, Nye E, et al. 2012. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small-cell lung cancer. Cell 149: 642–655. [DOI] [PubMed] [Google Scholar]
- Lamba S, Russo M, Sun C, Lazzari L, Cancelliere C, Grernrum W, Lieftink C, Bernards R, Di Nicolantonio F, Bardelli A. 2014. RAF suppression synergizes with MEK inhibition in KRAS mutant cancer cells. Cell Rep 8: 1475–1483. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. 2014. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26: 207–221. [DOI] [PubMed] [Google Scholar]
- Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, Yoshimura A, Reindl W, Sipos B, Akira S, et al. 2011. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 19: 456–469. [DOI] [PubMed] [Google Scholar]
- Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, Der CJ, Counter CM. 2005. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7: 533–545. [DOI] [PubMed] [Google Scholar]
- Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, Asthana S, Neel D, Yan JJ, Lu X, et al. 2015. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet 47: 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, Li J, Peng B, Fleming JB, Wang H, et al. 2012. KrasG12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell 21: 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J. 2011. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell 22: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. 2017. PARP inhibitors: Synthetic lethality in the clinic. Science 355: 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. 2008. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci 105: 20380–20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. 2009a. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137: 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. 2009b. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell 136: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Masson K, Jaffe JD, Silkworth W, Ross NT, Scherer CA, Scholl C, Frohling S, Carr SA, Stern AM, et al. 2012. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proc Natl Acad Sci 109: 2860–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Weissmueller S, Morris JPt, Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, et al. 2016. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 534: 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, Sircoulomb F, Medrano M, Fedyshyn Y, Koh JL, et al. 2012. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov 2: 172–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, Virtanen C, Bradner JE, Bader GD, Mills GB, et al. 2016. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell 164: 293–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM, et al. 2016. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353: 1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLornan DP, List A, Mufti GJ. 2014. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med 371: 1725–1735. [DOI] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T. 2009. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature 462: 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS, Downward J. 2013. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov 3: 548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie SA, Willumsen BM, Weber MJ, Wolfman A. 1993. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science 260: 1658–1661. [DOI] [PubMed] [Google Scholar]
- Morgan-Lappe SE, Tucker LA, Huang X, Zhang Q, Sarthy AV, Zakula D, Vernetti L, Schurdak M, Wang J, Fesik SW. 2007. Identification of Ras-related nuclear protein, targeting protein for xenopus kinesin-like protein 2, and stearoyl-CoA desaturase 1 as promising cancer targets from an RNAi-based screen. Cancer Res 67: 4390–4398. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, et al. 2006. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441: 106–110. [DOI] [PubMed] [Google Scholar]
- Papke B, Der CJ. 2017. Drugging RAS: Know the enemy. Science 355: 1158–1163. [DOI] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. 2015. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162: 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K, Politi K, Beverly LJ, Varmus HE. 2008. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci 105: 5242–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Heynen GJ, Germano G, Willems SM, Evers B, Vecchione L, Gambino V, Lieftink C, Beijersbergen RL, Di Nicolantonio F, et al. 2015. PTPN11 Is a central node in intrinsic and acquired resistance to targeted cancer drugs. Cell Rep 12: 1978–1985. [DOI] [PubMed] [Google Scholar]
- Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G, Guerra C, Santamaria D, Barbacid M. 2010. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small-cell lung carcinoma. Cancer Cell 18: 63–73. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. 2011. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer 11: 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. 1994. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370: 527–532. [DOI] [PubMed] [Google Scholar]
- Rosenbluh J, Mercer J, Shrestha Y, Oliver R, Tamayo P, Doench JG, Tirosh I, Piccioni F, Hartenian E, Horn H, et al. 2016. Genetic and proteomic interrogation of lower confidence candidate genes reveals signaling networks in β-catenin-active cancers. Cell Syst 3: 302–316 e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MB, Der CJ, Wang-Gillam A, Cox AD. 2015. Targeting RAS-mutant cancers: Is ERK the key? Trends Cancer 1: 183–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N, Clevers H. 2014. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev 24: 68–73. [DOI] [PubMed] [Google Scholar]
- Sarthy AV, Morgan-Lappe SE, Zakula D, Vernetti L, Schurdak M, Packer JC, Anderson MG, Shirasawa S, Sasazuki T, Fesik SW. 2007. Survivin depletion preferentially reduces the survival of activated K-Ras-transformed cells. Mol Cancer Therapeut 6: 269–276. [DOI] [PubMed] [Google Scholar]
- Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S, et al. 2009. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137: 821–834. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. 2015. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao DD, Tsherniak A, Gopal S, Weir BA, Tamayo P, Stransky N, Schumacher SE, Zack TI, Beroukhim R, Garraway LA, et al. 2013. ATARiS: Computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Res 23: 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, et al. 2014. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Winslow MM, Magendantz M, Ouyang C, Dowdle J, Subramanian A, Lewis TA, Maglathin RL, Tolliday N, Jacks T. 2011. Selective killing of K-ras mutant cancer cells by small molecule inducers of oxidative stress. Proc Natl Acad Sci 108: 8773–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. 2009. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Sweeney MF, Yu M, Burger A, Greninger P, Benes C, Haber DA, Settleman J. 2012. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell 148: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolander A, Yamamoto K, Huber BE, Lapetina EG. 1991. Association of p21ras with phosphatidylinositol 3-kinase. Proc Natl Acad Sci 88: 7908–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et al. 2013. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaargaren M, Bischoff JR. 1994. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap. Proc Natl Acad Sci 91: 12609–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckel M, Molina-Arcas M, Weigelt B, Marani M, Warne PH, Kuznetsov H, Kelly G, Saunders B, Howell M, Downward J, et al. 2012. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res 22: 1227–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen AG, Esposito D, Bagni RK, McCormick F. 2014. Dragging Ras back in the ring. Cancer Cell 25: 272–281. [DOI] [PubMed] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. 2017. Defining a cancer dependency map. Cell 170: 564-576.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. 2014. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514: 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent S, Chen R, Sayles LC, Lin C, Walker RG, Gillespie AK, Subramanian A, Hinkle G, Yang X, Saif S, et al. 2010. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J Clin Invest 120: 3940–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74: 205–214. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ngo VN, Marani M, Yang Y, Wright G, Staudt LM, Downward J. 2010. Critical role for transcriptional repressor Snail2 in transformation by oncogenic RAS in colorectal carcinoma cells. Oncogene 29: 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, Sabatini DM. 2017. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168: 890–903 e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne PH, Viciana PR, Downward J. 1993. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature 364: 352–355. [DOI] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE. 2015. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348: 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, et al. 2007. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 447: 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Liu D, Li L, Wempe MF, Guin S, Khanna M, Meier J, Hoffman B, Owens C, Wysoczynski CL, et al. 2014. Discovery and characterization of small molecules that target the GTPase Ral. Nature 515: 443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Stockwell BR. 2008. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol 15: 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, et al. 2011. Pancreatic cancers require autophagy for tumor growth. Genes Dev 25: 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, Von Hoff DD, Maitra A, Kimmelman AC. 2014. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov 4: 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. 2012. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149: 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TL, Fellmann C, Lee CS, Ritchie CD, Thapar V, Lee LC, Hsu DJ, Grace D, Carver JO, Zuber J, et al. 2014. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov 4: 1182–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, Roper J, Chio II, Giannopoulou EG, Rago C, et al. 2015. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science 350: 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. 2008. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135: 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Settleman J, Kyriakis JM, Takeuchi-Suzuki E, Elledge SJ, Marshall MS, Bruder JT, Rapp UR, Avruch J. 1993. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature 364: 308–313. [DOI] [PubMed] [Google Scholar]
- Zhang W, Nandakumar N, Shi Y, Manzano M, Smith A, Graham G, Gupta S, Vietsch EE, Laughlin SZ, Wadhwa M, et al. 2014. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal 7: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Adjei AA. 2014. The clinical development of MEK inhibitors. Nat Rev Clin Oncol 11: 385–400. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Aref AR, Cohoon TJ, Barbie TU, Imamura Y, Yang S, Moody SE, Shen RR, Schinzel AC, Thai TC, et al. 2014. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov 4: 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]