Abstract
Recent studies point to a diverse assemblage of prokaryotic cognates of the eukaryotic ubiquitin (Ub) system. These systems span an entire spectrum, ranging from those catalyzing cofactor and amino acid biosynthesis, with only adenylating E1-like enzymes and ubiquitin-like proteins (Ubls), to those that are closer to eukaryotic systems by virtue of possessing E2 enzymes. Until recently E3 enzymes were unknown in such prokaryotic systems. Using contextual information from comparative genomics, we uncover a diverse group of RING finger E3s in prokaryotes that are likely to function with E1s, E2s, JAB domain peptidases and Ubls. These E1s, E2s and RING fingers suggest that features hitherto believed to be unique to eukaryotic versions of these proteins emerged progressively in such prokaryotic systems. These include the specific configuration of residues associated with oxyanion-hole formation in E2s and the C-terminal UFD in the E1 enzyme, which presents the E2 to its active site. Our study suggests for the first time that YukD-like Ubls might be conjugated by some of these systems in a manner similar to eukaryotic Ubls. We also show that prokaryotic RING fingers possess considerable functional diversity and that not all of them are involved in Ub-related functions. In eukaryotes, other than RING fingers, a number of distinct binuclear (chelating two Zn atoms) and mononuclear (chelating one zinc atom) treble clef domains are involved in Ub-related functions. Through detailed structural analysis we delineated the higher order relationships and interaction modes of binuclear treble clef domains. This indicated that the FYVE domain acquired the binuclear state independently of the other binuclear forms and that different treble clef domains have convergently acquired Ub-related functions independently of the RING finger. Among these, we uncover evidence for notable prokaryotic radiations of the ZF-UBP, B-box, AN1 and LIM clades of treble clef domains and present contextual evidence to support their role in functions unrelated to the Ub-system in prokaryotes. In particular, we show that bacterial ZF-UBP domains are part of a novel cyclic nucleotide-dependent redox signaling system, whereas prokaryotic B-box, AN1 and LIM domains have related functions as partners of diverse membrane-associated peptidases in processing proteins. This information, in conjunction with structural analysis, suggests that these treble clef domains might have been independently recruited to the eukaryotic Ub-system due to an ancient conserved mode of interaction with peptides.
Introduction
Protein modification via covalent attachment of Ubiquitin (Ub) and related proteins (Ubls) plays a vital role in regulation of protein–protein interactions, signaling and protein stability.1 The enzyme cascade directing the conjugation of Ub/Ubls to amino groups in substrate proteins or lipids consists of the E1, E2, and E3 components.2 These components, respectively, charge the carboxyl group of the Ub/Ubl in an ATP-dependent manner, transfer it via transthiolation to an active cysteine, and finally conjugate it to the amino group on the substrate by the formation of an isopeptide linkage.3 Two distinct, structurally unrelated classes of E3 ligases have been identified: the HECT ligases and the RING ligases.4,5 HECT domains contain a conserved cysteine residue, which participates in a further thiotransfer of the Ub/Ubl from the E2 ligase, prior to its conjugation to the substrate protein.6 In contrast, the RING ligase acts as an adaptor protein facilitating the transfer of the Ub/Ubl from the E2 ligase directly to the substrate protein.7 The RING finger often co-occurs in the same polypeptide with other domains facilitating substrate interaction or as a subunit of a large protein complex, whose other components, such as the F-box proteins, mediate interactions with the target protein.8
The RING finger displays the treble-clef fold: a small, remarkably versatile structural scaffold found in a wide range of functional contexts across cellular life.9 The treble-clef fold is comprised of three substructures: an N-terminal “lateral flap”, a central β-hairpin, and a C-terminal α-helix (Fig. 1A).9–11 The classical treble-clef domains are stabilized primarily by means of a Zn2+ cation chelated by two cysteines from the tip of the lateral flap and another pair from the beginning of the C-terminal helix. These cysteines and the chelated metal might be lost in certain versions such as the U-box and some types of HNH/EndoVII-fold nucleases on account of secondary acquisition of alternative stabilizing interactions.12,13 The Zn-chelating treble-clef domains can further be divided into two general classes (Fig. 1A): those that retain the ancestral single Zn2+ ion chelation site are termed mononuclear, whereas those that have acquired a second Zn-chelation site are termed binuclear treble-clefs.9 The RING domain falls in the latter category, which also includes the chromatin peptide-binding PHD domain,14,15 the phosphatidylinositol 3-phosphate phospholipid (PI3P)-interacting FYVE domain,16 the diaglycerol-binding C1 domain,17 the ubiquitin-binding zinc-finger UBP domain,18 and the B-box, AN1, MYND, ZZ and UPF1 domains.19–22 In these binuclear versions, the second Zn-chelating site invariably involves a pair of cysteines or a cysteine–histidine dyad from the β-hairpin and another pair from either the end of the C-terminal helix or a further extension downstream of it.
Fig. 1.
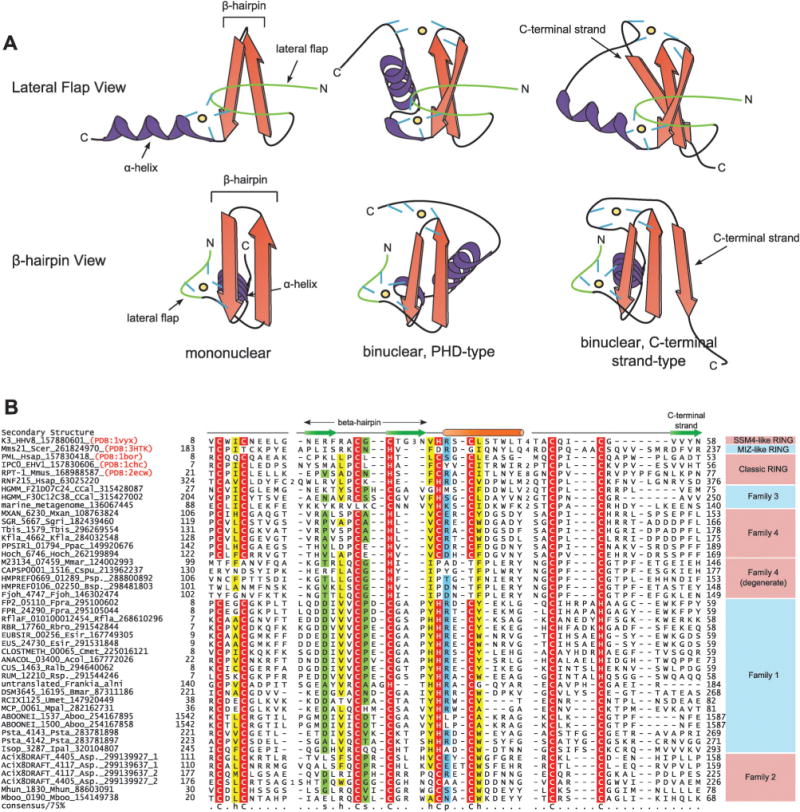
(A) Cartoon representations of mononuclear and PHD-type and C-terminal strand-containing binuclear treble clef domains. Two views are provided: one with the line of sight perpendicular to the lateral flap (top) and one with the line of sight perpendicular to the β-hairpin. β-Strands are depicted as arrows colored in orange with the arrowheads at the C-terminal end, while α-helices are depicted as coils, colored in purple. The lateral flap structure is colored in green. Zinc ion coordinating residues are depicted as short lines, colored in blue. The relative spatial location of zinc ions are marked with a black circle shaded in yellow. (B) Multiple sequence alignment of RING finger with a special emphasis on prokaryotic versions. Proteins are annotated by their gene names, species abbreviations and Genbank index (gi) numbers and are further grouped by their familial associations, shown to the right of the alignment. Secondary structure assignments are shown above the alignment, where the green arrow represents the β-strand and the orange cylinder the α-helix. Secondary structure was derived from a combination of crystal structures and alignment based predictions. Poorly conserved inserts are replaced by the corresponding number of residues. The alignment was colored based on 75% consensus and the coloring scheme and consensus abbreviations are as in Fig. 4. Species abbreviations are as follows: Aboo: Aciduliprofundum boonei; Acol: Anaerotruncus colihominis; Asp.: Acidobacterium sp.; Bmar: Blastopirellula marina; Bsp.: Bacteroides sp.; CCal: Candidatus Caldiarchaeum; Cmet: Clostridium methylpentosum; Cspu: Capnocytophaga sputigena; EHV1: Equid herpesvirus 1; Esir: Eubacterium siraeum; Fjoh: Flavobacterium johnsoniae; Fpra: Faecalibacterium prausnitzii; HHV8: Human herpesvirus 8; Hoch: Haliangium ochraceum; Hsap: Homo sapiens; Ipal: Isosphaera pallida; Kfla: Kribbella flavida; Mboo: Methanoregula boonei; Mhun: Methanospirillum hungatei; Mmar: Microscilla marina; Mmet: marine metagenome; Mmus: Mus musculus; Mpal: Methanocella paludicola; Mxan: Myxococcus xanthus; Ppac: Plesiocystis pacifica; Psp.: Prevotella sp.; Psta: Pirellula staleyi; Ralb: Ruminococcus albus; Rbro: Ruminococcus bromii; Rfla: Ruminococcus flavefaciens; Rsp.: Ruminococcus sp.; Scer: Saccharomyces cerevisiae; Sgri: Streptomyces griseus;Tbis: Thermobispora bispora; Umet: uncultured methanogenic archaeon RC-I.
Due to their presence in ancient proteins such as the ribosomal proteins S14 and L2411 and the HNH/EndoVII-like nucleases, the mononuclear treble clef domains are inferred to have been already present in the Last Universal Common Ancestor of Life (LUCA). However, several distinct clades of both mononuclear and binuclear treble-clef domains appear to have radiated in eukaryotes.23 In particular, the mononuclear versions furnished several eukaryote-specific nucleic acid-binding Zn-finger domains, such as the GATA, the nuclear hormone receptor, LIM, THAP and TRASH domains, the Ub-binding A20 domain and the ARF GTPase activating protein (GAP) domain, which is related to GATA.24 While prokaryotes do not have a comparably large expansion of mononuclear treble-clef clades, some diversity is seen in the DNA-binding domains of HNH proteins and the TraR-like transcription factors, YacG-like proteins, the ClpX N-terminal domain and the type-II thymidine kinase Zn-binding domain.9,24 In contrast, the majority of binuclear treble-clefs (e.g. B-box, MYND, ZZ, C1, FYVE, PHD and RING) were only known from eukaryotes.23,25 AN1 and UBP-type binuclear treble clefs were previously observed in prokaryotes; however, they were not considered to have a widespread role in those organisms.26,27 It was hence believed that most of the major binuclear versions emerged early in eukaryotic evolution and rapidly diversified concomitant with the expansion of quintessentially eukaryotic systems. This proposal was generally supported by their primary functional roles in these eukaryotic systems, such as chromatin structure and dynamics (e.g. PHD, ZZ and MYND), the ubiquitin system (RING and B-box),28 the cytoskeleton (ZZ and MYND),29,30 and cellular lipid-membranes and lipid-based signaling (C1 and FYVE).31–35
In our earlier work on the origin of the ubiquitin system we had used contextual information from conserved geneneighborhoods to show that a phylogenetically diverse group of bacteria possess systems combining genes encoding E1s, E2s, JAB domain peptidases and Ub-like proteins.36–38 These were inferred to function as prokaryotic cognates of the eukaryotic Ub-conjugation system and this proposal has received some support from recent work on archaeal systems related to Ub-conjugation.39,40 However, these neighborhoods all lacked the cognate E3 RING-like domain.36 Very recently, a gene encoding a putative treble clef-containing RING domain was reported in a newly-sequenced archaeal genome,41 suggesting a possible deeper origin for the RING domain. Furthermore, this remarkable archaeal gene was found to be adjacent to genes encoding the other core components of the Ub-system: the Ub modifier, the E1 and E2 ligases, and a JAB domain peptidase41 suggesting that it could indeed be a direct precursor of the eukaryote-type Ub-system.
This and other new genomic data raise questions regarding the early evolutionary history of what were considered to be purely eukaryotic binuclear treble-clef domains and the emergence of Ub-related functions in the treble clef fold. Hence, we systematically analyzed public sequence repositories to explore the diversity of RING fingers and other treble-clef domains in prokaryotes. In addition to confirming that the aforementioned archaeal sequence contains a bona fide RING finger, we uncovered other representatives of this domain sporadically distributed across phylogenetically diverse bacteria and archaea. Some of these members, similar to the recent finding in archaea, are found in gene neighborhoods containing homologs of the core components of the eukaryotic ubiquitin system. Additionally, we characterized the prokaryotic members of the UBP clade of binuclear treble-clef domains and use comparative genome contextual analyses to predict a role for the family in bacterial redox signaling pathways. We also present evidence that the AN1-like and B-box-like binuclear treble clef domains have clear prokaryotic origins. Comparison of the shared structural features of these binuclear treble-clef clades suggests that multiple versions of the binuclear treble clefs had emerged in prokaryotes and transferred to the progenitors of eukaryotes prior to the Last Eukaryotic Common Ancestor (LECA). These findings have several implications for understanding the recruitment of these domains to diverse eukaryotic roles, particularly the provenance of the Ub-modification system.
Results and discussion
Contextual evidence points to a potential modifier function for the YukD-family of Ubls in bacteria
In an attempt to extend the known network of functional associations for prokaryotic Ubls, we ran sensitive sequence profile searches using previously identified prokaryotic Ubl domains36,42 as seeds for the PSI-BLAST program.43 As a result we uncovered previously unidentified members of the YukD family of Ubls44 in the planctomycetes Pirellula staleyi (Psta_4149; gi: 283781904) and Isosphaera pallida (Isop_1340; gi: 320102883) and the actinobacterium Frankia alni (FRAAL0857; gi: 111220329). Examination of the gene neighborhoods for these proteins revealed the presence of prokaryotic homologs of components of the Ub-system, namely, E1 and E2 ligases and JAB-like peptidases (Fig. 2), similar to previously described gene neighborhoods for prokaryotic Ubls.36 Given that such conserved geneneighborhoods, especially those shared by phylogenetically distant prokaryotes, are indicative of a close functional interaction between the gene products,53,54 we reasoned that these YukD-like Ubls could be conjugated to target proteins through enzymatic ligase activity of the E1 and E2 enzymes and C-terminally processed or deconjugated by the action of the JAB peptidase. Consistent with this proposal, we detected a conserved AG motif near the C-terminus of these newly detected YukD-like Ubls (see ESI†) comparable to the flexible C-terminal tails with small residues that are important for the ligation reaction of both the Ubl49,50 and the structurally unrelated Pup modifiers.51,52
Fig. 2.
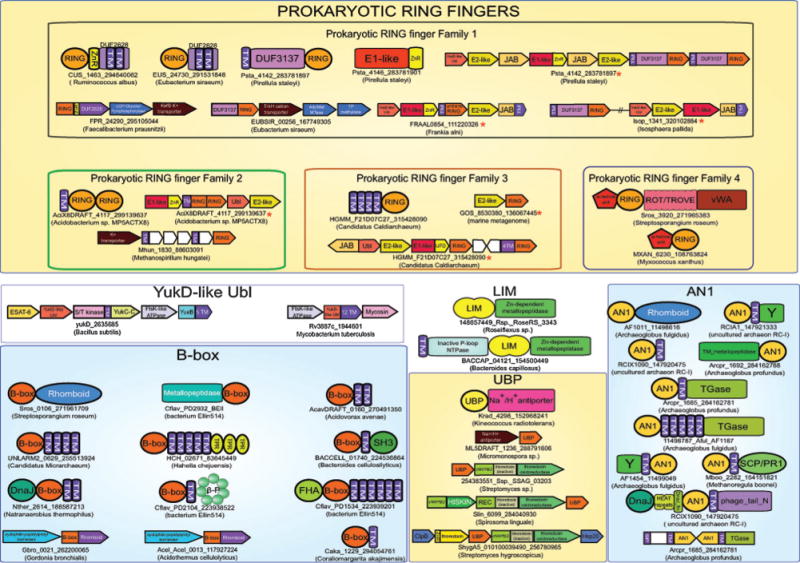
Architectures and operons are grouped according to the treble-clef domain or ubiquitin domain that is contained in them. Operons that contain other proteins or domains involved in the ubiquitin system pathway are marked with a red asterisk. Genes that are not translated in the database are marked with an “untrans” prefix. Note that the Isosphaera RING domain does not co-occur with the ubiquitin pathway genes, but is in a distinct genome location with respect to the latter. Architectures and operons are labeled by the gene names, gis and species name (in brackets). Genes in conserved gene neighborhoods are shown as boxed arrows with the arrow head pointing in the 3′ direction. Standard abbreviations are used for most domains. Non-standard abbreviations include: Y: uncharacterized conserved domain, β-P: WD40-like betapropeller repeats, HISKIN: histidine kinase, REC: receiver, TM: transmembrane helix, and TGase: transglutaminase, ZnR: zinc ribbon.
This observation was surprising because: (1) members of the YukD family have hitherto not been contextually linked to prokaryotic cognates of the eukaryotic Ub-system and (2) early experiments with Bacillus subtilis YukD did not find evidence for protein modification activity.44 Hence, we re-investigated the known members of the YukD family in light of the above observations. We had previously reported that the YukD-like Ubl family is associated in conserved gene neighborhoods with members of the ESAT-6 export pathway (also called Type VII secretion system or ESX) of Gram-positive bacteria, suggesting a role for YukD in regulating this export system.36 This prediction was borne out by subsequent experiments in firmicutes that have shown that the YukD (EsaB in S. aureus) protein regulates the secretion of effector molecules via the ESAT-6 secretion system.45 In actinobacteria, the YukD-like Ubl is fused to the N-terminus of a transmembrane protein of the ESAT-6 pumping pathway (typified by the Rv3887c protein in Mycobacterium tuberculosis), and is in the neighborhood of a gene encoding a subtilisin-like serine peptidase (mycosin; Fig. 2). Cleavage of regulatory components by mycosins, such as MycP1, has been shown to modulate the ESAT-6/ESX secretion system.46 In the firmicutes, the YukD-like Ubls are standalone versions, almost always co-occurring in predicted operons with two genes: (1) a gene encoding a membrane protein containing an N-terminal intracellular domain, which displays the same fold as the serine/threonine/tyrosine kinases, and a C-terminal α-helical extracellular domain (Fig. 2). (2) A gene coding for a 5-TM membrane protein (e.g. Bacillus subtilis protein YueB), analogous to the transmembrane transporter of the actinobacteria.
The gene coding for the protein with the kinase fold domain is usually located in the same relative position and orientation as the mycosin gene in the actinobacterial operons. While sharing a common fold, the kinase domain is not closely related to conventional protein kinases that phosphorylate peptides. Instead, it is reminiscent of the ATP-dependent ligases, which contain a divergent version of the protein kinase fold, and catalyze the formation of peptide-bond like linkages by activating the carboxyl group through phorphorylation. Such ligases catalyze the non-ribosomal formation of peptide linkages during the biosynthesis of metabolites like the siderophores, vibrioferrin and achromobactin.47,48 Hence, by analogy to Pup conjugation, which involves a functionally comparable peptide ligase,48 it is possible that the YukD-like Ubl of firmicutes is conjugated to the multi-TM protein of the ESAT-6/ESX system, by the predicted peptide ligase action of the kinase domain. This could conceivably be further regulated by interactions of the extracellular domain associated with the kinase domain with other molecules under specific conditions. In actinobacteria, instead of ligation, regulation could involve cleavage of the YukD-like Ubl domain that comes fused to the ESAT-6/ESX system transmembrane protein by the mycosin peptidase. A possible precedent for this proposal is provided by cleavage of the Ubl domains found in the tail assembly related proteins of the lambdoid bacteriophages by the associated JAB domain peptidase.36 Hence, our observations suggest that regulatory systems deploying YukD-like Ubls might span the entire spectrum, including forms which are not conjugated, those which are conjugated via an apparatus resembling the eukaryotic Ub-system and also perhaps via a distinct kinase-related peptide ligase.
Identification and characterization of novel prokaryotic RING-like domains
To better understand the conjugation apparatus of the newly identified YukD-like Ubls we further systematically investigated their gene-neighborhoods and domain architectures. In the Pirellula staleyi gene neighborhood, two paralogous genes were observed immediately downstream of the genes for the above-mentioned components, with single N-terminal transmembrane helices and C-terminal cysteine-rich globular regions (Fig. 2). Initial iterations of sequence profile-based searches initiated with this globular region identified several related proteins present across a range of phyletically diverse prokaryotes including actinobacteria, firmicutes, planctomycetes, and a single euryarchaeon. Two additional methanogenic euryarchaeal sequences were also recovered in further transitive searches with these sequences (Fig. 1B). Continued iterative searching with this domain detected a significant relationship with RING finger domains e.g. the cellulose synthase RING-like domain in various plants including Shorea parvifolia (detected gi: 254554078, iteration: 3, e-value = 3 × 10−3). In parallel, iterative HMM searches on this globular domain were initiated using the JACKHMMER program (http://hmmer.janelia.org), with results confirming the relationships observed above. For example, a search initiated with the euryarchaeal Aciduliprofundum boonei homolog identified homology with the RING domain in the Arabidopsis lyrata RHA1a protein (gi: 297809395, iteration: 2, e-value: 1.9 × 10−4) and the RING domain in the Rattus norvegicus TTC3 protein (gi: 157817021, iteration: 4, e-value: 2.5 × 10−6). Reciprocal JACKHMMER searches initiated with the abovementioned sequences from methanogenic euryarchaea also established their membership in this family. Using the Pirellula staleyi RING finger domain, as query in TBLASTN searches, we also recovered an unannotated gene coding for a protein with an N-terminal TM segment and a C-terminal RING finger domain in the YukD operon in Frankia alni.
Further searches initiated with the E2 domain encoded in the Pirellula gene neighborhood identified two additional closely related E2-like domains in Acidobacteria sp. MP5ACTX8. Examination of their neighboring genes revealed the presence of an E1 gene related to those observed in Pirellula staleyi and Frankia alni, along with paralogous genes encoding an N-terminal transmembrane helix and a large cysteine-rich region (gis: 299139637, 299139927, Fig. 2). JACKHMMER searches initiated with this region recovered significant hits mapping to eukaryotic RING domains in two non-overlapping locations (e = 10−4−10−5), suggesting the presence of two copies of the domain and also detected two novel methanogenic euryarchaeal sequences containing a single copy of the domain (Fig. 1B). New profile searches with PSI-BLAST and HMMSEARCH (from the HMMER3 package) run with profiles or PSSMs including the above-detected prokaryotic RING domains resulted in the identification of an additional family of bacterial RING domains from several actinobacteria, myxobacteria and bacteroidetes (e.g. MXAN_6230; gi: 108763824). Concurrent with these discoveries, a paper was recently published which described a single RING domain-containing gene in the newly-sequenced archeaon Candidatus Caldiarchaeum subterraneum, proposed to be representative of a novel division of archaea.41 This RING domain was also notably linked in a gene neighborhood encoding a Ubl, E1 and E2 ligases, and a JAB domain peptidase, the first neighborhood in an archaeon to contain the complete basic eukaryotic Ub conjugation machinery. Searches initiated with this sequence recovered two other paralogous RING finger proteins from Caldiarchaeum subterraneum, other related RING fingers from other poorly characterized marine prokaryotes, and multiple eukaryotic RING domains with significant e-values. These searches suggested that these RING fingers displayed an even closer affinity for their eukaryotic counterparts than the above identified set of prokaryotic RING homologs.
We constructed a multiple alignment of all prokaryotic RING-like sequences and examined it for concordance with the known features of the version of the treble-clef domain found in the eukaryotic RING fingers (Fig. 1B). Of primary interest was the conserved presence of the eight metal ion-coordinating residues in the anticipated locations in all but one of the prokaryotic versions. Interestingly, many of the identified prokaryotic RING domains have a C4HC3 coordinating residue arrangement, in contrast with the canonical C3HC4 arrangement observed in eukaryotes. Another variation observed in certain prokaryotic RING domains is a C4HC2H configuration, with a longer insert present between the final pair of coordinating residues (Fig. 1B). Yet other prokaryotic forms display only cysteine residues in the metalcoordinating positions. Finally, the RING fingers identified in bacteroidetes (Fig. 1B, see above) showed a loss of two pairs of Zn-chelating residues, namely those from the lateral flap and those from these C-terminal helices. However, they retain the two pairs from the central hairpin and the C-terminal extension (Fig. 1A and B). The conservation patterns help in delineating the prokaryotic RING fingers into four distinct clades (Fig. 1B). The diversity observed in the key coordinating residues of prokaryotic RING domains is reminiscent of the diversity observed in the catalytic residues of prokaryotic E1 and E2 domains, which are linked in conserved gene neighborhoods to other components of the predicted prokaryotic Ubl modification pathways.36–38 Thus, new genomic evidence indicates that RING fingers had already diversified in prokaryotes, were widely disseminated across diverse groups, and that at least a subset of these diverse forms primarily participated in the context of the prokaryotic cognates of the Ub-system. This suggests that the E3 function of RING fingers appears to have first emerged in prokaryotes. The versions identified here are primarily found in free-living bacteria. It should be stressed that these newly detected RING finger domains are distinct from the RING and U-box domains previously found in intracellular parasitic and symbiotic bacteria.55,56 These latter versions appear to be more recent lateral transfers from host eukaryotes that are deployed by the symbiotic or parasitic bacteria in the host cell to modify its behavior.57,58 They show no genome context associations with E2s or E1s and depend on host E1s and E2s for their activity.
We then attempted to exploit the identification of RING fingers in prokaryotes to decipher the emergence of E3 and other Ub-related functions among treble-clef domains. In eukaryotes, not just the RING finger, but also few mononuclear treble-clef domains (e.g. A20 and C4DM) and multiple binuclear domains (e.g. AN1, UBP, B-box and to a certain extent the MYND finger) are involved in functions related to the Ub-system.59–65 This raised the question as to whether the treble-clefs acquired these Ub-related functions in their common ancestor with the RING finger, or convergently on multiple occasions. Presence of multiple, distinct clades of prokaryotic RING fingers, with distinct patterns of metal chelation, alongside other binuclear treble clef domains hinted that the primary diversification of at least some of these domains might have occurred in prokaryotes. To better characterize the situation and understand the implications of the prokaryotic treble-clef domains for the emergence of functions related to the Ub-system we systematically analyzed: (1) the structural features characteristic of different clades of treble-clef domains; (2) their interactions with proteins and other biomolecules; (3) the phyletic distributions and contextual associations of prokaryotic treble-clef domains.
Structural distinctions among binuclear treble clef domains
The binuclear treble-clef domains, with the exception of the FYVE and IBR domains (see below), share a similar second metal-binding site C-terminal to the α-helix of the core trebleclef domain (Fig. 1A). The location of the second metal ion is largely spatially conserved between different binuclear versions, lying approximately in the middle of the angle formed by the central β-hairpin and the C-terminal α-helix.9,11 Further, the first residue of this last coordinating pair is always located at the end of the C-terminal helix (Fig. 1A). Together, these features argue for a common ancestry for most binuclear treble clef domains, namely the RING, B-box, TFIIH-p44 Zn-finger, ZZ, C1, UBP, PHD, AN1, and MYND domains.
Variations in the configuration of the final coordinating pair, in conjunction with C-terminal extensions, unite multiple distinct domains into structurally similar groups that might reflect further higher-order clades within this monophyletic assembly of binuclear treble clefs. The most conspicuous structural embellishment to the conserved core of the binuclear treble-clef domains is the presence of a C-terminal extended region that stacks back with the central β-hairpin, resulting in an effectively 3-stranded structure.10 This is observed in the RING, B-box, UBP, ZZ, TFIIH-p44 ZnF and C1-type treble clefs (Fig. 1A). As noted previously, this element has been moved to the N-terminus along with one of the Zn-chelating residues due to a postulated circular permutation event in the C1 domains.9 This strand appears unique to these binuclear domains; while some mononuclear treble-clef domains like the THAP and certain members of the HNH clade have extended regions proximal to the core treble clef scaffold,13,66,67 fundamental differences in their stacking, arrangement of the additional strands, and interactions with other structural elements strongly suggest that these are independent innovations. Presence of this C-terminal strand is also related to the positioning of the final metal coordinating pair. In the versions with this strand, the second residue in the pair is invariably found near the N-terminus of the strand. Notably, even in the circularly-permutated C1 domain, this residue is retained in the same position, providing further evidence that the C1 domain is indeed a member of the above assemblage of treble clef domains. These features together indicate that all the versions with this C-terminal strand are likely to form a monophyletic group within the binuclear treble clef domains.
Within this monophyletic assemblage, RING fingers (including the U-box) are distinguished from the rest by a distinctive “squiggle” comprised of half a helical turn (encompassing both final pairs of Zn-chelating residues), at the junction between the C-terminal strand and the preceding α-helix of the core treble-clef domain.12 The UBP domain is also clearly distinguished from the rest by several additional features: (1) the C-terminal additional strand is part of an extended β-meander that combines with the β-hairpin of the core treble clef to form a five-stranded β-sheet. (2) The two residues in the 3rd Zn-chelating pair (i.e. those at the beginning of the C-helix of the core treble-clef) are separated by a pronounced insert that forms an “overflow” of variable length. (3) The 4th pair (i.e. the terminal) of Zn-chelating residues straddles a complete helical turn, which does not form a separate squiggle as in the RING fingers, but forms a further turn of the C-terminal α-helix in the core treble clef. The remaining binuclear treble clef domains, such as the PHD, AN1 and MYND fingers, which lack the C-terminal strand, also show distinctive clade-specific differences in the configuration of the last pair of Zn-chelating residues. In all of these the second residue of the terminal Zn-chelating dyad is not associated with the beginning of a strand but occurs as part of an extension in the form of a further helical segment or a coil.9,11 The PHD finger, in particular, displays considerable C-terminal structural diversity in the secondary structure of the C-terminal extension that houses the second residue from the coordinating pair: it may be part of an extended coil region or part of an extensive α-helical elaboration.35 Thus, there appears to have been a limited overall constraint on the context of the terminal coordinating residue in these binuclear treble-clef domains, with some of the differences probably reflecting lineage-specific, local functional adaptations.
The remaining two binuclear domains, IBR and FYVE,24 do not appear to be specifically related to the other binuclear treble clefs. Of these, the IBR domain has two Zn2+ ions chelated by 4 pairs of chelating residues, but these do not occupy a position comparable to the other treble-clef domains.68 Specifically, the 2nd and 3rd pairs of chelating residues assume a configuration similar to that seen in the Zn-ribbon domains.69 Hence, the very relationship of the IBR to the treble clef domains is dubious and is not considered further in this article. While the FYVE domain coordinates two Zn2+ ions, it was previously noted to have a distinct pattern of arrangement of these coordination sites.16 This pattern was described as an overlapping doublet of treble-clef domains arising from duplication, wherein the first treble clef contributes a pair of cysteines to the second treble-clef.9,11 Even though the FYVE domain closely resembles the LIM fingers, which always occur as duplicated treble clef domains,70 a simple duplication cannot produce the pattern observed in the former domain because the lateral flap of the second treble clef is nested within the β-hairpin of the first treble clef. The most parsimonious explanation for the nesting observed in the FYVE domain requires the following steps: (1) a starting duplicated pair of mononuclear treble clef domains similar to the LIM fingers. (2) An internal circular permutation involving the second strand of the β-hairpin of the first of the duplicated copies and the lateral flap of the second copy. This permutation would have proceeded via a partial internal duplication after the initial complete duplication (Fig. 3A). Thus, the FYVE domain appears to have convergently acquired a structurally distinct binuclear state relative to all other bona fide binuclear treble clefs.
Fig. 3.

(A) A scheme for the possible origin of the FYVE domain. (1) The precursor mononuclear domain. (2) Duplication of the precursor gives rise to a LIM-like intermediate. (3) Partial duplication of the LIM-like intermediate. (4) Circular permutation event gives rise to the FYVE domain. Metal-ion coordinating residue pairs are denoted by “C” and pairs which coordinate the same zinc ion are joined by black lines. In step 3 the linkages which were retained in the FYVE domain are colored in red. β-Strands are colored in blue and numbered in each internal duplicated treble clef domain (for ease of display, the lateral flap is here represented by two smaller β-strands). The conserved core α-helix is colored in brown and denoted with the letter “H”. In step 4, a differential coloring scheme is employed to emphasize the respective origins of the observed structural components. (B) Interaction modes of treble clef domains with their partner proteins. Cartoon representation of the core treble clef scaffold, with coloring scheme similar to (A) but with the lateral flap colored in green and the variable C-terminal region colored in grey. Identified interaction partners are labeled by arrows with the name of the treble clef domain and its cognate binding partner separated by “↔”. General regions corresponding roughly to the binding pockets mentioned in the text are denoted by dashed circles shaded in grey. The surface which stacks binding partners against the exposed β-strand is depicted by a dashed line, colored in grey. Zinc ion residues are labeled and shown as black circles, shaded in yellow. Approximate locations of binuclear coordinating residues are shown as red lines.
These structural comparisons indicate that the treble clef domains with Ub-related functions (even within the binuclear treble clef domain) are not particularly close. While RING and UBP belong to the same higher order group of binuclear treble clefs with a C-terminal strand, the version of the fold in AN1 and MYND lacks this feature. Further, the RING and UBP domains themselves notably differ in placement of the last Zn-chelating residue. Similarly, the FYVE and C1 domains, both of which are involved in lipid or membrane-related functions, are also not structurally close binuclear treble clefs and have independently acquired a binuclear state.
Classification of the interaction sites of binding partners on the treble clef scaffold
While early studies have noticed certain commonalities in the interactions of the distinct treble clef fold domains with their functional partners,9,11 a considerable amount of new structural data has accumulated since then. We reasoned that a systematic survey and classification of the patterns of interaction of these domains could provide an independent means of assessing the emergence of Ub-related and other functions in this structural scaffold. Such analyses have previously been employed in uncovering trends in binding tendencies of other small domains which act as scaffolds facilitating a wide range of interactions.42,71 Hence, we comprehensively investigated the interactions of the treble clef scaffold using all available structures in the PDB database, where these domains are complexed with their physiologically relevant binding partners. Results of this analysis indicated that there are four primary interaction sites on the treble clef scaffold which are utilized by several distinct domain families, in addition to certain sites that have more restricted usage patterns (an overview of these findings is depicted in Fig. 3B; detailed views of each interaction are available in the ESI†). Two of the primary interaction sites take the form of pockets of variable depths: the first is formed at the interface of the lateral flap and the central β-hairpin and the second is formed by the α-helix combining with variable C-terminal extensions and the central β-hairpin (Fig. 3B and ESI†). The third major interaction region is formed by the exposed back “face” of the C-terminal α-helix of the core treble clef when oriented with its C-terminus pointing left. The fourth major interaction surface is via extended interactions along the length of the first (ascending strand) of the central β-hairpin (Fig. 3B and ESI†).
Each of these major sites tends to favor specific types of interacting partners. The pocket formed at the interface between the lateral flap and β-hairpin is almost exclusively utilized by mononuclear treble clef domains for interaction with nucleic acid or nucleotides: RNA by S14, DNA by THAP and MutM,67,72 and TDP and ADP by the Zn-binding domain of thymidine kinase73,74 (Fig. 3B and ESI†). However, in the mononuclear A20 domain the same pocket is utilized to recognize Ub.75 The second binding pocket is mainly used by mononuclear fingers and tends to recognize nucleic acids or accepts the terminal tails of peptides in the extended configuration (Fig. 3B and ESI†). In the mononuclear GATA, HNH, and NHR Zn-finger domains this region interacts with DNA,76–78 while in the MutM finger (related to the so-called TRASH domain) it interacts with the HhH domain found to its immediate C-terminus in the same polypeptide.72,79 The only binuclear treble clef to utilize this binding pocket is the UBP domain in which the pocket is used to accommodate the C-terminal tail of Ub18 (Fig. 3B and ESI†).
An interesting dichotomy is observed between the two remaining major binding sites: the “back” face of the α-helix is mainly used by mononuclear treble clef domains for nucleic acid binding, whereas interaction along the length of the β-hairpin is predominantly utilized for binding peptides (Fig. 3B and ESI†). The mononuclear GATA, NHR and HNH use this α-helix as a major DNA-binding site in conjunction with the above-described second binding pocket.76–78 The archaeo-eukaryotic ribosomal protein L24AE uses the same helical interface to bind the large subunit ribosomal RNA.80 The mononuclear A20 domain utilizes this site for Ub-binding by interacting along the length of the C-terminal strand of Ub.75 As in the case of the second pocket discussed above, the UBP domain is the only binuclear treble clef that utilizes this region, and binds Ub via this helix18 (Fig. 3B and ESI†). The 4th major interaction region is one of the most widely used across both mononuclear and binuclear treble clefs. Typically, it is used to bind peptides in the extended conformation, such that they form an antiparallel strand stacking with the first strand of the treble clef β-hairpin. Less frequently, the same region is also used to bind nucleic acids and lipids. Examples of this binding mode among mononuclear fingers include: (1) the LIM-interacting domain with the LIM domain;81 (2) a peptide from the L14 ribosomal protein by L24AE;80 (3) the SspB-tail region of the SspB adaptor protein with the ClpX treble clef;82 (4) DNA by the THAP domain;67 (5) 16S RNA by the ribosomal protein S1480 (Fig. 3B and ESI†). Among the binuclear forms, the PHD finger uses this interface to bind the tails of histone H3,83 while it uses the first pocket to accommodate the methylated or unmethylated H3K4 side chain (Fig. 3B and ESI†).15,84 This binding mode is also retained in the degenerate version of the PHD domain (PHD-X or the ZF-CW) that has lost the lateral flap.85 The binuclear MYND finger similarly binds peptides, such as those from the corepressor SMRT, in a similar configuration.86 Interestingly, the FYVE domain also interacts with its cognate lipid by binding it along the first strand of the β-hairpin87 (Fig. 3B and ESI†). The THAP domain similarly uses the same sites in recognition of its DNA partner.67 In the binuclear treble-clefs with the C-terminal strand, its packing with the first strand of the central β-hairpin is equivalent to the above-described peptide-binding mode. Thus, the C-terminal strand effectively “blocks” this binding mode in those treble clefs which possess it. In any given treble clef domain the same ligand might be contacted via more than one of these conserved interaction regions (e.g. the PHD finger).83
Beyond these major interaction surfaces there are two others which are used less often among the currently characterized treble clef domains (Fig. 3B): (1) the “front” face of the C-terminal α-helix of the treble clef and (2) the exposed “front” face of the lateral flap. Unlike the above described sites, the mode of interaction via these surfaces is decidedly non-uniform, beyond the shared general location. The first of these sites is utilized by the RING finger for the interaction with its E2 partner, with the helix being buried into the corresponding E2 pocket.88 The same region is used by the PHD finger to recognize acetylated peptides in histone tails, but in this case the ligand is merely accepted in a groove towards the N-terminus of the helix.83 The front face of the flap is utilized by the C1 domain to bind lipids, but in this case the lipid is inserted between the two extended elements of the lateral flap17 (Fig. 3B and ESI†). It is likely that this site is also required for interactions of the RING finger with Ub.37
These observations suggest that the four major interaction sites might have emerged early in the evolution of the treble-clef fold and predated the origin of the binuclear form. These sites appear to have been retained across the diversity of treble clef domains (mononuclear or binuclear) for interactions with biochemically distinct substrates. Emergence of the C-terminal strand in a subset of the binuclear treble clefs appears to have precluded utilization of a favored site, thereby fostering the use of alternative, atypical sites in the RING and C1 domains. This survey also indicates that the interaction modes of the different treble clef domains with Ub-related functions are not shared. In particular the binuclear treble clefs, UBP and RING, appear to have very distinct Ub-interaction modes. The mononuclear A20 likewise differs from both these binuclear forms in engaging Ub. Similarly the lipid/membrane interaction modes of the C1 and FYVE are completely unrelated to each other (Fig. 3). These observations, together with those pertaining to the structural differences between different treble-clef domains, indicate that Ub-interaction and Ub-related functions are likely to have convergently evolved on several independent occasions in the treble-clef fold.
Identification and functional characterization of prokaryotic binuclear treble clef domains
Given that evidence from structural analysis points to independent acquisition of Ub-related functions among structurally related and distant treble clef folds, we explored the prokaryotic versions of these domains to better understand the basis for this functional convergence. Hence, we systematically assessed the phyletic patterns of treble clef domains, with an emphasis on the binuclear forms among prokaryotes. In addition to prokaryotic RING domains, prokaryotic versions of the binuclear AN1, B-box, and UBP domains are known or were detected in this study (ESI†). We also investigated the prokaryotic versions of the mononuclear LIM domains, which tend to occur as pairs, and were hitherto predominantly observed in eukaryotes. For each of these, we analyzed domain architectures and genomic contexts to better decipher their functions in prokaryotes.
Prokaryotic RING fingers: evidence for Ub-related and Ub-independent roles
Of the four distinct families of prokaryotic RING domains (excluding those transferred from eukaryotes to endosymbiotic or endoparasitic bacteria) the first is found in several firmicutes, planctomycetes, and archaea such as Aciduliprofundum boonei and Methanocella paludicola. The second family is more restricted and found in Acidobacteria sp. and the methanogenic euryarchaea, Methanospirillum hungatei and Methanoregula boonei (Fig. 1B). The third family is comprised of the versions prototyped by the RING fingers found in the recently described genome of Caldiarchaeum subterraneum and is otherwise found in uncharacterized marine prokaryotes. The fourth family is currently known from actinobacteria and myxobacteria and bacteroidetes (ESI†).
As noted above, several members of the first three families are found in gene neighborhoods tightly linked to genes encoding other components of the Ub-system (Fig. 2A, marked with an asterisk). Additionally, some of these prokaryotic RING finger domains might be found in genomes encoding Ub-system components but in locations distant from these genes (e.g. in Isosphaera pallida and paralogs in Caldiarchaeum subterraneum). In bacteria, the Ubl associated with these RING proteins is always a member of the YukD family. The E1s found in these bacterial predicted operons are more distant from the eukaryotic versions and closer to other bacterial versions. All the components encoded by the Caldiarchaeum subterraneum operon are closer to their eukaryotic counterparts rather than the bacterial forms. The E1-like domain of this neighborhood contains a large insert within the core structure in a position observed only in eukaryotic E1-like domains (previously defined as insert location no. 2)38 to the exclusion of previously observed prokaryotic E1-like domains.38 Additionally, the Caldiarchaeum subterraneum E1-like domain contains the complete complement of core conserved residues observed in the eukaryotic versions, including both CxxC motifs and the ExxK motif found in the terminal α-helix, a set of motifs which are only very sporadically conserved across previously known prokaryotic E1-like domains which are linked to other Ubl pathway domains36,38 (ESI†). Perhaps most striking is the previously unreported fusion of a C-terminal UFD domain to the E1-like domain (Fig. 2) in the Caldiarchaeum subterraneum E1 cognate. The UFD domain is a circularly-permutated Ubl domain which is crucial in eukaryotic Ubl systems for mediating the presentation of the E2 domain to the active site of the E1 domain during Ub/Ubl conjugation reactions.89,90 While fusions to various catalysis-contributing C-terminal domains, including rhodanese and CCTBP, have previously been observed in prokaryotes,38 this is the first instance of fusion to the UFD domain which is essential for eukaryotic Ubl signaling. The core active site residues of the E2 domains encoded by prokaryotic genomes, also encoding one of the above three families of RING domains, display an “HPN” motif just upstream of the catalytic cysteine residue (ESI†). This signature is primarily seen in eukaryotic E2 lineages and is thought to play a role in stabilization and formation of the oxyanion hole during catalysis.37 This signature is variable in all other prokaryotic E2 domains, excluding the subset associated with the RING finger domains and the prokaryotic E2 family A which bears a cognate HXN signature.37 Other prokaryotic forms contain different residues conserved at a distinct set of locations on the E2 domain, which are predicted to play similar functional roles to the HPN motif. The above observations considerably extend the diversity of the prokaryotic Ub-system cognates relative to their eukaryotic counterparts. We now have bacterial, archaeal and bacteriophage cognates of the Ub-system, which have E1s or JAB peptidases alone39 or both E1s and E2s along with a JAB peptide and now those that might have a RING finger E3 (Fig. 2).36 In particular the E2s and in some cases the E1s of the prokaryotic systems with the RING finger E3 are closer to their eukaryotic counterparts. Thus, precursors of the eukaryotic system are likely to have emerged within this spectrum of diversification of systems related to Ub-conjugation that occurred in prokaryotes.
In several prokaryotes, members of all four families also occur in genomic contexts independently of Ub-system components. In some of these organisms, such as firmicutes and certain euryarchaea, there is no evidence for the encoding of any E2-like enzymes in their genomes. This suggests that in some prokaryotes the RING domains might have a distinct function that might be unlinked to an Ub-conjugation-like system. Nevertheless, both versions found in Ub-system-type operons and those occurring independently of it show a common domain architectural feature, i.e. linkage to TM segments (Fig. 2). In firmicutes, members of the first prokaryotic RING family are fused to a well-conserved TM module overlapping with the PFAM model DUF2628 (Fig. 2). This TM module is widely distributed in bacteria and also found linked to cell-wall binding and transport related domains71,91,92 (ESI†). These firmicutes proteins also display a Zn-ribbon domain immediately C-terminal to the RING domain (Fig. 2). In planctomycetes, members of the first prokaryotic RING finger family are fused to a single N-terminal TM helix and to a widely-distributed uncharacterized globular bacterial domain (Fig. 1; overlaps with PFAM model DUF3137). This domain is almost always fused either to a single TM helix or a sulfate permease domain, suggesting that it might function as a low molecular weight solute sensor (Fig. 2 and ESI†). The RING domains with fused TM domains may also be found in gene neighborhoods tightly linked to genes encoding transporters (Fig. 2). These associations suggest that a major fraction of the prokaryotic RING domains might have functions related to either regulation or modification of membrane-associated proteins, irrespective of whether they function with an associated Ub-conjugation-like system.
Though the fourth family of prokaryotic RINGs shows sequence features close to eukaryotic RING fingers (Fig. 1), it deviates from the other families in currently showing no clear cut gene-neighborhood or domain-architectural associations with Ub-system-like components. Furthermore, no E2 domains are encoded by any of the genomes which code for these RING finger proteins. Most of these RING fingers occur as part of a large polypeptide, wherein the RING finger is the second domain. The RING finger is flanked at the N-terminus by a small α-helical domain and at the C-terminus by a large α-helical domain, which in turn is followed by a vWA domain. Profile–profile comparisons and secondary structure predictions indicate that the large α-helical domain is related to the ROT/TROVE module (Fig. 2), which is comprised of around nine bihelical repeats arranged in the form of a toroidal structure. The ROT/TROVE module binds RNA, and most frequently occurs at the N-terminus of a vWA domain in proteins such as the animal Ro and Deinococcus Rsr,93–95 in an architecture mirroring the arrangement seen in the above proteins with the RING finger. Indeed, there are homologs of the above proteins in firmicutes, fusobacteria and fungi that have the remaining domains but lack the RING finger domain, suggesting that they have an organization similar to the classical Ro-like proteins (ESI†). The RING finger hence appears to have been inserted in between the N-terminal α-helical domain and the ROT/TROVE-like module in a subgroup within this family. In bacteria, Rsr has been shown to bind 23S rRNA and 3′ ends of misfolded RNAs and present them for processing or degradation.94,96 They also bind small non-coding RNAs called Y RNAs which negatively regulate their association with the 23S rRNA. By analogy it is possible that these proteins might be regulators of RNA stability or processing. Hence, it appears likely that this family of prokaryotic RING fingers does not have a role in connection to the Ub-system but functions as a regulatory component of a possible RNA-processing complex.
Thus, evidence from contextual information suggests that RING fingers in prokaryotes are functionally diverse, with both Ub-system associated roles as well as independent roles that might be related to regulatory interactions with membrane-associated and RNA-associated proteins.
Prokaryotic UBP domains participate in cyclic nucleotide and redox signaling
Prokaryotic UBP domains are particularly prevalent in actinobacteria, but are also found in several lineages of cyanobacteria, proteobacteria, chloroflexi, acidobacteria, bacteroides, and euryarchaeota (ESI†). In eukaryotes, the UBP domain binds the tails of Ub and typically occurs as part of large proteins combined with other domains such as the RING finger, the ubiquitin C-terminal hydrolase, histone deacetylase and Sirtuin domains.18 All prokaryotic UBP domains occur as standalone versions, with the exception of a few actinobacterial representatives, which are fused to a transmembrane Na+/H+ antiporter domain. Interestingly, genomic contexts reveal that prokaryotic UBP domains show no detectable linkages to any proteins related to the Ub-system. However, they did show well conserved gene neighborhoods that point to other functional associations. In actinobacteria a common association is with a gene encoding a Na+/H+ antiporter, which in conjunction with the above-noted fusions in this bacterial lineage points to a role of the UBP domain in regulation of ion transport via this membrane protein (Fig. 1). A much stronger operonic association is seen with a gene encoding a previously uncharacterized thioredoxin oxidoreductase with a Rossmann fold. This oxidoreductase domain is additionally fused to an inactive thioredoxin domain (i.e. lacking the canonical CxxxC motif which functions in redox reactions97) and to either a cNMP-binding domain (cNMPBD) or a receiver domain of the two component system. A subset of the versions fused to the cNMPBD is also accompanied by another gene in the predicted operon, which encodes an active thioredoxin domain fused to an N-terminal zinc ribbon domain (Fig. 2). Those versions fused to the receiver domain are accompanied by another gene in the operon that encodes a histidine kinase fused to a cNMPBD (Fig. 2 and ESI†). These associations suggest that prokaryotic UBP domains are involved in a signaling pathway that combines a redox process with cyclic nucleotides (either cAMP or cGMP) sensed by the cNMPBD. A straight-forward interpretation of the domain architectures and operonic associations is that the sensing of cNMP helps, either directly or via a two component relay directed by the histidine kinase and receiver domain, to regulate the activity of the thioredoxin oxidoreductase. Given the tight operonic linkage to the UBP domains it is likely to be an additional player in this signaling system.
Sequence analysis of the thioredoxin oxidoreductase and prokaryotic UBP domain provided further evidence in support of their role in the redox process. The thioredoxin oxidoreductase domain in these gene neighborhoods lacks the usual CxxC motif that mediates the reduction of the corresponding CxxxC motif in active thioredoxin domains;98 instead, these oxidoreductase domains contain another conserved cysteine residue downstream of this region. The prokaryotic UBP domains have a key, highly conserved sequence signature that is absent in all eukaryotic UBP domains (Fig. 4 and ESI†). This is a CD signature occurring one residue downstream of the third Zn-coordinating residue pair at the beginning of the C-terminal α-helix of the core treble clef domain. This observation indicates that the signature is uniquely related to the function of the prokaryotic UBP domains. Examination of the structure of the UBP domain indicates that the side chain of the cysteine in this signature is solvent exposed and clearly unrelated to Zn-coordination or stabilizing interactions within the structure (PDB: 2IDA). Given the association with the thioredoxin oxidoreductase, we propose that this conserved cysteine in the UBP domain, together with the conserved cysteine in the oxidoreductases domain forms part of a redox relay pair, that in some cases might additionally involve a thioredoxin domain protein encoded in the same operon. Thus, we predict that the prokaryotic UBP domain is a key component of a signaling system that coordinates a cNMP-dependent signal with regulation of redox potential. In light of this, it appears plausible that the UBP domains associated with the Na+/H+ antiporter potentially regulate ion transport in a redox-dependent fashion.
Fig. 4.
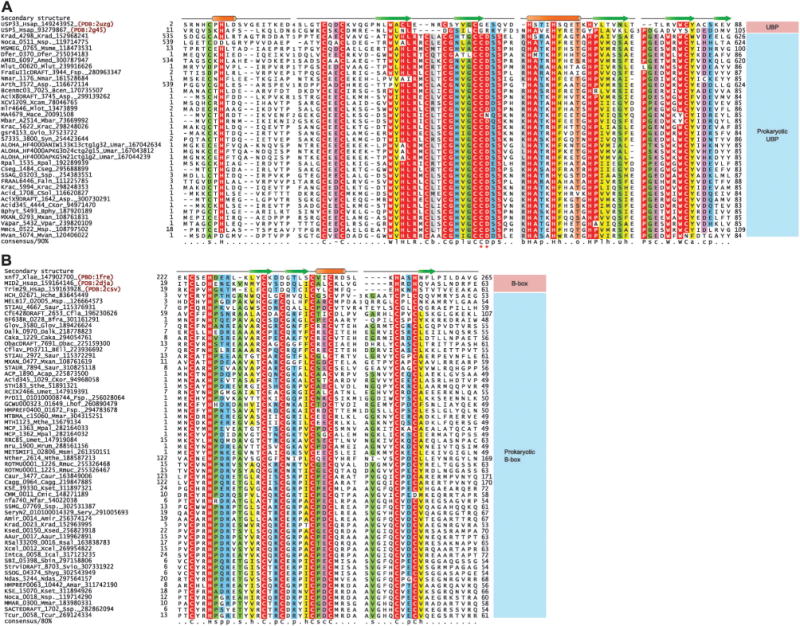
(A) Multiple sequence alignment of prokaryotic UBP and (B) B-box domains. Protein nomenclature and secondary structure assignments are as in Fig. 1. The highly conserved CD motif seen in prokaryotic UBP domains are marked with asterisks. Residue coloring is based on 90% consensus for the UBP domain and 80% consensus for the B-box domain. The coloring scheme and consensus abbreviations are as follows: h, hydrophobic (ACFILMVWY); l, aliphatic (LIV) and a, aromatic (FWY) residues shaded yellow; b, big residues (LIYERFQKMW), shaded gray; s, small residues (AGSVCDN) and u, tiny residues (GAS), shaded green; p, polar residues (STEDKRNQHC) shaded blue; –, acidic residues (DE), shaded magenta, o, alcohol (ST) group containing residues shaded orange, zinc coordinating residues and absolutely conserved residues are shaded red. Species abbreviations are as follows: Aaur: Arthrobacter aurescens; Acap: Acidobacterium capsulatum; Amar: Aeromicrobium marinum; Amed: Amycolatopsis mediterranei; Amir Actinosynnema mirum; Asp.: Acidobacterium sp.; Asp.: Arthrobacter sp.; BEll: bacterium Ellin514; Bcen: Burkholderia cenocepacia; Bfra: Bacteroides fragilis; Bphy: Burkholderia phytofirmans; CKor: Candidatus Koribacter; CSol: Candidatus Solibacter; Cagg: Chloroflexus aggregans; Caka: Coraliomargarita akajimensis; Caur: Chloroflexus aurantiacus; Cfla: Chthoniobacter flavus; Cmic: Clavibacter michiganensis; Cseg: Caulobacter segnis; Dalk: Desulfatibacillum alkenivorans; Dfer: Dyadobacter fermentans; Faln: Frankia alni; Fsp.: Frankia sp.; Fsp.: Fusobacterium sp.; Glov: Geobacter lovleyi; Gvio: Gloeobacter violaceus; Hche: Hahella chejuensis; Hsap: Homo sapiens; Ical: Intrasporangium calvum; Krac: Ktedonobacter racemifer; Krad: Kineococcus radiotolerans; Ksed: Kytococcus sedentarius; Kset: Kitasatospora setae; Lhof: Leptotrichia hofstadii; Mace: Methanosarcina acetivorans; Mbar: Methanosarcina barkeri; Mlot: Mesorhizobium loti; Mlut: Micrococcus luteus; Mmar: Methanothermobacter marburgensis; Mmar: Mycobacterium marinum; Mpal: Methanocella paludicola; Mrum: Methanobrevibacter ruminantium; Msme: Mycobacterium smegmatis; Msmi: Methanobrevibacter smithii; Msp.: Marinobacter sp.; Msp.: Mycobacterium sp.; Mthe: Methanothermobacter thermautotrophicus; Mvan: Mycobacterium vanbaalenii; Mxan: Myxococcus xanthus; Ndas: Nocardiopsis dassonvillei; Nfar: Nocardia farcinica; Nmar: Nitrosopumilus maritimus; Nsp.: Nocardioides sp.; Nthe: Natranaerobius thermophilus; Obac: Opitutaceae bacterium; Rmuc: Rothia mucilaginosa; Rpal: Rhodopseudomonas palustris; Rsal: Renibacterium salmoninarum; Saur: Stigmatella aurantiaca; Sbin: Streptomyces bingchenggensis; Sery: Saccharopolyspora erythraea; Shyg: Streptomyces hygroscopicus; Ssp.: Streptomyces sp.; Sthe: Symbiobacterium thermophilum; Svio: Streptomyces violaceusniger; Syn: Synechococcus sp.; Tcur: Thermomonospora curvata; Umar: uncultured marine; Umet: uncultured methanogenic archaeon; Vpar: Variovorax paradoxus; Xcam: Xanthomonas campestris; Xcel: Xylanimonas cellulosilytica; Xlae: Xenopus laevis.
The above observations strongly suggest that, despite their structural relationship, the prokaryotic UBPs are not related to the Ub-systems and acquired this function only in eukaryotes.
Prokaryotic B-box-like domains in membrane and proteolysis-related functions
In the current study we uncovered a widespread, previously uncharacterized family of prokaryotic treble clef domains related to the eukaryotic B-box domains (Fig. 4). This family is found in several bacterial lineages such as actinobacteria, chloroflexi, proteobacteria, verrucomicrobia, bacteroidetes, and among archaea, primarily in methanogenic euryarchaeotes. These domains display the typical spacing of metal-chelating residues as seen in the B-box domains, with the first pair displaying the characteristic CxxH signature. Profile–profile comparisons with these bacterial proteins preferentially recovered the eukaryotic B-box domains with much lower p-values than any other treble-clef domains (p < 10−8 with HHpred). Together, these observations suggest that these domains represent prokaryotic cognates of the B-box domain. The dominant domain architectural theme for these B-box-like domains, which is seen across diverse prokaryotes, is a fusion to the N-terminus of an integral membrane serine peptidase domain of the rhomboid family (Fig. 2). In addition to this, certain prokaryotic versions of the B-box-like domain are also found fused to an unrelated membrane-associated peptidase domain of the Zn-dependent metallopeptidase superfamily (Fig. 2). Versions which are not linked to these membrane-associated peptidases are always linked to TM segments. Additionally, these membrane proteins might also show other globular domains (Fig. 2), such as the DNAJ, FHA and SH3 domains and WD40 and tetratricopeptide repeats (TPRs). Of these, the DNAJ domain recruits the HSP70 chaperone protein to regulate protein folding or stability.99 The FHA domain binds phosphopeptides, whereas the SH3 domain, WD40 and TPRs mediate other protein–protein or protein–peptide interactions.100,101 In several bacteria the gene encoding the rhomboid-associated B-box-like domain is found in a predicted operon with a gene for a peptidyl–prolyl isomerase of the cyclophilin superfamily (Fig. 2). Together, the architectural associations suggest that the prokaryotic B-box-like domain primarily functions at the cell membrane, probably in proteolytic processing, folding or stability of membrane-associated proteins. Remarkably, this situation is paralleled by the prokaryotic versions of another binuclear treble clef domain, the AN1 domain (see below).
Involvement of prokaryotic AN1 domains in membrane-associated proteolysis
Prokaryotic members of the AN1 domain family were previously postulated to be derived from eukaryotic AN1-like domains based on the limited availability of completely-sequenced archaeal genomes available at the time of their original discovery.26 The current diversity of available genomes enables a more realistic assessment of the distribution of this domain, and indicates that AN1 domains are widely present across diverse euryarchaeota and also the so-called thaumarchaeota including Nitrosopumilus maritimus and Cenarchaeum symbiosum (see ESI†). This distribution argues for an archaeal origin for the AN1 domain. A strong theme emerges in the domain architectural linkages of the archaeal AN1 domains, namely the association with several structurally unrelated membrane-associated peptidase domains (Fig. 2). The most prevalent architecture is the fusion of an AN1 domain to the N-terminus of the rhomboid family serine peptidase domain in several methanogenic and halophilic archaea. Additionally, the AN1 domain is found at the N-termini of membrane-linked transglutaminase-like thiol peptidases of the papain-like fold102 and at the C-termini of integral membrane Zn-dependent metallopeptidases. Beyond these associations with peptidases, the AN1 domain is also found linked to TM segments (Fig. 2), DNAJ and SCP/PR1 domains. Thus, there is a striking parallel between the architectures of the prokaryotic B-box-like and AN1 domains, with the two domains often occurring as mutually exclusive alternatives in combinations with a similar set of domains. Thus, both these treble clef domains are likely to perform a similar function related to membrane-associated proteolytic processing of polypeptides and in regulating protein folding or stability.
In conclusion, while there is no evidence directly linking the prokaryotic B-box and AN1 domains to cognates of the Ub-system, they do appear to have a role in regulating protein stability and proteolytic degradation. It is possible that both these domains present peptides for processing by associated peptidase domains in prokaryotes. It is conceivable that in eukaryotes this original functional role of the B-box and AN1 domains was reused for regulating protein stability via the Ub-system. Indeed, certain eukaryotic AN1 proteins might have retained their ancestral role in relation to protein stability as suggested by recent studies on the eukaryotic AN1 protein AIRAP that functions as a heat-shock protein.103 Likewise, the eukaryotic ZFAND1 protein, which combines a Ubl domain with an AN1 domain, is part of the endoplasmic reticulum associated protein degradation (ERAD) system.8
Prokaryotic LIM domains
LIM domains are predominantly known from eukaryotes and primarily function in protein– protein interactions (see above). While they are mononuclear domains, they are unique in almost always occurring as pairs, unlike the other mononuclear domains involved in protein– protein and protein–nucleic acid interactions in both prokaryotes and eukaryotes. Given this unique organization, and the possible significance of such paired treble-clef domains in the emergence of the interlocked structures typified by the FYVE domain, we systematically investigated the prokaryotic history of the LIM domains. As a result we were able to uncover the previously underappreciated spread of prokaryotic LIM domains. They are found in representatives of chloroflexi, actinobacteria, verrucomicrobia, lentisphaerae, bacteroidetes and firmicutes (ESI†). All prokaryotic LIM domains display a fusion to a previously uncharacterized C-terminal Zn-dependent metallopeptidase domain (Fig. 2). Additionally, several prokaryotic versions also show a fusion to an inactive P-loop NTPase domain at their N-terminus (Fig. 2). Furthermore, most of the prokaryotic versions possess a single TM at their N-terminus. The prokaryotic LIM domains resemble their eukaryotic counterparts in occurring as a closely spaced pair of treble clef domains, wherein the C-terminus of the first repeat is shortened as it immediately leads into the N-terminus of the second one. Strikingly, the domain architectures of the prokaryotic LIM proteins resemble the above-described situation for the prokaryotic AN1 and B-box-like domains. This suggests that, like them, they are likely to regulate protein processing at the cell-membrane. This is interesting because, not just two structurally distinct binuclear treble clefs, but also the paired mononuclear domain appear to share a similar function in prokaryotes. This also indicates that the precursors of multiple treble clef domains that were previously known to be predominantly eukaryotic in the distribution had their origins in a similar functional context in prokaryotes.
Evolutionary considerations and general conclusions
Elucidating the origin of the core eukaryotic Ub-system
The remarkable affinity of the Caldiarchaeum Ub-system genes to their eukaryotic counterparts was proposed to support an origin for the eukaryotic Ub-system via the archaeal partner during symbiogenetic eukaryogenesis.41 Caldiarchaeum has been shown to be phylogenetically close to the thaumarchaeota, which share several distinct features with eukaryotes. These include histones, the tubulin precursor FtsZ, cognates of the ESCRT-II and ESCRT-III complexes, the VPS4 ATPase and SMC ATPases.104,105 While some of these components are also seen in euryarchaea and fewer in classical crenarchaea and korarchaea, the configuration closest to the predicted archaeal precursor of the eukaryotes is observed in the thaumarchaea (ESI†). In light of this evidence, and of the closeness of the Caldiarchaeum Ub-system components in terms of sequence similarity, domain architecture, and configuration and conservation of active site residues, it is conceivable that eukaryotes indeed acquired their Ub-system from such an archaeal progenitor. However, it should be noted that such an Ub-system is absent in other currently known archaea, including the thaumarchaea. Hence, alternative explanations could be possible. Most trivially one could speculate about horizontal “back-transfers” of the Ub-system from eukaryotes to prokaryotes. In our view this alternative should be considered rather implausible. First, given that eukaryotes lack operonic linkage of functionally related genes, such a scenario would require the individual transfer of 5 unrelated genes, namely those for Ub/Ubl ligase trio, the UB and the JAB peptidase followed by reassembly into an operon in archaea. This is a rather unlikely event given the near independent probabilities of each gene being transferred from a eukaryote and it being functionally irrelevant in the recipient archaeon without its partners. Second, as a free-living hyperthermophilic organism,41 Caldiarchaeum has much lower contact with eukaryotes from which it could have acquired such genes, unlike endoparasitic or symbiotic bacteria. Furthermore, in the latter organisms there is only evidence for acquisition of E3s from the host, rather than the whole Ub-system.
Our findings reported in this article have considerable significance in elucidating the possible events in this regard. The discovery of multiple bacterial operons that like the Caldiarchaeum system also contain the entire Ub/Ubl ligase trio, albeit more distantly related to their eukaryotic counterparts, suggests that the Caldiarchaeum system is merely one of a larger range of such systems that are present in prokaryotes. This should be considered in light of the following observations: (1) the earlier discovery of even more distant systems with E1s, E2s, Ubls and JAB-peptidases in bacteria36–38 and the report of Sampylation in haloarchaea that is only dependent on an E1-like enzyme.39 (2) The earlier discovery of E1s, JAB peptidases and Ubls involved in a diverse set of peptide-ligation-like and Ubl-cleavage-like reactions in prokaryotic biosynthesis of cofactors, amino acids and secondary metabolites such as siderophores and antibiotics and bacteriophage tail assembly.36,38,106–108 Together, they strongly suggest that systems resembling eukaryotic Ub-conjugation systems to different degrees were put together in prokaryotes during the diversification of such biosynthetic and regulatory pathways. For example, the cysteine, molybdopterin, thiamin, thiouridine and siderophore biosynthesis systems merely contain Ubls, E1 and JAB peptidases in adenylation and sulfur transfer reactions. The more complex systems, such as those we reported earlier and add upon here, including an E2 component, are likely to serve as regular Ub/Ubl-conjugation-like systems. Finally, there are those with RINGs that are likely to be close to the eukaryotic systems in every sense (Fig. 2).41 Our discovery of several novel prokaryotic RING domains, which appear unrelated to the Ub-system, suggests that the original diversification of these domains probably occurred in prokaryotes, with a subset of them being recruited as E3s right in the prokaryotic Ub/Ubl-related systems. Together, this lends strong support for a primarily prokaryotic origin for the complete Ub-system in the form of an operonic assembly linking all the key components that was acquired by the eukaryotic progenitor. Given that the Ubls of the newly detected bacterial systems with RING fingers belong to the YukD family, it appears likely the that eukaryote-type Ub itself emerged from the YukD family.
The evidence from this study, along with our earlier reported observations, indicate that such operons are present across phylogenetically distant prokaryotes, and often missing in close relatives of the forms that display such systems. Hence, these prokaryotic Ub/Ubl-related systems are apparently highly mobile and widely disseminated through lateral transfer, analogous to the restriction–modification and secondary metabolite biosynthesis gene clusters36,48,109 Therefore, we cannot be certain if the eukaryotic Ub-system indeed emerged from a Caldiarchaeum-like system in the archaeal symbiont during eukaryogenesis. Indeed, such systems might be present in as yet un-sampled bacteria suggesting that it is not unlikely that eukaryotes acquired such a system from the primary bacterial symbiont or even via an independent lateral transfer of the operon from yet another prokaryote. Finally, the persistent association of the gene encoding the JAB peptidase with genes coding for other components of a Ubl conjugation system suggests a piecemeal origin for the eukaryotic proteasomal complex. The ancestral form of the lid complex with the JAB peptidase is likely to have emerged from the predicted Ubl-conjugation systems such as those described here. Its association with the preexisting core proteasomal apparatus containing the NTN peptidases, AAA+ ATPases, and proteasomal chaperone is likely to have strengthened the functional link between the two systems. While the core proteasomal apparatus is of archaeal origin,110 it is also present in various bacteria111 of which some such as Frankia possess the complete core Ub-system. Hence, it is possible that this connection developed either in bacteria or archaea, and was merely retained in eukaryotes which vertically inherited their core proteasomal complex from the archaea.
The early diversification of the binuclear treble clefs and LIM domains and the independent recruitment of treble clef domains to Ub-related functions
This study also provides different lines of evidence for a prokaryotic origin for multiple treble clef domains that were considered to be predominantly eukaryotic in their provenance. In addition to the RING finger, the presence in prokaryotes of the B-box, the UBP and AN1 fingers suggests that these had already differentiated from each other in prokaryotes, and were probably transferred to eukaryotes early in their evolution. Given that the B-box and UBP are predominantly in bacteria, RING in both archaea and bacteria, and AN1 predominantly in archaea, it is conceivable that the LUCA already possessed at least one binuclear treble clef. This is a novel inference; previously only RNA-associated versions (e.g. S14) and, perhaps DNA-associated versions (e.g. HNH nuclease) were proposed as having been present in LUCA.9 It also indicates that even within the binuclear domains the two versions, i.e. those with the C-terminal strand extension and those without it, had already diversified in bacteria. In eukaryotes, there are several distinct treble clef domains performing Ub-related functions, namely RING, UBP, AN1, B-box, A20, C4DM and to a certain extent the MYND domains. Strikingly, evidence from structural features, ligand-interaction modes, and contextual functional inference for the prokaryotic homologs suggests that these domains are likely to have independently acquired their Ub-related functions. Even among the binuclear domains, including the structurally related RING, B-box and UBP domains, the evidence points to independent acquisition of Ub-related roles. In a similar vein, though the RING finger participates in modification of the ε-amino group of a lysine, and the PHD finger in recognizing a modified or unmodified ε-amino group of a lysine they appear to have convergently acquired this capability. Likewise, structural evidence points to convergence in acquisition of membrane or lipid-related functions among different treble-clefs, such as the FYVE and C1 domains. Thus, we have a remarkable case of independent emergence of similar functions in the same fold on multiple occasions.
This raises the question as to why such convergence might have occurred. The structural features of the treble clef fold point to its versatility in being used as an interface for interactions.9 The inferences regarding the functions of the prokaryotic binuclear treble clefs and LIM domains suggest more specific possibilities. The several prokaryotic B-box, AN1 and LIM domains are inferred to function as accessory domains in close conjunction with peptidase domains. This suggests that their radiation might have involved acquisition of peptide binding capability, in particular, for presentation of peptides to the linked peptidases. This might have provided a pre-adaptation that allowed them to be reused in a comparable context in the Ub-system of the eukaryotes, as well as in other contexts such as chromatin and cytoskeleton, where recognition of specific peptides was important. All prokaryotic binuclear treble clefs, especially the RING, B-box-like and AN1 domains, and the mononuclear LIM fingers are inferred to have a membrane-related role. Furthermore, the JAB peptidases found in operonic associations with the prokaryotic RING fingers also often show C-terminal TM segments (Fig. 2). Some bacterial E2s are fused to TM segments and extracellular TPR repeats, for example Desulfotalea psychrophila DP0803, gi: 51244655. Even those YukD-like Ubls, which function independently of the trio of ligases, and certain versions of the structurally unrelated polypeptide modifier Pup, have membrane-associated functions. This suggests that the diversification of different treble clef domains presenting peptides to peptidases and the prokaryotic Ub system cognates closest to those in eukaryotes happened in the context of the inner surface of the prokaryotic cell-membrane. This ancestral membrane-linked role might have brought them together during early eukaryotic evolution and allowed different treble clef domains to acquire Ub-related functions. In favor of this suggestion, it may be pointed out that components of the Ub-system, AN1 fingers and rhomboid-like peptidases function together as part of the membrane-associated ERAD system in eukaryotes.8 Indeed, other ancient roles of the Ub-system in eukaryotes, such as in modification of membrane lipids during autophagy and vesicular trafficking are also consistent with this proposed ancestral membrane-linked function. This proximity to the membrane might have also favored the emergence of lipid-interacting treble clef domains, such as the C1 and the FYVE domain. These domains are currently known only from eukaryotes, but it cannot be ruled out that there might be as yet undetected prokaryotic counterparts or that this function emerged early in eukaryotes from the membrane-linked treble clefs acquired from bacteria. Indeed, we favor the nested or overlapping treble-clef units in FYVE emerging from a LIM-like duplicated form, and C1 originating from a binuclear, membrane-associated version with the additional strand.
In conclusion, the observations presented here could provide a framework for future experimental studies that might better help in understanding the biochemistry of treble clef domains. Importantly, investigations on the novel cyclic nucleotide and thioredoxin oxidoreductase related pathway dependent on the UBP domain might help in uncovering a major unexplored theme in the function of treble domains—regulation of redox potential-dependent interactions. A systematic experimental study of the coordination between different treble clef domains and the linked peptidase domains might lead to a better understanding of the ancestral biochemistry of these domains. Finally, experimental studies on those prokaryotic RING fingers which are apparently unlinked to Ub-related systems might provide new insights on alternative functions of this domain that are apparently not known in eukaryotes.
Materials and methods
All sequence profile searches were performed against the non-redundant (nr) database of protein sequences housed at the National Center for Biotechnology Institute (NCBI, NIH, Bethesda, MD). PSI-BLAST43 searches were iterated until convergence using a threshold expectation value (e-value) of 0.01 for inclusion in construction of a position-specific-scoring matrix after each iteration. Profile-based iterative HMM searches were performed using the JACKHMMER program from the recently-released HMMER3 package (http://hmmer. janelia.org).112 Profile–profile comparisons were performed using the HHPRED program.113 Multiple sequence alignments were constructed with the output from the Kalign program subjected to manual correction based on PSI-BLAST high-scoring pair results, JPRED2-generated secondary structure predictions,114 and, where applicable, inspection of solved structures. Protein structures were visualized using the Swiss-PDB program.115 Transmembrane helices were predicted using the TMHMM116 and the TMpred programs (http://www.ch.embnet.org/software/TMPRED_form.html). Single-linkage clustering of similar protein sequences was performed using the BLASTCLUST program ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html, length and score threshold parameters for clustering were empirically determined. Gene neighborhoods were retrieved through a custom script which accesses neighboring genes from whole genome shotgun sequences or completely sequenced genomes. All large scale analyses were performed with the TASS software package (Anantharaman, V., Balaji, S., Aravind, L., unpublished results).
Supplementary Material
Acknowledgments
Work by LMI and LA is supported by the intramural funds of the National Library of Medicine at the National Institutes of Health, USA.
Footnotes
References
- 1.Hochstrasser M. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Glickman MH, Ciechanover A. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 4.Ardley HC, Robinson PA. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 6.Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T, Noel JP. Mol Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 7.Joazeiro CA, Weissman AM. Cell (Cambridge, Mass) 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 8.Venancio TM, Balaji S, Iyer LM, Aravind L. Genome-Biology. 2009;10:R33. doi: 10.1186/gb-2009-10-3-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grishin NV. Nucleic Acids Res. 2001;29:1703–1714. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aravind L, Iyer LM, Koonin EV. Cell Cycle. 2003;2:123–126. doi: 10.4161/cc.2.2.335. [DOI] [PubMed] [Google Scholar]
- 11.Krishna SS, Majumdar I, Grishin NV. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aravind L, Koonin EV. Curr Biol. 2000;10:R132–R134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D, Iyer LM, Aravind L. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misra S, Hurley JH. Cell (Cambridge, Mass) 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang G, Kazanietz MG, Blumberg PM, Hurley JH. Cell (Cambridge, Mass) 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 18.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. Cell (Cambridge, Mass) 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 19.Kadlec J, Guilligay D, Ravelli RB, Cusack S. RNA. 2006;12:1817–1824. doi: 10.1261/rna.177606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spadaccini R, Perrin H, Bottomley MJ, Ansieau S, Sattler M. J Mol Biol. 2006;358:498–508. doi: 10.1016/j.jmb.2008.01.071. [DOI] [PubMed] [Google Scholar]
- 21.Legge GB, Martinez-Yamout MA, Hambly DM, Trinh T, Lee BM, Dyson HJ, Wright PE. J Mol Biol. 2004;343:1081–1093. doi: 10.1016/j.jmb.2004.08.087. [DOI] [PubMed] [Google Scholar]
- 22.Borden KL, Lally JM, Martin SR, O’Reilly NJ, Etkin LD, Freemont PS. EMBO J. 1995;14:5947–5956. doi: 10.1002/j.1460-2075.1995.tb00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravind L, Iyer LM, Koonin EV. Curr Opin Struct Biol. 2006;16:409–419. doi: 10.1016/j.sbi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Int J Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV. J Mol Biol. 1999;289:729–745. doi: 10.1006/jmbi.1999.2827. [DOI] [PubMed] [Google Scholar]
- 27.Wang F, Xiao J, Pan L, Yang M, Zhang G, Jin S, Yu J. PLoS One. 2008;3:e4027. doi: 10.1371/journal.pone.0004027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews JM, Bhati M, Lehtomaki E, Mansfield RE, Cubeddu L, Mackay JP. Curr Pharm Des. 2009;15:3681–3696. doi: 10.2174/138161209789271861. [DOI] [PubMed] [Google Scholar]
- 29.Hnia K, Zouiten D, Cantel S, Chazalette D, Hugon G, Fehrentz JA, Masmoudi A, Diment A, Bramham J, Mornet D, Winder SJ. Biochem J. 2007;401:667–677. doi: 10.1042/BJ20061051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francke F, Buck F, Bachner D. Biochem Biophys Res Commun. 2005;334:1292–1298. doi: 10.1016/j.bbrc.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 31.Colon-Gonzalez F, Kazanietz MG. Biochim Biophys Acta, Mol Cell Biol Lipids. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Corvera S. Science’s STKE. 2000;2000 doi: 10.1126/stke.2000.37.pe1. pe1. [DOI] [PubMed] [Google Scholar]
- 33.Hayakawa A, Hayes S, Leonard D, Lambright D, Corvera S. Biochem Soc Symp. 2007;74:95–105. doi: 10.1042/BSS0740095. [DOI] [PubMed] [Google Scholar]
- 34.Wurmser AE, Gary JD, Emr SD. J Biol Chem. 1999;274:9129–9132. doi: 10.1074/jbc.274.14.9129. [DOI] [PubMed] [Google Scholar]
- 35.Chakravarty S, Zeng L, Zhou MM. Structure (London) 2009;17:670–679. doi: 10.1016/j.str.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer LM, Burroughs AM, Aravind L. GenomeBiology. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burroughs AM, Jaffee M, Iyer LM, Aravind L. J Struct Biol. 2008;162:205–218. doi: 10.1016/j.jsb.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burroughs AM, Iyer LM, Aravind L. Proteins. 2009;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong YJ, Jeong BC, Song HK. Biochem Biophys Res Commun. 2011;405:112–117. doi: 10.1016/j.bbrc.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, Takai K, Takami H. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burroughs AM, Balaji S, Iyer LM, Aravind L. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Ent F, Lowe J. FEBS Lett. 2005;579:3837–3841. doi: 10.1016/j.febslet.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Burts ML, DeDent AC, Missiakas DM. Mol Microbiol. 2008;69:736–746. doi: 10.1111/j.1365-2958.2008.06324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, Cox JS. Cell Host Microbe. 2010;7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmelz S, Kadi N, McMahon SA, Song L, Oves-Costales D, Oke M, Liu H, Johnson KA, Carter LG, Botting CH, White MF, Challis GL, Naismith JH. Nat Chem Biol. 2009;5:174–182. doi: 10.1038/nchembio.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. Mol BioSyst. 2009;5:1636–1660. doi: 10.1039/b917682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkinson KD, Audhya TK. J Biol Chem. 1981;256:9235–9241. [PubMed] [Google Scholar]
- 51.Cerda-Maira FA, Pearce MJ, Fuortes M, Bishai WR, Hubbard SR, Darwin KH. Mol Microbiol. 2010;77:1123–1135. doi: 10.1111/j.1365-2958.2010.07276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iyer LM, Burroughs AM, Aravind L. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huynen M, Snel B, Lathe W, 3rd, Bork P. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. Proc Natl Acad Sci U S A. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kubori T, Hyakutake A, Nagai H. Mol Microbiol. 2008;67:1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 56.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 57.Gohre V, Spallek T, Haweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S. Curr Biol. 2008;18:1824–1832. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- 58.Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB. Nature. 2007;448:370–374. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen MD, Bycroft M. Protein Sci. 2007;16:2072–2075. doi: 10.1110/ps.072967807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isobe T, Uchida C, Hattori T, Kitagawa K, Oda T, Kitagawa M. Biochem Biophys Res Commun. 2006;339:367–374. doi: 10.1016/j.bbrc.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 61.Ju D, Wang X, Xu H, Xie Y. Mol Cell Biol. 2008;28:1404–1412. doi: 10.1128/MCB.01787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linnen JM, Bailey CP, Weeks DL. Gene. 1993;128:181–188. doi: 10.1016/0378-1119(93)90561-g. [DOI] [PubMed] [Google Scholar]
- 63.Massiah MA, Matts JA, Short KM, Simmons BN, Singireddy S, Yi Z, Cox TC. J Mol Biol. 2007;369:1–10. doi: 10.1016/j.jmb.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 64.Mrosek M, Meier S, Ucurum-Fotiadis Z, von Castelmur E, Hedbom E, Lustig A, Grzesiek S, Labeit D, Labeit S, Mayans O. Biochemistry. 2008;47:10722–10730. doi: 10.1021/bi800733z. [DOI] [PubMed] [Google Scholar]
- 65.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 66.Campagne S, Saurel O, Gervais V, Milon A. Nucleic Acids Res. 2010;38:3466–3476. doi: 10.1093/nar/gkq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC. Nat Struct Mol Biol. 2010;17:117–123. doi: 10.1038/nsmb.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beasley SA, Hristova VA, Shaw GS. Proc Natl Acad Sci U S A. 2007;104:3095–3100. doi: 10.1073/pnas.0610548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krishna SS, Aravind L. J Struct Biol. 2010;172:294–299. doi: 10.1016/j.jsb.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perez-Alvarado GC, Miles C, Michelsen JW, Louis HA, Winge DR, Beckerle MC, Summers MF. Nat Struct Biol. 1994;1:388–398. doi: 10.1038/nsb0694-388. [DOI] [PubMed] [Google Scholar]
- 71.Balaji S, Aravind L. Nucleic Acids Res. 2007;35:5658–5671. doi: 10.1093/nar/gkm558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. EMBO J. 2000;19:3857–3869. doi: 10.1093/emboj/19.15.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Birringer MS, Claus MT, Folkers G, Kloer DP, Schulz GE, Scapozza L. FEBS Lett. 2005;579:1376–1382. doi: 10.1016/j.febslet.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 74.Welin M, Kosinska U, Mikkelsen NE, Carnrot C, Zhu C, Wang L, Eriksson S, Munch-Petersen B, Eklund H. Proc Natl Acad Sci U S A. 2004;101:17970–17975. doi: 10.1073/pnas.0406332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flick KE, Jurica MS, Monnat RJ, Jr, Stoddard BL. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- 77.Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 78.Rastinejad F, Perlmann T, Evans RM, Sigler PB. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 79.Fromme JC, Verdine GL. J Biol Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 80.Selmer M, Dunham CM, Murphy FVT, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 81.Deane JE, Mackay JP, Kwan AH, Sum EY, Visvader JE, Matthews JM. EMBO J. 2003;22:2224–2233. doi: 10.1093/emboj/cdg196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park EY, Lee BG, Hong SB, Kim HW, Jeon H, Song HK. J Mol Biol. 2007;367:514–526. doi: 10.1016/j.jmb.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 83.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Nature. 2010;466:258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He F, Umehara T, Saito K, Harada T, Watanabe S, Yabuki T, Kigawa T, Takahashi M, Kuwasako K, Tsuda K, Matsuda T, Aoki M, Seki E, Kobayashi N, Guntert P, Yokoyama S, Muto Y. Structure (London) 2010;18:1127–1139. doi: 10.1016/j.str.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y, Chen W, Gaudet J, Cheney MD, Roudaia L, Cierpicki T, Klet RC, Hartman K, Laue TM, Speck NA, Bushweller JH. Cancer Cell. 2007;11:483–497. doi: 10.1016/j.ccr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 88.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Cell (Cambridge, Mass) 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 89.Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lois LM, Lima CD. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mishima M, Shida T, Yabuki K, Kato K, Sekiguchi J, Kojima C. Biochemistry. 2005;44:10153–10163. doi: 10.1021/bi050624n. [DOI] [PubMed] [Google Scholar]
- 92.Freund C, Dotsch V, Nishizawa K, Reinherz EL, Wagner G. Nat Struct Biol. 1999;6:656–660. doi: 10.1038/10712. [DOI] [PubMed] [Google Scholar]
- 93.Bateman A, Kickhoefer V. BMC Bioinformatics. 2003;4:49. doi: 10.1186/1471-2105-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wurtmann EJ, Wolin SL. Proc Natl Acad Sci U S A. 2010;107:4022–4027. doi: 10.1073/pnas.1000307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Koonin EV, Aravind L. Trends Biochem Sci. 2000;25:223–224. doi: 10.1016/s0968-0004(00)01577-2. [DOI] [PubMed] [Google Scholar]
- 96.Chen X, Wurtmann EJ, Van Batavia J, Zybailov B, Washburn MP, Wolin SL. Genes Dev. 2007;21:1328–1339. doi: 10.1101/gad.1548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Atkinson HJ, Babbitt PC. PLoS Comput Biol. 2009;5:e1000541. doi: 10.1371/journal.pcbi.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chivers PT, Laboissiere MC, Raines RT. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 99.Qiu XB, Shao YM, Miao S, Wang L. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hammet A, Pike BL, McNees CJ, Conlan LA, Tenis N, Heierhorst J. IUBMB Life. 2003;55:23–27. doi: 10.1080/1521654031000070636. [DOI] [PubMed] [Google Scholar]
- 101.Andrade MA, Perez-Iratxeta C, Ponting CP. J Struct Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 102.Iyer LM, Koonin EV, Aravind L. Cell Cycle. 2004;3:1440–1450. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- 103.Rossi A, Trotta E, Brandi R, Arisi I, Coccia M, Santoro MG. J Biol Chem. 2010;285:13607–13615. doi: 10.1074/jbc.M109.082693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hobel CF, Albers SV, Driessen AJ, Lupas AN. Biochem Soc Trans. 2008;36:94–98. doi: 10.1042/BST0360094. [DOI] [PubMed] [Google Scholar]
- 105.Reeve JN, Bailey KA, Li WT, Marc F, Sandman K, Soares DJ. Biochem Soc Trans. 2004;32:227–230. doi: 10.1042/bst0320227. [DOI] [PubMed] [Google Scholar]
- 106.Godert AM, Jin M, McLafferty FW, Begley TP. J Bacteriol. 2007;189:2941–2944. doi: 10.1128/JB.01200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lehmann C, Begley TP, Ealick SE. Biochemistry. 2006;45:11–19. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xi J, Ge Y, Kinsland C, McLafferty FW, Begley TP. Proc Natl Acad Sci U S A. 2001;98:8513–8518. doi: 10.1073/pnas.141226698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Roberts RJ. Crit Rev Biochem Mol Biol. 1976;4:123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- 110.Koonin EV, Wolf YI, Aravind L. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lupas A, Zuhl F, Tamura T, Wolf S, Nagy I, De Mot R, Baumeister W. Mol Biol Rep. 1997;24:125–131. doi: 10.1023/a:1006803512761. [DOI] [PubMed] [Google Scholar]
- 112.Eddy SR. Genome Inf Ser. 2009;23:205–211. [PubMed] [Google Scholar]
- 113.Soding J, Biegert A, Lupas AN. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cuff JA, Barton GJ. Proteins: Struct, Funct, Genet. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 115.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 116.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


