Abstract
ADP-ribosylation—the transfer of ADP-ribose (ADPr) from NAD+ onto target molecules—is catalyzed by members of the ADP-ribosyltransferase (ART) superfamily of proteins, found in all kingdoms of life. Modification of amino acids in protein targets by ADPr regulates critical cellular pathways in eukaryotes and underlies the pathogenicity of certain bacteria. Several members of the ART superfamily are highly relevant for disease; these include the poly(ADP-ribose) polymerases (PARPs), recently shown to be important cancer targets, and the bacterial toxins diphtheria toxin and cholera toxin, long known to be responsible for the symptoms of diphtheria and cholera that result in morbidity. In this Review, we discuss the functions of amino acid ADPr modifications and the ART proteins that make them, the nature of the chemical linkage between ADPr and its targets and how this impacts function and stability, and the way that ARTs select specific amino acids in targets to modify.
ADP-ribosylation is an ancient post-translational modification found in viruses, bacteria and eukaryotes1. It is catalyzed by members of the ART superfamily of proteins, which transfer ADPr from nicotinamide adenine dinucleotide (NAD+) onto substrates via N-, O-, or S-glycosidic linkages on target molecules. Enzymatically active ARTs utilize a protein fold called the ART fold to catalyze these linkages at the 1″ position of the nicotinamide ribose via an SN1-like reaction mechanism2. Although most ARTs modify proteins (specifically at nucleophilic amino acids) some modify other substrate molecules, including tRNAs3,4 polynucleic acids5–9, and small-molecule antimicrobials10,11. ADP-ribosylation of target proteins by ARTs plays important functions in cells. ART-mediated ADP-ribosylation of target proteins modifies their function by various mechanisms, including altering their enzymatic activity1,12, changing their binding partners, and regulating their stability by functioning as signals for ubiquitylation13–16. ADP-ribosylation also functions as a signaling scaffold for the recruitment of binding proteins to ADPr-modified targets17–22. Importantly, ADPr modifications are reversible23–26, and the duration of the signaling that results from ADP-ribosylation of targets is dependent on the kinetics of turnover at the modification site27–29. Therefore, it is critical to understand how and where targets are modified by ARTs.
In this Review, we discuss the function of amino acid ADPr (aa-ADPr) modifications and the ART family of proteins that make them. To better understand the turnover of the modifications on targets, we discuss the nature and stability of the chemical linkage between ADPr and its targets, as well as how the enzymes responsible for its catalysis select specific sites in targets to modify. For the sake of simplicity, we limit our discussion to bacterial and human ARTs, with a specific focus on the PARP (also known as ARTDs, ADP-ribosyltransferases diphtheria toxin-like) and ectoART (also known as ARTCs) human subfamilies, as they contain multiple ARTs whose functions are better understood than other ARTs. Other ART proteins exist in lower Eukaryotes that are absent in humans, and for more information on these we suggest ref. 30.
The ART fold
The defining feature of ART proteins is the ART fold, which is found within the ART catalytic domains of all ARTs from viral ARTs to human. This protein fold is poorly conserved at the sequence level but is structurally well conserved, consisting of a split β-sheet with each half containing three strands1,2,31. The key amino acid–NAD+ binding interactions are shown in Figure 1 and are described in greater detail below. The ART fold is one of two known NAD+ binding protein folds, the other being the Rossman fold, which, in contrast to the ART fold, cannot catalyze ADPr transfer1. There are other important differences between the Rossman fold and the ART fold. The Rossman fold is not specific for NAD+ and binds several mononucleotides including NADP+ and FAD32, and proteins containing the Rossman fold, such as oxidoreductases, bind NAD+ in a relatively flat configuration, ideal for redox chemistry. The ART fold is highly specific for NAD+, and binds the molecule in a bent confirmation that induces strain on the pyridinium N-glycosidic bond, lowering the activation energy for glycosidic bond cleavage. A critical interaction in the ART fold is the interaction of the amide of nicotinamide with the main chain of an amino acid (Fig. 1). This interaction engenders specificity for the nicotinamide of NAD+ and is exploited by most NAD+-competitive ART inhibitors. In addition to this interaction, there are three other highly conserved amino acids in the ART fold that are critical for NAD+ binding and catalysis. The identify of these key catalytic amino acids organizes the ARTs into two major families in bacteria and relates the eukaryotic ARTs to their bacterial ancestors1.
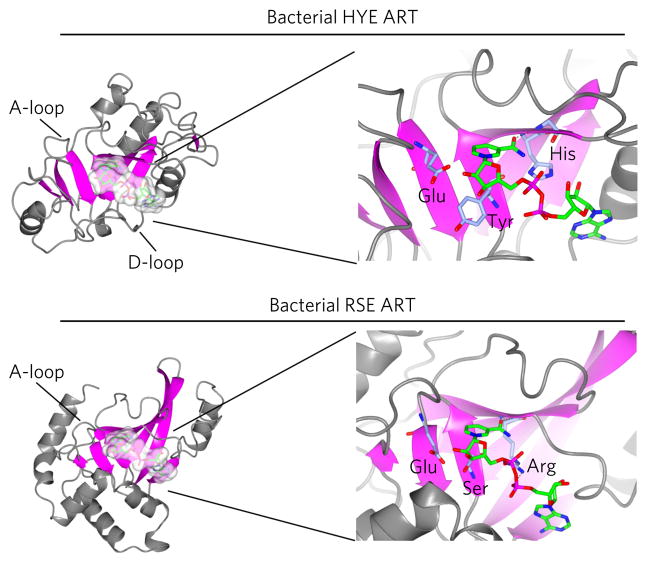
Figure 1. Structure of the HYE and RSE ART folds.
Shown are the crystal structures of the bacterial HYE ART ExoA (PDB ID 2ZIT; upper panel) and the structure of RSE ART Iota toxin (PDB ID 4H03; lower panel). The β-sheets of the ART fold are shown in magenta. NAD+ (shown in green) binds in a similar orientation in both types of bacterial ARTs. The conserved catalytic amino acids in both the bacterial HYE (His440, Tyr470 and Glu553) and RSE (Arg295, Ser338 and Glu380) ARTs are shown in purple.
The ART superfamily
Nearly all ART proteins contain additional protein domains outside of the ART catalytic domain. These domains play important roles in targeting the ARTs to their substrates and to specific cellular locations, and in some cases are able to sense specific cellular conditions such as DNA damage19,33,34. Although domains play important roles in ART function the key feature used to categorize the ART superfamily is the presence of certain catalytic amino acids within the ART fold1,35. ARTs are divided into two families based on the presence of three conserved amino acids required for NAD+ binding and enzymatic activity: histidine–tyrosine–glutamate found in bacterial HYE ARTs and arginine–serine–glutamate found in bacterial RSE ARTs (Fig. 1)1. In bacterial HYE ARTs, the histidine forms a hydrogen bond with the 2-OH of the adenosine ribose and the NH2 of the nicotinamide amide, the tyrosine pi stacks with the nicotinamide ring, and the glutamate is thought to stabilize the furanosyl oxocarbenium intermediate. The founding member of HYE ARTs is diphtheria toxin from Corynebacterium diphtheria1,35. In RSE ARTs, the arginine forms electrostatic interactions with the diphosphate backbone of NAD+, the serine makes hydrogen bonds with the several positions in the nicotinamide ribose, and the glutamate plays the same catalytic role as in HYE ARTs (Fig. 1).
The founding member of RSE ARTs is cholera toxin (CT) from Vibrio cholerae35. Despite the differences in the core catalytic amino acids between HYE ARTs and RSE ARTs, the confirmation of NAD+ in these pockets is generally the same, and both family members are able to catalyze ADP-ribosylation of target proteins (Fig. 1). This is, in large part, dictated by the interaction of the amide of nicotinamide with the backbone of a glycine in bacterial HYE ARTs and the backbone of an arginine in bacterial RSE ARTs (Fig. 1). In addition to these critical amino acids, ART folds from bacterial HYE and RSE ARTs and their eukaryotic homologs contain additional features that are important for function: the ADP-ribosylating turn-turn (ARTT) loop, also referred to as the acceptor or the A-loop, is found in all RSE and HYE ARTs, and the donor loop, also known as the D-loop in bacterial HYE ARTs, is found in all HYE ARTs but absent in RSE ARTs36. Several studies have demonstrated that the A-loop in bacterial RSE ARTs plays a critical role in targeting and amino acid selectivity37, and the D-loop plays important roles in NAD+-binding and catalysis38. A more detailed discussion of the role of these loops in ART function is described below.
In humans, there are two ART families that are homologs of these bacterial ARTs: poly-ADP-ribosepolymerases (PARPs) and ectoARTs. PARPs are related to the HYE ARTs and thus have also been referred to as ADP-ribosyltransferase diphtheria-toxin like (ARTDs)35. There are 17 PARP family members in humans; interestingly, only six PARPs (PARP1–6) contain the HYE residues, whereas the majority of PARP family members (PARP6–8, 10–12, and 14–16) contain a hydrophobic amino acid in the third position of the conserved catalytic amino acid residues35. The functional significance of this difference is unclear, but it is likely related to catalytic activity (more on this below). EctoARTs derive from the RSE bacterial ART lineage and are also referred to as ADP-ribosyltransferase cholera-toxin like (ARTCs)39. Other aspects of the ART fold are better conserved in human ARTs. Human PARPs contain both A- and D-loops, similarly to their bacterial HYE ART homologs, and all human ARTCs contain only the A-loop, like their bacterial RSE ART homologs39.
ART functions
ADP-ribosylation, or more specifically mono(ADP-ribosylation) (MARylation) is thought to have originally evolved in bacteria as a defense mechanism against viruses, other bacterial species and antimicrobial molecules (Fig. 2a)1,30. Ancestral ARTs targeted viral RNAs and DNAs and the proteins that regulate their replication and function, and even evolved the ability to target small-molecule antimicrobials such as rifamycin for MARylation10,11. Later, specific species of pathogenic bacteria evolved the ability to secrete ADP-ribosylating proteins called bacterial toxins (Fig. 2a). These secreted bacterial ARTs act as pathogenicity factors by entering their eukaryotic host cells and targeting key regulatory proteins for MARylation, including eukaryotic elongation factor 2 (eEF2), actin, Rho and other G proteins (Fig. 3a)1,29,40. MARylation of these targets leads to disastrous cellular consequences—inhibition of global protein synthesis in the case of eEF2 MARylation and overproduction of cAMP in the case of G-protein MARylation29. These aberrant MARylation events are the underlying causes of diphtheria and cholera, respectively, which when left untreated can be fatal.
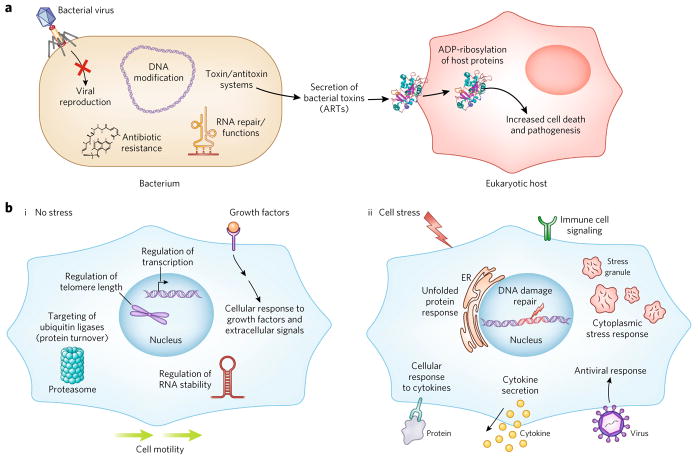
Figure 2. Key cellular functions of ARTs in bacteria and humans.
(a) Bacterial ART proteins play important cellular-defense functions that protect against viruses and antimicrobial molecules. They also play less defined roles in DNA modification, RNA repair and tRNA function. Their best understood functions involve their roles as toxins, whereby they act as pathogenicity factors in the host–pathogen response. (b) In eukaryotes, ARTs play important non-stress functions in the cell (i). They regulate telomere length, transcription, RNA stability and cell motility, and target proteins for degradation via the proteasome. They also play roles in the cellular response to extracellular signals such as growth factors. Under stress conditions (ii), eukaryotic ARTs regulate the unfolded protein response, the DNA damage response, the cytoplasmic stress response, and the cellular response to viruses. They also play important roles in immune cell signaling and activation, and regulate both the cellular response to cytokines and the expression and secretion of cytokines. Please note that not all ART functions are described.
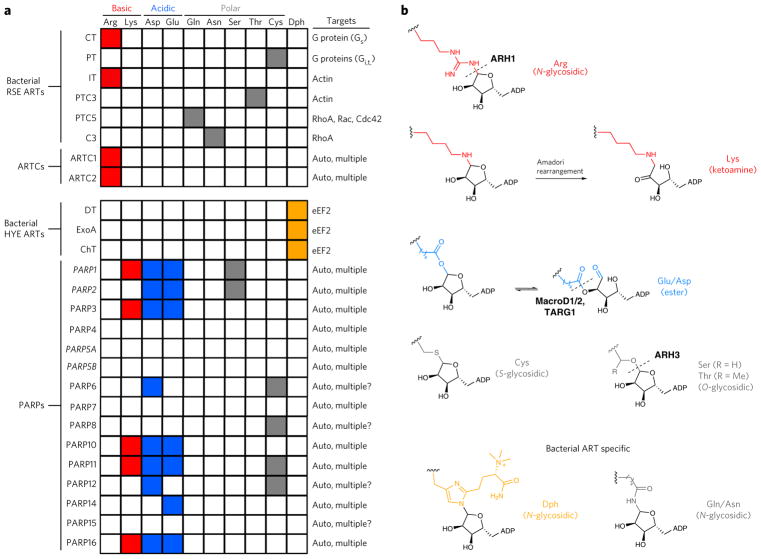
Figure 3. Amino acid targeting in bacterial ARTs, ARTCs and PARPs.
(a) In general, bacterial ARTs have a single target protein and target a single amino acid with high specificity. Eukaryotic ARTCs modify multiple protein targets, but appear to specifically modify arginine. PARPs also have multiple targets and have been shown to ADP-ribosylate multiple types of amino acids. PARPs shown in italic catalyze PARylation. PT, pertussis toxin; CT, cholera toxin; ChT, cholix toxin; DT, diphtheria toxin; IT, iota toxin; ARTC, ADP-ribosyltransferase cholera-toxin like. (b) Types of aa-ADPr bonds generated by bacterial ARTs and human ARTCs and PARPs. ADP, ADP-ribose. Dph, diphthamide. Known amino acid–specific hydrolyzing enzymes; sites of hydrolysis are shown as dotted lines.
ART bacterial toxins and their evolution in response to host immune systems are a classic example of host–pathogen genetic conflict1. Both the toxins and the eukaryotic host cellular innate immune system obtain often compensatory mutations in order to ‘gain the upper hand’. This genetic pressure is thought to have been an evolutionary driving force in the diversification of ART proteins1. Perhaps as a result, the bacterial toxins found key amino acid residues in their host proteins to MARylate, resulting in highly stringent site and target specificity. This exquisite specificity has made studying them more straightforward than Eukaryotic ARTs and, as a result, there is considerable accumulated knowledge about the chemical nature of the aa-ADPr conjugation site, and in some cases the mechanisms that define site specificity for bacterial ARTs.
Eukaryotes express more ART family members than prokaryotes, with humans expressing 21 ARTs; 17 PARPs and 4 ARTC proteins. human ARTs tend to be larger and more complex, and have more diverse domains outside the catalytic domain than bacterial ARTs1,35. This diversity in protein domain architecture facilitates the ability to integrate multiple cellular signals into a specific response-ADP-ribosylation of target proteins, an important feature of many eukaryotic ARTs. Together, these factors result in an expansion of functions for the human ART proteins relative to their bacterial relatives and, in many cases, provide more subtle control over the pathways regulated by the ARTs (Fig. 2b).
The eukaryotic ART targeting domains and signals sequences include nuclear localization signals, ER targeting sequences, DNA and RNA binding domains, protein–protein interaction domains, ubiquitin binding domains, and even ADPr binding domains, to list a few34. These protein domains target the PARPs to all internal cellular compartments, including the nucleus, cytoplasm, endoplasmic reticulum, and multiple membrane compartments33. In contrast, human ARTCs are primarily ecto-enzymes and are targeted to the plasma membrane, with their catalytic domains facing the extracellular space41.
The original cellular defense functions found in bacterial ARTs are still present in human PARPs and ARTCs. Multiple human ARTs play important antiviral functions in innate immunity, and several PARPs play additional functions in the regulation of acquired immunity and activation of immune cells (Fig. 2b, ii)42–45. In addition, PARPs play vital roles in regulating cellular stress responses related to the quality control of DNA, RNA and protein—specifically, the DNA damage response, the cytoplasmic stress response, and the unfolded protein response, respectively12,19,21,46. PARPs also play roles in the general, nonstress, nondefense pathways in cells by modulating gene expression, telomere length, transcriptional regulation, translation, mRNA stability, cell signaling, cell division and cell motility17,20,21,33,34,47–49. Furthermore, they also play important roles in regulating protein stability by targeting proteins for ubiquitylation by directly PARylating them13–16. Ironically, or perhaps through design, many of these pathways are ones that are targeted by the bacterial toxins for MARylation during infection.
Eukaryotic ARTs also evolved the ability to generate more elaborate ADPr modifications. Several PARPs can add multiple ADPr residues to the same amino acid site of modification in a process called poly(ADP-ribosylation) (PARylation). Multiple ADPr moieties are linked together via glycosidic bonds, resulting in a polymer that has characteristics of both nucleic acids and polysaccharides. These polymers can be quite large; PAR as long as 30 residues has been found in cells21. Due to its size and high negative charge density, PAR can function as a highly dynamic protein-recruiting scaffold. Several examples of this function exist, including assembly of DNA repair complexes during DNA damage, regulation of NF-κB signaling, cajal body function, mitotic spindle pole assembly, and cytoplasmic stress granule assembly18,47,50. Three high-affinity PAR binding domains have been identified: the tryptophan–tryptophan–-glutamate (WWE) domain, the poly(ADP-ribose)-binding zinc finger (PBZ) domain, and ‘macro’ domains. In addition, a lower affinity PAR-binding domain was identified called the PAR binding motif49. Macro domains also bind MAR, and examples have been found in bacterial MAR hydrolases that utilize Macro domains to bind and hydrolyze ADP-ribosylated proteins26. This suggests that protein binding was an early step in the evolution of ADPr function in ARTs and that, in addition to modifying the function of protein targets, ADP-ribosylation functions as an important signaling molecule. Collectively, ADPr binding domains are found in over 800 proteins, demonstrating the importance of MAR and PAR binding to eukaryotic life34.
How PARylating PARPs gained the ability to polymerize PAR and PARylate amino acids is no simple feat, as the chemical linkage between some aa-ADPr and ADPr-ADPr are substantially different. Counterintuitively, all PARPs that can generate PAR contain a glutamate in the third position of the HYE catalytic amino acids, which in bacterial ARTs does not impart PAR activity but is required for ADP-ribosylation. The presence of a glutamate in the third position of the HYE catalytic amino acids is necessary, but not sufficient, for PAR activity in PARPs. PARPs that contain a hydrophobic amino acid instead of a glutamate in the third position of the catalytic amino acids exclusively catalyze MARylation. Mutation of this amino acid to a glutamate in PARP10 (ref. 51) and PARP16 (ref. 52) does not confer PARylation activity and, in fact, abolishes MARylation activity. Given the importance of glutamate in stabilizing the furanosyl oxocarbenium intermediate, it is curious why these PARPs have evolved away from having a glutamate at this position.
The amino acid–ADPr linkage
ART-mediated ADP-ribosylation of protein targets has clearly evolved a variety of functions across many kingdoms of life1. Compared to bacterial ARTs, which for the most part target a single protein and a single amino acid in that target protein in a species-specific manner, eukaryotic ARTs appear to modify multiple targets and, for the PARPs, multiple amino acids in those targets50,52,53. One important question is whether the identity of the amino acid that is linked to ADPr impacts the function of the modification. For example, it is possible that the turnover, and hence the duration of the ADPr signal and/or the recognition by binding proteins and ADP-ribose hydrolases, is impacted by the nature of the amino acid attachment. Identifying the specific amino acids that are modified by specific ARTs is an important step for understanding the function of the proteins and their modifications.
More is known about bacterial-ART-generated aa-ADPr linkages because they are more homogeneous, and the amount of target protein modified with ADPr is much higher than that with eukaryotic ARTs. Bacterial HYE ARTs (diphtheria toxin (DT), exotoxin A (ExoA) and cholix toxin (ChT)) only target a unique, post-trans-lationally modified amino acid, termed diphthamide, found in eEF21,35. In contrast, as a family, bacterial RSE ARTs exhibit broader amino acid targeting and have been shown to modify arginine (for example, iota toxin), cysteine (for example, pertussis toxin), and threonine (for example, PTC3 toxin), as well as glutamine (for example, PTC5 toxin) and asparagine (for example, C3 toxin), which are not considered nucleophilic1. The amino acid preference for ADPr transfer mediated by these bacterial RSE ARTs is governed by interactions between the incoming amino acid nucleophile and the A-loop (which will be further discussed below)37. Despite their ability to modify multiple types of amino acids, each bacterial RSE ART family member only modifies a single protein target and only a single amino acid on that target (Fig. 3a)1,30.
Human ARTCs predominately modify arginines, similarly to their most closely related bacterial RSE ART ancestor, CT (Fig. 3a,b)39. Unlike their bacterial HYE ART ancestor diphtheria toxin, PARPs do not modify diphthamide, but instead modify several nucleophilic amino acids, predominately glutamate and aspartate (to form an ester bond); they have also been shown to modify serine, cysteine, and lysine (Fig. 3a,b)52,54–58. PARPs modify multiple amino acids on a single target, a feature that distinguishes them from bacterial ARTs52. The ADP-ribosylation of acidic amino acids is a unique feature of PARPs, and recent work has shown that the glutamate and aspartate can undergo a 1′–2′ transfer, which has important implications for how it is recognized and removed (Fig. 3b)59. Lysine-ADPr can undergo an Amadori rearrangement to form a stable ketoamine (Fig. 3b). It should be noted, however, that lysine can react with free ADPr, calling into question the relevance of lysine ADP-ribosylation60.
The importance of turnover
The turnover of ADPr or PAR at a specific site in target proteins has important implications for its function. For example, the duration of the cellular effects of bacterial toxins on their host proteins depends on the sustained modification of the target proteins29, and the DNA damage repair signal sent by PARP1 is dependent on a balance of sustained PARP1 activation and the regulation of PARG, the hydrolase that degrades PAR and reverses the ADPr signal49. Many factors affect the turnover of ADPR modifications, and most of these are likely dependent on the specific biology of the targets and the pathways affected. However, one key factor that can be pathway and target independent is the amino acid linkage. The specific aa-ADPr linkage could impact the turnover and function of the modification in several manners. Some chemical linkages may be inherently more stable and less likely to be reversed, although this is unlikely to seriously impact function. In addition, the recognition of some linkages by ADPr hydrolases or binding proteins may be impacted by the amino acid to which they are attached (Fig. 3a,b).
Thus far, enzymes that remove ADPr from arginine (ARH1)61, serine (ARH3)62,63 and acidic amino acids (MacroD1, D2, and TARG1)24,26 have been identified, suggesting that ADP-ribosylation at these amino acids is dynamic and that these amino acids are likely bona fide targets of ARTs in the cell (Fig. 3b). By contrast, enzymes that specifically cleave the lysine–ADPr or the cysteine–ADPr bond have not yet been identified, although the Nudix hydrolase NUDT16 (refs. 64,65) or the ectonucleotide pyrophosphatase (ENPP1)66, which can cleave the phosphodiester bond in protein-bound ADPr, might be responsible for their removal in cells. It is not well-studied whether specific aa-ADPr linkages impact recognition by specific ADPr hydrolases or alter the ability of specific ADPr hydrolases to remove the modification.
Target recognition and amino acid selection
Another critical question regarding the ARTs involves target recognition. How do the ARTs recognize their protein targets, and, specifically, how do they know which amino acid residue to modify? Important lessons can be learned by combining what we’ve discovered from bacterial and eukaryotic ARTs.
As mentioned above, bacterial ART toxins tend to target a single protein in host cells (Fig. 3). For example, the bacterial RSE ARTs CT and iota toxin (IT) MARylate a single arginine in the G protein Gs and actin, respectively1. Interestingly, there are instances in which bacterial RSE ARTs from different species can target distinct amino acids on the same protein target. IT MARylates Arg177 in actin, whereas the Photorhabdus luminescens toxin C3 (PTC3) MARylates Thr148 in actin67,68. The HYE ARTs (ExoA, DT, and ChT) are unique among the bacterial ART family because they only MARylate diphthamide, which is exclusively found in eEF2 (ref. 35).
Like eukaryotic ARTs, most bacterial ARTs contain protein domains outside of their catalytic domains. However, in contrast to eukaryotic ARTs, data suggest that these do not appear to play important roles in substrate specificity. Instead, site selectivity is thought to be primarily determined by features within the catalytic domain and by interactions these features make with the target. For bacterial RSE ARTs, amino acids in the A-loop play an essential role in target recognition and amino acid selection. Structural studies demonstrate that Tyr375 and Glu378 in the A-loop of IT interact with its substrate actin: Tyr375 is oriented toward Arg177 (human actin number), the site of IT-mediated MARylation, and Glu378 forms a hydrogen bond with Arg177 in actin69. Mutation of Glu378 to a glutamine (E378Q) abolished MARylation of actin; E378Q IT was still able to hydrolyze NAD+ at the glycosidic bond (NADase), demonstrating that this position is not required for the generation of the furanosyl oxocarbenium intermediate but rather for the transfer of ADPr specifically to Arg177 in actin69. The bacterial RSE ART C3 has a glutamine (Gln217) at an equivalent position in the A-loop, and structural studies demonstrate that Gln217 forms a hydrogen bond with Asn41 (site of C3-mediated MARylation) in RhoA70. Mutation of Gln217 to a glutamate (Q217E) abolishes MARylation of RhoA at Asn41; interestingly, however, Q217E C3 was able to MARylate an arginine in various substrates70. Together, these results demonstrate that the glutamate–glutamine in the A-loop motif directs which amino acid is targeted for MARylation.
Similarly to RSE ARTs, amino acids in the A-loop of bacterial HYE ARTs (DT, ExoA, and ChT) are important for target recognition. Double mutation of Glu546 and Arg551 to alanine (E546A R551A) in the HYE ART ExoA abolished MARylation of diphthamide in eEF2 without affecting NADase activity, and structural studies reveal that Glu546 and Arg551 in the A-loop interact with amino acids near diphthamide, further supporting a role for these amino acids in substrate recognition38. It does not appear that these amino acids, or any other amino acids in the A-loop, interact directly with diphthamide.
The D-loop of the HYE ART catalytic domain also appears to play an important role in substrate specificity. For example, several amino acids in the D-loop of ExoA play critical roles in MARylation activity and NAD+ binding. Structural studies show that Asp461 in the D-loop forms a hydrogen bond with the amino acid substrate diphthamide; however, mutation of Asp461 to an alanine does not affect MARylation activity38. Therefore, it is unknown which amino acid, if any, in the D-loop contributes to amino acid selection.
In humans, ARTCs are thought to MARylate arginines exclusively in protein targets, and thus exhibit specific amino acid selectivity in their targets, similar to their RSE bacterial ART ancestors24,71. Several ARTC family members contain a glutamate in the A-loop at the same position of Glu378 (IT numbering); therefore, it is likely that this glutamate in the A-loop of ARTCs is required for amino acid selection39. Unlike CT and IT, ARTCs appear to be more promiscuous, i.e., they have multiple protein targets and modify more than one arginine in those targets. Several ARTC family members can also MARylate arginines in peptide targets. For example, ARTC1 (also known as ART1) can MARylate multiple arginines in the immunemodulatory cationic peptides LL-37 and human neutrophil peptide-1 (HNP-1)71,72.
A large component of the substrate specificity of PARPs involves defined targeting domains outside of the catalytic domain described above. One of the best-described examples is the ankyrin repeat clusters (ARCs) in PARP5A and PARP5B (also known as tankyrase 1 and 2, respectively) that play a critical role in targeting these proteins to their substrates. PARylation of PARP5A and PARPB targets is a signal for their ubiquitylation and subsequent degradation by the proteasome, an important function for PAR in cells13–16. PARP5A and PARP5B contain multiple ARCs that bind to protein targets containing a tankyrase binding motif (TBM; R-x-x-G-[D/E]-G-no P-[D/E). In many cases, protein targets contain multiple TBMs, and therefore bind to multiple ARCs in PARP5A and PARP5B. One of the best-characterized substrates of PARP5A and PARP5B is axin, which, upon PARP5A/B-mediated PARylation in cells, is ubiquitylated and then targeted for proteasomal-mediated degradation13–16. Structural studies demonstrated that the two TBMs in axin bind to two ARCs (ARC2-3) in PARP5A and PARP5B73. Disruption of this interaction by mutation of a glycine in the TBM in axin abolishes proteasomal-mediated degradation of axin and ADP-ribosylation by PARP5A73. Sequence alignments demonstrate that TBMs are found in many proteins, of which several have been validated as targets of PARP5A/B74.
Another example involves the DNA-binding PARPs (PARP1, PARP2, and PARP3). The zinc finger domains in PARP1 recruits PARP1 to sites of DNA damage and leads to activation of ART activity49. Activation results in extensive auto-PARylation, and the PAR polymer is thought to recruit other DNA repair proteins (for example, XRCC1). Interestingly, target specificity of PARP1 can also be regulated by its binding proteins. Histone PARylation factor 1 (HPF1) binding to PARP1 or PARP2 causes these PARPs to PARylate serines in histones and other protein targets54,57. Interestingly, the serine–ADPr and ADPr–ADPr linkages in PAR are both O-glycosidic, acetal linkages, suggesting that PARP1 could preferentially transfer ADPr onto hydroxyl nucleophiles under certain conditions.
Aside from targeting through the protein-targeting domains, little is known about how PARPs select specific amino acids to modify, although one possibility is that the protein-targeting domains constitute the major selectivity mechanism of PARPs. In this model, the noncatalytic domains target the PARPs to their substrates, and proximity to the catalytic domain determines which amino acids are ADP-ribosylated (Fig. 4). This could explain the seemingly poor amino acid selectivity of PARPs relative to other bacterial ARTs. For example, PARP1 has been shown to modify dozens and dozens of proteins and multiple amino acids, including acidic amino acids glutamate, aspartate and serine, as well as the basic amino acid lysine54,56,75. The targets of MARylating PARPs appear to be even more broad. A recent study showed that multiple MARylating PARPs (for example, PARP6, PARP8, PARP10, PARP11, and PARP12) auto-MARylate on glutamate, aspartate, lysine, and cysteine52. Because these chemical linkages can be quite different, and many of the site identifications for PARPs appear to be dependent on the technique used to identify them, it is unclear which of the amino acids are the true sites of ADP-ribosylation in the cell. Combining recently developed strategies for finding the direct targets of MARylating PARPs with mass spectrometry methods to identify the aa-ADPr sites will help figure out the family-member-specific amino acid targeting preferences across multiple targets50,76. Indeed, a recent study combining a chemical–genetics strategy to identify targets of the nuclear PARPs (PARP1, PARP2, and PARP3) with a mass spectrometry method to identify aa-ADPr sites revealed potential family-member-specific consensus sequences50. These include: E*-P/E or E*-X-P (PARP1), E-E* (PARP2) and E*-X-A, E*-X-X-X-K, A-X-X-E* (PARP3). Additional studies, including mutagenesis, are required to evaluate the biological relevance of these sequences and to confirm that they constitute bona fide consensus sequences for these PARPs.
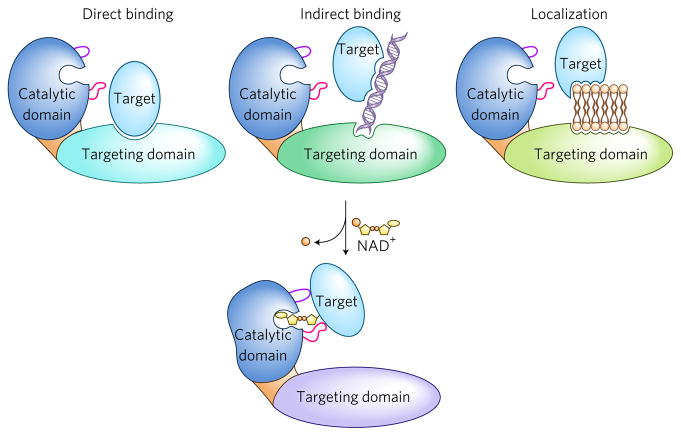
Figure 4. Mechanisms of substrate targeting.
Two-step model for substrate targeting. Step 1: targeting domains found in ART proteins bring the ART catalytic domain in close proximity to the target protein. Targeting can occur by direct binding of the targeting domain to the target protein, by indirect binding through other molecules including nucleic acids, or by concentrating the ART to specific organelles to which the targets are sequestered, including membranes. Step 2: structural features within the catalytic domains including the A-loop (purple) and D-loop (magenta), play additional roles in substrate targeting including amino acid selection.
Features within the catalytic domain of PARPs could also dictate target and amino acid specificity. Though in bacterial HYE ARTs, the A- and D-loops play important roles in target specificity, this possibility has not yet been examined in their homologs, the human PARPs. In PARPs the amino acids within these loops are not well conserved, and the length of the loops are highly variable among PARP family members52. Though the significance of these differences are not well understood, a recent study demonstrated that swapping the A-loop in PARP1 (37 amino acids) for the A-loop in PARP16 (13 amino acids) results in a decrease in PARP1 PARylation activity; conversely, swapping the A-loop in PARP16 for the A-loop in PARP1 did not affect PARP16 auto-MARylation activity52. These results suggest that exact composition and/or length of the A-loop in PARP1 is critical for amino acid recognition or catalysis (or both). Whether this is true for all PARPs remains to be determined.
In summary, although both bacterial and eukaryotic ARTs contain protein domains outside of their catalytic domains, bacterial ARTs appear to rely primarily on features within their catalytic domains for substrate selectivity. In contrast, a large component of eukaryotic ART target selectivity involves noncatalytic protein domains. It is important to note that key experiments with bacterial ARTs examining the importance of the targeting domains for substrate selectivity have not been performed and, similarly, the importance of specific features of the catalytic domain, specifically the A- and D-loop, on substrate selectivity for eukaryotic ARTs have not been systematically explored. In light of the available data, we favor a model in which target selectivity by all ARTs is determined by a combination of targeting domains and features within the catalytic domains. This model is described in Figure 4.
Concluding remarks
The ART family of proteins has evolved many important cellular functions, and members are found in all kingdoms of life. As a protein family with clear disease relevance, PARPs have been identified as important therapeutic targets for cancer, and are likely good targets for modulation of the immune system. Bacterial toxins are important disease proteins that have a broad impact on human health, and bacterial ARTs with antiviral and antimicrobial functions could be interesting targets for viral and bacterial infections, respectively. Yet, despite the importance and the promise of this interesting family of proteins, little is known about the function of ARTs and ADPr, particularly in humans.
What are the key questions in the field? There are many, of course, but the current priorities are the following: (1) What are the specific activating signals of the ARTs and what are their protein targets? (2) What are the key amino acid targets of the PARPs and how do they impact the function of the modification? (3) How does the aa-ADPr linkage impact the recognition and binding affinity of ADPr binding proteins and hydrolases. Importantly, we are finally in a position to start answering some of these questions, owing to technological advances and interest, and the answers will provide critical information that will allow us to better evaluate the therapeutic potential of ARTs and ADPr modifications in human disease and how to best exploit ADP-ribosylation for the treatment of disease.
Finally, one oddity about the field of ADP-ribosylation is the intellectual and, in fact, physical divide between bacterial ART scientists and those that study eukaryotic ARTs. Despite the common interest and expertise, there is currently very little interaction between these scientists, as we do not share reagents, we do not attend each other’s meetings, and we are essentially considered separate fields of biology. This makes little sense and is a major obstacle for progression in a field of biology that is already technically challenging and difficult. Bacterial and eukaryotic ARTs clearly share common mechanistic features, and the tools and knowledge necessary to study them and the modifications they make have significant overlap. It’s time for the two sides to get together.
Footnotes
Competing financial interests
The authors declare competing financial interests: details accompany the online version of the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aravind L, Zhang D, de Souza RF, Anand S, Min Iyer L. In: Endogenous ADP-Ribosylation. Koch-Nolte F, editor. Springer International Publishing; 2015. pp. 3–32. [Google Scholar]
- 2.Bazan JF, Koch-Nolte F. Sequence and structural links between distant ADP-ribosyltransferase families. Adv Exp Med Biol. 1997;419:99–107. doi: 10.1007/978-1-4419-8632-0_12. [DOI] [PubMed] [Google Scholar]
- 3.Culver GM, et al. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1″-2″cyclic phosphate. Science. 1993;261:206–208. doi: 10.1126/science.8392224. [DOI] [PubMed] [Google Scholar]
- 4.Spinelli SL, Kierzek R, Turner DH, Phizicky EM. Transient ADP-ribosylation of a 2′-phosphate implicated in its removal from ligated tRNA during splicing in yeast. J Biol Chem. 1999;274:2637–2644. doi: 10.1074/jbc.274.5.2637. [DOI] [PubMed] [Google Scholar]
- 5.Lyons B, et al. Scabin, a novel DNA-acting ADP-ribosyltransferase from Streptomyces scabies. J Biol Chem. 2016;291:11198–11215. doi: 10.1074/jbc.M115.707653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takamura-Enya T, et al. Mono(ADP-ribosyl)ation of 2″-deoxyguanosine residue in DNA by an apoptosis-inducing protein, pierisin-1, from cabbage butterfly. Proc Natl Acad Sci USA. 2001;98:12414–12419. doi: 10.1073/pnas.221444598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano T, et al. Purification and molecular cloning of a DNA ADP-ribosylating protein, CARP-1, from the edible clam Meretrix lamarckii. Proc Natl Acad Sci USA. 2006;103:13652–13657. doi: 10.1073/pnas.0606140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jankevicius G, Ariza A, Ahel M, Ahel I. The toxin-antitoxin system DarTG catalyzes reversible ADP-ribosylation of DNA. Mol Cell. 2016;64:1109–1116. doi: 10.1016/j.molcel.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munnur D, Ahel I. Reversible mono-ADP-ribosylation of DNA breaks. FEBS J. 2017;284:4002–4016. doi: 10.1111/febs.14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dabbs ER, et al. Ribosylation by mycobacterial strains as a new mechanism of rifampin inactivation. Antimicrob Agents Chemother. 1995;39:1007–1009. doi: 10.1128/aac.39.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baysarowich J, et al. Rifamycin antibiotic resistance by ADP-ribosylation: Structure and diversity of Arr. Proc Natl Acad Sci USA. 2008;105:4886–4891. doi: 10.1073/pnas.0711939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jwa M, Chang P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nat Cell Biol. 2012;14:1223–1230. doi: 10.1038/ncb2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 14.DaRosa PA, et al. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature. 2015;517:223–226. doi: 10.1038/nature13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, et al. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl) ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou ZD, Chan CHS, Xiao ZC, Tan EK. Ringfinger protein 146/Iduna is a poly(ADP-ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase. Cell Adh Migr. 2011;5:463–471. doi: 10.4161/cam.5.6.18356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 18.Chang P, Coughlin M, Mitchison TJ. Interaction between Poly(ADPribose) and NuMA contributes to mitotic spindle pole assembly. Mol Biol Cell. 2009;20:4575–4585. doi: 10.1091/mbc.E09-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vyas S, Chang P. New PARP targets for cancer therapy. Nat Rev Cancer. 2014;14:502–509. doi: 10.1038/nrc3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 21.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 22.Ahel I, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 23.Barkauskaite E, Jankevicius G, Ahel I. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol Cell. 2015;58:935–946. doi: 10.1016/j.molcel.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Jankevicius G, et al. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol. 2013;20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenthal F, et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol. 2013;20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 26.Sharifi R, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Haan L, Hirst TR. Cholera toxin: a paradigm for multi-functional engagement of cellular mechanisms (Review) Mol Membr Biol. 2004;21:77–92. doi: 10.1080/09687680410001663267. [DOI] [PubMed] [Google Scholar]
- 28.Ueda K, Hayaishi O. ADP-ribosylation. Annu Rev Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- 29.Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch-Nolte F, editor. Endogenous ADP-Ribosylation. 1. Vol. 384. Springer International Publishing; 2015. [Google Scholar]
- 31.de Souza RF, Aravind L. Identification of novel components of NAD-utilizing metabolic pathways and prediction of their biochemical functions. Mol Biosyst. 2012;8:1661–1677. doi: 10.1039/c2mb05487f. [DOI] [PubMed] [Google Scholar]
- 32.Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J Mol Biol. 1973;76:241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 33.Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat Commun. 2013;4:2240. doi: 10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bock FJ, Chang P. New directions in poly(ADP-ribose) polymerase biology. FEBS J. 2016;283:4017–4031. doi: 10.1111/febs.13737. [DOI] [PubMed] [Google Scholar]
- 35.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Pinto AF, Schüler H. Comparative structural analysis of the putative mono-ADP-ribosyltransferases of the ARTD/PARP family. Curr Top Microbiol Immunol. 2015;384:153–166. doi: 10.1007/82_2014_417. [DOI] [PubMed] [Google Scholar]
- 37.Sun J, Maresso AW, Kim JJP, Barbieri JT. How bacterial ADP-ribosylating toxins recognize substrates. Nat Struct Mol Biol. 2004;11:868–876. doi: 10.1038/nsmb818. [DOI] [PubMed] [Google Scholar]
- 38.Jørgensen R, Wang Y, Visschedyk D, Merrill AR. The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep. 2008;9:802–809. doi: 10.1038/embor.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laing S, Unger M, Koch-Nolte F, Haag F. ADP-ribosylation of arginine. Amino Acids. 2011;41:257–269. doi: 10.1007/s00726-010-0676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung DW, Collier RJ. Enzymatically active peptide from the adenosine diphosphate-ribosylating toxin of Pseudomonas aeruginosa. Infect Immun. 1977;16:832–841. doi: 10.1128/iai.16.3.832-841.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seman M, Adriouch S, Haag F, Koch-Nolte F. Ecto-ADP-ribosyltransferases (ARTs): emerging actors in cell communication and signaling. Curr Med Chem. 2004;11:857–872. doi: 10.2174/0929867043455611. [DOI] [PubMed] [Google Scholar]
- 42.Todorova T, Bock FJ, Chang P. Poly(ADP-ribose) polymerase-13 and RNA regulation in immunity and cancer. Trends Mol Med. 2015;21:373–384. doi: 10.1016/j.molmed.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwata H, et al. PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation. Nat Commun. 2016;7:12849. doi: 10.1038/ncomms12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozaki T, et al. Mitochondrial damage elicits a TCDD-inducible poly(ADPribose) polymerase-mediated antiviral response. Proc Natl Acad Sci USA. 2017;114:2681–2686. doi: 10.1073/pnas.1621508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atasheva S, Frolova EI, Frolov I. Interferon-stimulated poly(ADP-ribose) polymerases are potent inhibitors of cellular translation and virus replication. J Virol. 2014;88:2116–2130. doi: 10.1128/JVI.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leung AKL, et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bock FJ, Todorova TT, Chang P. RNA regulation by poly(ADP-ribose) polymerases. Mol Cell. 2015;58:959–969. doi: 10.1016/j.molcel.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Todorova T, Bock FJ, Chang P. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat Commun. 2014;5:5362. doi: 10.1038/ncomms6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupte R, Liu Z, Kraus WL. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31:101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson BA, et al. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 2016;353:45–50. doi: 10.1126/science.aaf7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleine H, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 52.Vyas S, et al. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daniels CM, Ong SE, Leung AKL. The promise of proteomics for the study of ADP-ribosylation. Mol Cell. 2015;58:911–924. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gibbs-Seymour I, Fontana P, Rack JGM, Ahel I. HPF1/C4orf27 is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol Cell. 2016;62:432–442. doi: 10.1016/j.molcel.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bilan V, Leutert M, Nanni P, Panse C, Hottiger MO. Combining higher-energy collision dissociation and electron-transfer/higher-energy collision dissociation fragmentation in a product-dependent manner confidently assigns proteomewide ADP-ribose acceptor sites. Anal Chem. 2017;89:1523–1530. doi: 10.1021/acs.analchem.6b03365. [DOI] [PubMed] [Google Scholar]
- 57.Bonfiglio JJ, et al. Serine ADP-ribosylation depends on HPF1. Mol Cell. 2017;65:932–940.e6. doi: 10.1016/j.molcel.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leidecker O, et al. Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat Chem Biol. 2016;12:998–1000. doi: 10.1038/nchembio.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morgan RK, Cohen MS. A clickable aminooxy probe for monitoring cellular ADP-ribosylation. ACS Chem Biol. 2015;10:1778–1784. doi: 10.1021/acschembio.5b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cervantes-Laurean D, Jacobson EL, Jacobson MK. Glycation and glycoxidation of histones by ADP-ribose. J Biol Chem. 1996;271:10461–10469. doi: 10.1074/jbc.271.18.10461. [DOI] [PubMed] [Google Scholar]
- 61.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 62.Fontana P, et al. Serine ADP-ribosylation reversal by the hydrolase ARH3. eLife. 2017;6:e28533. doi: 10.7554/eLife.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abplanalp J, et al. Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase. Nat Commun. 2017;8:2055. doi: 10.1038/s41467-017-02253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daniels CM, Thirawatananond P, Ong SE, Gabelli SB, Leung AKL. Nudix hydrolases degrade protein-conjugated ADP-ribose. Sci Rep. 2015;5:18271. doi: 10.1038/srep18271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palazzo L, et al. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem J. 2015;468:293–301. doi: 10.1042/BJ20141554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palazzo L, et al. ENPP1 processes protein ADP-ribosylation in vitro. FEBS J. 2016;283:3371–3388. doi: 10.1111/febs.13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandekerckhove J, Schering B, Bärmann M, Aktories K. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 1987;225:48–52. doi: 10.1016/0014-5793(87)81129-8. [DOI] [PubMed] [Google Scholar]
- 68.Lang AE, et al. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science. 2010;327:1139–1142. doi: 10.1126/science.1184557. [DOI] [PubMed] [Google Scholar]
- 69.Tsurumura T, et al. Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex. Proc Natl Acad Sci USA. 2013;110:4267–4272. doi: 10.1073/pnas.1217227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogelsgesang M, Aktories K. Exchange of glutamine-217 to glutamate of Clostridium limosum exoenzyme C3 turns the asparagine-specific ADP-ribosyltransferase into an arginine-modifying enzyme. Biochemistry. 2006;45:1017–1025. doi: 10.1021/bi052253g. [DOI] [PubMed] [Google Scholar]
- 71.Stevens LA, Levine RL, Gochuico BR, Moss J. ADP-ribosylation of human defensin HNP-1 results in the replacement of the modified arginine with the noncoded amino acid ornithine. Proc Natl Acad Sci USA. 2009;106:19796–19800. doi: 10.1073/pnas.0910633106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Picchianti M, et al. NAD-dependent ADP-ribosylation of the human antimicrobial and immune-modulatory peptide LL-37 by ADP-ribosyltransferase-1. Innate Immun. 2015;21:314–321. doi: 10.1177/1753425914536242. [DOI] [PubMed] [Google Scholar]
- 73.Morrone S, Cheng Z, Moon RT, Cong F, Xu W. Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc Natl Acad Sci USA. 2012;109:1500–1505. doi: 10.1073/pnas.1116618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pollock K, Ranes M, Collins I, Guettler S. Identifying and validating tankyrase binders and substrates: a candidate approach. Methods Mol Biol. 2017;1608:445–473. doi: 10.1007/978-1-4939-6993-7_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- 76.Carter-O’Connell I, et al. Identifying family-member-specific targets of mono-ARTDs by using a chemical genetics approach. Cell Rep. 2016;14:621–631. doi: 10.1016/j.celrep.2015.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]