Abstract
Recent work suggests that genes encoding complement proteins that are active in the innate immune system may confer risk for schizophrenia by disrupting typical synaptic pruning in late adolescence. Alterations in the complement pathway may contribute to aberrant cortical thinning in schizophrenia prodromes and reduced prefrontal cortical thickness in chronic schizophrenia patients; however, this theory needs to be translated to humans. We conducted a series of analyses in a sample of adult Swedish twins enriched for schizophrenia (N=129) to assess the plausibility of a relationship between complement gene expression and cortical thickness that could go awry in the etiology of schizophrenia. First, we identified that peripheral mRNA expression levels of two complement genes (C5, SERPING1) made unique contributions to the variance in superior frontal cortical thickness among all participants. Vertex-wise maps of the association between gene expression levels and thickness across the cortex suggested that this relationship was especially strong with SERPING1 in the superior frontal region, consistent with the pattern of disruption in cortical thickness observed in schizophrenia. Additional analyses identified that these genes are expressed in the human superior frontal cortex, that heritable genetic factors influence SERPING1 gene expression levels, and that these associations are observed regardless of case status. These findings provide initial evidence linking the complement system with cortical thinning in humans, a process potentially involved in the pathogenesis of schizophrenia.
Introduction
Investigations of individuals at clinically high risk for psychosis have found that the onset of the disorder is preceded by reductions in cortical gray matter thickness, particularly in the superior frontal cortex (Cannon et al, 2015). Post-mortem studies suggest that this may be influenced by excessive reduction of synaptic connections between cortical neurons (Bennett, 2011; Glausier and Lewis, 2013). This could lead to reduced integrity of the superior frontal cortex, disrupt cognitive processes that involve this region (Paus, 2005) such as source memory (Dobbins and Wagner, 2005), and ultimately contribute to the development of psychosis (Cannon, 2015). However, potential mechanisms underlying reductions in cortical synaptic density have not been clearly established. As both risk for schizophrenia (Sullivan et al, 2003) and measures of superior frontal cortical thickness (Goldman et al, 2013) are strongly influenced by genetic factors, examining genes associated with these phenomena may yield insight into underlying pathological processes of the disorder.
The strongest genomic region implicated in genome-wide association studies of risk for schizophrenia is the major histocompatibility complex locus (Ripke et al, 2014), which encodes many genes in signaling pathways involved in the immune response to pathogens (Horton et al, 2004). One immune pathway associated with this locus that has been increasingly linked with both synaptic pruning and risk for schizophrenia is the complement pathway (Sekar et al, 2016). This pathway is known to ‘tag’ both pathogens and damaged human cells for elimination by macrophage cells (Mayilyan et al, 2008), and emerging evidence from in vitro and rodent studies suggests that complement proteins also tags synapses for elimination (ie, synaptic pruning) in the developing visual cortex (Chu et al, 2010; Schafer et al, 2012). Synaptic pruning is a neurodevelopmental process that may peak during adolescence (Tamnes et al, 2010), coinciding with the typical onset of schizophrenia (Cannon et al, 2015). It is possible that overactivation of the complement pathway during this period could increase risk for schizophrenia via exacerbated synaptic elimination. In support of this theory, Sekar et al (2016) identified schizophrenia-associated genetic variants within the complement system that increased complement activation in mice, disrupting synaptic pruning. Though synaptic pruning cannot be directly measured in living humans, structural MRI measures of cortical thickness may be a somewhat sensitive proxy (Bourgeois and Goldman-Rakic, 1994; Glausier and Lewis, 2013). A link between the complement pathway and cortical thickness would provide a first step towards assessing the plausibility of a connection between genetic variants in the complement system and cortical thickness, with potential implications for understanding schizophrenia.
We assessed whether complement gene mRNA expression levels in peripheral blood leukocytes correlated with superior frontal cortical thickness in a sample of adult twins from Sweden enriched for schizophrenia cases. Further analyses were conducted to assess the plausibility of the proposed pathway, including mapping correlations between complement gene expression and thickness across the entire cortex, assessing complement gene expression in post-mortem human superior frontal cortical samples, examining whether heritable genetic factors influence variance in complement expression, and determining the influence of case status.
Materials and methods
Participants
The Swedish Twin Registry was used to identify a national cohort of twins born between 1940 and 1975 (see Supplementary Information). In total, 147 individuals who were interviewed, underwent an MRI scan, and provided blood samples that were assayed for gene expression were included in the present study. Fifteen participants were excluded due to MRI quality (see below) and three participants were excluded due to missing BMI data, yielding a total sample of 129 participants. Across the sample, 43% were male (n=56), average age was 49.1 years (95% CI (29.2, 69.0)), and average BMI was 26.4 (17.0, 35.7). Twenty-two individuals were diagnosed with schizophrenia, 21 were unaffected schizophrenia co-twins, 13 were diagnosed with bipolar disorder, 13 were unaffected bipolar co-twins, and 60 were control twin pairs. As this theory is based on studies of psychosis (Sekar et al, 2016) and information about psychotic features was not available for bipolar participants, bipolar participants and their co-twins were included with controls for all primary analyses. This approach parallels Sekar et al (2016), in which bipolar cases and controls were grouped when testing for and identifying increased C4 expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. To assess the influence of this grouping, secondary analyses were conducted with schizophrenia and bipolar cases and co-twins grouped together and with bipolar cases and co-twins excluded entirely (see Supplementary Information). Results did not change appreciably.
RNA Microarray
Gene expression was assayed from peripheral blood mononuclear cells (PBMCs) using the Illumina Human WG6 v3.0 chip. RNA was extracted from PBMCs isolated from a 10 ml blood sample (ABI Tempus System). RNA aliquots of 100 ng/μl were analyzed by the UCLA Biological Samples Processing Core. Technical replicates were run on all samples using the Illumina Human WG6 v3.0 chip, and correlations between these samples for each person were evaluated. All samples had a useable quantity of RNA, and all subjects had good correlations between their technical replicate samples (ICCs: 0.902–0.998). Samples were preprocessed using the Illumina Bead Studio package. Technical replicates were averaged to produce one vector per person, and samples were log-transformed to ensure normalization of the sample distributions. The Illumina chip indexed a total of 24 526 markers. After rank normalization and background subtraction, 5967 probes were excluded based on low signal strength (not significantly greater than 0 at a Bonferroni-corrected α<0.05).
Gene Selection and Quality Control
The Gene Ontology (GO) Consortium (geneontology.org) was used to identify a priori genes involved in the complement system. A total of 28 unique human genes were annotated with the term ‘regulation of complement activation’, and mRNA levels of 22 of the 28 genes identified in GO were available from the chip assay after quality control (see Supplementary Information). Outliers (>2 SD) gene expression values were winsorized (ie extreme values set to a value two standard deviations from the mean expression level) using the R package ‘robustHD’ (Alfons, 2016). Winsorization did not significantly influence results (see Supplementary Information).
Magnetic Resonance Imaging
MRIs were acquired on a 1.5-T scanner (GE Healthcare, Little Chalfont, Buckinghamshire, UK) using a 3D T1-weighted IRSPGR sequence acquired sagitally (TE=6 ms, flip angle=35°, and no interslice gap) in the Department of Clinical Neuroscience, Karolinska Institutet. The matrix size was 256 × 256 × 256 pixels, which corresponded to a resolution of 1 mm3. All MRI images were processed using FreeSurfer version 5.3 (http://surfer.nmr.mgh.harvard.edu/) (see Chung et al, 2015). Unrecoverable artifacts (eg motion, poor contrast) in scans led to the exclusion of 14 subjects (4 cases, 2 co-twins, 8 controls). An additional control was excluded due to abnormally large ventricles (potential undiagnosed neurological condition).
Statistical Analyses
Sample characteristics are compared by case status in Table 1. Expression levels did not differ by case status for any complement genes (see Supplementary Information). See Supplementary Information for a flow chart of analyses (Supplementary Information Figure S2).
Table 1. Demographic Characteristics of the Sample.
| SZ (n=22) | SZ co-twin (n=21) | Control (n=86) | Effect size | df | p | |
|---|---|---|---|---|---|---|
| Age (years) | 50.29 (31.65, 68.92) | 48.77 (28.76, 68.77) | 48.86 (28.53, 69.18) | F=0.19 | 2126 | 0.826 |
| Sex (% male) | 14 (63.6%) | 11 (52.4%) | 31 (36.0%) | χ2=6.25 | 2 | 0.044 |
| BMI | 27.43 (19.48, 35.38) | 25.30 (14.70, 35.89) | 26.37 (17.01, 35.73) | F=1.12 | 2126 | 0.328 |
| Smoking (% yes) | 11 (50%) | 7 (33.3%) | 23 (26.7%) | χ2=4.40 | 2 | 0.111 |
| Antipsychotics (% yes)a | 14 (63.6%) | 1 (7.1%) | 3 (4.3%) | χ2=42.42 | 2 | 0.001 |
| Zygosity (% MZ) | 8 (36.4%) | 7 (33.3%) | 44 (51.2%) | χ2=3.10 | 2 | 0.212 |
| Superior frontal thickness (mm3)b | 2.64 (2.21, 3.07) | 2.68 (2.37, 2.99) | 2.64 (2.31, 2.98) | F=0.43 | 2 | 0.654 |
95% confidence intervals listed in brackets.
Missing data: co-twin (n=14), control (n=69).
See Supplementary Table 3 for superior frontal thickness of schizophrenia and bipolar cases, their co-twins, and controls.
Prescreening
There was not sufficient power to account for expression levels of all 22 complement genes simultaneously in a model predicting superior frontal cortical thickness. As we did not have an external replication sample for this data set, we needed to nominate predictors for further investigation in a simultaneous model. To protect against Type I error, a parameter reduction selection approach was taken at a univariate level to identify transcripts that were most consistently associated with superior frontal cortical thickness across 1000 subsets of the data. Subsets were created by randomly selecting 85% of participants (N=105) without replacement (ie, resampling). The association between expression levels of a gene and superior frontal cortical thickness was examined in each subset, accounting for age, gender, BMI and diagnostic status as covariates and family as a random factor in a linear mixed model. Associations (β) below the prescreening threshold of p<0.05 were recorded as ‘1’, and those above were counted as ‘0’. We tallied the number of associations, and anticipated chance associations in approximately 50/1000 subsets. Analyses were conducted using the linear and nonlinear mixed effects models (nlme) package in R (Pinheiro and Bates, 2000), and degrees of freedom were estimated using the Satterthwaite option. Assumptions of linear mixed models were not rejected (see Supplementary Information).
Simultaneous Model
The transcripts selected in the univariate approach could account for shared or unique variance in superior frontal cortical thickness. To assess this, transcripts identified in the prescreening phase were entered simultaneously into a linear mixed model predicting superior frontal cortical thickness in the entire sample (N=129), accounting for the same covariates listed above. Gene expression variables were z-scored to promote interpretability of regression statistics. Model performance was compared with a covariate-only model, and covariation between transcripts was assessed using the vif.mer package (source code available at https://github.com/aufrank/R-hacks/blob/master/mer-utils.R). The influence of smoking, antipsychotic use, zygosity, and estimated total intracranial volume on model fit was assessed. For all hypothesis tests, a threshold of p<0.05 was used. The change in R2 between the full and covariate-only models was approximated using the multi-model inference (MuMIn) package (Barton, 2016).
Spatial Distribution
Relationships between each transcript of interest and cortical thickness were tested vertex-wise (ie, models predicting thickness at each vertex across the cortex, accounting for the same covariates) in the full sample (N=129). p-Values were mapped onto the cortical surface bilaterally at each vertex. Monte Carlo simulations were used across the surface and synthesized with a cluster-forming threshold of p<0.05 (two-sided) (Hagler et al, 2006; Hayasaka and Nichols, 2003). To visualize effects below the stringent statistical threshold, associations were mapped onto the cortical surface with and without multiple comparison corrections.
Post-mortem Human Cortex
To assess if these genes are expressed in neural tissue, we determined whether there was non-zero gene transcription of the nominated complement genes within the superior frontal cortex (bi-hemispheric) ROI using six non-psychiatric post-mortem microarray expression data sets obtained from the open-access Allen Human Brain Atlas (Hawrylycz et al, 2012) (see Supplementary Information). Because multiple microarray probes exist for the selected complement genes, we chose the probe that most reliably reflected ‘ground truth’ expression (C5: A_23_P71855; SERPING1: A_23_P139123), assessed with RNA-Seq by Miller et al (2014). A gene was marked as expressed above baseline within a tissue sample if a two-sided t-test comparing expression to the corresponding background was significant at p<0.01 and the difference between the background subtracted signal and the background was greater than 2.6 standard deviations away from the background mean (see data documentation at http://human.brain-map.org). Because microarray measures are typically used to gauge relative expression (Okoniewski and Miller, 2006), we also examined available RNA-Seq data to assess whether an absolute measure of transcript expression was above zero (Wang et al, 2009) (see Supplementary Information).
Heritability
For those mRNA probes found to be related to superior frontal cortical thickness, we used Mx to implement structural equation modeling comparing gene expression covariance in monozygotic (MZ) (n=34 pairs) to dizygotic (DZ) twin pairs (n=50 pairs) to assess the relative contribution of additive genetics (A), dominant genetics (D), common environment (C), and unique environment (E) (Plomin et al, 2013) (see Supplementary Information). To maximize power, all participants with available gene expression data were used for heritability estimates (N=190), regardless of availability of imaging data (see Zheutlin et al, 2016). We tested the significance of genetic and shared environmental contributions to each model with χ2 difference tests (ie, directly comparing models in which each of these parameters was present versus absent). Heritability of superior frontal cortical thickness was also assessed in all participants with usable structural MRI data (N=132), which included 22 MZ and 26 DZ twin pairs.
Case Status
The influence of case status (schizophrenia, co-twin, control) on the observed associations between complement gene expression levels and superior frontal cortical thickness was further assessed by including an interaction term between gene expression levels and case status predicting superior frontal cortical thickness.
Results
Prescreening
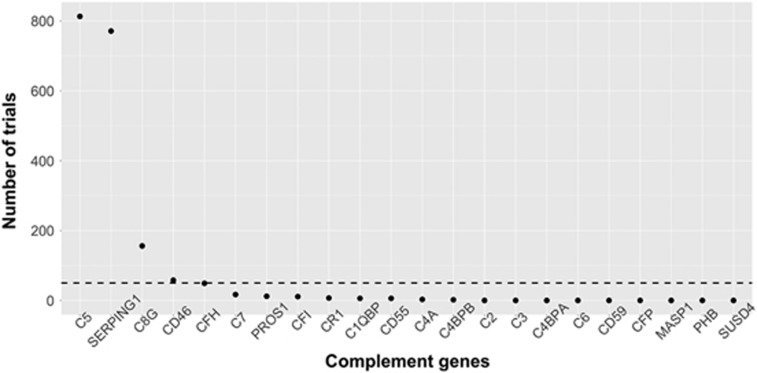
Associations within small samples are more likely to be driven by sample-specific characteristics, and thus may not always generalize to independent data sets. To reduce the likelihood of this possibility, we used a prescreening approach to nominate complement transcripts that were most consistently associated (threshold p<0.05) with superior frontal cortical thickness in 1000 random subsets (85%) of the sample (Figure 1). Expression levels of two genes were most consistently associated with superior frontal cortical thickness: C5 (813/1000) and SERPING1 (771/1000). The gene with the next highest number of associations, C8G, exhibited over 60% fewer associations (156/1000). A more stringent threshold of p<0.01 was used to further assess the strength of these associations. Again, SERPING1 (185/1000) and C5 (138/1000) exhibited substantially more associations with superior frontal cortical thickness than C8G (2/1000). C5 and SERPING1 were selected for further examination.
Figure 1.
Number of resampled subsets (85% of sample) in which each gene was associated with superior frontal thickness below the prescreening threshold (p<0.05), out of 1000 subsets. Dashed line represents number of expected associations due to chance with this threshold.
Simultaneous Model
The transcripts selected in the univariate approach could account for shared or unique variance in superior frontal cortical thickness. To assess this, C5 and SERPING1 were entered into the same linear mixed model predicting superior frontal cortical thickness in the entire sample (N=129). The model that included C5 and SERPING1 gene expression predicted superior frontal cortical thickness significantly better than a model that only included covariates, likelihood ratio χ2 (2)=13.344, p=0.001 (Table 2). Expression levels of C5 and SERPING1 accounted for an additional 7.4% of the variance in superior frontal cortical thickness when considering fixed covariates alone (conditional R2) and an additional 0.5% of the variance when considering both fixed and random (ie, family ID) covariates (marginal R2) (Nakagawa and Schielzeth, 2013). Both transcripts were significant in the model, suggesting that collinearity is not a major concern; this was corroborated by low VIF values of 1.02 (SERPING1) and 1.04 (C5). An increase in one standard deviation in C5 gene expression was associated with a decrease in 0.033 mm3 of superior frontal cortical thickness (95% CI: −0.058, −0.008). An increase in one standard deviation in SERPING1 gene expression was associated with a decrease in 0.032 mm3 of superior frontal cortical thickness (95% CI: −0.057, −0.007). Model fit was not significantly improved by the inclusion of C8G (runner-up in prescreening), smoking, antipsychotic use, zygosity, or estimated total intracranial volume (see Supplementary Information).
Table 2. Linear Mixed-Effects Model Predicting Superior Frontal Cortical Thickness (N=129).
| B | SE | df | T | p | |
|---|---|---|---|---|---|
| Sex (female) | 0.016 | 0.028 | 80 | 0.57 | 0.574 |
| Age | −0.009 | 0.001 | 41 | −6.31 | <0.001 |
| BMI | −0.006 | 0.003 | 41 | −2.27 | 0.029 |
| Case status (SZ) | 0.007 | 0.036 | 41 | 0.20 | 0.847 |
| Case status (co-twin) | 0.040 | 0.035 | 41 | 1.15 | 0.259 |
| C5 (z-scored) | −0.033 | 0.013 | 41 | −2.62 | 0.012 |
| SERPING1 (z-scored) | −0.032 | 0.013 | 41 | −2.50 | 0.016 |
| Intercept | 3.223 | 0.099 | 80 | 32.64 | <0.001 |
AIC=−133.713; R2m=0.390; R2c=0.576.
Spatial Distribution
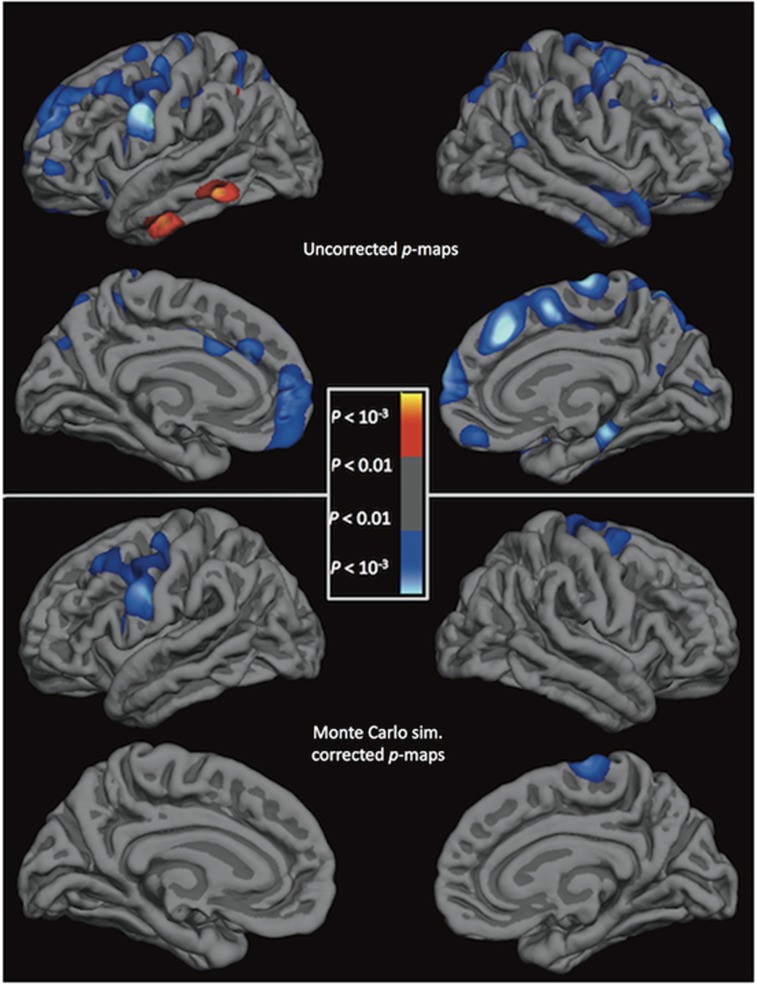
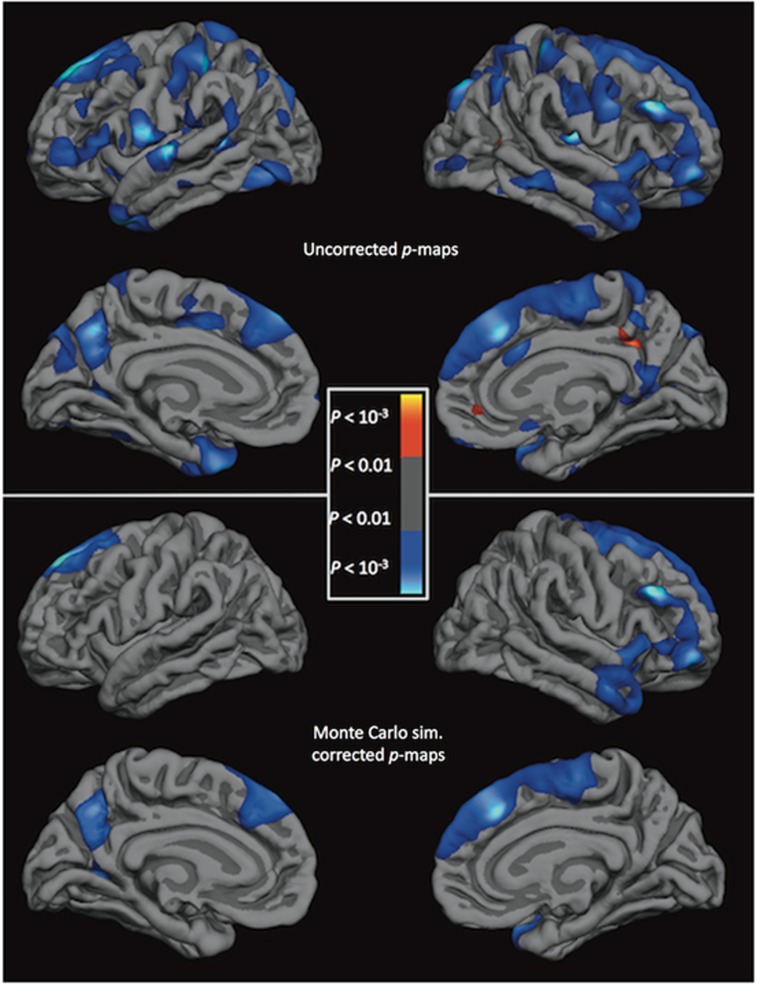
C5 and SERPING1 could be associated with cortical thickness throughout the brain or predominantly with the superior frontal cortex. To assess this, relationships between expression levels and cortical thickness were tested vertex-wise in the full sample. Significant negative correlations with SERPING1 gene expression levels and cortical thickness (Figure 2) survived the whole-brain analysis with cluster-based multiple correction in clusters covering the superior frontal cortex (cluster size=11.7 mm2, clusterwise p=0.017) and precuneus (7.1 mm2, p=0.040) in the left hemisphere and the superior frontal cortex (8.6 mm2, p<0.001), rostral middle frontal cortex (46.3 mm2, p=0.004), and insula (38.9 mm2, p=0.001) in the right hemisphere. Significant negative correlations with C5 gene expression levels and cortical thickness (Figure 3) survived correction in clusters covering the precentral gyrus in both the left (54.4 mm2, p<0.001) and right (22.9 mm2, p=0.014) hemispheres. We also display the uncorrected statistical significance maps of the relationship between cortical thickness and SERPING1 and C5 expression, with a nominal p<0.05 threshold. In addition to the regions previously identified, SERPING1 levels exhibited a negative correlation with thickness in the precentral and postcentral gyrii, and across the temporal gyrii in the right hemisphere (Figure 2). C5 levels were additionally negatively correlated with thickness in the middle frontal cortex in the left hemisphere and the superior frontal cortex in both hemispheres, and were positively correlated with the middle and inferior temporal gyrii in the left hemisphere (Figure 3).
Figure 2.
Statistical brain atlases plotting the association between SERPING1 gene expression levels and cortical thickness across the cortex. Cooler colors represent a negative association between gene expression levels and cortical thickness. Sim., simulation.
Figure 3.
Statistical brain atlases plotting the association between C5 gene expression levels and cortical thickness across the cortex. Cooler colors represent a negative association between gene expression levels and cortical thickness. Sim., simulation.
Post-mortem Human Cortex
Not all genes expressed in peripheral blood leukocytes are expressed in the human brain, nor is gene expression consistent across the human brain (Sullivan et al, 2006). We examined whether SERPING1 and C5 were expressed in post-mortem superior frontal cortical samples contained in the Allen Human Brain Atlas (Hawrylycz et al, 2012). Using microarray data, the mean percentage of samples with non-zero expression of complement genes in superior frontal cortex compared with background expression levels was 97% and 93% for SERPING1 and C5, respectively (Table 3). RNA-Seq data confirmed non-zero expression levels of SERPING1 and C5 in all available samples (see Supplementary Information).
Table 3. Number of Superior Frontal Cortical Samples from Each Donor with Microarray Complement Gene Expression Levels Above Baseline and Average Gene Expression across the Sample.
| Superior frontal samples (#) | SERPING1 (# samples expressed) | SERPING1 expression ratio | C5 (# samples expressed) | C5 expression ratio | |
|---|---|---|---|---|---|
| Donor 10021 | 26 | 26 | 1 | 25 | 0.96 |
| Donor 9861 | 38 | 30 | 0.79 | 29 | 0.76 |
| Donor 12876 | 12 | 12 | 1 | 10 | 0.83 |
| Donor 14380 | 15 | 15 | 1 | 15 | 1 |
| Donor 15496 | 19 | 19 | 1 | 19 | 1 |
| Donor 15697 | 19 | 19 | 1 | 19 | 1 |
| Average | 0.97 | 0.93 |
Heritability
Variance in gene expression and cortical thickness can be predominantly influenced by inherited genetic factors, common environmental factors, or both factors. Heritability models were used to assess the relative influence of these factors on variance in SERPING1 and C5 expression. The best fitting model for SERPING1 included a genetic component of 46%, indicating moderate heritability of variance in expression levels (Table 4). For C5, the best fitting model included a shared environment component of 64%, indicating moderate influence of the shared environment. The best fitting model for superior frontal thickness included an additive genetic (A) component of 65%, indicating high heritability. Heritability models for C5 and SERPING1 did not exhibit good fit to the data, but were best described by the selected models (see Supplementary Information).
Table 4. Heritability Estimates for Complement Gene Expression.
| Phenotype | Illumina probe | Model | Heritability |
|---|---|---|---|
| C5 | ILMN_1746819 | CE | 0.6355 (C) |
| SERPING1 | ILMN_1670305 | AE | 0.4669 (A) |
| Superior frontal cortical thickness | − | AE | 0.6547 (A) |
Note: C=shared environment; A=additive genetics (primary influence on gene expression levels identified by heritability analyses).
Case Status
There was not a significant main effect of case status (Table 2) or an interaction of case status with C5 or SERPING1 (data not shown) predicting superior frontal cortical thickness in the simultaneous model.
Discussion
We have provided preliminary evidence that complement gene expression may be associated with superior frontal cortical thickness in humans. Expression level of two nominated complement genes (C5, SERPING1) contributed uniquely to prediction of superior frontal cortical thickness across diagnostic groups. Vertex-wise analyses suggested that SERPING1 expression was most strongly associated with this region of the cortex. Evidence for moderate heritability of variance in SERPING1 expression levels and confirmation that these genes are expressed in the human superior frontal cortex further support the plausibility of a mechanistic link between genetic variants influencing the complement system and cortical thickness in humans.
The transcripts associated with superior frontal cortical thickness in our sample included both an inhibitor (SERPING1) and an activator (C5) of the complement system. SERPING1 encodes the plasma protease C1 inhibitor (C1-INH), a potent anti-inflammatory protein that inhibits activation of the complement system by binding to pro-inflammatory complement proteins (eg C1q, C3b) (Davis et al, 2010). Intriguingly, C1q and C3b have been implicated in synaptic pruning (Orsini et al, 2014). The observed negative association between SERPING1 mRNA levels and cortical thickness is counterintuitive, and could reflect an unmeasured compensatory process. In contrast, C5 encodes the C5 protein, which is cleaved into two active components: C5a, a potent inflammatory protein that recruits immune cells (Woodruff et al, 2010) and C5b, which initiates the formation of the membrane attack complex leading to cell rupture and death (Orsini et al, 2014).
The observed associations between C5 and SERPING1 expression and superior frontal cortical thickness cannot address causality, but represent a first step towards assessing whether complement-mediating synaptic pruning may occur in the human prefrontal cortex. To further test this theory, the assumptions underlying these preliminary analyses need to be assessed and expanded upon. First, the correlation between expression levels of SERPING1 and C5 and superior frontal thickness could reflect a chance fluctuation in expression levels, a third variable effect, or the trace of earlier development. Gene expression levels in PBMCs tend to be relatively consistent within individuals across a period of months (Radich et al, 2004). However, longitudinal assessments of complement gene expression are needed. Complement expression and cortical thickness could both be influenced by an unmeasured third variable, which future studies could assess (see Supplementary Information). Alternatively, complement expression in middle age may reflect individual differences in complement activity earlier in development. If complement proteins interact differently with specific cell types during neurodevelopment, they could differentially influence thickness in distinct cortical regions. In adulthood, associations between expression levels and thickness in those regions could still be observed cross-sectionally. From this perspective, it is intriguing that our SERPING1 association maps correspond closely with cortical maps of increased rates of thinning in adolescents who contract schizophrenia (Cannon et al, 2015). This association could be further elucidated with longitudinal studies of complement expression and cortical thickness in adolescents and additional investigation of these complement proteins in neurodevelopment.
A second assumption is that complement expression levels in PBMCs reflect complement expression levels in microglia. Microglia are particularly similar in function to peripheral monocytes, and gene expression patterns between the two may be relatively consistent (Saijo and Glass, 2011) (see Supplementary Information). However, peripheral immune activation can affect brain function via other pathways, including stimulation of the vagus nerve by peripheral immune proteins, transport of immune proteins across the blood brain barrier, and peripheral lymphocytes entering the brain (O’Connor et al, 2014). Additional work is needed to investigate these pathways.
Third, this theory suggests that the complement-mediated synaptic pruning contributes to reductions in cortical gray matter thickness in adolescence. However, the extent to which adolescence is a period of synaptic pruning, and if this contributes to observed cortical thinning (Gogtay et al, 2004), is debated (see Supplementary Information). Other neurodevelopmental changes that could contribute to cortical gray matter loss include myelination and gains in white matter volume (Catts et al, 2013). Additionally, the majority of research on the role of complement in synaptic pruning has been focused on the developing visual cortex (Chu et al, 2010; Schafer et al, 2012; Sekar et al, 2016). Further work is needed to confirm that the complement system is involved in synaptic pruning in the human prefrontal cortex during adolescence.
Fourth, this theory suggests that dysregulation of the complement system could result in an accelerated rate of cortical synaptic pruning, causing vulnerable individuals to cross below a critical threshold of synaptic density needed to support cognitive functioning and leading to psychosis (Cannon, 2015). The lack of influence of case status suggests that the association between complement gene expression and cortical thickness could reflect normative brain development, rather than a disease or medication effect. However, case status differences may be more apparent during adolescence, in which pruning is likely actively taking place. Longitudinal studies in adolescents at different levels of risk for schizophrenia may clarify whether complement system activity is associated with greater changes in superior frontal thickness in at-risk individuals.
To increase the likelihood of identifying reproducible signal in our relatively small sample, we used a resampling procedure to identify individual complement genes associated with superior frontal cortical thickness in many random sample replicates. As a second check for Type I error, we conducted a parallel approach in which we prescreened genes using 1000 bootstrapped samples. These analyses yielded the same conclusion (see Supplementary Information). The spatial analyses may also be considered a reliability check (see Supplementary Information). Though not a focus of our hypotheses, C5 expression levels correlated with thickness of the precentral gyrus and survived correction in the spatial analysis, suggesting that this is likely a robust association. Replication in independent samples is needed to confirm and further assess these associations.
These analyses are vulnerable to the inherent limitations of data acquisition techniques and data availability. For example, we were not able to account for some factors that may influence gene expression and microarray analyses (eg, time of blood draw, inflammatory and fasting status, sample cell type composition) (Whitney et al, 2003). We could not follow-up on identified complement targets in PBMCs or confirm expression with quantitative PCR in the Swedish sample. We did not have information on time of blood draw, isolation and storage of samples, sample quality, or cortical thickness for the Allan Brain Atlas post-mortem samples. The slightly older imaging equipment (ie 1.5T scanner, 1 mm3 resolution) used may have very slightly influenced measures of cortical thickness (Gronenschild et al, 2012; Han et al, 2006).
This study identified initial evidence suggesting that expression levels of SERPING1, a complement system gene, are associated with superior frontal cortical thickness in humans. These findings are a first step towards establishing the plausibility of a pathway linking the complement system with cortical thickness in humans. Further research is needed to confirm and expand upon the observed associations and assess whether these pathways may play a role in the etiology of schizophrenia. If so, this could provide novel targets for the prevention of schizophrenia.
Funding and disclosure
TDC is a consultant to the Los Angeles County Department of Mental Health and Boehringer Ingelheim Pharmaceuticals. DMA and KA’s work was funded by NSF Graduate Research Fellowships (DGE-1122492). The remaining authors declare no potential conflict of interest.
Acknowledgments
We thank the study participants, research staff members, and the Swedish Board of Health and Welfare. We also thank Avram Holmes, David Glahn, and Sebastian Therman for their comments on drafts of the manuscript. We thank the National Institute of Mental Health (RO1 MH052857, Principal Investigator: TDC) and the Stockholm County Council (ALF 20100305) for financial support.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Alfons A (2016). Robust methods for high-dimensional data. CRAN Repos at https://cran.r-project.org/web/packages/robustHD/robustHD.pdf.
- Barton K (2016). Package ‘MuMIn.’ CRAN Repos at https://cran.r-project.org/web/packages/MuMIn/MuMIn.pdf.
- Bennett MR (2011). Schizophrenia: susceptibility genes, dendritic-spine pathology and gray matter loss. Prog Neurobiol 95: 275–300. [DOI] [PubMed] [Google Scholar]
- Bourgeois J, Goldman-Rakic PS (1994). Synaptogenesis in the prefrontal cortex of rhesus monkeys. [DOI] [PubMed]
- Cannon TD (2015). How schizophrenia develops: cognitive and brain mechanisms underlying onset of psychosis. Trends Cogn Sci 19: 744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Chung Y, He G, Sun D, Jacobson A, van Erp TGM et al (2015). Progressive reduction in cortical thickness as psychosis develops: a multisite longitudinal neuroimaging study of youth at elevated clinical risk. Biol Psychiatry 77: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catts VS, Fung SJ, Long LE, Joshi D, Vercammen A, Allen KM et al (2013). Rethinking schizophrenia in the context of normal neurodevelopment. Front Cell Neurosci 7: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Jin X, Parada I, Pesic A, Stevens B, Barres B et al (2010). Enhanced synaptic connectivity and epilepsy in C1q knockoutmice. Proc Natl Acad Sci USA 107: 7975–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Jacobson A, He G, van Erp TG, McEwen S, Addington J et al (2015). Prodromal symptom severity predicts accelerated gray matter reduction and third ventricle expansion among clinically high risk youth developing psychotic disorders. Mol Neuropsychiatry 1: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AE, Lu F, Mejia P (2010). C1 inhibitor, a multi-functional serine protease inhibitor. Thromb Haemost 104: 886–893. [DOI] [PubMed] [Google Scholar]
- Dobbins IG, Wagner AD (2005). Domain-general and domain-sensitive prefrontal mechanisms for recollecting events and detecting novelty. Cereb Cortex 15: 1768–1778. [DOI] [PubMed] [Google Scholar]
- Glausier JR, Lewis DA (2013). Dendritic spine pathology in schizophrenia. Neuroscience 251: 90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC et al (2004). Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci USA 101: 8174–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman AL, Pezawas L, Doz P, Mattay VS, Fischl B, Verchinski BA et al (2013). Widespread reductions of cortical thickness in schizophrenia and spectrum disorders and evidence of heritability. Arch Gen Psychiatry 66: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenschild EHBM, Habets P, Jacobs HIL, Mengelers R, Rozendaal N, van Os J et al (2012). The effects of FreeSurfer version, workstation type, and Macintosh operating system version on anatomical volume and cortical thickness measurements. PLoS ONE 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler DJ, Saygin AP, Sereno MI (2006). Smoothing and cluster thresholding for cortical surface-based group analysis of fMRI data. Neuroimage 33: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S et al (2006). Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage 32: 180–194. [DOI] [PubMed] [Google Scholar]
- Hawrylycz M, Lein E, Guillozet-Bongaarts A, Shen E, Miller J, Al E (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka S, Nichols TE (2003). Validating cluster size inference: random field and permutation methods. Neuroimage 20: 2343–2356. [DOI] [PubMed] [Google Scholar]
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK et al (2004). Gene map of the extended human MHC. Nat Rev Genet 5: 889–899. [DOI] [PubMed] [Google Scholar]
- Mayilyan KR, Weinberger DR, Sim RB (2008). The complement system in schizophrenia. Drug News Perspect 21: 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Menon V, Goldy J, Kaykas A, Lee CK, Smith KA et al (2014). Improving reliability and absolute quantification of human brain microarray data by filtering and scaling probes using RNA-Seq. BMC Genomics 15: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Schielzeth H (2013). A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol 4: 133–142. [Google Scholar]
- O’Connor TG, Moynihan JA, Caserta MT (2014). Annual research review: the neuroinflammation hypothesis for stress and psychopathology in children—developmental psychoneuroimmunology. J Child Psychol Psychiatry Allied Discip 55: 615–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoniewski MJ, Miller CJ (2006). Hybridization interactions between probesets in short oligo microarrays lead to spurious correlations. BMC Bioinformatics 7: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini F, De Blasio D, Zangari R, Zanier ER, De Simoni M-G (2014). Versatility of the complement system in neuroinflammation, neurodegeneration and brain homeostasis. Front Cell Neurosci 8: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T (2005). Mapping brain maturation and cognitive development during adolescence. Trends Cogn Sci 9: 60–68. [DOI] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DMMixed-Effects Models in S and S-PLUS. Springer: New York; (2000). [Google Scholar]
- Plomin R, DeFries J, Knopik V, Neiderhiser JBehavioral Genetics, Vol. 6. Worth Publishers: New York, 2013.
- Radich JP, Mao M, Stepaniants S, Biery M, Castle J, Ward T et al (2004). Individual-specific variation of gene expression in peripheral blood leukocytes. Genomics 83: 980–988. [DOI] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA et al (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K, Glass CK (2011). Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol 11: 775–787. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R et al (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74: 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N et al (2016). Schizophrenia risk from complex variation of complement component 4. Nature 530: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM (2006). Evaluating the comparability of gene expression in blood and brain. Am J Med Genet—Neuropsychiatr Genet 141 B: 261–268. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC (2003). Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 60: 1187–1192. [DOI] [PubMed] [Google Scholar]
- Tamnes CK, Østby Y, Fjell AM, Westlye LT, Due-Tønnessen P, Walhovd KB (2010). Brain maturation in adolescence and young adulthood: regional age-related changes in cortical thickness and white matter volume and microstructure. Cereb Cortex 20: 534–548. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nature Rev Genet10: 57–63.. [DOI] [PMC free article] [PubMed]
- Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA et al (2003). Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA 100: 1896–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff TM, Ager RR, Tenner AJ, Noakes PG, Taylor SM (2010). The role of the complement system and the activation fragment C5a in the central nervous system. NeuroMolecular Med 12: 179–192. [DOI] [PubMed] [Google Scholar]
- Zheutlin AB, Viehman RW, Fortgang R, Borg J, Smith DJ, Suvisaari J et al (2016). Cognitive endophenotypes inform genome-wide expression profiling in schizophrenia. Neuropsychology 30: 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.