Abstract
The transcription factor PU.1 is often impaired in patients with acute myeloid leukemia (AML). Here, we used AML cells that already had low PU.1 levels and further inhibited PU.1 using either RNA interference or, to our knowledge, first-in-class small-molecule inhibitors of PU.1 that we developed specifically to allosterically interfere with PU.1-chromatin binding through interaction with the DNA minor groove that flanks PU.1-binding motifs. These small molecules of the heterocyclic diamidine family disrupted the interaction of PU.1 with target gene promoters and led to downregulation of canonical PU.1 transcriptional targets. shRNA or small-molecule inhibition of PU.1 in AML cells from either PU.1lo mutant mice or human patients with AML-inhibited cell growth and clonogenicity and induced apoptosis. In murine and human AML (xeno)transplantation models, treatment with our PU.1 inhibitors decreased tumor burden and resulted in increased survival. Thus, our study provides proof of concept that PU.1 inhibition has potential as a therapeutic strategy for the treatment of AML and for the development of small-molecule inhibitors of PU.1.
Keywords: Hematology, Therapeutics
Keywords: Drug therapy, Leukemias, Transcription
Introduction
Acute myeloid leukemia (AML) is a cancer of the hematopoietic system characterized by the abnormal clonal proliferation of immature cells following various genetic and epigenetic alterations. Despite efforts to discover new therapeutic options, survival of patients with AML remains poor, with a 5-year overall survival rate of 25% (1) and the worst outcomes for individuals over 60 years of age, who represent the vast majority of AML patients. In this age group in particular, clinical outcomes have not significantly improved in the past 4 decades. AML is a very heterogeneous genetic disease characterized by recurrent genetic mutations that often occur in combination in individual patients (approximately 30 mutations recur in patients at a frequency of >1%), and, on average, patients with AML carry a combination of 3 to 5 driver mutations (2). One of the major challenges facing currently ongoing “precision oncology” efforts is the low frequency of a larger number of individual mutations and their combinatorial occurrence. Instead of targeting specific genetic aberrations, an alternate strategy for AML treatment would be to target more commonly dysregulated pathways that are implicated in various AML subtypes and in larger subsets of patients.
In the past 15 years, increasing evidence has shown the critical importance of PU.1, a lineage-restricted transcription factor of the hematopoietic system in AML. Functionally critical decreases in PU.1 levels have been described in FLT3–internal tandem duplication (FLT3-ITD) (3), RUNX1-ETO (4), and promyelocytic leukemia (5), representing 24%, 7%, and 13% of all AMLs, respectively (cancer.sanger.ac.uk) (6, 7). Additionally, PU.1 loss-of-function heterozygous mutations or deletions have been described in AML and are found in approximately 10% of MLL-translocated AMLs (8–10). Homozygosity of a single nucleotide variant in an upstream regulatory element (URE) of PU.1, lowering PU.1 expression, has been described in AML with complex karyotype (11), and a study on highly purified stem cells from patients with AML showed reduced PU.1 levels in at least 40% of examined cases (12).
PU.1 is highly conserved between humans and mice, and its functions have been studied using a number of genetically engineered mouse models. PU.1 is important for myeloid and lymphoid differentiation (13, 14), as well as hematopoietic stem cell (HSC) maintenance (15, 16). Its role in AML development has been firmly established through mouse models with reduced, but not completely absent, PU.1 expression. Homozygous knockout of an enhancer (URE) located –14 kb upstream of PU.1 leads to an 80% decrease in PU.1 expression and development of stem cell–derived AML between 3 and 8 months of age (12, 17). Enhancer haplodeficiency of PU.1 is not sufficient to induce leukemia by itself; however, it leads to myeloid bias in (preleukemic) stem cells and MDS and AML development in combination with cooperating events (18).
Overall, disruption of PU.1 expression or activity is present in more than 50% of patients with AML and is associated with a specific transcriptional and epigenetic program (19, 20). Thus, targeting PU.1 in AML could be an appealing option for treatment. In the past, strategies to rescue PU.1 expression in AML cells have been explored. Overexpression of PU.1 is sufficient to trigger neutrophil differentiation in acute promyelocytic leukemia (APL) and leads to differentiation and apoptosis of various primary AML samples (5, 21). However, elevation of PU.1 levels or activity is difficult to achieve pharmacologically.
In this study, we used the inverse strategy. As complete loss of PU.1 leads to stem cell failure (15), we hypothesized that AML cells may be more vulnerable to further PU.1 inhibition in comparison with normal hematopoietic cells. We used 2 alternative approaches to test this hypothesis: RNA interference and newly developed PU.1 inhibitors.
We have recently reported proof of principle for the ability to inhibit PU.1 by novel heterocyclic diamidines, which are derivatives of clinically tested compounds such as furamidine (22, 23). DNA recognition by PU.1 requires specific binding in the DNA major groove at consensus sites harboring a 5′-GGAA/T-3′ motif that typifies target sites for the ETS family. Selectivity for PU.1 is conferred through additional contacts with the minor groove of adjacent AT-rich tracks (24). We initiated a development and screening effort to find optimized compounds that would recognize a larger number of base pairs adjacent to a core ETS site as more specific PU.1 inhibitors. The PU.1 inhibitors we identified target the minor groove and lead to inhibition of PU.1 binding in the major groove via an allosteric mechanism.
Using RNA interference as well as our small-molecule inhibitors, we show that PU.1 inhibition is effective at inhibiting AML cell growth, including in murine and human cell lines and in primary AML patients’ cells in vitro and in vivo, and thus represents what we believe to be a fundamentally new strategy for the treatment of AML.
Results
PU.1 knockdown decreases cell growth and clonogenic capacity and increases apoptosis of murine and human AML cells.
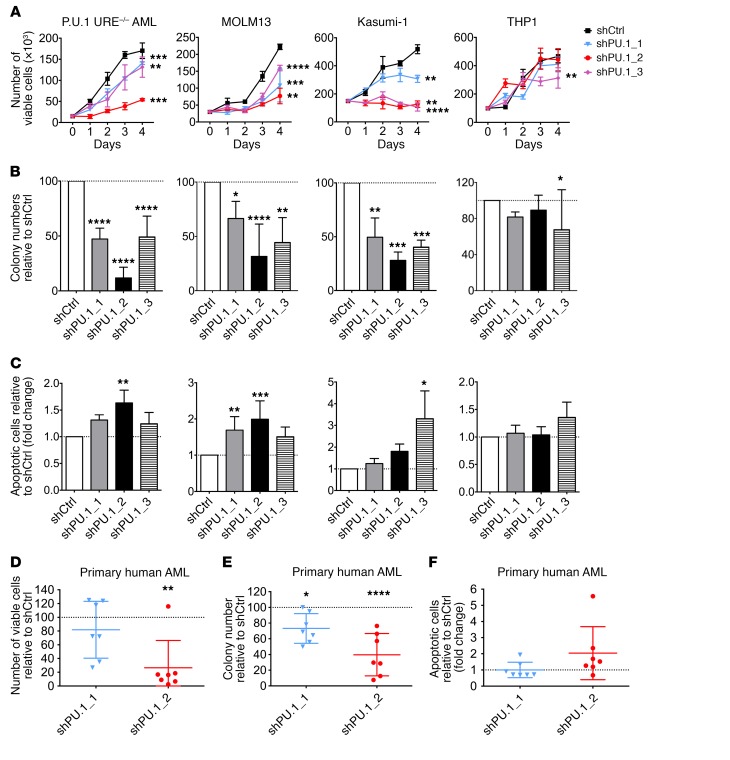
To determine whether PU.1 inhibition may be a suitable strategy in AML, we used an established model of AML driven by reduced PU.1 levels, PU.1 URE–/– AML, in which PU.1 expression is reduced to approximately 20% of normal levels by disruption of an upstream enhancer (URE) (12, 17). The PU.1 URE–/– AML cell line has been established from a leukemic mouse with homozygous deletion of the URE of the PU.1 gene, which has been previously described (17). We selected 3 shRNAs that decreased PU.1 expression in mouse and human cells (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI92504DS1). Knockdown of PU.1 in PU.1 URE–/– AML cells by the 3 different shRNAs led to significantly decreased cell growth and colony formation (Figure 1, A and B). Likewise, the percentage of apoptotic cells was substantially increased upon shRNA-mediated PU.1 knockdown in PU.1 URE–/– AML cells (Figure 1C). The degree of inhibition of growth and clonogenicity, as well as apoptosis induction, were greater with the shRNA PU.1_2, leading to more efficient PU.1 knockdown (Supplemental Figure 1A). Knockdown of PU.1 in an immature murine hematopoietic cell line with normal levels of PU.1 (BaF3) did not have significant effects on proliferation or apoptosis (Supplemental Figure 1, C–F).
Figure 1. PU.1 knockdown decreases cell growth and clonogenicity and increases apoptosis of murine and human AML cells.
(A) Cell proliferation assay of PU.1 URE–/– AML (n = 4), MOLM13 (n = 3), Kasumi-1 (n = 3), and THP1 (n = 3) cells after transduction with shPU.1_1, shPU.1_2, or shPU.1_3. Results from 1 representative experiment are shown. (B) Clonogenic capacities of PU.1 URE–/– AML (n = 4), MOLM13 (n = 6), Kasumi-1 (n = 3), and THP1 (n = 4) cells after transduction with shPU.1_1, shPU.1_2, or shPU.1_3. Fold change compared with shCtrl is shown. (C) Apoptosis induction in PU.1 URE–/– AML (n = 3), MOLM13 (n = 7), Kasumi-1 (n = 3), and THP1 (n = 3) cells after transduction with shPU.1_1, shPU.1_2, or shPU.1_3. Fold change of annexin-V+DAPI– cells compared with shCtrl is shown. (D–F) Primary human AML cells were transduced with PU.1 shRNAs, sorted (GFP+), and plated in semisolid media; colony numbers and viable and apoptotic cell numbers were assessed after 14 days of culture. Error bars indicate the mean ± SD, and each AML sample is represented by an individual dot. The percentage (D and E) and fold change (F) compared with vehicle (dotted line) are shown. (D and E) Number of viable cells and clonogenic capacities and (F) apoptosis induction (annexin-V+DAPI–) after transduction with shPU.1_1 and shPU.1_2 (n = 7 each). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 1-way ANOVA.
We next investigated the effect of PU.1 knockdown on human leukemic cell lines with different PU.1 levels (Supplemental Figure 1G). MOLM13 and Kasumi-1 cell lines harbor anomalies associated with low PU.1 levels [FLT3-ITD mutation for MOLM13 and t(8;21) for Kasumi-1] (3, 4), while THP1 cells have higher PU.1 levels. Decreased PU.1 led to a strong inhibitory effect on the growth and clonogenic capacity of MOLM13 and Kasumi-1 cells, whereas it had no or very modest effects on THP1 cell growth (Figure 1, A and B). Accordingly, the apoptotic fraction was increased following PU.1 knockdown in MOLM13 and Kasumi-1 cells, but we observed no major effect in THP1 cells (Figure 1C). To confirm that this phenotype was correlated with PU.1 levels, we overexpressed PU.1 in MOLM13 cells upon PU.1 knockdown with shPU.1_2 and found a rescue of the phenotype, with increased clonogenic capacity and decreased apoptosis (Supplemental Figure 1, H–J).
To determine whether PU.1 inhibition has an effect on primary cells from AML patients, we seeded mononuclear cells (MNCs) from AML patients in semisolid media for 2 weeks and assessed the number of colonies, the number of viable cells, and the proportion of apoptotic cells. Knockdown of PU.1 significantly decreased the number of viable cells (mean decrease of 18% for shPU.1_1 and 74% for shPU.1_2) (Figure 1D), as well as the colony-forming capacity of primary human AML cells (mean decrease of 27% for shPU.1_1 and 60% for shPU.1_2, compared with shCtrl) (Figure 1E). At the same time, the proportion of apoptotic cells increased on average by 2-fold upon knockdown of PU.1 in primary human AML cells with the more efficient shRNA (shPU.1_2) (Figure 1F).
Taken together, these data show that inhibition of PU.1 decreases cell growth and clonogenic capacity and increases apoptosis, in murine as well as human AML, and thus provide proof of concept for PU.1 inhibition as a possible therapeutic strategy in AML.
Development of small-molecule PU.1 inhibitors abrogating DNA binding by PU.1.
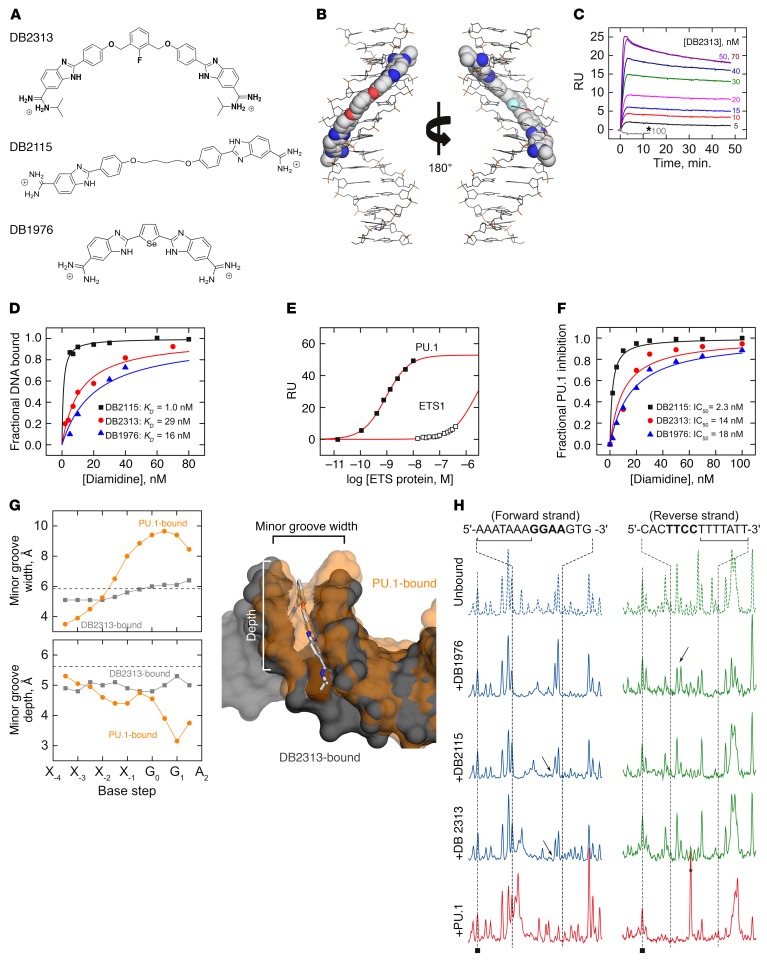
Classical heterocyclic diamidines, typified by furamidine (DB75), are highly selective and potent ligands for AT-rich DNA, a feature responsible for their success as inhibitors of kinetoplast DNA in trypanosomiasis (25, 26). As with other ETS family transcription factors, DNA site recognition by PU.1 involves contact with the major groove at the 5′-GGAA-3′ consensus, as well as an indirect readout of the minor groove backbone, where sequence-dependent geometry confers selectivity among ETS paralogs. PU.1 is distinguishable from other paralogs by the former’s strong preference for AT-rich sequences upstream of the 5′-GGAA-3′ consensus (27). We screened a number of designed AT-targeting heterocyclic diamidines and found that they efficiently competed with PU.1 for DNA binding. DNA footprinting experiments verified that the compounds targeted the flanking sequences found in cognate PU.1 sites, using the λB motif of the murine Igλ2-4 enhancer (5′-ATAAAAGGAAGTG-3′) (28) as a model. On the basis of the general structural features of our initial PU.1 inhibitors (23), we designed and prepared a focused library of larger derivatives for screening using the solution and cellular analytical methods (vide infra). DB2115 and DB2313 were selected as the 2 best compounds (Figure 2A). Both compounds harbor the amidine-benzimidazole-phenyl platform that provides excellent recognition for expanded AT sites. The concept for the units of these compounds is that the amidine cation imparts solubility, interacts favorably with phosphate groups that are close in the narrow DNA minor groove, and can hydrogen bond with acceptor groups on AT base pairs (carbonyl of T, T=O, and N3 of A) at the floor of the minor groove. The benzimidazole-NH points into the minor groove and can also H-bond with AT base pair acceptors. In addition, all 3 groups (amidine, benzimidazole, and phenyl) make excellent van der Waals contacts with the walls of the minor groove and edges of AT base pairs at the floor of the groove. The compounds also have the appropriate curvature to match the DNA minor groove in AT sequences. Together, they make an excellent AT recognition module. Linking 2 of these modules, as in DB2115 and DB2313, provides a molecule for expanded and very strong AT site recognition. A docked structure shows that DB2313, for example, inserted itself precisely and deeply into the DNA minor groove along the AT-track of the λB motif, interacting with the DNA base pair edges at the minor groove floor over almost 10 consecutive base pairs (Figure 2B). Thus DB2313 and DB2115, along with DB1976, which is the best compound from our initial studies of PU.1 inhibition (23), were evaluated in more detail.
Figure 2. Expanded heterocyclic diamidines target the DNA minor groove and inhibit PU.1 binding by an allosteric mechanism.
(A) Chemical structures of the heterocyclic diamidines. (B) Model of DB2313 docked to the track (5′-AAATAAAA-3′) upstream of the 5′-GGAA-3′ ETS core consensus in the λB motif. (C) Representative SPR sensorgrams for the interaction of DB2313 with the 5′ AT-rich binding site of the λB promoter DNA sequence. Note the lack of binding by DB2313 to an alternative site specific to the ETS homolog ETS1 (5′-GCCGGAAGTG-3′), even at high concentrations (100 nM, asterisk). (D) Comparison of the binding affinities for the λB promoter DNA sequence with different compounds. RU values from the SPR sensorgrams, as in B, were converted to r (r = RU/RUmax, moles of compound bound/mole of promoter DNA) and are plotted against the unbound compound concentration. (E) Specificity of the λB motif for PU.1. Under identical solution conditions, ETS1 bound negligibly at concentrations that saturated the target in the case of PU.1. (F) Normalized PU.1 inhibition resulted from biosensor SPR experiments. The plots represent the amount of PU.1-DNA complex inhibition as a function of the added compound concentration. (G) Perturbations of DNA minor groove width and depth by bound DB2313 or PU.1. The base steps marked “Xi” denote the bases 5′ to the ETS consensus (G0G1AA). Dashed lines indicate the expected values of B-form DNA. Aligned structures of the DB2313-bound (gray) and PU.1-bound (orange) DNA, rendered as van der Waals surfaces, show the mutually incompatible minor groove conformations induced by the diamidine and protein. (H) DNase I footprints of compound binding to the λB motif. The subsite at which the compounds bind is marked by a bracket. Arrows indicate distinct perturbations to the drug-induced DNA structure among the compounds as detected by DNase I. As a reference, the PU.1-bound footprint is also shown (red); note the DNase I–hypersensitive band (asterisk) in the reverse strand that is diagnostic of site-specific ETS-DNA complexes.
We measured the affinity of these heterocyclic diamidines for the λB motif, a typical native DNA-binding site for PU.1, following our biosensor surface plasmon resonance (SPR) procedure (29, 30). Briefly, duplex DNA harboring the λB motif was immobilized on a streptavidin-functionalized sensor chip via a 5′-end–labeled biotin. The affinity of each compound for the λB motif was determined from the concentration-dependent increase in the SPR sensorgrams at steady state, as shown in Figure 2C for DB2313. The specificity of the compounds was probed by substituting the λB motif with a model cognate site (5′-GCCGGAAGTG-3′, termed SC1) that is recognized by ETS1. As shown by DB2313, a very large excess of compound, at concentrations far in excess of those used to saturate the λB motif, failed to bind the SC1 site to detectable levels (gray sensorgram in Figure 2C). With the λB motif as a target, all 3 compounds bound their target DNA tightly, with 1:1 stoichiometry and dissociation constants in the 10–8 to 10–9 M range (Figure 2D).
To define the potential of the diamidines to inhibit cognate PU.1 binding, we loaded the immobilized λB site with the purified, recombinant PU.1 ETS domain (Figure 2E). In the absence of diamidine, PU.1 formed a 1:1 complex with the immobilized λB motif with a dissociation constant of 5.4 nM, in agreement with solution fluorescence anisotropy measurements (31). Control titrations with the ETS domain of ETS1, which we have shown to be structurally superimposable with PU.1, showed a 3-log selectivity of the λB site for PU.1 and attested to the specificity of our SPR methodology. Immobilized λB DNA was loaded to greater than 95% capacity and then titrated with graded concentrations of each compound (32). Displacement of PU.1 was detected by the drop in SPR signal as a function of the compound concentration. All 3 compounds efficiently inhibited PU.1 binding, with IC50 values in the 10–8 to 10–9 M range (Figure 2F). The rank order of the IC50 values (DB2115 < DB2313 < DB1976) corresponded with the rank order of affinities, suggesting that the 3 compounds inhibited PU.1 binding to the λB motif via the same mechanism.
To gain structural insight into the mechanism by which the diamidines inhibit PU.1 binding to the λB site, we performed a molecular dynamics simulation of the DB2313-λB complex. The λB-bound complex showed the expected binding mode, including the typical water-mediated contacts between the 2 diamidiniumsand the DNA backbone (Supplemental Figure 2) (33). Comparison of the equilibrated DB2313-DNA complex with the PU.1-DNA co-crystal structure (34) showed that the bound compound induced the double helix into a conformation that was incompatible with PU.1 binding. The co-crystal structure of the PU.1-DNA complex shows that extensive perturbations of the DNA groove structure occur in PU.1 binding. Specifically, compression of the minor groove in both width and depth at the AT-rich flanking sequences is required for indirect readout of the DNA backbone by PU.1. Crucially, insertion of DB2313 gives rise to a minor groove profile, in terms of groove width and depth, that deviates strongly from the requirement of PU.1 for binding (Figure 2G). The incompatible effects of DB2313 and PU.1 on DNA structure therefore suggested that denial of PU.1 occupancy through conformational perturbations of the target DNA represents a major component of the efficacy of our compounds as PU.1 inhibitors. We confirmed experimentally that the diamidines induced incompatible DNA conformations for PU.1 binding by probing the drug-bound DNA with DNase I, a minor groove probe that is highly sensitive to local DNA structure (Figure 2H). Although each diamidine induced a unique DNase I footprint, as would be expected from their different structures, all were highly dissimilar to the characteristic footprint produced by PU.1. In summary, computational and experimental evidence revealed mutually exclusive effects on DNA conformation as a fundamental mechanism of PU.1 inhibition by the identified diamidines.
Functional inhibition of PU.1 by heterocyclic diamidines.
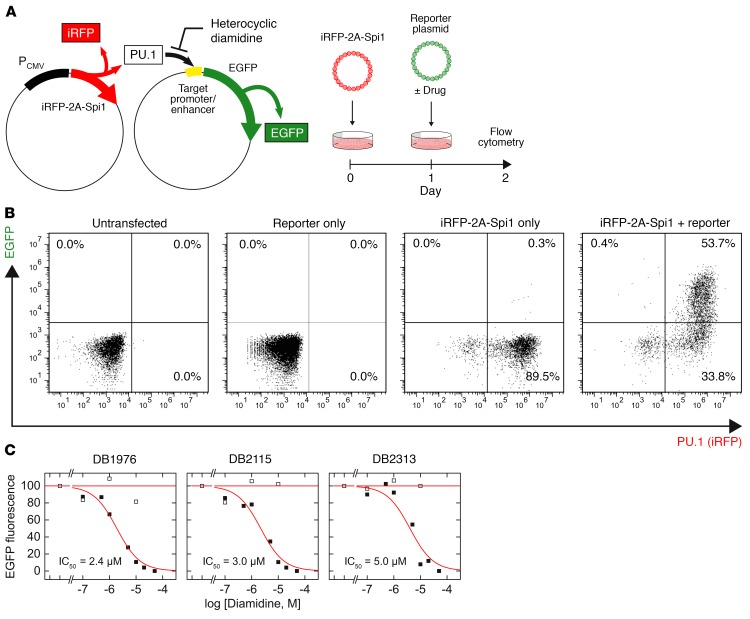
Having established the biophysical basis of PU.1 inhibition by DB2313, DB2115, and DB1976, we proceeded to define their cellular properties and potential to inhibit PU.1-dependent gene transactivation. To assess the potential for functional inhibition of PU.1 transactivation by the 3 diamidines, we tested their effects on the expression of a cellular EGFP reporter under the control of a minimal PU.1-dependent promoter harboring a triple tandem copy of the same λB enhancer site used in the SPR screen (Figure 3A). This cell-based system was designed such that ectopic PU.1 and reporter expression can be independently tracked by fluorescence to filter out any background activation of the reporter. In PU.1-negative HEK293 cells, which express several endogenous ETS paralogs including ETS2, GABPA, and members of the ELF subfamily (Human Protein Atlas), transfection of the λB-based reporter was silent in flow cytometry, indicating specificity of the λB enhancer to PU.1 (Figure 3B). All 3 compounds inhibited PU.1-dependent transactivation of the reporter in a dose-dependent manner, with similar IC50 values between 2 and 5 μM (Figure 3C). The relatively high IC50 values should be considered in light of the strong CMV promoter used to drive the expression of ectopic PU.1. To establish the specificity of diamidine action to the inhibition of PU.1, we tested whether the diamidines could inhibit transactivation at an analogous minimal promoter in which the λB enhancer sites were replaced by the SC1 sequence. Although PU.1 can bind the SC1 site, it is not found in native PU.1 target genes (35) and is therefore functionally nonspecific. None of the diamidines significantly inhibited transactivation at the SC1-based enhancer (Figure 3C), consistent with the lack of PU.1 binding to the SC1 site we observed by SPR (Figure 2C). Thus, heterocyclic diamidines appeared to maintain their strong AT selectivity in live cells and specifically inhibited PU.1 activity at functional DNA sites associated with PU.1 target genes.
Figure 3. Cellular properties and functional inhibition of PU.1 by expanded diamidines.
(A) Ectopic PU.1 activity, as indicated by an iRFP marker, in live HEK293 cells was measured through expression of a fluorescent EGFP reporter, transfected with or without compound, under the control of a minimal PU.1-dependent, λB-based promoter. (B) Flow cytometric analysis of the PU.1 marker (iRFP) and EGFP reporter, in which the upper right quadrant represents PU.1-dependent reporter activation. Note the lack of EGFP reporter activation by endogenous non-PU.1 ETS paralogs, indicating specificity of the λB reporter. (C) Inhibition of cellular EGFP fluorescence by the compounds. Black symbols represent the λB-based reporter, which was sensitive to diamidine inhibition in a concentration-dependent manner, and white symbols represent the mutated SC1-based reporter, which was insensitive. Curves are fits of the data to the Hill equation. The IC50 values for all compounds ranged between 2 μM and 5 μM.
Small-molecule PU.1 inhibitors decrease cell growth and the clonogenic capacity of AML cells and lead to increased apoptosis.
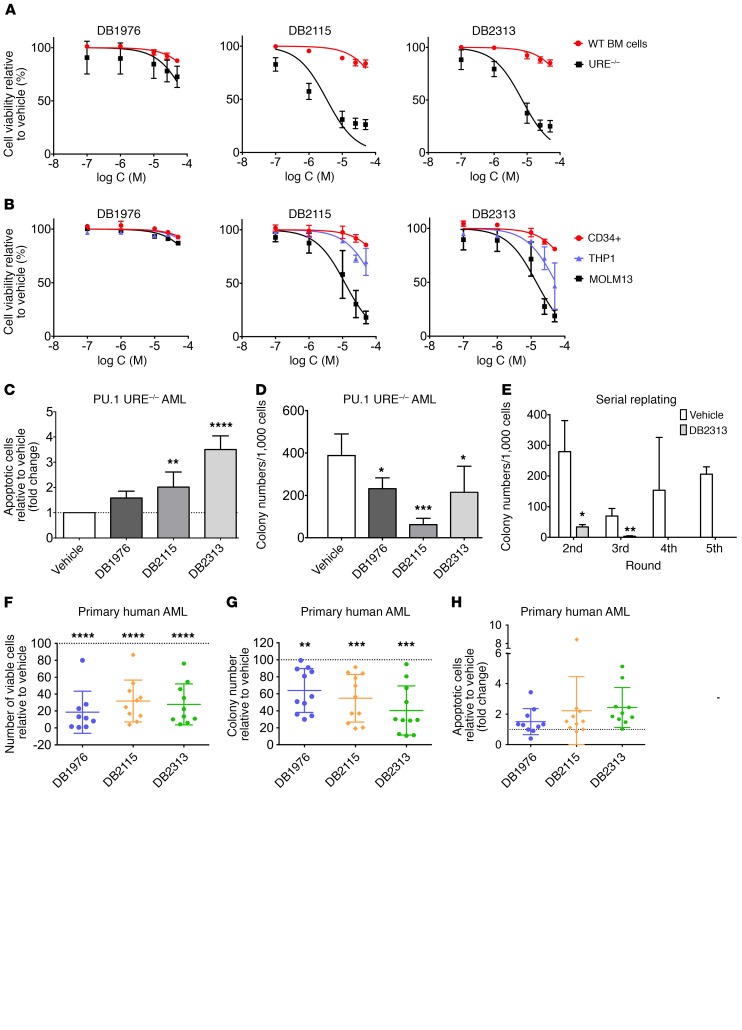
To determine whether our PU.1 inhibitors had functional effects on AML cells, we treated murine PU.1 URE–/– AML and human MOLM13 cell lines with compounds at different concentrations and compared them directly with the effects on murine WT bone marrow (BM) cells or human CD34+ cord blood cells, respectively. Treatment with compounds led to a profound decrease in the growth of PU.1 URE–/– AML cells (IC50 for DB2115: 3.4 μM < IC50 for DB2313: 7.1 μM < IC50 for DB1976: 105 μM), while showing little effect on normal hematopoietic cells at similar concentrations (calculated IC50 for DB2115: 192 μM; DB2313: 240 μM; DB1976: 334 μM) (Figure 4A). We made similar observations with human cells, with strong inhibitory effects on MOLM13 cells (PU.1 low), substantially lesser effects on THP1 cells (PU.1 high), and negligible effects on healthy CD34+ cells (Figure 4B). We confirmed the inhibitory effect on the proliferation of PU.1 URE–/– AML and MOLM13 cells during a treatment time course and observed no effect on AML cells with high PU.1 levels (THP1) (Supplemental Figure 3A). To confirm that the drug effects were related to PU.1 levels, we lentivirally expressed PU.1 in MOLM13 cells and observed a rescue of the cytotoxic effects of the compounds with increased cell viability upon treatment with DB1976, DB2115, or DB2313 (Supplemental Figure 3B).
Figure 4. Small-molecule PU.1 inhibitors decrease cell growth and increase apoptosis of AML cells.
(A) Cell viability of PU.1 URE–/– AML cells and WT BM cells after treatment with increasing concentrations of vehicle or small molecules (n = 3). (B) Cell viability of human CD34+, THP1, and MOLM13 cells after treatment with increasing concentrations of vehicle or small molecules (n = 3). (C) Apoptosis induction (annexin-V+PI–) in PU.1 URE–/– AML cells after 48 hours of treatment with DB1976 (n = 6), DB2115 (n = 6), or DB2313 (n = 3). Fold change compared with vehicle is shown. (D) Clonogenic capacities of PU.1 URE–/– AML cells after treatment with DB1976 (n = 5), DB2115 (n = 3) and DB2313 (n = 4). (E) Serial replating capacity of PU.1 URE–/– AML cells after treatment with DB2313 (n = 3). (F–H) Primary human AML cells were plated in semisolid media containing DB1976, DB2115, or DB2313; colony numbers, viable cell numbers, and apoptotic cells were assessed after 14 days of culture. Error bars represent the mean ± SD, and each AML sample is represented by an individual dot. The percentage (F and G) or fold change (H) compared with vehicle (dotted line) is shown. (F) Number of viable cells (n = 10) and (G) clonogenic capacity (n = 11) after treatment. (H) Apoptotic cell (annexin-V+PI–) fraction after treatment (n = 10). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 1-way ANOVA (C, D, and F–H) or 2-tailed Student’s t test (E).
Like our observations with PU.1-directed shRNAs, treatment with pharmacological PU.1 inhibitors led to a 1.6-, 2-, and 3.5-fold increase in apoptotic cells with DB1976, DB2115, and DB2313, respectively, in murine PU.1 URE–/– AML cells (Figure 4C), and we observed similar effects in human MOLM13 cells (Supplemental Figure 3C).
PU.1 inhibitors also significantly decreased the colony-forming capacity of PU.1 URE–/– AML and MOLM13 cells, but not of THP1 cells (Figure 4D and Supplemental Figure 3D). To assess the effect of PU.1 inhibitors on the long-term clonogenic capacity (in vitro self-renewal) of PU.1 URE–/– AML cells, we performed serial replating assays. Strikingly, we found that treatment with DB2313 led to a significant decrease in clonogenicity in the second and third rounds of plating and a complete disruption of clonogenic capacity in the fourth and higher rounds of plating (Figure 4E).
We also explored the effects of the small-molecule PU.1 inhibitors on primary human AML cells and treated 13 AML patients’ samples (Supplemental Table 1) with PU.1 inhibitors. PU.1 inhibitors led to a significant decrease in the number of viable cells (mean decrease of 81% for DB1976, 68% for DB2115, and 72% for DB2313) (Figure 4F) and clonogenic capacity (mean decrease of 36% for DB1976, 45% for DB2115, and 60% for DB23313) compared with vehicle-treated cells (Figure 4G). The apoptotic cell fraction increased on average by 1.5-fold with DB1976, 2.2-fold with DB2115, and 2.5-fold with DB2313 (Figure 4H). The apoptosis-inducing effect was also confirmed in short-term liquid culture for a few samples (Supplemental Figure 3E). Of note, we observed that PU.1 inhibition had an effect on the majority of samples, which harbored various genetic and cytogenetic anomalies.
Taken together, treatment with PU.1 inhibitors leads to decreased cell viability, colony formation, and increased apoptosis in PU.1lo-induced AML cell lines as well as in a majority of primary AML cell samples from patients.
Inhibitors show on-target PU.1-inhibitory activity in AML cells.
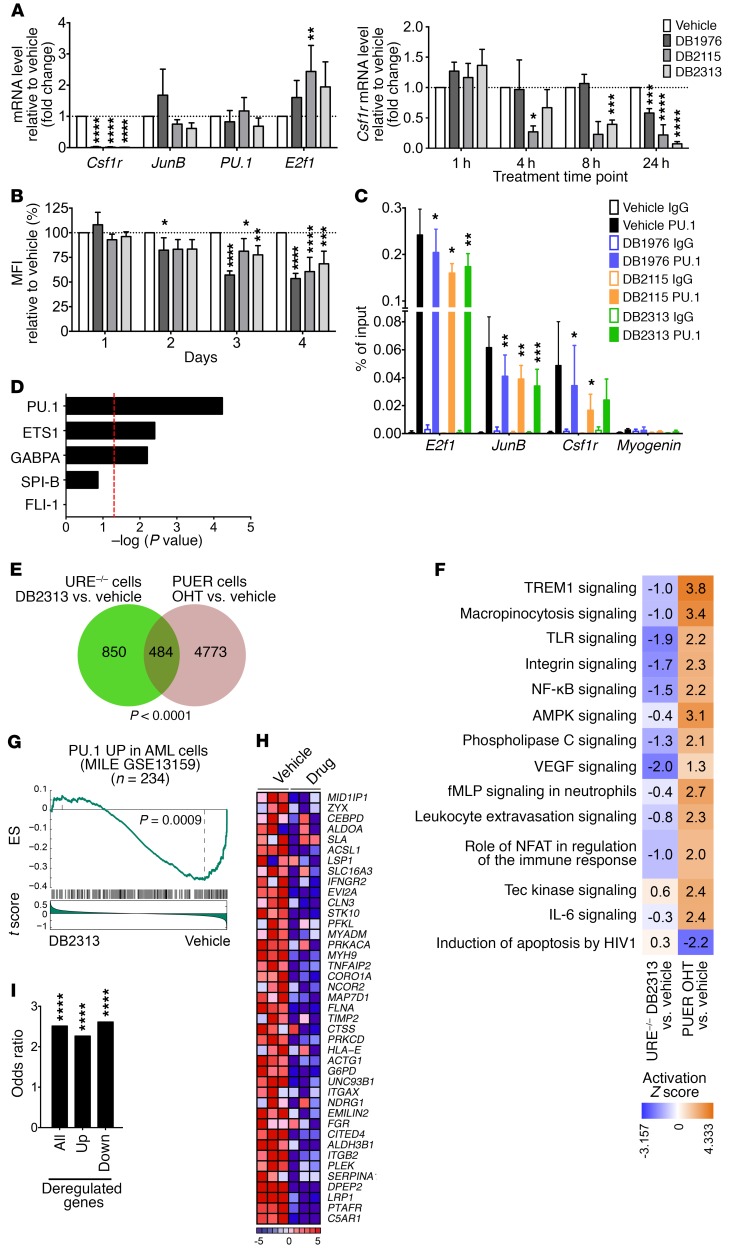
To assess the on-target activity of our inhibitors in AML cells, we measured transcript levels of well-known PU.1 targets in PU.1 URE–/– AML cells. It has been shown that PU.1 positively regulates CSF1R and JUNB and autoregulates itself, whereas it represses E2F1 (12, 16, 36, 37). In line with this, we found a decrease in Csf1r, Junb, and PU.1 transcript expression and an increase in E2f1 expression upon treatment with DB2115 or DB2313 (Figure 5A, left). As Csf1r is one of the most sensitive PU.1 targets in myeloid cells, we assessed its expression at different time points (1 h, 4 h, 8 h, and 24 h) after drug treatment. Interestingly, Csf1r expression significantly decreased as early as 4 hours after treatment (Figure 5A, right), in line with a direct effect of the drugs on PU.1 transcriptional activity. Furthermore, treatment of BM MNCs isolated from a PU.1-GFP–knockin reporter mouse model (38) led to a decrease in GFP reporter expression after treatment with DB1976, DB2115, or DB2313, further confirming a direct effect of the compounds on PU.1 transactivation (Figure 5B and Supplemental Figure 4A) and consistent with PU.1 positive autoregulation (36). ChIP assays revealed that treatment of AML cells with DB1976, DB2115, or DB2313 decreased PU.1 occupancy on E2f1, Junb, and Csf1r promoters, confirming that the compounds were directly interfering with PU.1 binding to chromatin in vivo (Figure 5C).
Figure 5. Inhibitors show on-target PU.1 inhibitory activity in AML cells.
(A) qRT-PCR analysis of PU.1 target genes after PU.1 URE–/– AML cell treatment (n = 3–7), normalized to Gapdh. Fold change compared with vehicle is shown. (B) Mean fluorescence intensity (MFI) of BM MNCs isolated from PU.1/GFP-knockin mice (38) after treatment (n = 5). Fold change compared with vehicle is shown. (C) Quantitative ChIP assays of PU.1 occupancy after treatment of PU.1 URE+/–Msh2–/– AML cells (n = 5). Myogenin was used as a negative control. (D–I) Transcriptome analysis of PU.1 URE–/– AML cells after a 24-hour treatment with DB2313 (n = 3) versus vehicle (n = 3). (D) Differentially expressed genes upon treatment were tested for enrichment of genes directly regulated by PU.1, or regulated by the other ETS transcription factors using Ingenuity Knowledge Base (generated with the use of IPA). Dotted line represents the significance threshold (–log P value >1.3). (E and F) Comparative analysis of deregulated genes in PU.1 URE–/– AML cells after treatment and in PUER cells after PU.1 induction (GEO GSE13125). (E) Venn diagram shows significant overlap between the 2 data sets. (F) Deregulated canonical pathways between the data sets. Colored squares indicate the activation Z score. (G) GSEA enrichment plot of PU.1 positively regulated genes (regulon) in AML cells (from the MILE AML network, as determined by the ARACNe algorithm) against the global list of differentially expressed genes upon treatment, ranked by the drug response (as measured by t score of DB2313 vs. vehicle). (H) Heatmap of leading-edge genes showing row-normalized relative expression. (I) Enrichment of PU.1 binding at promoters of deregulated genes in PU.1 URE–/– AML cells upon treatment. Publicly available PU.1 ChIP-seq data from PUER cells (GSE63317) were used for this analysis. Up, upregulated; Down, downregulated. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 1-way ANOVA (A–C), hypergeometric test (E), Fisher’s exact test (I), or according to ref. 57 (G).
To gain insight into the genome-wide transcriptional effects following treatment of AML cells with our inhibitors, we performed gene expression analysis. We found dysregulation of 1,648 transcripts (of 34,472 total) by at least 1.2-fold after DB2313 treatment of PU.1 URE–/– AML cells, with 867 probe sets upregulated and 781 probe sets downregulated. We found highly significant enrichment of known genes directly downstream of PU.1 (Figure 5D). Interestingly, enrichment of genes regulated by other ETS family transcription factors, such as ETS1, GABPΑ, SPI-B, or FLI-1, was much lower or nonsignificant, suggesting a preferential inhibitory effect of our compounds on PU.1 binding, with selectivity even within the ETS family. We confirmed the dysregulation of some known PU.1 target genes such as Ly96, Clec5a, Cdkn1a, Itgb2, Fcgr3, and Gfi1 by quantitative reverse transcription PCR (qRT-PCR) (Supplemental Figure 4B). Top canonical pathways and biological functions with significant enrichment identified by Ingenuity Pathway Analysis (IPA) included “hematological system development and function,” “cell death and survival,” “cellular development,” and “cellular growth and proliferation” (Supplemental Figure 4C), consistent with known functions of PU.1.
We compared the differentially expressed genes upon PU.1 inhibitor treatment of PU.1 URE–/– AML cells with publicly available data in which the PU.1-regulatory transcriptional network had been identified by tamoxifen-mediated induction of PU.1 expression in engineered PU.1-null immature hematopoietic cells (PUER cells) (Gene Expression Omnibus [GEO] GSE13125) (39). Of the 1,334 genes dysregulated after PU.1 inhibitor treatment of PU.1 URE–/– AML cells, 36% (484 cells) overlapped with previously identified canonical PU.1 targets in PUER cells (Figure 5E), which was highly significant (P < 0.0001). Furthermore, comparative pathway analysis revealed a significant inverse correlation between PU.1 URE–/– AML cells after treatment and PU.1 overexpression in PUER cells, suggesting that inhibitor treatment antagonizes canonical PU.1-regulated pathways (Figure 5F). Gene set enrichment analysis (GSEA) confirmed that DB2313 treatment leads to a change in expression of genes regulated by PU.1 in HSCs from the PU.1 URE–/– mouse model (12) (Supplemental Figure 4D). Moreover, we found an enrichment in genes regulated by transcription factors of the AP1 family, which are known PU.1 targets (Supplemental Figure 4E).
Furthermore, we performed interactome analysis of 3 independent large series of primary AML patients’ samples including those from The Cancer Genome Atlas (TCGA), the MILE (Microarray Innovations in LEukemia) study, and GEO GSE1159 to identify the PU.1 regulon specifically in AML cells using the ARACNe algorithm (40–42), and then tested the activity of PU.1 using this gene set in the DB2313-induced transcriptional signature using GSEA. In these analyses, the PU.1 “regulon” generated by the ARACNe algorithm was partitioned in a positive and a negative component on the basis of a positive or negative Spearman correlation (P < 0.05) between expression levels of PU.1 and the target gene across all samples of the data set used to generate the network. Strikingly, for each of these patient cohorts, we found that the PU.1 positive regulon (“PU.1 UP,” genes upregulated when PU.1 activity was high) was enriched under vehicle-treated conditions, consistent with a negative effect of DB2313 on PU.1 transcriptional activity in AML cells (Figure 5, G and H, and Supplemental Figure 4, F–I).
In addition, we performed a comparative analysis with publicly available PU.1 ChIP-sequencing (ChIP-seq) data (GEO GSE63317) (20) and found significant enrichment of PU.1 binding in the promoters of genes deregulated after treatment with PU.1 inhibitor (Figure 5I). Up- and downregulated genes were similarly affected, consistent with an interference of our drugs with PU.1-chromatin interaction and independent of downstream transactivating or repressive mechanisms.
We performed PU.1 ChIP-seq experiments and analyzed PU.1-bound sites in the vehicle-treated sample and PU.1 sites in the PU.1 inhibitor–treated (DB2313-treated) sample in PU.1 URE–/– cells. Comparison of PU.1 ChIP-seq of vehicle versus DB2313 treatment as the background showed a strong enrichment of PU.1 motifs, while the opposite comparison showed enrichment of motifs that are not related to ETS transcription factors (data not shown). This is in line with better PU.1 binding under control conditions. The most common motif in the PU.1 ChIP was AAAGAGGAAGTG, and the respective position weight matrix (PWM) found by Homer was present in 41% of peaks in the vehicle sample versus only 16% of the peaks in the DB2313-treated sample, which is again in line with PU.1 inhibitor treatment causing decreased binding of PU.1 to its canonical target sites. When we looked specifically at the peaks with a PU.1 motif in the vehicle-treated samples and subtracted peaks with a PU.1 motif in the DB2313-treated samples, we arrived at 268 bona fide PU.1 sites that were “lost” upon drug treatment. A complete list of these peaks/sites is provided in Supplemental Table 2. Examples of peaks from this analysis are shown in Supplemental Figure 5. Overall, the results from the ChIP-seq analyses are consistent with a genome-wide inhibitory effect of DB2313 on PU.1 binding to its canonical sites in AML cells.
PU.1 inhibitors have limited effects on normal hematopoiesis.
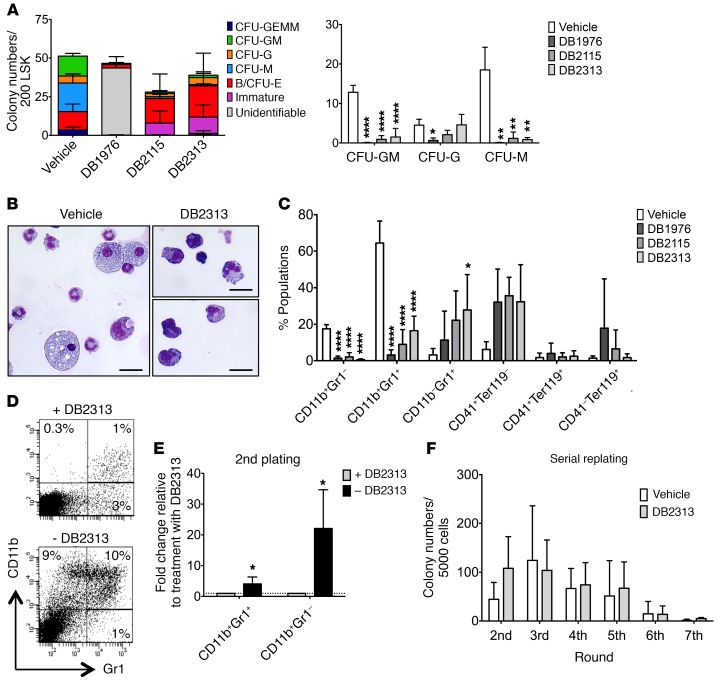
In order to determine the effect of our PU.1 inhibitors on normal hematopoietic differentiation, we sorted immature Lin–Sca1+c-Kit+ (LSK) cells from WT mice and treated them with either DB1976, DB2115, or DB2313. After treatment with PU.1 inhibitors, the total numbers of formed colonies were only slightly reduced, however, we saw a substantial reduction of the more mature myelomonocytic colony types (CFU-granulomonocyte [CFU-GM], CFU-granulocyte [CFU-G], and CFU-monocyte [CFU-M]) and a relative increase in the number of burst or CFU-erythroid (B/CFU-E), and small immature cell colonies (Figure 6A), consistent with the known important role of PU.1 in myelomonocytic differentiation. Of note, the initial compound DB1976 appeared to show toxicity on LSK cells, with a majority of colonies unidentifiable and a majority of dead cells, contrary to what we observed with the 2 newer compounds (DB2115 and DB2313), which we therefore prioritized for further investigation. Overall, the numbers of viable cells per colony were reduced upon treatment with PU.1 inhibitor (Supplemental Figure 6A), again consistent with an effect of PU.1 inhibition on more mature, differentiating cells. Consistently, cytomorphologic analysis revealed almost no macrophages, significantly fewer mature granulocytes, and an increase in immature cells, characterized by a higher nucleocytoplasmic ratio and basophilic cytoplasm after treatment (Figure 6B). This was confirmed by flow cytometric analysis, which revealed a reduction in mature monocytes (CD11b+Gr1–) and granulocytes (CD11b+Gr1+) and a slight increase in immature granulocytes (CD11b–Gr1+) and CD41+ cells, but no significant difference in erythrocyte (Ter119+CD41–) generation (Figure 6C and Supplemental Figure 6B). To further explore the granulocyte population and pinpoint the exact stage of maturation at which the small molecules impact differentiation, we used a combination of markers described by Guibal et al. (43) and further separated the cell populations into the myeloblast/promyelocyte-metamyelocyte/mature stages (Supplemental Figure 6C). After treatment, we observed a slight increase in the proportion of cells at the myeloblast/promyelocyte stage (stage 1), a strong increase of metamyelocytes (stage 2), and a decrease in more mature cells (stage 3) (Supplemental Figure 6D), indicating that our compounds primarily inhibit later stages of granulocytic differentiation.
Figure 6. PU.1 inhibitors decrease granulomonocytic differentiation in a reversible manner.
(A–C) WT LSK cells were plated in semisolid media containing PU.1 inhibitors (25 μM for DB1976, 700 nM for DB2115, and 330 nM for DB2313). (A) Number of CFU-G, CFU-M, CFU-GM, CFU-GEMM, B/CFU-E, and immature colonies after treatment (left). Detailed histograms of CFU-GM, CFU-G, and CFU-M numbers (right) (n = 3). (B) Morphological appearance of cytospun and May-Grünwald Giemsa–stained cells after colony formation assay with vehicle or DB2313 treatment. Scale bars: 20 μm. (C) FACS analysis showing the percentage of CD11b+Gr1–, CD11b+Gr1+, CD11b–Gr1+, CD41+Ter119–, CD41+Ter119+, and CD41–Ter119+ cells after colony formation assays (n = 4). (D and E) Cells from a first round of colony formation assays treated with DB2313 were replated in the presence (+DB2313) or absence (–DB2313) of DB2313. (D) Representative FACS plots. (E) Proportion of CD11b+Gr1– and CD11b+Gr1+ cells formed, with or without DB2313 treatment, in the replating (n = 3). Fold change compared with replating with DB2313 treatment is shown. (F) Serial replating assay with D2313 continuous treatment (n = 3). *P < 0.05, **P < 0.01, and ****P < 0.0001, by 1-way ANOVA (A and C) or 2-tailed Student’s t test (E).
As we had observed a reduction in mature myelomonocytic colonies upon treatment with PU.1 inhibitors, but persistence or even slight increases in more immature cells (CFU-granulocyte/erythrocyte/macrophage/megakaryocyte [GEMM] and immature colonies), we wanted to test whether these immature cells were still functional after drug removal. We focused on the compound with the lowest IC50 and the least effect on WT cells (DB2313). Interestingly, the production of mature granulocytes and monocytes increased significantly by 4-fold and 22-fold, respectively, after removing the treatment in the second plating, showing that the effects of PU.1 inhibition on granulocyte/monocyte (G/M) generation are reversible (Figure 6, D and E). In addition, we performed serial replating assays with continuous treatment with PU.1 inhibitors and found no significant differences in terms of serial replating capacity, indicating that treatment with PU.1 inhibitors does not significantly affect the long-term colony-forming potential or in vitro self-renewal capacity of immature hematopoietic stem and progenitor cells (Figure 6F).
We also explored the effect of the inhibitors on lymphoid cells. We treated B cells in liquid culture for 3 days and found that the compounds had no effect on the apoptotic fraction (Supplemental Figure 6G). To assess T cells, we performed culturing on OP9-Delta1 cells and found that compound treatment led to slightly delayed, but not completely blocked, T cell differentiation (Supplemental Figure 6, E and F). Finally, we assessed PU.1 inhibitor–treated BM cells in congenic transplantation assays and found no changes in T cell numbers and only modestly decreased B cell numbers after 20 weeks (Supplemental Figure 6H).
Taken together, our results indicate that treatment with our PU.1 inhibitors leads to effects on normal hematopoiesis that are consistent with fundamental roles of PU.1 function during hematopoiesis in myelomonocytic lineages, with limited effects on T and B cell lymphopoiesis. The effects on myeloid cells are reversible upon treatment discontinuation and seem to primarily affect more mature cells. However, in future studies, it will be of interest to perform longer-term in vivo treatment with further optimized compounds (once available) for a more comprehensive and definitive evaluation of the more subtle effects on normal hematopoiesis.
Treatment with PU.1 inhibitors decreases leukemia progression in vivo.
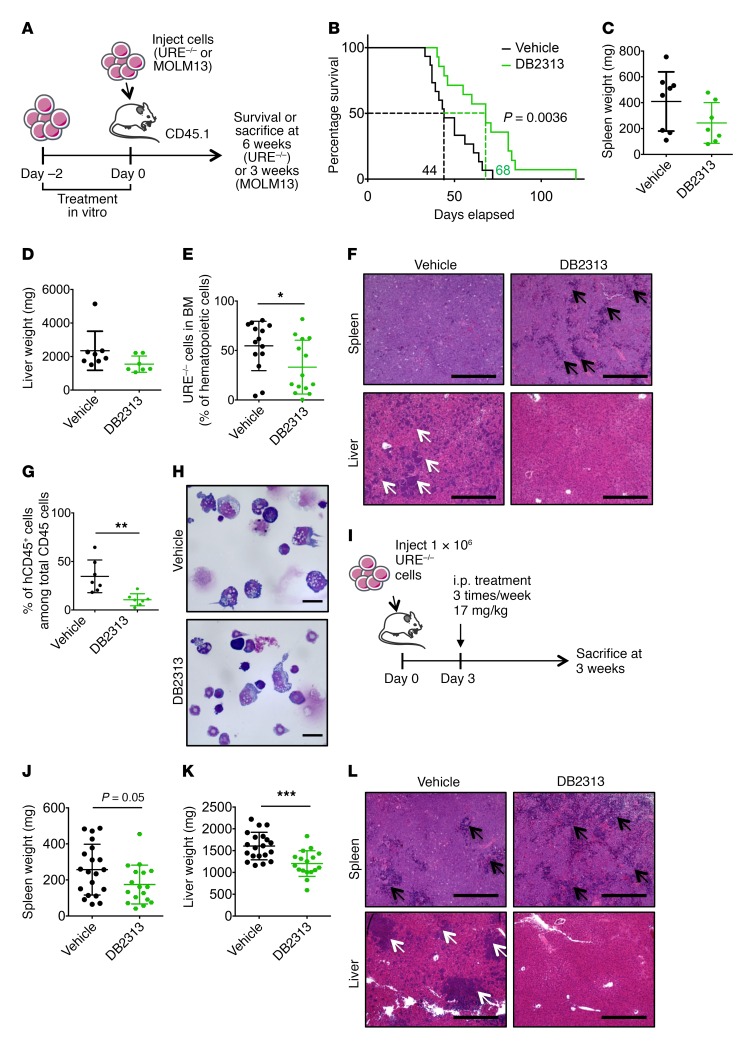
To assess the effect of PU.1 inhibitors on the growth of AML cells in vivo, we treated PU.1 URE–/– AML cells with DB2313 for 2 days and injected 2 × 105 viable cells into sublethally irradiated recipient mice (Figure 7A). Mice that received vehicle-treated AML cells succumbed to leukemia, with a median latency of 44 days, whereas mice that received DB2313-treated AML cells survived significantly longer (P = 0.0036), with a median latency of 68 days (Figure 7B). Assessment of tumor burden 6 weeks after transplantation showed a decrease in hepatoplenomegaly after treatment; mean splenic weight was 410 mg for vehicle-treated mice and 243 mg for DB2313-treated mice (Figure 7C); mean liver weight was 2,347 mg for vehicle-treated mice and 1,548 mg for DB2313-treated mice (Figure 7D). AML cell engraftment in the BM was significantly decreased after treatment with DB2313, with a mean chimerism of 55% for the vehicle-treated group and 33% for the DB2313-treated group (Figure 7E and Supplemental Figure 7A). Histological analyses revealed severe blast infiltration, with disruption of the splenic architecture and complete loss of the red pulp in the vehicle-treated group, and significantly less pronounced effects in the DB2313-treated group. Likewise, infiltration of the liver by leukemic blasts was substantially reduced in the DB2313-treated group compared with that detected in the vehicle-treated group (Figure 7F). When we performed the same experiment with human MOLM13 AML cells, we observed a similarly significant decrease in leukemic burden in the BM after treatment, with a mean chimerism of 35% for the vehicle-treated group and 11% for the DB2313-treated group (Figure 7, A, G, and H).
Figure 7. Treatment with PU.1 inhibitors leads to decreased tumor burden and increased survival in vivo.
(A) Experimental scheme for PU.1 URE–/– AML and MOLM13 cells transplants after in vitro treatment. (B–F) Transplantation of PU.1 URE–/– AML cells. (B) Kaplan-Meier survival analysis of transplanted mice (n = 15, vehicle group; n = 14, DB2313 group; 2 independent experiments). Dotted lines indicate median survival. (C and D) Spleen and liver weights 6 weeks after transplant (n = 8 mice, vehicle group; n 7, DB2313 group). (E) Chimerism of PU.1 URE–/– AML cells in the BM 6 weeks after transplantation (n = 28 mice total; 2 independent experiments). (F) Histological analysis of H&E-stained spleen and liver. Black arrows indicate the remaining red pulp in the spleen; white arrows indicate blast infiltration in the liver. Scale bars: 400 μm. (G and H) Transplantation of human MOLM13 cells. (G) Chimerism of MOLM13 cells (hCD45+) in the BM 3 weeks after transplantation (n = 7 mice/group). (H) Morphological appearance of May-Grünwald Giemsa–stained BM cells 3 weeks after transplantation. Scale bars: 20 μm. (I) Experimental scheme for PU.1 URE–/– AML cell transplantation followed by i.p. treatment with vehicle or DB2313. (J and K) Spleen and liver weights 3 weeks after transplantation (n = 20 mice, vehicle group; n = 17, DB2313 group; 3 independent experiments). (L) Histological analysis of H&E-stained spleen and liver. Black arrows indicate the remaining red pulp in the spleen; white arrows indicate blast infiltration in the liver. Scale bars: 400 μm. Error bars represent the mean ± SD, and each mouse is represented by an individual dot. *P < 0.05, **P < 0.01, and ***P < 0.001, by log-rank (Mantel-Cox) test (B) or 2-tailed Student’s t test (E, G, J, and K).
Further, we performed in vivo treatment by i.p. injections of DB2313 following transplantation of PU.1 URE–/– AML cells (Figure 7I). The effect on AML cells in the BM was modest, with a mean chimerism of 47% for the vehicle-treated group versus 37% for the DB2313-treated group, which was likely due to the low concentration of this prototype compound in the serum (Supplemental Figure 7, B and C). However, we observed decreased splenomegaly (mean weight of 257 mg for vehicle-treated mice and 174 mg for DB2313-treated mice) and hepatomegaly (mean weight of 1,604 mg for vehicle-treated mice and 1,203 mg for DB2313-treated mice) (Figure 7, J and K), reduced leukemic infiltration in the spleen (Figure 7L and Supplemental Figure 7D), and no detectable leukemic infiltration in the liver after DB2313 treatment (Figure 7L).
Taken together, although our compounds represent tool compounds that require further optimization, these data provide proof of concept for the antileukemic effects of PU.1 inhibitors in vivo, including decreased tumor burden and increased survival.
Discussion
During the past decade, increasing evidence has shown the importance of PU.1 dosage in leukemia development. Our study provides proof of concept that targeting PU.1, a commonly deregulated transcription factor in AML, has antileukemic effects.
Given the findings from Iwasaki et al. (15) showing that complete loss of PU.1 in HSCs leads to their exhaustion, we explored in this work inhibition of PU.1 activity as a therapeutic strategy in AML, with the hypothesis that leukemic cells with an already dysregulated PU.1 transcriptional network would be more susceptible to further PU.1 inhibition than normal cells. Our approach is similar in principle to what has been described recently for casein kinase 1A1, a tumor-suppressor gene located on chromosome 5q that is frequently heterozygously deleted in myelodysplastic syndrome. Haploinsufficiency of CSNK1A leads to HSC expansion, whereas complete deletion leads to HSC failure. Cells with already reduced levels of CSNK1A were more sensitive to CSNK1A pharmacological inhibition than were cells with normal levels (44).
We used 2 methods to decrease PU.1 activity: RNA interference and new PU.1 inhibitors. Only a few drugs directly targeting dysregulated transcription factors have been described so far, with one recent example being an inhibitor targeting a protein-protein interaction, RUNX1 binding to CBFβ-SMMHC, which is an oncogenic transcription factor fusion found in inv(16) AML (45).
The inhibitors reported here do not bind the PU.1 protein directly but instead prevent binding of PU.1 to cognate DNA sites, the requisite molecular event for gene regulation, via an allosteric mechanism. Specifically, occupancy by the compounds in the minor groove induces perturbations in DNA conformation that are transmitted to the orthosteric site of PU.1 in the major groove. Dissociation of DNA-bound PU.1 by DB2313 and distinctive DNA footprints of the compounds demonstrated that the bound conformations of PU.1 and the compounds with the same DNA sequence are not compatible. The compounds are derivatives of the heterocyclic diamidine family, some members of which (e.g., DB75 [furamidine] and derivatives for trypanosomiasis) have been established in the context of other clinical applications (46, 47).
Decreasing PU.1 activity and levels in human and murine PU.1lo-induced leukemic cells led to decreased cell growth and clonogenicity. We found that this phenotype was due to increased apoptosis after PU.1 inhibition. We also found inhibitory effects in primary human samples harboring various genetic alterations, suggesting that such a strategy could be applied to a larger fraction of patients with AML. Additionally, treatment with DB2313 increased survival and reduced tumor burden in vivo. Even though these inhibitors are early tool compounds and need further optimization for improved bioavailability in vivo, the fact that compounds from the same family have reached the clinic for a different indication offers encouragement for further development.
The reduction in PU.1 binding to chromatin, combined with changes in PU.1 target gene expression and the PU.1-dependent “regulon” in AML cells, demonstrates the on-target effect of the compounds. Of note, our compounds have a preference for PU.1-binding sites, which are characterized by highly AT-rich flanking sequences, a feature that is not shared by most other ETS transcription factors. And while the effect of our compounds does not seem to be exclusively restricted to PU.1-binding sites, we found significant selectivity for PU.1, which was sufficient to target AML cells and prevent unspecific toxicity. Consistent with known PU.1 function in normal hematopoiesis and myelomonocytic differentiation, we identified a transient decrease in the generation of terminally differentiated cells, especially from the myelomonocytic lineages, after treatment of immature stem and progenitor cells with PU.1 inhibitors. However, removal of the compounds led to restoration of normal hematopoietic differentiation, indicating that normal stem and progenitor cells are not significantly affected by treatment with PU.1 inhibitors.
In summary, our study provides proof of principle for inhibition of the transcription factor PU.1 as a therapeutic strategy in the treatment of AML. Furthermore, we present small molecules of the heterocyclic diamidine family as first-in-class PU.1 inhibitors that act via allosteric inhibition of PU.1-chromatin interaction and have antileukemic efficacy in vitro and in vivo. Our findings show that therapeutic interference with transcription factor–chromatin binding can be achieved with small molecules through a minor groove–targeting strategy in principle and support the further development of PU.1 inhibitors as potential therapeutics.
Methods
Cells.
THP1, MOLM13, BaF3, Kasumi-1, HEK293, and 293T cells were purchased from ATCC. MOLM13 and Kasumi-1 cells were cultured in RPMI 1640 medium supplemented with 20% heat-inactivated FBS and 1% penicillin/streptomycin (P/S). THP1, BaF3, and HEK293 cells were cultured in RPMI 1640 medium completed with 10% FBS and 1% P/S. BaF3 cells were supplemented with 10 ng/ml IL-3. 293T cells were maintained in DMEM supplemented with 10% FBS and 1% P/S. PU.1 URE–/– and PU.1 URE+/–Msh2–/– leukemia cells (17, 18) were cultured in M5300 medium (STEMCELL Technologies) supplemented with 10% FBS, 15% WEHI supernatant, 15% BHK supernatant, and 1% P/S. PU.1 URE–/– and PU.1 URE+/–Msh2–/– cell lines were established from leukemic URE–/– and URE+/–Msh2–/– mice, respectively.
Mouse cells from peripheral blood (PB), BM, spleen, or liver were isolated, treated with ACK buffer (pH 7.4) to lyse red blood cells, and cultured or stained for FACS analysis. Cells were maintained in Iscove’s Modified Dulbecco’s Media (IMDM) (Gibco, Thermo Fisher Scientific) supplemented with 20% FBS, 20 ng/ml recombinant mouse SCF (rmSCF) (Miltenyi Biotec), 10 ng/ml rmIL-3 (Gemini), 10 ng/ml rmIL-6 (Gemini), 20 ng/ml rmTPO (Miltenyi Biotec), and 1% P/S.
Human MNCs from BM or PB obtained from healthy or leukemic patients were isolated by density ultracentrifugation using Ficoll-Hypaque (GE Healthcare) and cultured in IMDM medium supplemented with 2% FBS, cytokines (10 ng/ml rhIL-3 [Preprotech], 25 ng/ml recombinant human IL-6 [rhIL-6], 50 ng/ml rhSCF, 50 ng/ml rhFLT3 ligand, and 100 ng/ml rhTPO [all from Gemini]), 40 μg/ml hLDL, and 1% P/S. All cell lines and primary cells were maintained in an incubator at 37°C and 5% CO2.
Small molecules.
The syntheses of DB1976 (23), DB2115 (48), and DB2457 (49) have been previously reported. The synthesis of DB2313 and the experimental details, along with characterization data, are provided in the supplemental materials.
After synthesis, DB1976, DB2115, DB2313, and DB2457 were dissolved in sterile water and stored at –20°C.
Mice.
PU.1 URE–/–, PU.1 URE+/–Msh2–/–, and PU.1 heterozygous mice with knockin of a GFP reporter into exon 1 of PU.1 have been previously described (17, 18, 38). Nonobese, diabetic severe combined immunodeficiency IL-2 receptor γ–null (NSG) mice were housed in the animal facility.
Patients’ characteristics.
See the Supplemental Methods and Supplemental Table 1.
Lentiviral vector and transduction.
shRNA sequences for knockdown of PU.1 have been previously described and validated (shPU.1_1 [ref. 50], and shPU.1_2 [ref. 51]); shPU.1_3 and shPU.1_4 were newly designed (shPU.1_3: 5′-AGATGCACGTCCTCGATAC-3′; shPU.1_4: 5′-TCCAGTTCTCGTCCAAGCACAA-3′). These sequences recognize both mouse and human sequences. Control (shCtrl) or shPU.1 sequences were cloned under an H1 promoter and inserted into the lentiviral vector pRRLsin-PGK-eGFP-WPRE (provided by Hana Raslova, Institut Gustave Roussy, Villejuif, France). PU.1 overexpression was performed using the lentiviral vector PCAD-PU.1-IRES-GFP or GFP alone as previously described (18). All viruses were produced using the 293T cell line. Cells were transduced by spin infection at 1,000 g for 60 minutes at 37°C in the presence of Polybrene. Seventy-two hours after transduction, cultures were sorted for GFP+ cells and used for further studies.
Molecular dynamics simulations.
See the Supplemental Methods.
Biosensor-SPR assays for compound binding affinity and PU.1-DNA complex inhibition by compound.
See the Supplemental Methods.
DNA footprinting.
See the Supplemental Methods.
Cell proliferation assays.
To evaluate the effect of the small molecules on cell viability, cells were cultured in the presence of vehicle (water) or different compounds. Cells were plated at a density between 0.5 × 105 and 2 × 105 in 100 μl or 500 μl of media and manually counted every 24 hours for 4 days using a trypan blue exclusion assay. For IC50 determination, cell viability was first assessed using a resazurin cell viability assay (Cell Titer Blue; Promega) after 48 hours of culture and confirmed by manual counting using a trypan blue exclusion assay. If not further specified, drugs for the treatment of mouse cells were used at the following concentrations: 25 μM for DB1976, 700 nM for DB2115, and 330 nM for DB2313. For the treatment of human cells, the same concentrations were used, with the exception of DB2313, which was used at a concentration of 660 nM.
Colony formation assays and serial replating assays.
For human cell line colony assays, 1,000 or 3,000 cells were plated per milliliter of methylcellulose in HSC002SF or HSC003 methylcellulose (R&D Systems). For mouse cell colony assays, 1,000 cells per milliliter of methylcellulose were plated in HSC007 methylcellulose (R&D Systems) and scored after 7 days. For human primary AML cell colony assays, 150,000 mononuclear cells were plated per milliliter of methylcellulose in HSC003 media and the colonies scored after 12 to 14 days. For LSK cells, 200 cells per 500 μl methylcellulose were plated in HSC007. Vehicle or inhibitors were added to the methylcellulose at the following concentrations: 25 μM for DB1976, 700 nM for DB2115, and 330 nM for DB2313 for murine cells and 660 nM for DB2313 for human cells. For serial replating of cell lines, the same numbers of cells initially plated were replated. For serial replating of primary LSK cells, 5,000 cells per 500 μl methylcellulose were replated.
Lymphoid cell culture.
See the Supplemental Methods.
Flow cytometric analysis and cell sorting.
See the Supplemental Methods.
Cytomorphology.
See the Supplemental Methods.
Western blot analysis.
See the Supplemental Methods.
Inhibition of the PU.1-DNA complex in live cells (reporter assay).
The functional inhibition of PU.1 transactivation by heterocyclic diamidines in live cells was measured using a fluorescent EGFP reporter. For this assay, a PU.1 expression plasmid was cloned by inserting a fragment encoding full-length PU.1 fused to an infrared RFP (iRFP) (52) reporter between the NheI/BamHI sites of pcDNA3.1(+). The fusion was linked by a sequence encoding a self-cleaving 2A peptide (53). Cultured HEK293 cells, which do not express PU.1, were transfected with the PU.1 expression plasmid for 24 hours. Cells were then retransfected, in the absence or presence of compounds, with an EGFP-based reporter under the control of a minimal promoter that was downstream from a synthetic enhancer harboring 5 tandem repeats of the λB motif (23). After an additional 24 hours, cells were trypsinized and counted by flow cytometry using an Accuri C6 instrument (BD) on 2 spectrally isolated channels for EGFP (488/511 nm) and iRFP (640/>670 nm).
Molecular modeling and docking.
See the Supplemental Methods.
ChIP.
PU.1 URE+/–Msh2–/– cells (10 × 106) were treated with vehicle, 50 μM DB1976, 1.4 μM DB2115, or 660 nM DB2313 for 8 hours. ChIP experiments were performed as previously described (54, 55), except that Dynabeads Protein A (Thermo Fisher Scientific) were used for the pulldown. Briefly, cells were cross-linked using a 1% formaldehyde solution for 15 minutes at room temperature. Samples were sonicated for 10 minutes to generate fragments below 1 kb. For immunoprecipitation, 5 μg PU.1 antibody (Santa Cruz Biotechnology; sc-352) or normal rabbit IgG (Santa Cruz Biotechnology; sc-3888) was used. DNA fragments were purified using a QIAquick PCR Purification Kit (QIAGEN) and subjected to qPCR amplification. The percentage of input was calculated using the comparative cycle threshold method. The primers used are listed in Supplemental Table 3.
qRT-PCR and microarray experiments.
RNA was extracted using an RNeasy Mini or Micro Kit (QIAGEN) and, for qRT-PCR, reverse transcription of RNA was performed using an iScript cDNA Synthesis Kit (Bio-Rad). qPCR reactions were performed on a ViiA7 instrument using SYBR Green PCR Master Mix (both from Life Technologies, Thermo Fisher Scientific). Expression levels of genes were normalized to Gapdh. The primers used are listed in Supplemental Table 3. For microarrays, RNA was isolated from PU.1 URE–/– AML cells after 24 hours of treatment with vehicle or DB2313 (330 nM). After checking RNA quality using a 2100 Bioanalyzer (Agilent Technologies), RNA was labeled with the Affymetrix GeneChip WT Terminal Labeling Kit, hybridized to Affymetrix 2.0 Mouse GeneST microarrays, and scanned by an Affymetrix GeneChip Scanner 3000 7G system using standard protocols. Microarray data are available in the NCBI’s GEO database (GEO GSE77651). Raw data were normalized using Expression Console software (Affymetrix), and differentially expressed genes were determined using a fold-change of 1.2 and a P value of 0.1 after analysis with Transcriptome Analysis Console software (Affymetrix). Pathway analyses were performed using IPA (QIAGEN). A –log (P value) cutoff of 1.3 was used as the significance threshold. GSEA was performed with gene sets from the Molecular Signatures Database (56, 57) and from the PU.1 URE–/– HSC signature database (12).
Integrative analysis with published data sets.
For comparative analysis, we used the NCBI GEO data set GSE13125 (39), derived from a PU.1 overexpression system with tamoxifen-inducible “PUER” cells. Comparative analysis between differentially expressed genes in this data set and in PU.1 URE–/– AML cells after treatment was performed using IPA. For GSEA, we used the NCBI GEO data set GSE5654, derived from PU.1 URE–/– HSC (12). To identify genes directly regulated by PU.1, we used the PU.1 ChIP-seq data obtained from GSE63317 (GSM1545739) (20). Among the genes dysregulated in PU.1 URE–/– after DB2313 treatment, we identified those with PU.1 occupancy in their promoters, defined as being within 1 kb upstream of the transcription start site (TSS). To determine the significance of correlation of differential gene expression upon PU.1 activation with the presence of PU.1 binding in the gene promoter, we performed a Fisher’s exact test using UCSC Genes (mm9) as a reference (https://genome.ucsc.edu/cgi-bin/hgGateway?db=mm9).
Interactome analyses (ARACNe Networks).
See the Supplemental Methods.
ChIP-seq analysis.
See the Supplemental Methods.
BM transplantation.
See the Supplemental Methods.
Determination of serum compound concentrations.
See the Supplemental Methods.
In vivo leukemia models.
NSG mice were sublethally irradiated (2.0 Gy) and transplanted via retro-orbital vein injection with PU.1 URE–/– AML cells or via tail vein injection with MOLM13 cells. For pretreatment experiments, AML cells were treated in vitro with vehicle or DB2313, viable cells were counted after 2 days of treatment, and 2 × 105 cells (PU.1 URE–/– AML) or 1 × 106 cells (MOLM13) per mouse were transplanted. Mice were monitored for signs of leukemia. For survival experiments, live BM aspiration was performed in mice 6 weeks after transplantation of PU.1 URE–/– cells. Otherwise, mice were sacrificed at 6 weeks (PU.1 URE–/– AML group) or 3 weeks (MOLM13 group). For i.p. treatment with PU.1 URE–/– AML cells, mice were transplanted with 1 × 106 cells per mouse. Mice were divided into vehicle-treated and DB2313-treated (17 mg/kg) groups. Treatment started 3 days after transplantation and continued for 3 weeks, with 3 injections per week. Mice were sacrificed 3–4 weeks after transplantation.
After sacrifice, BM, spleens, and livers were harvested from the mice, and engraftment was assessed by FACS using CD45.1, CD43, and c-Kit antibodies for PU.1 URE–/– AML cells, and CD45.1 and human CD45 antibodies for MOLM13 cells.
Statistics.
Data are presented as the mean ± SD of independent experiments, unless otherwise specified. Statistical analyses were performed with GraphPad Prism 7.0 (GraphPad Software) using 1-way ANOVA with Tukey’s post-test correction for multigroup comparisons or a 2-tailed Student’s t test for 2-group comparisons, unless otherwise specified in the figure legends. A P value of less than 0.05 was considered significant. For survival analyses, a log-rank test was used to determine statistical significance.
Study approval.
Human samples were collected after informed consent was obtained from patients and upon approval of the IRB of Albert Einstein College of Medicine (protocol 2008-842). All mouse experiments were approved by the IACUC of Albert Einstein College of Medicine (protocol 2013-1202).
Author contributions
IAD and US designed the study and experiments. AP and WDW designed the biosensor-SPR experiments. GMKP designed the DNA footprinting and (with HMK) the reporter experiments. GMKP, KH, AP, and WDW designed and executed the molecular simulations. DWB designed the novel compound syntheses, which were performed by AK and AAF. IAD, JL, HMK, SRN, KM, AP, and GMKP performed experiments. BB analyzed Chip-seq data sets. IAD and JC analyzed microarray data sets. LAC, TIT, BW, IM, AV, EG, AAI, and AAF analyzed and interpreted data. IAD, JL, US, AP, WDW, DWB, and GMKP wrote the manuscript.
Supplementary Material
Acknowledgments
We thank A. Skoultchi, K. Gritsman, J.B. Micol, and all members of the Steidl laboratory for helpful suggestions and discussions, and M. Ferreira for technical assistance. We also thank D. Sun from the Einstein Stem Cell Isolation and Xenotransplantation Facility (funded through New York Stem Cell Science [NYSTEM] grant C029154); D. Reynolds and W. Tran from the Einstein Genomics Core Facility; P. Schultes from the Department of Cell Biology at Albert Einstein College of Medicine; and S. Wang from the Georgia State University Mass Spectrometry facility for their expert technical support. We thank the Albert Einstein Cancer Center Shared Resources (NIH grant P30CA013330) and the Herbert Irving Cancer Center Shared Resources (NIH grant P30CA013696). This work was supported by NIH grants R01CA166429 and R01CA217092 (to US), R01GM111749 (to WDW and DWB), and R21HL129063 (to GMKP); a Translational Research Program (TRP) grant from the Leukemia & Lymphoma Society (to US); and a research grant from the Taub Foundation for Myelodysplastic Syndromes (MDS) Research (to US). IAD was partially supported by a postdoctoral fellowship from the Association pour la Recherche sur le Cancer. JC was supported by the Einstein Training Program in Stem Cell Research from the Empire State Stem Cell Fund through New York State Department of Health contract C30292GG. US is the Diane and Arthur B. Belfer Scholar in Cancer Research of the Albert Einstein College of Medicine and is a Research Scholar of the Leukemia & Lymphoma Society. AF is supported by NIH grant R35CA210065.
Version 1. 10/30/2017
Electronic publication
Version 2. 12/01/2017
Print issue publication
Version 2. 11/20/2017
Fixed author's name in authorship note.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(12):4297–4313. https://doi.org/10.1172/JCI92504.
Contributor Information
Iléana Antony-Debré, Email: ileana_antony@hotmail.com.
Ananya Paul, Email: apaul@gsu.edu.
Joana Leite, Email: joana.dasilvaleite@einstein.yu.edu.
Kelly Mitchell, Email: kelly.mitchell@phd.einstein.yu.edu.
Hye Mi Kim, Email: hkim127@student.gsu.edu.
Tihomira I. Todorova, Email: tihomira.todorova@einstein.yu.edu.
Kenneth Huang, Email: khuang8@student.gsu.edu.
Arvind Kumar, Email: rasaayan@yahoo.com.
Boris Bartholdy, Email: bbarthol@einstein.yu.edu.
Swathi-Rao Narayanagari, Email: swathi-rao.n@einstein.yu.edu.
Jiahao Chen, Email: jiahao.chen@phd.einstein.yu.edu.
Alberto Ambesi-Impiombato, Email: ambesia@outlook.com.
Ioannis Mantzaris, Email: imantzar@montefiore.org.
Evripidis Gavathiotis, Email: evripidis.gavathiotis@einstein.yu.edu.
Amit Verma, Email: amit.verma@einstein.yu.edu.
Britta Will, Email: britta.will@einstein.yu.edu.
W. David Wilson, Email: wdw@gsu.edu.
Gregory M.K. Poon, Email: gpoon@gsu.edu.
Ulrich Steidl, Email: ulrich.steidl@einstein.yu.edu.
References
- 1. Facts 2014-2015. Leukemia & Lymphoma Society. https://www.lls.org/sites/default/files/file_assets/facts.pdf Accessed September 28, 2017.
- 2.Cancer Genome Atlas Research Network, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizuki M, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101(8):3164–3173. doi: 10.1182/blood-2002-06-1677. [DOI] [PubMed] [Google Scholar]
- 4.Vangala RK, et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003;101(1):270–277. doi: 10.1182/blood-2002-04-1288. [DOI] [PubMed] [Google Scholar]
- 5.Mueller BU, et al. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 2006;107(8):3330–3338. doi: 10.1182/blood-2005-07-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimwade D, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 8.Bonadies N, Pabst T, Mueller BU. Heterozygous deletion of the PU.1 locus in human AML. Blood. 2010;115(2):331–334. doi: 10.1182/blood-2009-03-212225. [DOI] [PubMed] [Google Scholar]
- 9.Lavallée VP, et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 2015;47(9):1030–1037. doi: 10.1038/ng.3371. [DOI] [PubMed] [Google Scholar]
- 10.Mueller BU, et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100(3):998–1007. doi: 10.1182/blood.V100.3.998. [DOI] [PubMed] [Google Scholar]
- 11.Steidl U, et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest. 2007;117(9):2611–2620. doi: 10.1172/JCI30525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steidl U, et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet. 2006;38(11):1269–1277. doi: 10.1038/ng1898. [DOI] [PubMed] [Google Scholar]
- 13.McKercher SR, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15(20):5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 14.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 15.Iwasaki H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106(5):1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staber PB, et al. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell. 2013;49(5):934–946. doi: 10.1016/j.molcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbauer F, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36(6):624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 18.Will B, et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med. 2015;21(10):1172–1181. doi: 10.1038/nm.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staber PB, et al. The Runx-PU.1 pathway preserves normal and AML/ETO9a leukemic stem cells. Blood. 2014;124(15):2391–2399. doi: 10.1182/blood-2014-01-550855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sive JI, Basilico S, Hannah R, Kinston SJ, Calero-Nieto FJ, Göttgens B. Genome-scale definition of the transcriptional programme associated with compromised PU.1 activity in acute myeloid leukaemia. Leukemia. 2016;30(1):14–23. doi: 10.1038/leu.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durual S, et al. Lentiviral PU.1 overexpression restores differentiation in myeloid leukemic blasts. Leukemia. 2007;21(5):1050–1059. doi: 10.1038/sj.leu.2404645. [DOI] [PubMed] [Google Scholar]
- 22.Munde M, Poon GM, Wilson WD. Probing the electrostatics and pharmacological modulation of sequence-specific binding by the DNA-binding domain of the ETS family transcription factor PU.1: a binding affinity and kinetics investigation. J Mol Biol. 2013;425(10):1655–1669. doi: 10.1016/j.jmb.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munde M, et al. Structure-dependent inhibition of the ETS-family transcription factor PU.1 by novel heterocyclic diamidines. Nucleic Acids Res. 2014;42(2):1379–1390. doi: 10.1093/nar/gkt955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham TH, et al. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res. 2013;41(13):6391–6402. doi: 10.1093/nar/gkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soeiro MN, Werbovetz K, Boykin DW, Wilson WD, Wang MZ, Hemphill A. Novel amidines and analogues as promising agents against intracellular parasites: a systematic review. Parasitology. 2013;140(8):929–951. doi: 10.1017/S0031182013000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW. Antiparasitic compounds that target DNA. Biochimie. 2008;90(7):999–1014. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei GH, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29(13):2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisenbeis CF, Singh H, Storb U. PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin lambda 2-4 enhancer. Mol Cell Biol. 1993;13(10):6452–6461. doi: 10.1128/MCB.13.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen B, Tanious FA, Wilson WD. Biosensor-surface plasmon resonance: quantitative analysis of small molecule-nucleic acid interactions. Methods. 2007;42(2):150–161. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 30. Nanjunda RM, Munde M, Liu M, Wilson WD. Real-time monitoring of nucleic acid interactions with biosensor surface plasmon resonance. In: Wanunu Y, ed. Methods for Studying DNA/Drug Interactions. Boca Raton, FL: CRC Press; 2011:92–122. [Google Scholar]
- 31.Stephens DC, Kim HM, Kumar A, Farahat AA, Boykin DW, Poon GM. Pharmacologic efficacy of PU.1 inhibition by heterocyclic dications: a mechanistic analysis. Nucleic Acids Res. 2016;44(9):4005–4013. doi: 10.1093/nar/gkw229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang S, Poon GMK, Wilson WD. Quantitative Investigation of Protein–Nucleic Acid Interactions by Biosensor Surface Plasmon Resonance. In: Leblanc BP, Rodrigue S, eds. DNA-Protein Interactions. New York, NY: Humana Press; 2015:313–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei D, Wilson WD, Neidle S. Small-molecule binding to the DNA minor groove is mediated by a conserved water cluster. J Am Chem Soc. 2013;135(4):1369–1377. doi: 10.1021/ja308952y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kodandapani R, et al. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380(6573):456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 35.Szymczyna BR, Arrowsmith CH. DNA binding specificity studies of four ETS proteins support an indirect read-out mechanism of protein-DNA recognition. J Biol Chem. 2000;275(37):28363–28370. doi: 10.1074/jbc.M004294200. [DOI] [PubMed] [Google Scholar]
- 36.Chen H, et al. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene. 1995;11(8):1549–1560. [PubMed] [Google Scholar]
- 37.Zhang DE, Hetherington CJ, Chen HM, Tenen DG. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14(1):373–381. doi: 10.1128/MCB.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Back J, Allman D, Chan S, Kastner P. Visualizing PU.1 activity during hematopoiesis. Exp Hematol. 2005;33(4):395–402. doi: 10.1016/j.exphem.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Weigelt K, Lichtinger M, Rehli M, Langmann T. Transcriptomic profiling identifies a PU.1 regulatory network in macrophages. Biochem Biophys Res Commun. 2009;380(2):308–312. doi: 10.1016/j.bbrc.2009.01.067. [DOI] [PubMed] [Google Scholar]
- 40.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37(4):382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 41.Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7 Suppl 1:S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margolin AA, Wang K, Lim WK, Kustagi M, Nemenman I, Califano A. Reverse engineering cellular networks. Nat Protoc. 2006;1(2):662–671. doi: 10.1038/nprot.2006.106. [DOI] [PubMed] [Google Scholar]
- 43.Guibal FC, et al. Identification of a myeloid committed progenitor as the cancer-initiating cell in acute promyelocytic leukemia. Blood. 2009;114(27):5415–5425. doi: 10.1182/blood-2008-10-182071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider RK, et al. Role of casein kinase 1A1 in the biology and targeted therapy of del(5q) MDS. Cancer Cell. 2014;26(4):509–520. doi: 10.1016/j.ccr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Illendula A, et al. Chemical biology. A small-molecule inhibitor of the aberrant transcription factor CBFbeta-SMMHC delays leukemia in mice. Science. 2015;347(6223):779–784. doi: 10.1126/science.aaa0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das BP, Boykin DW. Synthesis and antiprotozoal activity of 2,5-bis(4-guanylphenyl)furans. J Med Chem. 1977;20(4):531–536. doi: 10.1021/jm00214a014. [DOI] [PubMed] [Google Scholar]
- 47.Wenzler T, Boykin DW, Ismail MA, Hall JE, Tidwell RR, Brun R. New treatment option for second-stage African sleeping sickness: in vitro and in vivo efficacy of aza analogs of DB289. Antimicrob Agents Chemother. 2009;53(10):4185–4192. doi: 10.1128/AAC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Chai Y, Kumar A, Tidwell RR, Boykin DW, Wilson WD. Designed compounds for recognition of 10 base pairs of DNA with two at binding sites. J Am Chem Soc. 2012;134(11):5290–5299. doi: 10.1021/ja211628j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo P, et al. The thiophene “sigma-hole” as a concept for preorganized, specific recognition of G.C base pairs in the DNA minor groove. Chemistry. 2016;22(43):15404–15412. doi: 10.1002/chem.201603422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon H, Boss JM. PU.1 binds to a distal regulatory element that is necessary for B cell-specific expression of CIITA. J Immunol. 2010;184(9):5018–5028. doi: 10.4049/jimmunol.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lausen J, Liu S, Fliegauf M, Lübbert M, Werner MH. ELA2 is regulated by hematopoietic transcription factors, but not repressed by AML1-ETO. Oncogene. 2006;25(9):1349–1357. doi: 10.1038/sj.onc.1209181. [DOI] [PubMed] [Google Scholar]
- 52.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29(8):757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE. 2011;6(4):e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carvajal LA, Hamard PJ, Tonnessen C, Manfredi JJ. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 2012;26(14):1533–1545. doi: 10.1101/gad.184911.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12(4):1015–1027. doi: 10.1016/S1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 56.Matthews CP, et al. Dominant-negative activator protein 1 (TAM67) targets cyclooxygenase-2 and osteopontin under conditions in which it specifically inhibits tumorigenesis. Cancer Res. 2007;67(6):2430–2438. doi: 10.1158/0008-5472.CAN-06-0522. [DOI] [PubMed] [Google Scholar]
- 57.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.