Abstract
Retroviral vectors, including those derived from gammaretroviruses and lentiviruses, have found their way into the clinical arena and demonstrated remarkable efficacy for the treatment of immunodeficiencies, leukodystrophies, and globinopathies. Despite these successes, gene therapy unfortunately also has had to face severe adverse events in the form of leukemias and myelodysplastic syndromes, related to the semi-random vector integration into the host cell genome that caused deregulation of neighboring proto-oncogenes. Although improvements in vector design clearly lowered the risk of this insertional mutagenesis, analysis of potential genotoxicity and the consequences of vector integration remain important parameters for basic and translational research and most importantly for the clinic. Here, we review current assays to analyze biodistribution and genotoxicity in the pre-clinical setting and describe tools to monitor vector integration sites in vector-treated patients as a biosafety readout.
Keywords: gene therapy, gammaretroviral, lentiviral, vector, insertional mutagenesis, regulatory authorities, clinical translation, AAV
Main Text
Therapeutic approaches involving the permanent genetic engineering of human cells are progressively moving from a niche application in rare genetic disorders toward an extensive clinical use for a wide spectrum of diseases. In particular, ex vivo gene therapy based on autologous infusion of hematopoietic cells permanently engineered by retroviral/lentiviral vectors has emerged as one of the most promising approaches for the cure of hematological diseases and tumors.1 The awareness of being on the verge of a potential epochal advance in medical science is calling researchers and institutions to provide efficient tools to evaluate the outcome of gene therapies at pre-clinical and clinical levels. This, in turn, demands a thorough revision of some state of the art technologies to understand the informative potentials of the current assays and also to shed light on the caveats and limitations that need to be taken into account and addressed in the near future. In the first section, we review the main experimental requirements and available assays for pre-clinical evaluation of retroviral vector platforms as well as gene therapy strategies in general. Specifically, we describe the main steps of in vivo biodistribution studies and the assays to evaluate the genotoxic potential of a vector according to current recommendations by regulatory agencies. The second section describes the current technologies for vector integration site analysis in treated patients directly at the clinical trial stage. The discussion is focused on recent advances in insertion site (IS) retrieval protocols and how they can be efficiently combined with next-generation sequencing platforms. We evaluate the impact of these analyses on gene therapy safety assessment and on the collection of relevant basic biological information. We also emphasize the bioinformatic structural requirements and point out the risks associated with “overdatafication,” i.e., conversion of processes potentially affected by technical biases and turning them into biological data, which leads to misinterpretation of results. Overall, this article provides the reader with an up-to-date overview of the current technologies for the pre-clinical and clinical evaluation of gene therapy products based on permanent modification of target cells.
The Pre-clinical Work Package
Before a new retroviral vector can be applied to the first patients, regulating agencies, e.g., the European Medicines Agency (EMA) in Europe or the Food and Drug Administration (FDA) in the United States, require sponsors and investigators to provide sufficient safety data. Pre-clinical assessment of pharmacodynamics and toxicology of the new drug is covered by guidelines published by the responsible authorities.2, 3 These directives are often relatively vague in their formulation, as too much specificity would require recurrent modification of the guidelines in the attempt to keep pace with the developments in the field. In general, the guidelines inform initiators of new trials what information must be gathered, but often not how. Consequently, this section discusses the main aspects of how to assess the relevant data regarding genotoxicity. We focus on hematopoietic gene therapy and how some tests can be combined to address more than one issue at a time, potentially saving money, time, and animal lives.
First of all, the earlier contact with the responsible regulatory authority is established, the better. The standard recommendations are often similar, but not entirely the same everywhere, even when the gene therapeutic application will be applied in more than one country or across continents. One has to make sure that the experimental design will later satisfy the national and the international regulatory agencies, especially in the framework of multicenter, international trials. The contact persons assist the applicant throughout the process of pre-clinical data assessment. Closer examination of the guidelines points out that the majority of biological effects of an advanced therapy medicinal product (ATMP), which in hematopoietic gene therapy is the genetically modified autologous stem cell, is believed to be a result of the vector particle, the vector backbone, and the transgene. What first seems obvious can be important regarding the legal status of the ATMP. The guideline presumes that there will be no immunological reaction toward the transplanted cells and thereby relieves one from testing for this effect. However, regulations require testing for the presence, integrity, and persistence of the genetic information delivered to the cells, especially when using integrating vectors. In case the transgene might elicit an immune reaction, this has to be addressed as well. Sometimes, data for a similar vector type already have been acquired in the course of pre-clinical tests and might be supportive but rarely sufficient to allow immediate translation of a new vector into the clinics. The respective document of the EMA closes the introductory statements with the notion that “any decision about the adequacy of the pre-clinical data can only be made on a case-by-case basis.”2 Therefore, it is important to personally discuss these issues with the authorities.
Many investigators and sponsors might wonder if the vectors share a similar basic design and are intended to treat the same disease, is there also a similar set of pre-clinical assays? The answer is often yes. In the experimental design, pharmacodynamics and biodistribution can often be combined and will be discussed below. The other arm of investigation concerns genotoxicity, for which either certain in vivo or in vitro tests can be performed. In the European guideline, it is specifically stated that standard genotoxicity tests are not generally required. Nevertheless, in the light of adverse events due to insertional mutagenesis, the EMA published a reflection paper on the management of the risks associated with integrating vector technologies.4 The manuscript adequately explains the most important aspects about insertional mutagenesis and which pre-clinical tests can be used to assess it. It describes that integration studies might be needed, in case pediatric patients are treated and if the vector system has the capacity to permanently integrate into the genome. This is actually the most common case for retroviral vectors in current clinical trials. The situation can differ, however, with the cell type to be treated. The risk for insertional mutagenesis after gene transfer into a post mitotic tissue might be lower compared to the treatment of multipotent stem cells.5, 6, 7 Nevertheless, regulators often require an analysis of the integration spectrum in the relevant target cell. The paper further exemplifies what information is expected when the insertion site profile of a new vector is analyzed. The investigators are encouraged to report any relevant aberration of the clonal composition of a graft and the presence of integration clusters. In light of what the field learned from clonal skewings in previous gene therapy trials,8, 9, 10, 11 these considerations surely make sense. However, the reflection paper does not specifically state how to interpret the results from such studies or more precisely, what is regarded as safe. Pre-clinical integration studies are not predictive for the subsequently treated patients, but they can help to understand and adequately judge risk factors like integration hotspots and vector associated deregulation of neighboring genes.
In addition to integrating retroviral vectors, further vector systems including adeno-associated viral vectors have entered the clinical arena with impressive success. As these vectors lack an intrinsic integrase activity they are defined as non-integrating vectors, which mediate long-term transgene expression in post-mitotic or slow proliferating tissue, such as adult healthy liver, muscle, or brain from episomes. Nevertheless, vector DNA might become integrated at DNA damage sites. These events are rare (1 × 10−4 to 1 × 10−5) as indicated by comprehensive LAM-PCR-based genome analyses of clinical samples from adeno-associated virus (AAV) gene therapy trials and occur in a random fashion.12, 13 Interestingly, integration sites were not detected in oncogenic regions reported by Nault et al.12, 14 However, integration site analyses and functional tests ought to be implemented in analyses of non-integrating vectors given the limited number of patients and the currently short observation period.
Biodistribution and Pharmacodynamics
Proof of efficacy is crucial to every pre-clinical application. This point is often addressed by standard flow cytometric measurements or antibody-based detection assays. However, it is not enough to show efficient gene transfer alone, but one must also show that the therapy has an effect. Beyond the proof of concept in vitro, the integrity of the transgene and its therapeutic effect have to be demonstrated in a relevant animal model, unless otherwise justified. In line with the case-by-case character of the evaluation procedure, which animal model is relevant, or how many mice should be used for the analysis is not specified. This somehow depends on the effect to be observed and thus careful biometric planning should be applied. Often, proof-of-concept studies have been performed and published, so that the type of readout is known in advance. One has to be sure about the kind of response to be expected (dichotomous, continuous, or survival) and calculate the number of animals required to support an experimental conclusion.15, 16 It is recommended to perform the in vivo experiment as a dose escalation study, meaning transductions with increasing multiplicities of infection. Ideally, this will allow the documentation of the minimal dose required for a therapeutic effect and the possibility to observe putative signs of toxicity at high dosages. Later, before the first patient is treated, a recommendation of dosage is necessary and could be supported by the conclusion of these studies.
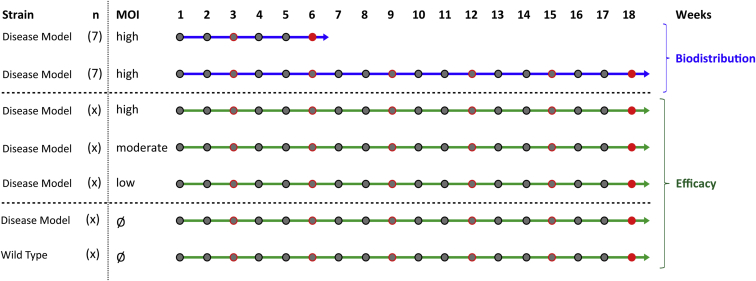
Regarding the experimental design of pharmacodynamics experiments, one usually starts with a cell culture experiment proving the effect of the genetic modification. Afterward, it is necessary to document, whether the treatment impedes with any normal function of the cell. Regarding hematopoietic stem cells (HSCs), the ability of these cells to differentiate normally and achieve long-term (>12 weeks) repopulation of the immune system has to be analyzed. It is wise to combine the experiments for pharmacodynamics with biodistribution studies, because the animals used in the efficacy analysis can also serve as backup for a vector screen by qPCR in different organs and vice versa. As displayed in Figure 1, we propose a time line of 6–18 weeks for these experiments. For both arms of the analysis, animal weight should be recorded weekly. Regular blood draws with documentation of white and red blood cells as well as platelet count should be performed every three weeks. The fate of the vector should be analyzed in both short- and long-term experimental settings. After 6 weeks, 10 different organs (brain, lung, liver, heart, kidney, gonads, spleen, thymus, bone marrow, and peripheral blood) can be isolated and examined for the presence of vector DNA by a qPCR copy number analysis. It is advantageous, if one can also provide flow cytometric data to analyze for donor cells (e.g., CD45.1/CD45.2 alloantigen) and present the combined measurements as a regression curve. One typically finds more engrafted cells in the hematopoietic organs (e.g., thymus, spleen, and bone marrow) with a correlating higher copy number. It is, however, normal to also observe a certain amount of cells in the lung after systemic injection, and these cells might remain there until the end of the analysis. At this time point, it is also possible to show normal differentiation by conventional surface-marker expression (granulocytes, monocytes, B cells, and T cells). This analysis is again repeated after 18 weeks to monitor the persistence of gene-modified cells in the hematopoietic organs and to provide evidence that gene-modified cells did not accumulate in non-hematopoietic organs, like the lung.
Figure 1.
Pre-clinical Pharmacodynamics and Biodistribution for Hematopoietic Gene Therapy
In blue, biodistribution includes observations of short- (6 weeks) and long-term effects (18 weeks). Body weight should be monitored weekly (gray dots), and blood should be taken every 3 weeks (red circles). At the time of end analysis (red dot), 10 different organs should be harvested and screened for vector presence by qPCR, sometimes in combination with flow cytometry. For pharmacodynamic studies (in green), the efficacy of the treatment and the lineage distribution can be monitored in peripheral blood over time and in the bone marrow at necroscopy. Different vector doses can be compared and analyzed with respect to wild-type and mock-treated animals.
For the efficacy and general toxicity studies, we recommend performing a dose escalation of at least three different multiplicities of infection (MOI) (low [1–5], moderate [10–30], and high [50–200]) during transduction. At any time (early and late), one can show that the gene modification follows a dose response. Combined with lineage stainings in flow cytometry, this is very convincing data to support the conclusion that the therapeutic effect does not interfere with hematopoiesis and the differentiation capacity of the cells. One should also include a non-treatment group and wild-type animals in the efficacy study to allow correlation of the observed effect with physiologic conditions. After 18 weeks, a full necroscopy should be performed. All macroscopic abnormalities and a thorough histopathologic analysis of several organs should be documented. Deciding which tissue types to analyze largely depends on the estimates regarding which organs possibly could be affected by a toxic impact of the treatment.
Genotoxicity
As mentioned previously, from the guideline’s point of view, general genotoxicity assays are not required for gene therapy.2 This is largely due to the nature of the available assays, of which none is predictive for the subsequent situation in the patient. Even the attempts to perform xenotransplantations of CD34-positive hematopoietic stem and progenitor cells into immune deficient mice, with the aim to humanize the mouse model, cannot overcome the principal limitations opposing a predictive value for subsequent patients. Whereas animal experiments are sensible for pharmacodynamics and biodistribution, the readout in terms of genotoxicity is debated controversially.17 Hence, the guidelines do not require any specific assay. In reality, however, regulators may request supporting data about the vector performance regarding the integration profile and insertional mutagenesis, especially in light of previous severe adverse events. In this chapter, we only describe possible in vitro and in vivo assays, which were practically useful. For a better description of the multitude of possible assays, please refer to the reviews of Rothe et al.18, 19
For analysis of genotoxicity in vivo, we propose using either the applicable disease model or a transplantation of human CD34-positive cells into immune-deficient mice. The purpose of this experiment is to determine the natural integration profile of the vectors and to possibly observe any kind of clonal imbalance after prolonged time. Either mice are monitored for an extended period (e.g., 12 months) or serial transplantation of the bone marrow can be accomplished to stress the hematopoietic compartment after 6 months. In any case, one should document the weight of the animals every week for the first 4 weeks and every 4 weeks thereafter. Animals developing a full blown leukemia rapidly lose weight and show signs of anemia, which often can be observed as pale feet. At the time of end analysis, a complete necroscopy should be performed. Any macroscopic abnormalities (thymus, lymph nodes, spleen, or liver) or findings in the histopathology (spleen, thymus, bone marrow, or enlarged lymph nodes) should be documented.20 Regarding insertion site analysis, it is highly recommended to keep an aliquot of the transfused material as a pre-transplantation (pre-TX) sample. With this sample, three important components of an insertion site analysis can be assessed. (1) The overall integration profile (frequency of intragenic insertions, distance to transcriptional start site, and to CpG islands), (2) any clusters of integrations, and (3) the occurrence of potential dangerous insertions near reported proto-oncogenes.21 Analysis of the pre-TX sample enables a comparison of the normal insertion site spectrum to that of repopulated long-term or even secondary transplanted animals. A conclusion could be that no relevant deviation from the polyclonal composition is observed over time. Details about a proper insertion site analysis, both for pre-clinical genotoxicity analysis and, even more importantly, for the patient follow-up after gene therapy, will be discussed in separate paragraph below.
Care must be taken during planning and interpretation of these mouse studies. The number of transplanted HSCs in mice is different from that in humans.22 The amount of long-term repopulating cells in the graft in combination with restrictions in the current linear amplification mediated (LAM)-PCR methodology can result in a reduced clonality even without an obvious selection for growth promoting insertions.23 This effect might even be aggravated when humanized mouse models are used. The stem cell nature and engraftment of cultured HSCs can strongly vary due to different culture conditions, leading to reduced clonality of gene modified cells, which is not caused by the gene therapy vector.24 The recently developed mouse models expressing human hematopoietic cytokines or with a mutation in c-kit might overcome this disadvantage due to better mimicking the niche for human HSCs or improved engraftment potential.25 Assuming a good engraftment, a possible outcome of a successful mouse genotoxicity study might conclude that no insertions in or near known proto-oncogenes are selected. In conjunction with physiologically normal health parameters and unobtrusive histopathologic findings, a careful statement regarding safety seems reasonable.
Another possibility to test the genotoxicity of a new vector in vivo is the use of a tumor prone mouse model like Cdkn2a−/−.26, 27 Here, even subtle differences in design of new self-inactivating (SIN)-lentiviral vectors can be uncovered. The normal endpoint in this case is always leukemia (also for the control mice) and the differences in median survival allow comparison of a test vector against other backbones with known mutagenic potential. We would recommend side-by-side comparison of mock transduced cells as a negative control, a vector with a strong viral promoter as a positive control and the new test vector. This model is extremely helpful to describe differences of genotoxic risks associated to a new vector. However, this strategy has the same deficiency as the other assays, in that it is not believed to be predictive for the outcome of future gene therapy in humans. Although, the test detected a genotoxic potential of SIN-lentiviral vectors, no adverse event has been observed with these vehicles in the clinic. Hence, the conclusion of such a vector comparison might be that a new vector is less prone to elicit enhancer mediated dysregulation or even aberrant splicing of genes compared to earlier vector generations.
Another useful test is the in vitro immortalization (IVIM) assay, first described by Modlich et al. in 2006.28 The assay is based on an observation by the lab of Jenkins and Copeland, who screened for enhancer mediated upregulation of proto-oncogenes like Mecom or Prdm16.29 Vector insertions into the Mecom and Prdm16 loci were involved in the severe adverse event of the first CGD gene therapy and were clinically relevant.30 For an IND (investigational new drug) application, a comparison of a new vector against a strong insertional mutagen like the LTR-driven gammaretroviral vector RSF91 can be performed. This vector contains promoter/enhancer sequences from spleen focus forming virus (SFFV) located in the long-terminal repeat (LTR) sequences. When murine lineage negative cells are transduced at high multiplicity of infections (starting cell number ≥ 1 × 105 cells and vector copy number above 3) and expanded for 2 weeks, the incidence of a replating phenotype at low-seeding densities (100 cells/well) is nearly 100%. Mock-transduced cells normally do not grow under these conditions. In contrast, clones from RSF91-transduced samples can be expanded even further and cultivated indefinitely. The incidence of positive plates can be quantified and reported together with the fitness of potentially immortalized clones. This fitness score is the frequency of cell growth on a 96-well plate. As cell numbers and proliferation behavior are monitored during the bulk culture, the effect of the transduction and possibly also the transgene on cell viability can also be analyzed. This assay can reproducibly measure the clinically most relevant factor of malignant insertional mutagenesis, i.e., the cis-activation of neighboring genes close to the integration site. Often, however, SIN-lentiviral vectors with weaker internal promoters do not reach a critical threshold for upregulation of neighboring genes in the IVIM assay. It remains to be determined whether more sensitive tests are needed to quantify differences in vector architecture or if the fact that no adverse event has been described in humans with SIN-lentiviral vectors is a biological truth reflected by the current setup of the assay. Of note, the IVIM assay measures myeloid transformation by insertional upregulation of Evi1 or Prdm16. In light of severe adverse events due to lymphoid leukemias that originate from lymphoid lineages, further assay systems, e.g., the assay described by Zhou et al. that specifically scores the LMO2 proto-oncogene activation, complement the IVIM.31
Integration Site Analysis
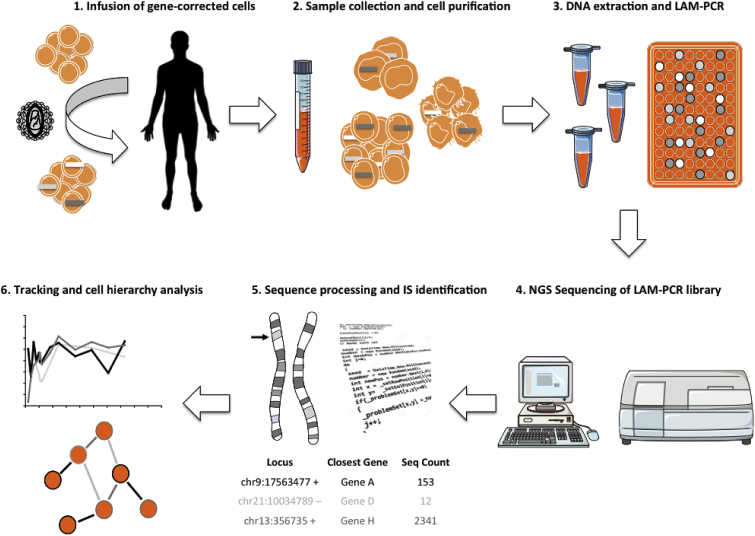
For the analysis of pre-clinical mouse studies and the follow-up of treated patients, the insertion sites and the relative frequency of transduced cell clones can be analyzed (Figure 2). All current techniques for integration site retrieval are based on in vitro enrichment of fragments containing vector-genome junctions combined with sequencing and bioinformatic processing for the retrospective identification of insertion sites. In the early days, LM (linker mediated)-PCR products were shotgun cloned into competent bacteria. The generated colonies were then picked and sequenced using the Sanger method. On average, 96 wells were analyzed per sample and the resulting sequences were manually processed and blasted on the genome of reference. In the clinical settings, this approach allowed the identification of hundreds of integration sites per patient and has been a powerful tool for the characterization of insertional mutagenesis events.32, 33, 34 Nonetheless, this technique was expensive, extremely time consuming and did not allow a comprehensive real time monitoring of the patients “integrome.” The arrival of next-generation sequencing (NGS) technologies provided a tool to perform high-throughput integration site analysis in a cost and time effective manner.35, 36, 37, 38, 39, 40 Importantly, together with the identification of vector insertions, these technologies allowed measurement of the relative abundance of each integration site according to sequencing read counts as a surrogate marker of the relative clonal size.
Figure 2.
Tracking Clonal Fate Dynamics In Vivo by Insertional Barcoding
The activity of collecting and analyzing integration sites for molecular tracking studies is based on the combination of in vitro cellular and molecular protocols and deep in silico data processing. Once LAM-PCR and bioinformatic pipelines are in place, the whole procedure may require 1–3 months from sample collection to the final data delivery depending on the availability of sequencing facilities in-house or to the outsourcing of NGS processing. Steps 2 to 4 are critical, as they may introduce contamination and collision events significantly affecting downstream analyses. Step 5 requires careful tuning of the bioinformatic pipeline in order to provide a proper set of filtered data as input to the final step 6, where extrapolation of biological information occurs.
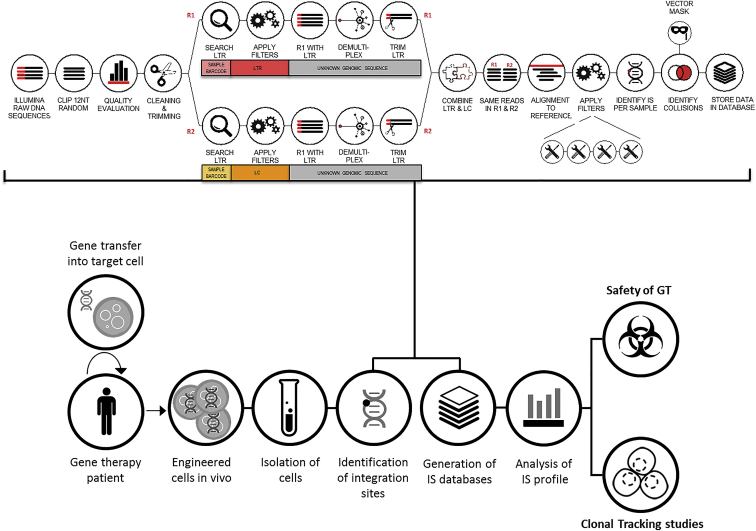
To date, LAM (linear amplification mediated)-PCR combined with Illumina (MiSeq or HiSeq) sequencing platform represent the state of the art for integration site retrieval. The LAM-PCR is a well-established technique and its protocol has been described in detail.41 To be effectively combined with the Illumina sequencing platform, LAM-PCR products must be subjected to an additional round of PCR to add Illumina adapters. These adapters are designed with a sequence complimentary to the two surface-bound amplification primers on the Illumina flow cells together with a sequence specific for the vector or linker cassette proximal to the binding site of the primers used for the last LAM-PCR step. It is advisable to add a 12-bp random sequence between these two segments for efficient cluster recognition upon Illumina sequencing and 6- to 8-bp barcode sequence for the sample identification in order to pool different samples in a single library. After Illumina sequencing, customized pipelines have been designed to process raw sequence data. The key steps involve data quality processing, barcode assignment to respective samples, trimming of vector and linker cassette sequences and mapping of the resulting sequences on the reference genome (Figure 3). The original localization of the vector insertion corresponds to the chromosomal position right at the end of the vector LTR sequence. The design of these pipelines has to take into account a series of technical issues that could affect the bona fide identification of vector integration sites: (1) a proper recognition of a complete LTR sequence before trimming is critical to assure that what is going to be mapped is not the result of a rearrangement occurring upon the genomic fragmentation step of LAM-PCR. (2) The barcode identification should be precise enough in order to allow a proper demultiplexing of the sequences and correctly assign a given mapped integration site to a specific sample. (3) The mapping procedure should allow efficient recognition of the variability of the alignment of a single insertion event around a small genomic window (usually an area of ±3 bp around the LTR end point, but could be even larger). These events are not rare occurrences and the technical reasons leading to these events remain to be clarified. The pipeline should provide a tool to distinguish between the identification of an individual integration site or of a cluster of independent insertional events mapping in the same narrow genomic area. (4) Sequences mapping to the same integration site should be pooled together and information regarding the sequence counts associated to each insertion should be stored for the analysis of clonal size.
Figure 3.
Schematic Representation of the Pipeline for IS Identification and Mapping for Safety and Clonal Tracking Studies on Samples for Gene Therapy Patients
Schematic representation of the pipeline for IS identification and mapping for safety and clonal tracking studies on samples from GT patients. Details of the main steps included in the informatic pipelines for Illumina paired-end sequence data processing of standard LAM-PCR products are reported in the workflow on the top of the figure (adapted from Leonardelli et al.44). The sequential order of the tasks and the nature of the filters considered for the bona fide identification of IS vary according to the pipelines developed by the different groups working in the field. LTR, long terminal repeat; LC, linker cassette; IS, insertion site; GT, gene therapy.
Once the integration sites are properly identified, the following analytical steps include the annotation of genomic features, like the presence of RefSeq or non-coding RNAs in the insertion site neighborhood. Other features, like chromatin conformation and gene expression levels can be linked to the insertion site analysis. However, one must remember that, in many cases, these characteristics are variable and cell-type specific. Therefore, inclusion of these features in the analytical pipeline should be carefully tuned according to the specific target cell of the study in order to avoid misinterpretation of the results.36 In this regard a valuable analytical approach should involve a thorough phenotypic characterization followed by a comprehensive insertional analysis of the cell product isolated from the target patient population before infusion. These data should then be cross-linked with information available on literature on chromatin accessibility of target cells (e.g., assay for transposase accessible chromatin with high-throughput sequencing [ATAC-seq], chromatin immunoprecipitation sequencing [ChIP-seq]) to create a baseline reference of the insertional profile of the engineered cells. This will provide a better understanding of the reasons driving in vitro integration preferences versus in vivo selection of insertion sites. Once a list of integration sites, each associated to its sequence count and neighboring features, has been created, a series of additional filtering steps are required to clean the results before further downstream analyses: (1) it is not uncommon to detect identical integration sites in two independent samples. Generally, it is thought that this could result from contamination occurring upon wet processing or during sequencing. Indeed, although one can assume that integration sites can be shared among different cell types of the same individual as the result of a differentiation from a common ancestor, it is questionable that a given insertion event could have occurred in the very same genomic position in cells from distinct individuals. As NGS becomes more and more applied to clinical and pre-clinical studies, the occurrence of so-called “collision” events has potentially increased. This is mainly due to the increased depth and intensity of the sequencing coupled with the requirements for wider multiplexing capacity for the simultaneous analysis of multiple samples. The bioinformatic processing pipeline design could disregard collision events from more than one independent individual or filters based on sequence count differences among independent patients could be applied in order to assign a specific insertion to a given individual. Importantly, collisions are often detected even among independent sequencing runs that are performed months apart from each other. Therefore, bioinformatic tools are required to compare the results of each new sequencing run with a previously generated insertion site database. Implementation of more complex barcoding strategies at the LAM-PCR level will most likely reduce the impact of these occurrences in the future. (2) Another filtering level could be needed for performing specific analyses. For example, studies based on the sharing of identical insertions found in different cell types or detected longitudinally in the same individual could be deeply affected by the purity of the isolated cell populations. To reduce the impact of biological contamination on the interpretation of the results from NGS, sequence reads-based filters could be applied to eliminate poorly represented insertions (e.g., removing integration sites with 3 or fewer sequencing reads). Then an additional filter could assign specific integrations only to a specific lineage or time point according to a preponderant sequence count as compared to other samples. In this case, identical integration will be considered as shared only when bearing comparable sequence counts among samples. This method is generally considered reasonable, but it is not exempt from technical flaws, particularly when dealing with studies involving longitudinal cell tracking as will be pointed out next.
The combination of NGS and insertion site retrieval technologies has significantly increased the amount of information available for the molecular tracking of gene-corrected cells infused into the patients. The level of detail reached by these analyses not only allows addressing issues regarding the safety of gene therapy but also sheds light on more general concepts of the biology and dynamics of gene-corrected cells in patients. These molecular tracking studies are based on the concept that each transduced cell that has successfully engrafted in an individual has become unequivocally marked by a unique integration site. Importantly, this permanent mark also will be inherited by the progeny of the target cells upon in vivo duplication/differentiation. Therefore, this “insertional barcoding” allows tracing the in vivo fate of infused cells. Each clone is identified by a univocal chromosomal localization and is commonly named by the closest gene flanking the relative integration site. The size of each clone is measured by means of its sequence counts and reported as the frequency of sequencing reads over the total. The population diversity can be estimated for each time point by combining the richness of integration sites and the evenness of their sequence count distribution. The survival of individual clones can be assessed by longitudinal resampling of the same integration sites overtime. If a clone expands significantly at the expense of the others, the sequence counts of the insertion site marking this clone will increase and as a consequence diversity of the relative population will decrease. Therefore, these analyses represent a powerful tool to monitor the stability of the population of genetically engineered cells.35, 39, 40 Despite the fact that sequencing read counts can provide a generally reliable estimate of population clonality, it should be kept in mind that they are the byproducts of extensive molecular processing of vector-genome junctions, which are exponentially amplified by several rounds of PCR. Thus, unless a consistent and self-evident clonal expansion is in place, the single clone size measurement could be affected by PCR-related biases, and the interpretation of these results should be taken with caution, particularly when performed on single clones and single time points. In this regard, different approaches have been developed based on fragmentation of the genomic DNA by sonication and enumeration of shear sites that belong to each IS sequence.42 Although theoretically valuable, the use of this technology is limited by the fact that the number of cells that compose a given clone could far exceed the number of fragments of different lengths generated upon fragmentation in proximity of a given IS, which results in saturation of the available diversity of shear sites. A more efficient way to overcome the quantification biases introduced by the PCR steps would be to tag genomic fragments with random barcodes as unique molecular identifiers prior to exponential amplification. Others and we have shown that inclusion of a string of random nucleotides upon adaptor ligation improves the IS quantification potential.43, 44
Insertional barcoding can be also exploited to study the hierarchical relationships between different cell types through the sharing of identical insertion sites. This type of analysis is based on the concept that if two or more clones belonging to different lineages share the same integration site, they should have derived from a common progenitor. In the context of gene therapy clinical trials based on the autologous gene correction of HSCs, the observation of identical insertions among CD34-positive cells, myeloid cells, and lymphoid cells is commonly considered a testimony of the efficient engraftment of multipotent progenitors and the level of multilineage sharing of integration sites in each individual cell type is generally correlated with its precursor potential.39, 40 An efficient application of this type of analysis was recently shown for insertional tracking of the hierarchical relationships between human T cell subtypes.45
To correctly implement integration site analysis as a molecular tracking tool for the study of in vivo biology and dynamics of engineered cells, data derived from NGS must be carefully processed according to the filtering procedure mentioned above with the aim of limiting false positives. Nonetheless, an overapplication of these filters may lead to significantly masking the biological information that lies beneath the tracking of integration sites. For example, in longitudinal studies, identical integration sites are often detected at multiple time points but carry a very different sequence read count at each follow-up. This could be the result of a variability intrinsic to the LAM-PCR procedure, of the known effects of subsampling (only a few milliliters of blood are collected at each time point), or of a true temporal fluctuation in clonal size. Similarly, it is common knowledge that clone size may vary at various stages of differentiation. Indeed, it is not unexpected to find differences between the sequence counts of an integration site belonging to a precursor (e.g., a progenitor in the bone marrow) versus a more differentiated lineage that may have gone through a physiological clonal burst (e.g., an effector T cell). Thus, in this context, applying a filter merely based on discarding insertion sites with low sequencing reads or assigning a given integration only to a specific lineage based on the uneven distribution of sequence counts, could lead to significant flaws in the analysis. The application of extensive bioinformatic filters should be justified in terms of overcoming technical issues and with a view toward the potential biological meaning of the collected data. Therefore, for molecular tracking studies, it is generally advisable to test the consistency of the results over different layers of data filtering and, when possible, exploit a weighted system designed to employ the entire integration site dataset.
Overall, the advent of NGS technologies has significantly increased the analytical power of insertion site retrieval, bringing the number of detectable integration sites from a few hundred to tens of thousands, while lowering the time and costs required for these studies. Although this represented a notable technical advance in the assessment of safety and efficacy of gene therapy, researchers that approach this technology should interpret their results with caution. Indeed, two considerations should be made on the future direction of these studies: (1) putting a strong effort in retrieving more integration sites by progressively increasing the intensity of sequencing may not necessarily be rewarding. This is because, like in many other NGS applications, the assumption that “a bigger dataset equals more information” is not always true. One should remember, for example, that the common insertion sites associated with oncogenic events as well as the general insertional profile of the different integrating vector platforms were all highlighted with good efficiency already in the early studies based on Sanger sequencing with a few hundred integration sites available and that many of the following high-throughput studies based on NGS were basically confirmatory of these results. Thus, much of the effort should now be advanced from the mere generation of bigger datasets to instead the formulation of new, sound and biologically relevant questions that NGS combined with insertion site retrieval could help to address (e.g., studying how target cell states affect insertional distribution and how this might affect in vivo IS selection). (2) Indeed, the sudden availability of a large amount of information within a relatively small time window and with cost-effective experiments has led to the common trend of overinterpretation of the results. Researchers are now more tempted to spend their resources for the acquisition of big datasets with the risk that many inferences are forced through a retrospective association of the data that fit a certain hypothesis without a strong knowledge of the statistical and technical constraint inherent to the analysis of NGS information. A typical case of potential overinterpretation in the field of integration site analyses involves the investigations of specific genomic areas surrounding an integration site with slightly overrepresented sequencing reads. If more than one insertion is found proximal to a locus where a specific genomic feature is present, investigators could be immediately tempted to retrospectively infer that a perturbation of this feature has occurred and that this event could have led to an increased fitness of the observed clone. Often, however, not only the gene closest to the integration but also surrounding genes might be affected. Furthermore, the overrepresentation of an integration site in terms of sequencing reads could be merely the result of LAM-PCR biases.23 Additionally, the increased resolution of integration site analysis by NGS has made it much more likely to “fish out” more than one insertion occurring into a locus of particular interest. The definition of common insertion sites, however, is affected by the increased sequencing depth. Hence, the probability of finding different insertions in the same genomic area is also higher. Unfortunately, this aspect is often overlooked. Conversely, the results of these observations are often overemphasized and rewarded with attention by the scientific community, although in many cases they represent only circumstantial evidence if not merely secondary epiphenomenon. Statistical and mathematical approaches are currently being designed to overcome these issues and will be of paramount importance for the accurate interpretation of NGS-based integration site analysis data, allowing specific events of interest to be properly highlighted.
Conclusions
In this article, the most important aspects regarding pre-clinical safety testing as well as analytical methods for patient follow-up during or after trials were discussed. We addressed issues involving regulatory aspects for pre-clinical evaluation of new vectors and suggestions for the careful interpretation of deep sequencing data. A significant number of new gene therapy trials are currently entering the clinical arena, thus the number of patients treated with gene therapy will continue to rise. The increase in treated individuals goes hand in hand with the responsibility for thorough safety testing before and after the vector is used in patients. Regarding the risk factors associated with integrating viral vectors, the most dramatic progress was made after in-depth analyses of the processes that caused severe adverse events helped us to better understand and prevent these incidents. We hope and anticipate that current safety testing will further improve and become predictive for humans.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB738, Cluster of Excellence REBIRTH [EXC 62/1], SFB670, and KFO 286), the Bundesministerium für Bildung und Forschung (BMBF, IFB-Tx, and PidNet), and the European Union (FP7 project CELLPID and SCIDNET). The work of L. Biasco was supported by the Gene Therapy Program of Dana-Farber/Boston Children’s Cancer and Blood Disorders Center (Boston, MA) and the UCL Great Ormond Street Institute of Child Health, Faculty of Population Health Sciences (London, UK). The authors thank M. Morgan for critically reading the manuscript.
References
- 1.Naldini L. Ex vivo gene transfer and correction for cell-based therapies. Nat. Rev. Genet. 2011;12:301–315. doi: 10.1038/nrg2985. [DOI] [PubMed] [Google Scholar]
- 2.European Medicines Agency. (2008). Guideline on the non-clinical studies required before first clinical use of gene therapy medicinal products. Committee for the medicinal products for human use (CHMP). http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/09/WC500003942.pdf.
- 3.FDA (2013). Guidance for industry: preclinical assessment of investigational cellular and gene therapy products. U.S. Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research. https://www.fda.gov/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/CellularandGeneTherapy/ucm376136.htm.
- 4.Aiuti A., Cossu G., de Felipe P., Galli M.C., Narayanan G., Renner M., Stahlbom A., Schneider C.K., Voltz-Girolt C. The committee for advanced therapies’ of the European Medicines Agency reflection paper on management of clinical risks deriving from insertional mutagenesis. Hum. Gene Ther. Clin. Dev. 2013;24:47–54. doi: 10.1089/humc.2013.119. [DOI] [PubMed] [Google Scholar]
- 5.Bartholomae C.C., Arens A., Balaggan K.S., Yáñez-Muñoz R.J., Montini E., Howe S.J., Paruzynski A., Korn B., Appelt J.U., Macneil A. Lentiviral vector integration profiles differ in rodent postmitotic tissues. Mol. Ther. 2011;19:703–710. doi: 10.1038/mt.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newrzela S., Cornils K., Li Z., Baum C., Brugman M.H., Meyer J., Hartmann S., Hansmann M.L., Fehse B., von Laer D. Resistance of mature T cells to oncogene transformation. Blood. 2008;112:2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- 7.Rittelmeyer I., Rothe M., Brugman M.H., Iken M., Schambach A., Manns M.P., Baum C., Modlich U., Ott M. Hepatic lentiviral gene transfer is associated with clonal selection, but not with tumor formation in serially transplanted mice. Hepatology. 2013;58:397–408. doi: 10.1002/hep.26204. [DOI] [PubMed] [Google Scholar]
- 8.Hacein-Bey-Abina S., von Kalle C., Schmidt M., Le Deist F., Wulffraat N., McIntyre E., Radford I., Villeval J.L., Fraser C.C., Cavazzana-Calvo M., Fischer A. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 9.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 10.Stein S., Ott M.G., Schultze-Strasser S., Jauch A., Burwinkel B., Kinner A., Schmidt M., Krämer A., Schwäble J., Glimm H. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 11.Braun C.J., Boztug K., Paruzynski A., Witzel M., Schwarzer A., Rothe M., Modlich U., Beier R., Göhring G., Steinemann D. Gene therapy for Wiskott-Aldrich syndrome--long-term efficacy and genotoxicity. Sci. Transl. Med. 2014;6:227ra33. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- 12.D'Avola D., López-Franco E., Sangro B., Pañeda A., Grossios N., Gil-Farina I., Benito A., Twisk J., Paz M., Ruiz J. Phase I open label liver-directed gene therapy clinical trial for acute intermittent porphyria. J Hepatol. 2016;65:776–783. doi: 10.1016/j.jhep.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Kaeppel C., Beattie S.G., Fronza R., van Logtenstein R., Salmon F., Schmidt S., Wolf S., Nowrouzi A., Glimm H., von Kalle C. A largely random AAV integration profile after LPLD gene therapy. Nat Med. 2013;19,:889–891. doi: 10.1038/nm.3230. [DOI] [PubMed] [Google Scholar]
- 14.Nault J.C., Datta S., Imbeaud S., Franconi A., Mallet M., Couchy G., Letouzé E., Pilati C., Verret B., Blanc J.F. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47:1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- 15.Dupont, W.D., and Plummer, W.D., Jr. (2014). PS: power and sample size calculation. http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize.
- 16.Zodpey S.P. Sample size and power analysis in medical research. Indian J. Dermatol. Venereol. Leprol. 2004;70:123–128. [PubMed] [Google Scholar]
- 17.Gonin P., Buchholz C.J., Pallardy M., Mezzina M. Gene therapy bio-safety: scientific and regulatory issues. Gene Ther. 2005;12(Suppl 1):S146–S152. doi: 10.1038/sj.gt.3302629. [DOI] [PubMed] [Google Scholar]
- 18.Rothe M., Modlich U., Schambach A. Biosafety challenges for use of lentiviral vectors in gene therapy. Curr. Gene Ther. 2013;13:453–468. doi: 10.2174/15665232113136660006. [DOI] [PubMed] [Google Scholar]
- 19.Rothe M., Schambach A., Biasco L. Safety of gene therapy: new insights to a puzzling case. Curr. Gene Ther. 2014;14:429–436. doi: 10.2174/1566523214666140918110905. [DOI] [PubMed] [Google Scholar]
- 20.Li Z., Modlich U., Anjali M. Leukemia diagnosis in murine bone marrow transplantation models. Methods Mol. Biol. 2009;506:311–329. doi: 10.1007/978-1-59745-409-4_21. [DOI] [PubMed] [Google Scholar]
- 21.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy K.F. Marrow frequency of rat long-term repopulating cells: evidence that marrow hematopoietic stem cell concentration may be inversely proportional to species body weight. Blood. 2003;101:3431–3435. doi: 10.1182/blood-2002-10-3026. [DOI] [PubMed] [Google Scholar]
- 23.Brugman M.H., Suerth J.D., Rothe M., Suerbaum S., Schambach A., Modlich U., Kustikova O., Baum C. Evaluating a ligation-mediated PCR and pyrosequencing method for the detection of clonal contribution in polyclonal retrovirally transduced samples. Hum. Gene Ther. Methods. 2013;24:68–79. doi: 10.1089/hgtb.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haemmerle R., Phaltane R., Rothe M., Schröder S., Schambach A., Moritz T., Modlich U. Clonal dominance with retroviral vector insertions near the ANGPT1 and ANGPT2 genes in a human xenotransplant mouse model. Mol. Ther. Nucleic Acids. 2014;3:e200. doi: 10.1038/mtna.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosgun K.N., Rahmig S., Mende N., Reinke S., Hauber I., Schäfer C., Petzold A., Weisbach H., Heidkamp G., Purbojo A. Kit regulates HSC engraftment across the human-mouse species barrier. Cell Stem Cell. 2014;15:227–238. doi: 10.1016/j.stem.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Montini E., Cesana D., Schmidt M., Sanvito F., Bartholomae C.C., Ranzani M., Benedicenti F., Sergi L.S., Ambrosi A., Ponzoni M. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cesana D., Ranzani M., Volpin M., Bartholomae C., Duros C., Artus A., Merella S., Benedicenti F., Sergi Sergi L., Sanvito F. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol. Ther. 2014;22:774–785. doi: 10.1038/mt.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modlich U., Bohne J., Schmidt M., von Kalle C., Knöss S., Schambach A., Baum C. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du Y., Jenkins N.A., Copeland N.G. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott M.G., Schmidt M., Schwarzwaelder K., Stein S., Siler U., Koehl U., Glimm H., Kühlcke K., Schilz A., Kunkel H. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 31.Zhou S., Fatima S., Ma Z., Wang Y.-D., Lu T., Janke L.J., Du Y., Sorrentino B.P. Evaluating the safety of retroviral vectors based on insertional oncogene activation and blocked differentiation in cultured thymocytes. Mol. Ther. 2016;24:1090–1099. doi: 10.1038/mt.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deichmann A., Hacein-Bey-Abina S., Schmidt M., Garrigue A., Brugman M.H., Hu J., Glimm H., Gyapay G., Prum B., Fraser C.C. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aiuti A., Cassani B., Andolfi G., Mirolo M., Biasco L., Recchia A., Urbinati F., Valacca C., Scaramuzza S., Aker M. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J. Clin. Invest. 2007;117:2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarzwaelder K., Howe S.J., Schmidt M., Brugman M.H., Deichmann A., Glimm H., Schmidt S., Prinz C., Wissler M., King D.J. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J. Clin. Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G.P., Berry C.C., Malani N., Leboulch P., Fischer A., Hacein-Bey-Abina S., Cavazzana-Calvo M., Bushman F.D. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115:4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biasco L., Ambrosi A., Pellin D., Bartholomae C., Brigida I., Roncarolo M.G., Di Serio C., von Kalle C., Schmidt M., Aiuti A. Integration profile of retroviral vector in gene therapy treated patients is cell-specific according to gene expression and chromatin conformation of target cell. EMBO Mol. Med. 2011;3:89–101. doi: 10.1002/emmm.201000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaspar H.B., Cooray S., Gilmour K.C., Parsley K.L., Adams S., Howe S.J., Al Ghonaium A., Bayford J., Brown L., Davies E.G. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 2011;3:97ra79. doi: 10.1126/scitranslmed.3002715. [DOI] [PubMed] [Google Scholar]
- 38.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt M., Schwarzwaelder K., Bartholomae C., Zaoui K., Ball C., Pilz I., Braun S., Glimm H., von Kalle C. High-resolution insertion-site analysis by linear amplification−mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 42.Berry C.C., Gillet N.A., Melamed A., Gormley N., Bangham C.R.M., Bushman F.D. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28:755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Firouzi S., López Y., Suzuki Y., Nakai K., Sugano S., Yamochi T., Watanabe T. Development and validation of a new high-throughput method to investigate the clonality of HTLV-1-infected cells based on provirus integration sites. Genome Med. 2014;6:46. doi: 10.1186/gm568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leonardelli L., Pellin D., Scala S., Dionisio F., Basso-Ricci L., Cittaro D., Di Serio C., Aiuti A., Biasco L. Computational pipeline for the identification of integration sites and novel method for the quantification of clone sizes in clonal tracking studies. Mol. Ther. 2016;24:S212. [Google Scholar]
- 45.Biasco L., Scala S., Basso Ricci L., Dionisio F., Baricordi C., Calabria A., Giannelli S., Cieri N., Barzaghi F., Pajno R. In vivo tracking of T cells in humans unveils decade-long survival and activity of genetically modified T memory stem cells. Sci. Transl. Med. 2015;7:273ra13. doi: 10.1126/scitranslmed.3010314. [DOI] [PubMed] [Google Scholar]