Abstract
Hypoxia and dysregulated metabolism are defining features of solid tumors. How cancer cells adapt to low O2 has been illuminated by numerous studies, with “reprogrammed” metabolism being one of the most important mechanisms. This metabolic reprogramming not only promotes cancer cell plasticity, but also provides novel insights for treatment strategies. As the most studied O2 “sensor,” hypoxia-inducible factor (HIF) is regarded as an important regulator of hypoxia-induced transcriptional responses. This minireview will summarize our current understanding of hypoxia-induced changes in cancer cell metabolism, with an initial focus on HIF-mediated effects, and will highlight how these metabolic alterations affect malignant phenotypes.
Keywords: cancer, glucose, glutamine, hypoxia, lipid metabolism, metabolism, IDH, ROS
Introduction
Molecular oxygen (O2) is a key nutrient required for aerobic metabolism to maintain intracellular bioenergetics and as a substrate in numerous organic and inorganic reactions. Hypoxia, defined by a deficiency in the amount of tissue O2 levels, occurs in a variety of physiological as well as pathological conditions. Significant research interest in variable O2 availability in cancers can be traced to the early 20th century, when Otto Warburg found that unlike most normal tissues, cancer cells preferentially “ferment” glucose to pyruvate and then lactate even in the presence of sufficient O2 to support mitochondrial metabolism (the “Warburg effect”). Although the underlying mechanisms of this observation were not clear and numerous subsequent studies demonstrated certain limitations in this theory (especially the hypothesis that cancer cells develop a defect in mitochondria that leads to impaired aerobic respiration), it opened a new territory of investigating how cancer cells rewire metabolism to adapt to changes in O2 levels. When facing hypoxia, cells modulate a number of conserved molecular responses, including those regulated by hypoxia-inducible factors (HIFs),3 endoplasmic reticulum (ER) stress responses, mechanistic target of rapamycin signaling, autophagy, and others. These processes promote altered metabolism to match O2 supply. In this minireview, we will discuss hypoxia responses according to HIF-dependent and -independent pathways and how they impact progression of solid tumors.
HIF-dependent metabolic reprogramming
The HIF hydroxylase system
The HIF system itself has been reviewed extensively elsewhere (1, 2). Initially identified as a transcriptional regulator bound to a hypoxia-response element (HRE) of the erythropoietin (EPO) gene to promote EPO production, it readily became apparent that this pathway operated much more widely, and it is currently recognized as a key modulator of the transcriptional response to hypoxic stress. Briefly, HIFs are heterodimeric transcription factors consisting of α and β subunits. Three HIFα isoforms exist in the mammalian genome (1α, 2α, and 3α), of which HIF1α and HIF2α are the best characterized. HIFα subunits heterodimerize with stable HIF1β (or ARNT), recognize, and then bind to HREs ((G/A)CGTG) throughout the genome to regulate downstream gene expression. Hypoxia-inducible behavior is conferred by the HIFα subunits, the protein abundance and transcriptional activity of which are regulated by O2-dependent prolyl and asparaginyl hydroxylation. In well-oxygenated environments, HIFα subunits are hydroxylated by prolyl hydroxylases (PHDs) and factor-inhibiting HIF (FIH), and in turn they are targeted for proteosomal degradation by an E3 ubiquitin ligase, the von Hippel-Lindau protein (pVHL) complex. In hypoxic conditions, PHD and FIH activities are diminished, and HIFα proteins are stabilized, which consequently induces transcription of thousands of genes supplying adaptive functions, such as vascular endothelial growth factor (VEGF), carbonic anhydrase IX (CAIX), EPO, and others.
HIF regulation of glucose metabolism
Glycolysis
Because O2 serves as an electron acceptor in oxidative phosphorylation, a central adaptation to hypoxia is a shift toward non-oxidative forms of carbon metabolism and ATP production, such as anaerobic glycolysis (Fig. 1A). This process is altered in cancer or other rapidly-dividing cells because of the high demand for glycolytic intermediates for macromolecular synthesis. HIF1α contributes to this shift by promoting the expression of genes at almost every step in central carbon metabolism, such as hexokinases (HK1 and HK2), phosphofructokinase 1 (PFK1), and phosphoglycerate kinase 1 (PGK1) (3). Glucose transporters GLUT1 and GLUT3 required for initial glucose internalization are also transcriptionally regulated by HIF1α (4, 5), which guarantees adequate glucose uptake and rapid energy production that compensates for its low efficiency. In addition, an end product of glycolysis, i.e. lactate, is efficiently removed from the cell through the action of HIF1α-inducible plasma membrane monocarboxylate transporter 4 (MCT4) (6). Taken together, HIF impacts both the glycolytic pathway itself and ancillary processes that support it.
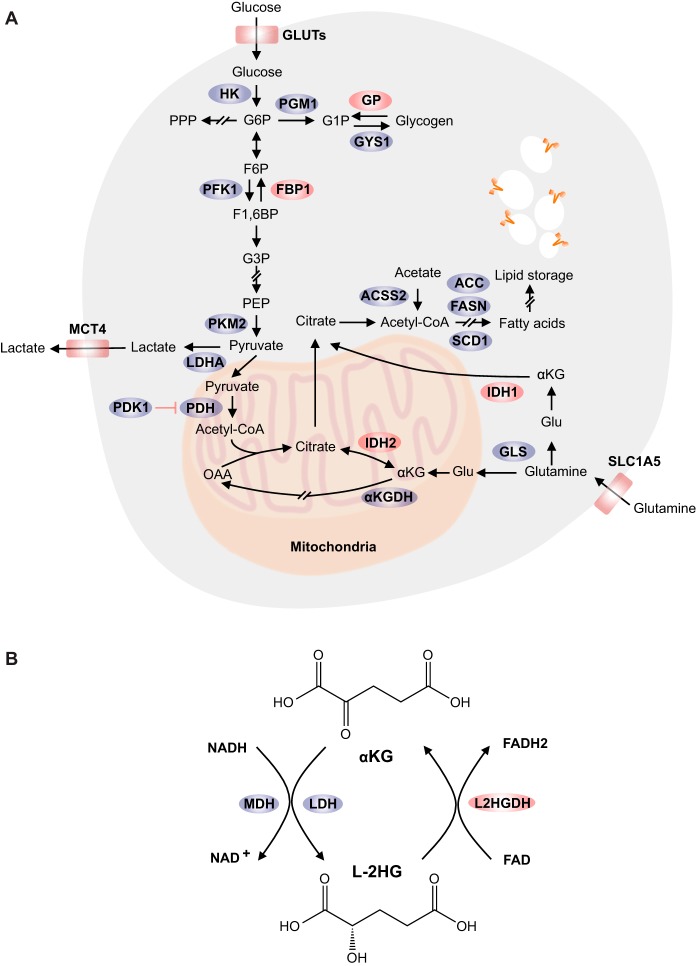
Figure 1.
Metabolic reprogramming in cancer cells under hypoxia. A, low O2 levels promote glucose uptake, glycolysis, lactate secretion, glutamine import, and glutaminolysis. Glucose can be stored as glycogen macromolecules, which are essential to maintain ATP levels and redox balance in hypoxic conditions. In addition, glycolytic enzymes FBP1 and PKM2 regulate HIFα activity through direct physical interaction. Moreover, cells exhibit increased rates of reductive carboxylation upon O2 deficiency. For lipid metabolism, hypoxic cells generate citrate from glutamine, and acetyl-CoA from acetate to compensate for decreased supply of these molecules from the TCA cycle. SCD1 activity is affected directly by changes in O2 availability. However, hypoxia results in increased lipid uptake, which counteracts the effects of SCD1 inhibition. The following genes are regulated by HIF: GLUTs (including GLUT1 and GLUT3), HK, PGM1, GYS1, PFK1, FBP1, PKM2, LDHA, PDK1, PDH, and MCT4. Also ACSS2, ACC, FASN, SCD1, IDH, and αKGDH protein expression/activity is directly or indirectly regulated by low O2 levels. One-headed arrows indicate single-direction reactions, and two-headed arrows represent those that are reversible. In addition, arrows with slashes indicate multistep reactions. B, enzymatic and cofactor requirements for l-2HG metabolism. The following abbreviations and names are used in figure: GLUT, glucose transporter; HK, hexokinase; PPP, pentose phosphate pathway; PGM1, phosphoglucomutase 1; GYS1, glycogen synthase 1; GP, glycogen phosphorylase; G6P, glucose 6-phosphate; G1P, glucose 1-phosphate; F6P, fructose 6-phosphate; F1,6BP, fructose-1,6-bisphosphatase; FBP1, fructose-1,6-bisphosphatase 1; G3P, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate; PKM2, pyruvate kinase M2; LDHA, lactate dehydrogenase A; MCT4, monocarboxylate transporter 4; PDH, pyruvate dehydrogenase; PDK1, pyruvate dehydrogenase kinase 1; OAA, oxaloacetate; IDH, isocitrate dehydrogenase; αKG, α-ketoglutarate; αKGDH, α-ketoglutarate dehydrogenase; Glu, glutamate; GLS, glutaminase; SLC1A5, solute carrier family 1 (neutral amino acid transporter), member 5; ACSS2, acyl-CoA synthetase short-chain family member 2; ACC, acetyl-CoA carboxylase; FASN, fatty-acid synthase; SCD1, stearoyl-CoA desaturase 1; MDH, malate dehydrogenase; L2HGDH, l-2-hydroxyglutarate dehydrogenase.
Interestingly, although it is well-known that HIF activates genes encoding glucose metabolic enzymes, recent evidence indicates that some enzymes in turn affect HIF activity through direct physical interaction. One important example is pyruvate kinase, which catalyzes the transfer of a phosphate group from phosphoenolpyruvate to adenosine diphosphate (ADP), yielding one molecule each of pyruvate and ATP. As the last step in glycolysis, pyruvate kinase is a key enzyme that determines glycolytic flux. HIF1α induces transcription of the PKM gene (7), which encodes PKM1 and PKM2 isoforms. It has been extensively reported that cancer cells replace PKM1 with the less active PKM2, thereby reducing pyruvate production and redirecting glycolytic carbon to biosynthetic pathways to support cell growth (8). Importantly, Luo et al. (7) showed that PKM2, but not PKM1, interacts directly with HIF1α subunits and promotes transactivation of HIF target genes by enhancing HIF1α binding and p300 recruitment to HREs, at least partially explaining the function of PKM2 in cancer cells. This observation has more recently been extended to macrophages (9): lipopolysaccharide, which initiates immune responses found in endotoxemia, induces PKM2 expression, enhancing the “Warburg effect” and shifting glucose metabolism away from oxidative phosphorylation toward a glycolytic program. This metabolic adaptation helps macrophages meet the increased demand for biosynthetic precursors required for mounting an immune response. Similarly, PKM2 interacts with HIF1α in activated macrophages to promote IL-1β production (9).
Another important regulatory enzyme is the gluconeogenic fructose-1,6-bisphosphatase 1 (FBP1). Li et al. (10) found FBP1 is uniformly depleted in over 600 clear cell renal cell carcinoma (ccRCC) tumors examined, in which pVHL is lost and HIF activity is constitutively maintained at high levels. Importantly, FBP1 opposes tumor growth not only though its enzymatic function by antagonizing glycolytic flux and inhibiting the “Warburg effect,” but also by inhibiting nuclear HIF function via direct interaction with the HIFα “inhibitory domain” (10). These two examples demonstrate complex regulation between HIF and its transcriptional targets and provide potential alternative therapeutic strategies (apart from targeting HIF directly) in tumors addicted to HIF signaling. Finally, whether and how other metabolic enzymes affect HIF activity is under active investigation.
Glycogen metabolism
Intracellular glucose is stored in the form of glycogen, a macromolecule essential for energy supply and glucose homeostasis (Fig. 1A) (11). Glycogen accumulation has been described in various cancer cells, although its abundance varies greatly across tumor types (12). Along with lipid deposition, glycogen accumulation contributes to the “clear cell” phenotype in a subset of breast, kidney, and ovary cancers (13). As with glycolysis, genes involved in glycogen biosynthesis have been identified as HIF targets in both normal and cancer cells, including phosphoglucomutase 1 (PGM1), protein phosphatase 1 regulatory subunit 3C (PPP1R3C), glycogen synthase 1 (GYS1), UTP–glucose-1-phosphate uridylyltransferase (UGP2), and 1,4-α-glucan-branching enzyme (GBE1) (14–17). The induction of these proteins correlates with significant increase in glycogen buildup in cells exposed to hypoxia. In addition, glycogenolysis allows these “hypoxia-preconditioned” cells to confront and survive glucose deprivation.
Several studies investigated the role of glycogen metabolism in the cancer setting. In a glioblastoma model, hypoxia induces GYS1 expression and subsequent glycogen accumulation (17). Interestingly, however, the glycogen degradation enzyme glycogen phosphorylase (PYGL) is also regulated by hypoxia. The authors demonstrated that GYS1 increases rapidly in hypoxia, and then declines, whereas PYGL climbs more slowly and reaches maximal levels at a later time point. This expression pattern is consistent with cellular glycogen levels, which are initially elevated and then gradually decline. In addition, PYGL knockdown leads to a p53-dependent induction of senescence and impaired tumorigenesis (17). Mechanistically, PYGL depletion is associated with glycogen deposition as well as increased reactive oxygen species (ROS), potentially due to a reduction in glucose carbon shuttling to the pentose phosphate pathway and impaired NADPH production. Furthermore, Lee et al. (18) show that the glycogen phosphorylase inhibitor CP-320626 reduces proliferation and increases apoptosis in pancreatic tumor cells, probably by limiting glucose oxidation, as well as de novo nucleic acid and fatty acid synthesis. These two studies provide a rationale to target glycogen degradation pathways in cancer cells, although more complete underlying mechanisms need to be defined. Finally, HIF-dependent regulation of glycogen synthesis has also been described in human ccRCC lines RCC4 and 786-O, which are characterized by pVHL loss and constitutive HIF activation (15). However, it remains to be determined whether and how this observation plays a role in ccRCC tumorigenesis.
HIF regulation of glutamine metabolism
Glutamine is the most abundant amino acid in the circulation and is an essential precursor for the synthesis of proteins, fatty acids, nucleotides, and many other important molecules (19). Glutamine metabolism has been reviewed extensively (20) and is briefly outlined here. Upon cellular entry via transporters such as SLC1A5, glutamine is converted by glutaminases to generate glutamate. Glutamate plays a variety of important roles in eukaryotic cells (Fig. 1A). For example, it is required for glutathione synthesis, the major cellular antioxidant, and is also the source of amino groups for other non-essential amino acids such as proline, alanine, aspartate, serine, and glycine. In addition, glutamate can be converted to α-ketoglutarate (α-KG), which is either oxidized by α-ketoglutarate dehydrogenase (αKGDH) to succinate and enters a “forward” tricarboxylic acid (TCA) cycle to generate ATP or reductively carboxylated by isocitrate dehydrogenase (cytosolic IDH1 or mitochondrial IDH2) to produce isocitrate and citrate by a “reverse” TCA cycle.
Under hypoxia, Wise et al. (21) demonstrated that glutamine becomes a major source of citrate through reductive carboxylation via wild-type IDH2 activity in the glioblastoma cell line SF188. Consequently, hypoxic cells are unable to proliferate when they are either glutamine-starved or rendered IDH2-deficient. This metabolic reprogramming might be partly through HIF, as constitutive HIF activation recapitulates the preferential reductive metabolic pathway even in normoxic conditions (21). In addition, IDH1 is also essential for HIF-mediated production of citrate at 1% O2, and citrate produced via IDH1-dependent reductive carboxylation is utilized for de novo lipogenesis, required for maintaining cell growth in hypoxic conditions (22–24). This shift appears to be due to proteolysis of the E1 subunit of the αKGDH complex, named “oxoglutarate dehydrogenase 2” (OGDH2) (25). Sun and Denko (25) found that the E3 ubiquitin ligase SIAH2 destabilized OGHD2 in hypoxia in an HIF-dependent manner, which results in diminished αKGDH activity and decreased glutamine oxidation. In addition, a ubiquitination-resistant OGDH2 336KA mutant impedes tumor growth in vivo, accompanied by a reduction in glutamine incorporation into lipids (25), highlighting this pathway as an essential mechanism to rewire glutamine fate in HIF-stabilized tumor cells. Finally, in VHL−/− ccRCC cells, glutamine is also utilized to generate aspartate for de novo pyrimidine biosynthesis via reductive carboxylation and glutathione for redox balance (26). When glutaminase is inhibited, nucleoside depletion and increased ROS levels lead to DNA replication stress, which sensitizes cells to poly(ADP-ribose) polymerase inhibition in vitro and in vivo. Taken together, these studies exemplify alternative fates of glutamine in an HIF-dependent manner to support cancer cell proliferation and viability under hypoxic stress or as a consequence of genetic mutations.
HIF regulation of lipid synthesis
Lipid synthesis provides essential building blocks and signaling molecules for tumor growth. Thus, despite a high-energy cost, de novo lipogenesis is clearly increased in most cancer cells (27–29). In this process, acetyl-CoA and NADPH together generate free fatty acid chains through stepwise enzymatic reactions via acetyl-CoA carboxylase (ACC) and fatty-acid synthase (FASN) (Fig. 1A). These fatty acids are then: 1) used to generate phospholipids, the basic units of lipid bilayers making up cell and organelle membranes; 2) incorporated into neutral lipids, such as triglycerides and cholesterol esters, for storage; 3) utilized to produce essential signaling molecules, such as sphingosine 1-phosphate, lysophosphatidic acid, diglyceride, and inositol 1,4,5-trisphosphate, which control inflammation, cell migration, and survival in cancer; and 4) employed to modify proteins, among others.
In hypoxic conditions, glucose-derived pyruvate entry into the TCA cycle is inhibited, as well as subsequent citrate and acetyl-CoA production. Therefore, cells must shift to alternative carbon sources to generate acetyl-CoA for fatty acid synthesis. These sources include citrate synthesis from reductive carboxylation of glutamate (reviewed above) and acetyl-CoA production through metabolizing acetate via cytoplasmic acetyl-CoA synthetase (ACSS2) (30). Indeed, based on 13C-tracing and mass spectrometry, cells cultured in O2-replete conditions produce more than 90% acetyl-CoA from glucose and glutamine-derived carbon. However, at 1% O2, acetate instead appears to be the primary hypoxia-induced contributor to acetyl-CoA in multiple cell lines (31). In addition, hypoxia enhances ACSS2 expression, increasing the use of acetate to sustain lipid biomass production and imparting a competitive growth advantage under microenvironmental stress (32). Exactly how ACSS2 is up-regulated in hypoxia remains unclear, although it has been suggested that sterol regulatory element-binding protein 2 (SREBP2) mainly controls ACSS2 expression, whereas HIF signaling enhances the up-regulation of ACSS2 by SREBP2 (32).
Multiple genes involved in lipid metabolism have been reported to be strongly induced under hypoxia. One example is the regulation of SREBP1 via phosphorylation of AKT followed by HIF1α activation (33). Of note, SREBP1 is a major transcriptional regulator of lipid metabolic genes, such as FASN. Moreover, some lipid metabolic genes are direct HIF transcriptional targets. For example, in hypoxic breast cancer MCF-7 cells and glioblastoma U87 cells, induction of fatty acid-binding protein 3 (FABP3), FABP7, and adipose differentiation-related protein (ADRP, also known as perilipin2, PLIN2) is an HIF1α-dependent but HIF-2α-independent event. These proteins are essential for lipid droplet (LD) formation, which helps to maintain ROS levels in these cells (34). Furthermore, PLIN2 up-regulation in ccRCC is HIF2α-dependent, which promotes lipid storage and contributes to “clear cell” phenotypes (35). Interestingly, PLIN2-mediated LD formation maintains integrity of the ER, suppressing cytotoxic ER stress responses that otherwise result from elevated protein synthetic activity characteristic of ccRCC cells. Therefore, PLIN2 depletion results in apoptosis and further sensitizes these cells to ER stress, identifying it as a targetable vulnerability created by HIF2α/PLIN2 signaling in this common renal malignancy.
HIF and redox stress
Because of its role as a potent electron acceptor, O2 has the tendency to form highly reactive oxygen species. Mitochondrial O2 metabolism is the dominant source of superoxide production via the mitochondrial electron transport chain (ETC). Several studies found HIFα subunits can be stabilized by mitochondrial ROS (36–39). Reciprocally, suppressing the production of ROS by ETC seems to be a fundamental function of HIF.
Under hypoxic conditions, HIF actively regulates mitochondrial function and subsequent ROS production through multiple mechanisms. First, HIF1α uncouples glycolysis and oxidative mitochondrial metabolism by transcriptionally up-regulating pyruvate dehydrogenase kinase 1 (PDK1) (40, 41). PDK1 phosphorylates and inhibits the function of mitochondrial pyruvate dehydrogenase (PDH), which transforms pyruvate into acetyl-CoA, subsequently used in the TCA cycle to carry out cellular respiration (Fig. 1A). Enforced PDK1 expression in hypoxic HIF1α-null cells attenuates ROS generation and protects these cells from hypoxia-induced apoptosis (40). At the same time, accumulating pyruvate is metabolized by lactate dehydrogenase A (LDHA) to generate lactate, which is then exported from the cell by MCT4, as discussed previously. Importantly, both LDHA and MCT4 are also HIF1α targets (42). Taken together, HIF1α-dependent transcriptional regulation coordinately impedes the entry of substrates for mitochondrial respiration and toxic ROS production. Second, HIF1α attenuates mitochondrial function by down-regulating several ETC components. For example, components of cytochrome c oxidase (COX; complex IV), the final ETC enzyme, are regulated in an HIF1α-dependent manner (43). HIF1α reciprocally manipulates COX4 subunit expression by activating transcription of the gene encoding COX4-2, while at the same time enhancing COX4-1 protein degradation by increasing the abundance of LON, a mitochondrial protease (43). The replacement of COX4-1 with COX4-2 allows improved adaption to hypoxia, with reduced ROS production. In addition, HIF1α increases mitochondrial NDUFA4L2 (NADH dehydrogenase 1α subcomplex, 4-like 2), which attenuates mitochondrial oxygen consumption through reduced complex I activity, limiting intracellular ROS production under low O2 conditions (44). Furthermore, HIF1α suppresses succinate dehydrogenase subunit B, a component of mitochondrial complex II (45), but whether this contributes to ROS level regulation is unclear. Finally, HIF influences mitochondrial function through effects at the whole-organelle level. In ccRCC cells lacking VHL, HIF1α negatively regulates c-Myc activity and subsequent mitochondrial biogenesis, O2 consumption, and ROS production (46). Moreover, the HIF target BNIP3 contributes to reduced mitochondrial mass through increased mitophagy (47). Taken together, these adaptive pathways underlying HIF activation attenuate ROS production and provide protective mechanisms for cancer cell growth. Whether they are “Achilles heels” and how we can utilize them against cancer are under investigation.
HIF, miR-210, and metabolism
The microRNAs (miRNAs) are small, non-coding RNA molecules ∼22 nucleotides in length that are evolutionarily conserved (48). Targeting most protein-coding transcripts, miRNAs are involved in nearly all physiological and pathological processes, including cancer cell metabolism. In recent years, more than 50 hypoxia-regulated miRNAs have been identified. Among them, miR-210 exhibits a robust and consistent up-regulation under hypoxia in virtually all cell types through an HIF1α-dependent regulation (49) and influences intracellular metabolism in both normal and malignant cells. For example, first, miR-210 represses the iron–sulfur cluster assembly proteins (ISCU1 in cytosol and ISCU2 in mitochondria) (50), which promote the assembly of [4Fe-4S] and [2Fe-2S] iron–sulfur clusters, which are incorporated into enzymes responsible for mitochondrial respiration, such as those in ETC complexes I, II, and III (51). Therefore, miR-210 expression decreases activity of complex I, oxygen consumption, and ATP generation (50). Moreover, an miR-210 antagonist decreases clonogenic survival in hypoxic MCF7 and HeLa cells (52). In both cases, miR-210 suppression increases ROS production. Second, the transcript coding for one of ETC components, SDHD, subunit D of succinate dehydrogenase complex, is a bona fide miR-210 target. SDHD knockdown mimics miR-210 mediated mitochondrial dysfunction. In addition, SDHD targeting also seems to stabilize HIF1α, which creates a positive feedback loop (53). Taken together, miR-210 serves as an important mediator shifting mitochondrial oxidative phosphorylation to glycolysis downstream of HIF. Whether other miRNAs are involved in more metabolic processes needs to be further investigated.
HIF-independent regulation of intracellular metabolic reactions
As discussed above, under low O2 tension, HIF coordinately regulates a variety of adaptive pathways to enable cancer cell growth, including but not limited to glucose, glutamine, lipid, and redox metabolism. However, O2 availability also affects cellular responses through HIF-independent mechanisms, the understanding of which will help to more completely describe how cancer cells adapt to hypoxic microenvironment.
l-2-Hydroxyglutarate (l-2HG) production and function
With the discovery of oncogenic mutations in isocitrate dehydrogenase enzymes (IDH1 and IDH2), the enzymatic product d-2HG is broadly appreciated as an oncometabolite that inhibits αKG and a large family of αKG-dependent dioxygenases, including histone demethylases and methylcytosine dioxygenases of the TET family, causing epigenetic dysregulation and a block in cellular differentiation (54). Although intensive efforts have been made to investigate the cellular functions of IDH-mutant derived d-2HG, and a selective, potent inhibitor of mutant IDH2, AG-221, was shown to be effective in IDH2 mutation-positive acute myeloid leukemia (55, 56), its mirror-image enantiomer l-2HG is less understood. Interestingly, recent studies found that hypoxia dramatically induces production of l-2HG in both normal and malignant cells (57, 58).
In different types of cancer cell lines, hypoxia substantially increases 2HG production 5–25-fold, detailed analysis specifies a selective accumulation of the l-enantiomer of 2HG in these cells (57). Surprisingly, this reaction is not through IDH1/2, but LDHA and malate dehydrogenase (MDH1 and MDH2) via their “promiscuous” catalytic activity (57). Further study confirmed these results by showing that purified LDH and MDH enzymes catalyze stereospecific reduction of αKG to l-2HG and that acidic reaction conditions dramatically enhance LDH- and MDH-mediated production of l-2HG in vitro and in cells. Mechanistically, hypoxia-induced acidic pH enhances LDHA-mediated reduction of αKG by driving equilibrium toward a protonated form of αKG that binds more stably to the LDHA enzyme (59). In normal cells, including human pulmonary arterial endothelial and smooth muscle cells, hypoxia also directs the production of l-2HG (58), highlighting it as a fundamental mechanism for hypoxia adaptation. Functionally, acid-enhanced production of l-2HG leads to stabilization of HIF1α in normoxic conditions (59). In addition, like d-2HG, l-2HG also inhibits the Jumonji family histone lysine demethylase KDM4C, resulting in aberrant accumulation of trimethylated histone 3 lysine 9 (H3K9me3) (57). Interestingly, a recent report demonstrated that tumor hypoxia reduces TET enzyme activity, which is independent of hypoxia-associated alterations in TET expression, proliferation, metabolism, HIF activity, or ROS production, and it is dependent directly on O2 shortage (60). This effect results in hypermethylation at gene promoters (including those suppressing glycolysis), which accounts for up to half of the hypermethylation events in cells. However, whether and how much hypoxia-mediated l-2HG plays a role in this process needs to be investigated in more detail. Furthermore, mitochondrial l-2HG is oxidized and converted back to αKG by l-2HG dehydrogenase (L2HGDH) (61), and αKG and l-2HG form a redox couple linked to NADH/NAD+ and FADH2/FAD (Fig. 1B). Therefore, 2HG may provide cells with a reservoir of reducing equivalents, and the αKG-2HG exchange maintains cellular redox balance. Consistent with this idea, a recent study in hematopoietic stem cells reported that loss of the mitochondrial complex III subunit Rieske iron–sulfur protein results in impaired mitochondrial respiration, increased NADH/NAD+ ratio, and increased 2HG levels, accompanied with DNA and histone hypermethylation (62). Because L2HGDH activity is dependent on active complex III (63), loss of complex III activity both increases NADH/NAD+ and impairs L2HGDH activity to favor 2HG production.
How HIF proteins contribute to l-2HG production seems to be context-dependent. In lung fibroblasts, whereas VHL knockdown and HIFα stabilization increase total 2HG production in normoxia, HIF1α depletion also elevates l-2HG in hypoxia, indicating HIF1α is sufficient but not necessary for l-2HG accumulation in these cells (58). Consistent with this, in SF188 cancer cells, HIF1A knockdown in hypoxia does not decrease but slightly increases l-2HG levels (57). In addition, HIF1α enhances hypoxia-induced l-2HG production in VHL-deficient RCC4 cells, as HIF1α depletion partly reverses hypoxia-mediated l-2HG increments (57). However, in murine CD8+ T lymphocytes, Hif1α−/− abolished 2HG accumulation under hypoxia, and l-2HG constitutes more than 90% of the 2HG pool, suggesting l-2HG production is dependent on HIF1α in this case (64).
Alternative effect of hypoxia on lipid metabolism
O2 deficiency affects lipid metabolism not only through HIF-dependent pathways, as discussed above, but also via direct effects on enzymes involved in this process. Stearoyl-CoA desaturase-1 (SCD1) is a key enzyme catalyzing the rate-limiting step in the formation of monounsaturated fatty acids. This process conducts electron flow from NADPH to the terminal electron acceptor molecular O2, rendering O2 a critical factor in the formation of double bonds in stearoyl-CoA. Indeed, Kamphorst et al. (65) found that consistent with impaired SCD1 activity, hypoxic cancer cells display decreased C18:1 production, as well as the C18:1/C18:0 ratios. Furthermore, SCD1 enzymatic activity is required to maintain cell viability in Tsc2−/− mouse embryonic fibroblasts with O2 deprivation (66). This is accompanied by elevated toxic ER stress, which might be caused by an unmet requirement of ER membrane quality in the conditions of increased protein synthesis by dysregulated mTORC1. This mechanism has been confirmed in multiple systems by demonstrating that SCD1 inhibition increases ER stress in vitro and in vivo (67, 68) and can be employed as a strategy to treat cancers.
Conclusion
Although our understanding of metabolic alterations in cancer has improved dramatically over recent years, whether and how hypoxia affects these pathways is still not entirely clear. As a typical feature of solid tumors, hypoxia is usually associated with chemoresistance, radioresistance, and poor prognosis. Defining how cancer cells cope with hypoxic stress is therefore critical for identifying potential new therapeutic targets. As discussed above, metabolic rewiring is one of the major hypoxia-induced responses, encompassing HIF-dependent and HIF-independent pathways. These adaptive responses collectively enable cell growth and survival, which at the same time could be an “Achilles heel” of these cells. Indeed, in cell culture and pre-clinical mouse model systems, targeting these processes results in cell death and tumor regression. However, will metabolic therapy be clinically viable? A therapeutic window may be difficult to achieve, and cancer cells could shift to adaptive pathways during long-term exposure to certain treatments. Therefore, future research needs to focus on how to specifically target cancer cells and improve efficacy, such as combining metabolic intervention with standard chemotherapy, radiotherapy, and immunotherapy.
This is the fourth article in the Thematic Minireview Series “Redox metabolism and signaling.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HIF
- hypoxia-inducible factor
- ER
- endoplasmic reticulum
- HRE
- hypoxia-response element
- 2HG
- 2-hydroxyglutarate
- ROS
- reactive oxygen species
- MDH
- malate dehydrogenase
- LDH
- lactate dehydrogenase
- ccRCC
- clear cell renal cell carcinoma
- TCA
- tricarboxylic acid
- FIH
- factor inhibiting HIF
- αKGDH
- α-ketoglutarate dehydrogenase
- α-KG
- α-ketoglutarate
- PHD
- prolyl hydroxylase
- ETC
- electron transport chain
- LD
- lipid droplet
- FASN
- fatty-acid synthase
- ACC
- acetyl-CoA carboxylase
- PDH
- pyruvate dehydrogenase
- EPO
- erythropoietin
- VHL
- von Hippel-Lindau
- miRNA
- microRNA
- SDHD
- subunit D of succinate dehydrogenase.
References
- 1. Nakazawa M. S., Keith B., and Simon M. C. (2016) Oxygen availability and metabolic adaptations. Nat. Rev. Cancer 16, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee K. E., and Simon M. C. (2015) SnapShot: hypoxia-inducible factors. Cell 163, 1288–1288 [DOI] [PubMed] [Google Scholar]
- 3. Denko N. C. (2008) Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 8, 705–713 [DOI] [PubMed] [Google Scholar]
- 4. Maxwell P. H., Dachs G. U., Gleadle J. M., Nicholls L. G., Harris A. L., Stratford I. J., Hankinson O., Pugh C. W., and Ratcliffe P. J. (1997) Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl. Acad. Sci. U.S.A. 94, 8104–8109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen C., Pore N., Behrooz A., Ismail-Beigi F., and Maity A. (2001) Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J. Biol. Chem. 276, 9519–9525 [DOI] [PubMed] [Google Scholar]
- 6. Ullah M. S., Davies A. J., and Halestrap A. P. (2006) The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through an HIF-1α-dependent mechanism. J. Biol. Chem. 281, 9030–9037 [DOI] [PubMed] [Google Scholar]
- 7. Luo W., Hu H., Chang R., Zhong J., Knabel M., O'Meally R., Cole R. N., Pandey A., and Semenza G. L. (2011) Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong N., Ojo D., Yan J., and Tang D. (2015) PKM2 contributes to cancer metabolism. Cancer Lett. 356, 184–191 [DOI] [PubMed] [Google Scholar]
- 9. Palsson-McDermott E. M., Curtis A. M., Goel G., Lauterbach M. A., Sheedy F. J., Gleeson L. E., van den Bosch M. W., Quinn S. R., Domingo-Fernandez R., Johnston D. G., Jiang J.-K., Jiang J.-K., Israelsen W. J., Keane J., Thomas C., et al. (2015) Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li B., Qiu B., Lee D. S., Walton Z. E., Ochocki J. D., Mathew L. K., Mancuso A., Gade T. P., Keith B., Nissim I., and Simon M. C. (2014) Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature 513, 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adeva-Andany M. M., González-Lucán M., Donapetry-García C., Fernández-Fernández C., and Ameneiros-Rodríguez E. (2016) Glycogen metabolism in humans. BBA Clin. 5, 85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zois C. E., Favaro E., and Harris A. L. (2014) Glycogen metabolism in cancer. Biochem. Pharmacol. 92, 3–11 [DOI] [PubMed] [Google Scholar]
- 13. Rousset M., Zweibaum A., and Fogh J. (1981) Presence of glycogen and growth-related variations in 58 cultured human tumor cell lines of various tissue origins. Cancer Res. 41, 1165–1170 [PubMed] [Google Scholar]
- 14. Pescador N., Villar D., Cifuentes D., Garcia-Rocha M., Ortiz-Barahona A., Vazquez S., Ordoñez A., Cuevas Y., Saez-Morales D., Garcia-Bermejo M. L., Landazuri M. O., Guinovart J., and del Peso L. (2010) Hypoxia promotes glycogen accumulation through hypoxia inducible factor (HIF)-mediated induction of glycogen synthase 1. PLoS ONE 5, e9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pelletier J., Bellot G., Gounon P., Lacas-Gervais S., Pouysségur J., and Mazure N. M. (2012) Glycogen synthesis is induced in hypoxia by the hypoxia-inducible factor and promotes cancer cell survival. Front. Oncol. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shen G.-M., Zhang F.-L., Liu X.-L., and Zhang J.-W. (2010) Hypoxia-inducible factor 1-mediated regulation of PPP1R3C promotes glycogen accumulation in human MCF-7 cells under hypoxia. FEBS Lett. 584, 4366–4372 [DOI] [PubMed] [Google Scholar]
- 17. Favaro E., Bensaad K., Chong M. G., Tennant D. A., Ferguson D. J., Snell C., Steers G., Turley H., Li J.-L., Günther U. L., Buffa F. M., McIntyre A., and Harris A. L. (2012) Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab. 16, 751–764 [DOI] [PubMed] [Google Scholar]
- 18. Lee W.-N., Guo P., Lim S., Bassilian S., Lee S. T., Boren J., Cascante M., Go V. L., and Boros L. G. (2004) Metabolic sensitivity of pancreatic tumour cell apoptosis to glycogen phosphorylase inhibitor treatment. Br. J. Cancer 91, 2094–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mayers J. R., and Vander Heiden M. G. (2015) Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem. Sci. 40, 130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altman B. J., Stine Z. E., and Dang C. V. (2016) From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wise D. R., Ward P. S., Shay J. E., Cross J. R., Gruber J. J., Sachdeva U. M., Platt J. M., DeMatteo R. G., Simon M. C., and Thompson C. B. (2011) Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U.S.A. 108, 19611–19616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Metallo C. M., Gameiro P. A., Bell E. L., Mattaini K. R., Yang J., Hiller K., Jewell C. M., Johnson Z. R., Irvine D. J., Guarente L., Kelleher J. K., Vander Heiden M. G., Iliopoulos O., and Stephanopoulos G. (2011) Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gameiro P. A., Yang J., Metelo A. M., Pérez-Carro R., Baker R., Wang Z., Arreola A., Rathmell W. K., Olumi A., López-Larrubia P., Stephanopoulos G., and Iliopoulos O. (2013) In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 17, 372–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mullen A. R., Wheaton W. W., Jin E. S., Chen P.-H., Sullivan L. B., Cheng T., Yang Y., Linehan W. M., Chandel N. S., and DeBerardinis R. J. (2011) Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun R. C., and Denko N. C. (2014) Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 19, 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okazaki A., Gameiro P. A., Christodoulou D., Laviollette L., Schneider M., Chaves F., Stemmer-Rachamimov A., Yazinski S. A., Lee R., Stephanopoulos G., Zou L., and Iliopoulos O. (2017) Glutaminase and poly (ADP-ribose) polymerase inhibitors suppress pyrimidine synthesis and VHL-deficient renal cancers. J. Clin. Invest. 127, 1631–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Currie E., Schulze A., Zechner R., Walther T. C., and Farese R. V. Jr. (2013) Cellular fatty acid metabolism and cancer. Cell Metab. 18, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beloribi-Djefaflia S., Vasseur S., and Guillaumond F. (2016) Lipid metabolic reprogramming in cancer cells. Oncogenesis 5, e189–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Röhrig F., and Schulze A. (2016) The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 [DOI] [PubMed] [Google Scholar]
- 30. Luong A., Hannah V. C., Brown M. S., and Goldstein J. L. (2000) Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 275, 26458–26466 [DOI] [PubMed] [Google Scholar]
- 31. Kamphorst J. J., Chung M. K., Fan J., and Rabinowitz J. D. (2014) Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schug Z. T., Peck B., Jones D. T., Zhang Q., Grosskurth S., Alam I. S., Goodwin L. M., Smethurst E., Mason S., Blyth K., McGarry L., James D., Shanks E., Kalna G., Saunders R. E., et al. (2015) Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 27, 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Furuta E., Pai S. K., Zhan R., Bandyopadhyay S., Watabe M., Mo Y.-Y., Hirota S., Hosobe S., Tsukada T., Miura K., Kamada S., Saito K., Iiizumi M., Liu W., Ericsson J., and Watabe K. (2008) Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 68, 1003–1011 [DOI] [PubMed] [Google Scholar]
- 34. Bensaad K., Favaro E., Lewis C. A., Peck B., Lord S., Collins J. M., Pinnick K. E., Wigfield S., Buffa F. M., Li J.-L., Zhang Q., Wakelam M. J., Karpe F., Schulze A., and Harris A. L. (2014) Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 9, 349–365 [DOI] [PubMed] [Google Scholar]
- 35. Qiu B., Ackerman D., Sanchez D. J., Li B., Ochocki J. D., Grazioli A., Bobrovnikova-Marjon E., Diehl J. A., Keith B., and Simon M. C. (2015) HIF2-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 5, 652–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chandel N. S., McClintock D. S., Feliciano C. E., Wood T. M., Melendez J. A., Rodriguez A. M., and Schumacker P. T. (2000) Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1α during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 275, 25130–25138 [DOI] [PubMed] [Google Scholar]
- 37. Guzy R. D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K. D., Simon M. C., Hammerling U., and Schumacker P. T. (2005) Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 1, 401–408 [DOI] [PubMed] [Google Scholar]
- 38. Mansfield K. D., Guzy R. D., Pan Y., Young R. M., Cash T. P., Schumacker P. T., and Simon M. C. (2005) Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-α activation. Cell Metab. 1, 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klimova T., and Chandel N. S. (2008) Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ. 15, 660–666 [DOI] [PubMed] [Google Scholar]
- 40. Kim J.-W., Tchernyshyov I., Semenza G. L., and Dang C. V. (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 41. Papandreou I., Cairns R. A., Fontana L., Lim A. L., and Denko N. C. (2006) HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 42. Keith B., Johnson R. S., and Simon M. C. (2011) HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 12, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fukuda R., Zhang H., Kim J.-W., Shimoda L., Dang C. V., and Semenza G. L. (2007) HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122 [DOI] [PubMed] [Google Scholar]
- 44. Tello D., Balsa E., Acosta-Iborra B., Fuertes-Yebra E., Elorza A., Ordóñez Á., Corral-Escariz M., Soro I., López-Bernardo E., Perales-Clemente E., Martínez-Ruiz A., Enríquez J. A., Aragonés J., Cadenas S., and Landázuri M. O. (2011) Induction of the mitochondrial NDUFA4L2 protein by HIF-1α decreases oxygen consumption by inhibiting complex I activity. Cell Metab. 14, 768–779 [DOI] [PubMed] [Google Scholar]
- 45. Dahia P. L., Ross K. N., Wright M. E., Hayashida C. Y., Santagata S., Barontini M., Kung A. L., Sanso G., Powers J. F., Tischler A. S., Hodin R., Heitritter S., Moore F., Dluhy R., Sosa J. A., et al. (2005) A HIF1α regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 1, 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang H., Gao P., Fukuda R., Kumar G., Krishnamachary B., Zeller K. I., Dang C. V., and Semenza G. L. (2007) HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of c-MYC activity. Cancer Cell 11, 407–420 [DOI] [PubMed] [Google Scholar]
- 47. Zhang H., Bosch-Marce M., Shimoda L. A., Tan Y. S., Baek J. H., Wesley J. B., Gonzalez F. J., and Semenza G. L. (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 283, 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Ha M., and Kim V. N. (2014) Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 [DOI] [PubMed] [Google Scholar]
- 49. Bertero T., Rezzonico R., Pottier N., and Mari B. (2017) Impact of microRNAs in the cellular response to hypoxia. Int. Rev. Cell Mol. Biol. 10.1016/bs.ircmb.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 50. Chan S. Y., Zhang Y.-Y., Hemann C., Mahoney C. E., Zweier J. L., and Loscalzo J. (2009) MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron–sulfur cluster assembly proteins ISCU1/2. Cell Metab. 10, 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rouault T. A., and Tong W. H. (2008) Iron–sulfur cluster biogenesis and human disease. Trends Genet. 24, 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Favaro E., Ramachandran A., McCormick R., Gee H., Blancher C., Crosby M., Devlin C., Blick C., Buffa F., Li J.-L., Vojnovic B., Pires das Neves R., Glazer P., Iborra F., Ivan M., et al. (2010) MicroRNA-210 regulates mitochondrial free radical response to hypoxia and Krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS ONE 5, e10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Puisségur M. P., Mazure N. M., Bertero T., Pradelli L., Grosso S., Robbe-Sermesant K., Maurin T., Lebrigand K., Cardinaud B., Hofman V., Fourre S., Magnone V., Ricci J. E., Pouysségur J., Gounon P., et al. (2011) miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 18, 465–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Losman J.-A., and Kaelin W. G. Jr. (2013) What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27, 836–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yen K., Travins J., Wang F., David M. D., Artin E., Straley K., Padyana A., Gross S., DeLaBarre B., Tobin E., Chen Y., Nagaraja R., Choe S., Jin L., Konteatis Z., et al. (2017) AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 7, 478–493 [DOI] [PubMed] [Google Scholar]
- 56. Shih A. H., Meydan C., Shank K., Garrett-Bakelman F. E., Ward P. S., Intlekofer A. M., Nazir A., Stein E. M., Knapp K., Glass J., Travins J., Straley K., Gliser C., Mason C. E., Yen K., et al. (2017) Combination targeted therapy to disrupt aberrant oncogenic signaling and reverse epigenetic dysfunction in IDH2- and TET2-mutant acute myeloid leukemia. Cancer Discov. 7, 494–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Intlekofer A. M., Dematteo R. G., Venneti S., Finley L. W., Lu C., Judkins A. R., Rustenburg A. S., Grinaway P. B., Chodera J. D., Cross J. R., and Thompson C. B. (2015) Hypoxia induces production of l-2-hydroxyglutarate. Cell Metab. 22, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oldham W. M., Clish C. B., Yang Y., and Loscalzo J. (2015) Hypoxia-mediated increases in l-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 22, 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Intlekofer A. M., Wang B., Liu H., Shah H., Carmona-Fontaine C., Rustenburg A. S., Salah S., Gunner M. R., Chodera J. D., Cross J. R., and Thompson C. B. (2017) l-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat. Chem. Biol. 13, 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thienpont B., Steinbacher J., Zhao H., D'Anna F., Kuchnio A., Ploumakis A., Ghesquière B., Van Dyck L., Boeckx B., Schoonjans L., Hermans E., Amant F., Kristensen V. N., Peng Koh K., Mazzone M., et al. (2016) Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature 537, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rzem R., Van Schaftingen E., and Veiga-da-Cunha M. (2006) The gene mutated in l-2-hydroxyglutaric aciduria encodes l-2-hydroxyglutarate dehydrogenase. Biochimie 88, 113–116 [DOI] [PubMed] [Google Scholar]
- 62. Ansó E., Weinberg S. E., Diebold L. P., Thompson B. J., Malinge S., Schumacker P. T., Liu X., Zhang Y., Shao Z., Steadman M., Marsh K. M., Xu J., Crispino J. D., and Chandel N. S. (2017) The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat. Cell Biol. 19, 614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Engqvist M. K., Esser C., Maier A., Lercher M. J., and Maurino V. G. (2014) Mitochondrial 2-hydroxyglutarate metabolism. Mitochondrion 19, 275–281 [DOI] [PubMed] [Google Scholar]
- 64. Tyrakis P. A., Palazon A., Macias D., Lee K. L., Phan A. T., Veliça P., You J., Chia G. S., Sim J., Doedens A., Abelanet A., Evans C. E., Griffiths J. R., Poellinger L., Goldrath A. W., and Johnson R. S. (2016) S-2-hydroxyglutarate regulates CD8(+) T-lymphocyte fate. Nature 540, 236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kamphorst J. J., Cross J. R., Fan J., de Stanchina E., Mathew R., White E. P., Thompson C. B., and Rabinowitz J. D. (2013) Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. U.S.A. 110, 8882–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Young R. M., Ackerman D., Quinn Z. L., Mancuso A., Gruber M., Liu L., Giannoukos D. N., Bobrovnikova-Marjon E., Diehl J. A., Keith B., and Simon M. C. (2013) Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 27, 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Peter A., Weigert C., Staiger H., Machicao F., Schick F., Machann J., Stefan N., Thamer C., Häring H.-U., and Schleicher E. (2009) Individual stearoyl-CoA desaturase 1 expression modulates endoplasmic reticulum stress and inflammation in human myotubes and is associated with skeletal muscle lipid storage and insulin sensitivity in vivo. Diabetes 58, 1757–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. von Roemeling C. A., Marlow L. A., Wei J. J., Cooper S. J., Caulfield T. R., Wu K., Tan W. W., Tun H. W., and Copland J. A. (2013) Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin. Cancer Res. 19, 2368–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]