Abstract
In metazoan cell nuclei, hundreds of large chromatin domains are in close contact with the nuclear lamina. Such lamina-associated domains (LADs) are thought to help organize chromosomes inside the nucleus and have been associated with gene repression. Here, we discuss the properties of LADs, the molecular mechanisms that determine their association with the nuclear lamina, their dynamic links with other nuclear compartments, and their proposed roles in gene regulation.
Introduction: the nuclear lamina and genome organization
In metazoan cell nuclei the inner nuclear membrane is lined by the nuclear lamina (NL), a fibrous layer consisting primarily of type V intermediate filament proteins named lamins. Early electron microscopy revealed the tight apposition of a layer of condensed chromatin adjacent to the NL (Fawcett, 1966) (Figure 1A). Later, DNA fluorescence in situ hybridization (FISH) demonstrated that specific genomic loci are preferentially located at the nuclear periphery, often when these loci exhibit low transcriptional activity (reviewed in Lanctot et al., 2007). During the past decade, genome-wide mapping methods have identified genomic regions that are in close contact with the NL, termed Lamina-Associated Domains (LADs).
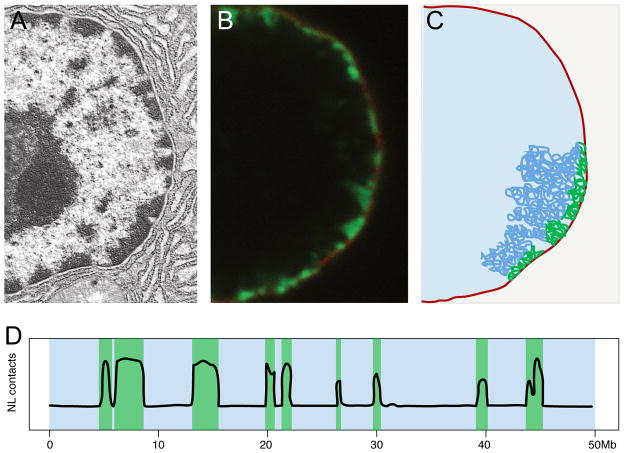
Figure 1. NL-associated heterochromatin.
A. Electron micrograph of part of a mouse cell nucleus. Densely stained chromatin is closely associated with the NL, but is also present around nucleoli and in patches elsewhere in the nucleus. Image provided by Kenneth M. Bart. B. Labeling of DNA–NL contacts by co-expression of Dam-Lamin B1 and a GFP-tagged m6A-tracer protein that binds to adenine-methylated DNA (green) in a cultured human cell. Image by Jop Kind. A single confocal section is shown. Lamin B1 is shown in red. C. Cartoon model illustrating how a chromosome (blue) is associated with the NL through multiple LADs that jointly form a heterochromatin layer (green). Only one chromosome is depicted. D. Schematic representation of a DamID track of interactions with the NL along part of a mammalian chromosome, illustrating the size range, relative sharply defined edges, and broad distribution of LADs. LADs are highlighted in green, inter-LAD regions in blue.
LADs are of particular interest for two broad reasons. First, their NL-anchoring helps to establish interphase chromosome topology and thus the overall genome spatial organization. Second, most of the several thousands of genes in LADs are expressed at very low levels, suggesting a role in gene repression.
Here, we summarize the features of LADs, the dynamics of their interactions with the NL, and recent progress in identifying the molecular mechanisms underlying their interactions with the NL. We also discuss current insights into the functional significance of LADs with respect to transcriptional regulation, and the links with other nuclear compartments. We conclude by raising key questions for future investigation.
Definition and characteristics of LADs
LADs are defined as genomic regions that make molecular contact with the NL. They have been identified primarily using the DamID technology, in which bacterial DNA adenine methyltransferase (Dam) is tethered to a NL protein (typically Lamin B1) leading to adenine methylation of DNA regions that contact the NL protein (Pickersgill et al., 2006). This modification can be visualized by microscopy (Figure 1B, C) or mapped genome-wide (Figure 1D). LADs can also be mapped by chromatin immunoprecipitation (ChIP) (Handoko et al., 2011), but this has been technically challenging for reasons that are only partially understood (Gesson et al., 2016; Lund et al., 2015).
LADs have been mapped in D. melanogaster, C. elegans, and mammalian cell lines (Guelen et al., 2008; Ikegami et al., 2010; Peric-Hupkes et al., 2010; Pickersgill et al., 2006). Mouse and human cells have ~1,000 – 1,500 LADs, typically 10 kb – 10 Mb in size (~0.5 Mb median), that are distributed along all chromosomes (Figure 1D). They cover more than one-third of the mouse and human genome in individual cell types, making LADs one of the most prominent features of the epigenome. In D. melanogaster, which has a much more compact genome, LADs are about 5-fold smaller. However, the average gene number per LAD is similar in flies and mammals, suggesting that LAD organization has co-evolved with gene spacing (van Bemmel et al., 2010). In C. elegans, NL-interacting chromatin domains are enriched at the distal parts of each chromosome (Gonzalez-Aguilera et al., 2014; Ikegami et al., 2010).
LADs correspond to heterochromatin at the nuclear periphery
As expected from the observed tight association of condensed chromatin with the NL, LADs possess several molecular features typical of heterochromatin (Table 1). Most genes in LADs are transcriptionally silent or express at low levels (Guelen et al., 2008; Peric-Hupkes et al., 2010). Furthermore, they overlap with regions that replicate late during S-phase (Guelen et al., 2008; Peric-Hupkes et al., 2010; Pope et al., 2014). LADs also have a low overall gene density, and include most “gene deserts”, defined as gene-free genomic regions >1 Mb. LADs are enriched for histone modifications H3K9me2 and H3K9me3 typical of heterochromatin (Guelen et al., 2008; Wen et al., 2009). The facultative heterochromatin mark H3K27me3 is also enriched at LAD boundaries of some cell types (Guelen et al., 2008; Harr et al., 2015). Human pericentromeric heterochromatin is also typically included in LADs, as are a subset of telomeric regions (Guelen et al., 2008). Finally, LADs are not enriched in cytosine methylation. In fact, in colorectal cancer cell genomes, large DNA hypomethylated regions show a strong overlap with LADs (Berman et al., 2011) but this association is not yet understood.
Table 1.
Features of mammalian LADs
| LAD | inter-LAD | |
|---|---|---|
| gene density | low* | high |
| gene expression | low | high |
| Hi-C compartment | B | A |
| replication timing | late | early |
| retroelements | LINE* | SINE |
| sequence A/T content | high* | low |
| histone marks | H3K9me2, H3K9me3, (H3K27me3) | aH3K4me1, H3K4me3, H3K27ac, other active marks |
| pericentric heterochromatin | frequent | rare |
| nucleolus association | frequent | infrequent |
more pronounced in cLADs compared to fLADs {Meuleman, 2013}
Constitutive and facultative LADs
DamID in various mammalian cell types has shown that some LADs are cell-type invariant (constitutive LADs, cLADs), while others interact with the NL in only certain cell types (facultative LADs, fLADs) (Meuleman et al., 2013; Peric-Hupkes et al., 2010). cLADs largely coincide with AT-rich isochores, are rich in LINE (long interspersed nuclear element) and poor in SINE (short interspersed nuclear element) repetitive elements, and form the most gene-poor subset of LADs. cLAD genomic positions and sizes, but not their actual DNA sequences, are strongly conserved between mouse and human (Meuleman et al., 2013). We speculate that cLADs collectively may form a structural “backbone”, tethering chromosomes to the NL at specific positions and thereby guiding the overall folding of interphase chromosomes.
fLADs make up over half of all LADs, are more gene-dense than cLADs, and their positions are less conserved between mouse and human (Meuleman et al., 2013). During differentiation of mouse cells, hundreds of genes change position relative to the NL, demonstrating the flexibility and dynamics of genome intranuclear spatial organization. Detachment from the NL is frequently accompanied by gene activation while attachment to the NL is frequently accompanied by gene inactivation (Peric-Hupkes et al., 2010; Robson et al., 2016). Below we will discuss the causality relationships between gene repression and NL interactions in more detail.
An extreme form of LAD relocation occurs in the retinal rod cells of nocturnal animals (Solovei et al., 2009). In fully differentiated rod cells, most of the heterochromatin normally located at the nuclear periphery coalesces in the center of the nucleus. LINE elements were also found in this central area in rod cells, suggesting that even most cLADs move to the nuclear center.
Single-cell dynamics of LADs
Chromatin within interphase nuclei is mobile, but the motion of an individual chromosome locus is usually confined to a short range of <1 μm (reviewed in Chuang and Belmont, 2007). Similar observations have been made for LADs in human cells. DNA regions contacting the NL were labeled in vivo through the inducible expression of a Dam-LaminB1 fusion protein and then tracked in live cells throughout a single interphase through their tagging by a fluorescent m6A-binding protein. LADs dynamically interact with the NL, but over a period of hours move only within a layer < 1 μm thick (Figure 1B), corresponding to the heterochromatin adjacent to the NL as seen by electron microscopy (Kind et al., 2013). Thus LADs form the layer of condensed chromatin lining the nuclear envelope but are mobile and only intermittently contact the NL (Figure 1C).
When m6A-tagged LADs were tracked through mitosis, many LADs that were associated with the NL in the mother cell instead were located deep in the nuclear interior of the daughter cells (Kind et al., 2013). This suggests LAD nuclear position is partially randomized after each mitosis. Certain genes appear to move to and from the nuclear periphery in a circadian rhythm (Zhao et al., 2015), but this is probably not a general principle.
More detailed views have emerged from single-cell DamID maps obtained from over one hundred individual G1 cells: certain LADs interact with the NL in almost every cell, while others do so in only a fraction of the cells (Kind et al., 2015). Each LAD has its own characteristic NL contact frequency. Interestingly, LADs contacting the NL in most cells tend to coincide with cLADs and have extremely low gene content (<1 gene/Mb). Thus a subset of LADs, predominantly cLADs and comprising ~15% of the genome, may act as specialized genomic regions that robustly anchor chromosomes to the NL.
Multivalent interactions
From a molecular perspective, LADs are huge: a typical LAD of 500 kb consists of ~2500 nucleosomes. How do such large chromatin domains interact with the NL? Single-cell DamID at a resolution of 100kb has shown that contacts with the NL typically involve continuous stretches of chromatin up to several Mb in length (Kind et al., 2015), suggesting multivalent interactions. Given that the NL and chromatin are both polymeric structures, multivalent interaction is perhaps not surprising.
At a larger scale, LADs on the same chromosome likely bind cooperatively to the NL. LADs on chromosomes with high LAD densities interact more robustly with the NL than those with low LAD densities (Kind et al., 2015). Visualization of H3K9me2 domains on a single human chromosome showed that these domains, presumably LADs, formed a chromosomal surface touching the NL (Chen et al., 2014b). Concerted interactions by multiple LADs may stably secure chromosomes to the NL.
DNA sequences that target genomic regions to the NL
To what extent are NL interactions encoded in the DNA sequence? Several studies useded microscopy to assay the autonomous targeting of stably integrated LAD-derived DNA sequences to the nuclear periphery. LADs at the human IgH and Cyp3a loci contain several DNA regions of 4–32 kb that each conferred NL association when integrated ectopically into a single genomic location. In one instance this was narrowed down to a simple repeat of GA dinucleotides (Zullo et al., 2012); however, this sequence motif is not generally enriched in LADs (Guelen et al., 2008). Dissecting an ~200 kb human genomic region encompassing the beta-globin (HBB) locus identified three peripheral targeting regions ranging in size from 6–32 kb (Bian et al., 2013). Each of these regions could redirect a ~110kb HBB region from pericentric heterochromatin to the NL; the largest was capable of targeting to the nuclear periphery by itself. A similar analysis of two fLADs identified many fragments (ranging from 0.9 kb to tens of kb) that could promote peripheral localization (Harr et al., 2015). Because the integration sites were not mapped in this study, it is unclear whether these elements are sufficient for peripheral targeting or rather require a specific context. Nevertheless, the observation again that multiple non-overlapping regions in a LAD exhibit peripheral targeting activity supports the model that LAD – NL interactions are typically multivalent.
Chromatin proteins and modifications mediating NL interactions
How LAD-derived sequences drive NL interactions remains enigmatic, but may be linked to their chromatin state. Significant progress has been made towards the identification of proteins that facilitate the anchoring of LADs to the NL. Several reports indicate that H3K9 methylation has a prominent role. In C. elegans, transgene repeats adopt a heterochromatic state and are positioned at the nuclear periphery. H3K9 methyltransferases MET-2 and SET-25 are required for this peripheral localization (Towbin et al., 2012). Depletion of both enzymes relocated a transgene array to the nuclear interior, and caused a partial loss of NL interactions along most chromosomes. In mammalian cells disruption or inhibition of G9a, a methyltransferase mediating H3K9me2 deposition, weakened NL-LAD association (Bian et al., 2013; Chen et al., 2014b; Harr et al., 2015; Kind et al., 2013).
Analysis of the ~ 1 Mb chromosome region encompassing the human HBB locus suggests that both H3K9me2 and H3K9me3 modifications contribute in different LAD regions to anchoring this chromosome region to the NL. Depletion of the two H3K9me3 methyltransferases Suv39H1 and Suv39H2 caused NL detachment of human HBB BAC transgenes, while inhibition of G9a caused NL detachment of BAC transgenes containing a LAD region several hundred kb distant from the HBB locus (Bian et al., 2013). Only the triple Suvar39H1, Suvar39H2, and G9a knockdown could peel the endogenous HBB locus and most of an adjacent, ~ 1 Mb LAD from the NL, indicating that these enzymes promote NL association in a redundant manner (Bian et al., 2013).
A study in C. elegans uncovered a mechanistic link between H3K9 methylation and the NL. A small protein named CEC-4 was found to be anchored in the inner nuclear membrane and to bind mono-, di- and tri-methylated H3K9 through its chromodomain. Loss of CEC-4 detached a heterochromatic transgene array from the NL and reduced genome-wide NL contacts similarly to a MET-2/SET-25 double mutant (Gonzalez-Sandoval et al., 2015). CEC-4 does not have obvious orthologs in mammals, but PRR14 is a human protein that may play a somewhat similar LAD-tethering role (Poleshko et al., 2013). PRR14 contains both a NL targeting domain and a separate domain that binds HP1α, a heterochromatin protein known to bind H3K9me2/3. PRR14 loads onto chromosomes immediately after mitosis via its interaction with HP1α, while later in interphase it requires Lamin A/C for its localization at the NL. Depletion of PRR14 caused a wrinkled NL morphology and partial loss of H3K9me3 at the nuclear periphery, although it has not yet been investigated whether LAD – NL interactions are disrupted.
The histone mark H3K27me3 may also be partially linked to LADs. Knockdown of EZH2, the methyltransferase responsible for deposition of this mark, caused reduced NL interactions of an ectopically integrated LAD fragment (Harr et al., 2015). Further studies are needed to confirm a direct role of H3K27me3 in specific LAD – NL interactions. Perhaps related to this histone modification is the reported role of the DNA-binding factor YY1 (Ying-Yang 1). Artificial tethering of YY1 to a reporter locus caused local accumulation of H3K27me3 and relocation of the locus to the nuclear periphery, while knockdown of YY1 shifted several LADs to the nuclear interior. It is puzzling, however, that the consensus recognition motif of YY1 is globally enriched in DNA outside LADs (Harr et al., 2015).
Because the small protein Barrier-to-autointegration (BAF/BANF1) can interact both with chromatin and with LEM-domain containing proteins that are part of the NL, BAF also has been suggested to link chromatin to the NL (Jamin and Wiebe, 2015). BAF preferentially associates with LADs, yet knockdown of BAF surprisingly caused no detectable decrease or a slight increase in LAD – NL interactions (Kind and van Steensel, 2014; Zullo et al., 2012), suggesting that BAF does not help to anchor LADs to the NL during interphase. However, in several organisms BAF is important for the assembly of the nuclear envelope onto chromosomes at the end of mitosis (Jamin and Wiebe, 2015).
LADs, NL substructure and internal Lamin A/C
Super-resolution microscopy has shown that the NL is not homogeneous. Mammalian lamins B1, B2, A and C form distinct, interwoven fibrous networks at the sub-micrometer scale (Shimi et al., 2015; Xie et al., 2016). Genome-wide DamID maps of lamins B1, B2 and A (C was not tested), however, are highly similar (Kind and van Steensel, 2014; Meuleman et al., 2013). Perhaps each lamin interacts with the same LADs, although with minor variations in contact frequencies.
While in most cell types B-type lamins are essentially confined to the NL, A-type lamins are also found throughout the nuclear interior (Gesson et al., 2014). Thus it is surprising that DamID maps of Lamin A and B are so similar. While this could mean that internal Lamin A does not interact with the genome, m6A-tracer experiments point to DNA – Lamin A contacts in the nuclear interior (Kind and van Steensel, 2014). It is therefore tempting to speculate that LADs are bound by the internal Lamin A pool when they are located internally. Some ChIP studies have suggested Lamin A/C binding sites in inter-LAD regions (Gesson et al., 2016; Lund et al., 2015), which could also represent interactions that occur in the nuclear interior. Further investigation is required to understand why such signals have so far not been observed in DamID maps.
NL proteins involved in NL – LAD interactions
Which NL proteins form the contact points for LADs? Lamins are obvious candidates because they interact in vitro both with DNA and chromatin (Gruenbaum and Foisner, 2015). Depletion of the single B-type lamin in Drosophila causes detachment of various genes from the NL (Kohwi et al., 2013; Shevelyov et al., 2009), while the only lamin gene in C. elegans, LMN-1, contributes to the sequestration of a heterochromatic transgene array at the nuclear periphery (Mattout et al., 2011). Surprisingly, deletion of all lamins in mouse embryonic stem cells has no or marginal effects on genome-wide contacts with the NL as determined by genomic interactions with emerin (Amendola and van Steensel, 2015; Zheng et al., 2015). Genetic studies in mouse suggest this result may be explained by redundant roles of Lamin A/C and the Lamin B receptor (LBR). LBR is inserted in the inner nuclear membrane, is part of the NL, and can interact with heterochromatin components such as HP1 and H4K20me3 (Olins et al., 2010). In post-mitotic cell types in which both LBR and Lamin A/C were knocked out, heterochromatin was found to coalesce in the nuclear interior (Solovei et al., 2013). Retinal rod cells in wild-type mice, which express neither Lamin A/C nor LBR, also show this central aggregation of heterochromatin, while ectopic expression of LBR in these rod cells causes heterochromatin to relocate to the nuclear periphery. Similarly, olfactory receptor genes in mouse olfactory sensory neurons (OSNs) move to the nuclear interior as a consequence of physiological downregulation of LBR (Clowney et al., 2012). LBR also contributes to tethering of the mammalian X chromosome to the nuclear periphery during X chromosome inactivation (Chen et al., 2016).
Besides LBR, the inner nuclear membrane harbors dozens of other Nuclear Envelope Transmembrane proteins (NETs) (Schirmer et al., 2003; Wong et al., 2014) and the depletion or overexpression of some of these NETs can alter chromosome localization relative to the nuclear periphery (Robson et al., 2016; Zuleger et al., 2013). While this chromosome repositioning could be indirect, NET39 contains a nucleoplasmic protein domain that was found to mediate anchoring of the Ptn gene, suggesting a direct role for NET39 in chromosome-tethering (Robson et al., 2016).
Emerin is a well-studied NET that can interact with LADs (Amendola and van Steensel, 2015; Guelen et al., 2008; Zheng et al., 2015). Emerin has been suggested to tether chromatin to the NL given that it interacts with NL proteins as well as chromatin components such as BAF (Berk et al., 2013). Exposure of epidermal stem cells to mechanical strain causes redistribution of emerin from the inner nuclear membrane to the endoplasmic reticulum, followed by partial replacement of H3K9me2/3 by H3K27me3 and changes in chromosome positioning inside the nucleus (Le et al., 2016). These effects on chromatin modifications can be mimicked by emerin knockdown, suggesting that emerin is part of a mechanosensory pathway signaling to heterochromatin.
Thus, several NL proteins have been linked to LAD positioning. Lamins, LBR, and emerin are widely expressed and may be redundant components of a scaffold to which LADs are anchored. Tissue specific interactions of fLADs may be in part regulated by NETs, which generally are restricted to specific cell types (Wong et al., 2014), through mechanisms still unknown.
Counterforces: tethering of inter-LAD regions to the nuclear interior
Just as LAD sequences are preferentially positioned near the NL, inter-LADs are preferentially positioned away from the NL. Throughout the nuclear interior RNA polymerase II has been found to cluster in distinct foci, termed “transcription factories”, and FISH studies revealed the transcription-dependent association of specific genes with these structures (Edelman and Fraser, 2012). Some active genes also localize preferentially near other substructures in the nuclear interior, including splicing factor speckles, PML bodies and Cajal bodies (reviewed in Mao et al., 2011) (Figure 2A).
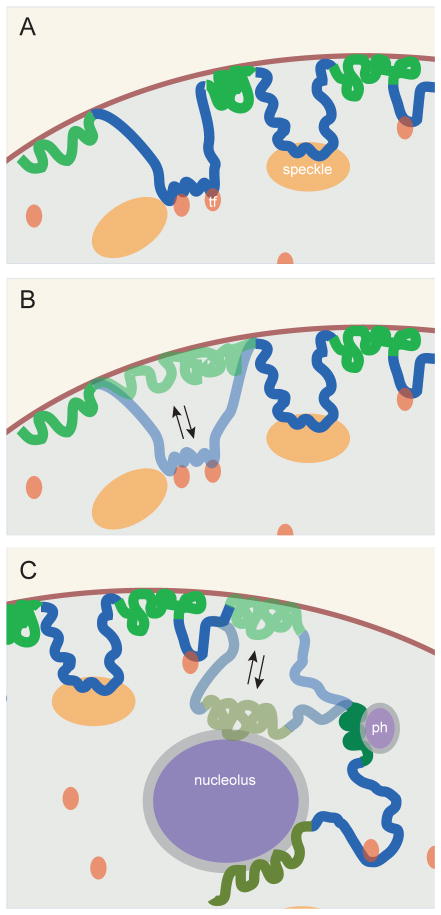
Figure 2. Dynamic compartmentalization of chromosomal domains.
A. Besides anchoring of LADs (green) to the NL, other regions (blue) may be tethered to nuclear structures that are permissive for transcription (orange), such as transcription factories (tf) or splicing factor speckles (speckles). B. Some LADs (semi-transparent green) contact the NL erratically (i.e., in a subset of cells) and may become transcriptionally active when associated with a permissive compartment (semi-transparent blue). C. Some LADs are apparently stochastically distributed between the NL, nucleoli and pericentromeric heterochromatin (ph), which are all repressive environments.
Mechanisms targeting genomic loci to these intra-nuclear compartments appear to compete with NL-anchoring mechanisms. For example, tethering of a viral transcriptional activation peptide to a transgene array located at the nuclear periphery caused relocation of this array to the nuclear interior (Chuang et al., 2006). Similar detachment from the NL was reported for genes located in fLADs when an activation domain was targeted to their promoter (Therizols et al., 2014). In both cases, relocalization to the nuclear interior was also induced by a synthetic peptide that caused chromatin decondensation but not gene activation (Chuang et al., 2006; Therizols et al., 2014), indicating that a change in chromatin state rather than transcription per se is responsible for the detachment from the NL. Possibly this involves erasure of H3K9me2 or H3K9me3, histone marks involved in NL association (see above). NL association of genes may also be counteracted by DNA-binding factors that drive clustering of multiple genomic loci in the nuclear interior (de Wit et al., 2013). Hence, the relative positioning of a gene relative to the NL may reflect the final balance reached between multiple targeting signals. Cell-to-cell variation in this balance may underlie some of the apparent stochasticity of LAD–NL interactions (Figure 2B).
Gene recruitment of genes to interior nuclear compartments may even involve a force-generating molecular mechanism. Live-cell microscopy revealed that relocation of the transgene array to the nuclear interior after tethering of the transcriptional acidic activator occurred through long-range, directed movements; both actin and nuclear myosin 1c were implicated in this transgene repositioning (Chuang et al., 2006). Directed movement of Hsp70 transgenes to nuclear speckles upon heat-shock, in some cases accompanied by chromatin stretching, also suggested a force-generating mechanism (Khanna et al., 2014). These results raise the interesting possibility that active recruitment of inter-LAD regions to the nuclear interior, together with attachment of neighboring LADs to the NL, could pull on the intervening chromosome region, direct effecting its intranuclear positioning and chromatin compaction. Through this “tug-of-war” mechanism, anchoring a LAD to the NL could have long-distance effects on the functional activity of neighboring inter-LAD chromosomes regions.
LAD borders
LADs tend to have sharply defined borders. In mammals these borders are enriched for active promoters, CpG islands, and binding of CTCF (Guelen et al., 2008). The latter is a DNA-binding protein implicated in the establishment of chromatin domain boundaries and the formation of large chromatin loops (Merkenschlager and Nora, 2016). Whether these elements actively establish LAD borders remains to be tested. An intriguing feature of LAD border elements is that they are generally located 5–10 kb outside LADs (Guelen et al., 2008), raising the interesting possibility that they may act through the above-mentioned tug-of-war mechanism. Active promoters at LAD borders could associate with transcription factories or other intra-nuclear compartments, thereby pulling the edges of LADs away from the NL. Similarly, CTCF binding sites, enriched just outside of LADs, may prevent NL interactions through formation of loops with more distal CTCF sites in inter-LADs (Handoko et al., 2011).
Other mechanisms should be considered as well. For example, active promoters and CTCF sites are usually marked by euchromatic histone modifications such as H3K4 methylation (Barski et al., 2007) that may antagonize spreading of heterochromatic marks mediating NL interactions. These models are not mutually exclusive.
LADs are repressive chromatin domains
Of the several thousands of genes located in LADs in a given cell type, the majority are expressed at very low or undetectable levels (Guelen et al., 2008). Increases in gene – NL contacts during differentiation are generally accompanied by reduced gene activity. Conversely, genes that move away from the NL during differentiation often become active (Lund et al., 2013; Peric-Hupkes et al., 2010; Pickersgill et al., 2006; Robson et al., 2016). During differentiation of mouse embryonic stem cells into neuronal precursor cells, about a third of all genes that detach from the NL remain inactive. These genes, however, were more prone to activation in a subsequent differentiation step, suggesting that release from the NL may “unlock” these genes for activation at a later stage (Peric-Hupkes et al., 2010).
Does this mean that LADs repress transcription, or are LADs a result of gene inactivity? One study addressed the first possibility by inserting thousands of identical reporter genes into random genomic locations. On average these reporter genes were on ~5–6 fold less active when inserted into LADs versus inter-LADs (Akhtar et al., 2013). Additionally, deletion of G9a, the histone methyltransferase responsible for H3K9 di-methylation in LADs, caused preferential upregulation of genes in LADs in mouse embryonic stem cells (Yokochi et al., 2009). Together these results indicate that LADs form a repressive environment that is in part controlled by H3K9me2.
Not all genes in LADs, however, are inactive; ~5–10% are expressed at high levels (Guelen et al., 2008; Peric-Hupkes et al., 2010). It remains to be elucidated how such genes ‘escape’ the repressive effects of the LAD environment.
Role of NL contacts in gene repression
Are NL contacts or their heterochromatic state key to the repressive effect of LADs? Considering the cell-to-cell variation in LAD positioning, it is difficult to imagine that NL contacts are sufficient for robust gene inactivation. Nevertheless, the NL contact frequency of genes as determined by single-cell DamID correlated inversely with their cell population average expression levels (Kind et al., 2015). Furthermore, sequential ChIP-DamID experiments indicated that interactions of individual genes with the NL are accompanied by reduced levels of H3K36me3, a histone mark linked to transcriptional elongation activity (Kind et al., 2013). This observation suggests an increased leakiness of the transcriptional repression of genes contained in LADs when they are not actually associated with the nuclear lamina. However, these data are only correlative. Insights into the causal relationship between NL association and gene expression have come from artificial tethering experiments and perturbation of NL proteins.
NL tethering experiments suggest reduced gene expression through NL contact. In one report, an integrated reporter gene showed an ~2–3-fold reduction in expression after NL-tethering using a lac operator array (Reddy et al., 2008). A similar study found only a marginal reduction in expression of a reporter gene near the tethering site, while a subset of endogenous flanking genes on the same chromosome showed dampened expression (Finlan et al., 2008). In a third study, tethering of a reporter gene array to the NL did not change the kinetics of reporter activation after induction by the strong VP16 acidic transcriptional activator (Kumaran and Spector, 2008). Finally, in Drosophila the tethering of Lamin C caused NL association and substantial repression of two reporter genes, but the magnitude of the repression depended on the integration site as well as the reporter gene (Dialynas et al., 2010). Together, these results indicate that proximity to the NL can cause reduction in gene expression, but only of a subset of genes.
If interactions with the NL are important for repression of transcription in the natural context, then disruption of the NL might be expected to de-repress transcription. In Drosophila, testis-specific gene clusters that are normally repressed and associated with the NL in somatic tissues are upregulated and detached from the NL upon depletion of Lamin (Shevelyov et al., 2009). During Drosophila neuroblast differentiation, the hunchback (Hb) gene is silenced and moves to the nuclear periphery. Depletion of Lamin prevents this relocation and results in incomplete silencing of Hb and, as a consequence, extension of neuroblast competence (Kohwi et al., 2013). In Drosophila fat bodies, genes contributing to the immune response are enriched in LADs, and an age-related reduction in Lamin expression was linked to loss of heterochromatin and derepression of these genes (Chen et al., 2014a). Reduction of Lamin in fat bodies in young animals recapitulated this derepression, while forced Lamin expression reversed the aging phenotype of chronic inflammation tied to loss of fat body heterochromatin and immune gene derepression in older animals. Thus, in Drosophila there are several indications that the NL contributes to repression of genes in LADs.
In contrast, in C. elegans a global reduction of NL interactions by depletion of the anchoring protein CEC-4 resulted in upregulation of only one single gene, while depletion of H3K9 methylation caused upregulation of many more genes (Gonzalez-Sandoval et al., 2015). NL contacts thus seemed to be of lesser importance for gene repression than H3K9 methylation itself, although effects of residual NL tethering by other proteins cannot be ruled out. However, deletion of CEC-4 did impair artificially induced differentiation into muscle cells (Gonzalez-Sandoval et al., 2015), suggesting that NL anchoring may be required to stabilize gene expression programs during particular differentiation steps. In mammalian cells depletion of NL components can lead to changes in gene expression (e.g., Amendola and van Steensel, 2015; Kim et al., 2011; Solovei et al., 2013), but so far these changes do not appear to be specific for LADs and therefore may be mostly unrelated to NL contacts. Together, these results illustrate that NL contacts may contribute to a varying degree to gene repression, depending on the species, cell type and genomic loci involved.
How NL contacts might promote repression is unknown. One possible model is that the NL harbors enzymatic activities that modify chromatin to induce a repressive state. For example, the histone deacetylase HDAC3 is activated by its interaction with emerin (Demmerle et al., 2012). By such mechanisms transient contact of a gene with the NL could lead to changes in the chromatin state that may last longer than the actual contact, but as yet there is no experimental evidence to support this. High-resolution FISH or live-cell imaging of the transcription activity of single genes in LADs may reveal whether genes having variable interactions with the NL become more active after they detach from the NL, or alternatively whether their repressed state is not dependent on permanent NL contact.
An alternative model posits that direct gene–NL contacts are not key to repression, but rather that anchoring of a locus to the NL results in its sequestration inside the peripheral layer of heterochromatin, which may be viewed as a separate three-dimensional compartment. This results in removal of the locus from transcriptionally active nuclear compartments such as transcription factories or nuclear speckles. The heterochromatic environment may also actively inhibit transcription, for example due to the high local concentration of repressive enzymatic activities such as H3K9 methyltransferases (Towbin et al., 2012), or because the heterochromatic layer restricts access of certain transcription regulators (Yao et al., 2011). This repressive effect may persist without actual NL contacts, as long as the locus remains embedded in the heterochromatin layer (Figure 2B, C).
Additional mechanisms may be involved in case of specific loci, as illustrated by the recently discovered role of LBR in X chromosome inactivation in female mouse cells (Chen et al., 2016). Xist is a non-coding RNA that normally coats the inactive X (Xi) and is crucial for its silencing. Mutational analysis and microscopy studies indicate that LBR binds to Xist and thereby helps to recruit Xi to the NL early in the inactivation process. This in turn promotes spreading of Xist along Xi, leading to stable silencing.
Links with nucleolus-associated chromatin and pericentromeric heterochromatin
Electron microscopy images show that nucleoli typically are surrounded by a layer of heterochromatin that is roughly similar in appearance to the heterochromatin adjacent to the NL (Figure 1A). Indeed, several studies indicate that this perinucleolar heterochromatin is closely related to LADs. Purification and mapping of DNA sequences associated with nucleoli has resulted in the genome-wide identification of nucleolus-associated domains (NADs) (Nemeth et al., 2010; van Koningsbruggen et al., 2010). These NADs overlap partially with LADs, and by FISH microscopy some NADs were indeed found to be located near the NL in a subset of cells. DamID and photoactivation labeling experiments showed that both nucleolus- and NL-associated loci could switch positions after mitosis (Kind et al., 2013) (van Koningsbruggen et al., 2010). Thus, at least a subset of LADs is variably positioned at either the NL or in close association with nucleoli (Figure 2C).
A third class of heterochromatin is formed by pericentromeric sequences, which in certain cell types and in some species aggregate in so-called chromocenters. Microscopy and DamID studies have shown preferential positioning of centromeres at the nuclear periphery, albeit with substantial centromere-to-centromere and cell-to-cell variation (Guelen et al., 2008; Solovei et al., 2004). 4C experiments have shown that pericentromeric heterochromatin is preferentially located near other LADs in nuclear space, particularly in differentiated cells (Wijchers et al., 2015). This may reflect their embedding in the peripheral layer of heterochromatin, together with many other LADs. It may also indicate that LADs occasionally associate with pericentric heterochromatin, away from the NL (Bian et al., 2013; Ragoczy et al., 2014).
A picture thus emerges (Politz et al., 2016) of large heterochromatic domains that can associate with the NL (in which case they are called LADs), with nucleoli (NADs), or with pericentric heterochromatin (Figure 2C). There is substantial overlap between these domain types, but they are not identical (Ragoczy et al., 2014). Some heterochromatic genomic regions such as cLADs appear to be preferentially positioned at the NL (Kind et al., 2015), others distribute nearly randomly over all three compartments, and yet others (particularly on small chromosomes) show a preference for nucleoli (Nemeth et al., 2010; Ragoczy et al., 2014; van Koningsbruggen et al., 2010). Interestingly, disruption of nucleoli causes a shift of associated genomic loci towards the NL (Ragoczy et al., 2014), while deletion of the regions that conferred NL association of the HBB transgenes, resulted in a shift towards the pericentromeric compartment (Bian et al., 2013). These results indicate that the distribution across these different heterochromatin compartments is determined in a competitive manner involving specific targeting elements. This balance and the cell-to-cell variation may depend on the cell type.
If embedding of a gene in any heterochromatin compartment is sufficient for its repression, then association with the NL, nucleoli or pericentromeric heterochromatin, or aggregates of heterochromatin within the nuclear interior, may all have the same repressive outcome. Hence, even if a LAD is not always located at the NL it could still be completely silenced. However, it cannot be ruled out that the NL compartment is special and cannot be fully compensated by association with other heterochromatin compartments. Systematic analysis of the dynamic partitioning of the genome over the major heterochromatic and euchromatic compartments, by integrated genomics and microscopy approaches, should shed light on this matter.
LADs, TADs and Compartment B
Chromatin conformation capture technologies such as 4C, 5C and Hi-C also have revealed a global domain organization of metazoan genomes. At the megabase scale, genomes are partitioned into domains that segregate into two major compartments, named A and B (Lieberman-Aiden et al., 2009; Simonis et al., 2006). Compartment A mostly includes active genes, while compartment B is predominantly transcriptionally inactive. A NL contact frequency map in human cells showed a nearly identical genome partitioning as the compartment A/B map, with LADs corresponding to B-type domains and inter-LADs corresponding to A-type domains (Figure 3) (Kind et al., 2015). This strong overlap indicates that LADs tend to interact with each other and are spatially separated from most of the transcriptionally active part of the genome. The simplest interpretation is that compartment B is mostly located at the NL, while compartment A is primarily located in the nuclear interior. However, considering the strong overlap of LADs, NADs and PADs, compartment B may also correspond to the union of all heterochromatin compartments, including at the NL, nucleoli and centromeres. Recent modeling of single-cell Hi-C mapping data suggest that this is indeed the case (Stevens et al., 2017).
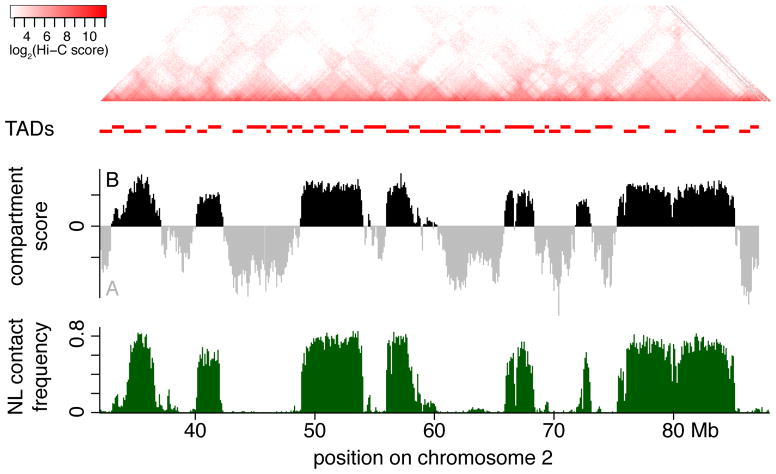
Figure 3. LADs compared to domains as identified by Hi-C.
Comparison of a NL contact frequency profile and Hi-C data in human KBM7 cells (see Data Analysis). Note the remarkably strong similarity of NL contact frequencies to the compartment A/B profile, and the partial similarity to TAD structure.
Hi-C mapping at higher resolution has revealed roughly 2,000 topologically associated domains (TADs), which are sharply defined genomic domains that exhibit preferential intra-domain contacts and are relatively isolated from their neighboring domains (Dixon et al., 2012; Nora et al., 2012) (Figure 3). Like LADs, TADs are frequently demarcated by CTCF binding sites at their borders (Merkenschlager and Nora, 2016). Numerous examples can be found of LADs that coincide with TADs and vice versa, but there are also many instances where they do not seem to overlap (Dixon et al., 2012; Nora et al., 2012). The relationship of TADs with LADs may appear less clear-cut due to the fact that TADs are often nested structures (i.e., subTADs within TADs), which makes their unambiguous annotation difficult and dependent on the algorithm and thresholds applied. Application of a TAD detection algorithm that takes this hierarchical structure into account indicated that high-level TADs (meta-TADs), which largely coincide with the compartment A/B domains, show a better overlap with LADs than low-level TADs (Fraser et al., 2015).
LADs do not appear to be linked to nuclear pore complexes
The nuclear membrane is punctuated by nuclear pore complexes (NPCs), which are large protein complexes that mediate transport between the nucleus and cytoplasm. In yeast, which does not have a NL, there is extensive evidence that NPCs can interact with specific genes as well as telomeres (Ibarra and Hetzer, 2015; Randise-Hinchliff and Brickner, 2016). In metazoans, the picture is more complex. Many nuclear pore proteins (Nups) are not only part of the NPC but also roam the nuclear interior, making interpretation of ChIP and DamID maps of these proteins challenging. Here we focus on a handful of reports in which Nup-interacting loci were confirmed by microscopy to be preferentially located at the nuclear periphery, and thus presumably associated with NPCs.
In C. elegans, a heat-shock promoter was found to target to the NPC upon its activation (Rohner et al., 2013). In D. melanogaster, DamID mapping with a modified Nup that was tightly anchored to the NPC primarily identified small interaction sites within long, moderately active genes (Kalverda et al., 2010). Some overlap of these sites with the insulator protein Su(Hw) was also reported (Kalverda and Fornerod, 2010), but the functional relevance is unclear. In mouse embryonic stem cells, DamID mapping identified Nup153 as binding near the transcriptional start site (TSS) of developmental genes, with additional experiments suggesting that Nup153 mediates repression of these genes through recruitment of the polycomb-repressive complex 1 (PRC1) (Jacinto et al., 2015). In stark contrast, in two human cell lines Nup93 and Nup153 showed a tendency to interact with super-enhancer regions that are preferentially located near the nuclear periphery. Depletion of Nup153 and Nup93 resulted in transcriptional changes of the genes associated with these enhancers (Ibarra et al., 2016).
To date, these results cannot easily be combined into a coherent picture. Nevertheless, they indicate that the genomic elements that interact with NPCs are generally excluded from LADs and are also not located near LAD borders, despite the close proximity of NPCs to the NL.
Outlook and future challenges
The extensive characterization of LADs has provided many insights, but there are still many unresolved issues; here we highlight several:
Heterogeneity of LADs and anchoring molecules
It is likely that LADs are heterogeneous in their chromatin composition, their regulatory functions, and the way they interact with the NL. We expect that multiple mechanisms target fLADs in cell-type specific manners to the NL. Even within individual LADs multiple anchoring mechanisms may act redundantly (Bian et al., 2013; Harr et al., 2015; Zullo et al., 2012). Screens and sensitive high-resolution mapping methods will be needed to efficiently unravel this complexity.
NL-targeting DNA sequences
A related question is whether LAD organization is encoded in the DNA sequence. For example, is it merely the high AT-content of DNA that drives cLAD formation, or are there more complex sequence elements with tethering functions hidden in cLADs? Do specific sequence motifs target fLADs to the NL in cell type dependent manners? New approaches may be needed to systematically identify such elements and compare their features. Likewise, deletion and ectopic insertion of LAD border sequences may provide important mechanistic insights into mechanisms that confine LADs. It is likely that all of these sequence elements act in concert, and hence the local context must be carefully characterized when studying the effects of individual elements. It will also be important to investigate whether genome rearrangements, as frequently found in cancer, affect NL interactions and downstream regulatory events.
Gene regulation in LADs
LADs harbor hundreds of transcriptionally inactive genes, but we should not assume that their inactive state is always due to the repressive effects of the LAD environment or the NL interactions. Many of these genes may be inactive simply because an essential tissue-specific activating transcription factor is not expressed. So far, few mammalian genes have been demonstrated to be regulated by their LAD chromatin state. One test would be to transplant a candidate gene to a neutral chromatin environment (e.g., an episomal vector) in the same cell type; if this restores expression then the gene must have been repressed in the LAD environment. Systematic identification of genes that are naturally repressed in LADs is a first step towards elucidating the underlying mechanisms of the repressive effects of LADs. Also interesting are LAD-embedded genes that appear not to be repressed: do they have specific regulatory elements that render them insensitive to the heterochromatic environment?
Structure of LADs and NL contacts
The macromolecular structures of LADs and of the LAD - NL interaction interface are completely unknown. How does LAD chromatin interact with the NL? If we could take an instant, high-resolution snapshot of a LAD, how long are the chromatin stretches that associate with the NL at that time, and how are they folded? Initial super-resolution microscopy experiments have suggested that patches of LAD chromatin are embedded in pockets in the NL (Kind et al., 2015), but much more work is needed to elucidate these interactions and their dynamics. Addressing these questions will require electron microscopy, super-resolution light microscopy in live cells and molecular probing methods.
Chromosome architecture
Anchoring of LADs to the NL likely contributes to the overall folding of interphase chromosomes. cLADs may be of particular interest, as they generally have the highest NL contact frequencies (Kind et al., 2015) and thus form the most stable anchors. Systematic deletion of cLADs along a chromosome, followed by microscopy and Hi-C analysis of the resulting changes in the spatial organization of the chromosome, should address this hypothesis. Tethering of chromosome regions to the NL may also provide an anchor point by which a local spreading of chromatin compaction can “reel” in neighboring chromatin away from “active” nuclear compartments in the nuclear interior.
Mechanical roles and mechanotransduction
Anchoring of the NL to chromosomes through LADs may also strengthen the NL itself and stiffen the nucleus, thus enhancing resistance of the nucleus to physical stress (Bustin and Misteli, 2016). Furthermore, LAD–NL attachments may be entry points for mechanotransduction across the nuclear envelope (Osmanagic-Myers et al., 2015). For example, small displacements of a bead attached to the cell surface were shown to lead to near instantaneous chromatin stretching inside the nucleus and an increase in transcription, proportional to the magnitude of stretching and detectable within seconds after the force application (Tajik et al., 2016). The LINC complex is of particular interest because it spans the space between the inner and outer nuclear membrane and directly connects the cytoskeleton to the NL (Chang et al., 2015). Whether and how LINC conveys mechanical signals to LADs may become an exciting area of future research.
Other nuclear functions
LADs show a strong overlap with domains of late DNA replication (Guelen et al., 2008; Peric-Hupkes et al., 2010; Pope et al., 2014). Does this merely reflect the heterochromatic nature of LADs or do NL interactions contribute to the control of DNA replication timing? Another nuclear function that may be modulated by NL interactions is DNA double strand break (DSB) repair. It was reported that artificial tethering of a DSB site to the NL causes a shift from repair by homologous recombination to repair by non-homologous end-joining (Lemaitre et al., 2014). Future studies may include more extensive comparisons of DSB repair kinetics and fidelity inside and outside of natural LADs.
Laminopathies
A wide range of human disorders have been linked to mutations in lamins and other NL proteins. Detailed reviews of these laminopathies can be found elsewhere, e.g. (Robin and Magdinier, 2016). Interestingly, loss of the peripheral heterochromatin layer has been observed by electron microscopy in cells from patients with autosomal recessive mandibuloacral dysplasia (Filesi et al., 2005) or Hutchinson–Gilford Progeria Syndrome (Shumaker et al., 2006), which are both caused by mutations in Lamin A. To what extent are laminopathies explained by altered LAD - NL interactions? Detailed genome-wide mapping of changes in NL interactions and gene expression as a consequence of laminopathy mutations may help to address this question. However, because the pathology of these disorders is extremely complex and involves specific sets of cell types, model systems must be chosen carefully. Answers to the fundamental questions as outlined above may contribute towards the understanding of laminopathies and possibly other human disorders.
Data analysis
Hi-C and single-cell DamID data in Figure 3 are from (Kind et al., 2015). Compartment scores as in (Kind et al., 2015) were provided by Geoffrey Fudenberg. The TAD track was kindly generated by Robin van der Weide using TADtool (Crane et al., 2015) with parameter settings window size = 590000 and cutoff = 29.98104.
Acknowledgments
We thank members of our laboratories, David Gilbert, Juan Carlos Rivera Mulia, Jian Ma, Elzo de Wit, V.K. Prasanth, Supriya Prasanth, Jie Chen and anonymous reviewers for helpful comments; Kenneth M. Bart and Jop Kind for providing images in Figures 1A and B, respectively; Geoffrey Fudenberg and Robin van der Weide for help with Figure 3. We apologize to colleagues whose work we could not discuss due to space constraints. BvS and ASB are supported by the NIH “4D Nucleome” U54 grant DK107965, NIH R01 GM58460 (ASB) and ERC Advanced Grant 293663 “ChromatinPrinciples” (BvS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LF, van Lohuizen M, van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Amendola M, van Steensel B. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep. 2015;16:610–617. doi: 10.15252/embr.201439789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berk JM, Tifft KE, Wilson KL. The nuclear envelope LEM-domain protein emerin. Nucleus. 2013;4:298–314. doi: 10.4161/nucl.25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2011;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Khanna N, Alvikas J, Belmont AS. beta-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J Cell Biol. 2013;203:767–783. doi: 10.1083/jcb.201305027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M, Misteli T. Nongenetic functions of the genome. Science. 2016;352:aad6933. doi: 10.1126/science.aad6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol. 2015;208:11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, Chow A, Cerase A, McDonel P, Guttman M. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- Chen H, Zheng X, Zheng Y. Age-associated loss of lamin-B leads to systemic inflammation and gut hyperplasia. Cell. 2014a;159:829–843. doi: 10.1016/j.cell.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yammine S, Shi C, Tark-Dame M, Gondor A, Ohlsson R. The visualization of large organized chromatin domains enriched in the H3K9me2 mark within a single chromosome in a single cell. Epigenetics. 2014b;9:1439–1445. doi: 10.4161/15592294.2014.971633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Belmont AS. Moving chromatin within the interphase nucleus-controlled transitions? Semin Cell Dev Biol. 2007;18:698–706. doi: 10.1016/j.semcdb.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Bouwman BA, Zhu Y, Klous P, Splinter E, Verstegen MJ, Krijger PH, Festuccia N, Nora EP, Welling M, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- Demmerle J, Koch AJ, Holaska JM. The nuclear envelope protein emerin binds directly to histone deacetylase 3 (HDAC3) and activates HDAC3 activity. J Biol Chem. 2012;287:22080–22088. doi: 10.1074/jbc.M111.325308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas G, Speese S, Budnik V, Geyer PK, Wallrath LL. The role of Drosophila Lamin C in muscle function and gene expression. Development. 2010;137:3067–3077. doi: 10.1242/dev.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman LB, Fraser P. Transcription factories: genetic programming in three dimensions. Curr Opin Genet Dev. 2012;22:110–114. doi: 10.1016/j.gde.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat. 1966;119:129–145. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- Filesi I, Gullotta F, Lattanzi G, D’Apice MR, Capanni C, Nardone AM, Columbaro M, Scarano G, Mattioli E, Sabatelli P, et al. Alterations of nuclear envelope and chromatin organization in mandibuloacral dysplasia, a rare form of laminopathy. Physiol Genomics. 2005;23:150–158. doi: 10.1152/physiolgenomics.00060.2005. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J, Ferrai C, Chiariello AM, Schueler M, Rito T, Laudanno G, Barbieri M, Moore BL, Kraemer DC, Aitken S, et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol Syst Biol. 2015;11:852. doi: 10.15252/msb.20156492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesson K, Rescheneder P, Skoruppa MP, von Haeseler A, Dechat T, Foisner R. Atype lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 2016;26:462–473. doi: 10.1101/gr.196220.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesson K, Vidak S, Foisner R. Lamina-associated polypeptide (LAP)2alpha and nucleoplasmic lamins in adult stem cell regulation and disease. Semin Cell Dev Biol. 2014;29:116–124. doi: 10.1016/j.semcdb.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C, Ikegami K, Ayuso C, de Luis A, Iniguez M, Cabello J, Lieb JD, Askjaer P. Genome-wide analysis links emerin to neuromuscular junction activity in Caenorhabditis elegans. Genome Biol. 2014;15:R21. doi: 10.1186/gb-2014-15-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A, Towbin BD, Kalck V, Cabianca DS, Gaidatzis D, Hauer MH, Geng L, Wang L, Yang T, Wang X, et al. Perinuclear Anchoring of H3K9-Methylated Chromatin Stabilizes Induced Cell Fate in C. elegans Embryos. Cell. 2015;163:1333–1347. doi: 10.1016/j.cell.2015.10.066. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol. 2015;208:33–52. doi: 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Benner C, Tyagi S, Cool J, Hetzer MW. Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 2016;30:2253–2258. doi: 10.1101/gad.287417.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015;29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Egelhofer TA, Strome S, Lieb JD. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010;11:R120. doi: 10.1186/gb-2010-11-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto FV, Benner C, Hetzer MW. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015;29:1224–1238. doi: 10.1101/gad.260919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamin A, Wiebe MS. Barrier to Autointegration Factor (BANF1): interwoven roles in nuclear structure, genome integrity, innate immunity, stress responses and progeria. Curr Opin Cell Biol. 2015;34:61–68. doi: 10.1016/j.ceb.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Fornerod M. Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle. 2010;9:4812–4817. doi: 10.4161/cc.9.24.14328. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Khanna N, Hu Y, Belmont AS. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol. 2014;24:1138–1144. doi: 10.1016/j.cub.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015;163:134–147. doi: 10.1016/j.cell.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Kind J, van Steensel B. Stochastic genome-nuclear lamina interactions: modulating roles of Lamin A and BAF. Nucleus. 2014;5:124–130. doi: 10.4161/nucl.28825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152:97–108. doi: 10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Le HQ, Ghatak S, Yeung CY, Tellkamp F, Gunschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol. 2016;18:864–875. doi: 10.1038/ncb3387. [DOI] [PubMed] [Google Scholar]
- Lemaitre C, Grabarz A, Tsouroula K, Andronov L, Furst A, Pankotai T, Heyer V, Rogier M, Attwood KM, Kessler P, et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014;28:2450–2463. doi: 10.1101/gad.248369.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Oldenburg AR, Delbarre E, Freberg CT, Duband-Goulet I, Eskeland R, Buendia B, Collas P. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res. 2013;23:1580–1589. doi: 10.1101/gr.159400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund EG, Duband-Goulet I, Oldenburg A, Buendia B, Collas P. Distinct features of lamin A-interacting chromatin domains mapped by ChIP-sequencing from sonicated or micrococcal nuclease-digested chromatin. Nucleus. 2015;6:30–39. doi: 10.4161/19491034.2014.990855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout A, Pike BL, Towbin BD, Bank EM, Gonzalez-Sandoval A, Stadler MB, Meister P, Gruenbaum Y, Gasser SM. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr Biol. 2011;21:1603–1614. doi: 10.1016/j.cub.2011.08.030. [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, Nora EP. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu Rev Genomics Hum Genet. 2016;17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Langst G. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins AL, Rhodes G, Welch DB, Zwerger M, Olins DE. Lamin B receptor: multi-tasking at the nuclear envelope. Nucleus. 2010;1:53–70. doi: 10.4161/nucl.1.1.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanagic-Myers S, Dechat T, Foisner R. Lamins at the crossroads of mechanosignaling. Genes Dev. 2015;29:225–237. doi: 10.1101/gad.255968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Poleshko A, Mansfield KM, Burlingame CC, Andrake MD, Shah NR, Katz RA. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 2013;5:292–301. doi: 10.1016/j.celrep.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Scalzo D, Groudine M. The redundancy of the mammalian heterochromatic compartment. Curr Opin Genet Dev. 2016;37:1–8. doi: 10.1016/j.gde.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T, Telling A, Scalzo D, Kooperberg C, Groudine M. Functional redundancy in the nuclear compartmentalization of the late-replicating genome. Nucleus. 2014;5:626–635. doi: 10.4161/19491034.2014.990863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randise-Hinchliff C, Brickner JH. Transcription factors dynamically control the spatial organization of the yeast genome. Nucleus. 2016;7:369–374. doi: 10.1080/19491034.2016.1212797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Robin JD, Magdinier F. Physiological and Pathological Aging Affects Chromatin Dynamics, Structure and Function at the Nuclear Edge. Front Genet. 2016;7:153. doi: 10.3389/fgene.2016.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson MI, de Las Heras JI, Czapiewski R, Le Thanh P, Booth DG, Kelly DA, Webb S, Kerr AR, Schirmer EC. Tissue-Specific Gene Repositioning by Muscle Nuclear Membrane Proteins Enhances Repression of Critical Developmental Genes during Myogenesis. Mol Cell. 2016;62:834–847. doi: 10.1016/j.molcel.2016.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol. 2013;200:589–604. doi: 10.1083/jcb.201207024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Shevelyov YY, Lavrov SA, Mikhaylova LM, Nurminsky ID, Kulathinal RJ, Egorova KS, Rozovsky YM, Nurminsky DI. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc Natl Acad Sci U S A. 2009;106:3282–3287. doi: 10.1073/pnas.0811933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell. 2015;26:4075–4086. doi: 10.1091/mbc.E15-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Solovei I, Schermelleh L, During K, Engelhardt A, Stein S, Cremer C, Cremer T. Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma. 2004;112:410–423. doi: 10.1007/s00412-004-0287-3. [DOI] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O’Shaughnessy-Kirwan A, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017 doi: 10.1038/nature21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater. 2016;15:1287–1296. doi: 10.1038/nmat4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- van Bemmel JG, Pagie L, Braunschweig U, Brugman W, Meuleman W, Kerkhoven RM, van Steensel B. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS One. 2010;5:e15013. doi: 10.1371/journal.pone.0015013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers PJ, Geeven G, Eyres M, Bergsma AJ, Janssen M, Verstegen M, Zhu Y, Schell Y, Vermeulen C, de Wit E, et al. Characterization and dynamics of pericentromere-associated domains in mice. Genome Res. 2015;25:958–969. doi: 10.1101/gr.186643.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong X, Luperchio TR, Reddy KL. NET gains and losses: the role of changing nuclear envelope proteomes in genome regulation. Curr Opin Cell Biol. 2014;28:105–120. doi: 10.1016/j.ceb.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Xie W, Chojnowski A, Boudier T, Lim JS, Ahmed S, Ser Z, Stewart C, Burke B. Atype Lamins Form Distinct Filamentous Networks with Differential Nuclear Pore Complex Associations. Curr Biol. 2016;26:2651–2658. doi: 10.1016/j.cub.2016.07.049. [DOI] [PubMed] [Google Scholar]
- Yao J, Fetter RD, Hu P, Betzig E, Tjian R. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 2011;25:569–580. doi: 10.1101/gad.2021411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci U S A. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sifakis EG, Sumida N, Millan-Arino L, Scholz BA, Svensson JP, Chen X, Ronnegren AL, Mallet de Lima CD, Varnoosfaderani FS, et al. PARP1- and CTCF-Mediated Interactions between Active and Repressed Chromatin at the Lamina Promote Oscillating Transcription. Mol Cell. 2015;59:984–997. doi: 10.1016/j.molcel.2015.07.019. [DOI] [PubMed] [Google Scholar]
- Zheng X, Kim Y, Zheng Y. Identification of lamin B-regulated chromatin regions based on chromatin landscapes. Mol Biol Cell. 2015;26:2685–2697. doi: 10.1091/mbc.E15-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleger N, Boyle S, Kelly DA, de las Heras JI, Lazou V, Korfali N, Batrakou DG, Randles KN, Morris GE, Harrison DJ, et al. Specific nuclear envelope transmembrane proteins can promote the location of chromosomes to and from the nuclear periphery. Genome Biol. 2013;14:R14. doi: 10.1186/gb-2013-14-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]