Abstract
Human noroviruses (NoV) are a leading cause of gastroenteritis globally, yet host factors required for NoV infection are poorly understood. We identified host molecules essential for murine NoV (MNoV) induced cell death including CD300lf as a proteinaceous receptor. CD300lf is essential for MNoV binding and replication in cell lines and primary cells. Additionally, Cd300lf−/− mice are resistant to MNoV infection. Expression of CD300lf in human cells breaks the species barrier restricting MNoV replication. The crystal structure of the CD300lf ectodomain revealed a potential ligand binding cleft composed of residues critical for MNoV infection. Therefore, the presence of a proteinaceous receptor is the primary determinant of MNoV species tropism while other components of cellular machinery required for NoV replication are conserved between humans and mice.
Main text
Noroviruses (NoV) are non-enveloped positive-sense RNA viruses (1, 2). Due to the strict species tropism of viruses in the NoV genus and the lack of robust replication of human noroviruses (HNoV) in animal models, murine norovirus (MNoV) has emerged as a model system to uncover basic mechanisms of NoV biology in vitro and in vivo (1, 3–8). MNoVs can establish persistent enteric infection enabling studies of the interplay between viral persistence, resident enteric microorganisms, and the host immune system (4–6). The capacity of HNoV and MNoV to bind cells, and the susceptibility of humans to HNoV, have been linked to expression of cell surface and secreted carbohydrates (1, 9–13), while another member of the Caliciviridae utilizes a proteinaceous receptor (14, 15). Host factors including receptor(s) required for NoV infection and pathogenesis have largely defied molecular identification; their discovery would aid in understanding mechanisms of NoV replication, vaccination, species tropism, and enteric viral persistence. To identify host molecules required for MNoV infection, we undertook an unbiased forward genetic approach (Fig. S1).
MNoV replicates and induces cell death in murine macrophage-like cells, including the microglial BV2 cell line, allowing identification of genes essential for MNoV replication using CRISPR-Cas9 technology. We introduced four independent genome-wide subpools of single guide RNAs (sgRNAs) of the murine Asiago library into BV2 cells stably expressing Cas9 and then infected the cells with MNoV strains that either cause acute systemic infection (MNoVCW3) or persistent enteric infection (MNoVCR6) in mice (16–18). sgRNA sequences from the surviving cells were sequenced and analyzed using the STARS gene-ranking algorithm (Fig. 1A) (17). sgRNAs targeting Cd300lf (CLM-1, CMRF35, MAIR-V, LMIR3), a cell surface immunoglobulin (Ig) domain-containing molecule within a family of proteins involved in binding lipids, were most significantly enriched for both MNoV strains (Fig. 1A and Table S1–3) (19). We generated two independent BV2ΔCD300lf clones; the growth of MNoVCW3 and MNoVCR6 was abolished in both clones while the replication of other viruses was unaffected (Fig 1B and S2). We also validated several additional molecules predicted to be important for MNoV-induced cell death (Fig. 1A and S3). Taken together, these data provide a systematic overview of the molecules required for NoV replication in these cells.
Fig. 1. CRISPR screen identifies CD300lf as necessary for MNoV infection.
(A) A heat map showing enrichment of genes in the two indicated conditions. Genes are color coded based upon their STARS score. (B) Wild-type BV2 cells or two independently-derived CD300lf-deficient clones (clone 1 and clone 2) transduced with an empty vector or a vector expressing CD300lf were challenged with an MOI of 0.05 of MNoVCW3 (top) or MNoVCR6 (bottom) infection and virus production was assessed by plaque forming units (PFU). Shown is mean ± SEM for data pooled from three independent experiments. L.O.D., limit of detection.
We selected CD300lf for further analysis because of its cell surface expression and the importance of viral receptors for conferring permissiveness for viral replication and species tropism. Transfection of MNoVCW3 RNA into BV2ΔCD300lf cells was sufficient to rescue MNoVCW3 production demonstrating, that CD300lf is essential for viral entry (Fig. 2A). Preincubation of cells with a polyclonal antibody targeting CD300lf (α-CD300lf) blocked MNoVCW3-induced cytopathic effects in BV2 cells (Fig. 2B). Similarly, incubating MNoVCW3 with recombinant CD300lf ectodomain (sCD300lf) neutralized MNoVCW3-induced cytopathic effects while Annexin V or phosphoserine treatment had no effect (Fig. 2C and S4). Similar results were obtained for infection of the B-cell line M12, bone marrow-derived dendritic cells (BMDC), and bone marrow-derived macrophages (BMDM) indicating the essential role of CD300lf in multiple cell types (Fig. 2D). Taken together these data indicate that interactions between MNoV and CD300lf are essential for MNoV infection.
Fig. 2. CD300lf is an MNoV receptor.
(A) Indicated BV2 cells were transfected with MNoVCW3 RNA and harvested 12 hours post-transfection. Viral production was measured by plaque assay. ns = not statistically significant. Shown is mean ± SEM for data pooled from three independent experiments. (B) BV2 cells were incubated with either α-CD300lf or an isotype control prior to infection with MNoVCW3 at an MOI 5.0. Cell viability was measured 24 hours post-infection. Shown is mean ± SEM for data pooled from three independent experiments. (C) MNoVCW3 was incubated with either the recombinant, soluble ectodomain of CD300lf (sCD300lf) or a control protein prior to infection of BV2 cells. Cellular viability was assayed 24 hours post-infection. Shown is mean ± SEM for data pooled from three independent experiments. (D) MNoVCW3 infection was inhibited by either cellular pretreatment of α-CD300lf or viral pretreatment with sCD300lf. Infection was measured by FACS for intracellular MNoV NS1/2 expression and inhibition relative to an isotype control or control protein, respectively. Shown is mean ± SEM for data pooled from three independent experiments. (E) MNoVCW3 binding assay in complete media to indicated BV2 cell lines as assayed by bound MNoV genomes via qPCR. Representative binding assay when cells or virus were preincubated with α-CD300lf or sCD300lf, respectively. Analyzed by one-way ANOVA with Tukey’s multiple comparison test; three independent experiments performed in triplicate. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. (F) MNoVCW3 binding assay done in PBS plus 10% fetal bovine serum (FBS) or derivatives as indicated. Analyzed by one-way ANOVA with Tukey’s multiple comparison test; three independent experiments performed in triplicate. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
To directly test if CD300lf functions as a binding receptor for MNoV, we analyzed the attachment of MNoVCW3 to BV2 cells on ice. BV2ΔCD300lf cells were impaired in MNoVCW3 binding (Fig. 2E). Additionally, treating BV2 cells with α-CD300lf, or preincubating virus with sCD300lf, reduced MNoV binding (Fig. 2E). In BV2ΔCD300lf cells, binding of MNoVCW3 was not further inhibited with α-CD300lf or by premixing virus with sCD300lf (Fig. 2E). Taken together, these data indicate that CD300lf mediates viral binding and is a functional receptor for MNoV.
Previous reports have suggested carbohydrates facilitate the binding of MNoV and HNoV to host cells and control the susceptibility of humans to HNoV infection (1, 9–11). Therefore, we assessed the relative contribution of carbohydrates to MNoV attachment and infection. Surprisingly, mice deficient in Fut2, which controls histo-blood group antigen (HBGA) secretor status, had similar MNoV loads compared to wild-type, littermate controls (Fig. S5A). Also, treating cells with the mannosidase I inhibitor kifunensine significantly reduced cell surface carbohydrates, but did not significantly alter MNoVCW3 binding to cells (Fig. S5B and S5C). These data suggest that MNoV binding and infection is dependent upon Cd300lf but not on protein-associated glycans or secretor status within the sensitivity of these assays.
In addition to CD300lf, we discovered that efficient MNoV binding to cells requires a cofactor present in serum (Fig. 2F). The cofactor is present in delipidated serum and is resistant to proteinase K and heat denaturation (95°C; Fig. 2F). Size fractionation of serum indicates that the cofactor is present in fractions with an average molecular weight of less than 5,000 Da (Fig 2F). Thus, the serum cofactor is consistent with a small non-proteinaceious heat stable molecule that interacts with MNoV and/or CD300lf to facilitate cellular binding.
Next, we sought to assess the physiologic relevance of MNoV and CD300lf interactions. MNoV isolated from the spleens of MNoVCW3-infected mice remained sensitive to sCD300lf inhibition, indicating that sCD300lf neutralizes MNoV regardless of the source of the virus (Fig. 3A). To test the role of CD300lf and MNoV interactions in vivo, we incubated MNoVCW3 with either sCD300lf or a control protein prior to infection of Stat1−/− mice, which succumb to MNoVCW3 infection. In a dose-dependent manner, sCD300lf preincubation was able to protect from MNoV-induced lethality (Fig. 3B and 3C). Finally, we generated Cd300lf−/− mice to test the in vivo role of CD300lf in MNoV infection. Cd300lf−/− mice were resistant to MNoVCR6 infection compared to littermate controls (Figure 3D). These data demonstrate that CD300lf is the primary physiological receptor for MNoV in vivo.
Fig. 3. CD300lf is a physiological MNoV receptor.
(A) MNoVCW3 harvested from the spleens of BL6/J mice or MNoVCW3 derived from BV2 cells was preincubated with sCD300lf prior to plaque assay. Inhibition is relative to a control protein. Shown is mean ± SEM for data pooled from three independent experiments. (B–C) Survival of Stat1−/− mice from challenge with (G) 103 PFU or (H) 105 PFU of MNoVCW3 preincubated with either sCD300lf or control protein. Analyzed by log-rank test; N=11 mice per cohort combined from three independent experiments. (D) Cd300lf−/− and littermate control mice were challenged with 106 PFU of MNoVCR6 perorally. Fecal shedding of MNoV genomes was monitored for 21 days post-challenge. N=9–10 mice per cohort combined from two independent experiments. Analyzed by repeated measures ANOVA; shown is mean ± SEM. ***P<0.001.
MNoV replicates in murine dendritic cells, macrophages, and B cells, but not in epithelial cells or human cells due to a restriction at viral entry (20, 21). Therefore, we tested if expression of murine CD300lf was sufficient to confer susceptibility of HeLa cells to MNoV. As expected, HeLa cells transfected with a control plasmid were unable to support MNoV replication (Fig. 4A). In contrast, HeLa cells expressing murine but not human CD300lf were susceptible to MNoV (Fig. 4A). These data indicate that CD300lf expression is sufficient for MNoV growth in human cells and suggest that differences between human and murine CD300lf could contribute to MNoV species restriction.
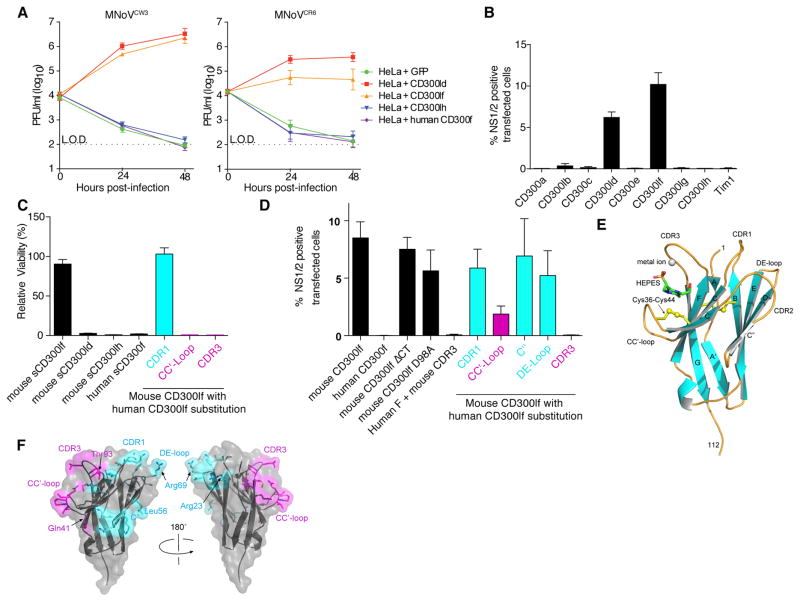
Fig. 4. Structure guided mapping identifies the CC′-loop and CDR3 of CD300lf critical for MNoV infection.
(A). HeLa cells transiently transfected with indicated constructs were infected with either MNoVCW3 (left) or MNoVCR6 (right) at an MOI of 0.05. Viral growth was measured by plaque assay at the indicated time points. Shown is mean ± SEM for data pooled from three independent experiments. (B) HeLa cells transiently transfected with indicated CD300 constructs or Tim1 were infected with MNoVCW3 at an MOI of 5.0 and analyzed for expression of CD300/Tim1 (FLAG) and MNoV NS1/2. Shown is mean ± SEM for data pooled from three independent experiments. (C) Recombinant ectodomains of indicated CD300 molecules (10 μg/ml) were preincubated with MNoVCW3 MOI 5 prior to infection of BV2 cells. Cellular viability was assayed 24 hours post-infection. Shown is mean ± SEM for data pooled from three independent experiments. (D) HeLa cells transiently transfected with indicated CD300 constructs were infected with MNoVCW3 at an MOI of 5.0 and analyzed for expression of CD300 (FLAG) and MNoV NS1/2. CD300lfΔCT, CD300lf with truncation of cytoplasmic domain. Shown is mean ± SEM for data pooled from three independent experiments. (E) Ribbon diagram of murine CD300lf ectodomain with bound metal ion and HEPES. The β-sheets (cyan) are lettered as a canonical V-type Ig domain with the positions of the CDR equivalent loops indicated. The disulfide bonds are shown in yellow. (F) Mapping of mutational results onto the CD300lf surface. Displayed in cyan are murine CD300lf residues replaced by human equivalent residues in CDR1, C″, and DE-loop that had no effect on infection. The CC′-loop and CDR3 mutations that diminished viral infection are shown in magenta.
In mice there are eight CD300 family members (22). Therefore, we sought to determine whether other CD300 molecules are capable of functioning as an MNoV receptor. After transfection into HeLa cells only CD300lf and CD300ld conferred susceptibility to MNoV in HeLa cells while the expression of other CD300 family members or the unrelated phosphatidylserine binding protein Tim1 was unable to support MNoV infection (Fig. 4B and S6) (23). We further confirmed that expression of CD300lf and CD300ld but not CD300lh was sufficient for viral replication in human cells (Fig. 4A). However, in contrast to murine CD300lf the recombinant ectodomains of murine CD300ld, CD300lh, or human CD300lf failed to neutralize viral infection (Figure 4C). Additionally, BV2ΔCD300ld cells are susceptible to MNoV infection suggesting that CD300ld can act as an MNoV receptor when ectopically expressed, but it is not a universal requirement (Fig. S7). However, we cannot exclude the possibility that CD300ld may play a role in viral tropism in some circumstances in vivo. Experiments in Cd300lf−/− mice indicate that this putative role is not essential for intestinal infection or shedding.
We next sought to define the mechanism for MNoV entry via CD300lf and to determine how MNoV discriminates between mouse and human CD300lf proteins. Importantly, the intracellular domain of CD300lf was not required to make HeLa cells susceptible, indicating that species tropism is determined by the ectodomain (CD300lfΔCT, Fig. 4D). We determined the crystal structure of the CD300lf ectodomain at 1.6Å resolution (Fig. 4E and Table S4). Interestingly, density corresponding to a bound HEPES molecule, present in the purification buffers, and a coordinated metal were visible in a surface cleft formed between the CDR3 loop and the β-hairpin turn that connects the C-C′ β-strands. CD300lf has been reported to mediate the phagocytosis of apoptotic cells through the calcium-dependent binding of lipids (24–26). HEPES has chemical resemblance to a phospholipid headgroup (Fig. S8). Our structure also reveals a metal coordinated primarily by CD300lf Asp98 and two CDR3 loop carbonyl oxygens. While mutation of murine CD300lf Asp98 has been shown to disrupt apoptotic cell surface binding, this mutant still allows MNoVCW3 infection of HeLa cells, suggesting that viral entry can occur in the absence of bound metal (Fig. 4D and S8) (25, 27). The Ig-domains of murine and human CD300lf share 59% sequence identity, and structurally the largest variation occurs in CDR3 and the CC′-loop (Fig. S9 and S10) (27). Interestingly, individual substitutions of human CC′-loop and CDR3 into murine CD300lf diminished and abolished MNoVCW3 infection of HeLa cells, respectively (Fig. 4D and 4F). Additionally, purified recombinant proteins harboring CC′-loop and CDR3 human sequences failed to neutralize MNoV infection (Fig. 4C and S11). However, the reciprocal CDR3 mutation (murine CDR3 into human CD300lf) was not sufficient for MNoV infection (Fig. 4D). Independently comparing murine CD300lf and CD300lh also indicated that the CD300lf CC′-loop and CDR3 sequences are necessary but not sufficient for receptor utilization by MNoV (Fig. S12). These data provide a framework for understanding how MNoV discriminates between CD300 family members.
Our work establishes that CD300lf is a functional MNoV receptor that mediates binding to the cell surface and is both necessary and sufficient for viral entry and replication in vitro and in vivo. Because MNoV serves as a model system for understanding how viruses persist and shape the immune system, the modulation of receptor availability either genetically or chemically may foster understanding of immunomodulation, persistence, and tropism of MNoV. This work also enables the future study of MNoV replication in human cells, which may uncover novel mechanisms of viral replication and pathogenesis and allow a direct identification and mechanistic dissection of the cellular factors required for NoV replication across species. Additionally, our work has implications for understanding HNoV infections. HNoV binds to HBGA and susceptibility to HNoV is correlated with host HBGA status (12, 13). These reports are the foundation for the hypothesis that glycans are HNoV receptors (9). However, HBGA alone cannot, to date, explain species tropism or the entry barrier for HNoV. In contrast, our data indicate that murine CD300lf is sufficient to explain tropism for MNoV and more broadly suggest the possibility that other NoVs utilize proteinacious receptors in addition to small molecule cofactors that are present in serum or other biological fluids. It is intriguing that CD300 molecules can bind a range of host lipids and, additionally, bacterial products (22, 28). It may therefore be that NoV cell and tissue tropism is determined in a combinatorial fashion by proteinaceous receptors interacting with permissiveness cofactors present at different sites.
Supplementary Material
Acknowledgments
We would like to thank S. Karst and M. Diamond for providing valuable reagents and S. Handley and C. Desai for their helpful discussion and figure generation. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, Mo., for the use of the Genome Engineeringand iPSC Center. CD300 constructs, cell lines, and mice are available from Dr. Herbert W. Virgin under a material transfer agreement with Washington University. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. Drs. Herbert Virgin, Daved Fremont, Robert Orchard, Craig Wilen, and Christopher Nelson are inventors on a patent application (US Provisional Application 62/301,965) held/submitted by Washington University entitled “Receptor for Norovirus and Uses Thereof”. The atomic coordinates are deposited in the Protein Data Bank under accession code 5FFL. The Siteman Cancer Center is supported in part by NCI Cancer Center SupportGrant #P30 CA091842, Eberlein, PI.” This work was supported by NIH grants U19 AI109725 (H.W.V.), 1F31CA177194 (B.T.M.), and 5T32CA009547 (M.T.B.).
References
- 1.Karst SM, Wobus CE, Goodfellow IG, Green KY, Virgin HW. Advances in norovirus biology. Cell host & microbe. 2014 Jun 11;15:668. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009 Oct 29;361:1776. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones MK, et al. Enteric bacteria promote human and murine norovirus infection of B cells. Science. 2014;346:755. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kernbauer E, Ding Y, Cadwell K. An enteric viral infection can functionally replace the beneficial cues provided by commensal bacteria. Nature. 2014;516:94. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldridge MT, et al. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science. 2015 Jan 16;347:266. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nice TJ, et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015 Jan 16;347:269. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman KL, Leon JS. Norovirus immunology: Of mice and mechanisms. European journal of immunology. 2015 Oct;45:2742. doi: 10.1002/eji.201545512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taube S, et al. A mouse model for human norovirus. mBio. 2013;4 doi: 10.1128/mBio.00450-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutson AM, Atmar RL, Estes MK. Norovirus disease: changing epidemiology and host susceptibility factors. Trends in Microbiology. 2004 Jun;12:279. doi: 10.1016/j.tim.2004.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis. 2002 May 1;185:1335. doi: 10.1086/339883. [DOI] [PubMed] [Google Scholar]
- 11.Taube S, et al. Murine noroviruses bind glycolipid and glycoprotein attachment receptors in a strain-dependent manner. Journal of virology. 2012 May;86:5584. doi: 10.1128/JVI.06854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindesmith L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003 May;9:548. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 13.Kambhampati A, Payne DC, Costantini V, Lopman BA. Host Genetic Susceptibility to Enteric Viruses: A Systematic Review and Metaanalysis. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2015 Oct 26; doi: 10.1093/cid/civ873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ossiboff RJ, Zhou Y, Lightfoot PJ, Prasad BV, Parker JS. Conformational changes in the capsid of a calicivirus upon interaction with its functional receptor. Journal of virology. 2010 Jun;84:5550. doi: 10.1128/JVI.02371-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ossiboff RJ, Parker JS. Identification of regions and residues in feline junctional adhesion molecule required for feline calicivirus binding and infection. J Virol. 2007 Dec;81:13608. doi: 10.1128/JVI.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strong DW, Thackray LB, Smith TJ, Virgin HW. Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. Journal of virology. 2012 Mar;86:2950. doi: 10.1128/JVI.07038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature Biotechnology. 2015 doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nice TJ, Strong DW, McCune BT, Pohl CS, Virgin HW. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. Journal of virology. 2013 Jan;87:327. doi: 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung DH, et al. CMRF-35-like molecule-1, a novel mouse myeloid receptor, can inhibit osteoclast formation. Journal of immunology. 2003 Dec 15;171:6541. doi: 10.4049/jimmunol.171.12.6541. [DOI] [PubMed] [Google Scholar]
- 20.Wobus CE, et al. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS biology. 2004 Dec;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward VK, et al. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proceedings of the National Academy of Sciences of the United States of America. 2007 Jun 26;104:11050. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borrego F. The CD300 molecules: an emerging family of regulators of the immune system. Blood. 2013 Mar 14;121:1951. doi: 10.1182/blood-2012-09-435057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi N, et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 2007 Dec;27:927. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izawa K, et al. The receptor LMIR3 negatively regulates mast cell activation and allergic responses by binding to extracellular ceramide. Immunity. 2012 Nov 16;37:827. doi: 10.1016/j.immuni.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 25.Tian L, et al. p85alpha recruitment by the CD300f phosphatidylserine receptor mediates apoptotic cell clearance required for autoimmunity suppression. Nature communications. 2014;5:3146. doi: 10.1038/ncomms4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi SC, et al. Cutting edge: mouse CD300f (CMRF-35-like molecule-1) recognizes outer membrane-exposed phosphatidylserine and can promote phagocytosis. Journal of immunology. 2011 Oct 1;187:3483. doi: 10.4049/jimmunol.1101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marquez JA, et al. The crystal structure of the extracellular domain of the inhibitor receptor expressed on myeloid cells IREM-1. Journal of molecular biology. 2007 Mar 23;367:310. doi: 10.1016/j.jmb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 28.Cannon JP, O’Driscoll M, Litman GW. Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics. 2012 Jan;64:39. doi: 10.1007/s00251-011-0562-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.