Abstract
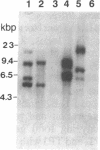
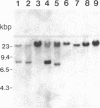
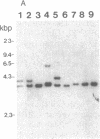
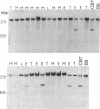
Friend murine leukemia virus (F-MuLV) induces erythroleukemia when inoculated into newborn BALB/c or NIH/Swiss mice. We have molecularly cloned F-MuLV host cell DNA junction fragments from an erythroleukemia cell line induced by F-MuLV to identify cellular genes involved in the leukemogenic process. One particular proviral integration site, Fli-1, is rearranged in 75% (9/12) of independently isolated erythroleukemia cell lines derived from either BALB/c or NIH/Swiss mice inoculated at birth with F-MuLV. Other hematopoietic neoplasms induced by F-MuLV, including myeloid (granulocytic) and lymphoid tumors, did not show rearrangements of the Fli-1 locus. Similarly, none of 35 erythroleukemia cell lines induced by the Friend virus complexes (FV-A and FV-P) was rearranged at the Fli-1 locus. In contrast, no rearrangements were detected at the Sfpi-1 locus, a preferred site of integration in either FV-P- or FV-A-induced leukemias. Using recombinant inbred mice, the Fli-1 locus was situated on mouse chromosome 9 close to the cellular protooncogene c-ets-1. DNA and RNA analysis suggests, however, that Fli-1 is different from ets-1. Thus, Fli-1 appears to define a distinct locus specifically involved in the induction of erythroid leukemias by F-MuLV.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartholomew C., Morishita K., Askew D., Buchberg A., Jenkins N. A., Copeland N. G., Ihle J. N. Retroviral insertions in the CB-1/Fim-3 common site of integration activate expression of the Evi-1 gene. Oncogene. 1989 May;4(5):529–534. [PubMed] [Google Scholar]
- Ben David Y., Prideaux V. R., Chow V., Benchimol S., Bernstein A. Inactivation of the p53 oncogene by internal deletion or retroviral integration in erythroleukemic cell lines induced by Friend leukemia virus. Oncogene. 1988 Aug;3(2):179–185. [PubMed] [Google Scholar]
- Chow V., Ben-David Y., Bernstein A., Benchimol S., Mowat M. Multistage Friend erythroleukemia: independent origin of tumor clones with normal or rearranged p53 cellular oncogenes. J Virol. 1987 Sep;61(9):2777–2781. doi: 10.1128/jvi.61.9.2777-2781.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Cuypers H. T., Selten G., Quint W., Zijlstra M., Maandag E. R., Boelens W., van Wezenbeek P., Melief C., Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984 May;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Duesberg P. H., Bister K., Moscovici C. Genetic structure of avian myeloblastosis virus, released from transformed myeloblasts as a defective virus particle. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5120–5124. doi: 10.1073/pnas.77.9.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung Y. K., Lewis W. G., Crittenden L. B., Kung H. J. Activation of the cellular oncogene c-erbB by LTR insertion: molecular basis for induction of erythroblastosis by avian leukosis virus. Cell. 1983 Jun;33(2):357–368. doi: 10.1016/0092-8674(83)90417-8. [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S., Fichelson S., Sola B., Bordereaux D., Hampe A., André C., Galibert F., Tambourin P. Frequent c-fms activation by proviral insertion in mouse myeloblastic leukaemias. Nature. 1987 Sep 17;329(6136):259–261. doi: 10.1038/329259a0. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Lavigueur A., Maltby V., Mock D., Rossant J., Pawson T., Bernstein A. High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Mol Cell Biol. 1989 Sep;9(9):3982–3991. doi: 10.1128/mcb.9.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H., Miller C., Koeffler H. P., Battifora H., Cline M. J. Rearrangement of the p53 gene in human osteogenic sarcomas. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7716–7719. doi: 10.1073/pnas.84.21.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau-Gachelin F., Tavitian A., Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988 Jan 21;331(6153):277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- Morishita K., Parker D. S., Mucenski M. L., Jenkins N. A., Copeland N. G., Ihle J. N. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988 Sep 9;54(6):831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- Mowat M., Bernstein A. Linkage of the Fv-2 gene to a newly reinserted ecotropic retrovirus in Fv-2 congenic mice. J Virol. 1983 Sep;47(3):471–477. doi: 10.1128/jvi.47.3.471-477.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat M., Cheng A., Kimura N., Bernstein A., Benchimol S. Rearrangements of the cellular p53 gene in erythroleukaemic cells transformed by Friend virus. Nature. 1985 Apr 18;314(6012):633–636. doi: 10.1038/314633a0. [DOI] [PubMed] [Google Scholar]
- Mushinski J. F., Potter M., Bauer S. R., Reddy E. P. DNA rearrangement and altered RNA expression of the c-myb oncogene in mouse plasmacytoid lymphosarcomas. Science. 1983 May 20;220(4599):795–798. doi: 10.1126/science.6687762. [DOI] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- O'Donnell P. V., Fleissner E., Lonial H., Koehne C. F., Reicin A. Early clonality and high-frequency proviral integration into the c-myc locus in AKR leukemias. J Virol. 1985 Aug;55(2):500–503. doi: 10.1128/jvi.55.2.500-503.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A., Oliff I., Schmidt B., Famulari N. Isolation of immortal cell lines from the first stage of murine leukemia virus-induced leukemia. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5464–5467. doi: 10.1073/pnas.81.17.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A., Ruscetti S. A 2.4-kilobase-pair fragment of the Friend murine leukemia virus genome contains the sequences responsible for friend murine leukemia virus-induced erythroleukemia. J Virol. 1983 Jun;46(3):718–725. doi: 10.1128/jvi.46.3.718-725.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A., Signorelli K., Collins L. The envelope gene and long terminal repeat sequences contribute to the pathogenic phenotype of helper-independent Friend viruses. J Virol. 1984 Sep;51(3):788–794. doi: 10.1128/jvi.51.3.788-794.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Savage P. D., Jones C., Silver J., Geurts van Kessel A. H., Gonzalez-Sarmiento R., Palm L., Hanson C. A., Kersey J. H. Mapping studies and expression of genes located on human chromosome 11, band q23. Cytogenet Cell Genet. 1988;49(4):289–292. doi: 10.1159/000132680. [DOI] [PubMed] [Google Scholar]
- Schiff R. D., Oliff A. The pathophysiology of murine retrovirus-induced leukemias. Crit Rev Oncol Hematol. 1986;5(3):257–323. doi: 10.1016/s1040-8428(86)80041-5. [DOI] [PubMed] [Google Scholar]
- Shibuya T., Mak T. W. Isolation and induction of erythroleukemic cell lines with properties of erythroid progenitor burst-forming cell (BFU-E) and erythroid precursor cell (CFU-E). Proc Natl Acad Sci U S A. 1983 Jun;80(12):3721–3725. doi: 10.1073/pnas.80.12.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J., Kozak C. Common proviral integration region on mouse chromosome 7 in lymphomas and myelogenous leukemias induced by Friend murine leukemia virus. J Virol. 1986 Feb;57(2):526–533. doi: 10.1128/jvi.57.2.526-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola B., Fichelson S., Bordereaux D., Tambourin P. E., Gisselbrecht S. fim-1 and fim-2: two new integration regions of Friend murine leukemia virus in myeloblastic leukemias. J Virol. 1986 Nov;60(2):718–725. doi: 10.1128/jvi.60.2.718-725.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tsichlis P. N., Shepherd B. M., Bear S. E. Activation of the Mlvi-1/mis1/pvt-1 locus in Moloney murine leukemia virus-induced T-cell lymphomas. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5487–5491. doi: 10.1073/pnas.86.14.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Strauss P. G., Hu L. F. A common region for proviral DNA integration in MoMuLV-induced rat thymic lymphomas. 1983 Mar 31-Apr 6Nature. 302(5907):445–449. doi: 10.1038/302445a0. [DOI] [PubMed] [Google Scholar]
- Villemur R., Monczak Y., Rassart E., Kozak C., Jolicoeur P. Identification of a new common provirus integration site in gross passage A murine leukemia virus-induced mouse thymoma DNA. Mol Cell Biol. 1987 Jan;7(1):512–522. doi: 10.1128/mcb.7.1.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein Y., Ihle J. N., Lavu S., Reddy E. P. Truncation of the c-myb gene by a retroviral integration in an interleukin 3-dependent myeloid leukemia cell line. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5010–5014. doi: 10.1073/pnas.83.14.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]