ABSTRACT
The regulation of latency is central to herpesvirus biology. Recent transcriptome-wide surveys have uncovered evidence for promiscuous transcription across the entirety of the Kaposi's sarcoma-associated herpesvirus (KSHV) genome and postulated the existence of multiple viral long noncoding RNAs (lncRNAs). Next-generation sequencing studies are highly dependent on the specific experimental approach and particular algorithms of analysis and therefore benefit from independent confirmation of the results. The antisense-to-latency transcript (ALT) lncRNA was discovered by genome-tiling microarray (Chandriani et al., J Virol 86:7934–7942, 2010, https://doi.org/10.1128/JVI.00645-10). To characterize ALT in detail, we physically isolated this lncRNA by a strand-specific hybrid capture assay and then employed transcriptome sequencing and novel reverse transcription-PCR (RT-PCR) assays to distinguish all RNA species in the KSHV latency region. These methods confirm that ALT initiates at positions 120739/121012 and encodes a single splice site, which is shared with the 3′-coterminal K14-vGPCR/ORF74 mRNA, terminating at 130873 (GenBank accession number GQ994935), resulting in an ∼10,000-nucleotide transcript. No shorter ALT isoforms were identified. This study also identified a novel intron within the LANA 5′ untranslated region using a splice acceptor at 127888. In summary, ALT joins PAN/nut1/T1.1 as a bona fide lncRNA of KSHV with potentially important roles in viral gene regulation and pathogenesis.
IMPORTANCE Increasing data support the importance of noncoding RNAs (ncRNAs), including microRNAs (miRNAs) and lncRNAs, which have been shown to exert critical regulatory functions without coding for recognizable proteins. Defining the sequences of these ncRNAs is essential for future studies aiming to functionally characterize a specific ncRNA. Most lncRNA studies are highly dependent on high-throughput sequencing and bioinformatic analyses, few studies follow up on the initial predictions, and analyses are at times discordant. The manuscript characterizes one key viral lncRNA, ALT, by physically isolating ALT and by a sequencing-independent assay. It provides for a simple assay to monitor lncRNA expression in experimental and clinical samples. ALT is expressed antisense to the major viral latency transcripts encoding LANA as well as the viral miRNAs and thus has the potential to regulate this key part of the viral life cycle.
KEYWORDS: long noncoding RNA, lncRNA, Kaposi's sarcoma-associated herpesvirus, KSHV, antisense-to-latency transcripts, ALT, LANA, gene expression, RNA enrichment, latency, herpesvirus
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8 causes Kaposi's sarcoma (KS), a cancer of endothelial cells that is a signature of the worldwide AIDS epidemic. In addition, KSHV instigates primary effusion lymphoma (PEL) and the plasmablastic variant of multicentric Castleman disease (MCD). These are rare but aggressive proliferative diseases of B cells that carry a poor prognosis even with treatment (1–3). There are no vaccines against KSHV. Standard therapies for KS, PEL, and MCD rely on cytotoxic therapy and improvement of immune system function in the context of HIV or organ transplantation. KSHV persists and replicates in the nucleus of infected cells, where its 165- to 170-kb double-stranded DNA (dsDNA) genome (4) is associated with histones and organized into transcriptionally active and inactive gene blocks, as inferred by different types of chromatin modifiers (5–10). Like other herpesviruses, KSHV undergoes a latent stage of its life cycle during which no viral progeny are produced, but the virus can reactivate and enter lytic replication. During latency, KSHV transcription is limited (11, 12), but reactivation to lytic replication initiates a cascade of RNA polymerase II-dependent transcription that spans the entire genome (13, 14).
Long noncoding RNAs (lncRNAs) belong to a diverse subclass of RNA molecules that lack obvious protein-coding potential and are classified by their relatively large size (>200 nucleotides [nt]) compared to smaller RNAs, such as microRNAs (miRNAs). Increasing evidence suggests that lncRNAs play key roles in cell differentiation, pathogenesis of cancer and other diseases, regulation of gene expression, and interactions between virus and host during infection (15–17). By virtue of their large size and numerous secondary and tertiary interactions, lncRNAs can form complex structures and serve as molecular scaffolds for protein complexes. In addition, single-stranded regions of lncRNAs can base pair with cDNA or RNA, allowing for silencing of specific mRNAs or for targeting of protein complexes to specific genomic loci. A notion that has gained prominence recently is that some lncRNAs can serve as sponges for miRNAs (15–17). More than two dozen human lncRNAs have been associated with human cancers, as the expression of these lncRNAs in tumors is dysregulated in a manner similar to that seen with protein-coding genes and miRNAs (16, 17). In addition, the expression of certain lncRNAs has been correlated with the degree of cancer prognosis and shown to play a role in metastasis, invasiveness, cell proliferation, and regulation of tumor suppressor proteins (16–18).
Like the human cells it infects, KSHV encodes a number of lncRNAs (13, 19–24). One of these viral lncRNA transcripts, polyadenylated nuclear (PAN) RNA, has been known for 2 decades (22, 25). Its origin and regulation has been well characterized (22, 25–32). In situ studies showed that PAN RNA is either not expressed or expressed at low basal levels during latency (33, 34) and then is induced to higher levels during lytic reactivation; however, PAN RNA is either the most abundant or among the most abundant transcripts during de novo infection (35, 36). PAN RNA has been reported to inhibit the expression of immune response genes such as interleukin-4 and gamma interferon, to interact with several viral and cellular proteins, to act as a global regulator of viral genes by interacting with cellular transcriptional regulators and chromatin-modifying proteins, and to potentially encode small peptides from noncanonical open reading frames (ORFs) (13, 26–29, 37–39). Although PAN RNA is one of the best-studied lncRNAs in KSHV, its complete repertoire of interactions and functions is still not fully understood (40). In addition to PAN RNA, some functional insights have been reported for three other KSHV lncRNAs: T3.0, T1.2, and T1.5 (the numbers indicate the transcript length in kilobase pairs). T3.0 and T1.2, which share a transcription start site (TSS) and are antisense to the mRNA of RTA (replication and transcription activator; ORF50), do not inhibit RTA (41). T1.5, a transcript near one of the two origins of lytic replication (ori-Lyt), is an immediate-early gene, and its transcription is required for viral replication (24, 42). Although these three lncRNAs do not have canonical ORFs, all have been reported to be ribosome associated (13, 41). In support of the potential to produce small peptides (<100 amino acids), a portion of T1.5 is found in the cytoplasm by Northern blotting and by RNA fluorescence in situ hybridization (FISH) (21), whereas T3.0 produces at least three small peptides (41), one of which induces its antisense transcript RTA (43). PAN RNA and T1.5 are also packaged within virions and expressed in the early stages of primary infection, suggesting an important role in establishing a successful infection (35, 36). Two other lncRNAs, ALT (antisense-to-latency transcripts) and T6.1, have been consistently observed in transcriptome sequencing (RNA-seq) surveys. Evidence for additional, less prominent viral lncRNAs can also be found as a by-product of mRNA transcriptome mapping studies (13, 19–21, 23, 44). The exact origin and regulation of many of these potential lncRNAs are not well defined at this time.
There is evidence of permissive transcription surrounding and across the ori-Lyt loci in KSHV, including lncRNAs T1.5, T6.1, and ALT, the microRNA cluster, and several protein-coding transcripts (13, 19, 23, 24, 45, 46). This feature is analogous to other DNA viruses, where transcription is sometimes seen as a means to keep ori-Lyt from becoming engulfed in repressive chromatin. In the case of Epstein-Barr virus (EBV), latent transcripts LMP2A and LMP2B cross the terminal repeat regions (47), which are fused and serve as the origin of replication for the circular plasmid during latency. Similar evidence for KSHV transcripts traversing the latent origin of replication, which, like in EBV, is comprised of fused terminal repeats, is missing due to the difficulty of mapping transcripts across these regions, although the mRNAs for the orthologous KSHV latent membrane proteins K1 and K15 originate proximal to the latent origin (48–51).
Of the more than a dozen potential lncRNAs in the KSHV genome that have been proposed on the basis of transcriptome-wide studies, most were detected in lytically reactivated cells. Many are not consistently detected across independent studies, and their transcriptional start and termination sites remain ill-defined. Unlike lncRNA PAN, the more recently reported and less abundant KSHV lncRNAs tend to run opposite of known coding regions and are thus antisense to known lytic mRNAs. The detailed functional characterization of these novel lncRNAs is dependent upon first creating a detailed map of their boundaries. We sought to address this gap in our understanding of KSHV by studying the transcription of one new KSHV lncRNA in detail and confirm its expression by means other than next-generation (NextGen) sequencing. We selected the KSHV lncRNA ALT (19), which lies opposite the well-characterized KSHV latency locus transcripts.
RESULTS
An updated map of known and potential KSHV mRNAs, miRNAs, and lncRNAs.
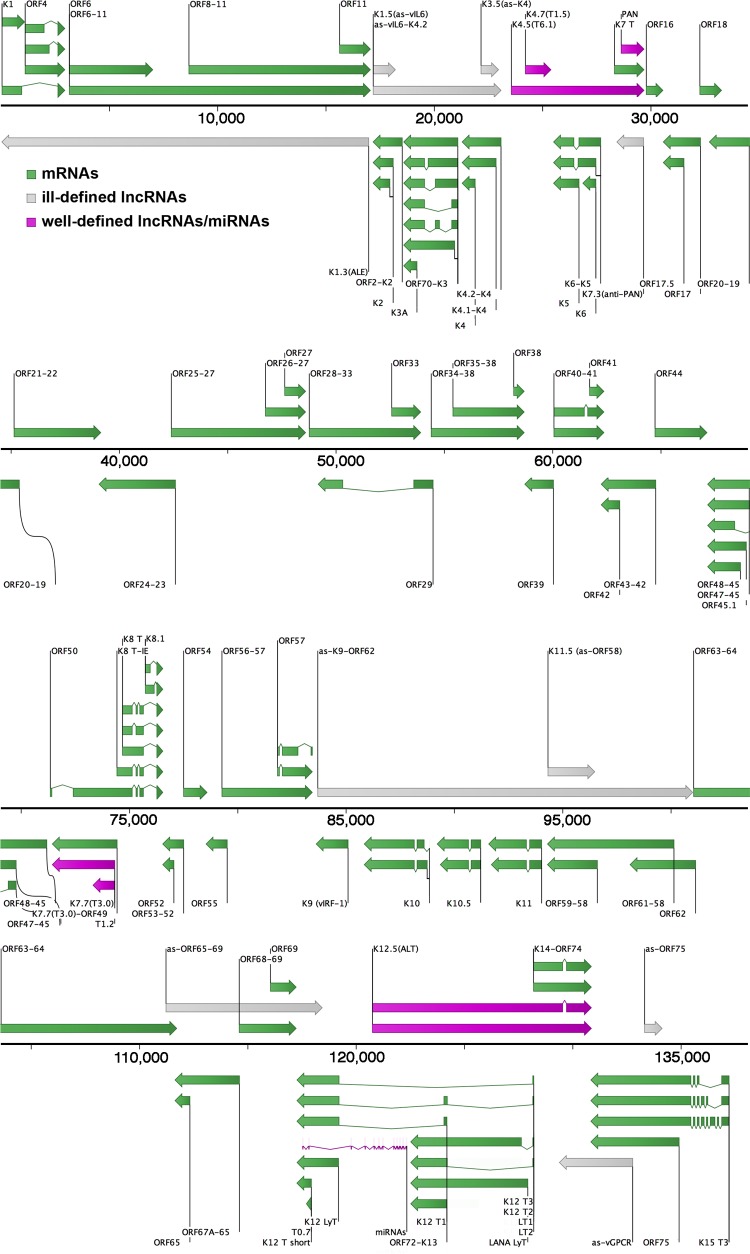
Arias et al. published KSHV 2.0, which mapped poly(A)-containing viral RNAs and assessed their translational status using ribosomal profiling (13), while Dresang et al. combined a tiling microarray with liquid chromatography and tandem mass spectrometry to explore the protein-coding capacity of the transcriptome (20). A high-throughput transcriptional mapping approach was taken by Majerciak et al. (21), whereas Chandriani et al. focused on tiling microarray supported by Northern blotting or 3′ rapid amplification of cDNA ends (RACE) (19). From these studies at least 16 potential lncRNAs have been reported (13, 19–21), although there are discrepancies between the different approaches (Table 1). Seven potential lncRNAs found by tiling microarray (19, 20) were not detected by subsequent RNA-seq studies (13, 21, 59, 60). We sought to integrate these and prior studies (1, 13, 19–21, 50, 59–61) into an updated, comprehensive map of KSHV transcription annotations (Fig. 1).
TABLE 1.
Potential lncRNAs in KSHV
| Descriptive name and categorya | Alias(es)a | Genome coordinatesb |
Direction | Size (kb) | Genome-wide screenc |
||||
|---|---|---|---|---|---|---|---|---|---|
| Start | End | A | B | C | D | ||||
| Undefined end(s) | |||||||||
| as-K1/ORF4-11 | ALE; K1.3; K1/11-AS | ∼17000 | 21 | L | ∼17.0 | + | + | + | |
| as-vIL6/ORF2 | K1.5 | ∼17200 | 18215 | R | ∼1.0 | + | + | ||
| as-vIL6/K4.2 | K2/K4.2-AS | ∼17200 | ∼23000 | R | ∼5.8 | + | |||
| as-K4s | K3.5 | ∼22100 | 22979 | R | ∼0.9 | + | |||
| as-PAN/K7 | K7.3; anti-PAN | ∼29700 | 28413 | L | ∼1.3 | + | |||
| as-K9/ORF62 | ∼83700 | ∼101000 | R | ∼17.3 | + | ||||
| as-ORF58/59 | K11.5 | ∼94300 | 96779 | R | ∼2.5 | + | |||
| as-ORF65/69 | ∼111000 | ∼118500 | R | ∼7.5 | + | ||||
| as-vGPCR | ∼133000 | ∼129000 | L | ∼4.0 | + | ||||
| as-ORF7 | ∼133300 | ∼134100 | R | ∼0.8 | + | ||||
| Defined ends | |||||||||
| as-K5/K6 | K4.5; T6.1; K5/6-AS | 23542 | 29690 | R | 6.1f | + | + | + | |
| as-IR | K4.7; T1.5 | 24191 | 25394 | R | 1.2f | + | + | + | |
| PAN RNAd | nut1; T1.1 | 28611 | 29690 | R | 1.1 | + | + | + | |
| as-RTA | K7.7; T3.0; 50AS-L | 74334 | 71423 | L | 2.9 | + | + | ||
| as-RTAe | T1.2; 50AS-S | 74334 | 73302 | L | 1.0 | ||||
| as-miR/K13/72/LANA | ALT; K12.5 | 120739;121012 | 130871 | R | 10.1f; 9.9f | + | + | + | + |
Underlined names are those used in this study. One or more ends of undefined lncRNAs have not been determined. Abbreviations: as, antisense; vIL6, viral homolog to interleukin-6 (ORF K2); vGPCR, viral G protein-coupled receptor (ORF74); IR, internal repeats; PAN RNA, polyadenylated nuclear RNA; RTA, replication and transcription activator (ORF50); miR, microRNAs; LANA, latency-associated nuclear antigen (ORF73); ALE, antisense to left end; anti-PAN, antisense to PAN RNA; nut1, nuclear U-RNA-like transcript 1; ALT, antisense to latency transcripts.
Coordinates from each reference were extrapolated to the KSHV strain JSC1 genome (GQ994935.1).
References for genome-wide screens are the following: A, Arias et al. (13), RNA-seq and ribosomal profiling; B, Chandriani et al. (19), tiling microarray; C, Dresang et al. (20), tiling microarray; D, Majerciak et al. (21), RNA-seq.
Reported in reference 41.
Actual size may vary due to differences in length of interior repetitive regions.
FIG 1.
Updated map of known and potential KSHV mRNAs, miRNAs, and lncRNAs. Coordinates of RNA transcripts from the literature were extrapolated to the KSHV strain JSC1 genome (GQ994935.1) and visualized with CLC Bio's Genomics Workbench software. The boundaries of ill-defined lncRNAs have not been determined, and their existence has not been validated. See the .gff file at https://www.med.unc.edu/orfeome/downloads/annotated-target-genomes for a more detailed annotation, including CDS, TSS, pA signals, TTS, repeats, and origins of replication.
At a global level, our analysis of existing data sets found evidence of extensive overlapping and antisense transcription across the entire KSHV genome which was not previously appreciated, as very few of the initially defined open reading frames (ORFs) overlap in KSHV. Nearly the entire genome is transcribed during lytic replication. Several of the KSHV lncRNAs overlap not only mRNAs but also each other, either on the same strand or on opposite strands. For instance, lncRNAs T1.5 and PAN RNA completely overlap T6.1 on the same strand, while PAN RNA is antisense to the anti-PAN lncRNA (Fig. 1). A large number of KSHV lncRNAs are reported to have ill-defined boundaries due to limitations in the methods used for their initial discovery. lncRNAs whose 5′ and/or 3′ ends have not yet been determined were classified as ill-defined in Table 1 and colored gray in Fig. 1, while lncRNAs with well-defined boundaries across multiple independent studies were annotated as such (Table 1; colored purple in Fig. 1). Two lncRNAs, as-vIL6-K4.2 and as-K9-ORF62, were annotated solely based on a verbal description in the results section of a single report (13), while three others, as-ORF65-69, as-vGPCR, and as-ORF75, were mapped by estimating their coordinates based on a published figure (19).
KSHV lncRNA ALT is an early lytic transcript.
ALT is too large and not sufficiently abundant to be reliably assessed by Northern blotting. Conventional reverse transcription-PCR (RT-PCR) methods of detecting RNA are difficult for the ALT lncRNA due to the highly abundant latency transcripts in an overlapping region on the opposite strand (see Fig. 3). To improve sensitivity and specificity, we designed pools of biotinylated antisense oligonucleotides (ASOs) to capture strand-specific RNA by using streptavidin-coupled magnetic beads (62, 63). To assess this enrichment, we used RT-PCR of control RNAs and regions unique to ALT. This strategy is applicable to lncRNAs whose ends are not well known, since capture oligonucleotides can be designed antisense to any region with evidence of transcription.
FIG 3.
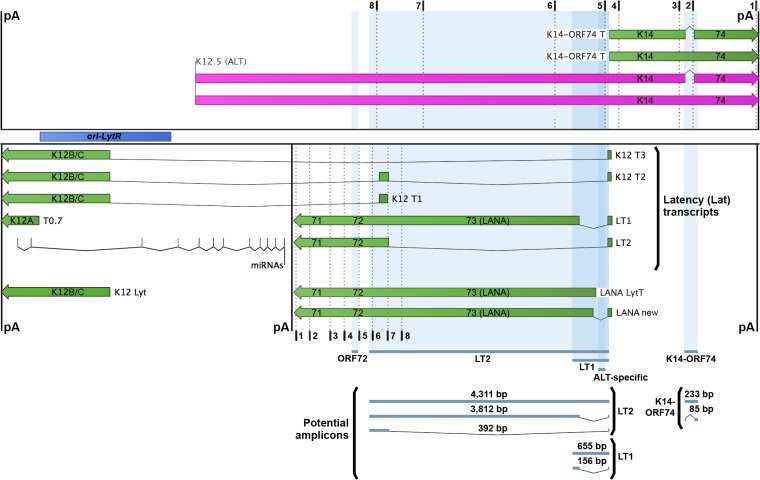
Schematic of the KSHV latency locus. The latency locus of KSHV was mapped with mRNAs in green and lncRNA ALT in purple. Dotted lines indicate two sets of biotinylated ASOs (numbered as shown in Table 2), one designed to enrich ALT and one to enrich the latency (Lat) transcripts. Shaded blue regions and lines below indicate potential amplicons. Polyadenylation (pA) signals are marked by black lines, and the right origin of lytic replication (ori-LytR) is shown by a dark blue bar.
To validate this approach, we used the well-characterized human lncRNA MALAT1 (metastasis-associated in lung adenocarcinoma transcript 1; also called NEAT2, for nuclear-enriched abundant transcript 2). First, we tested our ability to reverse transcribe full-length MALAT1, as lncRNAs are often difficult templates to extend due to their extensive length and secondary structure. We isolated total RNA from 2 million cells of the PEL cell line TREx BCBL1-RTA (64). Using oligo(dT) to initiate reverse transcription, we verified that three commercial reverse transcriptase (RT) enzymes could generate the full ∼6.6 kb of MALAT1 cDNA from its 3′-poly(A) tail to its 5′ end (as ascertained by primer MALAT#1). Importantly, of the three we tested, only ThermoScript RT also avoided false priming (65), which is critical to this study, as false priming obscures the strand origin for cDNA sequences and can lead to artifacts in stranded RNA-seq approaches. All oligonucleotide primer pairs for RT-PCR experiments amplified the appropriately sized products from genomic DNA (see Fig. 4B).
FIG 4.
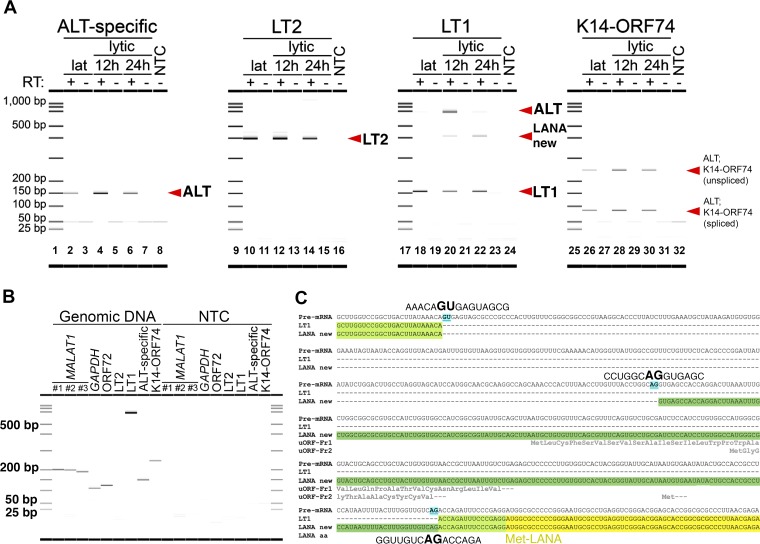
KSHV lncRNA ALT is expressed during the early lytic stage, and a new splice isoform of a LANA transcript is found. (A) Expression of RNAs during latency or 12 and 24 h postlytic reactivation, as detected by strand- or isoform-specific RT-PCR. The amplicons interrogated are listed above each digital gel image and are illustrated in Fig. 3. (B) Oligonucleotide primers designed for RT-PCR and listed in Table 2 were first tested on BCBL1 genomic DNA. All but the LT2 primers yielded an amplified product from DNA. A short extension time during PCR was intentionally chosen to avoid amplifying a full-length LT2 product from DNA to ensure these primers were specific for the spliced LT2 mRNA. (C) A new splice isoform (LANA new, whose longer 5′ UTR is highlighted in dark green) that contains the LANA CDS (yellow) and is expressed during the lytic stage was identified with an earlier splice acceptor site (blue) than Lat transcript LT1 (5′ UTR in light green). The amino acid sequences of three small peptides encoded by uORFs that are only present in LANA new are shown in gray text below the RNA sequence, while that of LANA is in black text.
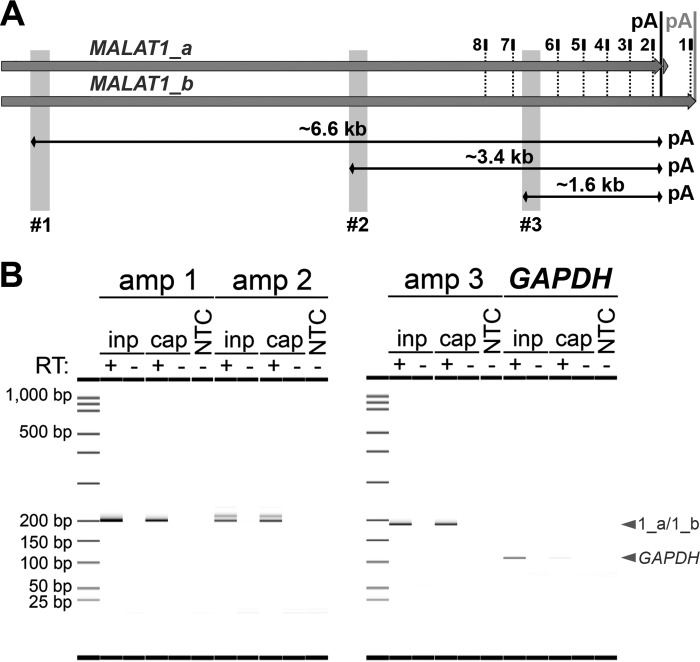
For strand-specific enrichment of MALAT1, we designed a pool of ASOs complementary to the 3′ end of MALAT1 transcripts starting near the transcription termination sequence (TTS) and spaced roughly every 200 nucleotides (Fig. 2A, dotted black lines, with numbering as shown in Table 2). Total RNA was incubated with the MALAT1-specific pool of biotinylated ASOs, and then ASO-bound RNA was isolated on streptavidin-coupled magnetic beads. RNA-ASO-bead complexes were washed on a magnetic stand to remove nonspecific interactors, and then the RNA of interest was eluted from ASO-bead complexes by high-temperature incubation. To assess the enrichment of MALAT1 transcripts, we performed RT-PCR on input or captured RNA using three MALAT1 amplicons (Fig. 2A, gray shading) and a negative-control amplicon for the highly abundant GAPDH mRNA. All RNA was incubated with or without RT, and no amplification was observed in the non-RT control samples (Fig. 2B). The RT-PCR products were visualized by a Caliper LabChip GX microfluidic instrument, which allows for high-accuracy sizing and quantification. All three MALAT1 regions were amplified from captured RNA at levels similar to input RNA, while GAPDH mRNA was significantly reduced in the captured RNA compared to the input (Fig. 2B). Here and in subsequent experiments, we added random hexamers to the RT reaction to increase consistency across captured lncRNAs of unknown size (Fig. 2B). These results establish that target RNAs, up to an estimated length of 6,600 nucleotides (nt), can be selectively enriched and converted to full-length cDNA using this method.
FIG 2.
Enrichment of human lncRNA MALAT1. (A) Biotinylated antisense oligonucleotides (ASOs; positions marked by dotted lines and numbered as shown in Table 2) and streptavidin-coupled magnetic beads were used to enrich the two major isoforms of lncRNA MALAT1 from TREx BCBL1-RTA cells at 12 h postlytic reactivation. Three amplicons (shaded gray) were used to assess the enrichment of RNA by RT-PCR. The polyadenylation (pA) signal for the minor isoform is marked by a gray vertical line, and the genome-encoded poly(A) tract for the major isoform (86) is marked by a black line. (B) MALAT1 was enriched as detected by all three amplicons (amp), while negative-control RNA GAPDH was not. All samples were run with or without RT. inp, RNA input; cap, ASO-captured RNA.
TABLE 2.
Primers used for RT-PCR assessment and enrichment of transcripts
| Name and category | Sequencea |
|---|---|
| RT-PCR of human transcripts | |
| MALAT1-Amp1-F204 | 5′-GAGAAAGGACTACAGAGCCCCG |
| MALAT1-Amp1-R204 | 5′-AAATGACGCAATTCTCCCTGCG |
| MALAT1-Amp2-F200 | 5′-CCATTGGGGAATAAGCATAACCCTG |
| MALAT1-Amp2-R200 | 5′-TGAGATGGACATTGCCTCTTCATTG |
| MALAT1-Amp3-F190 | 5′-TTCATTTCTGGTGGTGGGAGGG |
| MALAT1-Amp3-R190 | 5′-CCAAGGGAGGGGAGAGAGAAAC |
| GAPDH-F | 5′-ATGACATCAAGAAGGTGGTGAA |
| GAPDH-R | 5′-GCTGTTGAAGTCAGAGGAGAC |
| RT-PCR of KSHV transcripts | |
| ORF72-F | 5′-CCAGTTCACTTTGCTATGCCG |
| ORF72-R | 5′-GTGACGTCCGTCGCTAAGAC |
| ALT-specific-F | 5′-TAATCCGGGCGTGAGAAACA |
| ALT-specific-R | 5′-CCGTAAGGCACCCTTATCTTTG |
| 7307-LT2splice-F | 5′-AGCAGCAGCTTGGTCCGGCTG |
| 7209-LT2splice-R | 5′-GCATATGCGAAGTAAGAGATTGT |
| LT1splice-F | 5′-GGTCATCGCCAAGGTCACAT |
| LT1splice-R | 5′-GCTTGGTCCGGCTGACTTAT |
| K14-ORF74splice-F | 5′-TGGCCCAAACGGAGGATCCTAG |
| K14-ORF74splice-R | 5′-AGTTTCATTCCAGGATTCATCATC |
| Enrichment of human lncRNA MALAT1 | |
| MALAT1 even 8 | 5′-CACTGCAAGGTCTCATACAC-TEG-Biotin |
| MALAT1 even 7 | 5′-CAACACTCAGCCTTTATCAC-TEG-Biotin |
| MALAT1 even 6 | 5′-CCACTGGTGAATTCAACTGG-TEG-Biotin |
| MALAT1 even 5 | 5′-CCTTAGGATAATAGCGCTTT-TEG-Biotin |
| MALAT1 even 4 | 5′-CTTGCTTTTAACAGGCTTCT-TEG-Biotin |
| MALAT1 even 3 | 5′-ATAGAGCTACTTAGCTGTGG-TEG-Biotin |
| MALAT1 even 2 | 5′-CCTACTGAAGAGCATTGGAG-TEG-Biotin |
| MALAT1 even 1 | 5′-TAGGGCTTCTCAAAACACCA-TEG-Biotin |
| Enrichment of KSHV lncRNA ALT | |
| ALT even 8 | 5′-TATGGCAACTGCCAATAACC-TEG-Biotin |
| ALT even 7 | 5′-CCAGGAGATAATACACCAGA-TEG-Biotin |
| ALT even 6 | 5′-TCTCCAGAGTCTTCTCAAAG-TEG-Biotin |
| ALT even 5 | 5′-GGCACCCTTATCTTTGAAAT-TEG-Biotin |
| ALT even 4 | 5′-ACAGTGGGGGGTAAATTCTG-TEG-Biotin |
| ALT even 3 | 5′-AACGCGACACCAAAATGGGG-TEG-Biotin |
| ALT even 2 | 5′-ACAAGTAGTATAGGTGTTGT-TEG-Biotin |
| ALT even 1 | 5′-GTCACATCTGTGTACAAACC-TEG-Biotin |
| Enrichment of KSHV latency transcripts | |
| Lat even 8 | 5′-TAAGTTATGGGCGACTGGTC-TEG-Biotin |
| Lat even 7 | 5′-CAAATGCAAGTGCGGAGCGG-TEG-Biotin |
| Lat even 6 | 5′-CGCGGCTCAATTTCAAGAAT-TEG-Biotin |
| Lat even 5 | 5′-CGTGAGGCTTCTGAGCTTAC-TEG-Biotin |
| Lat even 4 | 5′-TGCAGCGCTGATAATAGAGG-TEG-Biotin |
| Lat even 3 | 5′-CCGGCTTGTATATGTGAAGG-TEG-Biotin |
| Lat even 2 | 5′-CCTATAGTGTCCTTGCTTAA-TEG-Biotin |
| Lat even 1 | 5′-TACATACATTCTACGGACCA-TEG-Biotin |
TEG, triethyleneglycol spacer for more efficient binding to magnetic beads.
As the name suggests, ALT lies on the opposite strand of the latency (Lat) transcripts (11, 30, 66), completely overlapping most of them (Fig. 3). In addition, ALT is on the same strand and is coterminal with a bicistronic lytic transcript containing ORF K14 (v-OX2, a homolog of cellular surface receptor OX2) and ORF74 (vGPCR, for viral G protein-coupled receptor) (19, 67). With such extensive overlapping transcription, careful placement of oligonucleotide primers is paramount when trying to detect, capture, and distinguish individual transcript isoforms in this region. To distinguish between the ALT lncRNA and the KSHV latency locus mRNAs, we used primers that span or lie within introns of the latency locus mRNAs and only permit amplification of transcripts from one strand or another (Fig. 3, blue shading). Two such strand-specific primer pairs (ALT specific or LT2, as listed in Table 2) were designed to only generate a single amplified product from one of the two opposing strands. The first, the ALT-specific primer pair (Fig. 3), lies completely within the introns of various latent LANA (latency-associated nuclear antigen) transcripts or upstream of a lytic transcript, labeled LANA LytT, on the bottom strand. It is also upstream of the K14-ORF74 bicistronic transcript on the top strand. Therefore, the ALT-specific primer pair only amplifies ALT cDNA. The second primer pair, the LT2 primer pair, has the potential to produce three amplicons, two from spliced latency transcripts and one from unspliced ALT lncRNA (Fig. 3). PCR amplification of cDNA from the Lat transcript isoform LT2 would result in a 392-bp amplicon, while relatively long amplicons would be produced from either a Lat isoform with a short intron (LT1; 3,812 bp) or the ALT lncRNA (4,311 bp). To ensure that the LT2 primer pair amplifies only the 392-bp product spanning the intron of isoform LT2, we varied the extension times during PCR on PEL cell DNA to determine conditions under which the two long amplicons are not amplified. Thus, we chose an extension time of 45 s, which underrepresents long products and yields a single LT2-specific product from cDNA (Fig. 4A and B).
Additionally, two other primer pairs (LT1 or K14-ORF74, as listed in Table 2 and Fig. 3) were designed to amplify multiple products of different sizes in order to differentiate between spliced or unspliced transcripts from either the same strand or opposite strands. The first, the LT1 primer pair (Fig. 3), spans an intron from Lat isoform LT1 and amplifies a product of 156 bp from that strand but a full-length amplicon of 655 bp from the ALT lncRNA on the opposite strand. The second, the K14-ORF74 primer pair (Fig. 3), spans an intron that is present in a subpopulation of the K14-ORF74 and ALT lncRNA transcripts (19), and it amplifies either a 233-bp product from unspliced RNA or an 85-bp spliced product. Finally, the ORF72 primer pair amplifies a single non-strand-specific product that can originate from either the Lat transcripts or the ALT lncRNA. All of these primer pairs gave the expected product using BCBL1 genomic DNA (Fig. 4B).
KSHV transcription expression is altered during lytic replication. Therefore, to determine the best conditions for maximal expression of ALT, we interrogated latent and lytic conditions. TREx BCBL1-RTA cells carry the potent inducible RTA/ORF50 expression plasmid, and upon addition of doxycycline, the cells synchronously initiate lytic transcription (64). Our laboratory has extensively characterized transcription from the latency locus in this cell line, and therefore we chose the time points of 12 and 24 h postinduction to assess lytic changes in transcription.
To investigate the expression levels of transcripts on both strands of the latency locus, we used oligo(dT) and random hexamers to reverse transcribe mRNA from either latent or lytic cells and then assessed transcripts using the strand- or isoform-specific primer pairs (Fig. 4A). Poly(A) RNA was quantified using the RNA HS assay on a Qubit 3.0 fluorometer, and the same mass of RNA was added to each RT reaction. Consistent with our prior studies (68), two LANA transcripts, LT1 and LT2, were expressed at steady levels during the latent and lytic stages, as seen with the Lat-specific LT2 primer pair (Fig. 4A, lanes 10, 12, and 14) or with the 156-bp LT1-specific amplicon from the LT1 primer pair (Fig. 4A, lanes 18, 20, and 22). Given the massive changes in cellular transcription during reactivation and the reproducibility of LT2 expression in this 24-h period, we used LT2 as a control for transcript expression. The ALT-specific primer pair (Fig. 4A, lane 4) and the LT1 primer pair (i.e., the 655-bp ALT-specific amplicon in Fig. 4A, lane 20) each amplified ALT, and we observed the highest expression of ALT relative to LT2 at 12 h postlytic reactivation. We were able to detect both the spliced and unspliced isoforms from the K14-ORF74 primer pair at all stages of the viral life cycle (Fig. 4A, lanes 26, 28, and 30), which supports the results from the ALT-specific primer pair. No product was observed in the non-RT negative-control samples (Fig. 4A, lanes 3, 5, 7, 11, 13, 15, 19, 21, 23, 27, 29, and 31) or the nontemplate PCR control (NTC) samples (Fig. 4A, lanes 8, 16, 24, and 32). The bands at or below 50 nucleotides represent single-stranded primers and double-strand primer-dimers, which are only visible due to the use of a high-sensitivity microfluidic device. It verifies that primer was added in excess in all reactions.
Using the LT1 primer pair, we noticed an unexpected amplicon of ∼400 bp that is absent from latent cells, appears at 12 h postreactivation, and is expressed at the highest level at 24 h postreactivation (Fig. 4A, lanes 18, 20, and 22). We found the same ∼400-bp amplicon in JSC1 and BC1 cells, confirming this is not a cell line-specific phenomenon. Further, we determined that this amplicon corresponds to a new splice isoform of a LANA-containing transcript. This isoform has a canonical splice acceptor site at genomic position 127888, which is upstream of the known acceptor site in latency transcript LT1 at position 127640. This yields a corresponding intron that is shorter than LT1, as shown in graphical form as LANA new (Fig. 3) and in a multiple-sequence alignment (Fig. 4C) using the online tool MUSCLE, version 3.8 (69). The intron of this new splice isoform lies entirely within the 5′ untranslated region (UTR), as with the intron of LT1, and therefore does not affect the LANA coding DNA sequence (CDS).
To improve the detection of the ALT lncRNA, we next sought to capture ALT prior to reverse transcription. Because ALT was expressed at its highest level at 12 h postreactivation (Fig. 4A, lanes 4 and 20), this time point was used for all subsequent experiments. To ensure the most complete lytic reactivation from doxycycline-inducible TREx BCBL1-RTA cells, we induced with doxycycline and added the histone deacetylase inhibitor valproic acid (VPA). Unlike other inducers, such as n-butyrate, 12-O-tetradecanoylphorbol-13-acetate (TPA), or vorinostat/suberanilohydroxamic acid (SAHA), VPA exhibits no toxicity in PEL even after prolonged incubation. We also added foscarnet/phosphonoformate (PFA) to abolish KSHV DNA replication, nuclear reorganization due to the formation of replication compartments, and premature cell lysis. This increases the chances of detecting authentic, transcriptionally regulated lytic lncRNAs, rather than fragmented, abortive transcription across replicating episomes. To establish strand specificity, we sought to enrich ALT and the Lat transcripts in separate ASO pools. We designed these pools of biotinylated ASOs clustered near the 3′ end of both sets of transcripts, avoiding repetitive regions (Fig. 3, dotted black lines, with numbering as shown in Table 2). For ALT, ASOs were distributed evenly throughout the predicted length of the lncRNA, while ASOs for Lat transcripts were grouped within a common region at the 3′ end to maximize the capture of all Lat transcripts.
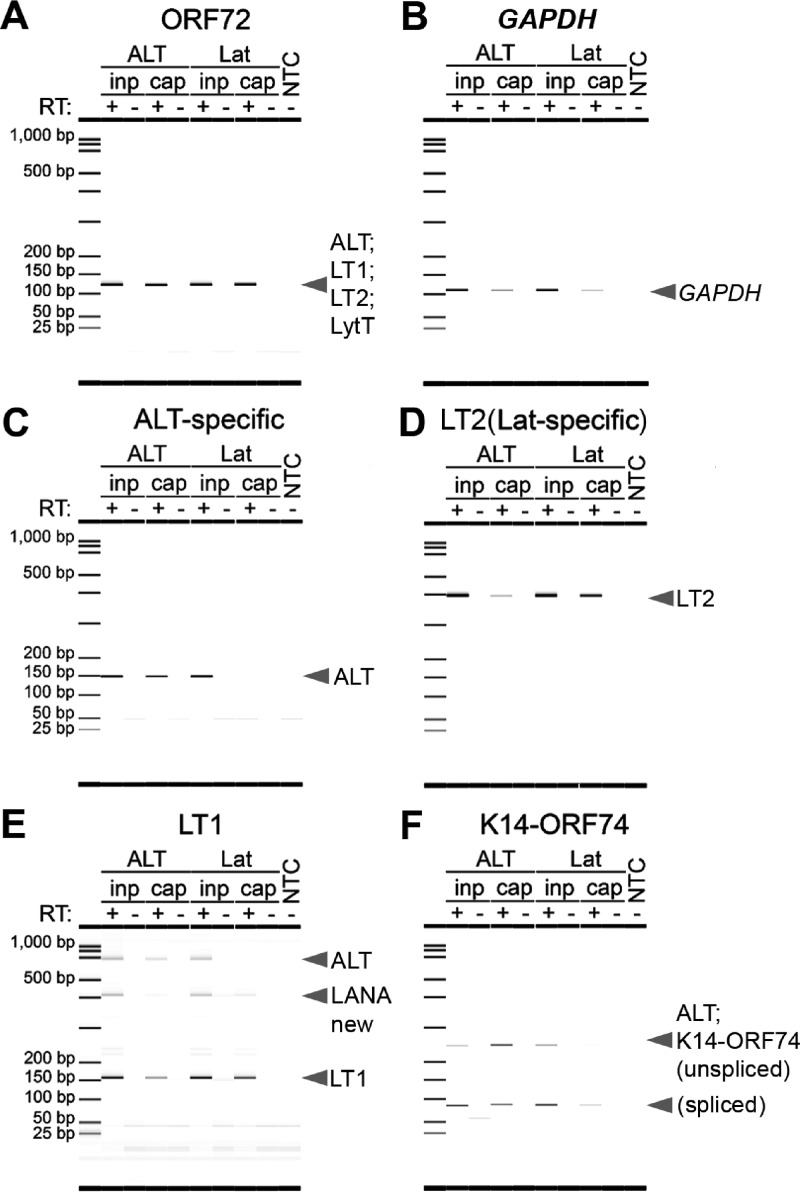
To assess the enrichment of ALT and Lat transcripts, we performed RT-PCR on input or captured RNA using the strand- or isoform-specific amplicons within the latency locus (Fig. 3, blue shading) and a negative-control amplicon within the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene. No amplification was observed in the negative-control samples incubated without RT (Fig. 5). Capture ASOs from both strands were effective, as the ORF72 primer pair, whose amplicon is not strand specific, equally amplified a single product in ALT- and Lat-captured RNA at levels similar to those of input RNA (Fig. 5A). The topography of the KSHV genome is highly repetitive and highly GC rich, leaving few targets for optimal design of ASOs. This, coupled with the low temperature at which ASO binding occurs, means that the expected outcome for this assay is enrichment, not isolation, of our target. Thus, we observe that some GAPDH is amplified in the captured RNA from each ASO pool (Fig. 5B), but at a much smaller amount than the intended target RNAs (e.g., ORF72) (Fig. 5A).
FIG 5.
ALT is enriched in a strand-specific manner. Strand- and isoform-specific RT-PCR was performed on ALT-captured, Lat-captured, or input RNA using the non-strand-specific primer pair ORF72 (A), negative-control GAPDH (B), the ALT-specific primer pair (C), the Lat-specific LT2 primer pair (D), or the isoform-specific LT1 (E) and K14-ORF74 (F) primer pairs, whose amplicons are illustrated in Fig. 3. ALT was amplified at 12 h postlytic reactivation in ALT-captured but not Lat-captured RNA, while latency transcripts were only amplified in Lat-captured RNA.
ALT was enriched in ALT-captured RNA compared to Lat-captured RNA (Fig. 5C, E, and F, left versus right side), as seen with either the ALT-specific primer pair (Fig. 5C), the 655-bp ALT-specific amplicon from the LT1 primer pair (Fig. 5E), or both K14-ORF74 amplicons (Fig. 5F). Conversely, the Lat transcripts were only amplified efficiently in Lat-captured but not ALT-captured RNA relative to input RNA (Fig. 5D and E, right versus left side), as seen with the Lat-specific LT2 primer pair (Fig. 5D) and the 156-bp Lat-specific amplicon from the LT1 primer pair (Fig. 5E). These data establish the presence of KSHV ALT by a direct, strand-selective approach independent of potential interpretation biases inherent to short-read sequencing and current heuristic, short-read mapping algorithms.
Identification of ALT 5′ and 3′ ends using RNA-seq.
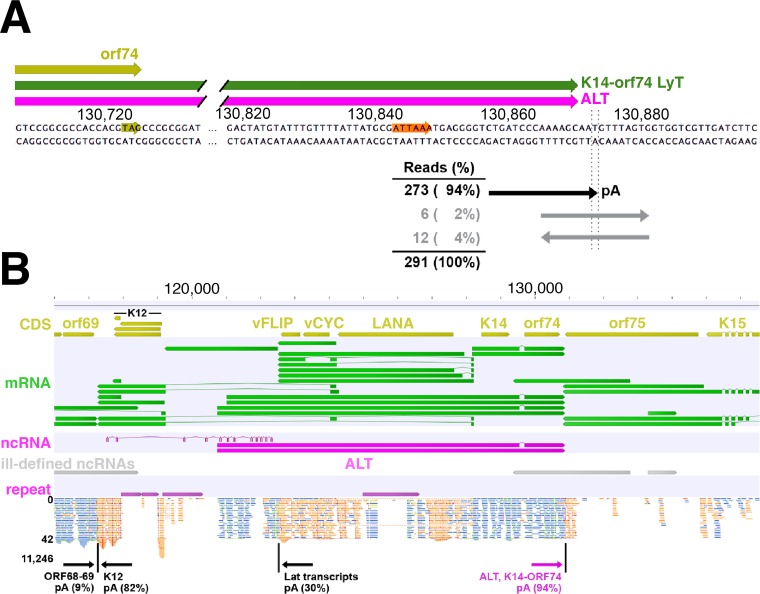
Having established the specificity of our enrichment method, we sought to define the ends of captured lncRNA by RNA-seq analysis. To this end, we created strand-specific RNA-seq libraries from TREx BCBL1-RTA cells 12 h postreactivation with or without ASO enrichment. We next aligned all sequence reads from ALT-enriched RNA to the KSHV strain JSC1 genome (GenBank accession number GQ994935.1). To identify 3′ ends and transcription termination sequences (TTS), we searched for genomic regions where an abundance of sequence reads contain a poly(A) tract that does not match the reference genome and identified the initial nonmatching adenine. We then determined the percentage of all reads in that genome position that contain adenine or any other nucleotide on either strand. As expected (11, 70), all poly(A)-containing reads in the latency locus data set aligned to known TTS of the Lat transcript, one at JSC1 genomic position 122510 (30% of all reads at that position), which is 3′ to ORF71, and one at position 117247 (82% of reads), which is 3′ to Kaposin/K12 (Fig. 6B).
FIG 6.
Identification of ALT 5′ and 3′ ends using RNA-seq analysis. (A) Strand-specific RNA-seq was performed on ALT-captured RNA from TREx BCBL1-RTA cells at 12 h postlytic reactivation, and reads were aligned to the KSHV strain JSC1 genome using BBMap. Nearly all reads downstream of the common ALT and K14-ORF74 pA signal (orange) contain ≥5 adenine residues starting at position 130873 (GQ994935.1), consistent with published reports, while the remainder span this position in either direction. (B) Reads in the latency locus from the top (blue) and bottom (orange) strands were visualized using Genomics Workbench. Reads align near the published 5′ end of the ALT lncRNA (19).
We identified a TTS downstream of the common ALT and K14-ORF74 polyadenylation (pA) signal. For this TTS shared by the coterminal ALT lncRNA and K14-ORF74 transcripts, 94% of all reads contain ≥5 adenine residues starting at position 130873 (Fig. 6A), while 6% of reads span this position in either direction. These results were obtained from RNA-seq analysis of ALT-enriched RNA, but similar percentages were observed at the same position in libraries from non-ASO-enriched RNA (all raw reads are available in the NCBI Sequence Read Archive under accession code SRP082699). As the two genome-encoded adenine residues upstream of position 130873 interfere with determining the exact start of this poly(A) tract, this result is consistent with the published TTS at positions corresponding to 130870 to 130872 in the JSC1 genome (13, 19, 21, 59, 60).
To examine the TSS of the ALT lncRNA, we mapped a continuous stretch of reads in the same strand orientation as the 5′ region of the ALT lncRNA (Fig. 6B). These reads covered the first ∼1,500 nucleotides of ALT but were not present in the region upstream of the published ALT TSS. Reads were clustered near the two reported 5′ ends of the ALT lncRNA, including the long predominant isoform starting at a position corresponding to 120739 in the JSC1 genome and the short isoform at 121012 (19). We also examined other properties of the ALT lncRNA. To explore the potential splicing of ALT, we used two different tools (71, 72) that predicted >200 potential splice donor and splice acceptor sites within the >10 kb of this lncRNA. Of note, each of these tools confirmed the known splice donor/acceptor pair at JSC1 genomic positions 129545 and 129694, respectively, which represents the only known intron shared by the K14-ORF74 and ALT lncRNA transcripts (19); however, we found no evidence in our RNA-seq data of intra-ALT splice sites.
Although the ends of ALT are clearly defined in our data, we note that there are a few interior areas that have poor read coverage, particularly in repetitive regions. Lack of coverage in the repetitive region within ALT is due to an inherent inability of the BBMerge algorithm to extend reads through repeats. Lack of coverage in other areas is likely due to reduced efficiency of cDNA initiation during reverse transcription or sequence-dependent reduced efficacy of sequencing chemistry used in the Illumina machine. In sum, our experiments confirmed the existence of the ALT lncRNA in KSHV by direct, strand-specific, sequencing-independent means (RT-PCR), uncovered a novel splice site in the 5′ UTR of LANA, and verified the termination and start site of the ALT lncRNA.
DISCUSSION
This work firmly establishes ALT as a bona fide KSHV lncRNA and thus provides the foundation and justification for further functional and genetic studies. The KSHV PAN lncRNA is so abundant that it can be identified by Northern blot analyses (22, 25). All other KSHV lncRNAs are either significantly less abundant or are inconsistently expressed and have not been verified by multiple, independent experimental approaches. With this work, ALT now has attained a status of significance and reproducibility similar to that of PAN RNA, and we provide a high-sensitivity, high-specificity, low-cost RT-PCR-based assay to distinguish ALT and LANA pri-miRNA expression patterns in KSHV.
The technical goal of this work was to validate a relatively fast and inexpensive method to enrich strand-specific RNAs that uses as few tiling oligonucleotides as possible. Other methods of enriching RNA require either a large pool of overlapping or closely spaced oligonucleotides tiled equally over the entire length of the target RNA or an oligonucleotide empirically determined to bind effectively (62, 73, 74). As several potential lncRNAs in KSHV are ≥5 kb (Table 1), adapting a tiled oligonucleotide method to enrich one or more of these viral lncRNAs can become prohibitively expensive.
We identified a new splice isoform (LANA new) originating from the latency cluster that is only expressed during lytic replication (Fig. 4A and C). Although most transcripts from the latency locus have high basal expression during latency and are not inducible by the lytic cycle regulator RTA or by chemical inducers of reactivation (11, 30, 66, 75), at least one other LANA-containing transcript, labeled LANA LytT in Fig. 3, is also inducible during the lytic stage (76). However, the LANA LytT transcript is controlled by a separate RTA-inducible promoter that is downstream of the constitutive latent promoter (76) and is located entirely within the intron of the LANA new isoform, suggesting that the LANA new transcript is controlled by yet another unidentified inducible lytic promoter. It is intriguing to speculate what effect alternative splicing of the LANA new isoform would have on translation of its CDS. Although the shorter intron of the LANA new splice isoform does not affect the LANA CDS, it contains a longer 5′ UTR than that of Lat transcript LT1 (Fig. 4C). The expanded 5′ UTR of this new LANA transcript isoform contains three additional upstream ORFs (uORFs), which are start and stop codons upstream of the main coding sequence. As uORFs generally decrease synthesis of downstream proteins (77), these additional uORFs in the LANA new isoform (Fig. 4C) can potentially lower the translation of the LANA protein in lytic cells relative to latent cells while allowing for continued expression of the KSHV miRNA cluster.
The complex and interconnected nature of the KSHV transcriptome bestows key characteristics for regulating gene expression that are still not fully understood. More than half of all KSHV ORFs are carried on polycistronic transcripts, >25% of all ORFs are carried by spliced mRNAs, and a few transcripts share promoters and/or pA signals (13, 21, 59, 60, 78, 79). The ALT lncRNA is coterminal with the bicistronic mRNA K14-ORF74 and includes the K14 and ORF74 CDS, but it is expected that K14 and ORF74 proteins are not translated from ALT, as these ORFs are >8 kb downstream of the ALT TSS. However, as microRNAs regulate gene expression by targeting 3′ UTRs for downregulation or degradation (79), the 3′ UTR common to ALT and the K14-ORF74 mRNA is likely subject to similar microRNA regulation. Some human miRNAs can downregulate the 3′ UTRs for K14 and ORF74 in latent PEL cells (60), and it is possible that these microRNAs have the same effect on ALT. It is speculated that grouping specific genes into bi- or polycistronic transcripts allows KSHV to maximize its coding capacity and level of gene regulation (78, 79). For example, splicing of one bicistronic mRNA that disrupts a large portion of the upstream ORF allows expression of the downstream protein (80). However, it is unknown what effect the lengthy 5′ UTR of ALT has upon its downstream CDSs K14 and ORF74, and it is possible that K14 and ORF74 proteins are exclusively translated from the two splice isoforms of K14-ORF74 bicistronic transcripts. The potential interactions and coregulatory functions of the ALT lncRNA with nearby or overlapping transcripts on both strands are numerous and poorly understood.
Finally, we provide a resource to the KSHV community (KSHV 2.1) by critically evaluating and integrating a series of mapping studies with an emphasis on viral ncRNAs. Just as more in-depth studies of the KSHV miRNAs (81–84) uncovered and highlighted the importance of structural variations (i.e., single-nucleotide variants with functional consequences, alternate strand incorporation, and alternate 5′ or 3′ end processing), this in-depth study of the KSHV lncRNAs affirms their existence and hints at their possible roles in viral gene expression, genome replication, and pathogenesis.
MATERIALS AND METHODS
Cell culture and reagents.
The KSHV-infected primary effusion lymphoma (PEL) cell line TREx BCBL1-RTA (64) was cultured in RPMI 1640 medium (Gibco), incubated at 37°C in 5% CO2, and supplemented with 10% tetracycline-free fetal bovine serum (Omega Scientific), l-glutamine (2 mM; Gibco), 100 g/ml streptomycin sulfate, and 100 U/ml penicillin G (Corning). Lytic reactivation of KSHV was induced with doxycycline (1 μg/ml) and valproic acid (1 mM) in the presence of foscarnet (phosphonoformate; 100 μM).
RNA isolation.
RNA was isolated from TREx BCBL1-RTA, JSC1, and BC1 cells. Doxycycline, valproic acid, and foscarnet were added when cultures reached the desired population of ≥10 million cells for total RNA or ≥100 million cells for poly(A) RNA. The culture was either harvested immediately or growth was continued for 12 or 24 h postinduction. Cells were pelleted by centrifugation at 300 × g for 5 min, and supernatants were removed. Cell pellets were resuspended in TRI Reagent (Molecular Research Center) and lysed according to the manufacturer's protocol, except an additional extraction was performed using acidic phenol-chloroform-isoamyl alcohol (125:24:1; Fisher). RNA pellets were dissolved in nuclease-free water and treated with 0.12 U/μl of TURBO DNase using a TURBO DNA-free kit (Life Technologies) according to the manufacturer's protocol. Removal of DNA was assessed by RT-PCR (described below), and RNA quality was determined using an RNA 6000 NanoChip on a Bioanalyzer instrument (Agilent). RNA was quantified using the RNA HS assay on a Qubit 3.0 fluorometer (Invitrogen). Poly(A) RNA was selected using Oligotex mRNA kits (Qiagen) according to the manufacturer's protocol, except that bead-bound RNA was first eluted twice with the same aliquot of nuclease-free water heated to 70°C and then eluted with an additional four aliquots of heated water. RNA in each eluted sample was quantified using Qubit 3.0. Poly(A) RNA was used to assess the expression of ALT and the Lat transcripts by RT-PCR (Fig. 4A) and to generate RNA-seq libraries from one set of biological replicates, while total RNA was used in all other RT-PCR or RNA-seq experiments.
Enrichment of specific RNAs by biotinylated ASOs.
Oligonucleotides antisense to the RNAs of interest were designed using the Stellaris online tool ChIRP (chromatin isolation by RNA purification) Probe Designer, based on work from the Chang laboratory (62, 63). ASOs were designed to bind to nonrepetitive regions of RNA, and biotin was added to the 3′ end along with a triethyleneglycol spacer to allow for more efficient binding to magnetic beads (Eurofins). ASO-mediated enrichment of poly(A) RNA was used to generate a single RNA-seq library (ALT03) (Fig. 6), while ASO-enriched total RNA was used in other RT-PCR experiments (Fig. 2B and 5). For enrichment, ≥2.0 μg poly(A) RNA from ≥70 million cells or ≥20.0 μg total RNA from ≥2 million cells was incubated with 100 pmol of pooled biotinylated ASOs in ChIRP hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris-HCl, pH 7.0, 1 mM EDTA, 15% formamide) (63) and RiboLock RNase inhibitor (Thermo Fisher Scientific) at 37°C for 2 h. ASO-bound RNA was then isolated by adding 100 pmol streptavidin-coupled magnetic beads in binding solution from a Dynabeads kilobaseBINDER kit (Invitrogen) and incubating on a rotator at room temperature for 2 h. Beads were washed and RNA was eluted in 10 mM Tris-HCl, pH 7.0, heated at 85°C for 2 min. The concentration of enriched RNA was determined using a Qubit 3.0 fluorometer (Invitrogen) as mentioned in “RNA isolation” above.
Assessment of RNA expression or ASO enrichment by RT-PCR.
Poly(A) RNA was used to assess the expression of ALT and Lat transcripts by RT-PCR (Fig. 4A), and ASO-enriched poly(A) RNA was used to generate the RNA-seq library ALT03 (Fig. 6). ASO-enriched or non-ASO-enriched (input) total RNA was used in RT-PCR experiments (Fig. 2B and 5). Total RNA, poly(A) RNA, or ASO-enriched RNA was reverse transcribed using ThermoScript reverse transcriptase (Invitrogen) with the following slight modifications to the supplier's protocol. The annealing step was performed with 90 to 140 ng of RNA and either with oligo(dT)20 or a mix of oligo(dT)20 and random hexamer primers at 75°C for 5 min. Reverse transcription was performed at 25°C for 10 min and 55°C for 15 min, and then heat inactivation was performed at 85°C for 5 min. The resulting cDNA was amplified using GoTaq Green master mix (Promega) and the pairs of oligonucleotide primers listed in Table 2. PCR primers were first tested on genomic DNA isolated from BCBL1 cells (Fig. 4B). For RT-PCR, cycling conditions included an initial denaturation step of 95°C for 30 s, amplification for 35 cycles (denaturation at 95°C for 20 s, annealing at 56°C for 20 s, and extension at 72°C for 30 s), and a final extension at 72°C for 1 min. Cycling conditions for genomic DNA were the same, except the extension time was 45 s. The reactions were run by capillary electrophoresis on a Caliper LabChip GX microfluidic instrument (PerkinElmer).
Preparation of RNA for high-throughput sequencing.
Strand-specific RNA-seq libraries were made using total RNA, poly(A) RNA, or ASO-enriched RNA (22 to 85 ng) from PEL cells 12 or 24 h postreactivation and the Ovation Universal RNA-seq system (NuGEN) with the following modifications to the supplier's protocol. cDNA was fragmented to an approximate size range of 200 to 500 bp using a TruSeq protocol (i.e., parameters included 5% acoustic factor, 175-W peak incident power, and 200 cycles per burst) with a modified treatment time of 40 s on a Covaris E220 focused ultrasonicator instrument. Human rRNAs were depleted using the provided NuGEN insert-dependent adapter cleavage primers. Libraries were amplified for 20 to 21 cycles. Final cDNA libraries were quantified using the dsDNA HS assay on a Qubit 3.0 fluorometer (Invitrogen), while the size distribution was determined using a high-sensitivity DNA chip on a Bioanalyzer instrument (Agilent). All libraries were paired-end sequenced using an Illumina HiSeq 2500 instrument for 50-bp or 100-bp read lengths.
RNA-seq data analysis.
Sequence reads were trimmed using bbduk.sh (ktrim = r, k = 23, hdist = 1) from the BBMap suite v.36.20 (https://sourceforge.net/projects/bbmap/) and the adapters.fa file included in the BBMap software that contains sequences for commonly used adapters from next-generation sequencing library preparation. Since the 50-bp reads used in this study do not overlap, we used a feature in the BBMerge program (included in the BBMap suite) that makes use of a tadpole (a read assembling program, also included in the BBMap suite). BBMerge tries to extend the 3′ end of each read via assembly using kmer frequencies. BBMerge was run on input read files (R1/R2) with these parameters (k = 31, extend2 = 200) to allow extension of reads up to 250 bp (Brian Bushnell, personal communication, and unpublished BBMerge manuscript). Reads (adapter-trimmed original or merged using BBMerge) were mapped with BBMap (with maxindel = 4000, intronlen = 10, ambig = random, qin = 33, sam = 1.3; the rest of the parameters were default) to the KSHV strain JSC1 genome (GQ994935). BBMap is able to generate alignment results directly as BAM files when SAMtools (v.1.3.1) software is available on the server. We obtained between 3 and 13 million sequence reads that aligned to the JSC1 genome.
We identified the 3′ ends of RNA transcripts in the latency locus of KSHV by using Integrative Genomics Viewer (IGV) v.2.3.79 (85). We first found genomic regions for which the majority of sequence reads contain ≥2 adenine residues that do not match the reference genome and form a poly(A) tract of ≥5 nucleotides, and we visually inspected each genomic position in that region to identify the initial nonmatching adenine. For that particular genomic position and its surrounding positions, we then used bam-readcount v.0.7.4 (https://github.com/genome/bam-readcount) to report the number and percentage of reads that contain each nucleotide base on either strand. We visualized 5′ ends by using Genomics Workbench software v.9.0.1 (CLC bio) to identify collections of sequence reads all in the same strand orientation without any upstream reads.
Accession number(s).
The sequence data have been deposited in the NCBI Sequence Read Archive under accession code SRP082699, and the annotated .gff file is available at https://www.med.unc.edu/orfeome/downloads/annotated-target-genomes.
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health (grant numbers DE018304 and AI107810 to D.P.D. and T32 CA009156 and T90 DE021986 to J.M.S). We acknowledge technical and scientific support from the UNC High-Throughput Sequencing Facility, which is supported by the University Cancer Research Fund (UCRF) and a Comprehensive Cancer Center Support grant (CA016086).
We thank Brian Bushnell of the DOE Joint Genome Institute for advice on optimal use of the BBMap suite of programs. We thank J. Jung for providing TREx BCBL1-Rta. We also thank Jedediah Seltzer and Carolina Caro-Vegas for critical readings of the manuscript and other members of the Dittmer laboratory for discussions.
We have no conflicts of interest to declare.
REFERENCES
- 1.Damania B, Cesarman E. 2013. Kaposi's sarcoma-associated herpesvirus, p 2080–2128. In Fields BN, Knipe DM, Howley PM (ed), Fields virology, 6th ed Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Dittmer DP, Damania B. 2013. Kaposi sarcoma associated herpesvirus pathogenesis (KSHV)–an update. Curr Opin Virol 3:238–244. doi: 10.1016/j.coviro.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaplan LD. 2013. Human herpesvirus-8: Kaposi sarcoma, multicentric Castleman disease, and primary effusion lymphoma. Hematol Am Soc Hematol Educ Program 2013:103–108. [DOI] [PubMed] [Google Scholar]
- 4.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A 93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen HS, Wikramasinghe P, Showe L, Lieberman PM. 2012. Cohesins repress Kaposi's sarcoma-associated herpesvirus immediate early gene transcription during latency. J Virol 86:9454–9464. doi: 10.1128/JVI.00787-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunther T, Grundhoff A. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog 6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilton IB, Simon JM, Lieb JD, Davis IJ, Damania B, Dittmer DP. 2013. The open chromatin landscape of Kaposi's sarcoma-associated herpesvirus. J Virol 87:11831–11842. doi: 10.1128/JVI.01685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu J, Yang Y, Turner PC, Jain V, McIntyre LM, Renne R. 2014. LANA binds to multiple active viral and cellular promoters and associates with the H3K4methyltransferase hSET1 complex. PLoS Pathog 10:e1004240. doi: 10.1371/journal.ppat.1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang H, Cho H, Sung GH, Lieberman PM. 2013. CTCF regulates Kaposi's sarcoma-associated herpesvirus latency transcription by nucleosome displacement and RNA polymerase programming. J Virol 87:1789–1799. doi: 10.1128/JVI.02283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, Jung JU. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog 6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol 72:8309–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fakhari FD, Dittmer DP. 2002. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J Virol 76:6213–6223. doi: 10.1128/JVI.76.12.6213-6223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. 2014. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 10:e1003847. doi: 10.1371/journal.ppat.1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YT, Kincaid RP, Arasappan D, Dowd SE, Hunicke-Smith SP, Sullivan CS. 2010. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA 16:1540–1558. doi: 10.1261/rna.1967910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cech TR, Steitz JA. 2014. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Merry CR, Niland C, Khalil AM. 2015. Diverse functions and mechanisms of mammalian long noncoding RNAs. Methods Mol Biol 1206:1–14. doi: 10.1007/978-1-4939-1369-5_1. [DOI] [PubMed] [Google Scholar]
- 17.Yang G, Lu X, Yuan L. 2014. LncRNA: a link between RNA and cancer. Biochim Biophys Acta 1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. 2013. Long noncoding RNAs and the genetics of cancer. Br J Cancer 108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandriani S, Xu Y, Ganem D. 2010. The lytic transcriptome of Kaposi's sarcoma-associated herpesvirus reveals extensive transcription of noncoding regions, including regions antisense to important genes. J Virol 84:7934–7942. doi: 10.1128/JVI.00645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dresang LR, Teuton JR, Feng H, Jacobs JM, Camp DG II, Purvine SO, Gritsenko MA, Li Z, Smith RD, Sugden B, Moore PS, Chang Y. 2011. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: insights on the detection and discovery of viral genes. BMC Genomics 12:625. doi: 10.1186/1471-2164-12-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majerciak V, Ni T, Yang W, Meng B, Zhu J, Zheng ZM. 2013. A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog 9:e1003749. doi: 10.1371/journal.ppat.1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun R, Lin SF, Gradoville L, Miller G. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A 93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor JL, Bennett HN, Snyder BA, Moore PS, Chang Y. 2005. Transcriptional analysis of latent and inducible Kaposi's sarcoma-associated herpesvirus transcripts in the K4 to K7 region. J Virol 79:15099–15106. doi: 10.1128/JVI.79.24.15099-15106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Li H, Chan MY, Zhu FX, Lukac DM, Yuan Y. 2004. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J Virol 78:8615–8629. doi: 10.1128/JVI.78.16.8615-8629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong W, Wang H, Herndier B, Ganem D. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci U S A 93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossetto CC, Pari GS. 2011. Kaposi's sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J Virol 85:13290–13297. doi: 10.1128/JVI.05886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossetto CC, Pari GS. 2012. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog 8:e1002680. doi: 10.1371/journal.ppat.1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossetto CC, Pari GS. 2014. PAN′s labyrinth: molecular biology of Kaposi's sarcoma-associated herpesvirus (KSHV) PAN RNA, a multifunctional long noncoding RNA. Viruses 6:4212–4226. doi: 10.3390/v6114212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossetto CC, Tarrant-Elorza M, Verma S, Purushothaman P, Pari GS. 2013. Regulation of viral and cellular gene expression by Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA. J Virol 87:5540–5553. doi: 10.1128/JVI.03111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J Virol 72:1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol 73:2232–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong W, Ganem D. 1997. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J Virol 71:1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staskus KA, Sun R, Miller G, Racz P, Jaslowski A, Metroka C, Brett-Smith H, Haase AT. 1999. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. J Virol 73:4181–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staskus KA, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson DJ, Ganem D, Haase AT. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol 71:715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purushothaman P, Thakker S, Verma SC. 2015. Transcriptome analysis of Kaposi's sarcoma-associated herpesvirus during de novo primary infection of human B and endothelial cells. J Virol 89:3093–3111. doi: 10.1128/JVI.02507-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J Virol 78:3601–3620. doi: 10.1128/JVI.78.7.3601-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borah S, Darricarrere N, Darnell A, Myoung J, Steitz JA. 2011. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog 7:e1002300. doi: 10.1371/journal.ppat.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell M, Kim KY, Chang PC, Huerta S, Shevchenko B, Wang DH, Izumiya C, Kung HJ, Izumiya Y. 2014. A lytic viral long noncoding RNA modulates the function of a latent protein. J Virol 88:1843–1848. doi: 10.1128/JVI.03251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell M, Kung HJ, Izumiya Y. 2014. Long non-coding RNA and epigenetic gene regulation of KSHV. Viruses 6:4165–4177. doi: 10.3390/v6114165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrad NK. 2016. New insights into the expression and functions of the Kaposi's sarcoma-associated herpesvirus long noncoding PAN RNA. Virus Res 212:53–63. doi: 10.1016/j.virusres.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y, Ganem D. 2010. Making sense of antisense: seemingly noncoding RNAs antisense to the master regulator of Kaposi's sarcoma-associated herpesvirus lytic replication do not regulate that transcript but serve as mRNAs encoding small peptides. J Virol 84:5465–5475. doi: 10.1128/JVI.02705-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Tang Q, Maul GG, Yuan Y. 2006. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J Virol 80:12171–12186. doi: 10.1128/JVI.00990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaber T, Yuan Y. 2013. A virally encoded small peptide regulates RTA stability and facilitates Kaposi's sarcoma-associated herpesvirus lytic replication. J Virol 87:3461–3470. doi: 10.1128/JVI.02746-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandriani S, Ganem D. 2010. Array-based transcript profiling and limiting-dilution reverse transcription-PCR analysis identify additional latent genes in Kaposi's sarcoma-associated herpesvirus. J Virol 84:5565–5573. doi: 10.1128/JVI.02723-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Komatsu T, Dezube BJ, Kaye KM. 2002. The Kaposi's sarcoma-associated herpesvirus K12 transcript from a primary effusion lymphoma contains complex repeat elements, is spliced, and initiates from a novel promoter. J Virol 76:11880–11888. doi: 10.1128/JVI.76.23.11880-11888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadler R, Wu L, Forghani B, Renne R, Zhong W, Herndier B, Ganem D. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J Virol 73:5722–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sample J, Liebowitz D, Kieff E. 1989. Two related Epstein-Barr virus membrane proteins are encoded by separate genes. J Virol 63:933–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowser BS, DeWire SM, Damania B. 2002. Transcriptional regulation of the K1 gene product of Kaposi's sarcoma-associated herpesvirus. J Virol 76:12574–12583. doi: 10.1128/JVI.76.24.12574-12583.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowser BS, Morris S, Song MJ, Sun R, Damania B. 2006. Characterization of Kaposi's sarcoma-associated herpesvirus (KSHV) K1 promoter activation by Rta. Virology 348:309–327. doi: 10.1016/j.virol.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 50.Schulz TF, Chang Y. 2007. KSHV gene expression and regulation. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 51.Wong EL, Damania B. 2006. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus K15 gene. J Virol 80:1385–1392. doi: 10.1128/JVI.80.3.1385-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu FX, Cusano T, Yuan Y. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol 73:5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenner RG, Alba MM, Boshoff C, Kellam P. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol 75:891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song MJ, Brown HJ, Wu TT, Sun R. 2001. Transcription activation of polyadenylated nuclear RNA by RTA in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J Virol 75:3129–3140. doi: 10.1128/JVI.75.7.3129-3140.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang PJ, Shedd D, Gradoville L, Cho MS, Chen LW, Chang J, Miller G. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J Virol 76:3168–3178. doi: 10.1128/JVI.76.7.3168-3178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conrad NK, Steitz JA. 2005. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J 24:1831–1841. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Massimelli MJ, Kang JG, Majerciak V, Le SY, Liewehr DJ, Steinberg SM, Zheng ZM. 2011. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5′ end to interact with viral ORF57 and cellular PABPC1. Int J Biol Sci 7:1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stubbs SH, Hunter OV, Hoover A, Conrad NK. 2012. Viral factors reveal a role for REF/Aly in nuclear RNA stability. Mol Cell Biol 32:1260–1270. doi: 10.1128/MCB.06420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bai Z, Huang Y, Li W, Zhu Y, Jung JU, Lu C, Gao SJ. 2014. Genomewide mapping and screening of Kaposi's sarcoma-associated herpesvirus (KSHV) 3′ untranslated regions identify bicistronic and polycistronic viral transcripts as frequent targets of KSHV microRNAs. J Virol 88:377–392. doi: 10.1128/JVI.02689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McClure LV, Kincaid RP, Burke JM, Grundhoff A, Sullivan CS. 2013. Comprehensive mapping and analysis of Kaposi's sarcoma-associated herpesvirus 3′ UTRs identify differential posttranscriptional control of gene expression in lytic versus latent infection. J Virol 87:12838–12849. doi: 10.1128/JVI.02374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain V, Plaisance-Bonstaff K, Sangani R, Lanier C, Dolce A, Hu J, Brulois K, Haecker I, Turner P, Renne R, Krueger B. 2016. A toolbox for herpesvirus miRNA research: construction of a complete set of KSHV miRNA deletion mutants. Viruses 8:E54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu C, Quinn J, Chang HY. 2012. Chromatin isolation by RNA purification (ChIRP). J Vis Exp 61:3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol 77:4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haddad F, Qin AX, Giger JM, Guo H, Baldwin KM. 2007. Potential pitfalls in the accuracy of analysis of natural sense-antisense RNA pairs by reverse transcription-PCR. BMC Biotechnol 7:21. doi: 10.1186/1472-6750-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarid R, Wiezorek JS, Moore PS, Chang Y. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol 73:1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nador RG, Milligan LL, Flore O, Wang X, Arvanitakis L, Knowles DM, Cesarman E. 2001. Expression of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor monocistronic and bicistronic transcripts in primary effusion lymphomas. Virology 287:62–70. doi: 10.1006/viro.2001.1016. [DOI] [PubMed] [Google Scholar]
- 68.Jeong J, Papin J, Dittmer D. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J Virol 75:1798–1807. doi: 10.1128/JVI.75.4.1798-1807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai X, Cullen BR. 2006. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J Virol 80:2234–2242. doi: 10.1128/JVI.80.5.2234-2242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dogan RI, Getoor L, Wilbur WJ, Mount SM. 2007. SplicePort–an interactive splice-site analysis tool. Nucleic Acids Res 35:W285–W291. doi: 10.1093/nar/gkm407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang M, Marin A. 2006. Characterization and prediction of alternative splice sites. Gene 366:219–227. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 73.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. 2011. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A 108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Talbot SJ, Weiss RA, Kellam P, Boshoff C. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 76.Matsumura S, Fujita Y, Gomez E, Tanese N, Wilson AC. 2005. Activation of the Kaposi's sarcoma-associated herpesvirus major latency locus by the lytic switch protein RTA (ORF50). J Virol 79:8493–8505. doi: 10.1128/JVI.79.13.8493-8505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Araujo PR, Yoon K, Ko D, Smith AD, Qiao M, Suresh U, Burns SC, Penalva LO. 2012. Before it gets started: regulating translation at the 5′ UTR. Comp Funct Genomics 2012:475731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng ZM. 2003. Split genes and their expression in Kaposi's sarcoma-associated herpesvirus. Rev Med Virol 13:173–184. doi: 10.1002/rmv.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu Y, Huang Y, Jung JU, Lu C, Gao SJ. 2014. Viral miRNA targeting of bicistronic and polycistronic transcripts. Curr Opin Virol 7:66–72. doi: 10.1016/j.coviro.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rimessi P, Bonaccorsi A, Sturzl M, Fabris M, Brocca-Cofano E, Caputo A, Melucci-Vigo G, Falchi M, Cafaro A, Cassai E, Ensoli B, Monini P. 2001. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi's sarcoma. J Virol 75:7161–7174. doi: 10.1128/JVI.75.15.7161-7174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gottwein E, Cai X, Cullen BR. 2006. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J Virol 80:5321–5326. doi: 10.1128/JVI.02734-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR. 2011. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manzano M, Shamulailatpam P, Raja AN, Gottwein E. 2013. Kaposi's sarcoma-associated herpesvirus encodes a mimic of cellular miR-23. J Virol 87:11821–11830. doi: 10.1128/JVI.01692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Umbach JL, Cullen BR. 2010. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J Virol 84:695–703. doi: 10.1128/JVI.02013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilusz JE, Freier SM, Spector DL. 2008. 3′ End processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]