Abstract
In this investigation a population of dogs at a rehoming center was monitored over a period of 2 years. Despite regular vaccination of incoming dogs against distemper, canine adenovirus type 2 (CAV-2), and canine parainfluenza virus (CPIV), respiratory disease was endemic. Tissue samples from the respiratory tract as well as paired serum samples were collected for analysis. The development of PCR assays for the detection of CPIV, canine adenovirus types 1 and 2, and canine herpesvirus (CHV) is described. Surprisingly, canine adenovirus was not detected in samples from this population, whereas 19.4% of tracheal and 10.4% of lung samples were positive for CPIV and 12.8% of tracheal and 9.6% of lung samples were positive for CHV. As reported previously, a novel canine respiratory coronavirus (CRCoV) was detected in this population (K. Erles, C. Toomey, H. W. Brooks, and J. Brownlie, Virology 310:216-223, 2003). Infections with CRCoV occurred mostly during the first week of a dog's stay at the kennel, whereas CPIV and CHV were detected at later time points. Furthermore, the evaluation of an enzyme-linked immunosorbent assay for detection of antibodies to CPIV and an immunofluorescence assay for detection of antibodies to CHV is described. This study shows that CPIV is present at kennels despite vaccination. In addition, other agents such as CHV and CRCoV may play a role in the pathogenesis of canine respiratory disease, whereas CAV-2 and canine distemper virus were not present in this population, indicating that their prevalence in the United Kingdom is low due to widespread vaccination of dogs.
Canine infectious respiratory disease (CIRD) is a disease complex that occurs in dogs usually housed in groups, for example, in rehoming centers, boarding kennels, or veterinary hospitals. The disease is characterized by a dry hacking cough, which in most cases is cleared within a short time; however, some dogs develop a severe bronchopneumonia that can be fatal. Although a fatal outcome is rare, CIRD poses a problem in kennels world-wide: it delays the rehoming of dogs and disrupts the training schedules of working dogs as well as leading to considerable treatment costs. Several studies of natural outbreaks of the disease have shown that the etiology is complex, with a variety of viruses and bacteria involved (1). The virus that has been most frequently reported to be present is canine parainfluenza virus (CPIV) (3, 18). Both canine adenovirus type 1 (CAV-1) and CAV-2 have been recovered from dogs suffering from CIRD, but CAV-2 has been more commonly associated with the disease (4, 19). Canine herpesvirus (CHV) has been isolated from dogs with CIRD, but its role remains uncertain (3, 14). Outbreaks of canine distemper virus (CDV) are usually characterized by more-severe respiratory symptoms and may be accompanied by gastrointestinal and central nervous system signs (1). Distemper can be controlled by vaccination, but it has been shown that a decrease in population immunity can lead to outbreaks (10). Serological tests have shown reoviruses and influenza virus to be prevalent in kennelled dogs, but their importance is unclear (2). The detection of a canine coronavirus similar to bovine coronavirus and associated with CIRD in dogs in the United Kingdom has been described recently (11).
Most studies of viral causes of naturally occurring CIRD were conducted more than 20 years ago. Since then vaccines against CAV, CDV, and CPIV have become more widely used, but despite this, outbreaks of CIRD frequently occur at many kennels.
This report describes a longitudinal study of viruses associated with endemic CIRD at a rehoming center. The disease affected the majority of dogs and in some cases led to death despite early use of antibiotic treatment.
MATERIALS AND METHODS
Study population and sample collection.
Dogs from a well-established rehoming kennel with a history of endemic respiratory disease were monitored for this study. Upon entry into the kennel, all dogs were vaccinated with KAVAK DA2 PiP69 (Fort Dodge Animal Health, Madison, Wis.), a live attenuated vaccine for distemper virus, canine adenovirus type 2, canine parainfluenza virus, and canine parvovirus. Also, a killed leptospirosis vaccine was used (KAVAK L; Fort Dodge Animal Health). The health status of each dog was assessed twice a day by a veterinary clinician, and respiratory signs were graded as follows: 1, no respiratory signs; 2, mild cough; 3, cough and nasal discharge; 4, cough, nasal discharge, and inappetence; 5, evidence of bronchopneumonia. The overall health status of the dogs was graded as follows: 1, good health; 2, poor health; 3, very poor health. The age, breed, and sex of the dogs were recorded.
A total of 211 dogs from the kennel population were euthanatized because they were unsuitable for rehoming due to behavioral problems or signs of severe (not exclusively respiratory) disease. For these dogs, a full postmortem examination was performed, and bronchoalveolar lavage (BAL) of the left apical lung lobe was carried out using 50 ml of Hanks balanced salt solution (Invitrogen, Paisley, United Kingdom). Furthermore, tracheal and lung samples were collected, and all samples were stored at −70°C until further use.
Serum samples were collected from 150 dogs on the day of entry into the rehoming kennel. For 105 dogs, a follow-up serum sample was available on day 7, and for 114 dogs, a serum sample was available on day 21 after entry. Serum samples were obtained from 13 dogs during week 4, and serum samples from 17 dogs were available during weeks 5 to 7 (day 30 to day 49; the average length of stay in that group was 38.7 days).
Virus strains.
Canine parainfluenza virus and canine adenovirus types 1 and 2 were kindly provided by H. Thompson, University of Glasgow. Canine herpesvirus strain T31 was isolated from the respiratory tract of a dog from the kennel population.
RNA and DNA extraction.
RNA was extracted from 170 tracheal samples and 106 lung samples by using TriReagent (Sigma, Poole, United Kingdom). Approximately 25 to 50 mg of homogenized tissue was used, and RNA was extracted as recommended by the manufacturer.
For extraction of RNA from the vaccine, the Seek Viral RNA kit (Talent, Trieste, Italy) was used by following the manufacturer's instructions for viral RNA extraction from biological fluids. The contents of one vial of KAVAK DA2 PiP69 were resuspended in 400 μl of distilled water, and 200 μl was used for RNA extraction.
DNA was extracted from 211 tracheal and 104 lung samples by using the Qiamp tissue kit (QIAGEN, Crawley, United Kingdom) according to the manufacturer's instructions.
Cloning and sequencing.
CHV strain T31 was obtained by isolation from the trachea of a study dog as described under “Virus isolation” below. Low-molecular-weight DNA was extracted from CHV-infected MDCK cells as described elsewhere (15), the DNA was digested by using EcoRI (Promega, Southampton, United Kingdom), and the fragments obtained were cloned into the EcoRI site of the pGEM-T easy vector (Promega). DNA sequencing and comparison of the sequences to published data in GenBank were used to identify clones that contained CHV DNA (data not shown). The nucleotide sequence of one clone was used to design CHV-specific primers for the region homologous to UL37 of herpes simplex virus type 1 (HSV-1). All PCR products that were to be used for cloning and sequencing were amplified by using Pfu polymerase (Promega) and purified by using the Qiaquick gel purification kit (QIAGEN) after separation on an agarose gel. The PCR products were cloned into the SmaI site of the pT7 blue2 vector (Novagen, CN Biosciences, Nottingham, United Kingdom) and sequenced by using the Thermo Sequenase fluorescent labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom) and Cy5-labeled primers.
PCR and reverse transcription-PCR (RT-PCR).
cDNA was synthesized by using random hexamers (Roche Diagnostics, Lewes, United Kingdom) and ImPromII reverse transcriptase (Promega).
The primer sequences for the amplification of CPIV, CHV, and CAV are given in Table 1. Primer pair I, as described previously by Frisk et al. (13), was used for amplification of the nucleoprotein gene of canine distemper virus. For all PCRs, Promega PCR buffer B without MgCl2 and a final primer concentration of 0.5 μM was used.
TABLE 1.
Primer sequences and positions
| Virus and primer | Sequence (5′ to 3′) | Nucleotide positions |
|---|---|---|
| Canine parainfluenza virusa | ||
| PNP1 | AGT-TTG-GGC-AAT-TTT-TCG-TCC | 120-140 |
| PNP2 | TGC-AGG-AGA-TAT-CTC-GGG-TTG | 786-766 |
| PNP3 | CGT-GGA-GAG-ATC-AAT-GCC-TAT-GC | 521-543 |
| PNP4 | GCA-GTC-ATG-CAC-TTG-CAA-GTC-ACT-A | 702-678 |
| Canine adenovirusb | ||
| CAV-F1 | TGT-CAA-CAA-GGT-TTT-GTC-TTT-T | 26876-26897 |
| CAV-R1 | TTT-TCA-AGG-GAG-GTG-CGT | 27129-27112 |
| CAV-VP1 | CTG-GGC-GGG-ATT-TAG-AGG-GTG-G | 18804-18825 |
| CAV-VP2 | CAA-GGG-CGT-GGG-CGG-AGT-TAG-A | 19507-19486 |
| Canine herpesvirusc | ||
| CHV1 | AAG-AGC-TCG-TGT-TAG-TGA-AAA-T | 1-22 |
| CHV2 | TAA-ACC-CGC-TGG-ATG-ATA-C | 494-476 |
| Canine herpesvirus glycoprotein B gened | ||
| GBN3 | TAA-TTC-ATA-TGT-CCC-CTT-TTT | 728-748 |
| GBN4 | GTC-CTG-TAT-CTT-CTA-ACT-CTG-CT | 2014-1992 |
Nucleotide positions are given for the nucleocapsid protein gene sequence of SV5 (GenBank accession number AF052755).
Nucleotide positions are given for the sequence of CAV-2 (GenBank accession number U77082).
Nucleotide positions in GenBank accession number AY582738.
Nucleotide positions according to GenBank accession number AF361073.
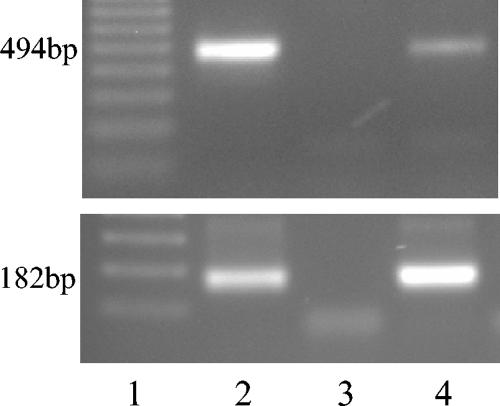
For CPIV and canine distemper virus, PCR conditions were as follows: initial denaturation at 95°C, followed by 35 cycles of 95°C for 1 min, 55°C for 40 s, and 72 °C for 1 min. A final elongation step was carried out at 72°C for 10 min. The expected size of the CPIV PCR product by use of primers PNP1 and PNP2 was 667 bp. For CPIV, 2 μl of the product of the PCR using PNP1 and PNP2 was subsequently used for a nested PCR using primers PNP3 and PNP4; the expected PCR product size was 182 bp (Fig. 1).
FIG. 1.
PCR for detection of canine herpesvirus (top) and nested RT-PCR for detection of canine parainfluenza virus (bottom). Lanes 1, molecular size marker (Sigma); lanes 2, positive control (DNA extracted from MDCK cells infected with canine herpesvirus or cDNA from MDCK cells infected with CPIV, respectively); lanes 3, DNA or cDNA sample obtained from uninfected MDCK cells; lanes 4, DNA or cDNA samples from lung tissues of dogs suffering from respiratory disease. Lanes 2 and 4 show a PCR band of the expected size of 494 bp for CHV or 182 bp for CPIV.
Primers CHV1 and CHV2 for detection of canine herpesvirus were designed to amplify a 494-bp fragment of a region homologous to HSV-1 UL37 (Fig. 1). The annealing step was carried out at 49°C; the MgCl2 concentration was 2.0 mM. All other conditions were as described above for CPIV. For sequencing of the partial glycoprotein B gene, primers GBN3 and GBN4 were used. The annealing step was performed at 45°C for 1 min, and extension was performed at 72°C for 3 min.
Primers CAV-F1 and CAV-R1, directed to the CAV-2 fiber gene, amplified a product of 254 bp. A final concentration of 2.0 mM MgCl2 and an annealing temperature of 52°C were used. Primers CAV-VP1 and CAV-VP2, directed to the CAV capsid protein gene, were designed to amplify a 704-bp product. The annealing temperature used was 60°C, and the MgCl2 concentration was 1.5 mM.
SNT.
Serum neutralization tests (SNT) for CPIV, CAV, and CDV were performed at the diagnostic laboratory of the College of Veterinary Medicine at Cornell University, Ithaca, N.Y.
To detect neutralizing antibodies to CHV, twofold serial serum dilutions from 1:2 to 1:32 were prepared in minimum essential medium (MEM) (Eagle) without serum (Sigma) and incubated with an equal volume of MEM containing 50 PFU of CHV for 1 h at 37°C in 96-well tissue culture plates. Following this, 5 × 103 MDCK cells were added per well. The plates were incubated for 1 week at 37°C. The titer was determined as the reciprocal of the highest serum dilution that showed complete inhibition of cytopathic effect (CPE).
CHV immunofluorescence test.
MDCK cells were infected with CHV and maintained until 50% of the cells showed signs of CPE. The cells were then seeded onto 10-well slides (VWR International, Lutterworth, United Kingdom) at 104 cells per well and incubated at 37°C overnight. The slides were then rinsed in phosphate-buffered saline (PBS), fixed in methanol-acetone solution (2 parts of methanol to 1 part of acetone) for 10 min at −20°C, and air dried. Slides were stored at −20°C.
For the immunofluorescence assay (IFA), the slides were rehydrated in PBS for 5 min at room temperature. Serum samples were diluted 1:30 in PBS and incubated on the slides for 1 h at 37°C. The slides were washed three times with PBS for 5 min and then incubated with a fluorescein isothiocyanate-labeled anti-dog immunoglobulin G antibody (1:128; Sigma) for 1 h at 37°C. After being washed as described above, the slides were mounted with 50% glycerol in PBS and analyzed under a fluorescent microscope.
CPIV ELISA.
For preparation of the CPIV enzyme-linked immunosorbent assay (ELISA) antigen, A72 cells were infected with CPIV at a multiplicity of infection of 0.01 PFU/cell in 75-cm2 tissue culture flasks and were maintained at 37°C until a CPE was visible. The culture medium was removed, and the cells were lysed by addition of 1.5 ml of lysis buffer (0.01 M Tris, 0.002 M EDTA, 0.2 M sucrose, 2% Triton X) as described by Blixenkrone-Møller et al. (6) and 25 μl of proteinase inhibitor (Sigma) per flask. The lysate was clarified by two consecutive centrifugation steps at 12,000 × g. The supernatant was stored at −80°C and used for ELISA at a dilution of 1:100. The same protocol was used to prepare a control antigen by using uninfected A72 cells. The plates were coated overnight with the CPIV and control antigens. All steps were performed as described previously for a coronavirus ELISA (11). The optical density (OD) was determined at 492 nm in an ELISA photometer. For each serum sample, the OD obtained for the negative-control antigen was subtracted from the OD obtained with the CPIV antigen. To evaluate the ELISA, 42 serum samples that had been tested by SNT at Cornell University were tested at a dilution of 1:100 in the CPIV ELISA. The mean OD obtained from 20 SNT-negative serum samples was 0.021, and the standard deviation was 0.026. The cutoff point was determined as the mean plus three times the standard deviation (an OD of 0.1). For 22 serum samples that had been identified as CPIV positive by SNT, the mean OD by the CPIV ELISA was 0.34 and the standard deviation was 0.19. The ODs of the 22 SNT-positive samples ranged from 0.11 to 0.71.
Virus isolation.
Virus isolation was attempted on canine adult lung fibroblasts (passages 3 to 7) and MDCK cells. Lung fibroblasts were maintained in MEM with 20% fetal calf serum, and MDCK cells were maintained in MEM with 5% fetal calf serum. Tracheal tissue samples (approximately 25 mg) were homogenized by using a scalpel and were mixed vigorously in 1 ml of MEM containing 100 U of penicillin/ml, 0.1 mg of streptomycin/ml, and 2.5 μg of amphotericin B (Sigma)/ml. The samples were centrifuged at 12,000 × g for 10 min, and the supernatant was used to inoculate cell cultures. After 30 min at 37°C, the supernatant was removed and maintenance medium was added to the cultures. The cultures were passaged up to three times if no cytopathic effect was visible. Virus isolation from 178 dogs was attempted by using either tracheal tissue samples (total number, 127) or BAL samples (total number, 115). For 64 dogs, virus isolation was attempted on both tracheal and BAL samples.
Statistical analysis.
The data were analyzed by using the chi-square test, and P values below 0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The CPIV partial nucleocapsid protein gene sequence has been assigned GenBank accession number AY581307. The accession number for the CHV glycoprotein B sequence is AY582737. The partial sequence of the CHV region homologous to HSV-1 UL37 has the accession number AY582738.
RESULTS
PCR analysis of samples from dogs with various clinical respiratory signs.
By RT-PCR, CPIV was detected in 33 out of 170 tracheal samples and in 11 out of 106 lung samples tested. Table 2 shows the distribution of CPIV in relation to clinical respiratory signs. CPIV RNA was detected in tracheal samples from dogs with all grades of CIRD as well as in samples from dogs without clinical symptoms. It was most frequently found in dogs suffering from mild respiratory disease. Lung samples were rarely found positive for CPIV RNA, and the positive samples were mostly from dogs without clinical respiratory disease.
TABLE 2.
Detection of CPIV and CHV by PCR in the tracheae and lungs of dogs in correlation with signs of clinical respiratory disease
| Symptoms of respiratory disease | No. of samples in which virus was detected/total samples (%)
|
|||
|---|---|---|---|---|
| CPIV
|
CHV
|
|||
| Trachea | Lung | Trachea | Lung | |
| None | 12/60 (20) | 7/33 (21.2) | 7/72 (9.7) | 1/46 (2.2) |
| Mild | 8/29 (27.6) | 3/23 (13) | 4/38 (10.5) | 1/14 (7.1) |
| Moderate | 12/61 (19.7) | 0/32 | 10/77 (13) | 4/28 (14.3) |
| Severe | 1/20 (5) | 1/18 (5.6) | 6/24 (25) | 4/16 (25) |
For detection of CHV, primers CHV1 and CHV2 were used, because they had been found to perform better than primers GBN3 and GBN4 in PCRs using DNA extracted from tissue samples (data not shown). In total, 211 tracheal samples were tested by PCR for CHV, and 27 were found to be positive, whereas 10 out of 104 lung samples were positive. Table 2 shows the distribution of CHV in relation to respiratory signs. CHV was more frequently detected in tracheal and lung samples taken from dogs with moderate to severe respiratory disease than in samples from dogs with no or mild clinical symptoms.
By using primer pair CAV-F1-CAV-R1, which is specific for CAV type 2, 29 tracheal and 34 lung samples were tested, and none were found to be positive. The same samples, as well as 66 additional tracheal samples, were subsequently tested with primers CAV-VP1 and CAV-VP2, which detect CAV-1 as well as CAV-2, but no positive samples were detected.
By RT-PCR, none of the 39 tracheal samples and 70 lung samples tested for canine distemper virus were found positive.
Prevalence of viruses over time.
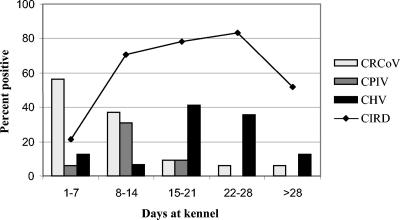
The presence of canine respiratory coronavirus (CRCoV) at this kennel and its prevalence in dogs with various clinical respiratory signs has been described previously (11). For comparative reasons, CRCoV data from these dogs were used in this report to study the prevalences of all viruses present in relation to the length of stay at the kennel. The prevalences of CPIV, CHV, and CRCoV in tracheal samples were analyzed in relation to the number of days the dogs had spent at the kennel before the samples were taken. For 149 tracheal samples, PCR results were available for all three viruses. CRCoV was detected most frequently within the first week of stay at the kennel, and its prevalence subsequently declined. The prevalence of CPIV showed a peak in the second week at the kennel but was rarely detected at earlier or later time points. In contrast, CHV was seldom present within the first 2 weeks but was frequently detected during weeks 3 and 4 (Fig. 2).
FIG. 2.
Prevalences of viruses at different time points as determined by PCR on tracheal samples. The x axis represents the number of days the dogs had stayed at the kennel before sampling. Each bar represents the percentage of dogs that were positive for the respective virus. The line represents the percentage of dogs that showed signs of respiratory disease. The total number of dogs tested at each time point was as follows: 1 to 7 days, n = 16; 8 to 14 days, n = 78; 15 to 21 days, n = 22; 22 to 28 days, n = 17; 29 days or more, n = 16.
Viral coinfections.
Of 149 dogs, 14 were infected with CPIV as well as with CRCoV. Seven of these dogs showed no clinical respiratory signs, three dogs each showed mild or moderate clinical signs, and one dog suffered from severe CIRD. One dog was positive for CHV and CPIV, and one dog was coinfected with CHV and CRCoV. In samples from two dogs, all three viruses could be detected; neither dog showed any sign of CIRD at the time of euthanasia.
Virus isolation from tracheal and BAL samples.
Canine herpesvirus was isolated from 8 out of 127 tracheal samples and from 6 out of 115 BAL samples tested. For two dogs a CHV isolate was obtained from the trachea as well as from the BAL fluid. Canine parainfluenza virus was recovered from one BAL specimen. Canine adenovirus was not isolated from any of these samples.
Sequence analysis of the CPIV PCR product.
The 667-bp PCR products amplified from two tracheal samples by using primer pair PNP1-PNP2 were cloned and sequenced. The sequences obtained were compared to each other and found to be 100% identical. Comparison with sequences available in GenBank revealed that both were 98.5% identical to the nucleocapsid protein gene sequence of simian virus 5 (SV5) strain W3A. For a further three PCR products, only the sequence amplified by the nested-PCR primer set PNP3-PNP4 was determined. In this 182-bp fragment, all five sequences compared were found to be identical and all showed the same four nucleotide changes relative to the sequence of SV5 strain W3A. Sequence analysis of the nested-PCR product obtained from the CPIV vaccine used at the kennel revealed that it was identical to the sequence of SV5 strain W3A.
Sequence analysis of canine herpesvirus strains.
The DNA sequence of a 1,287-bp fragment from the glycoprotein B gene was determined for 10 CHV isolates from the study population. All viral strains compared were identical in the sequenced region. Comparison to published sequences using the Fasta database search tool revealed 100% identity of the 10 sequences to the glycoprotein B sequence obtained from an Australian CHV isolate (GenBank accession number AF361073).
Serology.
On the day of entry into the kennel, all dogs had received a live attenuated vaccine against canine distemper virus, canine adenovirus, canine parainfluenza virus, and canine parvovirus as well as an inactivated Leptospira vaccine. A summary of the prevalence of viral antibodies is given in Table 3.
TABLE 3.
Prevalences of antibodies to viruses in serum samples from dogs at the time of entry into the kennel and at various time points afterwards
| Virus | No. of samples with antibodies/total samples (%) at:
|
||||
|---|---|---|---|---|---|
| Day 1 (entry) | Day 7 | Day 21 | Wk 4 | Wk 5 and later | |
| CPIV | 66/150 (44) | 72/105 (68.6) | 89/90 (98.9) | NDa | ND |
| CAV | 58/85 (68.2) | 24/27 (88.9) | 48/48 (100) | ND | ND |
| CDV | 13/24 (54.2) | 12/19 (63.2) | 11/11 (100) | ND | ND |
| CHV | 11/83 (13.3) | ND | 14/71 (19.7) | 6/13 (46.2) | 12/17 (70.6) |
| CRCoV | 37/123 (30.1) | 27/83 (32.5) | 113/114 (99.1) | ND | ND |
ND, not determined.
Presence of anti-CPIV antibodies on the day of entry and effect on the development of CIRD.
Forty-three dogs that were negative by CPIV ELISA on the day of entry were compared to 38 dogs that were positive. Out of the 43 dogs without antibodies, 11 remained healthy whereas 32 developed respiratory disease during the 21-day stay at the kennel. Of the 38 antibody-positive dogs, 16 remained healthy and 22 developed respiratory disease. The decreased prevalence of respiratory disease in antibody-positive dogs (57.9%) compared to antibody-negative dogs (74.4%) was shown not to be statistically significant (P = 0.115).
Comparison of a canine herpesvirus immunofluorescence assay to a serum neutralization test.
Fifty samples that had been tested by an IFA for antibodies to CHV were also tested by an SNT. Of 25 serum samples that had been negative by IFA, 22 were also found negative by SNT whereas 2 had an SNT titer of 2 and 1 had a titer of 8. Of 25 serum samples that had been positive by IFA, the result was confirmed by SNT for 24 whereas 1 serum sample was found to be negative. The neutralizing titers of the positive serum samples were as follows: 3 serum samples had a titer of 2, 4 had a titer of 4, 10 had a titer of 8, 3 had a titer of 16, and 5 had a titer of 32 or higher.
DISCUSSION
Canine infectious respiratory disease has a complex multifactorial etiology, and the pathogens involved may not be the same in every outbreak. This investigation sought to identify viruses present in dogs at a rehoming center with a history of endemic CIRD. The dog population was monitored over a 2-year period, which offered an opportunity to collect samples from dogs with various clinical respiratory signs as well as samples taken at a variety of time points after admittance to the kennel. Therefore, it was possible to associate the detection of viruses with respiratory signs as well as to gain insight into the chronological order of virus infections at this kennel.
In previous studies of respiratory disease in dogs, canine parainfluenza virus had been shown to be the most prevalent virus (3, 18). This investigation found that CPIV was present in the tracheae of almost 20% of dogs tested but was rarely detected in the lungs. There was no clear association between the presence of CPIV and the severity of respiratory symptoms. CPIV was also detected in the tracheae of a number of dogs that showed no clinical respiratory signs at the time of euthanasia, but it is possible that these dogs had an early stage of CIRD at which clinical signs would not yet have been manifest. CPIV infections occurred most frequently within the second week of stay at the kennel and decreased subsequently, presumably due to increasing antibody prevalence induced by vaccination. Many dogs demonstrated rapid seroconversion within 7 days after vaccination, which may indicate that they had previously been infected with or vaccinated against CPIV but had antibody levels below the detection limit of the ELISA on the day of entry. Respiratory disease was less frequent in dogs that were positive for CPIV antibodies upon entry into the kennel; however, the results were not statistically significant. Due to the high prevalence of respiratory disease in this population at the time of investigation, the number of dogs that remained healthy throughout the 3-week period was limited. Healthy dogs were also more likely to be rehomed during this period, so follow-up samples often could not be obtained.
Thus, CPIV was still present in this kennel despite the use of a vaccine. Almost all dogs developed anti-CPIV antibodies within 3 weeks after vaccination, but the infections occurred before the onset of an immune response. Regular vaccination of all dogs, however, has led to the build-up of a good population immunity, which may be responsible for the fairly low prevalence of CPIV in this population in comparison to the findings of similar studies of dogs with CIRD (3, 5). Comparison of the RNA sequences of the nucleoprotein genes of five CPIV RT-PCR products revealed that these sequences were identical to each other. It is not known how variable the nucleoprotein gene of CPIV is, because there were no sequences of actual CPIV isolates available in the GenBank database. However, all five sequences showed the same four nucleotide changes relative to that of simian virus 5 strain W3A, indicating that there may have been only one strain of CPIV present in this kennel. The same nucleotide changes were observed when the five sequences obtained from canine samples were compared to the sequence obtained from the CPIV vaccine strain. Thus, the detection of CPIV in tracheal and lung samples was not due to the presence of residual vaccine virus.
Surprisingly, CAV-2 was not present in this population, possibly due to the widespread use of vaccines against infectious canine hepatitis (caused by CAV-1). It has been shown that antibodies to CAV-1 confer cross-protection against CAV-2 (12). Canine distemper virus also was not detected in the study population, despite the fact that a proportion of the dogs showed severe respiratory symptoms. The serological data showed that the vaccine coverage was more than 50% of dogs entering the kennel. The higher prevalence of antibodies against CAV and CDV compared to anti-CPIV antibodies may indicate either that vaccines against the former viruses are used more frequently than vaccines against CPIV or that the immune response is longer lasting. The absence of CAV and CDV from this kennel could therefore be due to the frequent presence of antibodies or could reflect a very low prevalence of these viruses in the southeast of England.
Canine herpesvirus was the only virus that could be isolated consistently from this population. While CHV is well documented as a cause of fatal disease in puppies, it is considered to be less important in adult dogs. CHV infections occurred later than other viral infections in this population and were associated with more-severe respiratory signs. It has been shown previously that CHV is present in a variety of tissues in healthy dogs (7). It is therefore possible that CHV became reactivated during the course of CIRD due to stress caused by other viral and bacterial infections and hence was more frequently detected in dogs with severe clinical symptoms. Alternatively, dogs may have contracted CHV infections after entry into the kennel. One would expect to find a variety of CHV strains at the kennel if the presence of CHV was the result of activation of latent virus from different dogs. Analysis of 10 CHV strains obtained from dogs at the kennel by partial sequencing of the glycoprotein B gene revealed that all strains were 100% identical in the sequenced region; however, the sequence was also identical to a sequence obtained from an Australian isolate. It has been reported that isolates of canine herpesvirus world-wide show very low sequence variability (17). Further analysis by restriction digestion may be necessary to reveal potential variations in these CHV strains.
Only a small proportion of dogs were positive for antibodies to CHV on the day of entry. The number was too small to determine if the presence of antibodies led to decreased disease prevalence. Reading and Field (16) have reported the seroprevalence of CHV in the United Kingdom to be more than 80% by ELISA, but the majority of these dogs had titers of 2 or 4 when tested in an SNT. The lower sensitivity of the immunofluorescence assay used in this study compared to the SNT may explain the comparably low CHV seroprevalence detected. The IFA was used in this study because it was found to be much faster than the SNT. The serology results confirmed the finding by PCR that CHV infections occurred at a later time point than other viral infections in this population. Most dogs remained negative for anti-CHV antibodies during the first 3 weeks, but serum samples from dogs that had remained in the kennel for more than 3 weeks showed a dramatically increasing seroprevalence. Studies on experimental animals have shown that CHV causes only mild respiratory symptoms; however, in these experiments, other pathogens were not present. In naturally occurring outbreaks of CIRD, CHV may have an exacerbating effect on other viral or bacterial infections.
As previously reported, CRCoV was detected in this kennel and was mostly associated with mild respiratory symptoms. Almost all dogs tested were shown to develop antibodies to CRCoV within 3 weeks (11). In this report it was shown that the infection was most frequently detected in dogs that had stayed at the kennel for up to 2 weeks.
Only a small proportion of dogs (12%) had evidence of infections with several viruses at the same time. Coinfections with CRCoV and CPIV were detected most commonly, which may reflect the fact that infections with both viruses frequently occurred in the second week of stay at the kennel. Alternatively, infections with one virus may predispose the respiratory epithelium to entry of the other virus. Coinfections with CHV and other viruses were rare, since CHV infections appeared later than infections with CRCoV and CPIV.
Bacterial infections were also present in the study population, especially Bordetella bronchiseptica, which was most frequently detected in dogs with moderate respiratory symptoms (9), and Streptococcus equi subsp. zooepidemicus, which was associated with severe CIRD (8).
Conclusion.
Due to the widespread use of canine vaccines, canine adenovirus and canine distemper virus seem to have become less important in the etiology of CIRD, and the incidence of CPIV can be reduced by regular vaccination. In the kennel investigated, infections with CRCoV and CPIV may have initiated the disease, facilitating the entry of other microorganisms such as B. bronchiseptica, streptococci, and CHV.
The infectious agents involved in CIRD are diverse and evolving; the role of novel microorganisms therefore has to be assessed in order to enable development of appropriate diagnostic tools and efficacious vaccines.
Acknowledgments
We are most grateful to the Dogs Home Battersea for funding to J. Brownlie and for support and guidance.
We thank the veterinary nurses and clinicians at the Dogs Home Battersea for technical assistance.
REFERENCES
- 1.Appel, M., and L. N. Binn. 1987. Canine infectious tracheobronchitis short review: kennel cough, p. 201-211. In M. Appel (ed.), Virus infections of carnivores, 1st ed. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 2.Bibrack, B., U. Ackermann, and F. Benary. 1975. Serologic studies on the occurrence of virus infections in healthy dogs and in dogs with kennel cough. Zentbl. Veterinärmed. B 22:265-273. (In German.) [PubMed] [Google Scholar]
- 3.Binn, L. N., J. P. Alford, R. H. Marchwicki, T. J. Keefe, R. J. Beattie, and H. G. Wall. 1979. Studies of respiratory disease in random-source laboratory dogs: viral infections in unconditioned dogs. Lab. Anim. Sci. 29:48-52. [PubMed] [Google Scholar]
- 4.Binn, L. N., G. A. Eddy, E. C. Lazar, J. Helms, and T. Murnane. 1967. Viruses recovered from laboratory dogs with respiratory disease. Proc. Soc. Exp. Biol. Med. 126:140-145. [DOI] [PubMed] [Google Scholar]
- 5.Binn, L. N., E. C. Lazar, J. Helms, and R. E. Cross. 1970. Viral antibody patterns in laboratory dogs with respiratory disease. Am. J. Vet. Res. 31:697-702. [PubMed] [Google Scholar]
- 6.Blixenkrone-Møller, M., I. R. Pedersen, M. J. Appel, and C. Griot. 1991. Detection of IgM antibodies against canine distemper virus in dog and mink sera employing enzyme-linked immunosorbent assay (ELISA). J. Vet. Diagn. Investig. 3:3-9. [DOI] [PubMed] [Google Scholar]
- 7.Burr, P. D., M. E. Campbell, L. Nicolson, and D. E. Onions. 1996. Detection of canine herpesvirus 1 in a wide range of tissues using the polymerase chain reaction. Vet. Microbiol. 53:227-237. [DOI] [PubMed] [Google Scholar]
- 8.Chalker, V. J., H. W. Brooks, and J. Brownlie. 2003. The association of Streptococcus equi subsp. zooepidemicus with canine infectious respiratory disease. Vet. Microbiol. 95:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalker, V. J., C. Toomey, S. Opperman, H. W. Brooks, M. A. Ibuoye, J. Brownlie, and A. N. Rycroft. 2003. Respiratory disease in kennelled dogs: serological responses to Bordetella bronchiseptica lipopolysaccharide do not correlate with bacterial isolation or clinical respiratory symptoms. Clin. Diagn. Lab. Immunol. 10:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ek-Kommonen, C., L. Sihvonen, K. Pekkanen, U. Rikula, and L. Nuotio. 1997. Outbreak of canine distemper in vaccinated dogs in Finland. Vet. Rec. 141:380-383. [DOI] [PubMed] [Google Scholar]
- 11.Erles, K., C. Toomey, H. W. Brooks, and J. Brownlie. 2003. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology 310:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairchild, G. A., W. Medway, and D. Cohen. 1969. A study of the pathogenicity of a canine adenovirus (Toronto A26-61) for dogs. Am. J. Vet. Res. 30:1187-1193. [PubMed] [Google Scholar]
- 13.Frisk, A. L., M. Konig, A. Moritz, and W. Baumgartner. 1999. Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. J. Clin. Microbiol. 37:3634-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karpas, A., N. W. King, F. G. Garcia, F. Calvo, and R. E. Cross. 1968. Canine tracheobronchitis: isolation and characterization of the agent with experimental reproduction of the disease. Proc. Soc. Exp. Biol. Med. 127:45-52. [DOI] [PubMed] [Google Scholar]
- 15.McCance, D. J. 1996. DNA viruses: DNA extraction, purification and characterization, p. 192-193. In B. W. J. Mahy and H. O. Kangro (ed.), Virology methods manual, 1st ed. Academic Press, London, United Kingdom.
- 16.Reading, M. J., and H. J. Field. 1998. A serological study of canine herpes virus-1 infection in the English dog population. Arch. Virol. 143:1477-1488. [DOI] [PubMed] [Google Scholar]
- 17.Reubel, G. H., J. Pekin, K. Webb-Wagg, and C. M. Hardy. 2002. Nucleotide sequence of glycoprotein genes B, C, D, G, H and I, the thymidine kinase and protein kinase genes and gene homologue UL24 of an Australian isolate of canine herpesvirus. Virus Genes 25:195-200. [DOI] [PubMed] [Google Scholar]
- 18.Ueland, K. 1990. Serological, bacteriological and clinical observations on an outbreak of canine infectious tracheobronchitis in Norway. Vet. Rec. 126:481-483. [PubMed] [Google Scholar]
- 19.Wright, N. G., H. J. Cornwell, H. Thompson, A. Armitage, and I. Morrison. 1972. Canine adenovirus respiratory disease: isolation of infectious canine hepatitis virus from natural cases and the experimental production of the disease. Vet. Rec. 90:411-416. [DOI] [PubMed] [Google Scholar]