Abstract
We have identified a cytomegalovirus virion protein capable of modulating the rapid induction of an interferon-like response in cells that follows virus binding and penetration. Functional genomics revealed a role for the major cytomegalovirus structural protein, pp65 (ppUL83), in counteracting this response. The underlying mechanism involves a differential impact of this structural protein on the regulation of interferon response factor 3 (IRF-3). In contrast, NF-κB is activated independent of pp65, and neither STAT1 nor STAT3 becomes activated by either virus. pp65 is sufficient to prevent the activation of IRF-3 when introduced alone into cells. pp65 acts by inhibiting nuclear accumulation of IRF-3 and is associated with a reduced IRF-3 phosphorylation state. Thus, this investigation shows that the major structural protein of cytomegalovirus is committed to the modulation of the IRF-3 response, a primary mediator of the type I interferon response. By subverting IRF-3, the virus escapes throwing a central alarm devoted to both immediate antiviral control and regulation of the immune response.
Human cytomegalovirus (CMV) is a prominent opportunistic pathogen causing congenital disease as well as morbidity and mortality in immunocompromised hosts (reviewed in reference 50). Two characteristics of natural CMV infection stand out relative to other virus infections: primary infection takes many months to resolve and development of adaptive immunity is slow, even in fully immunocompetent individuals. This pattern suggests that CMV has the capacity to confound the priming of the immune process, and this has stimulated investigations into the means by which CMV interferes with antigen presentation (reviewed in reference 2). The infectious cycle of CMV starts with virus binding to the cell surface and is followed by fusion of the envelope with the plasma membrane with the release of viral structural components into the cell (reviewed in reference 43). Binding and penetration of virus or treatment with soluble envelope glycoprotein B (gB) or gH triggers a proinflammatory cellular response resembling an alpha/beta interferon (IFN-α/β) response (11, 13, 63, 76, 77, 80, 81). A virion- or virion glycoprotein-mediated activation of this IFN-like response is controlled through NF-κB (76-78) and may benefit viral infection, given the distribution of NF-κB sites on the viral genome (43). IFN-β gene transcription is induced by this IFN-like response in CMV-infected cells; however, IFN-β is only produced at low levels by infected cell cultures exposed to low multiplicities of infection (MOIs) (9, 55). IFN-β has not been detected in culture supernatants or virus preparations after infection at high MOIs or in purified virus stocks prepared by using low MOIs (references 41 and 81 and this report). Infection may also induce a novel IFN regulatory factor 3 (IRF-3)-related complex (9, 47, 53). In nonpermissive human peripheral blood mononuclear cells (PBMC) exposed to CMV, a response occurs via Toll-like receptor (TLR) signaling (15), suggesting that pattern recognition may contribute to this response.
IFNs are multifunctional cytokines with antiviral activity induced rapidly following infection (6, 60, 65). IFN-α and IFN-β are secreted and protect surrounding cells by signaling through the IFN-α/β receptor via Janus-activated kinase/signal transducer and activator of transcription (Jak/STAT) signaling to IFN-stimulated response element-regulated genes (17, 72). IFN-α/β has also been recognized as a major regulator of the adaptive immune response (6, 7). Induction of IFN-α/β expression may be mediated by a number of transcription factors such as NF-κB and AP-1; however, the activation of IRF-3 is believed to be the key signal to initiate IFN-β transcription and the IFN-α/β response (6, 34, 54). Inactive IRF-3 is retained in the cytoplasm of cells due to a strong nuclear export signal that dominates over nuclear import and thus serves as a gatekeeper of the IFN-α/β response (3, 54, 73). Virus infection or TLR signaling activates IRF-3 (26, 40, 54, 61, 68, 73). Upon activation, IRF-3 undergoes hyperphosphorylation, mediated by a virus-activated kinase pathway. Components of this pathway, IKKɛ and TBK1, have been identified (21, 61). Phosphorylated IRF-3 translocates to the nucleus and cooperates with the cellular acetyltransferases cyclic AMP-regulated enhancer binding protein (CBP) or p300 to mediate transcriptional activation of a subset of IFN response genes with a specific type of IFN-stimulated response element (17, 26, 33, 57, 72).
Although IFNs provide a rapid defense against a wide variety of RNA viruses, they are much less effective against DNA viruses, including many herpesviruses. Resistance has been attributed to functions encoded during viral infection, as reviewed by Katze et al. (34). Herpes simplex virus type 1 encodes functions that block IRF-3 as well as the effects of IFN (27, 37, 44, 46). Kaposi's sarcoma-associated herpesvirus encodes vIRF-1, which competes for p300 and prevents IRF-3 or IRF-1 function (4), and open reading frame 45 (ORF45), which targets IRF-7 (79). A number of other viruses encode functions that modulate IRF-3 activation (1, 22, 23, 67, 75). CMV encodes more than 160 gene products, some of which are needed for viral replication but many of which modulate diverse levels of the host response (1, 38, 42, 69). CMV is resistant to IFNs, based on clinical as well as in vitro studies (29). Virus infection of cultured cells is known to block IFN-stimulated Jak/STAT signaling as well as formation of the STAT-dependent IFN-stimulated gene factor 3 (41, 47). Two viral gene products (IRS1 and TRS1) block the activity of protein kinase R (14), one of the major effectors of IFN antiviral activity.
pp65 (ppUL83), the major constituent of both virions and noninfectious particles called dense bodies, localizes predominantly to the nucleus after virus penetration (58) and accumulates in both nucleus and cytoplasm as virus matures late in infection, where it may associate with a kinase (43). During infection pp65 is a major target of humoral (30) as well as cellular (CD4 and CD8 T-cell) immune responses (8, 35, 39, 74). Interestingly, pp65 is completely dispensable for productive infection of human fibroblasts (HFs) (59). Evidence has suggested that pp65 modulates antigen presentation (24, 48) and reduces the activation of NF-κB (12). Many other viral gene products encoded during replication have been shown to modulate viral and host cell processes (1, 42, 43, 69).
We have used a functional genomics approach to investigate the impact of pp65 on host cell transcription patterns at early times after CMV penetration of HFs. In a carefully controlled series of experiments comparing pp65 mutant to parental virus, we showed that pp65 dampened the virion-mediated IFN-like response (Abstr. Int. Herpesvirus Workshop, Regensburg, Germany, abstr. 1.10, 2001; Abstr. Eighth Int. Cytomegalovirus Workshop, Pacific Grove, California, abstr. 89A, 2001). Here we extend these results and describe the mechanism underlying this block. We find that pp65 subverts the activation of the transcription factor IRF-3 within early times after infection, thereby dampening the host antiviral response.
MATERIALS AND METHODS
Viruses and cells.
Primary human foreskin fibroblasts were grown and maintained as described (16). CMV strain AD169varDE (wild type [wt]) and RVAd65 (pp65 mutant virus derived from AD169varDE, a German variant of this widely used AD169 strain) (59), as well as strains AD169varATCC (American Type Culture Collection) (64), TownevarRIT3 (64), and Toledo (passage 10) were propagated, purified, and plaque assayed in HFs in complete medium as described (16). We employed a high MOI (4 PFU/cell) to give uniform infection levels of cells. RVAd65 and AD169varDE were confirmed to replicate to equivalent levels, as described when the mutant was first reported (59). Infections were carried out as previously described (16) with a parallel set of cultures evaluated as infection controls. RVAd65 and AD169varDE infected >98% of cells at an MOI of 4. Virus-free infected cell supernatants (wt or mutant-infected) collected at 4 or 6 h postinfection (hpi) after exposure to a high MOI inoculum failed to induce cellular gene expression or to interfere with replication of vesicular stomatitis virus (data not shown), consistent with previous observations (55). IFN-β is induced between 8 and 16 hpi at low MOIs with CMV (9, 55) but is not induced under high MOI conditions. UV inactivation, reducing plaque formation by >99.9999%, was carried out with 160 mJ/cm2 in a UV 1800 Stratalinker (Stratagene, La Jolla, Calif.) for 10 min with virus at a concentration of 4 × 107 PFU/ml in 4 ml. Inactivated virus was held for 1 h and diluted 10-fold in complete medium prior to use. CMV strain Towne pp65-expressing HFs transduced with MSCVpp65 or control cells transduced with empty LNCX vector were prepared and selected as described previously (66). pp65 expression was monitored by pp65 immunofluorescence staining, and the experiments shown in the present study were performed when pp65 was expressed in >95% of G418-selected HFs. PBMC from a CMV-seronegative adult donor were prepared by using Lymphoprep (Axis-Shield, Oslo, Norway) and suspended in complete medium prior to infection.
cDNA microarrays.
Polyadenylated RNA from 107 confluent HFs was purified by using Oligotex (QIAGEN, Valencia, Calif.) from Trizol (Invitrogen)-extracted total RNA. This RNA was prepared, reverse transcribed, labeled with Cy3-dUTP or Cy5-dUTP (Amersham, Little Chalfont, Buckinghamshire, United Kingdom) by random primed synthesis with DNA pol I Klenow (Amersham Life Science, Inc., Cleveland, Ohio), and hybridized to spotted, human cDNA microarrays as previously described (19). Sequence-verified human cDNA microarrays (HE and HG series, 31,000 spots; HD51 series, 17,000 spots) were produced at Stanford. Images were collected by using a GenePix 4000B microarray scanner, manually flagged to eliminate poor spots and analyzed by GenePix Pro 2.0 (Axon, Union City, Calif.) in combination with established methods (18, 25, 62). The data are available from the Stanford Microarray Database (SMD) (http://genome-www5.stanford.edu/MicroArray/SMD) Data were filtered for intensity (>150 pixels) and for regression correlation (>0.6) to remove dim spots. Cluster analysis was performed on a randomized seed of data (20). Background variability (normalized ratio of ± 1.4) was determined by hybridizing Cy3- and Cy5-cDNA from the same source to arrays (HE series). Significance analysis of microarrays (SAM) software was used as described in a one-class analysis (71). Expressed sequence tags (ESTs) and unnamed genes were evaluated by using BLAST (National Center for Biotechnology Information).
RNA blot analysis.
Total RNA (5 μg) was isolated and resolved by electrophoresis through 1% agarose, blotted, and hybridized by using biotinylated probes (interleukin-6 [IL-6], WARS, GBP-1, TAP-1, Mx1 [p78], ISG20, RANTES, Mip-1α, cig5, β-actin) derived from HF cell cDNA as previously described (16), with β-actin as a loading control with NorthernMax (Ambion). When used, actinomycin D (Sigma, St. Louis, Mo.) was added at a concentration of 5 μg/ml for 2 h at 4 hpi.
Antibodies, immunofluorescence microscopy, and immunoblot analysis.
Cells on 13-mm glass coverslips were fixed in ice-cold methanol for 20 min, rinsed, and blocked with phosphate-buffered saline containing 10% bovine serum albumin (Sigma). Mouse monoclonal antibodies to IRF-3 (SL12.1; BD Pharmingen, San Diego, Calif.) and pp65 (28-19 from William Britt, University of Alabama) and rabbit polyclonal antibodies to NF-κB (p50 subunit H-119), IRF-7 (H-246), STAT-1 (E-23), STAT-3 (H-190) (all from Santa Cruz Biotechnology, Santa Cruz, Calif.) were employed. Rabbit polyclonal antisera to IRF-3 from Santa Cruz Biotechnology used for localization in other reports (9, 12) exhibited only nonspecific immunofluorescence in our hands. Primary and Texas Red or fluorescein isothiocyanate-conjugated secondary antibodies (Vector Laboratories, Burlingame, Calif.) were in 2% bovine serum albumin. Nuclei were counterstained with Hoechst 44432 (Molecular Probes, Eugene, Oreg.). Localization was evaluated by epifluorescent microscopy. For immunoblot analysis, cells or isolated nuclei (4 × 105 cells or nuclei) were electrophoretically separated in 7.5 or 10% denaturing polyacrylamide gels, transferred, and probed with SL12.1 as described (67). Horseradish peroxidase-conjugated anti-mouse antibody (DakoCytomation Denmark A/S, Glostrup, Denmark) and an ECL Western blotting kit (Amersham) were employed. Nuclei were isolated following cell lysis in 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1% NP-40, and 0.5% deoxycholate. The IRF-3 phosphorylation state was determined at 4 hpi in buffer after collecting cells in the presence of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM EDTA, 1%NP-40, 1 mM sodium orthovanadate, 30 mM NaF, 0.1 mg of leupeptin/mg, and 1 mM phenylmethylsulfonyl fluoride.
IRF-3 nuclear translocation assay.
Cells in 12-well culture dishes were exposed to Superfect (QIAGEN)-loaded plasmid pcDNA3-EYFP (Clontech, Palo Alto, Calif.) for 2 h, according to the manufacturer's method. After treatment, the complete medium was added, and data were collected at 4 h (49).
RESULTS
pp65 modulation of host cell transcriptome.
Spotted human cDNA microarrays were used to assess the impact of CMV infection in the presence or absence of the major virion protein, pp65, on host cell transcript levels. We focused on very early times (1, 2, 3, and 4 hpi) following initial viral binding and penetration and employed a pair of viruses whose virions differ only in the presence of pp65 (UL83). Our experimental design employed two complementary microarray analyses to unveil the specific impact of pp65: (i) direct (or type I) analysis in which cDNA generated from the pp65 mutant was compared to cDNA from wt virus-infected cells, and (ii) indirect (or type II) analysis in which either mutant or wt virus-infected cell cDNA was compared to mock-infected cell cDNA collected at the same time point (19). These microarray approaches utilized replicates and cDNA synthesized directly from cellular mRNA template without any amplification. Data were subjected to a twofold change cutoff with clustering of data (20) as well as the stringent statistical criteria established by SAM (71).
Initially, we investigated the global impact of CMV strain AD169varDE infection on permissive HFs. Host transcript abundance in AD169varDE-infected cells was compared to mock-infected reference samples collected at the same time points and hybridized to 31,000-spot cDNA arrays. We observed changes in the pattern of gene expression at early times after infection that were consistent with previous reports (13, 63, 80, 81). When evaluated by using a twofold cutoff, 652 cDNAs were differentially expressed, with most being induced (584 cDNAs; 90% of spots) by virus infection at the 3 and/or 4 hpi time points. SAM analysis scored over 10,000 cDNAs as significantly altered by viral infection over this 4-h time course. Again, most (80%) were more abundant in virus-infected cells (http://genome-www5.stanford.edu/). Importantly, 565 (97%) virus-induced cDNAs identified by using a twofold cutoff criterion were included in this set. When genes represented by the cDNAs were grouped based on characteristics of regulation, an overwhelming majority (70%) of the 584 cDNAs represented IFN response-regulated genes, consistent with previous reports studying smaller data sets (13, 63, 80, 81). The extended lists of candidate cellular genes that respond to CMV infection can be found in the supplemental data (http://genome-www5.stanford.edu/MicroArray/SMD/).
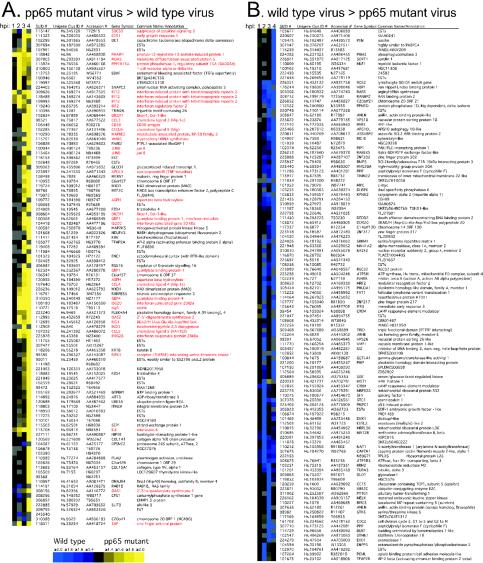
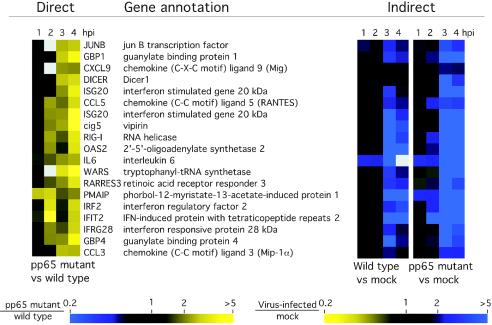
To investigate the influence of pp65 on CMV-mediated induction of cellular gene expression, we performed a direct microarray comparison of host transcript abundance in pp65-mutant and wt CMV-infected cells over a 4-h time course. When evaluated with a twofold cutoff, 220 cDNAs were differentially expressed at one time point or more over this time course. Cluster analysis placed these changes into two distinct categories that did not overlap (Fig. 1). A total of 101 cDNAs were induced more strongly by pp65 mutant virus (Fig. 1A, yellow), and 119 cDNAs were induced more strongly by wt virus (Fig. 1B, blue). The most dramatic differences in magnitude and numbers were observed at either 3 or 4 hpi. For example, at 4 hpi, 78 of the 101 cDNAs scored as more highly induced in pp65 mutant-infected cells, while 59 of the 119 cDNAs scored as more highly induced in wt virus-infected cells. Sixty-one annotated genes plus an additional 31 ESTs were represented in the set of 101 cDNAs that were more highly induced during mutant virus infection. Eighty annotated genes plus 36 ESTs were represented within the set of 119 cDNAs that were more strongly induced by wt virus infection with three cDNAs (CREM, PPIF, and ANLN) spotted in duplicate and giving similar patterns. When we sought common characteristics of host genes whose expression was coordinately regulated, 35 of the 61 genes (57%) induced more strongly by the mutant virus were identified as IFN-response genes (17, 26, 70). We could not find any recognizable pattern for the 80 genes induced more dramatically by wt virus infection, and so nothing more was done with this set. When the data from the time course were subjected to SAM analysis, which evaluates statistical consistency independent of the factor of change, 1,646 cDNAs were judged to be more strongly induced by mutant virus, and 1,604 cDNAs were induced more strongly by wt virus (http://genome-www5.stanford.edu/). The percentage of IFN-regulated genes increased as the stringency of cutoff was raised such that 92% of the genes meeting both the SAM significance and 2.0-fold cutoff criteria were in the IFN response group. Thus, our evaluation made use of several independent experimental strategies to show that pp65 mutant virus induced a response similar to that induced by a matched wt virus; however, the IFN-like component of this response was considerably stronger. When additional selection criteria were applied to this data set (twofold cutoff in two arrays), 19 cDNAs corresponding to 18 genes emerged as most strongly induced in pp65 mutant-infected cells (ISG20 was duplicated) (Fig. 2). All of these genes had been characterized as IFN-response genes (17, 70). We found similar results on smaller microarrays comparing mutant- and wt-infected cell cDNAs and also confirmed that mutant virus induced global IFN-like changes that were stronger than wt virus compared to mock infection at each time point (http://genome-www5.stanford.edu/). To illustrate how well this indirect analysis agreed with the direct analysis described above, Fig. 2 shows the primary data from both types of assay for the 18 genes that were most strongly induced by mutant virus. Thus, both direct and indirect microarray comparisons proved very powerful in showing the stronger stimulatory impact of the pp65 mutant over that of a matched pp65-expressing virus.
FIG. 1.
Cluster analysis (20) of genes regulated twofold or more in at least one time point (1, 2, 3, or 4 hpi) in a direct (type I) human cDNA microarray analysis comparing wt and pp65 mutant CMV. (A) Genes whose transcripts were more abundant upon infection with pp65 mutant RVAd65 than wt (pp65 mutant virus > wild-type virus). A total of 101 genes are represented in this panel. (B) Genes whose transcripts were more abundant upon infection with wt than pp65 mutant RVAd65 (wild-type virus > pp65 mutant virus). A total of 119 genes are represented in this panel. Each column of the microarray image represents a different time point, indicated at the top. Gray is used to indicate microarray spots that failed to pass the filtering criteria (see Materials and Methods). Red lettering denotes genes that are known to be IFN induced. Each row represents a different cDNA clone identified by a Stanford University identification number (SUID #). The Unigene cluster identification number (Clus ID #), GenBank accession number, gene symbol, and common gene name (or annotation) are also indicated for each clone. A color scale proportional to the factor of change is shown below panel A. Several cDNAs spotted in replicate on the microarrays (ISG20, 2 spots; dicer, 2 spots; MAX interacting protein 1, 2 spots; FZD-4, 2 spots; asparate β hydroxylase, 2 spots; GBP-1, 2 spots; JunB, 2 spots; C6orf37, 2 spots; and IFIT2, 3 spots) showed similar hybridization ratios. Complete data sets are available at http://genome-www5.stanford.edu.
FIG. 2.
Confirmation of pp65-induced genes by additional microarray experimental approaches. Cluster analysis of 19 genes for which transcripts were compared directly on a single microarray are shown at left. The transcripts shown here had a ratio of ≥2 for pp65 mutant virus-infected cell RNA versus wt virus-infected cell RNA in at least two time points (1, 2, 3, or 4 hpi) by using a direct analysis. A cluster analysis of the same genes following indirect analysis in which either wt or pp65 mutant virus infection was compared to mock infection at the same time point is shown at right. The color scales below the images indicate a factor of change ratio, shown with marks at 0.2-, 1-, 2-, and >5-fold spot intensity ratios. Ratios of mutant virus-infected to wt virus-infected cells are from a direct comparison (left), and ratios of either wt or pp65 mutant virus-infected cells to mock-infected cells are shown in the indirect comparisons (right). vs, versus.
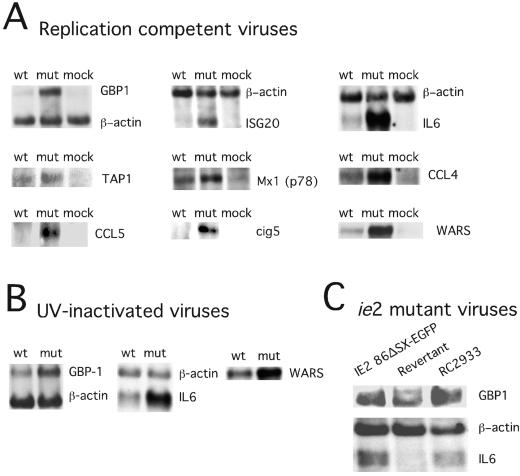
We confirmed the microarray data by RNA blot analysis of nine genes that were induced more dramatically by mutant than by wt virus (Fig. 3). To determine whether the difference in gene expression patterns reflected the delivery of virion pp65, we followed RNA levels of three genes (WARS, IL-6, and GBP-1) after exposure to UV-inactivated viruses. Both replication-competent and inactivated viruses induced similar RNA levels at 4 hpi (Fig. 3B) as expected (11, 13, 63, 76, 77, 80, 81), with the response to the pp65 mutant much stronger. These data implicated input virion pp65 as a modulator of cellular gene expression immediately following virus binding and penetration. We also found that newly synthesized pp65 made late during CMV infection (48 hpi) altered the expression of IFN-regulated genes GBP-1 and IL-6, mirroring the observations at the earlier time points (data not shown). To determine whether levels of pp65 in virions influenced the response, we investigated the behavior of two independently generated viral ie2 mutants, IE2 86ΔSX-EGFP, an AD169varATCC-based deletion mutant (56) and RC2933, a TownevarRIT3-based virus (J. Xu, D. Formankova, and E. S. Mocarski, unpublished data), that both fail to express late IE2 gene products (reference 56 and data not shown). Infection with either of these ie2 mutants resulted in a significantly reduced level of pp65 made late during the virus replication cycle (56), and this corresponded to a dramatic reduction in the incorporation of pp65 into virus particles (data not shown). Both of these mutants induced IL-6 and GBP-1 transcripts to higher levels than did the IE2 86ΔSX-EGFP revertant (Fig. 3C) or other control viruses (data not shown), which is consistent with the hypothesis that the amount of pp65 incorporated into virions influenced the IFN-like response to virus infection.
FIG. 3.
RNA blot analysis of selected IFN response genes. RNA blots (5 μg of total cell RNA per lane) from infected cells collected at 4 hpi at an MOI of 4 and hybridized with the indicated IFN response gene probes prepared as described in Materials and Methods. (A) Comparisons of wt-, pp65 mutant (mut)-, and mock-infected (mock) HFs for expression of GBP1, ISG20, IL-6, Tap-1, Mx-1 (p78), CCL4, CCL5, cig5, and WARS with a β-actin control are shown for some samples. (B) Comparisons of UV-inactivated wt and pp65 mutant (mut) virus-induced expression of GBP1, IL-6, and WARS with a β-actin control are shown for some samples. (C) Comparisons are shown of RNA collected 4 hpi from IE2 86ΔSX-EGFP-, revertant-, and RC2933-infected cells probed for GBP1, IL-6, and β actin.
pp65 control of the cytoplasmic localization and hypophosphorylation state of IRF-3.
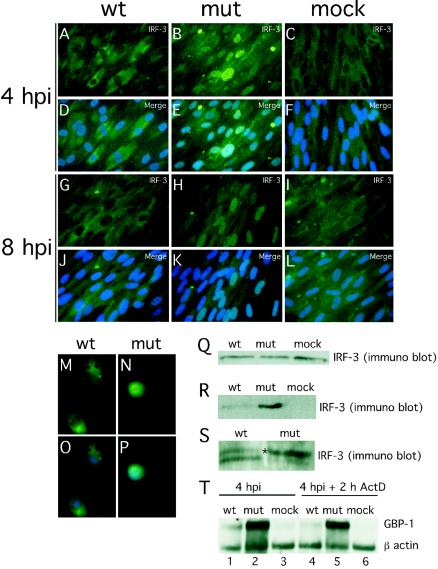
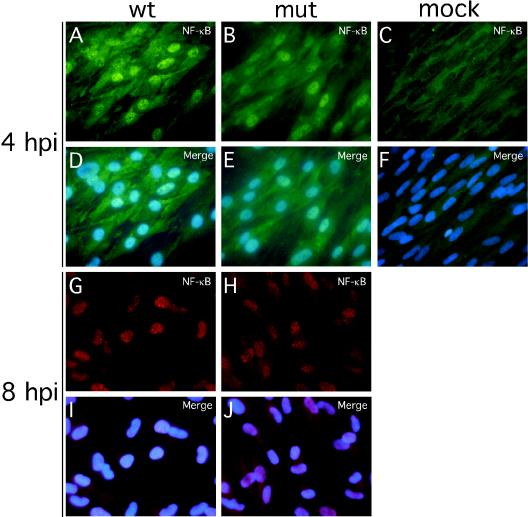
Binding sites for transcription factors (NF-κB, STATs, and IRFs) known to play crucial roles in the induction of IFN-response (54, 68) were contained in the promoter regions of the genes that responded differentially to wt and mutant virus infection. Upon activation, all of these factors translocate to and accumulate in the cell nucleus. We investigated the localization of IRF-3, NF-κB, STAT-1, STAT-3, and IRF-7 at 4 hpi under conditions leading to a uniform infection of >98% of cells (MOI of 4). Under these conditions an overwhelming majority of wt virus-infected HFs were pp65 antigen positive shortly after virus adsorption (data not shown), as expected from published reports (58). There was a dramatic difference in the localization of IRF-3 in wt (Fig. 4A and D) and mutant (Fig. 4B and E) virus-infected cells. IRF-3 remained localized to the cytoplasm following infection with wt virus, a pattern similar to mock-infected HFs (Fig. 4C and F), but localized to the nucleus in pp65 mutant virus-infected cells. Similar IRF-3 patterns were observed at 8 hpi in HFs (Fig. 4G to L) as well as in human PBMC at 4 hpi (Fig. 4M to P). In contrast to the differential impact we have seen on IRF-3 localization patterns, NF-κB translocated to the nucleus by 4 hpi and remained nuclear through 8 hpi, irrespective of whether mutant or wt virus was used (Fig. 5), results that are at variance with a recent report (12). In our hands, IRF-7 expression was not detectable by immunofluorescence analysis in either uninfected or infected cells (data not shown).
FIG. 4.
Impact of wt and pp65 mutant CMV infection on IRF-3 localization and phosphorylation state at early times after infection. (A to L) Immunofluorescence analysis of IRF-3 localization in HFs infected (MOI of 4) with wt virus at 4 (A and D) or 8 (G and J) hpi or with pp65 mutant virus (mut) at 4 (B and E) or 8 (H and K) hpi compared to mock-infected cells at 4 (C and F) or 8 (I and L) hpi. Overlays of Hoechst 44432-positive nuclei are shown in panels M to P (merge). Immunofluorescence analysis of IRF-3 localization in PBMC infected by wt (M and O) or mutant (N and P) virus (4 hpi with an MOI of 4). (Q to S) Immunoblot analyses of total (Q) and nuclear (R) IRF-3 levels in wt virus-, mutant virus- and mock-infected HFs, and immunoblot analysis of IRF-3 electrophoretic mobility forms revealing phosphorylation state in wt and mutant virus-infected cells (S). RNA stability assay (T) was performed with RNA extracted by wt CMV-, pp65 mutant CMV (mut)-, and mock-infected HFs (mock) in the absence (lanes 1 to 3) or presence of actinomycin D (2 h ActD; lanes 4 to 6). Infected-cell RNA was probed for GBP-1 and β-actin. Note that the monoclonal antibody we have employed in the experiments shown in Fig. 4 to 6 has been shown to give specific in situ immunofluorescence localization and immunoblot detection of IRF-3 (72; M. G. Wathelet, personal communication) and has been used for this purpose in several incisive studies (5, 23, 49, 67, 72).
FIG. 5.
Impact of wt and pp65 mutant CMV infection on NF-κB localization. Shown are immunofluorescence assays of NF-κB localization in HFs infected (MOI of 4) with wt virus at 4 (A and D) or 8 (G and I) hpi or with pp65 mutant virus (mut) at 4 (B and E) or 8 (H and J) hpi compared to mock-infected cells at 4 (C and F) hpi. Texas Red-conjugated secondary antibody was used to generate the results shown in panels G through J. Overlays of Hoechst 44432-positive nuclei are shown (merge).
In contrast to the activation pattern of NF-κB, both STAT-1 and STAT-3 remained cytoplasmic at 4 hpi with either wt or mutant virus (data not shown). The failure of STAT-1 to localize to the nucleus is consistent with earlier work that showed that STAT-1 remains inactive at early and late times during infection (41, 47) but is at variance with a recent report (12) showing that this transcription factor localizes to the nucleus shortly after exposure to virus.
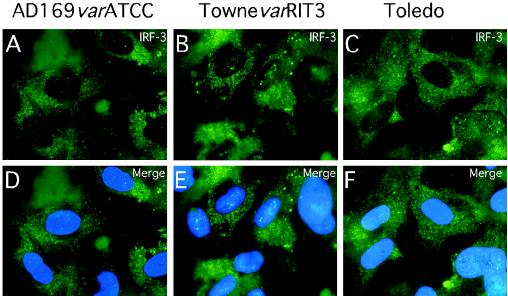
Overall, IRF-3 levels detected by immunoblotting were similar in wt virus-, mutant virus-, and mock-infected cells (Fig. 4Q). Consistent with the immunofluorescence results, nuclei fractionated from mutant virus-infected HFs contained higher levels of IRF-3 than nuclei from either wt- or mock-infected cells (Fig. 4R). Further analysis in the presence of phosphatase inhibitors revealed a slower migrating form of IRF-3 (Fig. 4S), suggesting a difference in phosphorylation states (72). Thus, pp65 appeared to counteract the hyperphosphorylation of IRF-3 that is associated with nuclear accumulation (54, 68, 73). Consistent with the IRF-3 results shown for AD169varDE, other pp65-expressing strains (AD169varATCC, TownevarRIT3, and Toledo) showed a cytoplasmic IRF-3 localization at 4 h after virus infection at high MOIs (Fig. 6). This result suggests that our observations were not dependent upon a single strain or strain variant. Given the behavior of CMV strain Toledo, which is fully virulent and expresses a complete set of CMV gene products (43), we believe the prevention of IRF-3 activation is likely to represent a normal activity of natural CMV strains.
FIG. 6.
Impact of infection with additional CMV strains and strain variants on IRF-3 localization at early times after infection. Shown are immunofluorescence analyses of IRF-3 localization in HFs infected (MOI of 4) with AD169varATCC (A and D), TownevarRIT3 (B and E), and Toledo (passage level 10; C and F). Overlays of Hoechst 44432-positive nuclei are shown (merge).
To determine whether RNA degradation played any role in the IRF-3-dependent activation of gene expression, we inhibited transcription with actinomycin D and compared RNA levels in pp65 mutant- and wt virus-infected cells. Both GBP-1 and IL-6 mRNAs were stable during a 2-h treatment at 4 hpi with no significant or differential effect on overall RNA stability (Fig. 4T; data not shown). Thus, it does not appear that CMV pp65 acts in a manner analogous to the virion host shut-off function (vhs/UL41) of herpes simplex virus (36).
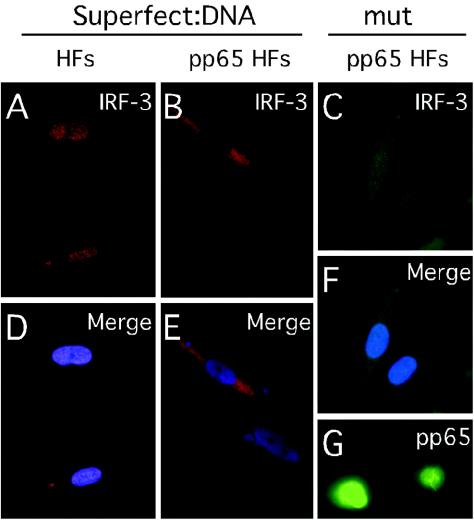
To determine whether pp65 was itself sufficient to prevent IRF-3 translocation independent of other virion structural proteins, IRF-3 localization was evaluated in retrovirus-transduced HFs that stably expressed pp65 (Fig. 7). We chose retrovirus vector-transduced cells for this study because this strategy does not in itself activate an IFN-like response that might interfere with interpretation of any experiments. We employed treatment with plasmid pcDNA3-EYFP-loaded Superfect liposomes to induce IRF-3 nuclear translocation in HFs independent of CMV infection, a method that activates IRF-3 (49), and compared empty LNCX vector-transduced HFs to pp65-transduced HFs. While nontransduced HFs (Fig. 7A and D) and HFs stably transduced with an empty LNCX vector (data not shown) supported nuclear accumulation of IRF-3, pp65-expressing HFs (Fig. 7G) exhibited cytoplasmic localization of IRF-3 following exposure to DNA-loaded liposomes (Fig. 7B and E). As expected, IRF-3 also remained predominantly cytoplasmic in pp65 HFs infected with pp65 mutant virus (Fig. 7C and F). Thus, pp65 alone was sufficient to prevent IRF-3 activation by a nonviral inducer.
FIG. 7.
Immunofluorescence analysis of IRF-3 localization in pp65-transduced cells. Immunofluorescence assays show IRF-3 localization 4 h after exposure to induction with pcDNA3-EYFP-loaded Superfect in control HFs (A) and pp65-transduced HFs (B) or at 4 hpi with pp65 mutant (mut) virus in pp65-transduced HFs (C). Overlays of IRF-3 stain with Hoechst 44432 show nuclei (D to F). Immunofluorescence assay shows pp65 localization in pp65-transduced HFs (G).
DISCUSSION
Many CMV gene products are committed to escape from the host immune response (1, 42, 69) through which this virus establishes a balance that sustains viral persistence and facilitates sporadic shedding throughout the life of the host. As a result of these tactics, CMV remains one of the most problematic opportunistic infections in immunocompromised hosts (50). Although a number of characterized immunomodulatory gene products help CMV to escape the effectors of the innate and adaptive immune response (1, 42, 69), few characterized functions target initiating events in the immune response. The first steps in this response include cell-intrinsic alarm signals such as apoptosis and activation of IFN-α/β (6). CMV-mediated suppressors of apoptosis, a cellular defense mechanism that helps prime the adaptive immune response via cross-presentation (2), is well documented and broadly conserved in CMVs (38, 42, 64); however, modulation of the IFN response by this virus is less well understood.
IRF-3 is the central transcription factor needed to initiate an IFN-α/β response, residing in the cytoplasm in an inactive form where it may be activated following virus infection and/or TLR signaling (54, 68, 70, 73). The importance of the IFN-α/β response in control of CMV infection can be gauged from the profound susceptibility of mice that lack IFN-α/β receptors (52) or that fail to support TLR signaling (28) in the activation of IRF-3. Both alphaherpesviruses (37, 46) and gammaherpesviruses (4, 79) encode gene products that prevent IRF-3 activation at very early times after infection. Although the tegument protein encoded by ORF45 of Kaposi's sarcoma-associated herpesvirus has not been studied in virus-infected cells and ORF45 is not the major tegument protein, its impact on IRF-7 phosphorylation and nuclear translocation (79) is most analogous to the impact we have described for pp65 on IRF-3. In the absence of pp65, induction of IRF-3 occurs immediately following entry into either HFs or PBMC. This is an effective strategy to interfere with IRF-3 activation in permissive cells that support productive infection as well as in nonpermissive cells where viral latency may be the outcome. In the presence of pp65, IRF-3 translocation to the nucleus and hyperphosphorylation are impeded. Our finding that the CMV pp65 modulates this key cellular mediator of the IFN-α/β response establishes that all three major subgroups of herpesviruses have the same impact but achieve it through different types of gene products. Because UL83 is conserved in characterized CMVs of rodents and primates, suppression of IRF-3 can be predicted to follow common pathways in this subset of betaherpesviruses. The importance of the murine CMV pp65 homolog is supported by evidence that M83 mutants are attenuated in immunocompetent mice, although these viruses replicate to normal levels in cell culture (45). Species specificity makes it impossible to evaluate the function of human CMV pp65 directly in an experimental host, making studies in the mouse critical to understanding the full role of this type of viral protein in infection.
While most pp65 appears to localize to the nucleus following virus entry (58), we do not yet know whether nuclear or cytoplasmic pp65 is responsible for the impact on IRF-3. Our data are consistent with two possible mechanisms that would interfere with known pathways of IRF-3 activation. Our data favor a model where pp65 in the nucleus would promote dephosphorylation and export of IRF-3, affecting the balance in nuclear-cytoplasmic shuttling (54), possibly also interfering with CPB/p300 and formation of the transcription factor DRAF1. This would allow nuclear export to dominate over import as it does in the absence of inducers (54). Alternatively, pp65 may interfere with the newly recognized kinases IKKɛ or TBK1 (21, 61) and prevent hyperphosphorylation, although this would most likely occur in the cytoplasm. Preliminary analyses have failed to detect a direct interaction between IRF-3 and pp65 by coimmunoprecipitation, suggesting that pp65 may modulate IRF-3 without forming a stable interaction.
The immediate impact of modulating IRF-3 activation and translocation to the nucleus would be the failure to activate IFN-stimulated genes (26, 54, 68, 70, 72). This would, in turn, allow virus to escape both direct antiviral effects as well as the amplification of innate and adaptive immunity that depends on genes that respond to IRF-3. The work we have presented suggests that the IRF-3 component of the virus-induced response, shown to be mediated via TLR2 signaling in PBMC exposed to CMV at MOIs ranging from 0.005 to 0.05 (15), appears to be inhibited when a high MOI is used, and this is likely due to the delivery of virion pp65 to cells. Interference with IRF-3 activation has downstream consequences and would modulate critical immune response cytokines, such as IL-12, IL-15, and IFN-γ, cytokines that control the intensity and quality of the innate and adaptive phases of the immune response (6, 7). The reported pp65-mediated alteration of antigen presentation (24, 48) may be another downstream consequence of IRF-3 modulation. Besides being introduced during viral entry, pp65 is an abundant late viral gene product which is distributed between the nucleus and cytoplasm and accompanies virion and dense body morphogenesis (43). Though not the main focus of this report, control of IRF-3 by pp65 is sustained at late times after infection. Activation of IRF-3 only occurs in mutant virus-infected cells at late times after infection, although the activation of IRF-3 responsive genes does not apparently suppress mutant virus replication (59). CMV may have a posttranscriptional mechanism to prevent full expression of IFN-β (13, 81) that may become partially compromised at low MOI (55). CMV is generally able to blunt the antiviral effects of IFN-α/β when applied exogenously so long as the virus is used at MOI that lead to uniform infection (29, 41, 47), and two gene products that block protein kinase R activity are known (14) to contribute to this resistance.
There is ample evidence that exposure of cells to either CMV or soluble CMV glycoproteins activates an IFN-like response (11, 13, 63, 76, 77, 80, 81) and that this activation is controlled through the NF-κB pathway (76-78). When pp65 mutant virus is used to infect cells, the broader and stronger IFN-like response to virion binding and penetration is likely due to the added impact of IRF-3 activation above and beyond NF-κB activation alone. While our work was under review by another journal, a report from Browne and Shenk (12) appeared and argued a role for pp65 in downregulating the virion-induced IFN response through a direct impact on NF-κB. The observations agreed with our original communication of microarray data but differed in the mechanism we have elucidated here. We have not observed modulation of NF-κB associated with pp65, a result that is in agreement with a range of studies on CMV (see works cited in reference 43, as well as references 76-78). We cannot provide an explanation for these divergent observations; however, we speculate that (i) virus strain differences, (ii) infection conditions, (iii) host cell variability, and/or (iv) virus preparation may have contributed. All studies on CMV rely on primary cells, which vary with source as well as with age. AD169 strain variants are in use in different laboratories; although they are often referred to by the same name, they are different (43, 51, 64). The pp65 mutant used here and by the Shenk group were both derived from the Plachter laboratory (J. Nelson, personal communication). RVAd65 was derived from an AD169 variant (59) that had been carried in German laboratories for several decades (M. Mach, personal communication). We obtained a parental stock and assigned the name AD169varDE to differentiate it from other AD169 variants in use (64). In the course of these studies we also examined the impact on IRF-3 of AD169varATCC, a commonly used virus (9, 11, 13, 51, 63, 76, 77, 80, 81) that has previously been used as a control (12). In our hands, matched mutant and control viruses revealed differences in IRF-3 localization but showed no differences in NF-κB localization to nuclei at 4 or 8 hpi. Aside from the possible contribution of viral strain variants and host cell variation, other factors in the infection and follow-up may have contributed to the different results. Finally, it is possible that use of adenovirus vectors (10), hemagglutinin-tagged pp65 (12), or some indirect impact of the IRF-3 activation state on NF-κB levels contributed to the differences in the observations. NF-κB has long been suggested to control expression of important viral genes (43, 77, 78) and has several NF-κB sites positioned near important regulatory genes. CMV benefits from activation of NF-κB, which follows the induction of mitogen-activated protein kinase as well as a phosphatidylinositol 3 kinase signaling cascade at early times after infection (31, 32). Thus, the virus appears to benefit from the activation of NF-κB but to suppress IRF-3 signals that would lead to a broader host IFN response.
An additional difference regarding the activation of IRF-3 by wt CMV (9, 12, 53) merits mention. We observed IRF-3 translocation to the nucleus by 4 hpi and continuing at least through 8 hpi, but this occurred only in mutant virus-infected cells. We employed a commercially available murine monoclonal antibody that has been widely applied to study IRF-3 (5, 23, 49, 67, 72) because it provides specific in situ immunofluorescence localization as well as a specific immunoblot detection of this protein (72; M. G. Wathelet, personal communication).
This study demonstrates the power of cDNA microarray analysis to dissect viral gene function, even in the absence of a significant growth defect. This functional genomics approach revealed dramatic differences in the magnitude of an IFN-type response to infection comparing wt and pp65 mutant virus immediately following entry when the virus-induced response was maximal. CMV mutants that incorporated reduced levels of pp65 into virions exhibited intermediate impact on this response, suggesting that the normally high level of pp65 in the virion tegument is present to blunt the cellular response to infection. The analysis we performed identified many genes that had been shown previously to be differentially induced by IRF-3 (26), although our data set did not overlap at all with IRF-3-repressed genes. As more information becomes available, our microarray standard (minimum information about a microarray experiment, or MIAME)-compliant, publicly accessible database (http://genome-www5.stanford.edu/) may yield additional insights into important targets of IRF-3 in the face of full NF-κB activation. Replicate comparisons and alternative data analysis criteria (cluster and SAM) allowed the retrieval of an informative and manageable number of genes that were functionally grouped in the same pathway.
CMV pp65 modulation of the IFN-α/β response provides further evidence for close coevolution of this virus in balance with the human host. Other transcription factors, notably NF-κB, may contribute to the cellular response independent of pp65 modulatory effects. Even in the absence of active IRF-3, CMV infection induces an IFN-like response. CMV exhibits a dose-dependent susceptibility to IFN-α/β inhibition in cell culture (29), consistent with achieving a standoff rather than completely overcoming the impact of IFN. This balance seems tipped in favor of the virus because IFNs-α/β showed little promise when administered to patients with the goal of controlling CMV infection and disease (50). Thus, the IRF-3 block characterized here may be a very important determinant in viral pathogenesis, potentially acting in concert with other less well understood effects on IFN-β expression (13, 81) and Jak/STAT signaling pathways (41).
Acknowledgments
We are grateful to Bodo Plachter, University of Mainz, for providing RVAd65, to Michael Mach, University of Erhlangen, for providing AD169varDE, to William Britt, University of Alabama, for providing antibody 28-19, to Deborah Spector, University of California San Diego, for providing the IE2 86ΔSX-EGFP mutant and the revertant viruses, and to Stanley Riddell, Fred Hutchinson Cancer Research Center, for providing MSCVpp65. We acknowledge the staff of the Stanford Microarray Database and Luciano Brocchieri, Department of Mathematics, Stanford University, for assistance with data analysis.
This work was supported by USPHS grants RO1 AI33852 and PO1 AI50153.
REFERENCES
- 1.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrode, G., and C. Davrinche. 2003. Dendritic cells and HCMV cross-presentation. Curr. Top. Microbiol. Immunol. 276:277-294. [DOI] [PubMed] [Google Scholar]
- 3.Au, W. C., P. A. Moore, W. Lowther, Y. T. Juang, and P. M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. USA 92:11657-11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 5.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biron, C. A., and G. C. Sen. 2001. Interferon and other cytokines. p. 321-351. In D. M. Knipe and P. M. Howley (ed.), Fields Virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 7.Biron, C. A. 2001. Interferons alpha and beta as immune regulators-a new look. Immunity 14:661-664. [DOI] [PubMed] [Google Scholar]
- 8.Bitmansour, A. D., S. L. Waldrop, C. J. Pitcher, E. Khatamzas, F. Kern, V. C. Maino, and L. J. Picker. 2001. Clonotypic structure of the human CD4+ memory T cell response to cytomegalovirus. J. Immunol. 167:1151-1163. [DOI] [PubMed] [Google Scholar]
- 9.Boehme, K. W., J. Singh, S. T. Perry, and T. Compton. 2004. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J. Virol. 78:1202-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowen, G. P., S. L. Borgland, M. Lam, T. A. Libermann, N. C. Wong, and D. A. Muruve. 2002. Adenovirus vector-induced inflammation: capsid-dependent induction of the C-C chemokine RANTES requires NF-kappa B. Hum. Gene Ther. 13:367-379. [DOI] [PubMed] [Google Scholar]
- 11.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Browne, E. P., and T. Shenk. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. USA 100:11439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Child, S. J., M. Hakki, K. L. De Niro, and A. P. Geballe. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 78:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Compton, T., E. A. Kurt-Jones, K. W. Boehme, J. Belko, E. Latz, D. T. Golenbock, and R. W. Finberg. 2003. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J. Virol. 77:4588-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courcelle, C. T., J. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Requirement for uracil-DNA glycosylase during the transition to late-phase cytomegalovirus DNA replication. J. Virol. 75:7592-7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diehn, M., G. Sherlock, G. Binkley, H. Jin, J. C. Matese, T. Hernandez-Boussard, C. A. Rees, J. M. Cherry, D. Botstein, P. O. Brown, and A. A. Alizadeh. 2003. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 31:219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 20.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 22.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert, M. J., S. R. Riddell, B. Plachter, and P. D. Greenberg. 1996. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature 383:720-722. [DOI] [PubMed] [Google Scholar]
- 25.Gollub, J., C. A. Ball, G. Binkley, J. Demeter, D. B. Finkelstein, J. M. Hebert, T. Hernandez-Boussard, H. Jin, M. Kaloper, J. C. Matese, M. Schroeder, P. O. Brown, D. Botstein, and G. Sherlock. 2003. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 31:94-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He, B., J. Chou, R. Brandimarti, I. Mohr, Y. Gluzman, and B. Roizman. 1997. Suppression of the phenotype of gamma(1)34.5- herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the alpha47 gene. J. Virol. 71:6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoebe, K., X. Du, P. Georgel, E. Janssen, K. Tabeta, S. O. Kim, J. Goode, P. Lin, N. Mann, S. Mudd, K. Crozat, S. Sovath, J. Han, and B. Beutler. 2003. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 24:743-748. [DOI] [PubMed] [Google Scholar]
- 29.Holmes, A. R., L. Rasmussen, and T. C. Merigan. 1978. Factors affecting the interferon sensitivity of human cytomegalovirus. Intervirology 9:48-55. [DOI] [PubMed] [Google Scholar]
- 30.Jahn, G., B. C. Scholl, B. Traupe, and B. Fleckenstein. 1987. The two major structural phosphoproteins (pp65 and pp150) of human cytomegalovirus and their antigenic properties. J. Gen. Virol. 68:1327-1337. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, R. A., S. M. Huong, and E. S. Huang. 2000. Activation of the mitogen-activated protein kinase p38 by human cytomegalovirus infection through two distinct pathways: a novel mechanism for activation of p38. J. Virol. 74:1158-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, R. A., X. Wang, X. L. Ma, S. M. Huong, and E. S. Huang. 2001. Human cytomegalovirus up-regulates the phosphatidylinositol 3-kinase (PI3-K) pathway: inhibition of PI3-K activity inhibits viral replication and virus-induced signaling. J. Virol. 75:6022-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juang, Y. T., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 35.Kern, F., T. Bunde, N. Faulhaber, F. Kiecker, E. Khatamzas, I. M. Rudawski, A. Pruss, J. W. Gratama, R. Volkmer-Engert, R. Ewert, P. Reinke, H. D. Volk, and L. J. Picker. 2002. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. J Infect. Dis. 185:1709-1716. [DOI] [PubMed] [Google Scholar]
- 36.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 84:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCormick, A. L., A. Skaletskaya, P. A. Barry, E. S. Mocarski, and V. S. Goldmacher. 2003. Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virology 316:221-233. [DOI] [PubMed] [Google Scholar]
- 39.McLaughlin-Taylor, E., H. Pande, S. J. Forman, B. Tanamachi, C. R. Li, J. A. Zaia, P. D. Greenberg, and S. R. Riddell. 1994. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J. Med. Virol. 43:103-110. [DOI] [PubMed] [Google Scholar]
- 40.Medzhitov, R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1:135-145. [DOI] [PubMed] [Google Scholar]
- 41.Miller, D. M., Y. Zhang, B. M. Rahill, W. J. Waldman, and D. D. Sedmak. 1999. Human cytomegalovirus inhibits IFN-alpha-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-alpha signal transduction. J. Immunol. 162:6107-6113. [PubMed] [Google Scholar]
- 42.Mocarski, E. S. 2002. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 10:332-339. [DOI] [PubMed] [Google Scholar]
- 43.Mocarski, E. S., Jr., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields Virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 44.Mohr, I., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 15:4759-4766. [PMC free article] [PubMed] [Google Scholar]
- 45.Morello, C. S., L. D. Cranmer, and D. H. Spector. 1999. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83). J. Virol. 73:7678-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Odeberg, J., B. Plachter, L. Branden, and C. Soderberg-Naucler. 2003. The human cytomegalovirus protein pp65 mediates accumulation of HLA-DR in lysosomes and destruction of the HLA-DRα chain. Blood 101:4870-4877. [DOI] [PubMed] [Google Scholar]
- 49.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In D. Knipe and P. Howley (ed.), Fields Virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 51.Patterson, C. E., and T. Shenk. 1999. Human cytomegalovirus UL36 protein is dispensable for viral replication in cultured cells. J. Virol. 73:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Presti, R. M., J. L. Pollock, A. J. Dal Canto, A. K. O'Guin, and H. W. I. Virgin. 1998. Interferon gamma regulates acute and latent murine cytomegalovirus infection and chronic disease of the great vessels. J. Exp. Med. 188:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reich, N. C. 2002. Nuclear/cytoplasmic localization of IRFs in response to viral infection or interferon stimulation. J. Interferon Cytokine Res. 22:103-109. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez, J. E., T. R. Loepfe, and N. S. Swack. 1987. Beta interferon production in primed and unprimed cells infected with human cytomegalovirus. Arch. Virol. 94:177-189. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez, V., C. L. Clark, J. Y. Yen, R. Dwarakanath, and D. H. Spector. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J. Virol. 76:2973-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, and S. Noguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 58.Schmolke, S., P. Drescher, G. Jahn, and B. Plachter. 1995. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J. Virol. 69:1071-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmolke, S., H. F. Kern, P. Drescher, G. Jahn, and B. Plachter. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 69:5959-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 61.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 62.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 66.Sun, Q., K. E. Pollok, R. L. Burton, L. J. Dai, W. Britt, D. J. Emanuel, and K. G. Lucas. 1999. Simultaneous ex vivo expansion of cytomegalovirus and Epstein-Barr virus-specific cytotoxic T lymphocytes using B-lymphoblastoid cell lines expressing cytomegalovirus pp65. Blood 94:3242-3250. [PubMed] [Google Scholar]
- 67.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 69.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 70.Toshchakov, V., B. W. Jones, P. Y. Perera, K. Thomas, M. J. Cody, S. Zhang, B. R. Williams, J. Major, T. A. Hamilton, M. J. Fenton, and S. N. Vogel. 2002. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat. Immunol. 3:392-398. [DOI] [PubMed] [Google Scholar]
- 71.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 73.Williams, B. R., and G. C. Sen. 2003. Immunology. A viral on/off switch for interferon. Science 300:1100-1101. [DOI] [PubMed] [Google Scholar]
- 74.Wills, M. R., A. J. Carmichael, K. Mynard, X. Jin, M. P. Weekes, B. Plachter, and J. G. Sissons. 1996. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 70:7569-7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yurochko, A. D., and E. S. Huang. 1999. Human cytomegalovirus binding to human monocytes induces immunoregulatory gene expression. J. Immunol. 162:4806-4816. [PubMed] [Google Scholar]
- 77.Yurochko, A. D., E. S. Hwang, L. Rasmussen, S. Keay, L. Pereira, and E. S. Huang. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-kappaB during infection. J. Virol. 71:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yurochko, A. D., M. W. Mayo, E. E. Poma, A. S. Baldwin, Jr., and E. S. Huang. 1997. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-kappaB promoters. J. Virol. 71:4638-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu, F. X., S. M. King, E. J. Smith, D. E. Levy, and Y. Yuan. 2002. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. USA 99:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]