Abstract
The initial stages of preprotein import into chloroplasts are mediated by the receptor GTPase Toc159. In Arabidopsis thaliana, Toc159 is encoded by a small gene family: atTOC159, atTOC132, atTOC120, and atTOC90. Phylogenetic analysis suggested that at least two distinct Toc159 subtypes, characterized by atToc159 and atToc132/atToc120, exist in plants. atTOC159 was strongly expressed in young, photosynthetic tissues, whereas atTOC132 and atTOC120 were expressed at a uniformly low level and so were relatively prominent in nonphotosynthetic tissues. Based on the albino phenotype of its knockout mutant, atToc159 was previously proposed to be a receptor with specificity for photosynthetic preproteins. To elucidate the roles of the other isoforms, we characterized Arabidopsis knockout mutants for each one. None of the single mutants had strong visible phenotypes, but toc132 toc120 double homozygotes appeared similar to toc159, indicating redundancy between atToc132 and atToc120. Transgenic complementation studies confirmed this redundancy but revealed little functional overlap between atToc132/atToc120 and atToc159 or atToc90. Unlike toc159, toc132 toc120 caused structural abnormalities in root plastids. Furthermore, when proteomics and transcriptomics were used to compare toc132 with ppi1 (a receptor mutant that is specifically defective in the expression, import, and accumulation of photosynthetic proteins), major differences were observed, suggesting that atToc132 (and atToc120) has specificity for nonphotosynthetic proteins. When both atToc159 and the major isoform of the other subtype, atToc132, were absent, an embryo-lethal phenotype resulted, demonstrating the essential role of Toc159 in the import mechanism.
INTRODUCTION
Protein import into chloroplasts is facilitated by multimeric translocon complexes in the outer and inner envelope membranes of chloroplasts called Toc and Tic, respectively (Keegstra and Cline, 1999; Chen et al., 2000; Hiltbrunner et al., 2001a; Jarvis and Soll, 2002). Identification of individual components of the translocation complexes was achieved biochemically using isolated pea (Pisum sativum) chloroplasts (Hirsch et al., 1994; Perry and Keegstra, 1994; Schnell et al., 1994; Wu et al., 1994). The core translocation complex of the outer envelope is formed by three components, Toc159, Toc34, and Toc75, with the stoichiometry 1 : 4–5 : 4 (Kessler et al., 1994; Seedorf et al., 1995; Bölter et al., 1998; Schleiff et al., 2003). Toc75 has a β-barrel structure, comprising 16 transmembrane β-strands, and forms the channel for preprotein conductance through the outer membrane (Hinnah et al., 2002). Toc159 and Toc34 belong to a unique class of GTPases and are proposed to mediate preprotein recognition (Hirsch et al., 1994; Kessler et al., 1994; Kouranov and Schnell, 1997; Sveshnikova et al., 2000). Toc34 is attached to the outer membrane by an 8-kD membrane anchor at its C terminus, and its GTP binding domain projects into the cytosol. Toc34 has been shown to interact directly with preproteins (Kouranov and Schnell, 1997; Sveshnikova et al., 2000) and to be important for the targeting of Toc159 to the Toc complex (Hiltbrunner et al., 2001b; Bauer et al., 2002; Smith et al., 2002; Wallas et al., 2003).
Toc159 was first identified as an 86-kD fragment because of its high sensitivity to proteolysis (Hirsch et al., 1994; Kessler et al., 1994; Perry and Keegstra, 1994; Bölter et al., 1998; Chen et al., 2000). It has a tripartite structure, consisting of a 52-kD, C-terminal membrane anchor domain (M-domain), a central GTP binding domain (G-domain), and an N-terminal acidic domain (A-domain) (Chen et al., 2000). Both the A-domain and the G-domain project into the cytosol (Bölter et al., 1998; Chen et al., 2000), and Toc159 has been shown to interact with preproteins during very early stages of protein import (Hirsch et al., 1994; Perry and Keegstra, 1994; Ma et al., 1996). Removal of the A- and G-domains abolishes detectable binding of preproteins at the chloroplast surface, which is consistent with its role as a receptor, although preprotein translocation can still occur (Chen et al., 2000). Recently, a soluble pool of Toc159 protein was detected in the cytosol (Hiltbrunner et al., 2001b), and it was proposed that Toc complex assembly is a dynamic process involving switching of Toc159 between a soluble form and an integral membrane form (Bauer et al., 2002; Smith et al., 2002). Toc159 might therefore be important for guiding preproteins from the cytosol to the translocon. However, the biological relevance of the soluble Toc159 pool has been questioned (Becker et al., 2004).
In Arabidopsis thaliana, both Toc-GTPases are encoded by small gene families (Jarvis et al., 1998; Bauer et al., 2000; Gutensohn et al., 2000; Hiltbrunner et al., 2001a; Jackson-Constan and Keegstra, 2001). Arabidopsis has four Toc159 homologs (atToc159, atToc132, atToc120, and atToc90) and two Toc34 homologs (atToc33 and atToc34). Several studies have attempted to address the reason for the existence of multiple Toc-GTPase isoforms in Arabidopsis. Characterization of an atToc159 knockout mutant, termed ppi2, led to the hypothesis that atToc159 might be a receptor with specificity for highly abundant, photosynthetic proteins (Bauer et al., 2000). In ppi2 plants, the differentiation of proplastids into chloroplasts is blocked, leading to a striking albino phenotype, although root plastids appear to develop normally (Bauer et al., 2000; Yu and Li, 2001). Photosynthetic genes were transcriptionally repressed in ppi2, and the accumulation of photosynthetic proteins was greatly reduced in the mutant, whereas nonphotosynthetic genes were unaffected (Bauer et al., 2000). Further evidence for the existence of an import pathway with preference for photosynthetic precursors was gained by detailed studies on the atToc33 knockout mutant ppi1 (Jarvis et al., 1998; Kubis et al., 2003). Using proteomics, transcriptomics, and in vitro import assays, ppi1 was shown to be specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins (Kubis et al., 2003). By extrapolation from these data, it was proposed that atToc132, atToc120, and atToc34 might be preferentially involved in the import of nonphotosynthetic proteins (Bauer et al., 2000; Kubis et al., 2003).
Whereas the function of atToc159 has previously been investigated using the ppi2 mutant (Bauer et al., 2000), similar molecular-genetic studies of the other three Arabidopsis Toc159 homologs had not been reported until very recently. We therefore conducted a comprehensive study of all four Arabidopsis Toc159 isoforms, using phylogenetics, gene expression studies, and knockout mutants for each component. In parallel with us, another laboratory independently conducted a similar study of the Arabidopsis Toc159 gene family and reached very similar conclusions (Ivanova et al., 2004). The results of Ivanova et al. (2004) are therefore discussed extensively throughout this report.
RESULTS
Phylogenetic Analysis of the Arabidopsis Toc159 Gene Family
In Arabidopsis, as mentioned above, four Toc159-related proteins are present (Bauer et al., 2000; Hiltbrunner et al., 2001a). All four proteins exhibit a characteristic tripartite structure, consisting of an N-terminal acidic domain (A-domain), a central GTP binding domain (G-domain), and a C-terminal membrane-anchor domain (M-domain) (Chen et al., 2000), although the A-domain is greatly reduced in atToc90. Sequence similarities vary between the domains, with the G- and M-domains displaying significantly higher sequence conservation than the A-domain. The two most similar proteins, atToc132 and atToc120, share 93.4% identity within the G-domains and 68.9% identity over their entire length. Amongst the other proteins, G-domain sequence identities range from 44.3% (between atToc159 and atToc90) to 58.1% (between atToc159 and atToc120); identities between the full-length proteins range from 30.5% (between atToc159 and atToc90) to 36.7% (between atToc159 and atToc120).
To look at the relatedness and evolution of the different Toc-GTPases, we constructed a phylogenetic tree using only the G-domain sequences of the different proteins because the G-domain is present and of a similar length in all proteins (Figure 1). In addition to the previously described Arabidopsis and pea proteins, Toc159- and Toc34-related proteins from the monocotyledonous species rice (Oryza sativa) and maize (Zea mays) were included as well as H-Ras p21, a GTPase from human, which was used as an outgroup. Although a similar phylogenetic study was conducted by Hiltbrunner et al. (2001a), the reliability of the predicted clades was not tested by bootstrap analysis, and monocotyledonous sequences were not included. In both studies, two major branches were observed, which correspond to the two main groups of Toc-GTPases: Toc159-related proteins (atToc159, atToc132, atToc120, atToc90, psToc159, osToc86-like_1, and osToc86-like_2 in Figure 1) and Toc34-related proteins (atToc33, atToc34, psToc34, zmToc34-1, and zmToc34-2 in Figure 1).
Figure 1.
Phylogenetic Analysis of Toc-GTPases from Arabidopsis and Other Species.
Amino acid sequences of the G-domains (equivalent to residues 828 to 1062 of psToc159) of Toc159 and Toc34 homologs from different species were aligned and used to produce a phylogenetic tree. Numbers of mutations are given above the clades, with bootstrap values below. Genes and accession numbers of the sequences used are as follows: atToc159, At4g02510; atToc132, At2g16640; atToc120, At3g16620; atToc90, At5g20300; atToc33, At1g02280; atToc34, At5g05000; psToc159, AAF75761; psToc34, Q41009; osToc86-like_1, AAG48839; osToc86-like_2, AAK43509; zmToc34-1, CAB65537; zmToc34-2, CAB77551; H-Ras p21, P01112. Species of origin of the sequences are indicated as follows: at, Arabidopsis thaliana; ps, Pisum sativum; os, Oryza sativa; zm, Zea mays. H-Ras p21 is from human.
The phylogenetic tree reflects the sequence identities observed within the Arabidopsis sequences. Arabidopsis atToc132 and atToc120 are within the same clade and, together with one of the rice Toc86-like proteins, form one subgroup of Toc159-related proteins. The other subgroup is comprised of atToc159, psToc159, and the other rice Toc86-like protein. All branches are supported by very high bootstrap values as indicated. The groupings imply that the Toc159 family diverged into two distinct subfamilies (atToc159-like and atToc132/atToc120-like) before species divergence occurred in a common ancestor of monocotyledonous and dicotyledonous species. This implies that there are at least two, fundamentally different subtypes of Toc159-related proteins in plants. By contrast, the data suggest that the Arabidopsis Toc34-related protein family formed later, after species divergence. Although atToc90 shows the lowest sequence identity to the other three Arabidopsis Toc159-related proteins, it clearly belongs to the Toc159 family.
Expression Profiles of the Arabidopsis Toc159 Homologous Genes
To gain an insight into the possible different functions of the four Arabidopsis Toc159-related proteins, we investigated their mRNA expression patterns in different tissues and at different developmental stages. Previous studies only compared three of the four genes (atTOC159, atTOC132, and atTOC120) in whole seedlings (Bauer et al., 2000) or mature leaves and roots (Yu and Li, 2001) by RT-PCR. We examined the expression patterns by comparative RNA gel blot analysis (Figure 2A). Identical RNA gel blots were probed in parallel using identically labeled cDNA probes for the four genes (see Methods). Quantification was performed using equivalent exposures of each blot, and the data were normalized for 28S rRNA (Figure 2B).
Figure 2.
Expression Profiles of the Four Arabidopsis Toc159 Homologous Genes.
(A) Fifteen-microgram samples of total RNA isolated from the indicated Arabidopsis tissues were analyzed by RNA gel blotting. RNA was isolated from wild-type seedlings grown in vitro for 10 d in the light (10d), from three different tissues of 28-d-old wild-type plants grown on soil (rosette leaves, young inflorescence tips, and roots), and from the four original Toc159 homolog knockout mutants (Figure 3A) grown in vitro for 10 d in the light. Filters were probed using 32P-labeled gene-specific probes of similar lengths with identical specific activities. rRNA (28S) was used as a loading control. The images shown were obtained after 10-d or 22-d exposures, as indicated, but quantifications were performed using identical exposures.
(B) Relative levels of expression of the atTOC159, atTOC132, atTOC120, and atTOC90 genes, normalized for 28S rRNA, are shown in the chart; y axis values between ∼55 and ∼95 have been removed to aid visualization.
The atTOC159 gene is the most regulated of the four: it is expressed highly in young, rapidly dividing photosynthetic tissues and at much lower levels in mature tissues and nonphotosynthetic tissues (Figure 2B). By contrast, the other three genes, atTOC132, atTOC120, and atTOC90, show relatively uniform levels of expression in all tested tissues (Figure 2B). Expression of atTOC132 is much lower than atTOC159 expression in most tissues but always higher (5- to 10-fold) than the expression of atTOC120. In 10-d-old seedlings, expression of atTOC159 is approximately eightfold higher than atTOC132 expression, which is in agreement with the data shown by Bauer et al. (2000). In roots, however, atTOC159 is downregulated, and expression of atTOC132 is actually higher than that of atTOC159, suggesting that atToc132 (and its close relative, atToc120) might be relatively more important than atToc159 in this tissue. These results are broadly consistent with those presented by Ivanova et al. (2004).
Overall, the expression profiles of atTOC159 and atTOC132/atTOC120 parallel those of atTOC33 and atTOC34, respectively (Kubis et al., 2003), which is consistent with the hypothesis that atToc159 is involved preferentially in the import of highly abundant, photosynthetic proteins and that atToc132 and atToc120 are involved preferentially in the import of nonphotosynthetic proteins (Bauer et al., 2000). Interestingly, atTOC90 is expressed at a uniformly high level throughout development (Figure 2B), suggesting that atToc90 may not exhibit similar substrate specificity.
Visible Phenotypes of Toc159 Homolog Knockout Mutants
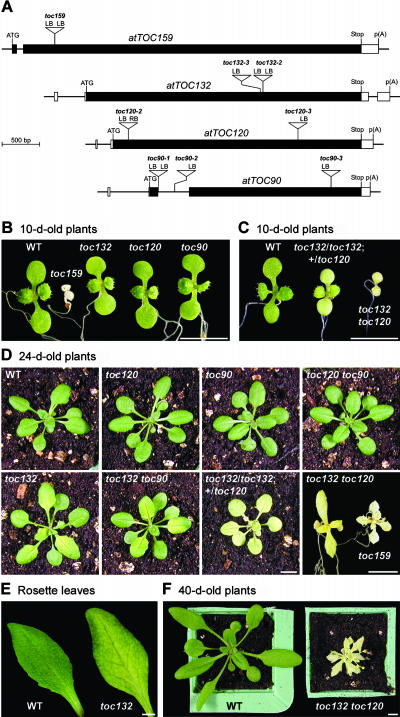
To directly address the hypothesis that different Toc159 isoforms are involved preferentially in import pathways with different preprotein recognition specificities, we identified Arabidopsis knockout mutants lacking each Toc159 isoform; the atToc159 knockout mutant ppi2 has been described previously (Bauer et al., 2000) and was kindly provided by Felix Kessler. Previously identified mutants affecting the import apparatus were referred to as ppi (Jarvis et al., 1998; Bauer et al., 2000; Constan et al., 2004) because the toc name had already been used to describe circadian clock mutants (Millar et al., 1995). However, the use of different names for corresponding mutants and proteins would become confusing when dealing with multiple components, and so the mutants described in this report are referred to using the names of the corresponding proteins (Schnell et al., 1997; Jackson-Constan and Keegstra, 2001); for the sake of consistency, the ppi2 mutant is referred to here as toc159. Because atToc132 and atToc120 knockout mutants (termed attoc132-1 and attoc120-1) were recently described by Ivanova et al. (2004), we refer to the primary, equivalent mutants described in this report as toc132-2 and toc120-2. The toc159, toc132-2, toc120-2, and toc90-1 mutants each contain a single-locus T-DNA insertion (Figure 3A, Table 1) and are null for the corresponding full-length mRNA as revealed by RNA gel blot analysis (Figure 2A). A shorter transcript was observed in the toc132 mutant (data not shown), which presumably corresponds to transcription of atTOC132 up to the T-DNA insertion site. A truncated protein encoded by this short transcript would lack part of its GTP binding domain and its entire membrane domain, and it is unlikely that it would be functional (Bauer et al., 2002; Lee et al., 2003).
Figure 3.
Visible Phenotypes of the Toc159 Homolog Knockout Mutants.
(A) Schematic diagrams showing the structure of the four Arabidopsis Toc159-related genes and the location of each T-DNA insertion. Protein-coding exons are represented by black boxes, and untranslated regions are represented by white boxes; introns are represented by thin lines between the boxes. T-DNA insertion sites are indicated precisely, but the insertion sizes are not to scale. ATG, translation initiation codon; stop, translation termination codon; p(A), polyadenylation site; LB, T-DNA left border; RB, T-DNA right border.
(B) Homozygous, single knockout mutant seedlings grown in vitro alongside the wild type.
(C) Pale and bleached toc132 toc120 double mutant seedlings grown in vitro alongside the wild type. The double mutants shown were homozygous for toc132 and heterozygous for toc120 (toc132/toc132; +/toc120; pale) or homozygous for both mutations (toc132 toc120; bleached).
(D) Mature (24-d-old) single and double mutants. Apart from the individual labeled toc132/toc132; +/toc120, all plants shown were homozygous for the indicated mutations. Plants were germinated in vitro and transferred to soil (or, in the case of toc132 toc120 and toc159, to medium containing 3% [w/v] sucrose) after 10 d of growth.
(E) Rosette leaves of the toc132 single mutant have a reticulate appearance.
(F) The toc132 toc120 double homozygote is able to survive to maturity on soil. The double mutant was allowed to establish itself on medium containing 3% (w/v) sucrose for 2 weeks before transfer to soil.
Bars = 5 mm.
Table 1.
Genetic Analyses of the Toc159 Homolog Knockout Mutants
| Resistant (if Plated on Selection)
|
|||||||
|---|---|---|---|---|---|---|---|
| Genotype | Selection | Greena | Albino | Total Resistant | Sensitive | Green:Albino | Resistant:Sensitive |
| toc159 (ppi2, Col-0) | None | 856 | 286 | –b | – | 3.00:1.00 | – |
| Kanamycin | 395 | 155 | 550 | 222 | 2.00:0.78 | 2.48:1.00 | |
| toc132, T3 | None | 444a | 0 | – | – | ∞ | – |
| Phosphinothricin | 1543a | 0 | 1543 | 487 | ∞ | 3.17:1.00 | |
| toc120, T5 | None | 393 | 0 | – | – | ∞ | – |
| Hygromycin | 446 | 0 | 446 | 165 | ∞ | 2.70:1.00 | |
| toc90, T4 | None | 489 | 0 | – | – | ∞ | – |
| Phosphinothricin | 732 | 0 | 732 | 215 | ∞ | 3.40:1.00 | |
Scoring of phenotypes was conducted after 10 d of growth in vitro, and so green plants here include toc132 homozygotes, which exhibit only slight chlorosis at the seedling stage.
Not determined.
The visible phenotypes of the original four mutants are shown in Figure 3. The strong albino phenotype of the toc159 mutant has been described previously and is consistent with the proposed role of atToc159 as the major receptor for photosynthetic protein import (Bauer et al., 2000). The toc132 mutant has a very slight pale phenotype in young plants (Figure 3B), which develops into a clear yellow-green, somewhat reticulate phenotype in mature plants (Figures 3D and 3E). By contrast, the toc120 and toc90 mutants were indistinguishable from the wild type throughout development (Figures 3B and 3D). To confirm these visible phenotypes, we identified additional, independent alleles of toc132, toc120, and toc90 (Figure 3A). The toc132-3 mutant had exactly the same yellow-green, reticulate phenotype as toc132-2 (see Supplemental Figure 1 online), and the toc120-3, toc90-2, and toc90-3 mutants all appeared exactly the same as wild-type plants (data not shown). Unless stated otherwise, all of the data presented in this article were derived using the originally identified mutant alleles (i.e., toc132-2, toc120-2, and toc90-1).
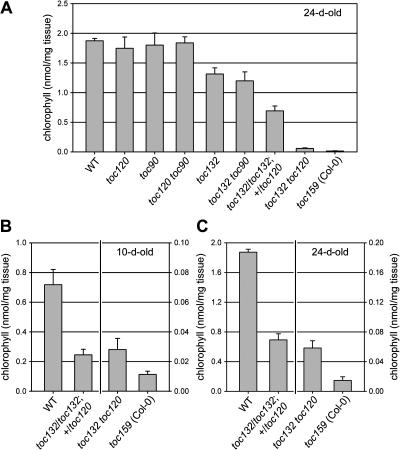
The phenotypes of the four original mutants were quantified by making chlorophyll measurements (Figure 4A). As expected, wild-type amounts of chlorophyll per unit of fresh weight were observed in the toc120 and toc90 mutants. By contrast, significantly reduced chlorophyll levels were present in 24-d-old toc132 plants (∼70% of the wild-type concentration), and only trace amounts of chlorophyll were present in toc159 plants of the same age (<1% of the wild-type concentration) (Figure 4A).
Figure 4.
Chlorophyll Accumulation in the Toc159 Homolog Knockout Mutants.
The chlorophyll contents of 10-d-old (B) and 24-d-old ([A] and [C]) plants are shown. The charts in (B) and (C) have a dual y axis, such that the right side of each chart is at 10-fold higher magnification than the left side. The data shown in (C) are also shown in (A). All values shown are means (±sd) derived from five independent samples.
Our observation that atToc120 knockout mutants display neither a visible phenotype nor a chlorophyll deficiency phenotype is entirely consistent with the results of Ivanova et al. (2004). However, whereas these authors reported that an atToc132 knockout mutant was indistinguishable from the wild type (Ivanova et al., 2004), we observed a clear, visible phenotype in two independent atToc132 knockout mutants (Figure 3; see Supplemental Figure 1 online) and a chlorophyll deficiency phenotype in the toc132-2 mutant (Figure 4). The reason for this apparent discrepancy is not clear. One possibility is that the phenotypic differences reflect the different genetic backgrounds of the mutants. Whereas the toc132-2 and toc132-3 mutants are of the Columbia-0 (Col-0) ecotype, the attoc132-1 mutant described by Ivanova et al. (2004) is of the Wassilewskija (Ws) ecotype, and it is well documented that Col-0 is genetically divergent from many other Arabidopsis ecotypes, including Ws (Barth et al., 2002). Another possibility is that the different observations reflect different plant growth conditions because we repeatedly observed that the toc132 mutant phenotype becomes clear only after a significant period of growth on soil.
Genetic Interactions between the Knockout Mutations
To assess the functional relationships amongst the Arabidopsis Toc159-related proteins, the four knockout mutants were crossed to each other in all pairwise combinations. For these experiments, a 7× Col-0 introgressed toc159 line was used (Table 1) because this mutant was originally of the Ws ecotype and all of the other mutants were of the Col-0 ecotype. All crosses were analyzed in the F2 and F3 generations for novel phenotypes and using appropriate single or double antibiotic selection media. To confirm the genotypes of putative double mutants, PCR analysis was performed using appropriate gene-specific and T-DNA–specific primers. The results of these various double mutant studies are discussed below and in the following sections.
In F2 families derived from crosses between toc90 and toc132 or toc120, individuals that were homozygous for both mutations could be identified in each case. The toc120 toc90 double mutant did not exhibit any new visible phenotypes and appeared exactly the same as wild-type plants (Figure 3D). This phenotypic normality was reflected in the unchanged chlorophyll levels of the double mutant (Figure 4A). Similarly, the toc132 toc90 double mutant displayed the same visible phenotype and chlorophyll concentration as the toc132 single mutant (Figures 3D and 4A). Thus, our data indicate that there is no significant functional overlap or redundancy between atToc90 and atToc120 or atToc132.
The toc132 toc120 Double Homozygote Appears Similar to toc159
When the F2 progeny of crosses between toc132 and toc120 were scored after 10 d of growth in vitro, all individuals could be placed into one of three discrete, phenotypic classes, termed green, pale, and bleached (Table 2); at this early stage of development, the visible phenotype of toc132 is slight and difficult to score (Figure 3B), and plants classified as green here also included toc132 homozygotes. No individuals falling outside of these three phenotypic classes were observed. All 18 pale individuals tested by PCR were found to be homozygous for toc132 and heterozygous for toc120 (genotype: toc132/toc132; +/toc120), whereas all five bleached individuals tested were found to be homozygous for both mutations (genotype: toc132/toc132; toc120/toc120). The ratio observed between these three phenotypes in the F2 generation (Table 2) supports the hypothesis that all pale and bleached individuals have these genotypes (the expected ratio is 13:2:1). The segregation ratios of the pale, bleached, and antibiotic resistance phenotypes in the F3 generation further confirmed the genotypes associated with these novel phenotypes (Table 2).
Table 2.
Genetic Analyses of Interactions between the toc132 and toc120 Mutations
| Resistant (if Plated on Selection)
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Selection | Greena | Pale | Bleached | Total Resistant | Sensitive | Green:Pale:Bleached | Resistant:Sensitive |
| toc132-2 × toc120-2, F2 | None | 556a | 88 | 9 | –b | – | 13.00:2.06:0.21 | – |
| toc132-2 × toc120-3, F2 | None | 317a | 53 | 25 | – | – | 13.00:2.17:1.03 | – |
| toc132-3 × toc120-2, F2 | None | 559a | 72 | 45 | – | – | 13.00:1.67:1.05 | – |
| toc132-2 × toc120-2, F3c | None | 108a | 190 | 55 | – | – | 1.00:1.76:0.51 | – |
| Phosphinothricin | 125a | 217 | 30 | 372 | 0 | 1.00:1.74:0.24 | ∞ | |
| Hygromycin | 0 | 229 | 76 | 305 | 109 | 0.00:2.00:0.66 | 2.80:1.00 | |
Scoring of phenotypes was conducted after 10 d of growth in vitro, and so green plants here include toc132 homozygotes, which exhibit only slight chlorosis at the seedling stage.
Not determined.
The data shown describe the progeny of pale F2 plants.
Our identification of a viable toc132 toc120 double homozygote conflicts with the results of Ivanova et al. (2004) because these authors reported that this genotype causes lethality during embryogenesis or early seedling growth. To further confirm our results, we crossed the toc132-3 and toc120-3 mutants with the original toc120-2 and toc132-2 mutants, respectively, and analyzed the resultant F2 populations. In both cases, green, pale, and bleached plants were once again observed at the expected frequencies (13:2:1; Table 2). Furthermore, when 20 pale plants and 25 bleached plants from the toc132-2 × toc120-3 F2 population were genotyped by PCR, all of the pale plants were found to be homozygous for the toc132 mutation and heterozygous for toc120, and all of the bleached plants were found to be doubly homozygous. We therefore conclude that the toc132 toc120 double homozygous genotype is not lethal in the Col-0 background. The reason for the discrepancy between our data and those of Ivanova et al. (2004) is not clear, but it is once again possible that the phenotypic differences reflect genetic dissimilarities between the Col-0 and Ws ecotypes (Barth et al., 2002) or different plant growth conditions.
The pale phenotype of toc132 toc120 heterozygotes (genotype: toc132/toc132; +/toc120) is clearly visible at the seedling stage, especially in the cotyledons (Figure 3C), and remains so throughout development (Figure 3D). This phenotype is significantly more severe than that of the toc132 single mutant, and this is reflected in the reduced chlorophyll levels of the double mutant (Figures 4A to 4C): the toc132 toc120 heterozygote contains ∼34 to 37% of the wild-type chlorophyll concentration throughout development. Although we occasionally observed some very subtle variegation of the leaves of toc132 toc120 heterozygous plants (data not shown), we never observed the strongly variegated phenotype reported by Ivanova et al. (2004). Again, this phenotypic difference may be because of the use of different plant growth conditions or genetic differences between the Col-0 and Ws ecotypes.
The toc132 toc120 double homozygote has a very strong visible phenotype that is almost as severe as that of the toc159 mutant (Figures 3B to 3D). Its chlorophyll concentration is only ∼10% of that present in the toc132 toc120 heterozygote (genotype: toc132/toc132; +/toc120) but still threefold to fourfold higher than that present in toc159 (Figures 4A to 4C). Thus, unlike toc159, the toc132 toc120 double homozygote is able to survive to maturity on soil (Figure 3F) if first allowed to establish itself on medium containing 3% sucrose.
The progression of phenotypic severity in homozygous toc132 mutants lacking either one (genotype: toc132/toc132; +/toc120) or both (genotype: toc132/toc132; toc120/toc120) atTOC120 alleles clearly demonstrates the existence of substantial functional overlap between atToc132 and atToc120.
The toc159 toc132 Double Homozygote Is Embryo Lethal
To investigate the functional relationships between atToc159 and the other three proteins, we crossed the toc159 (Col-0) mutant to toc132, toc120, and toc90. In the F2 generation of each cross, plants that were homozygous for toc132, toc120, or toc90 and heterozygous for toc159 were identified by scoring for the appropriate antibiotic resistance phenotypes and by conducting PCR analysis. The F3 progeny of these plants were then examined carefully for evidence of genetic interactions. Relatively normal frequencies of albino toc159 homozygotes were observed in the progenies of the toc120 and toc90 crosses (Table 3), and, when these double homozygotes were grown to maturity alongside toc159 single mutants (on medium containing 3% sucrose), no obvious effects of the toc120 or toc90 mutations on toc159 phenotypic severity could be observed (data not shown).
Table 3.
Genetic Analyses of Interactions between the toc159 Mutation and the Other Mutations
| Resistant (if Plated on Selection)
|
|||||||
|---|---|---|---|---|---|---|---|
| Genotype | Selection | Greena | Albino | Total Resistant | Sensitive | Green:Albino | Resistant:Sensitive |
| toc159 × toc132, F3b | Phosphinothricin | 1200a | 0 | 1200 | 0 | ∞ | ∞ |
| Kanamycin | 586a | 0 | 586 | 322 | ∞ | 1.82:1.00 | |
| toc159 × toc120, F3b | Hygromycin | 522 | 93 | 615 | 0 | 3.00:0.53 | ∞ |
| Kanamycin | 243 | 59 | 302 | 126 | 2.00:0.49 | 2.40:1.00 | |
| toc159 × toc90, F3b | Phosphinothricin | 546 | 117 | 663 | 0 | 3.00:0.64 | ∞ |
| Kanamycin | 462 | 117 | 579 | 139 | 2.00:0.51 | 4.17:1.00 | |
Scoring of phenotypes was conducted after 10 d of growth in vitro, and so green plants here include toc132 homozygotes, which exhibit only slight chlorosis at the seedling stage.
The data shown describe the progeny of F2 plants that were homozygous for toc132, toc120, or toc90 and heterozygous for toc159.
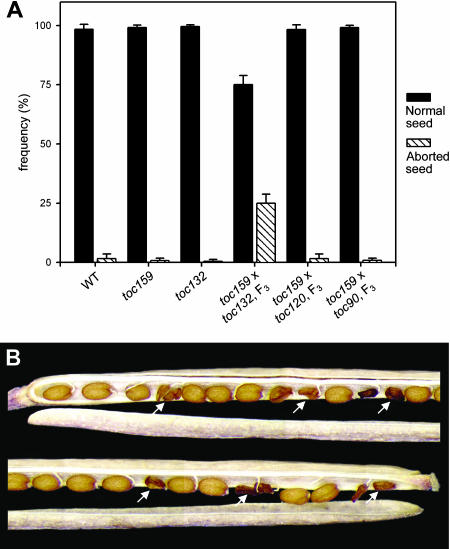
Interestingly, when the F3 progeny of the toc159 × toc132 crosses were examined, no albino plants were observed (Table 3). Because toc159 homozygotes would normally be expected to occur at a frequency of 25% in the F3 generation, the chances of observing no albinos in the total scored population of 2108 plants (Table 3), because of random chance alone, is essentially zero (χ2 = 702.667, df = 1, P value = 0.000). Thus, the data suggest that the toc159 toc132 double homozygous mutation is lethal during some early stage of development. Because plastids are known to play an important role during embryo development (Uwer et al., 1998; Apuya et al., 2001; Constan et al., 2004), we suspected that the defect might be occurring during embryogenesis. Consistent with this hypothesis, aborted seeds were observed at a frequency of exactly 25% in the siliques of F3 plants having the genotype +/toc159; toc132/toc132 (Figure 5). Taken together, these data provide strong evidence for the embryo lethality of the toc159 toc132 double homozygous genotype.
Figure 5.
Embryo Lethality of the toc159 toc132 Double Mutation.
(A) Three to four siliques from three different individuals (at least nine siliques in total) of the indicated genotypes were scored for the presence of aborted seeds. The F3 plants scored were all heterozygous for the toc159 mutation (introgressed into the Col-0 ecotype) and homozygous for the second mutation (Table 3). Error bars indicate sd.
(B) The appearance of aborted seeds within the silique of an F3 individual that was homozygous for toc132 and heterozygous for toc159.
In summary, our double mutant studies provided clear evidence for functional redundancy between the close relatives atToc132 and atToc120. No evidence for functional overlap between atToc159 and atToc120, or between atToc90 and any other protein, was observed. The lethality of the toc159 toc132 genotype can be interpreted in two different ways: (1) it reflects the disruption of import of a broad range of precursor types through two different import pathways and indicates minimal functional overlap between atToc159 and atToc132; (2) it reflects a severe or complete block in a single import pathway that is only partially disrupted in either single mutant and indicates significant functional redundancy between atToc159 and atToc132. Considering the sequence similarities between the atToc159, atToc132, and atToc120 proteins, and their relative levels of expression, the former explanation seems the most likely to be correct.
Transgenic Complementation Studies
To corroborate our findings on the functional relationships between atToc132, atToc120, and atToc90 and to differentiate between the two possible explanations for the embryo lethality of the toc159 toc132 genotype, we conducted transgenic complementation studies. All four Arabidopsis Toc159-related proteins were expressed under the control of the strong, constitutive, Cauliflower mosaic virus 35S promoter in two different mutant backgrounds: (1) the toc159 single mutant background; (2) the toc132 toc120 double mutant background. The atTOC90 cDNA used in these experiments (accession number AV548084) was recently shown to be truncated at the 5′ end by the identification of a slightly longer clone (accession number NM_122037). However, the former encodes a protein that is only 14 residues shorter (779 residues) than that encoded by the full-length clone (793 residues). Because this small, N-terminal deletion does not impinge upon the G-domain and because atToc159 mutants completely lacking the N-terminal A- or A+G-domains are active in vivo (Bauer et al., 2002; Lee et al., 2003), it seems unlikely that this mutation would strongly affect protein activity.
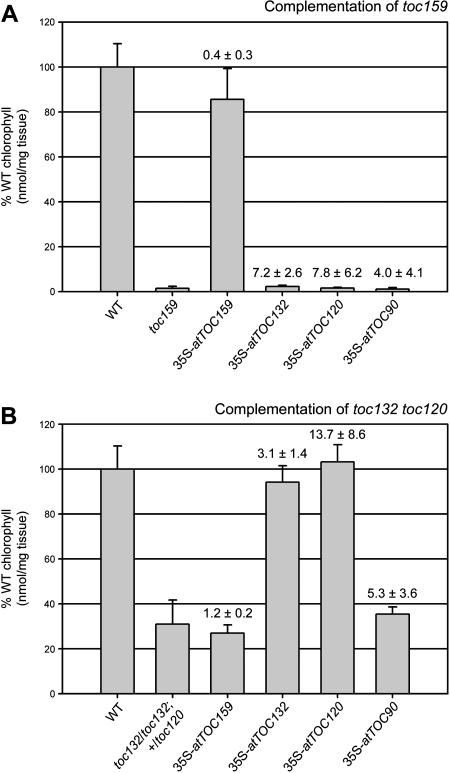
At least 10 independent transgenic lines were identified for each transgene/mutant combination. From these 10 lines, between four and six lines were selected for detailed analyses in each case. Lines were selected at random and were phenotypically representative of the entire set. In the selected lines, the degree of complementation of the mutant phenotypes was quantified by making chlorophyll measurements, and the extent of transgene overexpression, relative to the wild type, was assessed using semiquantitative RT-PCR (Figure 6).
Figure 6.
Transgenic Complementation of the toc159 Single Mutant and the toc132 toc120 Double Mutant.
Plants carrying the toc159 (A) or toc132 and toc120 (B) knockout mutations were stably transformed with four different T-DNA constructs: 35S-atTOC159, 35S-atTOC132, 35S-atTOC120, and 35S-atTOC90. Mutant complementation was quantified in the T3 generation by making chlorophyll measurements on plants with appropriate mutant genotypes (toc159/toc159 [A] and toc132/toc132; +/toc120 [B] or, occasionally, amongst the 35S-atTOC132 and 35S-atTOC120 transformants toc132/toc132; toc120/toc120 [B]) and that were either heterozygous or homozygous for the relevant transgene. The chlorophyll values shown are means (±sd) derived from measurements on four to six independent transformants; for each transformant, five independent measurements were made. Values are expressed as a percentage of the wild-type chlorophyll concentration. The extent of transgene overexpression in each transformant was estimated by conducting RT-PCR experiments using T2 plants that were either heterozygous or homozygous for the relevant transgene. Mean fold changes in expression (±sd), relative to the wild type, for each transgene/mutant combination are given textually above the corresponding chlorophyll data bars.
The results obtained from the toc159 complementation experiments were clear. All five selected 35S-atTOC159 lines displayed essentially wild-type chlorophyll concentrations (Figure 6A). These complemented plants were homozygous for the toc159 mutation (as revealed by segregation analysis and by PCR; data not shown) and were either heterozygous or homozygous for the 35S-atTOC159 transgene. Wild-type chlorophyll concentrations were achieved despite the fact that the transgene could only drive atTOC159 expression at ∼40% of the wild-type level (Figure 6A). By contrast, lines that were homozygous for the 35S-atTOC132, 35S-atTOC120, or 35S-atTOC90 transgenes, and the toc159 mutation, and which displayed significant levels of transgene overexpression, did not display significantly higher chlorophyll concentrations than untransformed toc159 homozygotes. Taking into account the data shown in Figure 2, the levels of transgene expression achieved with 35S-atTOC132 and 35S-atTOC120 are roughly equivalent to those achieved with 35S-atTOC159. Furthermore, when 35S-atTOC132 and 35S-atTOC120 lines were analyzed by immunoblotting using atToc132- and atToc120-specific antibodies (Ivanova et al., 2004), significant levels of protein overexpression were observed, indicating that both constructs were intact and fully active (see Supplemental Figure 2 online). Although the 35S-atTOC132 transformants did appear slightly larger than control toc159 plants (data not shown), the chlorophyll data clearly indicate that overexpression of atToc132 cannot compensate for the absence of atToc159 to any significant degree (Figure 6A).
The results obtained from the toc132 toc120 complementation experiments were also clear. All five selected 35S-atTOC132 lines and all four selected 35S-atTOC120 lines contained wild-type chlorophyll concentrations (Figure 6B). These complemented plants were homozygous for the toc132 mutation, either heterozygous or homozygous for the toc120 mutation, and either heterozygous or homozygous for the appropriate 35S transgene. These data confirm the previously drawn conclusion that atToc132 and atToc120 exhibit a high level of functional similarity. By contrast, toc132 toc120 heterozygotes (genotype: toc132/toc132; +/toc120) that were either heterozygous or homozygous for the 35S-atTOC159 or 35S-atTOC90 transgenes did not display significantly higher chlorophyll concentrations than untransformed toc132 toc120 heterozygotes (Figure 6B). Because the level of atTOC90 overexpression observed in these transgenic lines was good (5.3-fold on average; Figure 6B), these data strongly support the conclusion that atToc90 and atToc132/atToc120 do not share significant functional redundancy. However, because high levels of atTOC159 overexpression were not observed (presumably because atTOC159 is already expressed at very high levels in wild-type plants; Figure 2), the functional relationship between atToc159 and atToc132/atToc120 is more difficult to assess (data shown in Figure 6A are more informative in this regard). Nevertheless, individual lines that displayed 47 and 30% atTOC159 overexpression, relative to the wild type, did not contain significantly higher chlorophyll concentrations than untransformed toc132 toc120 heterozygotes, suggesting that the level of redundancy is minimal.
Taken together, these transgenic overexpression data confirm the conclusions drawn from the double mutant studies and indicate that the first hypothesis put forward to account for the lethality of the toc159 toc132 genotype is correct; that is, the severity of the phenotype of the double homozygote results from the disrupted import of a broad range of precursor types because of the fact that atToc159 and atToc132 operate preferentially in different import pathways.
Ultrastructure of Plastids in the Single and Double Mutants
To gain insight into the functional specialization of Toc159-related proteins implied by the phylogenetic, genetic, and transgenic data, we used transmission electron microscopy to characterize leaf and root plastid ultrastructure in the mutants (Figure 7). The severity of chloroplast ultrastructural defects broadly paralleled the severity of the visible and chlorophyll phenotypes of the mutants (Figures 3 and 4). In young plants, toc132 chloroplasts were similar to those in the wild type (data not shown), but the chloroplasts of the pale and bleached toc132 toc120 double mutants (genotypes: toc132/toc132; +/toc120 and toc132/toc132; toc120/toc120, respectively) were smaller with less developed thylakoids (Figure 7A). Remarkably, chloroplasts in the toc132 toc120 double homozygote were much more developed, with distinct thylakoids and grana, than those in the toc159 mutant (Figure 7A), despite the fact that two mutants look very similar (Figure 3). This latter observation is consistent with the hypothesis that atToc159 is relatively more important for the biogenesis of photosynthetic proteins and that atToc132 and atToc120 are relatively more important for the biogenesis of nonphotosynthetic proteins (Bauer et al., 2000).
Figure 7.
Ultrastructure of Plastids in the Toc159 Homolog Knockout Mutants at Different Developmental Stages.
(A) Plastids from the cotyledons of young (10-d-old) wild type, pale toc132 toc120 double mutant (genotype: toc132/toc132; +/toc120; arrowheads indicate five cytoplasmic inclusions, one of which contains a plastid [p]), bleached toc132 toc120 double homozygous (arrowheads indicate two cytoplasmic inclusions, one of which contains a mitochondrion [m]), and toc159 (two mitochondria and one plastid are indicated) plants are shown. All images are at the same magnification. Bar = 1 μm.
(B) Chloroplasts from the leaves of mature (28-d-old) wild type, toc132, and pale toc132 toc120 double mutant (genotype: toc132/toc132; +/toc120) plants are shown. The wild-type chloroplast is from an interveinal region; mutant chloroplasts are from veinal (v) and interveinal (iv) regions, as indicated. Arrowheads indicate five cytoplasmic inclusions. An enlargement of the indicated region (Z1) in the pale toc132 toc120 double mutant interveinal chloroplast is shown. All of the chloroplasts shown are at the same magnification; bar = 1 μm. The enlargement (Z1) is at eightfold higher magnification; bar = 0.1 μm.
(C) Plastids from the roots of 10-d-old wild type, toc159, and toc132 toc120 double homozygous plants are shown. Cytoplasmic inclusions are indicated (i). An enlargement of the indicated region (Z2) in the toc132 toc120 double mutant plastid is shown. All of the plastids are at the same magnification; bar = 1 μm. The enlargement (Z2) is at 5.3-fold higher magnification; bar = 0.1 μm.
In mature plants, chloroplasts in the toc132 single mutant and the pale toc132 toc120 heterozygote (genotype: toc132/toc132; +/toc120) were small with less developed thylakoids (Figure 7B). Interestingly, in both of these genotypes, the chloroplasts in cells surrounding the vascular bundles were more highly developed than those in the interveinal tissues of the mesophyll (Figure 7B). This observation most likely explains the reticulate appearance of toc132 leaves (Figure 3E). Arabidopsis cue1 mutants, which have defects in the phosphoenolpyruvate/phosphate translocator of the plastid inner envelope membrane, have a similar, but more severe, reticulate phenotype (Streatfield et al., 1999). The biosynthesis of aromatics is compromised in cue1, and the reticulate phenotype of the mutant can be rescued by feeding aromatic amino acids (Streatfield et al., 1999). It is possible that deficiencies in amino acid biosynthesis, caused by the inefficient chloroplast import of the necessary biosynthetic enzymes, are also responsible for the reticulate appearance of the toc132 mutant.
Particularly interesting observations were made when the root plastids of toc132 toc120 double homozygotes were examined. Whereas toc159 root plastids appeared relatively normal (Figure 7C) (Yu and Li, 2001), a high proportion of toc132 toc120 double homozygote root plastids were found to contain large cytoplasmic inclusions (Figure 7C), each surrounded by a normal double envelope membrane (Figure 7C). Of the 75 double mutant plastids examined, 31% contained at least one inclusion. By contrast, of the 49 wild-type and 45 toc159 root plastids examined, only 4 and 9%, respectively, contained cytoplasmic inclusions, and these tended to be smaller than those in the double mutant. Similar inclusions were also observed in the chloroplasts of toc132 toc120 plants (Figures 7A and 7B), which also exhibited other structural irregularities (i.e., they appeared elongated and flattened) (Figures 7A and 7B). It is possible that these structural defects are the result of the failure of toc132 toc120 plastids to import certain proteins, for example, cytoskeletal or division apparatus components. Alternatively, they may be the result of a specific response designed to increase the surface area of metabolically compromised plastids to facilitate the exchange of substances with the cytosol.
Taken together, these electron microscopy data are consistent with the hypothesis that atToc132 and atToc120 play a significant role in the import of nonphotosynthetic proteins.
Protein Import in the toc132/toc132; +/toc120 Double Mutant
To investigate whether the phenotypic differences between the toc132 toc120 double mutants and toc159 could be attributed to differential effects of the mutations on the import of different preproteins, import studies were performed using isolated chloroplasts. The pale toc132 toc120 double mutant (genotype: toc132/toc132; +/toc120) was chosen for these studies because it has a stronger phenotype than either single mutant and yet is sufficiently healthy to allow isolation of a reasonable yield of chloroplasts, which would not be feasible using the double homozygote. The F3 progeny of pale toc132 toc120 double mutants were plated on hygromycin plates, and hygromycin sensitive seedlings (corresponding to toc132 single mutants) were picked out by hand after 7 to 8 d of growth. Both pale and bleached toc132 toc120 seedlings (genotypes: toc132/toc132; +/toc120 and toc132/toc132; toc120/toc120, respectively) were used for chloroplast isolation, but, because of the greatly reduced growth of the double homozygotes, the yield of chloroplasts resulting from these was estimated to be <5% of the total.
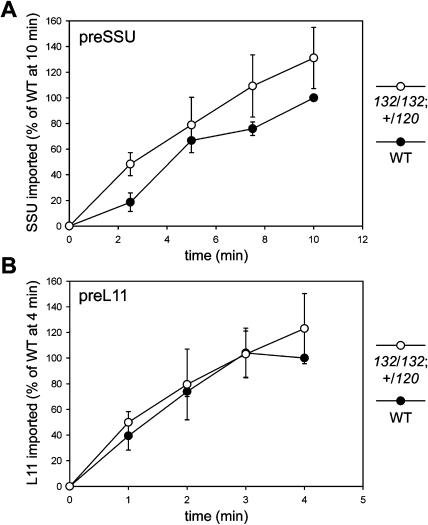
The photosynthetic preprotein, preSSU (precursor of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase [Rubisco]), and the nonphotosynthetic preprotein, preL11 (precursor of a 50S ribosomal protein), were selected for analysis because their import efficiencies were previously shown to be affected differentially by the ppi1 mutation (Kubis et al., 2003). However, using the wheat germ translation system for precursor synthesis, no defect in import was observed for either precursor in the double mutant chloroplasts (see Supplemental Figure 3 online). To eliminate the possibility that soluble factors in the wheat germ lysate were responsible for this negative result (Hiltbrunner et al., 2001b; Schleiff et al., 2002), we tested the same precursors again using the rabbit reticulocyte system for translation (Figure 8). Once again, however, no import deficiencies were detected, and our data show that double mutant chloroplasts import both precursors at a similar rate to wild-type chloroplasts in vitro.
Figure 8.
Comparison of Import Rates into Isolated Wild-Type and Mutant (toc132/toc132; +/toc120) Chloroplasts for Different Preproteins.
In vitro–translated, 35S-Met–labeled preSSU (A) and preL11 (B) were imported into wild-type and mutant (toc132/toc132; +/toc120) chloroplasts for the times indicated in the graphs. The amount of protein imported into chloroplasts was expressed as a percentage of the amount imported into wild-type chloroplasts at the final time point in each case. The data shown are means (±sd) of four (A) or three (B) independent experiments.
One possible explanation of these data is that the soluble forms of Toc159-related proteins act, in vivo, to increase the efficiency of import by facilitating the targeting of precursors from the cytosol to the translocation channel (Hiltbrunner et al., 2001b). Thus, in an in vitro import system that lacks these soluble components, base rates of import might still be observed, but the effects of the toc132 and toc120 mutations would no longer be apparent. Alternatively, it is possible that the atToc132 and atToc120 proteins are involved preferentially in the import of subsets of proteins that do not include either preSSU or preL11.
Analysis of the toc132 Chloroplast Proteome
In a further attempt to gain insight into the substrate preferences of the import pathway in which atToc132 (and atToc120) are involved, the chloroplast proteome of the toc132 mutant was analyzed. To do this, we employed the same procedure—involving difference gel electrophoresis (DIGE) and CyDye technology—that was used to analyze the ppi1 chloroplast proteome (Kubis et al., 2003). The degree of enrichment or depletion of toc132 chloroplast proteins, relative to the wild type, was determined by CyDyeDIGE analysis (see Supplemental Figure 4 online), and these data were then used to select proteins for identification by mass spectrometry (Table 4).
Table 4.
Proteins Identified by DIGE and Mass Spectrometry
| Spot No. | Gene No. | Fold Change in toc132a | Protein | MASCOT Scoreb |
|---|---|---|---|---|
| 1 | At1g32060 | 2.44 ± 0.05 | Phosphoribulokinase | 548 (Q) |
| 2c | At2g39730 | 1.95 ± 0.03 | Rubisco activase | 551 (Q) |
| 3c | At2g39730 | 1.86 ± 0.27 | Rubisco activase | 537 (Q) |
| 4 | At5g20720 | 1.81 ± 0.03 | Cpn21 chaperonin | 401 (Q) |
| 5 | At4g23100 | 1.80 ± 0.05 | γ-Glutamylcysteine synthetase | 134 (Q) |
| 6 | AtCg00120 | 1.74 ± 0.06 | ATPase synthase, α subunit | 74 (Q) |
| 7 | At5g35630 | 1.63 ± 0.17 | Glutamate-ammonia ligase | 98 (M) |
| 8 | At4g20360 | 1.59 ± 0.01 | Elongation factor, EF-Tu homolog | 83 (M) |
| 9 | At2g28000 | 1.51 ± 0.05 | Chaperononin 60α | 951 (Q) |
| 10 | AtCg00490 | – | Rubisco large subunit, LSU | 128 (M) |
| 11 | At5g38410 | – | Rubisco small subunit, SSU (Ats3B) | 90 (M) |
| 12 | At3g50820 | – | 33-kD subunit of the oxygen evolving complex, OE33 | 98 (Q) |
| 13 | At3g13470 | – | Chaperononin 60β | 92 (M) |
| 14 | At3g04790 | – | Ribose-5-phosphate isomerase | 124 (Q) |
| 15 | At5g09650 | – | Inorganic pyrophosphatase | 167 (Q) |
| 16 | At3g27850 | – | 50S ribsomal protein, L12-C | 91 (Q) |
| 17 | At4g24280 | – | Hsp70 homolog | 75 (M) |
| 18 | At2g04030 | – | Hsp90 homolog | 126 (M) |
| 19 | At3g62030 | – | Peptidyl prolyl isomerase | 161 (Q) |
| 20d | At3g58610 | 0.47 ± 0.20 | Ketol-acid reductoisomerase | 188 (Q) |
| 20d | At3g23940 | 0.47 ± 0.20 | Dihydroxy-acid dehydratase | 107 (Q) |
| 21 | At4g38970 | 0.43 ± 0.16 | Fructose-1,6-bisphosphate aldolase | 531 (Q) |
| 22 | At2g21330 | 0.40 ± 0.10 | Fructose-1,6-bisphosphate aldolase | 455 (Q) |
Values are means ± sd.
M indicates data acquired by peptide mass fingerprinting using a Micromass TofSpec2E MALDI Tof; Q indicates data acquired by liquid chromatography tandem mass spectrometry (LC-MS/MS) using a Micromass Qtof2.
These two spots correspond to the same protein and most likely differ as a result of posttranslational modification.
These two proteins were not resolved by two-dimensional gel electrophoresis and so were identified within the same spot.
The results clearly show that the perturbations that exist in the toc132 chloroplast proteome are distinct from those that were observed previously in the ppi1 chloroplast proteome. For example, photosynthetic proteins such as SSU, OE33, and ribose-5-phosphate isomerase—all of which were deficient in ppi1 chloroplasts—are present at normal levels in toc132. Furthermore, some photosynthetic proteins (e.g., Rubisco activase and the Calvin cycle enzyme phosphoribulokinase) are even slightly enriched in toc132 chloroplasts. However, no clear trend indicating a selective effect of the toc132 mutation on the accumulation of nonphotosynthetic proteins was observed because proteins depleted in the mutant included the Calvin cycle enzyme fructose-1,6-bisphospate aldolase in addition to obviously nonphotosynthetic proteins (the branched-chain amino acid biosynthetic enzymes ketol-acid reductoisomerase and dihydroxy-acid dehydratase). What is more, many nonphotosynthetic proteins are present at normal levels (e.g., the 50S ribosomal protein L12-C and the molecular chaperones Hsp70 and Hsp90) or slightly elevated levels (e.g., the ammonium fixation enzyme glutamate-ammonia ligase and the translation elongation factor EF-Tu) in toc132 chloroplasts. These data may indicate that atToc132 is not exclusively involved in the import of nonphotosynthetic precursors or reflect the continued presence of atToc120 protein in the toc132 mutant. Nevertheless, it is clear that the toc132 chloroplast proteome is quite different from the ppi1 chloroplast proteome, in that it is not specifically deficient in photosynthetic proteins.
The toc132 Transcriptome Response Is Distinct from the ppi1 Response
The expression of nuclear genes encoding plastidic proteins is regulated in response to complex signals emitted by the plastids themselves (Jarvis, 2003; Richly et al., 2003). We previously demonstrated that specific defects in the import and accumulation of photosynthetic proteins in the ppi1 mutant (which lacks a receptor, atToc33, with putative specificity for photosynthetic proteins) are correlated with the selective downregulation of the corresponding nuclear photosynthetic genes (Kubis et al., 2003). Because the transcriptome response of ppi1 reflected the characteristics of the ppi1 import defect, we used exactly the same procedure to characterize the toc132 transcriptome response. Thus, nylon filter DNA array technology (Richly et al., 2003) was employed to compare the mRNA expression of 3292 nuclear genes, most of them encoding plastidic proteins, in the toc132 mutant and the wild type; RNA gel blot analysis was previously used to confirm the accuracy of gene expression data generated using this method (Kubis et al., 2003). A total of 686 genes (∼20% of those analyzed) were found to be significantly differentially expressed in toc132 compared with the wild type (see Supplemental Table 1 online). Of those genes showing differential expression in toc132, ∼70% (486) were upregulated and ∼30% (200) were downregulated. A similar number of downregulated genes (161 of a total of 1461 differentially expressed genes) was observed in the ppi1 mutant (Kubis et al., 2003).
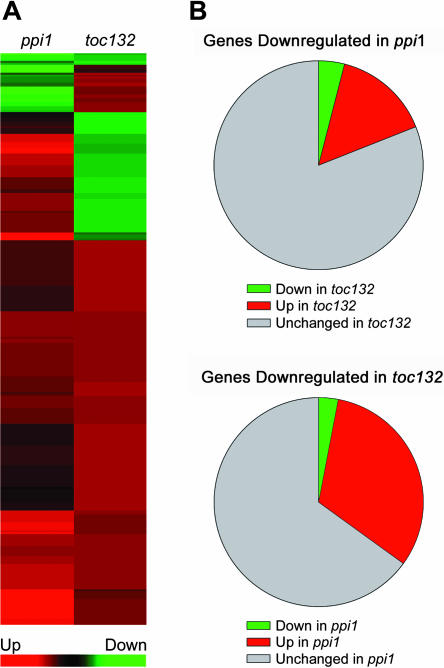
When we compared the toc132 transcriptome response with the previously described ppi1 response (Kubis et al., 2003) by cluster analysis, we observed that the responses were only partially overlapping (Figure 9A). Although the gene expression profiles of the two mutants were quite similar with respect to upregulated genes (Figure 9A), major differences were observed when only downregulated genes were considered (Figure 9B). Of all the genes showing downregulated expression in toc132 (200 genes), only 3% (six genes) were also downregulated in ppi1. Similarly, of all the genes showing downregulated expression in ppi1 (161 genes), only 4% (six genes) were also downregulated in toc132. Thus, toc132 does not exhibit the same specific downregulation of photosynthetic genes as was found in ppi1. Indeed, very few of the 200 genes that show reduced expression in toc132 encode known photosynthetic proteins (see Supplemental Table 1 online). Although these data do not provide a direct assessment of protein import in the toc132 mutant, they are nevertheless consistent with the hypothesis that atToc132 is involved preferentially in an import pathway with some specificity for nonphotosynthetic proteins.
Figure 9.
Effects of the toc132 and ppi1 Mutations on the Accumulation of Nuclear Transcripts Encoding Chloroplast Proteins.
(A) Nylon filter DNA array technology was used to characterize the ppi1 (Kubis et al., 2003) and toc132 (see Supplemental Table 1 online) nuclear chloroplast transcriptome responses. Hierarchical clustering of the expression profiles of the 288 genes that show significant differential expression, relative to the wild type, in both mutants (ppi1 and toc132) is shown. Colors indicate downregulated (green) or upregulated (red) gene expression relative to the wild type.
(B) The behavior of those genes that are downregulated in ppi1, relative to the wild type, in the toc132 mutant is shown in the top pie chart. Similarly, the behavior of those genes that are downregulated in toc132, relative to the wild type, in the ppi1 mutant is shown in the bottom pie chart. The top chart describes 161 genes, and the bottom chart describes 200 genes. Colors indicate downregulated (green), upregulated (red), or unchanged (gray) gene expression relative to the wild type.
DISCUSSION
Our phylogenetic analyses suggested that there are at least two distinct subtypes of Toc159-related proteins within plant cells—characterized by atToc159 and atToc132/atToc120, respectively—that differentiated before the divergence of monocotyledonous and dicotyledonous species (Figure 1). Accordingly, when the functional relationships between the four Arabidopsis proteins were tested genetically in double mutant studies (Figures 3 and 4, Table 2) and in transgenic complementation studies (Figure 6), atToc132 and aToc120 were shown to exhibit a high degree of functional redundancy. By contrast, the data indicated that there is very little functional overlap between atToc159 and atToc132 or atToc120 (Figure 6, Table 3; data not shown). Although toc132 toc120 double homozygotes exhibited a very severe phenotype, reminiscent of the toc159 phenotype, clear differences between the toc132 toc120 and toc159 phenotypes were observed (Figures 3, 4, and 7), implying functional differences between the two protein subtypes. Indeed, the fact that these two mutants each have such severe phenotypes (albino or near-albino) indicates that proteins of one subtype cannot completely compensate for the absence of protein(s) of the other subtype. Interestingly, the toc159 toc132 double homozygous mutation was found to cause embryo lethality (Figure 5, Table 3). Transgenic complementation studies (Figure 6) revealed that the strength of this genetic interaction is a reflection of functional nonredundancy between the atToc159 and atToc132 proteins and that lethality is most likely because of the disruption of import across a wide range of different precursor types.
Lethal effects of certain mutations affecting genes of chloroplast function have been reported previously. For example, inactivation of either plastidic glycyl-tRNA synthetase or chaperonin-60α resulted in developmental arrest during embryogenesis (Uwer et al., 1998; Apuya et al., 2001). The essential nature of plastids during embryo development can be attributed to their unique role as a site for the biosynthesis of important compounds (such as lipids, amino acids, purines, and pyrimidines) and, during later stages of embryogenesis, for photosynthesis. More recently, it was reported that a major component of the chloroplast protein import apparatus, Toc34, also plays an essential role in Arabidopsis (Constan et al., 2004). Whereas single knockout mutations affecting either atToc33 or atToc34 result in rather mild phenotypes (Jarvis et al., 1998; Constan et al., 2004), individuals that are homozygous for both mutations arrest during early embryo development (Constan et al., 2004). The essential nature of the Toc34 protein family indicates that this component plays a central role during protein import, rather than a peripheral or regulatory role as had previously been proposed. The fact that the toc159 toc132 genotype is also lethal indicates that the Toc159 protein family plays a similarly important role during import. It is interesting that both Toc-GTPases have been assigned to the so-called core Toc complex (Schleiff et al., 2003) because core components of the functionally similar Tom complex of mitochondria are also essential for viability (Baker and Schatz, 1991).
Specific defects in the expression and accumulation of photosynthetic proteins in ppi2 (toc159) led to the hypothesis that atToc159 is a receptor with specificity for photosynthetic proteins and, by extension, that atToc132 and atToc120 are receptors with specificity for nonphotosynthetic proteins (Bauer et al., 2000). The existence of a separate receptor to cope with the bulk flow of highly abundant, photosynthetic precursors, and thus prevent the fatal consequences that would result from outcompeted import of essential, nonphotosynthetic preproteins, is an attractive model. Although the inactivation of photosynthesis would perhaps not be expected to result in embryo lethality (Kanevski and Maliga, 1994), a complete block in the import of nonphotosynthetic proteins (involved in the biosynthesis of essential metabolites, for example) would because plastids are integral to cellular metabolism. Thus, if atToc132 and atToc120 are indeed receptors with preference for nonphotosynthetic proteins, the fact that toc132 toc120 double homozygotes are viable indicates that some other protein must be compensating for their absence in the double mutant. Because atToc159 is the most abundantly expressed isoform (Figure 2) and because no evidence for any detrimental effect of the toc90 mutation was observed (Figures 3 and 4, Table 3), the component most likely to be fulfilling this compensatory role is atToc159. The lethality of the toc159 toc132 genotype is certainly consistent with this hypothesis, and so we conclude that the two Toc159 subtypes, characterized by atToc159 and atToc132/atToc120, exhibit significant but incomplete functional specialization.
We recently proposed that atToc33 (the most abundant of two Toc34 isoforms in Arabidopsis) is preferentially involved in the import of photosynthetic proteins (Kubis et al., 2003). This hypothesis was based on the observation that the atToc33 knockout mutant, ppi1, is specifically deficient in the expression, chloroplast import, and accumulation of photosynthetic proteins. Interestingly, the ppi1 and toc132 transcriptome responses were extremely different with respect to downregulated genes (Figure 9B) because very few of the downregulated genes in toc132 encode known photosynthetic proteins (see Supplemental Table 1 online). Furthermore, when we used proteomics to characterize toc132 chloroplasts, we found that several of the photosynthetic proteins that were previously shown to be deficient in ppi1 were not depleted in toc132 (Table 4). We also observed pronounced structural defects in root plastids of the toc132 toc120 double homozygotes (defects that were not apparent in toc159 root plastids), and similar abnormalities were apparent in toc132 toc120 chloroplasts (Figure 7). Taken together, these data strongly support the hypothesis that atToc132 and atToc120 are receptors with some preference for nonphotosynthetic proteins.
The preferential involvement of the various Toc159 and Toc34 isoforms in different import pathways with substrate preferences has taken further significant support from the work of Ivanova et al. (2004). In addition to investigating functional relationships among atToc159, atToc132, and atToc120 in genetic experiments similar to some of those described in this report, these authors assessed physical associations between the different Toc isoforms directly and studied the binding of two different preproteins (one photosynthetic and one nonphotosynthetic) to the atToc132 receptor. An analysis of chloroplast membrane extracts by immunoaffinity chromatography revealed that atToc132 and atToc120 are present together in Toc complexes from which atToc159 is completely excluded. Furthermore, these atToc132/atToc120-containing complexes contained predominantly atToc34 and only small amounts of atToc33. By contrast, atToc159-containing complexes (from which atToc132 and atToc120 were excluded) contained predominantly atToc33, an observation that accounts nicely for the qualitatively similar phenotypes observed in the atToc159 and atToc33 knockout mutants ppi2 and ppi1 (Bauer et al., 2000; Kubis et al., 2003). When the binding of synthetic precursors bearing photosynthetic (preSSU) and nonphotosynthetic (prepyruvate dehydrogenase E1α) transit peptides was compared in pull-down assays, atToc132 was shown to exhibit a marked preference for the nonphotosynthetic transit peptide (Ivanova et al., 2004).
Given the uniformly high level of expression exhibited by the atTOC90 gene (Figure 2), it is surprising that we could find no evidence for a role of the atToc90 protein during chloroplast biogenesis: toc90 homozygotes were indistinguishable from the wild type with respect to appearance, rate of growth and development, and chlorophyll accumulation (Figures 3 and 4). Similarly, no obvious effect of the toc90 mutation was observed in the toc159, toc132, and toc120 backgrounds (Figures 3 and 4, Table 3), and the overexpression of atTOC90 was inactive in complementation studies (Figure 6), suggesting that the atToc90 protein does not share substantial functional redundancy with any of these components. Possible explanations of these data are that atToc90 is an evolutionary vestige that no longer plays an active role in protein translocation or that it plays a role unrelated to chloroplast protein import. Alternatively, atToc90 may play a nonessential or accessory role in import, perhaps acting to increase the efficiency of translocation under certain conditions that were not investigated during the course of this study.
We have identified new Arabidopsis knockout mutants lacking the atToc132, atToc120, and atToc90 proteins and characterized these mutants in detail along with the previously described atToc159 knockout mutant ppi2 (Bauer et al., 2000). In agreement with the results of Ivanova et al. (2004), our data provide clear evidence for (incomplete) functional specialization amongst the Toc159 protein family and are consistent with the notion that the closely related atToc132 and atToc120 proteins play a significant role in the import of nonphotosynthetic proteins. These new mutants will be useful tools for the further exploration of the substrate specificities of the Toc159-related receptors in the future. Given that chloroplasts isolated from the pale toc132 toc120 double mutant did not exhibit any evidence of an import defect in vitro, it is likely that further progress in this area will necessitate the use of in vivo approaches.
METHODS
Sequence Analysis
Amino acid sequences were aligned using ClustalW within the BioEdit program (Hall, 1999). Phylogenetic trees were calculated using PAUP* 4.0 b10 (Swofford, 2003). All analyses were performed using branch and bound searches, with the collapse option and furthest addition sequence selected. No weighting or ordering was imposed on the characters, and any gaps were treated as missing data. Indels were coded separately and appended to the sequence data matrix. Coding of indels was usually binary (deletions 0, insertions 1), but in places where more than one size occurred, the alternatives were coded as 2, 3, etc. Support for clades was estimated by means of nonparametric bootstrap analyses, as implemented in PAUP* 4.0, using 1000 replicates. The human c-H-Ras1 p21 proto-oncogene (accession number P01112) was used as an outgroup.
Plant Material and Growth Conditions
Arabidopsis thaliana plants, both wild type and mutants, used in this study were of the Col-0 ecotype. The ppi2 (toc159) mutant, which is of the Ws ecotype, was kindly provided by Felix Kessler (Bauer et al., 2000) and was subsequently introgressed into the Col-0 ecotype (see Results). Seeds were surface sterilized, and plants were grown as described previously (Aronsson and Jarvis, 2002). When necessary, the following antibiotics were included in the medium at the indicated concentrations: 50 μg/mL of kanamycin monosulphate (Melford Laboratories, Ipswich, UK) was used to select for toc159 and the Salk Institute Genomic Analysis Laboratory (SIGnAL) lines; 10 μg/mL of dl-phosphinothricin (Duchefa, Haarlem, The Netherlands) was used to select for toc132-2 and toc90-1; 15 μg/mL of hygromycin B (Duchefa) was used to select for toc120-2; and 110 μg/mL of gentamicin sulfate (Duchefa) was used to select for pCHF2 transformants. Plants grown on soil were kept under standard greenhouse conditions.
Identification of Knockout Mutants
The three new mutants that were the main focus of this study were obtained from the following sources: Syngenta, lines Garlic_667_D04 (toc132-2) and Garlic_1236_C11 (toc90-1); Csaba Koncz Laboratory, pool 286, line 28,567 (toc120-2). The additional, supportive mutants were obtained from SIGnAL, lines SALK_017374 (toc120-3), SALK_064469 (toc90-2), and SALK_119434 (toc90-3) and from Genomanalyse im Biologischen System Pflanze (GABI)-Kölner Arabidopsis T-DNA (Kat), line 394E01 (toc132-3). The mutants were isolated according to published procedures: Syngenta (Sessions et al., 2002); Csaba Koncz Laboratory (Ríos et al., 2002); SIGnAL (Alonso et al., 2003); GABI-Kat (Rosso et al., 2003).
Gene-specific and T-DNA–specific primers used to confirm the mutations were as follows: Toc132-F, 5′-GATGGGACTGAGTTTGTGGTTAGGTC-3′; Toc132-R, 5′-CAAAGCACATCAACGCTCAGCAAAATCCA-3′; Toc120-F, 5′-CTAACCAGGTTAGATTCTCGCCATCAC-3′; Toc120-R, 5′-GACGTGTTAAATTGACGAGCACTGTAGAG-3′; Toc90-F, 5′-TTCTTCTTTAGTCGGTTTATTGTGCGGTGGAG-3′; Toc90-R, 5′-AGGAGTGGAGAAATCAAAGAGAAAGCGAGGAG-3′; Syngenta T-DNA LB3, 5′-TAGCATCTGAATTTCATAACCAATCACGATACAC-3′; Syngenta T-DNA 104RB3, 5′-TAACAATTTCACACAGGAAACAGCTATGAC-3′; Koncz T-DNA LB FISH1, 5′-CTGGGAATGGCGAAATCAAGGCATC-3′; Koncz T-DNA RB HOOK2, 5′-TACTTTCTCGGCAGGAGCAAGGTGA-3′; SIGnAL T-DNA LBa1, 5′-TGGTTCACGTAGTGGGCCATCG-3′; GABI-Kat T-DNA LB, 5′-CCCATTTGGACGTGAATGTAGACAC-3′.
For the original three mutants, both T-DNA junctions were sequenced to verify the predicted location of the T-DNA with respect to each gene (Figure 3A); for the additional, supportive mutants, only one junction was sequenced in each case (Figure 3A). Complex T-DNA insertions were present in the toc132-2 and toc90-1 mutants because left border sequences were found at both ends of the insertion; however, no evidence for rearrangements of gene sequences or larger deletions was observed. Single-locus T-DNA insertion lines were identified for each of the original mutants (Table 1), and homozygotes derived from these lines were confirmed by PCR before further analysis.
Chlorophyll Quantification and Electron Microscopy
Chlorophyll determinations and transmission electron microscopy were performed as described previously (Porra et al., 1989; Constan et al., 2004). Transmission electron microscopy was performed at the Electron Microscope Laboratory, Faculty of Medicine and Biological Sciences, University of Leicester.
Identification of Double Mutants
Homozygous toc132, toc120, and toc90 plants and heterozygous toc159 (Col-0) plants were crossed to each other in all pairwise combinations, the resultant F1 plants were allowed to self-pollinate, and F2 seeds were obtained. After 10 d of growth in vitro, in the presence or absence of appropriate antibiotics, F2 plants were scored for antibiotic resistance and chlorotic phenotypes. After several more days of growth, genomic DNA was extracted (Edwards et al., 1991) from selected individuals and used for PCR genotyping. The PCR primers used were the gene-specific and T-DNA–specific primers given above.
Transgenic Complementation Experiments
The atTOC159, atTOC132, atTOC120, and atTOC90 open reading frames were expressed from a double-enhancer version of the 35S promoter, using plant transformation/expression vector, pCHF2 (Jarvis et al., 1998). For atTOC90, cDNA clone RZL46 g05 (accession number AV548084) was subcloned into pCHF2 as a SacI/end-filled Asp718 fragment into SacI/SmaI-cut pCHF2. The full-length atTOC120 open reading frame was amplified from Arabidopsis Col-0 genomic DNA using forward (5′-TCTCTGAGCTCTGTTTCGTGATTTTGGGTGAT-3′) and reverse (5′-TCAAAGTCGACAAACAAAAGTGAATTCCGACAAC-3′) primers and cloned into pCHF2 using SacI/SalI restriction sites introduced into the forward and reverse primers, respectively. The resulting atTOC120 complementation vector was confirmed by sequencing. The atTOC132 complementation vector was constructed from the truncated cDNA clone H3A2 (accession number W43632), which carries a 975-bp 5′ terminal deletion. Firstly, the truncated cDNA was subcloned as a SacI/XhoI fragment into a modified version of pCHF2 (carrying a single EcoRI site in the polylinker) cut with SacI/SalI. A 2-kb atTOC132 RT-PCR product, amplified using forward (5′-GTCTGAGCTCCTATCTCTAACTTCTGCGGTGGTG-3′) and reverse (5′-CAGGTGGATTCTTCTTGATG-3′) primers, was then cloned as a SacI/EcoRI fragment into the complementation vector to reconstitute the full-length atTOC132 cDNA. The PCR-amplified portion of the full-length atTOC132 cDNA was confirmed by sequencing. The full-length atTOC159 open reading frame, including the intron, was constructed from cDNA clone APZL25e08 (accession number AV523359), which carries a 5′ truncation, and a 3-kb PCR product amplified from Col-0 genomic DNA using forward (5′-CAAAGGTACCATGGACTCAAAGTCGGTTAC-3′) and reverse (5′-GTCTGAGCTCCTATCTCTAACTTCTGCGGTGGTG-3′) primers. The atTOC159 amplification product was ligated as an ApaI/HindIII fragment into the partial cDNA clone to reconstitute the full-length open reading frame, which was then confirmed by sequencing and cloned into pCHF2 as an Asp718/XbaI fragment.
The complementation constructs were introduced into Agrobacterium strain GV3101 (pMP90) and then used to transform toc159 heterozygotes or toc132 toc120 heterozygotes (genotype: toc132/toc132; +/toc120) by the floral dip method (Clough and Bent, 1998). Transgene overexpression was estimated by RT-PCR and, in some cases, by immunoblotting. RNA was extracted from 15-d-old T2 plants grown on medium containing gentamicin using the RNeasy plant mini kit (Qiagen, Valencia, CA) and treated with DNase I (DNA-free; Ambion, Austin, TX). RT-PCR was conducted using previously described procedures (Constan et al., 2004), using 18 cycles of amplification. PCR primers were selected to give ∼700-bp products and were as follows: atTOC159 forward, 5′-CAGTAGCAAAGCGGAAATGGACTCAAAG-3′; atTOC159 reverse, 5′-GCCACATCAACATGCACTGATTC-3′; atTOC132 forward, 5′-GATGGGACTGAGTTTGTGGTTAGGTC-3′; atTOC132 reverse, 5′-CTCTTGTTCTGTCTGTATGCC-3′; atTOC120 forward, 5′-AATTGGTTCGCAGGAGGGTC-3′; atTOC120 reverse, 5′-CTTTCAGTCTCTCCCTTCTC-3′; atTOC90 forward, 5′-TTCCGTGACCCTCATCAAGAAC-3′; atTOC90 reverse, 5′-GAGAAATCACTGTATCGCATGTCG-3′; eIF4E1 forward, 5′-AAACAATGGCGGTAGAAGACACTC-3′; eIF4E1 reverse, 5′-AAGATTTGAGAGGTTTCAAGCGGTGTAAG-3′. PCR products were resolved by agarose gel electrophoresis, stained with ethidium bromide, and quantified using Quantity One software (Bio-Rad, Hercules, CA); a λ DNA dilution series was used to confirm the accuracy of the quantification procedure. Data for the Toc159-related genes were normalized using equivalent data for the translation initiation factor gene eIF4E1 (Rodriguez et al., 1998). Total protein extracts were prepared from 10-d-old T3 plants as described previously (Aronsson et al., 2003) and were analyzed by staining with Coomassie Brilliant Blue R 250 (Fisher Scientific, Loughborough, UK) or by immunoblotting (Aronsson et al., 2003) using antibodies against atToc132 and atToc120 (Ivanova et al., 2004).
Complementation was estimated by making chlorophyll measurements using 10-d-old T3 plants. T3 families in the toc132 toc120 background were grown on medium containing phosphinothricin, hygromycin, and gentamicin, and only triply-antibiotic-resistant plants were used for chlorophyll quantifications. T3 families carrying the 35S-atTOC132, 35S-atTOC120, and 35S-atTOC90 constructs in the toc159 background were scored on medium containing gentamicin to identify 35S transgene homozygotes; chlorophyll measurements were then conducted using only 35S transgene/toc159 double homozygotes. T3 families carrying the 35S-atTOC159 construct in the toc159 background were scored phenotypically and by PCR to identify toc159 homozygotes; chlorophyll measurements were then conducted using only toc159 homozygotes carrying the 35S transgene.
Isolation of Arabidopsis Chloroplasts
Chloroplasts were isolated from 10-d-old wild-type and pale toc132 toc120 double mutant (genotype: toc132/toc132; +/toc120) plants grown in vitro as described previously (Aronsson and Jarvis, 2002; Kubis et al., 2003). Plant material was homogenized for 3 to 4 s (wild type) or 2 to 3 s (toc132 toc120) using a polytron. The yield and intactness of the chloroplasts were determined as described previously (Aronsson and Jarvis, 2002).
Protein Import into Chloroplasts
Template DNA for the in vitro transcription/translation of preproteins was amplified by PCR from cDNA clones using M13 primers. The preSSU and preL11 cDNA clones were as described previously (Aronsson and Jarvis, 2002). Transcription/translation was performed using a coupled reticulocyte lysate system (TNT T7 Quick for PCR DNA; Promega, Madison, WI) or a coupled wheat germ system (Promega), containing 35S-Met and T7 RNA polymerase, according to the manufacturer's instructions. Import reactions and quantifications were performed as described previously (Aronsson and Jarvis, 2002; Kubis et al., 2003).
Chloroplast Proteomics
Preparation of protein samples from wild-type and toc132 mutant chloroplasts, their labeling with complementary CyDyeDIGE fluors, separation, quantification, and identification was performed as described by Kubis et al. (2003). The following changes were applied: proteins within the gel-excised spots were first reduced, carboxyamidomethylated, and then digested to peptides using trypsin on a MassPrepStation (Micromass, Manchester, UK). The resulting peptides were applied to either matrix-assisted laser-desorption ionization time of flight MS (TofSpec2E; Micromass), for peptide mass fingerprinting, or LC-MS/MS. For LC-MS/MS, the liquid chromatographic separation was achieved with a PepMap C18 reverse phase, 180-μm i.d., 15-cm column (LC Packings, Amsterdam, The Netherlands), and the mass spectrometer was a Qtof2 (Micromass). Fragmentation data was used to search the National Center for Biotechnology Information database using the MASCOT search engine (http://www.matrixscience.com). Probability-based MASCOT scores were used to evaluate identifications. Only matches with P < 0.05 for random occurrence were considered significant (further explanation of MASCOT scores can be found at http://www.matrixscience.com).
RNA Isolation and RNA Gel Blot Analysis
RNA was isolated from plant material grown in vitro (10 d old) or on soil (28 d old) as described by Kubis et al. (2003). RNA gel electrophoresis, transfer to Hybond NX membrane (Amersham Pharmacia Biotech, Uppsala, Sweden), labeling of DNA fragments, and hybridization and washing of membranes at 65°C were all performed as described previously (Kubis et al., 2003). Band quantification was performed using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Identical filters were probed simultaneously with atTOC159, atTOC132, atTOC120, and atTOC90 probes. The four probes corresponded to cDNA fragments of the A-domain (A+G-domain in the case of atToc90) that were similar in length (786-bp atTOC132, 802-bp atTOC120, 874-bp atTOC90, and 910-bp atTOC159) and guanine/cytosine nucleotide content (39.15% atTOC120, 42.31% atTOC132, 43.36% atTOC90, and 46.37% atTOC159). The probes were labeled simultaneously under identical conditions using the same isotope and were shown to have identical specific activities by scintillation counting (<10% variation). Hybridization, washing, and exposure steps were performed simultaneously under identical conditions.
Cross-hybridization among the four genes can be excluded because probes were selected from the most variable region of the genes, where homology is low or restricted to short DNA stretches (in the case of atToc120 and atToc132), and the wash stringency used only allowed probe-target combinations with >83% homology to remain hybridized.
DNA Array Analysis
The 3292 GST array (Richly et al., 2003) was used for the transcriptome analysis of the toc132 mutant as described by Kubis et al. (2003). The data obtained for all significantly differentially regulated genes are given in Supplemental Table 1 online. Complete data are deposited at the GeneOmnibus Web site (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSM14832. Hierarchical clustering of the expression profiles of the 288 genes that show significant differential expression in both toc132 and ppi1 was performed using Genesis Software (version 1.1.3; Sturn et al., 2002).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AV523359, AV548084, NM_122037, P01112, and W43632.
Supplementary Material
Acknowledgments
We thank Penny Dudley for assisting in the identification of the toc120 mutant, Svenja Hester, Julie Howard, Christine Jackson, and Anthony Wardle for technical support, Natalie Allcock and Stefan Hyman for transmission electron microscopy, and Colin Ferris for help with the phylogenetic analysis. We thank Syngenta, SIGnAL, GABI-Kat, and Felix Kessler for providing the T-DNA lines. Funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation, and the seed were distributed by the Nottingham Arabidopsis Stock Centre. We are grateful to Felix Kessler and Danny Schnell for generously providing antibodies against the Arabidopsis Toc159-related proteins. This work was supported by a Universities UK Overseas Research Students award (to J.B.), a Royal Society/North Atlantic Treaty Organization postdoctoral fellowship (to S.K.), the Deutsche Forschungsgemeinschaft (to D.L.), the Royal Society Rosenheim Research Fellowship (to P.J.), and Biotechnology and Biological Sciences Research Council Grants 91/C12976, 91/P12928, and 91/C18638 (to P.J.). The GARNet proteomics facility at Cambridge was funded by the Biotechnology and Biological Sciences Research Council Investigating Gene Function Initiative.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Paul Jarvis (rpj3@le.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023309.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Apuya, N.R., Yadegari, R., Fischer, R.L., Harada, J.J., Zimmerman, J.L., and Goldberg, R.B. (2001). The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60α gene. Plant Physiol. 126, 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson, H., Combe, J., and Jarvis, P. (2003). Unusual nucleotide-binding properties of the chloroplast protein import receptor, atToc33. FEBS Lett. 544, 79–85. [DOI] [PubMed] [Google Scholar]
- Aronsson, H., and Jarvis, P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529, 215–220. [DOI] [PubMed] [Google Scholar]
- Baker, K.P., and Schatz, G. (1991). Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature 349, 205–208. [DOI] [PubMed] [Google Scholar]
- Barth, S., Melchinger, A.E., and Lübberstedt, T. (2002). Genetic diversity in Arabidopsis thaliana L. Heynh. investigated by cleaved amplified polymorphic sequence (CAPS) and inter-simple sequence repeat (ISSR) markers. Mol. Ecol. 11, 495–505. [DOI] [PubMed] [Google Scholar]
- Bauer, J., Chen, K., Hiltbunner, A., Wehrli, E., Eugster, M., Schnell, D., and Kessler, F. (2000). The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403, 203–207. [DOI] [PubMed] [Google Scholar]
- Bauer, J., Hiltbrunner, A., Weibel, P., Vidi, P.A., Alvarez-Huerta, M., Smith, M.D., Schnell, D.J., and Kessler, F. (2002). Essential role of the G-domain in targeting of the protein import receptor atToc159 to the chloroplast outer membrane. J. Cell Biol. 159, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, T., Jelic, M., Vojta, A., Radunz, A., Soll, J., and Schleiff, E. (2004). Preprotein recognition by the Toc complex. EMBO J. 23, 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölter, B., May, T., and Soll, J. (1998). A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide. FEBS Lett. 441, 59–62. [DOI] [PubMed] [Google Scholar]
- Chen, K., Chen, X., and Schnell, D.J. (2000). Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol. 122, 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Constan, D., Patel, R., Keesgtra, K., and Jarvis, P. (2004). An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 38, 93–106. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutensohn, M., Schulz, B., Nicolay, P., and Flügge, U.-I. (2000). Functional analysis of the two Arabidopsis homologues of Toc34, a component of the chloroplast protein import apparatus. Plant J. 23, 771–783. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Hiltbrunner, A., Bauer, J., Alvarez-Huerta, M., and Kessler, F. (2001. a). Protein translocon at the Arabidopsis outer chloroplast membrane. Biochem. Cell Biol. 79, 629–635. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner, A., Bauer, J., Vidi, P.A., Infanger, S., Weibel, P., Hohwy, M., and Kessler, F. (2001. b). Targeting of an abundant cytosolic form of the protein import receptor at Toc159 to the outer chloroplast membrane. J. Cell Biol. 154, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah, S.C., Wagner, R., Sveshnikova, N., Harrer, R., and Soll, J. (2002). The chloroplast protein import channel Toc75: Pore properties and interaction with transit peptides. Biophys. J. 83, 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, S., Muckel, E., Heemeyer, F., von Heijne, G., and Soll, J. (1994). A receptor component of the chloroplast protein translocation machinery. Science 266, 1989–1992. [DOI] [PubMed] [Google Scholar]
- Ivanova, Y., Smith, M.D., Chen, K., and Schnell, D.J. (2004). Members of the toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 15, 3379–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan, D., and Keegstra, K. (2001). Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol. 125, 1567–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, P. (2003). Intracellular signalling: The language of the chloroplast. Curr. Biol. 13, R314–R316. [DOI] [PubMed] [Google Scholar]
- Jarvis, P., Chen, L.-J., Li, H.-m., Peto, C.A., Fankhauser, C., and Chory, J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103. [DOI] [PubMed] [Google Scholar]
- Jarvis, P., and Soll, J. (2002). Toc, Tic, and chloroplast protein import. Biochim. Biophys. Acta 1590, 177–189. [DOI] [PubMed] [Google Scholar]
- Kanevski, I., and Maliga, P. (1994). Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 91, 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, F., Blobel, G., Patel, H.A., and Schnell, D.J. (1994). Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 266, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Kouranov, A., and Schnell, D.J. (1997). Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139, 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis, S., Baldwin, A., Patel, R., Razzaq, A., Dupree, P., Lilley, K., Kurth, J., Leister, D., and Jarvis, P. (2003). The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15, 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.H., Kim, S.J., Lee, Y.J., Jin, J.B., and Hwang, I. (2003). The M domain of atToc159 plays an essential role in the import of proteins into chloroplasts and chloroplast biogenesis. J. Biol. Chem. 278, 36794–36805. [DOI] [PubMed] [Google Scholar]
- Ma, Y., Kouranov, A., LaSala, S.E., and Schnell, D.J. (1996). Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 134, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J., Carre, I.A., Strayer, C.A., Chua, N.H., and Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163. [DOI] [PubMed] [Google Scholar]
- Perry, S.E., and Keegstra, K. (1994). Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. [Google Scholar]
- Richly, E., Dietzmann, A., Biehl, A., Kurth, J., Laloi, C., Apel, K., Salamini, F., and Leister, D. (2003). Co-variations in the nuclear chloroplast transcriptome reveal a regulatory master switch. EMBO Rep. 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos, G., et al. (2002). Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J. 32, 243–253. [DOI] [PubMed] [Google Scholar]
- Rodriguez, C.M., Freire, M.A., Camilleri, C., and Robaglia, C. (1998). The Arabidopsis thaliana cDNAs coding for eIF4E and eIF(iso)4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J. 13, 465–473. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53, 247–259. [DOI] [PubMed] [Google Scholar]
- Schleiff, E., Motzkus, M., and Soll, J. (2002). Chloroplast protein import inhibition by a soluble factor from wheat germ lysate. Plant Mol. Biol. 50, 177–185. [DOI] [PubMed] [Google Scholar]
- Schleiff, E., Soll, J., Küchler, M., Kühlbrandt, W., and Harrer, R. (2003). Characterization of the translocon of the outer envelope of chloroplasts. J. Cell Biol. 160, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell, D.J., Blobel, G., Keegstra, K., Kessler, F., Ko, K., and Soll, J. (1997). A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol. 7, 303–304. [DOI] [PubMed] [Google Scholar]
- Schnell, D.J., Kessler, F., and Blobel, G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266, 1007–1012. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., Waegemann, K., and Soll, J. (1995). A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 7, 401–411. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.D., Hiltbrunner, A., Kessler, F., and Schnell, D.J. (2002). The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP. J. Cell Biol. 159, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streatfield, S.J., Weber, A., Kinsman, E.A., Hausler, R.E., Li, J., Post-Beittenmiller, D., Kaiser, W.M., Pyke, K.A., Flugge, U.-I., and Chory, J. (1999). The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell 11, 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn, A., Quackenbush, J., and Trajanoski, Z. (2002). Genesis: Cluster analysis of microarray data. Bioinformatics 18, 207–208. [DOI] [PubMed] [Google Scholar]
- Sveshnikova, N., Soll, J., and Schleiff, E. (2000). Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proc. Natl. Acad. Sci. USA 97, 4973–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (2003). PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. (Sunderland, MA: Sinauer Associates).
- Uwer, U., Willmitzer, L., and Altmann, T. (1998). Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell 10, 1277–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallas, T.R., Smith, M.D., Sanchez-Nieto, S., and Schnell, D.J. (2003). The roles of Toc34 and Toc75 in targeting the Toc159 preprotein receptor to chloroplasts. J. Biol. Chem. 278, 44289–44297. [DOI] [PubMed] [Google Scholar]
- Wu, C., Seibert, F.S., and Ko, K. (1994). Identification of chloroplast envelope proteins in close physical proximity to a partially translocated chimeric precursor protein. J. Biol. Chem. 269, 32264–32271. [PubMed] [Google Scholar]
- Yu, T.S., and Li, H.-m. (2001). Chloroplast protein translocon components atToc159 and atToc33 are not essential for chloroplast biogenesis in guard cells and root cells. Plant Physiol. 127, 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.