SUMMARY
The reactivation of stalled DNA replication via fork regression invokes Holliday junction formation, branch migration, and the recovery of the replication fork after DNA repair or error-free DNA synthesis. The coordination mechanism for these DNA structural transitions by molecular motors, however, remains unclear. Here we perform single-molecule fluorescence experiments with Werner syndrome protein (WRN) and model replication forks. The Holliday junction is readily formed once the lagging arm is unwound, and migrated unidirectionally with 3.2 ± 0.03 bases/s velocity. The recovery of the replication fork was controlled by branch migration reversal of WRN, resulting in repetitive fork regression. The Holliday junction formation, branch migration, and migration direction reversal are all ATP dependent, revealing that WRN uses the energy of ATP hydrolysis to actively coordinate the structural transitions of DNA.
INTRODUCTION
DNA is constantly damaged (Vilenchik and Knudson, 2000). Various DNA lesions stall DNA replication, making the reactivation of stalled replication forks a major challenge for DNA replication (Dronkert and Kanaar, 2001; Nohmi, 2006; Prakash et al., 2005). In the initial stages of many reactivation pathways of stalled replication, the replication fork three-way junction is converted into a four-way junction (also called a Holliday junction or a chicken-foot structure) via the unwinding of the lagging and leading arms, followed by reannealing of the unwound parental strands and of the daughter strands, a process called replication fork regression (Atkinson and McGlynn, 2009; Fujiwara and Tatsumi, 1976; Higgins et al., 1976; Hotchkiss, 1974). Following the formation of a Holliday junction, the branchpoint of the Holliday junction migrates, making the damaged site accessible to the DNA repair machinery (Courcelle et al., 2003). Alternatively, the Holliday junction serves as a substrate for error-free DNA synthesis via the template-switching mechanism, or for DNA repair pathways via homologous recombination (Atkinson and McGlynn, 2009; Cox et al., 2000). In any case, eventual re-establishment of a functional replication fork is crucial for genome maintenance and cell survival.
The replication fork regression is executed by ATP-consuming molecular motors (Long and Kreuzer, 2009). In vitro biochemical studies at the ensemble level identified fork regression activities of diverse DNA helicases and translocases (Betous et al., 2012; Blastyak et al., 2007, 2010; Bugreev et al., 2011; Gari et al., 2008; Machwe et al., 2006; McGlynn and Lloyd, 2000; Yusufzai and Kadonaga, 2011), suggesting that fork regression is essential for various DNA repair pathways. However, it is still unknown how these molecular motors coordinate the conversion of a three-way junction to a Holliday junction, in order to initiate diverse DNA repair pathways. After DNA lesion repair or error-free DNA synthesis, fork regression should be reversed to recover the replication fork. Factors that mediate the reversal of the regressed forks have not yet been identified clearly. Recent single-molecule experiments based on the magnetic tweezers technique successfully demonstrated that the replication fork regression activity of UvsW helicase is utilized to restart the stalled replication of bacteriophage T4 holoenzyme (Manosas et al., 2012). Interestingly, they observed that the branch migration direction of UvsW was spontaneously reversed at random positions. This observation suggested that the recovery of replication forks might be controlled by stochastic reversal of the branch migration direction of UvsW. However, it still remains to be demonstrated whether this type of behavior is conserved in higher organisms. The mechanism of migration direction reversal also remains unclear.
Werner syndrome protein (WRN), whose mutation causes an autosomal recessive disease characterized by premature aging and predisposition to cancer, is a member of the RecQ helicase family that is highly conserved from bacteria to human (Bohr, 2008). Genetic and cellular studies collectively indicate important roles of WRN in preventing aberrant recombination events at sites of stalled replication forks (Sidorova, 2008). In vitro biochemical studies showed that WRN has fork regression and branch migration activities (Constantinou et al., 2000). To understand how WRN coordinates DNA structural changes during replication fork regression, we here performed single-molecule fluorescence-resonance energy transfer (FRET) experiments (Roy et al., 2008) with recombinant WRN (Rossi et al., 2010) and synthetic model replication forks. Our study reveals that WRN is a bidirectional motor that dynamically converts the DNA substrate between a replication fork and a Holliday junction.
RESULTS
Single-Molecule Observation of Replication Fork Regression
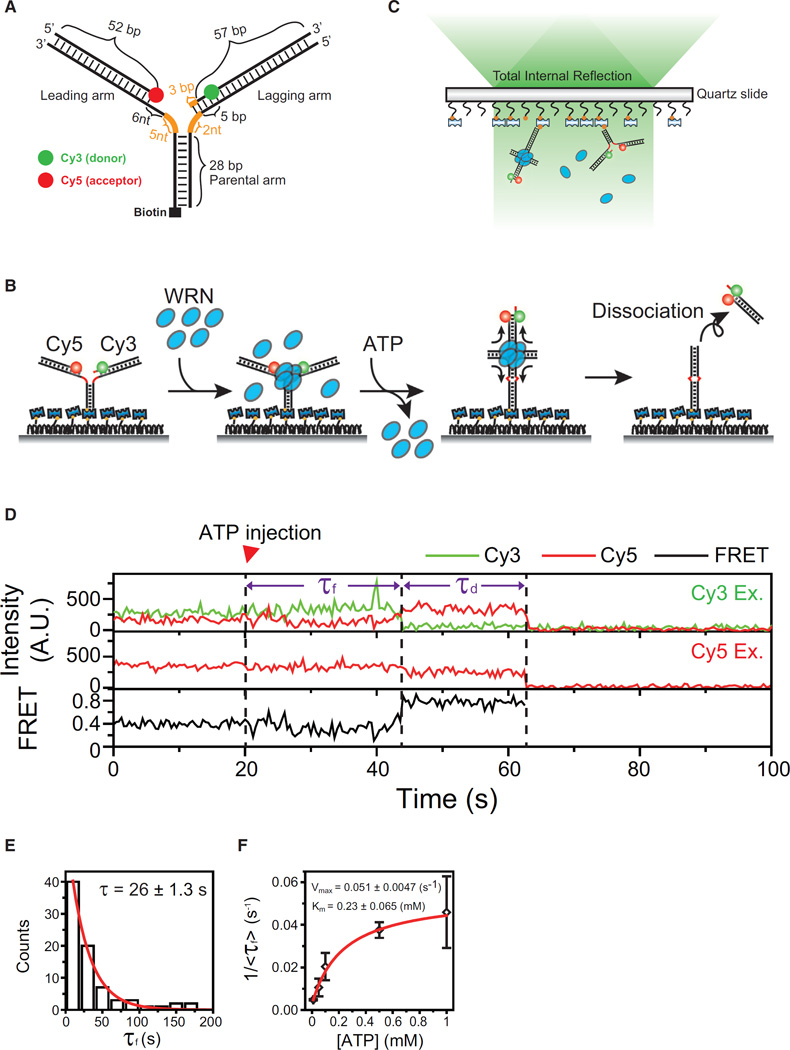
DNA lesions in the leading arm are more deleterious to DNA replication than those in the lagging strand (Atkinson and McGlynn, 2009). A DNA substrate was designed to mimic an intermediate structure formed when DNA synthesis terminates on the leading strand before the lagging strand (Figure 1A). It is composed of a 28-bp parental arm, a leading arm with a 52-bp duplex region and an 11-nt single-stranded gap at the junction, and a lagging arm with a 60-bp duplex and a 3-nt single-stranded gap at the junction (Figure 1A). To prevent spontaneous branch migration, we introduced heterologous nucleotides near the junction (orange, Figure 1A). A similar fork was shown to be a good substrate for the replication fork regression of WRN in vitro (Machwe et al., 2006, 2007). The DNA fork was labeled with Cy3 and Cy5 for FRET experiments, and biotin at the distal end of the parental arm for surface immobilization (Figure 1A).
Figure 1. Experimental Scheme.
(A) The design of a model replication fork. Orange lines represent heterologous bases.
(B) Experimental Procedures: (from left to right) immobilization of model replication forks, incubation with WRN (26 nM), flushing out of free WRN, initiation of fork regression by the delivery of imaging buffer containing ATP-Mg2+, and dissociation of the daughter duplex upon completion of fork regression.
(C) Scheme of single-molecule total internal reflection fluorescence microscopy.
(D) Example fluorescence intensity time traces of Cy3 (green) and Cy5 (red) at Cy3 excitation (top), that of Cy5 at Cy5 excitation (middle), and corresponding FRET time traces (bottom). Three events: ATP-Mg2+ injection, formation of a four-way junction, and dissociation of daughter strands are indicated by dashed lines. Time delays between the events are defined as τf and τd, as indicated.
(E) Distribution of τf in the presence of 1 mM ATP-Mg2+. The red line indicates a fit to a single-exponential function.
(F) ATP dependency of the four-way junction formation time. The inverse of average τf, <τf>, is plotted as a function of ATP concentration. Error bars are SD of three values obtained by averaging a dataset consisting of n > 30 data points, and each dataset was obtained at the same ATP concentrations but from independent measurements. The red line represents a fit to the Michaelis-Menten function.
See also Figure S1.
The Experimental Procedures is explained in Figure 1B. DNA forks were immobilized on a polymer-coated quartz surface via streptavidin-biotin interaction, and incubated with WRN. After flushing out unbound WRN, the fork regression reaction was initiated by delivery of imaging buffer containing 1 mM ATP-Mg2+. The whole reaction process was monitored using a total internal reflection FRET microscope equipped with the alternative laser excitation (ALEX) capability (Hohlbein et al., 2013; Lee et al., 2010b) (Figure 1C). In this experimental scheme, FRET appearance indicates the formation of a Holliday junction, whereas simultaneous disappearance of Cy3 and Cy5 signals represents the dissociation of the daughter duplex after the completion of fork regression (Figure 1B).
Figure 1D shows representative fluorescence intensity time traces of Cy3 (green) and Cy5 (red) at Cy3 excitation (top), that of Cy5 at direct excitation of Cy5 (middle), and corresponding FRET (bottom). More example traces and ensemble contour plot are shown in Figure S1. It is noticeable that an FRET jump occurs with a time delay (τf: the formation time of a Holliday junction) after the delivery of ATP-Mg2+. The high FRET state is maintained for a while (τd: the duration time of a Holliday junction) until Cy3 and Cy5 signals disappear simultaneously, indicating a successful completion of fork regression. Since the probability that both dyes photobleach at the same time is extremely low, the simultaneous disappearance of Cy3 and Cy5 signals is unlikely to be due to photobleaching. Under our standard experimental conditions (incubation of DNA with 26 nM WRN), about 14% of DNA molecules that had both Cy3 and Cy5 (145/1048) showed the fork regression activity. The efficiency could be increased by incubating DNA with higher concentration of WRN. Fork regression activity was not observed with non-hydrolyzable ATP analogs.
Lagging Strand Unwinding as a Rate-Limiting Step for the Formation of a Holliday Junction
First we asked why there was a time delay between the delivery of ATP-Mg2+ and the formation of a Holliday junction. The distribution of τf was well fitted to a single-exponential function, suggesting the existence of a single rate-limiting step for the formation of a Holliday junction (Figure 1E). The inverse of average τf over a range of ATP concentrations was well fitted to the Michaelis-Menten equation with a Km value of 0.23 ± 0.065 mM (Figure 1F), indicating that the Holliday junction formation is an active process requiring ATP hydrolysis.
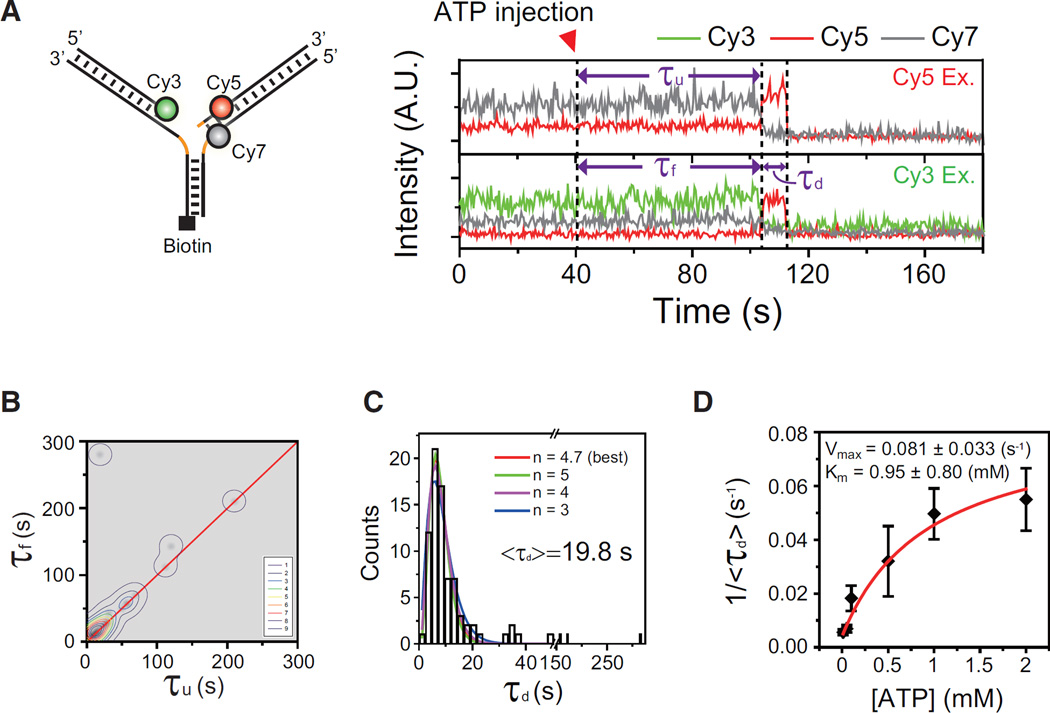
The conversion of a three-way junction into a Holliday junction would entail several steps, such as unwinding of the lagging and leading arms and pairing of the nascent daughter strands with concomitant reannealing of the parental strands. To determine which step is rate-limiting for the Holliday junction formation, we first studied the correlation between the unwinding of the lagging arm and the formation of the Holliday junction using a model replication fork labeled with Cy3, Cy5, and Cy7 (left, Figure 2A), and single-molecule three-color FRET (Lee et al., 2010b). In this labeling scheme, an FRET drop of the Cy5–Cy7 pair at Cy5 excitation represents the unwinding of the lagging arm, whereas an FRET jump of the Cy3–Cy5 pair at Cy3 excitation reports the formation of a Holliday junction. An example of fluorescence intensity time traces of Cy5, and Cy7 at Cy5 excitation (top), and those of Cy3, Cy5, and Cy7 at Cy3 excitation (bottom) are shown in Figure 2A. A time delay (τu) was observed between the delivery of ATP-Mg2+ and the unwinding of the lagging arm, and this time delay was comparable with the formation time of Holliday junction (τu ≈ τf). It means that after the initiation of the unwinding reaction, a Holliday junction was readily formed. The simultaneity of the lagging arm unwinding and the Holliday junction formation was generally observed in most replication fork regression events (Figure 2B); in 81% (21/26) of molecules, the Holliday junction was formed less than 2 s after the lagging arm unwinding.
Figure 2. Unwinding of the Lagging Arm as a Rate-Limiting Step for the Formation of a Holliday Junction.
(A) Single-molecule three-color FRET experiment to observe the correlation between the lagging arm unwinding and the four-way junction formation. Left: sample structure. Right: example intensity time traces of Cy3 (green), Cy5 (red), and Cy7 (gray) at Cy5 excitation (top) and Cy3 excitation (bottom). The unwinding time, τu, is defined as a time delay between the injection of 1 mM ATP-Mg2+ and the initiation of the lagging arm unwinding.
(B) Correlation plot of τf and τu. For clear visualization, each data point is represented as a gray-scaled 2D Gaussian distribution, and a contour plot of the population density is overlaid on the distribution. The red line is a linear function: y = x.
(C) Dwell time histogram of τd in the presence of 1 mM ATP-Mg2+. The red line represents a best fit to gamma distribution. Gamma distribution fitting with varying n values is shown in different colors.
(D) ATP dependency of the duration of a four-way junction. The inverse of average τd, <τd>, is plotted as a function of ATP concentration. Error bars are SD of three values obtained by averaging a dataset consisting of n > 30 data points, and each dataset obtained at the same ATP concentrations but from independent measurements. The red line represents a fit to the Michaelis-Menten function.
See also Figure S2.
Branch Migration Speed of WRN
Next, we investigated the relation between the high FRET dwell time, τd, and the branch migration time of WRN. As expected from a molecular motor that translocates with a well-defined step size, the distribution of τd was best fitted to a gamma distribution (tn−1 exp(−kt)) with n = 4.7 and k = 1.67 s (Figure 2C) (Park et al., 2010). This kind of distribution fit has been used to determine step sizes and speeds of various molecular motors. However, different gamma distributions also fitted our data reasonably well (Figure 2C), and we could not observe any correlation between n values and varying DNA length samples (Figure S2A). Furthermore, we observed rare, but non-negligible events with long τd that is not fitted to any gamma distribution (Figure 2C), indicating that WRN has a high heterogeneity in the reaction rate. It has been reported that heterogeneous reaction rates observed within an enzyme population (“static disorder”) causes the broadening of the dwell time distribution, hindering unique determination of the step size (Park et al., 2010). For this reason, we did not try to determine the kinetic step size of WRN using gamma distribution fits.
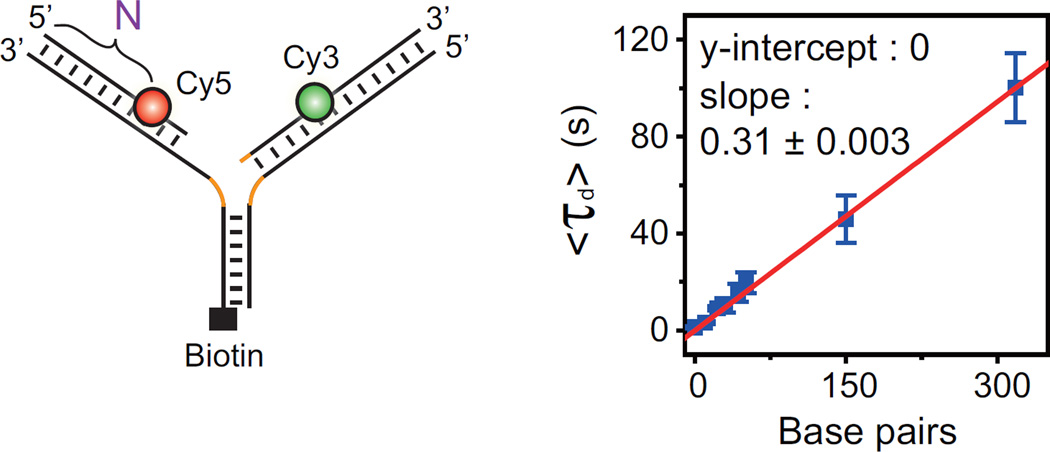
WRN uses ATP hydrolysis to translocate on DNA. To investigate the role of ATP in τd, the average of τd, <τd>, was determined over a range of ATP concentrations. The inverse of <τd> was fitted well to the Michaelis-Menten equation with a Km value of 0.95 ± 0.8 mM (Figure 2D). Finally, we prepared a series of model replication forks in which FRET probes are labeled at different positions (N = 0, 11, 21, 29, 40, 52, 150, and 319 bases from the ends of the lagging and leading arms) (Figure 3, left). In this way, each construct should migrate N bases for the completion of branch migration after the branchpoint arrives at the labeling position. <τd> was fitted well to a linear function (Figure 3, right), allowing us to determine the branch migration speed of WRN as 3.2 ± 0.03 bases/s. Representative traces at varying N are shown in Figure S2B.
Figure 3. Branch Migration Speed.
Length dependency of τd. Left: a series of model replication forks with FRET probes at different distances from the ends of the leading and lagging arms. The distance (N) means that Cy5 is labeled at the Nth base from the 5′ end of the leading daughter strand. The difference between Cy3 and Cy5 labeling positions is less than 3 nt. Right: <τd> at varying N. Error bars are SD of three values obtained by averaging a dataset consisting of n > 30 data points, and each dataset obtained from the same fork sample but independent measurement. The red line is a linear fit of the data with zero y-intercept. Orange lines represent heterologous bases.
See also Figure S2.
Repetitive Fork Regression of WRN
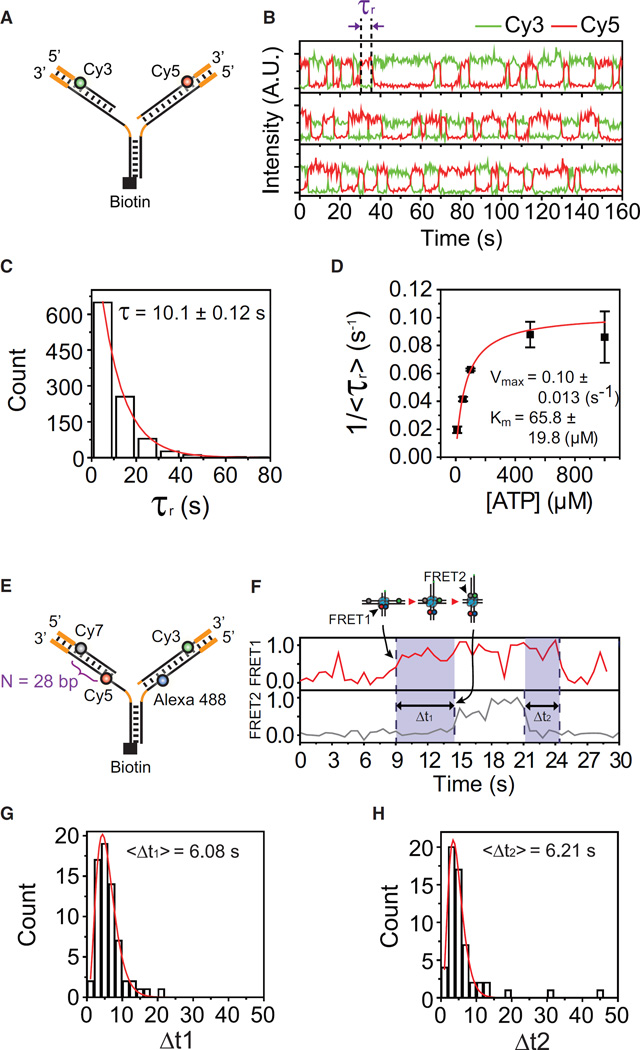
It has been shown that branch migration is greatly impeded when the molecular motors encounter heterologous sequences (Bugreev et al., 2011; Dennis et al., 2004; Kaplan and O’Donnell, 2004). To investigate how WRN handles heterologous sequences during fork regression, we prepared a model replication fork containing 15-bp heterologous regions at the distal ends of the leading and lagging arms (Figure 4A). Speculating that the branch migration machinery of WRN may be arrested at the heterologous region, the high FRET state was expected to be maintained until WRN eventually dissociates. Quite unexpectedly, however, we observed repetitive FRET jumps (Figure 4B). The repetitive behavior was observed among 89% of molecules that exhibited replication fork regression (134/150). For the remaining molecules, we could not tell whether FRET drops occurred or not due to photobleaching of fluorophores. The same repetitive behavior was observed using various model replication forks with different single-stranded regions regardless of the existence of non-homologous nucleotides flanking the junction (Figure S3). In contrast, the repetitive fork regression was not observed with a completely heterologous DNA fork (Figure S4), excluding the possibility that the FRET jumps represent simple juxtapositions of the leading and lagging arms by WRN. Interestingly, the dwell time of the high FRET state was fitted to a single-exponential function (Figure 4C) and shown to depend on ATP concentration (Figure 4D), indicating that the reversal of migration direction of WRN requires ATP hydrolysis. Since FRET probes were positioned near the heterologous sequences, the dwell time of the high FRET state is mainly determined by the time required for migration direction reversal. On the other hand, the low FRET dwell time is the sum of the branch migration time and the time required for the reactivation of fork regression.
Figure 4. Repetitive Fork Regression of WRN.
(A) Sample structure. Heterologous regions (15 bp) are added at the end of the leading and lagging arms (thick orange lines).
(B) Representative intensity time traces of Cy3 (green) and Cy5 (red) in the presence of 1 mM ATP-Mg2+. The reversal time, τr, is defined as the high FRET dwell time.
(C) Distribution of τr in the presence of 1 mM ATP-Mg2+. The red line represents a fit to a single-exponential function.
(D) ATP dependency of the branch migration reversal time. The inverse of average τr, <τr>, is plotted as a function of ATP concentration. Error bars are SD of three values obtained by averaging a dataset consisting of n > 200 data points, and each dataset obtained from the same ATP concentration but independent measurement. The red line is a fit to a Michaelis-Menten function.
(E) Sample structure for single-molecule four-color FRET experiment.
(F) Representative FRET time traces of FRET1 (FRET between Alexa 488 and Cy5) and FRET2 (FRET between Cy3 and Cy7) in the presence of 1 mM ATP-Mg2+. Time delay between FRET1 and FRET2 jumps in the regression phase (Δt1, n = 66) and that between FRET2 and FRET1 drops in the recovery phase (Δt2, n = 57) are clear.
(G and H) Distribution of Δt1 (G) and Δt2 (H) in the presence of 1 mM ATP-Mg2+.
See also Figures S3 and S4.
Because all experiments were performed in the absence of free WRN as shown in Figure 1B, we concluded that a single functional complex of fork regression machinery of WRN was driving the repetitive fork regression. At this point, the oligomeric state of the active WRN complex on replication forks remains to be determined (Compton et al., 2008; Huang et al., 2000; Xue et al., 2002). As a possible model, we speculated that WRN might switch its branch migration direction upon encounters with heterologous sequences and replication fork structures. To test this hypothesis, we prepared a DNA fork with an Alexa 488/Cy5 FRET pair at the proximal end and a Cy3/Cy7 FRET pair at the distal end (Figure 4E), and performed single-molecule four-color FRET experiments (Lee et al., 2010a). When all of the fluorophores interact with each other, six inter-fluorophore distances can be measured using single-molecule four-color FRET. In the current design, however, Alexa 488/Cy5 and Cy3/Cy7 FRET pairs are positioned far away, and they can be considered as two independent FRET pairs. Therefore, the Alexa 488/Cy5 FRET pair and the Cy3/Cy7 FRET pair are supposed to report the arrival/departure of the branchpoint at the proximal and distal ends, respectively. Figure 4F shows representative time traces of the Alexa 488/Cy5 FRET (FRET1) and the Cy3/Cy7 FRET (FRET2). Consistent with the progressive junction movement before migration direction switching, FRET1 appeared earlier than FRET2 in the regression phase, and disappeared later than FRET2 in the recovery phase. The time delay histograms in the regression phase (Δt1, n = 66) and the recovery phase (Δt2, n = 57) collected from more than 30 molecules were similar (Figures 4G and 4H), indicating symmetric operation of WRN in the regression and recovery phases. We would like to re-emphasize that the four-color FRET experiments were performed without free WRN in solution. Therefore, single WRN complex migrates the Holliday junction back and forth in a symmetric manner.
Branch Migration Reversal at Short Heterologous Sequences
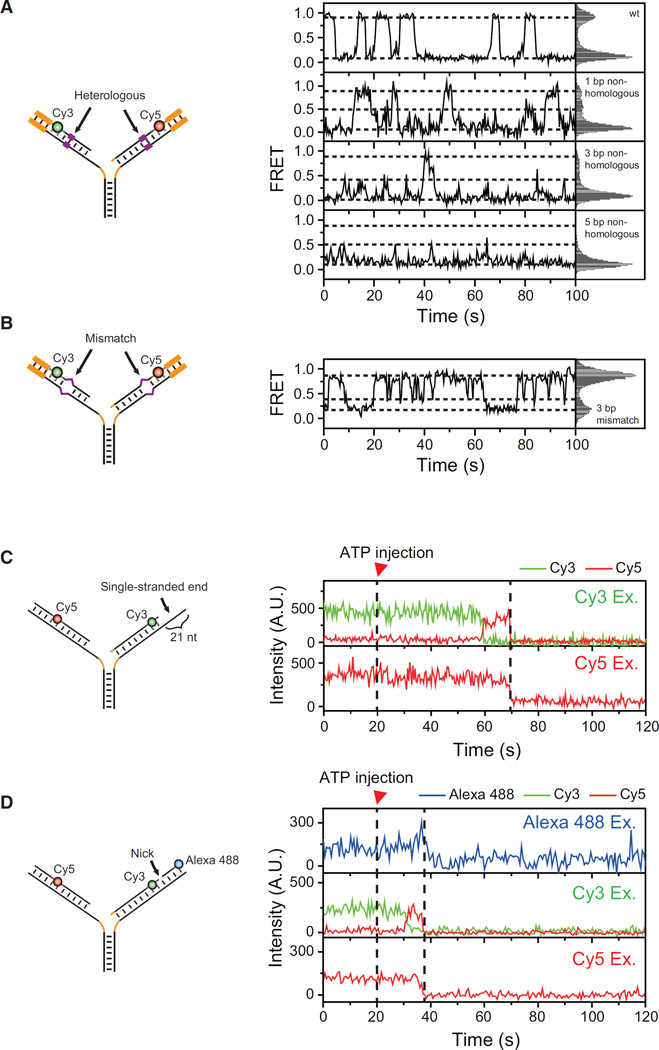
Short heterologous sequences are frequently encountered during branch migration, especially in mitotic recombination. Single-stranded breaks are a ubiquitous defect of the DNA helix. It has been shown that even a single heterologous base pair is able to block the spontaneous branch migration of a four-way junction (Biswas et al., 1998), whereas single-stranded breaks do not hinder spontaneous branch migration (Palets et al., 2010). Branch migration catalyzed by T7 gp4 was stalled by a single heterologous base pair (Rasnik et al., 2008). We investigated how these DNA structural features affected the branch migration reversal of WRN.
First, we introduced short heterologous sequences in the middle of the substrate that was used in Figure 4 (Figure 5A, left). The appearance of the new middle FRET state indicated that even a single heterologous base pair was a quite efficient inducer of branch migration reversal (Figure 5A, right, and Figure S4). However, when we introduced mismatches in the leading and lagging arms that were base-paired after fork regression (Figure 5B, left), branch migration reversal was not observed in the regression phase but only in the recovery phase (Figure 5B, right). Therefore, mismatches did not induce branch migration reversal. We also found that neither a long single-stranded region (Figures 5C and S5A) nor a nick (Figures 5D and S5B) induced branch migration reversal.
Figure 5. Branch Migration Reversal at Different DNA Structures.
(A) Branch migration reversal at short heterologous sequences. Left: a cartoon of sample structures. Short heterologous sequences (purple) are introduced in the middle of the leading and lagging arms. Right: representative FRET time traces (left panels) and corresponding FRET histograms (right panels) for different lengths of the heterologous regions. Dashed lines were added as an eye-guide. The histograms were made from more than 20 molecules.
(B) The same as (A) for the sample with 3-bp mismatches in the leading and lagging arms that are base-paired after fork regression.
(C) Branch migration through a long single-stranded region. Left: sample structure. Right: representative fluorescence intensity time traces of Cy3 (green) and Cy5 (red) at Cy3 excitation (top) and Cy5 excitation (bottom).
(D) Branch migration through a single-stranded break. Left: sample structure. Right: representative fluorescence intensity time traces of Alexa 488 (blue), Cy3 (green), and Cy5 (red) at Alexa 488 excitation (top), Cy3 excitation (middle), and Cy5 excitation (bottom).
See also Figures S4 and S5.
DISCUSSION
The repetitive translocation activities of various proteins have been reported (Abbondanzieri et al., 2008; Johnson et al., 2007; Myong et al., 2005, 2007; Yodh et al., 2009) and diverse biological functions of these repetitive motions have been suggested. We observed that WRN regresses replication forks in a repetitive fashion. Furthermore, it used the energy of ATP hydrolysis for all three steps required for repetitive fork regression: the formation of the Holliday junction, branch migration, and reversal of the migration. What is the biological function of the repetitive fork regression of WRN, which can be viewed as a seemingly wasteful behavior? During the reactivation of stalled replication forks via regression, a Holliday junction is a transient structure, which should be converted into a three-way junction for the reactivation of DNA replication after DNA repair or error-free synthesis. Although it has been reported that various ATPases with replication fork regression activity can also convert a Holliday junction into a functional replication fork (Bugreev et al., 2011; Machwe et al., 2011; Manosas et al., 2012), our study is a first demonstration that a single functional unit of WRN can dynamically interconvert a DNA substrate between a replication fork and a Holliday junction. In this way, we speculate that WRN provides time windows for the assembly of repair/lesion bypass complexes and for the restart of DNA replication after lesion repair.
Heterologous bases are expected to be rare during replication fork regression. What can be a biologically more relevant candidate to induce migration direction reversal? Firstly, it has been shown that T4 phage helicase UvsW switches its migration direction at random positions (Manosas et al., 2012). Although it was rare, we also observed that repetitive fork regression was still observed with fully homologous DNA substrates (Figure S5C), suggesting that WRN can also stochastically switch its branch migration direction at random positions. Secondly, it has been shown that protein road blocks hinder branch migration catalyzed by molecular motors (Achar et al., 2011). In eukaryotes, replication fork regression occurs in the context of chromatin. Therefore, it will be interesting to see whether WRN switches its migration direction at protein road blocks such as nucleosomes.
WRN has 3′–5′ directionality. Then, how can WRN migrate a Holliday junction in both directions? It is possible that the repetitive branch migration involves the microscopic dissociation and rebinding of a single WRN with an opposite orientation. It has been reported that multimers of WRN bind to a Holliday junction (Compton et al., 2008). Therefore, it is also possible that different WRN monomers in the WRN oligomer are activated during the regression and recovery phases. More studies are required to determine the oligomeric state of the active WRN and how they cooperate during repetitive replication fork regression.
EXPERIMENTAL PROCEDURES
Enzyme Preparation
Recombinant His6-tagged WRN protein (WRN) was overexpressed in Sf9 insect cells and purified as previously described (Orren et al., 1999).
Preparation of Short Replication Forks
DNA strands were purchased from Integrated DNA Technologies (IDT). For dye labeling, oligonucleotides and amine-reactive fluorophores were mixed with a 1:20 M ratio, and incubated overnight at room temperature in a reaction buffer (100 mM Na2B4O7 [pH 8.5]), followed by ethanol precipitation, and dissolution in a 10 mM Tris buffer (pH 8.0) with 50 mM NaCl. The labeling efficiency obtained from the comparison of the dye and oligonucleotide absorbances was better than 90%. The model replication forks were annealed sequentially. First, the leading and lagging arms were annealed separately by cooling the mixture of the parental and daughter strands from 90°C to 4°C with −1°C/min velocity in a buffer containing 10 mM Tris-HCl (pH 8.0) and 50 mM NaCl. Next, the leading and lagging duplexes were mixed and cooled down from 50°C to 4°C with −1°C/min velocity.
Preparation of Long Fork Samples
Long (>300 bp) DNA duplexes were prepared as follows. First, we obtained 309-bp double-strand DNA consisting of 303 bp of the 601 sequence (pIDTSMART-AMP: 601_and_flanking) and 6 bp of an EcoRI recognition sequence by PCR. PCR product was treated with EcoRI (R0101S, New England Biolabs) to generate a single-strand overhang. The cleaved product was ligated with a short DNA duplex that has a single-stranded overhang complementary to the EcoRI cut sequence using overnight incubation with T4 DNA ligase (M0202S, New England Biolabs) at 16°C. The ligated product was purified using denaturing PAGE.
Single-Molecule FRET Experiment
Quartz slides and glass coverslips were cleaned using piranha solution (mixture of sulfuric acid and hydrogen peroxide), coated with an 80:1 mixture of poly-ethylene glycol (PEG) and biotin-PEG. A sample chamber was made by assembling a quartz slide and a glass coverslip using double-sided tape. To immobilize replication forks, streptavidin and biotinylated replication forks were injected sequentially into the sample chamber. The immobilized fork was incubated with 26 nM WRN in a standard buffer (a Tris-HCl buffer [pH 8.0] containing 1 mM MgCl2, 100 ng/ml BSA, 1 mM DTT, and 1 mM trolox) for 5 min. Fork regression reaction was initiated by injecting the standard buffer containing 1 mM ATP-Mg2, unless otherwise indicated, using an automated syringe pump system (Fusion 100, Chemyx). The volume of the detection chamber was less than 25 µl, and a flow rate of 25 µl/s was used, rendering the buffer exchange time less than a second. The time resolution of the experiments was 1 s. To reduce the photobleaching, the imaging buffer also contained an enzymatic oxygen scavenger system (1 mg/ml glucose oxidase, 0.8% glucose, 0.04 mg/ml catalase). As excitation sources, 473-nm (Cobolt Blues 50, Cobolt), 532-nm (Compass 215M, Coherent), and 640-nm (Cube 640C, Coherent) lasers were used. Fluorescence signals were collected through a water-immersion objective (UPlanSApo 603, Olympus), separated using two dichroic mirrors (635dcxr and 740dcxr, Chroma) and a mirror (BB01-E02, Thorlabs), and imaged on an EM-CCD camera (Ixon DV897, Andor). Scattered laser light was filtered out using a long-pass filter for 535-nm (LP03-532RU-25, Semrock) and a notch filter for 640-nm (NF03-633 E-25, Semrock). Data acquisition, FRET trace extraction, and data analysis were done using homemade programs written in Visual C++ (Microsoft), IDL (ITT), and MATLAB (MathWorks), respectively.
Supplementary Material
Highlights.
The fork regression of WRN is readily activated after lagging arm unwinding
WRN migrates a Holliday junction with 3.2 ± 0.03 bases/s velocity
WRN reverses branch migration direction at short heterologous sequences
The branch migration reversal of WRN is an active process
In Brief.
By using single-molecule fluorescence measurements, Shin et al. show that the Holliday junction formation, branch migration, and migration direction reversal of WRN are all ATP dependent, revealing that WRN uses the energy of ATP hydrolysis to actively coordinate the structural transitions of DNA.
Acknowledgments
This work was supported by the Creative Research Initiatives (2009-0081562) to S.H., Priority Research Center Program through NRF (2009-0094050) to B.A., and funds from the Intramural Program of the National Institute on Aging, National Institutes of Health USA to V.A.B. Funding for open access charge: Creative Research Initiatives.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.str.2016.06.004.
AUTHOR CONTRIBUTIONS
S.S., J.L., S.Y., and S.H. designed the experiments. S.S., J.L., and S.Y. performed the experiments and analyzed the data. T.K. prepared the recombinant proteins. S.S., V.A.B., B.A., and S.H. wrote the manuscript.
REFERENCES
- Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achar YJ, Balogh D, Haracska L. Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc. Natl. Acad. Sci. USA. 2011;108:14073–14078. doi: 10.1073/pnas.1101951108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Yamamoto A, Hsieh P. Branch migration through DNA sequence heterology. J. Mol. Biol. 1998;279:795–806. doi: 10.1006/jmbi.1998.1769. [DOI] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A, Hajdu I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell. Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Rossi MJ, Mazin AV. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 2011;39:2153–2164. doi: 10.1093/nar/gkq1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD. The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J. Biol. Chem. 2008;283:24478–24483. doi: 10.1074/jbc.M803370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Donaldson JR, Chow KH, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Dennis C, Fedorov A, Kas E, Salome L, Grigoriev M. RuvAB-directed branch migration of individual Holliday junctions is impeded by sequence heterology. EMBO J. 2004;23:2413–2422. doi: 10.1038/sj.emboj.7600249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Tatsumi M. Replicative bypass repair of ultraviolet damage to DNA of mammalian cells: caffeine sensitive and caffeine resistant mechanisms. Mutat. Res. 1976;37:91–110. doi: 10.1016/0027-5107(76)90058-0. [DOI] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. USA. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J. Mol. Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- Hohlbein J, Craggs TD, Cordes T. Alternating-laser excitation: single-molecule FRET and beyond. Chem. Soc. Rev. 2013;43:1156–1171. doi: 10.1039/c3cs60233h. [DOI] [PubMed] [Google Scholar]
- Hotchkiss RD. Models of genetic recombination. Annu. Rev. Microbiol. 1974;28:445–468. doi: 10.1146/annurev.mi.28.100174.002305. [DOI] [PubMed] [Google Scholar]
- Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3’–>5’ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DL, O’Donnell M. Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol. Cell. 2004;15:453–465. doi: 10.1016/j.molcel.2004.06.039. [DOI] [PubMed] [Google Scholar]
- Lee J, Lee S, Ragunathan K, Joo C, Ha T, Hohng S. Single-molecule four-color FRET. Angew. Chem. Int. Ed. Engl. 2010a;49:9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee J, Hohng S. Single-molecule three-color FRET with both negligible spectral overlap and long observation time. PLoS one. 2010b;5:e12270. doi: 10.1371/journal.pone.0012270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Kreuzer KN. Fork regression is an active helicase-driven pathway in bacteriophage T4. EMBO Rep. 2009;10:394–399. doi: 10.1038/embor.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Karale R, Xu X, Liu Y, Orren DK. The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) Holliday junctions to functional replication forks. Biochemistry. 2011;50:6774–6788. doi: 10.1021/bi2001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Perumal SK, Croquette V, Benkovic SJ. Direct observation of stalled fork restart via fork regression in the T4 replication system. Science. 2012;338:1217–1220. doi: 10.1126/science.1225437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p) ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu. Rev. Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palets D, Lushnikov AY, Karymov MA, Lyubchenko YL. Effect of single-strand break on branch migration and folding dynamics of Holliday junctions. Biophys. J. 2010;99:1916–1924. doi: 10.1016/j.bpj.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Rasnik I, Jeong YJ, McKinney SA, Rajagopal V, Patel SS, Ha T. Branch migration enzyme as a Brownian ratchet. EMBO J. 2008;27:1727–1735. doi: 10.1038/emboj.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair (Amst) 2008;7:1776–1786. doi: 10.1016/j.dnarep.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilenchik MM, Knudson AG., Jr Inverse radiation dose-rate effects on somatic and germ-line mutations and DNA damage rates. Proc. Natl. Acad. Sci. USA. 2000;97:5381–5386. doi: 10.1073/pnas.090099497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ratcliff GC, Wang H, Davis-Searles PR, Gray MD, Erie DA, Redinbo MR. A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3’ – 5’ exonuclease and 5’-protruding strand endonuclease activities. Biochemistry. 2002;41:2901–2912. doi: 10.1021/bi0157161. [DOI] [PubMed] [Google Scholar]
- Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 2009;28:405–416. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT. Branching out with DNA helicases. Curr. Opin. Genet. Dev. 2011;21:214–218. doi: 10.1016/j.gde.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.