Abstract
The mating-type region of the fission yeast Schizosaccharomyces pombe comprises three loci: mat1, mat2-P and mat3-M. mat1 is expressed and determines the mating type of the cell. mat2-P and mat3-M are two storage cassettes located in a 17 kb heterochromatic region with features identical to those of mammalian heterochromatin. Mutations in the swi6+, clr1+, clr2+, clr3+, clr4+ and clr6+ genes were obtained in screens for factors necessary for silencing the mat2-P–mat3-M region. swi6+ encodes a chromodomain protein, clr3+ and clr6+ histone deacetylases, and clr4+ a histone methyltransferase. Here, we describe the cloning and characterization of clr2+. The clr2+ gene encodes a 62 kDa protein with no obvious sequence homologs. Deletion of clr2+ not only affects transcriptional repression in the mating-type region, but also centromeric silencing and silencing of a PolII-transcribed gene inserted in the rDNA repeats. Using chromatin immunoprecipitation, we show that Clr2 is necessary for histone hypoacetylation in the mating-type region, suggesting that Clr2 acts upstream of histone deacetylases to promote transcriptional silencing.
INTRODUCTION
Mutant screens aimed at identifying silencing factors acting in the mating-type region of fission yeast have identified several evolutionary conserved proteins required for the formation of heterochromatin. The mating-type region contains three loci: the expressed mat1 locus that determines the mating type of the cell and the silent storage cassettes mat2-P and mat3-M (1). Cells switch mating type by transposition of genetic information from mat2-P or mat3-M to mat1 (2,3). The mating-type region is localized on chromosome II, the centromere-proximal mat1 locus being separated from mat2-P by a ∼15 kb region called L and mat3-M being separated from mat2-P by a ∼11 kb region called ‘K’. The K-region contains sequences with strong similarity to the repeats found in centromeric regions, a 4.3 kb segment of the K-region displaying 96% similarity to the repeats in cen2 (4). Transcription and meiotic recombination are inhibited in the K-region by a heterochromatic structure spanning ∼17 kb of DNA and flanked by two inverted repeats IR-L and IR-R (5–7). The IR-L and IR-R repeats establish the borders between transcriptionally repressed heterochromatin and active euchromatin. The heterochromatic region shares many features with heterochromatin of Drosophila or mammals, including histone hypoacetylation, histone H3 lysine 9 methylation and association with a chromodomain protein, Swi6, bound to the modified histone H3 (6,8–11). The surrounding euchromatin on the other hand shares the features of active chromatin of higher eukaryotes with acetylated histones and histone H3 methylated at lysine 4 (6,10). Trans-acting factors required for silencing in the mating-type region were identified in four different mutant screens (12–15). These screens led to propose that Clr1, Clr2, Clr3, Clr4, Clr6 and Swi6 play a role in heterochromatin formation in Schizosaccharomyces pombe. Other factors such as Rik1 and Chp2 were identified by other approaches (13,16). Several of these trans-acting factors are also central to the formation of heterochromatic structures at the centromeres and telomeres of S.pombe (17–19). In addition, Clr3, Clr4, Swi6 and Chp2 are required for transcriptional silencing of PolII-transcribed genes inserted in the rDNA repeats (11,16). Clr3 and Clr6 belong to a family of histone deacetylases (15). Clr3 binds directly to the mating-type region and preferentially deacetylates lysine 14 of histone H3 (11). Clr4 is a methyltransferase that acts specifically on lysine 9 of histone H3 thereby forming a binding site for Swi6 (9,20,21). Moreover, it has recently been shown that a nucleosome remodeling factor, Hrp3 and an NAD+-dependent histone deacetylase, Sir2, are needed for transcriptional repression in the mating-type region (22,23).
Here, we describe the cloning and sequencing of the gene encoding the previously uncharacterized silencing factor Clr2. The clr2+ open reading frame (ORF) encodes a novel type of silencing factor with no obvious sequence homologs. We created a clr2 deletion allele that allowed us to investigate the contribution of Clr2 to transcriptional silencing at several chromosomal locations and we asked whether silencing is dependent on the concentration of Clr2 in the cell. Finally, we examined histone acetylation levels in clr2Δ cells, thereby gaining insight into the specific molecular defects caused by the lack of Clr2.
MATERIALS AND METHODS
Media
Media were prepared as in (24).
Cloning of clr2+
The strain PB1 (Table 1) was transformed with a genomic library in vector pDB248′ (25). Transformed cells were selected on PM-leu plates for complementation of the S.pombe leu1-32 mutation by the Saccharomyces cerevisiae LEU2 gene in pDB248′. After 6 days at 30°C Leu+ colonies were replica plated onto sporulation media and incubated at room temperature (20–25°C) for 3 days. The colonies were stained by iodine vapor, and dark-staining colonies were streaked on PM + FOA. A plasmid, pPB9, was recovered from a dark-staining FOA-resistant isolate. A 6 kb HindIII–XbaI fragment was subcloned from pPB9 into pDW232 (26) creating pPB11, which could rescue the Clr2 phenotype upon retransformation. The insert in pPB11 was sequenced.
Table 1. List of S.pombe strains.
| Strain | Genotype | Source |
|---|---|---|
| FY412 | h+, cc2(SphI)::ura4+, ura4-DS/E, leu1-32, ade6-M210 | (42) |
| FY498 | h+, imr1R(NcoI)::ura4+oriI, ura4-DS/E, leu1-32, ade6-M210 | (17) |
| FY597 | h90, mat3-M(EcoRV)::ura4+, ura4-DS/E, leu1-32, ade6-M210 | (17) |
| FY501 | h+, imr1L(NcoI)::ura4+oriII, ura4-DS/E, leu1-32, ade6-M210 | (17) |
| FY986 | h+, otr1L(dg1a/HindIII)::ura4+ oriII, ura4-DS/E, leu1-32, ade6-M210 | (17) |
| FY520 | h+, Ch16 m23::ura4+-TEL[72], ura4-DS/E, leu1-32, ade6-M210, (Ch16 ade6-M216) | (35) |
| FY611 | hA, Ch16 m23::ura4+-TEL[72], ura4-DS/E, leu1-32, ade6-M210, (Ch16 ade6-M216), swi6-S115 | (17) |
| KE78 | h90, clr2-E22, ura4-D18, ade6-M210 | (13) |
| PG9 | h90, mat3-M(EcoRV)::ura4+, ura4-D18, leu1-32, ade6-M216 | (12) |
| PG940 | h90, mat3-M(EcoRV)::ura4+, clr2-760, ura4-D18, leu1-32, ade6-M210 | (14) |
| PG970 | h90, mat3-M(EcoRV)::ura4+, clr2-785, ura4-D18, leu1-32, ade6-M210 | (14) |
| PG3088 | h90, mat3-M(EcoRV)::ura4+, clr2-E22, ura4-D18, leu1-32, ade6-M210 | This study |
| PG3089 | h90, mat3-M(EcoRV)::ura4+, clr4::LEU2, ura4-D18, leu1-32, ade6-M210 | This study |
| PG3090 | h+, imr1R(NcoI)::ura4+oriI, clr2-E22, ura4-DS/E, leu1-32, ade6-M210 | This study |
| PG3091 | h+, imr1R(NcoI)::ura4+oriI, clr4::LEU2, ura4-DS/E, leu1-32, ade6-M210 | This study |
| PG3092 | h+, cc2(SphI)::ura4+, clr2-E22, ura4-DS/E, leu1-32, ade6-M210 | This study |
| PG3093 | h+, cc2(SphI)::ura4+, clr4::LEU2, ura4-DS/E, leu1-32, ade6-M216 | This study |
| PG3094 | h+, leu1/Ylp2.4pUCura4+-7, clr2-E22, ura4-DS/E leu1-32 ade6-M216 | This study |
| PG3095 | h+, leu1/Ylp2.4pUCura4+-7, clr4::LEU2, ura4-DS/E leu1-32 ade6-M216 | This study |
| Hu393 | h+, leu1/Ylp2.4pUCura4+-7, ura4-DS/E leu1-32 ade6-M216 | (11) |
| Hu582 | h90, mat3-M(EcoRV)::ura4+, clr2::his7, his7-366, ura4-DS/E, leu1-32, ade6-M210 | This study |
| PB1 | h90, clr2-E22, mat3-M(EcoRV)::ura4+, ura4-D18, leu1-32, ade6-M216 | This study |
| PB65 | h90, rad1-1, lys1-1 | This study |
| PB101 | h90, ura4-D18, leu1-32, his7-366, ade6-M210 | This study |
| PB102 | h90, ura4-D18, leu1-32, his7-366, ade6-M216 | This study |
| PB140 | h90, mat3-M(EcoRV)::ade6+oriI, leu1-32, ade6-M210 | This study |
| PB141 | h90, mat3-M(EcoRV)::ade6+oriI, ura4-D18, leu1-32, ade6-M210 | This study |
| PB159 | h90, clr3-E36, mat3-M(EcoRV)::ade6+oriI, leu1-32, ade6-M210 | This study |
| PB169 | h90, clr4-S5, mat3-M(EcoRV)::ade6+oriI, leu1-32, ade6-M210 | This study |
| PB173 | h90, clr2-E22, mat3-M(EcoRV)::ade6+oriI, leu1-32, ade6-M210 | This study |
| PB180 | h90, swi6-S115, mat3-M(EcoRV)::ade6+oriI, leu1-32, ade6-M210 | This study |
| PB186 | h+N, clr2::his7+, ura4-D18, leu1-32, his7-366, ade6-M216 | This study |
| PB202 | h90, rik1::ura4+, mat3-M(EcoRV)::ade6+oriI, leu1-32, ade6-M210 | This study |
| PB250 | h90, clr1-6, mat3-M(EcoRV)::ade6+oriI, leu1-32, ade6-M210 | This study |
| PB254 | h90, clr2::his7+, ura4-D18, leu1-32, his7-366, ade6-M216 | This study |
| PJ32 | hA, cc2(SphI)::ura4+, clr2::his7, ura4-DS/E, leu1-32, his7-366, ade6-M210 | This study |
| PJ34 | hA leu1/Ylp2.4pUCura4+-7, clr2::his7 ura4-DS/E leu1-32, his7-366 ade6-M210 | This study |
| PJ42 | hA, imr1R(NcoI)::ura4+oriI, clr2::his7, ura4-DS/E, leu1-32, his7-366, ade6-M210 | This study |
hA, Mating type is ambiguous because the silent mating-type loci are deregulated in this background.
Deletion of clr2+
The primers PP45: 5′-CCCCTGCAGTTTGCTTTTATACCTCATTTATC-3′ and PP46: 5′-CCCCTGCAGTAGCTACTAAAAATGACAATAGC-3′ were used to amplify a 732 bp fragment upstream of the clr2 ORF. The primers PP47: 5′-GCGCCCGGGTAATAAGTATGGCAGATAAAATTAG-3′ and PP48: 5′-GCGCCCGGGACTCACATTACACTTTGTTCTC-3′ were used to amplify a 660 bp fragment downstream of the ORF. Restriction sites used for cloning are underlined. Both fragments were PCR amplified with 15 cycles of (2 min 94°C, 2 min 55.5°C, 1 min 72°C) using pPB11 as a template, and Taq (Perkin Elmer) as polymerase. The PCR products were cloned into pGEMR-T Easy Vector (Promega). The resulting plasmids were digested with PstI and XmaI, respectively. The PstI and XmaI fragments were cloned into plasmid pEA2 (27) PstI and XmaI site respectively. The resulting plasmid pPB48 was digested with KpnI and SphI and the 4.5 kb fragment was used to transform the PB101XPB102 diploid.
Generation of Clr2 overexpression plasmids
The 1611 bp clr2+ ORF was amplified by PCR from pPB11 (10 cycles of 2 min at 94°C, 2 min at 62.5°C, 1 min 30 s at 72°C) using the Taq polymerase (Perkin Elmer) and the primers PP49: 5′-CCCGGATCCATGCCTGCTATTACTTGTGTTTG-3′ and PP50 5′-CCCGGATCCTTATTACATTACAACTGCTGACACC-3′. The PCR product was cloned into pGEMR-T Easy Vector (Promega). The resulting plasmid was digested with BamHI and the 1.6 kb fragment was cloned into the pREP82X (28) BamHI site, giving plasmid pPB40. The BamHI fragment containing the clr2+ gene from pPB40 was cloned into pREP4X, pREP3X and pREP81X (28) producing the plasmids pPB63, pPB57 and pPB59.
Spot tests
Overnight cultures were diluted in steps of 10, and drops of 5 μl were applied to the plates.
Preparation of polyclonal antibodies against Clr2
A BamHI fragment comprising the clr2+ ORF was obtained from plasmid pPB40 and cloned into the BamHI site of vector pGEX-2T (Pharmacia Biotech) generating pPB46, where clr2+ is tagged in the C-terminus with glutathione S-transferase (GST). Escherichia coli DH5 was induced to express Clr2–GST. Cells were harvested and inclusion bodies were prepared using a protocol based on (29). The Clr2–GST protein was further purified on a 10% SDS–PAGE gel. The gel was stained with 4 M NaCH3CO2 and the white band corresponding to Clr2–GST was removed with a scalpel. The gel was fragmented mechanically and the Clr2–GST protein was electroeluted in an Isco, Model 1750, Electrophoretic Sample Concentrator O.N. at 1 W. Two rabbits were injected with Clr2–GSTp. Antibodies directed against Clr2 were purified in a column where Clr2–GST had been coupled to a matrix of Affigel 15 (BioRad). The antibodies were concentrated by centrifugation in a microcon 10 tube (Amicon) to a concentration of 0.8 mg/ml.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitations were performed according to (11). Antibodies were purchased from Upstate.
RESULTS
Cloning of the clr2+ gene
The clr2-E22 homothallic strain PB1 (Table 1) was transformed with an S.pombe genomic library. The phenotype of PB1 offers two strategies for cloning the clr2+ gene. First, PB1 colonies display an aberrant sporulation phenotype due to the simultaneous expression of mat2-P and mat3-M in haploid cells, an abnormal event that causes haploid sporulation and reduces the efficiency of mating. As S.pombe spore asci contain starch molecules that are stained black by iodine vapors, this sporulation phenotype can be assayed at the colony level: clr2-E22 colonies are stained gray by iodine vapors as compared to the black wild-type staining. Second, a ura4+ gene inserted near the mat3-M cassette in PB1 is derepressed by the clr2-E22 mutation. This renders PB1 sensitive to the drug 5-fluoroorotic acid [5-FOA; (30)]. Complementation of the clr2-E22 mutation can be expected to partially or totally restore the wild-type iodine staining and FOA resistance.
Out of 45 000 transformants, one transformant was obtained where the clr2-E22 phenotype described above was complemented. Restoration of a wild-type sporulation phenotype was confirmed by microscopy (Figure 1). The complementing plasmid was rescued from PB1 and its insert was sequenced, identifying an ORF of 1611 bp. This ORF corresponds to a previously uncharacterized gene in the Sanger Centre S.pombe Gene Database (locus C1B3.17, EMBL accession no. Z98598). The 1611 bp ORF is capable of encoding an acidic protein (pI = 5.6) of 537 amino acids (aa) with a predicted MW of 62 kDa (Swiss-Prot accession no. O13881). No obvious homologous protein was found when BLAST searches were performed at the National Center for Biotechnology Information (NCBI). No described motifs were identified using the Swiss Prot or Pfam domain databases. The physical distance between C1B3.17 and the rad1 gene is ∼11.2 kb (31). The genetic distance between rad1-1 (32) and clr2-E22 was determined by tetrad dissections following a cross between PB1 (clr2-E22) and PB65 (rad1-1). This distance was found to be 1.9 cM. A genetic distance of 1.9 cM in S.pombe is in good agreement with a physical distance of 11.2 kb, indicating C1B3.17 is clr2+ and not an extragenic suppressor. We found during the course of the linkage analysis that the clr2-E22 mutant strain is not sensitive to ultraviolet (UV) irradiation.
Figure 1.
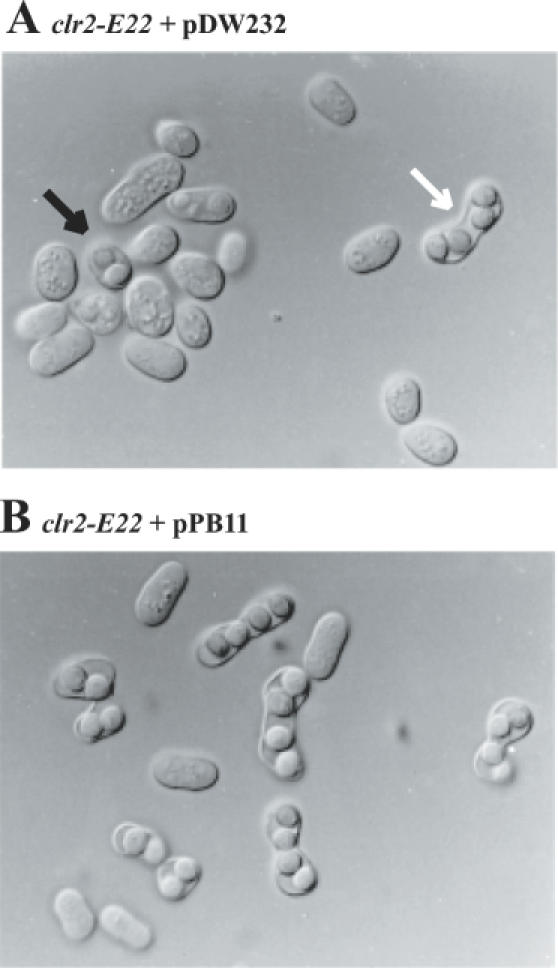
Complementation of the sporulation defect in a clr2-E22 mutant strain with cloned DNA. A black arrow points at a product of haploid meiosis and a white arrow points at a zygotic ascus in (A). Strain KE78 (clr2-E22) was transformed with vector DNA, pDW232 (A) or the clr2+-expressing plasmid pPB11 (B).
Analysis of clr2 mutant alleles
The clr2-E22 mutation (13) was sequenced as well as two other mutations in clr2 described in (14), clr2-760 and clr2-785. The clr2-E22 mutation is a 16 bp insertion at nucleotide 205 creating a frame shift. The clr2-760 and clr2-785 mutations are both non-sense mutations at nucleotide 968 or 903 respectively, introducing a stop instead of a tryptophan at codon 323 or 301. We conclude from our analysis of these truncated alleles that all or parts of the C-terminal 214 amino acids are necessary to make a functional Clr2 protein. The C-terminus of Clr2 also proved sensitive to other modifications since fusions to a green fluorescent protein (GFP) or hemagglutinin (HA) tag resulted in non-functional proteins (data not shown).
Deletion of clr2+
A construct was made with the S.pombe his7+ gene (27) flanked by sequences naturally found on the side of the clr2+ ORF. A linear fragment was used to transform a diploid (PB101 X PB102) homozygous for a his7-366 mutation in the endogenous his7 gene. Stable histidine phototrophic transformants were examined by tetrad dissection. A 2:2 segregation pattern was obtained for both the His+/His− and the Clr2+/Clr2− phenotypes, the Clr2− phenotype co-segregating with the His+ phenotype in 18 tetrads examined. A Southern blot prepared with one of the diploids and its offspring confirmed integration of the his7+ gene into the clr2+ locus (data not shown). The clr2Δ segregants being viable shows that clr2+ is not an essential gene. The 1611 bp clr2+ ORF was cloned into the pREP82X vector to place the ORF under the control of the weakest nmt1 (no message in thiamine) promoter (28). The resulting plasmid, pPB40, was able to rescue the sporulation defects of a strain where the entire reading frame of clr2+ had been deleted, thereby showing that the 1611 bp ORF encodes a functional Clr2 protein.
Transcriptional silencing in clr2Δ strains
Spontaneous or EMS-induced mutations in the clr2 gene were shown to affect transcriptional silencing in the mating-type region, in the centromeric imr repeats, and near telomeres (13,14,17). In order to determine whether the clr2 null mutation had a more severe silencing phenotype than these mutations, the clr2Δ deletion and the clr2-E22 mutation were combined with a ura4+ reporter gene inserted in the centromere or mating-type region (Figure 2). In addition, we decided to investigate whether Clr2 contributes to the more recently described silencing of PolII-transcribed genes inserted in the S.pombe ribosomal DNA (rDNA) (16). To this end, the clr2Δ and the clr2-E22 mutant alleles were each crossed into a strain containing an rDNA::ura4+ reporter. For comparison with a more extensively characterized silencing factor a clr4Δ (33) allele was also combined with each of the ura4+ reporter genes examined here. Expression of ura4+ was estimated by plating serial dilutions of cells on selective plates (Figure 2). clr2+ strains with the ura4+ gene inserted in the mating-type region [mat3-M(EcoRV)::ura4+], centromeric repeats [imr1R(NcoI)::ura4+], centromeric central core [cen2 (SphI)::ura4+] or rDNA (rDNA::ura4+) all grew poorly on plates lacking uracil and they formed colonies on plates containing FOA consistent with a transcriptional repression of the ura4+ gene at these locations (Figure 2A–D, top rows). In contrast, when either clr2 mutant allele or the clr4Δ allele was combined with the mat3-M(EcoRV)::ura4+ reporter, cells grew well on plates lacking uracil but they did not grow on plates containing FOA indicating the mat3-M(EcoRV)::ura4+ reporter is largely derepressed in the mutant backgrounds (Figure 2A, second to fourth rows). Similarly, all strains with a trans-acting mutation and a ura4+ reporter gene in the imr centromeric repeats [imr1R(NcoI)::ura4+] grew well on plates lacking uracil and poorly on plates containing FOA (Figure 2B, second to fourth rows). The few large FOA-resistant papilli present in these mutant strains probably contain DNA rearrangements in which the ura4+ gene is lost, as observed in other silencing mutants (16). Transcription is less stringently repressed in the central core of S.pombe centromeres than in the flanking repeats (17). Consistently, we observed a modest repression of cen2 (SphI)::ura4+ in our wild-type background (Figure 2C, first row). This weak repression was partially dependent on Clr2 and Clr4, the clr4Δ allele having a somewhat more pronounced effect than either the clr2Δ or clr2-E22 allele (Figure 2C, second to fourth rows). Finally, cells in which the clr2 or clr4 mutant alleles were combined with the rDNA::ura4+ reporter grew well on plates lacking uracil and poorly on plates containing FOA indicating rDNA::ura4+ is derepressed in the mutant backgrounds. In summary, we found that Clr2 has an effect on transcriptional silencing in the mating-type region, the imr repeats and the central core of the centromere and it also has a clear role in the repression of PolII-transcribed genes inserted in the S.pombe rDNA. The extent to which Clr2 represses a reporter gene at these locations is comparable to the repression exerted by Clr4.
Figure 2.
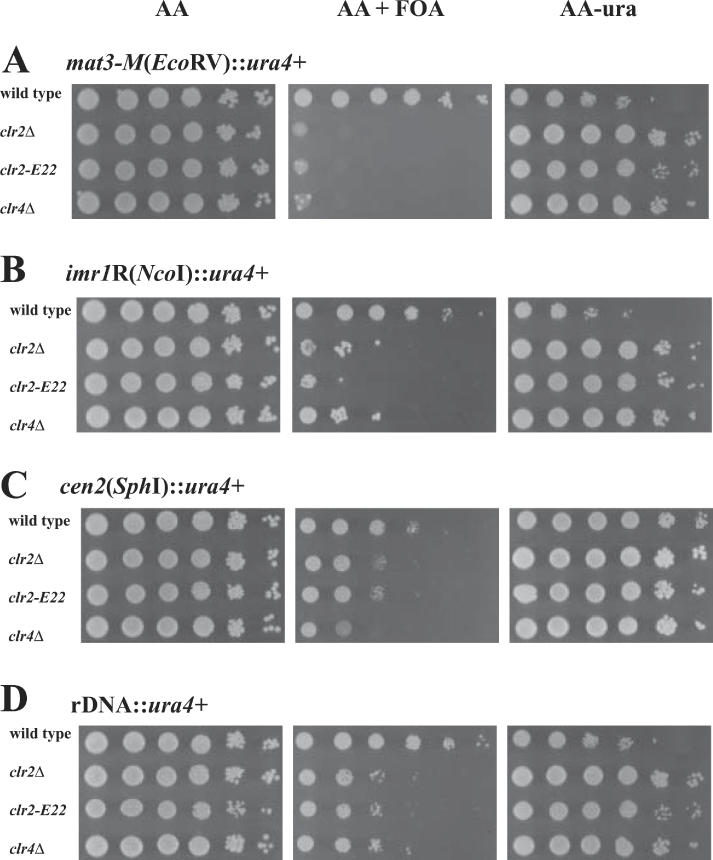
Clr2 is necessary for silencing in the mating-type region, centromere and the rDNA. Cells were serially diluted in steps of ten and spotted onto the indicated media. (A) The strains FY597 (wild type), Hu582 (clr2Δ), PG3088 (clr2-E22) and PG3089 (clr4Δ) have the ura4+ reporter gene inserted in the mating-type region [mat3-M(EcoRV)::ura4+]. (B) The strains FY498 (wild type), PJ42 (clr2Δ) PG3090 (clr2-E22) and PG3091 (clr4Δ) have the ura4+ reporter gene inserted in the repeats of centromere 1 [imr1R(NcoI)::ura4+]. (C) The strains FY412 (wild type), PJ32 (clr2Δ) PG3092 (clr2-E22) and PG3093 (clr4Δ) have the ura4+ reporter gene inserted into the central core of centromere 2 [cen2 (SphI)::ura4+]. (D) The strains Hu393 (wild type), PJ34 (clr2Δ), PG3094 (clr2-E22) and PG3095 (clr4Δ) have the ura4+ reporter gene inserted into the rDNA (rDNA::ura4+).
Analysis of the effect of Clr2 overexpression
It has been shown that overexpression of Swi6 increases transcriptional repression in the mating-type region (7,34). This finding prompted us to test whether overexpression of Clr2 would affect silencing. In order to monitor protein expression, we used antibodies produced against Clr2. The antibodies failed to recognize Clr2 on a western blot with a protein extract from a wild-type S.pombe strain. However, when Clr2 was overexpressed by transforming a wild-type strain with a plasmid where clr2+ is under the control of the weakest or the strongest nmt1 promoter a protein of the expected size of 62 kDa was detected (Figure 3).
Figure 3.
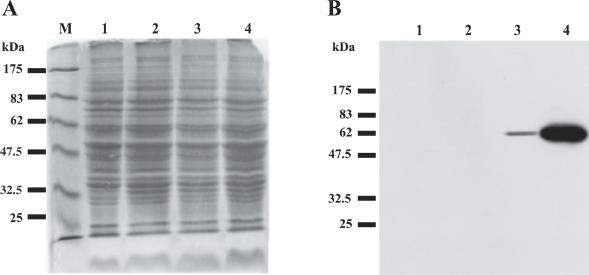
Antibodies against Clr2 recognize Clr2 from a total S.pombe protein extract when the protein is overexpressed. Protein extracts from S.pombe were (A) stained with Coomassie or (B) blotted and incubated with the Clr2p antibody. M, protein size markers; lane 1, PB254 (clr2Δ); lane 2, PB141 (clr2+) transformed with vector pREP82X; lane 3, PB141 (clr2+) transformed with plasmid pPB40 with the clr2+ gene under the control of the weakest nmt1 promoter; and lane 4, PB141 (clr2+) transformed with plasmid pPB63 where the clr2+ gene is under the control of the strongest nmt1 promoter.
The plasmids described above where clr2+ is under the control of the weakest or the strongest nmt promoter were transformed into strains carrying a ura4+ reporter gene inserted at two different positions in centromere 1 [imr1L(NcoI)] or [otr1L (HindIII)] (17), near a telomere, {Ch16 m23::ura4+-TEL[72]}, (35), or in the mating-type region [mat3-M(EcoRV)::ura4+], (12). Cell suspensions of the transformants were diluted and spotted onto selective plates where the expression of the reporter gene could be estimated. Overexpression of Clr2 had no detectable effect on the expression of the reporter genes at any of the chromosomal locations tested (data not shown). We also investigated whether overexpression of Clr2 could suppress mutations in other trans-acting factors necessary for silencing in the mating-type region. We found that overexpression of Clr2 did not restore the repression of the mat3-M(EcoRV)::ade6+ reporter gene (24) in a clr1-6, clr3-E36, clr4-S5, rik1::ura4, or swi6-S115 mutant background, indicating overexpression of Clr2 does not suppress mutations in these silencing factors. Furthermore, we investigated whether over-expression of Clr2 could restore telomeric silencing in a strain with a swi6-S115 mutation and found that this was not the case. These results all point to silencing not being sensitive to Clr2 dosage.
Deletion of clr2+ results in hyperacetylation of histones in the mating-type region
Silencing being affected in the mating-type region of clr2Δ cells suggested that the chromatin structure of that region might be altered in the mutant cells. Histones are hyopacetylated in the mating-type region of wild-type cells (11). We asked whether Clr2 was necessary for this low acetylation level by performing Chromatin Immunoprecipitations (ChIP) with antibodies directed against histone H3 acetylated at lysine 14 (α-H3AcK14), histone H4 acetylated at lysine 8 (α-H4AcK8), or histone H4 acetylated at lysine 12 (α-H4AcK12). Chromatin extracts were prepared from strains with a ura4+ reporter gene near the mat3-M cassette and a truncated ura4 allele, ura4-DS/E, at the endogenous ura4 locus (Figure 2). Figure 4 shows the result of the ChIP assays. The top panel displays results obtained with the wild-type strain (FY597), and the bottom panel displays results obtained with the clr2Δ strain (Hu582) (Table 1). The acetylation levels of mat3-M(EcoRV)::ura4+ were low in the wild-type strain consistent with previous observations (11). Deletion of clr2+ resulted in increased acetylation of the three lysine residues examined, the histones associated with the mat3-M(EcoRV)::ura4+ reporter gene in clr2Δ cells being as highly acetylated as those associated with the ura4-DS/E control. We conclude that Clr2 is necessary for the low histone acetylation level observed in the mating-type region.
Figure 4.
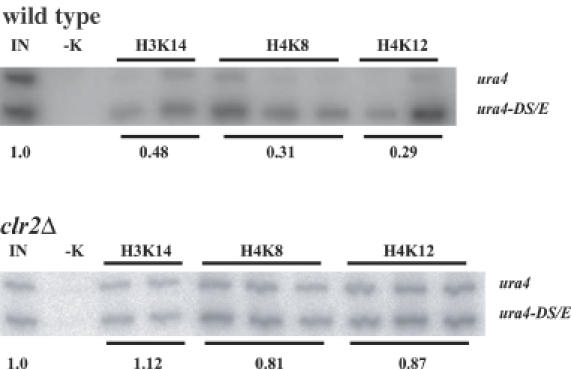
Histone acetylation levels in the mating-type region are elevated in the absence of Clr2. Radiolabeled PCR products amplifying a portion of the ura4 gene placed in the mating-type region (ura4+) or at its normal chromosomal location (ura4-DS/E) were obtained using total chromatin (IN) or immunoprecipitated chromatin. −K is a negative control where no antibody was added in the immunoprecipitation step. Antibodies recognizing specifically H3AcK14 (H3K14), H4AcK8 (H4K8) or H4AcK12 (H4K12) were used in the immunoprecipitations. Those were conducted in duplicate or triplicate as shown. The mean values of normalized ratios (ura4+/ura4-DS/E) are reported below the lanes for each antibody used. The strains used were FY597 (wild type) and Hu582 (clr2Δ).
DISCUSSION
Clr2 is a novel type of silencing factor
We found that the Clr2 silencing factor is encoded by a previously uncharacterized sequence orphan, designated as C1B3.17, in the S.pombe GeneBank at the Sanger Genome Sequencing Center. Most of the silencing factors that have been characterized in S.pombe are conserved in other organisms. Swi6 has homologs in the HP1 proteins found in Drosophila melanogaster and mammals (9,36). Clr4 also has homologs in Su(var)3-9p of D.melanogaster and Suv39h1 in mammals (20,21). Clr3 and Clr6 belong to a large conserved family of histone deacetylases (15,37). Other silencing factors in S.pombe contain recognizable motifs or domains. For example, Clr1 and Rik1 contain respectively three C2H2 Zinc fingers and a β-propeller domain of the UV-DDB-127 family, indicating these proteins might interact with DNA/RNA (G. Thon, unpublished data) (38,39). Chp2 contains a chromo- and chromoshadow domain (16). These similarities underscore the mechanistic conservation of heterochromatin formation in eukaryotes. Sequence orphans are present in the genomes of all organisms. In the S.pombe proteome there are 454 sequence orphans among 4965 proteins in total, or in other words ∼9% of the proteins encoded in its genome are unique to S.pombe (Sanger Genome Sequencing Center). Orphans might perform functions that are specific to the organism in which they are found, or they might perform conserved functions that do not require sequence conservation. Understanding their origin and evolution is a challenging task. Characterizing orphans, for whom a biological function has been uncovered, such as Clr2, will help understand the place of these proteins in evolution.
Clr2 is necessary for silencing at various chromosomal locations
We determined that clr2+ is not an essential gene. Mutated clr2 alleles were previously shown to alleviate silencing in the mating-type region and in the imr repeats of the centromere (13,14,17). Deletion of clr2+ caused silencing defects similar to those caused by deletion of the histone methyltransferase gene clr4+ at all chromosomal locations tested; the mating-type region, the imr repeats, the central core of centromere 2, and the rDNA (Figure 2), indicating that Clr2 is a general mediator of transcriptional silencing in S.pombe. The effects observed at the central core of centromere 2 [cen2 (SphI)::ura4+] are noticeably smaller than those observed at the other locations. Previous studies have shown that the central cores of S.pombe centromeres are associated with proteins not found in the flanking centromeric repeats such as Mis6, Sim4 and Cnp1 (40,41). Conversely, Swi6 is physically associated with centromeric repeats rather than with the central core (40). In spite of the undetectable association of Swi6 with central core sequences however, Swi6 and other heterochromatic proteins have some influence on the transcriptional repression occurring in central cores since a derepression of ura4+ placed within the central core of cen1 [TM1(NcoI)::ura4+] can be detected by growth assays in swi6Δ, rik1-304 or clr4-S5 mutant backgrounds (17). Our observations are consistent with Clr2 having an effect of the same magnitude as these other proteins involved in heterochromatin formation, whether the effect is directly exerted on the central core, or whether it is an indirect effect reflecting changes occurring in the neighboring heterochromatin.
Overexpression of Clr2 does not affect silencing
Transcriptional silencing in the mating-type region varies with the concentration of Swi6 or Clr4 in the cell. High dosage of the Swi6 protein strengthens silencing, whereas a high dosage of Clr4 alleviates silencing. Overexpression of other silencing factors such as Clr1 or Clr3 has no effect on mating-type silencing (7,20,34). Swi6 is a structural component of heterochromatin (34). If Clr2 also enhanced silencing when overexpressed it could be argued that Clr2 is a structural component of chromatin like Swi6. Here we report that overexpression of Clr2 does not affect the degree of silencing in the mating-type region, centromeres or telomeres. This indicates that the amount of Clr2 is not the limiting factor for heterochromatin formation and that Clr2 does not titrate other factors away from their point of action.
Clr2 promotes histone deacetylation in the mating-type region
In a strain with a clr2Δ deletion, the histones in the mating-type region are hyperacetylated compared to a wild-type strain (Figure 4). This indicates that histone deacetylases such as Clr3, Clr6 and Sir2 are not operating properly in the absence of Clr2, possibly because they are not recruited to the normally silent region. Alternatively, these enzymes might be properly recruited to elements nucleating heterochromatin formation, but heterochromatin might be unable to spread in the absence of Clr2. Histone hypoacetylation is not required solely for transcriptional silencing in the mating-type region, but it also mediates silencing at centromeres and in the rDNA repeats, which suggests that Clr2 affects silencing in these regions via increased histone acetylation levels as well. It is difficult to speculate about the precise mechanism of action of Clr2 since the protein sequence does not give any clues. Genetic selections using the S.pombe clr2 mutant strains described here might help identify functional homologs from other species, with unrelated or largely diverged sequences.
Acknowledgments
ACKNOWLEDGEMENTS
P.B. acknowledges a grant from the Swedish research council (2002-4948); G.T. acknowledges grants from the Novo Nordisk Foundation and from the Danish Research Council.
DDBJ/EMBL/GenBank accession no. Z98598
REFERENCES
- 1.Kelly M., Burke,J., Smith,M., Klar,A. and Beach,D. (1988) Four mating-type genes control sexual differentiation in the fission yeast. EMBO J., 7, 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beach D.H. and Klar,A.J. (1984) Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J., 3, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcangioli B. and Thon,G. (2004) Mating-type cassettes: structure, switching and silencing. In Egel,R. (ed.), The Molecular Biology of Schizosaccharomyces pombe. Springer, New York, NY, pp. 129–148. [Google Scholar]
- 4.Grewal S.I. and Klar,A.J. (1997) A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics, 146, 1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egel R., Willer,M. and Nielsen,O. (1989) Unblocking of meiotic crossing-over between the silent mating-type cassettes of fission yeast, conditioned by the pleiotropic mutant rik1. Curr. Genet., 15, 407–410. [Google Scholar]
- 6.Noma K., Allis,C.D. and Grewal,S.I. (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science, 293, 1150–1155. [DOI] [PubMed] [Google Scholar]
- 7.Thon G., Bjerling,P., Bunner,C.M. and Verhein-Hansen,J. (2002) Expression-state boundaries in the mating-type region of fission yeast. Genetics, 161, 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeppesen P. and Turner,B.M. (1993) The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell, 74, 281–289. [DOI] [PubMed] [Google Scholar]
- 9.Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- 10.Litt M.D., Simpson,M., Gaszner,M., Allis,C.D. and Felsenfeld,G. (2001) Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science, 293, 2453–2455. [DOI] [PubMed] [Google Scholar]
- 11.Bjerling P., Silverstein,R.A., Thon,G., Caudy,A., Grewal,S. and Ekwall,K. (2002) Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol., 22, 2170–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thon G. and Klar,A.J. (1992) The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics, 131, 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekwall K. and Ruusala,T. (1994) Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics, 136, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thon G., Cohen,A. and Klar,A.J. (1994) Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics, 138, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grewal S.I., Bonaduce,M.J. and Klar,A.J. (1998) Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics, 150, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thon G. and Verhein-Hansen,J. (2000) Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics, 155, 551–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allshire R.C., Nimmo,E.R., Ekwall,K., Javerzat,J.P. and Cranston,G. (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev., 9, 218–233. [DOI] [PubMed] [Google Scholar]
- 18.Ekwall K., Javerzat,J.P., Lorentz,A., Schmidt,H., Cranston,G. and Allshire,R. (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science, 269, 1429–1431. [DOI] [PubMed] [Google Scholar]
- 19.Ekwall K., Nimmo,E.R., Javerzat,J.P., Borgstrom,B., Egel,R., Cranston,G. and Allshire,R. (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell. Sci., 109, 2637–2648. [DOI] [PubMed] [Google Scholar]
- 20.Ivanova A.V., Bonaduce,M.J., Ivanov,S.V. and Klar,A.J. (1998) The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nature Genet., 19, 192–195. [DOI] [PubMed] [Google Scholar]
- 21.Rea S., Eisenhaber,F., O'Carroll,D., Strahl,B.D., Sun,Z.W., Schmid,M., Opravil,S., Mechtler,K., Ponting,C.P., Allis,C.D. et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- 22.Jae Yoo E., Kyu Jang,Y., Ae Lee,M., Bjerling,P., Bum Kim,J., Ekwall,K., Hyun Seong,R. and Dai Park,S. (2002) Hrp3, a chromodomain helicase/ATPase DNA binding protein, is required for heterochromatin silencing in fission yeast. Biochem. Biophys. Res. Commun., 295, 970–974. [DOI] [PubMed] [Google Scholar]
- 23.Shankaranarayana G.D., Motamedi,M.R., Moazed,D. and Grewal,S.I. (2003) Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol., 13, 1240–1246. [DOI] [PubMed] [Google Scholar]
- 24.Thon G., Bjerling,K.P. and Nielsen,I.S. (1999) Localization and properties of a silencing element near the mat3-M mating-type cassette of Schizosaccharomyces pombe. Genetics, 151, 945–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beach D., Piper,M. and Nurse,P. (1982) Construction of a Schizosaccharomyces pombe gene bank in a yeast bacterial shuttle vector and its use to isolate genes by complementation. Mol. Gen. Genet., 187, 326–329. [DOI] [PubMed] [Google Scholar]
- 26.Weilguny D., Praetorius,M., Carr,A., Egel,R. and Nielsen,O. (1991) New vectors in fission yeast: application for cloning the his2 gene. Gene, 99, 47–54. [DOI] [PubMed] [Google Scholar]
- 27.Apolinario E., Nocero,M., Jin,M. and Hoffman,C.S. (1993) Cloning and manipulation of the Schizosaccharomyces pombe his7+ gene as a new selectable marker for molecular genetic studies. Curr. Genet., 24, 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forsburg S.L. (1993) Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res., 21, 2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlow E. and Lane,D. (1999) Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 30.Boeke J.D., LaCroute,F. and Fink,G.R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet., 197, 345–346. [DOI] [PubMed] [Google Scholar]
- 31.Wood V., Gwilliam,R., Rajandream,M.A., Lyne,M., Lyne,R., Stewart,A., Sgouros,J., Peat,N., Hayles,J., Baker,S. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- 32.Sunnerhagen P., Seaton,B.L., Nasim,A. and Subramani,S. (1990) Cloning and analysis of a gene involved in DNA repair and recombination, the rad1 gene of Schizosaccharomyces pombe. Mol. Cell. Biol., 10, 3750–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Partridge J.F., Scott,K.S., Bannister,A.J., Kouzarides,T. and Allshrie,R.C. (2002) cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol., 12, 1652–1660. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama J., Klar,A.J. and Grewal,S.I. (2000) A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell, 101, 307–317. [DOI] [PubMed] [Google Scholar]
- 35.Nimmo E.R., Cranston,G. and Allshire,R.C. (1994) Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J., 13, 3801–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorentz A., Ostermann,K., Fleck,O. and Schmidt,H. (1994) Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene, 143, 139–143. [DOI] [PubMed] [Google Scholar]
- 37.Leipe D.D. and Landsman,D. (1997) Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res., 25, 3693–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuwald A.F. and Poleksic,A. (2000) PSI-BLAST searches using hidden markov models of structural repeats: prediction of an unusual sliding DNA clamp and of beta-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res., 28, 3570–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuzon C.T., Borgstrom,B., Weilguny,D., Egel,R., Cooper,J.P. and Nielsen,O. (2004) The fission yeast heterochromatin protein Rik1 is required for telomere clustering during meiosis. J. Cell. Biol., 165, 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Partridge J.F., Borgstrom,B. and Allshire,R.C. (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev., 14, 783–791. [PMC free article] [PubMed] [Google Scholar]
- 41.Pidoux A.L., Richardson,W. and Allshire,R.C. (2003) Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J Cell. Biol., 161, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allshire R.C., Javerzat,J.P., Redhead,N.J. and Cranston,G. (1994) Position effect variegation at fission yeast centromeres. Cell, 76, 157–169. [DOI] [PubMed] [Google Scholar]


