Abstract
The spread of modern humans across the globe has led to genetic adaptations to diverse local environments. Recent developments in genomic technologies, statistical analyses, and expanded sampled populations have led to improved identification and fine-mapping of genetic variants associated with adaptations to regional living conditions and dietary practices. Ongoing efforts in sequencing genomes of indigenous populations, accompanied by the growing availability of “-omics” and ancient DNA data, promises a new era in our understanding of recent human evolution and the origins of variable traits and disease risks.
Modern humans originated ~200,000 years ago in Africa. Over the past 100,000 years, humans spread across the globe into a variety of habitats, from tropical to arctic, from high altitudes to lowlands and even to toxic environments. After humans migrated out of Africa, they encountered and interbred with archaic populations such as Neandertals and Denisovans, resulting in introgression of archaic genomes into non-African modern human genomes (~1 to 6% of modern genomes) (1). Introgression within Africa also likely occurred (2) but is more challenging to quantify because ancient DNA (aDNA) does not preserve well in that region, and no archaic African genomes are currently available. Within the past 10,000 years, most human populations have transitioned from a hunting-gathering life-style to practicing agriculture and pastoralism, resulting in rapid population growth, increased population densities, and an increase in infectious diseases. The selection pressures for adapting to local environments and new diets have resulted in population- or region-specific genetic variants that influence variable phenotypes (such as height, innate immune response, lactose tolerance, fatty acid metabolic efficiency, and hemoglobin levels).
Establishing a complete picture of local human adaptation can be challenging because it involves identifying the genomic regions under selection, the phenotypes that selection is acting upon, and ideally, the external conditions driving the selection. Populations that have adapted to environments that severely challenge survival provide well-characterized cases for local adaptation. Here, we review several recent examples that illustrate the use of emerging data, such as aDNA and genome-wide association studies (GWAS), and the impact these adaptations have for disease risk (Fig. 1).
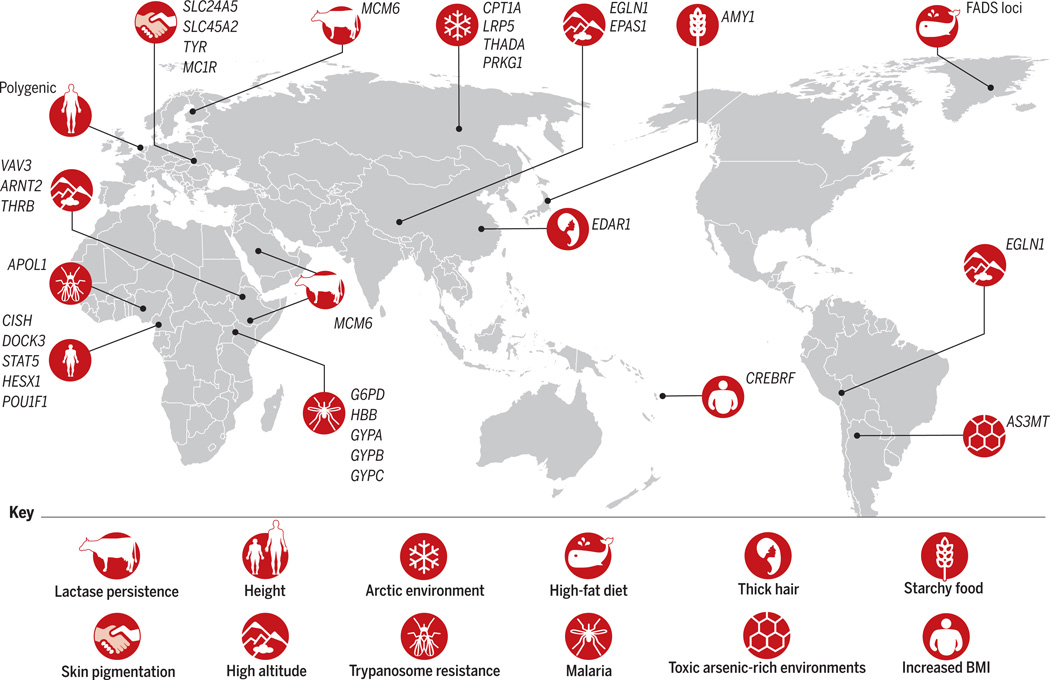
Fig. 1. Examples of human local adaptations, each labeled by the phenotype and/or selection pressure, and the genetic loci under selection.
[Adapted from (13)]
Adaptation to dairy consumption
The advent of cattle domestication in the Middle East and North Africa, ~10,000 years ago, lead to strong selection pressure for the ability to drink milk as adults. Variants near the LCT locus—coding for the lactase enzyme that metabolizes lactose, the main carbohydrate in milk—show some of the strongest signals of selection in the human genome (3). In most mammals and in most humans, the level of the lactase enzyme decreases after weaning [lactase nonpersistence (LNP)]. However, many populations that have traditionally practiced dairying maintain high levels of lactase into adulthood [lactase persistence (LP)].
A genetic variant associated with LP in Europeans was mapped to intron 13 of MCM6 upstream of LCT (4). Additional variants located within 100 base pairs (bp) of the European variant were identified in African populations (5). The LCT region was repeatedly reported as a target of a recent strong selective sweep (Fig. 2A) in Europeans and Africans, on the basis of numerous statistical tests including allele frequency comparisons between global populations [fixation index (FST) analyses] and extended haplotype homozygosity tests within [extended haplotype homozygosity (EHH) and integrated haplotype score (iHS)] and across [cross-population EHH (XP-EHH)] populations (Fig. 3) (3, 6, 7). Indeed, the homozygosity of African haplotypes containing the LP-associated variants extends on average nearly 2 Mb, but only ~2000 bp in ancestral haplotypes (5, 8). In vitro studies showed that these derived alleles enhance the expression of LCT (5, 9). The European LP-associated variant is estimated to be ~9000 years old, whereas the most common East African LP variant is ~5000 years old, which is consistent with archeological evidence for cattle domestication in the Middle East and east Africa (5). Sequencing of aDNA indicates that the European LP-associated allele was absent in early Neolithic Central Europeans and was at low frequency in late Neolithic Europeans (10), suggesting that LP spread recently (within the past ~4000 years) across Europe (11).
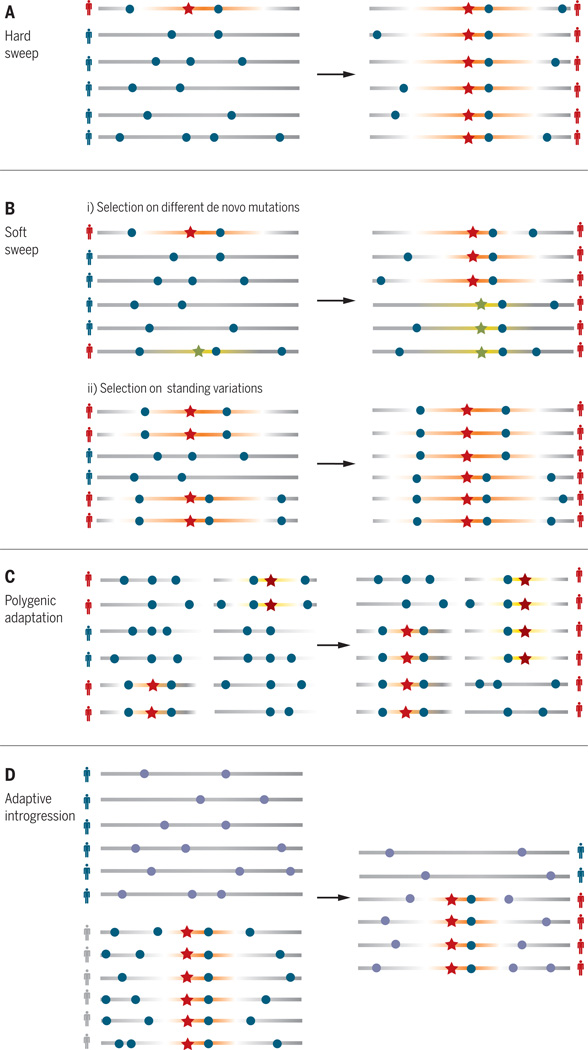
Fig. 2. Genomic signatures of local adaptation.
(A) A hard sweep. (B) A soft sweep acting on (i) multiple de novo beneficial mutations and (ii) standing variation. (C) Polygenic adaptation. (D) Adaptive introgression. Each horizontal bar represents a haplotype (a sequence of genetic variants on the same chromosome). The orange segment is graded to indicate the strength of linkage disequilibrium between beneficial (stars) and neutral (dots) variants.
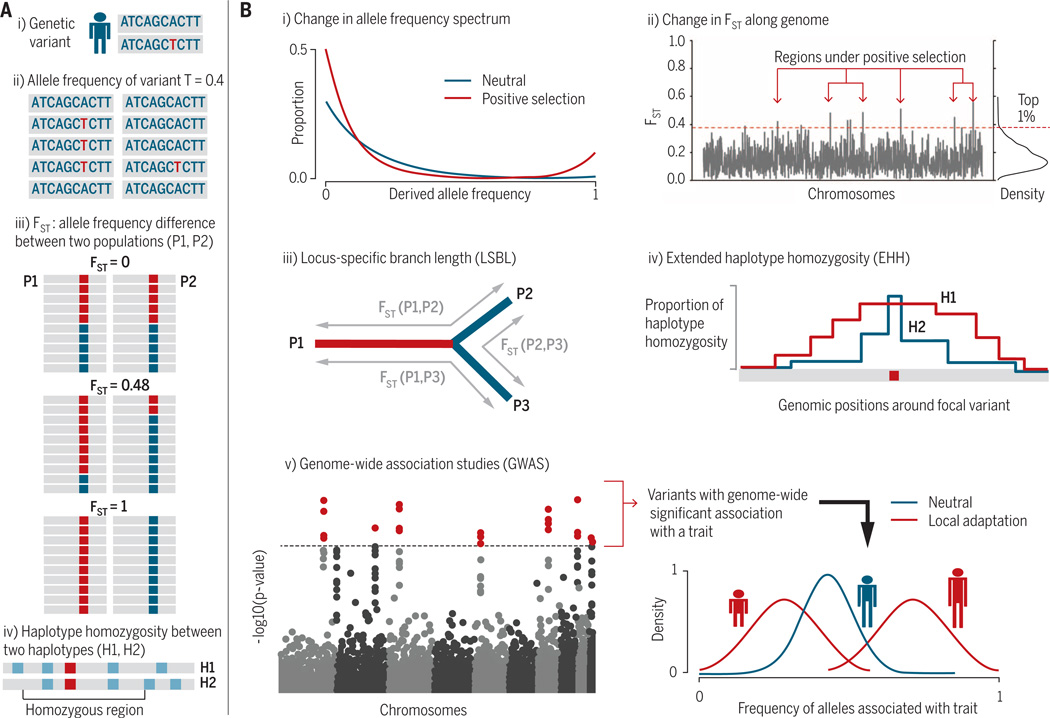
Fig. 3. Illustrations of basic concepts of population genetics and methods for detecting recent positive selection.
(A) Population genetics terminology. (i) A genetic variant (A/T, red) on the diploid chromosomes of an individual. (ii) The allele frequency of a variant in a population is the fraction of chromosomes containing the variant. In this case, allele frequency of variant T (red) = 0.4. (iii) FST measures proportion of genetic differentiation between populations. FST = 0 means the two populations have equal allele frequencies. FST =1 means the two populations are completely differentiated. (iv) Haplotype homozygosity is the shared stretches of variants between pairs of sampled haplotypes, typically measured around a focal variant (red square) and extending out to flanking variants (light blue). (B) Means of detecting positive selection. (i) Changes in the allele frequency spectrum. Positive selection (red) drives the alleles under selection and the hitchhiking alleles to high frequencies; at the same time, new mutations arise, creating an excess of rare alleles. (ii) FST along the genome. High FST values indicate loci enriched for targets of selection, with a cutoff (red dashed line) arbitrarily set to be the top 1%. (iii) LSBL, which quantifies branch lengths of a phylogenetic tree with three or more populations by using pairwise FST (17). At a locus, long branch length for one population (red) indicates a population-specific divergence. A similar statistic is the PBS. (iv) Haplotype homozygosity statistics [extended haplotype homozygosity (EHH) and integrated haplotype score (iHS)]. Overabundance of a long haplotype (H1, red) surrounding a focal variant indicates positive selection at the locus. The EHH statistic has also been extended to identify population-specific selective sweeps through cross-population comparisons (XP-EHH). (v) Results from GWAS, which identify genetic variants significantly associated with a specific trait (such as height), indicated by red dots on the Manhattan plot (left). Shifts in the frequency distribution of alleles associated with the trait (right) in specific populations (red) compared with the expectation under neutrality (blue) indicates that the trait is under selection. [Part of Fig. 3B is adapted from (27)]
The genetic adaptations resulting in LP are examples of convergent evolution in modern humans (causative genetic variants arose independently in geographically diverse populations owing to strong selective pressure for an adaptive phenotype). However, the identified LP-associated variants do not entirely explain the LP phenotype, particularly in western Africans and some central and southern Asian pastoralist populations. For example, the Fulani populations from Nigeria and Cameroon have the European variant at moderate frequency but lack the eastern African LP-associated variants and have a distinct haplotype with extended homozygosity near LCT, suggesting the presence of additional unknown functional variants (8). Furthermore, an outstanding question is the role that the human gut microbiome plays in LP.
Adaptation to an arctic environment
The Inuit populations in Alaska, Canada, and Greenland have adapted to a cold and dark Arctic environment and a marine diet rich in omega-3 polyunsaturated fatty acids. A recent study compared genomic diversity in Greenlandic Inuits with Europeans and Chinese using the population branch statistic (PBS) (Fig. 3B, iii) (12) and found that the most differentiated region encompasses a gene cluster that codes for fatty acid desaturase enzymes (FADS), which are important modulators of fatty acid composition. Two variants in the FADS region were significantly associated with short stature in the Inuits, possibly because of the influence of fatty acid composition on growth hormone regulation (12). These variants were also associated with height in a larger study of Europeans, but the variants were present at low frequencies and would have been hard to discover without studying the Inuit population, demonstrating why studies of indigenous populations can be informative for identifying variants of functional importance across ethnic groups (13).
Adaptation to tropical rainforests
Tropical rainforests are some of the harshest environments in the world, characterized by high temperature and humidity as well as the prevalence of parasites and other pathogens. Individuals living in tropical environments often have very short life spans, which directly affects reproductive success and, hence, can act as a selective pressure.
Short stature
One distinctive phenotype thought to be adaptive to a tropical rainforest environment is short stature (commonly referred to as a “pygmy” phenotype), which is defined as an average height of <150 cm in adult males. The short stature trait is an example of convergent evolution in rainforest hunter-gatherer (RFHG) populations across Africa, Asia, and South America. Selection for small body size may be due to limited food resources, resistance to heat stress, immune response, and/or a trade-off between early onset of reproduction and cessation of growth (14, 15). Perturbations in the GH1-IGF1 pathway have been implicated to play a role in short stature in RFHG populations in central Africa and southeast Asia (16).
In central African RFHG populations where admixture with neighboring populations is common, short stature is significantly correlated with ancestry and is highly heritable (14). Central African RFHG–specific genomic adaptations were identified through comparisons with other African populations by using methods such as locus-specific branch length (LSBL) (17) and XP-EHH (3) tests, which have high power to detect population-specific positive selection (Fig. 3B). These tests identified a 15-Mb region on chromosome 3 that shows signatures of strong positive selection (14). Several genes in this region are associated with short stature in Pygmies, including DOCK3, which is associated with height variation in non-Africans, and CISH, which is important in immune response but also inhibits human growth hormone receptor activity, indicating that short stature could be a product of selection acting on pleiotropic loci (14). A subsequent study discovered a ~200-kb haplotype containing HESX1 at high frequency in central African RFHGs but low frequency in other African populations; HESX1 is involved in the development of the anterior pituitary, where growth hormone is produced (18). This haplotype was not tagged in previous genotyping arrays, demonstrating the importance of including ethnically diverse populations in whole-genome sequencing studies (18). However, RFHG and neighboring agriculturalist populations in Uganda exhibit a distinct set of loci associated with short stature, which raises the possibility of convergent evolution of this trait across the RFHGs in Africa (15).
The studies discussed above suggest that short stature in central African RFHG is mostly driven by strong selection affecting a relatively small number of loci of moderate to strong effect. In contrast, hundreds of loci are associated with European height variation (19, 20). Detecting selection on a polygenic trait such as height is difficult because the causative variants can be ancient polymorphisms each of relatively small effect. Several recent methods use GWAS to identify trait-associated variants then test for allele frequency shifts between populations [either unweighted (20) or weighted by effect size (21)] greater than expected from neutral drift. Polygenic selection tests using GWAS data (Fig. 2C) have suggested weak selection influencing average height differences among European populations, with Northern Europeans generally being taller than Southern Europeans (20, 21). A recent aDNA analysis of Neolithic European populations suggests that the North-South European height gradient may reflect selection for shorter height in Early Neolithic migrants into southern Europe and admixture of taller steppe populations with northern Europeans (11). Although analysis of aDNA holds great promise for revealing human phenotypic history, this approach faces challenges for studies of indigenous populations such as RFHG because DNA is not well-preserved in tropical climates, and large-scale GWAS are not available.
Adaptation to endemic pathogens
Pathogenic environments are an important driver of local adaptation in humans (22), and nowhere is the challenge of survival from pathogens greater than in tropical rainforests. In particular, malaria, a mosquito-borne protozoan parasitic infection, is a major cause of mortality in sub-Saharan Africa. The most lethal malarial species, Plasmodium falciparum, causes >1 million child deaths in Africa each year (23). Tropical populations have long been known to have genetic variants that confer resistance to malaria, including the sickle cell, α+- and β+-thalassemia–causing alleles at the hemoglobin loci, as well as variants at ABO, GYPA, GYPB, GYPE, and G6PD [reviewed in (23)]. These variants are likely adaptive, as initially evidenced by the strong correlation between allele frequency and prevalence of malaria infection. However, several malaria-protective variants also cause common Mendelian diseases in hemi- or homozygotes [such as sickle cell anemia, G6PD deficiency, and thalassemia (23)] and are maintained through balancing selection.
Another example of adaptation to pathogens resulting in high frequencies of genetic variants associated with disease is at the APOL1 locus. The “G1” and “G2” genetic variants in the last exon of APOL1 are associated with chronic kidney disease, which is disproportionately common among individuals of African descent (24). These variants also confer resistance to human African trypanosomes (causing “African sleeping sickness”) by modifying the protein to lyse Trypanosoma brucei rhodesiense, a parasitic protozoa transmitted by the tsetse fly. The region flanking the G1 and G2 variants appear to be under recent positive selection, as indicated by extended haplotype homozygosity (25). G1 and G2 are common in some western African populations, but T.b. rhodesiense is currently common only in eastern and southern Africa, which raises questions about the evolutionary history of the APOL1 locus and the pleiotropic effects of G1 and G2 (25).
Adaptation to high altitude
Living at high altitude (>2500 m above sea level) in regions such as the Tibetan Plateau, the Andean Altiplano, and the Semien plateau of Ethiopia can be deadly because of an insufficient supply of oxygen to vital organs (hypoxia). However, populations living in these high-altitude regions for thousands of years have adapted and thrived, with varying physiological adaptations to hypoxic environments [reviewed in (26)].
Recent genome-wide scans of selection (such as LSBL, PBS, iHS, and XP-EHH) (Fig. 3B) uncovered genetic adaptations to high altitude by comparing Andean, Ethiopian, and Tibetan genomes with lowland populations with similar genetic ancestry [reviewed in (27)]. Signatures of positive selection were found repeatedly at genes involved in the hypoxia-inducible factor (HIF) pathway [reviewed in (28)] but on different haplotype backgrounds (for example, EGLN1 in Andean and Tibetan populations) owing to convergent evolution (29). One of the strongest signals of selection in the Tibetan populations is at EPAS1, a transcription factor influencing the HIF pathway. Sequence analysis suggests that the selected Tibetan haplotype may have originated from introgression of genomic DNA from Denisovans (Fig. 2D) (30).
Adaptation to toxic environments
Arsenic is acutely toxic to humans but is naturally present at high levels in groundwater across the globe (31). San Antonio de los Cobres (SAC), Argentina, is one such locale with high levels of arsenic yet has been settled by human populations for the past ~11,000 years. Arsenic metabolism involves methylating inorganic arsenic to monomethylarsonic acid (MMA) and subsequently to dimethylarsinic acid (DMA), which is less toxic. The fraction of arsenic metabolites (%DMA/%MMA) in urine indicates the efficiency of arsenic metabolism. Individuals in SAC show particularly low urinary excretion of MMA, suggesting a local adaptation to arsenic. A recent association study in SAC identified potential protective regulatory variants upstream of AS3MT, a gene involved in the arsenic methylation pathway. The variants are likely under positive selection because they are embedded in long haplotypes in the SAC population and are at higher frequency than in neighboring low-arsenic groups with similar genetic ancestry (31).
Adaptation to ultraviolet exposure
Variation in human skin color is one of the most striking examples of human phenotypic diversity. Unlike other primates, human skin is not covered by dense body hair and is the primary interface between our body and the environment. Ultraviolet radiation (UVR) exposure is an important driver of pigmentation evolution in humans, with selection pressure for darker skin at low latitudes for UVR protection and for lighter skin at higher latitudes, possibly to maintain vitamin D photosynthesis (32).
The earliest studies of the genetics of human pigmentation were based on candidate genes identified in model organisms and highly penetrant genetic variants of Mendelian disorders [for example, SLC24A5 in zebrafish color patterns, MC1R in mouse coat colors, and OCA1-4 in human oculocutaneous albinism (33)]. GWAS in European populations have included additional candidate loci associated with light skin, a subset of which (such as OCA2, TYRP1, TYR, SLC24A5, and SLC45A2) are under strong selection, as evidenced by multiple genome-wide scans of selection [reviewed in (33)]. Furthermore, a recent analysis of 230 ancient Eurasian genomes found that the allele associated with light skin pigmentation has likely reached fixation in modern Europeans from very low frequency during the Neolithic period due to strong selection pressure over the past ~4000 years (11).
Although many genetic variants associated with European light skin color are identified, little is known about the genetic basis of skin color in Asia and Africa. Indeed, Africans exhibit high variability in skin color (ranging from dark-skinned Nilotic pastoralists to light-skinned San hunter-gatherers), and the genetic basis of pigmentation in these populations has just recently begun to be explored (34).
Impact of local adaptation on common complex diseases
Genetic variants that were adaptive in the past can be maladaptive in modern environments. For example, the high prevalence of type 2 diabetes was proposed to be due to common variants that were adaptive for the efficient conversion of food into energy in the past when resources were scarce, but are maladaptive in modern urban environments (the “thrifty gene hypothesis”). This hypothesis remains controversial; an alternative hypothesis is that the disease-associated variants were never beneficial but became common through genetic drift (the “drifty gene hypothesis”) (35).
A recent study of Samoans provides functional evidence supporting the thrifty gene hypothesis. Over 80% of Samoans are overweight or obese [body mass index (BMI) > 26 kg/m2], which is among the highest prevalence in the world (36). By genotyping ~3000 Samoans, a missense variant in CREBRF was found to be associated with BMI and fasting glucose levels. This variant is under strong recent positive selection, as evidenced by extended haplotype homozygosity and high allele frequency in the Samoan population but not other populations. Functional expression experiments in adipose cells showed that the Samoan variant decreases energy use and increases adipose fat storage, suggesting that this variant may have been adaptive in the past by increasing tolerance to periods of starvation but is associated with risk for obesity and type 2 diabetes in modern populations.
Future directions
Unearthing human adaptation with aDNA
aDNA sequencing provides a direct historical record of genomic variation and provides new possibilities for inferring recent human evolutionary history of modern phenotypes. Studies based on comparison of modern human with Neandertal and Denisovan genomes have found evidence of adaptive archaic haplotypes in genes related to innate immune response, metabolism, and skin phenotypes in ethnically diverse modern human populations [reviewed in (1)]. Future studies of aDNA will be informative for reconstructing the origin of functional variants and inferring the strength of selection based on direct observation of changes in allele frequencies over time (37). However, current understanding of ancient adaptation events is limited by sparse aDNA data over broad geographic and temporal scales. Moreover, methods for studying ancient genotype variation tend to focus on ascertained variants in specific populations, particularly Europeans (1).
Adaptive potential of structural variation
Structural variants (SVs)—including duplications, copy number variants, deletions, insertions, and inversions—encompass a much larger fraction of the genome than single-nucleotide polymorphisms (SNPs) [~4.1 million to 5.0 million base pairs for SNPs compared with ~20 million base pairs for SVs (38)] and may consequently have substantial contributions to human adaptive evolution (39). For example, Perry et al. (40) observed higher copy numbers of AMY1, an enzyme that breaks down starches, in populations with high-starch diets. Further, a common inversion polymorphism at chromosome 17q21.31 is hypothesized to be under positive selection in Europeans because of increased fecundity of women carrying the inverted haplotype (41). However, SVs are extremely challenging to detect by using short-read sequencing technologies. The development of cost-effective high-throughput methods for obtaining long-sequence reads and novel computational approaches will facilitate identification and characterization of SVs in the future (42).
Challenges to detecting selection
Although numerous methods for detecting selection have been developed, there is a known lack of concordance across methods (43), partly because of the different selection time scales each method is capable of detecting (44). Strong positive selection on a de novo (newly arising) mutation is the classic process that many methods have been devised to detect, in which the haplotype containing the beneficial variant rapidly rises in frequency in the population, resulting in long stretches of identical haplotypes around the selected variant within the population (Figs. 2 and 3). There is some debate over the fraction of the human genome affected by selective sweeps within the past ~250,000 years, in part owing to complexities in accounting for background selection that purges deleterious genetic variation (45, 46) and the difficulty in detecting simultaneous selection on multiple beneficial haplotypes at a locus (“soft sweeps”) (Fig. 2B) (47). Although recent methods have increased the power to scan for soft sweeps (48, 49), their detection is still limited because of relatively weak genomic signals (50) and because regions flanking a hard sweep (the “shoulders”) can mimic a soft sweep (51, 52). Development of machine-learning methods, which can effectively “learn” the appropriate signals within complex data when given training examples, may be effective for detecting hard and soft selective sweeps (53).
Perhaps the largest challenge to detecting selection is the fact that most traits are polygenic. It is difficult to distinguish small frequency shifts over large numbers of independent loci (Fig. 2C) from neutral drift without knowing a priori which trait is under selection and the genetic variants that influence the trait. Incorporating the results of GWAS into selection tests (20, 21) is one promising avenue to identify cases of polygenic adaptation, although such integration will have to overcome issues common to genetic association tests, including ascertainment bias in the set of genotyped variants, population stratification, and a lack of observed heritability (“missing heritability”).
Integration of genetic, phenotypic, and functional data into adaptation studies
The best-characterized human adaptive variants are those related to Mendelian, or near Mendelian, traits for which the adaptive phenotype can be easily distinguished (for example, LP and skin pigmentation). Characterizing the functional variants that affect nonobvious or intermediate phenotypes—such as blood metabolite levels, gene expression across cell types, and epigenetic modifications—motivates the need for detailed phenotyping of global populations based on “-omics” profiling (such as transcriptomics, metabolomics, or epigenomics). Additionally, integration of genome-wide selection scans with GWAS helps improve the power of genotype-phenotype association analyses by localizing putative regions with a functional impact. This is particularly important in studies of indigenous populations, in which obtaining large sample sizes is challenging. Lastly, functional experiments in model organisms can directly establish the link between candidate adaptive variants and phenotypes. For example, a derived allele at the EDAR locus (EDAR370A) is significantly associated with hair thickness and dental morphology [reviewed in (27)] and is a target of recent strong positive selection in Asian populations (3, 54). Development of humanized mice containing EDAR370A replicated the hair thickness phenotype and also identified previously unknown phenotypic impact on mammary and eccrine glands (55). With the development of in vitro and in vivo technologies, such as tissue-specific cell lines, genome/RNA-editing (such as CRISPR technology), and induced pluripotent stem cells, it is increasingly possible to validate the function of adaptive variants in humans.
Conclusions
Selection pressures in response to regional conditions have influenced global human genomic diversity, as evidenced by the reviewed local adaptations to diverse physical environments, pathogen exposure, and dietary practices. None of these insights would be possible without global genome-wide population genetic data collected over the past 15 years. New insights into local human adaptation will require high-coverage whole-genome sequencing of ethnically diverse populations and detailed phenotyping. Extending current GWAS-based methods to detect polygenic adaptation in order to include more accurate models of genetic architecture that take into account nonadditive, epistatic, and gene-environment effects is an important future direction. The integration of high-quality genomic data from ancient and modern populations, detailed phenotype data, and advances in computational approaches will illuminate the mode and tempo of local adaptation as humans settled the globe.
Acknowledgments
We thank M. Rubel for her assistance with creating Fig. 1 in this manuscript and members of the Tishkoff laboratory for helpful scientific discussions. This work is funded by NIH grants 1R01DK104339-01 and 1R01GM113657-01 and NSF grant BCS-1317217 to S.A.T., NIH grant T32ES019851-02 to M.E.B.H. through the Center of Excellence in Environmental Toxicology at the University of Pennsylvania, and NIH grants LM009012 and LM010098 to Y.L. through J. H. Moore.
REFERENCES AND NOTES
- 1.Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E. Nat. Rev. Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsieh P, et al. Genome Res. 2016;26:291–300. doi: 10.1101/gr.196634.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabeti PC, et al. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enattah NS, et al. Nat. Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 5.Tishkoff SA, et al. Nat. Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voight BF, Kudaravalli S, Wen X, Pritchard JK. PLOS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williamson SH, et al. PLOS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranciaro A, et al. Am. J. Hum. Genet. 2014;94:496–510. doi: 10.1016/j.ajhg.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enattah NS, et al. Am. J. Hum. Genet. 2008;82:57–72. doi: 10.1016/j.ajhg.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plantinga TS, et al. Eur. J. Hum. Genet. 2012;20:778–782. doi: 10.1038/ejhg.2011.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathieson I, et al. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fumagalli M, et al. Science. 2015;349:1343–1347. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 13.Tishkoff S. Science. 2015;349:1282–1283. doi: 10.1126/science.aad0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarvis JP, et al. PLOS Genet. 2012;8:e1002641. doi: 10.1371/journal.pgen.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perry GH, et al. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3596–E3603. doi: 10.1073/pnas.1402875111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry GH, Dominy NJ. Trends Ecol. Evol. 2009;24:218–225. doi: 10.1016/j.tree.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Shriver MD, et al. Hum. Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachance J, et al. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lango Allen H, et al. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turchin MC, et al. Nat. Genet. 2012;44:1015–1019. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg JJ, Coop G. PLOS Genet. 2014;10:e1004412. doi: 10.1371/journal.pgen.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fumagalli M, et al. PLOS Genet. 2011;7:e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwiatkowski DP. Am. J. Hum. Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genovese G, et al. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko W-Y, et al. Am. J. Hum. Genet. 2013;93:54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beall CM. Integr. Comp. Biol. 2006;46:18–24. doi: 10.1093/icb/icj004. [DOI] [PubMed] [Google Scholar]
- 27.Lachance J, Tishkoff SA. Annu. Rev. Ecol. Evol. Syst. 2013;44:123–143. doi: 10.1146/annurev-ecolsys-110512-135833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bigham AW, Lee FS. Genes Dev. 2014;28:2189–2204. doi: 10.1101/gad.250167.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bigham A, et al. PLOS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huerta-Sánchez E, et al. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlebusch CM, et al. Mol. Biol. Evol. 2015;32:1544–1555. doi: 10.1093/molbev/msv046. [DOI] [PubMed] [Google Scholar]
- 32.Jablonski NG, Chaplin G. Proc. Natl. Acad. Sci. U.S.A. 2010;107(suppl. 2):8962–8968. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturm RA, Duffy DL. Genome Biol. 2012;13:248. doi: 10.1186/gb-2012-13-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beleza S, et al. PLOS Genet. 2013;9:e1003372. doi: 10.1371/journal.pgen.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speakman JR. Annu. Rev. Nutr. 2013;33:289–317. doi: 10.1146/annurev-nutr-071811-150711. [DOI] [PubMed] [Google Scholar]
- 36.Minster RL, et al. Nat. Genet. 2016;48:1049–1054. doi: 10.1038/ng.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickrell JK, Reich D. Trends Genet. 2014;30:377–389. doi: 10.1016/j.tig.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auton A, et al. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radke DW, Lee C. Brief. Funct. Genomics. 2015;14:358–368. doi: 10.1093/bfgp/elv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry GH, et al. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stefansson H, et al. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 42.Pendleton M, et al. Nat. Methods. 2015;12:780–786. doi: 10.1038/nmeth.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akey JM. Genome Res. 2009;19:711–722. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabeti PC, et al. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 45.Hernandez RD, et al. Science. 2011;331:920–924. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enard D, Messer PW, Petrov DA. Genome Res. 2014;24:885–895. doi: 10.1101/gr.164822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Messer PW, Petrov DA. Trends Ecol. Evol. 2013;28:659–669. doi: 10.1016/j.tree.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berg JJ, Coop G. Genetics. 2015;201:707–725. doi: 10.1534/genetics.115.178962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson BA, Petrov DA, Messer PW. Genetics. 2014;198:669–684. doi: 10.1534/genetics.114.165571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pritchard JK, Pickrell JK, Coop G. Curr. Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrider DR, Mendes FK, Hahn MW, Kern AD. Genetics. 2015;200:267–284. doi: 10.1534/genetics.115.174912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jensen JD. Nat. Commun. 2014;5:5281. doi: 10.1038/ncomms6281. [DOI] [PubMed] [Google Scholar]
- 53.Schrider DR, Kern AD. PLOS Genet. 2016;12:e1005928. doi: 10.1371/journal.pgen.1005928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grossman SR, et al. Science. 2010;327:883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 55.Kamberov YG, et al. Cell. 2013;152:691–702. doi: 10.1016/j.cell.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]