Significance
A reservoir of latently infected cells poses the greatest challenge to HIV-1 eradication. Efforts to develop strategies to eliminate the reservoir have been hampered, in part, by the lack of a precise understanding of the cellular and molecular nature of this reservoir. We describe a new method to analyze the replication-competent latent reservoir quantitatively and qualitatively. We find that over 50% of the replication-competent viruses in the reservoir form part of groups with identical env sequences. However, a negative correlation exists between integrated proviral clones and replication-competent viruses, such that the larger the proviral clone, the lower is its probability of representing a replication-competent virus.
Keywords: HIV, reservoir, replication-competent, culture, method
Abstract
HIV-1–infected individuals harbor a latent reservoir of infected CD4+ T cells that is not eradicated by antiretroviral therapy (ART). This reservoir presents the greatest barrier to an HIV-1 cure and has remained difficult to characterize, in part, because the vast majority of integrated sequences are defective and incapable of reactivation. To characterize the replication-competent reservoir, we have combined two techniques, quantitative viral outgrowth and qualitative sequence analysis of clonal outgrowth viruses. Leukapheresis samples from four fully ART-suppressed, chronically infected individuals were assayed at two time points separated by a 4- to 6-mo interval. Overall, 54% of the viruses emerging from the latent reservoir showed gp160 env sequences that were identical to at least one other virus. Moreover, 43% of the env sequences from viruses emerging from the reservoir were part of identical groups at the two time points. Groups of identical expanded sequences made up 54% of proviral DNA, and, as might be expected, the sequences of replication-competent viruses in the active reservoir showed limited overlap with integrated proviral DNA, most of which is known to represent defective viruses. Finally, there was an inverse correlation between proviral DNA clone size and the probability of reactivation, suggesting that replication-competent viruses are less likely to be found among highly expanded provirus-containing cell clones.
A reservoir of latently infected cells persists in HIV-1–infected individuals treated with antiretroviral therapy (ART) (1). This reservoir endures for the lifetime of the individual and presents the greatest barrier to an HIV-1 cure (2, 3). Although there is a growing understanding of the cellular and molecular nature of this compartment, many questions remain about the composition of the latent reservoir and the ability of current techniques to characterize it accurately (4, 5).
One of the challenges in studying the reservoir is that the majority (>90%) of integrated proviruses in CD4+ T-cell DNA are defective and cannot produce infectious virions (6–9). Comparatively few cells harbor the replication-competent proviruses that constitute the clinically relevant reservoir, and there are currently no markers to distinguish these cells from those cells bearing defective proviruses. An additional problem is that it is difficult to measure the size of the replication-competent reservoir accurately. The best available assay measures the size of the reservoir by limiting dilution cultures under conditions that favor latent virus outgrowth [quantitative viral outgrowth assay (QVOA)] (10, 11). However, this assay is typically performed on peripheral blood and can significantly underestimate the size of the replication-competent reservoir even within this compartment (7, 9).
Finally, the genetic characteristics of the reservoir have been studied primarily by sequencing integrated proviral DNA or cell-associated RNAs, many of which are defective and thus not representative of the replication-competent reservoir (12–16). Phylogenetic analyses of the replication-competent reservoir are usually limited because bulk outgrowth cultures produce species with little diversity, even when analyzed by ultra-deep sequencing (17–20).
Here, we report on a modified QVOA that includes a qualitative measure of the reservoir [qualitative and quantitative viral outgrowth assay (Q2VOA)]. We use the assay to describe the genetic and biological diversity of the replication-competent reservoir and examine the relationship between replicating and archived proviruses in CD4+ T cells from the same ART-suppressed individuals at two time points separated by 4–6 mo.
Results
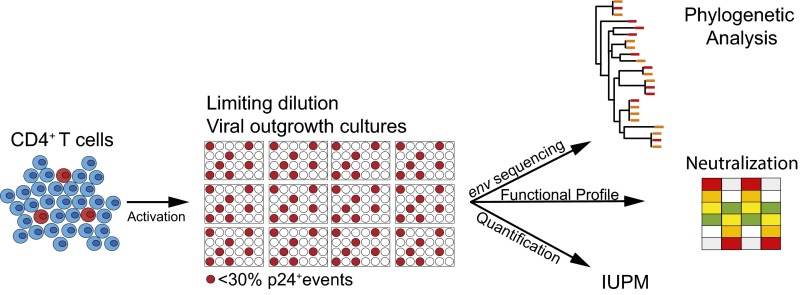
To investigate the genetic and phenotypic complexity of the replication-competent reservoir, we modified the QVOA protocol to increase the number of unique outgrowth cultures and sequenced the emerging viruses. Unlike QVOA, where multiple dilutions are assayed, Q2VOA is performed using a single predetermined dilution that produces less than 30% positive wells to maximize the total number of individual viruses that can be sequenced. Based on Poisson distribution, this technique produces cultures that are likely to contain single replication-competent proviruses (Fig. 1).
Fig. 1.
Quantitative and qualitative analysis of the replication-competent reservoir. Diagrammatic representation of the assay. CD4+ T cells are cultured at a limiting dilution under conditions whereby a single virus emerges from the latent reservoir in each positive well (red). The number of infectious units per million (IUPM) is determined directly from the number of p24-positive wells. Virus-containing supernatants from positive cultures are harvested for env sequencing and neutralization assays.
CD4+ T lymphocytes were isolated from each of four chronically infected individuals who had been virologically suppressed by combination ART for 4–22 y at two time points 4–6 mo apart (Table S1). We tested 0.40–1.44 × 108 CD4+ T lymphocytes from each ART-treated individual at each time point. On average, 13.5% of cultures were positive for p24. The number of cells yielding replication-competent viruses varied across individuals from 0.19 to 1.07 infectious units per million, which is similar to values obtained by others (3, 6) (Table 1).
Table S1.
Clinical characteristics of study subjects
| Study ID | Age | Sex | Year of HIV diagnosis | CD4 nadir | Years since HIV diagnosis | Years on ART | ART regimen |
| B106 | 27 | M | 2008 | 390 | 7 | 7 | TDF/FTC/RPV |
| B115 | 44 | M | 1993 | 200 | 22 | 22 | DRV/r, ABC, 3TC |
| B155 | 59 | M | 1993 | 444 | 22 | 15 | TDF/FTC/RPV |
| B199 | 49 | M | 2009 | 200 | 6 | 4 | TDF/FTC, RAL |
ABC, abacavir; DRV/r, darunavir/ritonavir; FTC, emtricitabine; ID, identification; M, male; RAL, raltegravir; RPV, rilpivirine; 3TC, lamivudine; TDF, tenofovoir disoproxil fumarate.
Table 1.
Q2VOA overall results and IUPM
| Months between time points | Total CD4+ cells tested | Wells tested | Positive wells (%) | IUPM | Total CD4+ cells tested | Wells tested | Positive wells (%) | IUPM | |
| Study ID | Time point 1 | Time point 2 | |||||||
| B106 | 4 | 39.6 × 106 | 132 | 31 (23.5) | 0.89 | 75.6 × 106 | 252 | 28 (11.1) | 0.39 |
| B115 | 4 | 57.6 × 106 | 192 | 40 (20.8) | 0.57 | 140 × 106 | 468 | 50 (10.7) | 0.38 |
| B155 | 6 | 75.6 × 106 | 252 | 69 (27.4) | 1.07 | 72 × 106 | 240 | 24 (16.7) | 0.35 |
| B199 | 4 | 43.2 × 106 | 144 | 24 (16.7) | 0.61 | 144 × 106 | 480 | 26 (5.4) | 0.19 |
ID, identification; IUPM, infectious units per million cells.
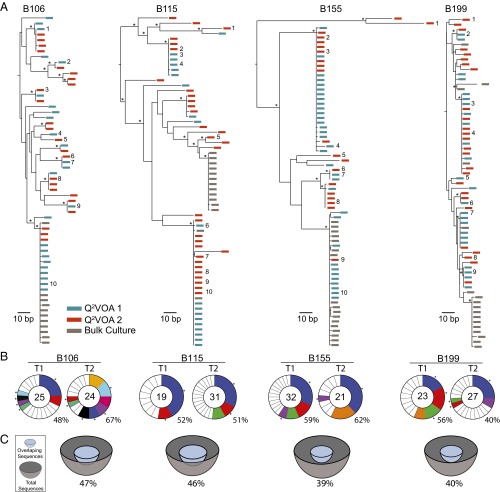
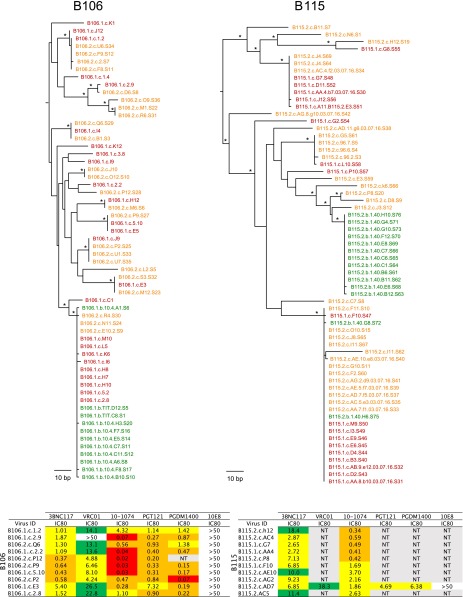
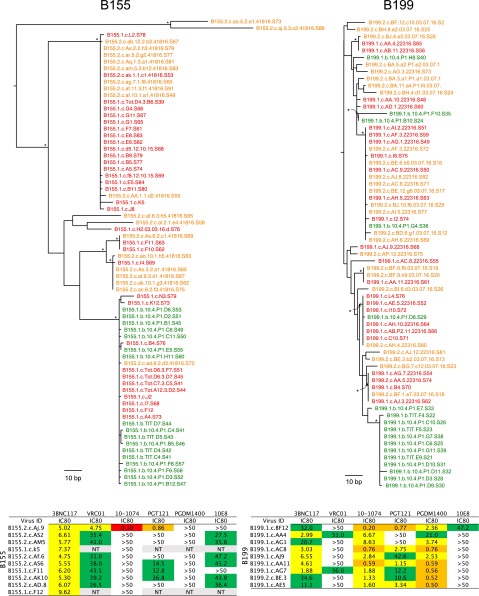
To characterize the cultured viruses molecularly, we produced cDNA from culture supernatants and sequenced the env gene using primers that resulted in a clonal prediction score of 94 of 100 (silicianolab.johnshopkins.edu/cps) (21). Thus, there was a high probability that identical env sequences represented identical full-length genomes. We obtained a total of 234 env sequences from Q2VOA, of which 13.7% were excluded from further analysis due to the presence of short reads (3.8%) or the presence of reads producing an inconclusive consensus (9.8%). The phylogenetic analysis of the remaining 202 env sequences showed that the four individuals were infected with epidemiologically unrelated clade B viruses (Fig. S1). Phylogenetic analysis of individual sequences revealed the existence of a diverse viral population composed of multiple (bootstrap-supported) clusters for each of the four individuals (Fig. 2A).
Fig. S1.
Maximum likelihood phylogenetic tree was constructed from viral env sequences from outgrowth culture supernatants as well as archived proviral DNA from all participants. Hypervariable (as defined in https://www.hiv.lanl.gov/content/sequence/VAR_REG_CHAR/) and other poorly aligned regions were excluded from the analysis. The tree was constructed using RAxML v. 8.0.22 (55) with a GTRGAMMA substitution model, with 1,000 bootstrap replicates and midpoint rooted.
Fig. 2.
Env sequences from outgrowth cultures. (A) Maximum likelihood phylogenetic trees of full-length env sequences of viruses from Q2VOA outgrowth cultures from four individuals. Viruses from time point 1 are green, viruses from time point 2 are red, and bulk culture SGA is gray. Asterisks indicate nodes with significant bootstrap values (bootstrap support ≥ 70%). Numbers next to sequences correspond to viruses assayed for neutralization in Fig. 6. (B) Pie charts depict the distribution of culture-derived env sequences from the two time points. The number in the inner circle indicates the total number of env sequences analyzed. White represents sequences isolated only once across both time points, and colored areas represent identical sequences that appear more than once. The size of the pie slice is proportional to the number of sequences in the clone. Clones found at both time points are the same color and denoted by asterisks. Percentages of identical sequences are displayed at the bottom right of each pie chart. (C) Representation of overlapping sequences between the two time points. The size of the hemisphere is proportional to the number of sequences. Light blue hemispheres represent overlapping sequences and gray hemispheres represent the total number of sequences. The percentage of overlap is indicated at the bottom of each hemisphere.
To compare the diversity of viruses obtained from a “bulk” culture with the diversity of viruses derived by Q2VOA, we performed single genome analysis (SGA) on bulk culture supernatants established at the same time from the same four individuals. In contrast to Q2VOA, bulk culture supernatants were mainly monotypic and showed much reduced overall diversity (Fig. 2A). Thus, when multiple infected cells are reactivated in a single culture, the strain or strains with the fastest growth kinetics, or the greatest fitness in culture, dominate.
Individual replication-competent viruses obtained from different Q2VOA cultures frequently encoded identical env sequences. When Q2VOA-derived viruses obtained at the two time points were compared for each subject, typically less than half (40–52%) of their env sequences were unique (Table 2). The majority of sequences were identical to at least one other independently derived replication-competent virus obtained from the same subject. For example, of a total of 49 env sequences isolated from the two time points from B106, only 21 were unique. The majority (22) were identical to at least one other sequence that appeared at one of the two time points. We will refer to these repeated sequences irrespective of whether they appear at one or both time points as “clones” because they must originate from at least two different CD4+ T cells. This finding does not necessarily imply that the viruses are integrated in the same location in the genome because it is possible that identical viruses can infect different CD4+ T cells. The size of these clones ranged from two to 10 members, with a mean of 3.69 when all four individuals and both time points were considered (Fig. 2 B and C and Table 2). Fifty-four percent of all replication-competent viruses emerging in Q2VOA cultures were derived from expanded clones (Fig. 2 B and C and Table 2).
Table 2.
Distribution of observed sequences in Q2VOA
| Nonclonal | Clonal | ||||||
| Study ID | T1 | T2 | Total (%) | T1 | T2 | Total (%) | Overlapping sequences between T1 and T2 (%) |
| B106 | 13 | 8 | 21/49 (42.9) | 12 | 16 | 28/49 (57.1) | 23/49 (46.9) |
| B115 | 9 | 15 | 24/50 (48.0) | 10 | 16 | 26/50 (52.0) | 23/50 (46.0) |
| B155 | 13 | 8 | 21/53 (39.6) | 19 | 13 | 32/53 (60.4) | 21/53 (39.6) |
| B199 | 10 | 16 | 26/50 (52.0) | 13 | 11 | 24/50 (48.0) | 20/50 (40.0) |
To determine whether the viral sequences obtained from the replication-competent reservoir remained stable over time, we compared the sequences from the two time points for each individual. Many branches in the phylogenetic trees contained sequences derived from both time points (Fig. 2 A–C and Table 2). The relationship between the sequences from both time points was formally assessed by determining their Genealogical Sorting Index (GSI), which quantitates the degree of phylogenetic association between sequences (23). GSI values, which range between 0 (complete interspersion) and 1 (complete monophyly), showed that for each of the four individuals, the sequences obtained by Q2VOA from both time points could not be segregated as distinct groups (Table 3). This finding demonstrates that the viral population emerging from the latent reservoir in four individuals was stable over the 4- to 6-mo time interval analyzed.
Table 3.
GSI and probability values for HIV env trees under the null hypothesis that Q2VOA-derived sequences from both visits are a single mixed group
| Q2VOA | ||||
| Visit 1 | Visit 2 | |||
| Study ID | GSI | P value | GSI | P value |
| B106 | 0.067 | 0.45 | 0.02 | 0.92 |
| B115 | 0.066 | 0.14 | 0 | 1 |
| B155 | 0.059 | 0.26 | 0.012 | 0.82 |
| B199 | 0.053 | 0.3 | 0 | 1 |
To examine the relationship between proviruses integrated into CD4+ T-cell DNA and the replication-competent viruses obtained by Q2VOA, we performed SGA on DNA isolated from primary CD4+ T lymphocytes from the same individuals at both time points. We obtained a total of 498 env sequences, of which 16.3% were excluded from further analysis due to the presence of hypermutated regions (2.4%), short reads (5.6%), or reads producing an inconclusive consensus (8.2%). The remaining 417 full-length env sequences, or 85–113 per individual, fell within the same four patient-specific clades as the Q2VOA-derived env sequences, indicating absence of sample mix-up or contamination (Fig. S1).
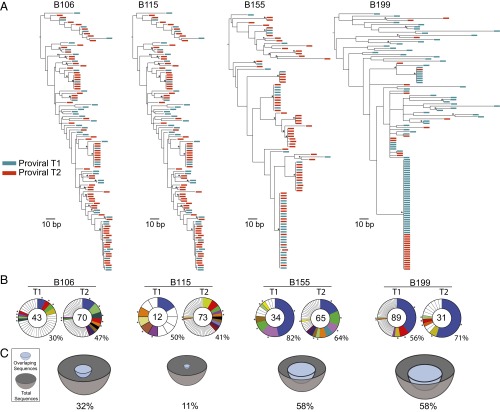
As previously reported for archived proviral DNA amplified from primary CD4+ T cells, we found unique sequences, as well as large groups of identical sequences, marking expanded cell clones (14, 15, 24, 25). For example, in B106, 67 of 113 env sequences obtained from the two time points were unique and the remaining 46 were members of clones. In addition, 36 of the 46 identical sequences overlapped between the two time points (Fig. 3 A and B and Table 4). Thus, like the Q2VOA-derived sequences, the archived proviral population was stable over the time interval analyzed.
Fig. 3.
Env sequences from archived proviral DNA. (A) Maximum likelihood phylogenetic trees of full-length env sequences derived by SGA from primary CD4+ T cells from four individuals. Viruses from time point 1 are green, and viruses from time point 2 are red. Asterisks indicate nodes with significant bootstrap values (bootstrap support ≥ 70%). (B) Pie charts depict the distribution of archived env sequences from the two time points. The number in the inner circle indicates the total number of env sequences analyzed. White represents unique sequences isolated only once across both time points, and colored areas represent identical sequences that appear more than once. The size of the pie slice is proportional to the number of sequences in the clone. Clones found at both time points are the same color and denoted by asterisks. Percentages of identical sequences are displayed at the bottom right of each pie chart. (C) Representation of overlapping sequences between the two time points. The size of the hemisphere is proportional to the number of sequences. Light blue hemispheres represent overlapping sequences and gray hemispheres represent the total number of sequences. The percentage of overlap is indicated at the bottom of each hemisphere.
Table 4.
Distribution of observed sequences in proviral DNA
| Nonclonal | Clonal | Overlapping | |||||
| Study ID | T1 | T2 | Total (%) | T1 | T2 | Total (%) | Sequences between T1 and T2 (%) |
| B106 | 30 | 37 | 67/113 (59.3) | 13 | 33 | 46/113 (40.7) | 36/113 (31.9) |
| B115 | 6 | 43 | 49/85 (57.6) | 6 | 30 | 36/85 (42.4) | 10/85 (11.8) |
| B155 | 6 | 23 | 29/99 (29.3) | 28 | 42 | 70/99 (70.7) | 58/99 (58.6) |
| B199 | 39 | 9 | 48/120 (40.0) | 50 | 22 | 72/120 (60.0) | 70/120 (58.3) |
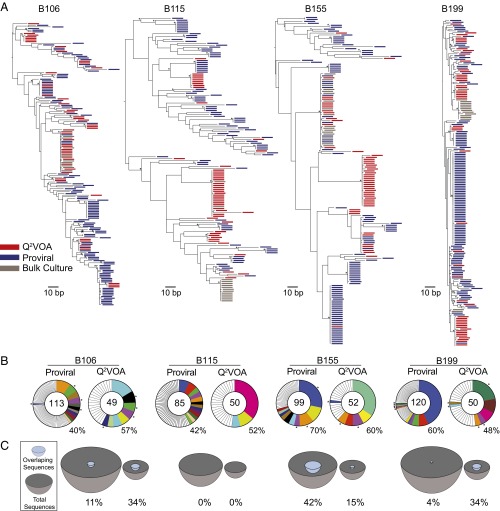
Archived proviral DNA sequences were then compared with replication-competent Q2VOA-derived sequences. Because both the proviral DNA and the replication-competent viral sequences for any given individual were overlapping at the two time points, we combined each of the two sets of sequences (Fig. 4). As might be expected, given that the sample size is limited, we found relatively limited overlap between the archived proviral sequences and Q2VOA culture-derived sequences in phylogenetic trees, with multiple large clusters composed of sequences isolated from only a single source (Fig. 4A). The 417 archived proviral sequences contained 50 expanded clones, 12 of which were also found in the replication-competent outgrowth cultures. The relative frequency of expanded clones from these two sources differed: Some greatly expanded clones identified in primary CD4+ T-cell DNA represented only a small fraction of clones identified in the outgrowth cultures, and rare archived clones were disproportionally abundant among the reactivated latent replication-competent viruses (Fig. 4B). This finding is best illustrated in subject B155, where the largest archived proviral clone, which comprised 28 of 99 total sequences, was found only once among 52 replication-competent viruses (Fig. 4 B and C). In contrast, in B199, one archived proviral sequence, which appeared only once in a total of 120 sequences derived from primary CD4+ T-cell DNA, was found in 11 of 50 Q2VOA culture-derived sequences emerging from the latent reservoir. B115 provided the clearest example of the discrepancy between proviral DNA and cultured viruses, with no instances of matching sequences between 85 proviral sequences and 50 outgrowth viruses (Fig. 4 B and C). Although this discrepancy is most likely due to the prevalence of defective archived proviral sequences (6–9), differences in proviral accessibility to polymerase may also contribute.
Fig. 4.
Comparison of env sequences from archived proviruses and replication-competent viruses. Sequences from the two time points were pooled for each patient. (A) Maximum likelihood phylogenetic trees of env sequences. Limiting dilution outgrowth viruses are red, bulk culture viruses are gray, and viral sequences amplified from primary CD4+ T cells are blue. Asterisks indicate nodes with significant bootstrap values (bootstrap support ≥ 70%). (B) Pie charts depicting the distribution of archived proviruses and culture-derived sequences. The numbers in the inner circles indicate the total number of env sequences analyzed. White represents sequences isolated only once, and colored areas represent identical sequences. The size of the pie slice is proportional to the number of sequences in the clone. Clones found in proviral DNA and outgrowth cultures are the same color and denoted by asterisks. Percentages of identical groups of sequences are displayed at the bottom right of each pie chart. (C) Representation of overlapping sequences between the two sources. The size of the hemisphere is proportional to the number of sequences. Light blue hemispheres represent overlapping sequences and gray hemispheres represent the total number of sequences. The percentage of overlap is indicated at the bottom of each hemisphere.
To determine the extent to which sequences from replication-competent viruses and archived proviruses were compartmentalized, we calculated their GSI. This analysis demonstrated that archived proviral and Q2VOA culture-derived sequences were significantly segregated for every single individual analyzed (Table 5). Thus, proviral sequences derived from primary CD4+ T-cell DNA do not provide an accurate representation of viruses comprising the replication-competent reservoir.
Table 5.
GSI and probability values for HIV env trees under the null hypothesis that Q2VOA- derived sequences and proviral-derived sequences are a single mixed group
| Q2VOA | Proviral | |||
| Study ID | GSI | P value | GSI | P value |
| B106 | 0.184 | <0.01 | 0.114 | <0.001 |
| B115 | 0.61 | <0.001 | 0.43 | <0.001 |
| B155 | 0.421 | <0.001 | 0.125 | <0.001 |
| B199 | 0.15 | <0.001 | 0.071 | <0.05 |
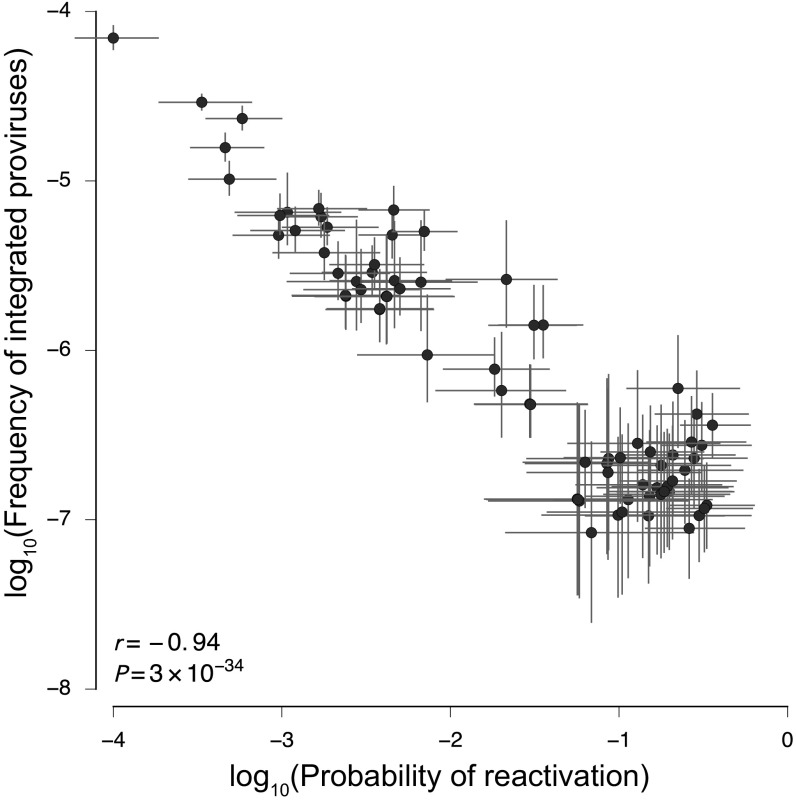
To understand the relationship between the two groups of sequences better, we analyzed them by a mathematical model that describes every sequence by two variables (SI Methods). The first (p) is the frequency of a sequence in the proviral compartment, and the second (r) is its probability of reactivation in the active viral culture. These parameters were extracted from the experimental data using Bayesian inference methods. The data show that clone size is negatively correlated with the activation probability (r = −0.94, p = 3.4 × 10−34) (Fig. 5). These results indicate that the larger the clone of archived proviral sequences, the lower is its probability of representing a replication-competent virus in the Q2VOA outgrowth culture.
Fig. 5.
Negative correlation between proviral clone size and probability of reactivation in culture. The frequency of integrated provirus (p) is negatively correlated with its probability of reactivation in culture (r). Bars denote interquartile ranges of posterior parameter estimates. The Pearson correlation is computed using median values for large clones. Clones with diverse frequencies and reactivation probabilities are observed in each patient.
Finally, to characterize the viruses obtained by Q2VOA further, we tested virus-containing supernatants selected to be representative of the diversity in the phylogenetic trees for sensitivity to a panel of broadly neutralizing antibodies (bNAbs) that are currently in clinical development. As might be expected, there was significant variation in the sensitivity profile of each individual’s replication-competent reservoir. In each case, there was at least one example of coexisting sensitive and resistant viruses for any one tested bNAb (Fig. 6A). For example, in B155, all but one of the 10 viruses tested were resistant to 10-1074, a bNAb that targets the base of the V3 loop and surrounding glycans (26). As might be expected, the 10-1074–susceptible virus segregated from the others in the phylogenetic tree (Fig. 6A and Fig. S2). When results from all individuals were combined, a large range in sensitivities was observed against the panel of bNAbs (Fig. 6B). In contrast, supernatants obtained from the bulk cultures showed bNAb sensitivity that appeared to be representative of a single viral cluster in any individual (Fig. 6A). Thus, Q2VOA differs from bulk cultures in that it enables measurement of the genetic and phenotypic diversity of the replication-competent reservoir. Finally, the observation that bNAb neutralization-sensitive and -resistant clones coexist in the reservoir provides an explanation for the observation that combination therapy will likely be required to maintain suppression (27, 28).
Fig. 6.
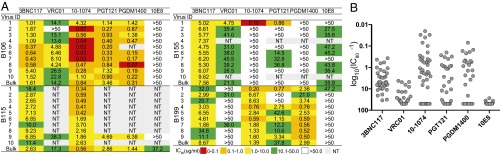
Sensitivity of latent viruses to bNAbs. (A) Table shows antibody concentration that inhibits infection by 80% (IC80) titers for selected outgrowth culture viruses (Fig. 2A) against a panel of bNAbs indicated at the top determined by TZM.bl assay. Viruses with low median tissue culture infectious dose (TCID50) titers could not be tested against the entire bNAb panel (red, IC80 of 0–0.1 μg/mL; orange, IC80 of 0.1–1.0 μg/mL; yellow, IC80 of 1.0–10 μg/mL; green, IC80 of 10–50 μg/mL). NT, not tested. (B) Graph shows combined IC80 titers across all four patients for each bNAb. Each dot represents one virus.
Fig. S2.
Maximum likelihood phylogenetic tree was constructed from viral env sequences from outgrowth culture supernatants for each participant. Viruses from time point 1 are red, viruses from time point 2 are orange, and bulk culture viruses are green. Asterisks indicate nodes with significant bootstrap values (bootstrap support ≥ 70%). Tables beneath each tree show concentration that inhibits response by 80% (IC80) titers for selected outgrowth culture viruses (red, IC80 of 0–0.1 μg/mL; orange, IC80 of 0.1–1.0 μg/mL; yellow, IC80 of 1.0–10 μg/mL; green, IC80 of 10–50 μg/mL). NT, not tested.
SI Methods
We applied Bayesian inference methods to estimate jointly the frequency of integrated proviruses and their reactivation probability, an approach enabled by the combined proviral sequencing and viral outgrowth assay data. We model the probability of measuring a certain number kDNA of integrated proviral sequences as binomial,
| [S1] |
Here, WDNA is the number of wells tested, CDNA is the number of cells per well, and p is the frequency of CD4+ T cells with integrated provirus. Similarly, the probability of observing kVOA-positive wells in the viral outgrowth assay is also binomial,
| [S2] |
As above, WVOA and CVOA are the number of wells and number of cells tested per well in the viral outgrowth assay, respectively. Here, r is the probability that an integrated provirus reactivates and grows out in the VOA.
We assumed the following prior distributions:
| [S3] |
where μ = max(kDNA/(WDNA × CDNA), 10−7) are weakly informative (54), intended simply to prevent pathological inferences. This approach yields conservative parameter estimates. For example, posterior distributions of reactivation probabilities for large clones are nonzero even if no reactivation events are observed. In models that include data from multiple visits, we also included an informative prior to take into account the expected decay of the latent reservoir in individuals on uninterrupted ART,
| [S4] |
Here, pi and ri are the frequency of integrated provirus and reactivation probability at visit i, respectively. Their product is therefore equivalent to the number of infectious units per million cells (IUPM), divided by 106. We used λ = −0.007 mo−1 as the IUPM decay rate and σ = 0.38 as the SD of its measurement, following previous estimates (3). The time between visits (in months) is t. We implemented all models in Stan (52) using the PyStan interface.
Discussion
A long-lived reservoir of HIV-1–infected cells that remain silent in infected individuals receiving ART is the primary impediment to HIV-1 cure (22, 29, 30). The reservoir is established early in infection (31) and has an estimated t1/2 of 44 mo (2, 3). Several mechanisms have been proposed to account for the persistence of this reservoir, including low-level viral replication, the long t1/2 of latently infected cells, and the proliferative expansion of these latently infected cells (5, 32, 33).
Arguments against persistent low-level viral replication include the observation that further intensification of ART has no measurable effect (34, 35). In addition, there is little evidence for viral evolution even after prolonged periods of ART (36). In contrast, the idea that the reservoir is maintained, at least in part, by the proliferation of latently infected cells is supported by the observation of increasing proportions of identical proviral sequences in circulating CD4+ T cells (15).
Archived proviral integration site analysis provided further support for clonal expansion of infected T cells (8, 24, 25). In all cases examined, a large proportion of the archived integrated proviruses were found to belong to expanded clones of CD4+ T cells that share a unique integration site. However, as expected from the observation that the vast majority of integrated proviruses are defective (7, 9), many of the proviruses found in clonally expanded T cells are also defective (8). The only exception was found in a singular individual with metastatic squamous cell carcinoma, which showed a large expanded clone of replication-competent virus (37). However, the integration site of the provirus in this individual was ambiguous and could not be mapped with certainty. Thus, the precise contribution of expanded cell clones bearing replication-competent proviruses to maintaining the latent reservoir is not defined well.
To examine the diversity of the latent but replication-competent reservoir, we developed a modification of current quantitative viral outgrowth protocols to include a qualitative measure of the reservoir of replication-competent proviruses. Q2VOA maximizes the yield of limiting dilution cultures to obtain viruses originating from a single cell. Consistent with the absence of viral evolution in individuals on suppressive ART, Q2VOA revealed that the latent replication-competent viral population is similar between time points 4–6 mo apart (12, 13, 38). This interval is relatively short in the life of an infected individual, and it will be interesting to evaluate the reservoir by Q2VOA over longer time intervals.
Fifty-four percent of the viruses emerging in Q2VOA belonged to expanded cell clones bearing the same env sequences, and these viruses have a high probability of also being identical in the rest of their genomes [prediction score of 94 of 100; available at silicianolab.johnshopkins.edu/cps (21)]. The isolation of identical sequences from multiple wells on different plates across two time points suggests that these replicating viruses do not accumulate significant mutations during 14 d in culture. The lack of immunological pressure under culture conditions may account for this finding.
There are several nonmutually exclusive explanations for the prevalence of expanded CD4+ T-cell clones carrying replication-competent viruses in the latent reservoir. Such cells could be clonally expanded as part of normal homeostatic processes, in response to antigen, or as a result of HIV-1 integration into and disruption or activation of genes that regulate cell division (24, 25). Alternatively, identical viral sequences could represent a burst of independent viral integration events into unrelated CD4+ T cells. Mapping the precise integration sites of the replication-competent proviruses is required to distinguish definitively between viral clones produced by CD4+ T-cell clonal expansion and by bursts of proviral integration.
Clonally expanded CD4+ T cells with shared proviral integration sites were detected in samples of 1–2 × 106 cells (8, 24, 25), which are far smaller than the samples assayed in Q2VOA. Only the largest of these previously described expanded cell clones were amenable to further characterization, and all 75 proviruses that were examined molecularly showed gross defects that rendered them replication-incompetent (8). The finding that large proviral clones are typically defective is entirely consistent with the observation that proviral clone size is inversely related to the probability of reactivation. The larger the archival proviral clone, the less likely it was to be represented in the replication-competent latent reservoir. This observation is also in agreement with the finding that most clonally expanded proviruses integrated into CD4+ T-cell DNA are defective (6–9).
The relative stability of clones in the replication-competent reservoir has significant implications for assessing efforts aimed at HIV-1 eradication. In particular, Q2VOA may be a sensitive assay with which to measure changes in the reservoir that result from immune-targeted interventions (39–42). Viruses isolated from the replication-competent latent reservoir demonstrated a broad range of neutralization sensitivity to bNAbs, and may also show differing levels of sensitivity to immune-based interventions. The coexistence of neutralization-sensitive and -resistant clones in the same patient may be important to consider in development of clinical strategies using bNAbs in the future. However, whether the viruses detected in Q2VOA represent species that are preferentially reactivated upon termination of ART in infected individuals remains to be determined.
In conclusion, we find clones of replication-competent viruses in the latent reservoir. The viral sequences of these clones show limited overlap with archived proviral sequences in primary CD4+ T cells, suggesting that the two compartments may be under different types of selective pressure in vivo. Finally, if the reservoir is maintained by CD4+ T-cell division, it may facilitate immunotherapy-directed efforts for HIV-1 cure because T-cell activation is thought to be associated with expression of HIV-1 antigens (39).
Methods
Study Participants.
The study was conducted under the approval of The Rockefeller University Institutional Review Board. Written informed consent was obtained from all participants under study protocol MNU-0628 at The Rockefeller University. Eligible participants were adults aged 18–65 y with HIV-1 infection and undetectable plasma HIV-1 RNA levels (<20 copies per milliliter) while on ART. Each participant underwent leukapheresis at two time points 4–6 mo apart.
Q2VOA.
Viral outgrowth cultures were performed as previously described (43, 44). Peripheral blood mononuclear cells (PBMCs) from virologically suppressed, chronically HIV-infected individuals on ART were obtained by leukapheresis. Briefly, leukapheresis products from each study participant at each time point were processed, and PBMCs were isolated by density centrifugation on Ficoll (Thermo Scientific). Cryopreserved PBMCs were then used to isolate total CD4+ T lymphocytes using negative selection by magnetic beads (Miltenyi). Purified CD4+ T lymphocytes were cultured at 0.3 × 106 cells per well in 24-well plates in 1 mL of RPMI 1640 (Gibco) supplemented with 10% (vol/vol) FBS (HyClone; Thermo Scientific), 1% penicillin/streptomycin (Gibco), 1 μg/mL phytohemagglutinin (Life Technologies), and 100 U/mL IL-2 (Peprotech) at 37 °C and 5% CO2. Each well also received 1 mL of medium containing 2.5 × 106 irradiated heterologous PBMCs. After 24 h, 1.5 mL of medium was discarded and 106 CD8+-depleted lymphoblasts from an HIV-negative donor were added to each well as target cells. At day 9, 1 mL of medium was removed and another 106 CD8+-depleted CD4+ T lymphoblasts were added to each well. At day 14, the supernatant of each well was tested for HIV-1 production using the Lenti-X p24 Rapid Titer Kit (Clontech) according to the manufacturer’s instructions. Under these conditions, using 0.3 × 106 donor cells, less than 30% of all cultures were positive for all individuals.
Bulk Cultures.
Bulk cultures were performed as previously described (45). Sequence analysis on bulk culture was also performed as previously described (28).
Q2VOA Sequence Analysis.
The supernatant from p24-positive cultures was extracted using the Qiagen MinElute Virus Spin kit QIAcube. The cDNA was produced using 1:10 diluted RNA with SuperScript III reverse transcriptase (Invitrogen Life Technologies) and the antisense primer env3out 5′–TTGCTACTTGTGATTGCTCCATGT–3′ followed by RNase H digestion (Invitrogen Life Technologies) for 20 min at 37 °C. The full gp160 env was amplified from 1:40 diluted cDNA using envB5out 5′–TAGAGCCCTGGAAGCATCCAGGAAG–3′ and envB3out 5′–TTGCTACTTGTGATTGCTCCATGT–3′ in the first round and the second-round nested primers envB5in 5′–TTAGGCATCTCCTATGGCAGGAAGAAG–3′ and envB3in 5′–GTCTCGAGATACTGCTCCCACCC–3′.
PCR assays were performed using High Fidelity Platinum Taq (Invitrogen) at 94 °C for 2 min; 94 °C for 15 s, 55 °C for 30 s, and 68 °C for 4 min × 35; and 68 °C for 15 min. Second-round nested PCR was performed using 1 μL of PCR1 product as a template and High Fidelity Platinum Taq at 94 °C for 2 min; 94 °C for 15 s, 57 °C for 30 s, and 68 °C for 4 min × 45; and 68 °C for 15 min. PCR2 products were checked using 1% 96-well E-Gels (Invitrogen). PCR bands with the expected HIV envelope size were quantified and subjected to library preparation using the Illumina Nextera DNA Sample Preparation Kit (Illumina) as described (46). Briefly, 10 ng of DNA per band was subjected to tagmentation, ligated to barcoded sequencing adapters using the Illumina Nextera Index Kit, and then purified using AmPure Beads XP (Agencourt). Ninety-six different purified samples were pooled into one library and then subjected to paired-end sequencing using Illumina MiSeq Nano 300 (Illumina) cycle kits at a concentration of 15 pM.
Sequence adapters were removed using Cutadapt v1.8.3. Read assembly for each virus was performed in three steps. First, de novo assembly was performed using Spades v3.6.1 to yield long contig files. Contigs longer than 255 bp were subsequently aligned to an HIV envelope reference sequence, and a consensus sequence was generated using Geneious 8. Finally, reads were realigned to the consensus sequence to close gaps, and a final read consensus was generated for each sequence. Sequences with double peaks (cutoff consensus identity for any residue <75%), with stop codons, or shorter than the expected envelope sizes were omitted from downstream analyses.
Proviral SGA.
DNA from 1 to 10 × 106 CD4+ cells from HIV-1–infected individuals was prepared as previously described (47). Briefly, cells were collected after magnetic isolation and lysed in Proteinase K buffer [100 mM Tris (pH 8), 0.2% SDS, 200 mM NaCl, 5 mM EDTA] and 20 mg/mL Proteinase K at 56 °C for 12 h. Genomic DNA was extracted by phenol chloroform precipitation. Aliquots of the resulting DNA were diluted and used as a template for full-length gp160 PCRs. All PCRs were performed using High Fidelity Platinum Taq (Invitrogen). The first PCR was performed at 94 °C for 2 min; 94 °C for 15 s, 58.5 °C for 30 s, and 68 °C for 4 min × 35; and 68 °C for 15 min. Second-round nested PCR was then performed using 1 μL of PCR1 product as a template and High Fidelity Platinum Taq at 94 °C for 2 min; 94 °C for 15 s, 61 °C for 30 s, and 68 °C for 4 min × 45; and 68 °C for 15 min. PCR2 products were checked using 1% 96-well E-Gels (Invitrogen). Bands with the expected size of the HIV-1 envelope obtained from diluted DNA samples that showed an amplification efficiency of less than 30% were subjected to library preparation and sequencing using the Illumina platform as described above. Hypermutants, sequences with double peaks (cutoff consensus identity for any residue <75%), sequences with stop codons, or sequences shorter than the expected envelope sizes were omitted from downstream analyses.
Viral Neutralization Testing by TZM.bl.
Supernatants from p24-positive Q2VOA wells were tested against a panel of bNabs using the TZM.bl neutralization assay, as previously described (48, 49). Neutralization assays were conducted in a laboratory meeting good clinical laboratory practice quality assurance criteria (laboratory of M.S.S.). Viruses with low median tissue culture infectious dose (TCID50) titers were only tested against 3BNC117 and 10-1074.
GSI.
The GSI (23) was used to calculate the degree of phylogenetic association of the env gene sequences. Phylogenies were inferred using PhyML version 3.0 (50). Multiple bifurcations with intervening zero-length branches were collapsed to polytomies using the di2multi method implemented in the ape package for R (51). These phylogram topologies were used to calculate the gsi values, and P values were determined using 10,000 replicate permutations (23).
Computational Analyses.
Bayesian inference was implemented in Stan (52) to estimate jointly the frequency of integrated proviruses and their reactivation probability. Data from proviral DNA sequencing and viral outgrowth assays were combined across multiple visits, incorporating a prior probability for the gradual decay of the reservoir (3, 53). We assumed a weakly informative prior for the frequency of integrated provirus and a uniform prior for the probability of reactivation. Parameters were estimated individually for viruses with identical sequences observed at least three times across both visits (large clones), whereas sequences observed less frequently were grouped together (small clones). The observed relationship between clone frequency and reactivation probability also holds in simpler models incorporating data from single visits only, and when clones that were not observed in both assays during the same visit are excluded. Additional details of the inference methods used are provided in SI Methods.
Acknowledgments
We thank all study participants who devoted time to our research. We also thank members of the M.C.N. laboratory for helpful discussions. J.C.C.L. is supported by Award 248676/2013-0 from the Brazil Conselho Nacional de Pesquisas “Ciencia sem Fronteiras.” Y.Z.C. is supported by Grant KL2TR001865, National Center for Advancing Translational Sciences (NCATS), and Grant UL1 TR000043, National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program. J.C.C.L. and Y.Z.C. are additionally supported by NIH/National Institute of Allergy and Infectious Diseases Grant U01 AI118536 (to M.C.). The contributions of J.P.B. and A.K.C. were supported by the Ragon Institute of MGH, MIT, and Harvard. E.F.K., G.H.L., and B.H.H. were supported by BEAT-HIV Delaney Collaboratory Grant UM1 AI126620. M.C.N. is a Howard Hughes Medical Investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KY113379–KY114054).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617789113/-/DCSupplemental.
References
- 1.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 3.Crooks AM, et al. Precise quantitation of the latent HIV-1 reservoir: Implications for eradication strategies. J Infect Dis. 2015;212(9):1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes SH, Coffin JM. What integration sites tell us about HIV Persistence. Cell Host Microbe. 2016;19(5):588–598. doi: 10.1016/j.chom.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: What, where and how to target them. Nat Rev Microbiol. 2016;14(1):55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- 6.Eriksson S, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2):e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn LB, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruner KM, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun TW, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 11.Chun TW, et al. In vivo fate of HIV-1-infected T cells: Quantitative analysis of the transition to stable latency. Nat Med. 1995;1(12):1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 12.Evering TH, et al. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog. 2012;8(2):e1002506. doi: 10.1371/journal.ppat.1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Josefsson L, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci USA. 2013;110(51):E4987–E4996. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Stockenstrom S, et al. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J Infect Dis. 2015;212(4):596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner TA, et al. An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol. 2013;87(3):1770–1778. doi: 10.1128/JVI.01985-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barton K, et al. Broad activation of latent HIV-1 in vivo. Nat Commun. 2016;7:12731. doi: 10.1038/ncomms12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Günthard HF, et al. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J Virol. 1999;73(11):9404–9412. doi: 10.1128/jvi.73.11.9404-9412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson JA, et al. Clonal sequences recovered from plasma from patients with residual HIV-1 viremia and on intensified antiretroviral therapy are identical to replicating viral RNAs recovered from circulating resting CD4+ T cells. J Virol. 2011;85(10):5220–5223. doi: 10.1128/JVI.00284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bui JK, Mellors JW, Cillo AR. HIV-1 virion production from single inducible proviruses following T-cell activation ex vivo. J Virol. 2015;90(3):1673–1676. doi: 10.1128/JVI.02520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SK, et al. Quantification of the latent HIV-1 reservoir using ultra deep sequencing and primer ID in a viral outgrowth assay. J Acquir Immune Defic Syndr. September 27, 2016 doi: 10.1097/QAI.0000000000001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laskey SB, Pohlmeyer CW, Bruner KM, Siliciano RF. Evaluating clonal expansion of HIV-infected cells: Optimization of PCR strategies to predict clonality. PLoS Pathog. 2016;12(8):e1005689. doi: 10.1371/journal.ppat.1005689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer S, Josefsson L, Coffin JM. HIV reservoirs and the possibility of a cure for HIV infection. J Intern Med. 2011;270(6):550–560. doi: 10.1111/j.1365-2796.2011.02457.x. [DOI] [PubMed] [Google Scholar]
- 23.Cummings MP, Neel MC, Shaw KL. A genealogical approach to quantifying lineage divergence. Evolution. 2008;62(9):2411–2422. doi: 10.1111/j.1558-5646.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- 24.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouquet H, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA. 2012;109(47):E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar K, et al. Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N Engl J Med. November 9, 2016 doi: 10.1056/NEJMoa1608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheid JF, et al. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature. 2016;535(7613):556–560. doi: 10.1038/nature18929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siliciano JM, Siliciano RF. The remarkable stability of the latent reservoir for HIV-1 in resting memory CD4+ T cells. J Infect Dis. 2015;212(9):1345–1347. doi: 10.1093/infdis/jiv219. [DOI] [PubMed] [Google Scholar]
- 30.Chun TW, Moir S, Fauci AS. HIV reservoirs as obstacles and opportunities for an HIV cure. Nat Immunol. 2015;16(6):584–589. doi: 10.1038/ni.3152. [DOI] [PubMed] [Google Scholar]
- 31.Chun TW, et al. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci USA. 1998;95(15):8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siliciano RF, Greene WC. HIV latency. Cold Spring Harb Perspect Med. 2011;1(1):a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barton K, Winckelmann A, Palmer S. HIV-1 reservoirs during suppressive therapy. Trends Microbiol. 2016;24(5):345–355. doi: 10.1016/j.tim.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallejo A, et al. The effect of intensification with raltegravir on the HIV-1 reservoir of latently infected memory CD4 T cells in suppressed patients. AIDS. 2012;26(15):1885–1894. doi: 10.1097/QAD.0b013e3283584521. [DOI] [PubMed] [Google Scholar]
- 35.Dinoso JB, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci USA. 2009;106(23):9403–9408. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joos B, et al. Swiss HIV Cohort Study HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci USA. 2008;105(43):16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonetti FR, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA. 2016;113(7):1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persaud D, et al. Slow human immunodeficiency virus type 1 evolution in viral reservoirs in infants treated with effective antiretroviral therapy. AIDS Res Hum Retroviruses. 2007;23(3):381–390. doi: 10.1089/aid.2006.0175. [DOI] [PubMed] [Google Scholar]
- 39.Halper-Stromberg A, et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 2014;158(5):989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruel T, et al. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat Commun. 2016;7:10844. doi: 10.1038/ncomms10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halper-Stromberg A, Nussenzweig MC. Towards HIV-1 remission: Potential roles for broadly neutralizing antibodies. J Clin Invest. 2016;126(2):415–423. doi: 10.1172/JCI80561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu CL, et al. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science. 2016;352(6288):1001–1004. doi: 10.1126/science.aaf1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van ’t Wout AB, Schuitemaker H, Kootstra NA. Isolation and propagation of HIV-1 on peripheral blood mononuclear cells. Nat Protoc. 2008;3(3):363–370. doi: 10.1038/nprot.2008.3. [DOI] [PubMed] [Google Scholar]
- 44.Laird GM, Rosenbloom DI, Lai J, Siliciano RF, Siliciano JD. Measuring the frequency of latent HIV-1 in resting CD4⁺ T cells using a limiting dilution coculture assay. Methods Mol Biol. 2016;1354:239–253. doi: 10.1007/978-1-4939-3046-3_16. [DOI] [PubMed] [Google Scholar]
- 45.Caskey M, et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522(7557):487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. Microbial evolution. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science. 2014;344(6191):1519–1522. doi: 10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147(1):95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005;Chapter 12:Unit 12 11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 51.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 52. Stan Development Team (2016) PyStan: The Python Interface to Stan, Version 2.9.0. Available at mc-stan.org.
- 53.Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 54.Gelman A, et al. A weakly informative default prior distribution for logistic and other regression models. Ann Appl Stat. 2008;2(4):1360–1383. [Google Scholar]
- 55.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]