Abstract
The last two decades have seen a worldwide resurgence in infections caused by Treponema pallidum subsp. pallidum, the syphilis spirochete. The syphilis spirochete’s well-recognized capacity for early dissemination and immune evasion has earned it the designation ‘the stealth pathogen’. Despite the many hurdles to studying syphilis pathogenesis, most notably the inability to culture and to genetically manipulate T. pallidum, in recent years, considerable progress has been made in elucidating the structural, physiologic, and regulatory facets of stealth pathogenicity. In this Review, we integrate this eclectic body of information to garner fresh insights into the highly successful parasitic lifestyles of the syphilis spirochete and related pathogenic treponemes.
Pathogenic treponemes cause venereal syphilis, yaws, endemic syphilis, and pinta—multi-stage, infections that, although similar, can be differentiated based on clinical, epidemiologic, and geographic criteria1,2. Only venereal syphilis (see Box 1 for a detailed description) is transmitted by sexual activity. The pathogenic treponemes are uncultivatable, slow-growing microorganisms with identical flat-wave morphologies. They poorly tolerate desiccation, elevated temperature, and ambient oxygen tension, traits that explain why efficient transmission requires close personal contact1,2. Landmark DNA-DNA hybridization studies3,4 showed that DNA from a venereal syphilis spirochete (Nichols strain) shared less than 5% homology with DNA from cultivatable treponemes but was indistinguishable from DNA of the yaws spirochete (Gauthier strain). This work led to the reclassification of the agents of venereal syphilis, endemic syphilis, and yaws as Treponema pallidum subspecies pallidum, endemicum, and pertenue, respectively. Genomic sequencing has established that these subspecies are clonal but form distinct genetic clusters with venereal syphilis strains, the most recently evolved group, likely diverging from the pertenue cluster several thousand years ago5. Genomic sequencing also has revealed that T. paraluiscuniculi (reclassified as Treponema paraluisleporidarum ecovar Cuniculus6), the cause of venereal spirochetosis in rabbits, shares a common ancestor with human pathogenic treponemes7. WHO estimates that ~90 million people in Africa, Asia and the Western Pacific currently are at risk for yaws8. Globally, ~11 million people acquired syphilis in 20089, with mother-to-child transmission occurring in nearly 2 million pregnancies10. T. pallidum is considered the most virulent subspecies because it routinely traverses blood-brain and maternal-fetal placental barriers11.
Box 1. Venereal syphilis in brief.
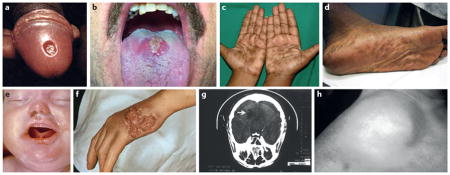
After an incubation period ranging from 9 to 90 days, during which spirochetes are already blood-borne, disease commences with the appearance of the hallmark ulcerative lesion, the chancre (A (http://phil.cdc.gov) and B (courtesy of Kevin Dieckhaus11)), at the site of inoculation. Mucocutaneous lesions of secondary syphilis (C (with permission from Reference 27) and D (courtesy of Adriana Cruz and Juan Salazar)) appear 4 to 10 weeks later. This stage is associated with the highest incidence of spirochetemia, the greatest treponemal burdens in blood and tissues, and the highest titers in serodiagnostic tests11. A pregnant woman with untreated secondary syphilis is at great risk of transmitting the disease to her fetus (E (http://phil.cdc.gov))28. Invasion of the central nervous system in as many as 40% of individuals with untreated early syphilis sets the stage for potentially devastating neurologic complications years later11. After weeks to months, the patient enters a period of latency during which, in the absence of a clinical relapse, the diagnosis can be made only by serologic testing. Latency is divided into early latent (infectious relapses and/or spirochetemia common) and late latent (relapses and/or spirochetemia unlikely) stages. Approximately 25% of patients will experience one or more secondary-like relapses during early latency, 90% of which occur in the first year. Approximately 30% of untreated patients will develop one of the recrudescent syndromes (‘benign’ gummas (F (http://phil.cdc.gov)), cardiovascular syphilis (G, aortic arch aneurysm), and neurosyphilis (H, right cerebellar infarct indicated by arrow, (http://phil.cdc.gov)) collectively designated tertiary syphilis11.
Venereal syphilis is acquired when treponemes are inoculated onto the mucosa or skin during sexual contact. Spirochetes directly penetrate mucous membranes or they enter through breaches in skin produced by sexual activity12. Attachment to host cells and the extracellular matrix is considered to be the crucial initial step of infection13,14. Once below the epithelium, the spirochetes multiply locally and disseminate through the lymphatics and the bloodstream12. T. pallidum’s flexuous, flat-wave morphology (Fig. 1A and B)15 enables it to penetrate tissues and vascular barriers throughout the body, while its periplasmic motility apparatus propels it forward via front-to-back undulations coordinated in response to poorly understood chemotactic signals16,17. Although it is not clear how T. pallidum benefits by invading deep visceral and musculoskeletal tissues, reaching and surviving in distal skin and mucosal sites enhances opportunities for subsequent transmission.
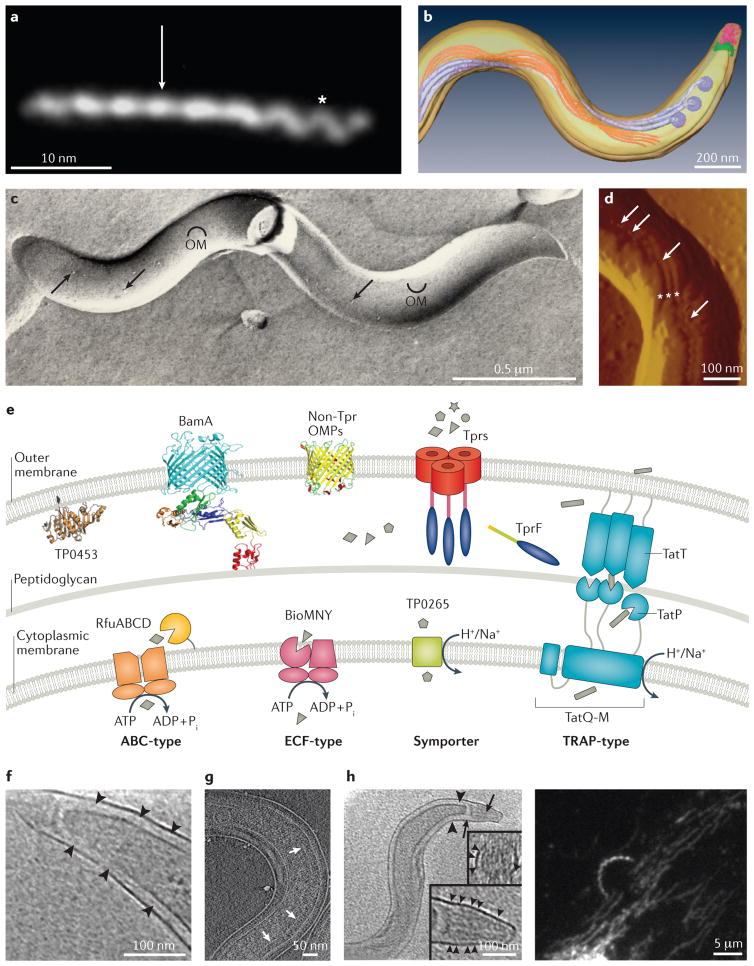
Figure 1. Morphology and cell envelope architecture of T. pallidum, the stealth pathogen.
A. Darkfield micrograph showing the flat wave morphology of T. pallidum. Asterisks and arrowhead indicate segments oriented 90 degrees from each other (with permission from Reference 15). B. Top view of a surface-rendered model of T. pallidum generated from cryoelectron tomograms showing the outer and cytoplasmic membranes (transparent yellow), flagellar motors (basal bodies, dark lavender), flagellar filaments (light lavender), cytoplasmic filaments (orange), cap (green), and cone (pink) (with permission from Reference 15). C. Freeze-fracture electron microscopy reveals scarce particles (integral membrane proteins) within the T. pallidum outer membrane. Convex and concave leaflets of the outer membrane are indicated; arrowheads indicate particles in the two leaflets (with permission from Reference 155). D. Scanning probe microscopy reveals rare particles on the T. pallidum surface (arrows), often located on the bulge in the outer membrane created by the underlying periplasmic flagella (asterisks) (with permission from Reference 91). E. Model for the molecular architecture of the T. pallidum cell envelope. Shown in the outer membrane are BamA (TP0326)59,64, a generic bipartite (that is, full-length) Tpr attached by its N-terminal portion to the peptidoglycan 59–61, a generic non-Tpr β-barrel, and TP0453 (p30.5), a structurally characterized lipoprotein attached to the outer membrane inner leaflet156. Tprs, such as TprF, lacking the MOSPC β-barrel forming domain, are located in the periplasm59. Substrates present in high concentration in the extracellular milieu probably traverse the outer membrane by simple diffusion through porins, such as TprC. Prototypic ABC-like transporters use a periplasmic substrate-binding protein (SBP), typically lipoproteins, and components with transmembrane and ATP-binding domains. The energy coupling factor (ECF)-type ABC transporters uses a transmembrane ligand-binding protein in place of a separate periplasmic SBP. To substitute for ATP hydrolysis, the symporters use a transmembrane permease that relies on energy from a chemiosmotic or electrochemical gradient that may be generated by the Rnf complex. The tripartite ATP-independent periplasmic (TRAP) transporters also lack ATP-binding modules and use transmembrane electrochemical gradients, but they are more complex. T. pallidum seems to have evolved a variation on the TRAP theme by also using an additional periplasmic component protein (TatT (TP0956)) containing a tetratricopeptide repeat (TPR) motif, giving rise to a newly described system denoted as a TPR protein-associated TRAP transporter (TPAT). The TPR protein TatT likely associates with the SBP TatP (TP0957) in a heterohexameric fashion to carry out ligand binding and uptake; structural analyses suggest that this complex may accommodate a chain-like hydrophobic molecule(s), such as a long-chain fatty acid(s). Uptake likely is facilitated by a putative membrane permease (TatQ-M (TP0958)) of the TPAT system101,102. F. Cryotomographic section of T. pallidum near the cell end showing the peptidoglycan layer (arrowheads) midway between the outer membrane and cytoplasmic membrane (with permission from Reference 15). G. Cryotomographic section of T. pallidum showing chemoreceptor arrays (arrows, with permission from Reference 91). H. Cryotomographic slice showing the cone-shaped structure at T. pallidum cell ends (with permission from Reference 15) along with darkfield micrograph of T. pallidum stably attached via its tip to the surface of a trophoblast cell. White arrowheads indicate fine fibers between the cone and the outer membrane.
Venereal syphilis can be thought of as a contest between the ability of T. pallidum, generally considered to be an extracellular pathogen, to avoid recognition and the adeptness of the host’s innate and adaptive immune responses to ‘track down’ the pathogen12. The spirochete’s fragile outer membrane lacks lipopolysaccharide (LPS), the highly proinflammatory glycolipid found in Gram-negative bacteria18. Although T. pallidum expresses numerous lipoproteins capable of activating macrophages and dendritic cells (DCs) through Toll-like receptor (TLR) 2-dependent signaling pathways19, they are predominantly below the surface13. The paucity of surface-exposed pathogen-associated molecular patterns (PAMPs) enables the bacterium to undergo repeated bouts of dissemination that are poorly detected by innate immunity and also explains the lack of systemic inflammatory symptoms characteristic of the disease11. The appearance of opsonic antibodies, which promote internalization, killing, and degradation of spirochetes within phagolysosomes, represents a turning point in the battle between host and pathogen19–21. However, by liberating lipopeptides for binding to TLRs lining the interior of the phagosome and antigenic peptides for presentation to resident and locally recruited T cells, bacterial clearance and killing becomes a double-edged sword. The ensuing inflammatory response causes tissue damage that gives rise to clinical manifestations11. Spirochetes replicating in tissues elicit a complex and variable inflammatory cell infiltrate consisting of macrophages, CD4+ and CD8+ T lymphocytes, and plasma cells, accompanied by varying degrees of endothelial cell swelling and proliferation. Production of IFN-γ by locally activated CD4+ and CD8+ T cells enhances the capacity of macrophages to internalize and degrade spirochetes but also bolsters their output of potentially tissue damaging, proinflammatory cytokines22–26.
Importantly, spirochetes are not easily cleared by opsonic antibodies; in fact, during secondary and early latent syphilis, viable bacteria circulate despite high titers of anti-treponemal antibodies27,28. How the pathogen burden is eventually decreased and latency established and maintained is a mystery. Passive immunization with sera from previously infected rabbits immune to rechallenge with T. pallidum suggests that antibodies play a critical role in suppressing spirochete burdens during latency29,30, although a contribution by cellular immunity cannot be ruled out19,31. Equally mysterious is where spirochetes reside during latency. Experiments in rabbits have suggested that T. pallidum may use hair follicles and nerves as protected niches32. T. pallidum’s capacity to survive for prolonged periods while hiding ‘in plain sight’ of humoral defenses represents its defining feature as a stealth pathogen33. In the remainder of this Review, we integrate an eclectic and sometimes puzzling body of information to garner fresh insights into a fundamental question of syphilis pathogenesis: how does T. pallidum make a living in its obligate human host?
Experimental roadblocks and new directions
T. pallidum was among the first major bacterial pathogens of humans to be identified, yet our knowledge of syphilis pathogenesis lags well behind that of other common bacterial infections34. One of the main reasons for this is the syphilis spirochete’s refractoriness to cultivation in vitro, despite intensive efforts dating almost from its discovery in 190635. In the 1940s, investigators defined incubations conditions and medium components that maintained treponemal motility and virulence for 6–8 days, although without replication36. In the early 1980s, reproducible increases (20- to 100-fold) in replication, with full preservation of motility and virulence, were obtained by cultivating treponemes under microaerophilic conditions in the presence of cottontail rabbit epithelial (Sf1Ep) cells37,38. Subsequent efforts to improve on these results by meticulously defining growth conditions and parameters yielded new information about T. pallidum growth requirements but only modest increases in replication without achieving continuous culture35,39. Although the genomic sequence confirmed that T. pallidum depends upon its host for virtually all of its nutritional requirements18, it did not yield obvious, untried solutions to the cultivation problem35. A contemporary bioinformatics-based analysis of the possible functions of the more than 400 ‘hypothetical’ proteins encoded in the T. pallidum genome40 may inform future attempts. Recent recognition of the importance of riboflavin uptake and flavin utilization for spirochete metabolism and energy generation may be pertinent as well41,42. Therefore, investigators today still rely upon intratesticular inoculation of rabbits to isolate T. pallidum strains from clinical samples and propagate them for experimentation43.
The once popular notion that T. pallidum’s poor surface antigenicity can be attributed to a pseudo-capsule of serum proteins and mucopolysaccharides44 gave way in the early 1990s to overwhelming evidence that the bulk of its immunogenic molecules are subsurface and that its delicate outer membrane forms the protective barrier13,45. Despite the formidable roadblock imposed by the cultivation barrier, compounded by the lack of a facile, inbred animal model for assessing protective immunity46, our understanding of this unorthodox bacterial outer membrane and its interactions with host defenses has increased substantially in recent years13,47. In parallel, there has been growing interest in the physiologic and regulatory foundations of stealth pathogenicity. Residence in a rich, relatively homeostatic environment has enabled T. pallidum to dispense with genes for de novo synthesis of nucleotides, fatty acids, vitamins, co-factors, amino acids, the tricarboxylic acid cycle, and oxidative phosphorylation18, resulting in the smallest of the spirochete genomes (~1.1 MB) and one of the smallest among pathogenic bacteria48. During the course of genomic reduction, the bacterium has undergone remarkable adaptations that enable it to acquire all of its required nutrients and optimize their usage within diverse niches, while coping with exogenous and endogenous stress.
The face of stealth-the outer membrane
Studying the properties and composition of the outer membrane of T. pallidum has been, and remains, arduous13,45. The quest for rare outer membrane proteins (OMPs) began in earnest with the discovery by freeze-fracture electron microscopy (Fig. 1C), recently confirmed by scanning probe microscopy (Fig. 1D), that T. pallidum contains ~100-fold fewer OMPs than Escherichia coli49,50. How then does T. pallidum meet its nutritional requirements and carry out its complex parasitic lifestyle with a minimalist outer membrane? A partial answer may lie with the bacterium’s slow (~30 h) rate of replication51, presumably an evolutionary ‘compromise’ between the density of OMPs needed for viability and the demands of stealth.
Tpr proteins
Little progress identifying rare OMPs was made until the genomic sequence revealed the existence of the T. pallidum repeat (Tpr) proteins, a 12 member paralogous family with sequence homology to the major outer sheath protein (MOSP) of Treponema denticola18,52,53, which is a known pore-forming protein and adhesin54. Of these, TprK (TP0897)21,52,55 has received the greatest attention because of its proposed role in immune evasion (see below), although its status as a bona fide OMP has been challenged56,57. Except for BamA (TP0326, originally known as TP9258), the T. pallidum genome does not encode proteins with a high degree of sequence homology to well-characterized OMPs of Gram-negative bacteria18. As an alternative to genome mining for OMP orthologs, we used a computational matrix to identify proteins predicted to adopt the hallmark conformation of an OMP, the β-barrel57. The two highest ranked Tprs, TprC/D (TprC (TP0117) and TprD (TP0131) are identical in the Nichols strain) and TprI (TP0620), possess all of the properties expected of a rare OMP59,60. The native proteins are low in abundance (~200 copies per cell), surface-exposed, and amphiphilic, whereas the folded recombinants form β-sheet-rich, heat-modifiable trimers that readily insert into artificial membranes. Similar to MOSP61, integration of TprC/D and TprI into liposomes results in permeability increases comparable to those produced by the archetypal porin, E. coli OmpF59,60. Formation of large, non-selective channels could explain how these rare proteins function cooperatively to meet the spirochete’s nutritional needs. With classical porins, the entire polypeptide forms the β-barrel62; a pivotal discovery is that TprC/D and TprI are bipartite (Fig. 1E)59,60. A C-terminal domain forms the β-barrel, while the N-terminal portion extends into the periplasm, anchoring the barrel to the murein sacculus. Consistent with this bipartite model, TprF, a truncated TprC/D/I ortholog of these Tprs lacking the C-terminal β-barrel domain, is periplasmic. Of note, these analyses provide a functional and topological template for the extended Tpr family61,63.
BamA and outer membrane biogenesis
Among the non-Tpr proteins predicted to form β-barrels57, BamA is of paramount importance. BamA is the essential central component of the β-barrel assembly machine (Bam) that catalyzes the insertion of newly exported OMPs into the outer membrane64,65. Like other members of the Omp85 superfamily, BamA has a dual domain architecture consisting of an outer membrane-inserted, C-terminal β-barrel and periplasmic polypeptide transport-associated (POTRA) repeats (in this case, five, Fig. 1E)64,65. As in other bacteria, BamA in T. pallidum is part of a complex (~400 kDa) whose subunits, unidentified thus far, assist in the insertion process64. Conceivably, the periplasmic chaperones Skp (TP0327) and SurA (TP1016), which ferry newly exported OMPs to the Bam apparatus66, function together with the POTRA repeats as a gatekeeper that regulates the protein composition and content of the outer membrane. A homology model based upon the solved structure of Neisseria gonorrhoeae BamA67 predicted that T. pallidum BamA contains a 16-stranded β-barrel in which short, weakly hydrogen-bonded β1 and β16 strands separate to allow lateral insertion of nascent OMPs into the outer membrane65. The BamA barrel has eight extracellular loops, with three (L4, L6 and L7) forming a dome that occludes the barrel’s extracellular opening, directing nascent OMPs laterally into the bilayer. Although much of the dome is poorly immunogenic, L4 contains a surface-exposed, immunodominant, opsonic epitope, a potential Achilles’ heel likely imposed by functional constraints on the β-barrel.
Adhesins
In vitro studies well predating the genomics era demonstrated that T. pallidum can bind to a variety of mammalian cell types68,69 as well as extracellular matrix proteins, particularly laminin and fibronectin68,70. Using bioinformatics to identify potential outer membrane proteins in combination with binding assays, potential adhesins for fibronectin (TP0155 and TP0483)71 and lamin (TP0751) were identified72,73. In addition to being an adhesin, TP0751 is a zinc-dependent metalloprotease (hence, was named “pallilysin”) that forms a complex with TP0751 (which contains a Von Willebrand factor type A domain) capable of degrading clots and extracellular matrix, which could facilitate both dissemination and attachment74,75. The lipoprotein TP0136 is a fibronectin-binding adhesin with sequence heterogeneity among T. pallidum strains (Table 1)76,77. Most recently, elegant gain of function experiements using B. burgdorferi as a surrogate genetic host have shown that the lipoprotein TP0435 (Tpp17) functions as a cytadhesin 78.
Table 1.
Candidate proteins potentially responsible for phenotypic differences between T. pallidum subsp. pertenue (TPE) and subsp. pallidum (TPA) strains*
| Protein | Name | Predicted function and comments | References |
|---|---|---|---|
| TP0009 | TprA | Potential porin (when C-terminal domain is present) | 59–61 |
| TP0117** | TprC | Porin | 59,60 |
| TP0131** | TprD | Porin | 59,60 |
| TP0133 | HP, immunogenic protein | 157,158 | |
| TP0134 | HP, putative outer membrane protein | Annotation | |
| TP0136** | Fibronectin-binding protein | 76,77 | |
| TP0314 | HP, not annotated in TPA | Annotation | |
| TP0316 | TprF | Potential porin (when C-terminal domain is present) | 59,60 |
| TP0326** | BamA | Originally Tp92; Outer membrane biogenesis | 58,64,65 |
| TP0433** | Arp | Acidic repeat protein, function unknown | Annotation |
| TP0462** | HP, putative lipoprotein | Annotation | |
| TP0488** | Mcp2-1 | Methyl-accepting chemotaxis protein | Annotation |
| TP0548** | Predicted rare outer membrane protein | 57 | |
| TP0619** | HP | Annotation | |
| TP0620** | TprI | Porin | 59,60 |
| TP0621 | TprJ | Potential porin | 59–61 |
| TP0733 | HP, outer membrane protein | Annotation | |
| TP0858 | Predicted outer membrane protein | 57 | |
| TP0865** | HP, putative outer membrane protein | Annotation | |
| TP0897** | TprK | Conflicting evidence regarding status as a bona fide outer membrane protein (see text) | 52,56,61 |
| TP0968 | HP | Annotation | |
| TP1031** | TprL | Potential porin | 59–61 |
Proteins with six or more amino acid replacements and/or major sequence differences between TPE and TPA strains according to Čejková et al.143
Candidates for phenotypic differences between TPA strains (Nichols, DAL-1, Chicago, SS14, and MexicoA).
HP, hypothetical protein.
Implications for limiting surface antigenicity
While there is consensus that resistance to antibody binding is the basis for immune evasion by T. pallidum12,55, much remains to be learned about how the pathogen accomplishes this impressive feat and, conversely, the counter-measures used by the host. Early immunolabeling experiments suggested that the outer membrane of T. pallidum is antigenically inert79,80. However, this simplistic notion is contradicted by biological assays showing that syphilitic infection does, in fact, induce antibodies that react with the bacterial surface19. Moreover, immunolabeling and opsonophagocytosis assays have revealed that T. pallidum populations are heterogeneous, consisting of antibody-binding and non-binding subpopulations, and that organisms that bind antibodies do so with markedly slow kinetics26,57,81. These observations likely explain the longstanding paradox that clearance and persistence of T. pallidum occur simultaneously26. At a given site, antibody binders would be slowly cleared, also ‘feeding’ inflammation, whereas immunoevasive non-binders would replicate locally and disseminate systemically.
How, then, does heterogeneity in surface antigenicity occur? With BamA, TprC/D, and TprI, the large majority of antibodies are directed against periplasmic domains60,64,65 and, are, therefore, ineffective for clearance. For those antibodies that are potentially opsonic, the extremely low copy numbers of the target proteins likely limits their accessibility, as evidenced by the sizable percentage of treponemes that fail to become surface-labeled or opsonized by high-titer recombinant antisera against TprC/D or BamA59,65. Similarly, recent studies suggest that the TP0750–TP0751 protease complex74,75 and TP043578 are expressed at low levels on the surfaces of some treponemes and, consequently, of limited availablity to antibody binding. Two distinct mechanisms for modulating antibody reactivity with Tprs have been proposed. TprK undergoes sequence and antigenic variation in seven hypervariable regions, B-cell epitopes proposed to be located in external loops, through gene conversion from a donor site located elsewhere on the chromosome55,82–86. Recent resequencing of the genome of strain SS14 has revealed sequence heterogeneity within the β-barrel-encoding portion of TprD (TP0131)87, suggesting that this OMP also undergoes antigenic variation. A second mechanism pertains to TprE, TprG and TprJ; changes in the number of G nucleotide repeats immediately upstream of their transcriptional start sites, due to slipped strand mispairing during DNA replication, influences the amount of message produced88. Unquestionably, much more investigation, including far better characterization of T. pallidum’s entire OMP repertoire and individual surface molecules, is needed before the pieces of this complex puzzle come together.
What lays beneath
Cell envelope structure
The architectural plan for the cell envelope of T. pallidum differs substantially from that of Gram-negatives89. Images obtained by conventional transmission electron microscopy of fixed, embedded organisms showed the outer and cytoplasmic membranes juxtaposed with the peptidoglycan layer sandwiched between90. However, visualization of the cell envelope in its native state by cryoelectron tomography15,91 revealed that the two membranes are actually well separated with a thin peptidoglycan layer creating two distinct zones within the periplasmic compartment (Fig. 1F). The lower, denser zone contains the periplasmic domains of integral proteins of the cytoplasmic membrane (various permeases), and the polypeptide moieties of lipoproteins, many of which are substrate-binding proteins (SBPs) for ABC transporters (Fig. 1E). Near the cell poles are chemoreceptor arrays containing the T. pallidum’s four methyl-accepting chemotaxis proteins (Mcp1 (TP0040), Mcp2 (TP0488), Mcp3 (TP0639) and Mcp4 (TP0640)) (Fig. 1G)91, sensors that bind exogenously derived ligands within the periplasm and relay chemotactic signals to the more distal flagellar motors92. A cone-like lattice at the cell tips, connected by fine fibrils to the outer membrane, could function as an organelle mediating end-on attachment (Fig. 1H)15,91. The proximity of this tip structure to the flagellar motors and chemoreceptor arrays theoretically enables close coordination between attachment, environmental sensing and directed motility. By providing receptors distributed along the length of the cell body, the aforementioned adhesins would enable organisms to gain an initial ‘foothold’ before becoming stably attached at their tips.
Transport across the cytoplasmic membrane
T. pallidum uses diverse ABC transporters and symporters (totaling ~5% of its genome) to transfer molecules required for cell viability from periplasm to cytosol (Fig. 1E, also see TransportDB, http://membranetransport.org/). Following passive diffusion of glucose across the outer membrane through nonselective porins, the methylgalactoside (Mgl) glucose-galacatose ABC transporter (consisting of TP0545 and TP0684-0686) is believed to mediate high-affinity transport across the cytoplasmic membrane93. Consistent with its lack of biosynthetic capacity, T. pallidum seems to possess a broad, though not easily understood, transporter repertoire for amino acids. One ABC transporter, MetI-MetN-MetQ (TP0119-TP0120-TP0821 (also known as TpN32)) is dedicated to the uptake of methionine94. The genome encodes three other putative SBPs with predicted specificities for oligopeptides (OppA (TP0585)), histidine (HisJ (TP0308)) and polar amino acids (TP0309), but their corresponding permeases and ATP-binding proteins have yet to be identified. Rounding out the T. pallidum’s amino acid requirements are putative symporters for aspartate and glutamate (TP0555 and TP0934), alanine and glycine (TP0414 and TP0998) and branched chain amino acids (TP0265). In addition, TP0144 functions as the SBP for a thiamine ABC transporter (TbpAPQ(TP0142-0144))95. BioMNY (TP0226-0228), a putative biotin importer, seems to be the bacterium’s only energy coupling factor (ECF)-type ABC transporter96.
Structural biology has become an essential tool for studying cell envelope constituents of T. pallidum. The crystal structure of TroA (TP0034), originally thought to be a rare OMP, revealed that it belonged to a newly discovered class of SBPs associated with transition-metal ABC transporters97. Crystallography and binding studies showed TmpC (TP0319) to be the SBP for the first ABC-type purine nucleoside transporter system (PnrABCDE (TP0319-323)) described in any bacterium98. Another lipoprotein, TP0655, was shown to be the SBP for a polyamine transporter (PotABCD (TP0652-0655)) with nanomolar binding affinities for putrescine and spermidine99. The X-ray structure of TP0298, also a lipoprotein, revealed it to be the SBP of a riboflavin transporter (RfuABCD (TP0298-0302))41 which collaborates with a dual function FAD pyrophosphatase-FMN transferase (Ftp (TP0796))42,100 to meet the T. pallidum’s prodigious requirements for flavin cofactors. A longstanding question has been how T. pallidum, a fatty acid auxotroph, obtains essential long-chain fatty acids (LCFAs). Recent structural analyses have led to the description of a multimeric lipoprotein complex (/TatT-TatP (TP0956–TP0957)) thought to comprise a tetratricopeptide repeat (TPR)-protein associated transporter that traffics LCFAs across the periplasm101,102.
Carbohydrate utilization and energy generation
Access to a plentiful supply of glucose in blood and interstitial fluids almost certainly explains why T. pallidum can rely on glycolysis as its primary means for generation of ATP (Fig. 2). In line with this notion is the lack of a complete pathway for gluconeogenesis and the inability to β-oxidize fatty acids and catabolize amino acids18. Telling in terms of the importance of glucose for driving T. pallidum’s ‘engine’ are classic studies showing rapid loss of motility upon glucose deprivation and rapid resumption of motility with it restored103. Whether T. pallidum needs alternative carbon sources is unclear. In contrast to Borrelia burgdorferi, the Lyme disease spirochete104, T. pallidum lacks genes for importing and utilizing exogenous glycerol, instead using glycerol phosphate dehydrogenase (GpsA (TP1009)) to convert dihydroxyacetone phosphate to glycerol-3-phosphate for phospholipid synthesis18. It also lacks phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) permeases (a ubiquitous means among bacteria for importing and regulating uptake of alternative sugars105), relying, instead, on hexokinase to phosphorylate glucose as the first step in glycolysis18. Bioinformatics, however, predict at least one additional ABC sugar permease (TP0075–TP0076), which uses two different SBPs (TP0074 and TP0737) and a nucleotide ATP-binding subunit, TP0804, an ortholog of E. coli MalK18. The functionality of this putative multiple sugar transporter is supported by the observation that, in addition to glucose, maltose and mannose can support replication of T. pallidum in vitro35.
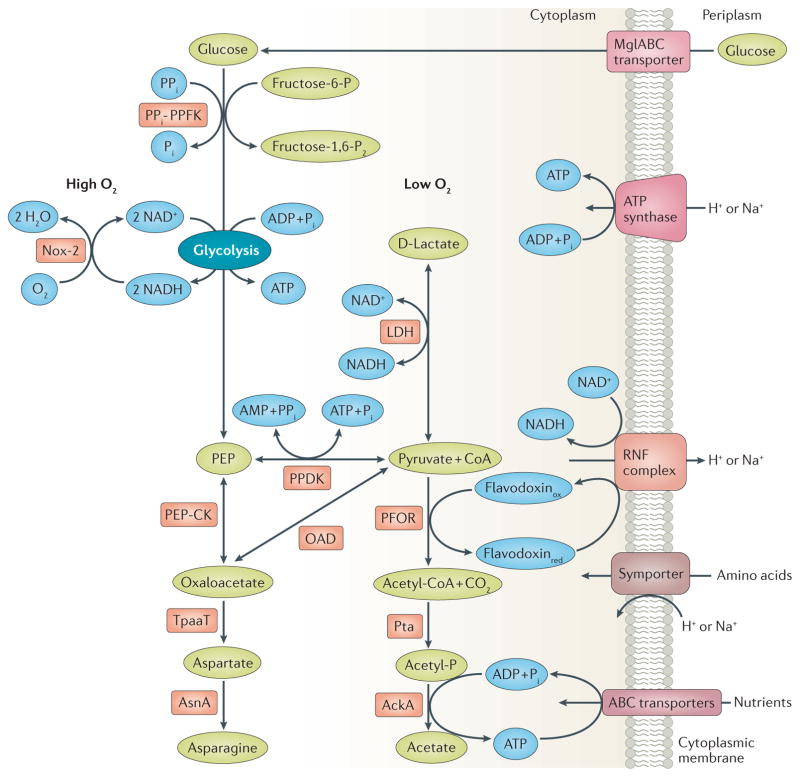
Figure 2. Energy generation, amino acid biosynthesis, and regeneration of NAD+ in T. pallidum.
T. pallidum’s glycolytic and pyruvate-to-acetate fermentation pathways are coupled to a putative Rnf pump, resulting in production of ATP by an ATP synthase; phosphoenolpyruvate (PEP) from glycolysis can be further metabolized to amino acids via PEP carboxykinase (PEP-CK). Metabolites, enzymes, and cofactors are shown in green, red, and blue, respectively. Abbreviations for enzymes are as follows: PPFK, phosphofructokinase; Nox, NADH oxidase; PPDK, pyruvate phosphate dikinase; OAD, oxaloacetate decarboxylase; TpaaT, aspartate aminotransferase; AsnA, asparagine synthetase; LDH, D-lactate dehydrogenase; PFOR, pyruvate-flavodoxin oxidoreductase; Pta, phosphate acetyl transferase; AckA, acetate kinase. Abbreviations for compounds/metabolites are as follows: Fru-6-P, fructose 6-phosphate; Fru-1, 6-P2, fructose 1, 6-bisphosphate; Pi, inorganic phosphate; PPi, inorganic pyrophosphate; CoA, coenzyme A; Acetyl-P; acetyl phosphate; RNF, a putative chemiosmotic pump with similarities to the Rhodobacter nitrogen fixation complex; Flavodoxinox and Flavodoxinred, oxidized and reduced forms of flavodoxin.
Some clever modifications in the conventional glycolytic pathway, shared with phylogenetically diverse organisms (eubacteria, protozoa, and plants106), enable T. pallidum to extract as much energy as possible from substrate level phosphorylation while enhancing metabolic versatility (Fig. 2). To spare ATP, T. pallidum uses a phosphofructokinase employing pyrophosphate (PPi-PPFK (TP0542)) instead of the ATP-dependent enzyme (ATP-PPFK)107; B. burgdorferi, also glycolysis-dependent, uses an orthologous enzyme108. Moreover, whereas ATP-PPFK is irreversible and a major point for allosteric regulation of standard glycolysis109, PPi-PFK is reversible and non-allosteric107. A second modification is the substitution of the reversible pyruvate phosphate dikinase (PPDK (TP0746)) for pyruvate kinase18,108. When coupled to adenylate kinase (TP0595), PPi-PPDK yields four ATPs per glucose compared with the two from pyruvate kinase in standard glycolysis110. The reversibility of the PPDK reaction, in concert with PEP carboxykinase (PEPCK (TP0122)), and oxaloacetate decarboxylase (OadA-OadB (TP0056–TP0057), creates a cycle for inter-conversion of pyruvate, PEP and oxaloacetate. PEP likely is central for T. pallidum to sense and regulate carbon and nitrogen flux (Box 2). T. pallidum can use oxaloacetate for its limited amino acid metabolism through conversion to aspartate by aspartate aminotransferase (TpaaT (TP0223)), with glutamate as the amine donor, and subsequently to asparagine by asparagine synthetase (AsnA (TP0556)). T. pallidum produces another ATP by converting pyruvate to acetate by pyruvate-flavodoxin oxidoreductase (PFOR (TP0939)), acetate kinase (AckA (TP0476)) and phosphate acetyl transferase (Pta (TP0094)). Importantly, whereas the more common pyruvate dehydrogenase reduces NAD+ to NADH, the electrons released by PFOR during oxidative decarboxylation of pyruvate are transferred to a low potential single electron carrier, probably flavodoxin (TP0925), which, as noted below, may then transfer them to a newly discovered pathway for chemiosmotic energy conservation.
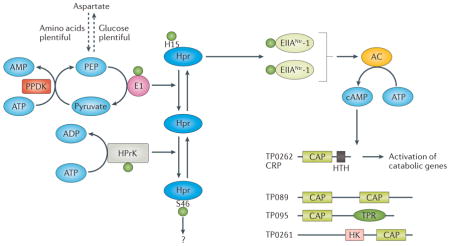
Box 2. A chimeric PTS for control of carbon and nitrogen metabolism.
Although T. pallidum has access to a continuous supply of nutrients, their availability and proportions likely vary in different niches. The phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) appears to be the spirochete’s primary mechanism for adjusting carbon and nitrogen utilization. Two general types of PTS have been described: a canonical PTS dedicated to carbohydrate utilization and a nitrogen-related PTS (PTSNtr) that is exclusively regulatory105,147. Because T. pallidum lacks PTS permeases, its PTS must be solely regulatory. However, it is also chimeric, combining elements derived from the sugar PTSs of Gram-negatives (Enzyme I (EI (TP0575)) and histidine phosphocarrier protein (Hpr (TP0589)) and Gram-positives (Hpr kinase (HprK (TP0591)) with the PTSNtr-derived components EIIANtr-1 (TP0085) and EIIANtr-2 (TP0755)105,148. The resulting hybrid seems capable of intricate metabolic control. Although EI and HprK both phosphorylate Hpr, they do so at different sites: E1 at His-15 (the catalytic site) and HprK at Ser-46 (the regulatory site), with PEP and ATP as the respective phosphoryl donors149,150. Since phosphorylation at one site inhibits phosphorylation at the other, the activities of EI and HprK seem counter-regulatory. Recalling the inter-conversion of PEP and aspartate through oxaloacetate described earlier, one can envision how cross-talk between the EI and HprK-Hpr pathways might regulate carbon-nitrogen flux; our proposed scheme is based on Saier’s postulate that the HprK-Hpr pathway is the direct regulator of glycolysis in treponemes151. When amino acids are relatively scarce, glycolysis would have to furnish substrate for their production in addition to meeting the cell’s energy needs. The resulting drain on PEP would diminish phosphorylation of Hpr at His-15 by EI, leading to enhanced glycolysis driven by HprK-Hpr. When amino acids are plentiful, the flow would be reversed, and aspartate would be converted to PEP, promoting the EI pathway and antagonizing phosphorylation of Hpr at Ser-46 by HprK.
In Gram-positives, Hpr(Ser)-P activates catabolite control protein A (CcpA), the transcription factor responsible for carbon catabolite repression105. T. pallidum does not contain this regulatory protein, so the effector molecule(s) influenced by the Hpr-HprK pathway is unknown. In E. coli, dephosphorylated EIIANtr broadly influences metabolism by regulating intracellular potassium levels through its interactions with the potassium transporter Trk152, which T. pallidum possesses. Interacting partners of phosphorylated EIIANtrs have not been identified in any bacterium153. Given the elements T. pallidum has appropriated from the sugar PTS, one possibility is that one or both phosphorylated EIIANtrs activate adenylate cyclase (TP0485)105. Although admittedly without precedent, it is important to note that TP0485 belongs the largest and most diverse group of adenylate cyclases, Class III, whereas the extensively studied E. coli enzyme belongs to Class I154. This has provocative implications given T. pallidum’s four predicted cAMP-binding proteins (TP0089, TP0261, TP0262, and TP0095) and the well-established link between production of cAMP and bacterial virulence154.
Besides depriving T. pallidum of the full energy content of glucose, the absence of oxidative phosphorylation creates two metabolic quandaries: (i) how to regenerate NAD+ from NADH in order to maintain glycolysis and (ii) how to generate the electrochemical potential across the cytoplasmic membrane that is needed to drive the flagellar motor and nutrient uptake by symporters. T. pallidum uses lactate dehydrogenase (LDH (TP0037)) and the flavoprotein NADH oxidase-2 (NOX-2 (TP0921)) to resolve the first dilemma (Fig. 2)111,112. Under low oxygen conditions, such as on mucosal surfaces, conversion of pyruvate to lactate by LDH predominates. In aerobic environments, NOX-2 uses NADH as the electron donor to reduce molecular oxygen to water. Once in the presence of oxygen, lactate generated under anaerobic conditions can be reclaimed as pyruvate, with NOX-2 regenerating NAD+ from the resulting NADH. Thus, NOX-2 serves a dual purpose: it promotes production of acetate from pyruvate and protects against oxygen toxicity. Reliance on NOX-2 likely accounts for the observation that T. pallidum, once considered an anaerobe because of its exquisite sensitivity to ambient oxygen concentrations, actually requires 3–5% O2 for optimal replication35,113, values closely matching those within mammalian tissues114. To resolve the second dilemma, with the added dividend of boosting energy production, T. pallidum is believed to couple the generation of an electrochemical gradient by a non-canonical flavin-dependent Rhodobacter nitrogen fixation (Rnf) Na+/H+ redox pump to ATP synthesis by one or both of its A-type ATP synthases (formerly annotated as V-type ATPases) (Fig. 2)42,115. Transfer of electrons from reduced flavodoxin, generated by PFOR, drives the electrogenic pump115 with NAD+, generated by LDH or NOX-2 depending upon oxygen availability, the ultimate electron acceptor.
Transition metals and redox stress
Transition metal uptake
Although lacking many well-characterized bacterial metalloproteins, genome mining and biochemical studies have clearly established that T. pallidum requires all three transition metals—(iron, manganese and zinc) to fulfill vital structural and catalytic functions116,117; in fact, the recent identification of multiple Fe-S cluster proteins indicates a greater need for iron than previously recognized. Characteristically, T. pallidum seems to use arcane strategies to wage the battle for transition metals117. It cannot synthesize siderophores and does not possess a TonB ortholog for energizing transport across the outer membrane. It can extract iron from surface-bound lactoferrin and transferrin118, but the receptors are unknown. TpD (TP0971), a 34-kDa lipoprotein avidly binds human lactoferrin but is periplasmic119, and, therefore, cannot be functionally analogous to TbpB, the Neisseria gonorrhoeae lipoprotein co-receptor for lactoferrin120. Ferriportin-mediated efflux of soluble Fe2+ from host cells,121 with subsequent diffusion across the outer membrane, might bybass the need for TonB-dependent uptake in some tissues. Whereas many pathogens possess highly redundant systems for transport of transition metals across the cytoplasmic membrane, T. pallidum seems to accomplish this task with just two ABC transporters: Tro (transport-related operon), which can import iron, manganese, and zinc, and the zinc-dedicated Znu116,122. Import of iron across the cytoplasmic membrane may be complemented by TP0972, a member of the iron permease Ftr1 superfamily119,123,124. A final metal-related oddity is T. pallidum’s ostensible lack of a global, transition metal-dependent transcriptional regulator. The tro operon encodes a zinc- and/or manganese-responsive DtxR-like repressor, TroR, which has no predicted binding sites outside of the tro promoter116,125.
Dealing with oxidative stress
Because T. pallidum lacks many of the anti-oxidant enzymes common to other bacteria, its defenses against oxidative stress were long obscure, even with the availability of the genomic sequence. As information on novel anti-oxidant proteins in bacteria has grown in recent years, it has been possible to elucidate T. pallidum’s redox defense mechanisms117,126. Instead of a classical superoxide dismutase, T. pallidum uses two iron-active center proteins, superoxide reductase (TP0823) and its reductant rubredoxin (TP0991), both acquired from hyperthermophilic anaerobes by a free-living ancestor,127 to protect against O2•−. The presence of these and other iron-containing proteins creates the need to carefully regulate intracellular iron stores to minimize iron-catalyzed production of protein-damaging and DNA-damaging hydroxyl radicals128. T. pallidum accomplishes this using the dps gene product TpF1 (TP1038), a dodecameric bacterioferritin with ferroxidase activity that binds soluble cytosolic Fe2+ and sequesters it as insoluble Fe3+ 129. For defense against peroxides, the bacterium depends upon a single robust, broad-spectrum, NADPH-dependent peroxiredoxin, the flavoprotein alkyl hydroperoxide reductase C (TpAhpC (TP0509))130. In other bacteria, AhpC-like peroxidredoxins are recycled by a dedicated flavoprotein disulfide reductase, alkyl hydroperoxide reductase F (AhpF). T. pallidum not only lacks this enzyme, its TpAhpC has diverged from other AhpCs to the extent that it cannot use AhpF as an electron donor; it relies instead on thioredoxin (TrX (TP0919)) and thioredoxin reductase (TrxR (TP0814))130. Both TpAhpC and TpTrx are expressed at very high levels in spirochetes freshly harvested from rabbit testes130, suggesting that peroxides are a major form of oxidative stress encountered during infection. TxR and TrxR presumably do double-duty defending against disulfide stress, although the low molecular weight, primary thiol redox buffer remains unidentified. Also unclear is how, without known redox regulatory proteins (for example, OxyR, SoxR, and PerR), the bacterium senses oxidative stress, although in vitro studies indicate it has the capacity to do so130. The degree of disulfide bonding between TpF1131 and Tpp17132 monomers may serve as non-transcriptional means for redox sensing in the cytosol and periplasm, respectively.
Regulation of gene expression
Unlike most pathogens and all environmental bacteria, T. pallidum does not contain two-component systems that mediate large shifts in gene expression in response to alterations in growth conditions18,133. On the other hand, in addition to a σ70 housekeeping sigma factor (RpoD (TP0493)), the T. pallidum genome encodes four annotated alternative sigma factors, σE (RpoE (TP0092)), σ54 (RpoN (TP1011)), σA (TP1012), and σ28 (RpoF (TP0700)), collectively indicating a capacity to re-direct gene transcription in response to specific environmental cues. While σ28 presumably transcribes flagellar genes, T. pallidum deviates from traditional flagellar biosynthesis pathways in that it lacks an ortholog for FlhDC, the master transcriptional regulator for class II flagellar genes, and its σ28 gene lacks an upstream binding site for σ54 134. Also notably absent is the alternative sigma factor (σ32) that mobilizes a heat shock response, likely accounting for T. pallidum’s sensitivity to supra-physiologic temperatures135. Furthermore, the genome encodes an unusual hybrid PTS system that putatively links carbon and nitrogen flux to gene regulation through the small nucleotide messenger cAMP (Box 2). The c-di-GMP signaling pathway is associated with a wide range of adaptive processes and behaviors in other pathogenic bacteria136. The presence of this pathway in T. pallidum is, therefore, a potentially important discovery. According to bioinformatics, T. pallidum contains two diguanylate cyclases for synthesizing c-di-GMP (TP0172 and TP0981)136, three phosphodiesterases for degrading it (TP0764, TP0877, and TP0912), and a putative effector protein (TP0086), which contains a c-di-GMP-binding PilZ domain40,136.
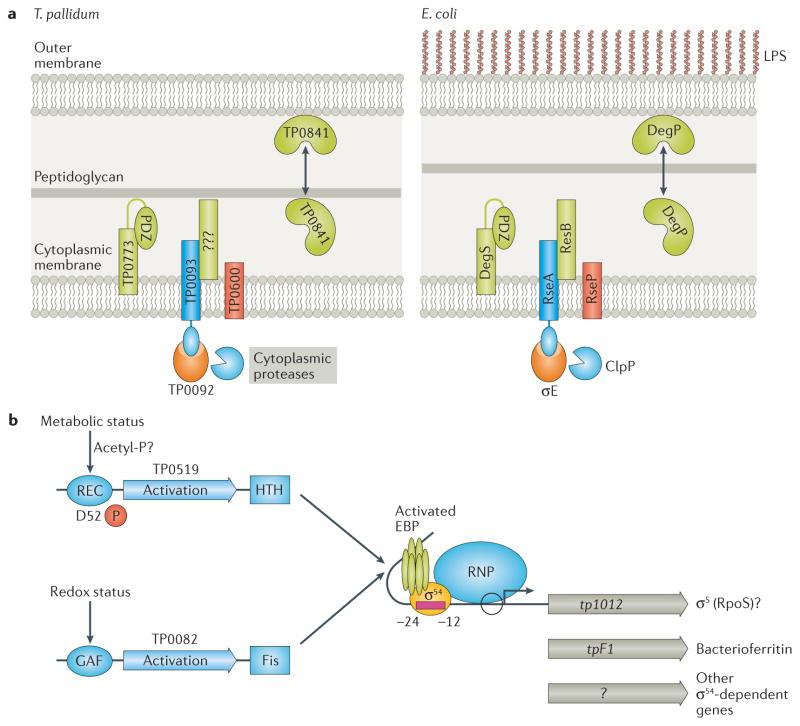
The susceptibility of the T. pallidum outer membrane to chemical and physical perturbations in vitro13,45 implies the need for a sensitive and robust system for sensing and responding to cell envelope stress in vivo. The σE (TP0092) pathway fulfills this function (Fig. 3A)137. tp0092 is highly transcribed during experimental syphilis,138 suggesting that σE-dependent genes are strongly induced by the inflammatory milieu in the rabbit testis. T. pallidum’s pathway for regulation of σE includes DegS (TP0773), RseP (TP0600), and RseA (TP0093) orthologs137. Notably absent is an identifiable ortholog for RseB, a negative regulator that binds to RseA and protects it from DegS-mediated cleavage137.
Figure 3. Proposed pathways for control of alternative sigma factors in T. pallidum.
A. σE cell envelope stress responses in T. pallidum and E. coli. Binding of misfolded OMPs to the PDZ domain of DegS (TP0773) initiates transduction of the stress signal across the cytoplasmic membrane. T. pallidum lacks a recognizable ortholog for RseB, a negative regulator that binds to RseA and protects it from DegS-mediated cleavage. B. σ54 (RpoN)-dependent gene expression. T. pallidum contains two enhancer binding proteins, the NtrC-like TP0519 and the NorR-like TP0082. Because TP0519 lacks a cognate histidine kinase, phosphorylation of TP0519 may occur through an acetyl-phosphate donor. Activation of TP0082 is predicted to occur following the binding of a small molecule to its GAF domain. tpF1 (tp1038), encoding a bacterioferritin, and tp1012, encoding an RpoS-like sigma factor, are predicted to be transcribed by σ54.
To initiate transcription, members of the σ54 family of alternative sigma factors recognize a unique -24/-12 type promoter and require hydrolysis of ATP by an enhancer-binding protein (EBP) that binds to an upstream sequence139,140. Further, interrogation of the T. pallidum genome for genes potentially transcribed by σ54 revealed tpf1 as the strongest candidate σ54-dependent gene18. The genome also encodes two EBPs, the nitrogen regulatory protein C (NtrC)-like TP0519 and the nitric oxide reductase regulator (NorR)-like TP0082 (Fig. 3B)18. TP1012, annotated as an ortholog of the Gram-positive housekeeping σ factor (σA)18, presents an interesting conundrum. It seems unlikely that T. pallidum would have two housekeeping sigma factors. However, TP1012 is also a distant relative of E. coli σS (RpoS); moreover, inspection of the tp1012 sequence identified an ATG translational start with an excellently placed σ54 promoter. Thus, T. pallidum might harbor an EBP-σ54-σS cascade (Fig. 3B) mirroring the virulence-related Rrp2-σ54-σS pathway in B. burgdorferi141,142.
Comparative Genomics
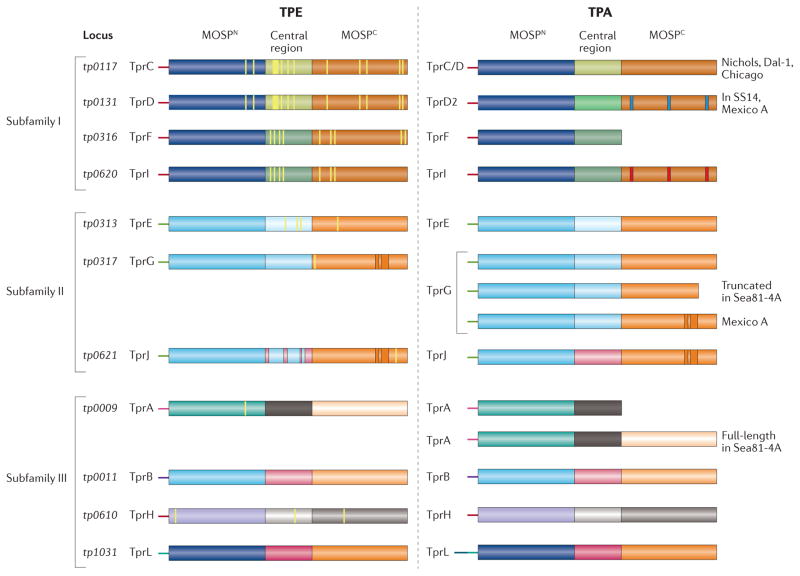
The overall level of genetic identity among the human pathogenic treponemes, > 99.8%, implies that differences in virulence, invasiveness, and tissue tropisms arise from changes in a relative handful of genetic loci5,143,144. Table 1 lists the strongest candidate proteins potentially responsible for phenotypic differences between pallidum and pertenue subspecies. Of the 22 genes listed, 15 encode proteins known or believed to reside in the cell envelope, with members of the Tpr family particularly well represented. Many of the amino acid substitutions in the Tprs are located in the C-terminal domains known or predicted to form outer membrane-inserted β-barrels (Fig. 4). Strikingly, the tprA and tprF loci, which typically encode frame-shifted truncated proteins in venereal syphilis spirochetes, encode full-length proteins in pertenue strains63,143,145; the pertenue TprF β-barrel is TprI-like whereas the TprA barrel is unique (Fig. 4). The list in Table 1 also contains most of the candidates for differences between subspecies pallidum strains. The Tpr repertoires among venereal syphilis spirochetes (Fig. 4) are, on the whole, very similar, although sequence variation in known or potential β-barrel forming domains does occur (e.g., TprC, TprD, and TprI ), along with insertions (e.g., TprG and TprJ), sporadic truncations (e.g., TprG in Seattle 81–4) or replacement of typically truncated proteins with full-length paralogs (for example, TprA, also in Seattle 81–4)145.
Figure 4. Domain architecture of the T. pallidum repeat (Tpr) family of proteins Tprs in T. pertenue (TPE) and T. pallidum (TPA) strains.
The Tprs are divided into three subfamilies based on sequence relatedness; within subfamilies I and II, members are closely related to each other, whereas members of subfamily III are heterogeneous52,63. Full-length Tprs consist of: (i) a variable extreme N-terminal stretch (~50 amino acids); (ii) MOSPN, a conserved N-terminal domain related to the corresponding domain in the N-terminus of the major outer sheath protein (MOSP) of T. denticola; (iii) a variable central region; and (iv) MOSPC, a conserved C-terminal β-forming domain related to the corresponding domain in T. denticola MOSP. The thin yellow lines denote amino acid variation in TPE paralogs relative to the corresponding paralogs in the TPA Nichols strain. Shown in umber are insertions in the MOSPC domains of TprG and TprJ relative to the corresponding paralog in the TPA Nichols reference strain. The central domain of TprJ in TPE contains sequences derived from the corresponding region of TprG. TprL in the TPA Nichols strain contains an extended variable N-terminal stretch. In TPE, TprA and TprF are full-length, bipartite proteins. The blue and red lines, respectively, are used to indicate sequence variation in TprD2 and TprI relative to TprC/D in TPA Nichols. With the exception of the Sea81-4 TPA strain, TprA is truncated, lacking the MOSPC domain; TprF is truncated in all TPA strains. TprK is not shown because of its hypervariability.
Structural homology modeling of BamA has afforded valuable insights into sequence differences revealed by genomics65. Mexico A-like strains have BamA β-barrels in which a single amino acid substitution in the immunodominant L4 surface loop markedly reduces reactivity of sera from patients infected with strains containing Nichols-like β-barrels65,146. Petrosova et al.146 made the seminal observation that the β-barrel in Mexico A TP0326 is identical to that in pertenue strains. They proposed that the pertenue TP0326 β-barrel was introduced into the subsp. pallidum genome as a result of recombination in an individual co-infected with yaws and venereal syphilis. Since this L4 epitope is a prime opsonic target65, the resulting strain theoretically would be less susceptible to pre-existing Nichols anti-L4 antibodies and, thus, capable of spreading in populations in which Nichols-like strains predominate.
Concluding remarks
Winston Churchill’s description of the Soviet Union as “a riddle, wrapped in a mystery, inside an enigma” could easily have been applied to T. pallidum. For decades, investigators obsessed with understanding protective immunity in syphilis pursued their Holy Grail, a syphilis vaccine, but were stymied by the ‘black box’ of the spirochete’s surface. Syphilologists now have in hand authentic OMPs and the experimental tools needed to establish authenticity for other candidates. These proteins also will enable clarification of the immunologic cat and mouse game that characterizes human syphilis and formulation of strategies to tease apart the virulence properties of the pathogenic treponemes. A relatively recent development in syphilis pathogenesis research has been the recognition that stealth pathogenicity has complicated and surprisingly obscure underpinnings that warrant intensive investigation. This Review underscores a theme now taking shape: the spirochete’s genomic reductionism, rather than being simply a matter of jettisoning unneeded genes, stems from a far more complicated evolutionary process, combining ultrastructural and metabolic parsimony with regulatory intricacy. Two forces appear to be at work: one is the need for T. pallidum to make the most of the nutrients it can usurp from its host, the other is the bacterium’s need to adapt to a diversity of micro-environments and stresses encountered during its journey within the human body. One of the great challenges to understanding these forces will be developing strategies to move beyond the static picture that has been an inadvertent byproduct of studying spirochetes extracted from a single milieu — the inflamed rabbit testis — to one that is dynamic and integrative.
Key points.
Pathogenic treponemes are clonal, uncultivatable, highly invasive bacteria that cause venereal syphilis, yaws, endemic syphilis, and pinta—multi-stage, infections that have many similarities but can be differentiated based on clinical, epidemiologic, and geographic criteria. Only Treponema pallidum subsp. pallidum is transmitted by sexual activity.
Key to the syphilis spirochete’s capacity for immune evasion and thus ‘stealth pathogenicity’ is its unusual outer membrane, which lacks lipopolysaccharide and contains an extremely low density of integral membrane proteins and a paucity of surface-exposed lipoproteins. Production of opsonic antibodies against low abundance surface antigenic targets is believed to be essential for control of syphilitic infection.
In recent years, considerable progress has been made in defining the syphilis spirochete’s repertoire of β-barrel-forming rare outer membrane proteins and the mechanisms by which the bacterium seems to limit exposure of surface molecules to the host’s antibody-mediated defenses.
During the course of genomic reduction, T. pallidum has undergone adaptations that enable it to acquire all of its required nutrients from its obligate human host and optimize their usage within various niches, while coping with exogenous and endogenous stress.
The T. pallidum genome encodes several alternative sigma factors and other regulatory molecules/pathways that collectively point to a previously unsuspected capacity to intricately regulate gene expression within diverse microenvironments.
Comparative genomics has enabled investigators to identify ‘hot spots’ for sequence variation that likely explain differences in virulence potential and tissue tropisms among the pathogenic treponemes; many of these are located in proteins known or predicted to reside at the host-pathogen interface.
Acknowledgments
The authors gratefully acknowledge support from NIH/NIAID grants AI26756 (JDR), AI56305 (MVN), and AI83640 (XFY); from Connecticut Children’s Medical Center (JDR), from the Ministry of Health of the Czech Republic grant NT11159-5/2010 (DS); and from the Grant Agency of the Czech Republic (P302/12/0574, DS). We also thank Chad Brautigam and Milton Saier for helpful discussions; Melissa Caimano for her many insightful comments and careful proofreading and editing; Morgan Ledoyt and Carson Karanian for assistance with figures; and Adriana Cruz, Juan Salazar, and Kevin Dieckhaus for providing images of syphilitic lesions.
Glossary terms
- Toll-like receptor (TLR) 2
A pattern recognition receptor that recognizes a variety of pathogen-associated molecular patterns, including bacterial lipoproteins.
- Opsonic antibodies
Antibodies directed against a pathogen’s surface-exposed epitopes that bind to Fc receptors on a phagocytic cell, triggering internalization through phagocytosis.
- Permease
A polytopic integral membrane protein that mediates energy-dependent uptake of small molecules across the plasma membrane of Gram-positives and the cytoplasmic membrane of Gram-negative bacteria.
- ABC Transporter
An ATP-binding cassette transporter couples hydrolysis of ATP to transport (usually import) of a substrate across the cytoplasmic membrane of Gram-negative bacteria and the plasma membrane of Gram-positive bacteria. Classical bacterial ABC transporters have a modular composition consisting of a substrate-binding protein, a dimeric membrane-bound permease, and a dimeric nucleotide binding protein with ATPase activity.
- Symporter
Transporter molecules that use the sodium or electrochemical gradient across the cytoplasmic membrane to drive the co-directional importation of substrates from the periplasmic to the cytosolic compartment.
- Auxotroph
An organism that has lost the ability to synthesize molecules required for growth. T. pallidum is considered an extreme auxotroph because of its very limited biosynthetic capacity.
- Two-component system
Two-component systems typically consist of a membrane-bound histidine kinase that senses a specific environmental stimulus and a cognate response regulator that mediates a cellular response, usually by activating and/or repressing differentially expressed genes.
- Housekeeping sigma factor
A sigma factor that binds to the catalytic core of RNA polymerase and recognizes promoters of genes required for core bacterial cell functions, such as maintenance and metabolism.
- Alternative sigma factor
A sigma factor that binds to the catalytic core of RNA polymerase, displacing the housekeeping sigma factor, re-directing transcription towards genes required to respond to a particular environmental stimulus, condition, or stress.
- Heat-shock response
The bacterial cell’s response to a sudden increase in temperature, involving differential gene expression regulated through the alternative sigma factor σ32.
Biographies
Justin D. Radolf is Professor of Medicine, Pediatrics, Immunology, Molecular Biology and Biophysics, and Genetics and Genome Sciences at UConn Health (UCH), Farmington, USA, and Senior Scientific Advisor to Connecticut Children’s Medical Center, Hartford, USA. He received his M.D. (1979) from the University of California, San Francisco, USA and completed postgraduate training in internal medicine (1982) at the Hospital of the University of Pennsylvania, Philadelphia, USA and infectious diseases (1986) at the University of California, Los Angeles, USA. In 1986, he joined the faculty in the Departments of Internal Medicine and Microbiology at the University of Texas Southwestern Medical Center, Dallas, USA and relocated to UCH in 1999. His research activities centre on the molecular pathogenesis and immunobiology of syphilis and Lyme disease.
Ranjit K. Deka is a senior scientist in the Microbiology Department at U.T. Southwestern Medical Center (Dallas, TX). He received his M.Sc. (1989) in biochemistry from Assam Agricultural University (Jorhat, India) and Ph.D. (1993) in protein biochemistry from the University of Glasgow (Scotland) where he studied the mechanisms of action of the shikimate pathway enzymes. His current discovery-driven research focuses on determining the structures and functions of T. pallidum periplasmic lipoproteins as an approach for discerning the roles of previously unknown metabolic pathways in syphilis pathogenesis.
Arvind Anand received his M.S. (2004) in zoology from Guru Nanak Dev University, Amritsar India and his Ph.D. (2010) degree in Molecular Biology and Biochemistry from the Institute of Microbial Technology, Chandigarh India. His dissertation involved cellular partners and protein-protein interactions of Vitreoscilla hemoglobin. His postdoctoral research at UConn Health (2010–2015) centered about structural and functional studies of rare outer membrane proteins of Treponema pallidum. Currently, he is a senior scientist at World Sciences Pvt. Ltd. Chandigarh, India.
David Šmajs is Vice-Chair and Professor, Department of Biology at the Faculty of Medicine, Masaryk University, Brno, Czech Republic. He received his M.D. (1993) and Ph.D. (1997) from Masaryk University and did postdoctoral training (1997–201) at the University of Texas Medical School at Houston, Texas, USA, and Baylor College of Medicine, Houston, Texas, USA, under the supervision of George M. Weinstock. His laboratory is focused on genetics and genomics of bacterial pathogens.
Michael V. Norgard is Professor and Chair of Microbiology at U.T. Southwestern Medical Center (Dallas, USA) where he holds the B. B. Owen Distinguished Chair in Molecular Research. He received his Ph.D. degree in 1977 from Rutgers Medical School (Newark, NJ) and performed postdoctoral work in genetic engineering from 1977–9 at the Roche Institute of Molecular Biology (Nutley, NJ). His research focuses on the membrane biology and physiology of Treponema pallidum as well as the strategic regulatory pathways that govern virulence expression by Borrelia burgdorferi, the Lyme disease spirochete.
X. Frank Yang is Associate Professor of Microbiology and Immunology at Indiana University (IU) School of Medicine, Indianapolis, USA. He received his Ph.D. (1997) in molecular biology from the University of Texas at Dallas, Texas, USA, studying signal transduction of Escherichia coli and did postdoctoral work at U.T. Southwestern Medical Center (Dallas, USA) (1998–2003) studying regulatory pathways controlling virulence gene expression in Borrelia burgdorferi. His laboratory at IU focuses on studying the mechanisms of gene regulation and host adaptation of spirochetal pathogens.
Footnotes
Conflicts of interest
The authors report no conflicts of interest.
References
- 1.Giacani L, Lukehart SA. The endemic treponematoses. Clin Microbiol Rev. 2014;27:89–115. doi: 10.1128/CMR.00070-13. This is a contemporary review of the origins, global epidemiology, and clinical features of the novenereal treponematoses and their continuing threat to worldwide health. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sena AC, Pillay A, Cox DL, Radolf JD. In: Manual of Clinical Microbiology. Jorgensen JH, et al., editors. ASM Press; Washington, D.C: 2015. pp. 1055–1081. [Google Scholar]
- 3.Miao R, Fieldsteel AH. Genetics of Treponema: relationship between Treponema pallidum and five cultivable treponemes. J Bacteriol. 1978;133:101–7. doi: 10.1128/jb.133.1.101-107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miao RM, Fieldsteel AH. Genetic relationship between Treponema pallidum and Treponema pertenue, two noncultivable human pathogens. J Bacteriol. 1980;141:427–9. doi: 10.1128/jb.141.1.427-429.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smajs D, Norris SJ, Weinstock GM. Genetic diversity in Treponema pallidum: implications for pathogenesis, evolution and molecular diagnostics of syphilis and yaws. Infect Genet Evol. 2012;12:191–202. doi: 10.1016/j.meegid.2011.12.001. This is a comprehensive review of the genetic and evolutionary relatioships among pathogenic treponemes based on genomic sequencing data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lumeij JT, Mikalova L, Smajs D. Is there a difference between hare syphilis and rabbit syphilis? Cross infection experiments between rabbits and hares. Vet Microbiol. 2013;164:190–4. doi: 10.1016/j.vetmic.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Smajs D, et al. Complete genome sequence of Treponema paraluiscuniculi, strain Cuniculi A: the loss of infectivity to humans is associated with genome decay. PLoS One. 2011;6:e20415. doi: 10.1371/journal.pone.0020415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitja O, et al. Global epidemiology of yaws: a systematic review. Lancet Glob Health. 2015;3:e324–31. doi: 10.1016/S2214-109X(15)00011-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. Global incidence and prevalence of selected curable sexually transmitted infections-2008. World Health Organization; Geneva: 2012. [Google Scholar]
- 10.Klausner JD. The sound of silence: missing the opportunity to save lives at birth. Bull World Health Organ. 2013;91:158–158A. doi: 10.2471/BLT.13.118604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radolf JD, Tramont EC, Salazar JC. In: Mandell, Douglas and Bennett's Principles and Practice of Infectious Diseases. Bennett JE, Dolin R, Blaser MJ, editors. Churchill Livingtone Elsevier; Philadelphia: 2014. pp. 2684–2709. [Google Scholar]
- 12.Radolf JD, Hazlett KRO, Lukehart SA. In: Pathogenic Treponema: Cellular and Molecular Biology. Radolf JD, Lukehart SA, editors. Caister Academic Press; Norfolk, UK: 2006. pp. 197–236. [Google Scholar]
- 13.Cameron CE. In: Pathogenic Treponema: Molecular and Cellular Biology. Radolf JD, Lukehart SA, editors. Caister Academic Press; Norwich, UK: 2006. pp. 237–266. [Google Scholar]
- 14.Lafond RE, Lukehart SA. Biological basis for syphilis. Clin Microbiol Rev. 2006;19:29–49. doi: 10.1128/CMR.19.1.29-49.2006. This is a complete, balanced review of the immunology, molecular biology, and pathogenesis of syphilis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izard J, et al. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol. 2009;191:7566–80. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas DD, et al. Treponema pallidum invades intercellular junctions of endothelial cell monolayers. Proc Natl Acad Sci U S A. 1988;85:3608–12. doi: 10.1073/pnas.85.10.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolgemuth CW. Flagellar motility of the pathogenic spirochetes. Semin Cell Dev Biol. 2015;46:104–12. doi: 10.1016/j.semcdb.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser CM, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–88. doi: 10.1126/science.281.5375.375. This landmark paper is an insightful, beautifully written presentation of the T. pallidum genome, one of the first bacterial genomes to be sequenced. [DOI] [PubMed] [Google Scholar]
- 19.Radolf JD, Lukehart SA. In: Pathogenic Treponema: Cellular and Molecular Biology. Radolf JD, Lukehart SA, editors. Caister Academic Press; Norfolk, UK: 2006. pp. 285–322. [Google Scholar]
- 20.Moore MW, et al. Phagocytosis of Borrelia burgdorferi and Treponema pallidum potentiates innate immune activation and induces gamma interferon production. Infect Immun. 2007;75:2046–62. doi: 10.1128/IAI.01666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukehart SA. Scientific monogamy: thirty years dancing with the same bug: 2007 Thomas Parran Award Lecture. Sex Transm Dis. 2008;35:2–7. doi: 10.1097/OLQ.0b013e318162c4f2. [DOI] [PubMed] [Google Scholar]
- 22.Abell E, Marks R, Jones EW. Secondary syphilis: a clinico-pathological review. Br J Dermatol. 1975;93:53–61. doi: 10.1111/j.1365-2133.1975.tb06476.x. [DOI] [PubMed] [Google Scholar]
- 23.van Voorhis WC, Barrett LK, Nasio JM, Plummer FA, Lukehart SA. Lesions of primary and secondary syphilis contain activated cytolytic T cells. Infect Immun. 1996;64:1048–50. doi: 10.1128/iai.64.3.1048-1050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salazar JC, et al. Treponema pallidum elicits innate and adaptive cellular immune responses in skin and blood during secondary syphilis: a flow-cytometric analysis. J Infect Dis. 2007;195:879–87. doi: 10.1086/511822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stary G, et al. Host defense mechanisms in secondary syphilitic lesions: a role for IFN-gamma-/IL-17-producing CD8+ T cells? Am J Pathol. 2010;177:2421–32. doi: 10.2353/ajpath.2010.100277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruz AR, et al. Immune evasion and recognition of the syphilis spirochete in blood and skin of secondary syphilis patients: two immunologically distinct compartments. PLoS Negl Trop Dis. 2012;6:e1717. doi: 10.1371/journal.pntd.0001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cruz AR, et al. Secondary syphilis in Cali, Colombia: new concepts in disease pathogenesis. PLoS Negl Trop Dis. 2010;4:e690. doi: 10.1371/journal.pntd.0000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shafii T, Radolf JD, Sanchez PJ, Schulz KF, Murphy FK. In: Sexually Transmitted Diseases. Holmes KK, et al., editors. McGraw Hill; New York: 2008. pp. 1577–1612. [Google Scholar]
- 29.Bishop NH, Miller JN. Humoral immunity in experimental syphilis. I. The demonstration of resistance conferred by passive immunization. J Immunol. 1976;117:191–6. [PubMed] [Google Scholar]
- 30.Perine PL, Weiser RS, Klebanoff SJ. Immunity to syphilis. I. Passive transfer in rabbits with hyperimmune serum. Infect Immun. 1973;8:787–90. doi: 10.1128/iai.8.5.787-790.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sell S, Norris SJ. The biology, pathology, and immunology of syphilis. Int Rev Exp Pathol. 1983;24:203–76. [PubMed] [Google Scholar]
- 32.Sell S, Salman J, Norris SJ. Reinfection of chancre-immune rabbits with Treponema pallidum. I. Light and immunofluorescence studies. Am J Pathol. 1985;118:248–55. [PMC free article] [PubMed] [Google Scholar]
- 33.Salazar JC, Hazlett KR, Radolf JD. The immune response to infection with Treponema pallidum, the stealth pathogen. Microbes Infect. 2002;4:1133–40. doi: 10.1016/s1286-4579(02)01638-6. [DOI] [PubMed] [Google Scholar]
- 34.Ho EL, Lukehart SA. Syphilis: using modern approaches to understand an old disease. J Clin Invest. 2011;121:4584–92. doi: 10.1172/JCI57173. This well written article presents a state-of-the-art summary of the global molecular epidemiology and pathogenesis of syphilis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norris SJ, Cox DL, Weinstock GM. Biology of Treponema pallidum: correlation of functional activities with genome sequence data. J Mol Microbiol Biotechnol. 2001;3:37–62. [PubMed] [Google Scholar]
- 36.Nelson RA., Jr Factors affecting the survival of Treponema pallidum in vitro. Am J Hyg. 1948;48:120–32. doi: 10.1093/oxfordjournals.aje.a119217. [DOI] [PubMed] [Google Scholar]
- 37.Fieldsteel AH, Cox DL, Moeckli RA. Cultivation of virulent Treponema pallidum in tissue culture. Infect Immun. 1981;32:908–15. doi: 10.1128/iai.32.2.908-915.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norris SJ. In vitro cultivation of Treponema pallidum: independent confirmation. Infect Immun. 1982;36:437–9. doi: 10.1128/iai.36.1.437-439.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norris SJ, Edmondson DG. Factors affecting the multiplication and subculture of Treponema pallidum subsp. pallidum in a tissue culture system. Infect Immun. 1986;53:534–9. doi: 10.1128/iai.53.3.534-539.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naqvi AA, Shahbaaz M, Ahmad F, Hassan MI. Identification of functional candidates amongst hypothetical proteins of Treponema pallidum ssp. pallidum. PLoS One. 2015;10:e0124177. doi: 10.1371/journal.pone.0124177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deka RK, Brautigam CA, Biddy BA, Liu WZ, Norgard MV. Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. MBio. 2013;4:e00615–12. doi: 10.1128/mBio.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deka RK, Brautigam CA, Liu WZ, Tomchick DR, Norgard MV. Evidence for posttranslational protein flavinylation in the syphilis spirochete Treponema pallidum: structural and biochemical insights from the catalytic core of a periplasmic flavin-trafficking protein. MBio. 2015;6 doi: 10.1128/mBio.00519-15. This report presents a biochemical/molecular analysis of a unique periplasmic flavin utilization pathway in T. pallidum and describes the spirochete's Rnf system for generating an electrochemical gradient across the cytoplasmic membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukehart SA, Marra CM. Isolation and laboratory maintenance of Treponema pallidum. Curr Protoc Microbiol. 2007;Chapter 12(Unit 12A):1. doi: 10.1002/9780471729259.mc12a01s7. [DOI] [PubMed] [Google Scholar]
- 44.Alderete JF, Baseman JB. Surface-associated host proteins on virulent Treponema pallidum. Infect Immun. 1979;26:1048–56. doi: 10.1128/iai.26.3.1048-1056.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radolf JD. Treponema pallidum and the quest for outer membrane proteins. Mol Microbiol. 1995;16:1067–73. doi: 10.1111/j.1365-2958.1995.tb02332.x. [DOI] [PubMed] [Google Scholar]
- 46.Silver AC, et al. MyD88 deficiency markedly worsens tissue inflammation and bacterial clearance in mice infected with Treponema pallidum, the agent of syphilis. PLoS One. 2013;8:e71388. doi: 10.1371/journal.pone.0071388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cameron CE, Lukehart SA. Current status of syphilis vaccine development: need, challenges, prospects. Vaccine. 2014;32:1602–9. doi: 10.1016/j.vaccine.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norris SJ, Weinstock GM. In: Pathogenic Treponema: Molecular and Cellular Biology. Radolf JD, Lukehart SA, editors. Norwich, U.K: 2006. pp. 19–38. [Google Scholar]
- 49.Radolf JD, Norgard MV, Schulz WW. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc Natl Acad Sci U S A. 1989;86:2051–5. doi: 10.1073/pnas.86.6.2051. This report describes the discovery of rare outer membrane proteins in T. pallidum by freeze-fracture electron microscopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker EM, Zampighi GA, Blanco DR, Miller JN, Lovett MA. Demonstration of rare protein in the outer membrane of Treponema pallidum subsp. pallidum by freeze-fracture analysis. J Bacteriol. 1989;171:5005–11. doi: 10.1128/jb.171.9.5005-5011.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magnuson HJ, Eagle H, Fleischman R. The minimal infectious inoculum of Spirochaeta pallida (Nichols strain) and a consideration of its rate of multiplication in vivo. Am J Syph Gonorrhea Vener Dis. 1948;32:1–18. [PubMed] [Google Scholar]
- 52.Centurion-Lara A, et al. Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J Exp Med. 1999;189:647–56. doi: 10.1084/jem.189.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gray RR, et al. Molecular evolution of the tprC, D, I, K, G, and J genes in the pathogenic genus Treponema. Mol Biol Evol. 2006;23:2220–33. doi: 10.1093/molbev/msl092. [DOI] [PubMed] [Google Scholar]
- 54.Ellen RP. In: Pathogenic Treponema: Molecular and Cellular Biology. Radolf JD, Lukehart SA, editors. Caister Academic Press; Norwich, UK: 2006. pp. 357–386. [Google Scholar]
- 55.Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. This review describes antigenic variation by T. pallidum in the context of well characterized mechanisms for varying surface antigenicity in other pathogens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hazlett KR, et al. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J Exp Med. 2001;193:1015–26. doi: 10.1084/jem.193.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cox DL, et al. Surface immunolabeling and consensus computational framework to identify candidate rare outer membrane proteins of Treponema pallidum. Infect Immun. 2010;78:5178–94. doi: 10.1128/IAI.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cameron CE, et al. Opsonic potential, protective capacity, and sequence conservation of the Treponema pallidum subspecies pallidum Tp92. J Infect Dis. 2000;181:1401–13. doi: 10.1086/315399. [DOI] [PubMed] [Google Scholar]
- 59.Anand A, et al. Bipartite topology of Treponema pallidum repeat proteins C/D and I: outer membrane insertion, trimerization, and porin function require a C-terminal β-barrel domain. J Biol Chem. 2015;290:12313–31. doi: 10.1074/jbc.M114.629188. This report describes the use of molecular, biophysical, and immunological methodologies to establish the bipartite topology of Tpr outer membrane proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anand A, et al. TprC/D (Tp0117/131), a trimeric, pore-forming rare outer membrane protein of Treponema pallidum, has a bipartite domain structure. J Bacteriol. 2012;194:2321–33. doi: 10.1128/JB.00101-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anand A, et al. The major outer sheath protein (Msp) of Treponema denticola has a bipartite domain architecture and exists as periplasmic and outer membrane-spanning conformers. J Bacteriol. 2013 doi: 10.1128/JB.00078-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Centurion-Lara A, et al. Fine analysis of genetic diversity of the tpr gene family among treponemal species, subspecies and strains. PLoS Negl Trop Dis. 2013;7:e2222. doi: 10.1371/journal.pntd.0002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Desrosiers DC, et al. TP0326, a Treponema pallidum beta-barrel assembly machinery A (BamA) orthologue and rare outer membrane protein. Mol Microbiol. 2011;80:1496–515. doi: 10.1111/j.1365-2958.2011.07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luthra A, et al. A homology model reveals novel structural features and an immunodominant surface loop/opsonic target in the Treponema pallidum BamA ortholog TP_0326. J Bacteriol. 2015;197:1906–20. doi: 10.1128/JB.00086-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Selkrig J, Leyton DL, Webb CT, Lithgow T. Assembly of β-barrel proteins into bacterial outer membranes. Biochim Biophys Acta. 2014;1843:1542–50. doi: 10.1016/j.bbamcr.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Noinaj N, et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature. 2013;501:385–90. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fitzgerald TJ, Johnson RC, Miller JN, Sykes JA. Characterization of the attachment of Treponema pallidum (Nichols strain) to cultured mammalian cells and the potential relationship of attachment to pathogenicity. Infect Immun. 1977;18:467–78. doi: 10.1128/iai.18.2.467-478.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hayes NS, Muse KE, Collier AM, Baseman JB. Parasitism by virulent Treponema pallidum of host cell surfaces. Infect Immun. 1977;17:174–86. doi: 10.1128/iai.17.1.174-186.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fitzgerald TJ, Repesh LA, Blanco DR, Miller JN. Attachment of Treponema pallidum to fibronectin, laminin, collagen IV, and collagen I, and blockage of attachment by immune rabbit IgG. Br J Vener Dis. 1984;60:357–63. doi: 10.1136/sti.60.6.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cameron CE, Brown EL, Kuroiwa JM, Schnapp LM, Brouwer NL. Treponema pallidum fibronectin-binding proteins. J Bacteriol. 2004;186:7019–22. doi: 10.1128/JB.186.20.7019-7022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cameron CE. Identification of a Treponema pallidum laminin-binding protein. Infect Immun. 2003;71:2525–33. doi: 10.1128/IAI.71.5.2525-2533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cameron CE, Brouwer NL, Tisch LM, Kuroiwa JM. Defining the interaction of the Treponema pallidum adhesin Tp0751 with laminin. Infect Immun. 2005;73:7485–94. doi: 10.1128/IAI.73.11.7485-7494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Houston S, Hof R, Honeyman L, Hassler J, Cameron CE. Activation and proteolytic activity of the Treponema pallidum metalloprotease, pallilysin. PLoS Pathog. 2012;8:e1002822. doi: 10.1371/journal.ppat.1002822. This report describes a biochemical/molecular characterization of TP0751 (pallilysin) as a zinc-dependent proteinase capable of degrading fibrin clots in addition to its role as a laminin-binding adhesin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Houston S, et al. The multifunctional role of the pallilysin-associated Treponema pallidum protein, Tp0750, in promoting fibrinolysis and extracellular matrix component degradation. Mol Microbiol. 2014;91:618–34. doi: 10.1111/mmi.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brinkman MB, et al. A novel Treponema pallidum antigen, TP0136, is an outer membrane protein that binds human fibronectin. Infect Immun. 2008;76:1848–57. doi: 10.1128/IAI.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ke W, Molini BJ, Lukehart SA, Giacani L. Treponema pallidum subsp. pallidum TP0136 protein is heterogeneous among isolates and binds cellular and plasma fibronectin via its NH2-terminal end. PLoS Negl Trop Dis. 2015;9:e0003662. doi: 10.1371/journal.pntd.0003662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan K, et al. Treponema pallidum lipoprotein TP0435 expressed in Borrelia burgdorferi produces multiple surface/periplasmic isoforms and mediates adherence. Sci Rep. 2016;6:25593. doi: 10.1038/srep25593. This is a ground breaking report in its use of Borrelia burgdorferi as a genetic surrogate for T. pallidum to characterize potential adhesins and surface antigenic targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Penn CW, Cockayne A, Bailey MJ. The outer membrane of Treponema pallidum: biological significance and biochemical properties. J Gen Microbiol. 1985;131:2349–57. doi: 10.1099/00221287-131-9-2349. [DOI] [PubMed] [Google Scholar]
- 80.Cox DL, Chang P, McDowall AW, Radolf JD. The outer membrane, not a coat of host proteins, limits antigenicity of virulent Treponema pallidum. Infect Immun. 1992;60:1076–83. doi: 10.1128/iai.60.3.1076-1083.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lukehart SA, Shaffer JM, Baker-Zander SA. A subpopulation of Treponema pallidum is resistant to phagocytosis: possible mechanism of persistence. J Infect Dis. 1992;166:1449–53. doi: 10.1093/infdis/166.6.1449. [DOI] [PubMed] [Google Scholar]
- 82.Centurion-Lara A, et al. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol Microbiol. 2004;52:1579–96. doi: 10.1111/j.1365-2958.2004.04086.x. [DOI] [PubMed] [Google Scholar]
- 83.LaFond RE, Molini BJ, Van Voorhis WC, Lukehart SA. Antigenic variation of TprK V regions abrogates specific antibody binding in syphilis. Infect Immun. 2006;74:6244–51. doi: 10.1128/IAI.00827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giacani L, et al. Antigenic variation in Treponema pallidum: TprK sequence diversity accumulates in response to immune pressure during experimental syphilis. J Immunol. 2010;184:3822–9. doi: 10.4049/jimmunol.0902788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reid TB, Molini BJ, Fernandez MC, Lukehart SA. Antigenic variation of TprK facilitates development of secondary syphilis. Infect Immun. 2014;82:4959–67. doi: 10.1128/IAI.02236-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giacani L, et al. Comparative investigation of the genomic regions involved in antigenic variation of the TprK antigen among treponemal species, subspecies, and strains. J Bacteriol. 2012;194:4208–25. doi: 10.1128/JB.00863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Petrosova H, et al. Resequencing of Treponema pallidum ssp. pallidum strains Nichols and SS14: correction of sequencing errors resulted in increased separation of syphilis treponeme subclusters. PLoS One. 2013;8:e74319. doi: 10.1371/journal.pone.0074319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giacani L, Lukehart S, Centurion-Lara A. Length of guanosine homopolymeric repeats modulates promoter activity of subfamily II tpr genes of Treponema pallidum ssp. pallidum. FEMS Immunol Med Microbiol. 2007;51:289–301. doi: 10.1111/j.1574-695X.2007.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hovind-Hougen K. In: Pathogenesis and Immunology of Treponemal Infection. Schell RF, Musher DM, editors. Marcel Dekker; New York: 1983. pp. 3–28. [Google Scholar]
- 91.Liu J, et al. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol. 2010;403:546–61. doi: 10.1016/j.jmb.2010.09.020. A materful use of cryoelectron microscopy and scanning probe microscopy to describe the native ultrastructure of T. pallidum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Briegel A, et al. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry. 2014;53:1575–85. doi: 10.1021/bi5000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deka RK, Goldberg MS, Hagman KE, Norgard MV. The Tp38 (TpMglB-2) lipoprotein binds glucose in a manner consistent with receptor function in Treponema pallidum. J Bacteriol. 2004;186:2303–8. doi: 10.1128/JB.186.8.2303-2308.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Deka RK, et al. Structural evidence that the 32-kilodalton lipoprotein (Tp32) of Treponema pallidum is an L-methionine-binding protein. J Biol Chem. 2004;279:55644–50. doi: 10.1074/jbc.M409263200. [DOI] [PubMed] [Google Scholar]
- 95.Bian J, Tu Y, Wang SM, Wang XY, Li C. Evidence that TP_0144 of Treponema pallidum is a thiamine-binding protein. J Bacteriol. 2015;197:1164–72. doi: 10.1128/JB.02472-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jaehme M, Slotboom DJ. Diversity of membrane transport proteins for vitamins in bacteria and archaea. Biochim Biophys Acta. 2015;1850:565–76. doi: 10.1016/j.bbagen.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 97.Lee YH, Deka RK, Norgard MV, Radolf JD, Hasemann CA. Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone. Nat Struct Biol. 1999;6:628–33. doi: 10.1038/10677. [DOI] [PubMed] [Google Scholar]
- 98.Deka RK, et al. The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum. J Biol Chem. 2006;281:8072–81. doi: 10.1074/jbc.M511405200. [DOI] [PubMed] [Google Scholar]
- 99.Machius M, et al. Structural and biochemical basis for polyamine binding to the Tp0655 lipoprotein of Treponema pallidum: putative role for Tp0655 (TpPotD) as a polyamine receptor. J Mol Biol. 2007;373:681–94. doi: 10.1016/j.jmb.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deka RK, Brautigam CA, Liu WZ, Tomchick DR, Norgard MV. The TP0796 lipoprotein of Treponema pallidum is a bimetal-dependent FAD pyrophosphatase with a potential role in flavin homeostasis. J Biol Chem. 2013;288:11106–21. doi: 10.1074/jbc.M113.449975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brautigam CA, Deka RK, Schuck P, Tomchick DR, Norgard MV. Structural and thermodynamic characterization of the interaction between two periplasmic Treponema pallidum lipoproteins that are components of a TPR-protein-associated TRAP transporter (TPAT) J Mol Biol. 2012;420:70–86. doi: 10.1016/j.jmb.2012.04.001. An extraordinary biochemical and structural analysis of a unique transporter in T. pallidum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deka RK, et al. Structural, bioinformatic, and in vivo analyses of two Treponema pallidum lipoproteins reveal a unique TRAP transporter. J Mol Biol. 2012;416:678–96. doi: 10.1016/j.jmb.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Austin FE, Cox CD. Lactate oxidation by Treponema pallidum. Current Microbiology. 1986;13:123–128. [Google Scholar]
- 104.Fraser CM, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–6. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 105.Gorke B, Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–24. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 106.Norris SJ, Fraser CM, Weinstock GM. Illuminating the agent of syphilis: the Treponema pallidum genome project. Electrophoresis. 1998;19:551–3. doi: 10.1002/elps.1150190415. [DOI] [PubMed] [Google Scholar]
- 107.Roberson RS, Ronimus RS, Gephard S, Morgan HW. Biochemical characterization of an active pyrophosphate-dependent phosphofructokinase from Treponema pallidum. FEMS Microbiol Lett. 2001;194:257–60. doi: 10.1111/j.1574-6968.2001.tb09479.x. [DOI] [PubMed] [Google Scholar]
- 108.Moore SA, Ronimus RS, Roberson RS, Morgan HW. The structure of a pyrophosphate-dependent phosphofructokinase from the Lyme disease spirochete Borrelia burgdorferi. Structure. 2002;10:659–71. doi: 10.1016/s0969-2126(02)00760-8. [DOI] [PubMed] [Google Scholar]
- 109.Fothergill-Gilmore LA, Michels PA. Evolution of glycolysis. Prog Biophys Mol Biol. 1993;59:105–235. doi: 10.1016/0079-6107(93)90001-z. [DOI] [PubMed] [Google Scholar]
- 110.Mertens E. ATP versus pyrophosphate: glycolysis revisited in parasitic protists. Parasitol Today. 1993;9:122–6. doi: 10.1016/0169-4758(93)90169-g. [DOI] [PubMed] [Google Scholar]
- 111.Higuchi M, Yamamoto Y, Kamio Y. Molecular biology of oxygen tolerance in lactic acid bacteria: Functions of NADH oxidases and Dpr in oxidative stress. J Biosci Bioeng. 2000;90:484–93. [PubMed] [Google Scholar]
- 112.Baker JL, Derr AD, Faustoferri RC, Quivey RG., Jr Loss of NADH oxidase activity in Streptococcus mutans leads to Rex-mediated overcompensation of NAD+ regeneration by lactate dehydrogenase. J Bacteriol. 2015 doi: 10.1128/JB.00383-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cox DL, et al. Effects of molecular oxygen, oxidation-reduction potential, and antioxidants upon in vitro replication of Treponema pallidum subsp. pallidum. Appl Environ Microbiol. 1990;56:3063–72. doi: 10.1128/aem.56.10.3063-3072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meyron-Holtz EG, Ghosh MC, Rouault TA. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science. 2004;306:2087–90. doi: 10.1126/science.1103786. [DOI] [PubMed] [Google Scholar]
- 115.Mayer F, Muller V. Adaptations of anaerobic archaea to life under extreme energy limitation. FEMS Microbiol Rev. 2014;38:449–72. doi: 10.1111/1574-6976.12043. [DOI] [PubMed] [Google Scholar]
- 116.Hazlett KR, et al. The Treponema pallidum tro operon encodes a multiple metal transporter, a zinc-dependent transcriptional repressor, and a semi-autonomously expressed phosphoglycerate mutase. J Biol Chem. 2003;278:20687–94. doi: 10.1074/jbc.M300781200. [DOI] [PubMed] [Google Scholar]
- 117.Gherardini FC, Boylan JA, Brett PJ. In: Pathogenic Treponema: Molecular and Cellular Biology. Radolf JD, Lukehart SA, editors. Caister Academic Press; Norwich, UK: 2006. pp. 101–126. [Google Scholar]
- 118.Alderete JF, Peterson KM, Baseman JB. Affinities of Treponema pallidum for human lactoferrin and transferrin. Genitourin Med. 1988;64:359–63. doi: 10.1136/sti.64.6.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Deka RK, et al. Crystal structure of the Tp34 (TP0971) lipoprotein of Treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin. J Biol Chem. 2007;282:5944–58. doi: 10.1074/jbc.M610215200. [DOI] [PubMed] [Google Scholar]
- 120.Noinaj N, et al. Structural basis for iron piracy by pathogenic Neisseria. Nature. 2012;483:53–8. doi: 10.1038/nature10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ward DM, Kaplan J. Ferroportin-mediated iron transport: expression and regulation. Biochim Biophys Acta. 2012;1823:1426–33. doi: 10.1016/j.bbamcr.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Desrosiers DC, et al. The general transition metal (Tro) and Zn2+ (Znu) transporters in Treponema pallidum: analysis of metal specificities and expression profiles. Mol Microbiol. 2007;65:137–52. doi: 10.1111/j.1365-2958.2007.05771.x. [DOI] [PubMed] [Google Scholar]
- 123.Brautigam CA, et al. Biophysical and bioinformatic analyses implicate the Treponema pallidum Tp34 lipoprotein (Tp0971) in transition metal homeostasis. J Bacteriol. 2012;194:6771–81. doi: 10.1128/JB.01494-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koch D, et al. Characterization of a dipartite iron uptake system from uropathogenic Escherichia coli strain F11. J Biol Chem. 2011;286:25317–30. doi: 10.1074/jbc.M111.222745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Posey JE, Hardham JM, Norris SJ, Gherardini FC. Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc Natl Acad Sci U S A. 1999;96:10887–92. doi: 10.1073/pnas.96.19.10887. This report describes the first molecular analysis of a transcriptional factor in T. pallidum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hazlett KR, et al. Contribution of neelaredoxin to oxygen tolerance by Treponema pallidum. Methods Enzymol. 2002;353:140–56. doi: 10.1016/s0076-6879(02)53044-5. [DOI] [PubMed] [Google Scholar]
- 127.Jenney FE, Jr, Verhagen MF, Cui X, Adams MW. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science. 1999;286:306–9. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 128.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–76. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Thumiger A, et al. Crystal structure of antigen TpF1 from Treponema pallidum. Proteins. 2006;62:827–30. doi: 10.1002/prot.20828. [DOI] [PubMed] [Google Scholar]
- 130.Parsonage D, et al. Broad specificity AhpC-like peroxiredoxin and its thioredoxin reductant in the sparse antioxidant defense system of Treponema pallidum. Proc Natl Acad Sci U S A. 2010;107:6240–5. doi: 10.1073/pnas.0910057107. This report presents a detailed biochemical analysis of T. pallidum's unique, robust enzymatic defense against peroxide stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Radolf JD, Borenstein LA, Kim JY, Fehniger TE, Lovett MA. Role of disulfide bonds in the oligomeric structure and protease resistance of recombinant and native Treponema pallidum surface antigen 4D. J Bacteriol. 1987;169:1365–71. doi: 10.1128/jb.169.4.1365-1371.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brautigam CA, Deka RK, Liu WZ, Norgard MV. Insights into the potential function and membrane organization of the TP0435 (Tp17) lipoprotein from Treponema pallidum derived from structural and biophysical analyses. Protein Sci. 2015;24:11–9. doi: 10.1002/pro.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Casino P, Rubio V, Marina A. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol. 2010;20:763–71. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 134.Anderson JK, Smith TG, Hoover TR. Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol. 2010;18:30–7. doi: 10.1016/j.tim.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stamm LV, Gherardini FC, Parrish EA, Moomaw CR. Heat shock response of spirochetes. Infect Immun. 1991;59:1572–5. doi: 10.1128/iai.59.4.1572-1575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guo MS, Gross CA. Stress-induced remodeling of the bacterial proteome. Curr Biol. 2014;24:R424–34. doi: 10.1016/j.cub.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Giacani L, Denisenko O, Tompa M, Centurion-Lara A. Identification of the Treponema pallidum subsp. pallidum TP0092 (RpoE) regulon and its implications for pathogen persistence in the host and syphilis pathogenesis. J Bacteriol. 2013;195:896–907. doi: 10.1128/JB.01973-12. This is the only report to use chromatin immunoprecipitation to define the cohort of genes controlled by an alternative sigma factor in T. pallidum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Studholme DJ, Buck M. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol Lett. 2000;186:1–9. doi: 10.1111/j.1574-6968.2000.tb09074.x. [DOI] [PubMed] [Google Scholar]
- 140.Bush M, Dixon R. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev. 2012;76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hubner A, et al. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A. 2001;98:12724–9. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2003;100:11001–6. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cejkova D, et al. Whole genome sequences of three Treponema pallidum ssp. pertenue strains: yaws and syphilis treponemes differ in less than 0.2% of the genome sequence. PLoS Negl Trop Dis. 2012;6:e1471. doi: 10.1371/journal.pntd.0001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Staudova B, et al. Whole genome sequence of the Treponema pallidum subsp. endemicum strain Bosnia A: the genome is related to yaws treponemes but contains few loci similar to syphilis treponemes. PLoS Negl Trop Dis. 2014;8:e3261. doi: 10.1371/journal.pntd.0003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Giacani L, et al. Complete genome sequence of the Treponema pallidum subsp. pallidum Sea81–4 strain. Genome Announc. 2014;2 doi: 10.1128/genomeA.00333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Petrosova H, et al. Whole genome sequence of Treponema pallidum ssp. pallidum, strain Mexico A, suggests recombination between yaws and syphilis strains. PLoS Negl Trop Dis. 2012;6:e1832. doi: 10.1371/journal.pntd.0001832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Peterkofsky A, Wang G, Seok YJ. Parallel PTS systems. Arch Biochem Biophys. 2006;453:101–7. doi: 10.1016/j.abb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 148.Pfluger-Grau K, Gorke B. Regulatory roles of the bacterial nitrogen-related phosphotransferase system. Trends Microbiol. 2010;18:205–14. doi: 10.1016/j.tim.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 149.Reizer J, Novotny MJ, Hengstenberg W, Saier MH., Jr Properties of ATP-dependent protein kinase from Streptococcus pyogenes that phosphorylates a seryl residue in HPr, a phosphocarrier protein of the phosphotransferase system. J Bacteriol. 1984;160:333–40. doi: 10.1128/jb.160.1.333-340.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Deutscher J, Kessler U, Hengstenberg W. Streptococcal phosphoenolpyruvate: sugar phosphotransferase system: purification and characterization of a phosphoprotein phosphatase which hydrolyzes the phosphoryl bond in seryl-phosphorylated histidine-containing protein. J Bacteriol. 1985;163:1203–9. doi: 10.1128/jb.163.3.1203-1209.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gonzalez CF, Stonestrom AJ, Lorca GL, Saier MH., Jr Biochemical characterization of phosphoryl transfer involving HPr of the phosphoenolpyruvate-dependent phosphotransferase system in Treponema denticola, an organism that lacks PTS permeases. Biochemistry. 2005;44:598–608. doi: 10.1021/bi048412y. [DOI] [PubMed] [Google Scholar]
- 152.Lee CR, Cho SH, Yoon MJ, Peterkofsky A, Seok YJ. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc Natl Acad Sci U S A. 2007;104:4124–9. doi: 10.1073/pnas.0609897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Deutscher J, et al. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78:231–56. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McDonough KA, Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Microbiol. 2012;10:27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Radolf JD, Bourell KW, Akins DR, Brusca JS, Norgard MV. Analysis of Borrelia burgdorferi membrane architecture by freeze-fracture electron microscopy. J Bacteriol. 1994;176:21–31. doi: 10.1128/jb.176.1.21-31.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Luthra A, et al. The transition from closed to open conformation of Treponema pallidum outer membrane-associated lipoprotein TP0453 involves membrane sensing and integration by two amphipathic helices. J Biol Chem. 2011;286:41656–68. doi: 10.1074/jbc.M111.305284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McKevitt M, et al. Genome scale identification of Treponema pallidum antigens. Infect Immun. 2005;73:4445–50. doi: 10.1128/IAI.73.7.4445-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Brinkman MB, et al. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J Clin Microbiol. 2006;44:888–91. doi: 10.1128/JCM.44.3.888-891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]