Abstract
The ATPase cycle of the chaperone Hsc70 is regulated by co-chaperones; Hsp40/DnaJ-related proteins stimulate ATP hydrolysis by Hsc70 and can bind unfolded polypeptides themselves. Conversely, various nucleotide exchange factors (NEFs) stimulate ADP-ATP exchange by Hsc70. We analyzed the purified Hsp40-related co-chaperones DJA1 (Hdj2) and DJA2 (Hdj3) and found that they had a distinct pattern of binding to a range of polypeptides. DJA2 alone could stimulate Hsc70-mediated refolding of luciferase in the absence of NEF, whereas DJA1 was much less active. The addition of the Bag1 NEF increased refolding by Hsc70 and DJA2, as did the newly characterized NEF Hsp110, but each NEF had a different optimal concentration ratio to Hsc70. Notably, the NEF HspBP1 could not increase refolding by Hsc70 and DJA2 at any concentration, and none of the NEFs improved the refolding activity with DJA1. Instead, DJA1 was inhibitory of refolding with DJA2 and Hsc70. All combinations of DJA1 or DJA2 with the three NEFs stimulated the Hsc70 ATPase rate, although Hsp110 became less effective with increasing concentrations. A chimeric DJA2 having its Hsc70-stimulatory J domain replaced with that of DJA1 was functional for polypeptide binding and ATPase stimulation of Hsc70. However, it could not support efficient Hsc70-mediated refolding and also inhibited refolding with DJA2 and Hsc70. These results suggest a more complex model of Hsc70 mechanism than has been previously thought, with notable functional divergence between Hsc70 co-chaperones.
The Hsp70 family of proteins are ATP-dependent molecular chaperones that assist the folding of polypeptides. Hsp70 chaperones have a typical structure divided into ATPase and substrate-binding domains that work in an ATP-driven substrate binding cycle. The mechanism of Hsp70 proteins has been well established in studies of the Escherichia coli homolog DnaK. In the ATP-bound state, an Hsp70 chaperone has low affinity for unfolded polypeptide. After hydrolysis of ATP, Hsp70 in the ADP-bound state binds substrate with high affinity. Exchange of ADP for ATP then reverts Hsp70 to its low polypeptide affinity state. Conversion of an Hsp70 between these two nucleotide states is controlled by different co-chaperone proteins. The Hsp40/DnaJ-related co-chaperones, including E. coli DnaJ, contain J domains that stimulate ATP hydrolysis by Hsp70, and consequently substrate binding. Nucleotide exchange factors (NEFs),2 such as GrpE in E. coli, trigger the dissociation of bound ADP from Hsp70 to allow the binding of ATP, resetting the cycle. The principles of this mechanism appear to be conserved in Hsp70 chaperones, including the major cytosolic form in humans, Hsc70 (HSPA8) (1, 2).
The DnaJ-related co-chaperones are also conserved between species. Type 1 J domain co-chaperones are homologous to DnaJ throughout their sequence and have the same domain architecture. Following their N-terminal J domains, they contain a linker sequence, zinc finger and central regions, and a C-terminal homodimerization region. Unfolded polypeptides are thought to be bound by the central region of these proteins; substrates are transferred to the Hsp70 partner upon J domain activation of ATP hydrolysis by the Hsp70 (1, 3). Although DnaJ is the only J domain co-chaperone of E. coli DnaK, humans have three cytosolic type 1 co-chaperones: DJA1 (DNAJA1, Hdj2, HSDJ), DJA2 (DNAJA2, Hdj3, HIRIP4), and DJA4 (DNAJA4, Hdj4). Comparison with the type 1 co-chaperone Ydj1 of Saccharomyces cerevisiae suggests that the human DJAs will have similar overall structures. DJA1 and DJA2 are constitutively expressed, whereas DJA4 is less highly expressed and may be specialized. In addition, eukaryotic type 2 J domain co-chaperones are known that diverge from DnaJ in the substrate-binding and C-terminal regions. The human type 2 member DJB1 (Hsp40, Hdj1) is expressed at low levels under normal conditions and is mainly expressed in the heat shock stress response. DJB1 also binds substrate more weakly than type 1 co-chaperones. Thus, the DJA1 and DJA2 co-chaperones are thought to be the major partners of human Hsc70 (3–10). The distinction between the human DJAs remains largely unexplored. Our recent work proposed a partial specialization between the human type 1 co-chaperones, perhaps to allow assistance of a wider range of substrates (11).
Interestingly, the NEF co-chaperones are divergent between E. coli and humans. The three types of human NEFs: Bag domain proteins, HspBP1, and Hsp110, are structurally unrelated to each other and to the single E. coli NEF GrpE. The C-terminal domain of Bag1 (C-Bag) was the first shown to have NEF activity for Hsc70, and homologous Bag domains have since been identified in several other human proteins. The mechanical action of C-Bag on Hsc70 appears to be equivalent to that of GrpE on DnaK, despite the difference in NEF structure (12–16). HspBP1 was subsequently found to have NEF activity. The structural effect of HspBP1 on Hsc70 is distinct from that of C-Bag (17–19). Most recently, Hsp110 (HSPH1, Hsp105α) was shown to be an NEF for Hsc70. Hsp110 represents a subclass of proteins structurally related to Hsp70; however, its ATPase activity and substrate binding are minimal. The Bag domain and HspBP1 NEFs are also thought to not bind substrate. A recent structural study suggests a large conformation shift within Hsp110 may be involved in its NEF function (20–27). Although the biochemical mechanisms of these NEFs have been recently revealed, little is known of the biological difference between them.
The important question of how Hsc70 mechanically assists polypeptide folding is still under investigation. Studies of the E. coli DnaK-DnaJ-GrpE system have provided the outline of a model (1, 28). Increases in the steady-state ATPase rate of DnaK require both DnaJ and GrpE to promote the ATP hydrolysis and nucleotide exchange steps. However, refolding of the model polypeptide luciferase is optimal at a particular ratio of DnaK to DnaJ and GrpE and, by implication, at the ATPase rate supported by that ratio. At this rate, the balance between nucleotide-dependent polypeptide binding and release by Hsc70 appears to be optimal for substrate folding. Other factors may be the binding of polypeptide by DnaJ and its transfer to DnaK.
The multiple DJAs and NEFs of human Hsc70 provide a fresh opportunity to examine this model. In the simplest formulation, an optimal Hsc70 ATPase rate for refolding should be supported by different co-chaperone combinations. The functions of DJA1 and DJA2 have been compared together with Hsc70 and the Bag1 NEF (6) but not with HspBP1 or Hsp110. We have reported that DJA1 was significantly less active than DJA2 in promoting the Hsc70- and C-Bag-mediated refolding of luciferase, although both had polypeptide binding and ATPase stimulatory properties (11), but differences between DJA1 and DJA2 with regard to the other NEFs are not known. Conversely, HspBP1 and Hsp110 have only been analyzed in combination with the biologically less relevant DJB1/Hsp40, like most data on Bag1 (13, 15, 18, 19, 22, 23). We therefore carried out a systematic analysis of the major DJA1 and DJA2 co-chaperones with Hsc70 and the Bag1, HspBP1, and Hsp110 NEFs.
EXPERIMENTAL PROCEDURES
Reagents and Proteins
Unless stated otherwise, all chemical reagents were from Sigma or BioShop Canada Inc. (Mississauga, Canada). Restriction enzymes and other recombinant DNA reagents were from New England Biolabs, Invitrogen, and Stratagene. Untreated rabbit reticulocyte lysate (RL) was from Green Hectares (Oregon, WI) and desalted into buffer G (100 mM KOAc, 20 mM Hepes-KOH, pH 7.5, and 5 mM MgOAc2) on a Hi-Prep 26/10 Fast Desalting column (GE Healthcare).
His-tagged human DJA1, DJA2, DJA4, Hsc70, and C-Bag were expressed in Rosetta 2 E. coli cells (Novagen) and purified as described (11). His-tagged human HspBP1 and Hsp110 (19, 23) were expressed in Rosetta 2 E. coli cells for 2 h at 37 °C and for 4 h at 30 °C, respectively. They were purified by chromatography on nickel-Sepharose HP (GE Healthcare) equilibrated in 500 mM NaCl, 20 mM imidazole, and 20 mM KH2PO4, pH 7.5, and eluted with 300 mM imidazole and 20 mM KH2PO4, pH 7.5, and then on a Mono Q 5/50 GL (GE Healthcare) equilibrated in 50 mM NaCl and 20 mM KH2PO4, pH 7.5, and eluted with a 50 to 600 mM NaCl gradient. HspBP1 was dialyzed into buffer G, and Hsp110 was further purified on a Superdex 200 Hi-Load 16/60 column (GE Healthcare) equilibrated in buffer G. DNA encoding the chimeric mutant DJA1–2 containing amino acids 1–95 of DJA1 and 96–412 of DJA2 was constructed by overlapping PCR and inserted into pPROEXHTa (Invitrogen), and the protein was purified similarly to DJA2. Where indicated, His tags were removed by digestion with His-tagged TEV protease 4 °C overnight followed by repurification on nickel-Sepharose.
Polypeptide Binding
Binding of various polypeptides to DJA1, DJA2, and DJA4 was tested as described (11). Plasmids encoding bovine phosphate carrier A (PiC) and mouse adenine nucleotide carrier 2 (ANT) were as described (29); those for rat citrate carrier (CiC) and bovine oxaloglutarate carrier (OGC) were from Vincenzo Zara (Lecce, Italy); those for human glucocorticoid receptor (GR), estrogen receptor α (ERα), progesterone receptor (PR), and mineralocorticoid receptor (MR) were from Theo Rein (Munich, Germany); and those for human cytochrome b5 and synaptobrevin 2 were from Stephen High (Manchester, UK). Purified DJA1, DJA2, and DJA4 were pre-bound on nickel-Sepharose in buffer containing 500 mM NaCl, 20 mM Hepes-KOH, pH 7.5, and 5 mM MgOAc2 for 30 min at 4 °C. Cell-free translations of the various polypeptides were performed with the TNT-coupled RL system with SP6 or T7 polymerase (Promega) supplemented with [35S]methionine (GE Healthcare and PerkinElmer Life Sciences), diluted 1:20 into buffer G containing 20 mM imidazole, 0.1% Triton X-100 and 2 mg/ml ovalbumin, and added to the immobilized DJA proteins. The final reactions contained 5 μM wild-type DJA protein and 5% translation mixture. After 5 min at room temperature, the binding reactions were terminated by the addition of 0.1 unit/μl apyrase. The protein complexes were recovered at 4 °C for 30 min and washed with buffer G containing 20 mM imidazole and 0.1% Triton X-100. The protein complexes were analyzed by SDS-PAGE and phosphorimaging quantitation.
Hsc70 Activation
Hsc70 activation by the co-chaperones was tested as described (11). To assay polypeptide refolding, firefly luciferase (Sigma) was denatured in buffer G containing 6 M guanidinium-HCl and 1 mM dithiothreitol for 10 min at room temperature. Refolding reactions were preassembled on ice, containing 4 μM Hsc70 and the indicated amounts of purified co-chaperones in buffer G supplemented with 39 mM NaCl and 2 mM ATP. Luciferase was diluted 1:100 into reactions to a final concentration of 5.4 nM and incubated at 30 °C. Control reactions contained 70% RL in the same buffer or treated with 0.1 unit/μl apyrase as a negative control. At the 60 min or indicated time point, the aliquots were diluted 2:25 into luciferase assay reagent (Promega), and luciferase activity was measured in an EG&G Berthold Lumat LB 9507 luminometer.
To test the ATPase activity of Hsc70, the reactions were preassembled on ice as described for the refolding assays, except that they were supplemented with 5 μCi/ml [α-32P]ATP (Perkin Elmer) in addition to 2 mM ATP, and the reactions were initiated by incubation at 30 °C. At various time points, the aliquots were removed and terminated with 37.5 mM EDTA. The samples were separated by thin layer chromatography on polyethyleneimine cellulose (Mallinckrodt Baker) developed in 0.5 M LiCl and 0.5 M formic acid. The ADP produced was determined by phosphorimaging quantitation, and the linear enzymatic rates (Vmax) were calculated by regression analysis.
RESULTS
Polypeptide Binding by DJAs
DJA1 and DJA2, as well as DJA4, are homologous to each other in their J domains and their central polypeptide-binding regions. However, sequence divergence between DJA1 and DJA2 (55% identity, 69% similarity) is almost as high as between each of them and Ydj1 (46–47% identity, 63–65% similarity), which would suggest functional differences between DJA1 and DJA2. These proteins were previously compared in their binding of two mitochondrial preproteins, which depend on the Hsc70-Hsp90 chaperone system for their targeting (11, 29). DJA1 bound the PiC and ANT ~2-fold better than DJA2, and DJA4 bound ANT better than PiC.
To obtain a more complete picture of DJA substrate binding characteristics, a wider range of polypeptides, which were potential chaperone substrates, was examined. The mitochondrial CiC and OGC, like PiC and ANT, belong to the structural family of metabolite transporters of the mitochondrial inner membrane. They all have six transmembrane domains and remain unfolded before insertion into the membrane (30). CiC and OGC preproteins also show involvement of the chaperone-Tom70 pathway in their mitochondrial import (31, 32). In contrast to the mitochondrial preproteins, the steroid hormone receptors are cytosolic and nuclear. Several of these proteins, including GR, ERα, PR and MR, respectively, are known to depend on the Hsc70-Hsp90 system to maintain them in a hormone-activable state (33, 34). Their hormone-binding domains appear to be the main chaperone binding sites and may be conformationally unstable in the absence of hormone. Certain transmembrane tail-anchored proteins such as synaptobrevin 2 are also assisted by Hsc70 during their insertion into the endoplasmic reticulum membrane. Other tail-anchored proteins such as cytochrome b5 appear to be less dependent on chaperones for insertion (35). The binding of the above polypeptides by the purified DJAs was therefore tested with our previously established method (11).
The various polypeptides were radiolabeled by cell-free translation in rabbit RL, a standard model cytosol for chaperone experiments. The labeled polypeptides were then co-precipitated with purified His-tagged DJA1, DJA2, or DJA4 and nickel-Sepharose. The existing chaperones in the RL compete for substrate binding with the DJAs but also help maintain solubility of the substrates; the radiolabeling allows accurate quantitation of the bound polypeptides by phosphorimaging analysis. The amount of bound polypeptide relative to input labeled material was determined, and the binding between the co-chaperones was compared. Substrate binding by DJA1, DJA2, and DJA4 relative to input is shown in Fig. 1. More CiC and OGC were bound by DJA1 than the established preprotein substrate PiC, over 80% of input in the case of OGC (Fig. 1). The amounts of steroid hormone receptors bound by DJA1 varied widely: 16% relative to input for PR, 25% for GR, 49% for ERα, and 66% for MR. The tail-anchored proteins cytochrome b5 and synaptobrevin 2 were bound poorly by DJA1, at less than 6% of input and close to background binding by the nickel-Sepharose control. So, the interaction of the mitochondrial preproteins and steroid hormone receptors with DJA1 seems to vary between the different proteins, whereas the tail-anchored proteins are bound poorly by DJA1.
FIGURE 1. Polypeptide binding by DJAs.
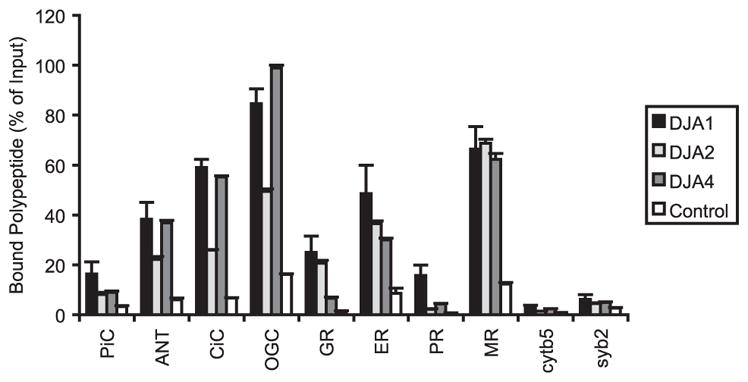
The indicated polypeptides were radiolabeled by cell-free translation and co-precipitated either with nickel-Sepharose alone or with purified His-tagged DJA1, DJA2, or DJA4 and nickel-Sepharose. Bound polypeptide was quantified by SDS-PAGE and phosphorimaging analysis. In these and all experiments, the error bars represent the standard deviations from the mean of at least three independent trials. The amount of polypeptide bound by negative control beads or by DJA1, DJA2, or DJA4 was plotted as a percentage of input translated material. cytb5, cytochrome b5; syb2, synaptobrevin 2.
Polypeptide binding by DJA2 and DJA4 was next analyzed. As observed before, PiC binding by DJA2 was approximately half that by DJA1, and binding of CiC and OGC by DJA2 was now also observed at approximately half relative to DJA1 binding, respectively (Fig. 1). DJA2 binding of GR and ERα was more similar to DJA1. Binding of PR by DJA2 was quite low, whereas binding of MR was essentially identical to DJA1. Very little cytochrome b5 and synaptobrevin 2 was bound by DJA2. In comparison, DJA4 binding also showed variation in the same range. Binding by DJA4 was comparable with DJA1 for CiC and OGC and progressively weaker for MR and ERα. GR and PR were bound most weakly by DJA4 at approximately one-fourth of DJA1 binding. Thus, the substrate binding profiles of DJA1 and DJA2 are clearly different. Overall, DJA1 shows moderately stronger binding of these polypeptides, but with exceptions (PR and MR). DJA4 may be more similar to DJA1 in its profile, although exceptions are also observed (GR and PR). The differences in binding are significant but mostly range within an order of magnitude. This is consistent with our proposed idea of partial specialization between the DJA co-chaperones.
Polypeptide Refolding by Hsc70, DJA2, and NEFs
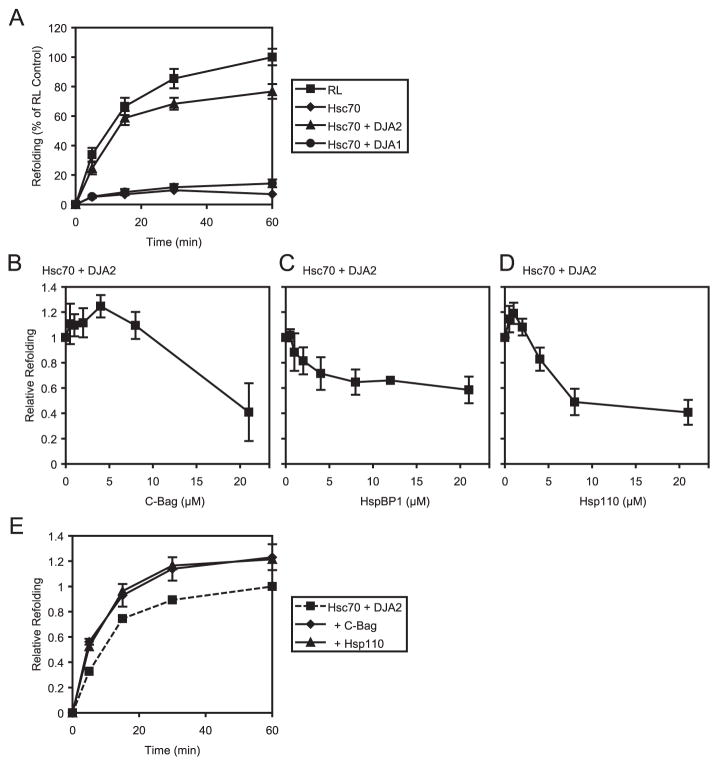
We recently reported that DJA2 was quite active in supporting the refolding of chemically denatured luciferase by purified Hsc70, whereas DJA1 had very poor activity (11). Those experiments were performed using 4 μM Hsc70, 4 μM of either DJA1 or DJA2, and 0.5 μM of the Bag1 NEF domain (C-Bag) (13, 14). The 8:1 ratio of Hsc70 to NEF had been thought to be optimal for Hsc70-mediated refolding, as well as for the E. coli DnaK-DnaJ-GrpE system (6). To compare DJA refolding functions with different NEFs, we first tested refolding in the absence of any NEF. The model cytosol RL was used as a positive control. Luciferase was denatured in guanidine and rapidly diluted 1:100 to 5.4 nM in RL or reactions containing purified proteins, supplemented with 2 mM ATP, and refolding at 30 °C was monitored by the luciferase enzymatic activity (11). As expected, RL provided efficient refolding of luciferase, whereas 4 μM Hsc70 alone could not (Fig. 2A). Remarkably, 4 μM Hsc70 and 4 μM DJA2 was relatively effective at refolding, to above 70% of the RL control by 60 min. This suggested that unlike for DnaK, an NEF may not be stringently required for Hsc70 function when an appropriate DJA partner is available. Consistent with earlier results, an equivalent amount of Hsc70 and DJA1 was inactive in refolding.
FIGURE 2. Polypeptide refolding by Hsc70, DJA2, and NEFs.
Luciferase was denatured in 6 M guanidine and diluted 1:100 into refolding reactions containing RL or the indicated combinations of Hsc70 and co-chaperones. Refolding at 30 °C was monitored by luciferase activity. A, activity upon refolding in RL or with 4 μM Hsc70 alone or with 4 μM DJA1 or DJA2, was monitored over time and plotted as a percentage of the activity at 60 min of the RL positive control. B–D, refolding activity after 60 min with 4 μM Hsc70, 4 μM DJA2, and the indicated amounts of C-Bag, HspBP1, and Hsp110 was plotted relative to that with Hsc70 and DJA2 alone. The y axis scales are identical. In this and all subsequent experiments, refolding after 60 min with 4 μM Hsc70 and 4 μM DJA2 was used as control reactions, with the amount of refolded luciferase activity set to 1. E, refolding activity over time with 4 μM Hsc70, 4 μM DJA2, and either 4 μM C-Bag or 1 μM Hsp110 was plotted relative to Hsc70 and DJA2 alone at 60 min. For comparison, the data for Hsc70 and DJA2 alone from A is replotted here.
Although an NEF may not be absolutely required for the Hsc70-DJA2 pair, it may still improve the efficiency of the refolding reaction. An increase in refolding of thermally denatured and in some cases chemically denatured luciferase had been reported for Bag1 and Hsp110 together with Hsc70 and DJB1/Hsp40 (13, 23). Therefore, chemically denatured luciferase refolding was tested with 4 μM Hsc70 and DJA2 as above, but with increasing concentrations of the C-Bag, HspBP1 and Hsp110 NEFs. The levels of refolding after 60 min relative to that of the Hsc70 and DJA2 control reaction without NEF was plotted (Fig. 2, B–D). Increasing amounts of C-Bag produced a moderate but significant increase in refolding that peaked at 1.24-fold above Hsc70 and DJA2 alone (Fig. 2B). The optimal C-Bag concentration for refolding was at 4 μM, equimolar to Hsc70 and DJA2. 21 μM inhibited refolding to ~0.4 of the Hsc70-DJA2 control.
HspBP1 had a very different behavior. Increasing HspBP1 amounts did not increase refolding by Hsc70-DJA2 at all, but progressively inhibited refolding to ~0.6 of the control (Fig. 2C). For Hsp110, the result was different again. 1 μM Hsp110 significantly raised refolding to 1.20-fold of the control (Fig. 2D), similar to the maximum effect of C-Bag. Higher levels of Hsp110 clearly inhibited refolding, to less than 0.5 of the control at 8 μM and above. To confirm these results, the time course of luciferase refolding at optimal concentrations of C-Bag (4 μM) and Hsp110 (1 μM) with Hsc70 and DJA2 was examined. The increase in refolding compared with Hsc70-DJA2 alone was evident throughout the time course and particularly at the early (5 min and 15 min) time points (Fig. 2E). So, both the Bag1 and Hsp110 NEF activities can boost refolding by Hsc70 and DJA2, although specific concentration ratios of these NEFs to Hsc70 seem to be required. As seen for HspBP1, NEF activity does not necessarily promote Hsc70-mediated refolding.
DJA1 Negatively Effects Hsc70-mediated Polypeptide Refolding
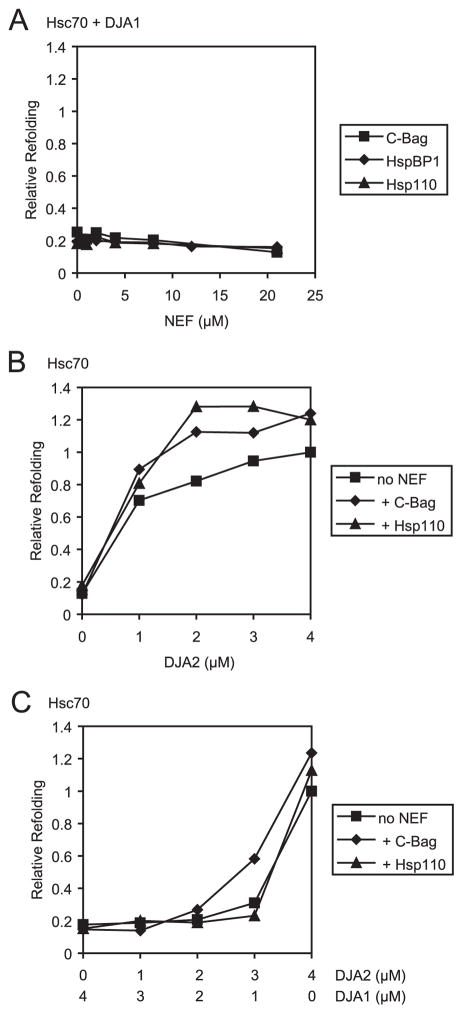
It was possible that one or more of the NEFs might have a greater effect on Hsc70-mediated refolding with DJA1, which had poor activity by itself. The refolding of chemically denatured luciferase was thus tested as above but with 4 μM each of Hsc70 and DJA1 and increasing amounts of C-Bag, HspBP1, and Hsp110. The levels of refolding after 60 min relative to that of Hsc70 and DJA2 control reactions were plotted. No amount of any of the NEFs could activate refolding by Hsc70 and DJA1 above 0.25 that of the Hsc70-DJA2 control (Fig. 3A). This clear difference between DJA1 and DJA2 suggests some mechanistic divergence, despite their sequence homology.
FIGURE 3. DJA1 inhibits polypeptide refolding.
Luciferase refolding was monitored as in Fig. 2, with the indicated combinations of Hsc70 and co-chaperones. A, refolding activity after 60 min with 4 μM Hsc70, 4 μM DJA1, and the indicated amounts of C-Bag, HspBP1, and Hsp110 was plotted relative to the Hsc70 and DJA2 control reactions. B, refolding activity after 60 min with 4 μM Hsc70, the indicated amounts of DJA2, and either no addition or 4 μM C-Bag or 1 μM Hsp110 was plotted relative to the Hsc70 and DJA2 control reactions. C, refolding activity after 60 min with 4 μM Hsc70, the indicated amounts of DJA2 and DJA1, and either 4 μM C-Bag or 1 μM Hsp110 was plotted relative to the Hsc70 and DJA2 control reactions.
The effect of DJA1 on an Hsc70-mediated refolding reaction might be neutral or inhibitory. In the first case, a refolding reaction with Hsc70 and DJA2 would remain unaffected by the addition of DJA1. In the latter case, DJA1 would interfere with the reaction. To address this question, luciferase refolding reactions having different amounts of DJA1 and DJA2 were examined. Reactions with 4 μM Hsc70 and a constant 4 μM total DJA proteins were analyzed (Fig. 3C), because optimization tests with 4 μM Hsc70 had previously shown maximum refolding at 4 μM DJA2 (Fig. 3B), whereas different concentrations of DJA1 in this range were equally ineffective with Hsc70 (not shown). In fact, we observed that DJA1 inhibited refolding when it was present. For example, reactions with 4 μM Hsc70 and 2 μM DJA2 alone produced significant refolding activity, 0.82 of the control, whereas further addition of 2 μM DJA1 reduced refolding to 0.25. The addition of C-Bag or Hsp110 NEF did not restore the refolding activity inhibited by DJA1. The addition of optimal concentrations of C-Bag (4 μM) and Hsp110 (1 μM) further increased refolding by Hsc70 and DJA2 alone (Fig. 3B), but the presence of DJA1 still blocked refolding (Fig. 3C). Indeed, refolding in the presence of 2 μM DJA1 was as poor when C-Bag or Hsp110 were added, as when the NEFs were absent. Thus, DJA1 is not only inactive in the Hsc70-mediated refolding tested here but appears to inhibit this particular function of DJA2 and Hsc70 and perhaps other functions. We further ruled out effects of the His tags on the inhibition of refolding. Hsc70 with its His tag removed behaved similarly to tagged Hsc70. It was activated to refold luciferase by DJA2, but not DJA1. C-Bag and Hsp110 further increased its refolding with DJA2, but not HspBP1. HspBP1, after removal of the His tag, did not enhance refolding by Hsc70-DJA2. Similarly, DJA1, after removal of the His tag, still could not activate Hsc70 to refold luciferase and was inhibitory of the Hsc70-DJA2 refolding reaction (not shown). Nontagged DJA2 was not isolated in sufficient amounts to be tested and may be more active than tagged DJA2.
NEFs Diverge in ATPase Stimulation of Hsc70
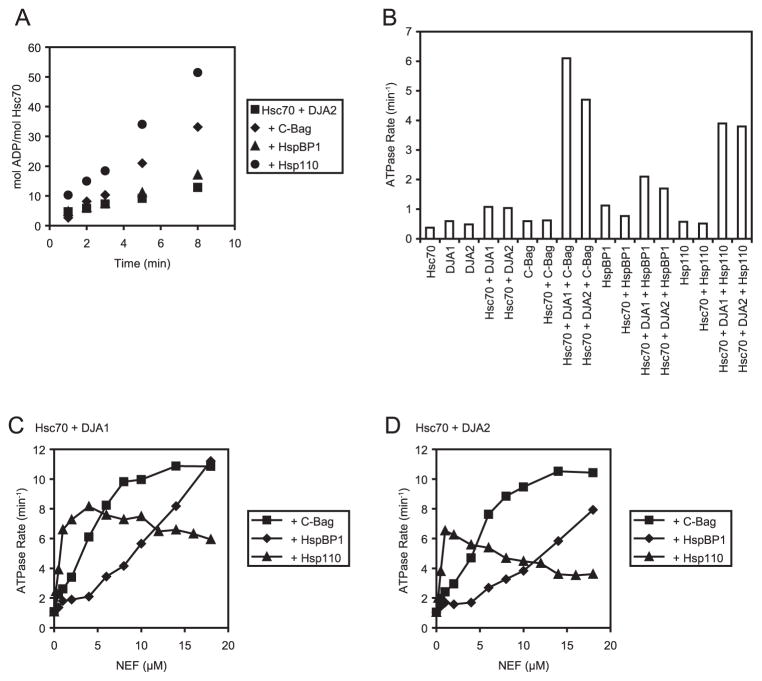
The results in Figs. 2 and 3 indicated that both C-Bag and Hsp110 were able to increase the refolding function of Hsc70 and DJA2 but not that with DJA1. With either of the DJA co-chaperones, the expected effect of an NEF would be to increase the overall ATPase rate of Hsc70. Earlier work found that the ADP-ATP exchange step can be rate-limiting for the steady-state Hsc70 (and DnaK) ATPase and that both an NEF and a DnaJ-related co-chaperone were required for ATPase stimulation (1, 36, 37). It was possible that increased refolding activity correlated with a specific ATPase rate of Hsc70; inability to reach this rate might explain the weaker refolding function of HspBP1 or, less probably, of DJA1. We therefore investigated the relationship between the Hsc70 ATPase rate and refolding activity at different concentrations of each NEF. As established (11), the reactions were set up similar to the refolding reactions but lacking denatured luciferase and containing [α-32P]ATP to monitor ADP production over time at 30 °C (Fig. 4A). Linear (steady-state) enzymatic rates were calculated, and representative rates are presented in Table 1.
FIGURE 4. NEFs diverge in Hsc70 ATPase stimulation.
ADP production in 30 °C reactions containing the indicated combinations of Hsc70 and co-chaperones was monitored by thin layer chromatography separation of radiolabeled ADP from ATP and phosphorimaging analysis. A, examples of ADP production over time in reactions with 4 μM Hsc70, 4 μM DJA2, and either 4 μM C-Bag, 4 μM HspBP1, or 1 μM Hsp110. Steady-state (linear) ATPase rates in Table 1 are based on similar experiments. B, Hsc70 ATPase rates were measured for reactions with the indicated combinations of 4 μM Hsc70, 4 μM DJA1 or DJA2, 4 μM C-Bag, 4 μM HspBP1, or 0.5 μM Hsp110. C and D, ATPase rates were measured with 4 μM Hsc70, 4 μM of either DJA1 or DJA2, and the indicated amounts of C-Bag, HspBP1, and Hsp110.
TABLE 1. Steady-state ATPase rates of Hsc70 in combination with the indicated co-chaperones.
Average enzymatic rates (min−1) and standard deviations from the mean are shown.
| NEF | 4 μM DJA1 | 4 μM DJA2 | 4 μM DJA1–2 | |
|---|---|---|---|---|
| 4 μM Hsc70 | 1.1 ± 0.1 | 1.0 ± 0.2 | 1.2 ± 0.1 | |
| 4 μM Hsc70 | 4 μM C-Bag | 6.1 ± 0.3 | 4.8 ± 0.2 | 5.5 ± 0.6 |
| 4 μM Hsc70 | 4 μM HspBP1 | 2.0 ± 0.2 | 1.7 ± 0.2 | 2.2 ± 0.1 |
| 4 μM Hsc70 | 1 μM Hsp110 | 8.0 ± 1.1 | 6.7 ± 0.3 | 6.6 ± 0.2 |
As expected, 4 μM Hsc70 with 4 μM of either DJA1 or DJA2 had a low enzymatic rate in the absence of NEF, 1.1 and 1.0 min−1, respectively (Table 1 and Fig. 4B). Low background rates were also observed for the co-chaperones in the absence of Hsc70, and the addition of DJA1 or DJA2 and C-Bag stimulated the Hsc70 ATPase activity. In the absence of Hsc70, the background ATPase rate of Hsp110 was not increased by either DJA1 or DJA2 (not shown). At 4 μM C-Bag and 4 μM DJA2, which provided maximum refolding function in our conditions, the Hsc70 ATPase rate was 4.8 min−1. In parallel Hsc70-DJA2 reactions, 4 μM HspBP1 produced less stimulation of the Hsc70 ATPase rate to 1.7 min−1, but 1 μM Hsp110 raised the rate more strongly to 6.7 min−1. The Hsc70 ATPase rates with the different NEFs were slightly higher with DJA1 than with DJA2 (Fig. 4B and Table 1). We previously observed that DJA1 was a somewhat better activator of the Hsc70 ATPase under saturating C-Bag concentrations (11), and our data now confirm this under conditions closer to that of the cell.
We next tested the Hsc70 ATPase rate at the same fixed concentrations of DJA2 and DJA1, over different concentrations of each NEF. The DJA1-stimulated Hsc70 ATPase rate increased steadily with C-Bag concentration until ~10 μM C-Bag, with higher concentrations causing only small increases in rate (Fig. 4C). A maximum rate of ~11 min−1 could be reached at high C-Bag levels. The C-Bag effect on Hsc70 with DJA2 was essentially the same, with a similar high rate observed above 10 μM C-Bag (Fig. 4D). HspBP1 also raised the Hsc70 ATPase rate with increasing concentration but was markedly less efficient than C-Bag. With DJA1, 10 μM HspBP1 was needed to achieve a rate of ~6 min−1, compared with 4 μM of C-Bag (Fig. 4C). At 18 μM HspBP1, the rate observed for DJA1 and Hsc70 was 10 min−1, whereas the maximum for the comparable DJA2 reaction was 8 min−1. Surprisingly, the behavior of Hsp110 was altogether different. As noted above, low concentrations of Hsp110 were strongly stimulatory of the Hsc70 ATPase, with DJA1 or DJA2. However, the maximum rate observed for DJA1-stimulated Hsc70 was just over 8 min−1 at 4 μM Hsp110, and higher amounts of the NEF caused a moderate reduction in the ATPase rate instead of a further increase (Fig. 4C). The effect with DJA2 was also clear; a peak rate of less than 7 min−1 was attained at 1 to 2 μM Hsp110, and higher concentrations reduced ATPase rates below this level (Fig. 4D). So, the three NEFs have quite different characteristics in their regulation of the Hsc70 ATPase.
In the refolding assays, the highest Hsc70-DJA2 function was observed between 2 and 8 μM C-Bag, with the highest point at 4 μM C-Bag (Fig. 2B). These conditions corresponded to DJA2-stimulated Hsc70 ATPase rates between 3 and 9 min−1, with the peak refolding activity ~5 min−1 (Fig. 4D). In comparison, refolding was stimulated by Hsp110 between a narrow concentration range of 0.5–2 μM, with maximum activity at ~1 μM. The ATPase rates supported by these amounts of Hsp110 were between 4 and 8 min−1, and peak refolding corresponded to just under 7 min−1. An Hsc70 ATPase rate ~6 min−1 could thus be the optimal rate for its refolding activity, regardless of the NEF being used. However, HspBP1 was also able to raise the Hsc70 rate with DJA2 to this range, at higher concentrations of the NEF ~14 μM but was still unable to increase the Hsc70 refolding function. Furthermore, 8 μM Hsp110 was strongly inhibitory of the refolding function, although the corresponding ATPase rate was close to 5 min−1 and not far from the proposed optimal rate. Finally, DJA1 appears generally similar to DJA2 in ATPase stimulation together with the NEFs despite its poor refolding function under all conditions. Therefore, for the DJA and the NEF co-chaperones, the ability to activate Hsc70 enzymatically to a certain level may be necessary for refolding function but is not in itself sufficient.
Activity of a DJA1-DJA2 Chimera
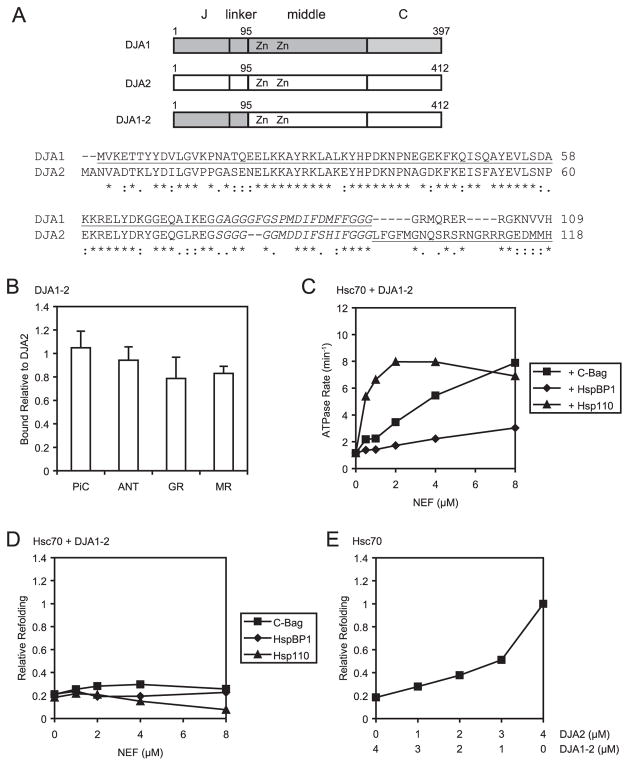
The unfavorable or negative effect of DJA1 on Hsc70-mediated refolding could not be explained by lack of ATPase stimulation (Fig. 4). Because DJA1 bound substrates somewhat more strongly than DJA2, it was possible that the stronger binding interfered with the refolding reaction, or some other feature of DJA2, such as coordination between its different domains, could be better adapted for this refolding reaction than in DJA1. To address these questions, we constructed a chimeric DJA that had the N-terminal J domain and linker of DJA1 (residues 1–95) and the substrate-binding zinc finger, central, and C-terminal regions of DJA2 (residues 96–412) (Fig. 5A). We reasoned that the chimeric DJA1–2 should have the same substrate binding properties of DJA2 and should stimulate the Hsc70 ATPase like DJA1. However, if DJA1 and DJA2 have different interactions between their respective N- and C-terminal regions, these interactions will be mismatched in DJA1–2. On gel filtration chromatography, purified DJA1–2 had a profile similar to those of DJA1 and DJA2, consistent with native homodimers (not shown). DJA1–2 was therefore tested for the properties of substrate binding, stimulation of the Hsc70 ATPase, and Hsc70-mediated refolding.
FIGURE 5. Activity of a DJA1-DJA2 chimera.
A, upper panel, schematic of the domain architecture of DJA1, DJA2 and the DJA1–2 mutant. The positions of the J domain, linker, middle domain with zinc fingers, and C-terminal domain are marked. DJA1–2 contains residues 1–95 of DJA1 and residues 96 – 412 of DJA2. Lower panel, CLUSTALW2 alignment of the N-terminal regions of DJA1 and DJA2. The linker sequences are in italics, and the sequence of DJA1–2 is underlined. B, binding of radiolabeled polypeptides to DJA2 and DJA1–2 was tested as in Fig. 1, and the amount bound by DJA1–2 was plotted relative to the amount bound by DJA2. C, Hsc70 ATPase rates were measured as in Fig. 4 for reactions with 4 μM Hsc70, 4 μM DJA1–2, and the indicated amounts of C-Bag, HspBP1, and Hsp110. D and E, luciferase refolding was monitored as in Fig. 2, with the indicated combinations of Hsc70 and co-chaperones. D, refolding activity after 60 min with 4 μM Hsc70, 4 μM DJA1–2, and the indicated amounts of C-Bag, HspBP1, and Hsp110, was plotted relative to the Hsc70 and DJA2 control reactions. E, refolding activity after 60 min with 4 μM Hsc70 and the indicated amounts of DJA2 and DJA1–2 was plotted relative to the Hsc70 and DJA2 control reactions.
Binding of the representative substrates PiC, ANT, GR, and MR to DJA1–2 was assayed as described in Fig. 1 and quantified relative to DJA2 binding (Fig. 5B). As expected, the levels of polypeptide binding were similar. Slightly less GR was bound by DJA1–2 than by DJA2, but it is not certain that this difference is significant. Next, the activity of the Hsc70 ATPase was tested as in Fig. 4. At 4 μM Hsc70 and 4 μM DJA1–2, a rate of 1.2 min−1 was observed, essentially the same as that with DJA1 and DJA2 (Table 1). The addition of various amounts of C-Bag, HspBP1, and Hsp110 NEF to the DJA1–2 and Hsc70 reaction boosted the ATPase rates (Fig. 5C), with a pattern closest to that of wild-type DJA1. The ATPase rates rose progressively with higher C-Bag concentrations, reaching 7.9 min−1 at 8 μM C-Bag. As observed with the wild-type DJAs, HspBP1 was less effective at stimulating the ATPase rates, with 8 μM HspBP1 supporting a rate of 3.0 min−1. Hsp110 stimulated the ATPase activity strongly at low concentrations, but peak ATPase rates of 8 min−1 were observed at 2 μM and 4 μM Hsp110. This was similar to the effect of Hsp110 on the DJA1 and Hsc70 reaction, which also peaked at 4 μM Hsp110 (Fig. 4C). For DJA2, maximum stimulation was observed at 1 μM Hsp110, and higher concentrations led to a decline in rate. Thus, DJA1–2 resembles DJA2 in its polypeptide binding characteristics and DJA1 in its ATPase stimulation in concert with NEFs.
DJA1–2 was next tested in the Hsc70-mediated refolding of luciferase. Reactions contained 4 μM Hsc70 and 4 μM DJA1–2 and as above were compared with control reactions containing 4 μM Hsc70 and DJA2 after 60 min of refolding. Interestingly, reactions with DJA1–2 showed little refolded protein, less than 0.25 of the DJA2-Hsc70 control (Fig. 5D). The addition of C-Bag NEF over a range of concentrations improved refolding with DJA1–2 only marginally; HspBP1 had no effect, whereas Hsp110 at higher concentrations appeared to reduce even the basal level of refolding. Thus, although the majority of DJA1–2 was derived from DJA2 and its J domain was clearly active, the mechanism required for the refolding reaction was disrupted.
To determine whether DJA1–2 had a neutral or negative effect on refolding, reactions with a constant level of 4 μM Hsc70 and different amounts of DJA1–2 and DJA2 were tested. As was done for DJA1 and DJA2 (Fig. 3C), DJA1–2 ranged from 4 to 0 μM, and DJA2 from 0 to 4 μM, with the total concentration of DJA remaining constant. Like DJA1, the DJA1–2 mutant appeared to inhibit DJA2-stimulated refolding (Fig. 5E), although not quite as strongly as DJA1. For example, 1 μM DJA1 inhibited refolding reactions with 3 μM DJA2 to less than 0.30 of the control (Fig. 3, B and C), and 1 μM DJA1–2 under the same conditions reduced refolding to 0.52. Overall, the results with the chimeric DJA1–2 mutant suggest that the complete refolding activity of DJA2 with Hsc70 requires more than the independent function of its domains, but also another feature such as interdomain coordination. The J domain and linker of DJA1–2, derived from DJA1, may be unable to properly interact with the rest of the protein. Furthermore, the coordination between the domains of wild-type DJA1 may be different enough from that within DJA2 that it cannot function well in the refolding reaction studied here. NEFs can improve the function of DJA2 with Hsc70 but cannot overcome the deficiency of DJA1 or DJA1–2.
DISCUSSION
The systematic investigation of the Hsc70 co-chaperones suggested three conclusions. First, DJA1 and DJA2 are functionally different. This difference cannot be simply explained by their polypeptide binding or Hsc70 stimulatory properties or preference for an NEF partner. Second, the Bag1 and Hsp110 NEFs are also functionally distinct from HspBP1. The appropriate ratios between Hsc70 and its co-chaperones may correlate with an optimal ATPase rate of Hsc70, but cycling at this rate is not sufficient for complete chaperone function. Third, our results suggest a more complex model of human Hsc70 function than is readily apparent in the E. coli DnaK system.
DJA1 and DJA2 are distinct in the characteristics of their separate domains: the stimulation of the Hsc70 ATPase by their J domains and the binding of a range of polypeptides by their central regions. These differences extend our earlier observation of variations between the DJAs, further supporting our proposal of partial specialization between the DJAs. However, the variation in domain properties, although significant, is within a limited range. This variation may contribute to their functional difference, as observed in the refolding with Hsc70 of guanidine-denatured luciferase, but seems insufficient to fully explain the difference. There may be an Hsc70 ATPase rate that is optimal for refolding when stimulated by DJA2 and either Bag1 or Hsp110, but DJA1 with the NEFs can also stimulate Hsc70 to the same extent. Because none of the NEFs can support refolding by DJA1 and Hsc70, the difference in refolding is not due to the preference of the DJAs for a specific NEF. The possibility that stronger polypeptide binding by DJA1 compared with DJA2 is the major hindrance to Hsc70-mediated refolding is argued against by the poor refolding ability of the chimeric DJA1–2 mutant, which has the substrate-binding domain of DJA2. So, there are likely to be other properties of the DJAs that are less well established but that contribute to their functional difference.
One such property may be coordination between the J domain, linker, and central regions of the DJAs. Such coordination may be important for the transfer of polypeptide from the DJA to Hsc70. This idea agrees with our results and may have a structural basis in the linker segment or domain surfaces of the DJAs. We hypothesize that the interdomain coordination of DJA1 is somewhat different from that of DJA2, such that the Hsc70-mediated refolding activity examined here is not supported. The ability of DJA1 to bind substrate, but not to transfer substrate productively to Hsc70, could explain the inhibition of refolding observed in our mixed DJA refolding experiments. This idea could also explain our observation that the DJA1–2 mutant inhibited the refolding mediated by Hsc70 and DJA2. Given that the DJA1–2 mutant has the J domain and linker of DJA1 and central region from DJA2, we might expect there to be a lack of coordination between the domains and hence impaired transfer of substrate to Hsc70. Nevertheless, the domain coordination in wild-type DJA1 might be arranged for the chaperoning of a range of substrates or polypeptide conformations different from that of DJA2. Indeed, we found that DJA1 may be more effective in the co-translational folding of luciferase (11), which is distinct from the refolding of mature protein studied here.
We expect that interdomain coordination involves specific structures within the DJAs. One possibility would be the Gly/Phe-rich linker between the J domain and central region, including an Asp-Ile-Phe tripeptide motif. Work with E. coli DnaJ and S. cerevisiae Ydj1 suggested that the Asp-Ile-Phe motif was important for the function of both co-chaperones, potentially for substrate transfer to the partner Hsp70s (38, 39). The linker in the DJAs is conserved only in the type 1 co-chaperones, suggesting a mechanism unique to this class and not found in type 2 proteins such as DJB1. Another possibility for interdomain coordination is contact between the J domains and the zinc finger regions of the DJAs. These structures are close together in the quaternary structure of the DJA1 homodimer (8) and presumably also in DJA2. J domains are thought to contact Hsc70 through a highly conserved surface (40), but other surfaces of the domain may form other interactions.
The Hsc70 NEFs are structurally very different from each other, so mechanistic differences were less unexpected. We propose that their biochemical properties observed here agree with their apparent biological roles. The Bag1 NEF is effective in promoting refolding, but at concentrations equimolar to Hsc70. In cells, Bag domains are typically found in combination with other functional domains, and these proteins seem to have more specialized roles. Bag1 and Bag2 are involved in regulating protein degradation, whereas Bag4 and Bag5 work in apoptotic signaling pathways (13, 16, 41–43). Such specialized proteins may be expressed at lower levels and would only reach concentrations equal to Hsc70 in localized complexes. HspBP1 is the least efficient as an NEF and is inhibitory of Hsc70 refolding function under all conditions tested. Biologically, HspBP1 promotes the degradation of proteins and blocks the anti-apoptotic function of stress-induced Hsp70 (44, 45). These functions are consistent with HspBP1 acting as an inhibitor of Hsc70. Such inhibition of Hsc70 may be useful for the cell when degradation as opposed to refolding becomes a better survival strategy. At low concentrations of NEF, Hsp110 is the strongest activator of the Hsc70 ATPase and promotes refolding at these same low concentrations. Thus, Hsp110 may be the general NEF for Hsc70 chaperoning function but with its expression levels tightly regulated. Hsp110 is inhibitory of both the refolding and ATPase activities of Hsc70 at higher concentrations. It is possible that Hsp110 at high concentrations binds substrates through its C-terminal region, competing with Hsc70. Overexpression of Hsp110 may be another method for the cell to switch Hsc70 from a folding to a degradation strategy.
The human Hsc70 system thus appears more complex than the canonical DnaK system of E. coli, not just in the range of co-chaperone proteins but in its actual biochemical and biological mechanism. This is seen most clearly in the ability of DJA1 and HspBP1 to activate the Hsc70 ATPase in a nonproductive manner. A comparison with the co-chaperones of Hsc70 (Ssa-type proteins) in S. cerevisiae agrees with this view (1, 2). Although this yeast has a single cytosolic DJA-type co-chaperone Ydj1, it has the three types of NEF in the cytosol. The Hsp110 orthologs Sse1 and Sse2 seem the most important biologically, and their combined deletion is lethal (23, 24). The single Bag-related protein Snl1 is membrane-anchored and seems to have a specialized function in the endoplasmic reticulum or nuclear membranes (41). The HspBP1 ortholog Fes1 is moderately important; its deletion causes temperature-sensitive growth, and it cannot fully substitute for Sse1/Sse2 deletion (17, 19, 24). These observations are consistent with the divergence between human Hsp110 as a general NEF, Bag proteins as functionally specialized, and HspBP1 as a distinct inhibitory factor. The human DJAs also seem to have diverged in their exact mechanisms.
Acknowledgments
We thank Melanie Bhangoo and Anna Fan for technical assistance.
Footnotes
This work was supported by a Canadian Institutes of Health Research operating grant and the Canadian Foundation for Innovation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: NEF, nucleotide exchange factor; ANT, adenine nucleotide translocator; CiC, citrate carrier; ERα, estrogen receptor α; GR, glucocorticoid receptor α isoform; MR, mineralocorticoid receptor; OGC, oxaloglutarate carrier; PiC, inorganic phosphate carrier; PR, progesterone receptor; RL, reticulocyte lysate.
References
- 1.Mayer MP, Bukau B. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young JC, Agashe VR, Siegers K, Hartl FU. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 3.Qiu XB, Shao YM, Miao S, Wang L. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minami Y, Hohfeld J, Ohtsuka K, Hartl FU. J Biol Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- 5.Terada K, Kanazawa M, Bukau B, Mori M. J Cell Biol. 1997;139:1089–1095. doi: 10.1083/jcb.139.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terada K, Mori M. J Biol Chem. 2000;275:24728–24734. doi: 10.1074/jbc.M002021200. [DOI] [PubMed] [Google Scholar]
- 7.Hafizur RM, Yano M, Gotoh T, Mori M, Terada K. J Biochem (Tokyo) 2004;135:193–200. doi: 10.1093/jb/mvh023. [DOI] [PubMed] [Google Scholar]
- 8.Borges JC, Fischer H, Craievich AF, Ramos CH. J Biol Chem. 2005;280:13671–13681. doi: 10.1074/jbc.M408349200. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Qian X, Sha B. Structure. 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Li J, Jin Z, Fu Z, Sha B. J Mol Biol. 2005;346:1005–1011. doi: 10.1016/j.jmb.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 11.Bhangoo MK, Tzankov S, Fan AC, Dejgaard K, Thomas DY, Young JC. Mol Biol Cell. 2007;18:3414–3428. doi: 10.1091/mbc.E07-01-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohfeld J, Jentsch S. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luders J, Demand J, Papp O, Hohfeld J. J Biol Chem. 2000;275:14817–14823. doi: 10.1074/jbc.275.20.14817. [DOI] [PubMed] [Google Scholar]
- 14.Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 15.Gassler CS, Wiederkehr T, Brehmer D, Bukau B, Mayer MP. J Biol Chem. 2001;276:32538–32544. doi: 10.1074/jbc.M105328200. [DOI] [PubMed] [Google Scholar]
- 16.Takayama S, Reed JC. Nat Cell Biol. 2001;3:E237–E241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 17.Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. FEBS Lett. 2002;531:339–342. doi: 10.1016/s0014-5793(02)03570-6. [DOI] [PubMed] [Google Scholar]
- 18.Raynes DA, Guerriero V., Jr J Biol Chem. 1998;273:32883–32888. doi: 10.1074/jbc.273.49.32883. [DOI] [PubMed] [Google Scholar]
- 19.Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Mol Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Oh HJ, Chen X, Subjeck JR. J Biol Chem. 1997;272:31636–31640. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- 21.Yamagishi N, Nishihori H, Ishihara K, Ohtsuka K, Hatayama T. Biochem Biophys Res Commun. 2000;272:850–855. doi: 10.1006/bbrc.2000.2864. [DOI] [PubMed] [Google Scholar]
- 22.Yamagishi N, Ishihara K, Hatayama T. J Biol Chem. 2004;279:41727–41733. doi: 10.1074/jbc.M407947200. [DOI] [PubMed] [Google Scholar]
- 23.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaner L, Sousa R, Morano KA. Biochemistry. 2006;45:15075–15084. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q, Hendrickson WA. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. J Biol Chem. 2008;283:8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- 28.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. Proc Natl Acad Sci U S A. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan AC, Bhangoo MK, Young JC. J Biol Chem. 2006;281:33313–33324. doi: 10.1074/jbc.M605250200. [DOI] [PubMed] [Google Scholar]
- 30.de Marcos-Lousa C, Sideris DP, Tokatlidis K. Trends Biochem Sci. 2006;31:259–267. doi: 10.1016/j.tibs.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Palmisano A, Zara V, Honlinger A, Vozza A, Dekker PJ, Pfanner N, Palmieri F. Biochem J. 1998;333:151–158. doi: 10.1042/bj3330151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zara V, Ferramosca A, Palmisano I, Palmieri F, Rassow J. J Mol Biol. 2003;325:399–408. doi: 10.1016/s0022-2836(02)01236-6. [DOI] [PubMed] [Google Scholar]
- 33.Pratt WB, Toft DO. Exp Biol Med (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 34.Picard D. Trends Endocrinol Metab. 2006;17:229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Abell BM, Rabu C, Leznicki P, Young JC, High S. J Cell Sci. 2007;120:1743–1751. doi: 10.1242/jcs.002410. [DOI] [PubMed] [Google Scholar]
- 36.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wawrzynow A, Banecki B, Wall D, Liberek K, Georgopoulos C, Zylicz M. J Biol Chem. 1995;270:19307–19311. doi: 10.1074/jbc.270.33.19307. [DOI] [PubMed] [Google Scholar]
- 38.Aron R, Lopez N, Walter W, Craig EA, Johnson J. Genetics. 2005;169:1873–1882. doi: 10.1534/genetics.104.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cajo GC, Horne BE, Kelley WL, Schwager F, Georgopoulos C, Genevaux P. J Biol Chem. 2006;281:12436–12444. doi: 10.1074/jbc.M511192200. [DOI] [PubMed] [Google Scholar]
- 40.Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young JC, Barral JM, Ulrich Hartl F. Trends Biochem Sci. 2003;28:541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Arndt V, Daniel C, Nastainczyk W, Alberti S, Hohfeld J. Mol Biol Cell. 2005;16:5891–5900. doi: 10.1091/mbc.E05-07-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai Q, Qian SB, Li HH, McDonough H, Borchers C, Huang D, Takayama S, Younger JM, Ren HY, Cyr DM, Patterson C. J Biol Chem. 2005;280:38673–38681. doi: 10.1074/jbc.M507986200. [DOI] [PubMed] [Google Scholar]
- 44.Alberti S, Bohse K, Arndt V, Schmitz A, Hohfeld J. Mol Biol Cell. 2004;15:4003–4010. doi: 10.1091/mbc.E04-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanimura S, Hirano AI, Hashizume J, Yasunaga M, Kawabata T, Ozaki K, Kohno M. J Biol Chem. 2007;282:35430–35439. doi: 10.1074/jbc.M707547200. [DOI] [PubMed] [Google Scholar]