Abstract
Dynamic reprogramming of metabolism is essential for T cell effector function and memory formation. However, the regulation of metabolism in exhausted CD8+ T (Tex) cells is poorly understood. We found that during the first week of chronic lymphocytic choriomeningitis virus (LCMV) infection, before severe dysfunction develops, virus-specific CD8+ T cells were already unable to match the bioenergetics of effector T cells generated during acute infection. Suppression of T cell bioenergetics involved restricted glucose uptake and use, despite persisting mechanistic target of rapamycin (mTOR) signaling and up-regulation of many anabolic pathways. PD-1 regulated early glycolytic and mitochondrial alterations and repressed transcriptional coactivator PGC-1α. Improving bioenergetics by overexpression of PGC-1α enhanced function in developing Tex cells. Therapeutic reinvigoration by anti-PD-L1 reprogrammed metabolism in a subset of Tex cells. These data highlight a key metabolic control event early in exhaustion and suggest that manipulating glycolytic and mitochondrial metabolism may enhance checkpoint blockade outcomes.
eTOC Blurb
Exhausted T cells progressively develop poor effector function in chronic infections and cancer and are a barrier controlling these diseases. Wherry and colleagues identify suppressed T cell metabolism as an early cellular perturbation driving exhaustion and implicate PD-1 and altered glycolytic, mitochondrial, and PGC-1α–driven metabolic pathways in the process.
Introduction
Upon viral infection, naïve virus-specific CD8+ T cells differentiate into effector T cells (Teff) that proliferate and produce antiviral effector proteins. To meet increased bioenergetic demands, these cells undergo metabolic reprogramming from quiescent mitochondrial oxidative phosphorylation (OXPHOS) to glycolysis (Buck et al., 2015). With these metabolic changes, glucose use is directed away from mitochondria, fueling less efficient cytoplasmic energy production but simultaneously allowing the generation of cellular building blocks necessary for proliferation and macromolecular synthesis to meet the demand for increased biomass. In addition, switching to glycolysis is directly linked to increased effector function (Chang et al., 2013; Ho et al., 2015). The change in metabolic lifestyle is thought to occur through T cell receptor (TCR)-linked, phosphoinositide 3-kinase (PI3K) and Akt-mediated mTOR signaling (Buck et al., 2015). After acute resolving infections, such as with the Armstrong (Arm) strain of lymphocytic choriomeningitis virus (LCMV), the virus-specific T cell response contracts and a pool of memory T cells (Tmem) is established (Wherry and Kurachi, 2015). The conversion to memory is characterized by a shift back to mitochondrial OXPHOS, fueled at least in part by fatty acid oxidation (O'Sullivan et al., 2014; van der Windt et al., 2012).
In contrast to acute resolving viral infections, virus-specific T cell function is compromised in persisting infections, such as in hepatitis C virus (HCV), human immunodeficiency virus (HIV) or infection with the chronic clone 13 strain of LCMV in mice as well as in cancer (Wherry and Kurachi, 2015). While there are similarities between the CD8+ Teff cell response in acute and chronic viral infections, virus-specific T cells in chronic infections can become progressively exhausted. Tex cells are defined by poor effector functions, high co-expression of multiple inhibitor receptors and an altered global transcriptional program compared to functional Teff or Tmem cells (Wherry and Kurachi, 2015). In addition, two subsets of Tex cells exist that are defined by high expression of the transcription factor T-bet and intermediate expression of inhibitory receptor PD-1 (T-betHiPD-1Int) or high Eomesodermin (Eomes) and high PD-1 expression (EomesHiPD-1Hi). Whereas both subsets are required for control of chronic infection, the PD-1Int subset functions as a progenitor pool giving rise to terminally differentiated PD-1Hi cells (Paley et al., 2012). Targeted blockade of PD-1 is effective in improving T cell function and reducing viral replication, mainly by reinvigorating the PD-1Int Tex cell subset (Blackburn et al., 2008). Inhibitory receptor blockade targeting immune checkpoints is also transforming human cancer therapy with impressive responses in multiple types of malignancies presumably due to reversal of T cell exhaustion (Wolchok and Chan, 2014).
Continued TCR signaling due to persisting antigen is thought to be a key driver of T cell exhaustion. One function of inhibitory receptors such as PD-1 is to attenuate signaling downstream of the TCR. The intracellular tail of PD-1 contains an immunotyrosine inhibitory motif (ITIM) and an immunotyrosine switch motif (ITSM), that can recruit phosphatases such as SHP-2, allowing dephosphorylation of key signal transducers (Chemnitz et al., 2004; Okazaki et al., 2001). Engagement of PD-1 by its ligand PD-L1 results in the formation of microclusters with the TCR (Yokosuka et al., 2012) and PD-1 inhibits proximal signaling molecules after TCR engagement (Sheppard et al., 2004). As a result, PD-1 can function as a rheostat to tune TCR signaling in tissues during infections (Honda et al., 2014; Okazaki et al., 2013). In addition, PD-1 ligation attenuates PI3K and Akt signaling inhibiting cell cycle at the G1 stage (Patsoukis et al., 2012). In vitro, engaging PD-1 or CTLA-4 on activated T cells reduces Akt and mTOR signaling, and suppresses glycolysis (Parry et al., 2005; Patsoukis et al., 2015). In vivo, a feedback loop between PD-1 and the transcription factor Foxo1 exists that desensitizes antigen-induced signaling via the Akt and mTOR pathway fostering survival of more terminally differentiated PD-1HiEomesHi Tex cells (Staron et al., 2014). These data suggest that the PD-1Int and PD-1Hi Tex cell subsets may have different metabolic requirements. Thus, previous data suggest a role for inhibitory receptor signaling in metabolism of Tex cells. However, the mechanisms responsible for the regulation of the metabolic lifestyle of CD8+ Tex cells, when these metabolic changes occur and the role of inhibitory receptors in this process in vivo remain to be fully defined.
Here, we examined these questions and demonstrate that functional metabolic derangement occurred early in the development of CD8+ T cell exhaustion. This early metabolic derangement included suppressed respiration, reduced glucose uptake, glycolysis and dysregulated mitochondrial energetics. Elevated mTOR activity and PD-1 signaling early during the development of T cell exhaustion contributed to these metabolic alterations. PD-1 repressed expression of the key metabolic regulator PGC1-α in CD8+ T cells early during chronic infection and retroviral (RV) expression of PGC1-α corrected some metabolic alterations in developing Tex cells and improved effector function. These data highlight a key metabolic control event that occurs early during the development of exhaustion. Metabolic dysregulation was maintained into established chronic infection and was controlled, at least in part, by PD-1 because blockade of PD-1:PD-L1 interactions resulted in metabolic reprogramming of PD-1Int Tex cells, but not the PD-1Hi subset. Thus, targeting Tex cell metabolism may constitute an important aspect of immunomodulatory therapies and could complement effects of blocking PD-1 or other checkpoints.
Results
Transcriptional analysis indicates early regulation of metabolism in Tex cells
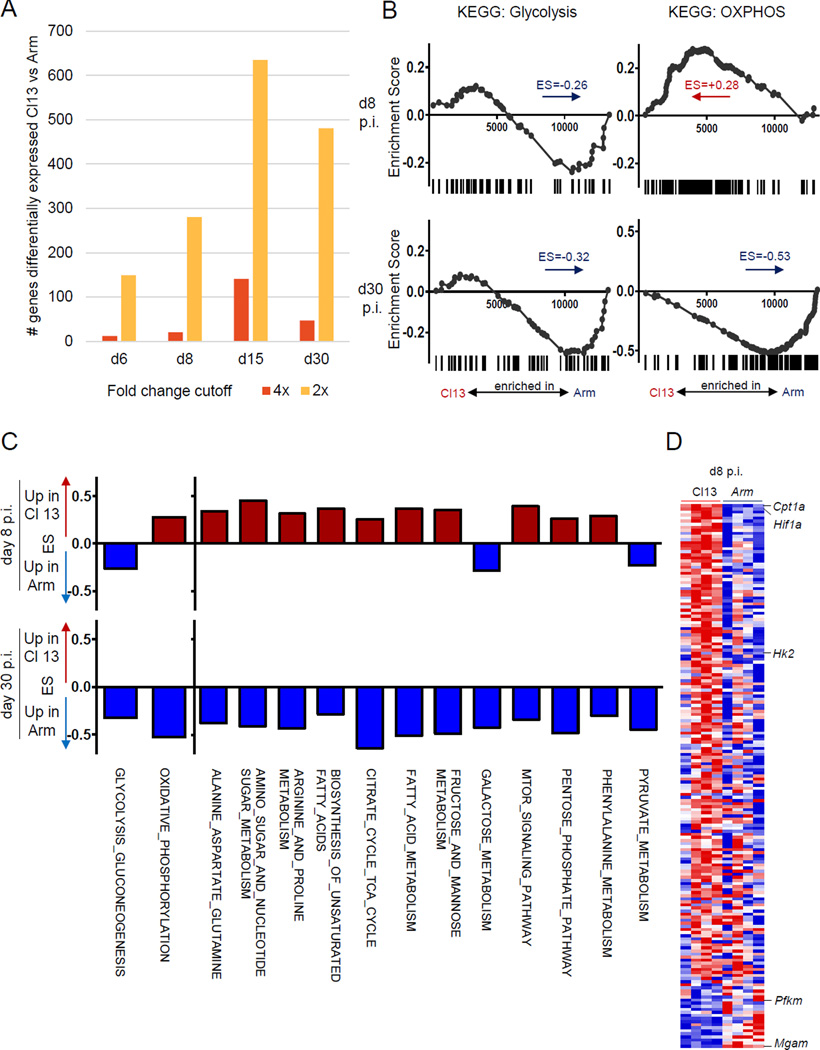
Transcriptional profiling revealed relatively few differences in LCMV-specific CD8+ T cells responding to acutely cleared Arm versus the chronic clone 13 strains of LCMV in early infection (d6 and d8 post infection (p.i.)) compared to later time points (d30+) (Fig. 1A) (Crawford et al., 2014; Doering et al., 2012). To examine whether metabolic pathways were regulated differently in CD8+ T cells prior to development of memory versus exhaustion, we first compared the gene-expression profiles of LCMV-specific CD8+ T cells from LCMV clone 13-infected mice to those from Arm-infected mice at the peak of the effector response and after development of Tmem and Tex cells (d8 versus d30 p.i.) by gene-set enrichment analysis (GSEA). Genes involved in glycolysis were enriched in CD8+ T cells from Arm infection (Fig. 1B). In contrast, genes involved in OXPHOS were enriched early during clone 13 infection (early Tex cells) (Fig. 1B). A broader analysis showed enrichment for other metabolic pathways in early clone 13 infection, including citrate cycle, fatty acid (FA) metabolism and amino acid metabolism (Fig. 1C). Leading edge analysis identified Cpt1a, the gene encoding a key enzyme fueling FA-driven mitochondrial OXPHOS, as highly enriched in clone 13 infection (Fig. 1D). In contrast, few metabolic pathways were enriched in LCMV-specific Teff from Arm infection. This pattern reversed at d30 p.i., when CD8+ T cell exhaustion was fully established, with broad upregulation of metabolic pathways in Tmem compared to Tex cells. These results pointed towards altered use of metabolic pathways in LCMV-specific T cells in evolving chronic infection, and suggested a role for dysregulated metabolism in establishing T cell exhaustion.
Figure 1. Altered regulation of metabolic genes early and late during clone 13 infection.
Microarray profiling of LCMV GP33-specific CD8+ T cells in LCMV Arm or clone 13 infection (GSE41867) analyzed for (A) number of differentially expressed genes (2-fold or 4-fold cutoff). (B) GSEA analysis for KEGG glycolysis and gluconeogenesis pathways and OXPHOS pathways at d8 or d30 p.i. Negative enrichment score (ES) indicates enrichment in Arm infection (C) GSEA of the indicated KEGG metabolic pathways at d8 and d30 p.i. (D) Heatmap of leading edge genes driving gene set enrichment of KEGG metabolic pathways at d8p.i. See also Fig. S1.
Glycolysis and oxidative phosphorylation are suppressed in early Tex cells
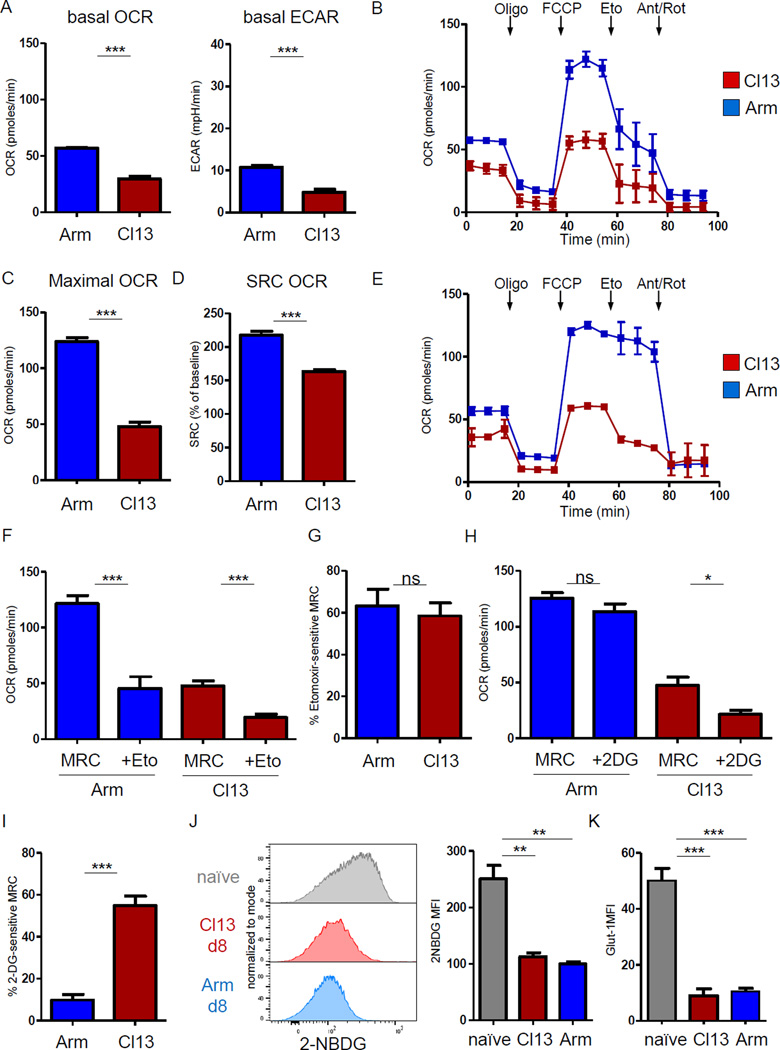
We next sought to test whether the transcriptional changes were linked functionally to the metabolism of LCMV-specific CD8+ T cells. At d8 p.i., Teff cells in Arm infection had a higher extracellular acidification rate (ECAR) than CD8+ T cells from clone 13 infection (Fig. 2A), in agreement with the enrichment for glycolytic pathway genes. The basal oxygen consumption rate (OCR) was also significantly reduced compared to Arm infection (Fig. 2A). Injection of the mitochondrial decoupler FCCP allowed evaluation of the maximal respiratory capacity (MRC) and spare respiratory capacity (SRC, i.e. the difference between MRC and baseline OCR; Fig. 2B, C). CD8+ T cells at d8 of clone 13 infection had reduced MRC and SRC (Fig. 2C, D). Thus, in addition to reduced respiration at baseline, early Tex cells displayed lower maximal respiration and SRC compared to Teff cells in Arm infection (Fig. 2D). These results suggested suppressed glycolysis and OXPHOS at the effector stage of clone 13 infection despite the transcriptional upregulation of several metabolic pathways.
Figure 2. Suppression of CD8+ T cell mitochondrial respiration and glycolysis early in chronic infection.
Metabolic flux profiling was performed on purified CD8+ T cells from mice d8 p.i. with LCMV Arm (blue) or clone 13 (red). (A) OCR and ECAR at baseline. (B) OCR obtained during mitochondrial stress test, performed by injection of Oligomycin (Oligo), mitochondrial decoupler FCCP, Cpt1a inhibitor Etomoxir (Eto) to interrogate FAO or ETC inhibitors Antimycin A/Rotenone (Ant/Rot). (C and D) MRC and SRC of CD8+ T cells from d8 p.i. was determined after FCCP injection. (E) Compared to (2B), 2-DG injection was used to assess glucose metabolism. (F) The effect of CPT1a inhibition on SRC and (G) the contribution of FA metabolism (i.e. Eto-sensitive SRC) is plotted for CD8+ T cells from Arm versus clone 13. (H) The effect of 2-DG injection on mitochondrial respiration and (I) the contribution of glucose (i.e. the 2-DG-sensitive SRC). (J) Glucose uptake measured by incorporation of glucose analogue 2-NBDG and Glut-1 expression (K) of naïve CD8+ T cells and LCMV-specific P14 T cells isolated on d8 p.i. of Arm versus clone 13 infection. Data are representative of 3 independent experiments (3–5 mice / experimental group). * indicates p<0.05, ** p<0.01, *** p<0.001 by unpaired Student’s T-test. Error bars are mean ± SEM. See also Fig. S2 and S3.
We next examined the relative contribution of FA versus glucose to cellular respiration. After mitochondrial decoupling, we blocked FA metabolism using the Cpt1a inhibitor etomoxir, as previously described for Teff and Tmem (O'Sullivan et al., 2014; van der Windt et al., 2012). Addition of etomoxir reduced respiration by ~60% in CD8+ T cells from both Arm and clone 13 infection (Fig. 2F,G), suggesting that at d8 p.i., both Teff and early Tex cells could use FA as an energy source, although effects of etomoxir on other targets cannot be fully excluded. In contrast, inhibition of glycolysis using 2-DG resulted in a modest reduction in OCR in Arm, but a ~50% decrease in respiration of CD8+ T cells from clone 13 infection (Fig. 2E, H, I). 2-DG inhibits glycolysis, by blocking early glycolytic enzymes (hexokinase and phosphoglucose-isomerase) and competing for glucose transporters. Thus, these results indicated that glucose was an important energy source fueling mitochondrial respiration that was rapidly depleted in early Tex cells during clone 13, but not Arm infection at d8 p.i. In agreement with this notion, a recently described glucose deprivation signature (Ho et al., 2015) was enriched in CD8+ T cells from clone 13 versus Arm infection at d8 p.i. (Fig. S2A). Glucose uptake by Teff cells in Arm peaks at d4 p.i. and declines by d8 p.i. around the time of antigen elimination (Fig. S2B). The ability to import glucose via glucose transporter-1 (Glut-1) is a limiting step for glucose use by T cells. Glucose uptake was low and linked to reduced Glut-1 on early Tex and Teff cells compared to naïve T cells (Fig. 2J, K). One interpretation of these observations was that while Teff cells in Arm infection have decreased their dependency on glucose mediated energy production associated with the clearance of infection and reduced TCR stimulation at d8 p.i., CD8+ T cells in clone 13 infection remained highly dependent on glucose because of persisting antigen and TCR signaling, but were unable to maintain effector metabolism due to glucose restriction.
We hypothesized that Glut-1 expression was limiting in early Tex cells. To test this idea, we transduced virus-specific TCR transgenic P14 cells specific for the LCMV GP33–41 epitope with a retrovirus (RV) expressing Slc2a1 encoding Glut-1 (Fig. S3). As expected, increased glucose uptake was observed in vitro upon Glut-1 overexpression (OE) (Fig. S3A, B). In vivo, while no major benefit was observed during Arm infection, Glut-1 OE during clone 13 infection resulted in a substantial numerical increase in Slc2a1-RV transduced P14 cells compared to control transduced cells (Fig. S3D, E). Glut-1 OE also resulted in a trend towards increased glucose uptake at d8 p.i. (p=0.053; Fig. S3E). These data suggested that RV-mediated Glut-1 expression could enhance the CD8+ T cell response during early clone 13 infection. However, the modest increase in glucose uptake at d8 p.i. suggested that other steps in the regulation of Glut-1 activity may also be important and that the benefit observed was either due to subtle increases in glucose uptake in vivo, or earlier events when Glut-1 OE may have a quantitatively larger impact on glucose uptake.
Metabolic dysregulation of early Tex cells involves mitochondrial depolarization
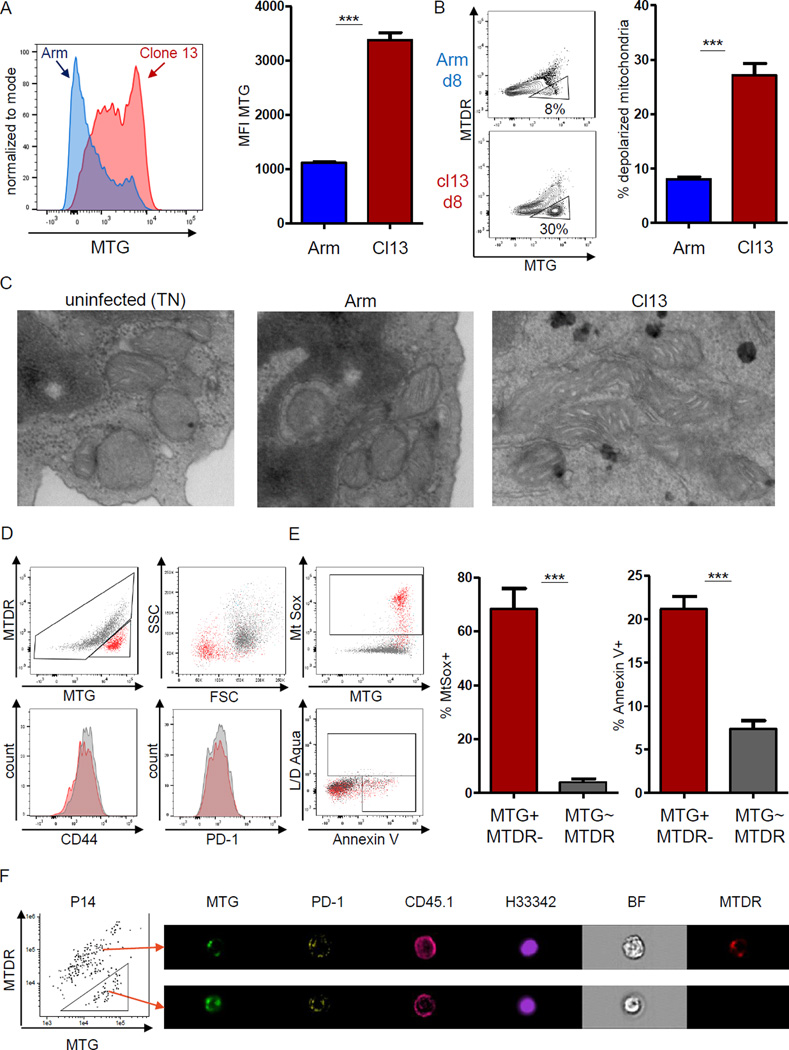
Mitochondrial respiration was significantly reduced in CD8+ T cells at d8 p.i. with clone 13 (Fig. 2). Glucose uptake in Arm infection correlated with mitochondrial mass in Teff cells (Fig. S2C). We thus asked if the reduced glucose uptake and mitochondrial respiration observed in early Tex cells were linked to mitochondrial mass. Early Tex cells had significantly higher mitochondrial mass as indicated by staining with Mitotracker Green (MTG) compared to Teff cells (Fig. 3A). These observations are in agreement with the transcriptional up-regulation of mitochondrial genes (Fig. 1), but appeared paradoxical when compared to the low mitochondrial energetics observed. Because mitochondrial membrane potential (Δψm) is required for production of ATP during OXPHOS, a low Δψm might contribute to insufficient mitochondrial function in CD8+ T cells in clone 13 infection. Thus, we combined a potential-dependent and a potential-independent mitochondrial dye to identify CD8+ T cells with depolarized mitochondria. Indeed, at d8 p.i., a significantly higher fraction of early Tex cells had depolarized mitochondria compared to Teff cells (Fig. 3B). Electron transmission microscopy of naïve CD8+ T cells, Teff and early Tex cells identified differences in the morphology of mitochondria in early Tex cells including fused mitochondria with cristae elongation and prominent electron-lucent mitochondrial matrix (Fig. 3C). Virus-specific CD8+ T cells with depolarized mitochondria expressed normal profiles of differentiation and exhaustion markers, such as CD44 and PD-1, but displayed reduced forward scatter suggesting smaller cell size (Fig. 3D). T cells with depolarized mitochondria exhibited increased mitochondrial reactive oxygen species (ROS) (Fig. 3E). Accumulation of ROS may inflict damage to cellular proteins and DNA, potentially leading to apoptosis (Belikov et al., 2015). To test this, we used a combination of Annexin V and LD Aqua staining that allows characterization of membrane perturbation, early apoptosis and overt cell death. We used the scatter properties identified in Fig. 3D to identify P14 cells with smaller cell size and assessed healthy (AnnexinV-LD−), early apoptotic (AnnexinV+LD−) and late apoptotic/necrotic (LD+) phenotypes (Fig. S4). We observed a significant frequency of small P14 cells undergoing cell death, however, ~33% of the cells analyzed showed no indication of apoptosis (Fig. S4). In contrast, when we analyzed the complete pool of LD- P14 cells, only 10–15% of cells with depolarized mitochondria stained AnnexinV+. Imagestream analysis revealed intact nuclear staining of these depolarized cells (Fig. 3F). These data suggested that depolarization of mitochondrial membrane potential results in increased ROS production, which may render a fraction of early Tex cells sensitive to death. Taken together, these results indicated that early during clone 13 infection, virus-specific CD8+ T cells displayed defects in regulation of mitochondrial metabolism including increased mitochondrial mass associated with accumulation of depolarized mitochondria and ROS production.
Figure 3. Depolarized mitochondria in virus-specific CD8+ T cells in early clone 13 infection.
Mitochondrial profiling of virus-specific CD8+ T cells at d8 after Arm (blue) or clone 13 (red) infection. Live, singlet CD8+ CD45.1+ P14 cells were analyzed (A) Mitotracker Green (MTG) staining of P14 cells at d8 p.i. (B) Virus-specific CD8+ T cells with depolarized mitochondria identified as positive for MTG and negative for Mitotracker DeepRed (MTDR) (triangle gate). (C) Electron microscopy of sorted P14 cells from uninfected, Arm or clone 13 infected mice (d8 p.i.). Cells with representative mitochondrial morphology are shown. (D) Representative plots of virus-specific CD8+ T cells at d8 p.i. of clone 13 with depolarized (red) versus regularly polarized mitochondria (grey). (E) ROS in P14 CD8+ T cells measured using MtSox. Cells were costained for Annexin V and amine-reactive L/D reagent. Gated on live singlet CD8+ CD45.1+ P14 cells, gates for MtSox and ROS are indicated on the left. (F) Amnis Imagestream analysis (60× magnification). Cells were gated on congenic CD45.1+ P14 and MTG versus MTDR staining was used to identify cells with depolarized mitochondria. Arrows indicate a representative cell staining with regular mitochondrial polarization (upper) versus depolarized mitochondria (lower). Nuclear integrity assessed by Hoechst 33342. Data are representative of 3 experiments (Imagestream: one experiment) with 3–5 mice in each experimental group. *** indicates p<0.001 by unpaired Student’s T-test. Error bars are mean ± SEM. See also Fig. S4
mTOR signaling contributes to mitochondrial depolarization in early Tex cells
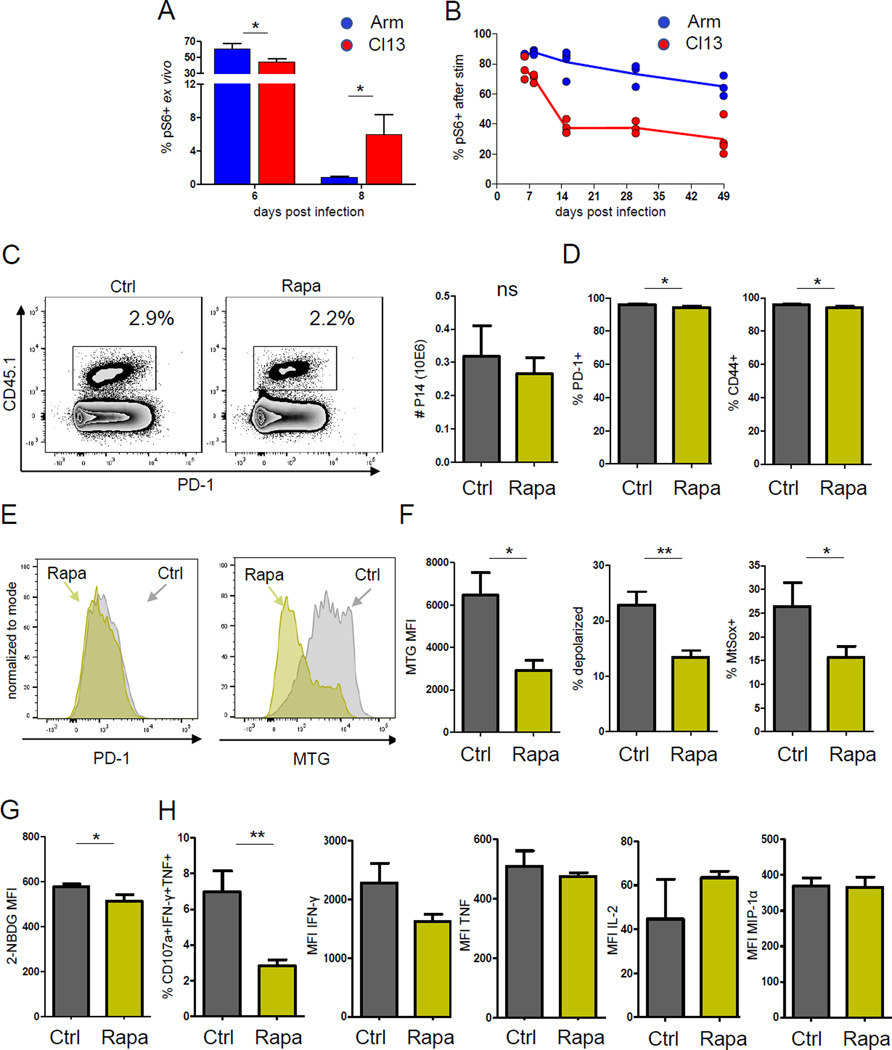
In a scenario of continuous metabolic demand such as what developing Tex cells experience, deprivation of glucose supply (e.g., involving low Glut-1) may result in reduced mitochondrial tricarboxylic acid (TCA) cycle substrate influx and compromised membrane potential. Thus, we hypothesized that mTOR signaling persisting past the normal effector stage in early Tex cells might compromise metabolic programming. We, therefore, interrogated the ability to phosphorylate ribosomal protein S6 downstream of mTOR upon antigen stimulation. Ex vivo phosphorylation of S6 in LCMV-specific CD8+ T cells was high at d6 p.i. with Arm or clone 13 (Fig. 4A). In Arm infection, ex vivo pS6 returned to baseline by d8 p.i. consistent with the control of infection. In contrast, during clone 13 infection, residual elevated ex vivo pS6 was observed at d8 p.i., likely reflecting ongoing in vivo signaling. We next tested the potential to induce new signaling via mTOR after peptide restimulation in vitro. mTOR activity was reduced during clone 13 compared to Arm infection especially after d8 p.i. (Fig 4A), suggesting lack of optimal induction of mTOR signaling upon restimulation. These results are in agreement with the recently described impairment of mTOR signaling in Tex cells in chronic infection (Staron et al., 2014). However, mTOR pathway genes were enriched at d8 p.i. of clone 13 infection (Fig. 1) and the direct ex vivo difference in pS6 at d8 p.i. suggested the possibility that ongoing residual, yet suboptimal, mTOR activity could play a role in the metabolic phenotype observed in early Tex cells. To test this idea we treated mice with 300 µg/kg of mTOR inhibitor rapamycin from d5–7 p.i. in clone 13 infection. This treatment mildly reduced the number of virus-specific T cells (Fig. 4B, C) as expected based on the role of mTOR in effector CD8+ T cell expansion (Araki et al., 2009; Pearce et al., 2009). Despite the impact on T cell expansion, only mild changes in typical T cell differentiation and exhaustion markers were observed in clone 13 infection (Fig. 4D). Similar treatment promoted development of memory precursors and reduced short-lived Teff in Arm infection (data not shown) as described (Araki et al., 2009). Short-term treatment with rapamycin during early clone 13 infection significantly reduced mitochondrial mass and depolarization (Fig. 4E, F), suggesting that persisting mTOR signaling was involved in the development of paradoxically large dysfunctional mitochondria in early Tex cells. These changes were also associated with a mild reduction in glucose uptake (Fig. 4G) and decreased polyfunctionality of virus-specific T cells (Fig. 4F). While limiting mTOR activity by rapamycin improved the mitochondrial fitness of early Tex cells, this metabolic benefit may come at the expense of reducing remaining effector function.
Figure 4. MTOR inhibition improves the mitochondrial phenotype during early clone 13 infection.
Role of mTOR in developing Tex cells. (A) Time course of ex vivo pS6 in P14 CD8+ T cells during LCMV Arm or clone 13 infection. (B) Time course of pS6 following 20min GP33 stimulation in vitro. Gated on live singlet CD8+ P14. (C) Clone 13 infected mice were treated daily from d5–8 p.i with 300µg/kg rapamycin (Rapa) or PBS (control) and analyzed on d8 p.i. Frequencies of CD45.1+ P14 cells are shown. Gated on live singlet CD8+ T cells (D) P14 CD8+ T cells analyzed for expression of PD-1 and CD44 (E) Representative histograms for PD-1 or MTG of P14 CD8+ T cells from control or Rapa treated mice. (F) Mitochondrial mass (MTG), depolarization (MTG+MTDR−) and ROS production (MtSox+) following Rapa treatment. (G) Glucose uptake by 2-NBDG fluorescence. (H) Functional profile of virus-specific T cells after GP33 peptide stimulation. Degranulating (CD107a+) P14 cells producing IFN-γ, TNF, IL-2, MIP1α, and frequency of polyfunctional cells is shown. Data are gated on live singlet CD8+CD45.1+ P14 cells. Data indicate n=3–5 mice / experimental group and are representative of 2 independent experiments. * indicates p<0.05, ** p<0.01 by unpaired Student’s T-test. Error bars are mean ± SEM
Mitochondrial and mTOR abnormalities persist in Tex cells in established chronic infection
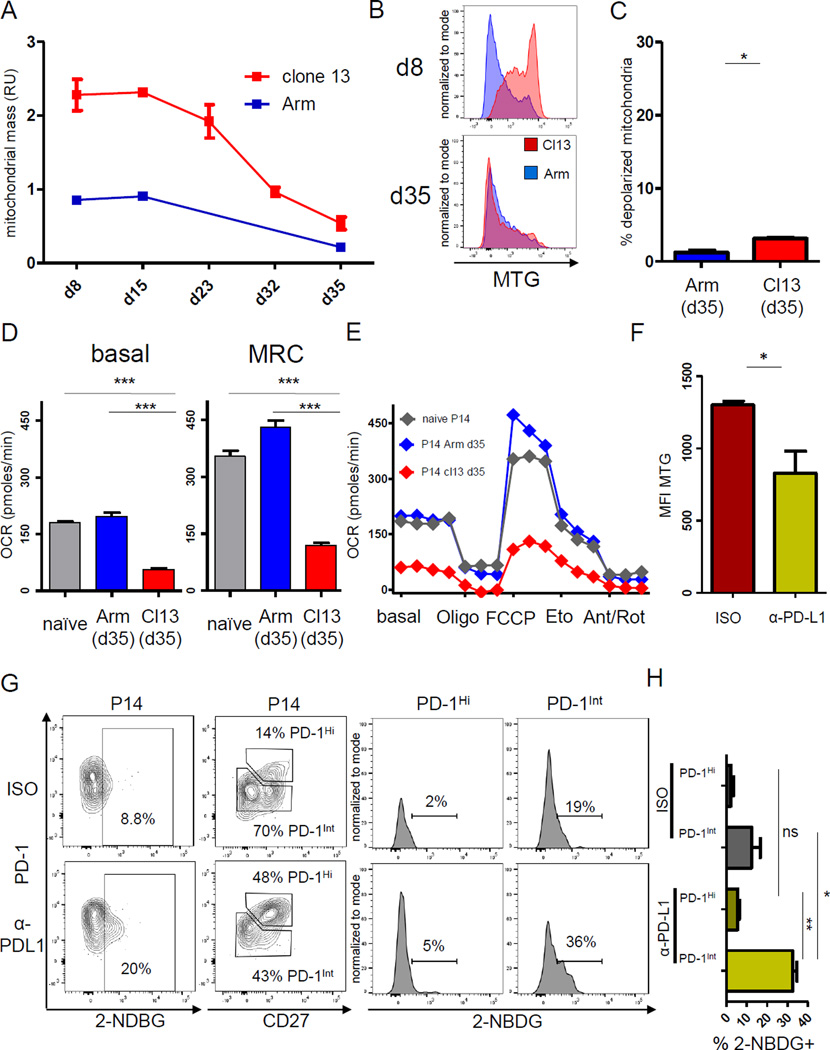
During the transition to full exhaustion after the effector phase in established chronic infection, virus-specific CD8+ T cells undergo major cellular changes, including downregulation of many metabolic genes (Fig. 1) (Wherry et al., 2007). We therefore investigated the connections between the metabolic phenotype in early versus established chronic infection. We observed a gradual reduction in mitochondrial mass over time, accompanied by a reduction in the frequency of depolarized mitochondria (Fig. 5A–C). However, compared to Tmem, Tex cells maintained a slightly greater mitochondrial mass and significant increase in the proportion of depolarized mitochondria. We also investigated the basal respiration and MRC of Tex cells in established chronic infection and observed reduced respiration compared to naïve T cells and Tmem (Fig. 5D, E), suggesting that the functional metabolic suppression observed early is maintained throughout chronic infection.
Figure 5. Mitochondrial mass, mTOR signaling and low mitochondrial bioenergetics during LCMV clone 13 infection.
Mitochondrial phenotype, respiratory function and mTOR signaling of P14 cells analyzed after d8 p.i. of LCMV Arm (blue) or clone 13 (Cl13) (red) infection. (A) Normalized mitochondrial mass indicated by relative units (RU) of MTG fluorescence, and (B) representative histograms of MTG on d8 and d35 p.i. are shown gated on live singlet P14 cells. (C) The fraction of depolarized mitochondria for P14 CD8+ T cells on d35 p.i. on similar scale as in Fig. 2. (D and E) LCMV-specific CD8+ T cells were sorted after d35 of Arm and clone 13 infection and mitochondrial stress test performed by injection of Oligo, FCCP, Eto, or Ant/Rot as in Fig 2. (F) C57Bl/6 mice were depleted of CD4+ T cells, P14 cells adoptively transferred and infected with clone 13. Mice were treated with PD-L1 blocking or isotype control antibody from d22–35 p.i. Mitochondrial mass was determined in P14 CD8+ T cells from control or treated mice by MTG. (G) Representative data for glucose uptake by P14 CD8+ T cells (left column) from isotype (upper) or anti-PDL1 (lower) treated mice by 2-NBDG fluorescence. Glucose uptake in PD-1Int and PD-1Hi subsets (gated by PD-1 versus CD27) of virus-specific CD8+ T cells (right panel). (H) Glucose uptake for the PD-1Int and PD-1Hi subsets in 5G is quantified. Kinetic data in 5A summarizes 5 experiments (3–5 mice / experimental group). Data in 5B is representative of 3 experiments (n=3 per group). Data in 5D–E represents pooled data from one experiment. Data in 5G-H represents an experiment with n=10 mice.* indicates p<0.05, *** p<0.001 by unpaired Student’s T-test. Error bars are mean ± SEM. See also Fig. S5
PD1hi and PD1int subsets of Tex cells differ metabolically
The pool of Tex cells contains two subsets defined by expression of PD-1 and T-box transcription factors; the PD-1Int T-betHi progenitor subset and the more terminally differentiated PD-1Hi EomesHi subset (Blackburn et al., 2008; Paley et al., 2012). We next tested whether the metabolic changes observed in early chronic infection were also found in PD-1Hi and PD-1Int Tex cell subsets in established chronic infection. Higher pS6 in the PD-1Hi subset suggested more mTOR activity (Fig. S5A, B). Conversely, we observed higher glucose uptake and a trend to lower mitochondrial mass in PD-1Int cells as well as an enrichment of pathways involved in glycolytic and OXPHOS metabolism compared to PD-1Hi cells (Fig. S5C–E). These observations demonstrated metabolic differences between Tex cell subsets defined by PD-1 expression, but may also point towards a possible role for PD-1 in the metabolic dysfunction of Tex cells.
PD-1 pathway blockade reverses metabolic depression in PD-1Int Tex cells
PD-1 blockade in tumor models is associated with transcriptional changes in metabolic genes (Gubin et al., 2014). We next tested whether the metabolic alterations in Tex cells might be reversed by transient PD-1 pathway blockade. To address this question, we performed PD-L1 blockade in established chronic infection. PD-L1 blockade resulted in an expansion of virus-specific T cells and lowered viral load (data not shown), as previously described (Barber et al., 2006). After PD-L1 blockade, reinvigorated Tex cells displayed reduced mitochondrial mass and increased glucose uptake (Fig 5F, G), suggesting increased metabolic fitness. The increased glucose uptake was largely confined to the PD-1Int Tex cell subset (Fig. 5G, H). These results suggested that targeting PD-1 partially relieves metabolic restrictions, allowing the PD-1Int progenitor population to meet the bioenergetic demands required for proliferation and reinvigoration.
PD-1 controls early Tex cell metabolism
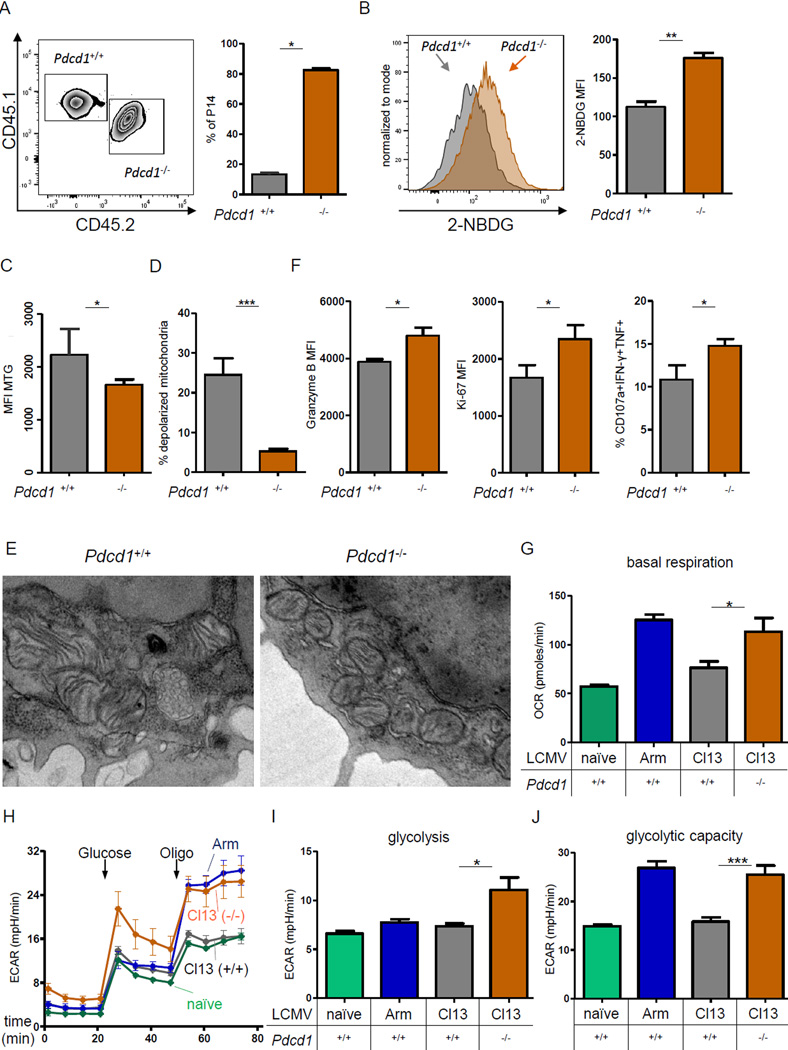
The origins of the metabolic defects that were already present in early Tex cells described above remained unclear. We therefore tested whether PD-1 had a causal role in establishing the early metabolic dysregulation in vivo in developing Tex cells. First, we employed a co-transfer model using a small number of adoptively transferred P14 cells that allows direct comparison of WT and PD-1 deficient cells using Pdcd1+/+ and Pdcd1−/− P14 cells in the same host (Odorizzi et al., 2015). This approach controls for alterations in viral load, inflammation or immunopathology that might occur in the absence of PD-1 (Frebel et al., 2012; Odorizzi et al., 2015). Congenically disparate Pdcd1+/+ and Pdcd1−/− P14 cells were adoptively transferred into WT recipient mice followed by clone 13 infection (Fig. 6A). In this model, Pdcd1−/− P14 cells preferentially expanded at d8 p.i. consistent with previous studies (Frebel et al., 2012; Odorizzi et al., 2015). The numerical advantage of Pdcd1−/− P14 cells was linked to a significant increase in glucose uptake (Fig. 6B) as well as reduced mitochondrial mass and fewer cells with depolarized mitochondria (Fig. 6C, D). Electron transmission microscopy of Pdcd1+/+ and Pdcd1−/− P14 cells highlighted a reduction in fused mitochondria, cristae elongation and translucent mitochondrial matrix in the absence of PD-1 in early Tex cells (Fig. 6E). Of note, Pdcd1−/− P14 cells displayed better polyfunctionality (Fig. 6F), suggesting that improved early metabolic fitness may prevent or delay development of exhaustion. These benefits of PD-1 genetic deficiency were only observed in early infection but not in established chronic infection (Fig. S5F), consistent with the temporary benefit of PD-1 genetic deficiency on other aspects of T cell exhaustion (Odorizzi et al., 2015). These results indicated that PD-1 acts as an upstream regulator of the metabolic phenotype in early Tex cells. To further interrogate the effects of genetic PD-1 ablation on metabolic function, we isolated purified CD8+ T cells from Pdcd1+/+ and Pdcd1−/− mice at d8 p.i. and performed metabolic flux analysis. Pdcd1−/− CD8+ T cells had significantly higher basal respiration at d8 p.i. with clone 13 (Fig. 6G). We interrogated the glycolytic metabolism further by addition of glucose to previously glucose-starved cells, followed by blockade of the mitochondrial ATP synthase with oligomycin, enforcing maximal cytoplasmic glycolysis (i.e. glycolytic capacity) (Fig. 6H). In early clone 13 infection, glycolysis and glycolytic capacity of Pdcd1−/− cells was substantially enhanced, with Pdcd1−/− cells resembling normal Teff cells isolated from Arm infection, while glycolytic capacity of Pdcd1+/+ cells from clone 13 infection was strongly impaired and not different from naïve non-glycolytic controls (Fig. 6I, J). Thus, these results indicated that PD-1 regulates metabolism, glycolysis and mitochondrial function of virus-specific CD8+ T cells early during clone 13 infection prior to development of full exhaustion.
Figure 6. PD-1 controls metabolic dysregulation of Tex cells in early and established chronic LCMV infection.
(A) Congenically distinct Pdcd1+/+ (CD45.1+CD45.2−) and Pdcd1−/− (CD45.1+CD45.2+) P14 CD8+ T cells were cotransferred into naïve (CD45.1−CD45.2+) mice, followed by infection with clone 13 (cl13). The frequency of Pdcd1+/+ versus Pdcd1−/− P14 cells at d8 p.i. is shown. (B–F) Cotransferred Pdcd1+/+ and Pdcd1−/− P14 CD8+ T cells analyzed for glucose uptake (B), mitochondrial mass (C), mitochondrial depolarization (D), and functional parameters (F) at d8 p.i. with cl13. (E) Representative electron microscopy on sorted cotransferred Pdcd1+/+ and Pdcd1−/− P14 cells. (G–J) Metabolic flux was performed on negatively selected, magnetic bead purified CD8+ T cells from naïve uninfected, Arm or cl13 infected WT or Pdcd1−/− mice at d8 p.i. (G) Basal respiration. (H) Glycolytic stress test measuring ECAR performed by injecting glucose to glucosestarved cells, followed by Oligo. (I) Basal glycolysis was calculated by the increase in ECAR post glucose injection for Pdcd1−/− clone 13 infection (J) Glycolytic capacity was determined after Oligo injection * indicates p<0.05, ** p<0.01, *** p<0.001 by unpaired (G–J) or paired (6A–F) Student’s T-test. Error bars are mean ± SEM. See also Fig. S5
Improving early metabolic fitness counteracts developing exhaustion
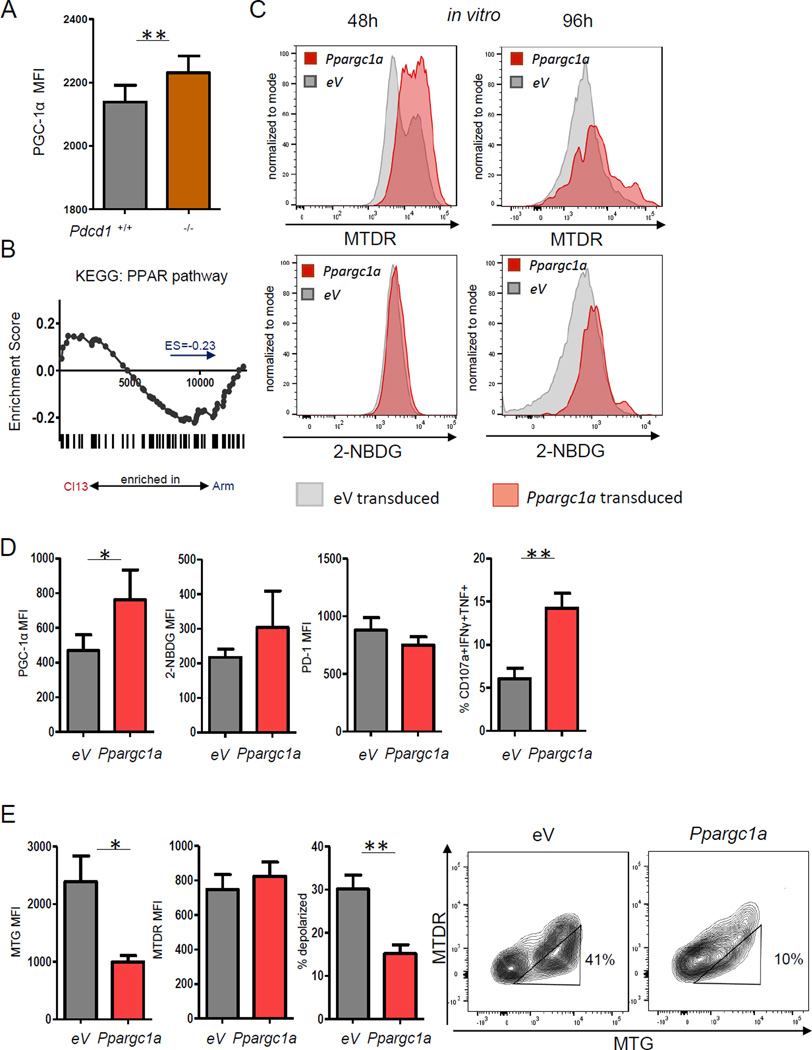
To further test the hypothesis that PD-1 regulation of metabolism contributes to exhaustion, we analyzed the expression of peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α) (Fig. 7A). PGC-1α is a key transcriptional regulator of genes controlling energy metabolism and mitochondrial biogenesis (Austin and St-Pierre, 2012). PGC-1α expression was reduced in Pdcd1+/+ compared to Pdcd1−/− Tex cells, suggesting negative regulation of PGC-1α by PD-1 as a mechanism of metabolic modulation. PGC-1α interacts with PPARγ and coordinates the expression of genes involved in mitochondrial function. Consistent with the increased respiration observed above (Fig. 2), PPAR target genes tended to be enriched in Teff from Arm infection compared to early Tex cells (Fig. 7B). We therefore tested whether increasing PGC-1α expression could improve the metabolic fitness in chronic infection. OE of PGC-1α by RV increased potential-dependent mitochondrial staining and glucose uptake in vitro (Fig. 7C). In vivo during clone 13 infection, Ppargc1a-transduced P14 cells had higher PGC-1α expression compared to control transduced early Tex cells (Fig. 7D, E). Of note, this overexpression significantly increased polyfunctionality (Fig. 7E). We also observed trends towards higher glucose uptake and lower PD-1 expression in early Tex cells after PGC-1α OE (Fig. 7E). Moreover, PGC-1α OE reversed the mitochondrial phenotype of early Tex cells and substantially reduced mitochondrial mass and the frequency of cells with depolarized mitochondria (Fig. 7F). Thus, PD-1 repressed PGC-1α and enhancing PGC-1α expression reversed metabolic perturbations associated with developing Tex cells. These data also suggested a causal link between the metabolic alterations and the development of T cell exhaustion.
Figure 7. PGC1α overexpression improves metabolism of Tex cells and restores function.
Expression of PGC1α was analyzed in developing Tex cells. (A) Cotransferred Pdcd1+/+ and Pdcd1−/− P14 CD8+ T cells were analyzed for PGC1α expression at d8 p.i. with clone 13. (B) GSEA of genes in the KEGG PPAR pathway at d8 p.i. with Arm or clone 13. (C) WT P14 cells were transduced with Ppargc1a RV and potential-dependent mitochondrial MTDR staining and glucose uptake analyzed at 48h and 96h post T cell activation. (D) RV-transduced P14 cells were transferred into mice at d1 of clone 13 infection and transduced P14 cells analyzed at d8 for PGC1α expression, glucose uptake, PD-1 expression and polyfunctionality. (F) Mitochondrial polarization analysis was performed at d8 p.i. with clone 13. Data indicate 3 independent experiments (n=3–5 mice/ group / experiment) * indicates p<0.05, ** p<0.01, by paired (A) or unpaired (D, E) Student’s T-test. Error bars are mean ± SEM.
Discussion
Changes in metabolic gene expression of Tex cells have been observed in previous microarray studies (Wherry et al., 2007), however it remained unclear when these changes occur and how they impact cellular metabolism. In this work, we identified functional alterations in metabolism that occurred early in chronic infection before major manifestations of exhaustion were established. For example, high mTOR activity, likely driven by persisting antigen signaling at the end of the first week of clone 13 infection, was connected to a cellular need for increased glycolysis and OXPHOS. However, early Tex cells could not meet this bioenergetic demand facing limited glucose uptake and use. This relative glucose deprivation occurred in the presence of increased anabolic signals and was controlled, at least in part, by the PD-1 pathway. This dysregulation skewed early Tex cells to an altered metabolic phenotype prone to mitochondrial depolarization and high ROS production. These defects could be partially reversed by improving glucose uptake, removing PD-1 signals or enforcing expression of the metabolic regulator PGC1α. Together these observations implicate distorted metabolism as an upstream driver of the pathways to T cell exhaustion.
Teff cells generated during acute infections switch to anabolic metabolism dominated by aerobic glycolysis (Buck et al., 2015). In the current work, we identified suppression of glycolysis and OXPHOS early during clone 13 infection that occurred despite the transcriptional up-regulation of several metabolic pathway genes, including those involved in OXPHOS. T cell commitment to exhaustion is thought to occur progressively over time (Angelosanto et al., 2012; Brooks et al., 2006; Crawford et al., 2014). Early, but not established Tex cells, have developmental plasticity and could develop into functional Tmem cells if isolated during the first week, but not later in infection (Angelosanto et al., 2012). One interpretation of these data is that the drivers of exhaustion develop gradually as chronic infection progresses. In the current study, early Tex cells had impaired glycolysis and respiration and this early metabolic dysregulation preceded major transcriptional, phenotypic and functional changes associated with established exhaustion. Restriction of glucose availability in the presence of persisting anabolic signaling (i.e. mTOR) appeared to be a major feature of early Tex cells. Perhaps to compensate for these defects in glycolysis, Cpt1a, a key regulator of FA oxidation (FAO) was upregulated in early Tex cells, consistent with a connection between PD-1, Cpt1a and FAO observed in vitro (Patsoukis et al., 2015). Blockade of Cpt1a reduced mitochondrial respiration by ~50% in early Tex cells suggesting that early Tex cells have access to alternate energy pathways when glucose use is limiting. The use of FA may endow Tex cells with a survival mechanism when faced with glucose restriction, but may be insufficient to fuel anabolic metabolism normally associated with robust TCR signaling, leading to restrictions on proliferative expansion and effector function. These findings support the notion that regulation of metabolism is an early driver, rather than a consequence, of exhaustion.
Our observations also implicate PD-1 as a major control pathway of metabolic dysregulation during early and established CD8+ T cell exhaustion. In vitro studies with activated effector-like T cells showed suppression of glycolysis by PD-1 and CTLA-4 (Parry et al., 2005; Patsoukis et al., 2015). Despite these connections between PD-1 and glycolysis, our results suggest that inhibition of glucose uptake is incomplete. In metabolic flux experiments, glucose addition prompted an increase in glycolysis in glucose-deprived early Tex cells whereas inhibition of residual glucose uptake by 2-DG reduced metabolic activity. In vitro approaches using activated Teff-like cells have highlighted more complete suppression of glycolysis in response to PD-1 signaling (Patsoukis et al., 2015). Differences between in vitro studies and the current data may be due to differences between in vitro activated Teff-like cells and in vivo generated Tex cells that are transcriptionally and phenotypically distinct from Teff cells (Doering et al., 2012; Wherry et al., 2007). Finally, while our results point to a strong connection between PD-1, metabolism and induction of T cell exhaustion, it should be pointed out that T cell exhaustion can occur even in the absence of PD-1 (Odorizzi et al., 2015), suggesting redundancy in pathways that can drive exhaustion.
Overexpression of Glut-1 in early Tex cells improved the T cell response during early clone 13, but not Arm infection, consistent with limited glucose uptake in developing Tex cells where TCR signaling and metabolic “stress” on the cell persists. In vivo, the uptake of 2-NBDG peaks at ~d3–4 p.i. with LCMV Arm and then declines. This early peak in glucose uptake is consistent with other data on in vitro activated Teff-like cells (Chang et al., 2013; Maciver et al., 2008; Patsoukis et al., 2015), but the decline in glucose uptake indicates a dynamic regulation in vivo that is distinct from short-term in vitro culture. Thus, the benefit of Glut-1 overexpression in developing Tex cells likely reflects either a role for improved glucose uptake early in the T cell expansion phase or residual low-level glucose uptake at later times. The incomplete reversal of metabolic defects and exhaustion by Glut-1 overexpression suggested that the metabolic restrictions underlying early Tex cells likely extend beyond this early glucose checkpoint. Indeed, our studies revealed PGC-1α, a central transcriptional regulator of oxidative metabolism, as a key node perturbed in developing Tex cells. PGC-1α expression was enhanced in the absence of PD-1 in early Tex cells. Overexpressing PGC-1α corrected the dysregulated mitochondrial phenotype and improved effector function, reversing hallmarks of exhaustion. Thus, in addition to previously described pathways such as Foxo1 (Staron et al., 2014), our results now indicate that PD-1 can influence metabolic programs via PGC-1α.
Activation of mTOR downstream of TCR-triggered PI3K and Akt signaling is central for the anabolic metabolism of Teff cells, whereas, limiting mTOR activity improves Tmem development in acute infection (Araki et al., 2009). Attenuating mTOR signaling in Tex cells can arise at least partly due to PD-1 and chronic TCR stimulation (Staron et al., 2014). This chronic stimulation results in a feedback loop in which Foxo1 sustains PD-1 expression and survival of terminal Tex cells (Staron et al., 2014). These observations are in agreement with the decline of ex vivo S6 phosphorylation during clone 13 infection observed here, and reduced pS6 following antigen stimulation in vitro. Our data also now indicate that the accumulation of paradoxically large depolarized mitochondria was due to persisting elevated or residual mTOR activity driving an anabolic mitochondrial program under conditions of nutrient restriction. One possible explanation for our findings is that in the setting of aberrant anabolic stimulation during clone 13 infection (e.g., persisting infection), mTOR signaling, although attenuated, remains stimulatory for mitochondrial biogenesis. However, this signaling occurs when glycolysis is already suppressed. Such a scenario may then cause intracellular depletion of TCA substrates, tempering the mitochondrial proton gradient, leading to mitochondrial membrane depolarization. In this model, restriction of mTOR signaling in early Tex cells by rapamycin may help adapt the nutrient demand to availability, preventing proton gradient breakdown and ROS production. In support of this notion, improving mitochondrial OXPHOS by overexpression or gain-of-function of lymphocyte expansion molecule (lem) increased virus-specific T cell function and viral control early during clone 13 infection (Okoye et al., 2015). Despite the advantages of rapamycin treatment for the metabolic phenotype of early Tex cells, its use is unlikely to be of overall benefit in a therapeutic setting, given its immunosuppressive effects (Powell et al., 2012). Thus, while limiting mTOR activity can benefit the mitochondrial fitness of developing Tex cells, this metabolic benefit may come at the expense of the remaining effector function.
While the characteristics of Tex cells have been defined in many settings, the early signals driving this state of T cell differentiation have remained poorly understood. The current study demonstrates metabolic control of T cell exhaustion starting early during chronic infection and implicates dysregulated metabolism as a driver of CD8+ T cell exhaustion. The PD-1 pathway constrained glucose uptake and use and repressed the key metabolic transcriptional co-activator PGC-1α, effectively suppressing glycolysis and OXPHOS in virus-specific T cells, but also compromising mitochondrial quality. These events occurred in a setting of high energetic demands leading to an altered metabolic phenotype prior to major manifestations of exhaustion. These results have implications for therapeutic strategies aiming at the reinvigoration of Tex cells in chronic infections and cancer.
Experimental Procedures
Mice, infections, adoptive transfer and treatments
Pdcd1−/− (Keir et al., 2007), LCMV GP33-specific TCR transgenic “P14”, Pdcd1+/+ P14 (Odorizzi et al., 2015) and C57BL/6 mice (Charles River) were housed at the University of Pennsylvania (Philadelphia, PA) and treated in accordance with protocols approved by the Institutional Animal Care and Use Committee. Infections with LCMV Armstrong (2 × 105 plaque-forming units (PFU) i.p.) or clone 13 (4 × 106 PFU i.v.) were performed as described (Odorizzi et al., 2015). Treatment with 300µg/kg of rapamycin or PBS was performed daily at d5–8 p.i. For antibody blockade, mice were injected with 200µg anti-CD4 (GK1.5; BioXcell) i.p. on day −1 and +1 of infection; treatment with 200µg anti-PDL1 (10F.9G2) antibody or isotype control was performed every three days between d22–35 p.i. For cotransfer experiments, 250 Pdcd1+/+ and Pdcd1−/− P14 cells were adoptively transferred as described (Odorizzi et al., 2015).
Retrovirus experiments
Slc2a1 (encoding Glut-1) or Ppargc1a (encoding PGC1α) cDNA was cloned into the MSCV-IRES-VEX plasmid. CDNA-expressing RVs were produced in 293T cells with MSCV and pCL-Eco plasmids using Lipofectamine 3000 (Invitrogen). For RV transduction, splenic P14 cells were enriched by CD8 negative selection (StemCell). After 24h of activation with anti-CD3 (1µg/ml; 2C11; eBioscience) and anti-CD28 (0.5µg/ml; 37.51; eBioscience) with recombinant human IL-2 (100U/ml), P14 cells were transduced with RV and polybrene (1µg/ml) by spin infection (2,000g for 60min at 30C). RV-transduced P14 cells were adoptively transferred into recipient infected mice, as described (Kurachi et al., 2014).
Gene set enrichment analysis (GSEA)
GSEA using Broad Institute software (http://www.broadinstitute.org/gsea/) was performed on microarray data from LCMV-specific CD8 T cells sorted at d6, 8, 15, 22 and 30 p.i. and from PD-1Hi and PD1Int Tex cells isolated on ~d30 p.i. with clone 13. These data are available in the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/gds) (GSE41867 and GSE41869). GSEA used gene sets from the Molecular Signature Database v.5.0 (Subramanian et al., 2005) including KEGG pathway gene lists obtained from http://www.genome.jp/kegg/pathway.html. Enrichment scores (ES) obtained by GSEA were used to compare different KEGG pathways at different time points of infection.
Seahorse Extracellular Flux Analysis
Seahorse experiments were performed on sorted virus-specific CD8 T cells or where cells were limiting, purified total CD8 T cells from infected mice (negative magnetic selection (Stem Cell)) using modified XF Cell Mito Stress kit or Glycolysis Stress kit (Seahorse Bioscience). Briefly, cells were plated on poly-D-lysine-coated 24- and 96- well polystyrene Seahorse plates, equilibrated for 1h at 37C and assayed for OCR (pMoles/min) and ECAR (mpH/min) after addition of oligomycin (1 µM), FCCP (1.5µM), etomoxir (200µM) or 2-DG (50mM) and antimycin A/rotenone (1µM/0.1µM). In some experiments, cells were equilibrated in glucose-free media and injection of glucose (10mM) was performed. SRC was defined as the difference between MRC and baseline OCR, expressed as % of baseline.
Flow cytometry and cell sorting
For mitochondrial studies, cells were incubated with 50nM MitoTracker Green (MTG) and/or 25nM MitoTracker DeepRed (MTDR) for 30min at 37C prior to staining. Mitochondrial superoxides were assessed using 5 µM MitoSox Red. Glucose uptake was assayed by exposing cells to 10µM 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-D-glucose (2-NBDG) at 37C. Reagents were obtained from Life Technologies. Staining for flow cytometry was performed as described (Odorizzi et al., 2015). Reagents were obtained from BD (Annexin V), BioLegend (CD3, CD4, CD8, CD19, CD39, CD44, CD45.1, CD45.2, CD71, CD98, CD127, CD160, PD-1), Ebioscience (CD8, CD27, CD38, CD39 CD62L, 2B4, Lag-3, KLRG1), R&D Systems (Glut-1) or Cell Signaling Technology (p-S6240/244). Tetramers were obtained from the NIH tetramer core. Dead cells were excluded based on LIVE/DEAD Fixable Dead Cell Stain (Life Technologies) or Zombie NIR (Biolegend). Annexin V staining was performed as per manufacturer’s protocol (BD). Data were collected on an LSRII (BD) and analyzed with FlowJo software (Tree Star). Cell sorting was performed using a FACSAria II (BD). For signaling experiments, cells were rested for 30–60min at 37°C in 10% RPMI, followed by restimulation with 1µM gp33 peptide for 20min. P-S6240/244 was detected using BD cytofix/cytoperm kit.
Polychromatic imaging cytometry
For Imagestream (Amnis) analysis, negative magnetic bead-purified (StemCell) CD8+ T cells were stained with antibodies and mitochondrial dyes as above. Hoechst 33342 staining (10mM) was performed for 30min at 37C. Single stained cells were used as compensation controls. Images were captured at 60× magnification and analysis was performed using IDEAS v6 software (Amnis).
Electron microscopy
Samples were purified by FACS and fixed by immersion in a 2% glutaraldehyde solution in 0.1M cacodylate buffer (pH 7.4). Processing and staining was performed as described (Picard et al., 2015). Thin sections (90µm) were mounted on filmed copper grids, stained with uranyl acetate and lead citrate and examined on a JEOL 1010 or FEI-Tecnai T12 electron microscope.
Statistical analysis
Prism (GraphPad Software) and JMP (SAS Inst. Inc) were used for statistical analysis. Paired and unpaired Student t test (as appropriate) were used in Fig. (2–7).
Supplementary Material
Highlights.
-
-
Early exhausted T cells exhibit repressed glycolytic and mitochondrial metabolism.
-
-
Metabolic skewing occurred despite mTOR-driven up-regulation of anabolic pathways.
-
-
PD-1 regulated bioenergetic insufficiencies early and late in infection
-
-
PGC-1α overexpression improved metabolism and T cell function
Acknowledgments
We thank J.-E. Lee and K. Foskett for help with flux analyses, M. Reuter for help with the Imagestream, D. Williams, for electron microscopy and J. Kurachi for technical support, M. Bohnert for mitochondrial expertise. Tetramers were obtained from the NIH tetramer core. B.B. was supported by German Research Foundation fellowship BE5496/1-1, G.M.D. by the Sidney Kimmel Foundation for Cancer Research (SKF-015-039), the Samuel and Emma Winters Foundation, and the NIH (P50CA121973 Skin Cancer SPORE), K.E.P. by a Robertson Foundation/Cancer Research Institute Irvington Fellowship and A.L.J. by a Philadelphia Foundation's Brody Family Medical Trust Fund fellowship. This work was supported by NIH grants AI105343, AI082630, AI095608, AI112521, AI115712, AI117718, AI108545, AI2010085 to E.J.W. This research was supported by the Parker Institute for Cancer Immunotherapy. E.J.W. has a patent licensing agreement on the PD-1 pathway.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
B.B., A.L.J and E.J.W. designed the study. B.B., A.L.J., M.K., P.M.O., K.E.P., J.A., E.S., L.M.M and M.A.P. performed experiments. B.B., A.L.J and E.J.W. analyzed the data. G.M.D. contributed reagents, B.B. and E.JW. wrote the paper, and all other authors edited the paper.
References
- Angelosanto JM, Blackburn SD, Crawford A, Wherry EJ. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol. 2012;86:8161–8170. doi: 10.1128/JVI.00889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S, St-Pierre J. PGC1alpha and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- Belikov AV, Schraven B, Simeoni L. T cells and reactive oxygen species. J Biomed Sci. 2015;22:85. doi: 10.1186/s12929-015-0194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Freeman GJ, Wherry EJ. Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc Natl Acad Sci U S A. 2008;105:15016–15021. doi: 10.1073/pnas.0801497105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, McGavern DB, Oldstone MB. Reprogramming of antiviral T cells prevents inactivation and restores T cell activity during persistent viral infection. J Clin Invest. 2006;116:1675–1685. doi: 10.1172/JCI26856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O'Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med. 2015;212:1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O'Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemnitz JM, Riley JL, Frauwirth KA, Braunstein I, Kobayashi SV, Linsley PS, Thompson CB, Parry RV. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Blood. 2004;104:726a–726a. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford A, Angelosanto JM, Kao C, Doering TA, Odorizzi PM, Barnett BE, Wherry EJ. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity. 2014;40:289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. Network Analysis Reveals Centrally Connected Genes and Pathways Involved in CD8(+) T Cell Exhaustion versus Memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frebel H, Nindl V, Schuepbach RA, Braunschweiler T, Richter K, Vogel J, Wagner CA, Loffing-Cueni D, Kurrer M, Ludewig B, Oxenius A. Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. J Exp Med. 2012;209:2485–2499. doi: 10.1084/jem.20121015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Antitumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T, Egen JG, Lammermann T, Kastenmuller W, Torabi-Parizi P, Germain RN. Tuning of antigen sensitivity by T cell receptor-dependent negative feedback controls T cell effector function in inflamed tissues. Immunity. 2014;40:235–247. doi: 10.1016/j.immuni.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir ME, Freeman GJ, Sharpe AH. PD-1 regulates self-reactive CD8+ T cell responses to antigen in lymph nodes and tissues. J Immunol. 2007;179:5064–5070. doi: 10.4049/jimmunol.179.8.5064. [DOI] [PubMed] [Google Scholar]
- Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol. 2014;15:373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver NJ, Jacobs SR, Wieman HL, Wofford JA, Coloff JL, Rathmell JC. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J Leukoc Biol. 2008;84:949–957. doi: 10.1189/jlb.0108024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi PM, Pauken KE, Paley MA, Sharpe A, Wherry EJ. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J Exp Med. 2015;212:1125–1137. doi: 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T, Chikuma S, Iwai Y, Fagarasan S, Honjo T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat Immunol. 2013;14:1212–1218. doi: 10.1038/ni.2762. [DOI] [PubMed] [Google Scholar]
- Okazaki T, Maeda A, Nishimura H, Kurosaki T, Honjo T. PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13866–13871. doi: 10.1073/pnas.231486598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye I, Wang LH, Pallmer K, Richter K, Ichimura T, Haas R, Crouse J, Choi O, Heathcote D, Lovo E, et al. T CELL METABOLISM The protein LEM promotes CD8(+) T cell immunity through effects on mitochondrial respiration. Science. 2015;348:995–1001. doi: 10.1126/science.aaa7516. [DOI] [PubMed] [Google Scholar]
- Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, Wherry EJ. Progenitor and Terminal Subsets of CD8(+) T Cells Cooperate to Contain Chronic Viral Infection. Science. 2012;338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, Linsley PS, Thompson CB, Riley JL. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Molecular and cellular biology. 2005;25:9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat Commun. 2015;6:6692. doi: 10.1038/ncomms7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsoukis N, Sari D, Boussiotis VA. PD-1 inhibits T cell proliferation by upregulating p27 and p15 and suppressing Cdc25A. Cell Cycle. 2012;11:4305–4309. doi: 10.4161/cc.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, McManus MJ, Csordas G, Varnai P, Dorn GW, 2nd, Williams D, Hajnoczky G, Wallace DC. Trans-mitochondrial coordination of cristae at regulated membrane junctions. Nat Commun. 2015;6:6259. doi: 10.1038/ncomms7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard KA, Fitz LJ, Lee JM, Benander C, George JA, Wooters J, Qiu YC, Jussif JM, Carter LL, Wood CR, Chaudhary D. PD-1 inhibits T-cell receptor induced phosphorylation of the ZAP70/CD3 zeta signalosome and downstream signaling to PKC theta. Febs Lett. 2004;574:37–41. doi: 10.1016/j.febslet.2004.07.083. [DOI] [PubMed] [Google Scholar]
- Staron MM, Gray SM, Marshall HD, Parish IA, Chen JH, Perry CJ, Cui G, Li MO, Kaech SM. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity. 2014;41:802–814. doi: 10.1016/j.immuni.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nature Reviews Immunology. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok JD, Chan TA. CANCER Antitumour immunity gets a boost. Nature. 2014;515:496–498. doi: 10.1038/515496a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T, Takamatsu M, Kobayashi-Imanishi W, Hashimoto-Tane A, Azuma M, Saito T. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. Journal of Experimental Medicine. 2012;209:1201–1217. doi: 10.1084/jem.20112741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.