Abstract
The wide spread of gene exchange and loss in the prokaryotic world has prompted the concept of ‘lateral genomics’ to the point of an outright denial of the relevance of phylogenetic trees for evolution. However, the pronounced coherence congruence of the topologies of numerous gene trees, particularly those for (nearly) universal genes, translates into the notion of a statistical tree of life (STOL), which reflects a central trend of vertical evolution. The STOL can be employed as a framework for reconstruction of the evolutionary processes in the prokaryotic world. Quantitatively, however, horizontal gene transfer (HGT) dominates microbial evolution, with the rate of gene gain and loss being comparable to the rate of point mutations and much greater than the duplication rate. Theoretical models of evolution suggest that HGT is essential for the survival of microbial populations that otherwise deteriorate due to the Muller’s ratchet effect. Apparently, at least some bacteria and archaea evolved dedicated vehicles for gene transfer that evolved from selfish elements such as plasmids and viruses. Recent phylogenomic analyses suggest that episodes of massive HGT were pivotal for the emergence of major groups of organisms such as multiple archaeal phyla as well as eukaryotes. Similar analyses appear to indicate that, in addition to donating hundreds of genes to the emerging eukaryotic lineage, mitochondrial endosymbiosis severely curtailed HGT. These results shed new light on the routes of evolutionary transitions, but caution is due given the inherent uncertainty of deep phylogenies.
Keywords: Horizontal gene transfer, prokaryotes, evolutionary transitions, microbial evolution, statistical tree of life
Pervasive horizontal gene transfer in microbial evolution and the statistical tree of life
As soon as several complete bacterial and archaeal genomes were sequenced in the mid to late 1990s, comparative and phylogenomic analyses have revealed a surprising complexity of microbial genome evolution 1– 6. These observations can be broadly summarized in the form of three major trends: i) bacterial and archaeal genomes have dramatically different gene compositions, with only a small set of core genes being universally conserved; ii) unexpected patterns of gene sharing have been detected, in particular many genes shared between hyperthermophilic archaea and bacteria; and iii) topologies of the phylogenetic trees for many bacterial genes were rarely fully compatible between each other or with the 16S ribosomal RNA (rRNA) tree, although at least some of these trees were highly reliable, indicating that the discrepancies could not be caused by methodological artifacts alone. Taken together, these findings appeared impossible to explain without invoking widespread horizontal gene transfer (HGT), prompting the concept of ‘lateral genomics’, which posits that the dominant process in microbial evolution is gene exchange between organisms rather than vertical descent along a tree 4, 7– 10. In its extreme form, lateral genomics denies the relevance of tree-like evolution and “tree thinking” in biology altogether 11, 12. This concept triggered an intense debate that continued for over a decade, with the entire spectrum of positions expressed forcefully, from complete dismissal of HGT as a consequential aspect of microbial evolution to an equally adamant denial of the importance of trees 13. A characteristic episode that might epitomize this entire extended discussion occurred in 2006. A computational pipeline that automatically produced a comprehensive ‘Tree of Life’ (TOL) from a concatenated sequence alignment of 31 universal proteins was hailed as a major advance of phylogenomics 14, only to be immediately debunked as a “tree of 1%”, i.e. one that (at best) accurately reflects the evolutionary history of only a small fraction of microbial genes 15.
Now, exactly 20 years after the comparison of complete microbial genomes became possible, where do we stand on the status of trees and HGT in microbial evolution? Not only phylogenies of individual genes but also the microbial TOL are clearly alive and apparently rather well. A remarkable testimony to the staying power of trees is the recent amendment to the microbial TOL, which now includes a major new branch discovered through metagenomics 16. However, the status of the TOL has changed irrevocably. Given the overwhelming evidence that the topologies of the phylogenetic trees of individual genes are rarely identical, the phylogeny of universal genes (let it be one gene, such as 16S rRNA, or multiple genes, such as those of ribosomal proteins) hardly can be considered an accurate representation of organismal evolution. The key question, then, is: does a tree of a universal gene reflect solely its own history or does it contain information on the evolution of other genes and, if so, how many genes and how much information? A phylogenomic study designed to address this question has revealed considerable orderliness among the topologies of several thousand trees in the microbial “phylogenetic forest” 17, 18. Specifically, the tree topologies of the (nearly) universal genes, which encode primarily translation system components (roughly, the notorious 1% of all analyzed trees), are highly consistent not only among themselves but also with trees of numerous other genes. The consensus topology of the nearly universal trees explains nearly 40% of the variance in the tree topologies across the “phylogenetic forest” 19. This tree-like signal of vertical inheritance is by far the strongest trend in microbial evolution because the remaining variance in tree topologies reflects the largely random gene exchange. Thus, the “tree of 1%” seems not to be a failed evolutionary hypothesis 20, 21 but rather an appropriate representation of the central current of microbial genome evolution, or a “statistical tree of life” (STOL) 22. The STOL provides the standard against which HGT can be identified—indeed, the very notion of horizontal gene flow becomes meaningless in the absence of such vertical standard—and, more generally, the framework for reconstruction of microbial genome evolution via gene gain and loss.
A more sophisticated argument against “tree thinking” has been that biased HGT, such that there exists a gradient of HGT rates from closely related to distant microbes, could mimic a tree pattern of evolutionary divergence 23. Subsequent simulation analyses differed with respect to whether this explanation was plausible 24– 26 or not 19 under realistic parameters of the evolutionary process. Testing this proposition depends on the subtleties of evolution modeling and is not easy.
Rapid dynamics of microbial evolution
Numerous comparisons of microbial genomes show that even genomes of organisms that are closely related in terms of the sequence similarity between universal genes (e.g. identical 16S rRNA sequences) often substantially differ in the gene repertoires 27, 28. Thus, information on the evolutionary dynamics of microbial genomes can be extracted from the patterns of gene presence-absence 29. The prominence of the vertical evolution trend in the “forest of life” (see above) justifies the use of phylogenetic trees of universal genes (species tree, for short) as a scaffold for evolutionary reconstruction. Briefly, all the genes in the pangenome of a species or another group of microbes (i.e. the entirety of the genes represented in the available isolates of the given group 30) are mapped to the leaves of the species tree. This mapping is used to reconstruct the evolutionary scenario for the pangenome, i.e. the history of gene gains, losses, and duplications. Initially, the reconstruction was performed using simple, intuitive maximum parsimony methods which identify the scenario with the minimum number of events 31– 33. At present, the approaches of choice are based on more sophisticated maximum likelihood algorithms that employ evolutionary birth-and-death models to derive statistical estimates for the number of different genomic events associated with each branch of the species tree 34– 37.
Application of the maximum likelihood approach to the reconstruction of evolution for diverse groups of closely related bacteria and archaea has revealed a striking picture of genomes in turmoil 38. Although the rates of gene gain, loss, and duplication greatly differ across the bacterial diversity, in the most dynamic groups, several gene gains and losses occur during the time the genome accumulates, on average, one nucleotide substitution per gene. Strikingly, the most common process of genome dynamics is actually loss of genes: for most (although not all) groups of microbes, evolutionary reconstructions indicate a twofold to threefold excess of losses over gains per nucleotide substitution. In the long term, excess of gene losses obviously would lead to genome degradation and eventually extinction, and indeed such is the fate of many groups of microbes, in particular parasites and symbionts 39, 40. In general, however, the gradual gene loss appears to be offset by episodic, massive gene gain that might accompany the emergence of major groups of prokaryotes 41 (see more below on such putative bursts of innovation). The same reconstructions indicate that in all analyzed groups of microbes, the rate of gene gain exceeds the gene duplication rate by at least an order of magnitude 38, 42. The primary source of gene gain is HGT, which accordingly is the principal route of evolutionary innovation in bacteria and archaea.
Taken together, the reconstructions of the dynamics of microbial genome evolution show that, in the microbial world, evolution primarily occurs not via the classic Darwinian process adopted by the Modern Synthesis of Evolutionary Biology, i.e. gradual accumulation of numerous, “infinitesimally small” changes (mutations) 43, 44, but rather by much bigger, at least gene-sized, leaps. In a sharp contrast to eukaryotes, in bacteria and archaea, the dominant feature of genome evolution is not gene duplication 45– 47 but rather evolution by extensive gene loss and gene gain via HGT.
Essentiality and evolvability of horizontal gene transfer in bacteria and archaea
Can microbes evolve without substantial HGT, simply via the competition of clonal populations? Apparently not, as population genetic modeling indicates that this evolutionary regime is unsustainable in the long term 48. Clonal populations typically deteriorate due to the action of the evolutionary mechanism known as Muller’s ratchet, i.e. gradual loss of fitness and eventual extinction caused by accumulation of slightly deleterious mutations via genetic drift 49, 50. Demise caused by Muller’s ratchet appears to be the typical fate of bacteria that are confined to intracellular parasitism or symbiosis, although the ratchet can be slowed down by lowering the mutation rate 51. However, such mechanisms cannot stop the ratchet altogether. The only route of actual escape from Muller’s ratchet appears to be gene acquisition via HGT resulting in either displacement of a mutated gene by a functional copy or gain of new genes that offsets the deleterious effects of accumulating mutations 48. Notably, the model shows that, thanks to the stochastic nature of the mutation process, protection from the ratchet is achievable despite the fact that environmental DNA that comes from dead microbes on average has a higher mutational load than the DNA of living recipient cells 48. Thus, in prokaryotes, HGT plays the same role of preventing mutational meltdown that in eukaryotes is played by meiotic sex 52.
Escape from Muller’s ratchet can be considered the most fundamental benefit of HGT in microbial evolution but it certainly is not the only one. Acquisition of new genes and whole operons appears to be the principal route of metabolic network expansion in microbes 42, 53. As the network grows, gain of a single enzyme is increasingly likely to provide access to a new nutrient leading to increased fitness 54.
Given the indispensability of HGT for the survival of microbial populations, a plausible hypothesis seems to be that HGT is evolvable, i.e. is an adaptive, selectable trait. However, whether or not this is the case is not an easy question because HGT might be considered a by-product of the presence of substantial amounts of DNA in the environment combined with genetic processes such as transformation and bacteriophage infection that leads to gene transduction 55. Diverse bacteria and archaea are competent for natural transformation that is mediated by dedicated DNA intake pumps 56. These pumps can be legitimately considered devices for utilization of environmental DNA as a source of nucleotides (simply put, food), with HGT being a fringe benefit. However, the demonstration that at least in some bacteria the ingested DNA is protected against degradation, thus preventing its use as a nucleotide source and conversely facilitating HGT, implies that, at least in part, natural competence evolved as a gene transfer mechanism 57. The long-known existence of DNA uptake signal sequences and proteins that bind them, which jointly comprise a discrimination mechanism allowing bacteria to preferentially take up DNA from closely related organisms, is another piece of evidence in support of the view of transformation as an evolved route of gene transfer, apart from the nutritional value of DNA 58, 59.
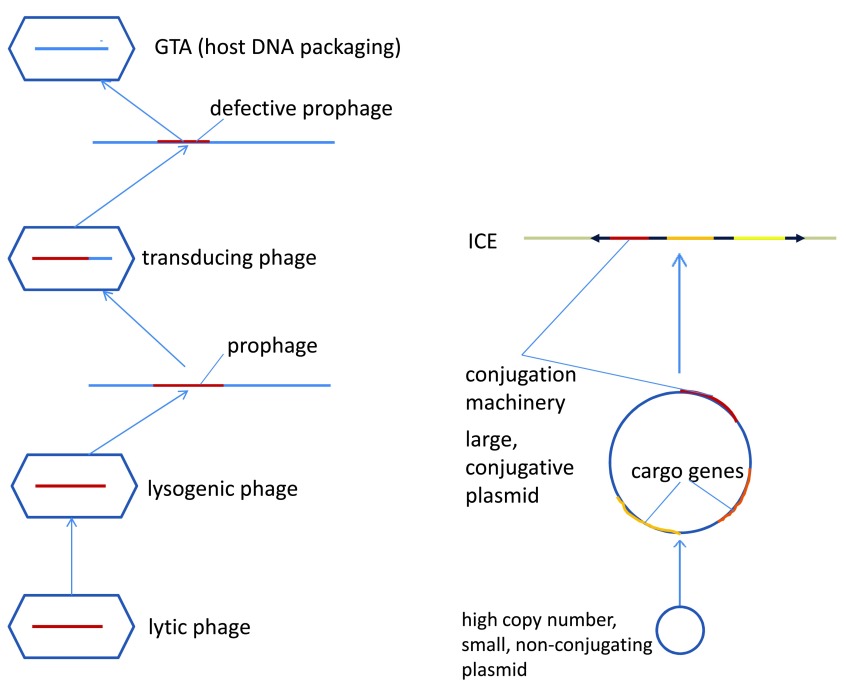
Bacterial and archaeal conjugation (prokaryotic sex) is a mechanism of genetic material transfer between microbial cells that combines features of selfish genetic elements and devices for gene transfer. Conjugative plasmids encode proteins required for autonomous replication, whereas integrative and conjugative elements (ICEs, or conjugative transposons) typically replicate while integrated into the host chromosome but have the ability to excise and form plasmid-like molecules 60, 61. Both types of elements are transferred by the conjugation molecular machinery (type IV secretion systems) and typically carry ‘cargo’ genes unrelated to the transposon life cycle. Thus, these selfish elements are at the same time vehicles for HGT that mediate microbial adaptation by introducing new genes into the recipient genomes 62.
Perhaps the most striking showcase for dedicated vehicles of HGT are the gene transfer agents (GTAs), defective prophages that form virus particles in which they package apparently random fragments of the bacterial chromosome rather than the phage genome 63, 64. The GTAs then infect other bacteria or archaea, and the transferred DNA integrates into the recipient genome. In marine bacterial communities, the rate of GTA-mediated gene transfer appears to be quite high and often involves distantly related organisms 65. Notably, the GTAs confer on their carriers the ability to donate rather than acquire genetic material. Such a capacity could be adaptive in the context of utilization of “public goods” by microbial communities. The wide spread of GTAs appears to present strong evidence of evolvability of HGT.
These examples highlight the apparent major route of evolution of HGT vehicles, through stepwise domestication and “enslavement” of selfish genetic elements, such as plasmids and viruses, whereby the hosts exploit the inherent ability of such elements to transfer genetic material ( Figure 1).
Figure 1. Domestication of selfish genetic elements en route to dedicated vehicles for horizontal gene transfer.
The figure depicts the hypothetical stages of the evolutionary paths from a lytic phage to a gene transfer agent and from a small, high copy number plasmid to an integrative and conjugative element. Abbreviations: GTA, gene transfer agent; ICE, integrative and conjugative element.
Horizontal gene transfer and evolutionary transitions
Several large-scale reconstructions of microbial genome evolution suggest that gene loss occurs in a roughly clock-like manner whereas gene gain tends to be episodic, occurring in bursts that involve acquisition of many genes over a short time 31, 38, 66. These observations prompted the hypothesis that emergence of new major groups of organisms often, perhaps typically, involves massive gene gain via HGT (in microbes) or extensive, in some cases, whole genome duplication (in eukaryotes) followed by gradual genome streamlining via gene loss in each of the lineages 41. The importance of massive HGT in at least two major evolutionary transitions, namely the origin of eukaryotes and the origin of the eukaryotic supergroup Archaeplastida (algae and plants), is beyond doubt 67. In these special cases, the sources of the hundreds if not thousands of transferred genes were mitochondria and chloroplasts, respectively, i.e. bacterial endosymbionts on the path to becoming eukaryotic organelles 68– 70. Can the model of punctuated gene gain be validated in a more general context? Comprehensive search of archaeal genomes for acquired bacterial genes has led to the conclusion that the origin of most, if not all, major archaeal clades was associated with and possibly caused by acquisition of hundreds or even thousands of bacterial genes 71, 72. The largest influx of bacterial genes was detected in mesophilic archaeal groups such as Halobacteria and Methanobacteria and apparently led to fundamental innovation, i.e. adaptation to new lifestyles and ecological niches. The conclusion on the acquisition of numerous bacterial genes at the roots of the major archaeal clades, as opposed to more uniform gain along the respective evolutionary lineages, has been reached by Nelson-Sathi and colleagues using an original statistical procedure for topological comparison of the phylogenetic tree of the (candidate) acquired bacterial genes and resident genes in the recipient group of archaea 72. A re-evaluation of these results using more traditional methods for reconstruction of gene gain and loss yielded results that were better compatible with piecemeal gene acquisition 73. Nevertheless, a more biologically oriented analysis seems to suggest that, at least for the origin of several groups of mesophilic archaea, acquisition of multiple bacterial genes has been the trigger of the lifestyle transition 74. Clearly, additional and probably extensive research with different methods is required to resolve this conundrum.
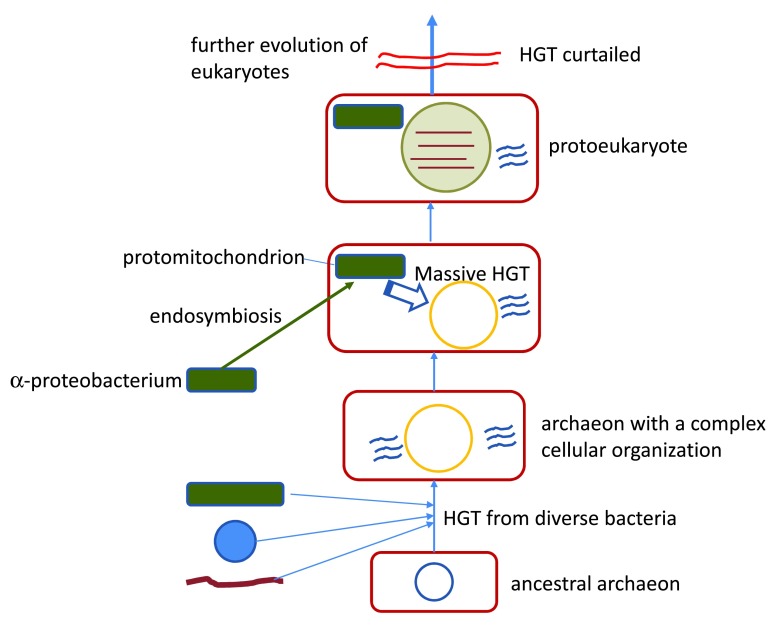
Two more recent, complementary studies have further addressed the question of episodic vs. continuous acquisition of genes via HGT in the context of symbiogenesis and early evolution of eukaryotes. One of these employed comprehensive comparison of the topologies of phylogenetic trees of eukaryotic genes of apparent bacterial and archaeal origin and arrived to the conclusion that eukaryotes acquired the majority of bacterial genes in the two major bursts associated with the origin of mitochondria and chloroplasts whereas subsequent, continuous acquisition of bacterial genes was limited in extent 75. The other analysis makes an attempt to go even further by directly comparing the phylogenetic distances from the closest bacterial homologs for the genes of apparent α-proteobacterial origin encoding proteins localized to the mitochondria and genes apparently derived from other bacteria 76. The conclusion is that the proto-mitochondrial genes are the closest homologs to their bacterial ancestors, hence probably the latest large bunch of bacterial genes acquired by eukaryotes.
The ‘mitochondria late’ conclusion superficially could be interpreted as an indication that the host of the proto-mitochondrion was an ‘archezoan’, a primitive amitochondrial eukaryote 77, 78. However, this does not appear to be a necessary implication of the actual observations. On the contrary, these findings seem to be fully compatible with the conclusions of Ku et al. on the scarcity of late, non-organellar HGT in eukaryotes 75 and with the earlier scenarios of eukaryogenesis, which proposed a complex archaeon with many acquired bacterial genes as the host of the proto-mitochondria 79, 80 ( Figure 2). Under this scenario, acquisition of the mitochondria precipitated the series of dramatic changes in the organization of the chimeric cell which led to the curtailment of HGT. Thus, the acquisition of numerous genes from the proto-mitochondrion comes across as the last major burst of HGT (other than the acquisition of chloroplast genes at the base of the Archaeplastida), although numerous lineage-specific acquisitions of relatively small but biologically consequential groups of bacterial genes as well as eukaryote-to-eukaryote transfers undoubtedly occurred at later stages 81– 83.
Figure 2. Eukaryogenesis and horizontal gene transfer.
The figure presents the ‘endosymbiotic’ model of eukaryogenesis under which the host of the protomitochondrial endosymbiont was a typical archaeon albeit one with a relatively complex intracellular organization and numerous genes captured from bacteria via HGT. Abbreviations: HGT, horizontal gene transfer.
Concluding remarks
Over a decade ago, the question has been asked whether the concept of HGT would soon ‘come of age’, causing a rather tense discussion 84, 85. These days, I believe, it is clear that the field has matured. There is no reasonable doubt anymore that HGT is a dominant process in microbial evolution that generally occurs at a high rate. Moreover, the relevance of ‘horizontal’ as applied to gene flow is validated by the strong evidence of the existence of a central vertical, tree-like trend in genome evolution. Thus, the focus of research has shifted towards the ‘how’s’ and ‘why’s’ of HGT and, in these directions, much more remains to be done than has been accomplished already.
Both theoretical models and tantalizing experimental clues suggest that HGT is essential for microbial survival and could be an evolvable, adaptive capacity mediated by dedicated vehicles originating from domesticated selfish elements. Yet this concept runs afoul of the distrust of ‘evolution of evolvability’ that is deeply ingrained among biologists. Indeed, much work remains to be done to make a compelling case for the evolvability of HGT. Somewhat similarly, albeit in a different area, there is accumulating evidence of a major role of HGT in evolutionary transitions. Yet these conclusions rest on the analysis of deep phylogenies which are inherently error prone and recalcitrant to definitive interpretation. Much like evolution itself, extensive HGT in the microbial world is a fact and not a ‘theory’. However, understanding the routes, causes, and consequences of horizontal gene flow as well as constructing the actual, quantitative theoretical framework of this pervasive process will keep many biologists busy for decades to come.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Olga Zhaxybayeva, Department of Biological Sciences, Dartmouth College, Hanover, NH, USA
Purificacion Lopez-Garcia, Unité d'Ecologie, Systématique et Evolution, Université Paris-Sud, Orsay, France
Funding Statement
The author’s research is supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
[version 1; referees: 2 approved]
References
- 1. Kolstø AB: Dynamic bacterial genome organization. Mol Microbiol. 1997;24(2):241–8. 10.1046/j.1365-2958.1997.3501715.x [DOI] [PubMed] [Google Scholar]
- 2. Koonin EV, Mushegian AR, Galperin MY, et al. : Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol Microbiol. 1997;25(4):619–37. 10.1046/j.1365-2958.1997.4821861.x [DOI] [PubMed] [Google Scholar]
- 3. Koonin EV, Mushegian AR, Rudd KE: Sequencing and analysis of bacterial genomes. Curr Biol. 1996;6(4):404–16. 10.1016/S0960-9822(02)00508-0 [DOI] [PubMed] [Google Scholar]
- 4. Doolittle WF: Phylogenetic classification and the universal tree. Science. 1999;284(5423):2124–9. 10.1126/science.284.5423.2124 [DOI] [PubMed] [Google Scholar]
- 5. Fraser CM, Eisen JA, Salzberg SL: Microbial genome sequencing. Nature. 2000;406(6797):799–803. 10.1038/35021244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koonin EV, Makarova KS, Aravind L: Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–42. 10.1146/annurev.micro.55.1.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aravind L, Tatusov RL, Wolf YI, et al. : Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14(11):442–4. 10.1016/S0168-9525(98)01553-4 [DOI] [PubMed] [Google Scholar]
- 8. Nelson KE, Clayton RA, Gill SR, et al. : Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399(6734):323–9. 10.1038/20601 [DOI] [PubMed] [Google Scholar]
- 9. Doolittle WF: Lateral genomics. Trends Cell Biol. 1999;9(12):M5–8. 10.1016/S0962-8924(99)01664-5 [DOI] [PubMed] [Google Scholar]
- 10. Martin W: Mosaic bacterial chromosomes: a challenge en route to a tree of genomes. Bioessays. 1999;21(2):99–104. [DOI] [PubMed] [Google Scholar]
- 11. Bapteste E, Susko E, Leigh J, et al. : Do orthologous gene phylogenies really support tree-thinking? BMC Evol Biol. 2005;5:33. 10.1186/1471-2148-5-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doolittle WF, Bapteste E: Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci U S A. 2007;104(7):2043–9. 10.1073/pnas.0610699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Malley MA, Boucher Y: Paradigm change in evolutionary microbiology. Stud Hist Philos Biol Biomed Sci. 2005;36(1):183–208. 10.1016/j.shpsc.2004.12.002 [DOI] [PubMed] [Google Scholar]
- 14. Ciccarelli FD, Doerks T, von Mering C, et al. : Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311(5765):1283–7. 10.1126/science.1123061 [DOI] [PubMed] [Google Scholar]
- 15. Dagan T, Martin W: The tree of one percent. Genome Biol. 2006;7(10):118. 10.1186/gb-2006-7-10-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hug LA, Baker BJ, Anantharaman K, et al. : A new view of the tree of life. Nat Microbiol. 2016;1(5): 16048. 10.1038/nmicrobiol.2016.48 [DOI] [PubMed] [Google Scholar]
- 17. Puigbò P, Wolf YI, Koonin EV: Search for a 'Tree of Life' in the thicket of the phylogenetic forest. J Biol. 2009;8(6):59. 10.1186/jbiol159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Puigbò P, Wolf YI, Koonin EV: Seeing the Tree of Life behind the phylogenetic forest. BMC Biol. 2013;11:46. 10.1186/1741-7007-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puigbò P, Wolf YI, Koonin EV: The tree and net components of prokaryote evolution. Genome Biol Evol. 2010;2:745–56. 10.1093/gbe/evq062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doolittle WF: Eradicating typological thinking in prokaryotic systematics and evolution. Cold Spring Harb Symp Quant Biol. 2009;74:197–204. 10.1101/sqb.2009.74.002 [DOI] [PubMed] [Google Scholar]
- 21. Martin WF: Early evolution without a tree of life. Biol Direct. 2011;6:36. 10.1186/1745-6150-6-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Malley MA, Koonin EV: How stands the Tree of Life a century and a half after The Origin? Biol Direct. 2011;6:32. 10.1186/1745-6150-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gogarten JP, Doolittle WF, Lawrence JG: Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19(12):2226–38. [DOI] [PubMed] [Google Scholar]
- 24. Andam CP, Gogarten JP: Biased gene transfer in microbial evolution. Nat Rev Microbiol. 2011;9(7):543–55. 10.1038/nrmicro2593 [DOI] [PubMed] [Google Scholar]
- 25. Andam CP, Williams D, Gogarten JP: Biased gene transfer mimics patterns created through shared ancestry. Proc Natl Acad Sci U S A. 2010;107(23):10679–84. 10.1073/pnas.1001418107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abby SS, Tannier E, Gouy M, et al. : Lateral gene transfer as a support for the tree of life. Proc Natl Acad Sci U S A. 2012;109(13):4962–7. 10.1073/pnas.1116871109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perna NT, Plunkett G, 3rd, Burland V, et al. : Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409(6819):529–33. 10.1038/35054089 [DOI] [PubMed] [Google Scholar]
- 28. Eppinger M, Mammel MK, Leclerc JE, et al. : Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A. 2011;108(50):20142–7. 10.1073/pnas.1107176108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tatusov RL, Koonin EV, Lipman DJ: A genomic perspective on protein families. Science. 1997;278(5338):631–7. 10.1126/science.278.5338.631 [DOI] [PubMed] [Google Scholar]
- 30. Tettelin H, Riley D, Cattuto C, et al. : Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11(5):472–7. 10.1016/j.mib.2008.09.006 [DOI] [PubMed] [Google Scholar]
- 31. Snel B, Bork P, Huynen MA: Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12(1):17–25. 10.1101/gr.176501 [DOI] [PubMed] [Google Scholar]
- 32. Mirkin BG, Fenner TI, Galperin MY, et al. : Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;3:2. 10.1186/1471-2148-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kunin V, Ouzounis CA: The balance of driving forces during genome evolution in prokaryotes. Genome Res. 2003;13(7):1589–94. 10.1101/gr.1092603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen O, Rubinstein ND, Stern A, et al. : A likelihood framework to analyse phyletic patterns. Philos Trans R Soc Lond B Biol Sci. 2008;363(1512):3903–11. 10.1098/rstb.2008.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Csurös M: Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26(15):1910–2. 10.1093/bioinformatics/btq315 [DOI] [PubMed] [Google Scholar]
- 36. Csurös M, Miklós I: Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol. 2009;26(9):2087–95. 10.1093/molbev/msp123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cohen O, Pupko T: Inference of gain and loss events from phyletic patterns using stochastic mapping and maximum parsimony --a simulation study. Genome Biol Evol. 2011;3:1265–75. 10.1093/gbe/evr101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Puigbò P, Lobkovsky AE, Kristensen DM, et al. : Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol. 2014;12:66. 10.1186/s12915-014-0066-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Merhej V, Georgiades K, Raoult D: Postgenomic analysis of bacterial pathogens repertoire reveals genome reduction rather than virulence factors. Brief Funct Genomics. 2013;12(4):291–304. 10.1093/bfgp/elt015 [DOI] [PubMed] [Google Scholar]
- 40. Merhej V, Raoult D: Rickettsial evolution in the light of comparative genomics. Biol Rev Camb Philos Soc. 2011;86(2):379–405. 10.1111/j.1469-185X.2010.00151.x [DOI] [PubMed] [Google Scholar]
- 41. Wolf YI, Koonin EV: Genome reduction as the dominant mode of evolution. Bioessays. 2013;35(9):829–37. 10.1002/bies.201300037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Treangen TJ, Rocha EP: Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7(1):e1001284. 10.1371/journal.pgen.1001284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Darwin C, Wallace A: On the Tendency of Species to form Varieties; and on the Perpetuation of Varieties and Species by Natural Means of Selection. Journal of the Proceedings of the Linnean Society of London. Zoology. 1858;3(9):45–62. 10.1111/j.1096-3642.1858.tb02500.x [DOI] [Google Scholar]
- 44. Dobzhansky T: Further Data on the Variation of the Y Chromosome in Drosophila Pseudoobscura. Genetics. 1937;22(3):340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lynch M, Conery JS: The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5. 10.1126/science.290.5494.1151 [DOI] [PubMed] [Google Scholar]
- 46. Ohno S: Evolution by Gene Duplication.Berlin, Heidelberg: Springer Berlin Heidelberg.1970. 10.1007/978-3-642-86659-3 [DOI] [Google Scholar]
- 47. Lynch M, Katju V: The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20(11):544–9. 10.1016/j.tig.2004.09.001 [DOI] [PubMed] [Google Scholar]
- 48. Takeuchi N, Kaneko K, Koonin EV: Horizontal gene transfer can rescue prokaryotes from Muller's ratchet: benefit of DNA from dead cells and population subdivision. G3 (Bethesda). 2014;4(2):325–39. 10.1534/g3.113.009845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muller HJ: The Relation of Recombination to Mutational Advance. Mutat Res. 1964;106(1):2–9. 10.1016/0027-5107(64)90047-8 [DOI] [PubMed] [Google Scholar]
- 50. Charlesworth D, Morgan MT, Charlesworth B: Mutation accumulation in finite outbreeding and inbreeding populations. Genet Res. 1993;61(1):39–56. 10.1017/S0016672300031086 [DOI] [Google Scholar]
- 51. Allen JM, Light JE, Perotti MA, et al. : Mutational meltdown in primary endosymbionts: selection limits Muller's ratchet. PLoS One. 2009;4(3):e4969. 10.1371/journal.pone.0004969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ku C, Nelson-Sathi S, Roettger M, et al. : Endosymbiotic gene transfer from prokaryotic pangenomes: Inherited chimerism in eukaryotes. Proc Natl Acad Sci U S A. 2015;112(33):10139–46. 10.1073/pnas.1421385112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Andersson JO: Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol. 2009;63:177–93. 10.1146/annurev.micro.091208.073203 [DOI] [PubMed] [Google Scholar]
- 54. Maslov S, Krishna S, Pang TY, et al. : Toolbox model of evolution of prokaryotic metabolic networks and their regulation. Proc Natl Acad Sci U S A. 2009;106(24):9743–8. 10.1073/pnas.0903206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bushman F: Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.2001. Reference Source [Google Scholar]
- 56. Claverys JP, Martin B, Polard P: The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev. 2009;33(3):643–56. 10.1111/j.1574-6976.2009.00164.x [DOI] [PubMed] [Google Scholar]
- 57. Johnston C, Polard P, Claverys JP: The DpnI/DpnII pneumococcal system, defense against foreign attack without compromising genetic exchange. Mob Genet Elements. 2013;3(4):e25582. 10.4161/mge.25582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Smith HO, Tomb JF, Dougherty BA, et al. : Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269(5223):538–40. 10.1126/science.7542802 [DOI] [PubMed] [Google Scholar]
- 59. Smith HO, Gwinn ML, Salzberg SL: DNA uptake signal sequences in naturally transformable bacteria. Res Microbiol. 1999;150(9–10):603–16. 10.1016/S0923-2508(99)00130-8 [DOI] [PubMed] [Google Scholar]
- 60. Frost LS, Koraimann G: Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 2010;5(7):1057–71. 10.2217/fmb.10.70 [DOI] [PubMed] [Google Scholar]
- 61. Carraro N, Burrus V: The dualistic nature of integrative and conjugative elements. Mob Genet Elements. 2015;5(6):98–102. 10.1080/2159256X.2015.1102796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Johnson CM, Grossman AD: Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu Rev Genet. 2015;49:577–601. 10.1146/annurev-genet-112414-055018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lang AS, Beatty JT: Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15(2):54–62. 10.1016/j.tim.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 64. Lang AS, Zhaxybayeva O, Beatty JT: Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol. 2012;10(7):472–82. 10.1038/nrmicro2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McDaniel LD, Young E, Delaney J, et al. : High frequency of horizontal gene transfer in the oceans. Science. 2010;330(6000):50. 10.1126/science.1192243 [DOI] [PubMed] [Google Scholar]
- 66. Wolf YI, Makarova KS, Yutin N, et al. : Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct. 2012;7:46. 10.1186/1745-6150-7-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Timmis JN, Ayliffe MA, Huang CY, et al. : Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5(2):123–35. 10.1038/nrg1271 [DOI] [PubMed] [Google Scholar]
- 68. Esser C, Ahmadinejad N, Wiegand C, et al. : A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21(9):1643–60. 10.1093/molbev/msh160 [DOI] [PubMed] [Google Scholar]
- 69. Martin W, Herrmann RG: Gene transfer from organelles to the nucleus: how much, what happens, and Why?. Plant Physiol. 1998;118(1):9–17. 10.1104/pp.118.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Martin W, Rujan T, Richly E, et al. : Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 2002;99(19):12246–51. 10.1073/pnas.182432999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nelson-Sathi S, Dagan T, Landan G, et al. : Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A. 2012;109(50):20537–42. 10.1073/pnas.1209119109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nelson-Sathi S, Sousa FL, Roettger M, et al. : Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature. 2015;517(7532):77–80. 10.1038/nature13805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Groussin M, Boussau B, Szollosi G, et al. : Gene Acquisitions from Bacteria at the Origins of Major Archaeal Clades Are Vastly Overestimated. Mol Biol Evol. 2016;33(2):305–10. 10.1093/molbev/msv249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lopez-Garcia P, Zivanovic Y, Deschamps P, et al. : Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol. 2015;13(7):447–56. 10.1038/nrmicro3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ku C, Nelson-Sathi S, Roettger M, et al. : Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524(7566):427–32. 10.1038/nature14963 [DOI] [PubMed] [Google Scholar]
- 76. Pittis AA, Gabaldon T: Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature. 2016;531(7592):101–4. 10.1038/nature16941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kurland CG, Collins LJ, Penny D: Genomics and the irreducible nature of eukaryote cells. Science. 2006;312(5776):1011–4. 10.1126/science.1121674 [DOI] [PubMed] [Google Scholar]
- 78. Poole A, Penny D: Eukaryote evolution: engulfed by speculation. Nature. 2007;447(7147):913. 10.1038/447913a [DOI] [PubMed] [Google Scholar]
- 79. Yutin N, Wolf MY, Wolf YI, et al. : The origins of phagocytosis and eukaryogenesis. Biol Direct. 2009;4:9. 10.1186/1745-6150-4-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Koonin EV, Yutin N: The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb Perspect Biol. 2014;6(4):a016188. 10.1101/cshperspect.a016188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Whitaker JW, McConkey GA, Westhead DR: The transferome of metabolic genes explored: analysis of the horizontal transfer of enzyme encoding genes in unicellular eukaryotes. Genome Biol. 2009;10(4):R36. 10.1186/gb-2009-10-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Richards TA, Talbot NJ: Horizontal gene transfer in osmotrophs: playing with public goods. Nat Rev Microbiol. 2013;11(10):720–7. 10.1038/nrmicro3108 [DOI] [PubMed] [Google Scholar]
- 83. Savory F, Leonard G, Richards TA: The role of horizontal gene transfer in the evolution of the oomycetes. PLoS Pathog. 2015;11(5):e1004805. 10.1371/journal.ppat.1004805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lawrence JG, Hendrickson H: Lateral gene transfer: when will adolescence end? Mol Microbiol. 2003;50(3):739–49. 10.1046/j.1365-2958.2003.03778.x [DOI] [PubMed] [Google Scholar]
- 85. Koonin EV: Horizontal gene transfer: the path to maturity. Mol Microbiol. 2003;50(3):725–7. 10.1046/j.1365-2958.2003.03808.x [DOI] [PubMed] [Google Scholar]