Abstract
Viral proteins mimic host protein structure and function to redirect cellular processes and subvert innate defenses1. Small basic proteins compact and regulate both viral and cellular DNA genomes. Nucleosomes are the repeating units of cellular chromatin and play an important role in innate immune responses2. Viral encoded core basic proteins compact viral genomes but their impact on host chromatin structure and function remains unexplored. Adenoviruses encode a highly basic protein called protein VII that resembles cellular histones3. Although protein VII binds viral DNA and is incorporated with viral genomes into virus particles4,5, it is unknown whether protein VII impacts cellular chromatin. Our observation that protein VII alters cellular chromatin led us to hypothesize that this impacts antiviral responses during adenovirus infection. We found that protein VII forms complexes with nucleosomes and limits DNA accessibility. We identified post-translational modifications on protein VII that are responsible for chromatin localization. Furthermore, proteomic analysis demonstrated that protein VII is sufficient to alter protein composition of host chromatin. We found that protein VII is necessary and sufficient for retention in chromatin of members of the high-mobility group protein B family (HMGB1, HMGB2, and HMGB3). HMGB1 is actively released in response to inflammatory stimuli and functions as a danger signal to activate immune responses6,7. We showed that protein VII can directly bind HMGB1 in vitro and further demonstrated that protein VII expression in mouse lungs is sufficient to decrease inflammation-induced HMGB1 content and neutrophil recruitment in the bronchoalveolar lavage fluid. Together our in vitro and in vivo results show that protein VII sequesters HMGB1 and can prevent its release. This study uncovers a viral strategy in which nucleosome binding is exploited to control extracellular immune signaling.
Keywords: Adenovirus, chromatin, innate immunity, HMGB1
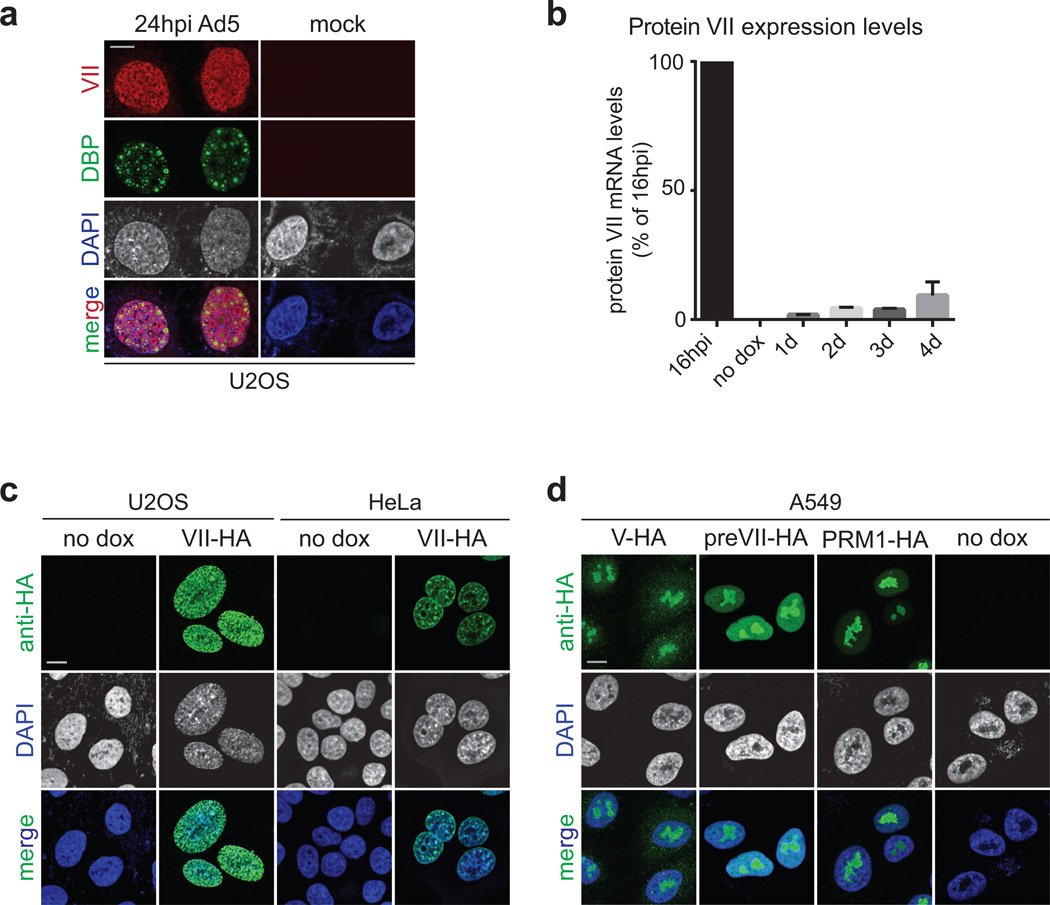
As viruses commandeer cellular functions to promote viral production, they induce numerous cellular changes. Manipulation of host chromatin is important for viral takeover of cellular functions1,8–11. Although there are known examples of viral control by manipulating gene expression2,9,12, an alternative strategy for immune evasion could exploit cellular chromatin to impact extracellular signaling. Genomes of DNA viruses are compacted and packaged into virus particles with small basic proteins encoded by host or virus. Adenoviruses encode protein VII, a small basic protein packaged with viral genomes3–5. We hypothesized that protein VII contributes to host chromatin manipulation. We investigated protein VII localization during infection, and found it present at both viral replication centers stained for viral DNA binding protein DBP (Fig 1a, Extended Data Fig 1a), and cellular chromatin stained for histone H1 and DAPI (Fig 1b). These observations suggest protein VII functions on both viral and host genomes. To determine protein VII’s impact on cellular chromatin, we generated cell lines with inducible expression. In multiple cell types we observed that protein VII accumulation altered nuclear DNA into a punctate appearance (Fig 1c and Extended Data Fig 1b–c). We tested whether other basic proteins produce similar effects on chromatin. Viral core protein V and the precursor of protein VII (preVII) localized to nucleoli and did not affect chromatin appearance (Extended Data Fig 1d). Human protamine PRM1, a basic protein involved in sperm DNA compaction13, also localized to nucleoli and did not affect chromatin appearance (Extended Data Fig 1d). Taken together, our data demonstrate that protein VII is sufficient to alter cellular chromatin and is distinct from other small basic proteins.
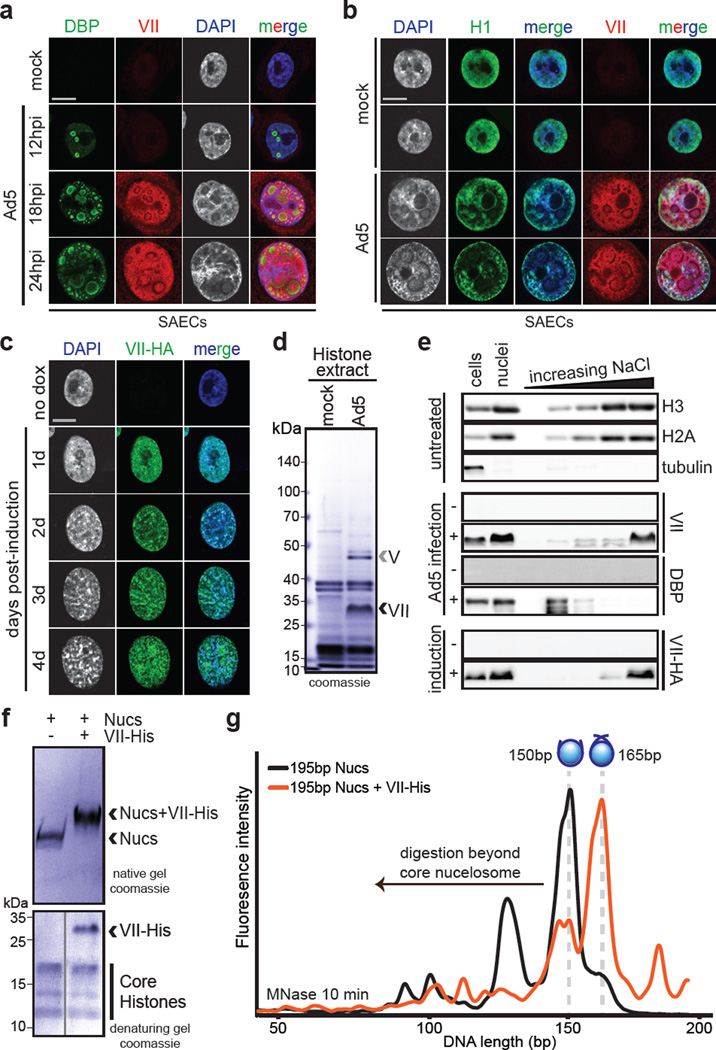
Figure 1. Protein VII is sufficient to alter chromatin and directly binds nucleosomes.
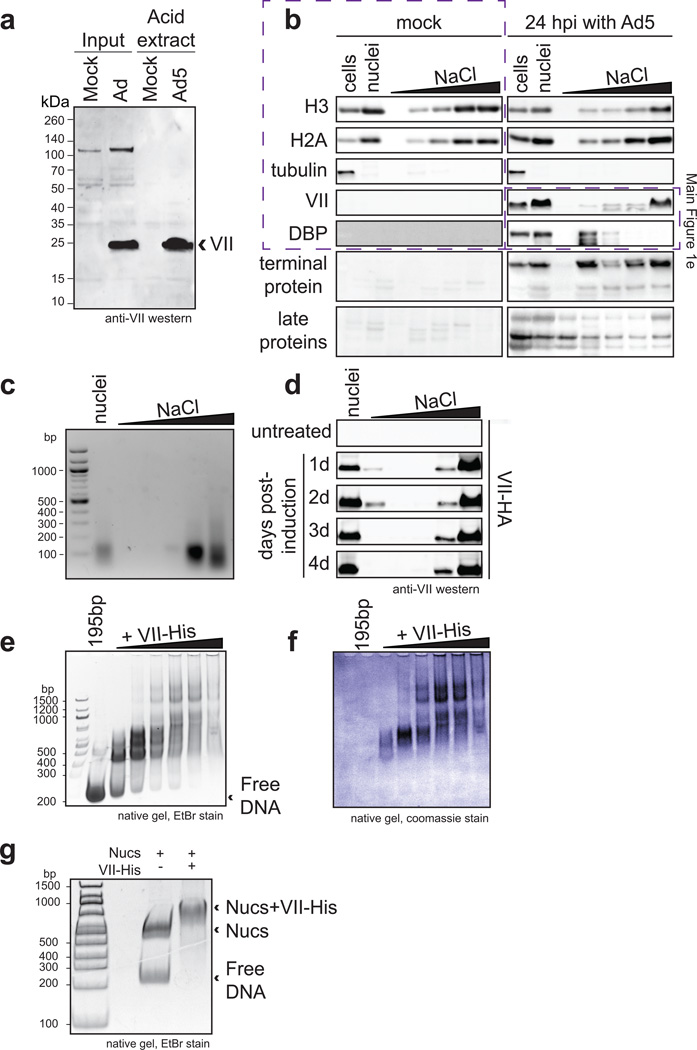
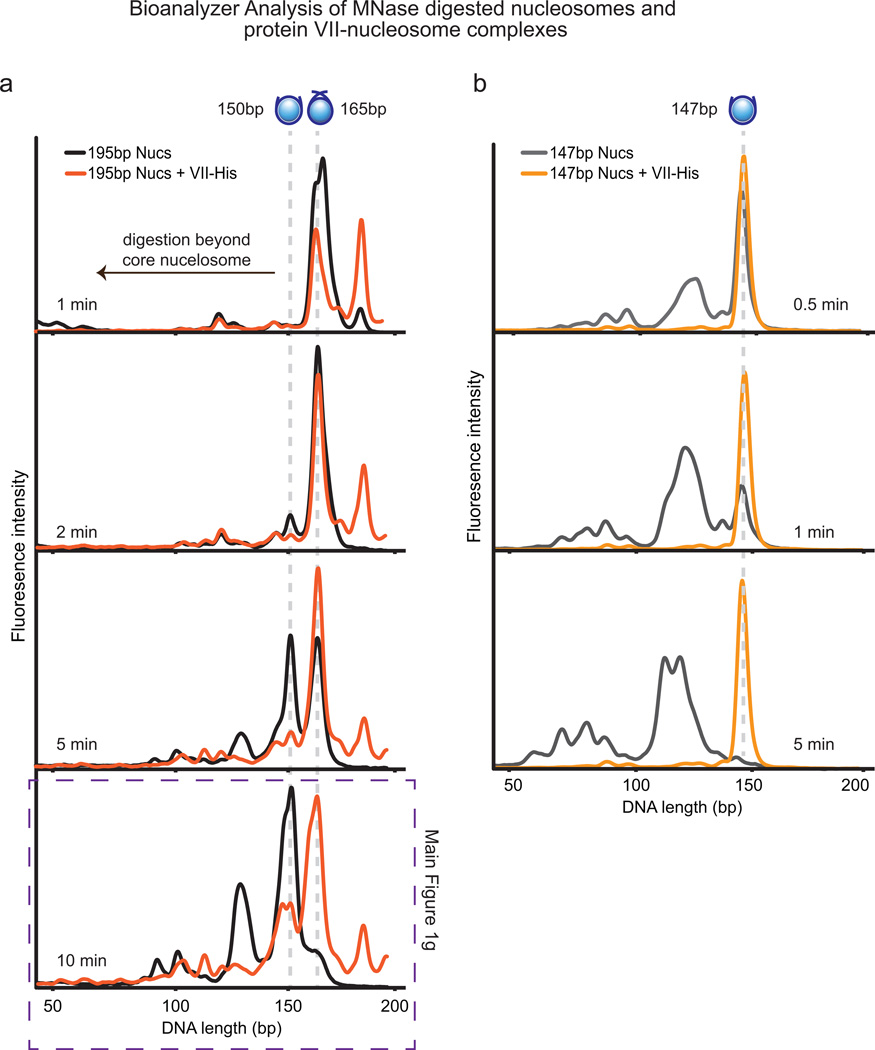
a–b Ad5-infected SAECs stained for protein VII (red) with DBP (a), or histone H1 (b), and DAPI (grey, blue in merge). c, Protein VII-HA induced cells over four days showing HA (green) and DAPI (grey, blue in merge). d, SDS-PAGE of histone extract from Ad5-infected cells showing protein V and protein VII. e, Western blot of chromatin fractionation from nuclei of Ad5-infected cells, induced for protein VII-HA, or untreated. f, Protein VII binds to nucleosomes. Protein bands from native gel stained with coomassie (top) were subjected to 2D analysis by SDS-PAGE (bottom). g, Protein VII protects nucleosome complexes from MNase digestion. Bioanalyzer curves represent nucleosomes alone (black) or protein VII-nucleosome complexes (orange).
To affect cellular chromatin at the nucleosome level during infection, we reasoned protein VII must be abundant and associated with histones. Acid extraction of histones14,15 from infected cells, revealed viral proteins VII and V isolated with cellular histones (Fig 1d), as verified by western blot (Extended Data Fig 2a) and mass spectrometry. Protein VII abundance was comparable to cellular histone levels (Fig 1d). We further analyzed association of protein VII with cellular chromatin by salt fractionation of nuclei16. We found protein VII with cellular histones and DNA in high salt fractions (Fig 1e and Extended Data Fig 2b–d). Ectopically expressed protein VII is also found in high salt fractions, in contrast to other viral proteins that elute at low salt (Fig 1e and Extended Data Fig 2b). These data suggest that protein VII is highly abundant and tightly associated with cellular chromatin.
We hypothesized protein VII interacts with chromatin by forming complexes with DNA, histones, or nucleosomes, and examined protein VII interactions in vitro. Purified recombinant protein VII binds to DNA5 (Extended Data Fig 2e–f). We reconstituted nucleosomes in vitro with recombinant histone proteins on 195 base pairs (bp) of DNA17. Protein VII changed nucleosome mobility upon native gel electrophoresis (Fig 1f and Extended Data Fig 2g). We analyzed native gel bands by denaturing SDS-PAGE, and confirmed complexes contained core histones with protein VII (Fig 1f, bottom). Unlike protamines13, protein VII forms complexes with nucleosomes but does not appear to replace histones. Next, we examined whether protein VII association with nucleosomes affects DNA wrapping using microccocal nuclease (MNase) digestion followed by DNA fragment analysis17. We found protein VII pauses nucleosomal DNA digestion at ~165bp, the point at which DNA strands crossover the nucleosome dyad (Fig 1g, Extended Data Fig 3a). In contrast, nucleosome digestion alone paused with core particles at ~150bp, suggesting protein VII encumbers DNA access. Unlike linker histone binding that is dependent on DNA length18, protein VII protects against MNase digestion on the nucleosome core particle of 147bp (Extended Data Fig 3b). Protein VII alone protects DNA from MNase digestion, as would be expected given its role in the viral core. Together, these data demonstrate protein VII binds directly to nucleosomes and limits DNA accessibility at the DNA entry/exit site.
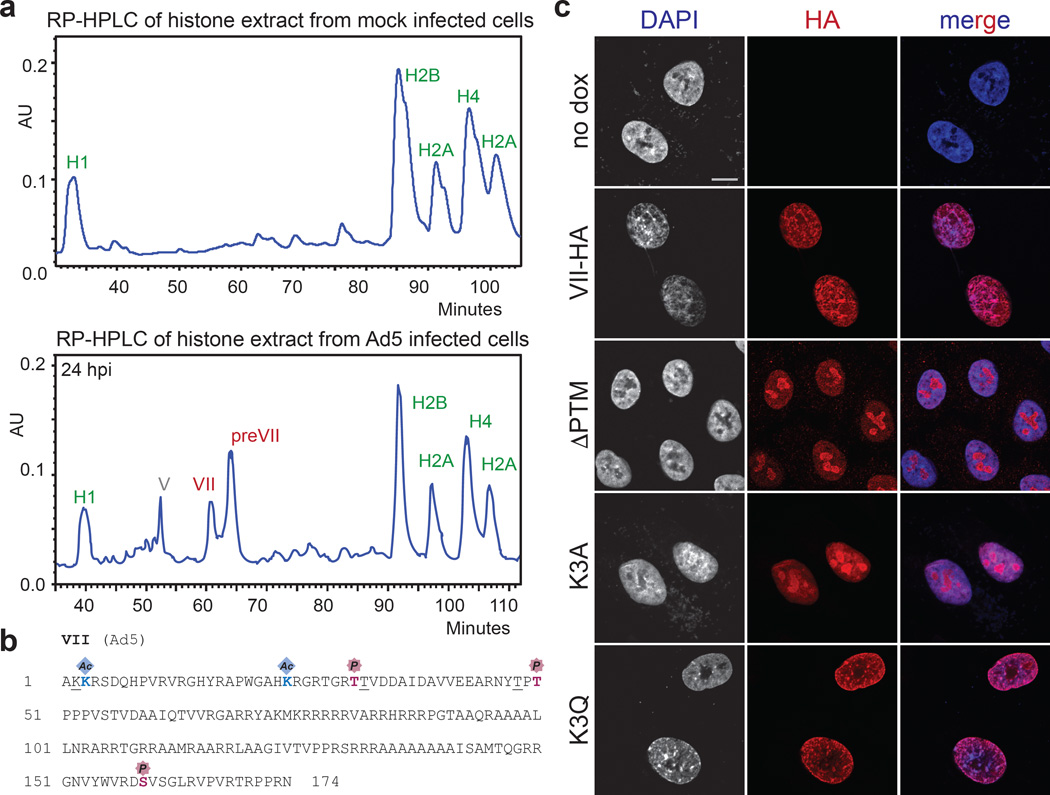
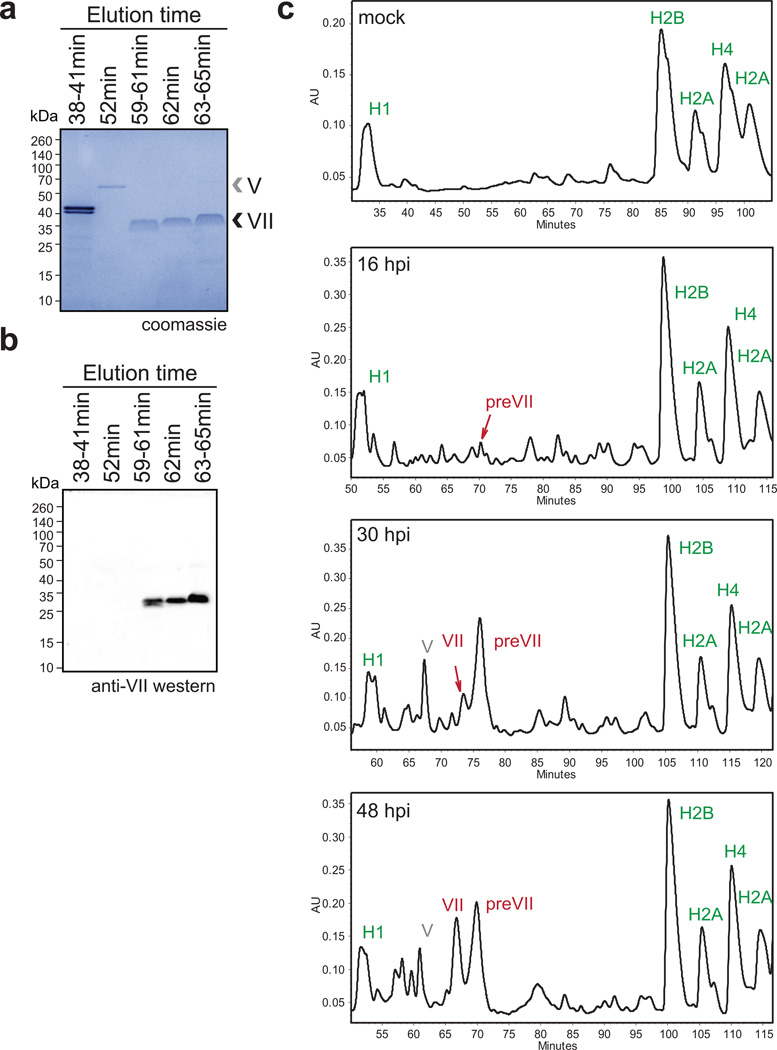
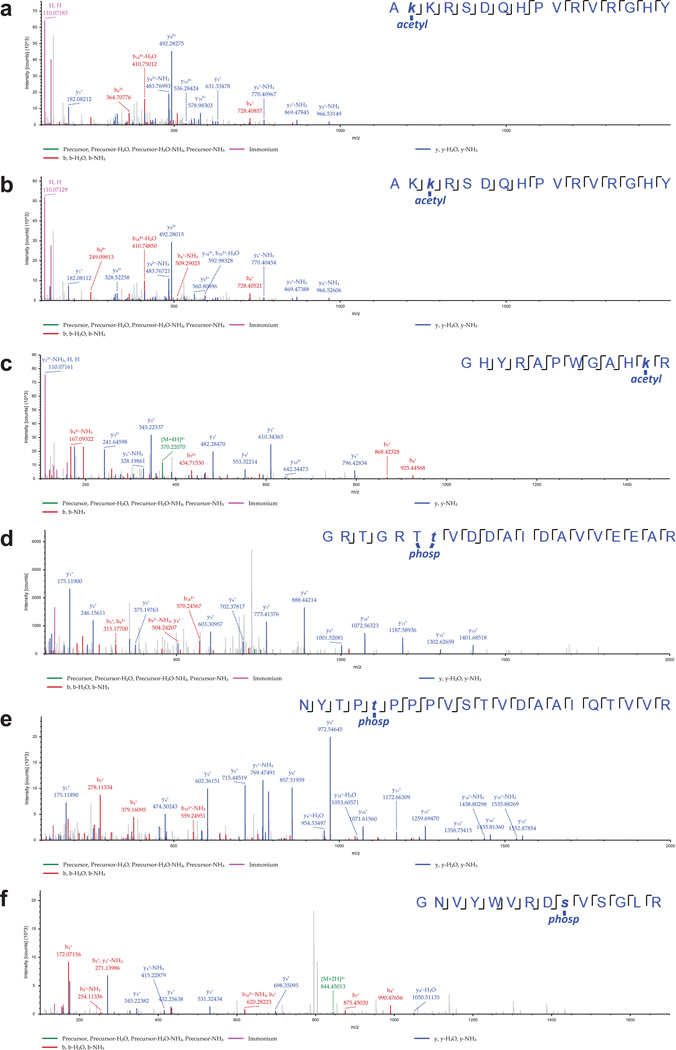
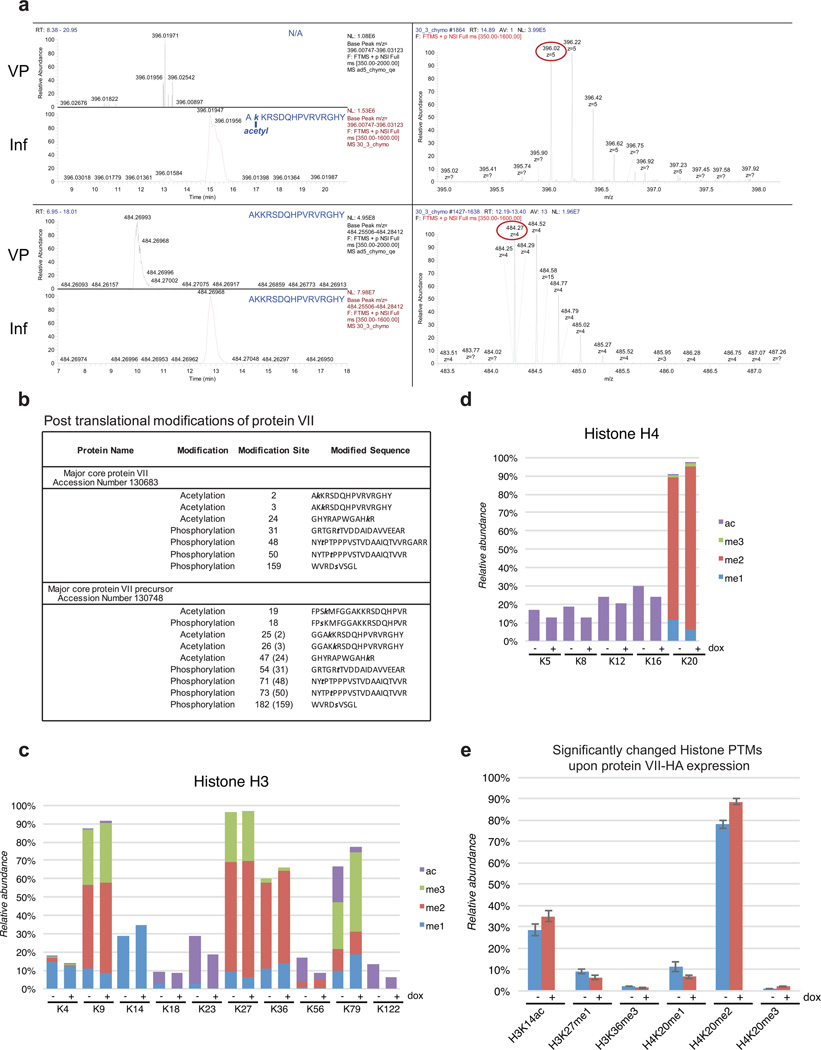
Post-translational modifications (PTMs) on histones are central to regulating chromatin structure14,19. Due to the histone-like nature of protein VII3, we hypothesized it is subject to post-translational modification similar to histones. Protein VII precursor was previously proposed to be acetylated by amino-terminal addition during protein synthesis20. We noted that protein VII contains conserved lysine residues within an AKKRS motif21, similar to the commonly modified canonical histone motif ARSK19. We therefore purified protein VII from histone extracts over an adenovirus infection time-course by reverse phase HPLC (Fig 2a, Extended Data Fig 4). Consistent with observations from histone extracts (Fig 1d), protein VII levels were comparable to endogenous histones. We digested purified protein VII and preVII with chymotrypsin to distinguish the two proteins, and analyzed peptides by tandem mass spectrometry. We identified several PTMs, with two acetylation sites and three phosphorylation sites as most abundant (Fig 2b, Extended Data Fig 5 and 6b). Interestingly, we identified acetylation sites on ectopically expressed protein VII but not on protein VII in virus particles (Extended Data Fig 6a). We speculate this provides a possible mechanism for distinguishing protein VII bound to cellular chromatin from protein destined for packaged virus. To investigate relevance of identified PTMs, we mutated modified sites in protein VII. An alanine-replacement mutant for all five PTM sites localized to nucleoli instead of cellular chromatin (Fig 2c). Results with individual point mutations suggest the K3 residue is important for chromatin localization, and employing glutamine as an acetylation mimic (K3Q) mirrored the pattern of wild-type protein (Fig 2c). Effects induced by protein VII are not due to global alteration of histone PTMs since only six PTMs on histones H3 and H4 showed minor but significant changes (Extended Data Fig 6c–e and SI Table 1). These data suggest protein VII modification plays critical roles during virus infection.
Figure 2. Post-translational modifications on protein VII contribute to chromatin localization.
a, RP-HPLC analysis of histone extracts. Viral proteins V, VII and preVII are indicated at 24 hpi. b, Primary sequence of protein VII with modified residues identified in infected cells. Underlined residues represent moieties that may also be modified in identified peptides (see ED). c, Immunofluoresence showing DAPI (grey, blue in merge) and protein VII (red) as wild-type or with alanine substitutions at PTM sites (ΔPTM), K3A or K3Q.
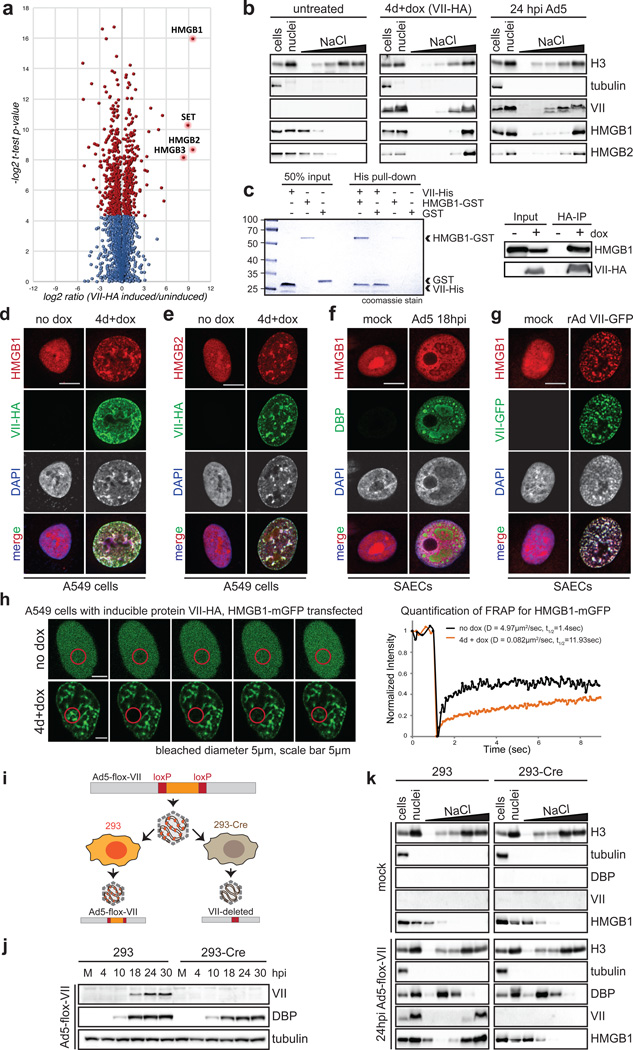
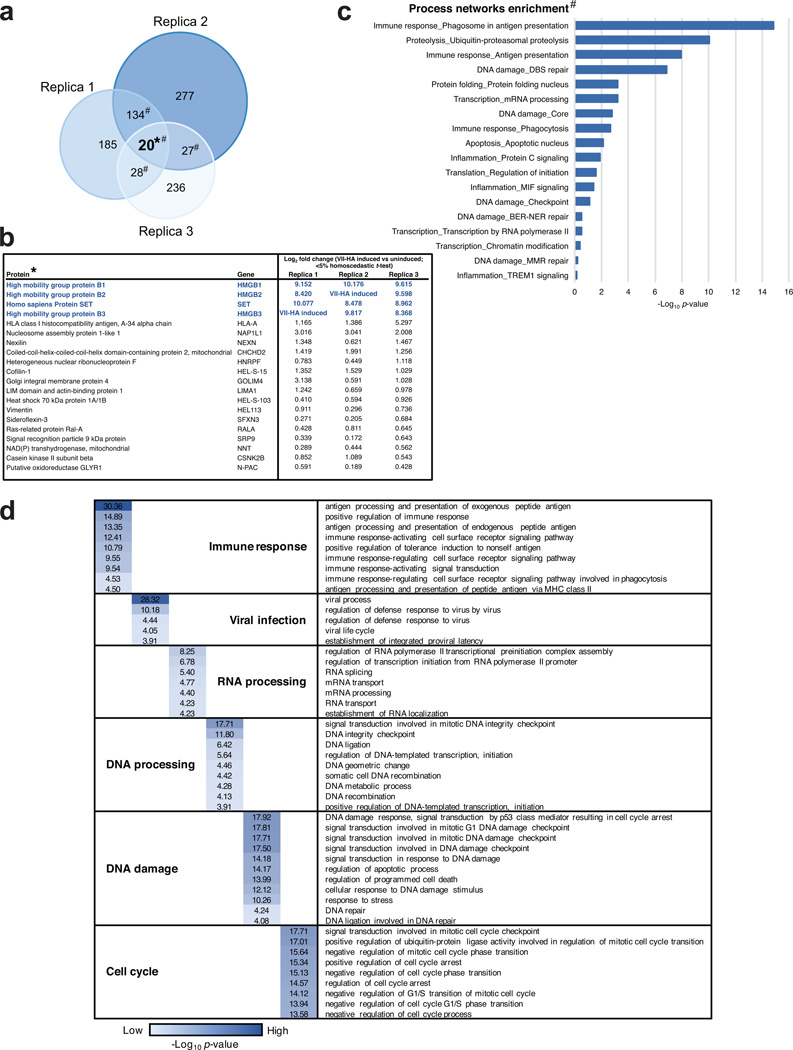
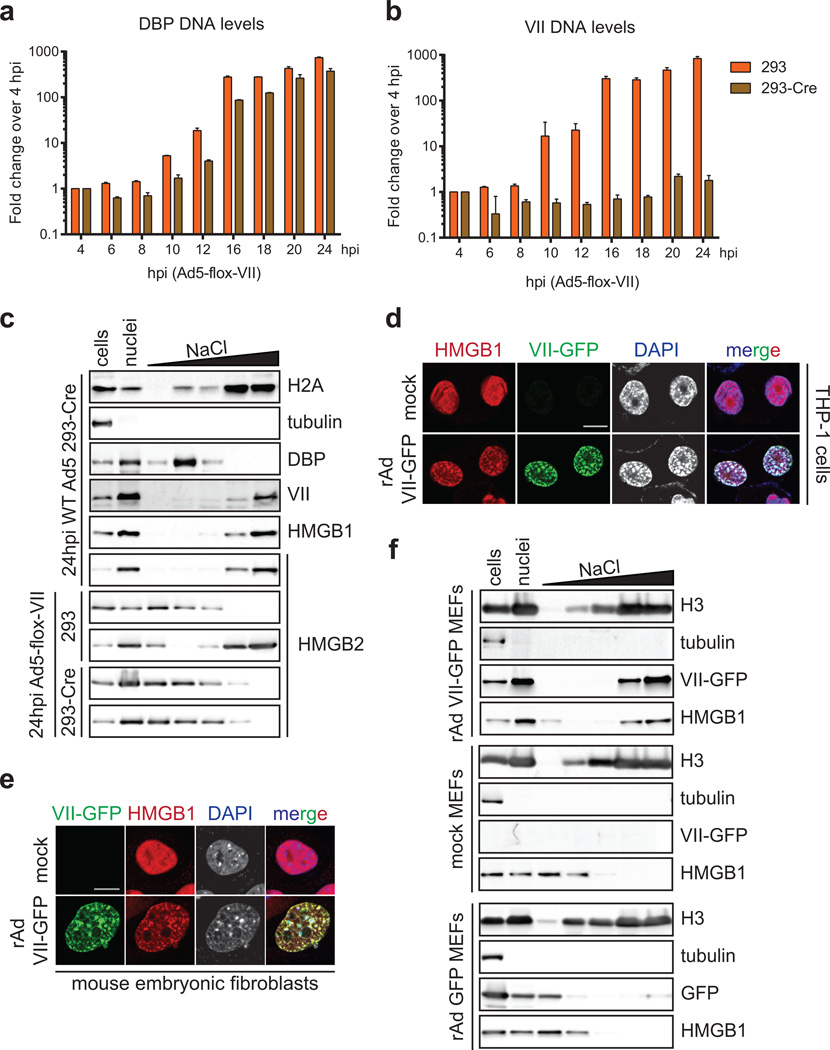
To determine whether protein VII manipulation of cellular chromatin is part of a strategy to counteract host defenses, we employed mass spectrometry to examine changes in protein composition of nuclear fractions. We compared the total chromatin proteome in the presence and absence of protein VII (Fig 3a and SI Table 2). We identified 20 proteins that changed significantly across three biological replicates (Extended Data Fig 7 and SI Table 2). The categories of proteins most significantly changed upon protein VII expression were related to immune responses (Extended Data Fig 7c). The top four proteins enriched in chromatin fractions by protein VII were SET/TAF-1, a protein previously shown to interact with protein VII22,23, and high mobility group box proteins HMGB1, HMGB2 and HMGB3 (Fig 3a). The HMGB proteins are alarmins with multiple functions as activators of immunity and inflammation6,7. HMGB1 is a nuclear protein normally only transiently associated with chromatin24,25. Cells also release HMGB1 as an extracellular danger signal that promotes immune responses after injury or infection26. We confirmed increased chromatin association of HMGB1 and HMGB2 by analysis of fractionated nuclei, upon protein VII expression and during adenovirus infection (Fig 3b). We verified these changes are not due to altered HMGB1 expression levels (Extended Data Fig 8a–b). We demonstrated direct binding of recombinant protein VII to HMGB1 in vitro and confirmed HMGB1 co-immunopreciptation with protein VII (Fig 3c). We visually observed reorganization of HMGB1 and HMGB2 distribution upon protein VII expression, and at late stages of infection (Fig 3d–f, Extended Data Fig 8c–e). We also showed reorganization of HMGB1 distribution by vector transduction to express protein VII-GFP (Fig 3g, Extended Data 8f). The effect of protein VII on HMGB1 is also conserved across human Ad serotypes (Extended Data Fig 8g). We further defined effects of protein VII on HMGB1 mobility by fluorescence recovery after photobleaching (FRAP) and found decreased HMGB1 diffusion (Fig 3h). We next investigated whether protein VII is necessary for chromatin retention of HMGB1 during virus infection. We used a replication competent adenovirus with loxP sites inserted on either side of the protein VII gene, allowing deletion of protein VII during infection of cells expressing Cre recombinase (Fig 3i–j, Extended Data Fig 9a–b). We fractionated nuclei from infected cells and found HMGB1 and HMGB2 were no longer retained in chromatin when protein VII was deleted (Fig 3k, Extended Data Fic 9c). Together, these data indicate that protein VII is necessary and sufficient to promote chromatin association and immobilization of HMGB1.
Figure 3. Protein VII directly binds HMGB1 and is necessary for retention of the alarmin in cellular chromatin.
a, Volcano plot for proteomics analysis of one representative biological replicate of high salt fraction. The y-axis represents −log2 statistical p-value and x-axis represents log2 protein fold-change between uninduced or protein VII expressing cells (homoscedastic two-tailed t-test, p<0.05 red dots; ntechnical=3). b, Nuclear fractionation shows HMGB1 and HMGB2 normally elute from nuclei at low salt concentrations but are retained in high salt fractions by protein VII-HA. c, Protein VII interacts with HMGB1 in pull-down of recombinant HMGB1-GST (left, coomassie-stained SDS-PAGE) and immunoprecipitation of HMGB1 (right, western blots). d–e, Protein VII expression alters localization of HMGB1 (d) and HMGB2 (e). Immunofluorescence shows protein VII-HA (green) co-localized with HMGB1 (d) and HMGB2 (e) in cellular chromatin, DAPI (grey, blue in merge). f, Same as (d) at 18 hpi with Ad5 DBP (green). g, Protein VII-GFP relocalizes HMGB1 (red) to chromatin with DAPI (grey, blue in merge). h, FRAP experiment with HMGB1-mGFP. Recovery of FRAP signal in time-course images (left) with quantification and diffusion coefficients (right). i, Schematic showing loxP strategy for deleting protein VII. j, Western blots comparing 293 and 293Cre cells infected with Ad5-flox-VII virus. k, Salt fractionation in nuclei from (j).
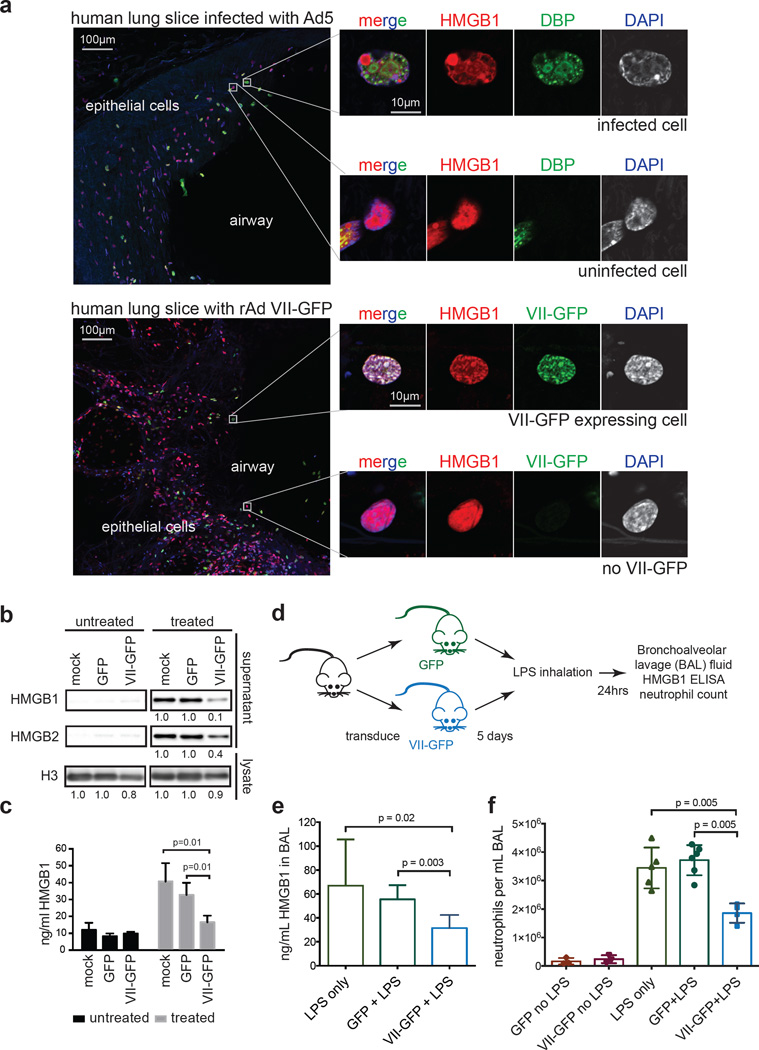
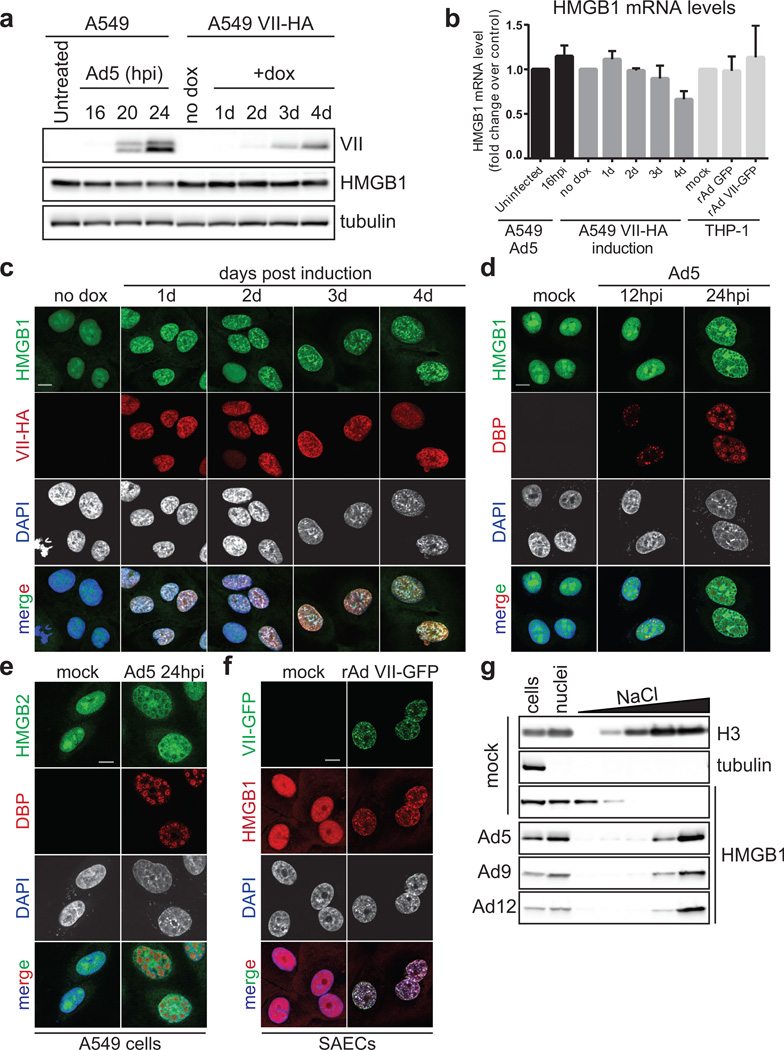
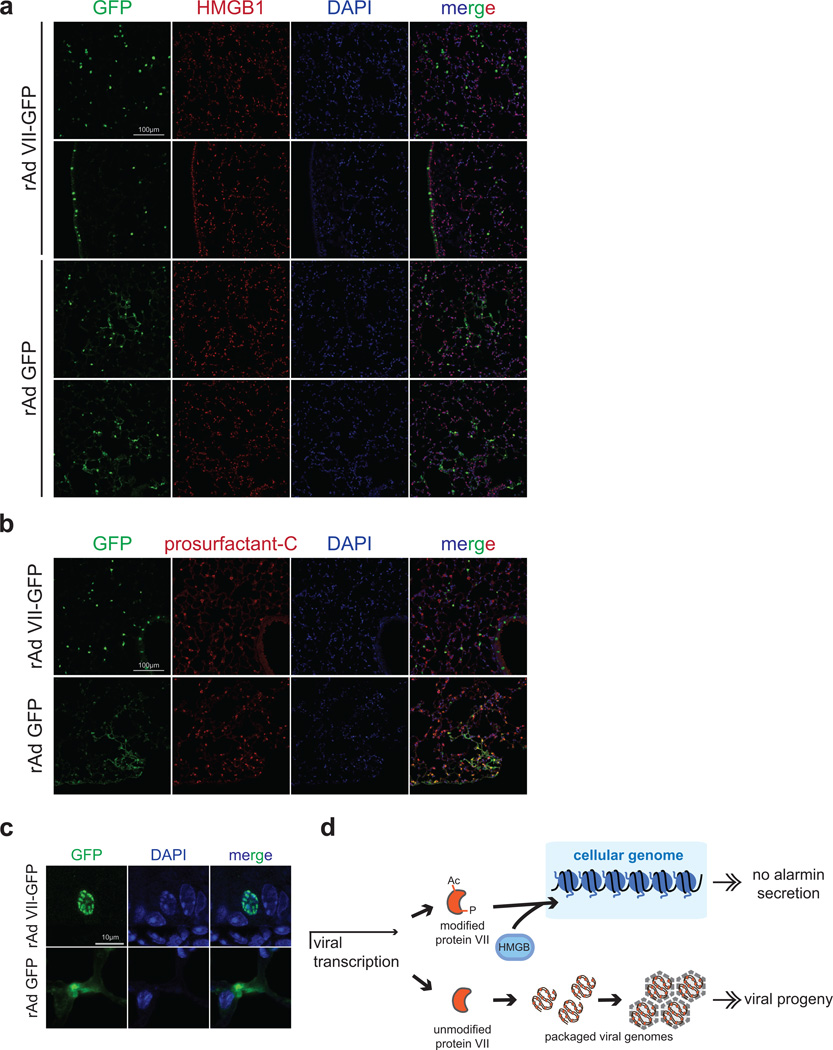
We hypothesized that protein VII retains HMGB1 in chromatin during natural infection to prevent cellular release and abrogate host immune responses. We therefore visualized endogenous HMGB1 during adenovirus infection in precision cut lung slices (PCLS)27 from human donors (Fig 4a). Consistent with cell culture experiments, we demonstrate that protein VII is sufficient to relocalize endogenous HMGB1. We then tested whether protein VII prevents HMGB1 release in cell culture and in vivo models. We expressed GFP or protein VII-GFP in macrophage-like THP-1 cells, and confirmed protein VII-GFP was sufficient to alter chromatin and HMGB1 localization (Extended Data Fig 9d). Cells were treated to stimulate inflammasomes, and HMGB1 analyzed in supernatants. Protein VII expression resulted in reduced levels of HMGB1 and HMGB2 in supernatants (Fig 4b–c). Subsequently, we employed a murine model of LPS induced lung injury28 to investigate protein VII’s impact on HMGB1 release and neutrophil recruitment in vivo (Fig 4d). We confirmed that protein VII was expressed in transduced mouse lungs (Extended Data Fig 10a–c) and retained mouse HMGB1 (Extended data Fig 9e–f). We exposed mice to inhaled LPS to induce HMGB1 release and neutrophil recruitment to alveoli. Bronchoalveolar lavage (BAL) fluid obtained 24 hours after LPS exposure showed that mice transduced to express protein VII had significantly less HMGB1 and fewer neutrophils than mice expressing GFP (Fig 4d–f). Together, these data suggest that protein VII functions in cellular chromatin to retain HMGB1 as a mechanism to blunt immune responses.
Figure 4. Protein VII prevents HMGB1 release.
a, Precision cut lung slices (PCLS) infected with Ad5 or transduced to express protein VII-GFP. Endogenous HMGB1 (red) is redistributed in cells with virus (DBP in top) and VII-GFP (bottom). b, Protein VII-GFP is sufficient to inhibit HMGB1 and HMGB2 release in THP-1 cells. Numbers indicate relative intensities of bands quantified with ImageJ. c, ELISA-based quantification of HMGB1 in supernatants from (b), error bars=±s.d., ntechnical=4, homoscedastic one-tailed t-test. d, Schematic for investigating protein VII in a mouse lung injury model. e, Expression of protein VII-GFP decreases HMGB1 in mouse BAL fluid as quantified by ELISA, error bars=±s.d., biological replicates: nLPS=4, nGFP+LPS=6, nVII-GFP+LPS=7, homoscedastic one-tailed (p=0.02) or two-tailed (p=0.003) t-test. f, Neutrophils in BAL fluid are significantly fewer in mice expressing protein VII-GFP, error bars=±s.d., biological replicates: nGFP+LPS=6, nVII-GFP+LPS=4, nLPS=5, nGFP=3, nVII-GFP=3 homoscedastic two-tailed t-test.
In summary, in addition to known roles on packaged viral DNA29,30, we show that protein VII interacts with cellular chromatin and binds nucleosomes. We suggest that protein VII PTMs contribute to chromatin localization, and that protein VII impacts chromatin-association of host proteins. Finally, we show that protein VII in cellular chromatin leads to sequestration of HMGB family members, contributing to abrogated immune responses (Extended Data Fig 10d). Our study reveals that chromatin retention of signaling molecules by a viral protein may represent a previously unrecognized immune evasion strategy.
Methods
Cells
Primary small airway epithelial cells (SAECs), U2OS, HeLa, 293, THP-1 and A549 cells were obtained from the American Tissue Culture Collection (ATCC) and grown according to the provider’s instructions. Cell lines were not authenticated or tested for mycoplasma. Acceptor cells for generation of inducible cell lines were kindly provided by E. Makeyev and used as previously reported31. Protein VII, preVII and V were cloned from genomic DNA isolated from HeLa cells infected with adenovirus type 5 and inserted into the inducible plasmid cassette with a C-terminal HA tag using restriction enzymes BsrGI and AgeI (primer sequences available upon request). Positive clones were selected in DH5α cells, sequenced, and transfected into A549, U2OS or HeLa acceptor cells along with plasmid expressing the Cre recombinase. Recombined clones were selected by puromycin resistance (1 µg/mL) and induced with doxycycline (0.2 µg/mL) to express the desired protein. Protein expression was verified by immunofluorescence and western blot. All figures shown are after 4 days of induction unless otherwise stated. Protein VII and preVII were also verified by HPLC purification and mass spectrometry analysis. Point mutations were generated by gene synthesis from Genewiz. 293-Cre cells were provided by from P. Hearing.
Viruses and infections
Wild-type adenovirus type 5 (Ad5), adenovirus type 9 (Ad9), adenovirus type 12 (Ad12), and recombinant adenovirus vectors expressing only GFP were propagated in 293 cells as previously described32. Recombinant adenovirus vector with VII-GFP replaced in the E1 region was a kind gift from D. Curiel33. Infections were carried out as described previously34 using a multiplicity of infection of 10 for primary cells and cell lines for Ad5 infections. Ad9 and Ad12 infections were carried out with a multiplicity of infection of 50 and 20, respectively. Ad5-flox-VII was generated by P. Hearing and also prepared using standard methods in 293 cells. LoxP sites were added flanking protein VII in the Ad5 genome resulting in protein VII deletion during infection of 293 cells expressing Cre recombinase.
Antibodies
Primary antibodies were purchased from Covance (HA MMS-101R), Abcam (H1 ab4269, H3 ab1791, HMGB1 ab18256, HMGB2 ab67282), Millipore (H2A 07-146, prosurfactin-C AB3786), and Santa Cruz (Ku86 sc5280, tubulin sc69969). The antibodies to DBP, adenoviral late proteins, terminal protein and protein VII were kind gifts from A. Levine35, J. Wilson32, R. Hay and L. Gerace, respectively. Secondary antibodies for immunoblotting were obtained from Jackson ImmunoResearch and secondary antibodies for immunofluorescence were obtained from Life Technologies.
Immunofluorescence
Cells were grown on glass coverslips in 24-well plates and either infected or induced with doxycycline (0.2 µg/mL). Cells were harvested for immunofluorescence at the indicated time points, washed in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 15 minutes and post-fixed with 100% ice-cold methanol for 5 minutes. Coverslips were then blocked and stained as previously described36 and mounted using ProLong Gold Antifade Reagent (Life Technologies). Immunofluorescence was visualized using a Zeiss LSM 710 Confocal microscope (Cell and Developmental Microscopy Core at UPenn) and ZEN 2011 software. Images were processed using ImageJ and assembled with Adobe CS6. All scale bars show in the figures are 10µm unless otherwise stated.
Immunoblotting
Western blot analysis was carried out using standard methods. Briefly, equal amounts of total protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Millipore) for at least 30 minutes at 30V. Membranes were stained with ponceau to confirm protein loading and blocked in 5% milk in TBST containing 0.1% azide. Membranes were incubated with primary antibodies overnight, washed for 30 minutes in TBST and incubated with secondary antibodies conjugated to horseradish peroxidase (Jackson Laboratories) for 1 hour. Membranes were washed again and proteins were visualized with Pierce ECL Western Blotting Substrate (Thermo Scientific) and detected using a Syngene G-Box.
Mice
All mice were housed in SPF conditions in an animal facility at the Children’s Hospital of Philadelphia. All studies in mice were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the Institutional Animal Care and Use Committee, Children's Hospital of Philadelphia Animal Welfare Assurance Number A3442-01. C57BL/6J male mice aged 8–10 weeks were used for experiments. Mice were sedated with ketamine and xylazine. Once sedated, mice underwent orotrachial intubation, as previously described37, with a 20G angiocatheter from BD (Franklin Lakes, NJ). Mice subsequently received 5e10 GC of recombinant adenovirus expressing VII-GFP or GFP purified by the Penn Vector Core as described above. Four days after infection, mice were exposed to aerosolized LPS, 3 mg/mL for 30 minutes as previously described38. One day after LPS exposure, BAL, and lung tissue were harvested as previously detailed39 and examined for HMGB1 content (ELISA, Chondrex 6010) and neutrophil count (hematoxylin and eosin stain kit EMD 65044/93). Immunostaining was carried out by the CHOP Pathology Core using standard methods. A minimum of four biological replicates were used for each condition studied. Mice were assigned a random number and color at the start of the experiment and were randomized. Technicians carrying out the experiments were blinded to the identity of the samples. Tissue samples were assigned a random study number such that the technician performing the analysis was blinded. Unblinding for the purpose of data analysis occurred only after all data had been collected.
Salt fractionation of nuclei
Salt fractionation of nuclei was adapted from established protocols16,40. Briefly, 2–4 × 107 cells were collected and resuspended in 2 mL of ice-cold buffer I (0.32 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 15 mM Tris, pH 7.5, 0.5 mM DTT, 0.1 mM PMSF and protease inhibitor cocktail from Roche). To dissolve the plasma membrane, 2 mL ice-cold buffer I supplemented with 0.1% IGEPAL were added and samples were incubated on ice for 10 minutes. The 4 mL of nuclei was layered on 8 mL of ice-cold buffer II (1.2 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 15 mM Tris, pH 7.5, 0.5 mM DTT, 0.1 mM PMSF and protease inhibitor cocktail from Roche) and centrifuged for 20 minutes at 10,000 × g and 4°C. The pelleted nuclei were resuspended in 400 µL buffer III (10 mM Tris pH 7.4, 2 mM MgCl2, 0.1 mM PMSF) supplemented with 5mM CaCl2 and the DNA was digested to mononucleosomes by addition of 1 unit of MNase (Sigma-Aldrich, N3755). The reaction was incubated at 37°C for 30 minutes and then stopped by addition of 25 µL of 0.1M EGTA. The samples were centrifuged for 10 minutes, 350 × g, at 4°C, and supernatants were set aside for western blot analysis. The pellet was resuspended in 400 µL of buffer IV (70 mM NaCl, 10 mM Tris pH 7.4, 2 mM MgCl2, 2 mM EGTA, 0.1% Triton X-100, 0.1 mM PMSF) with 80mM salt and rotated for 30 minutes at 4°C. The sample was centrifuged for 10 minutes at 350 × g, 4°C, and the supernatant collected for western blot analysis. This step was repeated for salt concentrations in buffer IV of 150 mM, 300 mM and 600 mM. The final pellet was resuspended in 400 µL ddH2O and all samples were analyzed together by western blot. An aliquot of each supernatant was set aside for DNA purification using a PCR purification kit (Qiagen) and analyzed by agarose gel electrophoresis. Alternatively, 4 × 107 cells were resuspended in 400 µL hypotonic buffer (10 mM HEPES pH = 7.9, 1.5 mM MgCl2, 10 mM KCl, 1:1000 PMSF, 0.5 mM DTT) and incubated on ice for 30 min. The cells were transferred to a 1 ml dounce tissue grinder and the cell membranes were gently disrupted with 40 strokes of a tight-fitting pestle. The samples were centrifuged for 5 min at 1,500g and 4°C. The pelleted nuclei were resuspended in 400 µL buffer III and the fractionation was continued as described above.
Preparation of salt fractions for mass spectrometry analysis
All chemicals used for preparation of mass spectrometry samples were of at least sequencing grade and purchased from Sigma-Aldrich (St Louis, MO), unless otherwise stated. Only the 600 mM salt fraction was used for LC-MS/MS analysis. The 0.1% TritonX-100 detergent was removed from samples prior MS analysis by precipitation using chloroform (CHCl3)-methanol (MeOH) precipitation41. The protein pellet from CHCl3-MeOH precipitation was resuspended in 6 M urea and 2 M thiourea in 50 mM ammonium bicarbonate. Samples were reduced with 10mM DTT for 1 hour at room temperature and the carbamidomethylated with 20 mM iodoacetamide for 30 minutes at room temperature in the dark. After alkylation proteins were digested first with endopeptidase Lys-C (Wako, mass spectrometry grade) for 3 hours, after which the solution was diluted 10 times with 20 mM ammonium bicarbonate. Subsequently, samples were digested with trypsin (Promega) at an enzyme to substrate ratio of approximately 1:50 for 12 hours at room temperature. The samples were acidified with 5% formic acid (FA) to a pH ≤ 3 and desalted using Poros Oligo R3 RP columns (PerSeptive Biosystems) packed in a P200 stage tip with C18 3M plug (3M Bioanalytical Technologies). Purified peptide samples were dried by lyophilization and stored at −20°C until further analysis. This procedure was carried out for three biological replicas.
Nano LC-MS/MS and analysis of salt fractions
Samples were loaded onto a 16 cm C18–AQ column (inner diameter 75 µm, 3 µm beads, Dr, Maisch GmbH, Germany) using an Easy nano-flow HPLC system (Thermo Fisher Scientific, Odense, Denmark). The nanoLC was coupled to an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher Scientific, San Jose, CA) via a nanoelectrospray ion source (Thermo Fisher Scientific, San Jose, CA). Peptides were loaded in buffer A (0.1% formic acid) and eluted with a 120 minute linear gradient from 2–30% buffer B (95% acetonitrile, 0.1% formic acid). After the gradient, the column was washed with 90% buffer B. Mass spectra were acquired using a data-dependent acquisition method with the TopSpeed set with 3-second cycle. Spectra were acquired in the Orbitrap analyzer with mass range of 350–1200 m/z and 120,000 resolution (200 m/z), with a maximum injection time of 50 msec and an AGC target of 5×10e5. Signals with 2–5 charges were selected for HCD fragmentation using a normalized collision energy of 27, a maximum injection time of 120 msec and an AGC target of 10,000. Fragments were analyzed in the ion trap. Raw MS files were analyzed by MaxQuant (v1.5.2.8)42 (http://www.maxquant.org). MS/MS spectra were searched against the UniProt-human database (Version June 2014, 59,345 entries). All used search parameters were default, with the exception of including the match between runs (1 minute window) and the iBAQ label-free quantification43. The search included variable modifications of methionine oxidation and N-terminal acetylation, and fixed modification of carbamidomethyl cysteine. Each iBAQ value was log2 transformed and subsequently normalized by the average protein abundance within each run. Biological process association analysis and process network enrichment were performed using the GeneGo's MetaCore pathways analysis package with false discovery rate (FDR) <5%; each GO term was ranked using p-value enrichment.
Purification of recombinant protein VII-His
Protein VII was cloned from genomic DNA isolated from adenovirus infected HeLa cells into a pET21a backbone to generate a C-terminal hexahistidine tag. Positive clones were selected in DH5α cells, sequenced, and transformed into BL21 (DE3) cells (NEB C2527I). The purification of insoluble protein VII-His was adapted from existing protocols to purify histone proteins from E. coli44,45. Briefly, BL21 cells were inoculated from overnight cultures and grown to an optical density of 0.5–0.6 OD260, induced with 0.1 mM IPTG (Sigma) and harvested after 4 hours at 37°C. Cells pellets were resuspended in a mild buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 1 mM PMSF, 5% glycerol, 2.5 µg/mL aprotinin, leupeptin and pepstatin) and disrupted by sonication using a Branson 250 sonifier. The lysate was then centrifuged at 27,000 × g for 20 minutes at 4°C. The supernatants were discarded and pellets were resuspended in a denaturing buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5% glycerol, 8 M urea). The suspension was centrifuged again to eliminate insoluble cell debris and the his-tagged protein was isolated using a cobalt resin (ThermoScientific 89964) according to the manufacturer’s instructions for denaturing conditions. The purified protein was then dialyzed against water and lyophilized. Purified protein was verified by western blot and mass spectrometry.
In vitro binding assays
HMGB1-GST (Abnova) or GST (Sigma) were combined with recombinant protein VII-His at equimolar ratios and incubated at 4°C for 1 hour. Complexes were then mixed with a cobalt resin (ThermoScientific 89964) to bind protein VII-His and any associated protein and washed three times in the binding buffer (50 mM Tris pH 8, 300 mM NaCl, 0.1% IGEPAL). The beads were then boiled in sample buffer, separated on a 4–12% NuPage gel and visualized by coomassie staining.
Nucleosome in vitro binding and MNase digestion assays
Gel shift and MNase digestion assays were carried out as previously described17,46,47. Briefly, nucleosomes were reconstituted by incubating purified recombinant histones with ‘601’ DNA of either 195 or 147bp over a series of dialysis. Recombinant protein VII-His was then combined with nucleosomes at various molar ratios, incubated at room temperature for 15 minutes, and analyzed by native gel electrophoresis. Complexes were also digested with MNase (Affymetrix) by addition of 1 unit per µg of DNA for 147bp nucleosome experiments and 0.1 unit per µg of DNA for 195bp nucleosome experiments, incubated at 22°C for varying amounts of time followed by the addition of EGTA and guanidine thiocyanate to stop the reaction. The DNA fragments were then purified using a MinElute PCR purification kit (Qiagen) and analyzed on an Agilent 2100 Bioanalyzer as previously described17.
Release assay of HMGB1 in THP-1 cells
THP-1 cells were seeded at a density of 2 × 105 cells per well in a 24-well plate, and stimulated into macrophage-like cells by addition of 10 ng/mL PMA for 48 hours. Cells were washed in PBS and transduced with recombinant adenovirus vectors expressing only GFP or protein VII-GFP such that > 90% of cells were GFP positive. At 48 hours post transduction, cells were washed and 200 µL of serum free RPMI was added. To stimulate the inflammasome, LPS (Sigma-Aldrich L2880) with a final concentration of 0.5 µg/mL was added to wells and incubated for 2 hours, then nigericin (Sigma-Aldrich N7143) was added with a final concentration of 10 µM for 1 hour. Supernatants were collected and proteins precipitated overnight at 4°C with a final concentration of 20% trichloroacetic acid (Sigma), washed with acetone, dried, and resuspended in 1× sample buffer with reducing agent (Invitrogen). For ELISA analysis, supernatants were harvested directly and HMGB1 content was detected by the manufacturer’s instructions (Chondrex 6010). Cells were also harvested by the addition of 1× sample buffer with reducing agent (Invitrogen) and boiled. Supernatants and lysates were analyzed together by western blot.
Acid extraction and Reverse Phase-HPLC for purification of protein VII and analysis of total histone PTMs
Histones were prepared for mass spectrometry analysis as detailed previously48. Nuclei were isolated and histones from infected cells were extracted by acid as previously described14. The pre-protein VII and protein VII variants were fractionated using an offline RP-HPLC. Briefly, ~100 µg proteins were resuspended in buffer A (0.1% trifluoroacetic acid (TFA) in HPLC grade water) and loaded onto a C18 5µm column (4.6 mm internal diameter × 250 mm, Vydac) using a Beckman Coulter (System Gold, Brea, CA) HPLC (Buffer A: 0.1% TFA, Buffer B: 95% acetonitrile, 0.08% TFA). The proteins were separated using a gradient from 30–45% buffer B in 100 minutes at a flow-rate of 0.2 mL/min. The fractions containing the proteins of interest were collected using an automatic fraction collector and individual peaks combined based on their UV signal. The fractions were subsequently dried by vacuum centrifugation and prepared for mass spectrometry (see below). Protein VII was purified from three biological replicates and analyzed as follows for MS.
Mass spectrometry analysis of protein VII PTMs
Sample preparation/protein VII
RP-HPLC purified samples of protein VII variants were reduced in 10 mM dithiothreitol (DTT) in 50 mM ammonium bicarbonate for 1 hour at 56 °C. After cooling to room temperature, samples were alkylated in 20 mM iodoacetamide in 50 mM ammonium bicarbonate for 30 minutes in the dark. Samples were digested with chymotrypsin or Arg-C, at an enzyme to substrate ratio of approximately 1:20 for 8 hours at 37°C. The samples were acidified to a final concentration of 5% formic acid to a pH ≤ 3 and desalted using P200 stage tip columns packed with C18 3M plug (3M Bioanalytical Technologies). Purified peptide samples were dried by lyophilization and stored at −20°C until further analysis.
Nano LC-MS/MS analysis of histone PTMs
The nanoLC-MS/MS analysis was performed as previously described48.
Nano LC-MS/MS analysis of protein VII peptides
The nanoLC-MS/MS analysis was performed in triplicate for each sample. Samples were loaded onto a 16 cm C18–AQ column (inner diameter 75 µm, 3 µm beads, Dr, Maisch GmbH, Germany) using an Easy nano-flow HPLC system (Thermo Fisher Scientific, Odense, Denmark). The nanoLC was coupled to an Orbitrap Velos Pro Mass Spectrometer (Thermo Fisher Scientific) via a nanoelectrospray ion source (Thermo Fisher Scientific, San Jose, CA). Peptides were loaded in buffer A (0.1% formic acid) and eluted with a 45 minute linear gradient from 2–30% buffer B (95% acetonitrile, 0.1% formic acid). After the gradient, the column was washed with 90% buffer B. Mass spectra were acquired using a data-dependent acquisition method with the Top15 most intense ions. Spectra were acquired in the Orbitrap analyzer with mass range of 350–1600 m/z and 60,000 resolution (400 m/z), with a maximum injection time of 10 msec and an AGC target of 10e6. Signals above 1000 count charges were selected for HCD fragmentation using normalized collision energy of 36, a maximum injection time of 100 msec and an AGC target of 50,000. Fragments were analyzed in the orbitrap.
Data processing of protein VII spectra
Raw mass spectrometer files were analyzed using Proteome Discoverer (v1.4, Thermo Scientific, Bremen, Germany). MS/MS spectra were converted to .mgf files and searched against the UniProt-adenovirus C serotype 5 database using Mascot (v2.5, Matrix Science, London, UK). Database searching was performed with the following parameters: precursor mass tolerance 10 ppm; MS/MS mass tolerance 0.05 Da; enzyme chymotrypsin (Promega) or Arg-C (Roche), with two missed cleavages allowed; fixed modification was cysteine carbamidomethylation; variable modifications were methionine oxidation, serine/threonine/tyrosine phosphorylation, lysine acetylation and methylation, asparagine and glutamine deamidation. Specifically, phosphorylation, acetylation, and methylation were searched separately, not as co-existing modifications. Peptides were filtered for <1% false discovery rate, Mascot ion score >20 and peptide rank 1.
Co-immunoprecipitation of protein VII-HA
A549 cells were induced to express protein VII with doxycycline for four days as described above. Approximately 4 × 107 cells were harvested and pelleted for each immunoprecipitation reaction. Cell pellets were resuspended in 500 µl of IC wash buffer with protease inhibitors (20 mM HEPES pH 7.9, 110 mM KOAc, 2 mM MgCl2, 150 mM NaCl, 0.1% Tween-20, 0.1% Triton X) and incubated on ice for 10 minutes with intermittent vortexing to disrupt cells. Samples were then incubated on ice for 1 hour with 5 µl of benzonase (Millipore) added to each sample to digest DNA to ~150bp, which was confirmed by DNA isolation and agarose gel analysis. Samples were then sonicated in a Diagenode Bioruptre for 30s on and 30s off for five rounds at 4°C and centrifuged at 14,000 g for 15 minutes at 4°C. Supernatants were then incubated rotating for 1 hour at 4°C with 30 µl of HA-conjugated magnetic beads (Thermo Scientific) and washed three times for five minutes in IC buffer. Isolated proteins were eluted with 100 µl of 2mg/ml HA peptide (Thermo Scientific) for 20 minutes rotating at 37°C and separated on and SDS-PAGE gel. For protein separation by SDS–PAGE the NuPAGE 1DE System was used (NuPAGE Novex 4–12% bis–tris 1.0 mm gels, Invitrogen, USA). Uninduced cells were used as a negative control. The immunopreciptation was carried out in biological triplicate and pull-down of protein VII-HA and HMGB1 was confirmed by western blotting standard techniques as described above.
Quantitative PCR
Genomic DNA was isolated using the PureLink Genomic DNA kit (Thermo Scientific). Quantitative PCR was performed using primers specific for viral DBP (5’ gccattgcgcccaagaagaa and 5’ ctgtccacgattacctctggtgat), protein VII (5’ gcgggtattgtcactgtgc and 5’ cacccaatacacgttgccc), and cellular tubulin (5’ ccagatgccaagtgacaagac and 5’ gagtgagtgacaagagaagcc). Values for DBP and VII were normalized internally to tubulin and to the 4 hour time point to control for any variation in virus input. RNA was isolated using the RNeasy Mini Kit (Qiagen) and reverse transcribed using the High Capacity RNA to cDNA Kit (Applied Biosystems). Quantitative PCR was performed using primers specific for HMGB1 (5’ taactaaacatgggcaaaggag and 5’ tagcagacatggtcttccac) and beta actin (5’ gcaccacaccttctacaatgag and 5’ ggtctcaaacatgatctgggtc). Quantitative PCR was performed using the standard protocol for Sybr Green (Thermo Scientific) and analyzed using the ViiA 7 Real-Time PCR System (Thermo Scientific).
Precision cut lung slice (PCLS) immunofluorescence
PCLS were obtained and prepared as previously described27,49. De-identified human lung tissue from donors was obtained from the National Disease Research Interchange (NDRI), Philadelphia, PA. Analysis of human samples was approved by the University of Pennsylvania Internal Review Board. Samples were infected with 108 pfu of Ad5 per slice or 109 GC of rAd VII-GFP for 24 hours. Samples were fixed in 4% PFA at room temperature for 15 minutes and washed three times in PBS. Samples were permiabilized with 0.5% Triton X and washed twice more in PBS. Samples were then incubated with 3% BSA and 0.03% Triton X in PBS for 1 hr to block. Primary antibodies (DBP or HMGB1) were incubated in the same buffer for 1 hr and then samples were washed three times in PBS with 3% BSA, incubated with secondary antibodies and DAPI for 1 hr, and washed three more times. Whole slices were mounted on slides with mounting solution and imaged by confocal microscopy.
Fluorescence recovery after photobleaching (FRAP)
Full-length HMGB1 was cloned from pcDNA3.1 Flag-hHMGB1 (Addgene 31609) into pEGFP-N1 containing a L221K mutation to prevent dimerization of GFP molecules50. A549 cells were induced to express protein VII for four days with doxycycline in glass-bottom dishes. Cells were then transfected with the construct that constitutively expresses HMGB1 with a monomeric GFP C-terminal tag. FRAP was carried out using standard methods on a Zeiss LSM 710 confocal microscope. Diffusion coefficients were calculated using the “simFRAP” algorithm (http://imagej.nih.gov/ij/plugins/sim-frap/index.html), a simulation based approach to FRAP analysis51.
Statistical analyses
Statistical details are reported in each figure legend. Statistical analyses were performed on at least three different biological replicates, unless otherwise stated in the figure legend. The sample size was chosen to provide enough statistical power to apply parametric tests (one- or two-tailed homoscedastic t-test). The t-test was considered as valuable statistical test since binary comparisons were performed and the number of replicates was limited. Furthermore, we applied the homoscedastic t-test assuming that the variance between the two datasets would remain homogeneous due to the use of the same cell lines in culture with and without protein VII expression. No samples were excluded as outliers (this applies to all proteomics analyses described in this manuscript). Proteins with p-value smaller than 0.05 were considered as significantly altered between the two tested conditions for two-tailed and one-tailed t-test. Data distribution was assumed to be normal but this was not formally tested. The nanoLC-MS analysis was performed in triplicate for each sample to determine technical variation.
Extended Data
Extended Data Figure 1. Adenovirus protein VII distorts chromatin.
a, Protein VII localizes to cellular chromatin and viral replication centers in U2OS similarly to SAECs in Fig 1a. b, Protein VII mRNA levels measured by quantitative PCR showing that after 4 days of induction in the A549 cell line, the level of protein VII transcripts is approximately 10% of that measured during infection at 16 hpi. Despite the low relative level, this amount of protein VII is sufficient to cause dramatic changes in the nucleus (error bars=±s.d., nbiological=3). c, Inducible cell lines of U2OS and HeLa expressing protein VII-HA show chromatin localization and distortion, similar to A549 cells in Fig 1c. d, Inducible A549 cell lines expressing viral protein V, the precursor for protein VII (preVII) or cellular protamine PRM1 with C-terminal HA tags. Although all 3 proteins possess a large number of charged residues, none are sufficient to distort cellular chromatin or increase nuclear size as observed with mature protein VII.
Extended Data Figure 2. Protein VII associates tightly with chromatin and binds DNA and nucleosomes in vitro.
a, Western blot analysis showing protein VII in histone extracts from infected HeLa cells at 24 hpi. b, Chromatin fractionation of lysates from A549 cells that were uninfected (mock) or infected for 24 hrs with Ad5. Viral and cellular proteins were detected by western blotting with various antibodies as indicated. c, Agarose gel analysis of DNA extracted from nuclear fractionation experiments indicating the size of DNA is between 100 – 200 bp and elutes predominantly in the higher salt fractions. d, Chromatin fractionation of cells induced to express protein VII indicating protein VII present in the highest salt fraction from the first day of induction. e–f, Recombinant protein VII-His binds DNA. Incubating increasing molar amounts of protein VII with 195bp DNA results in shifts by native gel electrophoresis indicating protein VII-DNA complex formation. Staining with either ethidium bromide (e) or coomassie (f) are shown to verify the presence of DNA and protein, respectively. g, Ethidium bromide staining shows DNA content of nucleosome shifts from gel in Fig 1f.
Extended Data Figure 3. Bioanalyzer examination of MNase digested nucleosomes and protein VII-nucleosome complexes.
a, 195bp Nucleosomes or protein VII-nucleosome complexes were incubated with MNase for the indicated times, the reaction was stopped, DNA extracted and analyzed. As in Fig 1g, nucleosomes are shown in black and protein VII-nucleosome complexes in orange. The presence of protein VII pauses digestion at 165bp, suggesting that protein VII is blocking access to the DNA. b, 147bp nucleosomes or protein VII-nucleosome complexes were incubated with MNase for the indicated times, the reaction was stopped, DNA extracted and analyzed. Graphs show nucleosomes in grey and protein VII-nucleosome complexes in orange. The presence of protein VII completely blocks digestion even after nucleosomes alone have been digested well beyond the core particle. In contrast to what would be expected for linker histones, protein VII protects the core nucleosome particle from digestion. These data indicate that protein VII may be masking the substrate for MNase through complex formation. This represents a unique mechanism of nucleosome binding and suggests a model for blocking DNA access in cellular chromatin during infection.
Extended Data Figure 4. Purification of protein VII from infected cells.
a, Coomassie stained SDS-PAGE analysis of fractions from RP-HPLC in Fig 2a. The bands in fraction 38–41 min correspond to histone H1. Protein VII and V, as indicated, were verified by mass spectrometry analysis (not shown). The slight upward shift of the protein VII bands in the later peak corresponds to the higher abundance of protein preVII, as seen by HPLC in Fig 2a. b, Western blot analysis of protein VII in HPLC fractions from (a). c, Time-course of infection followed by histone extraction and HPLC analysis. Mass spectrometry analysis verified peaks in each sample as indicated.
Extended Data Figure 5. Representative mass spectra.
Annotated MS/MS spectra of identified peptides of protein VII containing PTMs (a–c, acetylated peptides; d–f, phosphorylated peptides). The images represent the observed fragment ions collected using MS/MS collision induced dissociation (CID). Colored lines represent matches between observed and expected fragment ions of the given peptides. Specifically, green lines represent not fragmented precursor mass, blue lines represent matches with y-type fragments, red lines with b-type fragments, and yellow boxed masses represent fragments containing PTM neutral losses (e.g. ions that lost the phosphorylation during fragmentation).
Extended Data Figure 6. Acetylated VII spectra from virus particles and analysis of total histone PTM changes upon protein VII expression.
a, LC-MS analysis of unmodified and modified chymotryptic peptide AKKRSDQHPVRVRGHY. On the left, nanoLC–MS extracted ion chromatograms of protein VII peptides identified in the histone extracts of adenovirus infected cells (Inf) or viral particles (VP). The top left represents the modified form, while the bottom left the unmodified form. Non-modified forms were detected in both conditions for VII and VP, while the acetylated form was unique for the infected sample only (Inf). On the right, full MS spectrum of the modified (top) and unmodified (bottom) peptide. Circled mass represents the monoisotopic signal of the peptide. b, Summary of post-translational modifications detected on protein VII. Peptides shown were identified during infection at various time points with the mature protein VII in the top row and the precursor, preVII, in the bottom row. The numbers in brackets for preVII indicate the location of the same moiety in mature protein VII. Acetylation sites were detected in approximately 3% of peptides for mature protein VII and 2% of peptides in preVII. Phosphorylation was detected in approximately 1% of peptides for mature protein VII and preVII. c–d, Quantification of histone H3 (c) and H4 (d) PTMs in protein VII-HA induced (+dox) and uninduced (−dox) A549 cells from the analysis of crude histone mixtures (nbiological=3). Positions of PTMs are listed along the x-axis. Modification type is indicated by color as shown. y-axis represents the cumulative extent of PTMs as relative to the total histone H3 or H4, respectively. e, Breakdown of the histone marks (H3K14ac, H3K27me1, H3K36me3, H4K20me1, H4K20me2, and H4K20me3) found significantly different (nbiological=3) in terms of relative abundance between the protein VII-HA induced and uninduced states (<5% homoscedastic two-tailed t-test). Error bars represent ± standard deviation.
Extended Data Figure 7. Bioinformatic analysis of proteins enriched in the high salt fraction upon protein VII expression.
a, Venn diagram showing overlap between three biological replicates of high salt fraction proteins significantly enriched as compared to uninduced cells. b, Proteins found significantly enriched in the protein VII-HA induced state as compared to uninduced (<5% homoscedastic t-test) in all three biological replicates (VII-HA induced: proteins identified only in protein VII-HA induced condition). c–d, Classification of proteins significantly enriched in minimum two out of three biological replicates (protein VII-HA induced vs uninduced) according to process network enrichment and gene ontology (GO) biological process (GeneGo's MetaCore pathways analysis package; false discovery rate <5%); each GO term was ranked using p-value enrichment.
Extended Data Figure 8. Protein VII retains HMGB1 and HMGB2 in chromatin.
a, Western blot of adenovirus infected or doxycycline treated A549 cells showing the relative levels of protein VII expression. HMGB1 levels do not change upon infection or protein VII expression. Tubulin is shown as a loading control. b, Quantitative PCR analysis of mRNA transcripts of HMGB1 in various cell types as indicated (for A549, nbiological=3, for THP-1, nbiological=2, error bar=±s.d.). The levels of HMGB1 do not significantly change. c, Immunofluorescence analysis of a time-course of protein VII-HA (red) induction shown with HMGB1 (green) and DAPI (grey, blue in merge) in A549 cells. Expression of protein VII-HA results in a change to the HMGB1 distribution upon expression. d, HMGB1 (green) localization changes between 12 and 24 hpi of wild-type adenovirus in A549 cells, and adopts a pattern similar to protein VII as in Fig 1a. DBP (red) is shown as a marker of infection, DNA is stained with DAPI (blue in merge). e, Same as (d) showing HMGB2 adopts the same pattern as HMGB1 during Ad5 infection at 24 hpi. f, Multiple cells showing the same pattern of HMGB1 re-localization upon expressing VII-GFP as in Fig 3g. g, HMGB1 retention in the high salt fraction is conserved across Ad serotypes. Western blot analysis of HMGB1 from salt fractionated A549 cells infected with Ad5, Ad9 or Ad12 as shown.
Extended Data Figure 9. Protein VII is necessary and sufficient for chromatin retention of HMGB1 in human and mouse cells.
a–b, Replication of Ad5-flox-VII virus on 293 or 293-Cre cells. Quantitative PCR analysis of viral genomic DNA over a time-course of infection (a) shows the DBP gene is increasing exponentially in 293 and 293-Cre cells when infected with Ad5-flox-VII virus. In contrast, PCR for the protein VII gene (b) demonstrates deletion in 293-Cre cells (nbiological=2, error bar=±s.d.). c, Salt fractionation of 293-Cre cells infected with wild-type Ad5 indicating that the Cre recombinase does not interfere with the ability of protein VII to retain HMGB1 in the high salt chromatin fraction. Protein VII is also necessary for the chromatin retention of HMGB2. d, THP-1 cells transduced to express protein VII-GFP results in chromatin distortion and HMGB1 retention in chromatin. Immunofluorescence of transduced PMA-treated THP-1 cells showing protein VII-GFP (green), HMGB1 (red) and DNA (grey, blue in merge). e, Transduction to express protein VII-GFP is sufficient to relocalize mouse HMGB1 in mouse embryonic fibroblast (MEF) cells. f, Salt fractionation of mouse embryonic fibroblast cells transduced to express protein VII-GFP. Human Ad5 protein VII is sufficient to retain mouse HMGB1 in the high salt fraction in MEF cells. The control vector expressing GFP alone does not have this effect.
Extended Data Figure 10. Transduction of mouse lungs demonstrating expression of GFP or protein VII-GFP.
a, Sections of mouse lungs transduced to express protein VII-GFP or GFP co-stained for HMGB1. GFP signal shows multiple cell types transduced in both cases. Protein VII-GFP has a more distinct nuclear signal than GFP, which also appears cytoplasmic. Two sections for each condition are shown to indicate transduction efficiency. b, Same as (a) but co-stained for prosurfactant-C to mark type II pneumocytes. Some cells are positive for both, confirming multiple cell types transduced. c, Zoomed images of individual epithelial cells from mouse lungs showing the characteristic protein VII-GFP pattern colocalizing with DAPI in the nucleus. GFP only is mostly cytoplasmic. d, Schematic summarizing function of protein VII during infection. Newly synthesized protein VII late during infection can be post-translationally modified and binds to HMGB1, sequestering it on the cellular chromatin and preventing its release. Unmodified protein VII is packaged in viral progeny.
Supplementary Material
Acknowledgments
We thank members of the Weitzman Lab for insightful discussions and input, especially Robert Dilley and Bryan Simpson for generating reagents. We also thank R. Panetierri and C. Koziol-White for graciously providing precision cut lung slices. We are grateful to D. Curiel for generously sharing recombinant protein VII-GFP vectors and L. Gerace for anti-VII antibodies. We thank the Penn Vector Core for assistance in purifying recombinant vectors, the Penn CDB Microscopy Core for imaging and FRAP assistance, and the CHOP Pathology core for immunostaining of mouse lungs. We thank members of the Black, Garcia and Worthen labs for valuable technical help. We thank Craig Bassing, Igor Brodsky, Jorge Henao-Mejia, Rahul Kohli, Carolina Lopez, Adam Resnick, Sunny Shin, Kathy Spindler and Jonathan Weitzman for advice and critical reading of the manuscript. D.C.A. was supported in part by T32 CA115299 and F32 GM112414. N.J.P. was supported in part by T32 NS007180. N.S. was supported in part by funding from the American Cancer Society. Research was supported by grants from the National Institutes of Health (CA097093 to M.D.W., AI102577 and CA122677 to P.H., AI118891 and GM110174 to B.A.G., and GM082989 to B.E.B.), the Institute for Immunology of the University of Pennsylvania, and funds from the Children’s Hospital of Philadelphia (M.D.W.).
Footnotes
Author Contributions D.C.A. and M.D.W. conceived the project and designed experiments; D.C.A., C.H., N.S., J.P., N.J.P, and E.D.R. performed the experiments; D.C.A., C.H., and J.P. generated constructs and cell lines; K.K., R.C.M., S.S. and B.A.G. performed mass spectrometry analysis; P.O. and P.H. generated Ad5-flox-VII virus and provided 293-Cre cell line; D.C.A. and D.B. performed the FRAP experiments; A.J.P. and G.S.W. conducted all mouse experiments; B.E.B. and B.A.G. designed experiments and interpreted the data; D.C.A. and M.D.W. interpreted the data and wrote the manuscript and all authors were involved in editing the manuscript.
The authors declare no competing financial interests.
Accession codes
All proteomics raw files generated for this manuscript are collected into the public database Chorus (https://chorusproject.org/, Project number: 1047).
References
- 1.Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nature Reviews Microbiology. 2009;7:787–797. doi: 10.1038/nrmicro2222. [DOI] [PubMed] [Google Scholar]
- 2.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 3.Lischwe MA, Sung MT. A histone-like protein from adenovirus chromatin. Nature. 1977;267:552–554. doi: 10.1038/267552a0. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee PK, Vayda ME, Flint SJ. Adenoviral protein VII packages intracellular viral DNA throughout the early phase of infection. EMBO J. 1986;5:1633–1644. doi: 10.1002/j.1460-2075.1986.tb04406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vayda ME, Rogers AE, Flint SJ. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Research. 1983;11:441–460. doi: 10.1093/nar/11.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang R, et al. HMGB1 in health and disease. Mol. Aspects Med. 2014 doi: 10.1016/j.mam.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 8.Paschos K, Allday MJ. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends in Microbiology. 2010;18:439–447. doi: 10.1016/j.tim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marazzi I, et al. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari R, Berk AJ, Kurdistani SK. Viral manipulation of the host epigenome for oncogenic transformation. Nat Rev Genet. 2009;10:290–294. doi: 10.1038/nrg2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knipe DM, et al. Snapshots: chromatin control of viral infection. Virology. 2013;435:141–156. doi: 10.1016/j.virol.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari R, et al. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host and Microbe. 2014;16:663–676. doi: 10.1016/j.chom.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J. Biol. Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 14.Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Meth. Enzymol. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nature Protocols. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 16.Teves SS, Henikoff S. Salt fractionation of nucleosomes for genome-wide profiling. Methods Mol. Biol. 2012;833:421–432. doi: 10.1007/978-1-61779-477-3_25. [DOI] [PubMed] [Google Scholar]
- 17.Falk SJ, et al. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348:699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White AE, Hieb AR, Luger K. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci. Rep. 2016;6:19122. doi: 10.1038/srep19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Fedor MJ, Daniell E. Acetylation of histone-like proteins of adenovirus type 5. Journal of Virology. 1980;35:637–643. doi: 10.1128/jvi.35.3.637-643.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson CM, et al. Molecular evolution of human adenoviruses. Sci. Rep. 2013;3:1812. doi: 10.1038/srep01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyurcsik B, Haruki H, Takahashi T, Mihara H, Nagata K. Binding modes of the precursor of adenovirus major core protein VII to DNA and template activating factor I: implication for the mechanism of remodeling of the adenovirus chromatin. Biochemistry. 2006;45:303–313. doi: 10.1021/bi051248+. [DOI] [PubMed] [Google Scholar]
- 23.Haruki H, Okuwaki M, Miyagishi M, Taira K, Nagata K. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. Journal of Virology. 2006;80:794–801. doi: 10.1128/JVI.80.2.794-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 25.Sapojnikova N, et al. Biochemical observation of the rapid mobility of nuclear HMGB1. Biochim. Biophys. Acta. 2005;1729:57–63. doi: 10.1016/j.bbaexp.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Lu B, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488:670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koziol-White CJ, Damera G, Panettieri RA. Targeting airway smooth muscle in airways diseases: an old concept with new twists. Expert Rev Respir Med. 2011;5:767–777. doi: 10.1586/ers.11.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueno H, et al. Contributions of high mobility group box protein in experimental and clinical acute lung injury. Am. J. Respir. Crit. Care Med. 2004;170:1310–1316. doi: 10.1164/rccm.200402-188OC. [DOI] [PubMed] [Google Scholar]
- 29.Johnson JS, et al. Adenovirus protein VII condenses DNA, represses transcription, and associates with transcriptional activator E1A. Journal of Virology. 2004;78:6459–6468. doi: 10.1128/JVI.78.12.6459-6468.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karen KA, Hearing P. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. Journal of Virology. 2011;85:4135–4142. doi: 10.1128/JVI.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 31.Khandelia P, Yap K, Makeyev EV. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12799–12804. doi: 10.1073/pnas.1103532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozarsky KF, Jooss K, Donahee M, Strauss JF, Wilson JM. Effective treatment of familial hypercholesterolaemia in the mouse model using adenovirus-mediated transfer of the VLDL receptor gene. Nat. Genet. 1996;13:54–62. doi: 10.1038/ng0596-54. [DOI] [PubMed] [Google Scholar]
- 33.Orazio NI, Naeger CM, Karlseder J, Weitzman MD. The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection. Journal of Virology. 2011;85:1887–1892. doi: 10.1128/JVI.02134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le LP, et al. Core labeling of adenovirus with EGFP. Virology. 2006;351:291–302. doi: 10.1016/j.virol.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reich NC, Sarnow P, Duprey E, Levine AJ. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 36.Lilley CE, Chaurushiya MS, Boutell C, Everett RD, Weitzman MD. The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog. 2011;7:e1002084. doi: 10.1371/journal.ppat.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das S, MacDonald K, Chang H-YS, Mitzner W. A simple method of mouse lung intubation. J Vis Exp. 2013:e50318. doi: 10.3791/50318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeyaseelan S, Chu HW, Young SK, Worthen GS. Transcriptional profiling of lipopolysaccharide-induced acute lung injury. Infect. Immun. 2004;72:7247–7256. doi: 10.1128/IAI.72.12.7247-7256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nick JA, et al. Role of p38 mitogen-activated protein kinase in a murine model of pulmonary inflammation. J. Immunol. 2000;164:2151–2159. doi: 10.4049/jimmunol.164.4.2151. [DOI] [PubMed] [Google Scholar]
- 40.Zaret K. Micrococcal nuclease analysis of chromatin structure. Curr Protoc Mol Biol. 2005;Chapter 21(Unit 21.1) doi: 10.1002/0471142727.mb2101s69. [DOI] [PubMed] [Google Scholar]
- 41.Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 42.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 43.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka Y, et al. Expression and purification of recombinant human histones. Methods. 2004;33:3–11. doi: 10.1016/j.ymeth.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 45.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. Journal of Molecular Biology. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 46.Hasson D, et al. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulej K, Avgousti DC, Weitzman MD, Garcia BA. Characterization of histone post-translational modifications during virus infection using mass spectrometry-based proteomics. Methods. 2015;90:8–20. doi: 10.1016/j.ymeth.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper PR, Panettieri RA. Steroids completely reverse albuterol-induced beta(2)-adrenergic receptor tolerance in human small airways. J. Allergy Clin. Immunol. 2008;122:734–740. doi: 10.1016/j.jaci.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 50.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 51.Blumenthal D, Goldstien L, Edidin M, Gheber LA. Universal Approach to FRAP Analysis of Arbitrary Bleaching Patterns. Sci. Rep. 2015;5:11655. doi: 10.1038/srep11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.