Abstract
MicroRNAs (miRNAs) play an important role in the regulation of gene expression and are involved in many cellular processes including inhibition of viral replication in infected cells. In this study, three subtypes of influenza A viruses (pH1N1, H5N1 and H3N2) were analyzed to identify candidate human miRNAs targeting and silencing viral genes expression. Candidate human miRNAs were predicted by miRBase and RNAhybrid based on minimum free energy (MFE) and hybridization patterns between human miRNAs and viral target genes. In silico analysis presented 76 miRNAs targeting influenza A viruses, including 70 miRNAs that targeted specific subtypes (21 for pH1N1, 27 for H5N1 and 22 for H3N2) and 6 miRNAs (miR-216b, miR-3145, miR-3682, miR-4513, miR-4753 and miR-5693) that targeted multiple subtypes of influenza A viruses. Interestingly, miR-3145 is the only candidate miRNA targeting all three subtypes of influenza A viruses. The miR-3145 targets to PB1 encoding polymerase basic protein 1, which is the main component of the viral polymerase complex. The silencing effect of miR-3145 was validated by 3′-UTR reporter assay and inhibition of influenza viral replication in A549 cells. In 3′-UTR reporter assay, results revealed that miR-3145 triggered significant reduction of the luciferase activity. Moreover, expression of viral PB1 genes was also inhibited considerably (P value < 0.05) in viral infected cells expressing mimic miR-3145. In conclusion, this study demonstrated that human miR-3145 triggered silencing of viral PB1 genes and lead to inhibition of multiple subtypes of influenza viral replication. Therefore, hsa-miR-3145 might be useful for alternative treatment of influenza A viruses in the future.
Keywords: miRNA, miR-3145, silencing, inhibition, viral replication, influenza A virus
Introduction
Influenza A viruses are members of Orthomyxoviridae family. These enveloped viruses contain eight negative single strand RNA segments encoding for 11–12 viral proteins.1 Infection of influenza A viruses can affect the upper respiratory system and cause asymptomatic to severe symptoms including fever, sneezing, coughing, runny nose, nasal congestion, and diarrhea.1 Various subtypes of influenza A viruses can be classified by a combination of 17 different Hemagglutinin (HA) proteins and 10 various Neuraminidase (NA) proteins. The pH1N1, H5N1, and H3N2 subtypes cause serious worldwide public health concerns.2,3 The therapeutic approaches are limited to targeting neuraminidase or M2 ion channels, as well as vaccines, which have been restricted due to seasonal antigenic drift. Recent studies suggest that cellular microRNAs (miRNAs) are involved in the regulation of viral replication; therefore, miRNAs might be useful for an alternative treatment against influenza A virus.
MiRNAs are small non-coding RNAs approximately 22 nucleotides in length.4 MiRNAs are firstly transcribed as long hairpin RNAs called primary miRNAs (pri-miRNAs) which are continually cropped and trimmed to 60–100 nucleotides with a stem loop structure called precursor miRNAs (pre-miRNAs). The pre-miRNAs are then exported to cytoplasm by Exportin-5 protein. Cytoplasmic Dicer removes the loop structure of pre-miRNAs generating mature miRNA. Consequently, RNA-induced silencing complex (RISC) will assemble with miRNA duplexes and one strand of miRNA is removed by a helicase activity of the RISC while the remaining miRNA strand guides the RISC to a distinctive target mRNA via base pairing. MiRNAs regulate gene expression by mRNA degradation or translational repression, thus miRNAs play an important role in the regulation of many cellular processes including cell proliferation, apoptosis, and homeostasis.5 Moreover, cellular miRNAs also inhibit viral replications. For examples, hsa-miR-32 confines the accumulation of the primate foamy virus type 1 (PFV-1).6 In 2010, Song et al. reported that miR-323 miR-491 and miR-654 target PB1 genes of H1N1 influenza A viruses (A/WSN/33), leading to viral gene silencing and blocking viral replication.7 However, those miRNAs were proved to inhibit only one subtype (H1N1) of the influenza A virus in 1933, which may not represent the other subtypes or the recent strains of influenza A viruses infecting humans. Therefore, this study’s purpose is to identify human miRNA targets in multiple subtypes of influenza A viruses such as pH1N1, H5N1 and H3N2, then to validate the candidate miRNA on viral gene silencing.
Materials and methods
Prediction of candidate miRNAs targeting influenza viral genes
Three subtypes of influenza A viruses including pH1N1 (A/Thailand/104/2009) [accession no. GQ169381-5, GQ205443, GQ229379 and GQ259597], H5N1 (A/Thailand/NK165/2005) [accession no. DQ372591-8] and H3N2 (A/Thailand/CU-H1817/2010) [accession no. CY074963-70] were used to predict human miRNAs that target them. Two web-based programs, miRBase8–12 (www.mirbase.org) and RNAHybrid13 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) were used to screen the human miRNAs that target viral genomes based on the principles of miRNA-target recognition.14 Hybridization patterns between the miRNAs and their target mRNAs can be classified into 5′ canonical, 5′ seed, and 3′ compensatory. Therefore, criteria for the selection of miRNAs targeting influenza viral genes is based on effective hybridization patterns (5′ canonical, 5′ seed, or 3′ compensatory) and minimum free energy (MFE) for base pairing less than −17.5 kcal/mol. Details of the computational prediction method was described in previous work.15
Construction of a reporter vector
Total viral RNA was extracted by the Guanidium-isothiocyanate method and then reverse transcribed by random hexamers with SuperScript® III reverse transcriptase (Invitrogen, Waltham, Massachusetts, USA) following the manufacturer’s instruction. The targeting region fragment on viral PB1 genes was amplified by using NheI/PB1_ F and XhoI/PB1_R primers (Table 1). The thermal profile included an initial denaturation at 95℃ for 3 min, 40 cycles of amplification (95℃ for 15 s, 60℃ for 15 s, and 72℃ for 45 s) and final extension at 72℃ for 7 min. These primers were designed with restriction sites of NheI and XhoI for ligation with a pGL3MS2/Basic reporter vector containing firefly luciferase (FLuc) as a reporter gene. The pGL3MS2/Basic vector and PB1 PCR product were digested by restriction enzymes NheI and XhoI (Thermo Scientific, Waltham, MA, USA). The digested products were analyzed by agarose gel electrophoresis and purified by HiYield™ Gel Extraction kit (RBC Bioscience, New Taipei City, Taiwan). Then, the partial PB1 DNA fragment was ligated into XhoI and NheI sites at 3′-UTR of a firefly luciferase gene within a pGL3MS2/Basic vector by T4 DNA ligase (Thermo Scientific). This constructed vector was called pGL3MS2/Basic_PB1.
Table 1.
Primers for quantitative PCR (qPCR) and oligonucleotides for plasmid construction used in this study
| Primer/oligos | Sequence (5′ → 3′) | Strand | Application |
|---|---|---|---|
| PB1_ F | GACAGGGACATTTGAATTCAC | Sense | qPCR |
| PB1_R | GGTTTGATCCCACAGCTTCTT | Antisense | |
| GAPDH_F | GTGAAGGTCGGAGTCAACGG | Sense | qPCR |
| GAPDH_R | TCAATGAAGGGGTCATTGATGG | Antisense | |
| NheI/PB1_ F | TTGCTAGCGGATCCGACAGGGACATTTGAATTCAC | Sense | Reporter vector |
| XhoI/PB1_R | ACGGATCCTCGAGGTTTGATCCCACAGCTTCTT | Antisense | |
| miR-3145_TS | GATCCGTTCAACTCCAAACACTCAAAACTCATTGTTGAATGGAATGAG ATATTTTGAGTGTTTGGAATTGAATTTTTTGGAAA | Sense | Silencing vector |
| miR-3145_BS | AGCTTTTCCAAAAAATTCAATTCCAAACACTCAAAATATCTCATTCCA TTCAACAATGAGTTTTGAGTGTTTGGAGTTGAACG | Antisense | |
| shRNA-Luc_TS | GATCCGCGCTGCTGGTGCCAACCCTTCAAGAGAGGG TTGGCACCAGCAGCGCTTTTTTGGAAA | Sense | Silencing vector |
| shRNA-Luc_BS | AGCTTTTCCAAAAAAGCGCTGCTGGTGCCAACCCTCTCTT GAAGGGTTGGCACCAGCAGCGCG | Antisense |
Note: Underline indicates recognition sites of restriction enzymes.
Construction of silencing vectors
The pSilencer 3.0-H1 (Ambion®, Foster City, CA, USA) was used as a parent vector to construct the miRNA expression vector and positive silencing vector. For pSilencer_miR-3145 (encodes for hsa-miR-3145 short hairpin RNA), two oligonucleotides (1 µg/µL each) – miR-3145_TS (top strand) and miR-3145_BS (bottom strand) – were mixed in 1X annealing buffer then denatured at 90℃ for 3 min, followed by annealing at 37℃ for 1 h (Table 1). After that, the annealed fragment was ligated into BamHI and HindIII restriction sites within a pSilencer3.0-H1 according to the manufacturer’s protocol. For pSilencer_FLuc (encodes for short hairpin RNA against firefly luciferase reporter gene), two oligonucleotides – shRNA-Luc_TS and shRNA-Luc_BS (Table 1) – were annealed and then inserted into pSilencer3.0-H1 with the same protocol as described above.
Plasmids propagation
Plasmids (pGL3MS2/Basic, pGL3MS2/Basic_PB1, pSilencer_scramble, pSilencer_FLuc and pSilencer_miR-3145) were transformed into competent cells (E. coli strain DH5α) (RBC Bioscience, New Taipei City, Taiwan) by a heat shock protocol and then a colony was selected based on ampicillin-resistant characteristics. Nucleotide sequencing was performed in order to confirm the fidelity of each recombinant vector. The concentration of each plasmid was measured by NanoDrop 1000 spectrophotometer (Thermo Scientific).
Cell culture
Human lung adenocarcinoma epithelial cells (A549) were cultured in DMEM (Thermo Scientific) containing 10% heat-inactivated fetal bovine serum (Thermo Scientific) and 1% (v/v) antibiotic-antimycotic (Gibco, Waltham, Massachusetts, USA) at 37℃ under 5% CO2 with 95% air atmosphere.
Plasmid transfection for 3′-UTR reporter assay
In 3′UTR reporter assay, A549 cells were seeded at 8 × 103 cells/well in medium without antibiotic-antimycotic in 96 well-plates and incubated for 24 hours. For transfection into each well, three plasmids including 100 ng of a reporter vector (pGL3MS2/Basic_PB1 or pGL3MS2/Basic), 50 ng of a silencing vector (pSilencer_scramble or pSilencer_miR-3145 or pSilencer_FLuc) and 10 ng of a control vector (pCMV_RLuc) were diluted with Opti-MEM (Gibco, Waltham, MA, USA) and then co-transfected into the A549 cells by using 0.4 µL of Lipofectamine®2000 (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol. The transfected cells were incubated in a humidified atmosphere with 5% CO2 at 37℃ for 72 hours and then harvested. The Renilla luciferase activity obtained from pCMV_RLuc was used for normalization of the relative luciferase assay.
Dual Luciferase Assay
The dual luciferase assay was performed by using Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instruction. Briefly, cells were washed with 100 µL of phosphate buffer saline and then lysed by adding 20 µL of the Passive Lysis Buffer. The suspensions were then transferred into a white opaque 96-well plate (Costar®). After that, 100 µL of Luciferase assay reagent II (LARII) was added into each well. The emissions of firefly luciferase activity at 560 nm were measured by Synergy HT Microplate reader (BioTek, Winooski, VT, USA). After that, 100 µL of Stop and Glow reagent was added in order to stop firefly luciferase and then measure Renilla luciferase activity at 480 nm. The assay was performed in triplicates in at least three independent experiments. The relative luciferase activity was calculated using signal intensities of firefly luciferase from a reporter vector divided by Renilla luciferase from a control vector.
Plasmid transfection for silencing of viral genes
In a 24-well plate, A549 cells were seeded at 5 × 104 cells/well in DMEM medium without antibiotic-antimycotic. After 24 h of incubation, 600 ng of a silencing vector (pSlilencer_ scramble or pSlilencer_miR-3145) was diluted with Opti-MEM (Gibco, Waltham, Massachusetts, USA) and then individually transfected into the A549 cells by using 1.25 µL of Lipofectamine®2000 (Invitrogen, Waltham, Massachusetts, USA). Then influenza A virus subtypes pH1N1 (A/Thailand/104/2009), H3N2 (A/Thailand/CU-H187/2010) and H5N1 (A/Thailand/NK165/2005) were prepared by diluting the virus stocks with DMEM medium to yield the desired viral titer. After 24 h post-transfection, cell culture media were removed from each well and cells were washed with 1 mL of phosphate buffer saline. After that, 500 µL of each virus suspension (MOI = 0.1) was added into each well and then incubated in a humidified atmosphere with 5% CO2 at 37℃ for 1 h with occasional shaking every 15 min to allow the virus to adsorb into the cells. After adsorption, the virus suspension was removed from each well and then washed with 1 mL of phosphate buffered saline. Finally, 1 mL of complete DMEM medium was added into each well and cells were incubated in a humidified atmosphere with 5% CO2 at 37℃ for 48 h. After that, supernatant and cell lysate were collected for further analysis. The experiments were performed in triplicates under a biosafety level 3 (BSL3) laboratory, Faculty of Medicine, Chulalongkorn University. The protocol of this study was approved by Institutional Review Board (IRB No. 121/55), Faculty of Medicine, Chulalongkorn University.
Real-time RT PCR
Total RNA was extracted from infected cell lysates by RBC Real genomics total RNA extraction kit (RBC Bioscience, New Taipei City, Taiwan) and then reverse transcribed with random hexamers by SuperScript® III reverse transcriptase (Invitrogen, Waltham, Massachusetts, USA). The viral PB1 gene and internal control GAPDH gene were quantified by Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific). The primers used for amplifications of GAPDH and PB1 genes are summarized in Table 1. Real-time PCR amplification was carried out in Step One Plus™ Real-time PCR Systems (Applied Biosystems, Foster City, CA, USA). The thermal profile included one 3 min cycle at 95℃ and 45 cycles of amplification (95℃ for 15 s, 60℃ for 15 s, 72℃ for 45 s with fluorescence detection at the end of each cycle). After that, a melting curve analysis was performed by increasing the temperature from 60 to 95℃ with a temperature transition rate of 0.5℃/s while continuously collecting the fluorescent signal. The results were analyzed using StepOne™ Software v.2.2 analysis. The relative quantitation was calculated by comparative CT method to compare the viral gene silencing between cells transfected with pSlilencer_miR-3145 and pSlilencer_scramble control.
Statistical analysis
Data obtained from relative luciferase activity and relative gene expression between each group was analyzed by Student’s t-test. A P ≤ 0.05 was considered statistically significant.
Results
Prediction of candidate human miRNA targeting multiple subtypes of influenza A viruses
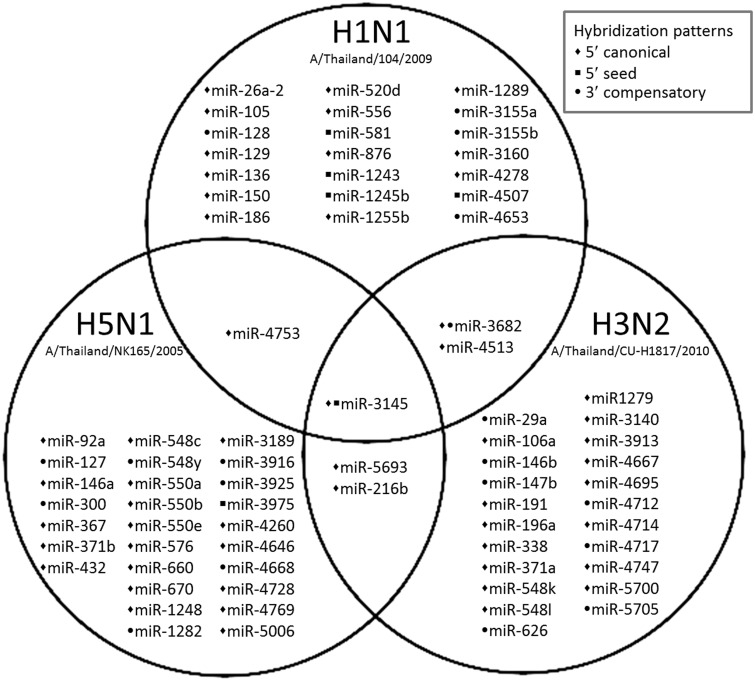
Viral genomes obtained from three subtypes of influenza A viruses including pH1N1 (A/Thailand/104/2009), H5N1 (A/Thailand/NK165/2005) and H3N2 (A/Thailand/CU-H1817/2010) were used to predict human miRNA targets based on the miRNA-target recognition patterns (5′ canonical or 5′ seed or 3′ compensatory) and minimum free energy for base pairing (less than -17.5 kcal/mol) criteria. Computational prediction revealed that there were 76 human miRNAs targeting theninfluenza A viruses used in this study. Seventy miRNAs were found to specifically target each subtype of influenza A virus, 21 of which targeted H1N1 subtype, 27 targeted H5N1 subtype and 22 targeted H3N2 subtype (Figure 1). There were 6 miRNAs that targeted multiple subtypes of influenza A viruses including hsa-miR-216 b (target on NA gene of H5N1 and H3N2 subtypes), hsa-miR-3682 (target on NS gene of pH1N1 and H3N2 subtypes), hsa-miR-4513(target on PA gene of pH1N1 and H3N2 subtypes), hsa-miR-4753 (target on PA gene of pH1N1 subtype and PB1 gene of H5N1 subtype), hsa-miR-5693 (target on PA gene of H5N1 and H3N2 subtypes) and hsa-miR-3145 (target on PB1 gene of pH1N1, H5N1 and H3N2 subtypes). The hybridization pattern and MFE of each predicted candidate miRNA are summarized in Table 2.
Figure 1.
Venn-Euler diagram shows a summary of candidate human miRNAs targeting influenza A viruses subtype pH1N1, H5N1, and H3N2. Bullets refer to different types of hybridization patterns between miRNAs and target viral RNA. The prediction showed that 76 human miRNAs target influenza A viruses, including 70 miRNAs that targeted specific subtype (21 for pH1N1, 27 for H5N1 and 22 for H3N2) and 6 miRNAs (miR-216b, miR-3145, miR-3682, miR-4513, miR-4753 and miR-5693) that targeted multiple subtypes of influenza A virus
Table 2.
Summary of candidate human miRNAs targeting multiple subtypes of influenza A viruses
| Human miRNAs | Viral Subtype | Viral gene (position) | Hybridization pattern | MFE (kcal/mol) | Pairing pattern |
|---|---|---|---|---|---|
| miR-216 b | H5N1 | NA (822–843) |  |
−31.3 | 5′canonical |
| H3N2 | NA (872–892) |  |
−30.1 | 5′canonical | |
| miR-3145 | pH1N1 | PB1 (1551–1570) | 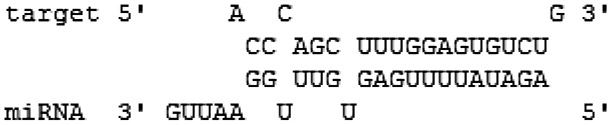 |
−18.2 | 5′seed |
| H5N1 | PB1 (1503–1541) | 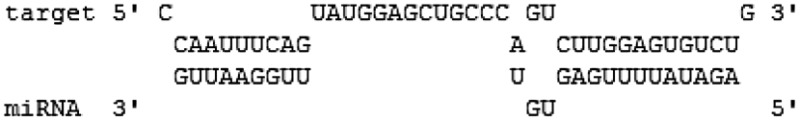 |
−20.7 | 5′canonical | |
| H3N2 | PB1 (1531–1549) |  |
−18.1 | 5′canonical | |
| miR-3682 | pH1N1 | NS (1011–1028) |  |
−19.1 | 5′canonical |
| H3N2 | NS (580–600) |  |
−22.6 | 3′compensatory | |
| miR-4513 | pH1N1 | PA (587–609) |  |
−32.3 | 5′canonical |
| H3N2 | PA (560–582) |  |
−32.3 | 5′canonical | |
| miR-4753 | pH1N1 | PA (1090–1115) |  |
−20.9 | 5′canonical |
| H5N1 | PB1 (541–564) |  |
−20.9 | 5′canonical | |
| miR-5693 | H3N2 | PA (1353–1370) |  |
−27.9 | 5′canonical |
| H5N1 | PA (1353–1370) |  |
−28.7 | 5′canonical |
Human miR-3145 targeting 3 subtypes of influenza A viruses
Interestingly, hsa-miR-3145 was the only candidate human miRNA targeting on PB1 gene of all three subtypes of influenza A viruses. The hybridization patterns between hsa-miR-3145 and its viral PB1 target in different subtypes of influenza A viruses were slightly different (Table 2). For pH1N1, the hybridization was classified as “5′seed” with the pairing energy or minimum free energy (MFE) at −18.2 kcal/mol. With H5N1, hsa-miR-3145 was hybridized with a “5′canonical” pattern with MFE at −20.7 kcal/mol. The PB1 of H3N2 targeted a “5′canonical” pattern with MFE at −18.1 kcal/mol. Moreover, alignment of PB1 genes obtained from various strains of human influenza A viruses revealed that PB1 target sites for the seed region of hsa-miR-3145 were conserved among various strains of the viruses (Figure 2). Implying that hsa-miR-3145 might be able to target PB1 of several strains of influenza A viruses. The silencing of PB1 expression by hsa-miR-3145 might have some effects on the replication of influenza A viruses. Therefore, hsa-miR-3145 was selected for further investigation.
Figure 2.
The alignment of viral PB1 gene revealed conserve target sites for seed region of hsa-miR-3145. Various strains in representative of four subtypes (pH1N1, H3N2, H5N1 and H7N9) of influenza A virus infecting humans were included in the alignment. The alignment was performed using Clastal W implemented in BioEdit software
Human miR-3145 targeted viral PB1 gene
To investigate the ability of hsa-miR-3145 to target viral PB1 gene, a 3′-UTR reporter assay was performed. According to the conserved target regions in PB1 genes among three viral subtypes, the pH1N1 virus was selected to be the representative for hsa-miR-3145 targeting validation. The pGL3MS2/Basic_PB1 reporter vector was constructed by adding a viral PB1 target region into 3′UTR of the firefly luciferase (FLuc) reporter gene in pGL3MS2/Basic vector. The pSilencer_scramble vector (a scrambled sequence that is not complemented by any gene of the human) and pSilencer_FLuc vector were constructed as negative silencing control vector and positive silencing control vector, respectively. The pSilencer_miR-3145 vector was constructed as a miR-3145 expression vector. The pCMV_RLuc vector was used for Renilla luciferase (RLuc) expression as a transfection control vector. The basis for using this assay is that the hybridization between miR-3145 and the viral PB1 target that linked with 3′-UTR of the firefly luciferase gene will trigger silencing of firefly luciferase activity compared to a control.
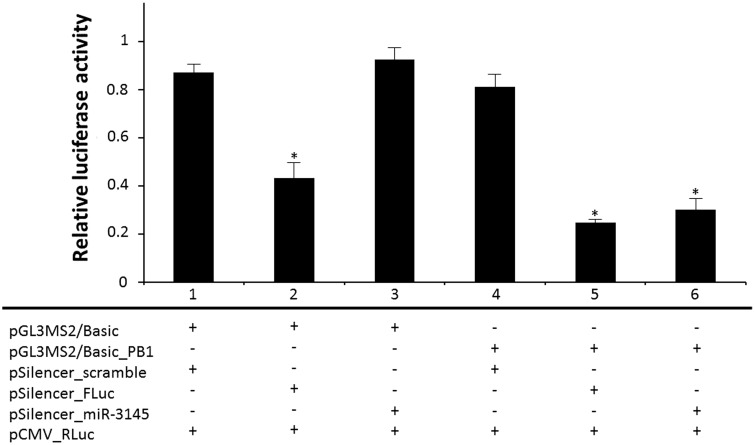
Basically, cells in each group were co-transfected with three plasmids including a reporter vector (pGL3MS2/Basic_PB1 or pGL3MS2/Basic), a silencing vector (pSilencer_scramble or pSilencer_FLuc or pSilencer_miR-3145) and a control vector (pCMV_RLuc). Each group was transfected with the same pCMV_RLuc control vector, but differed with respect to the types of reporter vector and silencing vector. Therefore, the transfected A549 cells were separated into six groups (Figure 3). In group 1, cells were transfected with pGL3MS2/Basic and pSilencer_scramble as the control group without target regions in the reporter vector and negative silencing control. In group 2, cells were transfected with pGL3MS2/Basic and pSilencer_FLuc was used as the positive silencing control due to the shRNA expression from pSilencer_FLuc specially targeting to FLuc reporter gene. As the results shown in Figure 3, the dual luciferase activity of group 2 was significantly lower (P = 0.0132) than those found in group 1, showing the effectiveness of the shRNA expression system. In group 3, pGL3MS2/Basic and pSilencer_miR-3145 were transfected into the cells in order to test whether miR-3145 triggered non-specific complementation with a firefly luciferase gene. Even the transfected cells could express hsa-miR-3145, but there was no target region in the reporter vector. As expected, the result revealed no significant difference (P = 0.3162) of the relative luciferase activity between group 3 and group 1, indicating that miR-3145 could not directly bind to the firefly luciferase gene. For group 4, cells were transfected with pGL3MS2/Basic_PB1 and pSilencer_scramble to test a silencing effect triggered by base complementation between an endogenous cellular miR-3145 and PB1 gene within 3′-UTR of the firefly luciferase gene. Results from group 4 showed no significant reduction (P = 0.3604) of the relative luciferase activity between group 4 and group 1, implying that the endogenous cellular miR-3145 was not expressed or was expressed at a very low level that was not sufficient to silence the reporter gene. In another positive silencing control in group 5, cells were transfected with pGL3MS2/Basic_PB1 and pSilencer_FLuc. The significant reduction of luciferase activity in group 5 compared to those found in group 1 and group 4 (P = 0.0026 and 0.0063 respectively) showed the effectiveness of positive silencing control. There was no significant difference of the relative luciferase activity observed between groups 2 and 5, implying that no additional silencing effect occurred when adding the viral PB1 target region into the 3′-UTR of the reporter gene. In group 6, pGL3MS2/Basic_PB1 and pSilencer_miR-3145 were transfected into the A549 cells. In this group, the A549 cells could overexpress hsa-miR-3145 to hybridize with a viral PB1 target region presented at 3′-UTR of the reporter gene. Interestingly, the relative luciferase activity in group 6 was significantly lower than those found in group 1 and group 4 controls (P = 0.0063 and 0.0115, respectively), indicating that hsa-miR-3145 triggered approximately a 63% reduction of the luciferase activity by targeting the viral PB1 segment within the 3′-UTR of the firefly luciferase reporter gene.
Figure 3.
The bar graph shows the relative luciferase activity among groups of transfected cells. Group 1, cells were transfected with pGL3MS2/Basic and pSilencer_scramble as the control group; group 2, cells were transfected with pGL3MS2/Basic and pSilencer_FLuc was used as the positive silencing control; group 3, cells were transfected with pGL3MS2/Basic and pSilencer_miR-3145 in order to test whether miR-3145 triggered non-specific complementary with a firefly luciferase gene; group 4, cells were transfected with pGL3MS2/Basic_PB1 and pSilencer_scramble to test a silencing effect triggered by base complementation between an endogenous cellular miR-3145 and the PB1 gene within 3′-UTR of the firefly luciferase gene; group 5, cells were transfected with pGL3MS2/Basic_PB1 and pSilencer_FLuc as another positive silencing control; group 6, cells were transfected with pGL3MS2/Basic_PB1 and pSilencer_miR-3145 to test the silencing effect of miR-3145. The pCMV_RLuc control vector was transfected into cells in each group. ‘*’ refers to significant differences from controls. Results revealed that the relative luciferase in groups 2, 5, and 6 were significantly lower (P < 0.05) than controls
Inhibition of influenza A virus replication triggered by hsa-miR-3145
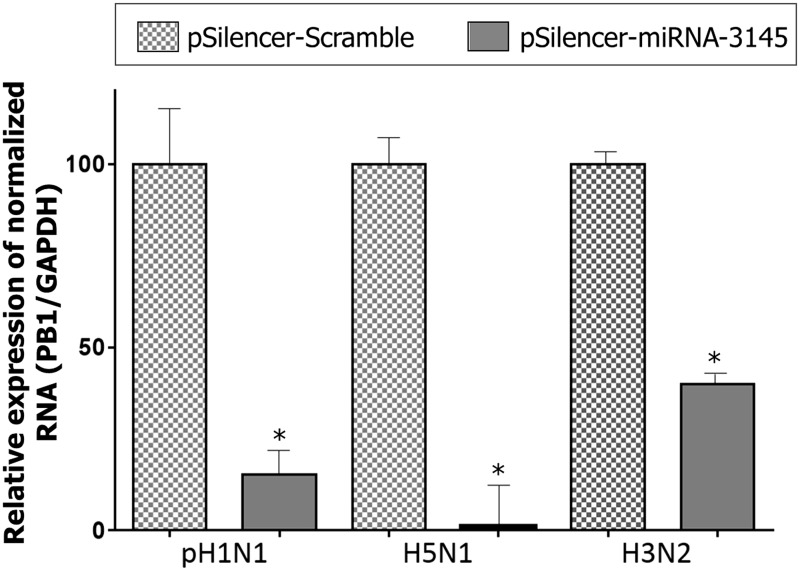
After 24 h of post-transfection with a silencing vector (pSilencer_scramble or pSilencer_miR-3145), the A549 cells were infected with pH1N1 influenza virus (MOI = 0.1). To investigate the antiviral activity of hsa-miR-3145, cell lysate was collected at 48 h post-infection followed by RNA extraction and cDNA synthesis. Expression level of viral PB1 gene was normalized by the amount of host GAPDH internal control gene, which was quantitated by real-time PCR. The infected cells with an overexpression of hsa-miR-3145 showed a significant reduction of viral PB1 RNA expression (P < 0.05) compared to cells transfected with pSilencer_scramble negative silencing control in all three subtypes of viral infection (Figure 4). For the pH1N1 subtype, the PB1 expression was decreased by approximately 84.62% (P = 0.0024) while that in the H5N1 subtype was decreased for 98.95% (P = 0.0004) and PB1 expression of H3N2 subtype was repressed 60.00% (P = 0.0001). Thus, the overexpression of hsa-miR-3145 seemed to inhibit the viral PB1 expression. The PB1 is one of the viral polymerase complexes (PB2, PB1 and PA) required for viral transcription and replication. Therefore, silencing of the viral PB1 gene led to inhibition of viral replication.
Figure 4.
Percentage of relative expression of viral PB1 gene normalized with GAPDH gene quantitated by real-time PCR. The result demonstrated the significant reduction (P < 0.05) of viral PB1 gene in the presence of miRNA-3145 compared to mock infection in all three subtypes of influenza A virus including pH1N1, H5N1 and H3N2. ‘*’ refers to significant differences from the control (mock infection)
Discussion
Although the computational tools of miRNA prediction are available, most of these tools can predict miRNAs target(s) only in some organisms, such as humans, mammals, fish, flies or worms.16,17 There are not many studies on predicting viral genes targeted by human miRNAs. The main category used in these tools is the complementary in the seed region of miRNA 5′ portion and their targets, leading to lots of false-positive and false-negative results.18 In this study, we used the categories based on the principle of miRNA target recognition.14 The hybridization patterns of miRNAs and their targets could be divided into three types; 5′ canonical, 5′ seed and 3′ compensatory. These patterns were used as the main criteria for the screening of candidate human miRNAs targeting various subtypes of influenza A virus. In addition, the pairing energy or minimum free energy (MFE) from the RNA Hybrid program was included in candidate miRNA selection. The criteria and prediction method used in this study was previously performed in order to identify miRNAs targeting HCV viral genes.15
In 2013, Zhang and colleagues reported that they identified miRNAs including hsa-miR-489, hsa-miR-325, hsa-miR-876-3p and hsa-miR-2117, which target H1N1 influenza A viral genes such as HA, PB2, MP and NS, respectively.19 Similarly, the hsa-miR-876 was also found to be pH1N1-targeting miRNA in our study. Previous studies indicated that human miRNAs can inhibit influenza virus replication. The hsa-miR-146a was reported to inhibit the replication of H1N1 and H3N2 subtypes of influenza A viruses.20 Ma et al. also verified that human miRNA let-7c can regulate the M1 expression of H1N1 influenza virus.21 Hsa-miR-26a, which was predicted to target the H1N1 influenza virus in our study, was also reported to inhibit the replication of the H1N1 subtype in MDCK cells.22 The hsa-miR-145 and hsa-miR-92a were suggested to target HA genes while hsa-miR-150 was reported to target PB2 genes of the H1N1 influenza A virus.23
However, most of the miRNAs targeting influenza viruses were differently identified in each study. This might be due to several factors including viral subtype, viral genome reference sequence, prediction software and miRNA selection criteria used in each individual study. According to the genetic variability of influenza A virus among different subtypes and strains, the aim of this study was to identify human miRNA targeting multiple subtypes of influenza A viruses infecting humans. Therefore, three subtypes of influenza A virus that infect humans, including pH1N1, H3N2 and H5N1, were analyzed. The result revealed that hsa-miR-3145 was the only human miRNA targeting influenza A viruses subtype pH1N1, H5N1, and H3N2. Interestingly, the target of this miRNA was the PB1 gene, which plays an important role in viral replication. Even the hybridizations between hsa-miR-3145 to three stains of influenza A viruses were slightly different, but the targets of miRNA seed region were conserved among various stains of the influenza A viruses as shown in the alignment of PB1 genes (Figure 2). H7N9 influenza A virus was recently reported to infect humans; therefore, the PB1 gene of H7N9 influenza A virus was also analyzed. Interestingly, H7N9 influenza A virus also contains conserved target sites for the seed region of hsa-miR-3145, implying that hsa-miR-3145 might be able to hybridize with PB1 gene of the H7N9 virus (Figure 2).
Similar to the study described by Song et al.,7 the 3′-UTR reporter assay was performed to validate the targeting and silencing ability of candidate miRNAs. For hsa-miR-3145 evaluation, the target site on viral PB1 gene was ligated to the 3′-UTR of luciferase reporter gene. It revealed that hsa-miR-3145 can target the viral PB1 and lead to the silencing activity.
Real-time RT-PCR was used to determine viral PB1 expression in A549 infected cells. The PB1 gene expression was significantly decreased in viral infected cells that were transfected with hsa-miR-3145 expression vector, indicating that hsa-miR-3145 triggered silencing of the PB1 gene. Interestingly, H5N1 subtype infected cells show a dramatic decrease in PB1 expression when compared to the pH1N1 and H3N2 infected cells. This may be caused by strong hybridization between the H5N1 PB1 target gene and hsa-miR-3145. The silencing of the PB1 gene by hsa-miR-3145 was proved in both 3′-UTR reporter assay and in vitro viral replication (Figures 3 and 4). The PB1 gene plays an important role in viral replication by encoding polymerase basic protein 1, which is the component of viral polymerase complex. When the PB1 gene was repressed, the viral replication would be inhibited, leading to the decrease in viral production. Thus, hsa-miR-3145 might have an antiviral effect against multiple subtypes of influenza A viruses.
The hsa-miR-3145 miRNA was first reported in the Melanoma miRNAome study by deep sequencing.24 Until now, its function or target has not been revealed. Unfortunately, the expression level of endogenous hsa-miR-3145 was very low in both uninfected and influenza viral-infected A549 cells (data not shown). This suggest that hsa-miR-3145 in the respiratory epithelial cell line is not constitutively expressed in normal conditions and not expressed in response to influenza viral infection. Therefore, further studies about induction mechanisms for endogenous hsa-miR-3145 expression might be useful for alternative antiviral approaches against influenza virus infection.
Conclusion
This study is the first report found that hsa-miR-3145 targeted multiple subtypes of influenza A viruses. The results revealed that hsa-miR-3145 targeted conserved regions in PB1 genes of influenza A viruses and triggered silencing of viral PB1. Thus, hsa-miR-3145 might be the best candidate for human miRNA inhibiting replication of influenza A viruses.
Authors’ contributions
KK performed in silico and in vitro analysis as well as drafted the manuscript. JM carried out viral infection and edit the manuscript. WP assisted in BSL3 process, data processing and graphic visualization. YP provided stock of viruses and revised the manuscript. SP took part in designing the study, data analysis, revision of the manuscript and coordination. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
We would like to appreciatively acknowledge the Department of Biochemistry and Research affairs, Faculty of Medicine, Chulalongkorn University, Thailand for their instrumental facilities. We would like to thank Prof. Jen-Ren Wang, Department of Medical Laboratory Science and Biotechnology, Center of Infectious Disease and Signaling Research, National Cheng Kung University, Taiwan for providing A549 cells. We also thank Assistant Prof. Nattanan Panjaworayan T-Thienprasert, Kasetsart University, Thailand for providing the original plasmids using in this study. Funding was supported by the Thailand Research Fund (TRF: RSA5680031); the Postdoctoral Fellowship, Ratchadapiseksompotch Fund (Faculty of Medicine, Chulalongkorn University); National Research University Project; Office of Higher Education Commission (WCU-007-HR-57); Centenary Academic Development Project (CU56-HR01); the Ratchadaphiseksomphot Endowment Fund of Chulalongkorn University (RES560530093), Research Chair Grant, National Science and Technology Development Agency (NSTDA) and Development and Promotion of Science and Technology Talents Project (DPST).
Declaration of conflicting interests
The authors hereby declare no personal or commercial conflict of interest with any aspect of this study.
References
- 1.Fleming DM, Chakraverty P, Sadler C, Litton P. Combined clinical and virological surveillance of influenza in winters of 1992 and 1993-4. BMJ 1995; 311: 290–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease C, Prevention. Update: Influenza A (H3N2)v transmission and guidelines – five states, 2011. MMWR Morbid Mortal Weekly Rep 2012; 60: 1741–4. [PubMed] [Google Scholar]
- 3.Tran TH, Nguyen TL, Nguyen TD, Luong TS, Pham PM, Nguyen vV, Pham TS, Vo CD, Le TQ, Ngo TT, Dao BK, Le PP, Nguyen TT, Hoang TL, Cao VT, Le TG, Nguyen DT, Le HN, Nguyen KT, Le HS, Le VT, Christiane D, Tran TT, Menno de J, Schultsz C, Cheng P, Lim W, Horby P, Farrar J, World Health Organization International. Avian Influenza Investigative Team Avian influenza A (H5N1) in 10 patients in Vietnam. New Engl J Med 2004; 350: 1179–88. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. microRNAs: tiny regulators with great potential. Cell 2001; 107: 823–6. [DOI] [PubMed] [Google Scholar]
- 5.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2007; 96 Suppl: R40–4. [PubMed] [Google Scholar]
- 6.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saïb A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science 2005; 308: 557–60. [DOI] [PubMed] [Google Scholar]
- 7.Song L, Liu H, Gao S, Jiang W, Huang W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol 2010; 84: 8849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucl Acid Res 2011; 39: D152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucl Acid Res 2014; 42: D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucl Acid Res 2006; 34: D140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths-Jones S. The microRNA Registry. Nucl Acid Res 2004; 32: D109–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucl Acid Res 2008; 36: D154–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 2004; 10: 1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol 2005; 3: e85–e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plakunmonthona S, T-Thienprasert NP, Khongnomnana K, Poovorawan Y, Payungporn S. Computational prediction of hybridization patterns between hepatitis C viral genome and human microRNAs. J Computat Sci 2014; 5: 327–31. [Google Scholar]
- 16.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003; 115: 787–98. [DOI] [PubMed] [Google Scholar]
- 17.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet 2005; 37: 495–500. [DOI] [PubMed] [Google Scholar]
- 18.Hamzeiy H, Allmer J, Yousef M. Computational methods for microRNA target prediction. Meth Mol Biol 2014; 1107: 207–21. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Li Z, Li Y, Liu Y, Liu J, Li X, Shen T, Duan Y, Hu M, Xu D. A computational method for predicting regulation of human microRNAs on the influenza virus genome. BMC Syst Biol 2013; 7: S3–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terrier O, Textoris J, Carron C, Marcel V, Bourdon JC, Rosa-Calatrava M. Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a. J General Virol 2013; 94: 985–95. [DOI] [PubMed] [Google Scholar]
- 21.Ma YJ, Yang J, Fan XL, Zhao HB, Hu W, Li ZP, Yu GC, Ding XR, Wang JZ, Bo XC, Zheng XF, Zhou Z, Wang SQ. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J Cell Molecul Med 2012; 16: 2539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Song L, Huang W. [MiR26a and miR939 regulate the replication of H1N1 influenza virus in MDCK cell]. Wei sheng wu xue bao (Acta Microbiol Sinica) 2010; 50: 1399–405. [PubMed] [Google Scholar]
- 23.He T, Feng G, Chen H, Wang L, Wang Y. Identification of host encoded microRNAs interacting with novel swine-origin influenza A (H1N1) virus and swine influenza virus. Bioinformation 2009; 4: 112–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark MS, Tyagi S, Nancarrow DJ, Boyle GM, Cook AL, Whiteman DC, Parsons PG, Schmidt C, Sturm RA, Hayward NK. Characterization of the Melanoma miRNAome by Deep Sequencing. PloS ONE 2010; 5: e9685–e9685. [DOI] [PMC free article] [PubMed] [Google Scholar]