Highlights
-
•
Knockdown of Zfp521 in BCL1 cell culture reduces viability and promotes apoptosis.
-
•
Genes expressed in B cells are down-regulated in cells with Zfp521 knockdown.
-
•
Cyclin D1 expression is increased in mouse tumors with Zfp521 over-expression.
Keywords: Zfp521, Pax5, Cyclin D1, Zfp423, Ebf1, Evi3
Abstract
Leukemia arises due to the dysregulated proliferation of hematopoietic progenitor cells. Errors in the multi-step commitment process result in excessive numbers of immature lymphocytes, causing malignant disease. Genes involved in the differentiation of lymphocytes are often associated with leukemia. One such gene, Zfp521, has been found to cause B-cell leukemia in mice when over-expressed. The role of Zfp521 in B-cell differentiation, and the mechanisms by which it leads to leukemic transformation, are unclear. In this study we report that Zfp521 knockdown causes apoptosis in a B-cell culture system and promotes down-regulation of genes acting at late stages of B-cell differentiation. We identify Pax5 and cyclin D1 as Zfp521 target genes, and suggest that excessive B-cell proliferation observed in mice with retroviral insertions near the Zfp521 gene is due to an up-regulation of cyclin D1 in B-cells. Overall, these results suggest links between dysregulated Zfp521 and B-cell survival.
1. Introduction
Leukemia originates due to errors in the hematopoietic differentiation of stem cells into mature lymphocytes [1]. Lymphocyte development requires a multi-step process whereby common lymphoid progenitors differentiate into B and T lineage-specific cells [2]. Developmental control of early B lineage cell differentiation is exerted by a regulatory network of key transcription factors [3], [4]. Sequential actions of these transcription factors results in the multi-step differentiation of mature B-cells from immature progenitors in the bone marrow [4].
Recombinant inbred (RI) mouse stains are a significant resource for leukemia gene discovery [5]. These mice harbor somatic viral insertions that alter the expression of tumor suppressor genes and proto-oncogenes [5], [6]. Sites of proviral insertion that are common to many animals within a strain or between multiple strains identify genomic locations where proto-oncogenes or tumor suppressor genes reside. The RI strain AKXD-27 possessed B-lineage lymphomas with a common proviral insertion site, designated as ecotropic viral integration site 3 (Evi3) [7]. Within the AKXD-27 strain 70% of mice exhibiting pre-B cell and B-cell tumors contained proviral insertions at the Evi3 locus; no rearrangements at the Evi3 locus were found in either T-cell or myeloid tumors [7], [8]. The high frequency of B-lineage lymphoma in mice with a proviral insertion at the Evi3 locus suggests that proviral insertion alters the expression of genes near the insertion site to promote B-lineage lymphoma.
Analysis of the genomic region surrounding the Evi3 retroviral integration site revealed that the virus had inserted upstream of a previously uncharacterized gene, which encodes a 30-zinc-finger protein with predicted DNA-binding and protein interaction domains [8], [9]. This gene was found to be up-regulated in tumors with retroviral insertions at Evi3, due to the strong viral promoters driving endogenous gene expression [9]. The human ortholog of this gene, EHZF, was identified due to its specific expression in CD34+ early hematopoietic progenitor stem cells [10]. Due to its zinc finger motifs, the gene at the Evi3 insertion site has been renamed Zfp521 in mice and ZNF521 in humans.
Although studies on the molecular function of Zfp521 have revealed a role in transcriptional regulation via chromatin remodeling [11], its place within the transcriptional network regulating B-cell differentiation remains unclear. To better understand the role of Zfp521 in B-lymphocytes, we developed a knockdown system in the lymphoblast cell line BCL1, which secretes IgD and IgM antibodies [12]. We assayed B-cell gene expression in this system, and found that certain genes which were up-regulated in B-cell tumors from AKXD-27 mice with Evi3 retroviral insertions [8] were conversely down-regulated in Zfp521 knockdown cells. Knocking down Zfp521 resulted in decreased cellular viability and increased cellular apoptosis. Using a cell viability rescue assay, we identified cyclin D1 as a potential mediator of increased B-cell proliferation in Zfp521 over-expressing leukemias, and propose a position for Zfp521 within the B-cell differentiation transcriptional regulatory network.
2. Materials & methods
2.1. Cell culture
Mouse lymphoblast cells (BCL1; ATCC® TIB-197) were cultured in RPMI 1640 medium supplemented with 2 mM L-glutamine (Lonza), 0.05 mM 2-mercaptoethanol (Sigma-Aldrich), 15% FBS, 5% penicillin, 5% streptomycin at 37 °C with 5% CO2.
2.2. Zfp521 shRNA constructs
shRNA constructs were purchased from OriGene (OriGene Technologies: SR422637). Four independent Zfp521 shRNA expression vectors with a CMV-Green Fluorescent Protein (GFP) marker were combined at equal concentration for transfection. A vector containing scrambled Zfp521 shRNA sequence and an empty vector lacking any shRNA sequence were used as controls.
2.3. Transfection
1 μg plasmid DNA was transfected into 1 × 105 BCL1 cells with FuGENE HD (Roche) in OptiMEM Media (Sigma). Transfection efficiency was calculated based on the GFP expression for each individual plasmid.
2.4. Viability assay
BCL1 were plated in triplicate at a density of 1 × 105 cells per well in 96-well plate. After 24 h, cells were transfected with shRNA plasmids or appropriate control plasmids and cultured for 1, 3 or 7 days. Cultured cells were incubated with CellTiter 96R Aqueous Non-Radioactive Cell Proliferation Assay (MTS) according to manufacturer’s instructions (Promega; no. G5421). Absorbance was recorded at 490 nm (Bio-Tek Powerwave HT Microplate Reader). Each assay was repeated with six technical replicates and three biological replicates.
2.5. Trypan blue stain
BCL1 cells were trypsinized in 1 ml trypsin (Sigma); cells were re-suspended in BCL1 media. An equal amount of cell suspension and trypan-blue solution (Sigma) were mixed together. Cells were visualized under light microscopy. Five different squares from a hemocytometer grid were counted to determine the total cell number and number of dead cells (stained blue). Each assay was repeated with three technical replicates and three biological replicates.
2.6. Caspase-3/7 assay
BCL1 cells were plated in triplicate at a density of 1 × 105 cells per well in 96-well plate. After 24 h, cells were transfected with Zfp521 shRNA or control plasmids as described. Caspase activity was assessed using Apo-ONE Homogenous Caspase-3/7 Assay (Promega; no: G7792) according to the manufacturer’s instructions. Absorbance was recorded at 490 nm. Wells with no cells were used a blank, and the average absorbance value of the blank was subtracted from the average absorbance value for each treatment condition. Fold change for each experimental condition was calculated by normalization to mock-transfected cells. Each assay was repeated with three technical replicates and three biological replicates.
2.7. Real-time quantitative PCR analysis
Total RNA was extracted with TRI reagent (Sigma), treated with DNaseI (Promega), reverse transcribed using random primers and prepared for real-time quantitative PCR (Promega GoTaq qPCR mix) according to manufacturer’s instructions. Primer sequences are listed in Supplemental Table 1. Reactions were performed in triplicate. Thermal cycling parameters were: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Expression levels were normalized to 18S RNA. Expression was analysed using the ΔΔCT method [13]. Results were analyzed by t test for statistical significance.
2.8. Site-directed mutagenesis
Site-directed mutagenesis primers were designed using software from New England BioLabs (http://www.neb.com/products/e0554-q5-site-directed-mutagenesis-kit). The Q5 site-directed mutagenesis kit (New England BioLabs; E0554S) was used following manufacturer’s instructions to create a stop codon within the Zfp521 sequence.
Forward primer: 5′-TGCACAGCTGAGACAGCTGCC-3′
Reverse primer: 5′-CAGCGTCCTCCTCCAACTC-3′
2.9. Rescue assay
BCL1 cells were plated in 24-well plates at a density of 1.0 × 106 cells per well. Cells were transfected with Zfp521 shRNA plasmid or control plasmids. The plates were incubated for 3 days, after which cells were transfected again with the plasmids listed below. An identical replicate control plate was mock-transfected without any addition of any plasmids. Plasmids transfected into knockdown cells: full-length Zfp521, mutant ΔZfp521, Zfp521Stop, Pax5 wild type, Pax5 V26G, Ebf1, and cyclin D1. Following the rescue transfection cells were subjected to the viability assay described above. The ratio of viable cells at day 7 post-transfection for rescued cells to cells without rescue was calculated for each transfection condition. A pairwise t test was performed to assess statistical significance.
3. Results
3.1. Generation of a Zfp521 B-cell knockdown system
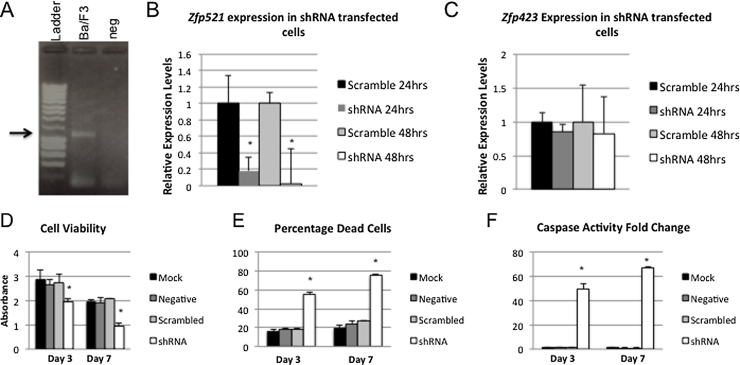
The majority of tumors found in AKXD-27 mice with over-expression of Zfp521 were of the B-lineage, at the Pro-B cell stage of differentiation [8]. Zfp521 expression has been reported in wild type mouse B-cells during early stages of B-cell differentiation [14], and we have detected Zfp521 expression in the IL-3 dependent murine pro-B-cell line Ba/F3 [15] (Fig. 1A), as well as in the mouse lymphoblast cell line BCL1 (Fig. 1B). To investigate the effects of reduced Zfp521 expression in B-cells, BCL1 cells were transfected with either a cocktail of four Zfp521 shRNA knockdown plasmids, a gene-specific scrambled control plasmid (scrambled Zfp521 target sequence), or an empty vector control. Mock-transfected cell were also analyzed as a further negative control. Zfp521 expression was assayed by qPCR in BCL1 cells 24 h and 48 h post-transfection. There was a significant decrease in Zfp521 expression in cells transfected with the knockdown plasmid cocktail as compared to cells transfected with the scrambled control plasmid at both 24 and 48 h post-transfection (Fig. 1B). We sought to confirm that our BCL1 cells demonstrated a specific knockdown of Zfp521 without affecting expression of its paralog Zfp423; therefore we measured the expression levels of Zfp423 in cells transfected with Zfp521 shRNA or the scrambled control plasmid. We found no difference in the expression levels of Zfp423 in cells with Zfp521 knockdown as compared to control plasmid transfected cells, confirming that the knockdown is specific to Zfp521 (Fig. 1C).
Fig. 1.
The Zfp521 BCL1 B-cell knockdown system. A. Zfp521 expression is present in the Pro-B cell line Ba/F3. Ladder = Bioline Hyperladder I, neg = no template control. B. Zfp521 expression is significantly reduced in BCL1 cells transfected with a specific shRNA as compared to mock-transfected cells at both 24 h (*p = 0.03; t test) and 48 h (*p = 0.006; t test). C. Zfp423 expression is unaffected in cells transfected with Zfp521 shRNA at either 24 h (p = 0.19; t test) or 48 h (p = 0.69; t test). D. The presence of viable cells is significantly reduced in cultures transfected with Zfp521 shRNA when compared to cells undergoing mock transfection at 3 days and 7 days post-transfection (*p < 0.05; t test), or transfection with vector containing no shRNA (negative) or scrambled shRNA sequence. E. The percentage of dead cells at days 3 and 7 post-transfection as identified by trypan blue staining is significantly increased in cells transfected with Zfp521 shRNA as compared to cells undergoing mock transfection (*p < 0.05; t test), or transfection with vector containing no shRNA (negative) or scrambled shRNA sequence. F. The fold-change in the levels of Caspase 3/7 activity in cells transfected with Zfp521 shRNA or control plasmids as compared to mock-transfected cells. A significant increase in Caspase 3/7 activity is detected in Zfp521 shRNA transfected cells compared to mock-transfected cells at day 3 and day 7 (*p < 0.05; t test) post-transfection.
3.2. Zfp521 knockdown cell phenotype
We employed an enzymatic cell viability assay to determine the number of live cells in our different transfection conditions. We found that Zfp521 knockdown cells had reduced viability as compared to other transfection conditions, with a significant decrease in cell viability by day 7 post-transfection (Fig. 1D). To confirm these findings, trypan blue staining was used to identify dead cells in each transfection condition. The percentage of dead cells was significantly increased in the Zfp521 knockdown transfection cells as compared to control transfection samples (Fig. 1E).
Because we detected reduced cell viability in Zfp521 knockdown cells, we sought to determine if the reduced viability was due to increased apoptosis. We measured caspase-3/7 activity at 24 h, 3 days, and 7 days post-transfection in Zfp521 knockdown and control plasmid transfected cells. The fold change in levels of apoptotic cells was increased in cells transfected with the Zfp521 shRNA plasmid cocktail as compared to mock transfection conditions, with a significant difference detected at day 7 post-transfection (Fig. 1F).
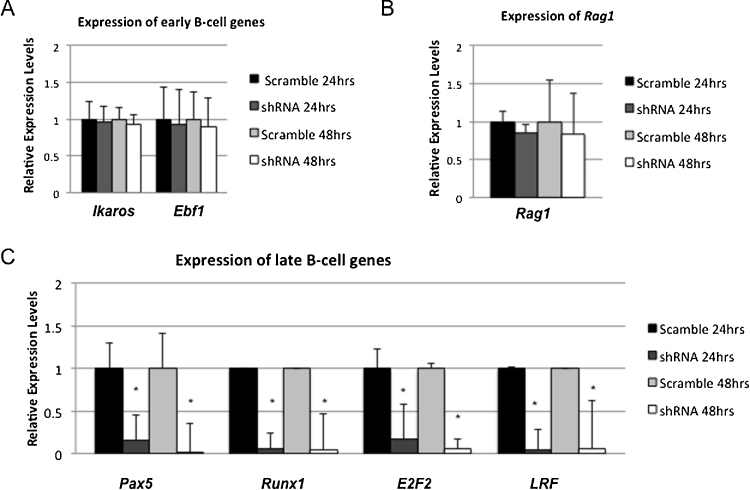
3.3. B-cell gene expression analysis
To determine the effect of the knockdown of Zfp521 on genes required for B-cell differentiation, we examined gene expression using quantitative RT-PCR in cells with Zfp521 knockdown as compared to control transfected cells. We analyzed the expression of B-cell transcription factors required prior to the Pro-B-cell stage of differentiation: Ikaros and Ebf1 [4]. Ikaros plays a critical role in regulating lymphocyte development, function, and homeostasis [16]. Early B-cell factor (EBF) is a transcription factor involved in the transcriptional regulation of many B-cell restricted genes, which is essential for B lymphocyte development [17]. There was no reduction in the expression of either gene in Zfp521 knockdown cells (Fig. 2A). Rag1 and Rag2 are lymphocyte-specific genes involved in V(D)J rearrangement of immunoglobulin (Ig) [18], which are expressed in early lymphoid progenitors [19]. No difference was observed in the expression of Rag1 in cells with Zfp521 knockdown as compared to control-transfected cells (Fig. 2B). We analyzed the expression of genes acting at later stages of B-cell differentiation: Pax5, Runx1, E2F2, and Lrf. Pax5 is expressed from early pro-B stage until the final stage of differentiation [20], and functions at the Pro-B-cell stage to activate genes crucial to B-cell lineage differentiation and repress genes required for commitment to other hematopoietic lineages [21]. Runx1 is expressed in early B-cell progenitor and in immature and mature B-cells [22]. Mice with deletions of E2F1 and E2F2 have inhibited B-cell differentiation, with development arrested at the pre-BII stage, indicating a requirement for E2F2 at the Large Pre-BII to Small Pre-BII differentiation transition [23]. Lrf regulates the lineage fate of mature B cells, and is highly expressed in lymphoma cell lines [24]. We found reduced Pax5, Runx1, E2F2, and Lrf gene expression levels in Zfp521 knockdown cells as compared to controls (Fig. 2C).
Fig. 2.
Expression of B-cell genes in Zfp521 knock-down cells. A. The expression of early B-cell genes was assayed by qPCR at 24 and 48 h post-transfection in BCL1 cells transfected with Zfp521 shRNA or scrambled control plasmid. B. The expression of the recombination gene Rag1 in scrambled control or Zfp521 shRNA transfections. C. The expression of late B-cell marker genes at 24 and 48 h post-transfection. Expression was significantly reduced in Zfp521 shRNA transfected cells as compared to cells transfected with scrambled plasmid for all genes reported in Panel D (*p < 0.05; t test). All panels show expression normalized to 18S RNA control.
3.4. Zfp521 knockdown rescue assay
To confirm that the reduced cell viability and increased apoptosis we saw in BCL1 knockdown cells was due to a loss of Zfp521 expression, we developed a rescue assay system in these cells. Cells were transfected with the Zfp521 knockdown shRNA cocktail or control plasmids as described above. On day 3 after the shRNA transfection, cells were transfected with a rescue construct. Cell viability was measured on day 7 after the rescue transfection. The ratio of viable cell number between cells receiving the rescue plasmid (denoted 7 + ) compared to control cells receiving no rescue plasmid (denoted 7) was calculated. We found that the addition of wild type Zfp521 resulted in a rescue of cell viability in cells originally transfected with the Zfp521 shRNA (Fig. 3A). The presence of caspase 3/7 activity was also measured at day 7 after the recue transfection. Cells that were not transfected with the Zfp521 rescue plasmid showed a significant increase in apoptosis following Zfp521 knockdown, however no such increase was noted in cells with Zfp521 rescue transfection (Fig. 3B).
Fig. 3.
Viability and apoptosis levels in rescue assay cell cultures. A. The ratio of cell viability measured on day 7 post-transfection for cells with Zfp521 wild type rescue plasmid added (day 7 + ) to cells without rescue plasmid added (day 7). A significant increase in viability was found in shRNA cells with Zfp521 wild type rescue plasmid added as compared to mock-transfected cells with rescue (*p < 0.05; t test). B. The fold-change in Caspase 3/7 activity in each condition as compared to mock-transfected cells. Cells without rescue are shown on the left, and cells with rescue plasmid added are shown in the right. A significant different was found between caspase activity levels in cells without Zfp521 rescue plasmid transfection to cells with rescue (*p < 0.05; t test). C. The ratio of cell viability measured on day 7 post-transfection for cells with ΔZfp521 plasmid added. No significant difference was detected in shRNA cells with ΔZfp521 plasmid added as compared to mock-transfected cells (p = 0.67; t test). D. The ratio of cell viability measured on day 7 post-transfection for cells with Zfp521Stop plasmid added. No significant difference was detected in shRNA cells with Zfp521Stop plasmid added as compared to mock-transfected cells (p = 0.15; t test). For all panels, the original transfection condition is shown in the legend.
Prior studies have shown that the final 4 zinc fingers of Zfp521 are required for protein-protein interactions with the B-cell transcription factor EBF1 [25]. A mammalian expression construct lacking the final 6 zinc fingers of Zfp521 has previously been demonstrated to modulate EBF1 activity [8]. This construct (ΔZfp521) did not rescue cell viability in cells transfected with the Zfp521 shRNA (Fig. 3C). The ΔZfp521 construct was generated through an internal restriction enzyme deletion of the wild type Zfp521 expression plasmid, and therfore has a reduced nucleotide length compared to the wild type Zfp521 expression plasmid. To confirm that the lack of rescue in Zfp521 knockdown cells was not an artifact of the ΔZfp521 construct, we generated another mutant Zfp521 expression construct through site-directed mutagenesis. This new construct (Zfp521Stop) is the same as the wild type Zfp521 plasmid with the exception of a single nucleotide substitution that generates a premature stop codon at amino acid 58 of Zfp521. The Zfp521Stop construct was also unable to rescue cell viability in our assay (Fig. 3D).
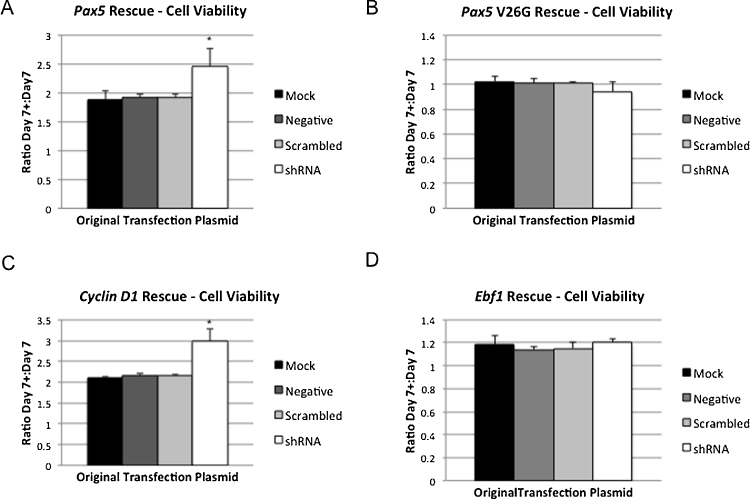
3.5. Zfp521 target gene rescue assay
We used our rescue assay system to examine potential downstream targets of Zfp521. We examined Pax5 because Pax5 expression levels are increased in lymphoid tumors from mice with retroviral insertions at the Evi3 site [8], supporting the hypothesis that Pax5 is downstream of Zfp521. To control for changes in viable cell number caused by general proliferative actions of the rescue candidates, we calculated the ratio of increase in viable cell number on day 7 for cells in each knockdown condition receiving the rescue plasmid (day 7 + ) as compared to cells without the addition of rescue plasmid (day 7). We found that the addition of wild type Pax5 resulted in a significant increase in cell viability in Zfp521 knockdown cells (Fig. 4A). To determine whether the DNA binding function of Pax5 was required for rescue, a construct containing a V26G mutation in Pax5 that lacks DNA binding ability was used [26]. The addition of mutant Pax5 showed no increase in viability of Zfp521 knockdown cells (Fig. 4B).
Fig. 4.
Cell viability measurements in rescue assays. A. The ratio of viable cells detected in cultures at day 7 post-transfection with wild type Pax5 rescue plasmid (7+) compared to cells mock transfected on day 7. A significant increase in viable cell number is seen in cells originally transfected with Zfp521 shRNA when compared to other transfection conditions (*p < 0.05; t test). B. The ratio of viable cells at day 7 post-transfection with Pax5 V26G mutant construct compared to cells without rescue. No significant differences were detected between any transfection groups. C. The ratio of viable cells in rescue assays with the addition of cyclin D1 plasmid as compared to cells with no rescue. A significant increase in viable cell number is seen in cells originally transfected with Zfp521 shRNA when compared to other transfection conditions (*p < 0.05; t test). D. The ratio of viable cells at day 7 post-transfection with Ebf1 plasmid compared to cells without rescue. No significant differences were detected between any transfection groups. For all panels, the original transfection condition is shown in the legend.
Notably, cyclin D1 has been identified as an indirect target of Zfp521 in chondrocytes, and depletion of Zfp521 in chondrocytes results in reduced cell proliferation [27]. We wished to examine whether the cell proliferation defects observed in Zfp521 knockdown cells might therefore be a consequence of perturbed cyclin D1 activity. Our rescue assay confirmed that the addition of cyclin D1 rescued cell viability specifically in the Zfp521 knockdown cells (Fig. 4C), suggesting that loss of Zfp521 disrupts cyclin D1 regulation in B-cells.
The B-cell transcription factor Ebf1 was found to be over-expressed in tumors with Zfp521 retroviral insertions [8], and is a putative binding partner for Zfp521 [28], [29]. Yet in our knockdown assay we found no significant alterations in Ebf1 expression levels. To further delineate the relationship between Ebf1 and Zfp521 we examined whether the introduction of Ebf1 expression would affect Zfp521 knockdown cell viability in our rescue assay. However, the addition of Ebf1 did not rescue cell viability defects (Fig. 4D), consistent with the finding that Ebf1 expression is not altered by the knockdown of Zfp521. This finding suggests that Ebf1 is not a downstream target of Zfp521.
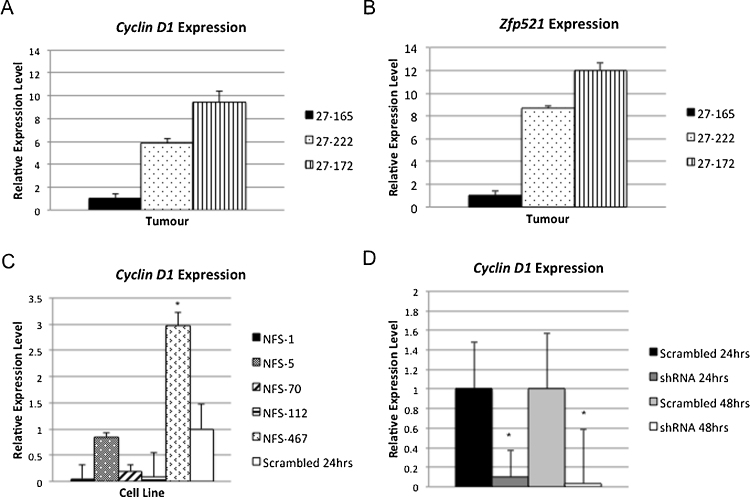
3.6. AKXD27 tumor expression analysis
Based on the finding that cyclin D1 rescued cell viability in Zfp521 knockdown cells, we examined whether AKXD27 B-cell tumors had perturbed expression of cyclin D1. We found that cyclin D1 expression is up regulated in tumors from AKXD27 mice with retroviral insertions at Evi3 as compared to a control tumor without an Evi3 retroviral insertion (Fig. 5A). Zfp521 over-expression is also detected in the tumors with retroviral insertions at the Evi3 locus (Fig. 5B). Furthermore, cyclin D1 expression is increased in a B-cell leukemia cell line (NSF-467) with a retroviral insertion at the Evi3 locus, when compared to B-cell lines without Evi3 retroviral insertions (Fig. 5C). We also confirmed that cyclin D1 expression is reduced in cells with Zfp521 knockdown compared to control cells, indicating that cyclin D1 is downstream of Zfp521 in B-cells (Fig. 5D).
Fig. 5.
Expression analysis in AKXD27 tumors and leukemia cell lines. A. cyclin D1 expression measured by qPCR in tumors from mice with retroviral insertions at Evi3 (27-222 and 27-172) is increased as compared to a control tumor (27-165). B. Zfp521 expression levels in the same AKXD27 tumors. C. cyclin D1 expression levels in leukemia cell lines. The NFS-467 cell line has a retroviral insertion at the Evi3 locus, and shows a significant increase in cyclin D1 expression as compared to BCL1 cells transfected with the Zfp521 scrambled shRNA (*p < 0.05; t test). D. The expression levels of cyclin D1 in BCL1 cells transfected with either the Zfp521 shRNA or scrambled shRNA at 24 and 48 h post-transfection. A decrease in cyclin D1 expression is shown in Zfp521 shRNA transfections compared to cells transfected with scrambled shRNA sequence (*p < 0.1; t test).
4. Discussion
Zfp521 encodes a transcription factor with an N-terminal transcriptional repressor motif [30] and 30 Kruppel-like-zinc finger domains [8], [9]. Due to the increased numbers of B-cells in mice with Zfp521 over-expression, we postulated that Zfp521 may function during B-cell development to regulate B-cell viability or proliferation. Additionally, the finding that mice with Zfp521 over-expression have an inappropriate B-cell surface marker profile, with upregulation of markers expressed from the pro-B stage of differentiation [8], raises the possibility that Zfp521 functions as part of the B-cell transcription factor network to promote differentiation events.
The results from our Zfp521 knockdown culture system are consistent with the above hypotheses. We discovered that BCL1 cell viability is reduced and apoptosis is significantly increased when Zfp521 is knocked down, suggesting a role for Zfp521 in B-cell survival. Our results support a role for Zfp521 in the regulation of genes expressed from the Pro-B-stage onwards. These results are consistent with the findings from a recent study reporting a role for Zfp521 in regulation of the pre-B-cell receptor genes BTK, BANK1, and BLNK [14]. Moderate levels of ZNF521 expression have also been noted in the human Raji (pre-B) cell line [10].
Retroviral insertion at the Evi3 locus causing Zfp521 over-expression is a cooperative event in the development of acute B-lineage leukemia in mice expressing an E2A-HLF chimeric protein associated with acute lymphoblastic leukemia t(17;19) [31]. The tumors in these mice are of the B-progenitor type, and express B220 and Cd19, similar to tumors found in AKXD27 mice with retroviral insertions at the Evi3 locus [8]. Further studies revealed ZNF521 overexpression in human leukemic cell lines with t(17;19) translocations [31], suggesting that the mechanism by which over-expression of Zfp521/ZNF521 causes B-cell leukemia may be conserved between mouse and human. Transgenic mice that over-express both the E2A-HLF fusion gene and Zfp521 showed increased proliferation of B-cell progenitors [31]. Our study suggests that the increased proliferation found in Zpf521 over-expressing Pro-B-cells is mediated by cyclin D1. Additionally, Zfp521 has been reported to regulate cyclin D3 expression in Pre-B-cells [14], suggesting that perturbations of Zfp521 expression disrupt the G1/S transition in the cell cycle.
The knockdown of Zfp521 in cultured hematopoietic progenitor cells has been reported to increase B-cell differentiation, as well as increase overall cell number in the culture [30]. The B-cell genes Ebf1, Pax5, and Mb1 were also up-regulated in these knockdown cultures [30]. It is possible that knocking down Zfp521 at the Pro-B-cell stage has an opposite effect to its knockdown in progenitor cells. Knockdown of Zfp521 at the Pre-B-cell stage also reduces cell proliferation [14], similar to our findings at the Pro-B-cell stage. Notably, in Pro-B-cell tumours with Evi3 retroviral insertions, Zfp521 over-expression occurs concomittantly with Ebf1, Pax5, and Mb1 over-expression, suggesting that Zfp521 has a positive effect on the transcription of these B-cell factors at the Pro-B-cell stage [8]. We therefore propose that Zfp521 functions within the B-cell transcriptional network to promote differentiation events subsequent to the Pro-B-cell stage of lineage commitment. Further study is needed to determine how perturbations of Zfp521 function at different stages of B-cell development disrupt the balance of gene expression patterns and cellular activities to culminate in leukemic transformation of B-lineage cells.
Conflict of interest
The authors declare that there are no conflicts of interest.
Acknowledgements
We thank John Murphy of University of Westminster for the kind gift of BCL1 cells. We thank Chris Thompson of the University of Manchester for access to qPCR equipment. We thank Shannon Kenney of the University of Wisconsin for Pax5 wild type and mutant expression plasmids. We thank Richard Pestell of Thomas Jefferson University for the cyclin D1 expression construct. We thank Mikael Sigvardsson of Lund University for the EBF expression plasmid. We thank Monica Justice of the University of Toronto for AKXD27 tumor tissues and NFS cell lines. S.A. was supported by a Kuwait Cultural Office. K.W. was supported by BBSRC CASE Ph.D. studentship BB/J500173/1.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.leukres.2016.03.013.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Shlush L.I., Minden M.D. Preleukemia the normal side of cancer. Curr. Opin. Hematol. 2015;22:77–84. doi: 10.1097/MOH.0000000000000111. [DOI] [PubMed] [Google Scholar]
- 2.Fuxa M., Skok J.A. Transcriptional regulation in early B cell development. Curr. Opin. Immunol. 2007;19:129–136. doi: 10.1016/j.coi.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Schebesta M., Heavey B., Busslinger M. Transcriptional control of B-cell development. Curr. Opin. Immunol. 2002;14:216–223. doi: 10.1016/s0952-7915(02)00324-2. [DOI] [PubMed] [Google Scholar]
- 4.Choukrallah M.A., Matthias P. The interplay between chromatin and transcription factor networks during B cell development: who pulls the trigger first? Front. Immunol. 2014;5:156. doi: 10.3389/fimmu.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mucenski M.L., Taylor B.A., Jenkins N.A., Copeland N.G. AKXD recombinant inbred strains: models for studying the molecular genetic basis of murine lymphomas. Mol. Cell. Biol. 1986;6:4236–4243. doi: 10.1128/mcb.6.12.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert D.J., Neumann P.E., Taylor B.A., Jenkins N.A., Copeland N.G. Susceptibility of AKXD recombinant inbred mouse strains to lymphomas. J. Virol. 1993;67:2083–2090. doi: 10.1128/jvi.67.4.2083-2090.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Justice M.J., Morse H.C., 3rd., Jenkins N.A., Copeland N.G. Identification of Evi-3, a novel common site of retroviral integration in mouse AKXD B-cell lymphomas. J. Virol. 1994;68:1293–1300. doi: 10.1128/jvi.68.3.1293-1300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hentges K.E., Weiser K.C., Schountz T., Woodward L.S., Morse H.C., Justice M.J. Evi3, a zinc-finger protein related to EBFAZ, regulates EBF activity in B-cell leukemia. Oncogene. 2005;24:1220–1230. doi: 10.1038/sj.onc.1208243. [DOI] [PubMed] [Google Scholar]
- 9.Warming S., Liu P., Suzuki T., Akagi K., Lindtner S., Pavlakis G.N. Evi3, a common retroviral integration site in murine B-cell lymphoma, encodes an EBFAZ-related Kruppel-like zinc finger protein. Blood. 2003;101:1934–1940. doi: 10.1182/blood-2002-08-2652. [DOI] [PubMed] [Google Scholar]
- 10.Bond H.M., Mesuraca M., Carbone E., Bonelli P., Agosti V., Amodio N. Early hematopoietic zinc finger protein (EHZF), the human homolog to mouse Evi3, is highly expressed in primitive human hematopoietic cells. Blood. 2004;103:2062–2070. doi: 10.1182/blood-2003-07-2388. [DOI] [PubMed] [Google Scholar]
- 11.Shen S., Pu J., Lang B., McCaig C.D. A zinc finger protein Zfp521 directs neural differentiation and beyond. Stem Cell Res. Ther. 2011;2:20. doi: 10.1186/scrt61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koganei S., Ito M., Yamamoto K., Matsumoto N. B-1a cell origin of the murine B lymphoma line BCL1 characterized by surface markers and bacterial reactivity of its surface IgM. Immunol. Lett. 2005;98:232–244. doi: 10.1016/j.imlet.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Hiratsuka T., Takei Y., Ohmori R., Imai Y., Ozeki M., Tamaki K. ZFP521 contributes to pre-B-cell lymphomagenesis through modulation of the pre-B-cell receptor signaling pathway. Oncogene. 2015 doi: 10.1038/onc.2015.385. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Tsapogas P., Breslin T., Bilke S., Lagergren A., Mansson R., Liberg D. RNA analysis of B cell lines arrested at defined stages of differentiation allows for an approximation of gene expression patterns during B cell development. J. Leukoc. Biol. 2003;74:102–110. doi: 10.1189/jlb.0103008. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida T., Georgopoulos K. Ikaros fingers on lymphocyte differentiation. Int. J. Hematol. 2014;100:220–229. doi: 10.1007/s12185-014-1644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boller S., Grosschedl R. The regulatory network of B-cell differentiation: a focused view of early B-cell factor 1 function. Immunol. Rev. 2014;261:102–115. doi: 10.1111/imr.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hikida M., Mori M., Takai T., Tomochika K., Hamatani K., Ohmori H. Reexpression of RAG-1 and RAG-2 genes in activated mature mouse B cells. Science. 1996;274:2092–2094. doi: 10.1126/science.274.5295.2092. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi H., Gregory S.C., Yokota T., Sakaguchi N., Kincade P.W. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- 20.Decker T., Pasca di Magliano M., McManus S., Sun Q., Bonifer C., Tagoh H. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Revilla I.D.R., Bilic I., Vilagos B., Tagoh H., Ebert A., Tamir I.M. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31:3130–3146. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukin K., Fields S., Lopez D., Cherrier M., Ternyak K., Ramirez J. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7869–7874. doi: 10.1073/pnas.1003525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F.X., Zhu J.W., Hogan C.J., DeGregori J. Defective gene expression, S phase progression, and maturation during hematopoiesis in E2F1/E2F2 mutant mice. Mol. Cell. Biol. 2003;23:3607–3622. doi: 10.1128/MCB.23.10.3607-3622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakurai N., Maeda M., Lee S.U., Ishikawa Y., Li M., Williams J.C. The LRF transcription factor regulates mature B cell development and the germinal center response in mice. J. Clin. Invest. 2011;121:2583–2598. doi: 10.1172/JCI45682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai R.Y., Reed R.R. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J. Neurosci. 1997;17:4159–4169. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raver R.M., Panfil A.R., Hagemeier S.R., Kenney S.C. The B-cell-specific transcription factor and master regulator Pax5 promotes Epstein-Barr virus latency by negatively regulating the viral immediate early protein BZLF1. J. Virol. 2013;87:8053–8063. doi: 10.1128/JVI.00546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Correa D., Hesse E., Seriwatanachai D., Kiviranta R., Saito H., Yamana K. Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev. Cell. 2010;19:533–546. doi: 10.1016/j.devcel.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang S., Akerblad P., Kiviranta R., Gupta R.K., Kajimura S., Griffin M.J. Regulation of early adipose commitment by Zfp521. PLoS Biol. 2012;10:e1001433. doi: 10.1371/journal.pbio.1001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiviranta R., Yamana K., Saito H., Ho D.K., Laine J., Tarkkonen K. Coordinated transcriptional regulation of bone homeostasis by Ebf1 and Zfp521 in both mesenchymal and hematopoietic lineages. J. Exp. Med. 2013;210:969–985. doi: 10.1084/jem.20121187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mega T., Lupia M., Amodio N., Horton S.J., Mesuraca M., Pelaggi D. Zinc finger protein 521 antagonizes early B-cell factor 1 and modulates the B-lymphoid differentiation of primary hematopoietic progenitors. Cell Cycle. 2011;10:2129–2139. doi: 10.4161/cc.10.13.16045. [DOI] [PubMed] [Google Scholar]
- 31.Yamasaki N., Miyazaki K., Nagamachi A., Koller R., Oda H., Miyazaki M. Identification of Zfp521/ZNF521 as a cooperative gene for E2A-HLF to develop acute B-lineage leukemia. Oncogene. 2010;29:1963–1975. doi: 10.1038/onc.2009.475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.