Abstract
Retinoic acid (RA) is one of the most potent inducers of differentiation of mouse embryonic stem cells (ESCs). However, previous studies show that RA treatment of cells cultured in the presence of a leukemia inhibitory factor (LIF) also result in the upregulation of a gene called Zscan4, whose transient expression is a marker for undifferentiated ESCs. We explored the balance between these two seemingly antagonistic effects of RA. ESCs indeed differentiated in the presence of LIF after RA treatment, but colonies of undifferentiated ESCs eventually emerged from these differentiated cells — even in the presence of RA. These colonies, named secondary colonies, consist of three cell types: typical undifferentiated ESCs expressing pluripotency genes such as Pou5f1, Sox2, and Nanog; cells expressing Zscan4; and endodermal-like cells located at the periphery of the colony. The capacity to form secondary colonies was confirmed for all eight tested ESC lines. Cells from the secondary colonies — after transfer to the standard ESC medium — retained pluripotency, judged by their strong alkaline phosphatase (ALP) staining, typical colony morphology, gene expression profile, stable karyotype, capacity to differentiate into all three germ layers in embryoid body formation assays, and successful contribution to chimeras after injection into blastocysts. Based on flow cytometry analysis (FACS), the proportion of Zscan4-positive cells in secondary colonies was higher than in standard ESC colonies, which may explain the capacity of ESCs to resist the differentiating effects of RA and instead form secondary colonies of undifferentiated ESCs. This hypothesis is supported by cell-lineage tracing analysis, which showed that most cells in the secondary colonies were descendents of cells transiently expressing Zscan4.
Keywords: retinoic acid, Zscan4, mouse embryonic stem cells, pluripotency
Introduction
Mouse embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of blastocysts and retain their pluripotent state in culture, with the capacity to differentiate into derivatives of all three embryonic germ layers in vitro and to contribute to chimeras after injection into blastocysts (Smith et al. 1988, Williams et al. 1988). ESCs readily differentiate after either removal of the leukemia inhibitory factor (LIF) or treatment with differentiation-inducing factors such as retinoic acid (RA), which induces differentiation of ESCs towards neural, endoderm, and mesoderm lineages (Colleoni et al. 2011, Edwards and McBurney 1983, Johannesson et al. 2009, Rohwedel et al. 1999).
Our previous studies further showed that the RA treatment of mouse ESCs resulted in six-fold upregulation of Zscan4 (a.k.a., Gm397) on the third day in RA conditions (Sharova et al. 2007). Zscan4 was originally identified for its specific expression in two-cell mouse embryos and in 1–5% of undifferentiated ESCs (Falco et al. 2007). It has also been shown that Zscan4 is a marker of undifferentiated ESCs and plays critical roles for telomere elongation and genome stability of ESCs (Amano et al. 2013, Zalzman et al. 2010). Therefore, its upregulation in RA conditions seemed contrary to the well-known differentiation-inducing effect of RA. The existence of alternative actions of RA is supported by findings that the pulse exposure to RA counteracts differentiation of ESCs even in the absence of LIF (Wang et al. 2008). Also, RA was shown to induce the development of pluripotent primordial germ cells (PGCs) and male gametes in 3D culture (Geijsen et al. 2004, Kerkis et al. 2007).
In this paper, we explore the balance of differentiating and counter-differentiating effects of RA in mouse ESCs and show that pluripotent colonies emerge in ESC cultures despite the continuous presence of RA. Besides standard pluripotent cells expressing Nanog and Pou5f1, these colonies (named secondary colonies) include an unusually high proportion of cells expressing Zscan4, as well as some endodermal-like cells on the periphery. Cell-lineage tracing analysis shows that most cells in the secondary colonies are descendents of cells transiently expressing Zscan4. We hypothesize that refraction of some ESCs to the differentiation-inducing effects of RA and to form secondary colonies could be related to the action of Zscan4.
Materials and Methods
Original MC1 (129.3) and MC2 (C57BL/6) ESC lines were obtained from the expanded frozen stock (vials with passages 5 and 3, respectively) at Johns Hopkins University, where they were derived; line dsRed was provided by Dr. Soriano. These cells were then expanded at NIA, frozen and later used to derive Zscan4-related clones: MC1-ZE-3 (Falco et al. 2007), MC2-ZE-18, and ZE-dsRed with Emerald reporter under control of Zscan4 promoter stably transfected in MC1, MC2, and dsRed ESCs, correspondingly. ESC line R1-Oct4-GFP with GFP-Oct4 reporter was provided by Dr. A. Nagy; C57BL/6 v26.2 was purchased from Open Biosystems (cat. #MES1393, Pittsburgh, PA); ES-D3-GL was purchased from ATCC (cat. #SCRC-1003, Manassas, VA); iPS-Ng-20D was provided by Dr. S. Yamanaka; and TGC 8-8 was provided by Dr. Hogan. Zscan4-cre-ERT2 was developed at NIA for lineage tracing of cells expressing Zscan4 (Zalzman et al. 2010). Standard ESC culture was done as described (Sharova et al. 2007). To generate secondary colonies, ESCs were seeded in six-well plates at clonal density 102/cm2 in standard ESC medium (DMEM, 15% FBS; 1000 u/ml LIF, ESGRO; 1mM sodium pyruvate; 0.1 mM NEAA, 2 mM glutamate, 0.1 mM beta-mercapto ethanol, and 50U/50μg per ml penicillin/streptomycin) supplemented with 50nM all trans retinoic acid (RA, Sigma, cat# R2625, St. Louis, MO) and cultured for 7 d with daily medium change. ALP staining was performed with 85R Sigma kit according to manufacturer protocol for microscopy. For flow cytometry analysis (FACS) of Emerald(+) cells, MC1-ZE-3 and MC2-ZE-18 cells were harvested and analyzed by Guava easyCyte(™) Mini System flow cytometer (Guava Technologies, Millipore, Billerica, MA).
For global gene expression profiling with microarrays, individual secondary colonies were manually picked on day 7, RNA extracted and processed as described (Sharova et al. 2007). For comparison, we used ESCs in standard culture conditions as well as cells derived from secondary colonies and then cultured 10 passages in standard conditions. Cy3-CTP-labeled sample targets (in two biological replications) were prepared with total RNA by Low RNA Input Fluorescent Linear Amplification Kit (Agilent, Santa Clara, CA) and hybridized to NIA Mouse 44K Microarray v3.0 (Agilent, design ID 015087) (Carter et al. 2005) together with Cy5-CTP labeled reference target, which was produced from mixture of Stratagene Universal Mouse Reference RNA and RNA from MC1 cells. For statistical analysis, we used NIA Array Analysis, which estimates the False Discovery Rate (FDR) to account for multiple hypothesis testing (Sharov et al. 2005). Microarray data are submitted to GEO/NCBI database, accession number GSE40495.
To visualize the expression of Pou5f1 and Nanog in relation to Zscan4-positive [Zscan4(+)] cells, MC1-ZE-3 ESCs were cultured in the presence of 50 nM RA for several days, fixed in 4% PFA and stained with POU5F1 and NANOG antibodies. For tracking Pou5f1 and Nanog expression in ESCs in the lineage of cells originated from Zscan4(+) cells, Zscan4-cre-ERT2 ESCs were cultured in the medium supplemented with RA and tamoxifen for 3 d and then in the RA medium without tamoxifen for four more days. ESCs were fixed on day 7 in 4% PFA and stained with lacZ, Pou5f1 and Nanog antibodies. Cre-recombinase in Zscan4-cre-ERT2 ESCs is under the control of the Zscan4 promoter. It translocates from the cytoplasm into the nucleus only in the presence of tamoxifen and excises a neomycin cassette from the LacZ ORF (Soriano 1999), leading to heritable constitutive lacZ expression in all cells that expressed Zscan4 at the time of exposure to tamoxifen. Zscan4-cre-ERT2 cells were fixed 4 min in tissue fixative (Millipore, Cat. #BG-5-C), washed with PBS and incubated 2 h at 37°C with X-Gal substrate (X-Gal Stock Solution, Millipore, Cat. #BG-3-G) diluted 1:40 in Tissue Stain Base Solution (Millipore, Cat. #BG-8-C). Immunohistochemistry was performed as described (Zalzman et al. 2010). Antibodies used: Oct3/4 – SantaCruz, Dallas, TX, mouse, 1:250 (sc5279), secondary – donkey-antiMouse 568(A10037, 1:800); Nanog – BD Biosciences, East Rutherford, NJ, mouse, 1:500 (560259), secondary – same as for Oct3/4; Gata4 – SantaCruz, goat, 1:1000 (sc1237), secondary – donkey-antiGoat 647(A21447, 1:400); β-gal – Abcam, Cambridge, MA, chicken, 1:500 or 1:1000 (ab9361), secondary – goat-antiChicken 488(A11039, 1:400); Zscan4 – GenScript, Piscataway, NJ, rabbit; 1:2000 (antiserum made for sequence CSTYHRHLRNYHRSD), secondary – donkey-antiRabbit 568 (A10042, 1:800). For flow cytometry analysis (FACS), MC1-ZE-3 cells were fixed in BD Cytofix/Cytoperm™ solution (BD Biosciences, cat #554722), permeabilized with BD Perm/Wash buffer (BD Biosciences, cat #554723), stained with specific antibodies (see above) and analyzed by Guava EasyCyte Mini System flow cytometer (Guava Technologies, Millipore).
For EB assay, ESCs were harvested on day 3 of culture, resuspended in EB medium (DMEM, 10% FBS; 1mM sodium pyruvate; 0.1 mM NEAA, 2 mM glutamate, 0.1 mM beta-mercapto ethanol, and 50U/50μg per ml penicillin/streptomycin) and plated in AggreWell™ 400 (Stemcell Technologies, Vancouver, BC), 1.2x106 cells/well to produce EB with ~1000 cells. On day 3 formed EB were transferred in Corning® Ultra-low attachment culture dishes (cat #3262, Corning, NY) and cultured for an additional four days. On day 7, floating EB were transferred to cell culture treated plates for attachment. The beating muscles and other differentiation types were scored on day 3 of attached EB culture. To assess the capacity to contribute to chimeras, ESCs were injected into blastocysts obtained from CD1 females (Charles River, Wilmington, MA, 8–12 wk old) which were superovulated by PMSG followed by hCG (Sigma) administration and mated with CD1 males. Two-cell embryos were collected by flushing oviducts using M2 medium (Millipore) and were cultured in KSOM (Millipore) drop for 3 d until they reached blastocyst stage at 37°C in 5% CO2. After cell injection, blastocysts were transferred to the uterus of recipient mice. Chimeras were evaluated by their coat color contribution. For karyotyping, ESCs were cultured in the standard medium with 30 ng/ml of colcemid (Gibco, Carlsbad, CA, #15212-012) for 4 h, treated with 0.55% KCl for 8 min at 37°C, and fixed in glacial acetic acid methanol (2:5). Prepared slides were stained with Giemsa (Gibco #10092-013) and analyzed under x100 magnification using Axiovert 200 (Zeiss, Oberkochen, Germany). Telomere length was estimated by qPCR as described earlier (Callicott and Womack 2006).
Results
Undifferentiated secondary colonies of ESCs emerged during retinoic acid supplementation
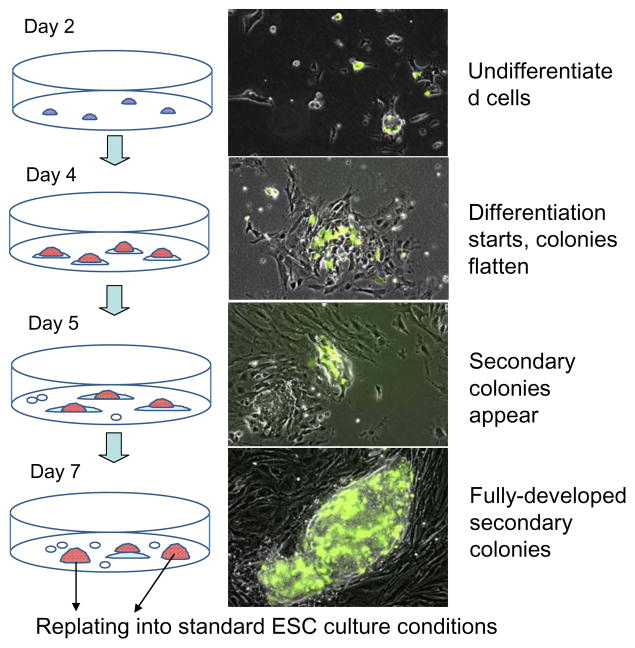
To explore the balance of opposite effects of RA – induction of differentiation and upregulation of undifferentiation-specific Zscan4 – on mouse ESCs, we first used cell line MC1-ZE-3 (Falco et al. 2007) containing an Emerald-Zscan4 reporter (Zscan4 promoter followed by Emerald ORF) to simplify the visualization of Zscan4-positive [Zscan4(+)] cells. Cells were plated at clonal density (at which growing colonies do not tend to coalesce with each other), and were then cultured in standard ESC medium supplemented with a low concentration of RA (50 nM, which is two-fold higher than the physiological plasma level) (Tzimas et al. 1996). We avoided higher concentrations of the RA, because they caused irreversible differentiation of ESCs. Initially ESCs showed signs of differentiation, such as flattening of colonies and reduction of both ALP staining and immunostaining for pluripotency-related transcription factors Pou5f1 and Nanog (Supplementary Figure S1A). However, by day 6–7, dome-shaped colonies similar to those in undifferentiated ESCs began to emerge among the seemingly differentiated cells (Fig. 1, Supplementary Figure S1A, B). These colonies were termed “secondary colonies” to distinguish them from untreated standard ESC colonies. The secondary colonies included many Zscan4(+) cells, detected by activation of the Emerald-Zscan4 reporter (green cells in Fig. 1). The majority of secondary colonies (~70%) also showed high expression of ALP, Pou5f1, and Nanog, comparable to control cells cultured without RA (Supplementary Figure S1A, B).
Figure 1.
Emergence of undifferentiated ESC colonies in retinoic acid conditions. Left panel: the experiment design, red cells are ALP-positive. Right panel: MC1-ZE-3 cells with Emerald-Zscan4 reporter (green) plated at clonal density show the emergence of secondary colonies.
When ESCs were cultured in medium with the same low concentration of RA (50 nM) but in the absence of LIF, no dome-shaped secondary colonies appeared within 8 d of culture, although a few groups of ALP-positive cells were present (Supplementary Figure S2A). The expression of pluripotency marker Pou5f1 also declined in these cells (not shown). This indicated that LIF was necessary for the formation of secondary colonies. Cells cultured in RA and in the absence of LIF had no self-renewal capacity, as they differentiated completely after replating in standard culture medium (Supplementary Figure S2B). Secondary colonies were also not observed when ESC cultures were plated at high density, even in the presence of LIF.
To test whether secondary colonies can form in other pluripotent cell lines, we used the same culturing method (i.e., clonal density, 50 nM RA, and LIF) for eight other pluripotent cell lines: six ESC lines (C57BL/6 V26.2, R1-Oct4-GFP, ES-D3-GL, MC2-ZE-18, ZE-dsRed, MC2); one embryonic germ cell (EGC) line (TGC 8-8); and one induced pluripotent stem cell (iPSC) line (iPS-Ng-20D). All cell lines tested formed secondary colonies by day 7. Strong ALP-staining is shown for cell lines C57BL/6 V26.2 and iPS-Ng-20D (Supplementary Figure S3A, B). In addition, R1-Oct4-GFP, which bears a GFP reporter under the control of the Pou5f1 promoter, showed a high expression of this pluripotency-related transcription factor, judged by the presence of GFP fluorescence (Supplementary Figure S3C). Expression of Zscan4 in the secondary colonies was confirmed in two cell lines with Emerald reporter under control of the Zscan4 promoter: MC2-ZE-18, ZE-dsRed (Supplementary Figure S3E, F).
To further test self-renewal capacity of cells derived from the secondary colonies, we replated them 10 times in the standard conditions (i.e., standard plating density, standard ES culture medium, 3 d in culture). After replating, these cells formed standard dome-shaped colonies with strong ALP-staining (Supplementary Figure S4). Secondary colonies can also be replated in RA conditions for at least three passages. However, continuous passaging of cells in RA conditions was not sustainable, as the proportion of differentiating cells increased progressively.
Secondary colonies included three sub-populations of cells: standard ESCs, Zscan4-positive cells, and endodermal-like cells
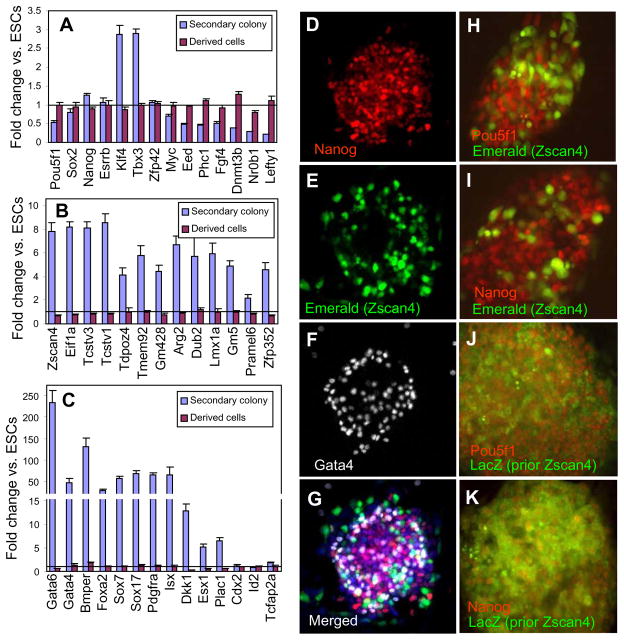
To explore molecular mechanisms involved in the formation of the secondary colonies, we used microarrays to measure gene expression in manually picked secondary colonies compared to standard ESCs. Expression of pluripotency-related genes in the secondary colonies could be either unchanged (e.g., Sox2, Esrrb, Zfp42); upregulated (e.g., Nanog, Klf4, Tbx3); or downregulated (e.g., Pou5f1, Eed, Phc1, Myc, Fgf4, Lefty1, Nr0b1, and Dnmt3b) compared to standard ESCs (Fig. 2A).
Figure 2.
Expression of genes in secondary colonies. Expression of (A) pluripotency related genes, (B) genes associated with Zscan4 expression in ESCs, and (C) differentiation-related genes in manually picked secondary colonies (MC1 cells in 50 nM retinoic acid on day 7) and in descendants of these cells cultured in standard ESC conditions for 10 passages (derived cells), based on microarray data; expression is normalized to undifferentiated standard ESCs (line = 1). Heterogeneity of gene expression: (D-G) expression of Nanog (immunostaining), Zscan4 (Emerald reporter), and Gata4 (immunostaining). Expression of Zscan4 in early secondary colonies (day 3, Emerald reporter) (H, I) and cumulative expression in late secondary colonies (day 7, lacZ staining in Zscan4-CreERT2) (J, K) combined with immunostaining for pluripotency-related factors Pou5f1 (H, J) and Nanog (I, K); lacZ staining in Zscan4-CreERT2 cells shows cells that previously expressed Zscan4.
Although Pou5f1 was downregulated ~2-fold in secondary colonies, its expression level still appeared sufficient to prevent the differentiation of cells. As expected, Zscan4 expression in secondary colonies was increased — by eight-fold compared to standard ESCs (Fig. 2B). Other genes that are known to be co-regulated with Zscan4 (Arg2, Dub2, Eif1a, Gm428, Gm5, Lmx1a, Pramel6, Tcstv1, Tcstv3, Tdpoz4, Tmem92, and Zfp352) (Amano et al. 2013, Macfarlan et al. 2012), also were increased in expression in the secondary colonies by 3- to 12-fold compared to standard ESCs (Fig. 2B). The secondary colonies also showed high expression of endoderm- and endothelial-related genes (e.g., Gata6, Gata4, Sox7, Sox17, Foxa2, Sox7, Sox17, Pdgfra, Isx, and Dkk1) (Fig. 2C), consistent with published reports on endoderm induction by RA (Johannesson et al. 2009). By contrast, genes associated with extra-embryonic lineages (Esx1, Plac1, Cited1, Cited2, and Hand1) were only slightly overexpressed in the secondary colonies. The gene expression changes were all reversed after passaging of cells derived from secondary colonies in standard culture condition (Fig. 2B, C purple bars).
Because expression of genes associated with the standard ESCs, Zscan4(+) cells, and cells differentiated from ESCs are known to be incompatible (Hamazaki et al. 2004, Macfarlan et al. 2012, Singh et al. 2007), we used immunocytochemistry to assess whether the secondary colonies were heterogeneous. Staining for Nanog (antibody, marker of standard ESCs), Zscan4 (Emerald reporter), and Gata4 (antibody, endoderm marker), showed that these three genes were indeed expressed in different subpopulations of cells (Fig. 2D–G). Early secondary colonies (day 3) showed a clear separation of Zscan4(+) cells (Emerald reporter, MC1-ZE-3 cell line) and Pou5f1(+) and Nanog(+) cells (Fig. 2H,I), in line with the characteristic findings that Zscan4(+) cells do not produce the POU5F1 or NANOG protein (Macfarlan et al. 2012).
FACS experiments with cells comprising secondary colonies stained for both Zscan4 and Gata4 confirmed that different subpopulations of cells express these genes, apart from a small proportion of cells (1.2–2.5%) that showed double staining. Cells with Gata4 expression (ca. 7%) were located mostly at the edges of colonies (Fig. 2F), similar to the pattern of cell segregation in ESCs (Niakan et al. 2010), whereas pluripotent cells expressing Nanog and Pou5f1 were located closer to the center of colonies (Fig. 2D, H, I). Cells with Zscan4 expression were scattered in a colony, although more cells seemed near the edges (Fig. 2E). Thus, secondary colonies included three subpopulations of cells: standard ESCs, Zscan4(+) cells, and endodermal-like cells.
Secondary colony-derived ESCs were pluripotent
The results thus far suggested that the secondary colonies that emerged from the seemingly differentiated cells contain pluripotent ESCs. To test this notion, we manually picked the secondary colonies with a micropipette and cultured them in the standard condition for 10 passages without RA. Morphologically, these ESCs became more like the standard ESCs. We then carried out in vitro cell differentiation assays. ESCs derived from the secondary colonies formed normal embryoid bodies in LIF(−) conditions and differentiated into derivatives of endoderm, ectoderm (neurons), and mesoderm (muscles, blood vessels) (Supplementary Figure S5). We observed no bias in the direction of differentiation compared to standard ESCs. Thus, although cells were exposed to the low dose of RA during the formation of the secondary colonies and showed a reduction of pluripotency markers Pou5f1 and Nanog, they regained their pluripotent phenotype after the removal of RA.
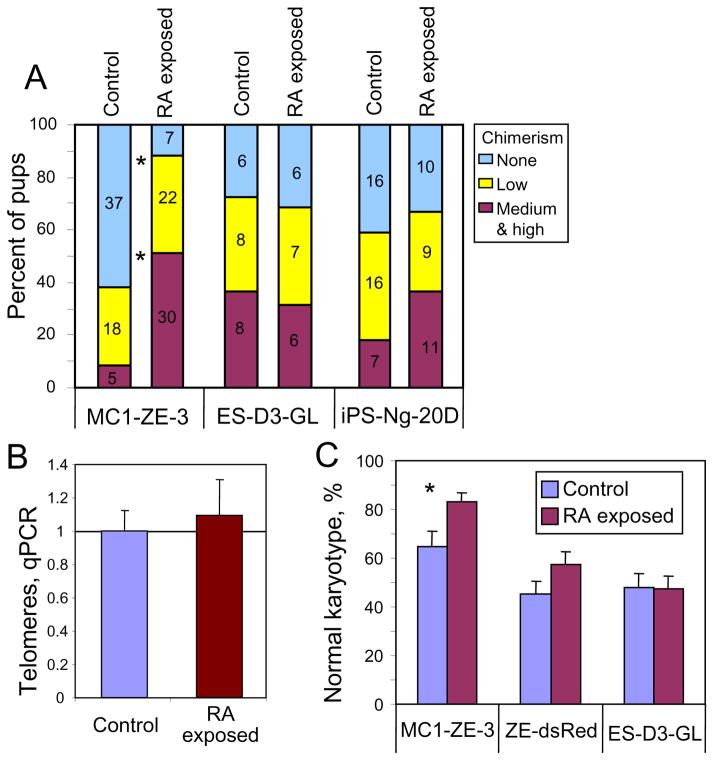
To further examine pluripotency in vivo, we injected the ESCs, derived from the secondary colonies and cultured for 10 passages in standard medium, into blastocysts and observed chimeras in the progeny from three tested cell lines: MC1-ZE-3, ES-D3-GL, and iPS-Ng-20D (Fig. 3A). Chimeras were formed in all three lines, which confirmed the pluripotent state of cells passaged through secondary colonies. In one cell line (MC1-ZE-3), chimerism was significantly higher compared to control ESCs (i.e., parental ESCs that underwent the same number of passages) (88.1% vs. 38.3%; χ2 = 31.7 p = 2×10−8) (Fig. 3A). However, in ES-D3-GL and iPS-Ng-20D cell lines, any change of chimerism was not significant (χ2 = 0.09, p = 0.76; χ2 = 0.43, p = 0.51, respectively). Known factors affecting the contribution of cells to chimeras include the length of telomeres and karyotype, both of which tend to deteriorate in ESCs after ca. 20 passages (Longo et al. 1997, Rebuzzini et al. 2008). Telomere length measured by qPCR appeared unchanged between MC1-ZE-3 cells derived from the secondary colonies and control ESCs (Fig. 3B), whereas the proportion of cells with normal karyotype was higher in MC1-ZE-3 cells exposed to RA than in control cells (χ2 = 6.248; p = 0.0124) (Fig. 3C). These results may explain their increased contribution to chimeras. In contrast, no difference in karyotype stability was observed in ZE-dsRed and ES-D3-GL cell lines.
Figure 3.
Contribution to chimeras and genome integrity of cells exposed to retinoic acid (RA) (i.e., from secondary colonies) and then cultured in standard conditions for 10 passages, as compared to control cells (i.e., parental ESCs that underwent the same number of passages). (A) Proportion of chimeric mice obtained from cell injection into blastocysts (number of pups shown); (B) telomere length assessed with qPCR; (C) karyotype of cells exposed to RA and in control cells; asterisk (*) indicates significant difference (p < 0.05).
Capacity of secondary colonies to resist the differentiation effects of RA is likely associated with the increased proportion of Zscan4-positive cells
The most unusual feature of secondary colonies is the upregulation of Zscan4, which is inferred from the intensive Emerald staining of secondary colonies formed by cell lines with Emereld-Zscan4 reporter (MC1-ZE-3, MC2-ZE-18, and ZE-dsRed, Fig. 1, Supplementary Figure S3) and from the results of microarray analysis (Fig. 2B). This motivated the study of whether the expression of Zscan4 was related to the capacity of secondary colonies to resist the differentiation effects of RA. First, we used FACS to check if the proportion of Zscan4(+) cells in secondary colonies was greater than in standard ESC culture (because this information cannot be inferred from microarrays or images of colonies). The proportion of Zscan4(+) (i.e., Emerald-positive) cells from line MC1-ZE-3 measured by FACS (Guava EasyCyte flow cytometer) was ~2-fold higher in RA conditions (i.e., in secondary colonies) than in controls without RA addition (Supplementary Figure S6A). Similar experiments with MC2-ZE-18 cells showed a >10-fold difference in Zscan4(+) cells between RA and control conditions (Supplementary Figure S6B). Stronger response of MC2-ZE-18 cells to RA treatment may result from a lower initial level of Zscan4 expression in this cell line. In both cell lines, the RA treatment resulted in the increase of the proportion of Zscan4(+) cells to 15–20% as compared to 1–5% in standard ESC cultures.
Although the proportion of Zscan4(+) cells has increased in secondary colonies, it did not approach 100%, and it is not clear how Zscan4(−) cells remained pluripotent. To analyze their individual capacity, we used lineage tracing to show that nearly all pluripotent cells without Zscan4 expression originated from cells that had transient expression of Zscan4 earlier. We used Zscan4-CreERT2 cells (Zalzman et al. 2010), which become permanently lacZ-positive after they express Zscan4 during treatment with tamoxifen. When tamoxifen was added to the medium with RA, the majority of cells in the secondary colonies showed strong lacZ staining, indicating that these cells were Zscan4(+) at some point during the tamoxifen/RA treatment (Supplementary Figure S6C).
To test whether Zscan4(+) cells later regained the expression of pluripotency-related transcription factors Pou5f1 and Nanog, we combined staining for lacZ with immunostaining for the transcription factors. In mature secondary colonies (on day 7), Pou5f1 and Nanog expression in Zscan4-cre-ERT2 ESCs widely overlapped with lacZ staining, which marked the cells that were Zscan4(+) 1–3 d earlier (Fig. 2J, K). This indicated reactivation of POU5F1 and NANOG proteins several cell cycles after Zscan4 was expressed, and confirmed that transiently Zscan4(+) cells remained pluripotent. When the secondary colonies were transferred to standard ESC medium, cells appeared mostly lacZ-positive (Supplementary Figure S6D), indicating that these cells are indeed descendants of Zscan4(+) cells in the secondary colonies.
Discussion
The major result of this study is that a low-dose (50 nM) of RA added to the standard medium did not cause total differentiation of mouse ESCs within 7 d. Instead, undifferentiated colonies (which we call “secondary”) appeared and grew, surrounded by differentiated cells. These colonies had typical undifferentiated morphology, ALP staining, and expressed pluripotency-related transcription factors. Formation of secondary colonies required both RA and LIF in the medium. In this respect our findings differ from a previous report that pulse-treatment with RA can support self-renewal of ESCs in medium without LIF (Wang et al. 2008). Cells derived from the secondary colonies had normal capacity for differentiation and showed a high contribution to chimeras after injection into blastocysts. The proportion of chimeras was not reduced compared to control cells of the same passage number and, in one cell line, it was even statistically higher than controls.
The peculiar feature of secondary colonies was the increased proportion of Zscan4(+) cells (15–20% compared to 1–5% in ESCs). Zscan4 is expressed in early preimplantation embryos, with a maximum at the late two-cell stage (Falco et al. 2007). In ESCs, it is co-expressed with several hundred other genes, the majority of which are also expressed in two-cell embryos (Akiyama et al. 2015, Amano et al. 2013, Macfarlan et al. 2012). Zscan4(+) cells show transient derepression of heterochromatin, which becomes actively transcribed (Akiyama et al. 2015). This can rationalize the upregulation of certain transposable elements (Macfarlan et al. 2012) and expression of the group of genes typical for the Zscan4(+) cells, because the majority of them (including Zscan4 itself) are ordinarily located in heterochromatin. Our previous experiments with induction and knockdown of Zscan4 showed that the gene is required to support the expression of many other genes typical of the Zscan4(+) cells (Nishiyama et al. 2012, Nishiyama et al. 2009). Thus, Zscan4 seems to play a key role in the activation of coregulated genes in heterochromatin.
The cell-lineage tracing experiments clearly showed that all pluripotent cells in secondary colonies once were Zscan4(+) during RA treatment. Thus, we think that Zscan4-related cell changes (e.g., derepression of heterochromatin and elongation of telomeres) may contribute to the resistance of secondary colonies to the differentiation-inducing effects of RA.
Regarding the increase of Zscan4(+) cells by RA, it is worth mentioning a possible involvement of Nr0b1. It has been shown that Nr0b1 is downregulated in RA conditions (Hosler et al. 1993) (see also Fig. 2C), and the downregulation of Nr0b1 results in a substantial increase of Zscan4c expression as well as in the upregulation of endoderm-related genes in ESCs (Fujii et al. 2015). Thus, Nr0b1(−/−) cells described in the previous report (Fujii et al. 2015) are possibly similar to the secondary colonies described in our study. However, it is not clear whether this is the only or the main mechanism that increases the proportion of Zscan4(+) cells in secondary colonies. For example, in our previous study, the knockdown of Nr0b1 via shRNA (90–95% reduction after 72 hr) did not result in the increase of Zscan4 expression (Nishiyama et al. 2012). Interestingly, the knockdown of Nr5a2 – an upstream gene of Nr0b1, did cause an increase in Zscan4 expression (Nishiyama et al. 2012), which seems to be consistent with experiments of Fujii et al.
Supplementary Material
Acknowledgments
We thank Dr. A. Nagy (Mount Sinai Hospital, Toronto, ON, Canada), Dr. S. Yamanaka (Kyoto University, Kyoto, Japan, and Gladstone Institute of Cardiovascular Disease, San Francisco, CA, USA), Dr. P. Soriano (School of Medicine, Mount Sinai, New York, USA), and Dr. B. Hogan (Duke University Medical Center, Durham, NC, USA) for providing transgenic cells. Also we thank current and past lab members for discussion and contribution to data published previously and integrated into the meta-analysis in this paper.
Funding
This research was supported entirely by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
References
- Akiyama T, Xin L, Oda M, Sharov AA, Amano M, Piao Y, Cadet JS, Dudekula DB, Qian Y, Wang W, Ko SB, Ko MS. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 2015;22:307–318. doi: 10.1093/dnares/dsv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, Hirata T, Falco G, Monti M, Sharova LV, Amano M, Sheer S, Hoang HG, Piao Y, Stagg CA, Yamamizu K, Akiyama T, Ko MS. Zscan4 restores the developmental potency of embryonic stem cells. Nat Commun. 2013;4:1966. doi: 10.1038/ncomms2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott RJ, Womack JE. Real-time PCR assay for measurement of mouse telomeres. Comp Med. 2006;56:17–22. [PubMed] [Google Scholar]
- Carter MG, Sharov AA, VanBuren V, Dudekula DB, Carmack CE, Nelson C, Ko MS. Transcript copy number estimation using a mouse whole-genome oligonucleotide microarray. Genome Biol. 2005;6:R61. doi: 10.1186/gb-2005-6-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni S, Galli C, Gaspar JA, Meganathan K, Jagtap S, Hescheler J, Sachinidis A, Lazzari G. Development of a neural teratogenicity test based on human embryonic stem cells: response to retinoic acid exposure. Toxicol Sci. 2011;124:370–377. doi: 10.1093/toxsci/kfr245. [DOI] [PubMed] [Google Scholar]
- Edwards MK, McBurney MW. The concentration of retinoic acid determines the differentiated cell types formed by a teratocarcinoma cell line. Dev Biol. 1983;98:187–191. doi: 10.1016/0012-1606(83)90348-2. [DOI] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Nishikawa-Torikai S, Futatsugi Y, Toyooka Y, Yamane M, Ohtsuka S, Niwa H. Nr0b1 is a negative regulator of Zscan4c in mouse embryonic stem cells. Sci Rep. 2015;5:9146. doi: 10.1038/srep09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijsen N, Horoschak M, Kim K, Gribnau J, Eggan K, Daley GQ. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- Hamazaki T, Oka M, Yamanaka S, Terada N. Aggregation of embryonic stem cells induces Nanog repression and primitive endoderm differentiation. J Cell Sci. 2004;117:5681–5686. doi: 10.1242/jcs.01489. [DOI] [PubMed] [Google Scholar]
- Hosler BA, Rogers MB, Kozak CA, Gudas LJ. An octamer motif contributes to the expression of the retinoic acid-regulated zinc finger gene Rex-1 (Zfp-42) in F9 teratocarcinoma cells. Mol Cell Biol. 1993;13:2919–2928. doi: 10.1128/mcb.13.5.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson M, Stahlberg A, Ameri J, Sand FW, Norrman K, Semb H. FGF4 and retinoic acid direct differentiation of hESCs into PDX1-expressing foregut endoderm in a time- and concentration-dependent manner. PLoS One. 2009;4:e4794. doi: 10.1371/journal.pone.0004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkis A, Fonseca SA, Serafim RC, Lavagnolli TM, Abdelmassih S, Abdelmassih R, Kerkis I. In vitro differentiation of male mouse embryonic stem cells into both presumptive sperm cells and oocytes. Cloning Stem Cells. 2007;9:535–548. doi: 10.1089/clo.2007.0031. [DOI] [PubMed] [Google Scholar]
- Longo L, Bygrave A, Grosveld FG, Pandolfi PP. The chromosome make-up of mouse embryonic stem cells is predictive of somatic and germ cell chimaerism. Transgenic Res. 1997;6:321–328. doi: 10.1023/a:1018418914106. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA, McMahon AP, Eggan K. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Sharov AA, Piao Y, Amano M, Amano T, Hoang HG, Binder BY, Tapnio R, Bassey U, Malinou J, Correa-Cerro LS, Yu H, Xin L, Meyers E, Zalzman M, Nakatake Y, Stagg C, Sharova L, Qian Y, Dudekula D, Sheer S, Cadet JS, Hirata T, Yang H, Goldberg I, Evans MK, Longo DL, Schlessinger D, Ko MSH. Systematic repression of transcription factors reveals limited patterns of gene expression changes in ES cells. Sci Rep. 2013;3:1390. doi: 10.1038/srep01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, Stagg C, Sharova L, Correa-Cerro LS, Bassey U, Hoang H, Kim E, Tapnio R, Qian Y, Dudekula D, Zalzman M, Li M, Falco G, Yang H, Lee S, Monti M, Stanghellini I, Islam MN, Nagaraja R, Goldberg I, Wang W, Longo DL, Schlessinger D, Ko MSH. Uncovering early response of gene regulatory networks in ES cells by systematic induction of transcription factors. Cell Stem Cell. 2009;5:420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebuzzini P, Neri T, Mazzini G, Zuccotti M, Redi CA, Garagna S. Karyotype analysis of the euploid cell population of a mouse embryonic stem cell line revealed a high incidence of chromosome abnormalities that varied during culture. Cytogenet Genome Res. 2008;121:18–24. doi: 10.1159/000124377. [DOI] [PubMed] [Google Scholar]
- Rohwedel J, Guan K, Wobus AM. Induction of cellular differentiation by retinoic acid in vitro. Cells Tissues Organs. 1999;165:190–202. doi: 10.1159/000016699. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Piao Y, Shaik N, Sullivan T, Stewart CL, Hogan BL, Ko MS. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev Biol. 2007;307:446–459. doi: 10.1016/j.ydbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Tzimas G, Collins MD, Burgin H, Hummler H, Nau H. Embryotoxic doses of vitamin A to rabbits result in low plasma but high embryonic concentrations of all-trans-retinoic acid: risk of vitamin A exposure in humans. J Nutr. 1996;126:2159–2171. doi: 10.1093/jn/126.9.2159. [DOI] [PubMed] [Google Scholar]
- Wang R, Liang J, Yu HM, Liang H, Shi YJ, Yang HT. Retinoic acid maintains self-renewal of murine embryonic stem cells via a feedback mechanism. Differentiation. 2008;76:931–945. doi: 10.1111/j.1432-0436.2008.00272.x. [DOI] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, Wersto RP, Ko MS. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.