Abstract
In this review, we provide an overview of protein synthesis in the yeast Saccharomyces cerevisiae. The mechanism of protein synthesis is well conserved between yeast and other eukaryotes, and molecular genetic studies in budding yeast have provided critical insights into the fundamental process of translation as well as its regulation. The review focuses on the initiation and elongation phases of protein synthesis with descriptions of the roles of translation initiation and elongation factors that assist the ribosome in binding the messenger RNA (mRNA), selecting the start codon, and synthesizing the polypeptide. We also examine mechanisms of translational control highlighting the mRNA cap-binding proteins and the regulation of GCN4 and CPA1 mRNAs.
Keywords: GCN4, translation elongation, translation initiation
A rapidly dividing yeast cell growing on rich medium is estimated to synthesize nearly 13,000 proteins per second (von der Haar 2008), limited by the availability of ribosomes (Shah et al. 2013). The average cell contains nearly 200,000 ribosomes (Warner 1999; Firczuk et al. 2013) and 15,000–60,000 messenger RNA (mRNA) molecules (with ∼1/3 encoding ribosomal proteins) (Warner 1999; Zenklusen et al. 2008). With levels ranging from 105 to 106 molecules per cell, translation elongation factors are among the most abundant proteins in the cell (Firczuk et al. 2013). Given the vast resources the yeast cell devotes to protein synthesis, a thorough understanding of protein synthesis is critical to understanding the biology of Saccharomyces cerevisiae. In addition to its critical role in synthesizing all of the proteins required for cell growth, the translation apparatus is also nimble and regulates both general and mRNA-specific protein synthesis in response to environmental cues.
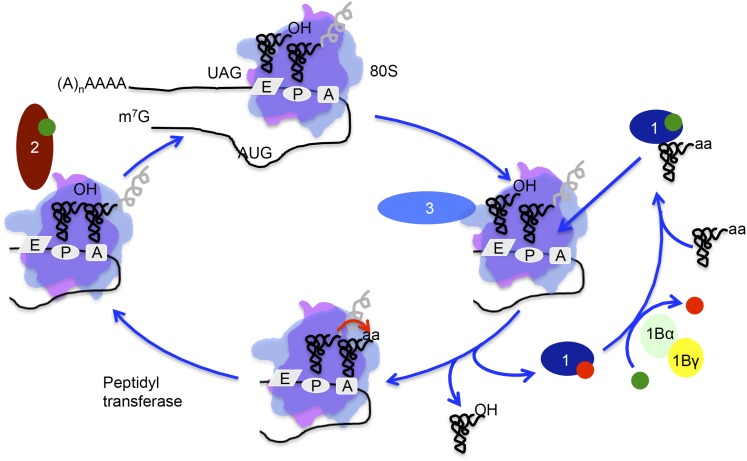
The basic mechanism of translating the nucleotide code of mRNA into the amino acid sequence of a protein, as performed by the ribosome, is well conserved throughout evolution. The process of protein synthesis can be subdivided into four major steps: initiation, elongation, termination, and ribosome recycling. During translation initiation, the small (40S) ribosomal subunit binds the specific initiator methionyl (Met)-transfer RNA (tRNA)iMet and an mRNA. The initiation step is completed when the small subunit selects a start codon and the large (60S) ribosomal subunit joins to form a functional ribosome. The elongation phase of protein synthesis refers to the codon-dependent addition of amino acids to the growing polypeptide chain. Finally, the termination steps involve release of the completed polypeptide chain from the ribosome, and recycling refers to dissociation of the ribosome and deacylated tRNA from the mRNA.
The initiation steps of protein synthesis have undergone the greatest changes during evolution. Whereas bacterial ribosomes locate translation start sites in part through base-pairing interactions between the ribosomal RNA (rRNA) in the ribosome and sequences immediately 5′ of the initiation codon (Kozak 2005; Laursen et al. 2005), eukaryotic ribosomes bind to the mRNA near the 5′ cap and scan in a 3′ direction inspecting the mRNA for start codons (Hinnebusch 2011). This change in initiation mechanisms between bacteria and eukaryotes is associated with a large increase in the number and complexity of factors required to facilitate protein synthesis. The three bacterial translation factors, IF1, IF2, and IF3 (Laursen et al. 2005; Schmeing and Ramakrishnan 2009), are replaced in yeast by 11 factors (Table 1). In contrast to the vastly different factor requirements for translation initiation in yeast vs. bacteria, the elongation and termination factors are structurally and/or functionally conserved with one exception between yeast and bacteria. The elongation factor eEF3 appears to be uniquely required in yeast as it is found neither in bacteria nor in higher eukaryotes (Belfield and Tuite 1993).
Table 1. Translation initiation factors.
| Factor | Subunit | Gene | Systematic name | Length (AA) |
|---|---|---|---|---|
| eIF1 | SUI1 | YNL244c | 108 | |
| eIF1A | TIF11 | YMR260c | 153 | |
| eIF2 | α | SUI2 | YJR007w | 304 |
| β | SUI3 | YPL237w | 285 | |
| γ | GCD11 | YER025w | 527 | |
| eIF2B | α | GCN3 | YKR026c | 305 |
| β | GCD7 | YLR291c | 381 | |
| γ | GCD1 | YOR260w | 578 | |
| δ | GCD2 | YGR083c | 651 | |
| ε | GCD6 | YDR211w | 712 | |
| eIF3 | a | RPG1/TIF32 | YBR079c | 964 |
| b | PRT1 | YOR361c | 763 | |
| c | NIP1 | YMR309c | 812 | |
| g | TIF35 | YDR429c | 274 | |
| i | TIF34 | YMR146c | 347 | |
| j | HCR1 | YLR192c | 265 | |
| eIF4A | TIF1 | YKR059w | 395 | |
| TIF2 | YJL138c | 395 | ||
| eIF4B | TIF3/STM1 | YPR163c | 436 | |
| eIF4E | CDC33 | YOL139c | 213 | |
| eIF4G | TIF4631 | YGR162w | 952 | |
| TIF4632 | YGL049c | 914 | ||
| eIF5 | TIF5 | YPR041w | 405 | |
| eIF5B | FUN12 | YAL035w | 1002 |
Over the last ≥15 years, molecular and biochemical studies have provided remarkable insights into the process of translation and the factors that assist the ribosome in producing proteins. Whereas the identity of most of the eukaryotic translation factors was established by biochemical studies conducted between the 1960s and 1980s, molecular investigations in yeast have provided novel insights into the functions and structure–function properties of the factors. In this review, we will focus on the initiation and elongation steps of protein synthesis, the functions of the translation factors, and the translational regulatory schemes in yeast. Due to space limitations, we will restrict our descriptions to the predominant scanning mechanism of translation initiation, and we will not provide a detailed description of the yeast ribosome nor of the tRNAs and complementary tRNA synthetases required for high-fidelity protein synthesis.
Mechanism of Translation Initiation
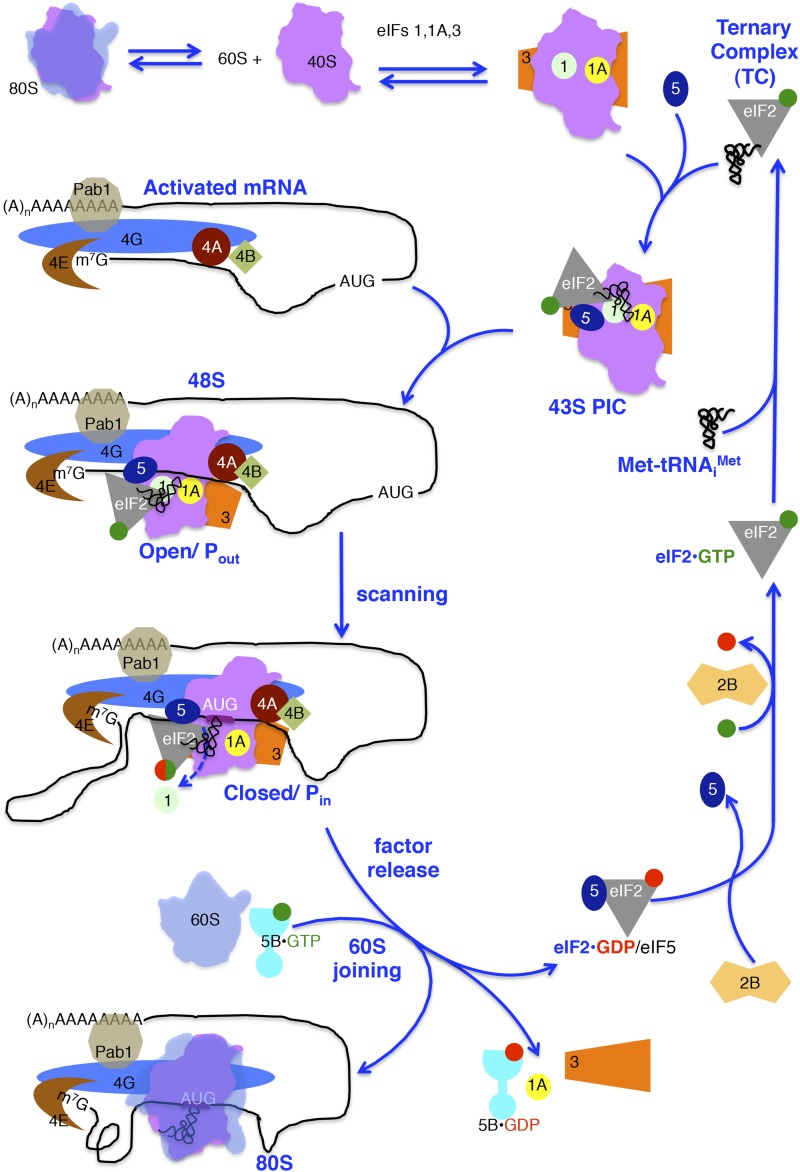
The most complex step of protein synthesis is translation initiation. In addition to the 40S and 60S ribosomal subunits, Met-tRNAiMet and 11 translation initiation factors consisting of 24 independent gene products (Table 1) are required to initiate translation on an mRNA. As detailed in the scheme in Figure 1, translation initiation factors function in an ordered fashion to assemble the 80S ribosomal complex that synthesizes proteins. First, the factor eIF2 binds GTP and Met-tRNAiMet forming a ternary complex (TC) that associates with the 40S ribosome along with the factors eIF1, eIF1A, eIF3, and perhaps eIF5 to form the 43S preinitiation complex (PIC). The eIF4 family of factors including the 7-methylguanosine (m7G) mRNA cap-binding protein eIF4E, the RNA helicase eIF4A, and the factors eIF4G and eIF4B, are thought to prepare the mRNA for binding to the 43S PIC to form a 48S PIC. Following binding near the 5′ end of the mRNA, the ribosomal complex scans down the mRNA in search of an AUG start codon. Selection of the translation start site is accompanied by completion of GTP hydrolysis by eIF2 and release of many of the initiation factors. The factor eIF5B, a second GTPase, promotes binding of the 60S subunit to form an 80S ribosome. Subsequent GTP hydrolysis by eIF5B leads to its release from the 80S monosome, which is poised to begin translation elongation.
Figure 1.
Pathway for yeast cytoplasmic translation initiation. Protein synthesis begins with the dissociation of ribosomal subunits and assembly of a 43S PIC. This is shown as consecutive steps in which eukaryotic initiation factors (eIFs) 1, 1A, and 3 bind to the 40S subunit first, followed by the eIF2–GTP (green circle)–Met-tRNAiMet ternary complex (TC) and eIF5. The 43S PIC binds an activated mRNA near the 5′ cap, forming a 48S complex. Activated mRNAs bear eIF4E at the 5′ cap, Pab1 bound to the poly(A) tail, bridged by eIF4G to form a loop along with eIF4A and eIF4B. During scanning, the 43S PIC in an open conformation, where Met-tRNAiMet is not fully base paired within the P site (Pout), moves in a 3′ direction along the 5′ UTR to the AUG codon. Either prior to or upon AUG recognition, GTP bound to TC is hydrolyzed to GDP+Pi (green and red hybrid circle), but Pi is not released until AUG recognition. Start codon selection is accompanied by release of eIF1, Pi loss from eIF2–GDP (red circle), release of eIF2 and eIF5, and reorganization of the 43S PIC to a closed state with Met-tRNAiMet in the Pin conformation and tightly bound to the complex. eIF5B–GTP promotes joining of the 60S subunit to the AUG-bound PIC. GTP hydrolysis and release of eIF5B–GDP and eIF1A forms the 80S complex with Met-tRNAiMet bound in the P site and a vacant A site ready for the elongation phase of protein synthesis. Recycling of eIF2 is accomplished by eIF2B displacing eIF5 from eIF2–GDP and then facilitating nucleotide exchange on eIF2. Met-tRNAiMet binds to eIF2–GTP reforming TC.
mRNA features in translation initiation
In addition to translation factors, mRNA features also contribute to formation of a translating 80S ribosome. While the most important feature of an mRNA is the open reading frame (ORF), other parts of the mRNA have significant impacts on protein synthesis. Nearly all yeast proteins are initiated with methionine encoded by an AUG codon. In addition, in almost all cases, protein synthesis starts at the first AUG codon from the 5′ end of the mRNA. To date, only a few exceptions to these rules have been identified, and interestingly several of the exceptional mRNAs are subject to translational regulation or encode proteins that are targeted to more than one subcellular compartment (Hinnebusch 2011).
Start codons and context nucleotides:
Translation initiation in yeast has generally been thought to be restricted to AUG codons. For example, when the AUG start codon of a CYC7 reporter gene was replaced by any of the nine single nucleotide near cognate codons (one mismatch from AUG), protein synthesis dropped to <0.5% of the AUG control (Clements et al. 1988). Likewise, all possible single nucleotide substitutions at the AUG start codon of a HIS4-lacZ reporter lowered expression to ≤2% of the AUG control (Donahue and Cigan 1988). However, the mRNAs encoding glycyl (Grs1) and alanyl (Ala1) tRNA synthetases initiate at both AUG and non-AUG codons (Chang and Wang 2004; Tang et al. 2004; Chen et al. 2008). Whereas the cytoplasmic synthetases initiate at an AUG start codon, the extended, mitochondrial enzymes initiate at upstream codons: UUG for Grs1 (Chen et al. 2008) and ACG for Ala1 (Tang et al. 2004).
Whole genome ribosomal profiling studies that mapped ribosome-protected mRNA fragments confirm the presence of ribosomes initiating translation at the non-AUG codons in the GRS1 and ALA1 mRNAs (Ingolia et al. 2009). While ribosome profiling studies have identified initiation at both AUG and non-AUG codons at short upstream open reading frames (uORFs) in the 5′ leader of yeast mRNAs, recent studies indicate that non-AUG codons rarely contribute to initiation of uORFs in vivo (Arribere and Gilbert 2013) and that sample processing procedures may have resulted in overrepresentation of some rarely used translation start sites (Gerashchenko and Gladyshev 2014).
The context of nucleotides around the start codon has been shown to be important in mammalian translation, but these flanking nucleotides appear to play a less significant role in yeast. Kozak defined an optimal sequence for start codon selection in mammalian cells as CC(A/G)CCAUG(G/A) (Kozak 2002, 2005). Within this context, the nucleotides at positions −3 and +4 relative to the A of the AUG codon were shown to be most important. Substitutions of pyrimidines at these positions lead to scanning ribosomes bypassing the AUG codon and thus leaky scanning to a start codon further 3′ in the mRNA. Three studies in yeast revealed only modest impacts of flanking nucleotides on AUG start codon selection. Studying the HIS4 gene, Cigan et al. (1988b) found that changing the preferred −3 A residue to C, G, or the least preferred U, reduced expression by only ∼3, 23, and 40%, respectively. Likewise, in studies of start codon context in derivatives of the CYC1 gene, Baim and Sherman (1988) found that U or C at the −3 position resulted in roughly a twofold increase in leaky scanning as compared to when a purine was at this position. Finally, based on a high throughput screen of start codon context nucleotides, Dvir et al. (2013) reported a significant, but modest (<30%), impact of a −3 purine on reporter gene expression. At odds with these studies, flanking nucleotides have been shown to be important in selection of the non-AUG start codon on the GRS1 mRNA (Chen et al. 2008) as well as in the selection of alternate AUG start codons on the MOD5 and CCA1 mRNAs (Werner et al. 1987; Slusher et al. 1991; Wolfe et al. 1994). In addition, the poor start codon context (C−3GUAUG) of the SUI1 gene encoding translation factor eIF1 in yeast impairs expression and enables autoregulation due to the role of eIF1 in start codon selection (Martin-Marcos et al. 2011).
mRNA leader length and secondary structure:
In general, yeast mRNAs have short and rather unstructured 5′ UTRs (Kertesz et al. 2010). A genome-wide analysis of transcription start sites in yeast enabled the characterization of the 5′ UTR for ∼80% of yeast genes (Nagalakshmi et al. 2008). These data revealed an average 5′ UTR length of 50 nucleotides with <5% of mRNAs having an AUG codon within 10 nucleotides of the 5′ end. Interestingly, mRNAs with short leaders, <12–20 nt, are subject to nonsense-mediated mRNA decay (NMD), apparently due to ribosomes bypassing the first start codon and initiating at downstream, out-of-frame sites (Arribere and Gilbert 2013).Thus, an AUG codon too close to the 5′ end of an mRNA is not readily recognized by the translating ribosome. In contrast, expanding the length of the 5′ UTR of a luciferase reporter mRNA from 43 nt to >1700 nt had no significant effect on relative luciferase expression (Berthelot et al. 2004). Thus, scanning ribosomes are thought to possess a high level of processivity at least on the relatively unstructured mRNAs present in yeast.
According to the scanning model of translation, secondary structure in the 5′ UTR could block translation by at least two different mechanisms. Secondary structure near the 5′ cap of the mRNA could prevent ribosome association with the mRNA, whereas secondary structure further down the 5′ UTR could prevent ribosome scanning. Several studies in yeast have demonstrated that insertion of stem-loop structures in the 5′ UTR interferes with translation (Baim and Sherman 1988; Cigan et al. 1988b; Abastado et al. 1991; Vega Laso et al. 1993; Berthelot et al. 2004; Sen et al. 2015). As expected, more stable stem-loop structures are more deleterious than weaker stem loops; however, the impact of cap-proximal vs. more distal secondary structure varies in the different published reports.
mRNA cap and poly(A) tail:
In addition to playing important roles in mRNA stability, the m7G-(5′)ppp(5′)-N cap and poly(A) tail contribute to the translation of an mRNA. All genomically encoded mRNAs in yeast are capped at their 5′ end with m7GTP. Capping occurs co-transcriptionally and is catalyzed by the enzymes Cet1, Ceg1, and Abd1, an RNA 5′ triphosphatase, a GTP-mRNA guanyltransferase, and an RNA guanine-7-methyltransferase, respectively (Shuman 2001). As described below, the cap structure is recognized by the translation factor eIF4E. At the 3′ end of the mRNA, the poly(A) tail is bound by the protein Pab1. Interestingly, Pab1 and eIF4E bind to separate sites on the translation factor eIF4G, and this binding has been shown to mediate mRNA circularization (Wells et al. 1998). The functional significance of mRNA circularization has not been resolved and it has been proposed to facilitate translation by helping shunt terminating ribosomes to the 5′ end of the same mRNA. Alternatively, mRNA circularization may serve a regulatory role to ensure translation of only intact (capped and polyadenylated) mRNAs.
Experiments in animal and plant cells demonstrated that the mRNA cap and poly(A) tail act synergistically to promote translation (Gallie 1991), and experiments using in vitro translation systems prepared from whole yeast cell extracts revealed a similar functional coupling between the cap and poly(A) tail (Iizuka et al. 1994; Tarun and Sachs 1995). Whereas the cap or poly(A) tail alone stimulated translation >20-fold compared to an mRNA lacking both features, the presence of both a cap and poly(A) tail enhanced translation an additional 2- to 8-fold (Tarun and Sachs 1995). As expected, cap-dependent translation is dependent on eIF4E, and poly(A) stimulation of translation is dependent on Pab1 (Otero et al. 1999).
Initiator methionyl-tRNA
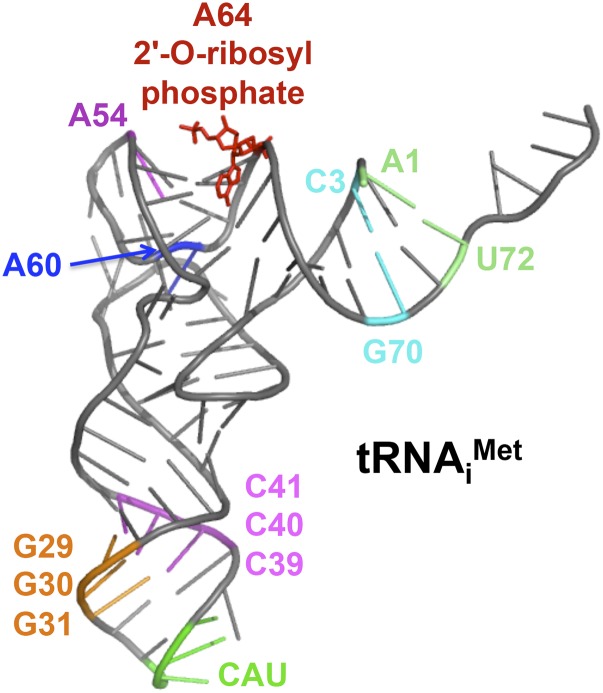
The tRNAiMet performs a unique role in protein synthesis. Distinct initiator and elongator tRNAs are used to incorporate methionine at the start codon vs. internal AUG codons in ORFs, respectively. Yeast contain four to five IMT genes encoding tRNAiMet and five EMT genes encoding elongator methionyl-tRNA (tRNAeMet) (Astrom et al. 1993). Whereas both sets of tRNAs contain a 5′-CAU-3′ anticodon, nucleotide and post-transcriptional modification differences restrict the function of the tRNAs to initiation vs. elongation. Swapping nucleotides between tRNAiMet and tRNAeMet has provided insights into the critical determinants for tRNAMet function in initiation vs. elongation. Functionally important features of tRNAiMet include: (1) A1:U72 and C3:G70 base pairs in the acceptor stem; (2) A54 and A60 in the T loop; and (3) three G:C base pairs in the anticodon stem (positions 29–31:39–41). In addition, the nucleotide A54 and an O-ribosyl phosphate modification of A64 restrict tRNAiMet from functioning in translation elongation (Figure 2) (von Pawel-Rammingen et al. 1992; Astrom et al. 1993; Astrom and Bystrom 1994).
Figure 2.
tRNAiMet features important for translation initiation. Features that enhance tRNAiMet function in initiation or restrict it from functioning in elongation are highlighted on the tertiary structure of yeast tRNAiMet (pdb 1YFG). Highlighted residues include A1:U72 and C3:G70 base pairs in the acceptor stem, residues A54 and A60 in the T loop, and a 2’-O-ribosyl phosphate modification on residue A64. Three consecutive G:C base pairs in the anticodon loop are important for the accuracy of start site selection. The anticodon 5′-CAU-3′ is depicted in green. Structure was generated using the PyMol Molecular Graphics System (version 1.7.6.6, Schrödinger).
Substitution of the A1:U72 base pair in tRNAiMet by G1:C72, as found in tRNAeMet, impaired yeast cell growth (von Pawel-Rammingen et al. 1992; Astrom et al. 1993), binding of Met-tRNAiMet to eIF2 (Farruggio et al. 1996; Kapp et al. 2006), and TC binding to the 40S ribosome (Kapp et al. 2006). Thus the identity of this base pair contributes both to TC formation and to later steps in the initiation pathway. Three consecutive G:C base pairs in the anticodon stem are important for tRNAiMet function in bacteria (Varshney et al. 1993; Mandal et al. 1996), and their critical role in eukaryotes has only recently been revealed (Dong et al. 2014). Disrupting the G31:C39 base pair in the anticodon loop altered the accuracy of translation start site selection in a manner that was sensitive to the presence of the A54 residue in the T loop (Dong et al. 2014). As the C3:G70 base pair in the acceptor stem likewise contributed to the accuracy of translation start site selection (Dong et al. 2014), and all of these mutations affected the binding of the eIF2–GTP–Met-tRNAiMet ternary complex to the 40S ribosome, albeit in distinct ways, these results indicate that conserved nucleotides of the tRNAiMet contribute to the accuracy of translation start site selection.
An additional important determinant in tRNAiMet is a 2-O-ribosyl phosphate modification of A64. In yeast strains lacking Rit1, the enzyme that catalyzes the modification, tRNAiMet can function in translation elongation (Astrom and Bystrom 1994). Interestingly, domain III of EF-Tu, and by analogy of the eukaryotic elongation factor eEF1A, contacts the T loop of the bound tRNA (Nissen et al. 1995). The O-ribosyl phosphate modification of position 64 in the T loop would be expected to sterically interfere with Met-tRNAiMet complex formation with eEF1A. Hence, it is thought that this modification restricts tRNAiMet function to initiation and thus prevents competition for methionyl-tRNA between translation initiation and elongation. Consistent with this idea, deletion of RIT1 exacerbated the growth defect in strains with mutations in eIF2 or tRNAiMet; and this growth defect was partially rescued by overexpression of tRNAiMet and further exacerbated by overexpression of eEF1A (Astrom et al. 1999).
While many nucleotides in tRNAs are post-transcriptionally modified (for example by methylation, conversion to pseudouridine, etc.), it is noteworthy that tRNAiMet appears to be especially sensitive to these modifications. Most of the 11 modifications of yeast tRNAiMet are nonessential; however, loss of the m1A58 modification destabilizes tRNAiMet and impairs translation initiation (Anderson et al. 1998, 2000). The Gcd10/Gcd14 complex catalyzes methylation of A58, and inactivation of GCD10 or GCD14 results in turnover of tRNAiMet by the Trf4/Rrp6 pathway (Kadaba et al. 2004).
Ternary complex formation
The translation factor eIF2 is responsible for binding Met-tRNAiMet to the 40S ribosome. A TC is formed between Met-tRNAiMet and the GTP-bound form of eIF2. The eIF2 is a heterotrimeric complex consisting of α (Sui2), β (Sui3), and γ (Gcd11) subunits. The yeast eIF2α (SUI2) and eIF2β (SUI3) genes were first discovered by Donahue et al. (1988) in a screen for mutations that suppress the histidine auxotrophy of his4-303 strains in which an AUU codon is substituted for the initiating AUG codon of the HIS4 gene. Spontaneous mutations in unlinked sui (suppressors of initiator codon) genes, including SUI2 (Cigan et al. 1989) and SUI3 (Donahue et al. 1988), enable translation to initiate at an in-frame UUG codon that normally encodes Leu as the third residue in HIS4 (Donahue et al. 1988). As discussed below, analysis of Sui− mutations in eIF2 and other translation factors has provided insights into the mechanism of start codon selection during translation initiation.
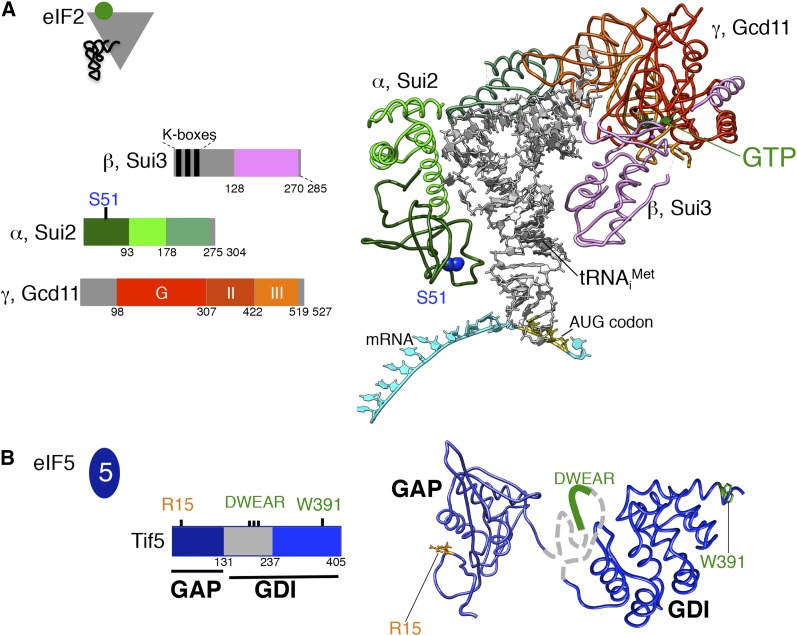
Structures of yeast (Dhaliwal and Hoffman 2003; Hussain et al. 2014; Llacer et al. 2015), archaeal (Schmitt et al. 2012), and mammalian eIF2α (Ito et al. 2004) revealed that the protein consists of three domains: an N-terminal OB-fold domain and a central α-helical domain that are connected through a flexible linker to a C-terminal α/β domain that binds to eIF2γ (Figure 3A) (Schmitt et al. 2012; Hussain et al. 2014; Llacer et al. 2015). A key mode of translational control in yeast and other eukaryotes involves phosphorylation of eIF2α. The yeast kinase Gcn2, which is conserved in all eukaryotes, phosphorylates the conserved Ser51 residue in a mobile loop of the OB-fold domain (Dever et al. 1992).
Figure 3.
Schematic and structural models of eIF2 and eIF5. (A) Structural model of the eIF2–GTP–Met-tRNAiMet ternary complex bound to an mRNA AUG codon (right) and cartoons depicting the eIF2 α, β, and γ subunit structural domains (left) using the same color schemes. The structural model is adapted from the structure of the yeast 48S complex (pdb 3JAP) with the 40S ribosome and other initiation factors omitted for clarity (Llacer et al. 2015). The eIF2α residue Ser51 (blue), GTP analog (green), Met-tRNAiMet (gray), and mRNA (cyan) with AUG codon (yellow) are indicated. (B) eIF5 domains and activities (left) and structural models (right) for the human GAP domain bearing R15 (pdb 2E9H) and the yeast CTD bearing W391 (pdb 2FUL) (Wei et al. 2006). Structures were drawn using Chimera software (University of California, San Francisco, UCSF).
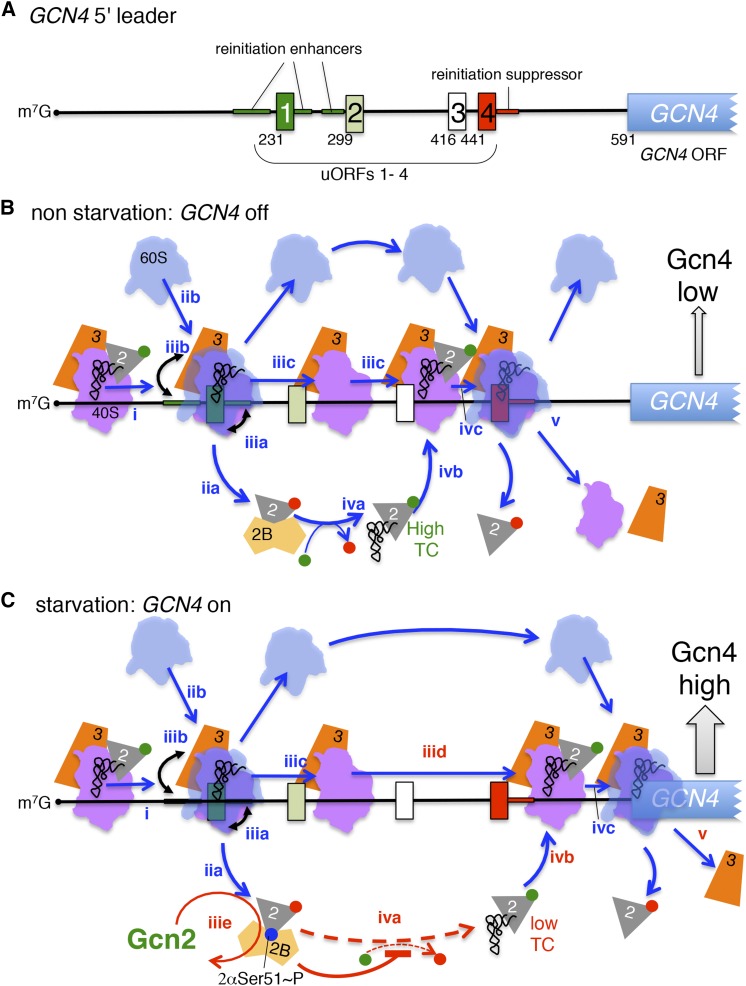
Whereas translation of the GCN4 mRNA in yeast is typically repressed by the presence of uORFs in the mRNA leader, phosphorylation of eIF2α enables ribosomes to bypass the inhibitory uORFs and initiate translation at the GCN4 ORF (reviewed in Hinnebusch 2005). Yeast lacking GCN2 are unable to grow under amino acid starvation conditions due to the failure to derepress GCN4 expression (Wek et al. 1989, 1990). Gcd− mutations, including mutations that impair eIF2 function (Williams et al. 1989), derepress GCN4 expression in the absence of GCN2, mimicking the effect of eIF2 phosphorylation in reducing TC assembly (Hinnebusch 2005).
The C-terminal half of yeast eIF2β shows significant sequence homology to archaeal aIF2β and consists of three elements: an N-terminal α-helix, a central helix–turn–helix domain, and a C-terminal zinc-binding domain (Figure 3A) (reviewed in Schmitt et al. 2010). The N-terminal α-helix, which is unstructured in the free form of aIF2β, binds to the backside of the aIF2γ GTP-binding (G) domain in the aIF2 complex (Sokabe et al. 2006; Yatime et al. 2007). Point mutations in this helix of yeast eIF2β, as well as in the docking site on yeast eIF2γ, disrupt eIF2 complex formation and confer Gcd− and Sui− phenotypes (Hashimoto et al. 2002; Borck et al. 2012). In the archaeal aIF2 complex, the C-terminal zinc-binding domain of aIF2β packs against the central α–β domain (Sokabe et al. 2006; Yatime et al. 2007). While the function of the zinc-binding domain has not been resolved, removal of this domain impairs RNA binding to isolated yeast eIF2β (Laurino et al. 1999) and confers a dominant Gcd− and recessive lethal phenotype (Castilho-Valavicius et al. 1992), while point mutations confer a dominant Sui− phenotype (Donahue et al. 1988; Castilho-Valavicius et al. 1992).
The N-terminal half of eIF2β is not present in the archaeal protein. Key features of this portion of eIF2β are three elements referred to as K-boxes K1, K2, and K3, each containing seven Lys residues and one Ser or Thr residue. Whereas deletion of any single or two K-boxes does not affect cell viability, removal of all three K-boxes is lethal (Asano et al. 1999; Laurino et al. 1999). Consistent with these findings, substituting Ala residues in place of the K3 Lys residues in a SUI3 allele lacking K1 and K2 was also lethal. In contrast, substituting Arg residues in place of the K3 Lys residues in the same allele was viable and had no impact on cell growth (Laurino et al. 1999). Thus, the positively charged character of at least one K-box is required for cell viability. Biochemical analyses revealed that removal of the K-boxes impairs mRNA, but not Met-tRNAiMet, binding to isolated eIF2 complexes (Laurino et al. 1999). Moreover, mutating the K-boxes in eIF2β impairs the binding of isolated eIF2β, as well as the eIF2 complex, with both the eIF2 GTPase stimulatory factor eIF5 and the catalytic ε-subunit of the eIF2 guanine-nucleotide exchange factor (GEF) eIF2B (Asano et al. 1999). A bipartite element consisting of acidic and aromatic amino acids is conserved at the C termini of eIF5 and eIF2Bε and mediates the K-box-dependent interaction with the N terminus of eIF2β (Asano et al. 1999).
The γ-subunit of eIF2, encoded by GCD11, was first identified based on the Gcd− phenotype of several mutants (Hannig et al. 1993). Interestingly, mutations in eIF2γ were independently isolated in a screen for Sui− mutants (Huang et al. 1997). Consistent with these findings, the GCD11-R510H mutant, originally isolated based on its ability to derepress GCN4 expression, also confers a Sui− phenotype (Dorris et al. 1995). The eIF2γ protein consists of three domains: an N-terminal GTP binding domain and β-barrel domains II and III (Figure 3A). Based on structural studies of the archaeal and yeast complexes, eIF2γ is the keystone of the eIF2 complex with separate docking sites for the eIF2α and eIF2β subunits (Schmitt et al. 2012; Hussain et al. 2014; Llacer et al. 2015). The incorporation of eIF2γ in the eIF2 complex is dependent on the apparently eIF2-specific chaperone Cdc123 (Perzlmaier et al. 2013). The amino acid sequence and structure of eIF2γ and aIF2γ show striking similarity to elongation factor EF-Tu from bacteria (Hannig et al. 1993; Schmitt et al. 2002; Roll-Mecak et al. 2004). Whereas EF-Tu binds diverse aminoacyl-tRNAs (aa-tRNAs) to the ribosomal A site, eIF2 specifically binds Met-tRNAiMet to the ribosomal P site. The structure of Phe-tRNA bound to EF-Tu revealed that the amino acid and acceptor stem of the tRNA bind in a pocket formed between the G domain and domain II. Supporting the notion that eIF2γ uses a similar pocket for Met-tRNAiMet binding, the slow-growth phenotype of the gcd11-Y142H mutant, which alters a residue in the proposed Met-tRNAiMet binding pocket, was partially suppressed by overexpression of tRNAiMet, and purified eIF2 complexes containing the mutant eIF2γ subunit showed defects in Met-tRNAiMet binding (Dorris et al. 1995; Erickson and Hannig 1996; Shin et al. 2011). Thus, at least the acceptor stem and amino acid binding site appears to be shared between eIF2 and EF-Tu. In contrast, a contact between the body of the tRNA, especially the T stem, and domain III of EF-Tu is apparently not conserved in eIF2γ (Nissen et al. 1995; Sanderson and Uhlenbeck 2007a,b; Shin et al. 2011; Schmitt et al. 2012; Hussain et al. 2014; Llacer et al. 2015). Instead, hydroxyl radical probing experiments and cryo-EM structures of 48S PICs indicated that domain III of eIF2γ projects toward, but does not contact, helix h44 on the subunit interface surface of the 40S ribosomal subunit (Shin et al. 2011).
Purified yeast eIF2 binds either GTP (Kd ∼1.7 µM) or GDP (Kd ∼0.02 µM) (Kapp and Lorsch 2004a). As for a number of G proteins, this 100-fold higher affinity for GDP relative to GTP introduces the requirement for eIF2B to recycle eIF2–GDP complexes to the functional eIF2–GTP form. Whereas eIF2–GTP complexes bind Met-tRNAiMet (Kd ∼9 nM) to form a ternary complex, eIF2–GDP binary complexes are defective for Met-tRNAiMet binding (Kd ∼150 nM) (Kapp and Lorsch 2004a). Thermodynamic coupling between GTP and Met-tRNAiMet binding to eIF2 results in a 10-fold increase in GTP binding affinity in the presence of Met-tRNAiMet (GTP Kd ∼0.2 nM) (Kapp and Lorsch 2004a). Consistent with this biochemical result, the slow-growth phenotype of an eIF2γ-K250R mutation, which impairs GDP and GTP binding to eIF2, is suppressed by overexpression of tRNAiMet (Erickson and Hannig 1996).
As noted above, conserved features of tRNAiMet contribute to ternary complex formation. However, the Met on Met-tRNAiMet appears to be the most important determinant for TC formation. Deacylation of Met-tRNAiMet decreases its affinity for binding to eIF2 by >10-fold (Kd ∼130 nM), comparable to the binding of Met-tRNAiMet to eIF2–GDP (Kapp and Lorsch 2004a). It is postulated that the thermodynamic coupling between eIF2 and the methionine residue on Met-tRNAiMet serves to ensure that translation initiates exclusively with Met.
Interestingly, the eIF2γ-K250R mutation in addition to weakening GDP binding enables cell survival in the absence of eIF2α (Erickson et al. 2001). The growth of the gcd11-K250R sui2Δ strain is further enhanced by overexpression of tRNAiMet (IMT4) and by overexpression of gcd11-K250R and SUI3 (Erickson et al. 2001). As weakening GDP binding to eIF2 enables elimination of eIF2α, these findings suggest that eIF2α plays a role in stimulating the eIF2B-catalyzed guanine nucleotide exchange on eIF2.
43S PIC formation
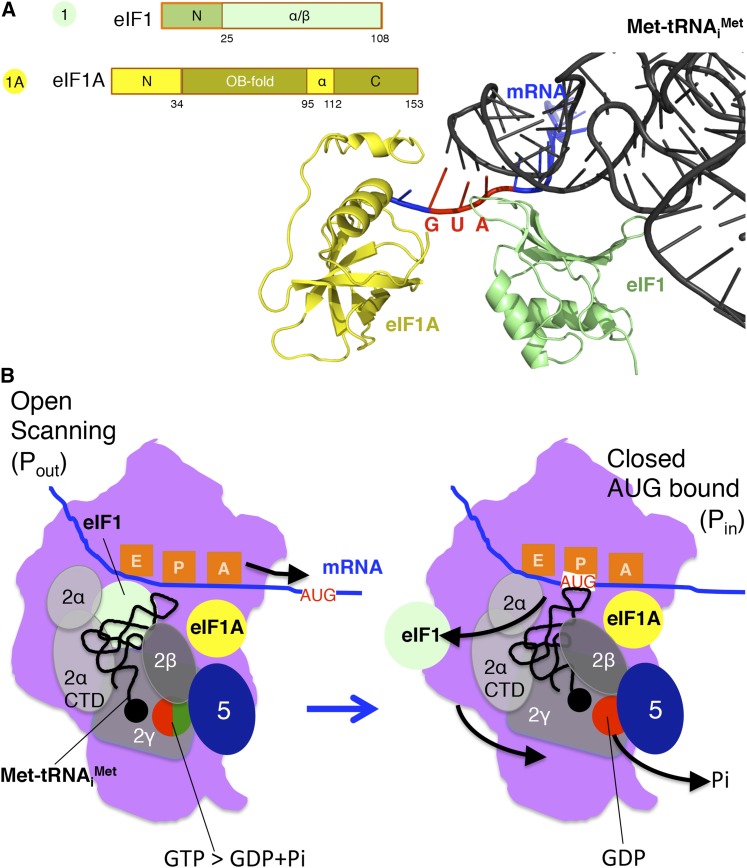
Binding of the eIF2 TC to the 40S subunit is facilitated by the factors eIF1 and eIF1A that bind directly to the 40S ribosome (Figure 1). The factor eIF1, encoded by SUI1, is a small (108 amino acid) protein that, based on structures of the yeast or the analogous Tetrahymena factor, binds to the platform of the 40S subunit near the P site (Figure 4, A and B) (Rabl et al. 2011; Hussain et al. 2014). The factor eIF1A, encoded by TIF11, is homologous to the bacterial factor IF1 (Battiste et al. 2000; Choi et al. 2000; Olsen et al. 2003; Hussain et al. 2014). Like IF1, eIF1A binds to the ribosomal A site and likely functions, in part, to prevent Met-tRNAiMet binding in the A site (Figure 4, A and B) (Carter et al. 2001; Hussain et al. 2014). Cryoelectron microscopy of the yeast 40S ribosome has revealed conformational changes accompanying the binding of eIF1 and eIF1A (Passmore et al. 2007; Hussain et al. 2014). In the absence of factors, the “latch” of the mRNA entry channel, composed of 18S rRNA helices h34 in the head and h18 in the body of the 40S subunit, is closed. Binding of eIF1 and eIF1A to the 40S subunit is accompanied by rotation of the head of the subunit (Hussain et al. 2014), perhaps providing access for the Met-tRNAiMet and TC and by weakening of the latch interactions to enable binding of mRNA (Passmore et al. 2007; Hussain et al. 2014). Interestingly, when only eIF1A is bound to the 40S subunit, the density corresponding to the latch is stronger than that observed in the apo-40S structure. As described below, this so-called “closed” complex in the absence of eIF1 is thought to be associated with selection of the translation start codon.
Figure 4.
Schematic and structural models of eIF1, eIF1A, and AUG codon selection. (A) Structural model (right) and schematics (left) of eIF1 (green) and eIF1A (yellow) bound to the 48S PIC (pdb 3JAP) along with Met-tRNAiMet (black) and mRNA (blue, AUG codon in red), but other factors and the ribosome are removed for clarity. Structure was generated using the PyMol Molecular Graphics System (version 1.7.6.6, Schrödinger). (B) Cartoon showing approximate positions of eIFs 1 and 1A with TC and eIF5 in the open scanning conformation (left) with Met-tRNAiMet not fully engaged in the P site (Pout), and factor movements (black arrows) induced by AUG codon recognition (right) and the transition to the closed complex (Pin) signaled by movement of eIF1 that triggers Pi release prior to eIF2–GDP–eIF5 release from the PIC.
Yeast eIF1 is composed of an ∼20-residue unstructured N-terminal tail (NTT) followed by an ∼88-residue folded α/β core (Figure 4A) (Reibarkh et al. 2008). The α/β core of eIF1 resembles similar domains in eIF2β, the N terminus of eIF5 (Figure 3C), and several ribosomal proteins (Reibarkh et al. 2008). In addition to binding the ribosome and regulating TC binding, eIF1 directly contacts eIF2β, the C-terminal domain of eIF5, and eIF3c. As these contacts have been mapped to distinct regions of eIF1, it is thought that eIF1 can simultaneously bind all three factors, and consistent with this idea, eIF1 can be found in a multifactor complex (MFC) with the eIF2 TC, eIF3, and eIF5 (Asano et al. 2000).
The 153-residue yeast eIF1A consists of a central OB-fold domain that resembles the bacterial factor IF1 (see Fekete et al. 2005). The core of eIF1A is buttressed on its C-terminal side by a helical region consisting of a long α2 helix and a short 310 helix. In addition, the factor has long unstructured N(∼25-residues)- and C(∼34 residue)-terminal tails (Figure 4A). In the 43S complex, the C-terminal tail (CTT) of eIF1A crosses through the P site (Hussain et al. 2014; Zhang et al. 2015). The Met-tRNAiMet is thus prevented from fully engaging the P site (Pin state) and instead is thought to be in a Pout state that is more conducive to scanning (Hinnebusch 2011, 2014). As discussed below, the CTT of eIF1A also interacts with the N-terminal domain (NTD) of eIF5 and with domain IV of eIF5B in subsequent steps of the initiation pathway.
Despite interacting with distinct sites, binding of eIF1 and eIF1A to the 40S subunit is thermodynamically coupled (Maag and Lorsch 2003). Moreover, both factors are required to achieve stable binding of the eIF2 TC in vitro (Algire et al. 2002). Consistent with these findings, mutations in the eIF1 core domain or in eIF1A that weaken their binding to the 40S ribosome likewise decrease the rate of TC binding in vitro and confer Gcd− phenotypes in vivo (Fekete et al. 2005; Cheung et al. 2007).
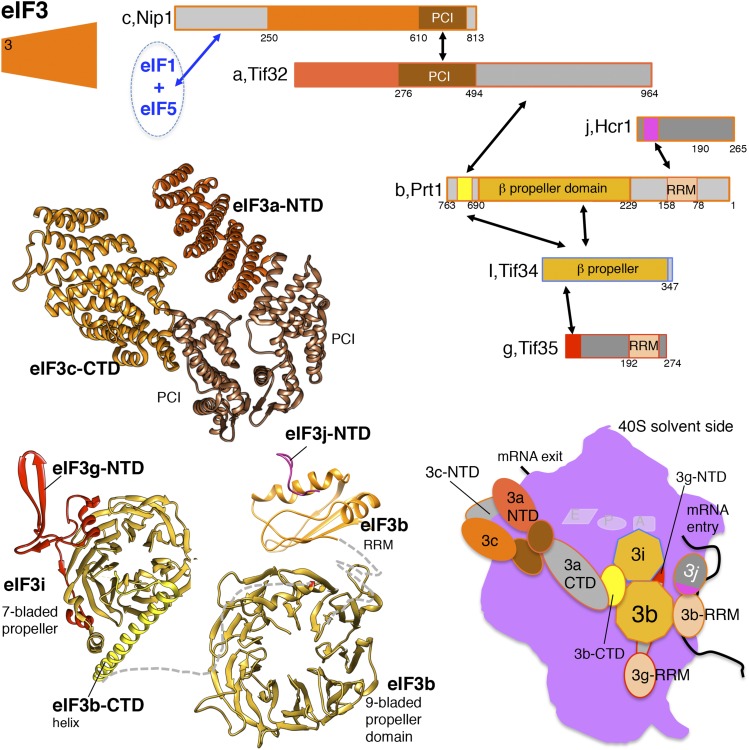
Whereas eIF1 and eIF1A are critical for TC binding to the 40S in the reconstituted yeast in vitro translation system, the factor eIF3 has been reported to stabilize TC binding by only approximately twofold (Kapp and Lorsch 2004b). Yeast eIF3 is composed of five essential subunits (a/Tif32, b/Prt1, c/Nip1, i/Tif34, and g/Tif35) and one nonessential subunit (j/Hcr1) (Figure 5) (note that the unusual nomenclature of the yeast eIF3 subunits is due to the presence of additional subunits in the mammalian factor that are not present in yeast eIF3). In addition, eIF5 (Tif5) purifies stoichiometrically with tagged forms of eIF3 from yeast (Phan et al. 1998). Extensive mapping studies of protein–protein interactions have provided insights into the structure of eIF3 and its interaction with other factors (reviewed in Valasek 2012). The eIF3b/Prt1 is thought to form the primary scaffold of the multisubunit complex. The N terminus of eIF3b/Prt1 contains an RNA recognition motif (RRM) that serves as a protein–protein interaction site for eIF3j/Hcr1 as well as for the C-terminal part of eIF3a/Tif32, which resembles eIF3j/Hcr1. The central part of eIF3b/Prt1 binds to eIF3c/Nip1, and the eIF3i/Tif34 and eIF3g/Tif35 subunits bind cooperatively to the C-terminal portion of eIF3b/Prt1. Finally, the N-terminal portion of the eIF3c/Nip1 subunit binds directly to eIF1 and to eIF5, which in turn binds the eIF2 TC (Figure 5). Thus, eIF3 and in particular the eIF3b subunit plays a central role in assembly of the 43S PIC. In contrast to eIF1, eIF1A, and the TC, which bind to the intersubunit face of the 40S subunit, cryo-EM studies revealed that the core of yeast eIF3 binds to the solvent-exposed face of the 40S with arm-like projections, including the PCI domains of eIF3a and eIF3c, that bind near the mRNA entry channel reaching around to the intersubunit face of the 40S (Erzberger et al. 2014; Aylett et al. 2015; Llacer et al. 2015). Consistent with this model of the eIF3–40S complex, yeast eIF3 subunits have been found to interact with 18S rRNA and ribosomal proteins on the solvent-exposed side of the 40S subunit (Valasek et al. 2003; Kouba et al. 2012a,b). The C terminus of eIF3a/Tif32 was shown to bind to a region of 18S rRNA encompassing helices h16–h18, and in two-hybrid assays this same portion of eIF3a/Tif32 bound to ribosomal proteins Rps2 and Rps3. These interactions place eIF3a near the mRNA entry channel of the 40S subunit.
Figure 5.
Schematic and structural models of eIF3. Schematics depict the eIF3 subunit organization and indicate major structural domains and protein–protein interactions (black arrows) within the eIF3 core complex. Structural models depicting these interactions are shown using Chimera software (UCSF) using pdb coordinates 4U1C (eIF3a/c), 4U1E (eIF3b-CTD/eIF3i/eIF3g-NTD), 4U1F (eIF3b β-propeller domain) (Erzberger et al. 2014), and 2KRB (eIF3b RRM/eIF3j peptide) (Elantak et al. 2010). The cartoon depicting eIF3 binding to the 40S solvent-exposed surface is based on cryo-EM reconstructions (Erzberger et al. 2014; Aylett et al. 2015; Llacer et al. 2015). The same color scheme is used for consistency between images.
Inactivation of a temperature-sensitive eIF3b/Prt1 mutant (Phan et al. 1998; Nielsen et al. 2004) or depletion of eIF3c/Nip1 (Phan et al. 1998; Valasek et al. 2004) impairs general translation in vivo and in vitro. Moreover, extracts from these strains exhibit a defect in binding Met-tRNAiMet to 40S subunits that was rescued by adding back the eIF3 complex (Phan et al. 1998). The Met-tRNAiMet and mRNA binding defects in extracts from the prt1-1 strain were also rescued by addition of an eIF3abc, but not an eIF3big, partial complex (Phan et al. 2001). It is noteworthy that the factors eIF5, eIF1, and eIF3j/Hcr1 co-purified with the eIF3abc partial complex, raising the possibility that these latter factors contributed to the complementing activity. These results uncover a functional specialization within the eIF3 complex and they also support previous studies in mammalian systems, indicating that Met-tRNAiMet binding to the 40S subunit is a prerequisite for the ribosome to bind to an mRNA (see Hinnebusch 2000).
In addition to the sequential assembly of the 43S complex with eIF1 and eIF1A binding to the 40S subunit prior to association of the TC, an en masse assembly of the 43S complex has also been proposed. A MFC consisting of eIF1, eIF2, eIF3, and eIF5 plus Met-tRNAiMet has been isolated from cells, and in crude cell extracts the MFC can be separated from the 40S ribosome (Asano et al. 2000). It has been proposed that preassembly of the MFC facilitates proper binding of Met-tRNAiMet to the 40S subunit. Consistent with this hypothesis, the protein–protein interactions required for MFC integrity, including the binding of eIF1 to eIF3 (Singh et al. 2004; Valasek et al. 2004), eIF5 to eIF1, eIF2β, and eIF3c (Singh et al. 2004, 2005; Valasek et al. 2004; Yamamoto et al. 2005) and eIF2 to eIF3a (Valasek et al. 2002; Nielsen et al. 2004), are also important for protein synthesis in vivo, and mutations that disrupt eIF3c interaction with eIF1 or eIF5 confer Sui− phenotypes (Valasek et al. 2004). While the in vivo data support the idea that MFC integrity is important for translation initiation, additional experiments are needed to define the function of the MFC. In particular, it is important to determine whether the MFC binds to the 40S en masse and serves as a more efficient means to bind Met-tRNAiMet to the 40S subunit. Alternatively, it has been proposed that the MFC might serve as a depot for the initiation factors that are critical for stable binding of Met-tRNAiMet to the 40S (Aitken and Lorsch 2012).
mRNA recruitment of the 43S PIC
The 5′ cap and 3′ poly(A) tail of mRNAs serve as binding sites for eIF4E and the poly(A) binding protein Pab1, respectively, that act synergistically to assist in recruiting additional translation initiation factors including eIF4G and the 43S PIC to near the 5′ end (Figure 1 and Figure 6A) (Tarun and Sachs 1995; Preiss and Hentze 1998). Yeast mRNA 5′ leader sequences are of variable length and can contain secondary structures that impede 43S binding and scanning to AUG initiation codons. As a consequence ATP-dependent RNA helicases such as eIF4A and Ded1 are recruited. Our understanding of the roles of factors in these key steps is outlined below.
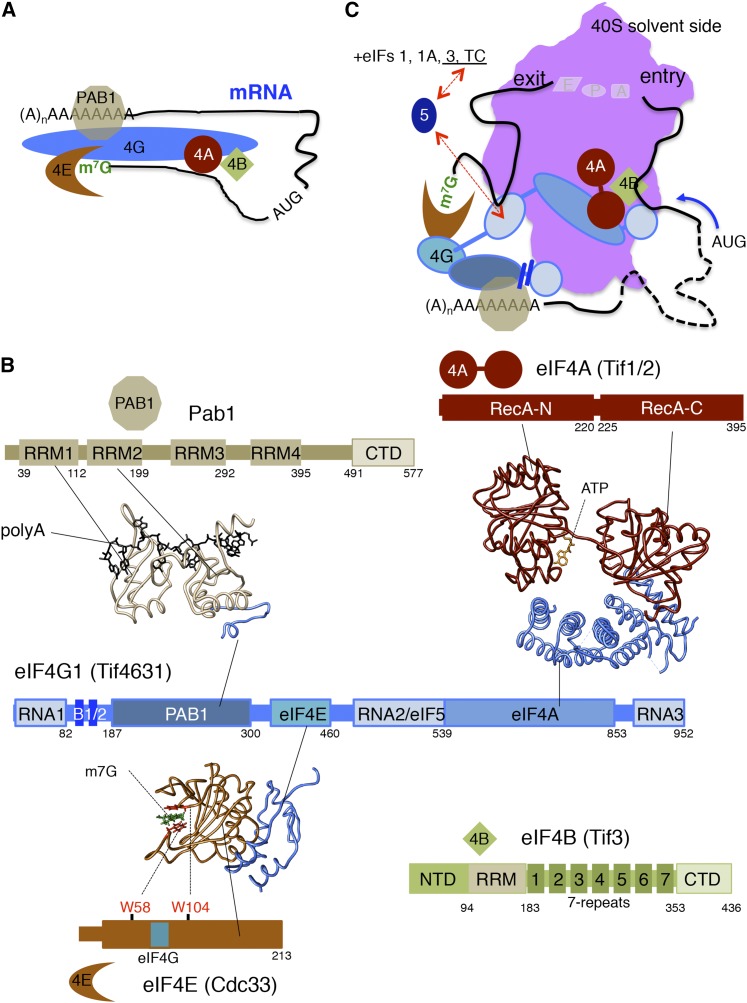
Figure 6.
Interactions among the m7G cap- and mRNA-binding factors. (A) Cartoon of mRNA recruitment step as in Figure 1. (B) Schematics of eIF4G (middle), Pab1 and eIF4A (top), and eIF4E and eIF4B (bottom). Factor binding domains on eIF4G are labeled, and structural models of the interacting factors are depicted. Structural models of human Pabp-poly(A)–eIF4G (Safaee et al. 2012), yeast eIF4A–eIF4G (Schutz et al. 2008), and yeast eIF4E–eIF4G (Gross et al. 2003) were drawn using Chimera software (UCSF). (C) Model for interactions of eIF4G domains with initiation factors and with the mRNA 5′ UTR on both the mRNA entrance and exit sides of the 40S ribosome to enhance mRNA binding to the ribosome.
eIF4E and mRNA 5′ cap recognition:
eIF4E has a compact single structural domain containing a cleft for mRNA 5′ cap binding. A pair of tryptophan residues (W58 and W104) form a 5′ cap-trapping sandwich (Altmann et al. 1988; Gross et al. 2003). A central region of eIF4G (eIF4G1393–460) binds to eIF4E on the opposite face to the 5′ cap interaction (Figure 6B). The eIF4E-4G binding interface overlaps with the surface important for binding 4E-binding proteins (4E-BPs) Caf20 and Eap1 that inhibit eIF4F assembly by competing with eIF4G to bind eIF4E (Altmann et al. 1997; Ptushkina et al. 1998; Cosentino et al. 2000).
While all mRNAs are capped and can bind eIF4E, they likely have differential affinity for eIF4E. Single-molecule FRET measurements with short model mRNAs calculated eIF4E affinity of 90 nM for an unstructured capped mRNA that was enhanced approximately fourfold by the presence of a modest secondary structure element 12 nt from the cap (O’Leary et al. 2013). Similarly, binding eIF4E to eIF4G enhances affinity for capped mRNA to ∼15–20 nM (Mitchell et al. 2010; O’Leary et al. 2013). Analysis of mRNAs bound to eIF4E in cells generally mirrors their levels in total RNA; however, >1000 mRNAs were enriched and a similar number were relatively depleted in eIF4E binding experiments (Costello et al. 2015). A simple conclusion is that eIF4E does not bind equally to all mRNAs in vivo.
The eIF4E-G113D (cdc33-1) temperature-sensitive (ts) mutant causes cell cycle arrest in G1 that was attributed to reduced translation efficiency of the CLN3 mRNA (Danaie et al. 1999). When Cln3 levels are elevated, cdc33-1 cells arrest randomly in the cell cycle rather than at G1, suggesting that translation of CLN3 mRNA, which bears an upstream ORF that contributes to its translational control (Polymenis and Schmidt 1997), becomes rate limiting for passage through G1 upon eIF4E inactivation. As cdc33 cells retain some protein synthesis activity at nonpermissive temperatures, the mutated factor may retain partial function (Altmann and Trachsel 1989). However, cell fluorescence studies suggest that when eIF4E is inactivated, the nuclear cap-binding protein Sto1 remains bound to mRNAs following their exit from the nucleus (Garre et al. 2012) and there is evidence that the nuclear cap complex, composed of Sto1 and Cbc2, may promote continued translation initiation, but with lower efficiency (Fortes et al. 2000).
eIF4G and Pab1 bring mRNA ends together:
eIF4G binds several translational components including eIF4E, eIF4A, Pab1, eIF5, 40S ribosomes, and mRNA (Figure 6, A–C). Although the two yeast eIF4G proteins (Table 1) are smaller than their mammalian counterparts, they share many of the same domains and interactions. Yeast eIF4G1 (Tif4631) and eIF4G2 (Tif4632) are 51% identical and appear to be functionally redundant. eIF4G1 expression levels are higher than eIF4G2 and poor growth phenotypes associated with tif4631∆ can be overcome by expressing eIF4G2 at higher levels from the stronger TIF4631 promoter (Clarkson et al. 2010).
eIF4G1 and 4G2 share an eIF4E interaction domain (eIF4G1393–460). This fragment adopts an α-helical structure on binding eIF4E (Figure 6B) (Gross et al. 2003) and bears a core conserved “YxxxFLL” motif that is critical for binding eIF4E. The tif4631-459 allele with both Leu residues (positions 457 and 458) mutated to alanine has greatly weakened eIF4E interaction and confers a ts phenotype in strains lacking eIF4G2, demonstrating that the eIF4E–eIF4G interaction is critical in vivo (Tarun and Sachs 1997). In vitro studies indicate eIF4G–eIF4E interactions both promote translation of mRNAs bearing a 5′ cap and suppress translation of uncapped mRNAs (Tarun and Sachs 1997; Mitchell et al. 2010).
eIF4G1 has three separate RNA-binding regions [termed RNA1(1–82), RNA2(492–539), and RNA3(883–952)] (Figure 6B) that likely contribute to the enhanced affinity for binding eIF4E to 5′-capped mRNAs (O’Leary et al. 2013), analogous to RNA-binding activities shown to promote mammalian eIF4G–eIF4E interactions (Yanagiya et al. 2009). In yeast, deletion of RNA1 or RNA3 is ts, when removed from the sole isoform of eIF4G, and further RNA motif removal is lethal (Berset et al. 2003). However, singly deleting RNA2 or RNA3 is lethal when combined with the tif4631-459 mutations that impair eIF4E binding (Park et al. 2011). This suggests that there is redundancy in the requirements for eIF4G–eIF4E and eIF4G–mRNA interactions for promoting translation initiation. In addition to the Pab1 binding region (eIF4G188–299), the adjacent RNA1, box 1 and box 2 conserved elements of eIF4G (Figure 6B) also promote binding to Pab1 and mRNA (Park et al. 2011).
The interaction between eIF4G1188–299 and Pab1 is primarily via the second of four RRMs in Pab1 (Figure 6B) (Kessler and Sachs 1998). eIF4G binding to Pab1 provides a further route for RNA recruitment. eIF4G can therefore bridge both the 5′ and 3′ ends of an mRNA as visualized as a “closed loop” (Wells et al. 1998) and supported by in vivo evidence (Preiss and Hentze 1998; Archer et al. 2015).
Closed loop promotes 43S ribosome recruitment:
The 5′ cap and poly(A) tail act synergistically to promote translation. eIF4G bridges these mRNA ends (Figure 1 and Figure 6A) and is implicated in recycling both 40S and 60S ribosome subunits to the start codon following termination. Using in vitro translation experiments and toe-printing techniques, 48S PIC association with the AUG start codon on a short mRNA was resistant to 5′ cap analog (m7GDP) in a manner that was dependent on both the 5′ cap and poly(A) tail as well as intact eIF4G, Pab1, and the termination factors eRF1 and eRF3 (Amrani et al. 2008). Further support for the eIF4E–4G–Pab1–mRNA closed loop comes from the capture of mRNAs bound to each factor from live cells and quantified by RT-PCR or high-throughput sequencing (Archer et al. 2015; Costello et al. 2015). However, it should be noted that the Pab1–eIF4G interaction is dispensable for cell growth unless the eIF4E–eIF4G interaction is also impaired (Tarun et al. 1997; Park et al. 2011). Thus, closed-loop assembly is not essential, but is rate enhancing for translation.
eIF4G–eIF5 interactions promote PIC recruitment:
eIF4E–eIF4G complexes play a role in ensuring that the AUG codon closest to the 5′ end is selected for translation. The 43S PIC is directed to bind an mRNA at the 5′ end. In mammalian cells, eIF4G binds the 43S complex via contacts with the c, d, and e subunits of eIF3 (Korneeva et al. 2000; LeFebvre et al. 2006; Villa et al. 2013). Neither eIF3d, eIF3e, nor the eIF4G domain used by mammals is conserved in yeast. Instead, a central region of eIF4G (eIF4GII residues 439–577) (He et al. 2003) encompassing RNA2 (Figure 6B) can bind to the eIF5 carboxyl terminal domain (CTD) (Asano et al. 2001) with high affinity (<15 nM) (Mitchell et al. 2010). Because the eIF5 CTD binds to both eIF3c/Nip1 and eIF2β (Yamamoto et al. 2005) it can recruit the 43S PIC to eIF4G-bound mRNAs near their 5′ end. It is noteworthy that eIF3, as well as eIF4 factors, are critical for recruitment of natural mRNAs to the PIC in the reconstituted in vitro system (Mitchell et al. 2010) and that depletion of eIF3 subunits in vivo causes a more dramatic impairment of mRNA binding to 40S subunits than does depletion of eIF4G (Jivotovskaya et al. 2006). Thus, in addition to stabilizing TC binding to the 43S PIC, eIF3 plays an important role in mRNA binding. As eIF3 binds directly to eIF5 (Asano et al. 1998; Phan et al. 1998), perhaps these factors cooperate with eIF4G to promote mRNA binding to the PIC.
eIF4A:
A C-terminal segment of eIF4G (eIF4G1542–883) interacts with the DEAD-box RNA helicase eIF4A (encoded by TIF1 and TIF2) (Figure 6B) (Dominguez et al. 1999; Neff and Sachs 1999). The α-helical HEAT repeats in this domain of eIF4G interact with both folded domains of eIF4A to form a stable complex where the ATPase and RNA-binding features of eIF4A are poised for action (Figure 6B) (Schutz et al. 2008). FRET measurements suggest that eIF4G HEAT domain binding to eIF4A shifts eIF4A from an “open” conformation with its two RecA homology domains separated, to one that is partially closed or “half open” as observed in the eIF4G/eIF4A co-crystal structure (Schutz et al. 2008; Andreou and Klostermeier 2014). This structural rearrangement is thought to stimulate eIF4A ATPase activity in the presence of RNA and facilitate PIC recruitment and movement along the mRNA in a 5′ to 3′ direction. Using model RNA templates and purified factors, it was shown that eIF4E/eIF4G both enhances ATPase activity and provides directional bias to eIF4A to unwind 5′ RNA duplexes (Rajagopal et al. 2012). The RNA1, RNA2, and RNA3 domains of eIF4G enhanced both of these activities, consistent with the ability of eIF4G to link different factors together (Figure 6), enhancing successive steps in translation initiation (Rajagopal et al. 2012).
The RNA helicase activity of eIF4A may be more important for creating a single-stranded stretch of mRNA for efficient 43S PIC recruitment to the mRNA 5′ end than for unwinding strong secondary structure elements within many 5′ leader sequences. Support for this idea comes from several genome-wide experiments. First, depletion of eIF4G reduced overall protein synthesis by only 75% and narrowed the range of translational efficiencies genome-wide (Park et al. 2011). The mRNAs most affected were not ones with longer 5′ UTRs, suggesting that eIF4G’s role in 43S PIC recruitment to mRNAs is more critical than its role in promoting scanning on long or structured 5′ UTRs (Park et al. 2011). Second, ribosome profiling of an eIF4A ts mutant found that the translational efficiency of most mRNAs were similarly affected by loss of eIF4A (Sen et al. 2015). As reporter mRNAs with 5′ UTRs of differing lengths and secondary structures were all affected by 30–50% in the eIF4A mutant, it was concluded that eIF4A is globally important for optimal initiation on all mRNAs (Sen et al. 2015).
eIF4B:
eIF4B, encoded by TIF3, enhances eIF4G–eIF4A complex formation and eIF4A helicase activity, and it also stimulates 43S recruitment to mRNA. Studies by Altmann and Trachsel (1989) demonstrated that yeast eIF4B is structurally divergent from its mammalian counterpart, but performs similar functions. TIF3 deletion causes both slow and cold-sensitive growth (Altmann et al. 1993). eIF4G interacts independently with both eIF4B and eIF4A and excess eIF4B suppresses ts mutations in the eIF4G HEAT domain and stabilizes eIF4G–eIF4A interactions, perhaps by altering the conformation of eIF4G in this complex (Park et al. 2013). In vitro measurements show that eIF4B stimulates RNA helicase activity of eIF4A, when eIF4G is also present. The eIF4A–eIF4G–eIF4B complex stimulated ATP hydrolysis and RNA unwinding by >12-fold over eIF4A alone, with eIF4B enhancing eIF4A RNA unwinding activity and eIF4G stimulating ATP hydrolysis (Andreou and Klostermeier 2014). FRET studies adding eIF4B to complexes including ATP, mRNA, and eIF4A/eIF4G promoted a switch from a half-open to a closed eIF4A conformation with juxtaposed RecA domains (Andreou and Klostermeier 2014). These structural transitions in eIF4A promoted by eIF4B and eIF4G are proposed to be important for efficient helicase activity during mRNA scanning (Andreou and Klostermeier 2014; Harms et al. 2014).
eIF4B is also implicated in 43S PIC recruitment. In vitro eIF4B significantly stimulated the recruitment of the 43S PIC to RPL41A and DAD4 mRNAs in the presence of eIF4F (Mitchell et al. 2010). In addition, eIF4B binds directly to eIF3 (Kd = 380 nM) (Mitchell et al. 2010) and interacts with single-stranded RNA (Kd = 2.2 µM) and the head region of the 40S subunit via an interaction with Rps20 (Kd = 360 nM) (Walker et al. 2013). Yeast eIF4B possesses an NTD, an RRM, which binds mRNA and stimulates RNA duplex annealing (Altmann et al. 1993; Niederberger et al. 1998), and a 20- to 26-residue module that is repeated seven times forming the “7-repeats” domain (Figure 6B). The 7-repeats are divergent from mammalian eIF4B, but are conserved among fungi (Zhou et al. 2014). The 7-repeats contribute to mRNA binding, as an allele missing both the NTD and 7-repeats fails to bind the 40S or to stimulate mRNA recruitment to the 43S PIC and impairs translation in vivo, whereas all of these activities are largely intact when only the RRM is missing (Walker et al. 2013). Interaction of eIF4B with Rps20, a protein within the 40S head region, may indicate that eIF4B helps load mRNAs into the 40S entry channel to promote scanning (Zhou et al. 2014). A speculative model for eIF4G domain interactions with multiple eIFs and the mRNA 5′ UTR to recruit the 43S PIC and promote scanning is shown in Figure 6C.
Ded1:
Ded1 is a second DEAD-box RNA helicase implicated in 43S PIC recruitment and scanning during translation initiation. DED1 is essential and is a close homolog of mammalian DDX3. Ded1, like eIF4A, contains two RecA-homology domains, but with a distinct NTD and CTD (Linder and Jankowsky 2011). In addition to translation initiation, Ded1 has roles in pre-mRNA splicing, mRNA export from the nucleus, and mRNA decay. While Ded1 is predominantly cytoplasmic (Chuang et al. 1997), it interacts with both nuclear and cytoplasmic mRNA 5′ cap complexes and moves between the nucleus and cytoplasm, suggesting that Ded1 is a general RNA chaperone (Senissar et al. 2014). Because Ded1 is an active helicase by itself, it is an excellent model for biochemical studies of RNA helicase activities (Iost et al. 1999; Yang and Jankowsky 2005). A recent comprehensive review of Ded1 provides a detailed summary of RNA helicase activities and wider roles of Ded1 and its homologs (Sharma and Jankowsky 2014).
Ded1 binds the RNA3 motif at the C terminus of eIF4G1 (Hilliker et al. 2011). While mutational inactivation of Ded1 inhibits translation initiation (Chuang et al. 1997), its overexpression rescues eIF4E ts alleles (de la Cruz et al. 1997). However, greater overexpression of Ded1 from an inducible GAL1 promoter represses translation, causing sequestration of eIF4E, eIF4G, and Pab1 in cytoplasmic granules (Hilliker et al. 2011). Ded1 can form very stable complexes with RNA in vitro (Liu et al. 2014) and it is important for resolving misfolded RNA structures and preventing higher-order structural contacts that would otherwise destabilize RNA architecture (Pan et al. 2014).
Ribosome profiling of DED1 mutants has shown differential requirements for Ded1 across different mRNAs. Inactivation of a ded1 cold-sensitive mutant impaired global translation rates, and greater than average reductions in translational efficiency were observed for only ∼600 genes (Sen et al. 2015). The Ded1 hyperdependent mRNAs had longer than average 5′ UTR lengths and greater propensity for secondary structure, thereby implicating Ded1 as critical for scanning through structured 5′ UTRs (Sen et al. 2015). Consistent with these findings, reporter mRNAs bearing long or structured 5′ UTRs exhibit heightened dependence on Ded1 (Berthelot et al. 2004; Sen et al. 2015).
AUG selection
Following binding of the 43S PIC near the 5′ end of the mRNA, it traverses in a 3′ direction inspecting for a start codon. Elegant experiments by Donahue and colleagues established that the anticodon of the Met-tRNAiMet in the 43S complex is primarily responsible for start codon selection. Mutation of the tRNAiMet anticodon from 5′-CAU-3′ to 5′-CCU-3′ enabled ribosomes to synthesize His4 when the HIS4 mRNA start codon was mutated from 5′-AUG-3′ to 5′-AGG-3′ (Cigan et al. 1988a). Moreover, insertion of a 5′-AGG-3′ codon in the HIS4 mRNA leader upstream and out-of-frame with the 5′-AGG-3′ codon at the HIS4 start site blocked His4 production (Cigan et al. 1988a). This latter result supports the model that the anticodon of Met-tRNAiMet in the scanning 43S complex inspects the mRNA in a base-by-base manner to select the translation start site.
This importance of codon–anticodon match in start codon selection was further supported by studies examining the kinetics and thermodynamics of 48S PIC formation in reconstituted yeast in vitro translation assays. Point mutations that altered the second or third positions of the AUG start codon on the mRNA dramatically lowered the affinity of Met-tRNAiMet binding in the 48S PIC. This binding defect was suppressed by mutations in the tRNAiMet anticodon that restored base-pairing interactions with the mRNA. As the start codon mutations in the mRNA mainly affected the on rate for Met-tRNAiMet binding, and not the off rate, it was proposed that 48S PIC formation is accompanied by a conformational change that locks in Met-tRNAiMet binding. Accordingly, in this closed state, Met-tRNAiMet is stably bound to the 40S subunit and fixed on the translation start codon of the mRNA (Kolitz et al. 2009).
In addition to the Met-tRNAiMet, translation factors play key roles in the transition of the 40S subunit from its open, scanning-competent state to the closed, scanning-arrested state following start codon selection (reviewed in Hinnebusch 2011, 2014). Genetic screens in yeast have provided key insights into the factors contributing to start codon selection. Spontaneous Sui− mutations that enhance initiation from a UUG codon were isolated in eIF1 (Yoon and Donahue 1992), all three subunits of eIF2 (Donahue et al. 1988; Cigan et al. 1989; Huang et al. 1997), and in eIF5 (encoded by TIF5) (Huang et al. 1997). In subsequent directed screens Sui− mutations have also been isolated in eIF1A (Fekete et al. 2007), eIF3 subunits (Valasek et al. 2004; Chiu et al. 2010; Elantak et al. 2010; Karaskova et al. 2012), and in 18S rRNA (Nemoto et al. 2010). In contrast to the Sui− mutations, which relax the stringency for start codon selection, a second class of mutations enhances start codon selectivity. The Ssu− (suppressor of Sui−) mutations block the ability of Sui− mutations to enhance initiation at a UUG codon in a mutant HIS4 allele (Asano et al. 2001; Fekete et al. 2007; Saini et al. 2010). In general, Sui− mutations are thought to block scanning and promote conversion of the 40S subunit to its closed, scanning arrested conformation (Pin). In contrast, the Ssu− mutations promote scanning and stabilize the open conformation of the 40S subunit (Pout).
Biochemical analyses of the Sui− and Ssu− mutant forms of initiation factors have provided insights into the mechanism of start codon selection. The dominant SUI5-G31R mutation in eIF5 was reported to alter the release of Pi from GTP following its hydrolysis by eIF2 (Saini et al. 2014), and the SUI3-2 (S264Y) mutation in eIF2β and the SUI4 (GCD11-N135K) mutation in eIF2γ were reported to increase the intrinsic (eIF5 independent) GTPase activity of eIF2 (Huang et al. 1997). Thus, it was proposed that premature GTP hydrolysis by eIF2 would enable release of Met-tRNAiMet to the P site in the absence of perfect codon–anticodon base pairing. The eIF5-G31R and eIF2β-S264Y mutations also stabilize 48S PICs at UUG codons in the presence of nonhydrolyzable GTP, indicating these mutations stabilize the closed Pin state of the 48S PIC (Martin-Marcos et al. 2014; Saini et al. 2014). The identification of Sui− mutations in the CTT and Ssu− mutations in the NTT of eIF1A likewise implicate these segments in stabilizing the Pout or Pin states of Met-tRNAiMet binding, respectively (Fekete et al. 2007; Saini et al. 2010).
Sui− mutations in eIF1 have been found to weaken eIF1 binding to the 40S subunit, consistent with their recessive phenotype and with the notion that eIF1 dissociation from the 48S complex is required for start codon selection (Cheung et al. 2007; Martin-Marcos et al. 2013). Moreover, overexpression of eIF1 suppresses Sui− mutations in other translation factors, indicating that eIF1 dissociation from the 48S complex is a key commitment step in start codon selection (Valasek et al. 2004; Cheung et al. 2007; Martin-Marcos et al. 2011; Martin-Marcos et al. 2013; Martin-Marcos et al. 2014). Cryo-EM structures of 48S PICs in open and closed states revealed a clash between eIF1 and Met-tRNAiMet in the Pin state (Llacer et al. 2015). This clash likely underlies the role of eIF1 in blocking initiation at non-AUG codons and evokes eIF1 release on AUG codon recognition (Figure 4, A and B). In addition to these genetic and structural studies, kinetic experiments have highlighted the critical gatekeeper function of eIF1 in regulating start codon selection. Recognition of the AUG start codon by the 48S PIC induces a conformational change that accelerates eIF1 dissociation, which in turn enables release of Pi from eIF2–GDP (Figure 4B) (Algire et al. 2005; Maag et al. 2005). Thus, eIF1 dissociation and the attendant release of Pi, and not simply GTP hydrolysis by eIF2 (Maag et al. 2005), is the irreversible step that commits the ribosome to initiate at the selected codon.
The 405-residue factor eIF5 folds into functionally distinct N- and C-terminal domains (Figure 3B). The N-terminal domain of eIF5 resembles eIF1 and like the C-terminal domain of eIF2β possesses a Zn-finger element (Conte et al. 2006). In contrast, the α-helical C-terminal domain of eIF5 folds into a HEAT repeat with structural similarity to the HEAT domains in the C terminus of eIF2Bε and in eIF4G (Wei et al. 2010). The N-terminal domain of eIF5 directly binds the G domain of eIF2γ (Alone and Dever 2006), and mutation of Arg15 in eIF5 confers a lethal phenotype and significantly impairs the ability of eIF5 to stimulate GTP hydrolysis by eIF2 (Das et al. 2001; Algire et al. 2005), supporting the notion that eIF5 functions as a GTPase activating protein (GAP) for eIF2. In accord with the gatekeeper function of eIF1, it is proposed that eIF1 release following start codon recognition enables eIF5 to move into the vacated space and thereby stimulate Pi release from eIF2–GDP+Pi (Nanda et al. 2009). Consistent with this proposed movement of eIF5 into the space previously occupied by eIF1, and thus closer to eIF1A, the SUI5-G31R mutation in eIF5 was found to strengthen eIF1A interaction with the PIC at a UUG codon (Maag et al. 2006). Moreover, mutations in the eIF1A CTT uncouple Pi release from eIF1 dissociation (Nanda et al. 2013). Thus the eIF5 N-terminal domain appears to be intimately involved in the structural rearrangements in the scanning ribosome upon start codon selection.
The C-terminal HEAT domain in eIF5 binds to eIF1, the NTT (K-boxes) of eIF2β, and to eIF3c/Nip1 (Yamamoto et al. 2005). Mutations in the eIF5 C-terminal domain that disrupted its interaction with both eIF1 and eIF2β conferred an Ssu− phenotype and destabilized the closed state of the 48S complex (Luna et al. 2012). Importantly, this mutation did not affect the ability of eIF5 to promote GTP hydrolysis by eIF2 (Luna et al. 2012), further strengthening the notion that GTP hydrolysis occurs prior to the step controlling start codon selection (Algire et al. 2005; Maag et al. 2005).
Taking into account the results from the various genetic and biochemical studies on the translation factors that participate in start codon selection, a model can be proposed wherein the factors eIF1, eIF1A, eIF3, eIF5, and the eIF2 TC are bound to the 40S subunit as it scans the mRNA (Figure 4B). In this open, scanning-competent complex the Met-tRNAiMet resides in the Pout state with eIF1 bound adjacent to the P site and eIF1A bound in the A site with its N- and C-terminal tails projecting into the P site. Both eIF1 and the eIF1A CTT prevent full accommodation of Met-tRNAiMet into the Pin state. When the scanning complex encounters a start codon, base-pairing interactions between the anticodon of Met-tRNAiMet and the start codon triggers entry of the Met-tRNAiMet to the Pin state. This movement is accompanied by displacement of eIF1 and movement of the eIF1A CTT toward eIF5. These factor movements trigger Pi release from eIF2, a critical commitment step in start codon selection (Algire et al. 2005), and conversion of the 43S PIC to its closed, scanning-arrested state (Hinnebusch 2011, 2014). In accord with this model, recent cryo-EM structures of 48S complexes in the open and closed states have revealed conformational changes in the 40S subunit as well as interaction of the eIF1A NTT with the Met-tRNAiMet–AUG codon duplex, and eIF2, eIF1A, and ribosomal proteins with the mRNA and start codon context nucleotides (Hussain et al. 2014; Llacer et al. 2015). Upon start codon selection, constriction of the mRNA channel and tightening of the P site are thought to block further scanning by the PIC.
Subunit joining
Following eIF1 and Pi release, the 48S PIC is in a closed conformation with Met-tRNAiMet fully accommodated in the P site. Accompanying these changes, eIF1A binding to the 48S complex becomes tighter. It is unclear when the eIF2–GDP complex, eIF5 and eIF3 dissociate from the PIC; however, it is clear that based on its binding site on the intersubunit face of the 40S subunit, eIF2 must dissociate prior to 60S subunit joining.
The factor eIF5B, encoded by FUN12 and an ortholog of the bacterial translation factor IF2, promotes 60S subunit joining (Choi et al. 1998; Pestova et al. 2000). eIF5B is 1002 amino acids in length and contains a GTP-binding domain near the center of the protein. Deletion of FUN12 severely impairs yeast cell growth and causes a loss of polysomes, consistent with a defect in translation initiation (Choi et al. 1998). Removal of the N-terminal ∼400 residues of eIF5B confers no growth defect in vivo and the truncated protein catalyzes subunit joining in vitro (Lee et al. 1999; Shin et al. 2002). The crystal structure of aIF5B, the archaeal ortholog of eIF5B, revealed a chalice-shaped protein with the G domain, domain II, and domain III, forming the cup of the chalice which is connected to the base, domain IV, by a long α-helix (Roll-Mecak et al. 2000; Kuhle and Ficner 2014a). Directed hydroxyl radical mapping studies of eIF5B–80S complexes placed domain II of yeast eIF5B near 18S rRNA helix h5 of the 40S subunit (Shin et al. 2009), consistent with results of a recent cryo-EM structure of eIF5B bound to an 80S initiation complex (Fernandez et al. 2013). This binding site is compatible with the eIF5B G domain binding to the GTPase activation center on the 60S subunit, similar to the binding sites of the bacterial translational GTPases IF2, EF-Tu, and EF-G on 70S ribosomes. Whereas the C-terminal domain of bacterial IF2, which corresponds to domain IV of eIF5B, directly binds the formylmethionine (fMet) on fMet-tRNAiMet, direct binding of eIF5B with Met-tRNAiMet has not been observed. However, based on the ribosomal binding site of eIF5B and the dimensions of aIF5B, domain IV of eIF5B is thought to project across the A site to interact with Met-tRNAiMet in the P site. This proposed contact of eIF5B with Met-tRNAiMet in the P site is consistent with cryo-EM structures of the initiation complex (Fernandez et al. 2013; Kuhle and Ficner 2014b) and with the instability of 48S PICs and decreased recognition of an inhibitory upstream AUG codon (“leaky scanning”) in yeast lacking eIF5B (Lee et al. 2002). It is proposed that eIF5B binding to the closed 48S complex at an AUG codon stabilizes Met-tRNAiMet binding following eIF2–GDP release and promotes 60S subunit joining. In the absence of eIF5B, the Met-tRNAiMet is not stably bound, causing some 48S complexes to dissociate from the mRNA and others to resume scanning to downstream start sites.
Domain IV of eIF5B binds to eIF1A (Choi et al. 2000) via interaction with the last five residues at the C terminus of eIF1A (Olsen et al. 2003; Acker et al. 2006; Fringer et al. 2007). Mutation of the eIF1A C terminus impairs subunit joining and full activation of eIF5B GTPase activity in vitro (Acker et al. 2006) and impairs yeast cell growth and eIF5B binding to 40S complexes in vivo. The growth and translation initiation defects of this eIF1A mutant are suppressed by overexpression of eIF5B, indicating that eIF1A helps recruit eIF5B to 40S complexes prior to subunit joining and thereby accelerates ribosomal subunit joining (Acker et al. 2009).
Both eIF1A and eIF5B are bound to the 80S ribosome following subunit joining, and GTP hydrolysis by eIF5B is required for their release (Shin et al. 2002; Fringer et al. 2007; Acker et al. 2009). Blocking eIF5B GTPase activity, either by inclusion of nonhydrolyzable GTP analogs or by mutation of the eIF5B G domain, does not impair subunit joining (Shin et al. 2002, 2007, 2009; Acker et al. 2009); however, it does impede eIF1A release from 80S ribosomes both in vivo and in vitro (Fringer et al. 2007; Acker et al. 2009). Mutations that disrupt the GTPase activity of eIF5B severely impair yeast cell growth (Shin et al. 2002, 2007, 2009). Suppressor mutations of these eIF5B mutants either restore the factor’s GTPase activity (Shin et al. 2007) or decrease the binding affinity of eIF5B for the 80S ribosome (Shin et al. 2002, 2009), consistent with GTP hydrolysis lowering eIF5B affinity for the ribosome. It is proposed that GTP hydrolysis by eIF5B alters the conformation of the 80S to promote eIF1A release (Acker et al. 2009). As the eIF5B suppressor mutants that bypass the requirement for GTP hydrolysis show enhanced levels of leaky scanning (Shin et al. 2002), GTP hydrolysis by eIF5B might serve as a checkpoint to ensure the fidelity of subunit joining (Shin et al. 2002). Following release of eIF5B and eIF1A, the ribosome is poised with Met-tRNAiMet in the P site and a vacant A site available to receive the first elongator aa-tRNA. It is possible that some initiation factors including eIF3 (Szamecz et al. 2008) remain associated with the ribosome through the first few steps of elongation.
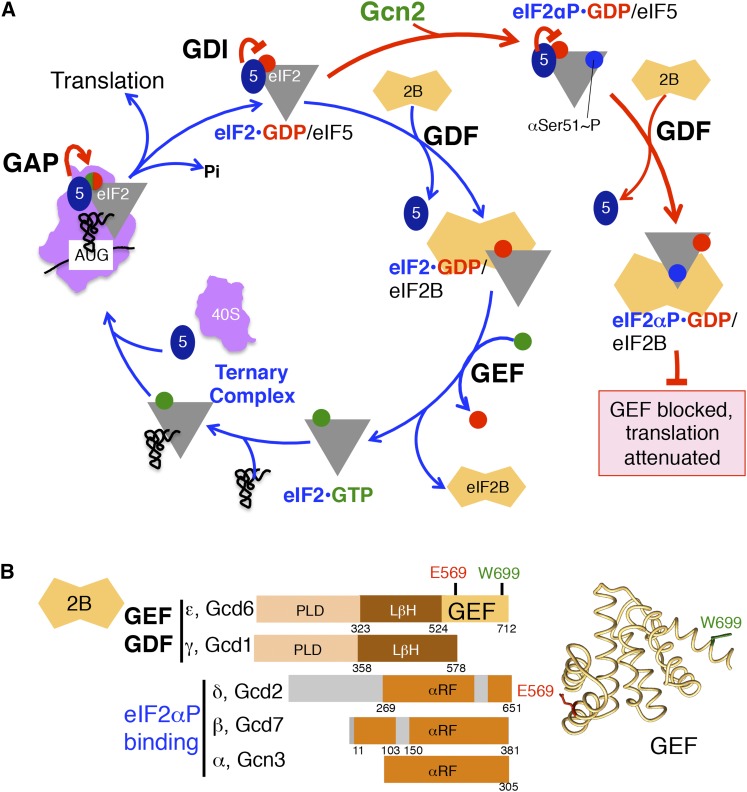
Recycling eIF2–GDP to eIF2–GTP
eIF2–GDP released from the 48S PIC following start codon selection must be converted to an active GTP-bound form to promote Met-tRNAiMet binding and continued rounds of translation initiation (Figure 1 and Figure 7A). This is an important step as phosphorylation of eIF2α converts eIF2 into an inhibitor of its GEF eIF2B, thereby lowering TC levels. eIF2 recycling was thought to be a single reaction involving eIF2B; however, eIF5 antagonizes eIF2B and must be displaced from eIF2 prior to nucleotide exchange (Figure 1 and Figure 7A) (Jennings and Pavitt 2014).
Figure 7.
Recycling and regulation of eIF2 by eIF2B. (A) Pathway of eIF2 nucleotide cycle and its regulation by eIF2α phosphorylation, adapted from Figure 1. GDI function of eIF5, GDF and GEF activities of eIF2B, and GAP function of eIF5 (5) are described in the text. Phosphorylation of eIF2α on Ser51 by GCN2 is represented by the blue circle; GDP, red circle; and GTP, green circle. (B) Schematics of eIF2B subunits and domain organization (left) and structure of the eIF2Bε GEF domain (right, pdb 1PAQ) (Boesen et al. 2004). Homologous domains are shown in identical color shades. PLD and LβH indicate the pyrophosphorylase-like and the left-handed β-helical domains, respectively (Reid et al. 2012). αRF indicates the α-helical domain followed by a Rossmann-like fold shared by the α-, β-, and δ-subunits. Structural models were drawn using Chimera software (UCSF).
GDP dissociation inhibitor function of eIF5:
In addition to its roles in PIC formation, AUG codon recognition and stimulation of eIF2–GTP hydrolysis, eIF5 functions as a GDP dissociation inhibitor (GDI) to prevent unregulated release of GDP from eIF2. eIF5 binds both eIF2–GDP and TC with identical high affinity (eIF2–GDP Kd = 23 ± 9 nM, TC Kd = 23 ± 5 nM) (Algire et al. 2005), and cells contain an abundant fraction of inactive eIF2–GDP/eIF5 complexes that are thought to be released from the 48S PIC following AUG recognition (Singh et al. 2006). eIF5 lowers the rate of spontaneous GDP release from eIF2 over a range of Mg2+ concentrations (Jennings and Pavitt 2010a), and this GDI activity requires the eIF5 CTD and the region linking it to the NTD. Thus, GDI and GAP activities of eIF5 are distinct (Jennings and Pavitt 2010a). Mutation of a conserved tryptophan (W391F) in the CTD, or seven substitutions within a conserved “DWEAR” motif in the linker region (termed L7A) (Figure 3B), eliminates GDI activity (Jennings and Pavitt 2010a,b). Though neither GDI mutation significantly alters growth of yeast on rich or minimal medium, they dramatically impair responses to eIF2 phosphorylation, indicating an important role of eIF5 in tight regulation of eIF2B GEF activity.
eIF2B displaces eIF5 from eIF2–GDP:
Because the CTD of eIF5 and the GEF domain of eIF2Bε (see Figure 3, C and D) share a common HEAT repeat structure (Boesen et al. 2004; Bieniossek et al. 2006) necessary for binding to eIF2β (Asano et al. 1999), binding of each factor to eIF2 is mutually exclusive (Jennings and Pavitt 2010b). eIF5 must dissociate from the stable eIF2–GDP/eIF5 complex to enable eIF2B GEF action; however, eIF2B itself can displace eIF5 (Jennings et al. 2013). In common with other G protein regulator nomenclature, eIF2B is a GDI displacement factor (GDF) (Figure 7A).
eIF2B consists of five subunits α–ε (encoded by GCN3, GCD7, GCD1, GCD2, and GCD6, respectively; Table 1) in equimolar stoichiometry (Figure 7B) (Cigan et al. 1993; Kito et al. 2007). The subunits are subdivided into two functionally and structurally distinct αβδ and γε subcomplexes (Figure 7B). The eIF2B GEF domain is within the γε subcomplex, lodged at the eIF2Bε C terminus (Pavitt et al. 1998; Gomez and Pavitt 2000; Boesen et al. 2004). This subcomplex is as effective as intact eIF2B for eIF5 displacement (GDF) activity; however, neither γ nor ε alone have GDF activity, suggesting that these subunits cooperate in GDF function (Jennings et al. 2013). Importantly, missense mutations in eIF2Bγ (gcd1-G12V and gcd1-L480Q) specifically impair GDF function (release of eIF5 from eIF2–GDP) and only affect nucleotide exchange function when eIF2–GDP is prebound by eIF5 (Jennings et al. 2013). As the gcd1 mutations confer slow-growth and Gcd− phenotypes, the GDF activity of eIF2B is critical in vivo.
eIF2B catalysis of guanine nucleotide exchange:
A long-established function of eIF2B is to recycle inactive eIF2–GDP complexes to functional eIF2–GTP complexes (Figure 7A) (Cigan et al. 1993). Similar to GEFs for other G proteins, eIF2B likely catalyzes exchange by decreasing the binding affinity of eIF2 for GDP (Sprang and Coleman 1998). Given the greater abundance of GTP vs. GDP in growing cells (Rudoni et al. 2001), release of GDP from eIF2 is likely sufficient to allow recharging of eIF2 with GTP. Yeast eIF2B was discovered through studies examining the translational control of the GCN4 mRNA. Mutations in the eIF2B subunits cause Gcd− phenotypes (Harashima and Hinnebusch 1986) and also confer slow-growth and reduced rates of translation initiation (Hannig et al. 1990; Foiani et al. 1991; Bushman et al. 1993a).
Yeast eIF2B promotes release of GDP from eIF2 (Vmax = 250.7 fmol of GDP released per minute, at 0°) at rates similar to values reported for mammalian eIF2B (Nika et al. 2000). eIF2Bε alone is ∼5- to 10-fold less effective than intact eIF2B at promoting nucleotide exchange, while eIF2Bγε subcomplexes have the same activity as intact eIF2B (Gomez and Pavitt 2000; Jennings et al. 2013). The GEF domain comprises the C-terminal ∼200 residues (eIF2Bε518–712) (Gomez and Pavitt 2000; Gomez et al. 2002) and adopts a HEAT repeats structure with conserved residues important for GEF activity on one face (Boesen et al. 2004). Conserved residue E569 is critical for GEF function (Boesen et al. 2004) and cell viability (Mohammad-Qureshi et al. 2007), and residue W699 is important for binding to eIF2β and γ (Figure 7D) (Mohammad-Qureshi et al. 2007).
Regulation of eIF2B activity:
eIF2α is phosphorylated at Ser51 by Gcn2 (Dever et al. 1992) causing inhibition of eIF2B GEF activity (Pavitt et al. 1998; Jennings et al. 2013). Genetic and biochemical experiments implicate the homologous eIF2Bα, β, and δ subunits in mediating translational control. Multiple missense mutations in each of these subunits impair the regulatory response to phosphorylated eIF2 (Vazquez de Aldana and Hinnebusch 1994; Pavitt et al. 1997) and deletion of GCN3, encoding nonessential eIF2Bα, blocks the Gcn2-dependent induction of GCN4 expression (Hannig and Hinnebusch 1988). The mutations cluster in two regions of sequence similarity shared by the subunits (Pavitt et al. 1997), and based on the structure of human eIF2Bα, the mutations are largely distributed across one surface of each subunit (Hiyama et al. 2009). Each subunit appears to independently contribute to the regulatory mechanism, rather than being redundant. Mutations in eIF2Bβ weaken interactions with phosphorylated eIF2α (Pavitt et al. 1997; Krishnamoorthy et al. 2001), and direct interactions between eIF2α and eIF2B are supported by allele-specific genetic interactions between eIF2α and eIF2Bβ (Dev et al. 2010) and by lysine-specific cross-links between eIF2α and eIF2Bδ (Gordiyenko et al. 2014). The current model for inhibition of eIF2B activity proposes that phosphorylation causes tight binding of eIF2α to an eIF2Bαβδ surface such that eIF2Bε can no longer bind productively to eIF2γ to promote nucleotide exchange. In addition, binding of one molecule of eIF2 to eIF2Bαβδ likely prevents simultaneous interaction of a second eIF2 molecule with the eIF2Bγε subunits (Pavitt et al. 1998; Krishnamoorthy et al. 2001; Jennings et al. 2013).
Recent evidence reveals that eIF2B is a dimer of pentamers and thus a decamer of 590 kDa bearing two copies of each subunit (Gordiyenko et al. 2014). The precise arrangement of these subunits is not yet clear. eIF2Bγ and eIF2Bε both share significant sequence homology with apparently functionally unrelated enzymes containing a pyrophosphorylase-like domain (PLD) and a left-handed β-helix (LβH) that are important for intersubunit interactions (Figure 7B) (Koonin 1995; Reid et al. 2012; Gordiyenko et al. 2014). The remaining three subunits, eIF2Bα, eIF2Bβ, and eIF2Bδ, also share sequence/structural similarity with each other and with closely related protein families in Archaea: ribose-1,5-bisphosphate isomerases, methylthioribose-1-phosphate isomerases, and a final group that are proposed archaeal eIF2B homologs (Dev et al. 2009). All of these proteins share a α-helical domain followed by a Rossmann-like fold (αRF) (Figure 7B). Lysine-specific cross-links indicate a strong network of connections among all the essential eIF2Bβ/γ/δ/ε subunits (Gordiyenko et al. 2014). Genetic observations suggest interactions between eIF2Bβ and eIF2Bγ (Dev et al. 2010) and between eIF2Bα and both eIF2Bγ and eIF2Bε (Bushman et al. 1993b). Although the complete eIF2B structure is not yet available, a recent structural study of eIF2Bβδ from the thermophilic filamentous fungus Chaetomium thermophilum (Kuhle et al. 2015) and modeling and biochemical studies of mammalian eIF2B subunits (Bogorad et al. 2014) suggest that the eIF2Bαβδ may form the dimer core of eIF2B.
A significant proportion of eIF2B forms higher order structures in cells referred to as a large “body” or “filament” (Campbell et al. 2005; Noree et al. 2010) that can diffuse through the cytoplasm (Taylor et al. 2010). In the absence of stress, GFP fusions of most translation factors exhibit a diffuse cytoplasmic localization, whereas eIF2B is present in both diffuse and localized forms (Campbell et al. 2005). Fluorescence recovery after photobleaching experiments showed that eIF2B is resident in the body, while eIF2 shuttles through it. eIF2 movement is reduced by translation inhibitors, eIF2 phosphorylation, or eIF2B mutation, suggesting that the eIF2B body is a major site of eIF2B function and may contribute to translational control (Campbell et al. 2005).
Mechanism of Translation Elongation, Termination, and Recycling
Relative to bacterial systems, many mechanistic and structural aspects of yeast elongation are well conserved. The elongation factors eEF1A and eEF2 in yeast (Table 2) are structural and functional homologs of the bacterial factors EF-Tu and EF-G, respectively, and the basic pathway of elongation is also conserved (Figure 8). An eEF1A–GTP–aa-tRNA TC binds to the A site of the ribosome. Codon recognition by the tRNA triggers GTP hydrolysis and release of eEF1A–GDP, which allows the aa-tRNA to be accommodated in the A site. The ribosomal peptidyl transferase center (PTC) positions the aa-tRNA in the A site and the peptidyl-tRNA in the P site to allow rapid peptide bond formation. Ratcheting of the ribosome following peptide bond formation moves the tRNAs into hybrid P/E and A/P states with the acceptor ends of the tRNAs in the E and P sites and the anticodon loops remaining in the P and A sites, respectively. Binding and GTP hydrolysis by eEF2–GTP promotes translocation of the tRNA anticodon loops into the E and P sites, respectively. The deacylated tRNA is released from the E site and the next eEF1A–GTP–aa-tRNA binds to the A site in a codon-dependent manner. The cycle continues until a nonsense codon is reached. Recycling of eEF1A–GDP to eEF1A–GTP between each cycle requires the GEF eEF1B. Distinctive features in yeast include the subunit composition of the GEF and the mode of interaction of its catalytic subunit with eEF1A, unique and functionally important post-translational modifications on several elongation factors, and most prominently, the requirement for the essential eukaryotic elongation factor 3 (eEF3). A comprehensive review of the structures of the yeast translation elongation factors and of many mutants of these factors was previously published (Taylor et al. 2007a). As described below, molecular analyses of translation elongation factors have provided additional insights into the accuracy of translation elongation (Valente and Kinzy 2003) and helped elucidate the function of post-translational modifications of elongation factors (Greganova et al. 2011). These genetic studies have been complemented by structural studies of the yeast elongation factors eEF1A and eEF1Bα (Andersen et al. 2000), eEF1Bγ (Jeppesen et al. 2003), eEF2 (Jørgensen et al. 2003), and eEF3 (Andersen et al. 2006). As the kinetic mechanisms of translation elongation have been extensively studied in bacteria (Wintermeyer et al. 2004), and also recently reviewed for eukaryotes (Rodnina and Wintermeyer 2009; Dever and Green 2012), this section will focus on insights obtained using the yeast system.
Table 2. Translation elongation and termination factors.
| Factor | Gene | Systematic name | Length (AA) |
|---|---|---|---|
| eEF1A (EF1α) | TEF1 | YPR080w | 458 |
| TEF2 | YBR118w | 458 | |
| eEF1Bα (EF1β) | EFB1/TEF5 | YAL003w | 206 |
| eEF1Bγ (EF1γ) | TEF4 | YKL081w | 412 |
| CAM1 | YPL048w | 415 | |
| eEF2 | EFT1 | YOR133w | 842 |
| EFT2 | YDR385w | 842 | |
| eEF3 | YEF3/TEF3 | YLR249w | 1044 |
| HEF3 | YNL014w | 1044 | |
| eIF5A | HYP2 | YEL034w | 157 |
| ANB1 | YJR047c | 157 | |
| eRF1 | SUP45 | YBR143c | 437 |
| eRF3 | SUP35 | YDR172w | 685 |
| Rli1 (ABCE1) | RLI1 | YDR091c | 608 |
Figure 8.
Model of yeast translation elongation. Starting at the top, an elongating ribosome contains a peptidyl-tRNA in the P site and a deacylated tRNA in the E site. eEF1A(1)–GTP (green circle) binds aa-tRNA for delivery to the cognate codon in the A site. The codon–anticodon match in the A site triggers conformational changes in eEF1A, GTP hydrolysis, and release of eEF1A–GDP (red circle), leaving the aa-tRNA in the A site. Guanine nucleotide exchange on eEF1A is catalyzed by the α-subunit of the eEF1Bαγ complex. Following ribosome-catalyzed formation of the peptide bind, the ribosomal translocase eEF2 (2)–GTP stimulates movement of the A-site peptidyl-tRNA to the P site and of the now deacylated tRNA in the P site to the E site. The fungal-specific and essential factor eEF3 (3) interacts with eEF1A and is proposed to assist in the release of the E-site tRNA to allow continued cycles of elongation.
aa-tRNA delivery by eEF1
The eEF1A–eEF1Bαγ complex in yeast, like the analogous EF-Tu–EF-Ts complex of bacteria, provides both aa-tRNA delivery (eEF1A) and GEF (eEF1Bαγ) functions. Consistent with their strong sequence and structural similarity, the canonical translation functions and kinetic mechanisms of G proteins eEF1A and EF-Tu are very similar. In addition, studies in yeast have illuminated roles for elongation factors outside translation elongation.
eEF1A:
Mutations in TEF2 (Sandbaken and Culbertson 1988), one of two genes encoding eEF1A (Table 2), and altered levels of eEF1A (Song et al. 1989) affect translation fidelity. Dominant TEF2 mutants selected to suppress nonprogrammed +1 ribosomal frameshifting (Sandbaken and Culbertson 1988) were found to have differential effects on +1 and −1 frameshifting and missense suppression when present as the only form of eEF1A in the cell (Dinman and Kinzy 1997; Plant et al. 2007). An N153T mutation in the guanine-specificity element of the GTP-binding domain increased the intrinsic GTPase activity of eEF1A, enhanced amino acid misincorporation rates in vitro (Cavallius and Merrick 1998), and promoted nonsense suppression while generally not affecting missense suppression in vivo (Carr-Schmid et al. 1999a). In support of the notion that eEF1A–GTP–aa-tRNA ternary complexes and termination factors compete for binding to the ribosomal A site, deleting the TEF2 gene to lower eEF1A levels decreased nonsense suppression in vivo (Song et al. 1989). Overall, the studies on eEF1A reveal a diversity of effects on A-site events when eEF1A activity is altered.
eEF1A is subject to multiple post-translational modifications, many of which occur across eukaryotes. In yeast, these include di- and trimethyl lysine, but do not include phosphoglycerol ethanolamine as observed in the mammalian factor (Cavallius et al. 1993). The functional role of these modifications, however, remains unclear as mutation of the methylation sites does not affect cell growth or general protein synthesis (Cavallius et al. 1997). The C-terminal lysine of yeast eEF1A is methyl esterified; however, the role of this modification remains unclear (Zobel-Thropp et al. 2000).
eEF1Bαγ and guanine nucleotide exchange:
Following delivery of aa-tRNA to the ribosomal A site, GTP hydrolysis by eEF1A results in release of an eEF1A–GDP complex. The eEF1B complex that catalyzes guanine nucleotide exchange on eEF1A is more complex than the larger, but functionally homologous, single polypeptide EF-Ts protein, the GEF for EF-Tu in bacteria. While both eEF1B and EF-Ts catalyze the release of GDP from the G protein, eEF1B has two subunits with the essential exchange function lodged in eEF1Bα. Unlike the two subunits in yeast eEF1B, other eukaryotic eEF1B is composed of three subunits, including the additional polypeptides eEF1Bβ in mammals or eEF1Bδ in plants. Whereas the critical function of eEF1Bα is dispensable when eEF1A is overexpressed, mutations impairing eEF1Bα alter the efficiency of nonsense and missense suppression in yeast (Carr-Schmid et al. 1999b; Plant et al. 2007). The eEF1Bα mutations likely result in a decreased pool of active eEF1A–GTP complexes, which in turn enables termination factors to effectively compete with eEF1A ternary complexes at stop codons, leading to decreased nonsense suppression. Despite their similar functions EF-Ts and eEF1Bα interact with their G-protein substrates in distinct manners. Whereas EF-Ts binds to domains I and III of EF-Tu (Kawashima et al. 1996), eEF1Bα interacts with domains I and II of eEF1A (Andersen et al. 2000, 2001). This different mode of binding may reflect an evolutionary difference in the cofactors of eEF1A, which include a conserved interaction with actin across eukaryotic species and, in yeast, an association with eEF3 (Anand et al. 2003, 2006). Notably, eEF1Bα competes with actin for binding to eEF1A (Pittman et al. 2009).
eEF2 and ribosomal translocation
eEF2 function:
The role of G protein eEF2 is functionally comparable to bacterial translocase EF-G: translocation of the mRNA and peptidyl-tRNA from the A site to the P site of the ribosome. Structurally, bacterial EF-G and S. cerevisiae eEF2 are highly conserved. Genetic analysis of yeast eEF2 has demonstrated the key role of the tip of domain IV of eEF2 that enters the ribosomal A site (Taylor et al. 2007b). Much analysis has focused on His699, the modified residue (see below) at the tip. Viable mutants that alter His699 or other residues within the tip of domain IV cause modest defects in translational fidelity, including increased programmed −1 frameshifting (Ortiz et al. 2006). Analysis of the site of binding of Sordarin, a natural product inhibitor of eEF2, via mutational and structural analyses, has demonstrated that the compound binds between domains I, III, and V of eEF2, matching the sites of Sordarin-resistant mutations (Justice et al. 1998; Soe et al. 2007). These findings highlight the movements of domains I and II relative to III, IV, and V during translocation. Interestingly, a P596H mutation in human eEF2 has been linked to the neurodegenerative disorder, dominant spinocerebellar ataxia. The yeast equivalent, eEF2-P580H, exhibits an increased rate of −1 programmed ribosomal frameshifting (Hekman et al. 2012). It is noteworthy that cells carefully maintain the levels of eEF2. Any mutations that inactivate eEF2, yet produce a stable protein, cause a dominant-negative phenotype, apparently due to reduced levels of active WT eEF2 in the cell (Ortiz and Kinzy 2005), suggesting negative autoregulation of eEF2 levels.
Post-translational modifications of eEF2:
Conserved His699 in eEF2 is post-translationally modified to diphthamide. Named for the source of the bacterial toxin that ADP-ribosylates the residue upon infection with Corynebacterium diphtheriae, studies in yeast have been instrumental in identifying and characterizing the seven gene products (DPH1-7) required for synthesis of diphthamide (Schaffrath et al. 2014). Interestingly, neither His699 (Kimata and Kohno 1994) nor any of the DPH genes except KTI11, which is also a subunit of the tRNA-modification complex Elongator, is essential in vivo. eEF2 is also subject to methylation. The enzyme Efm3, encoded by YJR129c and related to human FAM86A, trimethylates eEF2 on Lys509, causing an increase in ribosomal frameshifting and increased sensitivity to antibiotics including sordarin (Davydova et al. 2014; Dzialo et al. 2014). Efm2 also dimethylates Lys613 (Couttas et al. 2012). In addition to these methylations, Thr557 is phosphorylated by the kinase Rck2 (Teige et al. 2001); however, this residue is not essential for eEF2 activity in vivo (Bartish et al. 2007).
The unique elongation factor eEF3
Proposed role in promoting tRNA release from the E site:
The translation elongation factor 3 (eEF3) was first described in the 1970s as a novel activity required for protein synthesis in yeast (Skogerson and Wakatama 1976). Further analysis of eEF3 and its gene YEF3 showed that eEF3 is essential for fungal translation and cell viability, yet is not found in mammals (Dasmahapatra and Chakraburtty 1981; Hutchinson et al. 1984; Sandbaken et al. 1990). Biochemical studies suggested that eEF3 links the A- and E-site activities of the ribosome by facilitating release of deacylated tRNA from the E site, which enables eEF1A delivery of aa-tRNA to the A site (Triana-Alonso et al. 1995). This function of eEF3 is based, in part, on the proposed allosteric model of aa-tRNA binding to the three binding sites on the ribosome. However, as the role of the ribosomal E site and the allosteric model of elongation are not fully resolved (Petropoulos and Green 2012), the critical role for eEF3 in translation elongation remains unclear. Despite this uncertainty in eEF3 function, both genetic and biochemical studies have revealed physical links between eEF1A and eEF3 that are important for protein synthesis (Anand et al. 2003).
eEF3 binding site on ribosome:
The association of eEF3 with the ribosome is of fundamental interest because the requirement for eEF3 is defined by the source of ribosomes. Using eEF1A and eEF2 from yeast or mammals, the requirement for eEF3 is observed with yeast, but not with mammalian, ribosomes (Skogerson and Engelhardt 1977). The availability of both a high-resolution crystal structure of most of eEF3 and a cryo-EM reconstruction of eEF3 on the yeast ribosome revealed that eEF3 binds near the ribosomal E site (Andersen et al. 2006). eEF3 contains an N-terminal HEAT repeat domain as well as two ABC-type ATPase domains with a chromodomain-like insertion (residues 761–869) in the second ATP-binding domain. The chromodomain interacts through multiple sites with the ribosome. While mutation of ribosome-contacting residues in the chromodomain did not affect ribosome binding, they did impair cell growth, elongation rate, and ribosome-stimulated ATPase activity (Sasikumar and Kinzy 2014). Interestingly, many of the eEF3 mutations isolated in screens based on different eEF3 functions cause inhibition of the ATPase activity, indicating the central role of this activity to eEF3 function on the ribosome.
eEF3 related proteins in yeast and other eukaryotes:
While most work on eEF3 has focused on the YEF3 product, the form of the protein expressed under laboratory conditions, yeast HEF3 encodes a protein 84% identical to eEF3 (Maurice et al. 1998; Sarthy et al. 1998). While HEF3 cannot complement the lack of YEF3 when expressed from its own promoter, it can when expressed from the YEF3 promoter. As the HEF3 promoter is active under conditions of zinc deficiency, the inability of HEF3 to substitute for YEF3 may simply reflect the lack of expression of HEF3 under normal growth conditions. The role of the HEF3-encoded form of eEF3 remains unknown.
Traditionally, eEF3 has been viewed as a “fungal-specific” factor, since it was not found in mammals or plants. A partial eEF3-like protein has been described in Chlorella virus CVK2 (Yamada et al. 1993), and genes very similar to YEF3 were recently found in many single cell eukaryotes. Consistent with these findings, 2D gel electrophoresis and MS analysis identified a protein with sequence similarity to eEF3 in Phytophthora infestans (Ebstrup et al. 2005). While none of these eEF3-like proteins have been shown to function like eEF3, the identification of these apparent eEF3 orthologs in other eukaryotes raises questions as to the functional distribution and evolution of this essential yeast protein.
eIF5A promotion of peptide bond formation
The translation factor eIF5A was originally identified as an initiation factor based on its ability to promote methionyl–puromycin synthesis (Kemper et al. 1976; Schreier et al. 1977; Benne et al. 1978; Benne and Hershey 1978), an in vitro assay designed to monitor first peptide bond synthesis. However, this assay also monitors the peptidyl-transferase activity of the ribosome. Yeast eIF5A is encoded by TIF51A (HYP2) and TIF51B (ANB1). Whereas TIF51A is expressed in aerobically grown yeast, TIF51B expression is restricted to anaerobic conditions (Lowry et al. 1983; Schnier et al. 1991). It is not clear why yeast differentially express the two forms of eIF5A that differ at only 15 of their 157 residues.
eIF5A is post-translationally modified by the conversion of a lysine residue to hypusine (Wolff et al. 2007; Dever et al. 2014). An n-butylamine group is transferred from spermidine to the ε-amino group of Lys51 in eIF5A to generate deoxyhypusine. This reaction is catalyzed by deoxyhypusine synthase, encoded by the essential DYS1 gene (Kang et al. 1995; Sasaki et al. 1996; Park et al. 1998). In a second step, the enzyme deoxyhypusine hydroxylase, encoded by nonessential LIA1, hydroxylates the 2-position of the added moiety to generate hypusine (Park et al. 2006a).
Yeast eIF5A was found to interact with the translational machinery (Jao and Chen 2006; Zanelli et al. 2006; Saini et al. 2009), and depletion of eIF5A impaired protein synthesis and yeast cell growth (Kang and Hershey 1994; Saini et al. 2009; Henderson and Hershey 2011). Analysis of polyribosome profiles of yeast depleted of eIF5A or following inactivation of a temperature-sensitive eIF5A mutant revealed retention of polysomes in the absence of cycloheximide (CHX), indicating a defect in translation elongation (Gregio et al. 2009; Saini et al. 2009). However, it has also been reported that depletion of eIF5A causes a loss of polysomes, suggesting that eIF5A may have a function in translation initiation or perhaps a specialized function in translation elongation (Henderson and Hershey 2011). Consistent with the translation elongation defect observed in the eIF5A temperature-sensitive mutant, purified eIF5A stimulated tripeptide synthesis in reconstituted yeast in vitro translation assays (Saini et al. 2009). An eIF5A function in elongation is consistent with identification of an eIF5A mutant as a suppressor of nonsense-mediated mRNA decay (Zuk and Jacobson 1998), which depends on translation elongation (Peltz et al. 1992; Beelman and Parker 1994).
A prokaryotic ortholog of eIF5A termed EF-P was recently shown to stimulate translation of polyproline (Doerfel et al. 2013; Hersch et al. 2013; Ude et al. 2013), and a similar function was identified for yeast eIF5A (Gutierrez et al. 2013). Partial inactivation of a temperature-sensitive eIF5A variant confers reduced expression of reporter genes or authentic yeast ORFs containing homopolyproline stretches (Gutierrez et al. 2013), leading to defects in fertility and polarized cell growth (Li et al. 2014). The requirement for eIF5A for synthesis of polyproline sequences was also observed in vitro and shown to be dependent on the hypusine modification in eIF5A. Directed hydroxyl radical probing experiments revealed eIF5A binds near the ribosomal E site with the hypusine residue in the vicinity of the acceptor stem of the P-site tRNA and the peptidyl-transferase center of the ribosome. Thus, eIF5A is proposed to insert its hypusine residue into the active site of the ribosome and promote peptide bond formation especially for poor substrates like polyproline (Gutierrez et al. 2013).
Ribosomal frameshifting
The yeast system has proven a valuable genetic tool for dissecting the elongation pathway and in particular the fidelity of protein synthesis in vivo. The stoichiometry of structural and catalytic proteins produced by Ty retrotransposons of yeast is determined by a “programmed” +1 translational frameshift. For both Ty1 and Ty3 retrotransposons, a +1 ribosomal frameshifting event establishes the levels of Gag vs. Gag–Pol fusion proteins. While the precise mechanism of ribosomal frameshifting differs for Ty1 and Ty3, in both cases frameshifting is triggered by a codon that is decoded by a rare tRNA (Belcourt and Farabaugh 1990; Farabaugh et al. 1993). The L-A double-stranded RNA virus of yeast likewise encodes both Gag and Pol proteins with the catalytic Pol synthesized as a Gag–Pol fusion by −1 ribosomal frameshifting (Dinman et al. 1991). Mutations predicted to impair recycling of eEF1A–GDP to eEF1A–GTP and thus slow A-site tRNA binding enhance +1 ribosomal frameshifting that occurs on the P site, whereas the fungal eEF2-specific antibiotic sordarin as well as mutations that impair eEF2 function specifically inhibit +1 frameshifting (Dinman and Kinzy 1997; Harger et al. 2001, 2002). In contrast, mutations thought to impair GTP hydrolysis by eEF1A increase the probability of −1 frameshifting (Dinman and Kinzy 1997; Harger et al. 2001). These results are consistent with the notion that +1 frameshifting occurs by miscoding in the P site, while −1 frameshifting occurs on ribosomes in which both the A and P sites are occupied by tRNAs (Harger et al. 2002).
Ribosomal frameshifting is also used to control the production of cellular proteins in yeast. Polyamine-regulated +1 ribosomal frameshifting on the OAZ1 mRNA governs the synthesis of antizyme, a regulator of polyamine synthesis in cells (Palanimurugan et al. 2004; Kurian et al. 2011), and +1 frameshifting is also used to synthesize Est3, a subunit of telomerase (Morris and Lundblad 1997; Taliaferro and Farabaugh 2007), and Abp140 (Trm140), a tRNA methyltransferase (Asakura et al. 1998; D’Silva et al. 2011; Noma et al. 2011). Whereas +1 frameshifting can be triggered when the A site contains a stop codon or a sense codon decoded by a rare tRNA, −1 frameshifting relies on a slippery sequence that allows repairing of the A- and P-site tRNAs with the mRNA and on a downstream secondary structure that impedes forward movement of the ribosome. Computational approaches to identify slippery sites and potential downstream pseudoknot structures predict that as many as 10% of yeast genes, including several genes controlling telomere maintenance (Table 3, Advani et al. 2013), contain a frameshift signal (Jacobs et al. 2007; Belew et al. 2008) and that frameshifting on these sites impacts mRNA levels by activating the nonsense-mediated decay pathway (Belew et al. 2011).
Table 3. Selected examples of translationally controlled yeast mRNAs.
| mRNA | Mechanism of control | References |
|---|---|---|
| Initiation control | ||
| ASH1 | RNA structure elements in 3′ UTR and coding region repress translation of unlocalized mRNA | Chartrand et al. 2002; Irie et al. 2002; Olivier et al. 2005; Paquin et al. 2007; Deng et al. 2008 |
| BOI1, FLO8, GIC1, MSN1, NCE102, PAB1, TIF4632, YMR181c | eIF4G dependent, cap-independent element in 5′ UTR directs translation | Gilbert et al. 2007 |
| CLN3 | Contains uORF, expression affected by eIF4E activity | Polymenis and Schmidt 1997 |
| ENO1, FBA1, TPI1 | Translation induced by amino acid starvation, 5′ UTR dependent, A-rich region implicated for ENO1, TPI1 | Rachfall et al. 2011 |
| ERS1, STE12 | Caf20 dependent translational repression | Castelli et al. 2015; Park et al. 2006b |
| FOL1, MKK1, RPC11, TPK1, WSC3 | uORF represses translation | Zhang and Dietrich 2005 |
| GCN4 | Four uORFs regulate translation in response to amino acid levels via eIF2 phosphorylation | Hinnebusch 2005 |
| GRS1, ALA1 | Non-AUG initiation produces mitochondrial tRNA synthetase, leaky scanning to downstream in-frame AUG produces cytoplasmic enzyme | Chang and Wang 2004; Tang et al. 2004; Chen et al. 2008 |
| HAC1 | Long-range base pairing between intron and 5′ UTR sequences represses translation | Ruegsegger et al. 2001; Sathe et al. 2015 |
| INO2 | uORF represses protein expression in presence of inositol and choline | Eiznhamer et al. 2001 |
| MOD5 | Leaky scanning of first AUG directs tRNA modifying enzyme Mod5 to cytoplasm instead of mitochondria | Slusher et al. 1991 |
| POM34 | Translationally repressed by a complex involving Eap1, Asc1, Scp160, and Smy2 | Sezen et al. 2009 |
| SUI1 | Poor start codon context enables autoregulation of eIF1 levels | Martin-Marcos et al. 2011 |
| URE2 | 5′ cap-independent translation | Komar et al. 2003; Reineke and Merrick 2009 |
| YAP1 | uORF represses translation via a leaky scanning mechanism | Vilela et al. 1998; Zhou et al. 2001 |
| Elongation control | ||
| CPA1 | uORF stalls ribosomes and induces mRNA decay; regulated by arginine levels | Gaba et al. 2001; Gaba et al. 2005 |
| OAZ1 | Polyamine-regulated ribosome +1 frameshifting | Kurian et al. 2011 |
| TYA-TYB | Ribosome +1 frameshifting to produce Gag–Pol fusion protein | Belcourt and Farabaugh 1990; Farabaugh et al. 1993; Harger et al. 2001 |
| L-A | Ribosome −1 frameshifting to produce Gag–Pol fusion protein | Dinman et al. 1991 |
| EST3, TRM140 | Ribosome +1 frameshifting | Morris and Lundblad 1997; Asakura et al. 1998; Taliaferro and Farabaugh 2007; D’Silva et al. 2011; Noma et al. 2011 |
| EST1, EST2, STN1, CDC13 | Ribosome −1 frameshifting | Advani et al. 2013 |
| Termination control | ||
| BSC1–BSC6, IMP3, ZDS1, PDE2 | 3–25% read through of stop codon to create an extended protein | Namy et al. 2001; Namy et al. 2002; Namy et al. 2003; Beznoskova et al. 2015 |
Genes are grouped by publication/similar mechanism. Many examples given rely on a single publication and detailed mechanisms remain unknown.
Actin bundling and nontranslation functions of eEF1A
eEF1A has activities outside its canonical role in translation elongation, including functions in several steps of viral life cycles, apoptosis, actin bundling, and others in metazoans (Mateyak and Kinzy 2010). The interaction of eEF1A with actin was first identified in the slime mold Dictyostelium discoideum (Yang et al. 1990). Using a combination of genetic and/or biochemical approaches, this ability of eEF1A to bind actin and promote actin bundling was demonstrated to be conserved in both budding (Munshi et al. 2001) and fission yeast (Suda et al. 1999). Subsequent genetic studies in S. cerevisiae enabled an analysis of the functional interaction between these two highly abundant proteins in vivo. Utilizing both overexpression and mutational analysis, the actin-bundling activity of eEF1A was found to reside in multiple regions of the protein, in particular the N terminus and the C-terminal domain. Overexpression of eEF1A results in altered cell size, disorganization of the actin cytoskeleton, and accumulation of cells in the G1 phase of the cell cycle (Munshi et al. 2001), while a genetic screen designed to capitalize on these phenotypes identified a series of mutations in eEF1A that when coupled with an altered C terminus result in similar cellular effects (Gross and Kinzy 2005, 2007). The eEF1A mutant proteins, in particular the F309L and S405P mutants, reduced actin bundling in vitro, supporting the original biochemical data from the Condeelis laboratory (Yang et al. 1990), and caused defects in the cell cytoskeleton and morphology, revealing a critical biological role for this interaction. Most surprisingly, these eEF1A mutants showed translation defects at the level of initiation and increased phosphorylation of eIF2α (Perez and Kinzy 2014). Deletion of the eIF2 kinase GCN2 eliminated the initiation defect, but revealed an elongation defect. Taken together, these studies reveal linkages between the cellular cytoskeleton and translation and are consistent with early work highlighting this important interaction (Howe and Hershey 1984) .
Termination and recycling
Termination and ribosome recycling are linked processes critical to release the completed polypeptide and to provide a pool of 40S and 60S subunits for additional rounds of translation. Termination begins with recognition of one of the 3 stop codons in the A site by the release factor eRF1 (Sup45), which binds to the ribosome together with the GTP-bound form of factor eRF3 (Sup35). The eRF1, composed of three domains, functionally mimics a tRNA with the N-terminal domain recognizing the stop codon (Bertram et al. 2000), the central domain with its methylated GGQ motif promotes hydrolysis of the peptidyl-tRNA bond (Heurgue-Hamard et al. 2005), and the C-terminal domain interacts with eRF3. Recognition of all three stop codons by eRF1 is mediated by the YxCxxxF and TASNIKS motifs, as well as by other binding pockets/cavities in the N domain (Conard et al. 2012; Blanchet et al. 2015). The interaction between eRF1 and eRF3 is critical for stop codon recognition (Wada and Ito 2014), and GTP hydrolysis by eRF3 facilitates eRF1 discrimination of stop codons (Salas-Marco and Bedwell 2004) and accelerates peptide release (Eyler et al. 2013). Upon GTP hydrolysis eRF3 dissociates, leaving eRF1 in the A site. Binding of Rli1, an ABC-family ATPase, promotes eRF1-mediated hydrolysis of the aminoacyl bond linking the polypeptide to the peptidyl-tRNA (reviewed in Dever and Green 2012). Structural analysis of yeast eRF1 in complex with eRF3 or Rli1 on the ribosome revealed conformational changes in eRF1, while eRF3 and Rli1 were bound to the intersubunit space overlapping the binding sites for eEF1A and eEF2 (Preis et al. 2014). In complex with eRF3, eRF1 interacts with the stop codon; while in complex with Rli1, eRF1 no longer interacts with the stop codon but the GGQ motif is positioned toward the peptidyl-transferase center to promote hydrolysis. These structural changes reveal an uncoupling of stop codon recognition from peptide release by eRF1 (Preis et al. 2014). As would be expected, both eRF1 and eRF3 are essential for yeast viability; however, the N terminus of eRF3 can be deleted. This N-terminal prion domain of eRF3 is the basis of the [PSI+] aggregation of eRF3 (reviewed in Liebman and Chernoff 2012), resulting in impaired translation termination (Baudin-Baillieu et al. 2014).
Following release of the completed polypeptide, an 80S ribosome is bound to the mRNA with a deacylated tRNA in the P site base paired to the penultimate codon. ATP hydrolysis by Rli1 promotes release of the 60S subunit (Shoemaker and Green 2011). Depletion of Rli1 leads to aberrant reinitiation near the stop codon, leading to translation of 3′ UTR sequences (Young et al. 2015). In mammals, the protein Ligatin, or the complex of MCT-1 and DENR, promote release of mRNA and deacylated tRNA from the 40S subunit in the final step of recycling (Skabkin et al. 2010). The yeast orthologs of these 40S recycling factors [termed Tma64 (Ligatin), Tma20 (MCT-1), and Tma22 (DENR)] were previously identified as ribosome-associated proteins, and expression of human MCT-1 complemented translation defects in a strain lacking TMA20 (Fleischer et al. 2006). These results suggest that the mechanism of ribosome recycling is well conserved between yeast and mammals.
Translational Control in Yeast
Many studies have examined how protein synthesis is controlled in yeast. One premise is that certain mRNAs are efficiently translated only when the encoded proteins are required at a specific location (e.g., the growing bud tip) or time during the cell cycle, or in response to a specific stress. Where transcriptional control alone cannot provide a sufficiently rapid response or precise localization of proteins, translational regulation can provide the needed additional control. Progress has been made in uncovering the scope of translational controls in yeast, signal transduction pathways involved, and mRNA-specific elements that enable mRNAs to respond to the perceived demand. These studies show that translation is rapidly reprogrammed when cells experience changes in their external environment and that the translational adjustments are important for cells to adapt to the altered environment. Translational changes involve both global repression of translation of many mRNAs and activation of translation of specific stress-response genes. Yeast studies have provided molecular insights into mechanisms of control operating across eukaryotes. Indeed, the detailed understanding of GCN4 translation is now widely viewed as primary evidence to support the scanning mechanism of translation initiation. In the sections below, we first outline global approaches used to address translation changes and then describe translational control by the 4E-BPs as well as control of the GCN4 and CPA1 mRNAs via distinct mechanisms.
Global approaches to studying translation controls
Global translational activity in cell populations is often monitored by polysome profile analysis. “Freezing” ribosomes on mRNAs with CHX immediately prior to cell harvest provides a snapshot of translation. CHX binds to the ribosome E site, preventing tRNA release and trapping ribosome–mRNA complexes (Schneider-Poetsch et al. 2010). In recent years, formaldehyde cross-linking has also been used to preserve factor–ribosome interactions (Valasek et al. 2007), while for high-throughput sequencing applications, adding CHX at very high concentrations or only during cell lysis is now favored to avoid possible artifacts (Gerashchenko and Gladyshev 2014).
For polysome profile analyses, lysates are sedimented through sucrose gradients and then fractionated to generate an absorbance trace of rRNA that reveals a snapshot of the ribosome distribution (Pospisek and Valasek 2013). In actively growing cells, most ribosomes (85 ± 5%) are engaged in translation. mRNAs have a mean density of 0.64 ± 0.31 ribosomes per 100 nt coding region, with the density varying from <0.1 to >1.6 between mRNAs. Typically, longer ORFs have lower ribosome density (Arava et al. 2003). Stressed cells, or those with mutations that inhibit global translation initiation, exhibit an increased proportion of 80S ribosomes [monosomes and 80S couples (associated 40S and 60S subunits not bound to an mRNA)] relative to polysomes. Northern blotting or RT-PCR analysis of polysome gradient fractions have been used to study specific candidate genes, while microarray and high-throughput sequencing approaches such as ribosome footprint profiling (ribo-seq) have been used to assess changes in translation across multiple mRNAs simultaneously (Ingolia et al. 2009).
Stresses found to deplete bulk polysomes include the sudden withdrawal of glucose (Ashe et al. 2000; Arribere et al. 2011; Vaidyanathan et al. 2014) or amino acids (Smirnova et al. 2005); nutrient limitation that induces sporulation (Brar et al. 2012); temperature shift (Preiss et al. 2003); and the addition of cellular stress agents: hyperosmotic salt (Melamed et al. 2008), hydrogen peroxide (Shenton et al. 2006), fusel alcohols (Smirnova et al. 2005), or drugs: rapamycin (Preiss et al. 2003), calcofluor-white (Halbeisen and Gerber 2009), and chlorpromazine (Deloche et al. 2004). These studies have shown that there is widespread reprogramming of translation following stress with diminished ribosome association of some mRNAs hypersensitive to stress, while the translation of other mRNAs is relatively resistant to the stress. Moreover, the mechanisms of translational reprogramming are stress specific. For example, the global response to amino acid starvation is dependent on Gcn2 and eIF2α phosphorylation, while the response to hydrogen peroxide involves inhibiting both initiation via Gcn2 and translation elongation (Shenton et al. 2006), causing pausing at aspartic acid and serine codons according to ribo-seq experiments (Pelechano et al. 2015); however, antioxidant-response mRNAs become well translated (Shenton et al. 2006; Kershaw et al. 2015). The response to glucose depletion is rapid and does not require eIF2α phosphorylation, with almost all ribosomes being lost from mRNAs within 1–2 min (Ashe et al. 2000), perhaps via alterations in eIF4A function (Castelli et al. 2011).
Unexpectedly, some mRNAs, translationally activated 15 min after glucose withdrawal, share the same promoter sequence that binds Hsf1 (Zid and O’Shea 2014), suggesting that transcription and mRNA nuclear history might contribute to active translation during stress (Zid and O’Shea 2014). The fate of translationally repressed mRNAs following glucose depletion has also been analyzed. mRNAs enter cytoplasmic foci termed P bodies or stress granules or are degraded (Hoyle et al. 2007; Buchan et al. 2008; Arribere et al. 2011). P bodies and stress granules form with different kinetics and contain different protein components (Hoyle et al. 2007; Buchan et al. 2008) and repressed mRNAs enter P bodies at different times after stress (Simpson et al. 2014). After several hours of starvation, growth resumes with a distinct pattern of highly translated mRNAs, allowing enhanced synthesis of mitochondrial-targeted proteins (Vaidyanathan et al. 2014). Altered protein kinase A signaling contributes to the translational reprograming (Ashe et al. 2000; Tudisca et al. 2012; Vaidyanathan et al. 2014). Hence diverse cellular stress conditions can rapidly perturb global and mRNA-specific protein synthesis via multiple mechanisms to achieve stress-specific translational reprogramming. In addition to revealing translation changes under stress conditions, ribosomal profiling techniques have provided mechanistic insights into elongation through the use of distinct inhibitors (Lareau et al. 2014) as well as by identifying specific pauses during elongation (Pelechano et al. 2015).
Regulating eIF4E–eIF4G interactions by 4E-BPs
In higher eukaryotes, 4E-BPs play a prominent role in controlling eIF4E function and cellular translation (Richter and Sonenberg 2005). The 4E-BPs are a diverse set of proteins that share a common, albeit rather degenerate, motif YxxxxLΦ (where x is any residue and Φ is hydrophobic) that enables them to compete with eIF4G for binding to the surface of the cap-binding protein eIF4E. The 4E-BPs regulate a variety of cellular and developmental processes in higher eukaryotes (Kong and Lasko 2012). Yeast has two characterized 4E-BPs that contain the consensus eIF4E-binding motif, Caf20 (also called p20) and Eap1, which are 18 and 70 kDa, respectively (Altmann et al. 1997; Cosentino et al. 2000). Caf20 and Eap1 share no sequence similarity outside the eIF4E-binding motif, mutation of which abrogates eIF4E interactions (Altmann et al. 1997; Cosentino et al. 2000; Ibrahimo et al. 2006). Because mutations in eIF4E have differential impacts on the binding of eIF4G and Caf20, the eIF4E-interaction interfaces with eIF4G and Caf20 are likely overlapping, but distinct (Ptushkina et al. 1998). This potentially enables Caf20 to displace eIF4G from eIF4E. Recent structural studies show higher eukaryote 4E-BPs share similar eIF4E-binding properties (Peter et al. 2015). These data suggest the yeast proteins are parallels of the higher eukaryote 4E-BPs.
Both 4E-BPs are nonessential and deletion mutants display normal polysome profiles in optimum growth conditions (Cridge et al. 2010). The 4E-BPs play nonredundant roles in the adaptive growth response to nitrogen limitation as deletion mutants prevent pseudohyphal development and invasive growth of the Σ1278b strain (Ibrahimo et al. 2006), while S288c deletion strains are sensitive to growth on alternative nitrogen sources (Cridge et al. 2010). Translational repression upon cell treatment with the antipsychotic drug chlorpromazine or the oxidants cadmium and diamide are partially dependent upon Eap1 (Deloche et al. 2004; Mascarenhas et al. 2008), while caf20∆ displays synthetic growth phenotypes when combined with tif3∆ (eIF4B deletion) (de la Cruz et al. 1997) or the tif4631-459 allele that disrupts eIF4G1 binding to eIF4E (Hershey et al. 1999). These phenotypes are consistent with both 4E-BPs being translational repressors.
Targeted and global studies have been used to identify mRNA targets of Caf20 and Eap1 regulation. For example, the expression of STE12, GPA2, and CLN1 was shown to be translationally upregulated in caf20∆ cells during filamentous growth, and STE12 regulation was dependent on Caf20 (Park et al. 2006b). Eap1 (together with Scp160, Asc1, and Smy2) is involved in translational repression of the POM34 mRNA (Sezen et al. 2009). These data support the idea that the 4E-BPs can interact with and repress the translation of particular mRNA targets, possibly via interactions with other sequence-specific mRNA binding proteins, similar to examples from higher eukaryotes (Kong and Lasko 2012).
Microarray analysis of polysome-associated mRNAs identified >1000 genes whose polysome association was affected by deletion of either 4E-BP (Cridge et al. 2010). A computational analysis suggested that Caf20 binds mRNAs with structured 5′ UTRs (Cawley and Warwicker 2012). High-throughput sequencing of mRNAs associated with TAP-tagged Caf20 or Eap1 revealed that the two 4E-BPs bind mainly the same set (>1000) of longer than average mRNAs, suggesting that translation of these mRNAs is dampened or that their translation is poised for repression by 4E-BP binding (Costello et al. 2015). Most 4E-BP-bound mRNAs were also enriched in eIF4E interaction, as expected if the 4E-BPs repress translation via their interaction with eIF4E. However, some 4E-BP-bound mRNAs were not enriched for eIF4E, suggesting that the 4E-BPs can act independently of eIF4E (Costello et al. 2015). Over 100 mRNAs were found to interact with Caf20 independently of its ability to bind eIF4E (Castelli et al. 2015). The 3′ UTR of one mRNA tested (ERS1) directed Caf20-mediated repression of translation, and this regulation could be transplanted to a heterologous reporter (Castelli et al. 2015). In summary, both 4E-BPs can compete with eIF4G for binding to eIF4E on many mRNAs, resulting in impaired translational efficiency. However, the 4E-BPs may also act as repressors independently of eIF4E on some mRNAs.
Translationally regulated mRNAs
There are many examples of translationally controlled mRNAs in yeast. General features of translational control at the initiation phase include regulated recognition of uORFs by scanning ribosomes (GCN4), inhibition of scanning by mRNA secondary structure (HAC1), leaky scanning to initiate internally (MOD5 and GRS1), or translational repression during mRNA localization through recognition of mRNA structural elements by RNA-binding proteins (ASH1). In addition, the elongation and termination phases of translation can be regulated. For example, ribosome stalling (CPA1) as well as frameshifting (retrotransposons, L-A virus, and OAZ1) and stop codon read-through can lead to the production of alternate proteins. Programmed frameshifting can further alter the mRNA levels by triggering decay such as for EST1, EST2, STN1, and CDC13 (Advani et al. 2013).
In addition to the cap-dependent scanning model of translation initiation, an alternative cap-independent mode of translation has been proposed for some mRNAs in higher organisms (Jackson 2013). While this alternate mode of translation initiation is best characterized for viral mRNAs, some cellular mRNAs may also employ this alternate means of translation initiation (Gilbert 2010; Jackson 2013). In general, cap-independent initiation is thought to rely on special secondary structure elements in the mRNAs to recruit translation factors and/or the 40S subunit (or PIC). In experiments employing in vitro translation assays and mRNA electroporation experiments, the translation of several yeast mRNAs encoding proteins required for invasive growth was maintained when the canonical m7GTP cap was replaced by an ApppG cap structure (Gilbert et al. 2007). This cap-independent translation was dependent on eIF4G and was attributed to a poly(A) element in the 5′ UTR of the mRNAs (Gilbert et al. 2007). Similarly, the URE2 mRNA was reported to direct the synthesis of both full-length Ure2 and an N-terminally truncated Ure2 that initiates from an internal AUG codon (Komar et al. 2003). Interestingly, synthesis of the truncated Ure2 was maintained upon genetic inactivation of the mRNA cap-binding protein eIF4E and when sequences with high secondary structure were inserted in the 5′ UTR to block scanning ribosomes (Komar et al. 2003). This putative internal initiation of translation on the URE2 mRNA was enhanced in cells lacking the protein eIF2A (YGR054w) (Komar et al. 2005) and was found to depend on a 104-nt A-rich stem-loop element that includes the internal AUG start codon (Reineke et al. 2008; Reineke and Merrick 2009). Currently it is unclear under what conditions cap-independent translation will be important in yeast and whether the mRNA elements that support cap-independent translation might function as general translational “enhancers” to help the mRNAs compete for the translational apparatus by the conventional cap-dependent pathway (Gilbert 2010).
Selected examples of translationally controlled mRNAs and associated references are given in Table 3. Detailed discussions of GCN4 and CPA1 control mechanisms are presented below.
GCN4
Probably the best-characterized example of regulation of protein synthesis in yeast is GCN4. Environmental signals are transduced to modulate start codon selection on the GCN4 mRNA and limit production of the Gcn4 protein to specific cellular stress conditions. Gcn4 is a basic leucine zipper (bZIP) transcriptional activator of amino acid and related biosynthetic genes (Natarajan et al. 2001). GCN4 mRNA translation is controlled by a reinitiation mechanism that requires an interplay of sequences in its 5′ leader including uORFs with translation initiation factors and ribosomes. Under nonstarvation conditions the flow of ribosomes to the GCN4 AUG start codon is limited by up to ∼100-fold and GCN4 translation is repressed unless cells are starved (Hinnebusch 2005).
Following starvation for one or more amino acids, a signaling pathway, termed general amino acid control, is deployed, which activates Gcn2 to phosphorylate eIF2α (Dever et al. 1992) causing inhibition of eIF2B (Pavitt et al. 1998) and a reduction in eIF2 TC levels (Dever et al. 1995). The lower level of TC leads to reduced ribosome engagement with most mRNAs; however, paradoxically, more ribosomes reach the GCN4 AUG codon and Gcn4 levels increase by up to 10-fold (Albrecht et al. 1998). Because GCN4 translation is acutely sensitive to the levels of active TC, it has proved an extremely useful tool to probe the role of translation factors in the scanning mechanism of protein synthesis as well as many of the details of translational control by eIF2 kinases (Hinnebusch 2005; Hinnebusch 2011).
Translation reinitiation at uORFs represses GCN4 expression:
The 5′ leader of the GCN4 mRNA is unusually long (591 nucleotides) and contains four short uORFs (uORF1–uORF4), each encoding either a di- or tripeptide product (Figure 9A) (Hinnebusch 1984). Extensive mutational analyses of the 5′ leader have shown that the uORFs are essential for mediating both the repression under replete conditions and for induction of Gcn4 levels under stress (Mueller and Hinnebusch 1986). uORFs 1 and 4 are both critical and have opposing roles (Mueller et al. 1988). Exquisite genetic experiments coupled with biochemistry have established and tested a model for control (Figure 9 and described here).
Figure 9.
Translational regulation by reinitiation on GCN4 mRNA. (A) The GCN4 5′ leader sequences showing uORFs 1–4 and the start of the GCN4 ORF as filled boxes in their relative positions. The nucleotide positions of each AUG codon are shown relative to the transcription start site. The approximate location of reinitiation enhancer and suppressor sequences is indicated. (B) Reinitiation model in nonstarvation conditions with stepwise depiction of ribosomes and key factor interactions with the GCN4 leader sequence (cartoons as per Figure 1). Blue arrows depict the movement and blue numbered steps (i–v) are explained in the main text. Note: uORF spacing has been altered to accommodate the ribosome cartoons. As depicted, following uORF1 translation high TC levels enable reinitiation at uORF4 leading to ribosome disengagement from the mRNA and GCN4 expression is repressed. (C) Reinitiation model under amino acid starvation conditions. Initial steps through translation of uORF1 (blue numbered i–iiic) are unchanged from nonstarvation conditions. Subsequent steps (red numbered iiid–v) are altered by activation of the eIF2α kinase Gcn2 (step iiie) resulting in low levels of TC. Ribosomes traverse past uORF4 without initiating and then reacquire TC (step ivb). The scanning ribosome (step ivc) recognizes the GCN4 start codon and GCN4 expression is derepressed.
Ribosomes bind the GCN4 mRNA close to the 5′ cap and follow the normal scanning mechanism for ribosome recruitment and AUG recognition to translate uORF1 (Figure 9B, step i). Rather than completely dissociating following translation of the uORF1 tripeptide, a portion of ribosomes (estimated as ∼50%) remains attached to the mRNA. This aberrant termination/ribosome-release cycle permits reinitiation to occur at a subsequent downstream AUG codon. It is likely that only the 40S ribosome remains attached to the mRNA following termination at uORF1 and that the 40S retains some bound translation factors, including eIF3 (Szamecz et al. 2008; Munzarova et al. 2011; Peter et al. 2015). The 40S lacks eIF2, as this factor was released at AUG recognition (Figure 9B, step iia) and prior to 60S joining (Figure 9B, step iib). This release of eIF2 is important for the mechanism of control. As uORF1 encodes only a tripeptide, the ribosome has not cleared the AUG codon before encountering the stop codon. Reporter analyses suggest that reinitiation can be efficient when uORFs are shorter than 35 codons (Poyry et al. 2004; Rajkowitsch et al. 2004).
To reinitiate at a downstream ORF, the 40S subunit must resume migration along the 5′ leader sequence. The precise factor requirements for resumed ribosome movement are not clear. The eIF2–GTP–Met-tRNAiMet TC is not necessary for 40S movement, but is required for AUG recognition by the tRNAiMet anticodon. Ribosomes migrating downstream of uORF1 must reacquire TC before reinitiating translation. Under nonstarvation conditions, TC levels are not limiting and reinitiation is efficient. Translation of uORF4 (or uORF3) is sufficient to prevent almost all ribosomes from reaching the GCN4 AUG codon because translation of uORF4, also encoding a tripeptide, does not favor reinitiation. In contrast to uORF1, uORF4 favors release of translating ribosomes and prevents resumed scanning/reinitiation events. These differences in reinitiation properties following translation of uORF1 vs. uORF4 are determined by the sequences flanking the uORFs (Figure 9A), which for uORFs 1 and 2, favor 40S and eIF3 retention.
Enhancer elements both 5′ and 3′ of uORF1 act to promote retention of 40S ribosomes (Grant and Hinnebusch 1994; Grant et al. 1995). The 5′ reinitiation enhancer region (5′ RER) was shown to interact with eIF3a bound to the 40S (Szamecz et al. 2008). The 5′ RER contains four distinct elements, two of which are important for eIF3a binding (Munzarova et al. 2011). Based on its ribosome-binding properties, eIF3a bridges the mRNA interaction to the 40S protein Rps0A located near the 40S mRNA exit channel (Szamecz et al. 2008). The 5′ RER acts in concert with the AU-rich sequences immediately downstream of the uORF1 stop codon to promote reinitiation (Miller and Hinnebusch 1989; Grant and Hinnebusch 1994; Rajkowitsch et al. 2004). On encountering the uORF1 stop codon, the 3′ enhancer element interacts with the terminating ribosome mRNA entry channel retaining the 40S (Figure 9B, step iiia). The 5′ RER, which has emerged from the mRNA exit channel of the ribosome, contacts eIF3a (Figure 9B, step iiib) stabilizing eIF3 and the 40S on the GCN4 mRNA (Szamecz et al. 2008). To facilitate 40S migration downstream, presumably eIF1 and eIF1A are recruited (or retained) to promote ribosome movement in a 3′ direction (Figure 9B, step iiic) (Passmore et al. 2007).
Although uORFs 2 and 3 are not necessary for regulated control of GCN4, uORF2 functions similarly to uORF1 and promotes reinitiation by retaining eIF3–40S interactions. Thus uORF2 is suggested to act as a “fail safe” to catch any scanning ribosomes that do not initiate at uORF1 (Gunisova and Valasek 2014). Ribo-seq experiments identified two non-AUG initiating uORFs positioned upstream of uORF1 (Ingolia et al. 2009). Although translation of one these elements was confirmed with reporter constructs, these noncanonical uORFs are not necessary for GCN4 translational control (Zhang and Hinnebusch 2011).
Efficient recycling of eIF2–GDP to eIF2–GTP by eIF2B (Figure 9B, step iva) favors reacquisition of TC and eIF5 by 40S ribosomes engaged with the GCN4 mRNA (Figure 9B, step ivb) and thus will promote reinitiation at uORF3 or uORF4 (Figure 9B, step ivc). The sequences 3′ of uORF4 are GC rich and do not favor reinitiation (Grant and Hinnebusch 1994). Following translation of uORF4, ribosomes terminate and disengage from the mRNA, the GCN4 ORF is not translated, and Gcn4 levels are repressed (Figure 9B, step v).
Delayed reinitiation activates GCN4 translation in starved cells:
Under amino acid starvation, the mechanism of cap-dependent initiation at uORF 1 and resumed 40S movement are as described for nonstarvation cells (Figure 9C, steps iiia–c). An important distinction in starved cells is that following translation of uORF1 40S ribosomes migrate for a longer time and further along the GCN4 leader than in unstarved cells (Figure 9C, step iiid). This is caused by activation of the eIF2α kinase Gcn2, which phosphorylates eIF2α on Ser51 (Figure 9C step iiie). As eIF2α phosphorylation inhibits the activity of its GEF eIF2B (Figure 9C step iva), reduced TC levels and impaired global translation initiation ensue (Figure 7). This slows the rate of reacquisition of TC by ribosomes migrating along the GCN4 mRNA, enabling 40S bypass of uORFs 2–4 before TC binding (Figure 9C, step iiid). TC binding to the 40S in the interval between uORF4 and the GCN4 AUG codon, enables ribosomal scanning and recognition of the GCN4 start codon. Subsequent joining of the 60S subunit (Figure 9C, step ivc) permits translation of GCN4. As translation elongation proceeds on the GCN4 ORF, eIF3 will be lost (Figure 9C, step v). Hence, the balance between reinitiation at the uORF4 and GCN4 AUG codons facilitates control of GCN4 translation (Hinnebusch 2005, 2011). Importantly, this model of GCN4 translational control is supported by an extensive series of experiments employing mutated GCN4 leader sequences with altered secondary structure, codons, uORF lengths, and uORF spacings (Miller and Hinnebusch 1989; Abastado et al. 1991; Grant and Hinnebusch 1994; Grant et al. 1994, 1995). In addition, ribosomal toe printing on in vitro translated GCN4 mRNAs (Gaba et al. 2001) provide further support for the model and confirm the role of eIF2 levels in regulating reinitiation at uORFs 3 and 4 vs. the GCN4 start codon.
Gcn1 and Gcn20 sense amino acid levels to activate Gcn2:
GCN4 translation is regulated physiologically by eIF2α phosphorylation, primarily in response to amino acid limitation. Aminoacyl tRNA synthetases bind specific free amino acids and deacylated tRNAs to “charge” the latter for use in protein synthesis. When one or more amino acids becomes limiting, cells accumulate higher levels of deacylated tRNAs (Zaborske et al. 2009). Uncharged (deacylated) tRNAs directly bind and activate Gcn2 (Dong et al. 2000). Gcn2 is one of a family of protein kinases that phosphorylate eIF2α on Ser51 to regulate global protein synthesis across eukaryotes (Wek et al. 2006; Dever et al. 2007). In its basal state, the Gcn2 kinase is inactive, and in response to an activation signal, conformational changes and altered interactions within the multidomain protein enable its eIF2α kinase activity (Qiu et al. 2001, 2002; Padyana et al. 2005; Garriz et al. 2009; Lageix et al. 2014).
Gcn2 is a 190 kDa protein composed of a central kinase domain (KD) surrounded by domains necessary to regulate its function and interactions, including a Gcn1-binding domain and pseudokinase domain N terminal to the KD, and a histidyl-tRNA synthetase-related (HisRS) domain followed by a ∼160 residue CTD that binds 60S subunits (ribosome binding domain, RBD). Gcn2 is a dimer with multiple intermolecular interactions between monomers as well as intramolecular interactions between adjacent domains and longer range interactions between the KD and the RBD within Gcn2 monomers (Ramirez et al. 1992; Zhu et al. 1996; Qiu et al. 1998, 2001; Padyana et al. 2005; Garriz et al. 2009; Lageix et al. 2014). Binding of uncharged tRNA to the HisRS domain (Zhu et al. 1996; Dong et al. 2000) stimulates autophosphorylation of the activation loop in the KD on Thr882 and Thr887 (Romano et al. 1998) enabling phosphorylation of its only known substrate eIF2α (Dey et al. 2005, 2007; Padyana et al. 2005).
Inactivating mutations in Gcn1 or Gcn20, an ABC-type ATPase, prevent Gcn2 activation in response to amino acid starvation (Marton et al. 1993; Vazquez de Aldana et al. 1995). Both Gcn1 and Gcn20 bind elongating ribosomes (Marton et al. 1997) via the Gcn1 NTD, and this binding is required for Gcn2 activation (Sattlegger and Hinnebusch 2005). A Gcn2–Gcn1–Gcn20 complex forms by interactions between the Gcn1 central region, which is homologous to eEF3, and the Gcn20 NTD, as well as by the Gcn1 CTD binding the Gcn2 NTD (Sattlegger and Hinnebusch 2000; Kubota et al. 2001). A model for Gcn2 activation is based in part on Gcn1–Gcn20 homology with eEF3. It proposes that deacylated (uncharged) tRNA binding to the ribosomal A site is the amino acid starvation signal sensed by Gcn1–Gcn20. They pass deacylated tRNA from the A site to the Gcn2 HisRS domain, promoting Gcn2 activation and the chain of regulatory events that lead to GCN4 translation (Hinnebusch 2005).
Other regulators of Gcn2:
Gcn2 engages in cross-talk with other nutrient-sensing regulatory pathways. The TOR complex 1 (TORC1), important for controlling cell growth and rRNA synthesis among other targets (Martin et al. 2006), senses nitrogen and carbon sufficiency (Beck and Hall 1999; Loewith and Hall 2011). TORC1 deploys a complex phosphatase-signaling network involving Tap42 and Sit4 (Loewith et al. 2002) and indirectly mediates the inhibitory phosphorylation of Gcn2 on Ser577, providing a link between nitrogen and amino acid signaling (Cherkasova and Hinnebusch 2003). Inhibition of TORC1 activates Sit4 leading to dephosphorylation of Ser577 and constitutive activation of Gcn2 via enhanced binding of uncharged tRNAs to the Gcn2 HisRS domain (Garcia-Barrio et al. 2002; Kubota et al. 2003). Genome-wide studies confirm that Gcn4 targets are activated by rapamycin treatment, an allosteric TORC1 inhibitor (Staschke et al. 2010).
Snf1 senses low glucose levels in yeast and also promotes Gcn2 activity upon amino acid starvation (Cherkasova et al. 2010). In addition to nutrient-sensing kinases, the protein Yih1 regulates Gcn2 activity. Yih1 resembles the Gcn2 NTD and, when overexpressed, interacts with Gcn1 and prevents it from activating Gcn2 (Sattlegger et al. 2004, 2011). It is not yet clear how Yih1 interactions with Gcn1 are regulated to control Gcn2 activation.
Dephosphorylation of eIF2α to reset GCN4 control:
When amino acids are no longer scarce, high levels of GCN4 translation and Gcn2 activity are not required and eIF2α phosphorylation levels fall. The essential type 1 protein phosphatase Glc7 has a broad range of substrates including eIF2α (Wek et al. 1992). Typically, Glc7 is targeted to its substrates via interactions with dedicated regulatory (targeting) subunits (Cannon 2010). However an N-terminal extension unique to budding yeast eIF2γ (Gcd11) contains a PP1-docking motif that targets Glc7 to eIF2 to dephosphorylate eIF2α (Rojas et al. 2014). Glc7 may not be the sole eIF2 phosphatase. Like Glc7 mutants, PP2A/Sit4 and PP2C/Ptc2 mutants also increase eIF2α phosphorylation levels (Taylor et al. 2010), and Sit4 can interact with eIF2α (Cherkasova et al. 2010). Thus, multiple phosphatases may contribute to eIF2α dephosphorylation.
Arginine-regulated ribosome stalling controls CPA1 translation
CPA1 encodes the small subunit of carbamoyl phosphate synthetase, an enzyme that catalyzes a step in the synthesis of citrulline, an intermediate in the arginine biosynthesis pathway. CPA1 mRNA translation is regulated by a uORF, and by Arg, via a ribosome-stalling mechanism that is conserved across fungi (Hood et al. 2007). The CPA1 uORF, YOR302W, encodes a 25 amino acid arginine attenuator peptide (AAP). The peptide sequence, especially residues 6–23, is critical for translational repression by Arg (Werner et al. 1987; Delbecq et al. 2000; Hood et al. 2007). Many single missense mutations in the uORF eliminate Arg-controlled ribosome stalling (Delbecq et al. 2000). Of these, the D13N mutant has been used widely to inform mechanistic understanding (Wang et al. 1999; Gaba et al. 2001). In contrast to GCN4, most nucleotide sequences around the CPA1 uORF are not important (Delbecq et al. 1994).
In the absence of Arg, both the uORF and CPA1 are translated. The uORF AUG context is poor, ensuring that some scanning PICs bypass the uORF AUG and instead initiate translation at CPA1 (Werner et al. 1987). This was confirmed by ribosome toe print analyses using a uORF-regulated luciferase reporter and yeast translation extracts (Gaba et al. 2001), as well as via ribo-seq experiments (Ingolia et al. 2009). There is no evidence supporting ribosome reinitiation after uORF translation. The presence of high Arg concentrations induces ribosome stalling at the uORF stop codon, with 80S complexes retaining a P-site tRNA-linked nascent peptide within the ribosome exit tunnel. The stalled 80S prevents any PICs that leaky scan through the uORF AUG codon from progressing to the CPA1 AUG codon (Wang et al. 1999; Gaba et al. 2001). This ensures Cpa1 levels drop when Arg is abundant.
CPA1 control is not mediated by monitoring tRNA Arg charging levels and the Saccharomyces AAP sequence does not contain Arg residues (Wang et al. 1999). Hence it is mechanistically distinct from both GCN4 uORF control and the ribosome stalling associated with Trp attenuation in bacteria. Studies performed using the orthologous Neurospora crassa Arg-2 locus have helped inform the mechanism of peptide and Arg-induced stalling. Control requires Arg itself, which alters interactions between the P-site tRNA/nascent AAP and both rRNA and ribosomal proteins within the PTC and the peptide exit tunnel of the 60S. Although ribosome stalling naturally occurs at the end of the AAP sequence, it was shown that Arg-dependent stalling occurs during translation elongation as removing the stop codon to extend the peptide or transferring the AAP sequence to the middle of a reporter gene generated novel Arg-mediated ribosome-stalling contexts (Wang et al. 1998; Fang et al. 2004). Structural analysis of the 80S-bound stalled nascent peptide by cryo-EM revealed that residues 10–24 of the N. crassa AAP (equivalent to residues 11–25 of yeast AAP) forms an α-helix within the exit tunnel and that AAP makes a series of contacts with tunnel-exposed conserved 28S rRNA bases in the upper tunnel and with residues of Rpl4 and Rpl17 at the exit tunnel constriction point (Bhushan et al. 2010). Complementary analyses with photo-cross-linked amino acids suggest that Arg alters the AAP conformation within the exit tunnel, affecting its interactions with both Rpl4 and Rpl17 (Wu et al. 2012). The cryo-EM data also suggest that Arg stabilizes a distinct conformation of 28S rRNA residue A2062 such that the AAP-linked P-site tRNA conformation within the PTC could prevent eRF1 action (Bhushan et al. 2010). An in vitro translation puromycin release assay confirmed that Arg-mediated stalling inhibits peptidyl transfer activity, thereby preventing normal translation termination and ribosome recycling (Wei et al. 2012). How Arg interferes with PTC function is not yet clear.
As stalling occurs at the uORF stop codon, Cpa1 levels are further controlled by the NMD quality control pathway that recognizes mRNAs with aberrant premature stop codons (Kervestin and Jacobson 2012). NMD requires three proteins: Upf1, Nmd2, and Upf3. When 80S complexes translating the uORF stall in the presence of Arg, the CPA1 mRNA is destabilized, dependent on Upf1 (Messenguy et al. 2002) and Nmd2 (Gaba et al. 2005). Consistent with these findings, mutating each NMD gene enhances Cpa1 activity in the presence of Arg (Messenguy et al. 2002). Translation of the AAP and ribosome stalling over the uORF stop codon are necessary for NMD as mutations altering the AUG start codon or the D13N mutation in the AAP eliminate NMD (Gaba et al. 2005).
Perspective
The use of S. cerevisiae to study protein synthesis has provided novel insights into both the mechanism and regulation of translation that are shared among all eukaryotic organisms. Molecular genetic, biochemical, and structural studies in yeast have been especially useful in deciphering the functions of translation factors in recruiting an mRNA to the ribosome and in selecting the start codon by the scanning ribosome. Moreover, the elegant GCN4 translation control system in yeast has not only provided novel insights into the functions of a variety of translation factors, but this mechanism of gene-specific translational control has served as a paradigm for the integrated stress response in mammalian cells. In addition, studies of translation in yeast have led to the development of the ribosomal profiling technique to monitor genome-wide protein synthesis. By combining the new techniques of ribosomal profiling, high-resolution cryo-EM imaging, and single-molecule biochemistry with traditional, yet powerful, molecular genetic approaches, yeast is an ideal system to study protein synthesis and the translational control processes operating in all eukaryotes.
Acknowledgments
We thank Alan Hinnebusch and the anonymous referee for their insights to improve the manuscript. Work in the Pavitt laboratory is funded by United Kingdom Biotechnology and Biological Sciences Research Council grants BB/L000652/1, BB/L020157/1, and BB/M006565/1. Work in the Kinzy laboratory is supported by National Institutes of Health (NIH) grant GM57483. Work in the Dever laboratory is supported by the Intramural Research Program of the NIH.
Footnotes
Communicating editor: A. Hinnebusch
Literature Cited
- Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G., 1991. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol. 11: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acker M. G., Shin B. S., Dever T. E., Lorsch J. R., 2006. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem. 281: 8469–8475. [DOI] [PubMed] [Google Scholar]
- Acker M. G., Shin B. S., Nanda J. S., Saini A. K., Dever T. E., et al. , 2009. Kinetic analysis of late steps of eukaryotic translation initiation. J. Mol. Biol. 385: 491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advani V. M., Belew A. T., Dinman J. D., 2013. Yeast telomere maintenance is globally controlled by programmed ribosomal frameshifting and the nonsense-mediated mRNA decay pathway. Translation (Austin) 1: e24418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken C. E., Lorsch J. R., 2012. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19: 568–576. [DOI] [PubMed] [Google Scholar]
- Albrecht G., Mosch H. U., Hoffmann B., Reusser U., Braus G. H., 1998. Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273: 12696–12702. [DOI] [PubMed] [Google Scholar]
- Algire M. A., Maag D., Savio P., Acker M. G., Tarun S. Z., Jr, et al. , 2002. Development and characterization of a reconstituted yeast translation initiation system. RNA 8: 382–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire M. A., Maag D., Lorsch J. R., 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20: 251–262. [DOI] [PubMed] [Google Scholar]
- Alone P. V., Dever T. E., 2006. Direct binding of translation initiation factor eIF2γ-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2Bε. J. Biol. Chem. 281: 12636–12644. [DOI] [PubMed] [Google Scholar]
- Altmann M., Trachsel H., 1989. Altered mRNA cap recognition activity of initiation factor 4E in the yeast cell cycle division mutant cdc33. Nucleic Acids Res. 17: 5923–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M., Edery I., Trachsel H., Sonenberg N., 1988. Site-directed mutagenesis of the tryptophan residues in yeast eukaryotic initiation factor 4E. Effects on cap binding activity. J. Biol. Chem. 263: 17229–17232. [PubMed] [Google Scholar]
- Altmann M., Muller P. P., Wittmer B., Ruchti F., Lanker S., et al. , 1993. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 12: 3997–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M., Schmitz N., Berset C., Trachsel H., 1997. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 16: 1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Ghosh S., Mangus D. A., Jacobson A., 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453: 1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand M., Chakraburtty K., Marton M. J., Hinnebusch A. G., Kinzy T. G., 2003. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J. Biol. Chem. 278: 6985–6991. [DOI] [PubMed] [Google Scholar]
- Anand M., Balar B., Ulloque R., Gross S. R., Kinzy T. G., 2006. Domain and nucleotide dependence of the interaction between Saccharomyces cerevisiae translation elongation factors 3 and 1A. J. Biol. Chem. 281: 32318–32326. [DOI] [PubMed] [Google Scholar]
- Andersen C. B., Becker T., Blau M., Anand M., Halic M., et al. , 2006. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 443: 663–668. [DOI] [PubMed] [Google Scholar]
- Andersen G. R., Pedersen L., Valente L., Chatterjee I., Kinzy T. G., et al. , 2000. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Ba. Mol. Cell 6: 1261–1266. [DOI] [PubMed] [Google Scholar]
- Andersen G. R., Valente L., Pedersen L., Kinzy T. G., Nyborg J., 2001. Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Bα complex. Nat. Struct. Biol. 8: 531–534. [DOI] [PubMed] [Google Scholar]
- Anderson J., Phan L., Cuesta R., Carlson B. A., Pak M., et al. , 1998. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 12: 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J., Phan L., Hinnebusch A. G., 2000. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreou A. Z., Klostermeier D., 2014. eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. J. Mol. Biol. 426: 51–61. [DOI] [PubMed] [Google Scholar]
- Arava Y., Wang Y., Storey J. D., Liu C. L., Brown P. O., et al. , 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100: 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer S. K., Shirokikh N. E., Hallwirth C. V., Beilharz T. H., Preiss T., 2015. Probing the closed-loop model of mRNA translation in living cells. RNA Biol. 12: 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Gilbert W. V., 2013. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 23: 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Doudna J. A., Gilbert W. V., 2011. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol. Cell 44: 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura T., Sasaki T., Nagano F., Satoh A., Obaishi H., et al. , 1998. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene 16: 121–130. [DOI] [PubMed] [Google Scholar]
- Asano K., Phan L., Anderson J., Hinnebusch A. G., 1998. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 273: 18573–18585. [DOI] [PubMed] [Google Scholar]
- Asano K., Krishnamoorthy T., Phan L., Pavitt G. D., Hinnebusch A. G., 1999. Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 18: 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Clayton J., Shalev A., Hinnebusch A. G., 2000. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 14: 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Shalev A., Phan L., Nielsen K., Clayton J., et al. , 2001. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 20: 2326–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M. P., De Long S. K., Sachs A. B., 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11: 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrom S. U., Bystrom A. S., 1994. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell 79: 535–546. [DOI] [PubMed] [Google Scholar]
- Astrom S. U., von Pawel-Rammingen U., Bystrom A. S., 1993. The yeast initiator tRNAMet can act as an elongator tRNAMet in vivo. J. Mol. Biol. 233: 43–58. [DOI] [PubMed] [Google Scholar]
- Astrom S. U., Nordlund M. E., Erickson F. L., Hannig E. M., Bystrom A. S., 1999. Genetic interactions between a null allele of the RIT1 gene encoding an initiator tRNA-specific modification enzyme and genes encoding translation factors in Saccharomyces cerevisiae. Mol. Gen. Genet. 261: 967–976. [DOI] [PubMed] [Google Scholar]
- Aylett C. H., Boehringer D., Erzberger J. P., Schaefer T., Ban N., 2015. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat. Struct. Mol. Biol. 22: 269–271. [DOI] [PubMed] [Google Scholar]
- Baim S. B., Sherman F., 1988. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartish G., Moradi H., Nygard O., 2007. Amino acids Thr56 and Thr58 are not essential for elongation factor 2 function in yeast. FEBS J. 274: 5285–5297. [DOI] [PubMed] [Google Scholar]
- Battiste J. B., Pestova T. V., Hellen C. U. T., Wagner G., 2000. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell 5: 109–119. [DOI] [PubMed] [Google Scholar]
- Baudin-Baillieu A., Legendre R., Kuchly C., Hatin I., Demais S., et al. , 2014. Genome-wide translational changes induced by the prion [PSI+]. Cell Rep. 8: 439–448. [DOI] [PubMed] [Google Scholar]
- Beck T., Hall M. N., 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. [DOI] [PubMed] [Google Scholar]
- Beelman C. A., Parker R., 1994. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 269: 9687–9692. [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J., 1990. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62: 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belew A. T., Hepler N. L., Jacobs J. L., Dinman J. D., 2008. PRFdb: a database of computationally predicted eukaryotic programmed -1 ribosomal frameshift signals. BMC Genomics 9: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belew A. T., Advani V. M., Dinman J. D., 2011. Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 39: 2799–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield G. P., Tuite M. F., 1993. Translation elongation factor 3: A fungus-specific translation factor? Mol. Microbiol. 9: 411–418. [DOI] [PubMed] [Google Scholar]
- Benne R., Hershey J. W. B., 1978. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253: 3078–3087. [PubMed] [Google Scholar]
- Benne R., Brown-Luedi M. L., Hershey J. W. B., 1978. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J. Biol. Chem. 253: 3070–3077. [PubMed] [Google Scholar]
- Berset C., Zurbriggen A., Djafarzadeh S., Altmann M., Trachsel H., 2003. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA 9: 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E., 2004. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol. 51: 987–1001. [DOI] [PubMed] [Google Scholar]
- Bertram G., Bell H. A., Ritchie D. W., Fullerton G., Stansfield I., 2000. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beznoskova P., Wagner S., Jansen M. E., von der Haar T., Valasek L. S., 2015. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 43: 5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S., Meyer H., Starosta A. L., Becker T., Mielke T., et al. , 2010. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell 40: 138–146. [DOI] [PubMed] [Google Scholar]
- Bieniossek C., Schutz P., Bumann M., Limacher A., Uson I., et al. , 2006. The crystal structure of the carboxy-terminal domain of human translation initiation factor eIF5. J. Mol. Biol. 360: 457–465. [DOI] [PubMed] [Google Scholar]
- Blanchet S., Rowe M., Von der Haar T., Fabret C., Demais S., et al. , 2015. New insights into stop codon recognition by eRF1. Nucleic Acids Res. 43: 3298–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesen T., Mohammad S. S., Pavitt G. D., Andersen G. R., 2004. Structure of the catalytic fragment of translation initiation factor 2B and identification of a critically important catalytic residue. J. Biol. Chem. 279: 10584–10592. [DOI] [PubMed] [Google Scholar]
- Bogorad A. M., Xia B., Sandor D. G., Mamonov A. B., Cafarella T. R., et al. , 2014. Insights into the architecture of the eIF2Balpha/beta/delta regulatory subcomplex. Biochemistry 53: 3432–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck G., Shin B. S., Stiller B., Mimouni-Bloch A., Thiele H., et al. , 2012. eIF2γ mutation that disrupts eIF2 complex integrity links intellectual disability to impaired translation initiation. Mol. Cell 48: 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar G. A., Yassour M., Friedman N., Regev A., Ingolia N. T., et al. , 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Muhlrad D., Parker R., 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183: 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman J. L., Asuru A. I., Matts R. L., Hinnebusch A. G., 1993a Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 1920–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman J. L., Foiani M., Cigan A. M., Paddon C. J., Hinnebusch A. G., 1993b Guanine nucleotide exchange factor for eukaryotic translation initiation factor 2 in Saccharomyces cerevisiae: interactions between the essential subunits GCD2, GCD6, and GCD7 and the regulatory subunit GCN3. Mol. Cell. Biol. 13: 4618–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S. G., Hoyle N. P., Ashe M. P., 2005. Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body: implications for translation control. J. Cell Biol. 170: 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon J. F., 2010. Function of protein phosphatase-1, Glc7, in Saccharomyces cerevisiae. Adv. Appl. Microbiol. 73: 27–59. [DOI] [PubMed] [Google Scholar]
- Carr-Schmid A., Durko N., Cavallius J., Merrick W. C., Kinzy T. G., 1999a Mutations in a GTP-binding motif of eEF1A reduce both translational fidelity and the requirement for nucleotide exchange. J. Biol. Chem. 274: 30297–30302. [DOI] [PubMed] [Google Scholar]
- Carr-Schmid A., Valente L., Loik V. I., Williams T., Starita L. M., et al. , 1999b Mutations in Elongation Factor 1b, a guanine nucleotide exchange factor, enhance translational fidelity. Mol. Cell. Biol. 19: 5257–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A. P., Clemons W. M., Jr, Brodersen D. E., Morgan-Warren R. J., Hartsch T., et al. , 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501. [DOI] [PubMed] [Google Scholar]
- Castelli L. M., Lui J., Campbell S. G., Rowe W., Zeef L. A., et al. , 2011. Glucose depletion inhibits translation initiation via eIF4A loss and subsequent 48S preinitiation complex accumulation, while the pentose phosphate pathway is coordinately up-regulated. Mol. Biol. Cell 22: 3379–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli L. M., Talavera D., Kershaw C. J., Mohammad-Qureshi S. S., Costello J. L., et al. , 2015. The 4E-BP Caf20p mediates both eIF4E-dependent and independent repression of translation. PLoS Genet. 11: e1005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho-Valavicius B., Thompson G. M., Donahue T. F., 1992. Mutation analysis of the Cys-X2-Cys-X19-Cys-X2-Cys motif in the a subunit of eukaryotic translation factor 2. Gene Expr. 2: 297–309. [PMC free article] [PubMed] [Google Scholar]
- Cavallius J., Merrick W. C., 1998. Site-directed mutagenesis of yeast eEF1A. Viable mutants with altered nucleotide specificity. J. Biol. Chem. 273: 28752–28758. [DOI] [PubMed] [Google Scholar]
- Cavallius J., Zoll W., Chakraburtty K., Merrick W. C., 1993. Characterization of yeast EF-1a: non-conservation of post-translational modifications. Biochim. Biophys. Acta 1163: 75–80. [DOI] [PubMed] [Google Scholar]
- Cavallius J., Popkie A. P., Merrick W. C., 1997. Site-directed mutants of post-translationally modified sites of yeast eEF1A using a shuttle vector containing a chromogenic switch. Biochim. Biophys. Acta 1350: 345–358. [DOI] [PubMed] [Google Scholar]
- Cawley A., Warwicker J., 2012. eIF4E-binding protein regulation of mRNAs with differential 5′-UTR secondary structure: a polyelectrostatic model for a component of protein-mRNA interactions. Nucleic Acids Res. 40: 7666–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. J., Wang C. C., 2004. Translation initiation from a naturally occurring non-AUG codon in Saccharomyces cerevisiae. J. Biol. Chem. 279: 13778–13785. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Meng X. H., Huttelmaier S., Donato D., Singer R. H., 2002. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell 10: 1319–1330. [DOI] [PubMed] [Google Scholar]
- Chen S. J., Lin G., Chang K. J., Yeh L. S., Wang C. C., 2008. Translational efficiency of a non-AUG initiation codon is significantly affected by its sequence context in yeast. J. Biol. Chem. 283: 3173–3180. [DOI] [PubMed] [Google Scholar]
- Cherkasova V. A., Hinnebusch A. G., 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 17: 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V., Qiu H., Hinnebusch A. G., 2010. Snf1 promotes phosphorylation of the α subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol. Cell. Biol. 30: 2862–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y. N., Maag D., Mitchell S. F., Fekete C. A., Algire M. A., et al. , 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 21: 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W. L., Wagner S., Herrmannova A., Burela L., Zhang F., et al. , 2010. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell. Biol. 30: 4415–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. K., Lee J. H., Zoll W. L., Merrick W. C., Dever T. E., 1998. Promotion of Met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 280: 1757–1760. [DOI] [PubMed] [Google Scholar]
- Choi S. K., Olsen D. S., Roll-Mecak A., Martung A., Remo K. L., et al. , 2000. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol. 20: 7183–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang R. Y., Weaver P. L., Liu Z., Chang T. H., 1997. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science 275: 1468–1471. [DOI] [PubMed] [Google Scholar]
- Cigan A. M., Feng L., Donahue T. F., 1988a tRNAiMet functions in directing the scanning ribosome to the start site of translation. Science 242: 93–97. [DOI] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Donahue T. F., 1988b Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 2964–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Feng L., Donahue T. F., 1989. Yeast translation initiation suppressor sui2 encodes the α subunit of eukaryotic initiation factor 2 and shares identity with the human α subunit. Proc. Natl. Acad. Sci. USA 86: 2784–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Bushman J. L., Boal T. R., Hinnebusch A. G., 1993. A protein complex of translational regulators of GCN4 mRNA is the guanine nucleotide-exchange factor for translation initiation factor 2 in yeast. Proc. Natl. Acad. Sci. USA 90: 5350–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson B. K., Gilbert W. V., Doudna J. A., 2010. Functional overlap between eIF4G isoforms in Saccharomyces cerevisiae. PLoS One 5: e9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J. M., Laz T. M., Sherman F., 1988. Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 4533–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conard S. E., Buckley J., Dang M., Bedwell G. J., Carter R. L., et al. , 2012. Identification of eRF1 residues that play critical and complementary roles in stop codon recognition. RNA 18: 1210–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte M. R., Kelly G., Babon J., Sanfelice D., Youell J., et al. , 2006. Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry 45: 4550–4558. [DOI] [PubMed] [Google Scholar]
- Cosentino G. P., Schmelzle T., Haghighat A., Helliwell S. B., Hall M. N., et al. , 2000. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello J., Castelli L. M., Rowe W., Kershaw C. J., Talavera D., et al. , 2015. Global mRNA selection mechanisms for translation initiation. Genome Biol. 16: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttas T. A., Raftery M. J., Padula M. P., Herbert B. R., Wilkins M. R., 2012. Methylation of translation-associated proteins in Saccharomyces cerevisiae: identification of methylated lysines and their methyltransferases. Proteomics 12: 960–972. [DOI] [PubMed] [Google Scholar]
- Cridge A. G., Castelli L. M., Smirnova J. B., Selley J. N., Rowe W., et al. , 2010. Identifying eIF4E-binding protein translationally-controlled transcripts reveals links to mRNAs bound by specific PUF proteins. Nucleic Acids Res. 38: 8039–8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Silva S., Haider S. J., Phizicky E. M., 2011. A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA 17: 1100–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaie P., Altmann M., Hall M. N., Trachsel H., Helliwell S. B., 1999. CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E. Biochem. J. 340: 135–141. [PMC free article] [PubMed] [Google Scholar]
- Das S., Ghosh R., Maitra U., 2001. Eukaryotic translation initiation factor 5 functions as a GTPase- activating protein. J. Biol. Chem. 276: 6720–6726. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra B., Chakraburtty K., 1981. Purification and properties of elongation factor 3 from Saccharomyces cerevisiae. J. Biol. Chem. 256: 9999–10004. [PubMed] [Google Scholar]
- Davydova E., Ho A. Y., Malecki J., Moen A., Enserink J. M., et al. , 2014. Identification and characterization of a novel evolutionarily conserved lysine-specific methyltransferase targeting eukaryotic translation elongation factor 2 (eEF2). J. Biol. Chem. 289: 30499–30510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Iost I., Kressler D., Linder P., 1997. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94: 5201–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbecq P., Werner M., Feller A., Filipkowski R. K., Messenguy F., et al. , 1994. A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol. Cell. Biol. 14: 2378–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbecq P., Calvo O., Filipkowski R. K., Pierard A., Messenguy F., 2000. Functional analysis of the leader peptide of the yeast gene CPA1 and heterologous regulation by other fungal peptides. Curr. Genet. 38: 105–112. [DOI] [PubMed] [Google Scholar]
- Deloche O., de la Cruz J., Kressler D., Doere M., Linder P., 2004. A membrane transport defect leads to a rapid attenuation of translation initiation in Saccharomyces cerevisiae. Mol. Cell 13: 357–366. [DOI] [PubMed] [Google Scholar]
- Deng Y., Singer R. H., Gu W., 2008. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 22: 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev K., Santangelo T. J., Rothenburg S., Neculai D., Dey M., et al. , 2009. Archaeal aIF2B interacts with eukaryotic translation initiation factors eIF2α and eIF2Bα: implications for aIF2B function and eIF2B regulation. J. Mol. Biol. 392: 701–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev K., Qiu H., Dong J., Zhang F., Barthlme D., et al. , 2010. The β/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo. Mol. Cell. Biol. 30: 5218–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Green R., 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 4: a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., et al. , 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68: 585–596. [DOI] [PubMed] [Google Scholar]
- Dever T. E., Yang W., Astrom S., Bystrom A. S., Hinnebusch A. G., 1995. Modulation of tRNAiMet, eIF-2 and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2·GTP·Met-tRNAiMet ternary complexes. Mol. Cell. Biol. 15: 6351–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Dar A. C., Sicheri F., 2007. The eIF2α kinases, pp. 319–344 in Translational Control in Biology and Medicine, edited by Mathews M. B., Sonenberg N., Hershey J. W. B. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Dever T. E., Gutierrez E., Shin B. S., 2014. The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol. 49: 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey M., Trieselmann B., Locke E. G., Lu J., Cao C., et al. , 2005. PKR and GCN2 kinases and guanine nucleotide exchange factor eukaryotic translation initiation factor 2B (eIF2B) recognize overlapping surfaces on eIF2α. Mol. Cell. Biol. 25: 3063–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey M., Cao C., Sicheri F., Dever T. E., 2007. Conserved intermolecular salt bridge required for activation of protein kinases PKR, GCN2, and PERK. J. Biol. Chem. 282: 6653–6660. [DOI] [PubMed] [Google Scholar]
- Dhaliwal S., Hoffman D. W., 2003. The crystal structure of the N-terminal region of the α subunit of translation initiation factor 2 (eIF2α) from Saccharomyces cerevisiae provides a view of the loop containing serine 51, the target of the eIF2α-specific kinases. J. Mol. Biol. 334: 187–195. [DOI] [PubMed] [Google Scholar]
- Dinman J. D., Icho T., Wickner R. B., 1991. A -1 ribosomal frameshift in a double-stranded RNA virus forms a Gag-pol fusion protein. Proc. Natl. Acad. Sci. USA 88: 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J. D., Kinzy T. G., 1997. Translational misreading: mutations in translation elongation factor 1a differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA 3: 870–881. [PMC free article] [PubMed] [Google Scholar]
- Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., et al. , 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339: 85–88. [DOI] [PubMed] [Google Scholar]
- Dominguez D., Altmann M., Benz J., Baumann U., Trachsel H., 1999. Interaction of translation initiation factor eIF4G with eIF4A in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274: 26720–26726. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Cigan A. M., 1988. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol. Cell. Biol. 8: 2955–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Cigan A. M., Pabich E. K., Castilho-Valavicius B., 1988. Mutations at a Zn(II) finger motif in the yeast elF-2b gene alter ribosomal start-site selection during the scanning process. Cell 54: 621–632. [DOI] [PubMed] [Google Scholar]
- Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A. G., 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6: 269–279. [DOI] [PubMed] [Google Scholar]
- Dong J., Munoz A., Kolitz S. E., Saini A. K., Chiu W. L., et al. , 2014. Conserved residues in yeast initiator tRNA calibrate initiation accuracy by regulating preinitiation complex stability at the start codon. Genes Dev. 28: 502–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorris D. R., Erickson F. L., Hannig E. M., 1995. Mutations in GCD11, the structural gene for eIF-2γ in yeast, alter translational regulation of GCN4 and the selection of the start site for protein synthesis. EMBO J. 14: 2239–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir S., Velten L., Sharon E., Zeevi D., Carey L. B., et al. , 2013. Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc. Natl. Acad. Sci. USA 110: E2792–E2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzialo M. C., Travaglini K. J., Shen S., Roy K., Chanfreau G. F., et al. , 2014. Translational roles of elongation factor 2 protein lysine methylation. J. Biol. Chem. 289: 30511–30524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebstrup T., Saalbach G., Egsgaard H., 2005. A proteomics study of in vitro cyst germination and appressoria formation in Phytophthora infestans. Proteomics 5: 2839–2848. [DOI] [PubMed] [Google Scholar]
- Eiznhamer D. A., Ashburner B. P., Jackson J. C., Gardenour K. R., Lopes J. M., 2001. Expression of the INO2 regulatory gene of Saccharomyces cerevisiae is controlled by positive and negative promoter elements and an upstream open reading frame. Mol. Microbiol. 39: 1395–1405. [PubMed] [Google Scholar]
- Elantak L., Wagner S., Herrmannova A., Karaskova M., Rutkai E., et al. , 2010. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J. Mol. Biol. 396: 1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson F. L., Hannig E. M., 1996. Ligand interactions with eukaryotic translation initiation factor 2: role of the γ-subunit. EMBO J. 15: 6311–6320. [PMC free article] [PubMed] [Google Scholar]
- Erickson F. L., Nika J., Rippel S., Hannig E. M., 2001. Minimum requirements for the function of eukaryotic translation initiation factor 2. Genetics 158: 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger J. P., Stengel F., Pellarin R., Zhang S., Schaefer T., et al. , 2014. Molecular architecture of the 40SeIF1eIF3 translation initiation complex. Cell 158: 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler D. E., Wehner K. A., Green R., 2013. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J. Biol. Chem. 288: 29530–29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Spevak C. C., Wu C., Sachs M. S., 2004. A nascent polypeptide domain that can regulate translation elongation. Proc. Natl. Acad. Sci. USA 101: 4059–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Zhao H., Vimaladithan A., 1993. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell 74: 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farruggio D., Chaudhuri J., Maitra U. and U. L. RajBhandary, 1996. The A1•U72 base pair conserved in eukaryotic initiator tRNAs is important specifically for binding to the eukaryotic translation initiation factor eIF2. Mol. Cell. Biol. 16: 4248–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete C. A., Applefield D. J., Blakely S. A., Shirokikh N., Pestova T., et al. , 2005. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J. 24: 3588–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete C. A., Mitchell S. F., Cherkasova V. A., Applefield D., Algire M. A., et al. , 2007. N-and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO 26: 1602–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I. S., Bai X. C., Hussain T., Kelley A. C., Lorsch J. R., et al. , 2013. Molecular architecture of a eukaryotic translational initiation complex. Science 342: 1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firczuk H., Kannambath S., Pahle J., Claydon A., Beynon R., et al. , 2013. An in vivo control map for the eukaryotic mRNA translation machinery. Mol. Syst. Biol. 9: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer T. C., Weaver C. M., McAfee K. J., Jennings J. L., Link A. J., 2006. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 20: 1294–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G., 1991. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 3203–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P., Inada T., Preiss T., Hentze M. W., Mattaj I. W., et al. , 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6: 191–196. [PubMed] [Google Scholar]
- Fringer J. M., Acker M. G., Fekete C. A., Lorsch J. R., Dever T. E., 2007. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell. Biol. 27: 2384–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A., Wang Z., Krishnamoorthy T., Hinnebusch A. G., Sachs M. S., 2001. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 20: 6453–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A., Jacobson A., Sachs M. S., 2005. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20: 449–460. [DOI] [PubMed] [Google Scholar]
- Gallie D. R., 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5: 2108–2116. [DOI] [PubMed] [Google Scholar]
- Garcia-Barrio M., Dong J., Cherkasova V. A., Zhang X., Zhang F., et al. , 2002. Serine 577 is phosphorylated and negatively affects the tRNA binding and eIF2α kinase activities of GCN2. J. Biol. Chem. 277: 30675–30683. [DOI] [PubMed] [Google Scholar]
- Garre E., Romero-Santacreu L., De Clercq N., Blasco-Angulo N., Sunnerhagen P., et al. , 2012. Yeast mRNA cap-binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock. Mol. Biol. Cell 23: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriz A., Qiu H., Dey M., Seo E. J., Dever T. E., et al. , 2009. A network of hydrophobic residues impeding helix αC rotation maintains latency of kinase Gcn2, which phosphorylates the α subunit of translation initiation factor 2. Mol. Cell. Biol. 29: 1592–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko M. V., Gladyshev V. N., 2014. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 42: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. V., 2010. Alternative ways to think about cellular internal ribosome entry. J. Biol. Chem. 285: 29033–29038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. V., Zhou K., Butler T. K., Doudna J. A., 2007. Cap-independent translation is required for starvation-induced differentiation in yeast. Science 317: 1224–1227. [DOI] [PubMed] [Google Scholar]
- Gomez E., Pavitt G. D., 2000. Identification of domains and residues within the ε subunit of eukaryotic translation initiation factor 2B (eIF2Bε) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol. Cell. Biol. 20: 3965–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez E., Mohammad S. S., Pavitt G. D., 2002. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J. 21: 5292–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordiyenko Y., Schmidt C., Jennings M. D., Matak-Vinkovic D., Pavitt G. D., et al. , 2014. eIF2B is a decameric guanine nucleotide exchange factor with a γ2ε2 tetrameric core. Nat. Commun. 5: 3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. M., Hinnebusch A. G., 1994. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol. Cell. Biol. 14: 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. M., Miller P. F., Hinnebusch A. G., 1994. Requirements for intercistronic distance and level of eukaryotic initiation factor 2 activity in reinitiation on GCN4 mRNA vary with the downstream cistron. Mol. Cell. Biol. 14: 2616–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. M., Miller P. F., Hinnebusch A. G., 1995. Sequences 5′ of the first upstream open reading frame in GCN4 mRNA are required for efficient translational reinitiation. Nucleic Acids Res. 23: 3980–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greganova E., Altmann M., Butikofer P., 2011. Unique modifications of translation elongation factors. FEBS J. 278: 2613–2624. [DOI] [PubMed] [Google Scholar]
- Gregio A. P., Cano V. P., Avaca J. S., Valentini S. R., Zanelli C. F., 2009. eIF5A has a function in the elongation step of translation in yeast. Biochem. Biophys. Res. Commun. 380: 785–790. [DOI] [PubMed] [Google Scholar]
- Gross J. D., Moerke N. J., von der Haar T., Lugovskoy A. A., Sachs A. B., et al. , 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750. [DOI] [PubMed] [Google Scholar]
- Gross S. R., Kinzy T. G., 2005. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12: 772–778. [DOI] [PubMed] [Google Scholar]
- Gross S. R., Kinzy T. G., 2007. Improper organization of the actin cytoskeleton affects protein synthesis at initiation. Mol. Cell. Biol. 27: 1974–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunisova S., Valasek L. S., 2014. Fail-safe mechanism of GCN4 translational control–uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4 expression. Nucleic Acids Res. 42: 5880–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E., Shin B. S., Woolstenhulme C. J., Kim J. R., Saini P., et al. , 2013. eIF5A promotes translation of polyproline motifs. Mol. Cell 51: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbeisen R. E., Gerber A. P., 2009. Stress-dependent coordination of transcriptome and translatome in yeast. PLoS Biol. 7: e1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Hinnebusch A. G., 1988. Molecular analysis of GCN3, a translational activator of GCN4: evidence for posttranslational control of GCN3 regulatory function. Mol. Cell. Biol. 8: 4808–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Williams N. P., Wek R. C., Hinnebusch A. G., 1990. The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics 126: 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Cigan A. M., Freeman B. A., Kinzy T. G., 1993. GCD11, a negative regulator of GCN4 expression, encodes the γ subunit of eIF-2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 506–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima S., Hinnebusch A. G., 1986. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 6: 3990–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger J. W., Meskauskas A., Nielsen J., Justice M. C., Dinman J. D., 2001. Ty1 retrotransposition and programmed +1 ribosomal frameshifting require the integrity of the protein synthetic translocation step. Virology 286: 216–224. [DOI] [PubMed] [Google Scholar]
- Harger J. W., Meskauskas A., Dinman J. D., 2002. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 27: 448–454. [DOI] [PubMed] [Google Scholar]
- Harms U., Andreou A. Z., Gubaev A., Klostermeier D., 2014. eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res. 42: 7911–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto N. N., Carnevalli L. S., Castilho B. A., 2002. Translation initiation at non-AUG codons mediated by weakened association of eukaryotic initiation factor (eIF) 2 subunits. Biochem. J. 367: 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., von der Haar T., Singh C. R., Ii M., Li B., et al. , 2003. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol. Cell. Biol. 23: 5431–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekman K. E., Yu G. Y., Brown C. D., Zhu H., Du X., et al. , 2012. A conserved eEF2 coding variant in SCA26 leads to loss of translational fidelity and increased susceptibility to proteostatic insult. Hum. Mol. Genet. 21: 5472–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A., Hershey J. W., 2011. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 108: 6415–6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch S. J., Wang M., Zou S. B., Moon K. M., Foster L. J., et al. , 2013. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. MBio 4: e00180–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey P. E., McWhirter S. M., Gross J. D., Wagner G., Alber T., et al. , 1999. The Cap-binding protein eIF4E promotes folding of a functional domain of yeast translation initiation factor eIF4G1. J. Biol. Chem. 274: 21297–21304. [DOI] [PubMed] [Google Scholar]
- Heurgue-Hamard V., Champ S., Mora L., Merkulova-Rainon T., Kisselev L. L., et al. , 2005. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 280: 2439–2445. [DOI] [PubMed] [Google Scholar]
- Hilliker A., Gao Z., Jankowsky E., Parker R., 2011. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell 43: 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., 1984. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. USA 81: 6442–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes, pp. 185–243 in Translational Control of Gene Expression, edited by Sonenberg N., Hershey J. W. B., Mathews M. B. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hinnebusch A. G., 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75: 434–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., 2014. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83: 779–812. [DOI] [PubMed] [Google Scholar]
- Hiyama T. B., Ito T., Imataka H., Yokoyama S., 2009. Crystal structure of the α subunit of human translation initiation factor 2B. J. Mol. Biol. 392: 937–951. [DOI] [PubMed] [Google Scholar]
- Hood H. M., Spevak C. C., Sachs M. S., 2007. Evolutionary changes in the fungal carbamoyl-phosphate synthetase small subunit gene and its associated upstream open reading frame. Fungal Genet. Biol. 44: 93–104. [DOI] [PubMed] [Google Scholar]
- Howe J. G., Hershey J. W., 1984. Translational initiation factor and ribosome association with the cytoskeletal framework fraction from HeLa cells. Cell 37: 85–93. [DOI] [PubMed] [Google Scholar]
- Hoyle N. P., Castelli L. M., Campbell S. G., Holmes L. E., Ashe M. P., 2007. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Yoon H., Hannig E. M., Donahue T. F., 1997. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 11: 2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T., Llacer J. L., Fernandez I. S., Munoz A., Martin-Marcos P., et al. , 2014. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. S., Feinberg B., Rotwell T. C., Moldave K., 1984. Monoclonal antibody specific for yeast elongation factor 3. Biochemistry 23: 3055–3063. [DOI] [PubMed] [Google Scholar]
- Ibrahimo S., Holmes L. E., Ashe M. P., 2006. Regulation of translation initiation by the yeast eIF4E binding proteins is required for the pseudohyphal response. Yeast 23: 1075–1088. [DOI] [PubMed] [Google Scholar]
- Iizuka N., Najita L., Franzusoff A., Sarnow P., 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 7322–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S., 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I., Dreyfus M., Linder P., 1999. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274: 17677–17683. [DOI] [PubMed] [Google Scholar]
- Irie K., Tadauchi T., Takizawa P. A., Vale R. D., Matsumoto K., et al. , 2002. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 21: 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Marintchev A., Wagner G., 2004. Solution structure of human initiation factor eIF2α reveals homology to the elongation factor eEF1B. Structure 12: 1693–1704. [DOI] [PubMed] [Google Scholar]
- Jackson R. J., 2013. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol. 5: a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J. L., Belew A. T., Rakauskaite R., Dinman J. D., 2007. Identification of functional, endogenous programmed -1 ribosomal frameshift signals in the genome of Saccharomyces cerevisiae. Nucleic Acids Res. 35: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao D. L., Chen K. Y., 2006. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J. Cell. Biochem. 97: 583–598. [DOI] [PubMed] [Google Scholar]
- Jennings M. D., Pavitt G. D., 2010a eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 465: 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M. D., Pavitt G. D., 2010b eIF5 is a dual function GAP and GDI for eukaryotic translational control. Small GTPases 1: 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M. D., Pavitt G. D., 2014. A new function and complexity for protein translation initiation factor eIF2B. Cell Cycle 13: 2660–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M. D., Zhou Y., Mohammad-Qureshi S. S., Bennett D., Pavitt G. D., 2013. eIF2B promotes eIF5 dissociation from eIF2*GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev. 27: 2696–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen M. G., Ortiz P., Shepard W., Kinzy T. G., Nyborg J., et al. , 2003. The crystal structure of the glutathione S-transferase-like domain of elongation factor 1Bγ from Saccharomyces cerevisiae. J. Biol. Chem. 278: 47190–47198. [DOI] [PubMed] [Google Scholar]
- Jivotovskaya A. V., Valasek L., Hinnebusch A. G., Nielsen K. H., 2006. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 26: 1355–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen R., Ortiz P. A., Carr-Schmid A., Nissen P., Kinzy T. G., et al. , 2003. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Biol. 10: 379–385. [DOI] [PubMed] [Google Scholar]
- Justice M. C., Hsu M. J., Tse B., Ku T., Balkovec J., et al. , 1998. Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 273: 3148–3151. [DOI] [PubMed] [Google Scholar]
- Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G., et al. , 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. A., Hershey J. W., 1994. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J. Biol. Chem. 269: 3934–3940. [PubMed] [Google Scholar]
- Kang K. R., Wolff E. C., Park M. H., Folk J. E., Chung S. I., 1995. Identification of YHR068w in Saccharomyces cerevisiae chromosome VIII as a gene for deoxyhypusine synthase. Expression and characterization of the enzyme. J. Biol. Chem. 270: 18408–18412. [DOI] [PubMed] [Google Scholar]
- Kapp L. D., Lorsch J. R., 2004a GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 335: 923–936. [DOI] [PubMed] [Google Scholar]
- Kapp L. D., Lorsch J. R., 2004b The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73: 657–704. [DOI] [PubMed] [Google Scholar]
- Kapp L. D., Kolitz S. E., Lorsch J. R., 2006. Yeast initiator tRNA identity elements cooperate to influence multiple steps of translation initiation. RNA 12: 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaskova M., Gunisova S., Herrmannova A., Wagner S., Munzarova V., et al. , 2012. Functional characterization of the role of the N-terminal domain of the c/Nip1 subunit of eukaryotic initiation factor 3 (eIF3) in AUG recognition. J. Biol. Chem. 287: 28420–28434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Berthet-Colominas C., Wulff M., Cusack S., Leberman R., 1996. The structure of the Escherichia coli EF-Tu.EF-Ts complex at 2.5 A resolution. Nature 379: 511–518. [DOI] [PubMed] [Google Scholar]
- Kemper W. M., Berry K. W., Merrick W. C., 1976. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J. Biol. Chem. 251: 5551–5557. [PubMed] [Google Scholar]
- Kershaw C. J., Costello J. L., Castelli L. M., Talavera D., Rowe W., et al. , 2015. The yeast La related protein Slf1p is a key activator of translation during the oxidative stress response. PLoS Genet. 11: e1004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M., Wan Y., Mazor E., Rinn J. L., Nutter R. C., et al. , 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature 467: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S., Jacobson A., 2012. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S. H., Sachs A. B., 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y., Kohno K., 1994. Elongation factor 2 mutants deficient in dipthamide formation show temperature-sensitive cell growth. J. Biol. Chem. 269: 13497–13501. [PubMed] [Google Scholar]
- Kito K., Ota K., Fujita T., Ito T., 2007. A synthetic protein approach toward accurate mass spectrometric quantification of component stoichiometry of multiprotein complexes. J. Proteome Res. 6: 792–800. [DOI] [PubMed] [Google Scholar]
- Kolitz S. E., Takacs J. E., Lorsch J. R., 2009. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA 15: 138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A. A., Lesnik T., Cullin C., Merrick W. C., Trachsel H., et al. , 2003. Internal initiation drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells. EMBO J. 22: 1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A. A., Gross S. R., Barth-Baus D., Strachan R., Hensold J. O., et al. , 2005. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J. Biol. Chem. 280: 15601–15611. [DOI] [PubMed] [Google Scholar]
- Kong J., Lasko P., 2012. Translational control in cellular and developmental processes. Nat. Rev. Genet. 13: 383–394. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., 1995. Multidomain organization of eukaryotic guanine nucleotide exchange translation initiation factor eIF-2B subunits revealed by analysis of conserved sequence motifs. Protein Sci. 4: 1608–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva N. L., Lamphear B. J., Hennigan F. L., Rhoads R. E., 2000. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G–1. J. Biol. Chem. 275: 41369–41376. [DOI] [PubMed] [Google Scholar]
- Kouba T., Danyi I., Gunisova S., Munzarova V., Vlckova V., et al. , 2012a Small ribosomal protein RPS0 stimulates translation initiation by mediating 40S-binding of eIF3 via its direct contact with the eIF3a/TIF32 subunit. PLoS One 7: e40464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouba T., Rutkai E., Karaskova M., Valasek L., 2012b The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes. Nucleic Acids Res. 40: 2683–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M., 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299: 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M., 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy T., Pavitt G. D., Zhang F., Dever T. E., Hinnebusch A. G., 2001. Tight binding of the phosphorylated α subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 21: 5018–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H., Ota K., Sakaki Y., Ito T., 2001. Budding yeast GCN1 binds the GI domain to activate the eIF2α kinase GCN2. J. Biol. Chem. 276: 17591–17596. [DOI] [PubMed] [Google Scholar]
- Kubota H., Obata T., Ota K., Sasaki T., Ito T., 2003. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2α kinase GCN2. J. Biol. Chem. 278: 20457–20460. [DOI] [PubMed] [Google Scholar]
- Kuhle B., Ficner R., 2014a eIF5B employs a novel domain release mechanism to catalyze ribosomal subunit joining. EMBO J. 33: 1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhle B., Ficner R., 2014b Structural insight into the recognition of amino-acylated initiator tRNA by eIF5B in the 80S initiation complex. BMC Struct. Biol. 14: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhle B., Eulig N. K., Ficner R., 2015. Architecture of the eIF2B regulatory subcomplex and its implications for the regulation of guanine nucleotide exchange on eIF2. Nucleic Acids Res. 43: 9994–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian L., Palanimurugan R., Godderz D., Dohmen R. J., 2011. Polyamine sensing by nascent ornithine decarboxylase antizyme stimulates decoding of its mRNA. Nature 477: 490–494. [DOI] [PubMed] [Google Scholar]
- Lageix S., Rothenburg S., Dever T. E., Hinnebusch A. G., 2014. Enhanced interaction between pseudokinase and kinase domains in Gcn2 stimulates eIF2α phosphorylation in starved cells. PLoS Genet. 10: e1004326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L. F., Hite D. H., Hogan G. J., Brown P. O., 2014. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 3: e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurino J. P., Thompson G. M., Pacheco E., Castilho B. A., 1999. The β subunit of eukaryotic translation initiation factor 2 binds mRNA through the lysine repeats and a region comprising the C2-C2 motif. Mol. Cell. Biol. 19: 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen B. S., Sorensen H. P., Mortensen K. K., Sperling-Petersen H. U., 2005. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69: 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Choi S. K., Roll-Mecak A., Burley S. K., Dever T. E., 1999. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc. Natl. Acad. Sci. USA 96: 4342–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Pestova T. V., Shin B. S., Cao C., Choi S. K., et al. , 2002. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl. Acad. Sci. USA 99: 16689–16694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFebvre A. K., Korneeva N. L., Trutschl M., Cvek U., Duzan R. D., et al. , 2006. Translation initiation factor eIF4G–1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 281: 22917–22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Belda-Palazon B., Ferrando A., Alepuz P., 2014. Fertility and polarized cell growth depends on eIF5A for translation of polyproline-rich formins in Saccharomyces cerevisiae. Genetics 197: 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman S. W., Chernoff Y. O., 2012. Prions in yeast. Genetics 191: 1041–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Jankowsky E., 2011. From unwinding to clamping: the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12: 505–516. [DOI] [PubMed] [Google Scholar]
- Liu F., Putnam A. A., Jankowsky E., 2014. DEAD-box helicases form nucleotide-dependent, long-lived complexes with RNA. Biochemistry 53: 423–433. [DOI] [PubMed] [Google Scholar]
- Llacer J. L., Hussain T., Marler L., Aitken C. E., Thakur A., et al. , 2015. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell 59: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Hall M. N., 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., et al. , 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10: 457–468. [DOI] [PubMed] [Google Scholar]
- Lowry C. V., Weiss J. L., Walthall D. A., Zitomer R. S., 1983. Modulator sequences mediate oxygen regulation of CYC1 and a neighboring gene in yeast. Proc. Natl. Acad. Sci. USA 80: 151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R. E., Arthanari H., Hiraishi H., Nanda J., Martin-Marcos P., et al. , 2012. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Reports 1: 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D., Lorsch J. R., 2003. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 330: 917–924. [DOI] [PubMed] [Google Scholar]
- Maag D., Fekete C. A., Gryczynski Z., Lorsch J. R., 2005. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell 17: 265–275. [DOI] [PubMed] [Google Scholar]
- Maag D., Algire M. A., Lorsch J. R., 2006. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 356: 724–737. [DOI] [PubMed] [Google Scholar]
- Mandal N., Mangroo D., Dalluge J. J., McCloskey J. A., Rajbhandary U. L., 1996. Role of the three consecutive G:C base pairs conserved in the anticodon stem of initiator tRNAs in initiation of protein synthesis in Escherichia coli. RNA 2: 473–482. [PMC free article] [PubMed] [Google Scholar]
- Martin D. E., Powers T., Hall M. N., 2006. Regulation of ribosome biogenesis: Where is TOR? Cell Metab. 4: 259–260. [DOI] [PubMed] [Google Scholar]
- Martin-Marcos P., Cheung Y. N., Hinnebusch A. G., 2011. Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol. Cell. Biol. 31: 4814–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P., Nanda J., Luna R. E., Wagner G., Lorsch J. R., et al. , 2013. β-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNAMet recruitment and accuracy of AUG selection in vivo. J. Biol. Chem. 288: 27546–27562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P., Nanda J. S., Luna R. E., Zhang F., Saini A. K., et al. , 2014. Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy. RNA 20: 150–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton M. J., Crouch D., Hinnebusch A. G., 1993. GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol. 13: 3541–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton M. J., Vazquez de Aldana C. R., Qiu H., Chakraburtty K., Hinnebusch A. G., 1997. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2α kinase GCN2. Mol. Cell. Biol. 17: 4474–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas C., Edwards-Ingram L. C., Zeef L., Shenton D., Ashe M. P., et al. , 2008. Gcn4 is required for the response to peroxide stress in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 19: 2995–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak M. K., Kinzy T. G., 2010. eEF1A: thinking outside the ribosome. J. Biol. Chem. 285: 21209–21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice T. C., Mazzucco C. E., Ramanathan C. S., Ryan B. M., Warr G. A., et al. , 1998. A highly conserved intraspecies homolog of the Saccharomyces cerevisiae elongation factor-3 encoded by the HEF3 gene. Yeast 14: 1105–1113. [DOI] [PubMed] [Google Scholar]
- Melamed D., Pnueli L., Arava Y., 2008. Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA 14: 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Vierendeels F., Pierard A., Delbecq P., 2002. Role of RNA surveillance proteins Upf1/CpaR, Upf2 and Upf3 in the translational regulation of yeast CPA1 gene. Curr. Genet. 41: 224–231. [DOI] [PubMed] [Google Scholar]
- Miller P. F., Hinnebusch A. G., 1989. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes Dev. 3: 1217–1225. [DOI] [PubMed] [Google Scholar]
- Mitchell S. F., Walker S. E., Algire M. A., Park E. H., Hinnebusch A. G., et al. , 2010. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol. Cell 39: 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad-Qureshi S. S., Haddad R., Hemingway E. J., Richardson J. P., Pavitt G. D., 2007. Critical contacts between the eukaryotic initiation factor 2B (eIF2B) catalytic domain and both eIF2β and -2γ mediate guanine nucleotide exchange. Mol. Cell. Biol. 27: 5225–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. K., Lundblad V., 1997. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 7: 969–976. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G., 1986. Multiple upstream AUG codons mediate translational control of GCN4. Cell 45: 201–207. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Jackson B. M., Miller P. F., Hinnebusch A. G., 1988. The first and fourth upstream open reading frames in GCN4 mRNA have similar initiation efficiencies but respond differently in translational control to change in length and sequence. Mol. Cell. Biol. 8: 5439–5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi R., Kandl K. A., Carr-Schmid A., Whitacre J. L., Adams A. E., et al. , 2001. Overexpression of translation elongation factor 1α affects the organization and function of the actin cytoskeleton in yeast. Genetics 157: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzarova V., Panek J., Gunisova S., Danyi I., Szamecz B., et al. , 2011. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet. 7: e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., et al. , 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Hatin I., Rousset J. P., 2001. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2: 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Duchateau-Nguyen G., Rousset J. P., 2002. Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol. Microbiol. 43: 641–652. [DOI] [PubMed] [Google Scholar]
- Namy O., Duchateau-Nguyen G., Hatin I., Hermann-Le Denmat S., Termier M., et al. , 2003. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res. 31: 2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda J. S., Cheung Y. N., Takacs J. E., Martin-Marcos P., Saini A. K., et al. , 2009. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 394: 268–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda J. S., Saini A. K., Munoz A. M., Hinnebusch A. G., Lorsch J. R., 2013. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J. Biol. Chem. 288: 5316–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K., Meyer M. R., Jackson B. M., Slade D., Roberts C., et al. , 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21: 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff C. L., Sachs A. B., 1999. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol. Cell. Biol. 19: 5557–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto N., Singh C. R., Udagawa T., Wang S., Thorson E., et al. , 2010. Yeast 18 S rRNA is directly involved in the ribosomal response to stringent AUG selection during translation initiation. J. Biol. Chem. 285: 32200–32212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger N., Trachsel H., Altmann M., 1998. The RNA recognition motif of yeast translation initiation factor Tif3/eIF4B is required but not sufficient for RNA strand-exchange and translational activity. RNA 4: 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K. H., Szamecz B., Valasek L., Jivotovskaya A., Shin B. S., et al. , 2004. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 23: 1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nika J., Yang W., Pavitt G. D., Hinnebusch A. G., Hannig E. M., 2000. Purification and kinetic analysis of eIF2B from Saccharomyces cerevisiae. J. Biol. Chem. 275: 26011–26017. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., et al. , 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270: 1464–1472. [DOI] [PubMed] [Google Scholar]
- Noma A., Yi S., Katoh T., Takai Y., Suzuki T., et al. , 2011. Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA 17: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C., Sato B. K., Broyer R. M., Wilhelm J. E., 2010. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 190: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary S. E., Petrov A., Chen J., Puglisi J. D., 2013. Dynamic recognition of the mRNA cap by Saccharomyces cerevisiae eIF4E. Structure 21: 2197–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier C., Poirier G., Gendron P., Boisgontier A., Major F., et al. , 2005. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol. Cell. Biol. 25: 4752–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen D. S., E. M. Savner , A. Mathew, F. Zhang, T. Krishnamoorthy et al, 2003. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 22: 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz P. A., Kinzy T. G., 2005. Dominant-negative mutant phenotypes and the regulation of translation elongation factor 2 levels in yeast. Nucleic Acids Res. 33: 5740–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz P. A., Ulloque R., Kihara G. K., Zheng H., Kinzy T. G., 2006. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J. Biol. Chem. 281: 32639–32648. [DOI] [PubMed] [Google Scholar]
- Otero L. J., Ashe M. P., Sachs A. B., 1999. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 18: 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padyana A. K., Qiu H., Roll-Mecak A., Hinnebusch A. G., Burley S. K., 2005. Structural basis for autoinhibition and mutational activation of eukaryotic initiation factor 2α protein kinase GCN2. J. Biol. Chem. 280: 29289–29299. [DOI] [PubMed] [Google Scholar]
- Palanimurugan R., Scheel H., Hofmann K., Dohmen R. J., 2004. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 23: 4857–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Potratz J. P., Cannon B., Simpson Z. B., Ziehr J. L., et al. , 2014. DEAD-box helicase proteins disrupt RNA tertiary structure through helix capture. PLoS Biol. 12: e1001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N., Menade M., Poirier G., Donato D., Drouet E., et al. , 2007. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell 26: 795–809. [DOI] [PubMed] [Google Scholar]
- Park E. H., Walker S. E., Lee J. M., Rothenburg S., Lorsch J. R., et al. , 2011. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1•PABP mRNPs in vivo. EMBO J. 30: 302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. H., Walker S. E., Zhou F., Lee J. M., Rajagopal V., et al. , 2013. Yeast eukaryotic initiation factor 4B (eIF4B) enhances complex assembly between eIF4A and eIF4G in vivo. J. Biol. Chem. 288: 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Aravind L., Wolff E. C., Kaevel J., Kim Y. S., et al. , 2006a Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc. Natl. Acad. Sci. USA 103: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. H., Joe Y. A., Kang K. R., 1998. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273: 1677–1683. [DOI] [PubMed] [Google Scholar]
- Park Y. U., Hur H., Ka M., Kim J., 2006b Identification of translational regulation target genes during filamentous growth in Saccharomyces cerevisiae: regulatory role of Caf20 and Dhh1. Eukaryot. Cell 5: 2120–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore L. A., Schmeing T. M., Maag D., Applefield D. J., Acker M. G., et al. , 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell 26: 41–50. [DOI] [PubMed] [Google Scholar]
- Pavitt G. D., Yang W., Hinnebusch A. G., 1997. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 17: 1298–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt G. D., Ramaiah K. V., Kimball S. R., Hinnebusch A. G., 1998. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 12: 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Wei W., Steinmetz L. M., 2015. Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161: 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S. W., Donahue J. L., Jacobson A., 1992. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 5778–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez W. B., Kinzy T. G., 2014. Translation elongation factor 1A mutants with altered actin bundling activity show reduced aminoacyl-tRNA binding and alter initiation via eIF2α phosphorylation. J. Biol. Chem. 289: 20928–20938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perzlmaier A. F., Richter F., Seufert W., 2013. Translation initiation requires cell division cycle 123 (Cdc123) to facilitate biogenesis of the eukaryotic initiation factor 2 (eIF2). J. Biol. Chem. 288: 21537–21546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T. V., Lomakin I. B., Lee J. H., Choi S. K., Dever T. E., et al. , 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335. [DOI] [PubMed] [Google Scholar]
- Peter D., Igreja C., Weber R., Wohlbold L., Weiler C., et al. , 2015. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol. Cell 57: 1074–1087. [DOI] [PubMed] [Google Scholar]
- Petropoulos A. D., Green R., 2012. Further in vitro exploration fails to support the allosteric three-site model. J. Biol. Chem. 287: 11642–11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L., Zhang X., Asano K., Anderson J., Vornlocher H. P., et al. , 1998. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol. 18: 4935–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L., Schoenfeld L. W., Valášek L., Nielsen K. H., Hinnebusch A. G., 2001. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNAiMet. EMBO J. 20: 2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman Y. R., Kandl K., Lewis M., Valente L., Kinzy T. G., 2009. Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Bα. J. Biol. Chem. 284: 4739–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E. P., Nguyen P., Russ J. R., Pittman Y. R., Nguyen T., et al. , 2007. Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae. PLoS One 2: e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenis M., Schmidt E. V., 1997. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 11: 2522–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisek M., Valasek L., 2013. Polysome profile analysis–yeast. Methods Enzymol. 530: 173–181. [DOI] [PubMed] [Google Scholar]
- Poyry T. A., Kaminski A., Jackson R. J., 2004. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 18: 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis A., Heuer A., Barrio-Garcia C., Hauser A., Eyler D. E., et al. , 2014. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Reports 8: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T., Hentze M. W., 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392: 516–520. [DOI] [PubMed] [Google Scholar]
- Preiss T., Baron-Benhamou J., Ansorge W., Hentze M. W., 2003. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat. Struct. Biol. 10: 1039–1047. [DOI] [PubMed] [Google Scholar]
- Ptushkina M., von der Haar T., Vasilescu S., Frank R., Birkenhager R., et al. , 1998. Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J. 17: 4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Garcia-Barrio M. T., Hinnebusch A. G., 1998. Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions in the C-terminal ribosome-binding region and the protein kinase domain. Mol. Cell. Biol. 18: 2697–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Dong J., Hu C., Francklyn C. S., Hinnebusch A. G., 2001. The tRNA-binding moiety in GCN2 contains a dimerization domain that interacts with the kinase domain and is required for tRNA binding and kinase activation. EMBO J. 20: 1425–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Hu C., Dong J., Hinnebusch A. G., 2002. Mutations that bypass tRNA binding activate the intrinsically defective kinase domain in GCN2. Genes Dev. 16: 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J., Leibundgut M., Ataide S. F., Haag A., Ban N., 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331: 730–736. [DOI] [PubMed] [Google Scholar]
- Rachfall, N., I. Heinemeyer, B. Morgenstern, O. Valerius and G. H. Braus, 2011 5′TRU: identification and analysis of translationally regulative 5′ untranslated regions in amino acid starved yeast cells. Mol Cell Proteomics 10: M110.003350. [DOI] [PMC free article] [PubMed]
- Rajagopal V., Park E. H., Hinnebusch A. G., Lorsch J. R., 2012. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5′-overhangs. J. Biol. Chem. 287: 20301–20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L., Vilela C., Berthelot K., Ramirez C. V., McCarthy J. E., 2004. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 335: 71–85. [DOI] [PubMed] [Google Scholar]
- Ramirez M., Wek R. C., Vazquez de Aldana C. R., Jackson B. M., Freeman B., et al. , 1992. Mutations activating the yeast eIF-2α kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol. Cell. Biol. 12: 5801–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibarkh M., Yamamoto Y., Singh C. R., del Rio F., Fahmy A., et al. , 2008. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J. Biol. Chem. 283: 1094–1103. [DOI] [PubMed] [Google Scholar]
- Reid P. J., Mohammad-Qureshi S. S., Pavitt G. D., 2012. Identification of intersubunit domain interactions within eukaryotic initiation factor (eIF) 2B, the nucleotide exchange factor for translation initiation. J. Biol. Chem. 287: 8275–8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L. C., Merrick W. C., 2009. Characterization of the functional role of nucleotides within the URE2 IRES element and the requirements for eIF2A-mediated repression. RNA 15: 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L. C., Komar A. A., Caprara M. G., Merrick W. C., 2008. A small stem loop element directs internal initiation of the URE2 internal ribosome entry site in Saccharomyces cerevisiae. J. Biol. Chem. 283: 19011–19025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J. D., Sonenberg N., 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480. [DOI] [PubMed] [Google Scholar]
- Rodnina M. V., Wintermeyer W., 2009. Recent mechanistic insights into eukaryotic ribosomes. Curr. Opin. Cell Biol. 21: 435–443. [DOI] [PubMed] [Google Scholar]
- Rojas M., Gingras A. C., Dever T. E., 2014. Protein phosphatase PP1/GLC7 interaction domain in yeast eIF2γ bypasses targeting subunit requirement for eIF2α dephosphorylation. Proc. Natl. Acad. Sci. USA 111: E1344–E1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., Cao C., Dever T. E., Burley S. K., 2000. X-Ray structures of the universal translation initiation factor IF2/eIF5B. Conformational changes on GDP and GTP binding. Cell 103: 781–792. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., Alone P., Cao C., Dever T. E., Burley S. K., 2004. X-ray structure of translation initiation factor eIF2γ: implications for tRNA and eIF2α binding. J. Biol. Chem. 279: 10634–10642. [DOI] [PubMed] [Google Scholar]
- Romano P. R., Garcia-Barrio M. T., Zhang X., Wang Q., Taylor D. R., et al. , 1998. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol. Cell. Biol. 18: 2282–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudoni S., Colombo S., Coccetti P., Martegani E., 2001. Role of guanine nucleotides in the regulation of the Ras/cAMP pathway in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1538: 181–189. [DOI] [PubMed] [Google Scholar]
- Ruegsegger U., Leber J. H., Walter P., 2001. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107: 103–114. [DOI] [PubMed] [Google Scholar]
- Safaee N., Kozlov G., Noronha A. M., Xie J., Wilds C. J., et al. , 2012. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol. Cell 48: 375–386. [DOI] [PubMed] [Google Scholar]
- Saini A. K., Nanda J. S., Lorsch J. R., Hinnebusch A. G., 2010. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNAiMet binding to the ribosome. Genes Dev. 24: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini A. K., Nanda J. S., Martin-Marcos P., Dong J., Zhang F., et al. , 2014. Eukaryotic translation initiation factor eIF5 promotes the accuracy of start codon recognition by regulating Pi release and conformational transitions of the preinitiation complex. Nucleic Acids Res. 42: 9623–9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P., Eyler D. E., Green R., Dever T. E., 2009. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459: 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J., Bedwell D. M., 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 24: 7769–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbaken M., Lupisella J. A., DiDomineco B., Chakraburtty K., 1990. Protein synthesis in yeast: isolation and characterization of the structural gene encoding elongation factor 3. J. Biol. Chem. 265: 15838–15844. [PubMed] [Google Scholar]
- Sandbaken M. G., Culbertson M. R., 1988. Mutations in elongation factor EF-1a affect the frequency of frameshifting and amino acid misincorporation in Saccharomyces cerevisiae. Genetics 120: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson L. E., Uhlenbeck O. C., 2007a The 51–63 base pair of tRNA confers specificity for binding by EF-Tu. RNA 13: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson L. E., Uhlenbeck O. C., 2007b Directed mutagenesis identifies amino acid residues involved in elongation factor Tu binding to yeast Phe-tRNAPhe. J. Mol. Biol. 368: 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy A. V., McGonigal T., Capobianco J. O., Schmidt M., Green S. R., et al. , 1998. Identification and kinetic analysis of a functional homolog of elongation factor 3, YEF3 in Saccharomyces cerevisiae. Yeast 14: 239–253. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Abid M. R., Miyazaki M., 1996. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 384: 151–154. [DOI] [PubMed] [Google Scholar]
- Sasikumar A. N., Kinzy T. G., 2014. Mutations in the chromodomain-like insertion of translation elongation factor 3 compromise protein synthesis through reduced ATPase activity. J. Biol. Chem. 289: 4853–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe L., Bolinger C., Mannan M. A., Dever T. E., Dey M., 2015. Evidence that base-pairing interaction between intron and mRNA leader sequences inhibits initiation of HAC1 mRNA translation in yeast. J. Biol. Chem. 290: 21821–21832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattlegger E., Hinnebusch A. G., 2000. Separate domains in GCN1 for binding protein kinase GCN2 and ribosomes are required for GCN2 activation in amino acid-starved cells. EMBO J. 19: 6622–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattlegger E., Hinnebusch A. G., 2005. Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2α kinase GCN2 during amino acid starvation. J. Biol. Chem. 280: 16514–16521. [DOI] [PubMed] [Google Scholar]
- Sattlegger E., Swanson M. J., Ashcraft E. A., Jennings J. L., Fekete R. A., et al. , 2004. YIH1 is an actin-binding protein that inhibits protein kinase GCN2 and impairs general amino acid control when overexpressed. J. Biol. Chem. 279: 29952–29962. [DOI] [PubMed] [Google Scholar]
- Sattlegger E., Barbosa J. A., Moraes M. C., Martins R. M., Hinnebusch A. G., et al. , 2011. Gcn1 and actin binding to Yih1: implications for activation of the eIF2 kinase GCN2. J. Biol. Chem. 286: 10341–10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffrath R., Abdel-Fattah W., Klassen R., Stark M. J., 2014. The diphthamide modification pathway from Saccharomyces cerevisiae–revisited. Mol. Microbiol. 94: 1213–1226. [DOI] [PubMed] [Google Scholar]
- Schmeing T. M., Ramakrishnan V., 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461: 1234–1242. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Blanquet S., Mechulam Y., 2002. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 21: 1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E., Naveau M., Mechulam Y., 2010. Eukaryotic and archaeal translation initiation factor 2: a heterotrimeric tRNA carrier. FEBS Lett. 584: 405–412. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Panvert M., Lazennec-Schurdevin C., Coureux P. D., Perez J., et al. , 2012. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nat. Struct. Mol. Biol. 19: 450–454. [DOI] [PubMed] [Google Scholar]
- Schneider-Poetsch T., Ju J., Eyler D. E., Dang Y., Bhat S., et al. , 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnier J., Schwelberger H. G., Smit-McBride Z., Kang H. A., Hershey J. W. B., 1991. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 3105–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier M. H., Erni B., Staehelin T., 1977. Initiation of mammalian protein synthesis: purification and characterization of seven initiation factors. J. Mol. Biol. 116: 727–753. [DOI] [PubMed] [Google Scholar]
- Schutz P., Bumann M., Oberholzer A. E., Bieniossek C., Trachsel H., et al. , 2008. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc. Natl. Acad. Sci. USA 105: 9564–9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N. D., Zhou F., Ingolia N. T., Hinnebusch A. G., 2015. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 25: 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senissar M., Le Saux A., Belgareh-Touze N., Adam C., Banroques J., et al. , 2014. The DEAD-box helicase Ded1 from yeast is an mRNP cap-associated protein that shuttles between the cytoplasm and nucleus. Nucleic Acids Res. 42: 10005–10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezen B., Seedorf M., Schiebel E., 2009. The SESA network links duplication of the yeast centrosome with the protein translation machinery. Genes Dev. 23: 1559–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J. B., 2013. Rate-limiting steps in yeast protein translation. Cell 153: 1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Jankowsky E., 2014. The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit. Rev. Biochem. Mol. Biol. 49: 343–360. [DOI] [PubMed] [Google Scholar]
- Shenton D., Smirnova J. B., Selley J. N., Carroll K., Hubbard S. J., et al. , 2006. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 281: 29011–29021. [DOI] [PubMed] [Google Scholar]
- Shin B. S., Maag D., Roll-Mecak A., Arefin M. S., Burley S. K., et al. , 2002. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell 111: 1015–1025. [DOI] [PubMed] [Google Scholar]
- Shin B. S., Acker M. G., Maag D., Kim J. R., Lorsch J. R., et al. , 2007. Intragenic suppressor mutations restore GTPase and translation functions of a eukaryotic initiation factor 5B switch II mutant. Mol. Cell. Biol. 27: 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin B. S., Kim J. R., Acker M. G., Maher K. N., Lorsch J. R., et al. , 2009. rRNA suppressor of a eukaryotic translation initiation factor 5B/initiation factor 2 mutant reveals a binding site for translational GTPases on the small ribosomal subunit. Mol. Cell. Biol. 29: 808–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin B. S., Kim J. R., Walker S. E., Dong J., Lorsch J. R., et al. , 2011. Initiation factor eIF2 promotes eIF2-GTP-Met-tRNAiMet ternary complex binding to the 40S ribosome. Nat. Struct. Mol. Biol. 18: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C. J., Green R., 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. USA 108: E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S., 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66: 1–40. [DOI] [PubMed] [Google Scholar]
- Simpson C. E., Lui J., Kershaw C. J., Sims P. F., Ashe M. P., 2014. mRNA localization to P-bodies in yeast is bi-phasic with many mRNAs captured in a late Bfr1p-dependent wave. J. Cell Sci. 127: 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C. R., Yamamoto Y., Asano K., 2004. Physical association of eukaryotic initiation factor (eIF) 5 carboxyl-terminal domain with the lysine-rich eIF2β segment strongly enhances its binding to eIF3. J. Biol. Chem. 279: 49644–49655. [DOI] [PubMed] [Google Scholar]
- Singh C. R., Curtis C., Yamamoto Y., Hall N. S., Kruse D. S., et al. , 2005. Eukaryotic translation initiation factor 5 is critical for integrity of the scanning preinitiation complex and accurate control of GCN4 translation. Mol. Cell. Biol. 25: 5480–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C. R., Lee B., Udagawa T., Mohammad-Qureshi S. S., Yamamoto Y., et al. , 2006. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 25: 4537–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin M. A., Skabkina O. V., Dhote V., Komar A. A., Hellen C. U., et al. , 2010. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 24: 1787–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogerson L., Wakatama E., 1976. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc. Natl. Acad. Sci. USA 73: 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogerson L., Engelhardt D., 1977. Dissimilarity in protein chain elongation factor requirements between yeast and rat liver ribosomes. J. Biol. Chem. 252: 1471–1475. [PubMed] [Google Scholar]
- Slusher L. B., Gillman E. C., Martin N. C., Hopper A. K., 1991. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc. Natl. Acad. Sci. USA 88: 9789–9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova J. B., Selley J. N., Sanchez-Cabo F., Carroll K., Eddy A. A., et al. , 2005. Global gene expression profiling reveals widespread yet distinctive translational responses to different eukaryotic translation initiation factor 2B-targeting stress pathways. Mol. Cell. Biol. 25: 9340–9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soe R., Mosley R. T., Justice M., Nielsen-Kahn J., Shastry M., et al. , 2007. Sordarin derivatives induce a novel conformation of the yeast ribosome translocation factor eEF2. J. Biol. Chem. 282: 657–666. [DOI] [PubMed] [Google Scholar]
- Sokabe M., Yao M., Sakai N., Toya S., Tanaka I., 2006. Structure of archaeal translational initiation factor 2βγ-GDP reveals significant conformational change of the β-subunit and switch 1 region. Proc. Natl. Acad. Sci. USA 103: 13016–13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. M., Picologlou S., Grant C. M., Firoozan M., Tuite M. F., et al. , 1989. Elongation factor EF-1a gene dosage alters translational fidelity in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 4571–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang S. R., Coleman D. E., 1998. Invasion of the nucleotide snatchers: structural insights into the mechanism of G protein GEFs. Cell 95: 155–158. [DOI] [PubMed] [Google Scholar]
- Staschke K. A., Dey S., Zaborske J. M., Palam L. R., McClintick J. N., et al. , 2010. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J. Biol. Chem. 285: 16893–16911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda M., Fukui M., Sogabe Y., Sato K., Morimatsu A., et al. , 1999. Overproduction of elongation factor 1α, an essential translational component, causes aberrant cell morphology by affecting the control of growth polarity in fission yeast. Genes Cells 4: 517–527. [DOI] [PubMed] [Google Scholar]
- Szamecz B., Rutkai E., Cuchalova L., Munzarova V., Herrmannova A., et al. , 2008. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 22: 2414–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro D., Farabaugh P. J., 2007. An mRNA sequence derived from the yeast EST3 gene stimulates programmed +1 translational frameshifting. RNA 13: 606–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. L., Yeh L. S., Chen N. K., Ripmaster T., Schimmel P., et al. , 2004. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J. Biol. Chem. 279: 49656–49663. [DOI] [PubMed] [Google Scholar]
- Tarun S. Z., Jr, Sachs A. B., 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9: 2997–3007. [DOI] [PubMed] [Google Scholar]
- Tarun S. Z., Jr, Sachs A. B., 1997. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol. Cell. Biol. 17: 6876–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S. Z., Jr, Wells S. E., Deardorff J. A., Sachs A. B., 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. USA 94: 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. J., Frank J., Kinzy T. G., 2007a Structure and function of the eukaryotic ribosome and elongation factors, pp. 59–85 in Translational Control in Biology and Medicine, edited by Mathews M. B., Sonenberg N., Hershey J. W. B. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Taylor D. J., Nilsson J., Merrill A. R., Andersen G. R., Nissen P., et al. , 2007b Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 26: 2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. J., Campbell S. G., Griffiths C. D., Reid P. J., Slaven J. W., et al. , 2010. Fusel alcohols regulate translation initiation by inhibiting eIF2B to reduce ternary complex in a mechanism that may involve altering the integrity and dynamics of the eIF2B body. Mol. Biol. Cell 21: 2202–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Reiser V., Ruis H., Ammerer G., 2001. Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc. Natl. Acad. Sci. USA 98: 5625–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triana-Alonso F. J., Chakraburtty K., Nierhaus K. H., 1995. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 270: 20473–20478. [DOI] [PubMed] [Google Scholar]
- Tudisca V., Simpson C., Castelli L., Lui J., Hoyle N., et al. , 2012. PKA isoforms coordinate mRNA fate during nutrient starvation. J. Cell Sci. 125: 5221–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S., Lassak J., Starosta A. L., Kraxenberger T., Wilson D. N., et al. , 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339: 82–85. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan P. P., Zinshteyn B., Thompson M. K., Gilbert W. V., 2014. Protein kinase A regulates gene-specific translational adaptation in differentiating yeast. RNA 20: 912–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L., Nielsen K. H., Hinnebusch A. G., 2002. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 21: 5886–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L., Mathew A. A., Shin B. S., Nielsen K. H., Szamecz B., et al. , 2003. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 17: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L., Nielsen K. H., Zhang F., Fekete C. A., Hinnebusch A. G., 2004. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 24: 9437–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L., Szamecz B., Hinnebusch A. G., Nielsen K. H., 2007. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 429: 163–183. [DOI] [PubMed] [Google Scholar]
- Valasek L. S., 2012. ‘Ribozoomin’–translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr. Protein Pept. Sci. 13: 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L., Kinzy T. G., 2003. Yeast as a sensor of factors affecting the accuracy of protein synthesis. Cell. Mol. Life Sci. 60: 2115–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Lee C. P. and U. L. RajBhandary, 1993. From elongator tRNA to initiator tRNA. Proc. Natl. Acad. Sci. USA 90: 2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez de Aldana C. R., Hinnebusch A. G., 1994. Mutations in the GCD7 subunit of yeast guanine nucleotide exchange factor eIF-2B overcome the inhibitory effects of phosphorylated eIF-2 on translation initiation. Mol. Cell. Biol. 14: 3208–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez de Aldana C. R., Marton M. J., Hinnebusch A. G., 1995. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J. 14: 3184–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Laso M. R., Zhu D., Sagliocco F., Brown A. J., Tuite M. F., et al. , 1993. Inhibition of translational initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J. Biol. Chem. 268: 6453–6462. [PubMed] [Google Scholar]
- Vilela C., Linz B., Rodrigues-Pousada C., McCarthy J. E., 1998. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 26: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa N., Do A., Hershey J. W., Fraser C. S., 2013. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem. 288: 32932–32940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T., 2008. A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Syst. Biol. 2: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Pawel-Rammingen U., Astrom S., Bystrom A. S., 1992. Mutational analysis of conserved positions potentially important for initiator tRNA function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Ito K., 2014. A genetic approach for analyzing the co-operative function of the tRNA mimicry complex, eRF1/eRF3, in translation termination on the ribosome. Nucleic Acids Res. 42: 7851–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S. E., Zhou F., Mitchell S. F., Larson V. S., Valasek L., et al. , 2013. Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA 19: 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Fang P., Sachs M. S., 1998. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol. Cell. Biol. 18: 7528–7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gaba A., Sachs M. S., 1999. A highly conserved mechanism of regulated ribosome stalling mediated by fungal arginine attenuator peptides that appears independent of the charging status of arginyl-tRNAs. J. Biol. Chem. 274: 37565–37574. [DOI] [PubMed] [Google Scholar]
- Warner J. R., 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Wei J., Jia M., Zhang C., Wang M., Gao F., et al. , 2010. Crystal structure of the C-terminal domain of the ε subunit of human translation initiation factor eIF2B. Protein Cell 1: 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Wu C., Sachs M. S., 2012. The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol. Cell. Biol. 32: 2396–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Xue Y., Xu H., Gong W., 2006. Crystal structure of the C-terminal domain of S. cerevisiae eIF5. J. Mol. Biol. 359: 1–9. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Jackson B. M., Hinnebusch A. G., 1989. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl. Acad. Sci. USA 86: 4579–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Ramirez M., Jackson B. M., Hinnebusch A. G., 1990. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol. Cell. Biol. 10: 2820–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Cannon J. F., Dever T. E., Hinnebusch A. G., 1992. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2α kinase GCN2. Mol. Cell. Biol. 12: 5700–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Jiang H. Y., Anthony T. G., 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34: 7–11. [DOI] [PubMed] [Google Scholar]
- Wells S. E., Hillner P. E., Vale R. D., Sachs A. B., 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2: 135–140. [DOI] [PubMed] [Google Scholar]
- Werner M., Feller A., Messenguy F., Pierard A., 1987. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell 49: 805–813. [DOI] [PubMed] [Google Scholar]
- Williams N. P., Hinnebusch A. G., Donahue T. F., 1989. Mutations in the structural genes for eukaryotic initiation factors 2α and 2β of Saccharomyces cerevisiae disrupt translational control of GCN4 mRNA. Proc. Natl. Acad. Sci. USA 86: 7515–7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W., Peske F., Beringer M., Gromadski K. B., Savelsbergh A., et al. , 2004. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem. Soc. Trans. 32: 733–737. [DOI] [PubMed] [Google Scholar]
- Wolfe C. L., Lou Y. C., Hopper A. K., Martin N. C., 1994. Interplay of heterogeneous transcriptional start sites and translational selection of AUGs dictate the production of mitochondrial and cytosolic/nuclear tRNA nucleotidyltransferase from the same gene in yeast. J. Biol. Chem. 269: 13361–13366. [PubMed] [Google Scholar]
- Wolff E. C., Kang K. R., Kim Y. S., Park M. H., 2007. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids 33: 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Wei J., Lin P. J., Tu L., Deutsch C., et al. , 2012. Arginine changes the conformation of the arginine attenuator peptide relative to the ribosome tunnel. J. Mol. Biol. 416: 518–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Fukuda K., Tamura S., Furukawa P., Songari P., 1993. Expression of the gene encoding a translation elongation factor 3 homolog of chlorella virus CVK2. Virology 197: 742–750. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Singh C. R., Marintchev A., Hall N. S., Hannig E. M., et al. , 2005. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3, and eIF4G. Proc. Natl. Acad. Sci. USA 102: 16164–16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagiya A., Svitkin Y. V., Shibata S., Mikami S., Imataka H., et al. , 2009. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol. Cell. Biol. 29: 1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Demma M., Warren V., Dharmawardhane S., Condeelis J., 1990. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature 347: 494–496. [DOI] [PubMed] [Google Scholar]
- Yang Q., Jankowsky E., 2005. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry 44: 13591–13601. [DOI] [PubMed] [Google Scholar]
- Yatime L., Mechulam Y., Blanquet S., Schmitt E., 2007. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc. Natl. Acad. Sci. USA 104: 18445–18450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. J., Donahue T. F., 1992. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol. Cell. Biol. 12: 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. J., Guydosh N. R., Zhang F., Hinnebusch A. G., Green R., 2015. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell 162: 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborske J. M., Narasimhan J., Jiang L., Wek S. A., Dittmar K. A., et al. , 2009. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J. Biol. Chem. 284: 25254–25267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli C. F., Maragno A. L., Gregio A. P., Komili S., Pandolfi J. R., et al. , 2006. eIF5A binds to translational machinery components and affects translation in yeast. Biochem. Biophys. Res. Commun. 348: 1358–1366. [DOI] [PubMed] [Google Scholar]
- Zenklusen D., Larson D. R., Singer R. H., 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15: 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Hinnebusch A. G., 2011. An upstream ORF with non-AUG start codon is translated in vivo but dispensable for translational control of GCN4 mRNA. Nucleic Acids Res. 39: 3128–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Saini A. K., Shin B. S., Nanda J., Hinnebusch A. G., 2015. Conformational changes in the P site and mRNA entry channel evoked by AUG recognition in yeast translation preinitiation complexes. Nucleic Acids Res. 43: 2293–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Dietrich F. S., 2005. Identification and characterization of upstream open reading frames (uORF) in the 5′ untranslated regions (UTR) of genes in Saccharomyces cerevisiae. Curr. Genet. 48: 77–87. [DOI] [PubMed] [Google Scholar]
- Zhou F., Walker S. E., Mitchell S. F., Lorsch J. R., Hinnebusch A. G., 2014. Identification and characterization of functionally critical, conserved motifs in the internal repeats and N-terminal domain of yeast translation initiation factor 4B (yeIF4B). J. Biol. Chem. 289: 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Edelman G. M., Mauro V. P., 2001. Transcript leader regions of two Saccharomyces cerevisiae mRNAs contain internal ribosome entry sites that function in living cells. Proc. Natl. Acad. Sci. USA 98: 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Sobolev A. Y., Wek R. C., 1996. Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J. Biol. Chem. 271: 24989–24994. [DOI] [PubMed] [Google Scholar]
- Zid B. M., O’Shea E. K., 2014. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514: 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel-Thropp P., Yang M. C., Machado L., Clarke S., 2000. A novel post-translational modification of yeast elongation factor 1A. Methylesterification at the C terminus. J. Biol. Chem. 275: 37150–37158. [DOI] [PubMed] [Google Scholar]
- Zuk D., Jacobson A., 1998. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 17: 2914–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]