Summary
The MYC oncogene encodes MYC, a transcription factor that binds the genome through sites termed E-boxes (5′-CACGTG-3′), which are identical to the binding sites of the heterodimeric CLOCK-BMAL1 master circadian transcription factor. Hence, we hypothesized that ectopic MYC expression perturbs the clock by deregulating E-box-driven components of the circadian network in cancer cells. We report here that deregulated expression of MYC or N-MYC disrupts the molecular clock in vitro by directly inducing REV-ERBα to dampen expression and oscillation of BMAL1, and this could be rescued by knockdown of REV-ERB. REV-ERBα expression predicts poor clinical outcome for N-MYC-driven human neuroblastomas that have diminished BMAL1 expression, and reexpression of ectopic BMAL1 in neuroblastoma cell lines suppresses their clonogenicity. Further, ectopic MYC profoundly alters oscillation of glucose metabolism and perturbs glutaminolysis. Our results demonstrate an unsuspected link between oncogenic transformation and circadian and metabolic dysrhythmia, which we surmise to be advantageous for cancer.
Graphical Abstract
Introduction
The molecular clock regulates rhythmic gene expression and synchronizes cellular metabolism with food availability and the 24-hour sleep-wake cycle (Asher and Schibler, 2011; Bass and Takahashi, 2010; Sahar and Sassone-Corsi, 2012). Cancer cells are known for having a growth advantage from deregulated metabolism, such as through MYC-driven aerobic glycolysis or glutaminolysis that supports biosynthesis and cell growth, but it is unknown how or whether the MYC oncogenic transcription factor could disrupt and uncouple metabolism from the influence of the molecular clock (Cantor and Sabatini, 2012).
Oncogenic expression of the MYC family of transcription factors lies at the center of many human cancers (Dang, 2012; Eilers and Eisenman, 2008). The MYC (c-MYC) gene is frequently amplified in human cancers (Ciriello et al., 2013), and the highly related MYCN (N-MYC) is amplified in poor-prognosis neuroblastoma (Maris, 2010). MYC heterodimerizes with its partner MAX to bind E-box sequences or non-canonical sites in target gene promoters or enhancers and regulate transcription (Wolf et al., 2015). Aberrant expression of MYC results in upregulation of many target genes, particularly those involved in glycolysis, oxidative metabolism, glutamine metabolism, mitochondrial and ribosomal biogenesis, cell growth and cell cycle regulation (Eilers and Eisenman, 2008). Intriguingly, while MYC has not been previously implicated in control or regulation of circadian rhythm in cancer cells, E-box elements bound by MYC drive expression of metabolic genes, such as nicotinamide phosphoribosyltransferase (NAMPT) (Menssen et al., 2012), that are also bound by the central molecular clock heterodimeric transcription factor CLOCK-BMAL1, which controls rhythmic circadian gene expression (Dang, 2012; Hogenesch et al., 1998; Ramsey et al., 2009). The molecular clock is driven by a key feedback loop, in which CLOCK-BMAL1 induces PER and CRY proteins, which in turn form inhibitory complexes with CLOCK-BMAL1 (Sato et al., 2006). CLOCK-BMAL1 also induces REV-ERBα and REV-ERBβ (Bugge et al., 2012; Cho et al., 2012), which transcriptionally repress BMAL1 at retinoic acid receptor-related orphan receptor elements (ROREs), thereby constituting a second important interlocking feedback loop (Preitner et al., 2002).
Circadian rhythms are ~24-hour cycles that control metabolic gene expression and cell division as well as other physiological processes, such as blood pressure and body temperature, coupling food availability and sleep with cellular metabolism (Bass and Takahashi, 2010). In mammals, the central clock is located within the suprachiasmatic nucleus (SCN) of the brain that senses light and synchronizes peripheral clocks through humoral, neuronal, and behavioral cues (Bass and Takahashi, 2010). The peripheral clocks, which are present in nearly every cell in the body (Bass and Takahashi, 2010), operate independently from the central clock and can be entrained by feeding in addition to temperature and synchronization cues from the central clock.
Mutations of circadian genes occur in many cancers, but their functional consequences are not well understood (Savvidis and Koutsilieris, 2012). Two key circadian genes, PER1 and PER2, have been identified as putative tumor suppressors in salivary gland, gastro-intestinal system, breast tissue, and the immune compartment (Savvidis and Koutsilieris, 2012; Zhao et al., 2014). Loss of PER2 function predisposed genetically engineered mice to spontaneous lymphomagenesis, but mouse strain specificity was not documented (Fu et al., 2002). Hematologic malignancies such as diffuse large B-cell lymphoma, chronic lymphocytic leukemia, and acute myeloid leukemia show strong downregulation of the central circadian regulator BMAL1 (Taniguchi et al., 2009). Indeed, recent studies identify BMAL1 as a putative tumor suppressor (Savvidis and Koutsilieris, 2012; Yeh et al., 2014). Ovarian cancers have altered circadian gene expression and oscillation compared to normal tissue, but the functional significance and mechanism for these alterations were not established (Tokunaga et al., 2008). Still other studies found circadian gene expression alterations in cancers such as pancreatic, prostate, and glioma (Savvidis and Koutsilieris, 2012). Importantly, altered circadian rhythm may affect cancer treatment outcomes, as studies in mice and humans have identified circadian-dependent toxicity in more than 40 anti-cancer agents, including antimetabolites, mitosis inhibitors, alkylating agents, and cytokines (Levi et al., 2010). Thus, altered circadian gene expression occurs broadly in cancer; however, it is not known whether MYC could contribute to disruption of circadian gene expression.
Given that the target core consensus sequence of CLOCK-BMAL1 appears identical to those recognized by MYC, we have previously hypothesized that oncogenic MYC could disrupt the molecular clock in vitro through dysregulating E-box-driven components of the clock machinery and thereby perturb circadian influence of central bioenergetic metabolism (Dang, 2012). Here, we report that MYC directly activates expression of multiple repressors of the clock and disrupts the circadian oscillation of the BMAL1::luciferase reporter and BMAL1 (ARNTL) mRNA. Through loss-of-function studies, we found that REV-ERB induction by MYC functionally contributes to the repression and dampening of BMAL1 oscillation. Further, N-Myc also induces REV-ERBα (NR1D1), whose elevated expression suppresses BMAL1 and portends poor clinical outcome for neuroblastoma patients. We also found that ectopic expression of BMAL1 reduces the clonogenicity of neuroblastoma cell lines, supporting the putative tumor suppressive function of BMAL1 (Yeh et al., 2014). Importantly, we found that MYC not only disrupts the circadian clock, but it also profoundly perturbs circadian glucose metabolism and glutaminolysis, favoring MYC-induced biosynthesis for cell growth. Our findings indicate that MYC and N-MYC, which are frequently deregulated in human cancers, can disrupt the cellular molecular clock and significantly affect cellular metabolism.
Results
MYC disrupts circadian oscillation of BMAL1 levels in cultured cells
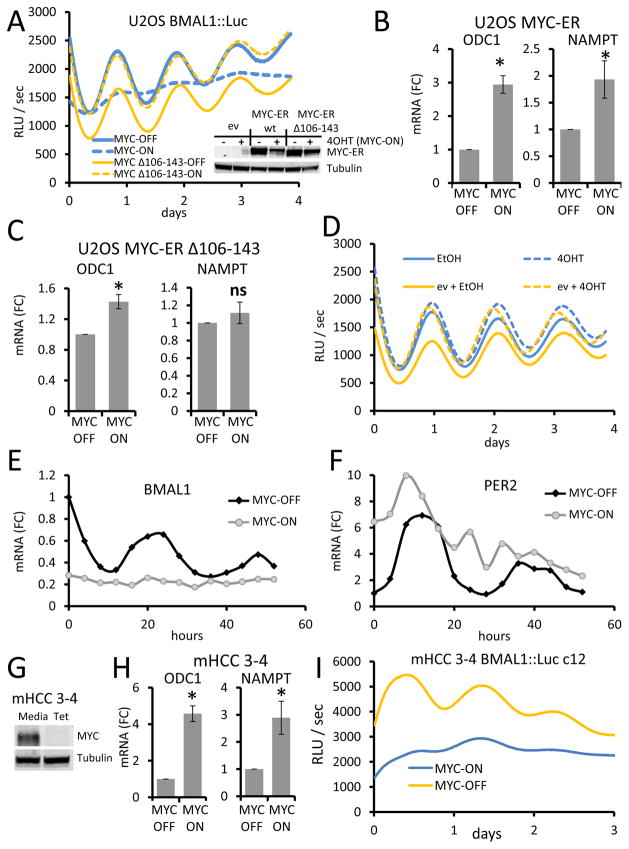
We sought to determine whether ectopic MYC expression could disrupt the cellular clock machinery. We used the human osteosarcoma cell line U2OS, which is a commonly used line to study circadian rhythm and has been engineered to stably express luciferase under the control of the BMAL1 promoter such that circadian oscillation of luciferase can be observed with the LumiCycle luminometer in real time after synchronizing the cells with dexamethasone (Baggs et al., 2009; Balsalobre et al., 2000). As U2OS cells have relatively low endogenous MYC (Walz et al., 2014), we stably expressed inducible wild-type MYC-ER™ (estrogen receptor Tamoxifen Mutant) or a mutant MYC-ER™ Δ106–143, which lacks the transactivation MYC Box II domain (Littlewood et al., 1995) (Figure 1A inset). When stimulated with 4-hydroxytamoxifen (4OHT, MYC-ON), MYC-ER translocated into nucleus and upregulated the MYC targets ornithine decarboxylase 1 (ODC1) and NAMPT, demonstrating the increased activity of MYC (Figure 1B) (Menssen et al., 2012; Walz et al., 2014). In contrast, cells expressing mutant MYC-ER Δ106–143 showed only slight upregulation of ODC1 and no upregulation of NAMPT in response to 4OHT stimulation (Figure 1C). While vehicle-treated (EtOH-treated, MYC-OFF) dexamethasone-synchronized U2OS MYC-ER cells showed 24-hour oscillation of luciferase, activation of MYC-ER with 4OHT profoundly ablated this oscillation (Figures 1A, S1A). By contrast, this oscillation was not diminished in MYC-ER Δ106–143-expressing cells treated with 4OHT (Figures 1A, S1A) or in cells which were uninfected or transduced with control empty vector (Figures 1D, S1B). Collectively, these observations documented that wild-type MYC activity dampens the oscillating circadian activity of the BMAL1 promoter.
Figure 1. MYC disrupts circadian rhythm.
A. U2OS BMAL1::Luc cells stably expressing either human wild-type MYC-ER™ or human MYC-ER™ Δ106–143 (as visualized by immunoblot, inset) were treated with 4-hydroxytamoxifen (4OHT, MYC-ON) to activate MYC-ER™ or ethanol (EtOH, MYC-OFF) control and synchronized with dexamethasone. BMAL1 promoter activity luminescence was continuously measured (every 10 min) in a LumiCycle™ luminometer. Data are representative of more than three experiments. B, C. Expression of ornithine decarboxylase (ODC) or NAMPT in U2OS MYC-ER™ (B) or MYC-ER™ Δ106–143 cells as determined by RT-PCR, normalized to Beta-2-microglobulin (β2M). Data are shown as means + SD (n = 4). mRNA (FC) = Fold Change. D. U2OS BMAL1::Luc cells expressing pBabe-Zeo empty vector (ev) or mock infected were cultured with 4OHT or ethanol, synchronized with dexamethasone, and monitored in LumiCycle™ luminometer. Data are representative of three or more experiments. E. Endogenous BMAL1 or (F) PER2 mRNA expression in synchronized U2OS MYC-ER cells was determined by RT-PCR, normalized to β2M. Data are representative of two or more experiments. G. Immunoblot of tetracycline regulated MYC in mHCC 3–4 cells. Tubulin serves as loading control. H. Expression of ODC and NAMPT in mHCC 3–4 cells as determined by RT-PCR. Data are shown as means + SD (n = 5). I. mHCC 3–4 BMAL1::Luc cell clone (c12) treated with tetracycline (tet, MYC-OFF) or control (−tet, MYC-ON) and synchronized with dexamethasone was monitored as in (A). RLU = relative light units. For B, C, and H, *p < 0.05 by Student’s t test of MYC-ON samples relative MYC-OFF samples.
Next, we collected RNA every four hours and assessed endogenous circadian gene expression by qPCR in synchronized U2OS cells. Endogenous BMAL1 (ARNTL) mRNA level was strongly decreased and oscillation significantly blunted by MYC activation (Figure 1E). The endogenous core circadian factor PER2 mRNA level was increased, but oscillation was also altered by MYC (Figure 1F). The fact that PER2 continued to oscillate with a disrupted phase is consistent with previously published results showing that static BMAL1 mRNA expression alters but does not eliminate PER oscillation (McDearmon et al., 2006). These data indicated that MYC affects not only the BMAL1 promoter-reporter, but also endogenous cycling of circadian genes.
To determine whether MYC-mediated disruption of circadian oscillation of BMAL1 promoter function could be restored in cancer, we used the mouse hepatocellular carcinoma cell line ‘mHCC 3–4’ derived from a liver tumor with a conditional c-MYC Tet-Off expression system (Shachaf et al., 2004; Xiang et al., 2015) (Figure 1G). Ectopic MYC predictably increased levels of Odc1 and Nampt mRNAs (Figure 1H). As previously done in U2OS cells (Baggs et al., 2009), we produced MYC-transformed mHCC 3–4 cells that stably express BMAL1::Luc. By LumiCycle luminometer analysis, circadian oscillation was restored with diminished ectopic MYC expression in this mouse liver cancer derived cell line in multiple clones (Figures 1I, S1C and S1D). Reduction of ectopic MYC expression also appeared to restore the expression of BMAL1 (Arntl) mRNA at later time points (Figure S1E) and reduced Per2 mRNA levels (Figure S1F). These observations collectively indicated that MYC overexpression alters circadian oscillation in different cell types and that diminished MYC expression in a MYC-driven tumor-derived liver cancer cell line could partially restore BMAL1 promoter activity oscillation.
MYC upregulates circadian repressor genes and downregulates BMAL1 expression
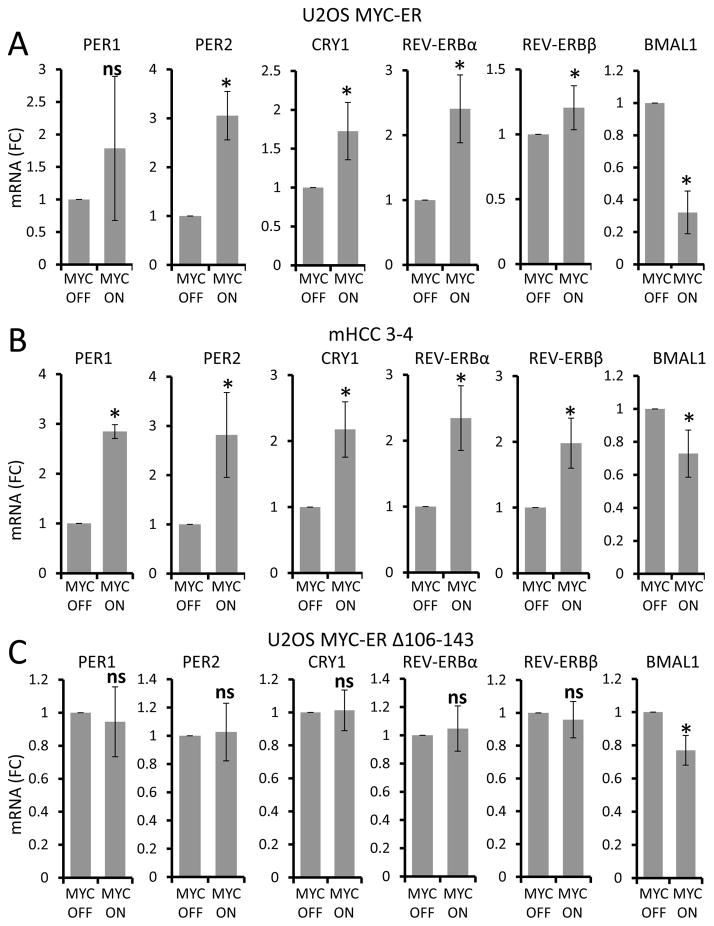
To delineate the mechanism by which MYC disrupts BMAL1 oscillation, we hypothesized that MYC transactivates E-box-driven BMAL1 repressors, such as PER and CRY, REV-ERBα (NR1D1) or REV-ERBβ (NR1D2) (Bass, 2012; Bass and Takahashi, 2010). We observed that MYC significantly upregulated levels of PER2, CRY1, REV-ERBα, and REV-ERBβ mRNAs in both U2OS MYC-ER (Figure 2A) and mHCC 3–4 (Figure 2B) cells. Some variation was observed between cell lines, as Per1 was upregulated by MYC in mHCC 3–4 but not in U2OS MYC-ER. In contrast to wild-type MYC, the inactive MYC-ER Δ106–143 mutant did not show significant upregulation of any circadian genes upon treatment with 4OHT, and slight downregulation of steady-state BMAL1 expression in U2OS cells (Figure 2C). To determine whether PER, CRY, REV-ERBα, and REV-ERBβ behave as direct MYC target genes, we analyzed publicly available MYC chromatin immunoprecipitation (ChIP) data from multiple cancer cell lines and U2OS cells with inducible MYC, and we also performed promoter studies with U2OS and mHCC (see Supplemental Experimental Information and Figures S2 and S3A–C). We found that each of these genes, with the exception of BMAL1 (ARNTL), could be induced and bound by MYC and hence behave as direct MYC targets in multiple cell lines. These observations suggested that MYC could induce the expression of negative regulators of BMAL1, which can in turn disrupt the molecular clock in vitro.
Figure 2. MYC upregulates circadian repressor genes.
A–C. Expression of clock factor genes (indicated above each graph) in (A) U2OS MYC-ER cells treated with either 4OHT (MYC-ON) or EtOH (MYC-OFF) control for 24 h. (B) mHCC 3–4 cells treated with (+Tet, MYC-OFF) or without (−Tet, MYC-ON) tetracycline for 24 h, or (C) U2OS MYC-ER™ Δ106–143 cells treated with either 4OHT (MYC-ON) or EtOH (MYC-OFF) control for 24 h. mRNA expression determined by RT-PCR was normalized to β2M expression. mRNA (FC) = Fold Change. Means and SDs from at least three experiments are shown. *p < 0.05 by Student’s t test of 4OHT (MYC-ON) samples relative to EtOH (MYC-OFF) samples or −Tet (MYC-ON) samples relative to +Tet (MYC-OFF) samples, ns = not significant.
REV-ERBa and β are necessary for disruption of circadian rhythm by MYC
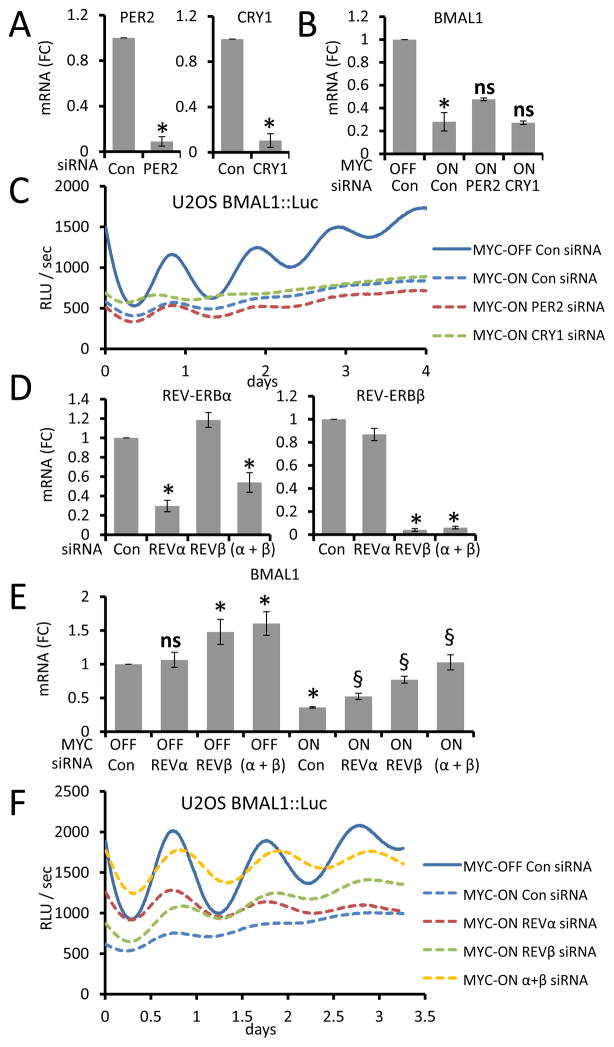
To delineate which BMAL1 repressors were responsible for disruption of circadian rhythm, we used siRNAs against the repressors to determine whether diminished expression could rescue MYC-mediated disruption of BMAL1-promoter circadian rhythm. PER and CRY negatively regulate circadian rhythm by repressing CLOCK-BMAL1 activity (Bass, 2012; Bass and Takahashi, 2010). We used siRNAs targeting CRY1 and PER2, which were consistently induced by MYC in multiple cell lines (Figure 2), in U2OS cells expressing MYC-ER to determine if they were responsible for the downregulation of BMAL1 and disruption of circadian oscillation by MYC. While we observed a significant reduction in the expression of each gene after siRNA treatment (Figure 3A), neither PER2 nor CRY1 knockdown significantly rescued either BMAL1 mRNA expression suppressed by MYC (Figure 3B) or the blunted BMAL1::Luc oscillation in MYC-ER-activated U2OS cells (Figure 3C). These data suggest that while PER2 and CRY1 behave as MYC targets, they may not be functionally responsible for disruption of BMAL1::Luc oscillation by MYC in U2OS cells. However, MYC’s induction of PER2 and CRY1 may still alter the molecular clock, because of their effects on BMAL1 protein activity rather than BMAL1 mRNA. Further, their reduction by siRNA to non-basal level could confound the interpretation of their roles in the suppression of BMAL1::Luc by MYC.
Figure 3. REV-ERBa and β are necessary for MYC disruption of BMAL1 promoter oscillation.
A. mRNA levels (normalized to expression of β2M) for PER2 or CRY1 determined by RT-PCR in U2OS MYC-ER cells transfected with either non-targeting siRNA (Con), siRNA against PER2, or CRY1 for 48 h with 4OHT (MYC-ON) being added to cells 24 hours after transfection. Means and SDs from at least three experiments are shown. *p < 0.05 for Con vs. siPER2 or siCRY1. mRNA (FC) = Fold Change. B. BMAL1 mRNA expression (normalized to expression of β2M) in U2OS MYC-ER transfected as described in (A) with siRNAs indicated at the bottom. *p < 0.05 by Student’s t test for MYC-ON vs. MYC-OFF. ns = not significant. C. U2OS MYC-ER cells were transfected with siRNAs as indicated in the figure, synchronized with dexamethasone and monitored using a LumiCycle™ luminometer. Data are representative of two or more experiments. D. mRNA levels (normalized to expression of β2M) for REV-ERBα or REV-ERBβ determined by RT-PCR in U2OS MYC-ER cells transfected with either non-targeting siRNA (Con), siRNA against REV-ERBα, REV-ERBβ, or both REV-ERBs (α+β) for 48 h with 4OHT (MYC-ON) being added to cells 24 hours after transfection. Means and SDs from at least three experiments are shown. *p < 0.05 for siREV vs. control. E. BMAL1 mRNA levels (normalized to expression of β2M) in U2OS MYC-ER transfected with the indicated siRNAs (bottom) as described in (D) and cultured with 4OHT (MYC-ON) or EtOH (MYC-OFF) after 24 hours. Means and SDs from at least three experiments are shown. *p < 0.05 by Student’s t test for siREV or Con MYC-ON vs. Con MYC-OFF. §, p< 0.05 siREV vs. Con MYC-ON. F. U2OS MYC-ER cells transfected with the indicated siRNAs for 48 hours, cultured with 4OHT (MYC-ON) or EtOH (MYC-OFF), synchronized with dexamethasone and monitored as in (C). Data are representative of two or more experiments. RLU = relative light units.
REV-ERBα is a transcriptional repressor that blunts BMAL1 expression and oscillation in U2OS cells (Preitner et al., 2002; Relogio et al., 2011). Further, overexpression of REV-ERBα in transgenic mouse liver suppressed BMAL1 levels and oscillation in vivo (Kornmann et al., 2007). REV-ERBα closely cooperates with REV-ERBβ to regulate gene expression and occupy promoter elements; hence, elimination of both α and β profoundly disrupted circadian rhythm while elimination of REV-ERBα alone only had mild effects on circadian oscillation (Bugge et al., 2012; Cho et al., 2012). Thus, to determine whether MYC disrupted circadian rhythm through REV-ERB genes, we targeted either REV-ERBα, REV-ERBβ, or both genes with siRNAs in U2OS MYC-ER (Figure 3D). While siRNA knockdown of either REV-ERBα or REV-ERBβ increased BMAL1 expression in MYC-activated cells, knockdown of both genes together led to a complete rescue of BMAL1 expression in the presence of MYC (Figure 3E). Note that REVERB knockdown in MYC-OFF cells also increased basal BMAL1 mRNA levels as expected (Figure 3E). Knockdown of REV-ERBα in mHCC 3–4 cells also led to a significant rescue in BMAL1 mRNA expression (Figure S3D). Importantly, knocking down both REV-ERB mRNAs substantially rescued the MYC-ER-mediated dampened expression and oscillation of BMAL1::Luc in U2OS cells, while individual REV-ERB gene knock down could partially rescue the dampening effects of MYC (Figure 3F). These data suggested that MYC’s disruption of circadian rhythm is largely carried out through MYC-mediated upregulation of REV-ERB transcription. In fact, we also observed that high MYC expression in a panel of human-derived T-ALL (T-cell acute lymphoblastic leukemia) lines correlates with increased REV-ERBα expression (Figures S3E and S3F).
N-MYC disrupts circadian gene expression and oscillation
It has been shown previously that c-MYC and N-MYC, encoded by the MYCN gene, have overlapping functions, targets, and binding motifs (Malynn et al., 2000). Given the functional similarities between MYC and N-MYC, we sought to determine whether N-MYC could also induce REV-ERBα expression like MYC. To study N-MYC, we used the human neuroblastoma cell lines Shep N-MYC-ER and SKNAS N-MYC-ER (Ushmorov et al., 2008; Valentijn et al., 2005). Both Shep and SKNAS lack amplified N-MYC, but when ectopic N-MYC-ER was activated with 4OHT (MYC-ON), the canonical Myc targets ODC1 and NAMPT were induced (Figures S4A–B). N-MYC activation also resulted in strong upregulation of REV-ERBα and downregulation of BMAL1 (Figures 4A, S4B). PER1, PER2, and CRY1 were similarly upregulated in the presence of 4OHT, suggesting that N-MYC perturbs circadian gene regulation like MYC (Figures S4A–B). These data suggest that inducible N-MYC similarly disrupts circadian gene expression in two different neuroblastoma cell lines.
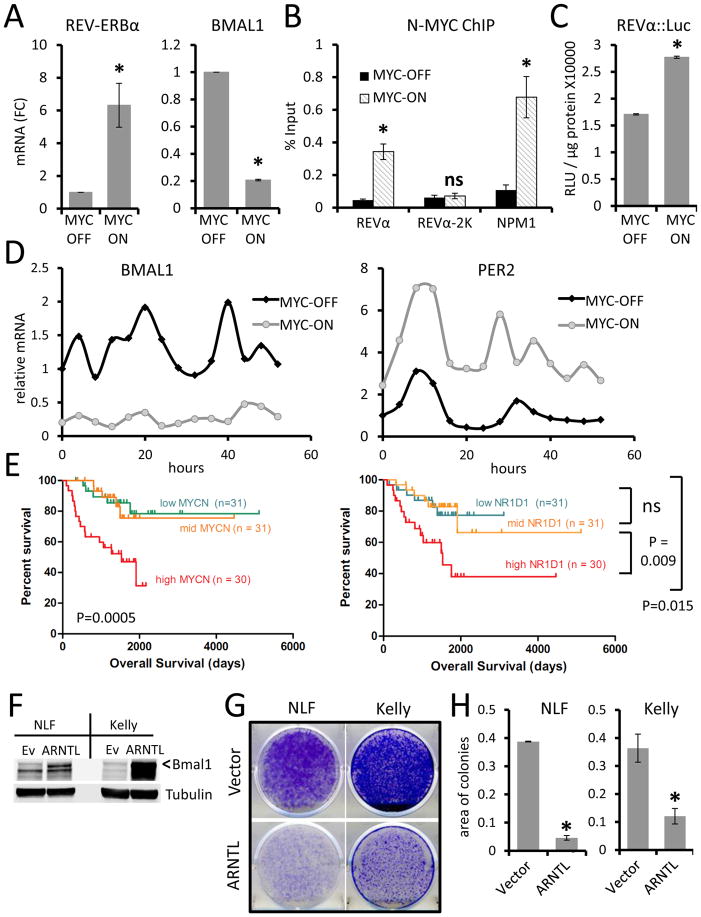
Figure 4. N-MYC directly upregulates REV-ERBa and disrupts circadian rhythm.
A. REV-ERBα and BMAL1 mRNA expression (normalized to expression of β2M) determined by RT-PCR in Shep N-MYC-ER expressing neuroblastoma cells with MYC-ON or MYC-OFF. mRNA (FC) = Fold Change. Means and SDs from at least three experiments are shown. *p < 0.05 by Student’s t test for MYC-ON vs. MYC-OFF. B. Chromatin immunopreciptitation (ChIP) analysis with anti-N-MYC antibody in MYC-ON vs. MYC-OFF in Shep N-MYC-ER cells. ChIP signals (% input) are shown for the Rev-erbα promoter E-box region (REVα, 81 bp region), 2Kb upstream of REV-ERBα transcription start site (REVα-2k), and the promoter of the canonical MYC target nucleophosmin 1 (NPM1). Means and SDs from technical triplicates are shown. *p < 0.05 by Student’s t test for MYC-ON vs. MYC-OFF. Data are representative of three or more experiments. C. Activity of the REV-ERBα promoter::luciferase construct (−1156 to +31 region) in Shep N-MYC-ER-expressing cells expressing under MYC-ON vs. MYC-OFF conditions. RLU: relative light unit. Means and SDs from triplicate samples are shown, and *p < 0.05 by Student’s t test for MYC-ON vs. MYC-OFF. Data are representative of three or more experiments. D. Time-series (every 4h) expression of BMAL1 (left panel) or PER2 (right panel) mRNA (relative to β2M) determined by RT-PCR in synchronized Shep N-MYC-ER-expressing cells with MYC-ON or MYC-OFF. Data are representative of two or more experiments. E. Kaplan-Meier survival analysis of patients with neuroblastoma expressing different levels of MYCN or (REV-ERBα) NR1D1 analysis grouped by tertiles based on individual gene expression. Log rank p-values shown. F. Ectopic expression of BMAL1 (ARNTL, indicated by <) in high MYCN neuroblastoma cell lines NLF and Kelly determine by immunoblotting Tubulin serves as loading control. G. Colony suppression assay of cells treated with 1mg/ml G418 showed that ectopic expression of ARNTL suppressed colony formation capacities of NLF and Kelly cells as compared with vector control (ev). H. Quantitation of colony suppression by ARNTL determined by area of colonies on culture plates measured by ImageJ. n = 3, error bars represent SD. *p <0.05 by Student’s t-test.
Corroborating the findings with the N-MYC-ER models, REV-ERBα expression also positively tracks with expression of N-MYC in three different neuroblastoma cell lines (Shep, NLF, and Kelly) with varying MYCN expression levels (Figure S4D). Like MYC in U2OS cells, N-MYC directly bound the promoter regions of REV-ERBα and NPM1 in Shep N-MYC-ER, but not with a region of DNA 2000 bases upstream from the REV-ERBα promoter start site (Figure 4B). Shep N-MYC-ER cells expressing a REV-ERBα::luciferase reporter also showed increased luciferase activity in the presence of 4OHT (Figure 4C), indicating that REV-ERBα is a direct target of N-MYC. Consistent with MYC upregulating REV-ERBα, Shep and SKNAS N-MYC-ER cells with active N-MYC-ER (MYC-ON) showed greatly diminished expression and disrupted oscillation of BMAL1 (Figures 4D, S4C), as well as increased amplitude and altered oscillation of PER2 (Figures 4D, S4C), consistent with results from U2OS and mHCC 3–4 cells. Importantly, this same trend was found in the three neuroblastoma cell lines (Figure S4D); we observed some circadian BMAL1 oscillation in the N-MYC single copy Shep cells, but not in the N-MYC amplified NLF or Kelly lines (Figure S4E).
Increased REV-ERBa and decreased BMAL1 expression correlate with high risk neuroblastoma, and BMAL1 functions as a tumor suppressor in MYCN amplified neuroblastoma cells
We analyzed gene expression from a collection of human primary neuroblastoma tumor samples to establish the potential clinical relevance of N-MYC regulation of REV-ERBα in human neuroblastoma (Wang et al., 2006). Consistent with previous findings, patients with the highest MYCN expressing neuroblastomas had the worst survival outcomes (Figure 4E). Intriguingly, elevated REV-ERBα alone also highly correlated with poor prognosis (Figure 4E), suggesting that REV-ERBα may play a role in the aggressiveness of neuroblastoma. REV-ERBα (NR1D1) expression was elevated in high-risk tumors, particularly those with MYCN amplification, compared to tumors classified as low or intermediate risk (Figure S5A) (Pugh et al., 2013). Conversely, BMAL1 (ARNTL) expression was lowest in the high-risk MYCN-amplified group (Figure S5B). Further, low BMAL1 or high REV-ERBα expression also correlated with poor prognosis in two different patient cohorts (Figures S5C–5E) (Kocak et al., 2013). Together, these data suggested that overexpressed N-MYC, as observed in cell lines and in primary patient samples, specifically upregulates REV-ERbα, which decreases BMAL1 expression and disrupts the molecular clock in neuroblastoma.
The decreased BMAL1 expression in high-risk neuroblastoma could be related to its putative tumor suppressor function (Yeh et al., 2014). To determine its tumor suppressive role, we overexpressed BMAL1 in NLF and Kelly neuroblastoma cell lines that have high N-MYC expression (Figure S4D) and assessed BMAL1’s effect on colony formation. BMAL1 was expressed by transient transfection of pCMV6-ARNTL (BMAL1), and the pCMV6 empty vector (Ev) served as control (Figures 4F–G). Cells were then selected with G418 and stained with crystal violet. As shown in Figure 4H, ectopic BMAL1 expression significantly suppressed colony formation in both NLF and Kelly cells, illustrating a tumor suppressive function of BMAL1.
MYC disrupts circadian oscillation of glycolysis and pAMPK and alters glutamine metabolism
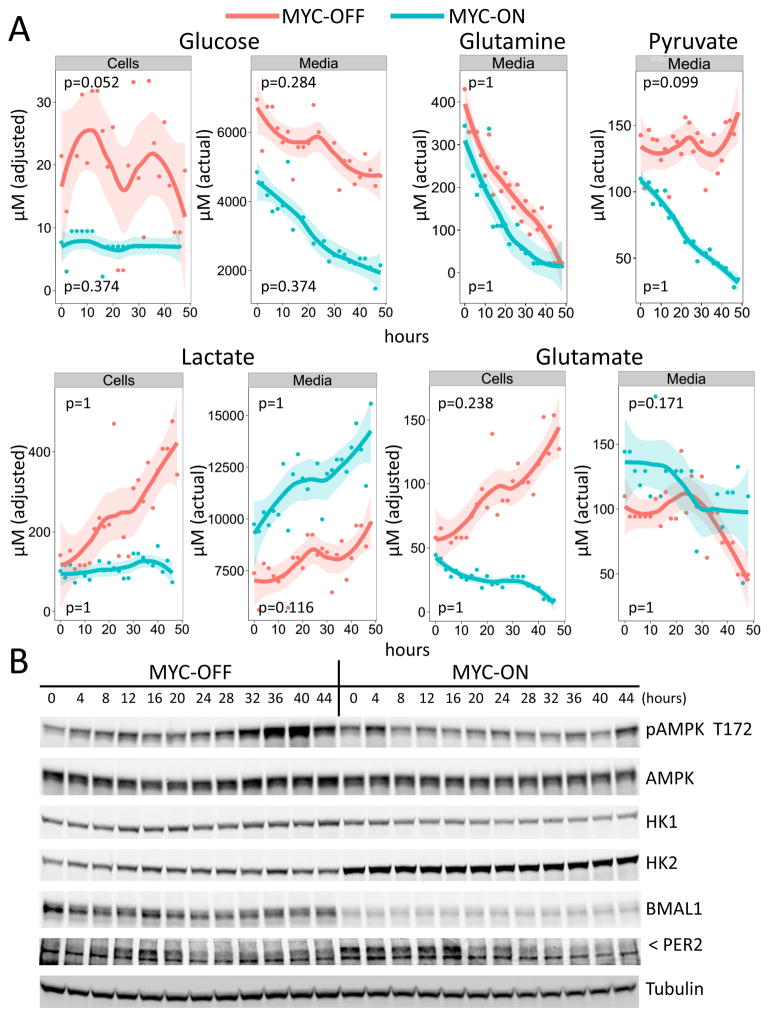
To determine whether MYC-mediated disruption of the circadian transcription factor network altered metabolism, we performed targeted metabolomics using NMR focusing on glucose, lactate, glutamine, glutamate and pyruvate levels in culture media and U2OS MYC-ER cells. In a time-series experiment at 2-hour intervals, media and cells from MYC-OFF (EtOH) or MYC-ON (4OHT) conditions were harvested for measurements of metabolites over a period of 48 hours, 24 hours after dexamethasone synchronization without changing the media. NMR peak intensities were quantified at each time point and were subjected to detrending and JTK-cycle analysis to identify oscillation among the selected metabolites (see Figure S6 for example NMR spectra) (Hughes et al., 2010; Weljie et al., 2006). As shown in Figure 5A, glucose was consumed at a higher rate in MYC-ON cells than MYC-OFF cells, which displayed phases of glucose consumption with slowed rates of loss from the media between 14 and 24 hours. Intriguingly, intracellular glucose levels oscillated in a circadian manner (JTK cycle time = 24 h; p-value = 0.052; ‘adjusted’ intracellular levels reflect concentrations in the extracts and not actual intracellular concentrations) peaking at about 12 and 36 hours. By contrast, while MYC-ON cells consumed glucose from the media at a higher rate, intracellular glucose levels were so low that circadian oscillation is not discernible, suggesting that glucose was consumed for biosynthesis as postulated (Murphy et al., 2013).
Figure 5. MYC suppresses circadian oscillation of glucose metabolism and alters glutamine metabolism in U2OS cells.
A. Cellular (Cells) or media metabolite (top labels) levels determined by NMR spectroscopy are shown for a time-series experiment with 2 h-intervals from U2OS with MYC-OFF (EtOH) or MYC-ON (4OHT), 24 hours after initial dexamethasone synchronization. Concentrations for metabolites represent the adjusted concentration for cells, based on extraction volume, or the actual concentration of media metabolites. For each metabolite profile, the time-series data were plotted using the JTK v2 algorithm after linear detrending and rhythmicity assessed via a non-parametric cosine function fit. The colored bands represent 95% confidence intervals from the locally weighted scatterplot smoothing function that generated the curves. P-values indicate the significance of the cosine wave fit to experimental data. Note that MYC-ON media 24 hour sample and MYC-ON cells 48 hour sample were excluded due to inconsistencies in sample measurements. B. Synchronized U2OS MYC-ER cells were harvested after treatment and lysates were processed for immunoblots of hexokinase 1 (HK1), hexokinase 2 (HK2), phospho-AMP kinase (pAMPK Thr172), AMP-kinase (AMPK), BMAL1, and PER2 (indicated by <). Tubulin serves as a loading control. Data are representative of two individual experiments.
Consistent with MYC-driven biosynthesis, MYC-ON U2OS cells had remarkably low intracellular lactate levels relative to MYC-OFF cells, while extracellular lactate levels in the media accumulated to high levels for MYC-ON cells compared with MYC-OFF cells (Figure 5A). It is notable that the apparent oscillation of media lactate levels in MYC-OFF cells paralleled changes in cellular glucose levels; however, the oscillation in the media did not reach statistical significance by JTK analysis. The significant disappearance of pyruvate from the medium of MYC-ON cells as compared with MYC-OFF cells (Figure 5A) supports the notion of MYC-driven biosynthesis, particularly since intracellular pyruvate levels were below the detection limit of this NMR approach. These patterns reveal a previously unsuspected circadian oscillation of glucose metabolism and its conversion to lactate in synchronized U2OS cells, which were disrupted by induction of MYC, suggesting that MYC-mediated disruption of the circadian clock perturbs metabolic oscillation.
We also examined glutamine and glutamate, and found that U2OS cells readily consumed glutamine, and the consumption was further heightened by the induction of MYC (Figure 5A). Intracellular glutamine levels were below detection limit in both MYC-ON and MYC-OFF cells (not shown). Notably, media glutamate levels were generally higher for MYC-ON cells compared to MYC-OFF cells. Intracellular glutamate decreased in MYC-ON cells while it accumulated in MYC-OFF cells, seemingly in a circadian fashion (which did not reach statistical significance by JTK analysis), suggesting glutamine and derived glutamate were consumed for biosynthesis in MYC-ON cells. These observations collectively illustrate that circadian fluctuation of key metabolites involved in glycolysis and glutaminolysis in U2OS cells was profoundly disrupted by MYC activation, which is known to drive biosynthesis for cell growth (Murphy et al., 2013).
To gain further insight into the bioenergetics of U2OS cells with different MYC activities, we analyzed phosphorylated AMPK (pAMPK T172) as a surrogate measure of cellular energy status. As shown in Figure 5B, pAMPK levels were higher, particularly at later time points, and oscillating in MYC-OFF cells as compared with MYC-ON cells. Oscillation of pAMPK levels was in-phase with intracellular glucose levels and appears to correlate with high intracellular lactate levels (glycolysis). pAMPK levels were at their lowest level when intracellular glucose levels were lowest (Figure 5B) in MYC-OFF cells. Intriguingly, the levels and oscillation of pAMPK were suppressed in MYC-ON cells suggesting that the bioenergetics status of MYC-driven U2OS cells was sustained by increased glutaminolysis and consumption of glucose. We also determined levels of hexokinases, HK1 and HK2, and found that both enzyme levels, particularly HK1, appear to oscillate (minimally) in phase with glucose consumption in MYC-OFF cells (Figure 5B). However, in MYC-ON cells, HK2, a known MYC target (Kim et al., 2007), was induced and ceased to oscillate. Note that these changes correspond to MYC-induced decreased BMAL and increased PER2 protein levels (Figure 5B). These observations demonstrate that, in addition to metabolic oscillation, the bioenergetic status of MYC-OFF U2OS cells also oscillated in a manner that is suppressed by ectopic MYC activity, which drives glycolysis and glutaminolysis for biosynthesis.
Discussion
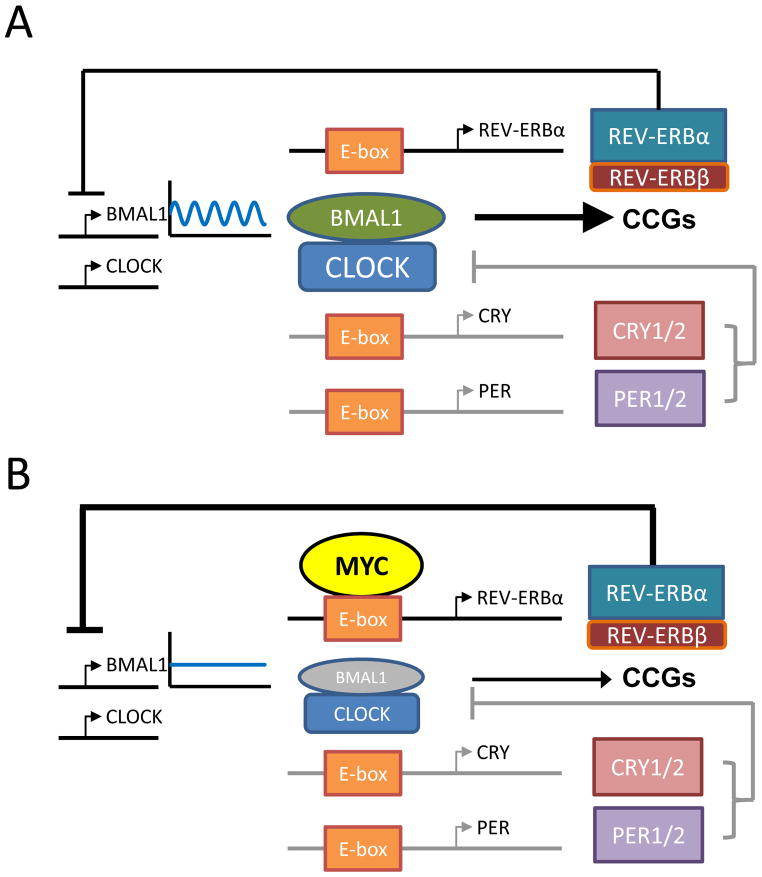
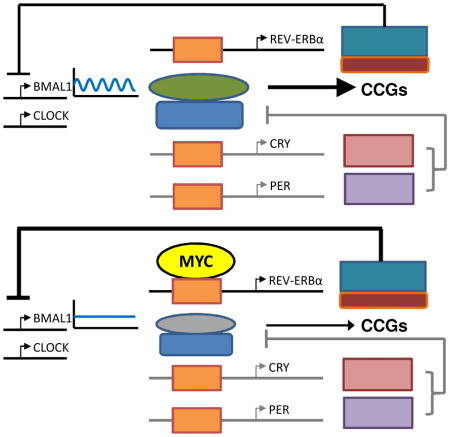
MYC and circadian rhythm are frequently altered in cancer (Sahar and Sassone-Corsi, 2009), but whether this altered circadian rhythm can be mechanistically linked to deregulated MYC transcriptional function in cancer has not been determined. Our work here documents the previously unsuspected role of oncogenic MYC in the disruption of the molecular clock and metabolic oscillation in vitro, providing a foundation for future in vivo studies. We found that oncogenic MYC disrupts circadian oscillation through upregulation of REV-ERBα and REVERBβ, which in turn dampen BMAL1 expression and oscillation (Figure 6); however, we have not excluded the possible roles of MYC-induced CRY and PER in MYC-mediated clock disruption. Specifically, it is notable that MYC increased PER2 mRNA and protein levels, while perturbing the circadian oscillation of PER2 expression (Figures 1F and 5B). We demonstrate that MYC directly bound to the promoter of REV-ERBα and upregulated its transcription, leading to lower levels of BMAL1 expression and loss of rhythmicity. These data are consistent with previous studies showing that ectopic REV-ERBα expression can dampen the oscillation of BMAL1, both in vitro and in vivo (Kornmann et al., 2007; Relogio et al., 2011). Our finding that MYC could occupy circadian E-boxes as previously proposed (Kondratov et al., 2006) is consistent with a study showing that the E-box binding helix-loop-helix leucine zipper transcription factor USF1 can also compete with CLOCK-BMAL1 to bind to circadian regulatory elements (Shimomura et al., 2013). However, we have not distinguished between E-box co-occupancy or competition between MYC and CLOCK-BMAL1.
Figure 6. Model of circadian rhythm disruption by MYC.
Summary diagram of normal molecular clock (top) molecular clock disrupted by Myc (bottom).
During the course of our studies, a recent report documented a role for MYC in disrupting the emergence of the circadian clock during differentiation of embryonic stem cells (Umemura et al., 2014). However, in these normal cells, MYC disruption of the circadian clock was dominated by a block in PER nuclear import rather than a direct effect on circadian E-boxes as we demonstrated in cancer cell lines. Our observations underscore the complexity of MYC-mediated transcription, which does not simply amplify expression of genes that are already expressed. Rather, MYC-mediated gene expression culminates from the intricate interplay between MYC and other transcription factors such as MIZ-1, which can result in time-dependent gene expression changes particularly when the molecular clock transcription factors are involved (Wolf et al., 2015).
While REV-ERBα was universally upregulated by MYC and N-MYC in all cell lines and databases we queried, REV-ERBβ upregulation was less consistent across these cell lines, despite also being predicted as a MYC target. Nonetheless, knockdown of both REV-ERBβ and REV-ERBα was required to substantially rescue BMAL1::luciferase amplitude and oscillation, which were inhibited by MYC. These observations are consistent with previous findings that REV-ERBα and REV-ERBβ cooperate closely to regulate transcription (Bugge et al., 2012; Cho et al., 2012), and, as such, both would have to be considered together when studying circadian gene oscillation.
Our observation that REV-ERBα expression induced by N-MYC correlates with poor prognosis for neuroblastoma contributes to the notion that REV-ERBα expression might be important in tumorigenesis. Notably, the REV-ERBα gene is co-amplified with ERBB2 in some breast cancers and may play an important role in the growth of the breast cancer cells (Kourtidis et al., 2010). REV-ERBα has not been previously recognized as an oncogene, but its suppression of BMAL1 could contribute to its tumorigenic potential. In particular, BMAL1 has been implicated as a tumor suppressor (Yeh et al., 2014) and our studies corroborate this notion and illustrate that low BMAL1 expression correlates with high-risk neuroblastoma and that ectopic expression of BMAL1 suppressed colony formation by two different high N-MYC neuroblastoma cell lines.
Given that circadian rhythm controls metabolic function, circadian oscillation therefore constrains metabolism at certain times of the day (Bass, 2012). Hence, perturbation of circadian gene expression, as is broadly found in cancer, could release the circadian restraints on cancer metabolism. MYC can elevate metabolic gene expression in transformed cells compared to normal cells (Dang, 2012), whose expression of metabolic genes is constrained by the molecular clock. We speculate that MYC induction of REV-ERBα, which in turn suppresses BMAL1, could lead to elevated, unconstrained expression of metabolic genes through clock disruption to provide an advantage to cancer cells. In this regard, we found for the first time that intracellular glucose metabolism in synchronized U2OS cells oscillated in a circadian fashion that was profoundly disrupted by MYC activation, which induced high levels of HK2 and increase glucose flux and lactate production. Our observations are consistent with a recent study showing circadian oscillation of NADH/NAD+ ratio in epidermal stem cells (Stringari et al., 2015). Significant changes in pAMPK, lactate, pyruvate, glutamine and glutamate in our studies are consistent with a role of activated MYC in driving biosynthesis, which correlates MYC’s ability to disrupt the circadian molecular clock. However, our current study does not define the interplay between MYC’s transcriptional disruption of the circadian clock machinery and its direct regulation of metabolic genes. Further studies will be required to parse the roles of MYC in genetic and metabolic regulation of circadian rhythm.
Because MYC and MYCN are amplified or their expressions are altered in at least 50% of cancers (Dang, 2012), our work suggests that a large group of cancers may have disrupted circadian molecular clocks and metabolism. While the potential tumorigenic advantage of an oncogene-disrupted circadian rhythm needs to be further established in the future through in vivo studies, we speculate that the differences between normal cellular circadian gene expression and the arrhythmic expression of circadian genes in cancer cells could potentially be exploited to optimize cancer therapy.
Abbreviated Experimental Procedures
Detailed Experimental Procedures are provided in the Supplemental Information. Standard molecular techniques such as PCR, immunoblotting, cell culture, transfection, and genomics studies are described in the Supplemental section.
Real-time luminescence monitoring
Lumicycle analysis of U2OS BMAL1::Luc cells was described previously (Baggs et al., 2009). Cell culture plates were cultured with beetle luciferin (Promega, Madison, WI, USA), sealed with high vacuum grease (Dow Corning, Midland, MI, USA), and measured for luminescence after dexamethasone synchronization every 10 minutes for at least 4 days on a Lumicycle™ luminometer, and presented as relative light units (RLU) per second (Actimetrics, Wilmette, IL, USA). The experiments were performed with technical triplicates and multiple biological replicates.
Time-series mRNA experiments
MYC was activated one day prior to collection. Experiment was performed as a ‘split timecourse’. 24 hours prior to the start of the experiment, half the cells were synchronized in dexamethasone, and at the start, the other half of the cells were synchronized. At each time point, two plates were harvested representing 0 and 24 hours +dex, 4 and 28, etc, to arrive at a 52 hour timecourse.
Generation of cell lines and clonogenic assay
Cells were transfected and selected as described in the Supplemental Information. For neuroblastoma clonogenic suppression, ARNTL or control expression vectors were transfected and cells were selected with G418 before staining with crystal violet. Plates were scanned and quantified with ImageJ, and representative images were uniformly contrasted.
siRNA
Human or mouse SMARTpool® siRNAs were purchased from Dharmacon (GE Healthcare, Lafayette, CO, USA). Cells were transfected with siRNAs using Lipofectamine 2000 or RNAiMAX (Life Technologies). Non-targeting siRNAs with same molar amount were used as negative control. For all experiments, a total of 60 nM siRNA were used. For experiments in which REV-ERBα and REV-ERBβ were used together in Figure 3, 30 nM of each siRNA were used.
Primary neuroblastoma survival data
For data in Figure 4, previously published data (Wang et al., 2006) for ninety-two human tumors was assessed for overall survival based on MYCN, NR1D1, and ARNTL expression (Figure S5C). Survival analyses were performed using the methods of Kaplan and Meier. Patients were divided into separate groups based on tertiles of individual mRNA expression. For overall-survival (OS), time was defined as the time from diagnosis until the time of death from disease or until the time of last contact if death did not occur. Patients who were alive were censored at the time last known alive. Log rank p-values < 0.05 were considered significant.
Cell and media extraction for NMR analysis
Snap frozen U2OS cell pellets were thawed and extracted. The aqueous fraction of an organic:aqueous extraction was dried and immediately prepared for NMR analysis as described in the Supplemental section using an Avance III HD 700 MHz NMR spectrometer from Bruker Biospin (Billerica, MA).
Quantitative analysis of metabolite oscillations
Output concentrations from the NMR analysis were imported into the R statistical framework (Team, 2015), and were noted to have significant temporal change in overall concentrations. Oscillations were assessed using the non-parametric JTK_CYCLE algorithm (Hughes et al., 2010), testing for period lengths between 22 and 26 hours corresponding to circadian events. The default cosine function was used to assess rhythmic patterns and significance.
Supplementary Material
Highlights.
The MYC and MYCN oncogenes disrupt the circadian molecular clock.
MYC directly activates negative regulators of BMAL1-CLOCK.
Constitutive activation of negative regulators, REV-ERBs, suspends the clock.
Clock suspension is associated with disruption of circadian metabolic oscillation.
Acknowledgments
We thank Michael Hogarty (University of Pennsylvania, Philadelphia, PA) for the Shep N-MYC-ER cells, Linda Valentijn (University of Amsterdam, Netherlands) for the SKNAS N-MYC-ER line, and Kathryn E. Wellen (University of Pennsylvania) for advice. We thank Brian Keith (University of Pennsylvania) for critical comments and suggestions on the manuscript. This work is partially supported by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) R01CA057341 (C.V.D), The Leukemia and Lymphoma Society LLS 6106-14 (C.V.D.), and the Abramson Family Cancer Research Institute. B.J.A, Z.E.S, and Z.E.W were supported by the NCI F32CA180370, F32CA174148, and F30CA200347, respectively. J.M.M contribution of neuroblastoma data from the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) project was supported by RC1MD004418.
Footnotes
The authors declare no competing financial interests.
Author Contributions
Conceptualization, B.J.A., A.L.H., C.V.D.; Methodology, B.J.A., A.L.H., A.V., D.I.B., M.C.S., M.A.L., D.W.F., J.B.H., A.M.W., C.V.D.; Formal Analysis, B.J.A., A.L.H., S.J.D., A.M.W.; Investigation, B.J.A., A.L.H., A.S., S.Y.K., Z.E.W., A.M.G., P.G-H., S.J.D., A.M.W.; Resources, Z.E.S., A.V., B.L., S.J.D., D.I.B., M.C.S., J.C.R., J.M.M., D.W.F., J.B.H., A.M.W., C.V.D.; Writing – Original Draft, B.J.A., A.L.H., C.V.D.; Writing – Review and Editing, B.J.A., A.L.H.,Z.E.S., Z.E.W., J.B.H., A.M.W., C.V.D.; Visualization, B.J.A., A.L.H., Z.E.S., S.J.D., A.M.W., C.V.D.; Supervision, C.V.D.; Funding Acquisition, B.J.A., C.V.D.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. Network features of the mammalian circadian clock. PLoS Biol. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of biological rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D, Roels F, Theissen J, Westermann F, Deubzer H, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell death & disease. 2013;4:e586. doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 2006;20:530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtidis A, Jain R, Carkner RD, Eifert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Res. 2010;70:1783–1792. doi: 10.1158/0008-5472.CAN-09-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, de Alboran IM, O’Hagan RC, Bronson R, Davidson L, DePinho RA, Alt FW. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Luscher B, Larsson LG, Hermeking H. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci U S A. 2012;109:E187–196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TA, Dang CV, Young JD. Isotopically nonstationary 13C flux analysis of Myc-induced metabolic reprogramming in B-cells. Metab Eng. 2013;15:206–217. doi: 10.1016/j.ymben.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relogio A, Westermark PO, Wallach T, Schellenberg K, Kramer A, Herzel H. Tuning the mammalian circadian clock: robust synergy of two loops. PLoS Comput Biol. 2011;7:e1002309. doi: 10.1371/journal.pcbi.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol Metab. 2012;23:1–8. doi: 10.1016/j.tem.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvidis C, Koutsilieris M. Circadian rhythm disruption in cancer biology. Mol Med. 2012;18:1249–1260. doi: 10.2119/molmed.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Shimomura K, Kumar V, Koike N, Kim TK, Chong J, Buhr ED, Whiteley AR, Low SS, Omura C, Fenner D, et al. Usf1, a suppressor of the circadian Clock mutant, reveals the nature of the DNA-binding of the CLOCK:BMAL1 complex in mice. Elife. 2013;2:e00426. doi: 10.7554/eLife.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C, Wang H, Geyfman M, Crosignani V, Kumar V, Takahashi JS, Andersen B, Gratton E. In vivo single-cell detection of metabolic oscillations in stem cells. Cell reports. 2015;10:1–7. doi: 10.1016/j.celrep.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Fernandez AF, Setien F, Ropero S, Ballestar E, Villanueva A, Yamamoto H, Imai K, Shinomura Y, Esteller M. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 2009;69:8447–8454. doi: 10.1158/0008-5472.CAN-09-0551. [DOI] [PubMed] [Google Scholar]
- Team, R.C. R: A language and environment for statistical computing. 2015 http://www.R-project.org.
- Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira J, Higashimoto M, Mashiko M, Ito K, Niikura H, Takenoshita S, Yaegashi N. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand. 2008;87:1060–1070. doi: 10.1080/00016340802348286. [DOI] [PubMed] [Google Scholar]
- Umemura Y, Koike N, Matsumoto T, Yoo SH, Chen Z, Yasuhara N, Takahashi JS, Yagita K. Transcriptional program of Kpna2/Importin-alpha2 regulates cellular differentiation-coupled circadian clock development in mammalian cells. Proc Natl Acad Sci U S A. 2014;111:E5039–5048. doi: 10.1073/pnas.1419272111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushmorov A, Hogarty MD, Liu X, Knauss H, Debatin KM, Beltinger C. N-myc augments death and attenuates protective effects of Bcl-2 in trophically stressed neuroblastoma cells. Oncogene. 2008;27:3424–3434. doi: 10.1038/sj.onc.1211017. [DOI] [PubMed] [Google Scholar]
- Valentijn LJ, Koppen A, van Asperen R, Root HA, Haneveld F, Versteeg R. Inhibition of a new differentiation pathway in neuroblastoma by copy number defects of N-myc, Cdc42, and nm23 genes. Cancer Res. 2005;65:3136–3145. doi: 10.1158/0008-5472.CAN-04-2469. [DOI] [PubMed] [Google Scholar]
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D, Seiser E, Jagannathan J, Shusterman S, Bansal M, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Analytical chemistry. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- Wolf E, Lin CY, Eilers M, Levens DL. Taming of the beast: shaping Myc-dependent amplification. Trends Cell Biol. 2015;25:241–248. doi: 10.1016/j.tcb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Stine ZE, Xia J, Lu Y, O’Connor RS, Altman BJ, Hsieh AL, Gouw AM, Thomas AG, Gao P, et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. The Journal of clinical investigation. 2015;125:2293–2306. doi: 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CM, Shay J, Zeng TC, Chou JL, Huang TH, Lai HC, Chan MW. Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int J Oncol. 2014;45:2101–2107. doi: 10.3892/ijo.2014.2627. [DOI] [PubMed] [Google Scholar]
- Zhao H, Zeng ZL, Yang J, Jin Y, Qiu MZ, Hu XY, Han J, Liu KY, Liao JW, Xu RH, et al. Prognostic relevance of Period1 (Per1) and Period2 (Per2) expression in human gastric cancer. International journal of clinical and experimental pathology. 2014;7:619–630. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.