Abstract
Flaviviruses include a highly diverse group of arboviruses with a global distribution and a high human disease burden. Most flaviviruses cycle between insects and vertebrate hosts; thus, they are obligated to use different cellular machineries for their replication and mount different mechanisms to evade specific antiviral responses. In addition to coding for viral proteins, the viral genome contains signals in RNA structures that govern the amplification of viral components and participate in triggering or evading antiviral responses. In this review, we focused on new information about host-specific functions of RNA structures present in the 3’ untranslated region (3’ UTR) of flavivirus genomes. Models and conservation patterns of RNA elements of distinct flavivirus ecological groups are revised. An intriguing feature of the 3’ UTR of insect-borne flavivirus genomes is the conservation of complex RNA structure duplications. Here, we discuss new hypotheses of how these RNA elements specialize for replication in vertebrate and invertebrate hosts, and present new ideas associating the significance of RNA structure duplication, small subgenomic flavivirus RNA formation and host adaptation.
Keywords: flaviviruses, viral RNA structures, dengue virus, RNA viruses evolution, sfRNAs, host-adaptation
Flaviviruses
The Flavivirus genus includes a large number of taxonomically recognized species, many of which are important human pathogens such as dengue, yellow fever, Japanese encephalitis, West Nile and other viruses that cause fever and encephalitis. Dengue virus (DENV) is the most important viral disease in humans transmitted by insects. It is responsible for about 390 million infections each year, without vaccines or antivirals available for its control. Yellow fever virus (YFV) is endemic in a number of African and South American countries, and causes 200,000 cases and 30,000 deaths in Africa even with effective vaccines available (http://www.who.int/csr/disease/yellowfev/YellowFeverBurdenEstimation_Summary2013.pdf). Other diseases caused by flaviviruses include West Nile encephalitis and Zika fever, which are considered emerging diseases with important outbreaks around the world [1].
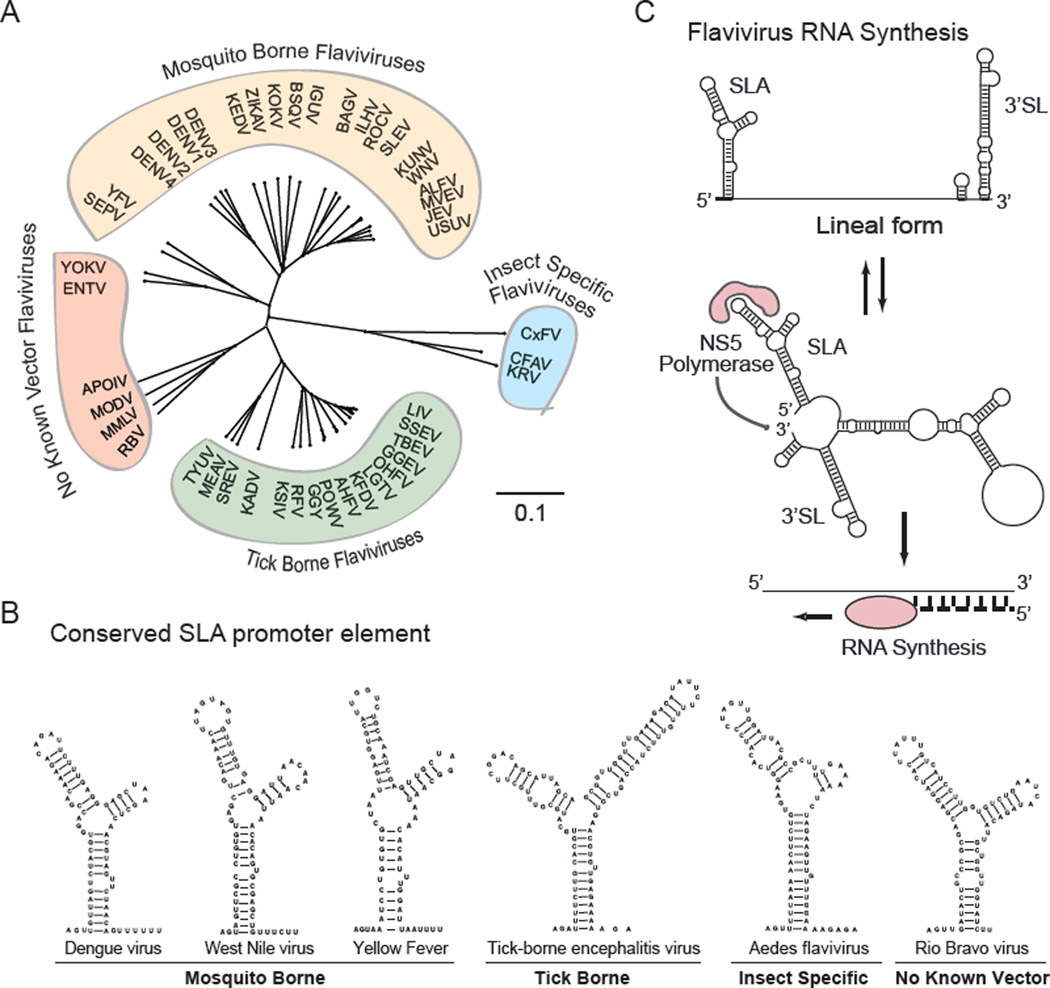
Despite the similar organization of flavivirus genomes and their mechanisms of replication, they possess differences in their host ranges and transmissibilities. In this regard, flaviviruses are divided into four large ecological groups: the mosquito borne group (MBFV), the tick borne group (TBFV), the vertebrate specific flavivirus group, referred to as no known vector viruses (NKFV), and the ones that have been only isolated from insects, which constitutes a growing group of viruses known as insect specific flaviviruses (ISFV) (for a recent review see [2]) (Figure 1A).
Figure 1.
Conserved Features and Mechanism of Flavivirus RNA Synthesis. a) Schematic representation of the distance tree of the four ecological groups of flaviviruses drawn using the neighbor joining method and jukes-cantor substitution model. b) Predicted RNA structure and sequence of SLA elements of different flaviviruses. c) General mechanism of viral RNA synthesis that involves the promoter element SLA at the 5’ end of the viral RNA, cyclization of the viral genome, and polymerase initiation at the 3’ end.
Flaviviruses are small, enveloped viruses with a single, positive-strand RNA genome of 10 to 12 kb. A type I cap structure is present at the 5’ end followed by the conserved dinucleotide 5’-AG-3’ [3]. The cap structure of flaviviruses contains a methyl group at the N7 position, and a second methyl group at the ribose 2’OH position of the first nucleotide, m7GpppAmpN2 [4, 5]. The 3’ end of the genome lacks a polyadenylate tail and terminates in a conserved 5’-CU-3’ [6]. The genome encodes a single open reading frame flanked by highly structured 5’ and 3’ untranslated regions (UTR). The 5’ UTRs are about 100 nucleotides long while the 3’ UTRs are in general between 400 and 700 nucleotides, while in some exceptional cases it can be over 900 nucleotides [2, 7, 8]. Although different flavivirus groups contain conserved RNA structures in the 5’ and 3’ UTRs, only two RNA elements are conserved in all flavivirus genomes. These are the Y shape stem-loop A structure (SLA) present at the 5’ end of the viral genome (Figure 1B), and the small hairpin 3’ stem-loop (sHP-3’ SL) located at the 3’ end of the viral RNA. These two essential RNA structures participate in the basic mechanism of viral RNA synthesis (Figure 1C). Sequence and structural features of the SLA as the promoter for viral polymerase binding and activation were first described in DENV and then extrapolated to other flaviviruses [9–11, 12]. The 3’ SL was the first RNA structure described in flaviviruses and it was originally observed in the genome of MBFVs [13–15]. Sequence conservation analysis within each flavivirus group indicates that both the SLA and the 3’ SL are the most conserved regions of the viral genomes (Figure 2). In addition, an important conserved feature in the genome of all flaviviruses is the presence of inverted complementary sequences that mediate genome cyclization, which allows the polymerase, bound to the promoter SLA, to reach the 3’ SL initiation site for RNA synthesis (for review see [16]). In this review, we will discuss the function of viral RNA elements that specialize for flavivirus replication in insect and mammalian hosts.
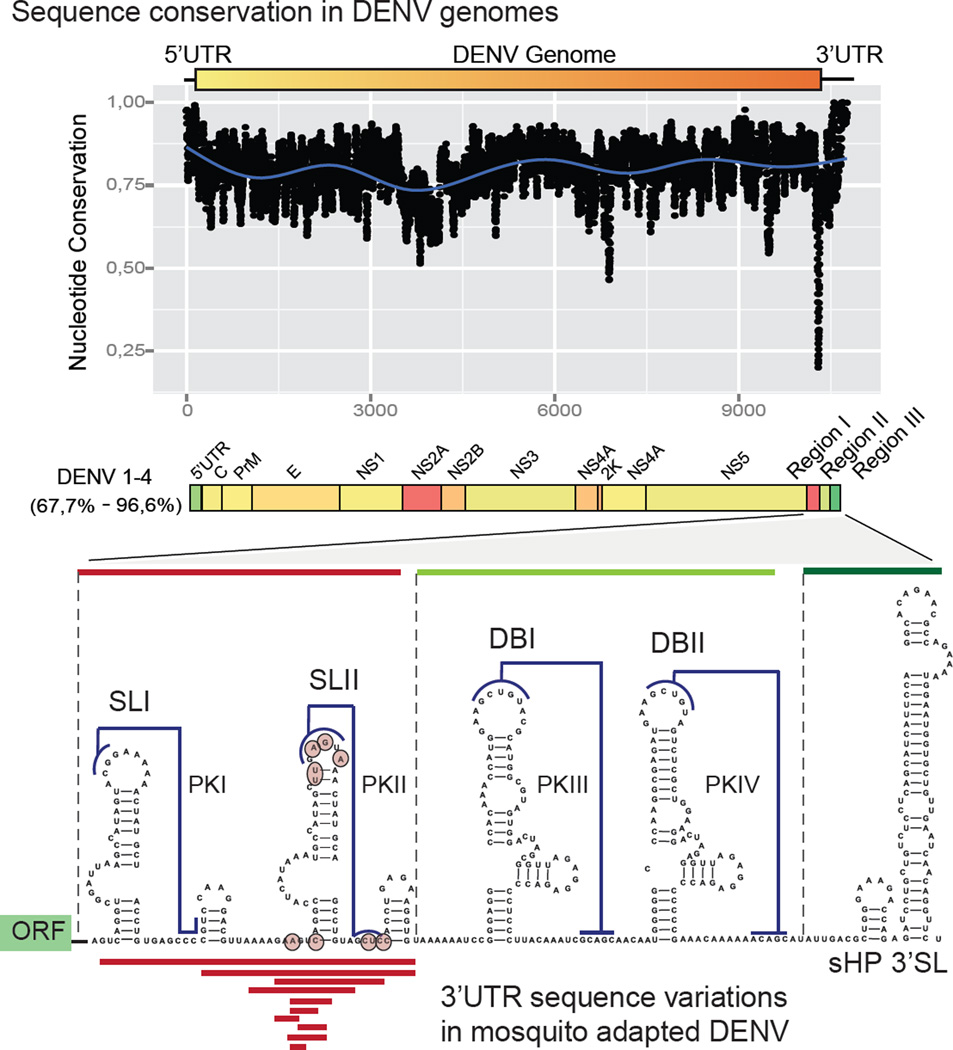
Figure 2.
Plot of Sequence Conservation of the Four DENV Serotypes. The 3’ UTR is divided into three regions according to the sequence variability (regions I, II, and III). Below, is a secondary structure model of the 3’ UTR of DENV2 predicted by conservation, stability and chemical probing; deletions (red lines) or point mutations (circles) rescued from mosquito cell-adapted populations are indicated.
Specialization of RNA Structures as a Strategy for Viral Adaptation to Mosquito or Human Cells
MBFV are a large group of viruses that cycle between Aedes or Culex mosquitoes and vertebrates. The process of jumping between such different hosts requires adaptation to different cellular machineries for viral replication and evasion of different types of antiviral responses. In general, RNA viruses have high capacity to adapt to different environments due to the genetic diversity of viral populations [17, 18]. However, it has been proposed that viruses that naturally alternate between different hosts evolve less rapidly than those that specialize in a single host [19]. This evolutionary constraint can be attributed to conflicting demands for efficient infection in different hosts [20].
A number of studies have investigated genetic variations that take place during flavivirus adaptation to vertebrate and invertebrate hosts (reviewed in [21, 22] and [23]). However, little is known about variations in functional RNA structures that act as cis-acting factors during flavivirus host adaptation. Several reports have documented mutations in flavivirus 3’ UTRs with differential impact on viral replication in insect or mammalian hosts [24–29]. For DENV a host specific requirement of the sHP present at the viral 3’ UTR has been reported [30]. In this regard, point mutations in the sHP abrogated infection in mosquito cells without affecting replication in mammalian cells. In a recent study, DENV restriction to replicate in vertebrate or invertebrate cells resulted in the selection of different viral populations with a hot spot for variations in the viral 3’ UTR [31]. Deep sequencing of viral populations revealed that mutations selected in mosquito cells mapped in a single RNA structure, named stem loop II (SLII, Figure 2). Selected mutations disrupting this RNA structure were associated with higher viral fitness in mosquito cells and in Aedes albopictus mosquitoes as compared with viruses with an intact SLII. In contrast, mutations that disrupted this RNA structure reduced viral replication in human cells [31]. Interestingly, cycles of disruption and reconstitution of the SLII structure were observed after host switch. These observations provide an example of a viral RNA structure that is under opposite selective pressures in mosquito and human cells and highlight a process of host specialization of a cis-acting RNA element.
The DENV2 3’ UTR is structurally divided into three regions: region I is located just downstream of the translation stop codon and contains a hypervariable sequence followed by two SLs similar in sequence and structure (SLI and SLII), with sequences involved in pseudoknot interactions (PKI and PKII, respectively); region II contains a duplication of a conserved structure known as dumbbell (DBI and DBII) also involved in pseudoknot interactions (PKIII and PKIV, respectively); and region III includes the essential terminal structure sHP-3’ SL (Figure 2). The functional significance of complex RNA structure duplication in the viral 3’ UTR is still unknown. Regarding the host-dependent variation of DENV SLII, it is important to mention that the sequence and structure of SLI remains intact during viral replication in mosquito or human cells, supporting the idea of a functional diversification of the duplicated SL elements.
The available data provides evidence of distinct functions of flavivirus RNA structures during infection in vertebrate and invertebrate hosts, resulting in different adaptation processes.
Functional Significance of Flavivirus RNA Structure Duplication
The presence of two SLs and two DBs is a conserved feature in most MBFV 3’ UTRs. Sequence similarities and defined structural blocks in the SLs and DBs support duplication as the origin of these RNA structures; however, recent functional studies provided evidence that each SL and each DB have distinct roles during viral replication [31–35]. In the case of SLI and SLII, studies using DENV2 indicate that while deletion or mutations of SLII provide a great replication advantage in mosquito cells, deletion of SLI marginally reduces viral replication, supporting different functions of the two structures. In contrast, in human cells, the two SLs appeared to play redundant functions. In this regard, while deletion of either structure slightly reduces viral replication, deletion of both results in a large decrease in viral fitness [31]. Regarding the DB elements, sequences involved in DB1 and DB2 formation were reported to play opposite roles in flavivirus genome cyclization. While DB1 bears nucleotides that are complementary to sequences present at the 5’ end of the genome that enhance long range RNA-RNA interactions and genome cyclization [34, 36], the sequence at the top-loop of DB2 competes with genome cyclization by forming a PK with nucleotides that overlap with 3’CS, one of the most conserved cyclization element among flaviviruses [32–34]. These observations support a diversification of function between the two SLs and between the two DBs.
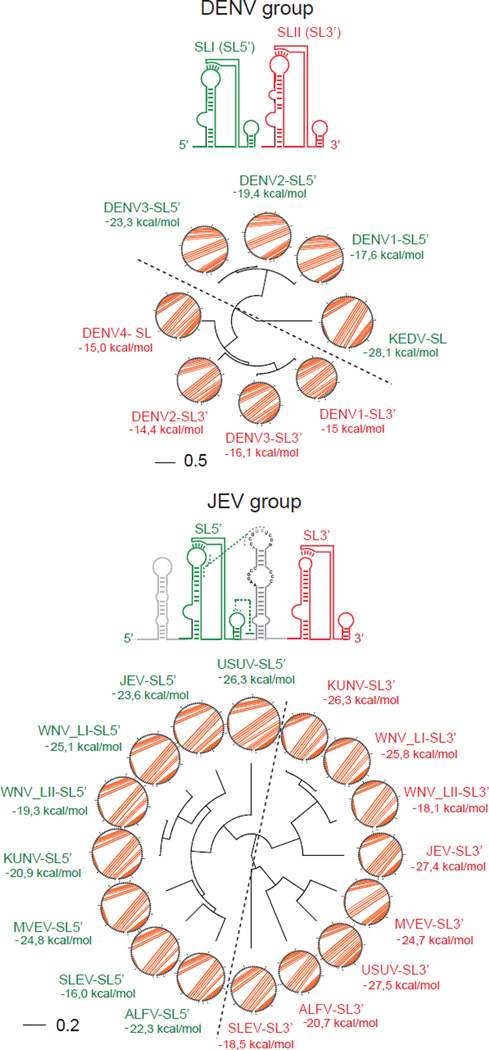
Interestingly, RNA sequence alignments and RNA structure comparison using RNAforester, which estimates RNA structural similarities based on the tree alignment model [37], shows that SLI from different DENVs are more closely related than SLI and SLII from the same virus (Figure 3). This observation was extended to all members of the Japanese encephalitis virus (JEV) group, supporting the idea that SL structure duplication occurred early during emergence of these viral groups, and then each of the duplicated element evolved, acquiring specific functions. To avoid confusion with the nomenclature of RNA structures from different groups, in Figure 3, the related SLI and SLII are also referred as SL5’ and SL3’, respectively (based on their location).
Figure 3.
Representation of Local Analysis of RNA Structure Similarities Using the RNAforester Algorithm. Top, comparison of the duplicated structures SLI (SL5’, indicated in green) and SLII (SL3’ indicated in red) of members of the DENV group. A fan dendrogram indicating the distance between these structures is shown with the corresponding circle plot with the sequence and arcs denoting base pairings. Bottom, a similar comparison of SL5’ and SL3’ structures are shown for members of the JEV group. Bar indicates number of nucleotide substitution per site
In the case of YFV, which naturally bears a single SL and likely has less capacity to tolerate mutations in this region, an interesting correlation between viral virulence and the integrity of the single SL was found [38]. In this case, sequence and structure comparison of wild and vaccine strains of YFV (performed to define determinants of viral attenuation) revealed that all vaccine strains, except 17DD, displayed a disrupted SL and PK structures (originally referred to as Region I). In the wild type strains, the SL structure was maintained, displaying several covariations. Interestingly, 17DD was the only vaccine strain that retained the SL-PK structure (also by co-variations) and was the most virulent of the vaccine strains in a mouse model [39]. This pioneering analysis of 3’ UTR structures provided an interesting correlation between SL-PK integrity and YFV pathogenesis.
More recently using DENV, the presence of a double SL-PK in the viral 3’ UTR was found to facilitate host switch from mosquito to humans [31]. In this regard, recombinant viruses with a single SL carrying mosquito adaptive mutations failed to replicate in human cells, while this was reverted by incorporation of a second SL. Based on these observations, it is possible that SL duplication serves as a mechanism for higher tolerance of mutations that are beneficial in one host but deleterious in the other, by maintaining one of the two structures intact during host switch.
In summary, experimental evidence supports the significance of RNA structure duplication in members of the MBFV group. Instead of a redundant function, as it was originally proposed, each of the duplicated RNA elements appear to play distinct roles during viral replication [31–34], and in some cases are directly linked to specific requirements in different host.
RNA Structure Duplication, Viral Fitness and sfRNA Production
During flavivirus infection, in addition to the full-length genomic viral RNA, other smaller RNA species accumulate, named subgenomic flavivirus RNAs (sfRNAs) [40–42]. Several recent studies provide interesting ideas on how the sfRNAs enhance flavivirus infections [42–48]. For a comprehensive description of the sfRNA functions, the reader is referred to recent reviews [49, 50].
The sfRNAs are products of partial degradation of viral RNAs. After removal of the cap structure present at the 5’ end of flavivirus genomes, the RNA is degraded by the host exoribonuclease XRN1 from the 5’ to the 3’ direction [42]. The biogenesis of sfRNAs is linked to highly structured RNA elements (xrRNAs) present at the viral 3’ UTR, which stall the XRN1 exonuclease activity [51–53]. The process results in the accumulation of small RNA molecules with nucleotide sequences corresponding to viral 3’ UTRs. In the case of JEV group members, the first of the duplicated SLs appears to be the main structure responsible for XRN1 stalling and sfRNA accumulation [42, 54]. In YFV, the single SL-PK (also known as SL-E) has been proposed to be the structure responsible for sfRNA formation [55]. For members of the DENV group, it is still uncertain which one of the two duplicated SL-PK structures is the main determinant for XRN1 arrest for sfRNA formation in vivo.
Interestingly, infection with flaviviruses carrying mutations that artificially disrupt SL-PK structures showed that RNA degradation by the XRN1 advances towards the 3’ end of the genome, but then stalls at the DB-PK structures, generating shorter sfRNA species [54]. This observation indicates that there are at least two types of structures in the MBFV 3’ UTRs capable of stalling the XRN1 activity (the SL-PK and the DB-PK). Based on the mechanism of sfRNA formation, it is intriguing the conserved duplication of RNA structures responsible for XRN1 arrest. Moreover, the process of DENV adaptation to mosquito cells specifically disrupts a SL-PK structure, which likely results in accumulation of different types of sfRNAs. These observations lead us to speculate about a possible link between host adaptation and sfRNA production (Figure 4, Key Figure). In addition, a recent study using different DENV isolates, proposed an association between the levels of sfRNA accumulation and the viral epidemiological fitness [48]. Based on the available data, it is possible that the variability observed at the 3’ UTR of different DENV isolates is a consequence of specific selective pressures in the mosquito.
Figure 4, Key Figure.
New Functions of RNA Structures Present at the 3’ UTR of Mosquito-Borne Flaviviruses. Host adaptation selects different dengue virus populations and the link between RNA structure duplication, host adaptation, and production of subgenomic flavivirus RNAs (sfRNAs) is described. Red lines under each population illustrate the change in frequency of the same viral variant.
Conserved RNA Structure Duplications in the 3’ UTRs of Flavivirus Genomes
To understand the biological significance of RNA structure duplication in the flavivirus genomes, the conservation of structural blocks of RNA elements found in each group of the Flavivirus genera was constructed and compared (Figure 5 and 6). Models of secondary structures of flavivirus 3’ UTRs including MBFVs, TBFVs, NKFVs, and ISFVs were elaborated using RNAalifold, RNAz and CentroidFold software [56–58]. When available, information from chemical or enzymatic probing was included in the predictions. The models described here also gather information from previous studies that identified a number of structural features shared by different flavivirus 3’ UTRs [35, 59–66].
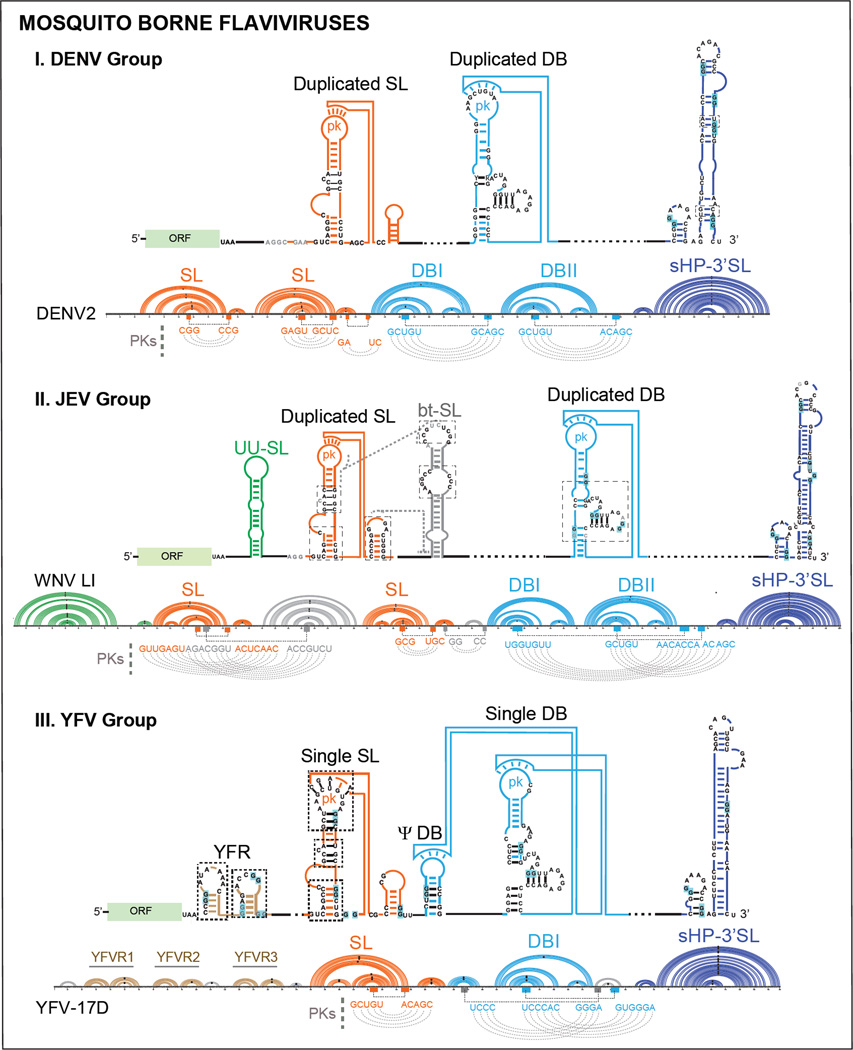
Figure 5.
Models of Conserved RNA Structural Elements of Mosquito Borne Flavivirus 3’ UTRs. Schematic representation of RNA structures and conserved RNA motifs for each group (DENV, JEV and YFV) are shown. Below the models, an example of one member of each group (DENV2, WNV-LI, and YFV-17D) is shown using an Arc plot secondary structure representation, including the nucleotide sequence. The color code for each structural motif is maintained in all the representations. Pseudoknot interactions (PKs) are indicated underneath each Arc plot. Abbreviations: SL: stem-loop, DB: dumbbell, sHP: small hairpin, 3’ SL: 3’ stem-loop, UU-SL: U rich stem-loop, bt-SL: between stem-loop, YFVR: yellow fever virus repeat, ORF: open reading frame.
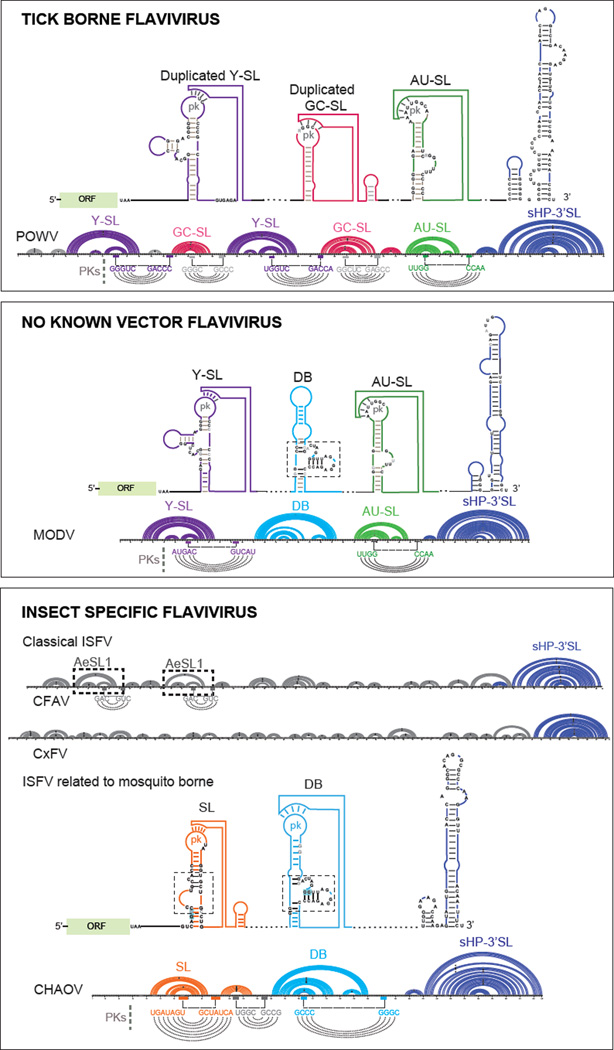
Figure 6.
Models of Conserved RNA Structural Elements of Tick-Borne, No-Known Vector, and Insect-Specific Flaviviruses. Schematic representation of RNA structures and conserved RNA motifs for each group are shown. Below the models, an example of one member of each group is shown using an Arc plot secondary structure representation including the nucleotide sequence: Powassan virus (POWV), Modoc virus (MODV) and for insect specific flavivirus examples the cellular fusion agent virus (CFAV), Culex flavivirus CxFV), and Chaoyang virus (CHAOV) are included. The color code for each structural motif is maintained in all the representations. Pseudoknots interaction (PKs) are indicated underneath each Arc plot. Abbreviations: Y-SL: Y-shape stem-loop, AU-SL: AU containing stem-loop, GC-SL: GC containing stem-loop, DB: dumbbell, sHP: small hairpin, 3’ SL: 3’ stem-loop, AeSL: Aedes stem-loop, ORF: open reading frame.
MBFVs
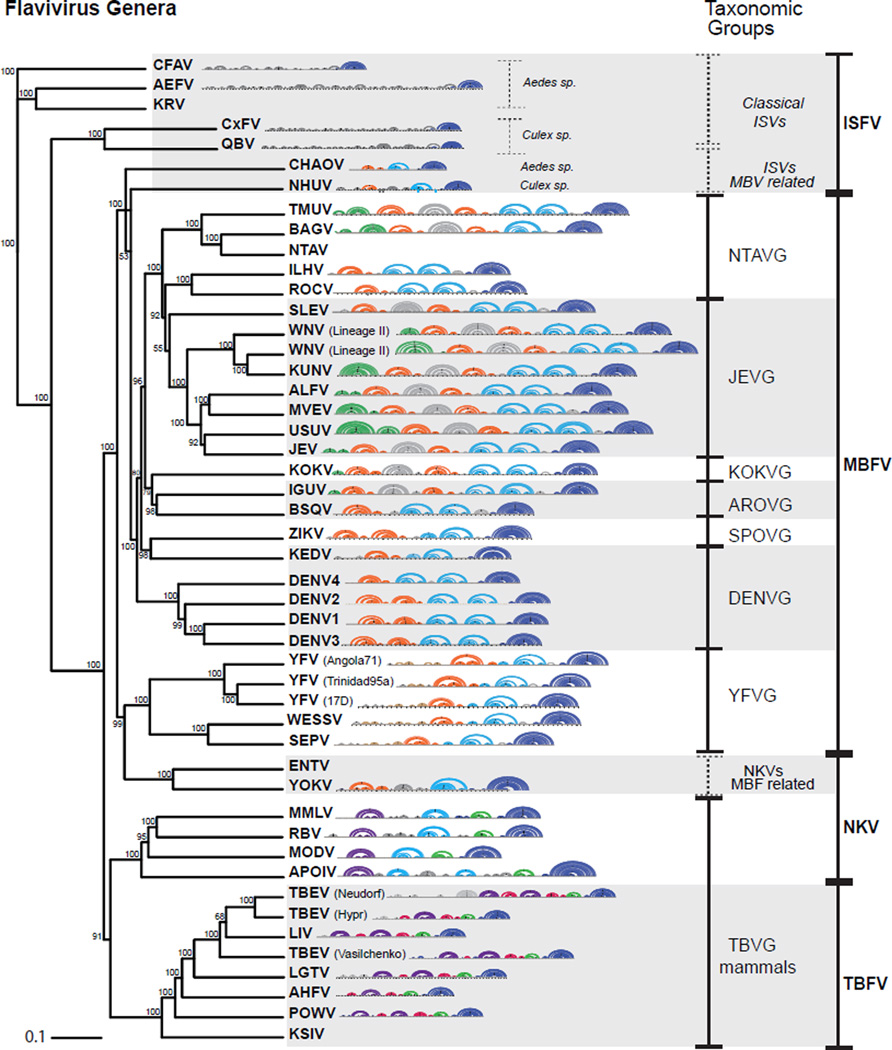
This large group of viruses transmitted by Aedes or Culex mosquitoes is divided into seven subgroups: the DENV group, JEV group, YFV group, Aroa virus group, Kokobera virus group, Ntaya virus group, and Spondweni virus group [67]. Most MBFV genomes display conserved structural blocks that were defined as SLs or DBs [31]. For clarity the names SLI and SLII are used to refer to the duplicated conserved elements present in region I of MBFV 3’ UTRs, included in the structures also known as xrRNA1 and xrRNA2, respectively [52–53].
DENV 3’ UTRs bear duplicated SLs and DBs (Figure 5). The exception is DENV4 that contains a single SL and a duplicated DB.
Models of RNA structures in the JEV group were constructed using sequences of the following viruses: WNV-LI, WNV-LII, JEV, Saint Louis encephalitis virus (SLEV), Alfuy virus (ALFV), Usutu virus (USUV), Kunjin virus (KUNV) and Murray Valley encephalitis virus (MVEV). They contain a more complex 3’ UTR as compared with that of members of the DENV group (Figure 5). In region I of the 3’ UTR, just downstream of the translation stop codon, four structural elements can be defined: a duplicated SL with conserved structural blocks similar to that observed in DENV, preceded and intercalated by two unrelated structures (Figure 5). The first structure is a U rich element (UU-SL) of variable length with low GC content. This RNA element is absent in some members of the group, such as in SLEV. The second structure is located between the two conserved SLs (bt-SL) and is present in all JEV group members analyzed (Figure 5). The loop sequence of this structure is predicted to form a tertiary interaction with a sequence located within the upstream SL [31]. When first described, the four RNA elements of region I of the JEV group were named SLI, SLII, SLIII and SLIV according to their position in the genome; however, only the originally described SLII and SLIV contain the structural blocks and sequences conserved in the duplicated SL elements found in all MBFVs [42]. The duplicated DB structures (DBI and DBII) are also present in most JEV group members. In this regard, although both DBs form pseudoknot interactions, the two loop sequences display contacts with sequences present downstream of DBII, showing a different arrangement as that observed for members of the DENV group (see WNV-LI arch PK structures, Figure 5).
Members of the YFV group, including YFVs, Sepik virus (SEPV) and Wesselbron virus (WSSV), contain single copies of the conserved SL and DB structures. However, they bear repetitions of hairpin structures specific to this group, named YFV repeats (YFVR). In this regard, three YFVR were observed in YFV isolates from West Africa, two repeats in isolates from East Africa and single structures in South American isolates [64, 68]. An additional RNA structure was identified with remnant elements of a DB, named pseudo dumbbell (ΨDB) (Figure 5). A similar arrangement of a ΨDB and DB structures was observed in Zika and Kedougou viruses, belonging to the Spondweni and the DENV groups, respectively. Interestingly, in certain YFV isolates from Brazil a duplication of the DB structure was found [69]. This observation places the question of whether an original YFV 3’ UTR contained a duplicated DB.
TBFVs
In this group the 3’ UTR of tick borne encephalitis virus (TBEV) vasilchenko, TBEV neudoerfl, TBEV hypr, Powassan virus (POWV), Louping ill virus (LIV), Langat virus (LGTV), and Alkhurma hemorrhagic fever virus (AHFV) were included in the analysis. The 3’ UTRs of these viruses show size heterogeneity and do not contain SLs or DBs as the ones described for MBFVs; instead they contain duplications of group-specific RNA structures in which conserved structural blocks were identified. Three different group-specific structures were defined (Figure 6). The first structure named Y shape stem-loop (Y-SL) [8] is often duplicated in different locations and its top loop sequence is involved in a PK structure with downstream sequences (Figure 6). The second structure also forms a stem-loop (referred to as GC-SL) that is duplicated in many TBFV members, with a loop containing a conserved GGC sequence involved in a PK interaction. The third structure forms a very stable stem-loop (referred to as AU-SL) with a loop containing a conserved AAUU sequence involved in a PK interaction. This last element is found in a single copy and is located just upstream of the conserved terminal sHP-3’ SL (Figure 6). Although TBFV members do not bear SL or DB duplications, they have group specific RNA structure duplications.
NKFVs
Members of this group were most commonly isolated from rodents and bats [70]. The sequences used for this analysis included the 3’ UTRs from Yokose virus (YOKV), Rio Bravo virus (RBV), Leucoencephalitis miotis virus (MMLV), Apoi virus (APOIV) and Modoc virus (MODV). The 3’ UTRs of these viruses contain in general single copies of conserved RNA structures that resemble that found in mosquito or tick borne viruses [71, 72]. Based on sequence and prediction comparisons at least three RNA structural elements were observed: one Y-SL and one AU-SL, similar to those described in TBFVs; and one DB element containing structural blocks and sequences of that conserved in MBFVs but with no evidence of PK formation (Figure 6). Many members of the NKFV group including MODV, MMLV, APOIV and RBV have been associated to both TBFVs and MBFVs, suggesting a common origin of these viruses (Figure 7). In addition, YOKV bears 3’ UTR structures resembling the SL and DB of MBFV and has been associated to the YFV group [73, 74]. It is possible that this virus evolved with MBFVs but lost the ability to replicate in insects.
Figure 7.
Phylogenetic Relationships between Members of the Flavivirus Genus with Arch Representations of Conserved Structures of the Viral 3’ UTRs. Phylogenetic tree using coding regions of flaviviruses employing a neighbor-joining method is shown. 3’ UTR structures are represented in arc plots using the following color code: SL structures in orange, DB and ΨDB in pale blue, YFVR in brown, UU-SL in green, bt-SL in gray, YSL in violet, GC-SL in pink, AU-SL in pale green and sHP-3’ SL in blue. Mosquito-borne flavivirus taxonomic groups: DENV group, JEV group, YFV group, Aroa virus group (AROVG), Kokobera virus group (KOKVG), Ntaya virus group (NTAVG), and Spondweni virus group (SPOVG).
ISFVs
A general property of the 3’ UTRs of these viruses is the presence of multiple direct sequence repeats [65], but the analysis presented here suggests the lack of high-order RNA structure duplication. For predicting formation of conserved RNA structures, this group was divided in two sub-groups: one that is distinct to all known flaviviruses, known as classical ISFVs, and the other one related to MBFVs [2, 75]. In the case of the classical ISVFs, including the cellular fusion agent virus (CFAV), Culex flavivirus (CxFV), Aedes flavivirus (AEFV) and Quang Binh virus (QBV), the 3’ UTRs display a variable number of short hairpins some of them duplicated with low potential for high order interactions. One duplicated structure with a potential PK interaction was predicted in some viruses isolated from Aedes mosquitoes (Ae-SL, Figure 6). The other members of the ISFV group, including Chaoyang virus (CHAOV) and Nhumirin virus (NHUV), contain RNA elements with conserved structural blocks similar to SLs and DBs of MBFVs but in single copies.
RNA secondary structure comparisons of flavivirus 3’ UTRs allows an association between RNA structure duplication and the ability of a virus to replicate in vertebrate and invertebrate hosts. MBFVs bear duplications of SLs and DBs, while TBFVs bear duplications of Y-SLs and GC-SLs. In contrast, in most of the single host flaviviruses NKFVs and ISFVs, the presence of secondary structure duplications is less evident, although they conserve stretches of repeated nucleotide sequences [8]. Common RNA structural modules for each region of the 3’ UTR of all available flavivirus genomes are shown in Figure 7. Hopefully, this unique and comprehensive analysis of RNA structures will stimulate further evolutionary and functional studies of this important group of insect-borne human viral pathogens.
Concluding Remarks
Models of common structural RNA elements present at the 3’ UTR of all flaviviruses have been revised to facilitate functional studies and as tools for comparisons between different viral groups. There is an evident association between flaviviruses that alternate between insects and mammalian hosts and conserved complex RNA structure duplications. Interestingly, the identification of opposite selective pressures on RNA structures in mosquito and human cells provide new ideas for understanding the need of duplicated elements.
A great deal of experimental information has been generated in the past few years that had led to an association between host specialization of RNA structures, sfRNA production, and viral fitness. These studies will be instrumental in generating new models that explain evolutionary mechanisms that enable flaviviruses to replicate in multiple hosts (see Outstanding Questions).
Understanding how host-virus interactions shape viral evolution will help to elucidate the factors that govern the emergence of new viruses and the expansion of already known RNA viral pathogens.
Outstanding Questions.
The SLII RNA structure is beneficial for DENV replication in human cells but detrimental in mosquito cells. What is the benefit of evolving conflicting requirements within a cis-acting RNA structure for viral replication in different hosts? Is this differential requirement the cause of RNA structure duplication as a mechanism for host adaptation?
What is the selective pressure on the viral SLII structure in infected human and mosquito cells? sfRNA accumulation requires an intact SL-PK structure. Is the requirement of specific sfRNA species in different hosts the cause of the cyclic disruption/reconstitution of SL-PK?
Why are the RNA structures that stall the XRN1 duplicated in mosquito-borne flaviviruses?
What is the impact of flavivirus 3’UTR variability on viral transmission?
Is the mechanism of RNA structure specialization observed during DENV replication in different hosts common to other arboviruses?
Trends.
Recent advances in molecular virology provide new hypotheses of how RNA structures in mosquito-borne flavivirus genomes mediate host adaptation, viral replication and evasion of antiviral responses.
Dengue virus RNA structures play different functions during infection in vertebrate and invertebrate hosts.
Conflicting requirements of viral RNA elements shape the composition of viral populations obtained in human or mosquito cells.
Viral RNA structures can modulate the type and extent of host antiviral responses.
Complex RNA structures present at the viral 3’ untranslated region (3’UTR) that stall the host exoribonuclease XRN1 and generate small viral-derived RNAs are duplicated in mosquito-borne flaviviruses.
Conservation of RNA structure duplication in the 3’UTR of insect-borne viruses is associated with mechanisms of host adaptation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Musso D, et al. Zika virus: following the path of dengue and chikungunya? Lancet. 2015;386:243–244. doi: 10.1016/S0140-6736(15)61273-9. [DOI] [PubMed] [Google Scholar]
- 2.Blitvich BJ, Firth AE. Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses. 2015;7:1927–1959. doi: 10.3390/v7041927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleaves GR, Dubin DT. Methylation status of intracellular dengue type 2 40 S RNA. Virology. 1979;96:159–165. doi: 10.1016/0042-6822(79)90181-8. [DOI] [PubMed] [Google Scholar]
- 4.Ray D, et al. West Nile virus 5'-cap structure is formed by sequential guanine N-7 and ribose 2'-O methylations by nonstructural protein 5. J Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y, et al. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wengler G. Terminal sequences of the genome and replicative-from RNA of the flavivirus West Nile virus: absence of poly(A) and possible role in RNA replication. Virology. 1981;113:544–555. doi: 10.1016/0042-6822(81)90182-3. [DOI] [PubMed] [Google Scholar]
- 7.Markoff L. 5' and 3' NCRs in Flavivirus RNA. Elsevier Academic Press; 2003. [Google Scholar]
- 8.Gritsun TS, Gould EA. Origin and evolution of 3'UTR of flaviviruses: long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. Adv Virus Res. 2007;69:203–248. doi: 10.1016/S0065-3527(06)69005-2. [DOI] [PubMed] [Google Scholar]
- 9.Filomatori CV, et al. A 5' RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodeiro MF, et al. Structural and functional studies of the promoter element for dengue virus RNA replication. J Virol. 2009;83:993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B, et al. Genetic interactions among the West Nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5' stem-loop of genomic RNA. J Virol. 2008;82:7047–7058. doi: 10.1128/JVI.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5'-terminal region of flavivirus genome RNA. Virology. 1988;162:290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 13.Grange T, et al. Stable secondary structures at the 3'-end of the genome of yellow fever virus (17 D vaccine strain) FEBS Lett. 1985;188:159–163. doi: 10.1016/0014-5793(85)80895-4. [DOI] [PubMed] [Google Scholar]
- 14.Brinton MA, et al. The 3'-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- 15.Mohan PM, Padmanabhan R. Detection of stable secondary structure at the 3' terminus of dengue virus type 2 RNA. Gene. 1991;108:185–191. doi: 10.1016/0378-1119(91)90433-c. [DOI] [PubMed] [Google Scholar]
- 16.Villordo SM, Gamarnik AV. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139:230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinhauer DA, et al. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122:281–288. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- 18.Borderia AV, et al. RNA virus population diversity: implications for inter-species transmission. Curr Opin Virol. 2011;1:643–648. doi: 10.1016/j.coviro.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins GM, et al. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- 20.Woolhouse ME, et al. Population biology of multihost pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 21.Ciota AT, Kramer LD. Insights into arbovirus evolution and adaptation from experimental studies. Viruses. 2010;2:2594–2617. doi: 10.3390/v2122594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coffey LL, et al. Factors shaping the adaptive landscape for arboviruses: implications for the emergence of disease. Future Microbiol. 2013;8:155–176. doi: 10.2217/fmb.12.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deardorff ER, et al. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS Pathog. 2011;7:e1002335. doi: 10.1371/journal.ppat.1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alvarez DE, et al. Role of RNA structures present at the 3'UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Men R, et al. Dengue type 4 virus mutants containing deletions in the 3' noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J Virol. 1996;70:3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tumban E, et al. Replacement of the 3' untranslated variable region of mosquito-borne dengue virus with that of tick-borne Langat virus does not alter vector specificity. J Gen Virol. 2011;92:841–848. doi: 10.1099/vir.0.026997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tajima S, et al. Characterization of the variable region in the 3' non-translated region of dengue type 1 virus. J Gen Virol. 2007;88:2214–2222. doi: 10.1099/vir.0.82661-0. [DOI] [PubMed] [Google Scholar]
- 28.Zeng L, et al. Identification of specific nucleotide sequences within the conserved 3'-SL in the dengue type 2 virus genome required for replication. J Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaney JE., Jr Dengue virus type 3 vaccine candidates generated by introduction of deletions in the 3' untranslated region (3'-UTR) or by exchange of the DENV-3 3'-UTR with that of DENV-4. Vaccine. 2008;26:817–828. doi: 10.1016/j.vaccine.2007.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villordo SM, Gamarnik AV. Differential RNA sequence requirement for dengue virus replication in mosquito and mammalian cells. J Virol. 2013;87:9365–9372. doi: 10.1128/JVI.00567-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villordo SM, et al. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Pathog. 2015;11:e1004604. doi: 10.1371/journal.ppat.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manzano M, et al. Identification of cis-acting elements in the 3'-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J Biol Chem. 2011;286:22521–22534. doi: 10.1074/jbc.M111.234302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sztuba-Solinska J, et al. Structural complexity of Dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res. 2013;41:5075–5089. doi: 10.1093/nar/gkt203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Borba L, et al. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J Virol. 2015;89:3430–3437. doi: 10.1128/JVI.02677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yun SI, et al. 3' cis-acting elements that contribute to the competence and efficiency of Japanese encephalitis virus genome replication: functional importance of sequence duplications, deletions, and substitutions. J Virol. 2009;83:7909–7930. doi: 10.1128/JVI.02541-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu ZY, et al. Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. J Virol. 2013;87:6804–6818. doi: 10.1128/JVI.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochsmann M, et al. Pure multiple RNA secondary structure alignments: a progressive profile approach. IEEE/ACM Trans Comput Biol Bioinform. 2004;1:53–62. doi: 10.1109/TCBB.2004.11. [DOI] [PubMed] [Google Scholar]
- 38.Proutski V, et al. Secondary structure of the 3'-untranslated region of yellow fever virus: implications for virulence, attenuation and vaccine development. J Gen Virol. 1997;78(Pt 7):1543–1549. doi: 10.1099/0022-1317-78-7-1543. [DOI] [PubMed] [Google Scholar]
- 39.Barrett AD, Gould EA. Comparison of neurovirulence of different strains of yellow fever virus in mice. J Gen Virol. 1986;67(Pt 4):631–637. doi: 10.1099/0022-1317-67-4-631. [DOI] [PubMed] [Google Scholar]
- 40.Urosevic N, et al. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J Gen Virol. 1997;78(Pt 1):23–29. doi: 10.1099/0022-1317-78-1-23. [DOI] [PubMed] [Google Scholar]
- 41.Lin KC, et al. Accumulation of a 3'-terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells. J Virol. 2004;78:5133–5138. doi: 10.1128/JVI.78.10.5133-5138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pijlman GP, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Moon SL, et al. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnettler E, et al. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang RY, et al. Japanese encephalitis virus non-coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet Microbiol. 2013;166:11–21. doi: 10.1016/j.vetmic.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 46.Schuessler A, et al. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol. 2012;86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bidet K, et al. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014;10:e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manokaran G, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke BD, et al. Functional non-coding RNAs derived from the flavivirus 3' untranslated region. Virus Res. 2015;206:53–61. doi: 10.1016/j.virusres.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 50.Charley PA, Wilusz J. Standing your ground to exoribonucleases: Function of Flavivirus long non-coding RNAs. Virus Res. 2015 Sep 11; doi: 10.1016/j.virusres.2015.09.009. pii: S0168-1702(0115)30062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapman EG, et al. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014;344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chapman EG, et al. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife. 2014;3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kieft JS, et al. New hypotheses derived from the structure of a flaviviral Xrn1- resistant RNA: conservation, folding, and host adaptation. RNA Biol. 2015 Nov 2;12(11):1169–1177. doi: 10.1080/15476286.2015.1094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Funk A, et al. RNA structures required for production of subgenomic flavivirus RNA. J Virol. 2010;84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silva PA, et al. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol. 2010;84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gruber AR, et al. RNAz 2.0: improved noncoding RNA detection. Pac Symp Biocomput. 2010;2010:69–79. [PubMed] [Google Scholar]
- 57.Hamada M, et al. Improving the accuracy of predicting secondary structure for aligned RNA sequences. Nucleic Acids Res. 2011;39:393–402. doi: 10.1093/nar/gkq792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olsthoorn RC, Bol JF. Sequence comparison and secondary structure analysis of the 3' noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA. 2001;7:1370–1377. [PMC free article] [PubMed] [Google Scholar]
- 60.Proutski V, et al. Secondary structure of the 3' untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 1997;25:1194–1202. doi: 10.1093/nar/25.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rauscher S, et al. Secondary structure of the 3'-noncoding region of flavivirus genomes: comparative analysis of base pairing probabilities. RNA. 1997;3:779–791. [PMC free article] [PubMed] [Google Scholar]
- 62.Wallner G, et al. The flavivirus 3'-noncoding region: extensive size heterogeneity independent of evolutionary relationships among strains of tick-borne encephalitis virus. Virology. 1995;213:169–178. doi: 10.1006/viro.1995.1557. [DOI] [PubMed] [Google Scholar]
- 63.Thurner C, et al. Conserved RNA secondary structures in Flaviviridae genomes. J Gen Virol. 2004;85:1113–1124. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- 64.Mutebi JP, et al. Genetic relationships and evolution of genotypes of yellow fever virus and other members of the yellow fever virus group within the Flavivirus genus based on the 3' noncoding region. J Virol. 2004;78:9652–9665. doi: 10.1128/JVI.78.18.9652-9665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gritsun DJ, et al. Molecular archaeology of Flaviviridae untranslated regions: duplicated RNA structures in the replication enhancer of flaviviruses and pestiviruses emerged via convergent evolution. PLoS One. 2014;9:e92056. doi: 10.1371/journal.pone.0092056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gritsun TS, Gould EA. Direct repeats in the flavivirus 3' untranslated region; a strategy for survival in the environment? Virology. 2007;358:258–265. doi: 10.1016/j.virol.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 67.Gubler DKG, Markoff L. Fields Virology. Lippincott-Raven; 2007. Flaviviruses. [Google Scholar]
- 68.Wang E, et al. Genetic variation in yellow fever virus: duplication in the 3' noncoding region of strains from Africa. Virology. 1996;225:274–281. doi: 10.1006/viro.1996.0601. [DOI] [PubMed] [Google Scholar]
- 69.Bryant JE, et al. Size heterogeneity in the 3' noncoding region of South American isolates of yellow fever virus. J Virol. 2005;79:3807–3821. doi: 10.1128/JVI.79.6.3807-3821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burns KF, Farinacci CJ. Virus of bats antigenically related to St. Louis encephalitis. Science. 1956;123:227. doi: 10.1126/science.123.3189.227. [DOI] [PubMed] [Google Scholar]
- 71.Charlier N, et al. Complete genome sequence of Montana Myotis leukoencephalitis virus, phylogenetic analysis and comparative study of the 3' untranslated region of flaviviruses with no known vector. J Gen Virol. 2002;83:1875–1885. doi: 10.1099/0022-1317-83-8-1875. [DOI] [PubMed] [Google Scholar]
- 72.Leyssen P, et al. Complete genome sequence, taxonomic assignment, and comparative analysis of the untranslated regions of the Modoc virus, a flavivirus with no known vector. Virology. 2002;293:125–140. doi: 10.1006/viro.2001.1241. [DOI] [PubMed] [Google Scholar]
- 73.Kuno G, Chang GJ. Characterization of Sepik and Entebbe bat viruses closely related to yellow fever virus. Am J Trop Med Hyg. 2006;75:1165–1170. [PubMed] [Google Scholar]
- 74.Gaunt MW, et al. Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J Gen Virol. 2001;82:1867–1876. doi: 10.1099/0022-1317-82-8-1867. [DOI] [PubMed] [Google Scholar]
- 75.Kenney JL, et al. Characterization of a novel insect-specific flavivirus from Brazil: potential for inhibition of infection of arthropod cells with medically important flaviviruses. J Gen Virol. 2014;95:2796–2808. doi: 10.1099/vir.0.068031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]