Abstract
Gene amplification at the chorion loci in Drosophila ovarian follicle cells is a model for the developmental regulation of DNA replication. Previously, we showed that the Drosophila homolog of the Myb oncoprotein family (DmMyb) is tightly associated with four additional proteins and that DmMyb is required for this replication-mediated amplification. Here we used targeted mutagenesis to generate a mutant in the largest subunit of the DmMyb complex, the Aly and Lin-9 family member, Myb-interacting protein 130 (Mip130). We found that mip130 mutant females are sterile and display inappropriate bromodeoxyuridine (BrdU) incorporation throughout the follicle cell nuclei at stages undergoing gene amplification. Whereas mutations in Dm-myb are lethal, mutations in mip130 are viable. Surprisingly, Dm-myb mip130 double mutants are also viable and display the same phenotypes as mip130 mutants alone. This suggests that Mip130 activity without DmMyb counteraction may be responsible for the Dm-myb mutant lethality. RNA interference (RNAi) to selectively remove each DmMyb complex member revealed that DmMyb protein levels are dependent upon the presence of several of the complex members. Together, these data support a model in which DmMyb activates a repressive complex containing Mip130 into a complex competent to support replication at specific loci in a temporally and developmentally proscribed manner.
Keywords: Drosophila, DmMyb, replication, chorion amplification
DNA replication in higher eukaryotes is developmentally regulated, with patterns of origin selection and cell-cycle behavior determined in a tissue-specific and temporal manner. Dramatic examples come from studies with Xenopus laevis and Drosophila melanogaster, where DNA replication in the early embryo occurs very rapidly from closely spaced origins in a seemingly non-sequence-specific manner (Bell 2002). In higher eukaryotes, the initiator complex, ORC, does not bind DNA with sequence specificity (Vashee et al. 2003; Remus et al. 2004), and this early promiscuous selection of DNA sites for replication initiation may reflect a “ground state” for the general initiation process. Later in development, the number of origins decreases and their spacing increases, and replication starts from discrete genomic locations in a cell type-specific manner. How such specialization evolved is generally not understood, though current speculation centers on the possible roles of chromosomal domains and gene expression patterns facilitating replication initiation site selection (Gilbert 2001). DNA amplification for the purpose of providing extra templates for transcription is a clear example of such interactions between gene expression and replication.
In Drosophila, a well studied replication program and DNA replication origin specification occurs at the chorion (eggshell) gene loci. At a precisely defined point in oogenesis, the follicle cells surrounding the developing oocyte cease widespread genomic replication and shortly thereafter begin repeated DNA-replication initiations from a small number of origins at four genomic locations that include the chorion loci on the X and third chromosomes (Calvi et al. 1998; Claycomb et al. 2004). Replication at these origins leads to amplification of the DNA surrounding the sites of initiation to meet the demand for high levels of eggshell protein transcripts and protein later in oogenesis (Orr-Weaver 1991). Such amplification is under the control of Cyclin E, a key regulator of the mitotic S phase (Calvi et al. 1998). Mutations in other key S-phase regulators (E2F1, E2F2, DP, and Rbf1) also selectively disrupt the amplification program (Royzman et al. 1999; Bosco et al. 2001; Cayirlioglu et al. 2001). As the process requires repeated rounds of DNA replication initiation, the basal factors that are required for universal genomic replication are also required for chorion gene amplification. These include members of the prereplicative complex (pre-RC; DmOrc2, Cdt1, Mcm6; Landis et al. 1997; Whittaker et al. 2000; Schwed et al. 2002) as well as the replication initiation regulators, Geminin, and Dbf4 (Landis and Tower 1999; Quinn et al. 2001). The key difference with DNA replication in other situations is that the amplification origins specifically escape, by an unknown mechanism, the normal control mechanisms that prevent re-replication within a cell cycle, while other potential replication initiation sites are kept inactive.
Drosophila contains a single Myb-related gene (Dm-myb) that appears to be most closely related to the vertebrate B-Myb (Ganter and Lipsick 1999; Simon et al. 2002). Dm-myb is essential (Katzen and Bishop 1996; Fitzpatrick et al. 2002; Manak et al. 2002), and studies of Dm-myb mutants suggest nontranscriptional roles for DmMyb in S phase and in the prevention of genomic instability, as well as transcriptional roles at the cyclin B promotor (Okada et al. 2002). Previously, we showed that a DmMyb-containing complex binds site-specifically to the two critical control elements required for chorion gene amplification, ACE3 and Ori-β, on the third chromosome (Beall et al. 2002). Reporter transgenes harboring deletions in regions of the DNA important for complex binding were defective for amplification, supporting the notion that the DmMyb complex is required for gene amplification in both cis and trans. Furthermore, somatic clones in the follicle cell layer surrounding the developing oocyte that were mutant for Dm-myb were defective for amplification. The characteristic thin eggshell phenotype of female-sterile mutations and loss of bromodeoxyuridine (BrdU) incorporation at the amplification foci were found in Dm-myb mutant patches despite proper DmOrc2 and Cdt1 localization at the amplification foci. Taken together, these results suggest that DmMyb is required for replication at the chorion origins at a step postprereplication complex (pre-RC) assembly. Because both DmMyb and one other complex member, Mip120, bind to ACE3 in vivo (as assayed by chromatin immunoprecipitation; Beall et al. 2002), and no “free” DmMyb could be detected biochemically, it seemed likely that DmMyb was acting at the chorion origins in association with the other complex members, Mip40, Caf1 p55, Mip120, and Mip130. These data left open important questions such as: What is DmMyb's function at the chorion origins and in replication in general? Does DmMyb always work in association with the four other complex members, or does DmMyb have functions that are independent of the DmMyb complex? Stated another way: While DmMyb's essential function(s) for viability are undetermined, we wanted to know if DmMyb's activity was executed coordinately with the other complex members.
To address the issue of DmMyb's dependence on the complex and to gain a better understanding of the function(s) of the DmMyb complex in vivo, we used targeted mutagenesis by homologous recombination (Rong and Golic 2000, 2001; Rong et al. 2002) to generate mutations in the largest subunit of the DmMyb complex, Mip130. Unlike with Dm-myb, we found that mutations in mip130 were viable. In addition, mip130 mutant females were sterile and showed BrdU incorporation throughout the follicle cell nucleus at stages normally undergoing chorion gene amplification, reminiscent of the phenotype observed with e2f2 mutants (Cayirlioglu et al. 2001). Most surprisingly, but certainly solidifying a critical genetic interaction between the members of the DmMyb complex, we found that Dm-myb mip130 double mutants were viable. This result suggests that in the absence of DmMyb, a Mip130 activity may be responsible for the observed lethality of Dm-myb mutants. RNA interference (RNAi) to selectively eliminate protein expression for each DmMyb complex member revealed that DmMyb protein levels were dependent upon several of the complex members. Together, our data suggest that DmMyb's primary function is as a member of the defined complex, and that members of the complex have both positive and negative roles in replication. Our current model is that DmMyb activates a repressive complex containing Mip130, normally required for silencing regions of the chromosome not targeted for replication, into a complex competent to support replication at specific loci, such as the chorion origins. Based upon other observations to be discussed, we posit that the activation/repression roles of the DmMyb complex function more generally for many DNA-related activities, including gene expression.
Results
mip130 mutants are viable and show decreased DmMyb stability
We used “ends in” gene targeting to generate mutant alleles in the largest subunit of the DmMyb complex, mip130 (Rong and Golic 2000, 2001; Rong et al. 2002). To do this, we engineered an I-SceI restriction endonuclease site and point mutations in the coding region of mip130 on either side of the I-SceI site to produce truncated forms of Mip130 upon targeted integration. The mutagenesis required step-wise strain construction: Transgenic animals carrying the mutant mip130 construct within a P element (that also harbored binding sites for FLP recombinase flanking the insert) were first generated by standard methods. The mip130 mutant was then obtained after crossing this P element-harboring line to animals that express both the I-SceI restriction enzyme and FLP recombinase. Expression of both FLP and I-SceI in an individual resulted in excision of the donor sequences from the P element and generated a double-stranded DNA break within mip130. The double-stranded DNA break stimulated homologous recombination between the donor DNA and the corresponding chromosomal target locus. The recombination resulted in two mutant copies of mip130 at the normal genomic locus that were separated by intervening sequences as outlined in Figure 1 (mip1301-723 + 1-36). We engineered point mutations in the donor construct such that the left copy of mip130 contained a new EcoRI site 2136 nucleotides from the start codon that could produce a 723-amino acid, C-terminal truncated protein. The right copy contained a new BamHI site 45 nucleotides from the start codon that could produce a 36-amino acid protein (of which the last 21 amino acids were not Mip130 sequence). Of two potential candidates, we found that one gave the correct restriction endonuclease pattern for a targeted insertion at the proper location on the X chromosome for mip130 (Fig. 1B).
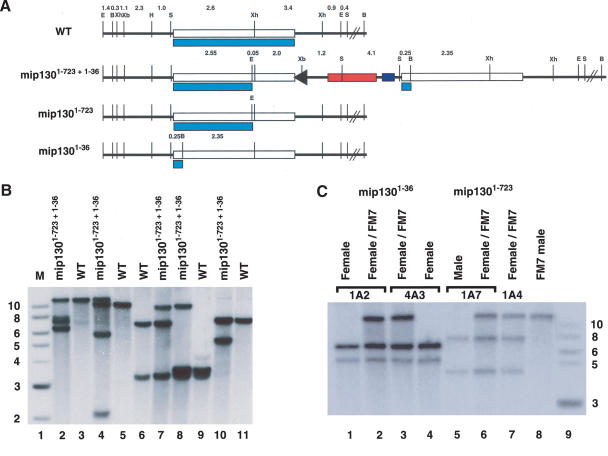
Figure 1.
Molecular verification of the mip130 mutant. (A) Restriction endonuclease map of the region surrounding the mip130 gene. (WT) Wild type. The normal chromosomal configuration without any mutations. (mip1301-723 + 1-36) The targeted mutation showing the duplication of mip130 and intervening P-element sequences. The arrowhead indicates the residual FLP-recombinase-binding site, the red box indicates the mini-white gene marker, and the blue box indicates the I-CreI restriction endonuclease site. (mip1301-723) The single-copy reduction event leaving the mip1301-723 allele at the locus. (mip1301-36) The single-copy reduction event leaving the mip1301-36 allele at the locus. Light blue boxes denote the Mip130 protein product that can be encoded by the locus. Distances in kilobases, between restriction endonuclease sites, are indicated at the top. (B) BamHI; (E) EcoRI; (H) HindIII; (S) SnaBI; (Xb) XbaI; (Xh) XhoI. (B) Southern blot of mip1301-723 + 1-36. Samples were analyzed in pairs for each enzyme, with digested DNA from mip1301-723 + 1-36/FM7 and wild type (WT) loaded side by side for direct comparison. Digests and expected fragments in kilobases are as follows: (lanes 2,3) EcoRI and BamHI (11.6 wild type and 7.6, 7.25, and 6.65 mutant); (lanes 4,5) EcoRI and XbaI (10.2 wild type and 12.2, 5.85, and 2.05 mutant); (lanes 6,7) XhoI (7.0 and 3.4 wild type and 9.9 mutant); (lanes 8,9) HindIII and XhoI (3.6 and 3.4 wild type and 9.9 mutant); (lanes 10,11) SnaBI (7.3 wild type and 5.8 mutant). The probe used was a 1.8-kb fragment derived from the mip130 gene spanning the internal XhoI site. M, molecular weight markers with sizes in kilobases, given at the left. (C) Southern blot of reduction alleles. Two independent lines for each allele (either mip1301-36 or mip1301-723) and wild type (FM7) were analyzed from the fly type indicated at the top. DNAs were digested with both EcoRI and BamHI, and fragments were detected with an internal mip130 probe spanning the XhoI site. Sizes expected in kilobases are as follows: mip1301-36 allele, 11.6 wild type and 6.65 and 4.95 mutant; mip1301-723 allele, 11.6 wild type and 7.25 and 4.35 mutant.
Because one gene copy at the mip1301-723 + 1-36 locus can encode a truncated Mip130 protein of substantial size, we reduced the tandem duplication to a single copy in order to generate a true null mutant. We expressed I-CreI endonuclease in the mip1301-723 + 1-36 heterozygous mutant animals to generate a double-stranded DNA break at the specific I-CreI-site in the locus (Fig. 1A). Repair of the break by recombination between the homologous regions flanking the double-stranded DNA break can eliminate the intervening sequences, including the marker gene, thereby providing a simple eye-color selection to detect reduction events. Reduction can lead to recovery of either mip130 allele, in addition to the wild-type configuration at the locus. We found the reduction to be very efficient, and we recovered several lines for each allele. Southern blot analysis confirmed that we had successfully isolated each allele (Fig. 1C).
Although we found that all alleles were viable (Table 1A,B), they were generally not healthy, appeared smaller than nonmutant siblings, and the mip1301-723 + 1-36 and mip1301-723 alleles displayed a mild rough-eye phenotype (data not shown). Interestingly, we found that there were significant differences in the alleles. The mip1301-723 allele was more severely affected than the mip1301-723 + 1-36 and mip1301-36 alleles, and we were unable to generate mip1301-723 mutant females for fertility tests and staining assays due to a very short adult lifespan. Based on these data and data to be presented below, we believe that we have generated an allelic series ranging from a null mutant (mip1301-36) to a weak neomorph (mip1301-723 + 1-36) to a strong neomorph (mip1301-723).
Table 1A.
mipl 30 mutant male viability
| Strain | Heterozygous female | FM7/FM7 female | FM7 male | Mutant male |
|---|---|---|---|---|
| mip 1301-723 + 1-36 | 33.7 ± 1.16 | 23.7 ± 1.8 | 21.9 ± 1.3 | 20.8 ± 1.1 |
| mip 1301-36 1A2 | 29.3 ± 2.1 | 21.4 ± 2.9 | 20.6 ± 0.8 | 28.8 ± 1.4 |
| mip 1301-723 1A7 | 34.8 ± 1.6 | 24.5 ± 1.4 | 24.9 ± 3.7 | 15.8 ± 3.3 |
| MH107 | 45.5 ± 5.7 | 28.9 ± 5.5 | 25.6 ± 1.0 | 0 ± 0 |
| myb mip 1301-723 + 1-36 #6 | 39.4 ± 2.7 | 26.3 ± 1.6 | 29.1 ± 3.9 | 7.3 ± 3.0 |
| myb mip 1301-723 + 1-36 #19 | 39.5 ± 2.0 | 30.1 ± 2.0 | 25.1 ± 3.2 | 5.3 ± 1.4 |
| myb mip 1301-36 1A2 | 32.9 ± 1.2 | 26.3 ± 1.9 | 26.3 ± 2.6 | 14.6 ± 2.3 |
| myb mip 1301-723 1A7 | 38.7 ± 2.5 | 32.6 ± 2.0 | 28.2 ± 3.4 | 0.6 ± 0.2 |
|
B. mip 130 mutant female viability
|
|
|
|
|
|---|---|---|---|---|
| Strain | Heterozygous female | Homozygous mutant female | FM7 male | Mutant male |
| mip 1301-723 + 1-36 | 27.8 ± 0.7 | 24.4 ± 0.7 | 26.6 ± 0.8 | 21.2 ± 1.4 |
| mip 1301-36 1A2 | 30.0 ± 2.4 | 25.7 ± 2.6 | 15.6 ± 2.1 | 28.8 ± 1.8 |
| myb mip 1301-723 + 1-36 #6 | 39.2 ± 3.3 | 17.6 ± 1.9 | 34.6 ± 3.0 | 8.6 ± 3.2 |
| myb mip 1301-36 1A2 | 28.3 ± 1.3 | 25.3 ± 1.1 | 22.4 ± 1.2 | 24.0 ± 0.6 |
(A) All progeny were counted from the following cross: mutant/FM7cKrGFP virgins × FM7cKrGFP males. Between 700 and 4500 progeny per strain were counted, and the values listed are the average percent viability ± standard deviation from at least three independent experiments. MH107 is a lethal mutation in Dm-myb. (B) All progeny were counted from the following cross: mutant/FM7cKrGFP virgins × mutant males. Between 1800 and 3000 progeny per strain were counted, and the values listed are the average percent viability ± standard deviation from at least three independent experiments.
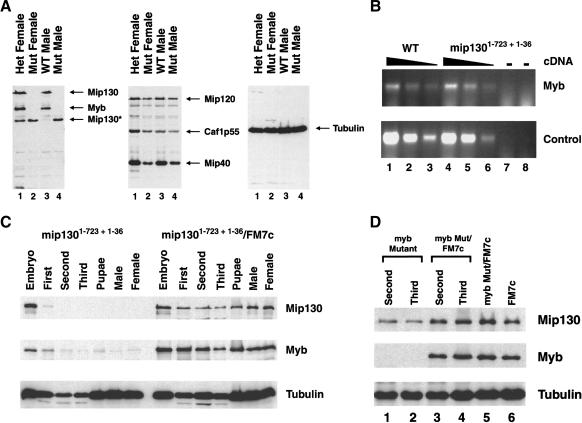
Immunoblot analysis of the three mip130 mutants showed that none produced a full-length Mip130 protein (Figs. 2A [left panel], 6D [below]). In fact, the mip1301-723 + 1-36 and mip1301-723 alleles produced a truncated Mip130 protein of a molecular weight as anticipated to be encoded by the mip1301-723 allele. Interestingly, we found that DmMyb protein levels were severely reduced in all mip130 mutants (Figs. 2A [left panel, lanes 2,4], 6D [below, both panels, lane 4]), whereas the levels of the other complex members were only modestly reduced (Fig. 2A, middle panel). Because low levels of DmMyb were detected in the mip1301-36 mutant (Fig. 6D [below], upper panel, cf. lanes 3 and 4), we speculate that in vivo, the truncated Mip130 protein present in the DmMyb complex in either the mip1301-723 + 1-36 or the mip1301-723 mutant affects DmMyb degradation most effectively. As we will show below, the phenotypic differences in the allelic series are not due to differences in residual DmMyb protein levels. We suspect that the allelic differences may be due to the expression levels of the truncated Mip130 protein and that the viability differences between alleles may in fact be the result of different levels of the truncated Mip130 protein in a particular cell type. In extracts derived from mutant adult males, ∼20% more of the truncated Mip130 protein was measured in mip1301-723 animals versus mip1301-723 + 1-36 animals (Supplementary Fig. 1), consistent with the most severely affected allele expressing the highest levels of the truncated Mip130 protein.
Figure 2.
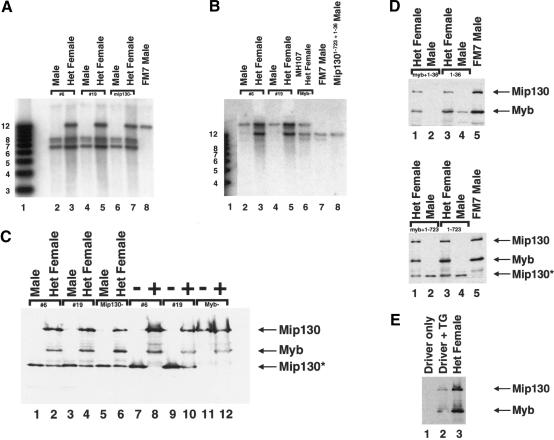
DmMyb levels are severely reduced in mip130 mutants. (A) Flies of the type indicated at the top of each lane were homogenized in sample buffer, and 0.5 fly per lane was examined for the presence of the protein indicated at the right of each panel by immunoblot analysis. Tubulin was used as a loading control (right panel). Mip130* represents the truncated Mip130 product produced by the mip1301-723 allele in mip1301-723 + 1-36 mutants. (B) RT-PCR analysis of RNA isolated from ovaries dissected from either mip1301-723 + 1-36 mutant or wild-type control females. Twofold serial dilutions of the input cDNA were analyzed for either Dm-myb transcripts or an unrelated transcript (control) as indicated on the right. (-) PCR performed on the isolated RNA prior to cDNA synthesis in order to ensure that there was no contaminating genomic DNA. (C) Progeny from the cross between mip1301-723 + 1-36/FM7cKrGFP (GFP-containing chromosome) females and mip1301-723 + 1-36 mutant males were sorted for GFP-containing (mip1301-723 + 1-36/FM7c) and non-GFP (homozygous mip1301-723 + 1-36 mutant) animals at the stages indicated at the top of the figure. Immunoblot analysis was performed with the antibodies indicated on the right. Tubulin was used as a loading control. (D) Same as in C except that the cross was between Dm-myb mutant MH107/FM7cAcGFP females and FM7c males.
Figure 6.
Dm-myb mip130 homozygous double mutants are viable. (A) Southern blot analysis of Dm-myb mip1301-723 + 1-36 double mutants: mip130 locus determination. Two independent lines (#6 and #19), the original mip1301-723 + 1-36 mutant, and wild type (FM7) were analyzed as indicated at the top. DNAs were digested with both EcoRI and BamHI, and fragments were detected with an internal mip130 probe spanning the XhoI site. Sizes expected, in kilobases, are as follows: 11.6 for wild type and 7.6, 7.25, and 6.65 for mutant. (B) Southern blot analysis of Dm-myb mip1301-723 + 1-36 double mutants: Dm-myb locus determination. Two independent lines (#6 and #19), the original Dm-myb mutant (MH107), wild-type (FM7), and mip1301-723 + 1-36 mutants were analyzed as indicated on the top. DNAs were digested with BamHI, and fragments were detected using a probe specific for Dm-myb. Sizes expected, in kilobases, are as follows: 10.4 wild type and 14.5 mutant. (C) Immunoblot analysis of Dm-myb mip1301-723 + 1-36 double mutants. Flies (lanes 1-6) and pupae (lanes 7-12) of the type indicated at the top were blotted for Mip130 and DmMyb as indicated at the right. (-) GFP-negative pupae representing homozygous mutant animals, (+) GFP-positive control siblings. (D) Immunoblot analysis of the Dm-myb mip1301-723 and Dm-myb mip1301-36 double mutants. Flies of the type indicated at the top of each panel were blotted for Mip130 and DmMyb as indicated at the right. (Upper and lower panels, lane 1) double mutant/FM7 female. (Lane 2) Homozygous double mutant male. (Lane 3) Heterozygous mip1301-36 1A2/FM7 (upper) or mip1301-723 1A7/FM7 (lower) female. (Lane 4) Homozygous mip1301-36 1A2 (upper) or mip1301-723 1A7 (lower) mutant male. (Lane 5) FM7 control male. (E) Immunoblot analysis of the mip1301-36 mutant complementation experiment. Transgenic lines (TG) containing the Mip130 cDNA under the control of Gal4 binding sites were used to complement the mip1301-36 mutant allele for female sterility. Gal4 was under the control of the e22c enhancer (driver), which expresses Gal4 in the ovary as well as at other stages in development. Females were homogenized in sample buffer, and 0.5 fly per lane was analyzed for Mip130 and DmMyb as indicated on the right. (Lane 1) Flies that were mip1301-36 homozygous mutant and carried the e22c driver only. (Lane 2) Flies that were mip1301-36 homozygous mutant and carried both the e22c driver and the mip130 cDNA transgene-1. (Lane 3) Heterozygous mip1301-36/FM7 sisters that carried the e22c driver only. The flies carrying both the driver and Mip130 transgene (lane 2) resulted in expression of both Mip130 and DmMyb.
Immunofluorescence analysis of mutant ovaries from mip1301-723 + 1-36 (data not shown) and mip1301-36 females also showed an absence of DmMyb protein staining in the follicle cell nuclei surrounding the developing oocyte (see Fig. 5A, below). RT-PCR analysis of RNA isolated from mip1301-723 + 1-36 mutant ovaries showed that the Dm-myb transcript levels were comparable to those from wild-type ovaries (Fig. 2B), suggesting that the absence of DmMyb protein in mip1301-723 + 1-36 mutants is due to protein instability and not the result of reduced transcription.
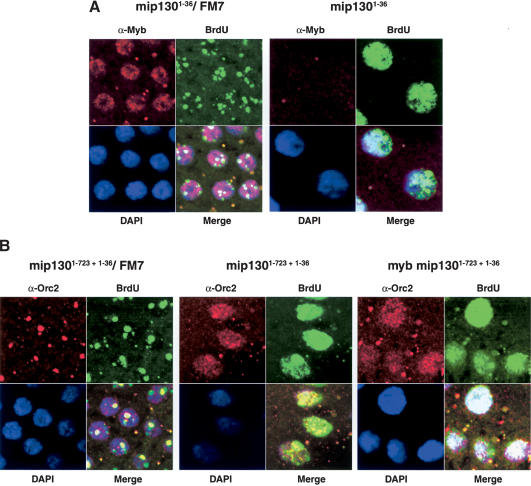
Figure 5.
BrdU incorporation occurs throughout the nucleus in mip130 mutants. (A) Stage 10 egg chambers from either mip1301-36/FM7a or mip1301-36 homozygous mutants. (Red) Anti-Myb; (green) anti-BrdU; (blue) DAPI. (B) Stage 10 egg chambers from mip1301-723 + 1-36/FM7 (left), mip1301-723 + 1-36 homozygous mutant (center), and Dm-myb mip1301-723 + 1-36 homozygous double mutants (right). (Red) Anti-Orc2; (green) anti-BrdU; (blue) DAPI.
Because we found that DmMyb protein levels were reduced in mip130 mutants, we were interested in determining at what stage in mip130 mutant development DmMyb protein levels decline. We created a mip1301-723 + 1-36 mutant strain in which the nonmutated X chromosome harbored a green fluorescent protein (GFP)-producing transgene so that mutant animals at any stage could be sorted from nonmutant animals. When heterozygous mutant females were crossed to mutant males, the absence of the GFP signal using a fluorescence microscope allowed for such manual sorting of homozygous mutant animals. We found that Mip130 levels severely dropped after the maternal stores were depleted following embryogenesis, and that there was a corresponding drop in DmMyb levels shortly thereafter (Fig. 2C). When we performed the reciprocal experiment, we found that with Dm-myb mutants, Mip130 levels were reduced but not eliminated (Fig. 2D). Together, these data suggest that DmMyb requires the presence of full-length Mip130, possibly in association with the other DmMyb complex members, for normal protein levels. The DmMyb protein itself may be unstable without Mip130, perhaps due to misfolding or perhaps due to targeted degradation when not in complex with the other members. The finding that Mip130 levels were not eliminated in Dm-myb mutant animals is also consistent with the presence of a Mip130-containing complex lacking DmMyb in such mutant strains.
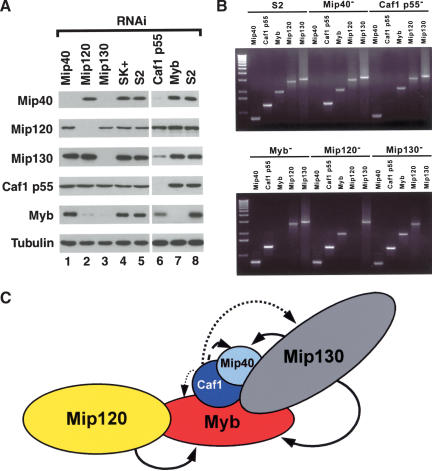
Because we saw such a dramatic decrease in DmMyb protein levels in mip1301-723 + 1-36 mutants, we were curious to examine the level of each DmMyb complex member when a specified member was removed. To this end, we used RNA-interference (RNAi) on Drosophila Schneider S2 cells and analyzed cell pellets after a 3-d incubation with the double-stranded RNA for the presence of each DmMyb complex member, using immunoblot analysis. During the RNAi analysis, no effects upon the growth rates of the RNAi-treated cells were detected (data not shown). We found that DmMyb was the most sensitive to the loss of another complex member, as depletion of Mip120, Mip130, or Caf1 p55 had dramatic effects upon DmMyb protein levels (Fig. 3A). Similar results were obtained using another Drosophila cell line (Kc), and separate experiments using two distinct, nonoverlapping double-stranded RNAs for Mip120 and Mip130 showed identical results (data not shown). The reduced protein levels in the RNAi experiments were caused by posttranscriptional mechanisms, because the only DmMyb complex member transcript affected in each RNAi experiment was that to which the double-stranded RNA was targeted (Fig. 3B). Depletion of Caf1 p55 affected all complex members except Mip120, consistent with its role as a platform for protein complex assembly in many other complexes. Conversely, Caf1 p55 appeared to be the least sensitive to loss of any DmMyb complex member, presumably due to its presence in a number of complexes in Drosophila. To summarize, we schematically illustrated the interactions between the DmMyb complex members in Figure 3C. The bold arrows indicate that loss of a given target protein had a profound effect upon the levels of the subunit to which the arrow is directed, and dotted lines depict very reproducible but smaller effects. We are uncertain as to the true contacts made by individual complex members; however, it seems that the levels of DmMyb and Mip40 are the most sensitive to another subunit loss. Furthermore, loss of Caf1 p55 and Mip130 affected the levels of several other complex members most significantly and therefore may be part of the “core” of the DmMyb complex. Previous biochemical fractionation experiments showed that DmMyb was always found in complex with the other four subunits, as measured by immunoblot analysis, through the final purification step (data not shown). Further fractionation experiments following the other complex members will be needed to explore the nature of other subcomplex assemblies.
Figure 3.
RNAi analysis of the Myb complex. (A) RNA-interference was performed on Drosophila S2 cells with the double-stranded RNAs targeted towards the transcript indicated above each lane. Cells were harvested after 3 d, and immunoblot analysis was performed on the cell pellets with the antibodies indicated on the left. Tubulin was used as a loading control. SK+, RNAi using a nonspecific control RNA against a fragment of the Blue-Script plasmid. S2, mock-treated cells. (B) RT-PCR analysis on RNA isolated from RNAi-treated cells as indicated above each line. Shown is an ethidium bromide-stained agarose gel containing PCR products from reactions performed with primers targeted to the specific transcript indicated above each lane. For each RNAi sample, selective depletion of the transcript for which the dsRNA was targeted was observed. (C) Schematic diagram of the stability of each protein after RNAi analysis. Arrow strength (solid > large dash > medium dash > small dash) is indicative of the amount that each protein was reduced in the absence of the indicated complex member.
mip130 mutant females display defects in chorion gene amplification
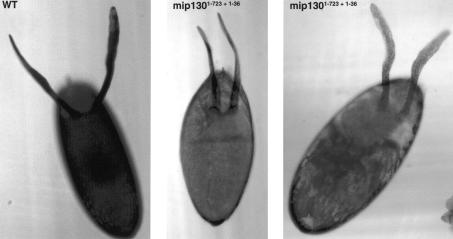
One of the hallmark features of some viable replication mutants is female sterility resulting from reduced chorion gene amplification that leads to thin eggshell production (Calvi and Spradling 1999). As with the viability results, we found a phenotypic range for the different mip130 mutants ranging from partial fertility for the null allele (mip1301-36) to complete infertility for the mip1301-723 + 1-36 allele (Table 2). Moreover, the “fertility” displayed by the mip1301-36 mutant females was severely impaired: Most individual mip1301-36 females had only one or two larvae crawling in vials after 7 d of egg laying, whereas the control heterozygous sisters had upwards of 100 larvae crawling in a vial from a single female. Eggs that were laid by mip130 mutant females were flaccid and contained thin eggshells, as exemplified by mip1301-723 + 1-36 in Figure 4. Often, the dorsal appendages were thin and fragile, another phenotype associated with defective chorion gene amplification. When we measured the amplification levels in mip1301-723 + 1-36 mutants, we found that the levels were reduced twofold compared to heterozygous mutant sisters (data not shown), consistent with the notion that the mip1301-723 + 1-36 mutant female sterility is in part due to problems with chorion gene amplification.
Table 2.
Summary of mip130 mutant female fertility
| Strain | Mutant/FM7 | Homozygous mutant |
|---|---|---|
| mip1301-723 + 1-36 | 100 ± 0 | 0 ± 0 |
| mip1301-36 1A2 | 98 ± 4 | a26 ± 19 |
| myb, mip1301-723 + 1-36 #6 | 90 ± 7 | 0 ± 0 |
| myb, mip1301-723 + 1-36 #19 | 100 ± 0 | 0 ± 0 |
| myb, mip1301-36 1A2 | 97 ± 5 | a21 ± 11 |
| mip1301-36 + driver | ND | a14 ± 4.9 |
| mip1301-36 + driver + TG-1 | ND | 91 ± 8.3 |
| mip1301-36 + driver + TG-2 | ND | 78 ± 9.8 |
Females scored were derived from the cross: mutant/FM7cKrGFP virgin × mutant male, except for the complementation experiment (below dotted line). See Materials and Methods for details of the complementation cross. At least 10 independent experiments, for a total of 100 females, were tested for each genotype listed, and the values listed represent the average percent fertility ± standard deviation. Fertility was determined by scoring for the presence of at least one larvae in each vial after 7 d at 25°C. aActual number of larvae per vial was approximately 0.5%-1% of wild type. (Driver) Ga14 expression under control of the e22c ovary enhancer. (TG) UAS mip130 transgene; (ND) not done.
Figure 4.
Absence of mip130 results in a thin chorion. Images shown are eggs laid by either wild-type (WT, left) or mip1301-723 + 1-36 mutant mothers (center and right). The wild-type (WT) egg appears very dark, due to the opacity of the thick chorion. The eggs laid by mip1301-723 + 1-36 mutant females are thin and often collapsed (center), and contain thin and fragile dorsal appendages. Images were taken using an Axiophot 373 light microscope.
In order to ensure that the mutant phenotypes that we were studying were due to loss of Mip130 only, we performed rescue-complementation experiments using the null, mip1301-36 allele, and transgenic animals carrying the mip130 cDNA under the control of the Gal4 transcription factor. We used the e22c “driver” that expressed Gal4 (and therefore, Mip130) at several developmental stages, including in the ovary. We found that homozygous mip1301-36 mutant females that contained both the e22c driver and mip130 cDNA were significantly more fertile than their sisters that contained the e22c driver only (Table 2). The complementation restored fertility to 80%-90% of wild-type levels. Importantly, expression of Mip130 resulted in a restoration of the expression of DmMyb (see Fig. 6E [below], cf. lanes 1 and 2).
In the follicle cells surrounding the Drosophila oocyte, genome-wide replication normally ceases prior to stage 10. Subsequently, DNA replication starts again, but at this time, DNA replication initiation occurs at only four loci. These amplification foci can be visualized as four spots of BrdU incorporation or as intense spots of DmOrc2 staining in follicle cell nuclei (Fig. 5B, mip1301-723 + 1-36/FM7; Calvi et al. 1998; Royzman et al. 1999). For both mip1301-723 + 1-36 and mip1301-36 mutants, the normal shut-off of DNA replication prior to stage 10 occurred (Supplementary Fig. 2). However, beginning at stage 10, when we looked at BrdU incorporation in mip130 mutant ovaries, we found that the patterns were very different from wild type: In most nuclei, BrdU incorporation occurred throughout the entire nucleus (Fig. 5A, mip1301-36 and Fig. 5B, mip1301-723 + 1-36) and was mirrored by a punctate but dispersed DmOrc2 staining pattern (Fig. 5B, mip1301-723 + 1-36; data not shown). Superimposed upon the general BrdU and DmOrc2 staining, many mip130 mutant nuclei displayed rather large and dense staining centers that are suspected to be the chorion amplification foci. This staining pattern was mosaic: In a small fraction of mip130 mutant nuclei, the BrdU and DmOrc2 staining patterns were near normal (data not shown). Even though the abnormal BrdU incorporation persisted into the latest stages in egg chamber development (data not shown), an extra complete endocycle was not manifest in mip130 mutants (Supplementary Fig. 2). As discussed below, such phenotypes are strikingly similar to those reported for e2f2 mutants (Cayirlioglu et al. 2001). These data provide evidence that Mip130 has a role as a negative regulator to keep genome-wide replication off in regions of the chromosome not normally targeted for replication.
Dm-myb mip130 double mutants are viable
Mutations in Dm-myb are lethal, and mutants die as third instar larvae/prepupae (Katzen and Bishop 1996; Manak et al. 2002). We were therefore surprised to find DmMyb protein levels nearly absent in some mip130 mutants, because mip130 mutants were viable. In order to determine whether low levels of DmMyb were responsible for mip130 mutant viability, we made double mutant animals that contained both a lethal mutation in Dm-myb and a mutation in mip130. Because both genes reside on the X chromosome, we generated recombinants that contained both mutations on a single X chromosome. Different lines were generated for each of the alleles of mip130 in combination with the lethal allele of Dm-myb. The presence of both the mutant mip130 and Dm-myb alleles was confirmed by Southern and immunoblot analyses, and representative examples are provided (Fig. 6A,B; data not shown). To our surprise, we found that all of the Dm-myb mip130 double mutant combinations were viable (Table 1A,B). Moreover, when combined with the Dm-myb mutation, the phenotypic differences in the mip130 allelic series were even more apparent. Given that the Dm-myb lethal mutation was constant in these strains and that the null double mutant was the most robust, the allelic differences likely result from the truncated Mip130 protein levels in the different alleles, as discussed (Supplementary Fig. 1). The double mutant animals were not healthy and displayed the same phenotypes as those observed for mip130 single mutants: Males were fertile, females were generally sterile (Table 2), and BrdU incorporation and anti-DmOrc2 staining in most follicle cell nuclei occurred throughout the nucleus (Fig. 5B, right panel, Dm-myb mip1301-723 + 1-36 double mutant). Summarizing the behavior of the various double mutants, it appears that mip130 is epistatic to Dm-myb. Based on the above data, we conclude that residual DmMyb activity is not responsible for the observed viability of mip130 mutants. Furthermore, as the Dm-myb mip130 double mutants were viable, it appears that loss of Mip130 works to suppress the normal requirement for DmMyb in critical but unknown cell types.
Discussion
The primary goal of this study was to explore a potential genetic interaction between Dm-myb and the largest subunit of the DmMyb complex that we had previously defined biochemically (Beall et al. 2002). To that end, we generated mutations in mip130 and uncovered an unexpected regulatory interplay between the members of the DmMyb complex. We found that mip130 mutants, although viable, showed severely reduced DmMyb protein levels. Moreover, RNAi against each member of the DmMyb complex revealed that DmMyb protein levels were critically dependent on several of the complex members. Thus, DmMyb and other members of the complex are interdependent upon each other for their steady-state protein levels and might be unfolded or otherwise marked for degradation when the complex loses integrity.
The above experiments, though of central importance in understanding the in vivo context in which the DmMyb protein operates, were reasonable extensions of our biochemical characterization of a tightly associated five-subunit complex containing the vast majority of the cellular DmMyb protein. Unanticipated based on any previous work, we found that Dm-myb mip130 double mutants were viable, suggesting that some function of Mip130 is responsible for the observed lethality of Dm-myb mutants. The genetic interaction between Dm-myb and mip130, together with the protein accumulation data, suggests that the primary function(s) carried out by DmMyb are in close association with the other complex members. The key observation that helped unravel this genetic complexity was that loss of Mip130 leads to un-scheduled DNA replication throughout the follicle cell nuclei. A simple hypothesis that can explain the present data is that DmMyb itself activates a repressive complex that contains Mip130 as an important member. By extension, Mip130, therefore, may function as a member of a complex that can serve as either a repressor or activator of chromosomal activity. Repression and activation carried out by the DmMyb complex must be vitally important only in some tissues, as follicle cell development is not required for female viability. In tissues critical for viability, the inability to activate the repressor is more deleterious (i.e., in Dm-myb mutants alone) than when both components are lost (i.e., in Dm-myb mip130 double mutants). Clearly in the ovarian follicle cells, proper development requires both components. However, before discussing our model in greater detail, we wish to first summarize other data and draw parallels to another constellation of DNA-binding factors that have a similar functional relationship to DmMyb and Mip130.
Our data showing that DmMyb is bound to ACE3 in vivo, DmMyb-binding sites are required in cis for amplification, the DmMyb complex is associated with DmORC, and Dm-myb mutant follicle cell clones show pre-RC assembly at the amplification foci without BrdU incorporation (Beall et al. 2002) suggest that DmMyb is required as a positive regulator for replication initiation at the chorion origins. Our observation that in mip130 mutants, BrdU incorporation occurs throughout the follicle cell nuclei at stages normally undergoing site-specific amplification suggests that Mip130 acts as an inhibitor of replication at regions of the genome not normally targeted for replication. Interestingly, these Dm-myb and mip130 mutant phenotypes are strikingly similar to those of e2f1 and e2f2 mutants. For e2f1i1 mutants that contain a mutation in the DNA-binding domain of E2F1, there is severely reduced or no BrdU incorporation in follicle cell nuclei at amplification stages (Royzman et al. 1999). In contrast, in e2f2 null mutants, genome-wide BrdU incorporation occurs throughout the follicle cell nucleus at amplification stages (Cayirlioglu et al. 2001). Thus E2F1 works as an activator of site-specific amplification, whereas E2F2 clearly participates in repression. Further drawing a parallel with the Dm-myb/mip130 data presented here, even though e2f1 null mutants are lethal at late larval/early pupal stages, e2f1 e2f2 double mutants survive to near adulthood (Frolov et al. 2001). The complicated genetic interaction between the Drosophila E2Fs suggests that the observed lethality of e2f1 mutants is in part due to unchecked E2F2 activity.
E2F1 has been primarily studied as a transcription factor, required for gene expression of some of the key proteins required for the G1/S transition. Nevertheless, E2F1 directly binds to ACE3 and is associated with DmOrc2 in ovary extracts (Bosco et al. 2001), providing evidence that E2F1 has roles at the chorion loci that are independent of transcription. These data together imply that both the E2F and DmMyb factors site-specifically coregulate replication at the ACE3 locus and may also carry out parallel roles in repression. Data to be presented elsewhere (P. Lewis and M. Botchan, in prep.) directly support these arguments: Fractionation of embryo extracts with a final affinity step using anti-Mip120 antibodies has shown that Mip120 and the other DmMyb complex members are in tight association with E2F2, DP1, Rbf1, Rbf2, and the histone deacetylase Rpd3. These biochemical results link the repressive activities of the Mip complex and E2F2, first brought to light by genetic and cytological experiments. A reasonable extrapolation from these data is that at ACE3, the E2F2 and Mip activities coordinate repression before stage10 and subsequently, E2F1 and DmMyb act coordinately to activate replication site-specifically during amplification. A proposed switch between E2Fs has been suggested previously (Frolov et al. 2001; Cayirlioglu et al. 2003). Although both the DmMyb and E2F complexes are cis-acting at the ACE3 enhancer (Bosco et al. 2001; Beall et al. 2002; E. Beall and M. Botchan, unpubl.) there are no direct data as yet that link the complexes to other specific chromosomal replication origin control elements.
Focusing solely on the Mips and DmMyb, in Figure 7 we illustrate how these factors might function as regulators of chromosomal activity. We posit that at some stage prior to stage 10 in egg chamber development, the DmMyb complex is bound to many sites in the genome and is required for inactivating potential replication initiation sites and perhaps inhibiting gene expression. In this configuration, DmMyb acts as a silent partner and is not required for repression. Through unknown mechanisms, the ACE3 locus is targeted as a site for replication initiation and is subsequently bound by members of the pre-RC. In a late step of origin activation, we suggest that DmMyb somehow converts an inactive replication origin to one that is competent for initiation, possibly through posttranslational modification of DmMyb. Such a switch in mammalian B-Myb transcriptional activity after phosphorylation has been suggested (Li and McDonnell 2002). In the absence of Mip130, as exemplified in the mip1301-723 + 1-36 strain, repression (both transcriptional and replicative) is relieved, pre-RC protein transcript levels may increase, and pre-RC assembly at chromosomal positions may no longer be restricted. This could lead to pre-RC association at sites spread through different regions along the entire chromosome. The model stipulates that neither DmMyb nor Mip130 targets the pre-RC to an origin, but rather suggests that DmMyb and Mip130 participate in origin activation and repression directly. We further emphasize that our data have shown a cis-acting function for the DmMyb complex only at ACE3, and it seems entirely likely that the DmMyb complex has functions both in cis and in trans in the developmental pathway under focus here.
Figure 7.
Model for how the DmMyb complex functions in transcription and/or replication. The DmMyb complex comprised of Mip40, Caf1 p55, DmMyb, Mip120, and Mip130 (blue and green balls) is bound to many sites along the chromosome and ensures a silenced transcriptional and/or replicative state. In this model, DmMyb is a silent partner of the repressive complex and does not play a role in repression. Through some unknown mechanism, a site is targeted for replication initiation, and is subsequently bound by members of the pre-RC (orange balls). In a late step of replication initiation, DmMyb (red) converts an inactive replication origin (purple) to one that is competent for initiation (yellow), possibly after appropriate posttranslational modification(s) of DmMyb have occurred. In this way, the lethal Dm-myb mutant phenotype results from the inability to activate the repressive complex containing Mip130. In double mutant animals, the loss of the activator (DmMyb) is compensated by the loss of the repressive complex (Mips), and pre-RCs can assemble at many sites along the chromosome that serve as sites of replication initiation.
Microarray analyses of e2f2 mutant follicle cells reveal an increase in transcription of several key replication factors, including DmOrc5, which may in part explain the genome-wide replication observed in e2f2 mutants (Cayirlioglu et al. 2003). Thus by analogy, we posit that the DmMyb complex has roles both in cis and in trans for gene expression (and other chromosomal activities), perhaps in some cases acting antagonistically with E2F family members (Fitzpatrick et al. 2002). It is clear that the DmMyb complex is not required for replication from every origin, as mutants lacking both DmMyb and Mip130 are viable. Rather it appears that the DmMyb complex is required for activating replication of a set of origins at certain developmental stages or tissues, such as in the follicle cells undergoing chorion gene amplification.
Certain facts about the members of the DmMyb complex also provide some hints about the mechanistic functions of the complex. Caf1 p55 is a member of several different complexes involved in histone metabolism in Drosophila, including the chromatin-remodeling complex NURF (Martinez-Balbas et al. 1998) and the chromatin assembly complex Caf 1 (Tyler et al. 1996). Mip130 is a member of a new family of proteins that is conserved from plants to humans and includes the Drosophila always early (aly) gene and the Caenorhabditis elegans SynMuvB gene, lin-9 (White-Cooper et al. 2000). Both the aly and SynMuvB genes encode chromatin-binding proteins and may have roles in DNA remodeling complexes for both transcription and DNA replication (White-Cooper et al. 1998; Solari and Ahringer 2000). Furthermore, Mip120 contains a C-terminal cysteinerich domain that is found in a number of chromatin-binding proteins including members of the polycomb group. Myb family members have been shown to interact with both histone acetyltransferases and deacetylases (Ganter and Lipsick 1999), and DmMyb is known to interact with the acetyltransferase p300 (Fung et al. 2003). The nucleosome structure surrounding eukaryotic origins is usually in a nuclease-sensitive state similar to that found at promoters and enhancers, and an ORC-dependent nucleosome configuration at ARS1, in Saccharomyces cerevisiae, is required for pre-RC assembly and origin firing (Lipford and Bell 2001). Together, these data support a role for chromatin modifying and/or remodeling complexes in promoting DNA replication initiation. Therefore, it is tempting to speculate that one function for the DmMyb complex is to regulate chromatin structure in such a way as to permit replication initiation and/or transcription through association of a chromatin-remodeling and/or modifying complex.
Materials and methods
Plasmid construction
For the mip130 donor element, 4.3 kb of DNA containing all of exon 2 and flanking genomic DNA (containing 485 bp upstream and 1007 bp downstream of the coding sequence) was amplified from genomic DNA in three pieces and subcloned into pHSX. Nucleotide changes were introduced in the PCR fragments as follows to generate the mip1301-723 and mip1301-36 alleles: The mip1301-723 allele genomic sequence was changed from 5′-CAGAATCTCGGCGGCGACTGCG-3′ to 5′-CAGAATTCTCGGCGGCGACTGC-3′ by introduction of a new EcoRI site as indicated, and the mip1301-36 allele genomic sequence was changed from 5′-GCAGGAGCTTCCGTATCCGACTCCGC-3′ to 5′-GCAGGATCCTTCCGTATCCGACTCCGC-3′ by introduction of a new BamHI site as indicated. An I-SceI site was introduced into the unique Xmn site between the new EcoRI and BamHI sites using PCR. The 4.3-kb DNA was flanked by NotI sites, the modified fragment was cloned into the pT2 transformation vector (Rong et al. 2002), and Drosophila transformants were made using standard methods.
Genetics
Targeting cross: We screened for targeting events by the loss of white+/white mosaicism induced by FLP recombinase as described (Rong and Golic 2001; Rong et al. 2002). Details of the crosses used can be found in the above references. Southern blotting was used to identify and verify the targeted mutant using standard methods.
I-CreI reduction to single alleles: Reduction was carried out using the following cross: mip1301-723 + 1-36/FM7 virgins were crossed to FM7w; 70I-CreI, Sb/TM6 males (Rong et al. 2002). FM7w/mip1301-723 + 1-36; 70I-CreI, Sb females were crossed to FM7w males. After 3 d, the parents were removed and the vials heat-shocked (36°C, 1h). Heterozygous female heat-shocked progeny were crossed to FM7w males, and the white-eyed, Sb+ progeny were selected as reduction allele candidates. Southern blotting was used to identify and verify that both alleles were recovered, using standard methods.
Generation of myb mip130 double mutants: The original mip130 mutant (mip1301-723 + 1-36), mip1301-36 allele line 1A2 and mip1301-723 allele line 1A7 were used to make double mutant lines as follows: Heterozygous mip130 mutant/FM7a females were crossed to MH107/Dp(1;Y)shi[+]1,y[+] males (Dm-myb null mutant in which the region on the X chromosome spanning the Dm-myb locus is also carried on the Y chromosome as a duplication; Katzen et al. 1998) and non-FM7 female progeny were crossed to FM7cKrGFP males. Single heterozygous FM7cKrGFP virgin progeny were used to generate stocks. Double mutants were identified using PCR with the following primer pairs: to detect the Dm-myb mutation (MH107), 5′-GC TAAGGACCAGAGAAAATGCG-3′ and 5′-CTTAATAGCA CACTTCGGCACG-3′; to detect the mip1301-36 allele, 5′-GAG CAAGTAATGAAGCGAAGG-3′ and 5′-CTCATCACGGCAT TCGAATAG-3′; to detect the mip1301-723 allele, 5′-CTAC GACTCACTAAACGCCAT-3′ and 5′-GTCAGTCAGAAAGC TATGCGG-3′. Double mutant lines were confirmed by Southern analysis.
Developmental time course: mip1301-723 + 1-36/FM7cKrGFP virgins were crossed to FM7cKrGFP males, and progeny were sorted at each stage using a Leica MZFLIII microscope. Samples were homogenized in sample buffer and analyzed using 9% SDS-PAGE followed by immunoblotting with affinity-purified anti-Myb, anti-Mip130, or anti-Tubulin (Sigma) antibodies.
Complementation: For the mip130 complementation, transgenic lines containing the Mip130 cDNA under the control of Gal4 with the use of the pUAST transformation vector were constructed. The “driver” strain (the source of Gal4) was y[1] w[*]; Pw[+mW.hs] = en2.4-GAL4e22c/SM5 (Bloomington). The cross for complementation was as follows: mip1301-36/FM7; P e22c/CyO; +/+ virgin females were crossed to mip1301-36; +/+; pUASTmip130/TM3 males. Two independent UAST-Mip130 lines were tested for complementation. Ten independent experiments in which 10 single-pair matings using females of the following genotype (for a total of 100 females) were scored for fertility: mip1301-36/mip1301-36; Pw[+mW.hs] = en2.4-GAL4e22c/+; pUASTMip130/+. As a control, mip1301-36/mip1301-36; Pw[+mW.hs] = en2.4-GAL4e22c/+; TM3/+ females were scored for fertility.
RNAi and RT-PCR analysis
Double-stranded RNA production: Gene-specific PCR products (∼700 bp) flanked by T7 polymerase-binding sites (5′-CGGCCAGTGAATTGTTTAATACGACTC) were used as templates in in vitro transcription reactions to produce dsRNA with the RiboMAX large-scale RNA production system (Promega). The dsRNAs were cleaned up using the RNeasy Mini Kit (QIAGEN), eluted in RNA annealing buffer (100 mM NaCl, 20 mM Tris at pH 8.0, 1 mM EDTA), and annealed by incubation at 68°C for 10 min followed by 30 min at 37°C. An aliquot of each dsRNA was analyzed on a 1% agarose gel to check the size and integrity of the dsRNA. Primers used to generate the specific PCR products were as follows: mip40, FlyBase accession CG15119, sense-primer 5′-14355765-14355747, antisense-primer 5′-14355044-14355065-3′; caf1 p55, FlyBase accession CG4236, sense-primer 5′-11051773-11051792-3′, antisense-primer 5′-11052588-11052569-3′; Myb, FlyBase accession CG9045, sense-primer 5′-15588042-15588023-3′, antisense-primer 5′-15587607-15587628-3′; mip120, FlyBase accession CG6061, sense-primer 5′-8575300-8575318-3′, antisense-primer 5′-8576076-8576058-3′; mip130, FlyBase accession CG3480, sense-primer 5′-1675099-1675117-3′, antisense-primer 5′-1675824-1675806-3′.
RNAi: Drosophila L2 cells were diluted to a final concentration of 1 × 106 cells/mL. For each RNAi experiment, 3 mL of cells were washed with serum-free medium, dsRNA (60 μg) added directly to the medium, and the cells were rocked for 2 h at 25°C. Three milliliters of medium supplemented with 10% FCS was added, and the cells were rocked at 25°C for 3 d.
RT-PCR and immunoblot analysis: For RT-PCR, ∼5 × 106 cells were collected and total RNA was isolated using the RNeasy Mini Kit (QIAGEN). RNA samples were treated with RQ1 Rnase-free DNAse (Promega) to eliminate contaminating genomic DNA, and subsequently cleaned up with the RNeasy mini Kit (QIAGEN). Total RNA (2 μg) was used for cDNA synthesis using Superscript One-Step RT-PCR with Platinum Taq (Invitrogen). RT-PCR was performed with gene-specific primers using standard methods. For immunoblot analysis, total protein (corresponding to ∼0.15 × 106 cells) were separated using 12% SDS-PAGE and transferred to nitrocellulose; proteins were detected using affinity-purified polyclonal rabbit antibodies against individual complex members as described (Beall et al. 2002). Anti-tubulin monoclonal antibodies were from Sigma.
Antibody staining and BrdU labeling of ovaries
Ovaries were fixed and processed as described (Beall et al. 2002). Staining was carried out with anti-BrdU monoclonal (Becton-Dickinson), affinity-purified anti-Myb (Beall et al. 2002), and/or anti-Orc2 (Austin et al. 1999) polyclonal antibodies followed by fluorophore-conjugated secondary antibodies (Molecular Probes). Ovaries were visualized with a Zeiss LSM510 confocal microscope.
Acknowledgments
We thank Kent Golic for providing plasmids and fly stocks to construct the targeted mutant, Jeff Sekelsky for advice in generating the mip130 mutant strains, John Manak and Joe Lipsick for helpful advice and providing the Dm-myb mutant, and Peter Lewis and Dirk Remus for helpful discussions on the manuscript. E.L.B. was supported by an NIH training grant (CA09041) to the Cancer Research Laboratory of the University of California at Berkeley. D.G. was supported by a fellowship from the Belgian American Educational Foundation (B.A.E.F.-75). This work was also supported by NIH Grant CA-30490.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1206604.
References
- Austin R.J., Orr-Weaver, T.L., and Bell, S.P. 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes & Dev. 13: 2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall E.L., Manak, J.R., Zhou, S., Bell, M., Lipsick, J.S., and Botchan, M.R. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833-837. [DOI] [PubMed] [Google Scholar]
- Bell S.P. 2002. The origin recognition complex: From simple origins to complex functions. Genes & Dev. 16: 659-672. [DOI] [PubMed] [Google Scholar]
- Bosco G., Du, W., and Orr-Weaver, T.L. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3: 289-295. [DOI] [PubMed] [Google Scholar]
- Calvi B.R. and Spradling, A.C. 1999. Chorion gene amplification in Drosophila: A model for metazoan origins of DNA replication and S-phase control. Methods 18: 407-417. [DOI] [PubMed] [Google Scholar]
- Calvi B.R., Lilly, M.A., and Spradling, A.C. 1998. Cell cycle control of chorion gene amplification. Genes & Dev. 12: 734-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P., Bonnette, P.C., Dickson, M.R., and Duronio, R.J. 2001. Drosophila E2F2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development 128: 5085-5098. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P., Ward, W.O., Silver Key, S.C., and Duronio, R.J. 2003. Transcriptional repressor functions of Drosophila E2F1 and E2F2 cooperate to inhibit genomic DNA synthesis in ovarian follicle cells. Mol. Cell Biol. 23: 2123-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J.M., Benasutti, M., Bosco, G., Fenger, D.D., and Orr-Weaver, T.L. 2004. Gene amplification as a developmental strategy: Isolation of two developmental amplicons in Drosophila. Dev. Cell 6: 145-155. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick C.A., Sharkov, N.V., Ramsay, G., and Katzen, A.L. 2002. Drosophila myb exerts opposing effects on S phase, promoting proliferation and suppressing endoreduplication. Development 129: 4497-4507. [DOI] [PubMed] [Google Scholar]
- Frolov M.V., Huen, D.S., Stevaux, O., Dimova, D., Balczarek-Strang, K., Elsdon, M., and Dyson, N.J. 2001. Functional antagonism between E2F family members. Genes & Dev. 15: 2146-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S.M., Ramsay, G., and Katzen, A.L. 2003. MYB and CBP: Physiological relevance of a biochemical interaction. Mech. Dev. 120: 711-720. [DOI] [PubMed] [Google Scholar]
- Ganter B. and Lipsick, J.S. 1999. Myb and oncogenesis. Adv. Cancer Res. 76: 21-60. [DOI] [PubMed] [Google Scholar]
- Gilbert D.M. 2001. Making sense of eukaryotic DNA replication origins. Science 294: 96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen A.L. and Bishop, J.M. 1996. myb provides an essential function during Drosophila development. Proc. Natl. Acad. Sci. 93: 13955-13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen A.L., Jackson, J., Harmon, B.P., Fung, S.M., Ramsay, G., and Bishop, J.M. 1998. Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes & Dev. 12: 831-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G. and Tower, J. 1999. The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development 126: 4281-4293. [DOI] [PubMed] [Google Scholar]
- Landis G., Kelley, R., Spradling, A.C., and Tower, J. 1997. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc. Natl. Acad. Sci. 94: 3888-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. and McDonnell, D.P. 2002. The transcription factor B Myb is maintained in an inhibited state in target cells through its interaction with the nuclear corepressors N-CoR and SMRT. Mol. Cell Biol. 22: 3663-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford J.R. and Bell, S.P. 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7: 21-30. [DOI] [PubMed] [Google Scholar]
- Manak J.R., Mitiku, N., and Lipsick, J.S. 2002. Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability. Proc. Natl. Acad. Sci. 99: 7438-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas M.A., Tsukiyama, T., Gdula, D., and Wu, C. 1998. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl. Acad. Sci. 95: 132-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Akimaru, H., Hou, D.X., Takahashi, T., and Ishii, S. 2002. Myb controls G(2)/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J. 21: 675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L. 1991. Drosophila chorion genes: Cracking the eggshell's secrets. Bioessays 13: 97-105. [DOI] [PubMed] [Google Scholar]
- Quinn L.M., Herr, A., McGarry, T.J., and Richardson, H. 2001. The Drosophila Geminin homolog: Roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes & Dev. 15: 2741-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D., Beall, E.L., and Botchan, M.R. 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 23: 897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y.S. and Golic, K.G. 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013-2018. [DOI] [PubMed] [Google Scholar]
- ____. 2001. A targeted gene knockout in Drosophila. Genetics 157: 1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y.S., Titen, S.W., Xie, H.B., Golic, M.M., Bastiani, M., Bandyopadhyay, P., Olivera, B.M., Brodsky, M., Rubin, G.M., and Golic, K.G. 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes & Dev. 16: 1568-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I., Austin, R.J., Bosco, G., Bell, S.P., and Orr-Weaver, T.L. 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes & Dev. 13: 827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwed G., May, N., Pechersky, Y., and Calvi, B.R. 2002. Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication. Mol. Biol. Cell 13: 607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A.L., Stone, E.A., and Sidow, A. 2002. Inference of functional regions in proteins by quantification of evolutionary constraints. Proc. Natl. Acad. Sci. 99: 2912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari F. and Ahringer, J. 2000. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr. Biol. 10: 223-226. [DOI] [PubMed] [Google Scholar]
- Tyler J.K., Bulger, M., Kamakaka, R.T., Kobayashi, R., and Kadonaga, J.T. 1996. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell Biol. 16: 6149-6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S., Cvetic, C., Lu, W., Simancek, P., Kelly, T.J., and Walter, J.C. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes & Dev. 17: 1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H., Schafer, M.A., Alphey, L.S., and Fuller, M.T. 1998. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development 125: 125-134. [DOI] [PubMed] [Google Scholar]
- White-Cooper H., Leroy, D., MacQueen, A., and Fuller, M.T. 2000. Transcription of meiotic cell cycle and terminal differentiation genes depends on a conserved chromatin associated protein, whose nuclear localisation is regulated. Development 127: 5463-5473. [DOI] [PubMed] [Google Scholar]
- Whittaker A.J., Royzman, I., and Orr-Weaver, T.L. 2000. Drosophila double parked: A conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes & Dev. 14: 1765-1776. [PMC free article] [PubMed] [Google Scholar]