Abstract
Diagnosis and therapeutic interventions in pathological conditions rely upon clinical monitoring of key metabolites in the serum. Recent studies show that a wide range of metabolic pathways are controlled by circadian rhythms whose oscillation is affected by nutritional challenges, underscoring the importance of assessing a temporal window for clinical testing and thereby questioning the accuracy of the reading of critical pathological markers in circulation. We have been interested in studying the communication between peripheral tissues under metabolic homeostasis perturbation. Here we present a comparative circadian metabolomic analysis on serum and liver in mice under high fat diet. Our data reveal that the nutritional challenge induces a loss of serum metabolite rhythmicity compared with liver, indicating a circadian misalignment between the tissues analyzed. Importantly, our results show that the levels of serum metabolites do not reflect the circadian liver metabolic signature or the effect of nutritional challenge. This notion reveals the possibility that misleading reads of metabolites in circulation may result in misdiagnosis and improper treatments. Our findings also demonstrate a tissue-specific and time-dependent disruption of metabolic homeostasis in response to altered nutrition.
Keywords: circadian rhythm, liver, metabolism, metabolomics, serum
Introduction
Circadian rhythms govern a large variety of behavioral, physiological, and metabolic processes (1–4). Recent advances reveal that a very large fraction of mammalian metabolism undergoes circadian oscillations (5–12). This notion is critical, and it raises awareness of the need for increased attention to the time of monitoring clinically relevant values in patients. Indeed, studies in humans show that levels of key markers oscillate significantly (13–18), possibly leading to false or misleading reads that may result in questionable therapeutic outcomes. Thus, a comprehensive comparative analysis of the circadian metabolome in the serum versus peripheral tissues is critical to decipher the circulating metabolites that constitute a specific signature of a given physiological state.
Circadian rhythms are under the control of clocks that ensure cyclic regulation of a large spectrum of cellular and molecular mechanisms. In mammals, the central clock is located in the suprachiasmatic nucleus (SCN)2 of the anterior hypothalamus. The SCN integrates external daily cues, such as the light-dark cycle, and operates as a synchronizer for a multitude of peripheral clocks located in most tissues (19–21). Peripheral clocks respond to nutritional cues and can be uncoupled from the SCN by restricted feeding (9, 22–24). Recent studies have shown that restriction of the time of feeding (9) as well as nutritional challenge by a high fat diet (HFD) (24–26) result in extensive modifications of liver metabolism. Furthermore, the liver clock displays a highly dynamic homeostasis associated with an elaborate reprogramming of its molecular gears under nutritional challenge (26). Accumulating evidence underscores the intimate interplay between the circadian clock and cellular metabolism (21, 26–29). Indeed, many metabolic pathways are under circadian control and, in turn, may feedback to the clock system to assist in circadian timekeeping (6, 7, 11, 30). Although transcriptomics studies have extensively illustrated a substantial fraction of the genome controlled by the molecular clock (18, 23, 31–36), analysis of the metabolome has lagged behind, mostly because of technical difficulties. The relatively recent use of technologies such as liquid chromatography-mass spectrometry (LC-MS) and the subsequent development of appropriate bioinformatics tools (37) have been valuable in starting to unraveling the contribution of the circadian clock to mouse and human metabolism in a number of tissues as well as in blood and saliva (13–16).
Analyses of serum metabolome in humans and mice have been performed in a variety of conditions (i.e. sleep deprivation, phase shifting, etc.) (14, 15, 17). Because serum is a biological sample most often harvested in human patients and is also a critical linker between peripheral tissues as well as between peripheral tissues and the brain, we wanted to understand how a high fat diet affects the circadian clock at the level of serum metabolites. Here we reveal that, unlike in the liver, the overall effect of a high fat diet on the serum circadian metabolome is a profound loss of rhythmicity. Our results demonstrate that monitoring the levels of metabolites in the serum is a poor predictor of the metabolic landscape of the liver. Moreover, we underscore the possibility that the uncoupling of peripheral clocks from the SCN, known to be detrimental for energy balance (38–48), may occur through metabolic information present in the serum. Finally, we have identified specific serum metabolites diagnostic of the risk of diabetes, obesity, and other metabolic disorders that are associated with nutritional challenge.
Experimental Procedures
Mice and Diets
Age-matched, male C57BL/6J mice (JAX, 00064) were maintained on a 12-h light/12-h dark cycle. Mice had access to water and pellet food ad libitum. Animal care and use were in accordance with guidelines of the institutional animal care and use committee at the University of California (Irvine, CA). At 6 weeks of age, animals were placed on a normal chow diet (Prolab RMH 2500) or an HFD (60% of calories from fat; Research Diets, D12492) for 10 weeks. Body weight was measured weekly. Animals were separated into individual cages 1 week before sacrifice. Liver and serum were harvested across the circadian cycle every 4 h. Serum was prepared from an abdominal/thoracic blood sample. All samples were stored at −80 °C until analysis.
Mass Spectrometry for Metabolomics Analysis
Metabolomics analysis was carried out by Metabolon, Inc. (Durham, NC) using a published methodology (49). Briefly, small molecules from the liver and serum (n = 5/group and zeitgeber (ZT)) of age-matched, adult, male mice were harvested at ZT0, ZT4, ZT8, ZT12, ZT16, ZT20, and ZT24, with ZT0 corresponding to lights on and ZT12 corresponding to lights off in the animal facility. Animals were housed on a 12-h light/12-h dark schedule. Livers and serums were harvested, and livers were rinsed briefly in PBS and immediately frozen in liquid nitrogen. Frozen samples were shipped to Metabolon. The metabolic profiling platform used for this kind of analysis by Metabolon combines three independent platforms: ultra-HPLC/MS/MS optimized for basic species, ultra-HPLC/MS/MS optimized for acidic species, and gas chromatography/mass spectrometry (GC/MS). Briefly, a comparable amount of liver and serum tissue from each replicate animal for normal chow (NC) and HFD was used. Liver samples were processed essentially as described previously (26, 50). For serum samples, the LC/MS portion of the platform was based on a Waters ACQUITY UPLC system and a Thermo-Finnigan LTQ mass spectrometer, which consisted of an electrospray ionization source and linear ion trap mass analyzer. The sample extract was split into two aliquots, dried, and then reconstituted in acidic or basic LC-compatible solvents, each of which contained 11 or more injection standards at fixed concentrations. One aliquot was analyzed using acidic positive ion-optimized conditions and the other using basic negative ion-optimized conditions in two independent injections using separate dedicated columns. Extracts reconstituted in acidic conditions were gradient-eluted using water and methanol, both containing 0.1% formic acid, whereas the basic extracts, which also used water/methanol, contained 6.5 mm ammonium bicarbonate. The MS analysis alternated between MS and data-dependent MS2 scans using dynamic exclusion. The samples destined for GC/MS analysis were redried under vacuum desiccation for a minimum of 24 h before being derivatized under dried nitrogen using bistrimethyl-silyl-trifluoroacetamide. The GC column was 5% phenyl, and the temperature ramp was from 40 to 300 °C in a 16-min period. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast scanning single-quadrupole mass spectrometer using electron impact ionization. The instrument was tuned and calibrated for mass resolution and mass accuracy on a daily basis. Metabolites were identified by automated comparison of the ion features in the experimental samples with a reference library of chemical standard entries that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as associated MS spectra and curated by visual inspection for quality control using software developed at Metabolon (51).
Adipokine Level Measurements
Analyses of IL-6, leptin, and adiponectin levels in serum and liver were carried out by Eve Technologies by multiplexing laser bead technology.
Western Blotting
Equal amounts of protein from whole liver cell lysates (30 μg) were separated on polyacrylamide gel and transferred to nitrocellulose membranes. After blocking, the membranes were incubated overnight, at 4 °C, with one of the following primary antibodies: anti-phospho-GSK3β (Ser9) (Cell Signaling Technology), anti-phospho-AKT (Ser473) (Cell Signaling Technology), anti-AKT (Cell Signaling Technology), and actin. All of the primary antibodies were used at a concentration of 1:1,000.
Statistics
Data from each experimental group were assessed through replicates (n = 5) for representative mean and image. Data represent mean ± S.E. of experiments. Experiments with two variables were analyzed by two-way analysis of variance followed by Tukey's post hoc test for multiple comparisons (GraphPad Prism version 5.0). For analysis of rhythmic metabolites, the nonparametric test JTK_CYCLE was used, incorporating a window of 20–28 h for the determination of circadian periodicity, as described previously (26, 50). Metabolites with p < 0.05 were considered significant (supplemental Files 1 and 2).
Results
Comparative Circadian Metabolomics
We used LC-MS metabolomics to analyze the relative abundance of metabolites in the mouse serum and liver throughout the circadian cycle under NC and HFD conditions. Wild-type mice were divided in two experimental groups, the first fed an NC diet and the second fed an HFD (60% of calories from fat (26)) for 10 weeks. Liver and serum were harvested across the circadian cycle every 4 h. Although the hepatic circadian metabolome and transcriptome have been previously reported to undergo extensive reprogramming following nutritional challenge (26), the degree and specificity to which circulating metabolites oscillate have not been determined.
Our metabolome analysis identified 362 known metabolites in the serum (Fig. 1A), belonging to major metabolic pathways. Although a large fraction of metabolites are present in both tissues (222 metabolites), 140 metabolites, corresponding to 38.6% of the total, were detected only in serum and not in the liver (supplemental Files 1 and 2). This indicates that either 1) their abundance is too low in the liver to be detected; 2) these metabolites are not typically made in or transported to the liver; or 3) due to technological variability, they were unable to be detected.
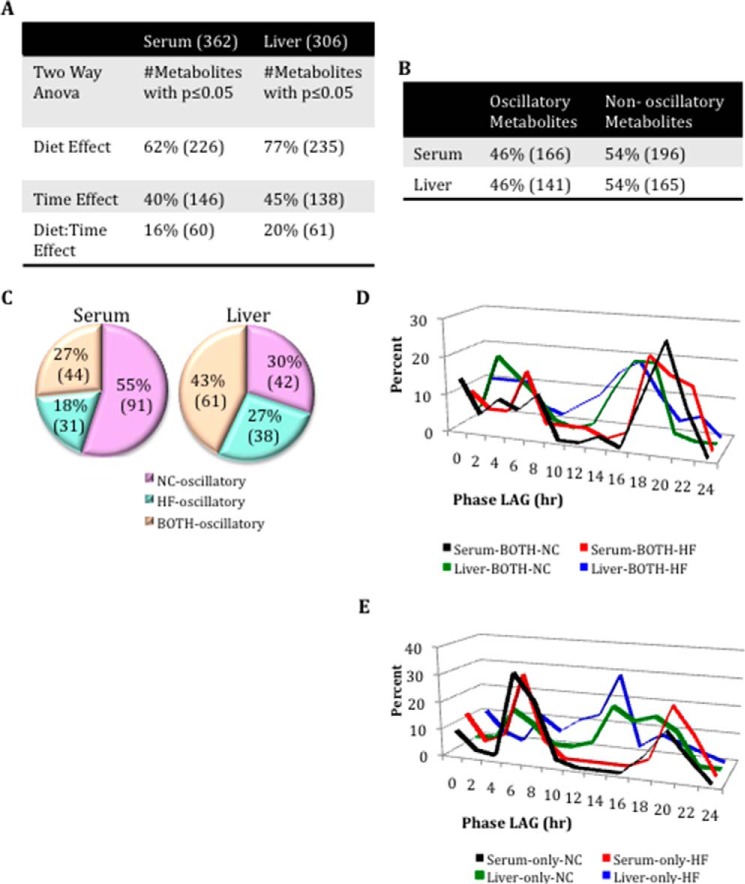
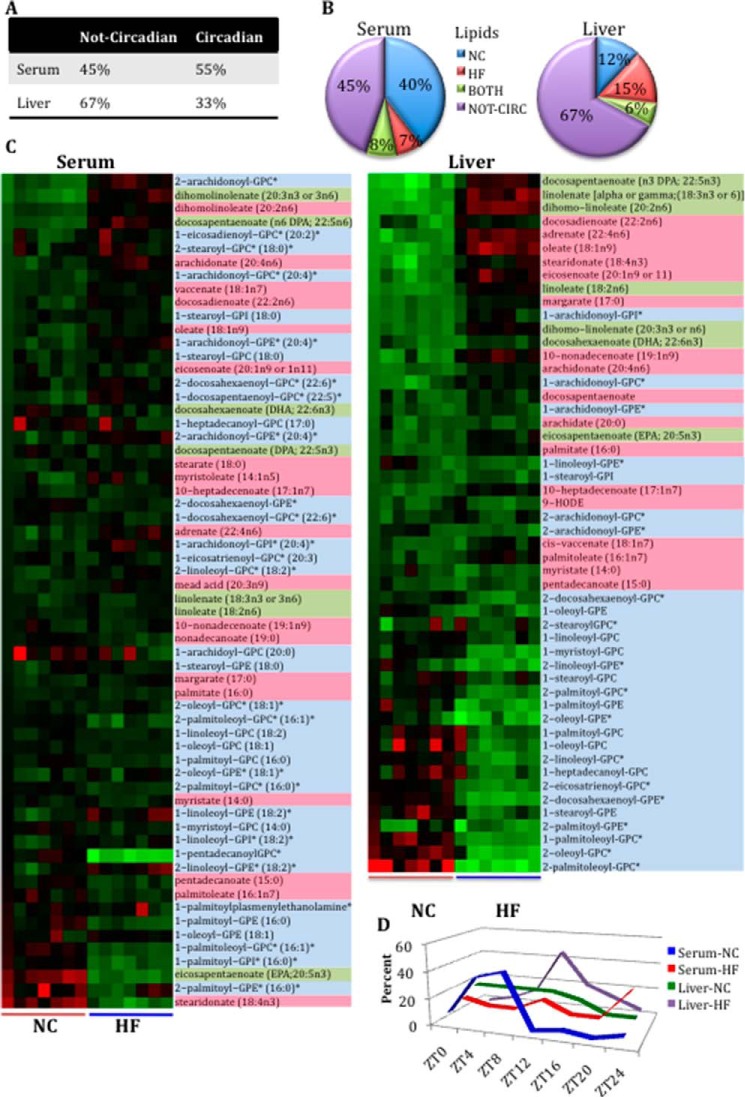
FIGURE 1.
General loss of oscillation in serum metabolites after HFD compared with liver. A, comparison of the number of serum and liver metabolites affected by diet or time. B, comparison of oscillating and not oscillating metabolites in serum and liver. C, comparison of the numbers of oscillatory metabolites only in NC, only in HF, or in both NC and HF groups (BOTH) for serum and liver (p < 0.05, JTK_cycle, and n = 5 biological replicates). D, phase graphs of serum and liver metabolites that oscillate in both feeding conditions. E, phase graphs of serum and liver metabolites that oscillate only in the NC or HFD.
In both tissues, a large number of metabolites (62% in serum, 77% in the liver) were affected by HFD. When analyzed by analysis of variance, 40% of serum metabolites and 45% of liver metabolites deviated in abundance throughout the circadian cycle. However, when analyzed for circadian oscillation specifically (p < 0.05, JTK_cycle (26)), the fraction of oscillating metabolites was the same. Specifically, 46% oscillated in the serum, and 46% cycled in the liver under a distinct feeding condition (i.e. NC, HFD, or both (BOTH)) (Fig. 1B). Strikingly, in serum, HFD induced an extensive disruption in the oscillation of metabolites that cycle in NC (55%). A smaller fraction of metabolites (27%) oscillated in both feeding conditions, and only 18% oscillated in HFD. This profile is in stark contrast to the situation in the liver, where 43% of metabolites oscillate in both feeding conditions, 30% oscillate only in NC (i.e. oscillation is lost under HFD), and 27% oscillate only in HFD (26) (Fig. 1C). Thus, although a similar number of metabolites oscillated in both feeding conditions in the liver, there was a 3-fold decrease in the number of metabolites oscillating in HFD in the serum relative to NC. Thereby, it appears that the serum metabolome is much more sensitive to nutritional challenge than the liver. Moreover, an analysis of the phase of oscillation in both tissues under NC and HFD revealed some important differences (see Figs. 4F, 5 (E and J), 6F, 7D, and 8 (E and J)). In serum, apart from the loss of oscillation in carbohydrates and cofactors in HFD, the remaining oscillatory metabolites were phase-delayed in HFD. Moreover, some variance was noted in the phase of oscillation for metabolites that oscillated in both diets. In particular, serum metabolites oscillating in both feeding conditions were phase-advanced in HFD (Fig. 1D), whereas, considering the phase of metabolites that oscillated only in NC or only in HFD, liver metabolites in HFD were somewhat phase-advanced compared with metabolites oscillating only in NC (26) (Fig. 1E). Also, we analyzed the relative abundance of major classes of metabolites at ZT0 and ZT12 for serum and liver (37). Significant decreases in serum were detected at ZT0 under HFD compared with NC diet for peptides and xenobiotics. Moreover, lower levels of peptides were also found at ZT12 compared with ZT0 under NC. Interestingly, some major classes of metabolites showed a diet effect in the liver. This is the case for amino acids, xenobiotics, and nucleotides, with a marked decrease in their content under HFD for both of the time points analyzed.
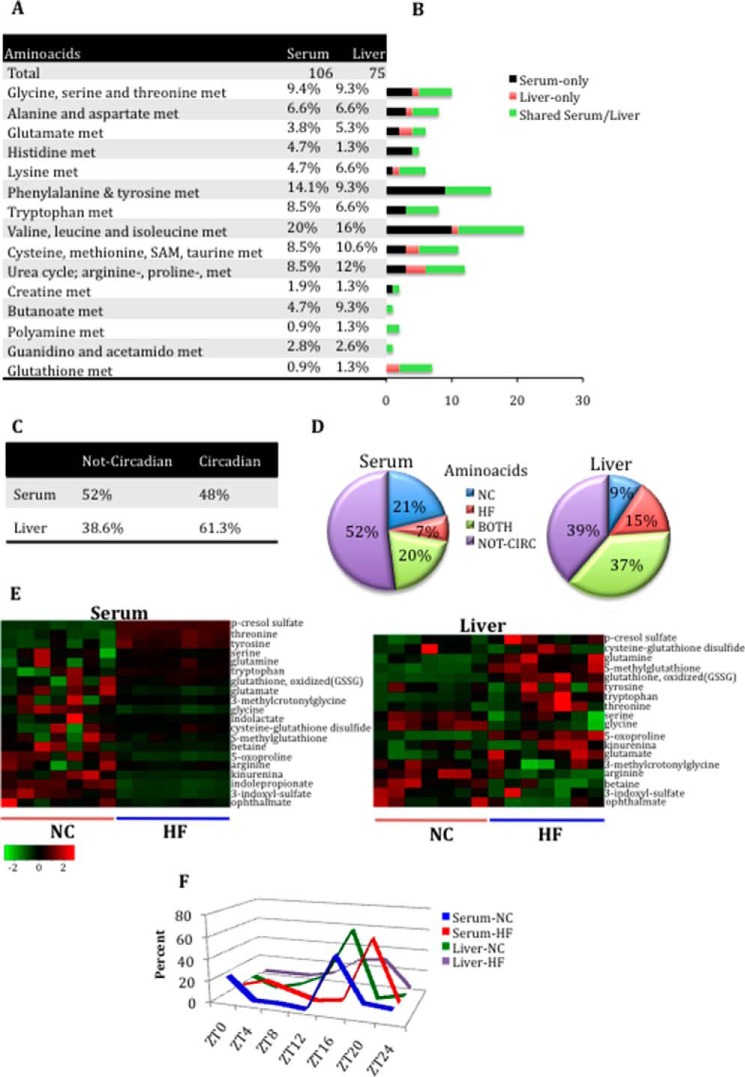
FIGURE 4.
Loss of oscillation in serum amino acid metabolites after HFD compared with the liver. A, amino acid metabolite composition in serum and liver. B, differences in amino acid metabolite composition between serum and liver. Black bars, number of metabolites identified only in serum. Red bars, number of metabolites identified only in the liver. Green bars, number of metabolites shared between serum and liver. C, number of amino acid metabolites oscillating or not oscillating in serum and liver. D, comparison of the numbers of oscillatory amino acid metabolites only in NC, only in HFD, or in both NC and HF groups for serum and liver (p < 0.05, JTK_cycle, and n = 5 biological replicates). E, heat maps of select metabolites in the serum and liver of animals on NC or HFD. F, phase profiles for amino acid metabolites oscillating for each feeding condition in serum and liver. The percentage of oscillatory metabolites that peak at a specific ZT in NC and HFD compared with the total number of oscillatory metabolites in that metabolic pathway is plotted.
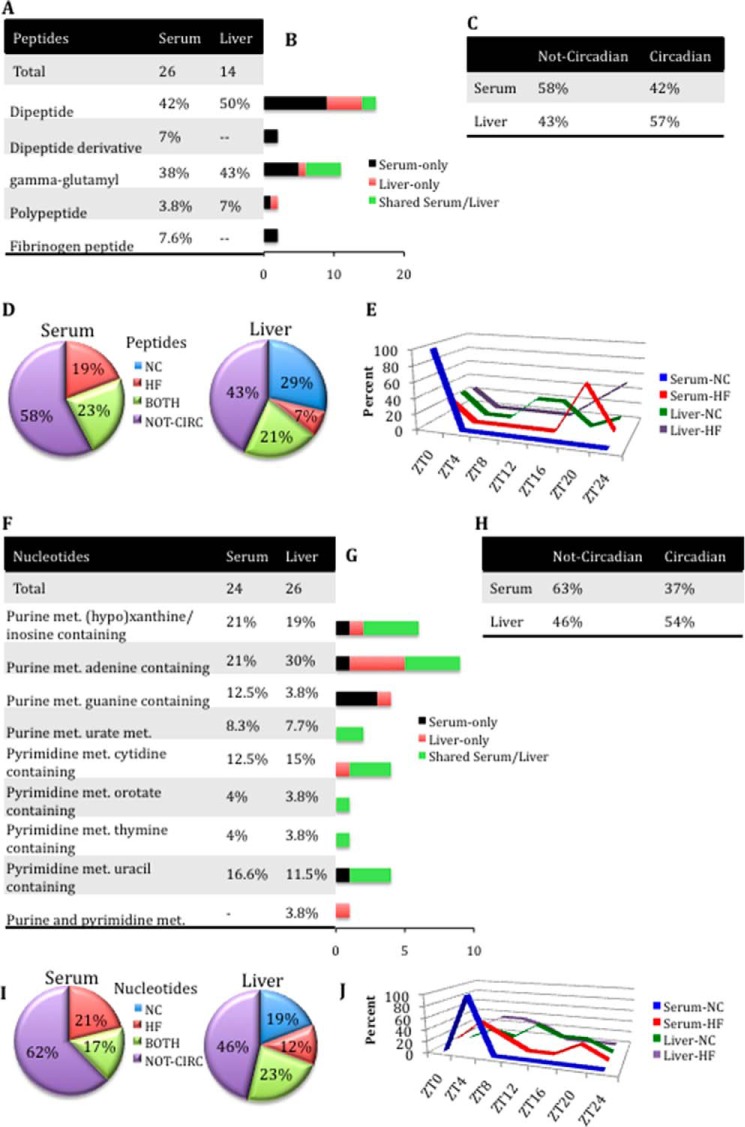
FIGURE 5.
Gain of oscillation in peptide metabolites in serum under HFD conditions compared with liver (A–E) and gain of oscillation in nucleotide metabolites in serum in HFD compared with liver (F–J). A, peptide metabolite composition in serum and liver. B, differences in peptide metabolite composition between serum and liver. Black bars, number of metabolites identified only in serum. Red bars, number of metabolites identified only in the liver. Green bars, number of common (shared) metabolites between serum and liver. C, number of peptide metabolites oscillating or not oscillating in serum and liver. D, comparison of the numbers of oscillatory peptide metabolites only in NC, only in HF, or in both NC and HF groups for serum and liver (p < 0.05, JTK_cycle, and n = 5 biological replicates). E, phase profiles for peptide metabolites oscillating for each feeding condition in serum and liver. The percentage of oscillatory metabolites that peak at a specific ZT in NC and HFD compared with the total number of oscillatory metabolites in that metabolic pathway is plotted. F, nucleotide metabolite composition in serum and liver. G, differences in nucleotide metabolite composition between serum and liver. Black bars, number of metabolites identified only in serum. Red bars, number of metabolites identified only in the liver. Green bars, number of common (shared) metabolites between serum and liver. H, comparison of number of nucleotide metabolites oscillating or not oscillating in serum and liver. I, comparison of the numbers of oscillatory nucleotide metabolites only in NC, only in HF, or in both NC and HF groups for serum and liver (p < 0.05, JTK_cycle, and n = 5 biological replicates). J, phase profiles for nucleotide metabolites oscillating for each feeding condition in serum and liver. The percentage of oscillatory metabolites that peak at a specific ZT in NC and HFD compared with the total number of oscillatory metabolites in that metabolic pathway is plotted.
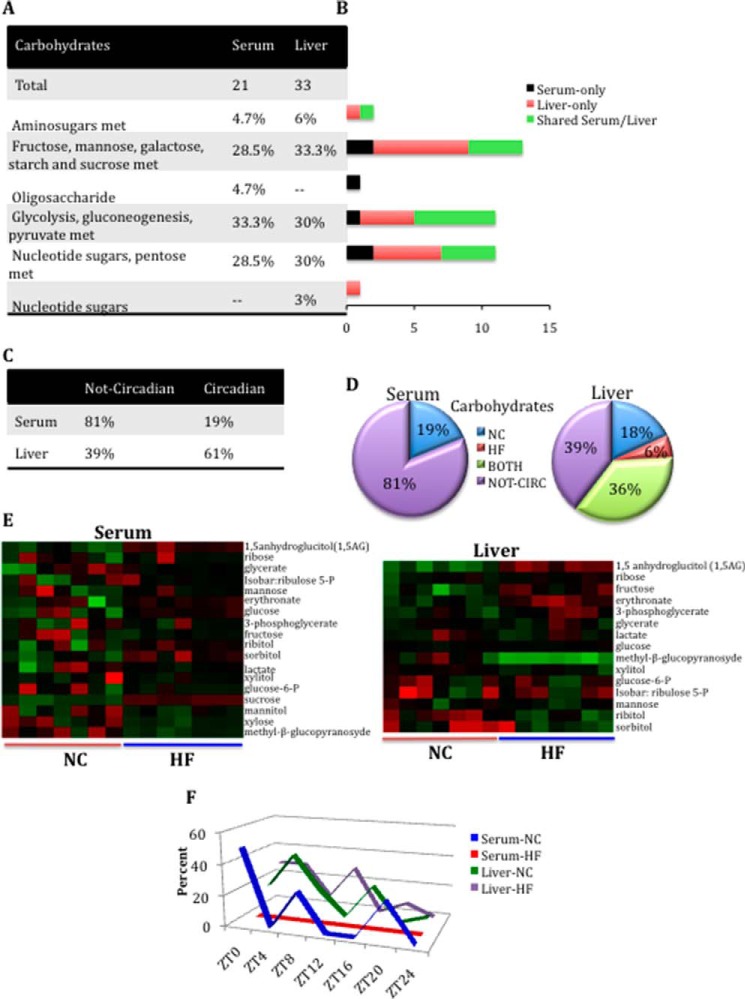
FIGURE 6.
Loss of oscillation in serum carbohydrate metabolites compared with liver in HFD. A, carbohydrate metabolite composition in serum and liver. B, differences in carbohydrate metabolite composition between serum and liver. Black bars, number of metabolites identified only in serum. Red bars, number of metabolites identified only in the liver. Green bars, number of common (shared) metabolites between serum and liver. C, comparison of carbohydrate metabolites oscillating or not oscillating in serum and liver. D, comparison of the numbers of oscillatory carbohydrate metabolites only in NC, only in HF, or in both NC and HF groups for serum and liver (p < 0.05, JTK_cycle, and n = 5 biological replicates). E, heat maps of select shared carbohydrate metabolites in the liver and serum under NC and HFD conditions. F, peak profiles for carbohydrate metabolites oscillating for each feeding condition in serum and liver. The percentage of oscillatory metabolites that peak at a specific ZT in NC and HFD compared with the total number of oscillatory metabolites in that metabolic pathway is plotted.
FIGURE 7.
Gain of cycling serum lipid metabolites compared with liver. A, percentage of lipid metabolites circadian or not circadian under normal chow or high fat diet conditions. B, comparison of the numbers of oscillatory lipid metabolites only in NC, only in HF, or in both NC and HFD groups for serum and liver (p < 0.05, JTK_cycle, and n = 5 biological replicates). C, heat maps of lipid metabolites oscillating and not oscillating in serum (left map) and liver (right map) samples under NC and HFD. Shown are lysolipids (light blue), long-chain fatty acids (pink), and essential fatty acids (green). D, phase profiles for lipid metabolites oscillating for each feeding condition in serum and liver. The percentage of oscillatory metabolites that peak at a specific ZT in NC and HFD compared with the total number of oscillatory metabolites in that metabolic pathway is plotted.
FIGURE 8.
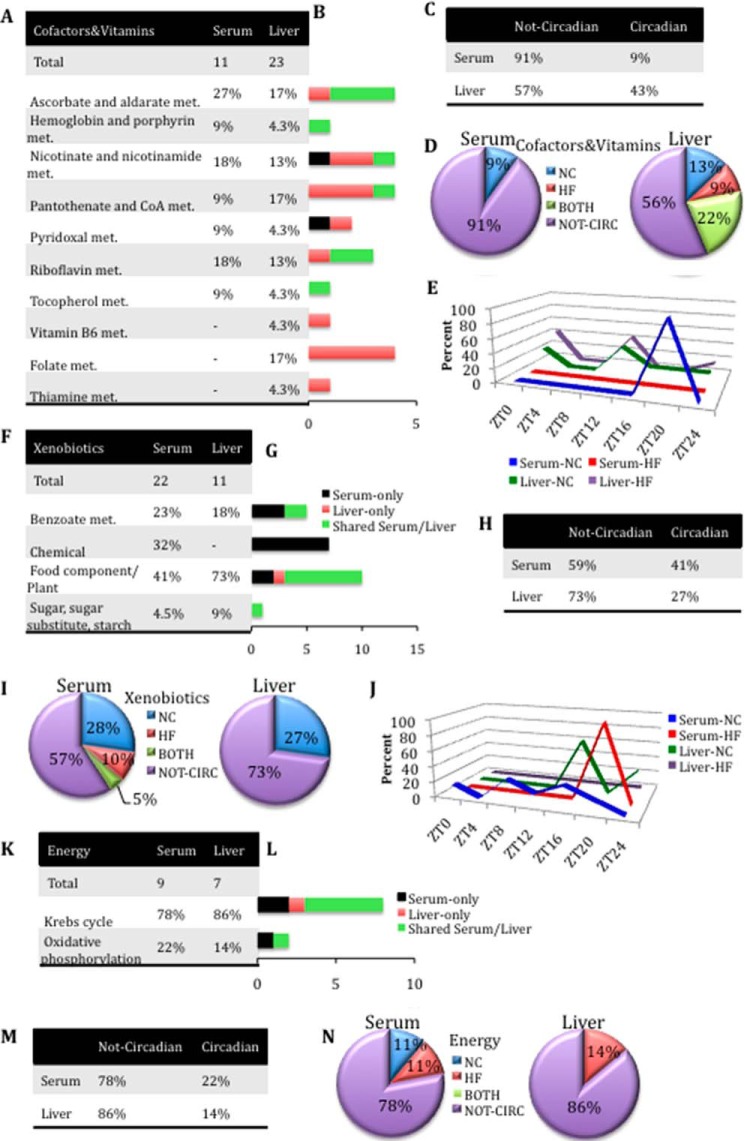
Loss of oscillation in cofactor and vitamin metabolites in serum in HFD compared with liver (A–E), gain of oscillation in xenobiotic metabolites in serum in NC and HFD compared with liver (F–J), and loss of oscillation in serum and liver of energy-related metabolites (K–N). A, cofactors and vitamin metabolite composition in serum and liver. B, differences in cofactors and vitamin metabolite composition between serum and liver. Black bars, number of metabolites identified only in serum. Red bars, number of metabolites identified only in the liver. Green bars, number of metabolites shared between serum and liver. C, number of cofactor and vitamin-related metabolites oscillating or not oscillating in serum and liver. D, comparison of the numbers of oscillatory cofactors and vitamin metabolites only in NC, only in HF, or in both NC and HF groups for serum and liver (p < 0.05, JTK_cycle, and n = 5 biological replicates). E, phase profiles for cofactor and vitamin metabolites oscillating for each feeding condition in serum and liver. The percentage of oscillatory metabolites that peak at a specific ZT in NC and HFD compared with the total number of oscillatory metabolites in that metabolic pathway is plotted. F, xenobiotic metabolite composition in serum and liver. G, differences in xenobiotic metabolites composition between serum and liver. Black bars, number of metabolites identified only in serum. Red bars, number of metabolites identified only in the liver. Green bars, number of metabolites shared between serum and liver. H, number of xenobiotic metabolites oscillating or not oscillating in serum and liver. I, comparison of the numbers of oscillatory xenobiotic metabolites only in NC, only in HF, or in both NC and HF groups for serum and liver (p < 0.05, JTK_cycle, and n = 5 biological replicates). J, phase profiles for xenobiotic metabolites oscillating for each feeding condition in serum and liver. The percentage of oscillatory metabolites that peak at a specific ZT in NC and HFD compared with the total number of oscillatory metabolites in that metabolic pathway is plotted. K, energy metabolite composition in serum and liver. L, differences in energy metabolite composition between serum and liver. Black bars, number of metabolites identified only in serum. Red bars, number of metabolites identified only in the liver. Green bars, number of metabolites shared between serum and liver. M, comparison of the numbers of energy metabolites oscillating or not oscillating in serum and liver. N, comparison of the numbers of oscillatory energy metabolites only in NC, only in HF, or in both NC and HF groups for serum and liver (p < 0.05, JTK_cycle, and n = 5 biological replicates).
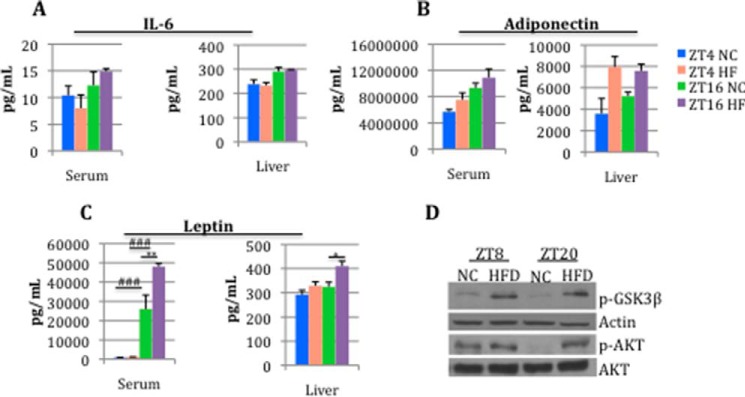
By analyzing a variety of parameters, we have previously shown that, as expected, HFD-fed mice develop an obese phenotype (26). Adipose tissue in mammals not only acts as storage for excess of nutrients; it also acts as an endocrine organ secreting adipokines that are involved in a wide range of functions (52–55). Specifically, obesity is associated with oxidative stress and inflammatory responses in adipose tissue (due to adipocytes hypertrophy and hyperplasia) with consequent increased levels of local and systemic pro-inflammatory cytokines. We analyzed the levels of IL-6, leptin, and adiponectin (56–59) in serum and liver at two different time points (ZT4 and ZT16) to monitor whether the obese status elicited by the high fat diet could affect the circadian secretion of these adipokines (Fig. 2, A–C). No changes or little change was observed in IL-6 and adiponectin levels between NC and HFD in both tissues. This could be due to various factors, such as the diet composition and/or the circadian changes occurring at other ZTs. Importantly, the levels of leptin at ZT16 were significantly higher in HFD as compared with NC in both serum and liver. This result not only indicates a temporal disruption of this adipokine; it also suggests a misreading of the signal for the brain of the body's energy stores (Fig. 2C). Moreover, because leptin is implicated in the etiology of insulin resistance (60–62), we extended our analysis by monitoring AKT and GSK3β. As expected, we found that HFD induced an increase in basal (non-insulin-stimulated) AKT phosphorylation at Ser473 and GSK3β inactivation (as measured by phosphorylation at Ser9) (Fig. 2D). Importantly, we also observed a complete loss of rhythmicity in AKT phosphorylation in the animals fed an HFD, whereas robust rhythmicity in phosphorylation of AKT is seen in NC-fed animals.
FIGURE 2.
HFD affects leptin levels in serum and liver. Serum and liver levels of IL-6 (A), adiponectin (B), and leptin (C) within ZT4 and ZT16 in NC and HFD (asterisks, diet effect; number symbols, time effect; two-way analysis of variance; *, p < 0.05; **, p < 0.01; ###, p < 0.001, Tukey's post hoc test; error bars, S.E.; n = 3 biological replicates). D, immunoblotting analyses of phospho-AKT (Ser473), AKT, and GSK3β (Ser9) of NC and 10-week HFD livers at ZT8 and ZT20.
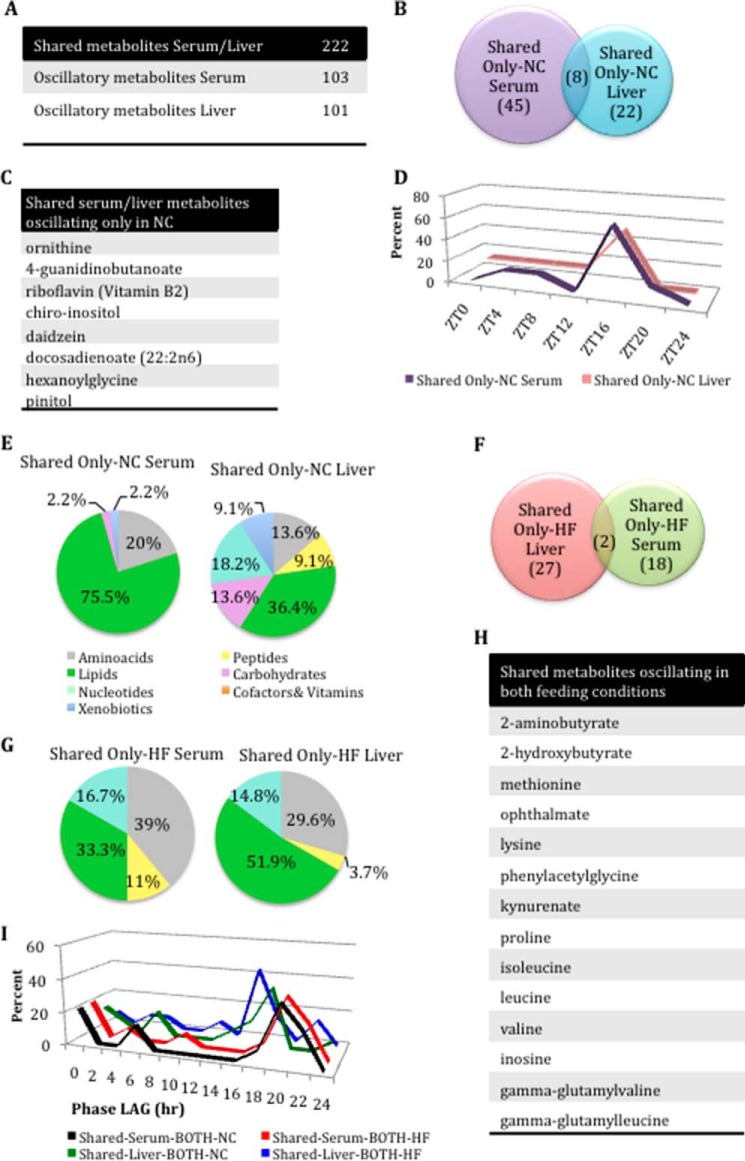
Next, in the pool of shared metabolites between serum and liver (Fig. 3A), we analyzed in detail the metabolites present in both tissues and oscillating only in NC (Fig. 3, B–E). Of the eight metabolites oscillating in both tissues under NC (Fig. 3, B and C), most were synchronous by showing a similar phase peak at ZT16 (Fig. 3D). Importantly, there was a different composition of the metabolites shared between serum and liver in NC. Specifically, 75.5% of the serum lipids oscillated in NC versus 36.4% in the liver. Another striking difference relates to nucleotide metabolites; 18% of the shared nucleotide oscillated in the liver, whereas none oscillated in the serum (Fig. 3E). A parallel analysis for oscillating metabolites only in HFD revealed that only two are shared between liver and serum (allantoin and 2-oleoylglycerophosphoethanolamine) (Fig. 3F).
FIGURE 3.
Phase alteration of shared serum and liver metabolites oscillating in both feeding conditions. A, number of shared metabolites oscillating in serum and liver. B, number of shared serum and liver metabolites oscillating only in NC. C, shared metabolites oscillating only in the NC conditions in both tissues. D, phase profiles for shared metabolites oscillating only in NC conditions in serum and liver. E, differences in metabolite composition between shared serum and liver metabolites that oscillate only in the NC conditions. F, number of shared serum and liver metabolites oscillating only in the HFD condition. G, differences in metabolite composition in shared serum and liver metabolites that oscillate only under HFD. H, shared metabolites oscillating in both feeding conditions. I, phase graphs of serum and liver metabolites that oscillate in common in under both feeding conditions (BOTH). Black line, shared serum metabolites that oscillate in the NC subset of the both category (i.e. oscillating under both feeding conditions but considering only NC conditions). Red line, shared serum metabolites oscillating in the HFD subset of the both category. Green line, shared liver metabolites oscillating in the NC subset of the both category. Blue line, shared liver metabolites in the HFD subset of the both category.
Another revealing difference relates to metabolites oscillating in HFD, where 52% of liver lipid metabolites cycled, whereas only 33% did so in serum (Fig. 3G). Interestingly, 14 metabolites are shared and oscillate under both feeding conditions in both tissues, and most of them belong to the amino acid pathway (Fig. 3H). Analysis of the phase of oscillation for these shared metabolites revealed that, unlike the liver, the serum metabolome is not phase-advanced under HFD (Fig. 3I).
Amino Acids and Peptides
A significant fraction of amino acid metabolites are common to serum and liver, although some unique profiles are distinctive of the two tissues. Specifically, a number of amino acids were found exclusively in the serum (40% of all serum amino acid metabolites), including sarcosine, N6-acetyllysine, phenylpyruvate, and creatinine (Fig. 4 (A and B) and supplemental Files 1 and 2). A smaller fraction of amino acid metabolites are unique to the liver (17% of all liver amino acid metabolites), including glutarate, hypotaurine, and S-adenosylmethionine. Comparison of the profiles reveals that fewer amino acids oscillate in the serum than in the liver (48% versus 61.3%) (Fig. 4C). In particular, we observed a 2-fold reduction in circadian amino acids affected by HFD in the serum as compared with the liver (Fig. 4D).
During the analysis of the amino acid pathway, we found that several amino acids, including glycine, serine, and threonine, display rhythmicity only under HFD in the serum. Conversely, metabolites belonging to the subpathway of the tryptophan metabolism, including tryptophan, indolelactate, indolepropionate, and kinurenine, completely lost oscillation in HFD (Fig. 4E and supplemental File 1). These metabolites, however, showed a significant decrease in levels, indicating that one of the effects of HFD is to influence the homeostasis of tryptophan metabolism. Moreover, HFD results in an increase of the overall levels of tyrosine and induces temporal regulation of tyrosine-related metabolites, such as cresol sulfates and 3-(4-hydroxyphenyl) lactate. Although HFD did not appear to regulate the circadian levels of other metabolites belonging to this pathway (phenol sulfates, 4-hydroxyphenylpyruvate), it decreased temporal aspects of their degradation. Interestingly, serotonin levels were significantly decreased in the serum under HFD compared with NC (supplemental File 1), in contrast to observations in human plasma following sleep deprivation (17).
The comparison between serum and liver amino acid metabolites revealed that some metabolites, including betaine, glutamate, glutamine, and 3-methylcrotonylglycine, are not oscillatory in serum, whereas they are robustly cyclic in the liver only in HFD. Similarly, amino acids of glutathione metabolism were devoid of oscillation in the serum (except for the ophthalmate and 5-oxoproline), in contrast to the liver, where they are highly oscillatory in both feeding conditions. In contrast, isobutyrylcarnitine, tyrosine, tryptophan, p-cresol sulfate, 3-indoxyl sulfate, and arginine were not cyclic in the liver but were cyclic in serum in NC or in HFD (Fig. 4E and supplemental Files 1 and 2). Comparison of the oscillation phase revealed that metabolites of both tissues tended to peak later under HFD than what was typically observed in NC. Interestingly, under NC, most of serum and hepatic amino acids peaked at ZT16 (Fig. 4F). Serum also showed an absence of fibrinogen cleavage peptides and dipeptide derivatives and a higher number of metabolites belonging to the dipeptide subpathway compared with the liver (Fig. 5 (A and B) and supplemental Files 1 and 2). Also, there was an increase in oscillating peptides in the serum compared with liver in HFD and a complete loss of rhythmicity for serum peptides that cycle only in NC as compared with liver (Fig. 5D and supplemental Files 1 and 2). No temporal similarity for the oscillation phase was found between the two tissues analyzed (Fig. 5E).
Nucleotides
Most nucleotide metabolites are shared between liver and serum. The liver is the primary organ of de novo nucleotide synthesis, although many tissues use salvage pathways to generate nucleotide levels sufficient for cellular functions (63). Several differences between liver and serum were, however, found, specifically in the content of the purine metabolites adenine and guanine (Fig. 5, F and G). Interestingly, there was a loss in nucleotides oscillating in the serum compared with the liver (37% versus 54%) (Fig. 5H). In particular, no oscillating nucleotide metabolites were found under NC conditions in the serum (Fig. 5I). Moreover, in the serum, nucleotide metabolites tended to peak mostly at ZT4 under both of the feeding conditions, whereas in the liver, they were phase-advanced under HFD (Fig. 5J).
Carbohydrates
Analysis of carbohydrate metabolites showed that 15 are shared between serum and liver. Almost one-third (29%) of the total serum carbohydrate metabolites were found exclusively in the serum, whereas 50% of total liver carbohydrates were found only in the liver (Fig. 6 (A and B) and supplemental Files 1 and 2). There was a striking difference in the number of carbohydrate metabolites whose levels changed in a circadian manner in the liver versus serum. Indeed, whereas only four metabolites (mannose, mannitol, sucrose, and xylose) oscillated in the serum (19%), 20 did so in the liver (61%) (Fig. 6C). Moreover, all circadian serum carbohydrates in NC lost their cycling profile in HFD, whereas in the liver, the oscillations were conserved also under nutritional challenge (Fig. 6, D and F). The cycling of carbohydrates in general appears to be more prominent in the liver, the primary site of both glucose uptake and glucose production. The loss of cycling under HFD of most carbohydrate metabolites in the serum reinforces the notion that the general effect of nutritional challenge is the disruption of homeostasis.
In addition to carbohydrates, another pathway that in the serum undergoes circadian disruption by HFD is glycolysis. Under nutritional challenge, all metabolites involved in glycolysis lost oscillation in the serum, including glucose 6-phosphate, lactate, glucose, and 3-phosphoglycerate, all of which remained oscillatory in the liver (Fig. 6E and supplemental Files 1 and 2). An intriguing example is sucrose (present in serum but not in the liver). Sucrose, which cycles in NC conditions, completely lost oscillation in HFD (supplemental File 1). A likely explanation could be that mice under HFD have a delay in glucose clearance compared with those in NC, mostly because of peripheral insulin resistance that results from higher levels of circulating free fatty acids.
Lipids
Many lipid metabolites are shared between serum and the liver. However, 33% of the lipid species found in the serum were not present in the liver (supplemental Files 1 and 2). Most of these belong to the subpathways of medium-chain fatty acids, monohydroxy fatty acids, branched-chain fatty acids, lysolipids, and metabolites in the carnitine metabolism pathway. Conversely, 18% of liver lipids are not found in the serum. Importantly, whereas more than half of the serum lipids oscillated across the circadian cycle, fewer did so in the liver (55% versus 33%) (Fig. 7A). Diets also differentially affect lipid metabolites in the liver and serum. In particular, under NC, a higher number of lipids oscillated in the serum compared with the liver (Fig. 7B). Conversely, there was a massive loss of lipid metabolites under HFD in the serum as compared with the liver. Strikingly, most metabolites of the lysolipid pathway (55%) cycled in NC but lost oscillation in HFD, in striking contrast to the situation in the liver, in which only 28% of the lysolipids were shown to have a diet effect (Fig. 7C and supplemental Files 1 and 2). Also, metabolites in the essential fatty acid and long chain fatty acid pathways oscillated in the serum only under NC, although they showed a non-cyclic trend in the liver under any diet condition. On the other hand, few lipids of the carnitine metabolism subpathway (e.g. myristate, carnitine, acetylcarnitine, and stearoylcarnitine) oscillated in the liver but not in the serum under HFD (supplemental Files 1 and 2). For a number of metabolites, there were also changes in the phase. Some serum lipid metabolites were phase-delayed in HFD; also, whereas most of the liver metabolites oscillated in NC between ZT0 and ZT12, most serum lipids peaked at ZT8 (Fig. 7D). Thus, circadian lipid profiles are profoundly affected by HFD and show loss of circadian oscillation in the serum.
Cofactors and Xenobiotics
The comparison between cofactors revealed the absence of vitamin B6, folate, and thiamine metabolites in the serum, whereas these metabolites were highly present in the liver (Fig. 8 (A and B) and supplemental Files 1 and 2). Interestingly, 91% of serum cofactor and vitamin-related metabolites were not oscillating throughout the circadian cycle compared with the liver (57%), whereas the remaining 9% oscillated only in NC conditions (Fig. 8, C and D). This difference was inverted for xenobiotic metabolites (Fig. 8, F–J). Indeed, 41% of serum xenobiotic metabolites were circadian versus only 27% in the liver (Fig. 8, H and I), where there was a complete loss of oscillation under HFD. Moreover, serum xenobiotic-related metabolites were phase-advanced in NC compared with those in the liver (Fig. 8J).
Energy Metabolism
Metabolites related to the Krebs cycle and oxidative phosphorylation showed no major changes along the circadian cycle (Fig. 8, K–N). Importantly, only two metabolites (22%) oscillated across the circadian cycle in the serum and only one in the liver (14%) (Fig. 8, M and N). These data suggest that these critical metabolites must maintain relatively stable levels throughout the circadian cycle and under nutritional challenge.
Serum Metabolites as Markers for Metabolic Disease
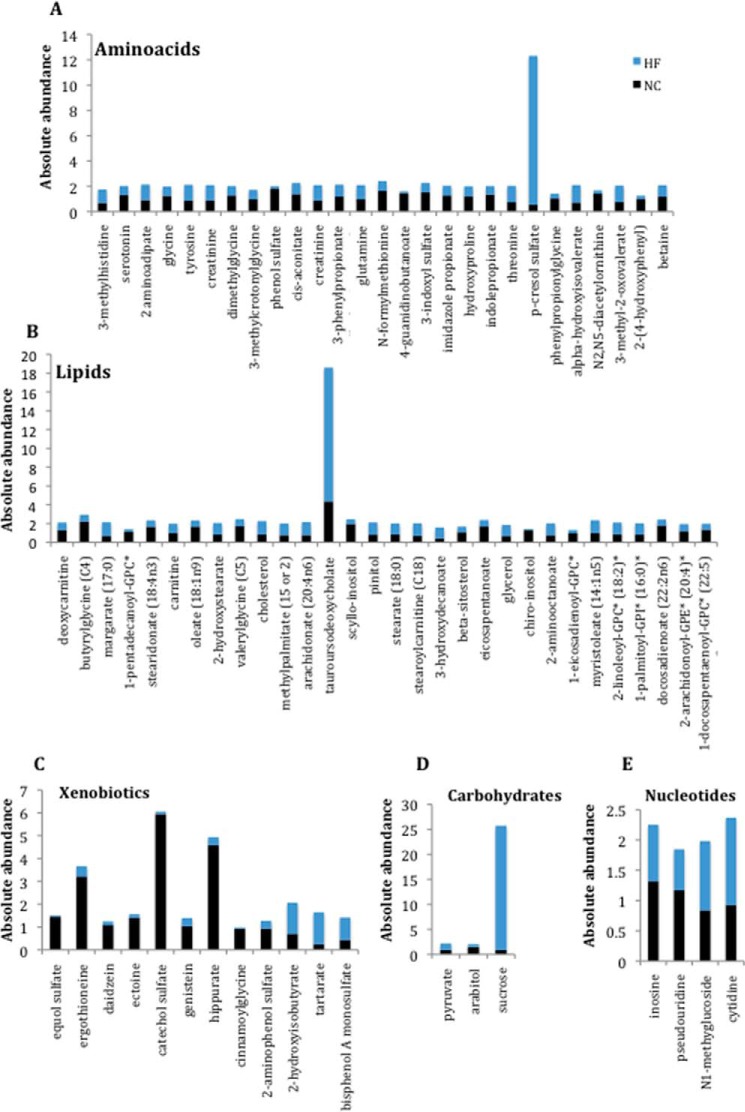
The comparison between serum and liver metabolomes showed that almost 39% of the total metabolites identified, across most of the metabolic pathways, were exclusively present in serum (supplemental File 1). Most of the metabolites found in the serum are known to be present in all tissues and organs of the body, including intestine, muscle, brain, epidermis, and spleen, indicating that a given metabolite may be not specific to serum but rather present in low abundance or is not found in the liver. Considering serum metabolites only, 60% of them undergo changes in response to the diet, independent of whether it is a decrease or an increase upon HFD (Figs. 1A and 9 (A–D) and supplemental File 1). Interestingly, many of these metabolites can be considered as markers for various diseases, including cancer, cardiovascular and renal diseases, and metabolic disorders. For example, in our study, we found altered levels of arachidonate, cholesterol, stearate, betaine, glycerol, sucrose, 2-aminoadipate, eicosapentaenoate, 3-methyl-2-oxovalerate, and oleate (Fig. 9, A and B). These metabolites have been shown to be associated with obesity, metabolic syndrome, or type II diabetes (64–71). Moreover, the levels of several metabolites associated with cardiovascular diseases and/or renal failure/dysfunctions were significantly changed, such as, for example, fatty acid, p-cresol sulfate, inosine, genistein, and daidzain (72–79) (Fig. 9, A–C and E). Serotonin, which has a key role in appetite control, was severely affected by HFD, possibly underscoring the tight link between this neurotransmitter and obesity (Fig. 9A). Peripheral serotonin could be involved in the obesity-induced adipose tissue inflammation in our mice (80–84).
FIGURE 9.
HFD affects the absolute abundance of several metabolites in serum. Shown is a representation of serum metabolites found to be affected by diet (p < 0.05).
Other examples of metabolites that we have found altered and are known to be associated with human diseases are β-sitosterol, hydroxyproline, N1-methyguanosine, cytidine, indoxyl sulfate, pyruvate, and creatinine, all of which have been reported to be involved in cancer (85–93) (Fig. 9, A, B, D, and E). Interestingly, our analysis showed higher levels of tyrosine, pyruvate, and glutamine, paralleling the abnormal concentration of these metabolites in patients affected by schizophrenia (94, 95) (Fig. 9, A and D). Finally, our analysis also revealed some metabolites used as markers for muscle damage, such as hydroxyproline and 3-methyl histidine (96, 97), which are more abundant in HFD (Fig. 9A).
Discussion
The relevance of blood in clinical tests lies in the fact that tissue lesions, organ dysfunction, and pathological states alter metabolite composition in the serum, providing valuable information for diagnosis. Here we have presented a high throughput, comparative analysis of the serum metabolome as compared with the liver, along the circadian cycle and under nutritional challenge. In addition to its intrinsic clinical value, this study provides insights on the organism-wide processes of communication among tissues that may take place in a time-specific manner. Considerable advances in a variety of biochemical analytical techniques have allowed significant progress in the deciphering the metabolome in a number of physiological conditions. Specifically, metabolic changes throughout the circadian cycle in serum of both humans and rodents have been analyzed. In addition, a number of studies have detected metabolites that vary throughout the circadian cycle in other tissues, including the liver (50, 98, 99). These studies show that many metabolites cycle in abundance throughout the 24-h cycle and that many of these oscillations are subject to disruption in the liver following high fat feeding (26). A number of studies support the notion that desynchrony between the central and peripheral clocks is disadvantageous for energy balance and homeostasis (46, 100), whereas normal rhythmicity can be altered by HFD administered ad libitum in some mouse models (24, 101). Also, phosphatidylcholine (18:0/18:1) has recently been shown to be a diurnal metabolite capable of integrating lipogenesis in the liver to the use of fatty acids peripherally (102). In addition, circulating metabolites whose levels are disrupted as a result of the loss of adipose tissue-specific clock function have been shown to alter energy balance by disrupting the rhythmic expression of orexigenic and anorexigenic peptides in the hypothalamus (103). To determine the extent and specificity to which diets affect the circadian metabolome in ways that might be disadvantageous to overall energy balance, we compared serum and liver metabolomes in mice made obese through a high fat diet. This comparison, the first of this type, has revealed that the serum is generally devoid of oscillation following nutritional challenge and that the remaining oscillatory events are not synchronized with the liver clock. A few previous studies have used a single time point (or non-circadian time points) to assess the metabolite profile of obese humans (104). Some similarities between our results and these studies exist, specifically when analyzing metabolites of the glycerophospholipid and lysolipid pathways. In both obese humans and our obese mice, the abundance of many of these metabolites was substantially increased, supporting the notion that enhanced lipolysis constitutes a signature of the obese state. Unlike the liver, where only a few lipid metabolites showed circadian oscillation, over half of the lipids showed a circadian oscillation in the serum, the majority of which lose their oscillation after high fat feeding. Indeed, whereas de novo lipogenesis occurs in the liver after chronic high fat feeding, loss of oscillating lipids in the serum probably reflects constitutive breakdown of adipose tissue, which occurs in the insulin-resistant state (105). Breakdown of adipose tissue causes the release of free fatty acids directly into the bloodstream, but it is not the only source of lipids in the blood. Indeed, short- and medium-chain fatty acids can be absorbed directly from the intestine, so it is possible that with the large increase in dietary lipids, oscillations were lost or could not be detected. Moreover, loss of rhythmicity in this metabolite group may also reflect the fact that other peripheral clocks become misaligned under HFD. Another potential indication of this desynchrony between tissues is the total loss of carbohydrate oscillations after HFD in the serum but not in the liver. One remarkable effect of HFD on the serum metabolome is that, unlike in the liver, where numerous metabolites take on de novo oscillation only after HFD feeding (26), most serum metabolites lost their rhythmicity. Obviously, this has profound implications for the likelihood of synchronicity across peripheral tissues and between peripheral tissues and the brain. Whereas the SCN responds to light and functions as the central pacemaker (4, 19, 20), clocks located in peripheral tissues can respond to other zeitgebers, such as nutrients during restricted feeding (9, 22, 47). These result in the uncoupling of peripheral clocks from the SCN, which has been shown to be highly disadvantageous for energy balance and to cause a variety of physiologic imbalances, as shown in both human and rodent studies (22–24, 39, 41, 45–47). Our results regarding the effect of HFD show some interesting similarities to the data of Davies et al. (17), who studied circadian metabolite profiles in humans after sleep deprivation. For example, whereas many amino acid-related metabolites generally have not been found to vary in a circadian fashion in humans, isoleucine and valine have both been shown to be rhythmic in human plasma, with both metabolites increasing in abundance after sleep deprivation. We have also found that rhythmicity of isoleucine and valine in mouse serum persists under HFD, although the overall levels of these amino acids and some of their related metabolites are significantly increased (supplemental File 1).
In addition, paralleling the effect of sleep deprivation, we observe increases in lysolipids under HFD. The increased abundance of lysolipids in the serum, as previously mentioned, may reflect breakdown of muscle and/or adipose tissue membranes. Thus, HFD and sleep deprivation appear to cause similar effects in terms of rhythmicity and abundance of several circulating metabolites.
Our study shows that HFD induces a misalignment between a peripheral tissue and the serum, where a significant fraction of circulating metabolites lose their rhythmicity under nutritional challenge. Also, peripheral clocks appear to respond in a highly tissue-specific manner to nutritional challenge. For example, unlike in the liver, a large number of circulating lipids are oscillatory only when mice are fed the normal chow diet.
Both nutritional challenge and disturbance of normal circadian patterns are risk factors for obesity (29). Also, metabolic disruptions elicited by HFD lead to reprogramming of the circadian clock in the liver (26) and presumably in other tissues and serum (this study), resulting in the uncoupling of peripheral clocks and SCN (29).
Thus, further investigations on the metabolome of other tissues along the circadian cycle and in response to different nutritional challenges will help in building a metabolic interconnective map of circadian metabolism. Indeed, loss of rhythmicity in fatty acids in the serum is likely to reflect the desynchronization of other peripheral clocks. A similar scenario seems to be present for carbohydrates, whose oscillation is lost under HFD only in serum and not in the liver. This result may suggest the presence of active lipogenesis along the circadian cycle under HFD. Indeed, it has been shown that excess of food intake is translated into altered expression levels of lipogenic genes (106). Thus, a diet rich in fat could stimulate the conversion of carbohydrates into lipids for subsequent storage in the adipose tissue. A relevant effect of HFD on the liver clock is the phase advance of a group of metabolites and transcripts (26, 28). This phenomenon appears to be inverted in serum, where in HFD, metabolites belonging to major metabolic pathways were generally phase-delayed. This type of misalignment across tissues may be responsible for lack of appropriate circadian communication, resulting in a loss of energy balance.
In conclusion, our study has profound implications for deciphering how circadian disruption is induced by nutrient challenge and its differential effect on serum or liver. A critical value of our findings relates to the application of this knowledge at the clinical level, by extending these high throughput metabolomics studies to personalized medicine in various physiological conditions.
Author Contributions
S. A. and P. S. C. designed the study; S. A. performed experiments, analyzed data, and wrote the paper; K. L. E.-M. helped in data collection and interpretation and reviewed drafts of the paper; N. J. C and P. B. were involved in the bioinformatics and statistical analysis; and P. S.-C. planned and supervised the research, reviewed drafts, and obtained funding for the research work.
Supplementary Material
Acknowledgments
We thank all members of the Sassone-Corsi laboratory for discussions, help, and support.
This work was supported by National Institutes of Health Grants GM081634 and AG033888, by INSERM, and by Merieux Research Grant MRG-53923. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This work contains supplemental Files 1 and 2.
- SCN
- suprachiasmatic nucleus
- HFD
- high fat diet
- NC
- normal chow
- ZT
- zeitgeber
- GSK
- glycogen synthase kinase.
References
- 1.Adamovich Y., Rousso-Noori L., Zwighaft Z., Neufeld-Cohen A., Golik M., Kraut-Cohen J., Wang M., Han X., and Asher G. (2014) Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 19, 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang W., Ramsey K. M., Marcheva B., and Bass J. (2011) Circadian rhythms, sleep, and metabolism. J. Clin. Invest. 121, 2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahar S., and Sassone-Corsi P. (2012) Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol. Metab. 23, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dibner C., Schibler U., and Albrecht U. (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549 [DOI] [PubMed] [Google Scholar]
- 5.Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L. P., and Sassone-Corsi P. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakahata Y., Sahar S., Astarita G., Kaluzova M., and Sassone-Corsi P. (2009) Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peek C. B., Affinati A. H., Ramsey K. M., Kuo H. Y., Yu W., Sena L. A., Ilkayeva O., Marcheva B., Kobayashi Y., Omura C., Levine D. C., Bacsik D. J., Gius D., Newgard C. B., Goetzman E., Chandel N. S., Denu J. M., Mrksich M., and Bass J. (2013) Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342, 1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peek C. B., Ramsey K. M., Marcheva B., and Bass J. (2012) Nutrient sensing and the circadian clock. Trends Endocrinol. Metab. 23, 312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokkan K. A., Yamazaki S., Tei H., Sakaki Y., and Menaker M. (2001) Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 [DOI] [PubMed] [Google Scholar]
- 10.Oike H., Oishi K., and Kobori M. (2014) Nutrients, Clock Genes, and Chrononutrition. Curr. Nutr. Rep. 3, 204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., Hong H. K., Chong J. L., Buhr E. D., Lee C., Takahashi J. S., Imai S., and Bass J. (2009) Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belden W. J., and Dunlap J. C. (2008) SIRT1 is a circadian deacetylase for core clock components. Cell 134, 212–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallmann R., Viola A. U., Tarokh L., Cajochen C., and Brown S. A. (2012) The human circadian metabolome. Proc. Natl. Acad. Sci. U.S.A. 109, 2625–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minami Y., Kasukawa T., Kakazu Y., Iigo M., Sugimoto M., Ikeda S., Yasui A., van der Horst G. T., Soga T., and Ueda H. R. (2009) Measurement of internal body time by blood metabolomics. Proc. Natl. Acad. Sci. U.S.A. 106, 9890–9895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasukawa T., Sugimoto M., Hida A., Minami Y., Mori M., Honma S., Honma K., Mishima K., Soga T., and Ueda H. R. (2012) Human blood metabolite timetable indicates internal body time. Proc. Natl. Acad. Sci. U.S.A. 109, 15036–15041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Psychogios N., Hau D. D., Peng J., Guo A. C., Mandal R., Bouatra S., Sinelnikov I., Krishnamurthy R., Eisner R., Gautam B., Young N., Xia J., Knox C., Dong E., Huang P., Hollander Z., Pedersen T. L., Smith S. R., Bamforth F., Greiner R., McManus B., Newman J. W., Goodfriend T., and Wishart D. S. (2011) The human serum metabolome. PLoS One 6, e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies S. K., Ang J. E., Revell V. L., Holmes B., Mann A., Robertson F. P., Cui N., Middleton B., Ackermann K., Kayser M., Thumser A. E., Raynaud F. I., and Skene D. (2014) Effect of sleep deprivation on the human metabolome. Proc. Natl. Acad. Sci. U.S.A. 111, 10761–10766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K., Mall C., Taylor S. L., Hitchcock S., Zhang C., Wettersten H. I., Jones A. D., Chapman A., and Weiss R. H. (2014) Mealtime, temporal, and daily variability of the human urinary and plasma metabolomes in a tightly controlled environment. PLoS One 9, e86223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schibler U., and Sassone-Corsi P. (2002) A web of circadian pacemakers. Cell 111, 919–922 [DOI] [PubMed] [Google Scholar]
- 20.Welsh D. K., Takahashi J. S., and Kay S. A. (2010) Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green C. B., Takahashi J. S., and Bass J. (2008) The meter of metabolism. Cell 134, 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vollmers C., Gill S., DiTacchio L., Pulivarthy S. R., Le H. D., and Panda S. (2009) Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 21453–21458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes M. E., DiTacchio L., Hayes K. R., Vollmers C., Pulivarthy S., Baggs J. E., Panda S., and Hogenesch J. B. (2009) Harmonics of circadian gene transcription in mammals. PLoS Genet. 5, e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E. A., Gill S., Leblanc M., Chaix A., Joens M., Fitzpatrick J. A., Ellisman M. H., and Panda S. (2012) Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohsaka A., Laposky A. D., Ramsey K. M., Estrada C., Joshu C., Kobayashi Y., Turek F. W., and Bass J. (2007) High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421 [DOI] [PubMed] [Google Scholar]
- 26.Eckel-Mahan K. L., Patel V. R., de Mateo S., Orozco-Solis R., Ceglia N. J., Sahar S., Dilag-Penilla S. A., Dyar K. A., Baldi P., and Sassone-Corsi P. (2013) Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bass J. (2012) Circadian topology of metabolism. Nature 491, 348–356 [DOI] [PubMed] [Google Scholar]
- 28.Eckel-Mahan K., and Sassone-Corsi P. (2013) Metabolism and the circadian clock converge. Physiol. Rev. 93, 107–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asher G., and Sassone-Corsi P. (2015) Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 [DOI] [PubMed] [Google Scholar]
- 30.Sahar S., Masubuchi S., Eckel-Mahan K., Vollmer S., Galla L., Ceglia N., Masri S., Barth T. K., Grimaldi B., Oluyemi O., Astarita G., Hallows W. C., Piomelli D., Imhof A., Baldi P., Denu J. M., and Sassone-Corsi P. (2014) Circadian control of fatty acid elongation by SIRT1 protein-mediated deacetylation of acetyl-coenzyme A synthetase 1. J. Biol. Chem. 289, 6091–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller B. H., McDearmon E. L., Panda S., Hayes K. R., Zhang J., Andrews J. L., Antoch M. P., Walker J. R., Esser K. A., Hogenesch J. B., and Takahashi J. S. (2007) Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 104, 3342–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmer S. L., Hogenesch J. B., Straume M., Chang H. S., Han B., Zhu T., Wang X., Kreps J. A., and Kay S. A. (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113 [DOI] [PubMed] [Google Scholar]
- 33.Ceriani M. F., Hogenesch J. B., Yanovsky M., Panda S., Straume M., and Kay S. A. (2002) Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22, 9305–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes M., Deharo L., Pulivarthy S. R., Gu J., Hayes K., Panda S., and Hogenesch J. B. (2007) High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb. Symp. Quant. Biol. 72, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., and Hogenesch J. B. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 [DOI] [PubMed] [Google Scholar]
- 36.Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., and Hogenesch J. B. (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 111, 16219–16224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel V. R., Eckel-Mahan K., Sassone-Corsi P., and Baldi P. (2012) CircadiOmics: integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat. Methods 9, 772–773 [DOI] [PubMed] [Google Scholar]
- 38.Salgado-Delgado R., Angeles-Castellanos M., Buijs M. R., and Escobar C. (2008) Internal desynchronization in a model of night-work by forced activity in rats. Neuroscience 154, 922–931 [DOI] [PubMed] [Google Scholar]
- 39.Salgado-Delgado R. C., Saderi N., Basualdo Mdel C., Guerrero-Vargas N. N., Escobar C., and Buijs R. M. (2013) Shift work or food intake during the rest phase promotes metabolic disruption and desynchrony of liver genes in male rats. PLoS One 8, e60052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suwazono Y., Dochi M., Sakata K., Okubo Y., Oishi M., Tanaka K., Kobayashi E., Kido T., and Nogawa K. (2008) A longitudinal study on the effect of shift work on weight gain in male Japanese workers. Obesity 16, 1887–1893 [DOI] [PubMed] [Google Scholar]
- 41.Knutsson A. (2003) Health disorders of shift workers. Occup. Med. (Lond.) 53, 103–108 [DOI] [PubMed] [Google Scholar]
- 42.Parkes K. R. (2002) Shift work and age as interactive predictors of body mass index among offshore workers. Scand. J. Work Environ. Health 28, 64–71 [DOI] [PubMed] [Google Scholar]
- 43.Karlsson B. H., Knutsson A. K., Lindahl B. O., and Alfredsson L. S. (2003) Metabolic disturbances in male workers with rotating three-shift work. Results of the WOLF study. Int. Arch. Occup. Environ. Health 76, 424–430 [DOI] [PubMed] [Google Scholar]
- 44.Fonken L. K., Workman J. L., Walton J. C., Weil Z. M., Morris J. S., Haim A., and Nelson R. J. (2010) Light at night increases body mass by shifting the time of food intake. Proc. Natl. Acad. Sci. U.S.A. 107, 18664–18669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antunes Lda C., Jornada M. N., Ramalho L., and Hidalgo M. P. (2010) Correlation of shift work and waist circumference, body mass index, chronotype and depressive symptoms. Arq. Bras. Endocrinol. Metabol. 54, 652–656 [DOI] [PubMed] [Google Scholar]
- 46.Antunes L. C., Levandovski R., Dantas G., Caumo W., and Hidalgo M. P. (2010) Obesity and shift work: chronobiological aspects. Nutr. Res. Rev. 23, 155–168 [DOI] [PubMed] [Google Scholar]
- 47.Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., and Schibler U. (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eberly R., and Feldman H. (2010) Obesity and shift work in the general population. Internet J. Allied Health Sci. Pract. 8, 10 [Google Scholar]
- 49.Evans A. M., DeHaven C. D., Barrett T., Mitchell M., and Milgram E. (2009) Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 81, 6656–6667 [DOI] [PubMed] [Google Scholar]
- 50.Eckel-Mahan K. L., Patel V. R., Mohney R. P., Vignola K. S., Baldi P., and Sassone-Corsi P. (2012) Coordination of the transcriptome and metabolome by the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 109, 5541–5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dehaven C. D., Evans A. M., Dai H., and Lawton K. A. (2010) Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminform. 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon H., and Pessin J. E. (2013) Adipokines mediate inflammation and insulin resistance. Front. Endocrinol. 4, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Travers R. L., Motta A. C., Betts J. A., and Thompson D. (2015) Adipose tissue metabolic and inflammatory responses to a mixed meal in lean, overweight and obese men. Eur. J. Nutr. 10.1007/s00394-015-1087-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Travers R. L., Motta A. C., Betts J. A., Bouloumie A., and Thompson D. (2015) The impact of adiposity on adipose tissue-resident lymphocyte activation in humans. Int. J. Obes. (Lond.) 39, 762–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fantuzzi G. (2005) Adipose tissue, adipokines, and inflammation. J. Allergy Clin. Immunol. 115, 911–919 [DOI] [PubMed] [Google Scholar]
- 56.Ellingsgaard H., Ehses J. A., Hammar E. B., Van Lommel L., Quintens R., Martens G., Kerr-Conte J., Pattou F., Berney T., Pipeleers D., Halban P. A., Schuit F. C., and Donath M. Y. (2008) Interleukin-6 regulates pancreatic alpha-cell mass expansion. Proc. Natl. Acad. Sci. U.S.A. 105, 13163–13168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S., Shin H. J., Ding E. L., and van Dam R. M. (2009) Adiponectin levels and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA 302, 179–188 [DOI] [PubMed] [Google Scholar]
- 58.Friedman J. M., and Halaas J. L. (1998) Leptin and the regulation of body weight in mammals. Nature 395, 763–770 [DOI] [PubMed] [Google Scholar]
- 59.Koch C. E., Lowe C., Pretz D., Steger J., Williams L. M., and Tups A. (2014) High-fat diet induces leptin resistance in leptin-deficient mice. J. Neuroendocrinol. 26, 58–67 [DOI] [PubMed] [Google Scholar]
- 60.Le Bacquer O., Petroulakis E., Paglialunga S., Poulin F., Richard D., Cianflone K., and Sonenberg N. (2007) Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J. Clin. Invest. 117, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yadav A., Kataria M. A., Saini V., and Yadav A. (2013) Role of leptin and adiponectin in insulin resistance. Clin. Chim. Acta 417, 80–84 [DOI] [PubMed] [Google Scholar]
- 62.Morton G. J., Gelling R. W., Niswender K. D., Morrison C. D., Rhodes C. J., and Schwartz M. W. (2005) Leptin regulates insulin sensitivity via phosphatidylinositol-3-OH kinase signaling in mediobasal hypothalamic neurons. Cell Metab. 2, 411–420 [DOI] [PubMed] [Google Scholar]
- 63.Brault J. J., and Terjung R. L. (2001) Purine salvage to adenine nucleotides in different skeletal muscle fiber types. J. Appl. Physiol. 91, 231–238 [DOI] [PubMed] [Google Scholar]
- 64.Steffen L. M., Vessby B., Jacobs D. R. Jr., Steinberger J., Moran A., Hong C. P., and Sinaiko A. R. (2008) Serum phospholipid and cholesteryl ester fatty acids and estimated desaturase activities are related to overweight and cardiovascular risk factors in adolescents. Int. J. Obes. (Lond.) 32, 1297–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suhre K., Meisinger C., Döring A., Altmaier E., Belcredi P., Gieger C., Chang D., Milburn M. V., Gall W. E., Weinberger K. M., Mewes H. W., Hrabé de Angelis M., Wichmann H. E., Kronenberg F., Adamski J., and Illig T. (2010) Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One 5, e13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du F., Virtue A., Wang H., and Yang X. F. (2013) Metabolomic analyses for atherosclerosis, diabetes, and obesity. Biomark Res. 1, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newgard C. B., An J., Bain J. R., Muehlbauer M. J., Stevens R. D., Lien L. F., Haqq A. M., Shah S. H., Arlotto M., Slentz C. A., Rochon J., Gallup D., Ilkayeva O., Wenner B. R., Yancy W. S. Jr., Eisenson H., Musante G., Surwit R. S., Millington D. S., Butler M. D., and Svetkey L. P. (2009) A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9, 311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang T. J., Ngo D., Psychogios N., Dejam A., Larson M. G., Vasan R. S., Ghorbani A., O'Sullivan J., Cheng S., Rhee E. P., Sinha S., McCabe E., Fox C. S., O'Donnell C. J., Ho J. E., Florez J. C., Magnusson M., Pierce K. A., Souza A. L., Yu Y., Carter C., Light P. E., Melander O., Clish C. B., and Gerszten R. E. (2013) 2-Aminoadipic acid is a biomarker for diabetes risk. J. Clin. Invest. 123, 4309–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menni C., Fauman E., Erte I., Perry J. R., Kastenmüller G., Shin S. Y., Petersen A. K., Hyde C., Psatha M., Ward K. J., Yuan W., Milburn M., Palmer C. N., Frayling T. M., Trimmer J., Bell J. T., Gieger C., Mohney R. P., Brosnan M. J., Suhre K., Soranzo N., and Spector T. D. (2013) Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 62, 4270–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z. H., Miyahara H., Takeo J., and Katayama M. (2012) Diet high in fat and sucrose induces rapid onset of obesity-related metabolic syndrome partly through rapid response of genes involved in lipogenesis, insulin signalling and inflammation in mice. Diabetol. Metab. Syndr. 4, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y. M., Liu Y., Liu Y. H., Wang X., Guan K., and Zhu H. L. (2015) Higher serum concentrations of betaine rather than choline is associated with better profiles of DXA-derived body fat and fat distribution in Chinese adults. Int. J. Obes. (Lond.) 39, 465–471 [DOI] [PubMed] [Google Scholar]
- 72.Nørrelund H., Wiggers H., Halbirk M., Frystyk J., Flyvbjerg A., Bøtker H. E., Schmitz O., Jorgensen J. O., Christiansen J. S., and Møller N. (2006) Abnormalities of whole body protein turnover, muscle metabolism and levels of metabolic hormones in patients with chronic heart failure. J. Intern. Med. 260, 11–21 [DOI] [PubMed] [Google Scholar]
- 73.Niwa T., Takeda N., and Yoshizumi H. (1998) RNA metabolism in uremic patients: accumulation of modified ribonucleosides in uremic serum: technical note. Kidney Int. 53, 1801–1806 [DOI] [PubMed] [Google Scholar]
- 74.Meijers B. K., Claes K., Bammens B., de Loor H., Viaene L., Verbeke K., Kuypers D., Vanrenterghem Y., and Evenepoel P. (2010) p-Cresol and cardiovascular risk in mild-to-moderate kidney disease. Clin. J. Am. Soc. Nephrol. 5, 1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crisafulli A., Altavilla D., Marini H., Bitto A., Cucinotta D., Frisina N., Corrado F., D'Anna R., Squadrito G., Adamo E. B., Marini R., Romeo A., Cancellieri F., Buemi M., and Squadrito F. (2005) Effects of the phytoestrogen genistein on cardiovascular risk factors in postmenopausal women. Menopause 12, 186–192 [DOI] [PubMed] [Google Scholar]
- 76.Yang Y., Nie W., Yuan J., Zhang B., Wang Z., Wu Z., and Guo Y. (2010) Genistein activates endothelial nitric oxide synthase in broiler pulmonary arterial endothelial cells by an Akt-dependent mechanism. Exp. Mol. Med. 42, 768–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qin Y., Shu F., Zeng Y., Meng X., Wang B., Diao L., Wang L., Wan J., Zhu J., Wang J., and Mi M. (2014) Daidzein supplementation decreases serum triglyceride and uric acid concentrations in hypercholesterolemic adults with the effect on triglycerides being greater in those with the GA compared with the GG genotype of ESR-β RsaI. J. Nutr. 144, 49–54 [DOI] [PubMed] [Google Scholar]
- 78.Lissin L. W., and Cooke J. P. (2000) Phytoestrogens and cardiovascular health. J. Am. Coll. Cardiol. 35, 1403–1410 [DOI] [PubMed] [Google Scholar]
- 79.Glew R. H., Okolie H., Huang Y. S., Chuang L. T., Suberu O., Crossey M., and VanderJagt D. J. (2004) Abnormalities in the fatty-acid composition of the serum phospholipids of stroke patients. J. Natl. Med. Assoc. 96, 826–832 [PMC free article] [PubMed] [Google Scholar]
- 80.Crane J. D., Palanivel R., Mottillo E. P., Bujak A. L., Wang H., Ford R. J., Collins A., Blümer R. M., Fullerton M. D., Yabut J. M., Kim J. J., Ghia J. E., Hamza S. M., Morrison K. M., Schertzer J. D., Dyck J. R., Khan W. I., and Steinberg G. R. (2015) Inhibiting peripheral serotonin synthesis reduces obesity and metabolic dysfunction by promoting brown adipose tissue thermogenesis. Nat. Med. 21, 166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tecott L. H., Sun L. M., Akana S. F., Strack A. M., Lowenstein D. H., Dallman M. F., and Julius D. (1995) Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature 374, 542–546 [DOI] [PubMed] [Google Scholar]
- 82.Lumeng C. N., and Saltiel A. R. (2011) Inflammatory links between obesity and metabolic disease. J. Clin. Invest. 121, 2111–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kato S. (2013) Role of serotonin 5-HT(3) receptors in intestinal inflammation. Biol. Pharm. Bull. 36, 1406–1409 [DOI] [PubMed] [Google Scholar]
- 84.Kim J. J., and Khan W. I. (2014) 5-HT7 receptor signaling: improved therapeutic strategy in gut disorders. Front. Behav. Neurosci. 8, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan B., Qiu Y., Zou X., Chen T., Xie G., Cheng Y., Dong T., Zhao L., Feng B., Hu X., Xu L. X., Zhao A., Zhang M., Cai G., Cai S., Zhou Z., Zheng M., Zhang Y., and Jia W. (2013) Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 12, 3000–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muti P., Awad A. B., Schünemann H., Fink C. S., Hovey K., Freudenheim J. L., Wu Y. W., Bellati C., Pala V., and Berrino F. (2003) A plant food-based diet modifies the serum β-sitosterol concentration in hyperandrogenic postmenopausal women. J. Nutr. 133, 4252–4255 [DOI] [PubMed] [Google Scholar]
- 87.Baskar A. A., Ignacimuthu S., Paulraj G. M., and Al Numair K. S. (2010) Chemopreventive potential of β-sitosterol in experimental colon cancer model: an in vitro and in vivo study. BMC Complement Altern. Med. 10, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blum R., and Kloog Y. (2014) Metabolism addiction in pancreatic cancer. Cell Death Dis. 5, e1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dang C. V. (2012) Links between metabolism and cancer. Genes Dev. 26, 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ganti S., Taylor S. L., Abu Aboud O., Yang J., Evans C., Osier M. V., Alexander D. C., Kim K., and Weiss R. H. (2012) Kidney tumor biomarkers revealed by simultaneous multiple matrix metabolomics analysis. Cancer Res. 72, 3471–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weinstein S. J., Mackrain K., Stolzenberg-Solomon R. Z., Selhub J., Virtamo J., and Albanes D. (2009) Serum creatinine and prostate cancer risk in a prospective study. Cancer Epidemiol. Biomarkers Prev. 18, 2643–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seidel A., Brunner S., Seidel P., Fritz G. I., and Herbarth O. (2006) Modified nucleosides: an accurate tumour marker for clinical diagnosis of cancer, early detection and therapy control. Br. J. Cancer 94, 1726–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Metwally N. S., Ali S. A., Mohamed A. M., Khaled H. M., and Ahmed S. A. (2011) Levels of certain tumor markers as differential factors between bilharzial and non-biharzial bladder cancer among Egyptian patients. Cancer Cell Int. 11, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang J., Chen T., Sun L., Zhao Z., Qi X., Zhou K., Cao Y., Wang X., Qiu Y., Su M., Zhao A., Wang P., Yang P., Wu J., Feng G., He L., Jia W., and Wan C. (2013) Potential metabolite markers of schizophrenia. Mol. Psychiatry 18, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Koopman K. E., Booij J., Fliers E., Serlie M. J., and la Fleur S. E. (2013) Diet-induced changes in the lean brain: hypercaloric high-fat-high-sugar snacking decreases serotonin transporters in the human hypothalamic region. Mol. Metab. 2, 417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paul G. L., DeLany J. P., Snook J. T., Seifert J. G., and Kirby T. E. (1989) Serum and urinary markers of skeletal muscle tissue damage after weight lifting exercise. Eur. J. Appl. Physiol. Occup. Physiol. 58, 786–790 [DOI] [PubMed] [Google Scholar]
- 97.Rathmacher J. A., Flakoll P. J., and Nissen S. L. (1995) A compartmental model of 3-methylhistidine metabolism in humans. Am. J. Physiol. 269, E193–E198 [DOI] [PubMed] [Google Scholar]
- 98.Dyar K. A., Ciciliot S., Wright L. E., Biensø R. S., Tagliazucchi G. M., Patel V. R., Forcato M., Paz M. I., Gudiksen A., Solagna F., Albiero M., Moretti I., Eckel-Mahan K. L., Baldi P., Sassone-Corsi P., Rizzuto R., Bicciato S., Pilegaard H., Blaauw B., and Schiaffino S. (2014) Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 3, 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Masri S., Rigor P., Cervantes M., Ceglia N., Sebastian C., Xiao C., Roqueta-Rivera M., Deng C., Osborne T. F., Mostoslavsky R., Baldi P., and Sassone-Corsi P. (2014) Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Froy O. (2010) Metabolism and circadian rhythms–implications for obesity. Endocr. Rev. 31, 1–24 [DOI] [PubMed] [Google Scholar]
- 101.Chaix A., Zarrinpar A., Miu P., and Panda S. (2014) Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 20, 991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu S., Brown J. D., Stanya K. J., Homan E., Leidl M., Inouye K., Bhargava P., Gangl M. R., Dai L., Hatano B., Hotamisligil G. S., Saghatelian A., Plutzky J., and Lee C. H. (2013) A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature 502, 550–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paschos G. K., Ibrahim S., Song W. L., Kunieda T., Grant G., Reyes T. M., Bradfield C. A., Vaughan C. H., Eiden M., Masoodi M., Griffin J. L., Wang F., Lawson J. A., and Fitzgerald G. A. (2012) Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 18, 1768–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim J. Y., Park J. Y., Kim O. Y., Ham B. M., Kim H. J., Kwon D. Y., Jang Y., and Lee J. H. (2010) Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC-Q-TOF MS). J. Proteome Res. 9, 4368–4375 [DOI] [PubMed] [Google Scholar]
- 105.Choi S. M., Tucker D. F., Gross D. N., Easton R. M., DiPilato L. M., Dean A. S., Monks B. R., and Birnbaum M. J. (2010) Insulin regulates adipocyte lipolysis via an Akt-independent signaling pathway. Mol. Cell Biol. 30, 5009–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kersten S. (2001) Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2, 282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.