Abstract
Influenza A virus is characterized by high genetic diversity.1–3 However, most of what we know about influenza evolution has come from consensus sequences sampled at the epidemiological scale4 that only represent the dominant virus lineage within each infected host. Less is known about the extent of intra-host virus diversity and what proportion is transmitted between individuals.5 To characterize those virus variants that achieve sustainable transmission in new hosts, we examined intra-host virus genetic diversity within household donor/recipient pairs from the first wave of the 2009 H1N1 pandemic when seasonal H3N2 was co-circulating. While the same variants were found in multiple members of the community, the relative frequencies of variants fluctuated, with patterns of genetic variation more similar within than between households. We estimated the effective population size of influenza A virus across donor/recipient pairs to be approximately 100–200 contributing members, which enabled the transmission of multiple lineages including antigenic variants.
Keywords: Influenza A virus, evolution, diversity, virus transmission, next generation sequencing
We have previously shown that pandemic H1N1 and seasonal H3N2 viruses—both present during the first wave of the H1N1 pandemic in Hong Kong6—have similar transmission potential in household settings, and that antigenic variants of H3N2 co-circulated with clades of H1N1/2009.6,7 In other parts of the world, and during the same time period, the unseasonal transmission of H3N2 was observed along with pandemic H1N1 virus.8 To characterize patterns of viral evolution at a finer-scale, and particularly the extent of virus genetic diversity that was transmitted among hosts, we performed whole genome deep sequencing on nasopharyngeal swabs collected from index cases with confirmed influenza along with their household contacts. Importantly, the household epidemiological information enabled us to assign donor/recipient pairs in suspected transmission events with relatively high confidence, compare these with unrelated pairs, and estimate spatio-temporal transmission chains.
The virus sample set was collected in July and August 2009 from 84 individuals (67 index patients and 17 other household members) living in Hong Kong; 16 patients were sampled twice, 2–4 days apart. We estimated intra-host virus diversity for each sample by mapping polymorphic sites onto the consensus genome assemblies to generate a list of single nucleotide variants (SNVs or minor variants) present at a frequency of at least 3%. Intra-host diversity was measured by the Shannon entropy, H, assuming site independence. Mean intra-host diversity was significantly higher (Wilcoxon rank-sum test p = 1.89e–12) for H3N2 (H = 33) than H1N1/2009 (H = 13). There was no significant Pearson correlation between high intra-host virus diversity and high viral titer7 (r = −0.3 for H1N1 and r = −0.16 for H3N2) for most of the genes, with the exception of PA and M for H1N1/2009 (Supplementary Table 1).
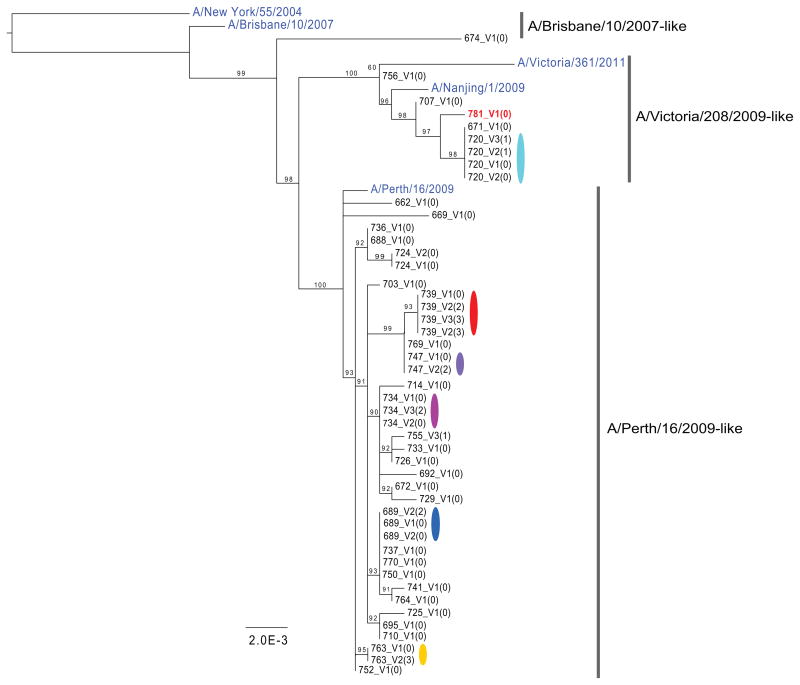
Phylogenetic analysis clustered whole genome consensus sequences by household for each group of patients diagnosed as infected with either H3N2 (Fig. 1) or H1N1/2009 (Supplementary Fig. 1). Comparisons of phylogenetic trees from each gene revealed no evidence for reassortment within this population during the time-frame of the study (data not shown). Three antigenic sublineages of H3N2 (A/Brisbane/10/2007-like, A/Victoria/208/2009-like, and A/Perth/16/2009-like) and three clades of H1N1/2009 (clades 3, 6 and 7) circulated in this population.6 Despite the relatively small population size, one case of mixed subtype infection was observed (patient 781_V1(0)), indicating that dual infection with seasonal and pandemic strains may not be a rare event.9
Figure 1. Maximum likelihood phylogenies of concatenated coding regions for H3N2.
M1/M2 and NS1/NS2 genes were represented as one segment for each covering the sequence between the first ATG to the last stop codon. Bootstrap support values are shown as percentages on nodes. Values below 50% were treated as equivocal and not shown. Public sequences downloaded from GenBank for use as out groups, or included within the diversity of the samples, are colored in blue. One patient, 781_V1(0), was infected with H1N1/2009 clade 7 after having been diagnosed with H3N2 strain A/Victoria/208/2009-like. Only the HA and NA from the H1N1/2009 could be unambiguously assembled from this individual (accession CY115455 and CY115458), while a whole genome was assembled for the H3N2.
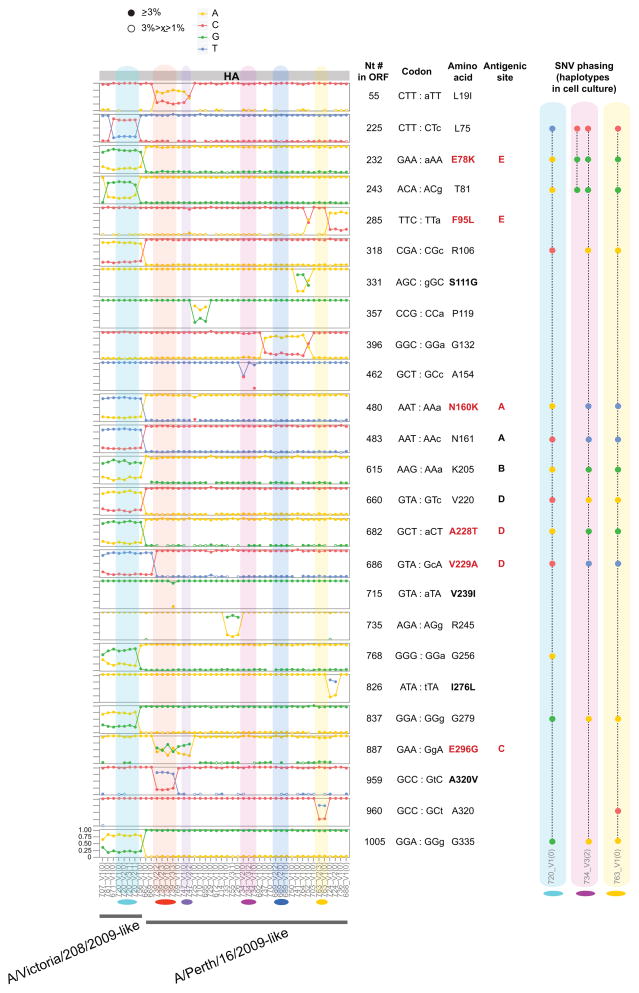
We compared SNVs across samples to determine if minor variants were shared within and between households. For both H3N2 (Fig. 2) and H1N1/2009 (Supplementary Fig. 2) we observed multiple positions in HA—including potential antigenic sites—where the minor variant nucleotide in one clade or lineage became the major nucleotide in another, with evidence of mixed infection at many other sites across the genome (Supplementary Figs. 3 and 4). For example, H3N2 households 707, 781, 671, 720 and 755 reveal a bimodal virus population that appears to have been transmitted intact in multiple transmission events. Overall, we tentatively estimated that approximately 66% of the H3N2-infected patients and 40% of the H1N1/2009-infected patients likely harbored mixed lineage infections (see Supplementary Table 2). To confirm these findings from the clinical specimens, we phased the SNVs into haplotypes by single molecule sequencing for 12 of the cell culture samples from 6 different households (Fig. 2 and Supplementary Fig. 2; Supplementary Tables 3–8). Notably, although the dominant haplotype indicates that each sample belongs to one major lineage, patients often carry a minor haplotype that resembles a separate lineage. This suggests that a number of the SNVs are not only de novo mutations that occurred in the index patient from a household, but are also shared across the community as a whole. We see a similar sharing of variant nucleotides when looking at global consensus sequences across seasons. Using HA consensus sequence data available in GenBank and human 2008 H3 sequences as a reference, we observed a shift of nucleotide frequency at some positions in subsequent seasons of H3N2 epidemics (Supplementary Fig. 5). This phenomenon is more pronounced for variants from the A/Victoria/208/2009-like lineage, in marked contrast to the decreasing trend observed for the A/Perth/16/2009-like lineage. However, no such trend was observed in pandemic H1N1 after the 2009 season. Additionally, frequency variations in H1N1/2009 are far less common than in H3N2. It is important to note that the A/Victoria/208/2009-like virus replaced the A/Perth/16/2009-like virus as the dominant lineage in recent years, leading in 2012 to a change of vaccine strain from A/Perth/16/2009-like virus to A/Victoria/361/2011-like virus (a phylogenetic subgroup of A/Victoria/208/2009). In contrast, pandemic H1N1 virus is antigenically stable and there was no change of vaccine strain after its introduction in humans in 2009. Overall, these data indicate that some viral lineages can be transmitted between individuals below current surveillance thresholds.
Figure 2. Comparison of HA minor variant frequencies across households.
Only polymorphic sites located in the HA1 domain are represented. Amino acid positions were numbered according to the first methionine (start codon) of the protein (and not according to the HA1 numbering schema). Site information for all segments is available in Supplementary Fig. 3. The x axis lists samples by position on the phylogenetic trees in Fig. 1; households with more than one member are colored. The y axis displays nucleotide frequencies with graph lines corresponding to 0, 25%, 75% and 100% frequency. ORF= open reading frame; Antigenic site= previously identified as corresponding to antigenic sites.16–19 Text in red highlights non-synonymous mutations located in antigenic sites. Closed circles represent minor variants found at a frequency 3% and higher, while open circles correspond to frequencies equal or higher than 1%, but below 3%. Boxes show how minor variant nucleotides are phased on the same molecules, representing haplotypes. These were determined from single molecule sequencing of cell culture viruses for 3 household pairs: 720_V1(0)/720_V2(1) (Supplementary Table 6), 734_V1(0)/734_V3(2) (Supplementary Table 7), 763_V1(0)/763_V2(3) (Supplementary Table 8).
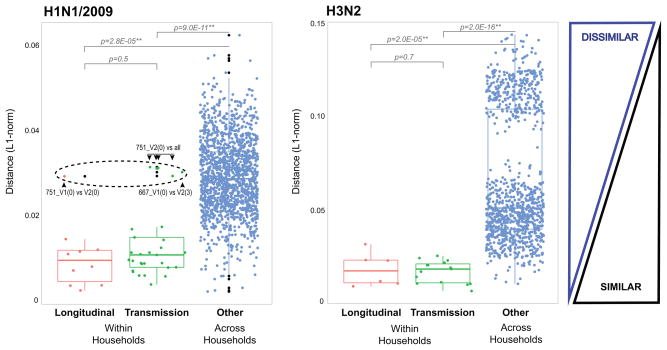
Since each virus sample collected will contain de novo mutations and potentially a mixed infection, we determined the similarity of the viral populations across the data set. To this end we calculated the genetic distance between samples by performing an all-versus-all pairwise comparison for each variant nucleotide position using an L1-norm (see Online Methods). We grouped pairwise comparisons by longitudinal pairs (same individual, sampled at two different visits), within households and across household pairs (Fig. 3). We determined that the median L1 genetic distances within household pairs or longitudinal pairs are significantly closer than any random pairing. This indicates that minor variants and their proportions can be used to infer inter-host transmission, even if a number of these correspond to co-infecting variants that are shared with individuals across households. Interestingly, for H1N1/2009 we see a number of “within household” pairs that are outliers (Fig. 3, dashed circle), providing further evidence of mixed infection. For example, variants present at a minor frequency in most of the samples from household 751 have become dominant in the visit 2 sample for the index case (751_V2(0)) (Supplementary Fig. 1). Although random sampling effects will impact mutational frequencies, such a profound increase in frequency is compatible with a selective advantage in that patient.
Figure 3. Box-plots of L1-norm pairwise genetic distance within and across households.
We used the L1-norm values obtained from the variant nucleotide analysis across all genes to compare overall genetic distance of longitudinal pairs and transmission pairs to every other possible sample pair combination. Each dot on the figure represents the genetic distance between a unique pair. The longitudinal pairs are represented by 16 individuals in 12 households who have been sampled at two different time points, 2–3 days apart. The transmission pairs are from 13 households where at least 2 members have been sampled; there is a total of 22 predicted donor and recipient pairs within households, and 22 more when including more than one time point per individual. The boxplots show the median of the distances; the bottom and top of each box represent the first and third quartiles. The lengths of the whiskers extend to 1.5 times the interquartile range. Outliers are marked by black dots. The dashed black circle in the H1N1/2009 plot marks the outliers. One of the H1N1/2009 pairs—household 751, index case (0), visit 1 and visit 2: 751_V1(0) and 751_V2(0)—had a pairwise genetic distance that was above the expected threshold (H1N1/2009, Longitudinal). When each of these was then used in within household pairwise comparisons (H1N1/2009, Transmission), the visit 2 sample appeared clearly as an outlier. The pairwise genetic distance between the index case in household 667 (667_V1(0)) and its other household member (667_V2(3)) also appeared as an outlier pair.
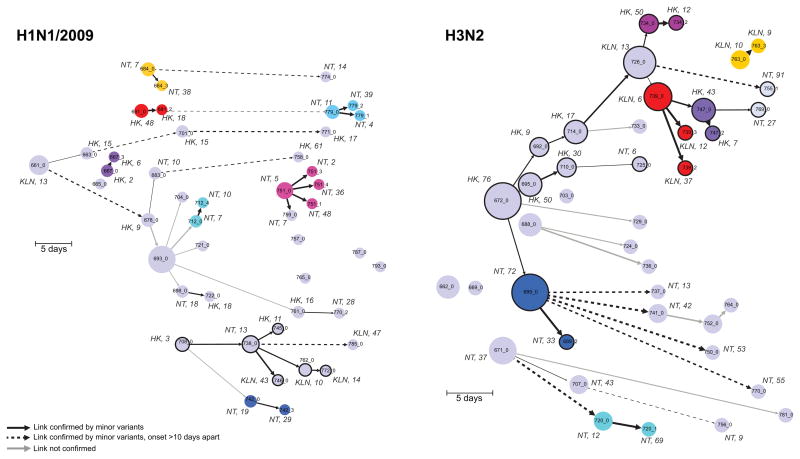
After excluding outliers and considering only a single sample (visit 1) per individual, there were 21 viable “within household” transmission pairs. To select other potential epidemic links within the community, we used the transmission and longitudinal pairs to identify outliers and determine a threshold of maximum genetic distance (after excluding outliers) (Fig. 3). Each pair was epidemiologically linked to a short transmission chain (see below). Using consensus sequences, we first inferred transmission networks across the population using a parsimony and graph-based algorithm.10,11 We then used minor variant data to highlight potential localized outbreaks (Fig. 4) with cross-region links (i.e. Hong Kong Island, Kowloon and New Territories). This network agrees with the fact that there is a high volume of population flow within Hong Kong each day, allowing ample opportunity for influenza transmission across regions.
Figure 4. Reconstruction of potential transmission pathways of H1N1/2009 and H3N2 outbreaks.
Transmission networks are inferred from the consensus whole genome sequences and date of onset. Each sample is a node on the graph and the directed edges indicate putative ancestries and transmissions. Time is represented on the x axis and shows the number of days since the first date of onset. A unique color is assigned to households with more than one member sampled. The size of the node is determined by the number of out degrees. A dashed line indicates a putative transmission link greater than 10 days. The weight of an edge is inversely proportional to the number of nucleotide differences between two samples (i.e. the thicker the edge, the smaller the number of differences). Nucleotide differences were separated into quartiles. H1N1/2009: 0–2 nt; 3–6 nt; 7–15 nt; 16–28 nt. H3N2: 0–5 nt; 6–9 nt; 10–19 nt; 20–45 nt. The links were confirmed by the genetic distances (L1-norm) and normalized for the edge weights. Circles with thick black edges are nodes within a chain of transmission with more than 2 individuals. Locality and age of the patient is indicated for a number of the nodes. HK: Hong Kong; NT: New Territories; KLN: Kowloon.
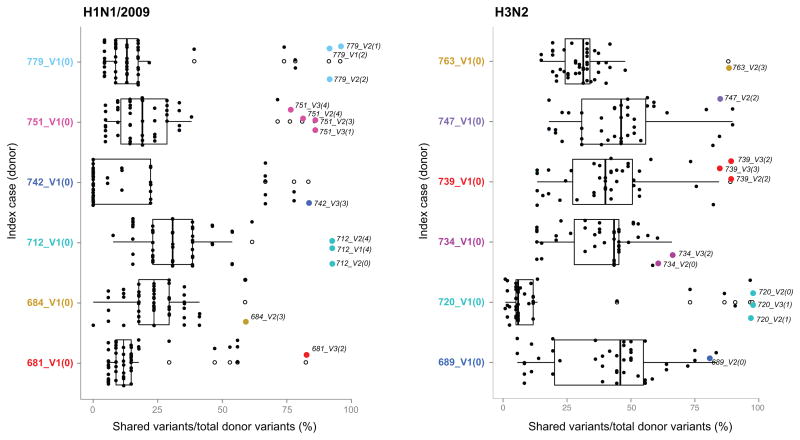
To further explore shared virus populations within households, we compared minor variants at each position in donor (index cases) and recipient transmission pair samples. Most variants found in the donor were shared with the potential recipient (Fig. 5, colored dots). The frequency of shared variants is much lower in pairs of unrelated samples (Fig. 5, black dots), although we find more shared variants in H3N2 than in H1N1/2009 pairs. We observed that the relative frequency of variants in the recipient is more often similar to that found in the donor, which is not the case for the same variants found in any other individual (Wilcoxon signed-rank test, p < 0.05), and implies the lack of a substantial genetic bottleneck at transmission. This in turn suggests that shared variants found in the recipient are not the result of de novo mutations but are more likely present in viruses that transmit between hosts and replicate.
Figure 5. Box-plots comparing shared variant frequencies within and across households.
We compared shared variant frequencies between samples from index cases and their household members (colored dots) or with any other sample (black dots). White boxes indicate interquartile ranges and white dots indicate outliers. Household members tend to share most of the variants found in the index case. Each H1N1/2009 household index case is compared to 54 other samples; each H3N2 household index case is compared to 46 other samples.
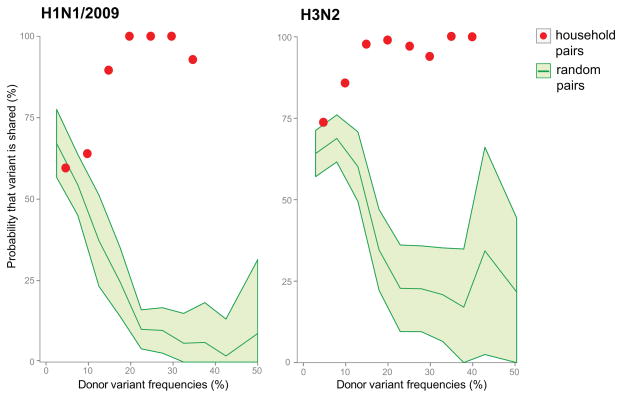
From the household transmission pairs we estimated the probability that multiple variants are transmitted between hosts. In particular, polymorphic sites with variants only detected in the donor and those detected in both donor and recipient samples were selected to determine the probability of transmission as a function of variant frequency. Accordingly, for H1N1/2009, a donor variant found at a frequency of 10% has a 64% chance of being transmitted to the recipient; for H3N2, a donor variant at 10% has an 86% chance of transmission (Fig. 6). Because of the limited sample size it was not possible to determine with confidence the probability of transmission for variants present at frequencies below 10%.
Figure 6. Probability of variant transmission as a function of relative frequency of the minor variants.
Variants that were only detected in the donor and those that were shared between donor and recipient samples were used in determining the probability of transmission. “Household pairs” (red dots) are comparisons between members of the same household. Each point is the proportion of shared variants over the total number of variants found in a window size of 10%. “Random pairs” (green shaded area) are 30 random donor/recipient pairs resampled 100 times to get a standard deviation estimate.
To infer the size of the virus population before and after transmission that is able to generate productive progeny, we estimated the effective population size, Ne, by modifying a version of the Wright-Fisher (WF) idealized population model for our data. Specifically, for the donor/recipient pairs we took the frequency of the shared minor variants, p; the frequency of the major nucleotide at that position, q; and then calculated the variance of the difference in donor/recipient frequencies to obtain a variance effective size. For this we obtained a mean of 192 viral particles (median: 124; mean standard deviation (SD) range: 114–276) for H1N1/2009 and a mean of 248 (median: 138; mean SD range 47–457) for H3N2. To confirm the scale of our estimates, we utilized a different method based on the Kullback-Leibler divergence (KLD) (see Online Methods).12 This gave a mean of 90 (median: 80; mean SD: 55) for H1N1/2009 and a mean of 114 (median 121; mean SD: 55) for H3N2. To estimate how many haplotypes would be present within these replicating populations, we used the phased SNV and reconstructed haplotype data and observed an average of 3 haplotypes for H1N1/2009 and 5 haplotypes for H3N2 transmitted across donor/recipient pairs (Supplementary Tables 3–8). The sample size is too small for the difference between H1N1/2009 and H3N2 to be significant. It is, however, theoretically possible that H3N2 has a higher Ne because the virus has been circulating in the human population since 1968 so that there is greater background genetic diversity and hence a greater diversity of lineages that can be transmitted among hosts. Crucially, these Ne and haplotype estimates suggest that multiple variants can be routinely transmitted between individuals, such that any transmission bottlenecks are fairly loose, and that a relatively small number of viral particles can initiate a productive infection with a number of variant strains that are co-transmitted.
In sum, we have analyzed minor variant dynamics in the transmission of influenza A virus within and across households during an epidemic and used that information to determine potential transmission events. The shared minor variant information between donors and recipients in transmission pairs was then utilized to estimate the number of viral particles that are able to infect and replicate in the recipient. The approach taken here could help define how prior immunity or other host factors, as well as virus subtype and strain, may affect transmission dose, of which our effective size estimates likely capture lower bounds. Indeed, this revealed the transmission of multiple variants, both from mixed infections and from within-host de novo haplotypes, indicating a relatively loose transmission bottleneck. Importantly, the shared variant data also suggest that there has been a single co-infection or super-infection event by two genetically distinct viruses during this epidemic, with this bimodal virus population then being transmitted intact in multiple subsequent transmission events. This is unsurprising in light of recent observations that natural selection can act on pools of virus variants linked by their co-localization in the same cell.13 In addition, this demonstrates that there are likely more cases of mixed lineages within infected patients than can be captured with standard consensus-based diagnostic assays. Such co-infections will obviously facilitate the occurrence of reassortment, and may help explain the frequent detection of reassortants between seasonal H3 viruses.14 Although similar observations have been made in animal studies,11,15 this is the first demonstration for influenza A virus in humans. Characterizing the genetic information of transmitted virions allows a better understanding of influenza virus transmission in humans, and provides more accurate information for modeling epidemics and disease control strategies.
Online Methods
Sample collection
Retrospective pooled specimens of nasal and throat swabs studied in our previous household influenza transmission investigations6,7 were subjected to next generation sequencing by HiSeq 2000 (Illumina). This data set comprises 102 virus samples (55 H1N1/2009 and 47 H3N2) collected from 84 individuals in Hong Kong over July and August 2009. There were multiple home visits and 16 individuals were sampled twice on 2 or 3 household visits (visit 1, V1; visit 2, V2; visit 3, V3), 2–4 days apart.
Sample preparation and sequencing
Multi-segment reverse-transcription PCR (M-RT-PCR)20 was used to amplify influenza-specific segments from total RNA, followed by sequence-independent, single-primer amplification (SISPA).21 Each RNA sample was subjected to 2 rounds of M-RT-PCR and these in turn were amplified by SISPA using different barcodes to control for barcode-specific amplification bias; these technical replicates were then pooled separately for 100 bp paired-ends sequencing on different lanes of a HiSeq 2000 sequencer (Illumina). Potential SISPA PCR duplicate sequence reads were removed with the ELVIRA package. SISPA barcoded reads were demultiplexed with a bespoke DNA Barcode Deconvolution software, and the demultiplexed reads were trimmed of M-RT-PCR primer sequences and low quality regions. Sequence reads were then de novo assembled using CLC Bio’s clc_novo_assemble program (Qiagen) and the resulting contigs were used to identify influenza virus reference segment sequences by performing BLASTN searches against complete influenza virus segments published at GenBank. CLC Bio’s clc_ref_assemble_long software (version 3.22.55705) was then used to map trimmed reads to the segments of the reference genome.
Phylogenetic analyses
All eight Influenza A coding sequences were concatenated into an alignment of 13,425 nucleotides (nt) for H3N2 and 13,392 nt for H1N1/2009. Coding sequences were concatenated in the order of the segment number on which they were encoded (PB2-PB1- PA-HA-NP-NA-M1-M2-NS1-NS2). All isolates were included except for 781_V1(0), which appeared to be a mix of H3N2 and H1N1/2009, encoding genes related to both H1N1/2009 and H3N2 strains. Other taxa not included in this study were used as outgroup taxa (A/California/04/2009 and A/New York/55/2004 for H1N1/2009 and H3N2, respectively). These were selected based on their position in widely sampled single gene phylogenies (data not shown). Two additional taxa—A/Brisbane/10/2007 and A/Nanjing/1/2009—were included in the H3N2 phylogeny to capture the full diversity of this part of the H3N2 tree. Maximum likelihood phylogenies were generated with RAxML22 using the GTR nucleotide substitution model, with among-site rate variation modeled using a discrete gamma distribution using four rate categories. Bootstrap support values were generated using 1,000 fast bootstrap replicates, and represented as percentages on nodes (values below 50% not shown).
Variant analysis
Minor variants were identified using the ELVIRA package, which applies statistical tests to minimize false positive SNV calls that can be caused by sequence specific errors (SSE) that may occur on Illumina platforms.23 This involves observing the forward and reverse reads of a SNV call. Based on a binomial distribution cumulative probability, we calculate the p-values. If both p-values are within a Bonferroni-corrected significance level (alpha = 0.05), the SNV call is accepted. A minimum minor allele frequency of 3% was used as the threshold and a minimum coverage of 200 reads for a given site (see Supplementary Table 9 for coverage average for each sample). This conservative cutoff was selected based on the same control sample that was sequenced in two different sequence runs, and then examining concordance (SNV found in both samples) and discordance (SNV found in only one of 2 samples) for different frequency thresholds. At 3%, 16/17 sites were concordant, while at 4% 14/14 sites were concordant. We chose the lower cut-off to gain more information, even if the error was higher. As a comparison, at 1%, only 32/62 sites were concordant and at 2%, 16/26.
Quantification of intra-host diversity
We used Shannon entropy to quantify the intra-host diversity of each sample through the relative frequencies of each single nucleotide variant using the short read (Illumina) data. This was done across all segments and assumes that all SNVs are independent of each other. We find that the entropy scores between H1N1/2009 and H3N2 are significantly different from each other (p = 1.27E–06).
Where P(i) is the relative frequency of a variant at position i.
Genetic distance across samples
The genetic distance between samples was estimated using three different methods: L1-norm, L2-norm and the Jensen-Shannon divergence (JSD) measure. For the L1-norm, we compare each sample against every other sample (all-versus-all pairwise comparison) at each variant nucleotide position:
Here dk is the distance measured at nucleotide position k between two samples.
n is the total number of possible nucleotide configurations (A, C, G, T).
p and q are vectors containing the relative frequencies of the different variant nucleotides observed (these are analogous to “alleles”).
Between two samples we observe a nucleotide position of a coding sequence (dk) and then sum over all positions to obtain D, the distance measured between two samples for a specific CDS; N is the length of the CDS.
This results in a single number that informs us of the distance (or dissimilarity) between two samples for each of the coding sequences. This was repeated across all segments.
We verified our analysis by comparing against two other distance measures. The L2-norm uses Euclidean distance and follows a similar procedure to the L1-norm with dk computed as such:
D is similarly calculated by summing over all values of dk.
For the third method, the JSD modifies the Kullback-Leibler divergence so that the resulting output is symmetric and will always have a finite value:
The JSD is calculated by:
where
A t-test was used to score significance between the three methods (data not shown). Since no significance was found, we used the L1-norm.
Estimating the virus effective population size (Ne)
We used a modified version of the Wright-Fisher idealized population model24 to estimate the effective population size of influenza A virus from the shared SNVs in our donor/recipient pairs. This model assumes the population does not grow or shrink, there are discrete generations, that every generation is “replaced” by offspring, and that each of the variant sites is independent (The parameter values used in the Wright-Fisher calculations can be found in the Supplementary Table 10). We then calculated a variance effective size, the size of a Wright-Fisher population with the same variance,
where is the variance effective population size for a given nucleotide position i, q is the major variant frequency of a donor j, and p is the minor variant frequency of j. For variants that were shared by all donors for a given strain with a frequency greater than 1% (we use this less conservative threshold so that we have more sites to include in our estimate and better resolution), we calculated the change in variant frequency between donor and recipients for all pairs,
with being the minor variant frequency of the recipient. The variance in this quantity appears in the effective size formula. For H1N1/2009, the size of j is 8 unique donor-recipient pairs with 21 shared variants. The equivalent values for H3N2 are j of 6 unique donor/recipient pairs with 81 shared variants.
To estimate the variance of the effective population size across all household pairs we included a standard deviation (SD) parameter defined by:
which is used in the following modified wright-fisher equations:
This ensures that E[pj ± ε] + E[qj ∓ ε] ≈ 1 and captures the mean standard deviation range and Δj (ε,ε′) is the change in frequencies at the j-th site between the donor an recipient.
To confirm the scale of our estimates, we employed a second method that utilizes Kullback-Leibler divergence, as previously used to measure Ebola virus transmission.12 This approach measures the distance from a true probability distribution, q, to a target probability distribution, p, which are our donor and recipient populations, respectively, and uses their similarity to estimate the number of times the donor distribution was sampled. As with the Wright-Fisher approach, this assumes independence between variant sites and will consequently return a lower bound estimate (N̂) on infectious dose size.
The number of shared variants between donor and recipient is represented by s. A variant has to be shared by both donor and recipient to be included. KL(qi|pi) is the Kullback-Leibler divergence from qi to pi, where qi is the set of nucleotide frequencies found in the donor at position i and pi is the set of nucleotide frequencies found in the recipient at the same site. This value is summed over the variant positions across all segments where a shared variant is discovered on both the donor and recipient. We calculated this for each donor/recipient pair for H1N1/2009 and H3N2.
Haplotype reconstruction by SMRT Sequencing
SNVs identified by Illumina sequencing were phased into haplotypes for six of our donor/recipient pairs (H1N1/2009 681_V1(0)/681_V3(2), 742_V1(0)/742_V3(3), 779_V1(0)/779_V2(1); H3N2: 720_V1(0)/720_V2(1), 734_V1(0)/734_V3(2), 763_V1(0)/763_V2(3)) using SMRT sequencing on the PacBio platform (Pacific Biosciences). DNA library preparation and sequencing was performed according to the manufacturer’s instructions and reflects the P6-C4 sequencing enzyme and chemistry, using 4-hour movie collection parameters. Each barcoded influenza M-RTPCR cDNA was assessed by Qubit analysis and DNA 12000 Agilent Bioanalyzer gel chip to quantify the mass and size distribution of the double-stranded cDNA present. After quantification, samples were pooled in batches of 2–3 samples per SMRTbell library preparation. The barcoded amplicon pools were then re-purified using a 1.8X AMPure XP purification step to assure removal of any damaged fragments and/or biological contaminant. After purification, ~100 ng of each of the purified, unsheared samples was taken into end-repair, which was incubated at 25°C for 5 minutes, followed by a second 1.8X Ampure XP purification step. Next, 0.75 μM of Blunt Adapter was added to the cDNA, followed by 1X template Prep Buffer, 0.05 mM ATP low and 0.75 U/μL T4 ligase to ligate (final volume of 47.5 μL) the SMRTbell adapters to the DNA amplicons. This solution was incubated at 25°C overnight, followed by a 65°C 10-minute ligase denaturation step. After ligation, the library was treated with an exonuclease cocktail to remove un-ligated DNA fragments using a solution of 1.81 U/μL Exo III 18 and 0.18 U/μL Exo VII, then incubated at 37°C for 1 hour. Two additional 1.8X Ampure XP purifications steps were performed to remove any adapter dimer or molecular contamination. Upon completion of library construction, samples were validated using another Agilent Bioanalyzer DNA 12000 gel chip as well as Qubit analysis. For all cases, the yield was sufficient and primer was annealed to the SMRTbell libraries for sequencing. The polymerase-template complex was then bound to the P6 enzyme using a ratio of 10:1 polymerase to SMRTbell at 0.5 nM for 4 hours at 30°C and then held at 4°C until ready for magbead loading, prior to sequencing. The magbead-loaded, polymerase-bound, SMRTbell libraries were placed onto the RSII machine at a sequencing concentration of 50 pM and configured for a 240-minute continuous sequencing run to allow for the maximum number of passes for consensus error-correction through the reads of insert protocol version 2.3.0. Sequencing was conducted to ample coverage using a single SMRTcell for each of the sample pools, where reads were rigorously filtered using a 10-pass, 95% single molecule CCS filter criteria to yield ~23,000–25,000 post-filtered reads per SMRTcell for each of the pooled sample sets. Continuous long read data with 21–26 single-molecule passes was generated and passed through the RS_ReadsOfInsert.1 pipeline version 2.3.0 using an ~99.2% accuracy cut-off to achieve higher quality CCS FASTA and FASTQ files for variant calling. Reads were aligned against the same reference genome used for the Illumina data. The alignment was performed with BLASR,25 using the default parameters. Reads that mapped against each segment were retrieved using SAMtools (version 1.2)26 and converted to FASTA format. We used the variant calls obtained from the Illumina reads and phased them with the PacBio reads to identify linked variants. The GenBank accession for the H1N1/2009 reference was CY111731 while that for the H3N2 reference was CY106640.
Supplementary Material
Acknowledgments
T.S. was a predoctoral trainee supported by NIH T32 training grant T32 EB009403 as part of the HHMI-NIBIB Interfaces Initiative. This research was supported with a grant from the Research Grants Council of the Hong Kong Special Administrative Region, China (Project No. T11-705/14N) (L.L.M.P, Y.G, J.S.M.P and B.J.C), federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services, under contract numbers HHS-N272201400006C (L.L.M.P, Y.G., and J.S.M.P.) and HHS-N266200700005C (B.J.C.), HHS-N272200900007C (E.G., X.L., R.A.H., T.B.S. and D.E.W.), from the National Institute of General Medical Science (NIGMS/NIH) under award number U54 GM088491 (E.G., R.R., J.V.D.) and U54 GM088558 (B.J.C.) and NHMRC Australia Fellowship AF30 (E.C.H). The data for this manuscript and its preparation was generated while D.E.W. was employed at JCVI. The opinions expressed in this article are the author’s own and do not reflect the views of the Centers for Disease Control, the Department of Health and Human Services, or the United States government.
Footnotes
Accession Codes
Sequence data have been deposited in the NCBI nucleotide and sequence read archive (SRA) databases. Accession numbers for the HA and NA genes are listed in the phylogenetic trees; the Illumina raw sequence reads appear in SRA as BioSamples SAMN01095441 to SAMN01095495 for H1N1/2009 and SAMN01095144 to SAMN01095190 for H3N2; The PacBio raw sequence reads for the 12 viral isolates appear in SRA under experiment accessions SRX1117304, SRX1117319, SRX1117320, SRX1117563-SRX1117566, SRX1117568-SRX1117572.
Author Contributions
All the authors read and approved the manuscript. L.L.M.P. and E.G. conceived and designed the experiments, supervised research, performed analyses and wrote the manuscript. T.S. analyzed the deep sequence data, performed the variant codon and clustering analyses, and wrote the manuscript. B.G. and R.R. supervised research on the inoculum size estimates and wrote the manuscript. X.L., R.H., D.E.W., B.Z., and R.S. performed the sample preparation and sequencing. T.B.S., A.T. and J.V.D. performed the bioinformatic analyses. M.B.R. performed phylogenetic analyses, E.C.H. performed phylogenetic analyses and wrote the paper. Y.G. and J.S.M.P. conceived and designed the experiments. B.J.C. conceived and designed the experiments and supervised research.
Competing financial interests:
Authors have no competing financial interests.
References
- 1.Bush RM, Fitch WM, Bender CA, Cox NJ. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol Biol Evol. 1999;16:1457–65. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- 2.Drake JW. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci U S A. 1993;90:4171–5. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A. 1999;96:13910–3. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viboud C, Nelson MI, Tan Y, Holmes EC. Contrasting the epidemiological and evolutionary dynamics of influenza spatial transmission. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120199. doi: 10.1098/rstb.2012.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fordyce SL, et al. Genetic diversity among pandemic 2009 influenza viruses isolated from a transmission chain. Virol J. 2013;10:116. doi: 10.1186/1743-422X-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poon LL, et al. Viral genetic sequence variations in pandemic H1N1/2009 and seasonal H3N2 influenza viruses within an individual, a household and a community. J Clin Virol. 2011;52:146–50. doi: 10.1016/j.jcv.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowling BJ, et al. Comparative epidemiology of pandemic and seasonal influenza A in households. N Engl J Med. 2010;362:2175–84. doi: 10.1056/NEJMoa0911530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghedin E, et al. Unseasonal transmission of H3N2 influenza A virus during the swine-origin H1N1 pandemic. J Virol. 2010;84:5715–8. doi: 10.1128/JVI.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee N, Chan PK, Lam WY, Szeto CC, Hui DS. Co-infection with pandemic H1N1 and seasonal H3N2 influenza viruses. Ann Intern Med. 2010;152:618–9. doi: 10.7326/0003-4819-152-9-201005040-00021. [DOI] [PubMed] [Google Scholar]
- 10.Jombart T, Eggo RM, Dodd PJ, Balloux F. Reconstructing disease outbreaks from genetic data: a graph approach. Heredity (Edinb) 2011;106:383–90. doi: 10.1038/hdy.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes J, et al. Transmission of equine influenza virus during an outbreak is characterized by frequent mixed infections and loose transmission bottlenecks. PLoS Pathog. 2012;8:e1003081. doi: 10.1371/journal.ppat.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emmett KJ, Lee A, Khiabanian H, Rabadan R. High-resolution Genomic Surveillance of 2014 Ebolavirus Using Shared Subclonal Variants. PLoS Curr. 2015;7 doi: 10.1371/currents.outbreaks.c7fd7946ba606c982668a96bcba43c90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Combe M, Garijo R, Geller R, Cuevas JM, Sanjuan R. Single-Cell Analysis of RNA Virus Infection Identifies Multiple Genetically Diverse Viral Genomes within Single Infectious Units. Cell Host Microbe. 2015;18:424–32. doi: 10.1016/j.chom.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westgeest KB, et al. Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011. J Virol. 2014;88:2844–57. doi: 10.1128/JVI.02163-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varble A, et al. Influenza a virus transmission bottlenecks are defined by infection route and recipient host. Cell Host Microbe. 2014;16:691–700. doi: 10.1016/j.chom.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu R, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–60. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitikoon P, et al. Pathogenicity and transmission in pigs of the novel A(H3N2)v influenza virus isolated from humans and characterization of swine H3N2 viruses isolated in 2010–2011. J Virol. 2012;86:6804–14. doi: 10.1128/JVI.00197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tharakaraman K, et al. Antigenically intact hemagglutinin in circulating avian and swine influenza viruses and potential for H3N2 pandemic. Sci Rep. 2013;3:1822. doi: 10.1038/srep01822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong Y, et al. Reassortant between human-Like H3N2 and avian H5 subtype influenza A viruses in pigs: a potential public health risk. PLoS One. 2010;5:e12591. doi: 10.1371/journal.pone.0012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou B, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83:10309–13. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djikeng A, et al. Viral genome sequencing by random priming methods. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–71. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura K, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 25.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.