Abstract
Regulation of mitochondrial biogenesis is essential for proper cellular functioning. Mitochondrial DNA (mtDNA) depletion and the resulting mitochondrial malfunction have been implicated in cancer, neurodegeneration, diabetes, aging, and many other human diseases. Although it is known that the dynamics of the mammalian mitochondrial genome are not linked with that of the nuclear genome, very little is known about the mechanism of mtDNA propagation. Nevertheless, our understanding of the mode of mtDNA replication has advanced in recent years, though not without some controversies. This review summarizes our current knowledge of mtDNA copy number control in mammalian cells, while focusing on both mtDNA replication and turnover. Although mtDNA copy number is seemingly in excess, we reason that mtDNA copy number control is an important aspect of mitochondrial genetics and biogenesis and is essential for normal cellular function.
Keywords: mitochondria, DNA copy number, DNA turnover, replication
Introduction
Mitochondria are ubiquitous organelles of eukaryotic systems that are essential for the aerobic production of ATP via oxidative phosphorylation (Attardi and Schatz, 1988). Mitochondria play an important role in apoptosis and are involved in signal transduction for cell proliferation (Wallace, 2008). These important functions demonstrate that mitochondria are essential for maintaining the health of an organism.
Mitochondria stand out as they own their own genomes. Although mitochondrial DNA (mtDNA) is quite diverse in the eukaryotic kingdom, the organization of the mammalian mitochondrial genome is significantly conserved (Clayton, 1992b). More importantly, mitochondria have a unique genetic system that includes multiple copies for each cell (Schon, 2000).
Altered mtDNA copy number regulation can result in disease. MtDNA depletion has been associated with infantile neurogenetic disorders that cause unexplained weakness, hypotonia, and developmental delays in early childhood (Macmillan and Shoubridge, 1996). Variation in mtDNA copy number was observed in multiple sclerosis patients (Blokhin et al., 2008). Also, a decrease of mtDNA copy number has been associated with renal cell carcinoma (Xing, 2008), liver disease (Morten et al., 2007), biliary atresia (Tiao et al., 2007), type 2 diabetes (Choi et al., 2001), cardiomyopathy (Bai and Wong, 2005), and breast cancer (Yu et al., 2007). Disease caused by excess mtDNA proliferation is less common, although it was reported as a compensatory effect of mtDNA deletions (Bai and Wong, 2005). Loss of mtDNA copy number control is associated with aging (Laderman et al., 1996) and is likely to be linked to either nuclear or mtDNA mutations.
Although tremendous progress has been made in recent years in many aspects of mitochondrial biology, there are many gaps in our understanding of mitochondrial genome regulation. How mtDNA copy number is regulated is of particular interest, due to the unique genetic system of mitochondria. It is a basic mechanistic question that is currently unanswered, and knowledge that is necessary for understanding how functional mitochondria are maintained. The goal of this review is to summarize current viewpoints of mtDNA regulation and to reveal new directions for future studies.
Mitochondrial genome and genetics
The human mitochondrial genome is a 16.6 kb circular structure of double-stranded DNA (Attardi and Schatz, 1988). The two strands are identified as either the heavy (H) strand or the light (L) strand by their varying densities in alkaline CsCl gradients. Thirteen genes code for polypeptides that are part of the oxidative phosphorylation system, with twelve of these genes being located on the H strand. Two rRNAs and twenty-two tRNAs are also encoded by the mitochondrial genome. The displacement loop (D loop) is a 1.1 kb noncoding control region that is important in replication and transcription. Other mitochondrial proteins are nuclear encoded, and they are synthesized in the cytoplasm, and then transported to the mitochondria.
Mitochondrial genetics are unique from the Mendelian genetic system observed in the nucleus in several important ways. First, mtDNA shows a maternal inheritance pattern. Paternal mtDNA is almost never passed on to progeny. Second, mitochondria are polyploid, with up to several thousand copies of their genome per cell. Also, replication and transcription are coupled in mitochondria, with unique machinery that carries out these processes. The mitochondrial genome and nuclear genome are maintained in separate cellular compartments, but they work together to ensure the normal functioning of the mitochondria (Pon et al., 1989).
The polyploidy aspect of mitochondrial genetics invokes the concepts of homoplasmy and heteroplasmy. Homoplasmy means that all the copies of the mitochondrial genome are the same, whereas heteroplasmy means that there is a mixture of two or more mitochondrial genotypes. When studying mitochondrial mutations, it is important to consider the degree of heteroplasmy of mutant versus wild-type DNA to identify when disease-causing thresholds are reached.
It is also important to note that mtDNA is not naked. It is packaged into a nucleoid structure with a group of protein factors. The nucleoid may also serve as a mitochondrial genetic unit as it has been shown that there is very little mtDNA exchange between individual nucleoids (Gilkerson et al., 2008). The mechanism of mtDNA replication with regards to nucleoid structure is an important area for future research to further elucidate the maintenance of heteroplasmy.
Nucleoid structure of mtDNA
The mitochondrial nucleoids are considered stable genetic structures of the mitochondria. In the past, it has been thought that individual mitochondrion might maintain their own genetic identity, but fusion and fission events that occur with great frequency allow for an exchange of nucleoids between mitochondria.
A mitochondrial nucleoid is made up of an aggregate of mtDNA genomes, and a group of protein factors such as mitochondrial transcription factor A (TFAM), mitochondrial single-strand binding protein (mtSSBP), and the helicase Twinkle (Wang and Bogenhagen, 2006). The number of nucleoids per mitochondrion varies depending on the tissue examined. Nucleoids give organization to mtDNA that is important during segregation and affects the degree of mtDNA heteroplasmy. It has been suggested that mitochondrial fusion and fission could influence a nucleoid’s access to proteins that are required for replication and transcription (Bogenhagen et al., 2008). Nucleoids are distributed at regular spatial intervals throughout the mitochondria, indicating a high degree of mtDNA regulation (Margineantu et al., 2002).
The nucleoid structure of mtDNA is similar to the nucleoid structure of bacterial DNA in several ways. An average nucleoid contains 5–7 copies of the genome and has a diameter of about 70 nm (Iborra et al., 2004), which is similar to the packing density of the E. coli genome (Woldringh, 2002). Also, both bacterial and mitochondrial nucleoids are anchored to the membrane. These similarities give even more strength to the endosymbiotic theory, which hypothesizes that mitochondria are the result of endocytosis of aerobic bacteria.
Recently, a layered nucleoid structure was proposed. In this model, replication and transcription take place in the core of the nucleoid, while translation and complex assembly occur in the peripheral region, tying the nucleoid to the inner membrane to allow gene expression to occur near ROS production sites (Bogenhagen et al., 2008). In bacteria, this sort of coupling of translation and membrane insertion that keeps the DNA against the cell membrane is called transertion (Woldringh, 2002). It has been shown that this same coupling probably occurs in yeast mitochondria (Bryan et al., 2002). The identification of a human mitochondrial transcription/translation coupler suggests that a similar mechanism likely regulates human mitochondrial nucleoids, as well (Wang et al., 2007).
MtDNA replication
It is well accepted that mtDNA replication does not coincide with the cell cycle and occurs independently of nuclear DNA replication (Bogenhagen and Clayton, 1977). However, all trans-acting factors associated with mtDNA replication are encoded by nuclear DNA, indicating that the nucleus plays an important role in regulating mtDNA copy number. Since transcription and replication are coupled in mitochondria, transcription-related proteins are also involved in replication (Clayton, 1992a).
DNA polymerase γ (POLG) is the only known DNA polymerase in animal mitochondria. It is a heterodimeric complex made up of a 120–140 kDa catalytic core and a 35–50 kDa accessory subunit (Olson et al., 1995). The accessory subunit is involved in RNA primer binding (Fan et al., 1999) and also binds dsDNA (Lim et al., 1999). In the past, it was thought that DNA repair and recombination in mitochondria was very limited, but POLG exhibits 3′ to 5′ exonuclease activity (Kunkel and Soni, 1988), as well as 5′-deoxyribose phosphate lyase activity (Longley et al., 1998), which shatters this notion (Kaguni, 2004). The components of POLG possess all activities necessary to carry out mtDNA replication and repair to some extent.
Mitochondrial single-stranded DNA-binding protein (mtSSBP) and Twinkle, the mitochondrial helicase, work together to achieve helix destabilization during replication. MtSSBP is a 13–15 kDa protein that acts as a 56 kDa tetramer; it has a high affinity for DNA and a DNA footprint of 8–17 nucleotides (Kaguni, 2004). Twinkle is stimulated by mtSSBP to achieve increased processivity and fidelity of POLG (Korhonen et al., 2003).
TFAM, which is an essential component of nucleoid structure, is necessary for transcription and replication initiation. In addition to providing structure to mtDNA, it is hypothesized that it regulates protein binding at the D-loop, the cis-regulatory region of the mitochondrial genome (Ghivizzani et al., 1994). Significantly, the amount of mtDNA has been shown to be directly proportional to total TFAM levels (Ekstrand et al., 2004). The human TFAM promoter region is known to be regulated by transcription factors Sp1 (Virbasius, 1994), nuclear respiratory factor 1 (NRF-1) (Virbasius, 1994), nuclear respiratory factor 2 (NRF-2) (Virbasius 1994), and hStaf/ZNF143 (Gerard et al., 2007). There is variability of TFAM promoter maps, but there are six conserved motifs across mammalian TFAM promoters (Gerard et al., 2007). The specific role of TFAM and the proteins associated with mtDNA replication are not fully understood and, therefore, worthy of further research.
The regulation of mtDNA replication is one of many factors implicated in mitochondrial biogenesis. Peroxisome proliferators-activated receptor gamma coactivator 1 alpha (PGC-1α) plays a key role in mitochondria biogenesis by inducing NRF-1, NRF-2, and TFAM expression (Wu, 1999) and by interacting with the nuclear hormone receptor PPARγ and thyroid hormone receptor (Puigserver et al., 1998). Thyroid hormone receptors regulate mitochondrial biogenesis by binding to thyroid hormone response element (TRE) sequences in the mitochondrial genome and thereby increasing mitochondrial transcription and replication (Weitzel et al., 2003). Weitzel et al. (2003) also hypothesizes that TREs possibly regulate intermediate factors, such as NRF-1 and PGC-1α, which control a second series of thyroid hormone target genes. PGC-1α overexpression results in the upregulation of genes involved in mitochondrial fatty acid oxidation and can cause massive mitochondrial proliferation in cardiac cells (Lehman, 2000). p38MAPK can activate PGC-1α (Knutti and Kralli, 2001) and its partner PPARα (Barger et al., 2001), suggesting that it has a role in mitochondrial biogenesis (Kelly and Scarpulla, 2004). Nitric oxide, a vasodilator has also been shown to increase mitochondrial biogenesis (Nisoli et al., 2003). Although mitochondrial biogenesis is controlled by many regulatory cascades in response to developmental and physiological stimuli that are still not fully understood, some shared key players as mentioned above in this process have been identified.
There are currently two models of the mechanism of mtDNA replication. The asynchronous displacement mechanism involves two unidirectional, independent origins. According to this model, the heavy strand is synthesized first until the light strand replication origin site is reached, and then light strand synthesis begins in the opposite direction (Clayton, 2003). Alternatively, the leading and lagging strand synthesis mechanism involves coupled, unidirectional mtDNA synthesis (Holt et al., 2000). It was further proposed that both mechanisms of mtDNA replication could occur in a cell and that alterations in the mechanism could result in a change in mtDNA copy number.
MtDNA turnover
When examining mtDNA copy number control, it is important to consider how long a particular mtDNA molecule is preserved within the mitochondria. Early work, using pulse-chase experiments, indicates that the synthesis and degradation rates of mtDNA are coordinated to maintain mtDNA copy number (Berk and Clayton, 1974). MtDNA turnover has been measured by detecting the percentage of 3H-methylthymidine uptake, which indicates new DNA synthesis, through separation by centrifugation. If the degradation rate slows down, the mtDNA synthesis rate decreases as well. A reduction in mtDNA turnover is evidence of an overall slowing of mitochondrial biogenesis, which could be correlated to reduced energy metabolism associated with aging (Litoshenko, 1984).
In the late 1960’s, Gross et al. (1969) investigated the turnover rates of mtDNA and mitochondrial phospholipids in rat tissues to try to determine if mitochondria are synthesized or destroyed as a whole. It was found that the half-life of mtDNA was highly regulated, but varied depending on the tissue. Rat heart, liver, and kidney had the highest mtDNA turnover rates; whereas brain had a significantly lower mtDNA turnover rate, in comparison. We now know that mitochondria undergo fission and fusion events, but what remains unclear is how mtDNA molecules are chosen to be replicated or degraded.
More recently, rapid and random mtDNA turnover has been reported (Kai et al., 2006). Surprisingly, rapid turnover is observed even in nonproliferating cells, which supports the idea that mtDNA replication is independent of the cell cycle. This indicates that mtDNA is not generated solely for the purpose of producing daughter cells. One hypothesis presented is that mtDNA is vulnerable to damage, and the cell needs to get rid of damaged copies of the genome. A recent report showed that herpes simplex virus could be a source of mtDNA damage that led to depletion (Saffran et al., 2007). Although preferential degradation of damaged mtDNA was not detected, mild cellular damage was suggested to increase the rate of the synthesis-degradation cycle (Kai et al., 2006).
There is almost no knowledge about how this synthesis-degradation relationship is regulated to control mtDNA turnover. However, loss of regulation by increased mitophagy, which results in increased mtDNA degradation, has been implicated in human diseases such as Parkinson’s disease (Dagda et al., 2008). Mitophagy normally targets mitochondria with abnormal organization to rid the cell of damaged mitochondria that could likely carry mutant mtDNA (Gomes and Scorrano, 2008). Mitophagy can be induced by nutrient deprivation or laser-induced photodamage of mitochondria (Kim et al., 2007). The mitochondrial permeability transition (MPT) appears to be involved in mitophagy. The opening of MPT pores leads to mitochondrial depolarization, followed by mitochondrial swelling and the eventual rupture of the outer membrane of the mitochondria (Kim et al., 2007). It has been shown that depolarized mitochondria move into autophagosomes and autolysosomes, which increase in number during nutrient deprivation (Rodriguez-Enriquez et al., 2006). Questions concerning the regulatory pathways that control mitophagy and how a mitochondrion is marked for degradation remain. Future studies that elucidate the mechanisms and regulation of mtDNA turnover will make it possible to recognize mitochondrial abnormalities that cause disease and eventually lead to cures.
Possible mechanisms of monitoring mtDNA copy number
Evidence supporting the hypothesis that mtDNA copy number is tightly regulated has recently accumulated. For example, the expression level of the mitochondrial gene ND5, which is essential for respiration, is dependent on the number of wild-type genes present (Bai et al., 2000). Also, during mammalian oocyte maturation, there appears to be an upregulation of mtDNA copy number, whereas a constant mtDNA copy number is maintained in primordial germ cells (Cao et al., 2007), indicating that mtDNA copy number regulation is crucial during early development.
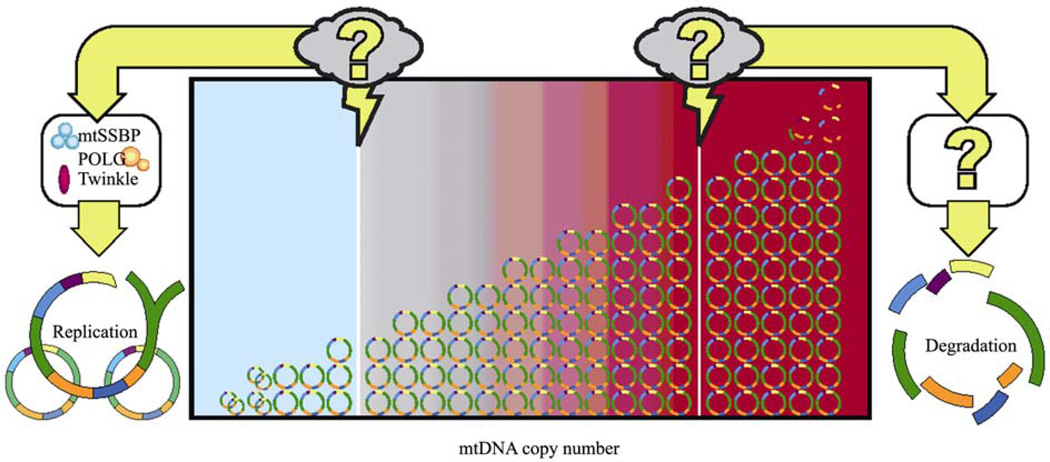
MtDNA copy number is not random; it is specific to tissue type and to developmental stage. However, it is unclear how mtDNA copy number is regulated. We propose the model that there is a lower threshold of mtDNA copy number that triggers an activation of replication by up-regulating the mitochondrial replication machinery, and a higher threshold of mtDNA copy number that triggers the machinery that leads to mtDNA degradation (Fig. 1). Together, these thresholds work to push the mtDNA copy number towards a middle range and regulate the amount of mtDNA in each cell. We do not know what these triggers are, or even what proteins must be activated in these processes. Further investigation of these areas will shed light on the mechanism of control of mtDNA copy number.
Fig. 1.
Threshold hypothesis of mtDNA copy number control. MtDNA copy number is regulated by thresholds. When the copy number reaches the lower threshold, unknown factors trigger the upregulation of the mtDNA replication machinery that instigates replication to push the mtDNA copy number back up. When the copy number reaches the higher threshold, unknown factors trigger the upregulation of proteins that lead to mtDNA degradation to push the mtDNA copy number back down.
There are several models for mtDNA copy number regulation. One idea is that the requirement for ATP dictates the amount of mtDNA, but there are no data to support this possibility. Another idea is that the availability of nucleotides could regulate mtDNA replication (Tang et al., 2000b). Another hypothesis says that mtDNA copy number is regulated by multiple replication origins (Tang et al., 2000a). Replication intermediate studies indicate that TFAM could affect mtDNA copy number by altering the mechanism of replication (Pohjoismaki et al., 2006). It will be important to look at all mtDNA binding proteins to determine limiting factors in mtDNA synthesis; perhaps one of these is released from an inhibitory complex. A review by Moraes (Moraes, 2001) discusses these models in more detail.
On the other hand, almost nothing is known about the mechanism of monitoring mtDNA copy number. MtDNA turnover rate and the proteins involved in the replication and degradation of mtDNA are obvious topics for future research. As we gain more knowledge about mtDNA synthesis and degradation, we will better understand how mtDNA copy number is regulated.
Perspectives
Mitochondrial gene expression and regulation is becoming increasingly relevant to human disease, as more mitochondrial disorders are identified. Mitochondrial dysfunction has been implicated in a range of diseases that includes diabetes, cancer, neurodegeneration, muscle atrophy, as well as aging. However, our knowledge of mtDNA copy number regulation and turnover is far from substantial.
As awareness and identification of mitochondrial disease heightens, more cellular models derived from human patients and animal models with mtDNA copy number abnormalities will appear. With the availability of those models, we expect that more trans-acting factors involved in mtDNA copy number control will be identified, which are both housekeeping genes and inducible factors of replication and turnover. MtDNA turnover and enzymes associated with the process will gain more attention. The isolation and resolution of the three-dimensional structure complexes involved in replication and turnover will be one of the biggest goals. The elucidation of both protein components and cis-acting elements within mtDNA responsible for copy number maintenance will certainly attract more attention. Determining the potential signaling pathways involved in mtDNA copy number monitoring as well as the regulation of mtDNA replication and turnover are top priorities. With the increasing understanding of mtDNA copy number regulation, a more comprehensive picture of mitochondrial genetics and biogenesis will emerge, which in turn will help us to develop new approaches to maintaining healthy mitochondria.
Acknowledgements
The work was supported by the National Institute of Aging/National Institution of Health, USA (No. AG025223 and AG024640) to YB.
References
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu. Rev. Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Bai RK, Wong LJ. Simultaneous detection and quantification of mitochondrial DNA deletion(s), depletion, and over-replication in patients with mitochondrial disease. J. Mol. Diagn. 2005;7:613–622. doi: 10.1016/S1525-1578(10)60595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Shakeley RM, Attardi G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell. Biol. 2000;20:805–815. doi: 10.1128/mcb.20.3.805-815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger PM, Browning AC, Garner AN, Kelly DP. p38 MAP kinase activates PPARα: A potential role in the cardiac metabolic stress response. J. Biol. Chem. 2001;276:44495–44501. doi: 10.1074/jbc.M105945200. [DOI] [PubMed] [Google Scholar]
- Berk AJ, Clayton DA. Mechanism of mitochondrial DNA replication in mouse L-cells: Asynchronous replication of strands, segregation of circular daughter molecules, aspects of topology and turnover of an initiation sequence. J. Mol. Biol. 1974;86:801–824. doi: 10.1016/0022-2836(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Blokhin A, Vyshkina T, Komoly S, Kalman B. Variations in mitochondrial DNA copy numbers in MS brains. J. Mol. Neurosci. 2008;35:283–287. doi: 10.1007/s12031-008-9115-1. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D, Clayton DA. Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell. 1977;11:719–727. doi: 10.1016/0092-8674(77)90286-0. [DOI] [PubMed] [Google Scholar]
- Bryan AC, Rodeheffer MS, Wearn CM, Shadel GS. Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics. 2002;160:75–82. doi: 10.1093/genetics/160.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Shitara H, Horii T, Nagao Y, Imai H, Abe K, Hara T, Hayashi J, Yonekawa H. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat. Genet. 2007;39:386–390. doi: 10.1038/ng1970. [DOI] [PubMed] [Google Scholar]
- Choi YS, Kim S, Pak YK. Mitochondrial transcription factor A (mtTFA) and diabetes. Diabetes Res. Clin. Pract. 2001;54(Suppl. 2):S3–S9. doi: 10.1016/s0168-8227(01)00330-8. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Transcription and replication of animal mitochondrial DNAs. Int. Rev. Cytol. 1992a;141:217–232. doi: 10.1016/s0074-7696(08)62067-7. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Structure and function of the mitochondrial genome. J. Inherit. Metab. Dis. 1992b;15:439–447. doi: 10.1007/BF01799602. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Mitochondrial DNA replication: What we know. IUBMB Life. 2003;55:213–217. doi: 10.1080/1521654031000134824. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: Implications for Parkinson’s disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genetics. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- Fan L, Sanschagrin PC, Kaguni LS, Kuhn LA. The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: Implications for a dual role as a primer recognition factor and processivity clamp. Proc. Natl. Acad. Sci. USA. 1999;96:9527–9532. doi: 10.1073/pnas.96.17.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard MA, Krol A, Carbon P. Transcription factor hStaf/ZNF143 is required for expression of the human TFAM gene. J. Biol. Chem. 2007;273:21998–22006. [Google Scholar]
- Ghivizzani SC, Madsen CS, Nelen MR, Ammini CV, Hauswirth WW. In organello footprint analysis of human mitochondrial DNA: Human mitochondrial transcription factor A interactions at the origin of replication. Mol. Cell. Biol. 1994;14:7717–7730. doi: 10.1128/mcb.14.12.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson RW, Schon EA, Hernandez E, Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J. Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim. Biophys. Acta. 2008;1777:860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- Gross NJ, Getz GS, Rabinowitz M. Apparent turnover of mitochondrial deoxyribonucleic acid and mitochondrial phospholipids in the tissues of the rat. J. Biol. Chem. 1969;244:1552–1562. [PubMed] [Google Scholar]
- Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- Kai Y, Takamatsu C, Tokuda K, Okamoto M, Irita K, Takahaski S. Rapid and random turnover of mitochondrial DNA in rat hepatocytes of primary culture. Mitochondrion. 2006;6:299–304. doi: 10.1016/j.mito.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutti D, Kralli A. PGC-1, a versatile coactivator. Trends Endocrinol. Metab. 2001;12:360–365. doi: 10.1016/s1043-2760(01)00457-x. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5' → 3' DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Soni A. Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-gamma. J. Biol. Chem. 1988;263:4450–4459. [PubMed] [Google Scholar]
- Laderman KA, Penny JR, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent functional alterations of mitochondrial DNA (mtDNA) from human fibroblasts transferred into mtDNA-less cells. J. Biol. Chem. 1996;271:15891–15897. doi: 10.1074/jbc.271.27.15891. [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase γ enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- Litoshenko AIa. Renewal of mitochondrial DNA in the liver of rats of different ages. Biull. Eksp. Biol. Med. 1984;97:299–301. [PubMed] [Google Scholar]
- Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5'-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc. Natl. Acad. Sci. USA. 1998;95:122448–122448. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macmillan CJ, Shoubridge EA. Mitochondrial DNA depletion: Prevalence in a pediatric population referred for neurologic evaluation. Pediatr. Neurol. 1996;14:203–210. doi: 10.1016/0887-8994(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Margineantu DH, Gregory, Cox W, Sundell L, Sherwood SW, Beechem JM, Capaldi RA. Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochondrion. 2002;1:425–435. doi: 10.1016/s1567-7249(02)00006-5. [DOI] [PubMed] [Google Scholar]
- Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- Morten KJ, Ashley N, Wijburg F, Hadzic N, Parr J, Jayawant S, Adams S, Bindoff L, Bakker HD, Mieli-Vergani G, Zeviani M, Poulton J. Liver mtDNA content increases during development: A comparison of methods and the importance of age- and tissue-specific controls for the diagnosis of mtDNA depletion. Mitochondrion. 2007;7:386–395. doi: 10.1016/j.mito.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Fancolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: The role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- Olson MW, Wang Y, Elder RH, Kaguni LS. Subunit structure of mitochondrial DNA polymerase from Drosophila embryos. Physical and immunological studies. J. Biol. Chem. 1995;270:28932–28937. doi: 10.1074/jbc.270.48.28932. [DOI] [PubMed] [Google Scholar]
- Pohjoismäki JL, Wanrooij S, Hyvärinen AK, Goffart S, Holt IJ, Spelbrink JN, Jacobs HT. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon LA, Vestweber D, Yang M, Schatz G. Interaction between mitochondria and the nucleus. J. Cell Sci. Suppl. 1989;11:1–11. doi: 10.1242/jcs.1989.supplement_11.1. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffran HA, Pare JM, Corcoran JA, Weller SK, Smiley JR. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 2007;8:188–193. doi: 10.1038/sj.embor.7400878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA. Mitochondrial genetics and disease. Trends Biochem. Sci. 2000;25:555–560. doi: 10.1016/s0968-0004(00)01688-1. [DOI] [PubMed] [Google Scholar]
- Tang Y, Manfredi G, Hirano M, Schon EA. Maintenance of human rearranged mitochondrial DNAs in long-term cultured transmitochondrial cell lines. Mol. Cell. Biol. 2000a;11:2349–2358. doi: 10.1091/mbc.11.7.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Schon EA, Wilichowski E, Vazquez-Memije ME, Davidson E, King MP. Rearrangements of human mitochondrial DNA (mtDNA): New insights into the regulation of mtDNA copy number and gene expression. Mol. Cell. Biol. 2000b;11:1471–1485. doi: 10.1091/mbc.11.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiao MM, Lin TK, Kuo FY, Huang CC, Du YY, Chen CL, Chuang JH. Early stage of biliary atresia is associated with signifcant changes in 8-hydroxydeoxyguanosine and mitochondrial copy number. J. Pediatr. Gastroenterol. Nutr. 2007;45:329–334. doi: 10.1097/MPG.0b013e3180cc2c0f. [DOI] [PubMed] [Google Scholar]
- Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respoiratory factors: A potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria as chi. Genetics. 2008;179:727–735. doi: 10.1534/genetics.104.91769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bogenhagen DF. Human mitochondrial DNA nuceloids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- Wang Z, Contey J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J. Biol. Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel JM, Iwen KA, Seitz HJ. Regulation of mitochondrial biogenesis by thyroid hormone. Exp. Physiol. 2003;88:121–128. doi: 10.1113/eph8802506. [DOI] [PubMed] [Google Scholar]
- Woldringh CL. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 2002;45:17–29. doi: 10.1046/j.1365-2958.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, de Andrade M, Swan GE, Wu X. Mitochondrial DNA content: Its genetic heritability and association with renal cell carcinoma. J. Natl. Cancer Inst. 2008;100:1104–1112. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R. Reduced mitochondrial DNA copy number is correlated with tumor progression in Chinese breast cancer patients. IUBMB Life. 2007;59:450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]