Introductory Paragraph
Genetic defects in myelin formation and maintenance cause leukodystrophies, a group of white matter diseases whose mechanistic underpinnings are poorly understood1,2. Hypomyelination and Congenital Cataract (HCC), one of these disorders, is caused by mutations in FAM126A, a gene of unknown function3. We show that FAM126A/Hyccin regulates the synthesis of phosphatidylinositol 4-phosphate (PI4P), a determinant of plasma membrane identity4–6. HCC patient fibroblasts exhibit reduced PI4P levels. FAM126A is an intrinsic component of the plasma membrane phosphatidylinositol 4-kinase complex that comprises PI4KIIIα and its adaptors TTC7 and EFR35,7. A FAM126A–TTC7 co-crystal structure reveals an all-α-helical heterodimer with a large protein-protein interface and a conserved surface that may mediate binding to PI4KIIIα. Absence of FAM126A, the predominant FAM126 isoform in oligodendrocytes, destabilizes the PI4KIIIα complex in mouse brain and patient fibroblasts. We propose that HCC pathogenesis involves defects in PI4P production in oligodendrocytes, whose specialized function requires massive plasma membrane expansion and thus generation of PI4P and downstream phosphoinositides8–11. Our results point to a role of FAM126A in supporting myelination, an important process in development and also following acute exacerbations in multiple sclerosis12–14.
Main Text
Phosphoinositides (PIPs) are low abundance anionic membrane phospholipids that play critical roles in many physiological processes15,16. At the plasma membrane, a major PIP is PI4P, which has direct signaling roles and in addition serves as the precursor of two other plasma membrane-enriched PIPs with major signaling functions in this membrane, PI(4,5)P2 and PI(3,4,5)P34,17. In oligodendrocytes and Schwann cells, these lipids regulate several steps in the biogenesis and maintenance of myelin, including the recruitment of myelin basic protein (MBP) to the plasma membrane by PI(4,5)P2 and the promotion of myelin growth by PI(3,4,5)P38–11.
Phosphorylation of phosphatidylinositol (PI) to generate PI4P at the plasma membrane is mediated by PI 4-kinase Type IIIα (PI4KIIIα, Stt4 in yeast)5,18,19. The properties, targeting mechanisms and regulation of this enzyme have only recently come into focus. Two factors required for its localization at the plasma membrane have been described5,7 and structurally characterized20: EFR3 and TTC7 (Efr3 and Ypp1 in yeast).
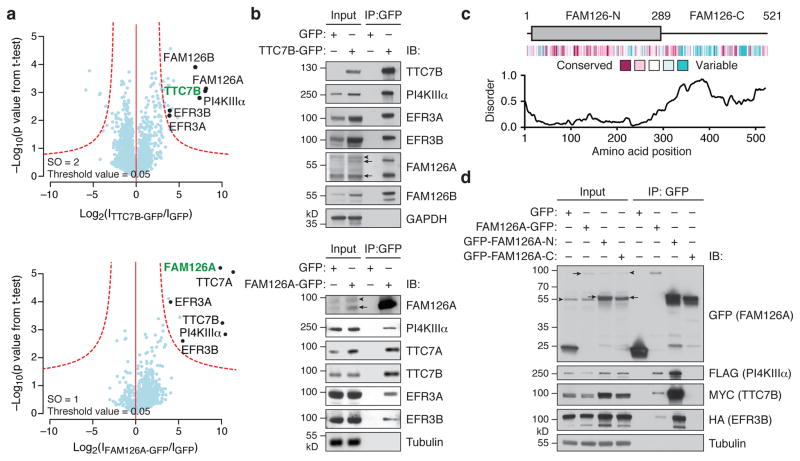
To identify additional regulators of PI4KIIIα, we used quantitative interaction proteomics. We immunoprecipitated either stably expressed TTC7B-GFP or GFP, followed by protease digestion and mass spectrometry analysis, to identify candidate TTC7B-binding proteins (Fig. 1a and Supplementary Table 1). These experiments identified the known TTC7B interaction partners PI4KIIIα, EFR3A, and EFR3B. Among the other hits, two of the most significantly enriched candidates were the paralogous FAM126A/hyccin and FAM126B, both of which were also identified in similar experiments using EFR3A-GFP or EFR3B-GFP as the bait (Supplementary Fig. 1a).
Figure 1. FAM126 is a novel component of the PI4KIIIα complex.
(a) Volcano plots of proteins associating with TTC7B-GFP vs. GFP only (top) and FAM126A-GFP vs. GFP only (bottom), from a label-free proteomics analysis of anti-GFP immunoprecipitates of HEK 293T cells stably expressing TTC7B-GFP, FAM126A-GFP, or GFP only. The logarithmic ratios of protein intensities are plotted against negative logarithmic p values of two-tailed Student’s t-test, equal variance, performed from n = 3 independent experiments. The red dotted line (significance, 0.05) separates specifically interacting proteins (top right portion of plot) from background. Selected top hits are indicated with black dots (bait is indicated in green), and all specific interactors are reported in Supplementary Table 1. (b) Immunoblot (IB) analysis of PI4KIIIα complex components in anti-GFP immunoprecipitates of HEK 293T cells stably expressing TTC7B-GFP (top), FAM126A-GFP (bottom), or GFP only. (c) Domain structure, evolutionary conservation, and predicted extent of disordered secondary structural elements of FAM126. (d) Immunoblot analysis of anti-GFP immunoprecipitates from HeLa cells transfected with 3xFLAG-PI4KIIIα, TTC7B-MYC, EFR3B-HA, and the indicated GFP-tagged construct. Arrows denote specific bands and arrowheads denote non-specific antibody bands.
Mutations in FAM126A that lead to loss of the FAM126A/hyccin protein cause a recessive leukoencephalopathy termed Hypomyelination and Congenital Cataract (HCC)3. Manifestations of this condition, which include progressive neurological impairment, mild to moderate cognitive defects, and peripheral neuropathy, stem from hypomyelination in the central and peripheral nervous systems3,21,22. No molecular functions or activities have been ascribed to FAM126A, and the cellular and molecular mechanisms of HCC pathogenesis are unknown. Thus, the identification of FAM126A as a potential interaction partner of TTC7 and EFR3 led us to investigate its role in PI4KIIIα complex formation and function as a first step toward understanding whether defects in phosphoinositide metabolism may cause HCC pathology.
We first explored and confirmed the interaction of FAM126A with TTC7, and more generally its association with the PI4KIIIα complex, in co-immunoprecipitation/immunoblot experiments (Fig. 1b). Two FAM126A immunoreactive species (58 and 47 kD) were enriched in TTC7B-GFP immunoprecipitates, corresponding to two predicted splice forms of FAM126A (Supplementary Fig. 1b). Accordingly, these two bands are absent in tissues from FAM126A knockout mice (Supplementary Fig. 1c). Given that the longer, 58 kD band represents the major form in mammalian brain (Supplementary Fig. 1d), we chose to focus on it in our subsequent studies. We then performed a reciprocal proteomics experiment using FAM126A-GFP-expressing cells to assess whether PI4KIIIα, TTC7, and EFR3 are the major interaction partners of FAM126A. Indeed, all of these proteins were the strongest hits in analogous quantitative proteomics experiments (Fig. 1a and Supplementary Table 1) and were highly enriched in immunoblot analysis of FAM126A-GFP immunoprecipitates (Fig. 1b).
Examination of the primary amino acid sequence of FAM126A revealed a highly conserved, structured N-terminal portion (FAM126A-N, residues 1–289) common to both splice forms and a poorly conserved C-terminal tail predicted to be disordered (FAM126A-C, residues 290–521) (Fig. 1c). Co-immunoprecipitation experiments of GFP-tagged full-length FAM126A, FAM126A-N, or FAM126A-C with differentially tagged PI4KIIIα, TTC7B, and EFR3B revealed an interaction of all of these components with full-length FAM126A and FAM126A-N (Fig. 1d). Note that the apparent more robust interaction of these proteins with FAM126A-N than with full-length FAM126A (Fig. 1d, lanes 6 and 7) reflects higher levels of FAM126A-N in the total lysate. Interestingly, overexpression of FAM126A-N led to a marked increase in levels of transfected TTC7B in total lysate (Fig. 1d, lane 3), suggesting a stabilizing interaction between these two proteins, as further confirmed by experiments described below.
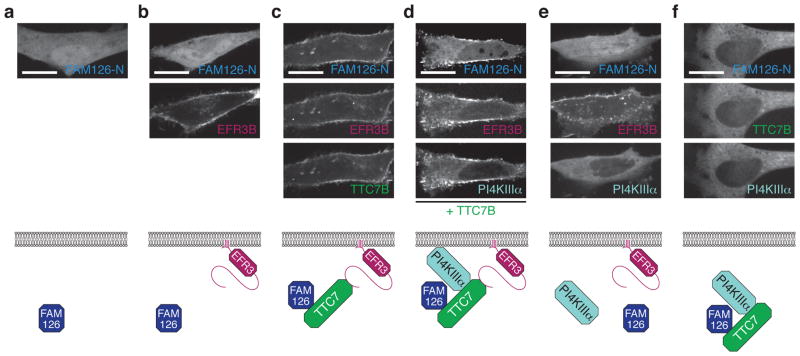
We next examined whether, as would be expected for a direct TTC7 interactor5, FAM126A-N localizes to the plasma membrane when co-expressed with other PI4KIIIα complex subunits. Using confocal microscopy, we found that GFP-tagged FAM126A-N and full-length FAM126A were localized to the cytosol in HeLa and COS-7 cells (Fig. 2a and Supplementary Fig. 2a). While co-expression of FAM126A-N with EFR3B, the membrane anchor for the PI4KIIIα complex (Fig. 2, bottom), did not change the FAM126A-N localization (Fig. 2b), co-expression of FAM126A-N or full-length FAM126A with both EFR3B and TTC7B resulted in a relocalization of FAM126A to the plasma membrane (Fig. 2c and Supplementary Fig. 2b). Further, co-expression of FAM126A-N, TTC7B, EFR3B, and PI4KIIIα resulted in colocalization of all four proteins at the plasma membrane in a manner that was dependent on the presence of TTC7B (Fig. 2d, e). Omission of EFR3B revealed a cytosolic colocalization (but nuclear exclusion) of TTC7B, FAM126A-N, and PI4KIIIα (Fig. 2f). Collectively, these data argue for a central role of TTC7B not only in bridging PI4KIIIα to EFR3B, the plasma membrane anchor, as reported previously5,7,20, but also in mediating the association of FAM126A to the plasma membrane, consistent with a direct interaction.
Figure 2. FAM126A is recruited to the plasma membrane in a complex with TTC7B, PI4KIIIα, and EFR3B.
Top, Confocal imaging of live HeLa cells transfected with GFP-FAM126A-N and the following additional plasmids: (a) none; (b) EFR3B-BFP; (c) EFR3B-BFP and TTC7B-mCherry; (d) EFR3B-BFP, PI4KIIIα-mCherry, and TTC7B-3xFLAG; (e) EFR3B-BFP and PI4KIIIα-mCherry; (f) TTC7B-BFP and PI4KIIIα-mCherry. Bottom, cartoon representation of the transfected constructs and interpretation of the results. Scale bars, 20 μm.
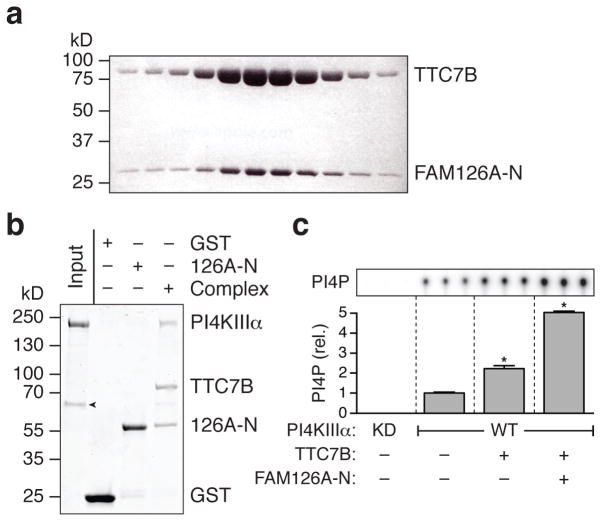
The localization experiments did not, however, allow us to conclusively determine whether, at the plasma membrane, FAM126A and PI4KIIIα can bind to TTC7B simultaneously, or, alternatively, whether FAM126A competes with PI4KIIIα for binding to TTC7B. To address this issue, we first confirmed the direct interaction between FAM126A-N and TTC7B by co-expression of the proteins in E. coli and analysis of the resulting complex by size-exclusion chromatography (Fig. 3a). The two proteins co-eluted as a heterodimer. Importantly, protein-protein interaction experiments with purified proteins showed that the TTC7B/FAM126A-N subcomplex interacts directly with PI4KIIIα (Fig. 3b), ruling out a competition between FAM126A and PI4KIIIα for TTC7 binding.
Figure 3. FAM126A binds directly to TTC7B, and both FAM126A and TTC7B bind to PI4KIIIα and stimulate its catalytic activity.
(a) TTC7B and FAM126A-N co-migrate as a heterodimer on a sizing column (Superdex 200), as shown by SDS-PAGE analysis of elution fractions. Proteins were visualized using Coomassie blue staining. (b) TTC7B/GST-FAM126A-N pulls down PI4KIIIα in a protein-protein interaction experiment with purified proteins. The GST-tagged proteins (GST, GST-FAM126A-N (126A-N), or the GST-FAM126A-N/TTC7B dimer (Complex)) were immobilized on glutathione-conjugated sepharose and incubated with purified PI4KIIIα. Proteins were visualized using Coomassie blue staining. The experiment was performed in triplicate. Arrowhead denotes the chaperone Hsp70, which co-purified with PI4KIIIα. (c) Kinase activity assay using purified PI4KIIIα sub-complexes. Wild-type (WT) or kinase-dead (KD) PI4KIIIα was produced either alone or in complex with TTC7B or TTC7B/FAM126A-N as indicated (Supplementary Fig. 3a), and in vitro lipid kinase activity assays were performed using PI-containing liposomes and γ32P-labeled ATP. The extent of PI4P formation was assessed by TLC and autoradiography. Two-tailed Student’s t-test with unequal variance: *, p = 0.0001 (n = 3 independent experiments). Error bars represent standard deviation.
Given that the TTC7B/FAM126A-N heterodimer bound tightly to PI4KIIIα, we next examined the effects of each of these proteins on the catalytic activity of PI4KIIIα. We purified wild-type or kinase-dead PI4KIIIα alone or in complex with either TTC7B only or both TTC7B and FAM126A-N (Supplementary Fig. 3a) and assessed the relative kinase activity in vitro by monitoring the formation of PI4P from phosphatidylinositol in the presence of γ32P-labeled ATP. The PI4KIIIα/TTC7B complex was about twice as enzymatically active as PI4KIIIα only, and the PI4KIIIα/TTC7B/FAM126A-N ternary complex was roughly five-fold more active (Fig. 3c). Thus both TTC7B and FAM126A appear to play a role in stabilizing the PI4KIIIα fold and/or stimulating the intrinsic enzymatic activity of the kinase.
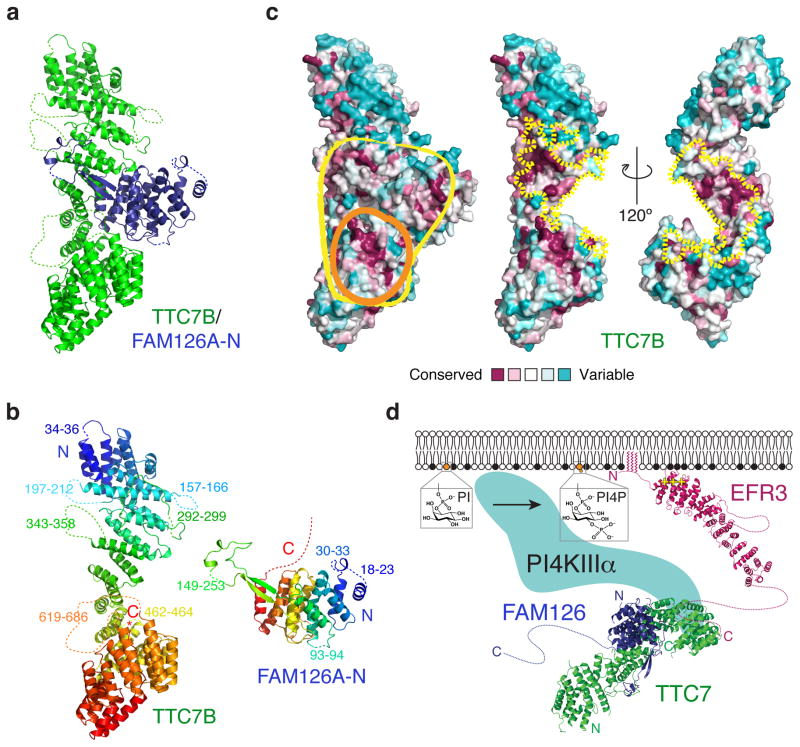
To obtain further insights into the role of FAM126A in the kinase complex, we determined the crystal structure of the TTC7B/FAM126A-N dimer at 2.9 Å resolution (Fig. 4a and Supplementary Fig. 3b). Both proteins are almost entirely alpha-helical. In TTC7B, as in its yeast homolog Ypp120 (PDBID 4N5C), the alpha helices are arranged into a superhelix, with the very C-terminal peptide inserted into the center of the superhelix (Fig. 4b). FAM126A-N is globular, except for a long hairpin (residues 121–165) that extends out and wraps around TTC7B (Fig. 4b). The proteins interact via an unusually large interface (~6000 Å2 total occluded surface area, where >2000 Å2 is considered large), indicative of a high-affinity, stable interaction as would be expected if FAM126A is an intrinsic part of the kinase complex (Fig. 4c).
Figure 4. Crystal structure of a TTC7B/FAM126A co-complex reveals an unusually large protein-protein interface and a conserved binding surface for PI4KIIIα.
(a) Ribbon diagram for the TTC7B/FAM126A-N complex. Note the FAM126A-N hairpin structure that wraps around TTC7B like an arm. Point mutations in FAM126A that underlie HCC are indicated in Supplementary Fig. 3c. (b) Ribbon diagrams for TTC7B and FAM126A-N colored from blue (N-terminus) to red (C-terminus). The “arm” in FAM126A is green. Disordered residues absent from the model are indicated by dotted lines. (c) Space-filling models of the TTC7B/FAM126A-N complex (left) and TTC7B (middle and right) colored by sequence conservation. Conserved surfaces on TTC7B/FAM126A-N (yellow outline) or TTC7B alone (orange outline) that may interact with PI4KIIIα are circled (left). The TTC7B surfaces at the interface with FAM126A-N are indicated by dotted yellow lines (middle and right). (d) Model for PI4KIIIα assembly at the plasma membrane. The EFR3 model is based on the structure of yeast Efr320 (PDBID 4N5A).
Interestingly, while the interacting surfaces in both proteins are well conserved in metazoans (Fig. 4c), the FAM126A-interacting surface of TTC7B is not conserved in fungi (including in S. cerevisiae Ypp1)20. Indeed, we have not been able to identify a fungal FAM126A homolog, and it is possible that FAM126 is not conserved across eukaryotic evolution but is a modification to the PI4KIIIα system present in higher eukaryotes only. Nevertheless, the overall organization of EFR3, TTC7 and PI4KIIIα within the complex is most likely conserved (Fig. 4d). Thus, as the yeast homolog of TTC7 interacts with the kinase via conserved surfaces in the C-terminal lobe20, we hypothesize that mammalian TTC7 interacts with PI4KIIIα in a similar way. We note that in the TTC7B/FAM126A-N subcomplex there is a conserved surface comprising the C-terminal portion of TTC7B and adjoining FAM126A residues, and we propose that these surfaces together form the binding site for PI4KIIIα in mammals (Fig. 4c).
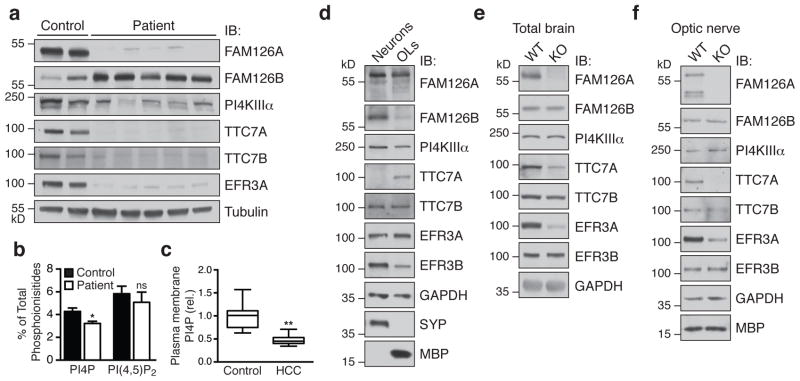
The structural data suggest a stabilizing function for FAM126A in the PI4KIIIα complex. To test this hypothesis, we first examined the levels of PI4KIIIα complex components in primary human skin fibroblasts from five HCC patients that are homozygous for either nonsense or missense mutations in FAM126A3,21,23. Cells from patients with nonsense mutations were completely devoid of the FAM126A/hyccin protein (Fig. 5a, lanes 3, 5, and 7), and cells from patients with missense mutations (L53P and C57R, see Supplementary Fig. 3c) exhibited greatly reduced levels of FAM126A (Fig. 5a, lanes 4 and 6), consistent with the misfolding and near-complete degradation of FAM126A in these cells. Indeed, immunoblot analysis revealed decreases in the levels of PI4KIIIα and its adaptors TTC7A, TTC7B, and EFR3A (EFR3B was not detectable in the fibroblasts) (Fig. 5a and Supplementary Fig. 4a), suggesting a general destabilization and degradation of the PI4KIIIα complex components in the absence of FAM126A. Accordingly, reintroduction of GFP-tagged FAM126A into the patient fibroblasts using a lentiviral vector partially rescued this phenotype (Supplementary Fig. 4b). While a compensatory increase in FAM126B, a paralog of FAM126A, was also observed in patient fibroblasts (Fig. 5a and Supplementary Fig. 4a), the approximately 10-fold lower expression of FAM126B mRNA in fibroblasts (Supplementary Fig. 4c) may explain why this increase is not sufficient to compensate for lack of FAM126A.
Figure 5. Loss of FAM126A leads to defects in the PI4KIIIα complex in HCC patient fibroblasts and FAM126A knockout mouse brain.
(a) Immunoblot (IB) analysis of PI4KIIIα complex components in control and HCC patient fibroblasts. (b) HPLC analysis of total 3H-inositol-labeled phosphoinositide content in control and HCC patient fibroblasts. Two-tailed Student’s t-test, unequal variance, *, p = 0.0019, n = 4 independent experiments. Error bars represent standard deviation. (c) Quantification of the plasma membrane PI4P pool in control and HCC patient fibroblasts by immunofluorescence using an anti-PI4P antibody. Two-tailed Student’s t-test, unequal variance, **, p = 0.0001. Box plot demarcates 25th and 75th percentile (middle line is median), and bars represent the minimum and maximum values (n = 19 cells for control, n = 23 cells for patient, from a total of four independent experiments). Similar results were seen in a second HCC patient cell line. (d) Immunoblot (IB) analysis of PI4KIIIα complex components in wild-type primary cortical neuronal and oligodendrocyte cultures. (e) and (f) Immunoblot (IB) analysis of PI4KIIIα complex components in total brain (e) or optic nerve (f) from male WT or FAM126A knockout mice at age P30. See Supplementary Fig. 4a for quantification of the immunoblot data from a and d–f.
HPLC analysis of total phosphoinositide content revealed a decrease in total cellular PI4P in HCC fibroblasts relative to control fibroblasts (Fig. 5b), and a specific decrease of the plasma membrane fraction of PI4P in HCC fibroblasts relative to control fibroblasts was confirmed by immunofluorescence (Fig. 5c and Supplementary Fig. 4d). Thus, FAM126A loss specifically affects PI4KIIIα complex assembly and PI4KIIIα-mediated PI4P synthesis at the plasma membrane.
Global knockout (KO) of PI4KIIIα in several model organisms leads to lethality5,24–26, while loss of FAM126A is permissive of life, although it causes defects in myelination in humans, leading to HCC3. To investigate the potential role of FAM126A in promoting PI4KIIIα function in myelination, we compared the expression levels of PI4KIIIα complex subunits in mouse primary cortical neurons and oligodendrocytes. We found that FAM126A was expressed in both oligodendrocytes and neurons, while its paralog FAM126B was expressed at much lower levels in oligodendrocytes relative to neurons (Fig. 5d and Supplementary Fig. 4a). Thus, in principle, a global loss of FAM126A could, in the brain, be more effectively compensated by FAM126B in neurons than in oligodendrocytes and thus more severely affect PI4KIIIα function in the latter cell type.
To support this possibility, we evaluated the levels of PI4KIIIα complex components in brain tissue from FAM126A KO mice27. Although these mice do not exhibit an obvious abnormal phenotype or specific defects in myelination as evaluated by light and electron microscopy analysis (Supplementary Fig. 5), immunoblot analysis of lysates from total brain and optic nerve (a pure white matter tract) revealed a selective decrease of TTC7A and EFR3A relative to wild-type (Fig. 5e,and Supplementary Fig. 4a). These are the isoforms of TTC7 and EFR3 whose levels are more highly expressed in oligodendrocytes relative to neurons, compared to their respective paralogs TTC7B and EFR3B (Fig. 5d and Supplementary Fig. 4a).
These results are further supported by immunoblot analysis of cellular fractions of different lineages that were generated from dissociated brain tissue by affinity purification from WT and FAM126A KO mice. Notably, there was a more pronounced decrease in PI4KIIIα, EFR3A/B, and TTC7A/B in cells of the oligodendroglial lineage compared to those of the neuronal lineage (Supplementary Fig. 4e). Taken together, these data connect FAM126A to PI4KIIIα function in the nervous system and implicate the white matter as a region potentially susceptible to PI4KIIIα dysfunction upon global loss of FAM126A, pointing to a mechanistic hypothesis for HCC disease pathogenesis. While it appears surprising that the lack of FAM126A in mice does not produce the neurological phenotype observed in humans, we note that the greater diameter of axons and thickness of the myelin sheath around such axons in humans implies a larger surface area of oligodendrocytes28,29. Thus, a greater demand on the machinery responsible for myelin biogenesis and maintenance in humans may explain their enhanced sensitivity to loss of FAM126A and impaired PI4P synthesis.
Interestingly, FAM126A expression was reported to be repressed by activated β-catenin30, and canonical Wnt/β-catenin/TCF signaling was recently shown to be a powerful negative regulator of oligodendrocyte differentiation31, a step that precedes the massive plasma membrane biogenesis that occurs in myelination. Our data suggest that activation of PI4KIIIα may be an important downstream consequence of the relief of Wnt/β-catenin/TCF signaling that occurs in oligodendrocyte development.
In sum, our results demonstrate that FAM126A is an intrinsic component of the PI4KIIIα complex and an important regulator of PI4P production at the plasma membrane, the first step in the synthesis of the bulk of the downstream PIPs PI(4,5)P2 and PI(3,4,5)P3. We further provide evidence for an important role of FAM126A/hyccin in oligodendrocytes, a cell type whose specialized function is to dramatically expand its plasma membrane. PI(4,5)P2 is implicated in myelin compaction8,9, and PI(3,4,5)P3 helps drive myelin growth10,11. Collectively, our studies point to impaired production of these PIPs as a mechanism through which absence of FAM126A/hyccin results in a hypomyelinating leukoencephalopathy in humans, and they demonstrate the critical importance of plasma membrane PIP homeostasis in myelin development and potentially14 also in remyelination after myelin loss.
Methods
Plasmids and cloning
TTC7B-mCherry, mCherry-PI4KIIIα (NCBI NP_477352.3), and EFR3B-HA were previously described5, and PM-mCherry was obtained from the De Camilli laboratory. 3xFLAG-PI4KIIIα was generated by subcloning PI4KIIIα into the p3xFLAG-CMV-10 vector (Sigma) using NotI and EcoRI. TTC7B-3xFLAG was generated by subcloning TTC7B into the p3xFLAG-CMV-14 vector (Sigma) using HindIII and XbaI. EFR3B-BFP and TTC7B-BFP were generated by subcloning EFR3B into the pTagBFP-N vector (Evrogen) using XhoI and EcoRI (for EFR3B) and XhoI (for TTC7B). TTC7B-MYC was generated by subcloning TTC7B into the pCMV-MYC-N vector (Clontech) using XhoI and NotI. FAM126A (isoform 1, NM_032581.2) was obtained from Origene and subcloned into the pEGFP-N1 vector (Clontech) using XhoI and BamHI to make FAM126A-GFP. GFP-FAM126A-N and GFP-FAM126A-C were generated by subcloning FAM126A1-289 and FAM126A289-521, respectively, into the pEGFP-C1 vector (Clontech) using KpnI and BamHI. For generation of stable HEK 293T cell lines, TTC7B-GFP, FAM126A-GFP, and GFP were subcloned into the pCDNA5/FRT or pCDNA5/FRT/TO vectors (Thermo Fisher) using NheI and NotI (FRT, TTC7B-GFP), BamHI and XhoI (FRT/TO, FAM126A), or EcoRV and NotI (FRT/TO, GFP). The coding sequence for TTC7B9-843 was cloned into a modified pCOLADuet-1 vector, which introduces an N-terminal dodeca-histidine-SUMO tag. The sequence for FAM126A2-308 was cloned into pGEX-6P-1 (GE Healthcare). The sequence for the 419-amino acid splice form of FAM126A (isoform 2, corresponding to NCBI XP_005249951.1) was synthesized as a gBlock (Integrated DNA Technologies) and cloned into the GFP-N1 vector using SalI and BamHI. This GFP-tagged alternate splice form, FAM126A1-419-GFP, was subcloned into the pLL3.7 lentiviral vector (Addgene plasmid #11795, gift from Michael Higley) downstream of the CAG promoter at the EcoRI restriction site using the InFusion system (Clontech). The lentiviral packaging vectors pMD2.G and psPAX2 (Addgene #12259 and 12260) were kindly provided by Didier Trono.
Antibodies
Working dilutions are listed in parentheses for all antibodies. An antibody to EFR3B (1:1000) was previously reported5. Antibodies to mouse TTC7A (1:200) and mouse TTC7B (1:200) were generated by immunization of rabbits with KLH conjugates of the following peptides: Ac-CEASSPVLPFSIIAREL-NH2 (TTC7A); Ac-RLETEIERCRSECQWERIPELC-NH2 (TTC7B) (Cocalico Biologicals, Inc.). Sera were affinity purified using the antigenic peptide immobilized on SulfoLink resin, following the manufacturer’s instructions (Thermo Fisher). An antibody to FAM126A (1:1000) was generated by immunization of rabbits with a purified FAM126A fragment (residues 2–308), and sera were affinity purified by incubation with a nitrocellulose membrane onto which the FAM126A(2–308) fragment had been adsorbed, followed by elution with mild acid. The sources for the other antibodies are as follows: CNPase (1:1000, clone 11-5B, MAB326R, EMD Millipore), EFR3A (1:1000, Ab2, Sigma), FAM126B (1:1000, Novus Biologicals), FLAG (1:2000, M2, Sigma), GAPDH (1:5000, 1D4, Proteus Biosciences), GFP (1:5000, Living Colors Monoclonal, Clontech), HA (1:5000, 3F10, Roche), MBP (1:1000, SMI 99, Covance), MYC (1:2000, rabbit polyclonal, EMD Millipore), NeuN (1:1000, clone A60, MAB377, EMD Millipore) human TTC7B (1:500, mouse IgG fraction, Sigma), PI4KIIIα (1:1000, Cell Signaling Technology), PI4P (1:100, Echelon), SYP (1:20,000, G95, Synaptic Systems), Tubulin (1:10,000, B-5-1-2, Sigma).
Cell culture
All cells were grown at 37 °C in a 5% CO2 atmosphere (except for Expi293 cells (obtained from Thermo Fisher), which were grown at 37 °C in an 8% CO2 atmosphere, rotating at 125 rpm on an orbital shaker) in cell culture medium (Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and penicillin/streptomycin) and were tested to be negative for mycoplasma. HEK 293T cell lines stably expressing TTC7B-GFP, FAM126A-GFP, or GFP were generated by transfection of Flp-In-293 cells (obtained from Thermo Fisher) with flippase (pOG44, Thermo Fisher) and either TTC7B-GFP, FAM126A, or GFP in the pCDNA5/FRT or pCDNA5/FRT/TO vector, followed by selection according to the manufacturer’s instructions (Thermo Fisher) using Hygromycin B (Sigma). Stable HEK 293T cell lines were maintained in cell culture medium supplemented with 10 μg/mL hygromycin B. Stable HeLa cell lines expressing EFR3A-GFP, EFR3B-GFP, or GFP were generated by transient transfection of HeLa cells (obtained from ATCC) followed by selection using G418 (Thermo Fisher), clonal isolation, and expansion. HCC patient and control primary human skin fibroblasts (obtained from the Cell Line and DNA Biobank from Patients Affected by Genetic Diseases, Gaslini Institute, Genova, Italy) were described previously21,27. Following ethical guidelines, the samples were obtained for analysis and storage with written informed consent. The consent was sought using a form approved by the Gaslini Institute Ethics Committee (Genova, Italy).
Transfection
HeLa and HEK 293T cells were transfected with appropriate plasmids using Fugene HD (Promega) according to the manufacturer’s instructions approximately 18–24 h prior to analysis. Expi293 cells were transfected with ExpiFectamine (Thermo Fisher) according to the manufacturer’s instructions.
Immunoprecipitation and immunoblots
HEK 293T cells stably expressing TTC7B-GFP, FAM126A-N-GFP, or GFP were harvested, resuspended in lysis buffer (150 mM NaCl, 20 mM Tris, 1 mM EDTA, 1% Triton X-100, pH 7.4, supplemented with protease inhibitors (cOmplete, EDTA-free (Roche)), sonicated briefly, and centrifuged for 10 min at 16,000 × g. The supernatant was immunoprecipitated in lysis buffer by addition of GFP-trap agarose (Chromotek) and rocking for 1 h at 4 °C. The resin was then isolated by centrifugation at 1000 × g, rinsed three times with lysis buffer, and analyzed either by SDS-PAGE, immunoblot (with detection by chemiluminescence), or mass spectrometry (see below). Original (uncropped) immunoblots are shown in Supplementary Fig. 6.
Pull-down analysis by MS
Resin containing purified immunoprecipitates was rinsed three times with wash buffer (150 mM NaCl, 50 mM Tris, pH 7.4) and then denatured for 30 min with urea (8 M) in 0.1 M Tris, pH 7.4, 1 mM DTT before alkylation and pre-digestion with Endoproteinase LysC (Wako Chemicals USA, Inc.). After incubation for 3 h, samples were diluted 4-fold with ammonium bicarbonate (25 mM) and further digested with trypsin (Promega) overnight. Digestions were stopped by addition of trifluoroacetic acid (TFA, 1 μL), and the resulting peptides were loaded and desalted on C18 Stage Tips.
LC-MS/MS analysis
Peptides were eluted from C18 Stage Tips with 60 μL of elution buffer (80% acetonitrile and 0.1% formic acid), and samples were dried down to 5 μL in a vacuum centrifuge. Peptides were then subjected to reversed phase chromatography on an Easy nLC 1000 system (Thermo Fisher Scientific) using a 50-cm column (New Objective) with an inner diameter of 75 μm, packed in-house with 1.9 μm C18 resin (Dr. Maisch GmbH). Peptides were eluted with an acetonitrile gradient (5–30% for 95 min at a constant flow rate of 250 nL/min) and directly electrosprayed into a mass spectrometer (Q Exactive; Thermo Fisher Scientific). Mass spectra were acquired on the spectrometer in a data-dependent mode to automatically switch between full scan MS and up to 10 data-dependent MS/MS scans. The maximum injection time for full scans was 20 ms, with a target value of 3,000,000 at a resolution of 70,000 at m/z = 200. The ten most intense multiple charged ions (z ≥ 2) from the survey scan were selected with an isolation width of 3Th and fragmented with higher energy collision dissociation (HCD) with normalized collision energies of 25. Target values for MS/MS were set to 1,000,000 with a maximum injection time of 120 ms at a resolution of 17,500 at m/z = 200. To avoid repetitive sequencing, the dynamic exclusion of sequenced peptides was set to 20 s.
MS data analysis
MS and MS/MS spectra were analyzed using MaxQuant (version 1.4.0.5), utilizing its integrated ANDROMEDA search algorithms32. Scoring of peptides for identification was carried out with an initial allowed mass deviation of the precursor ion of up to 6 ppm for the search for peptides with a minimum length of six amino acids. The allowed fragment mass deviation was 20 ppm. The false discovery rate (FDR) was set to 0.01 for proteins and peptides. Peak lists were searched against a local database for human proteome. Maximum missed cleavages were set to 2. The search included carbamidomethylation of cysteines as a fixed modification and methionine oxidation and N-terminal acetylation as variable modifications. All calculations and plots were performed as described33 using the R software package.
Imaging
For live-cell imaging, cells were grown in glass-bottom dishes (#1.5 thickness, MatTek Corporation). For immunofluorescence, cells were grown on cover slips (#1 thickness, neuVitro). Immunofluorescence labeling for PI4P was performed as previously described34. Imaging experiments were performed on a spinning disc confocal microscope, using the PerkinElmer UltraVIEW VoX system including a Nikon Ti-E Eclipse inverted microscope equipped with Perfect Focus, temperature-controlled stage, 14-bit EMCCD camera (Hamamatsu C9100-50), and spinning disc confocal scan head (CSU-X1, Yokogawa) controlled by Volocity software (PerkinElmer). All images were acquired through a 60× or 100× oil objective (1.4 NA, CFI Plan Apo VC). Blue fluorescence was excited with a 405 nm/50 mW diode laser (Melles Griot) and collected by a BP 445/60 filter. Green fluorescence was excited with a 488 nm/50 mW diode laser (Coherent) and collected by a BP 527/55 filter. Red fluorescence was excited with a 561 nm/50 mW diode laser (Cobolt) and collected by a BP 615/70 filter. Multicolor images were acquired sequentially. Image acquisition was performed using the Volocity software (PerkinElmer) and image analysis was performed using Fiji. For the quantification of plasma membrane PI4P immunofluorescence, samples were blinded, and average intensity maximum projections were generated. The cell outline was drawn manually, and the mean pixel intensity was recorded for each sample.
Protein expression (bacterial)
Plasmids containing the coding sequences for His12-SUMO-TTC7B9-843 and GST-FAM126A2-308 were co-transformed into E. coli BL21(DE3) cells. The cells were grown to an optical density (OD600) of ~0.7 at 37 °C and then shifted to 18 °C. Protein expression was induced at 18 °C by addition of isopropylthio-β-galactosidase (IPTG) to final concentration of 0.5 mM. The cells were harvested 20 h after induction and frozen at −80 °C.
Protein purification
For the TTC7B9-843/FAM126A 2-308 complex, cells were resuspended in lysis buffer (20 mM Tris (pH 8.0 at 22 °C), 200 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP), and 20 mM imidazole) supplemented with protease inhibitors (cOmplete, EDTA-free (Roche)), and lysed using a cell disruptor (Avestin). The complex was first isolated using Ni-NTA resin (QIAGEN). After elution, the His12-SUMO tag was cleaved off with SUMO protease. Protein was then bound to Glutathione-S-Sepharose 4B resin (GE Healthcare). Bound complex was eluted from the resin by cleavage with PreScission protease to remove the GST tags. Protein was further purified by size-exclusion chromatography on Superdex 200 (GE Healthcare) in buffer (20 mM Tris (pH 8.0 at 22 °C), 200 mM NaCl, 1 mM TCEP) and concentrated to 6 mg/mL. For protein-protein interaction experiments, GST-tagged FAM126A alone or complexed with TTC7B were isolated by Glutathione-S-Sepharose 4B resin and then eluted from the resin using glutathione. Protein was further purified by size-exclusion chromatography on Superdex 200 (GE Healthcare) in buffer (20 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM TCEP). Selenomethionine-substituted proteins for structure determination were prepared similarly to native proteins as previously described35.
Crystallization
Crystals of native and selenomethionine-substituted TTC7B9-843/FAM126A2-308 complexes were grown by the hanging drop method at 20 °C, mixing 1.35 μL each of protein solution (6 mg/mL) and mother liquor (0.1 M HEPES (pH 7.0–7.4), 4–5% polyethylene glycol (PEG) 8,000) and 0.3 μL of 0.2 M 3-(1-Pyridino)-1-propane sulfonate (NDSB-201). The crystals belong to space group P212121 with two copies of the complex in the unit cell.
Data collection and structure determination
Crystals were serially transferred into mother liquor supplemented with ethylene glycol from 10% to 30%, loop mounted, and flash-frozen in liquid nitrogen. Diffraction data were collected at the selenium anomalous edge. Data for both native and selenomethionine-substituted crystals were collected at the NE-CAT beamline 24ID-E at Advanced Photon Source (APS). The data were processed using HKL200036 (Supplementary Table 2). For phasing, we used the selenomethionine-substituted crystals in the SAD method37. Phasing was carried out using PHENIX38. A representative electron density map is shown in Supplementary Fig. 3b (top). We built the model of the complex using COOT39 and refined it against native data in PHENIX38 using positional, translation/libration/screw motion, and individual B-factor refinement options and secondary structure restraints (Supplementary Table 2 and Supplementary Fig. 3b (bottom). The final structure has good geometry (98.3 and 1.7% of residues are in allowed and generously allowed regions of the Ramachandran plot, respectively). The N-terminus of one of the two TTC7B molecules in the asymmetric unit is not ordered and was not modeled, and several loops were omitted in both copies of TTC7B (A: residues 33-36, 157-166, 197-212, 292-299, 343-358, 462-464, 619-686; B: 9-139, 154-170, 194-213, 286-318, 342-362, 460-464, 618-688) and FAM126A (A: residues 2-7, 18-23, 30-33, 93-94, 149-153, 288-308; B: 2-6, 16-24, 30-35, 49-52, 93-98, 114-120, 140-157, 288-308).
Protein expression (mammalian) for pull-down and kinase assay
Expi293 cells (Thermo Fisher) were maintained and transfected with ExpiFectamine (Thermo Fisher), according to the manufacturer’s instructions, with the following combinations of plasmids: (1) 3xFLAG-PI4KIIIα (wild-type); (2) 3xFLAG-PI4KIIIα (D1957A kinase-dead mutant); (3) 3xFLAG-PI4KIIIα (WT) and TTC7B-HA; or (4) 3xFLAG-PI4KIIIα (WT), TTC7B-HA, and GFP-FAM126A(1-289). Cells were harvested, resuspended in lysis buffer (250 mM NaCl, 20 mM Tris pH 8.0, 10% glycerol, 1 mM TCEP, supplemented with protease inhibitors (cOmplete, EDTA-free (Roche))), sonicated briefly, and centrifuged for 10 min at 16,000 × g. The supernatant was immunoprecipitated in lysis buffer by addition of M2 anti-FLAG agarose (Sigma) and rocking for 2 h at 4 °C. The resin was then isolated by centrifugation at 1000 × g, rinsed three times with lysis buffer, and protein complexes were eluted by incubation with 3xFLAG peptide (125 μg/mL in lysis buffer).
PI4KIIIα lipid kinase assay
The relative activity of the different PI4KIIIα preparations was assayed as follows. Liposomes containing C16-PI (Echelon Biosciences) were generated by sonication of a 1 mg/mL solution in kinase buffer (100 mM Tris-HCl pH 7.5, 50 mM EGTA, 100 mM MgCl2)) and incubated (at 80 μM) in a 50 μL reaction with 400 ng of PI4KIIIα or PI4KIIIα complex (equal amounts of PI4KIIIα in each sample was ensured by the use of both the BCA assay and SDS-PAGE/Coomassie staining), γ32P-labeled ATP (10 μCi), and cold ATP (50 μM) in kinase buffer for 5 min at 37 °C. The reaction was quenched by the addition of 700 μL of 2:1 chloroform:methanol containing 10 μg/mL Folch fraction (brain phosphoinositides) and 400 μL of 0.1 M HCl. The organic extracts were dried, resuspended in a small amount of 1:1 chloroform:methanol, and equal amounts were analyzed by thin-layer chromatography (mobile phase is 14:32:24:30:64 water:acetic acid:methanol:acetone:chloroform). PI4P was identified by comparison with PI3P generated in a parallel reaction using PI3K p110γ (Sigma) and quantified by autoradiography using a STORM 860 system (Molecular Dynamics).
Phosphoinositide analysis
PI4P and PI(4,5)P2 levels were quantified by 3H-inositol metabolic labeling for 72 h. Lipid extraction, deacylation, and HPLC separation by anion-exchange with detection using a radiometric detector were performed as previously described5.
Mouse husbandry and neuronal and oligodendrocyte cultures
To generate the FAM126A knockout mice, VelociGene technology was used to replace the exons 2 and 3 of FAM126A (Ensembl Accession Number: ENSMUSG00000028995; GenBank Accession number: NM_053090) and the intervening intron, corresponding to an 8.4-kb genomic region (chr5: 23,991,590–23,999,988; UCSC-genome Browser GRCm38/mm10, Dec11 Assembly) with a β-galactosidase(LacZ)/neomycin (NeoR) cassette as previously described27. The F2 offspring created in the mixed 129/C57BL/6 genetic background were backcrossed for ten generations into the C57BL/6 pure strain. Animal care and use was carried out in accordance with institutional guidelines (Animal Care and Use Committee of Istituto de Fisiologia Clinica, CNR, Pisa, Italy). Primary cortical neuronal and oligodendroglial cultures were generated as previously described40. Samples used for immunoblot analysis (Fig. 5) were harvested at DIV16 (neurons) and DIV5 (oligodendrocytes). No statistical method was used to predetermine samples size, and the experiments were not randomized.
Lentivirus production and rescue experiment
Lentiviral particles were purified as described herein. HEK 293T cells were transfected with either pLL3.7-FAM126A1-419-GFP or pLL3.7 empty vector in combination with the packaging plasmids pMD2.G and psPAX2 using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s instructions. Seventy-two hours after transfection, the viral supernatant was harvested, filtered through a 0.45 μm filter, and centrifuged in UV-sterilized tubes at 37693 × g for 90 min at 4 °C using an SW28.1 rotor (Beckman). The liquid was decanted, and 250 μL of OptiMEM (Thermo Fisher) was added to the tube and incubated at 4 °C overnight. The following day, the virus was gently resuspended, virus titer was measured by ELISA (p24 HIV ELISA kit, Cell BioLabs), and aliquots were generated and stored at −80 °C. HCC patient fibroblasts were infected with 0.5×1012 viral particles/mL in antibiotic-free media in the presence of polybrene (8 μg/mL, Sigma-Aldrich). After 24 h, the supernatant was replaced with fresh media. After an additional 6 d, the cells were rinsed, trypsinized, and lysates were generated and analyzed by immunoblot.
Quantitative RT-PCR analysis
Total RNA was isolated from control primary human skin fibroblasts using the RNeasy kit (Thermo Fisher) according to the manufacturer’s instructions and stored in nuclease-free water at −20 °C. Total RNA (2 μg) was reverse transcribed with an oligo(dT) primer using SuperScript polymerase (Thermo Fisher). cDNA was analyzed in triplicate by qRT-PCR amplification using SYBR Green Supermix on a Bio-Rad CFX96 Real-Time PCR Detection System. PCR amplification conditions were as follows: 95 °C (2 min) and 4 cycles of 95 °C (5 s) and 60 °C (30 s). Primer pairs (FAM126A: CACGAGTCGAGGTCCTGC and TCCTCCACAACCCCTTTCTC; FAM126B: TCCCCTCCTTATCCAAGCCT and ATGCTGACACAATGCCCCTT) were designed to amplify mRNA-specific fragments, and unique products were tested by melt-curve analysis. PCR efficiencies were 101.6% (slope of −3.285 in a ten-fold dilution series) for FAM126A and 96.7% (slope of −3.404 in a ten-fold dilution series) for FAM126B using the indicated primers. Data were analyzed using ΔΔCt, and values were normalized to the housekeeping gene ribosomal protein S26 (using CCGTGCCTCCAAGATGACAA and GCAATGACGAATTTCTTAATGGCCT as primers).
Immunoisolation of oligodendrocyte and neuronal lineage cells
For acute cell isolation, cells were isolated using the MACS cell selection kit (Miltenyi Biotec). Briefly, brains from male FAM126A KO and littermate WT control mice (on the pure C57BL/6 background) at postnatal day 8 were isolated, minced, and mechanically dissociated with a gentleMACS dissociator according to the manufacturer’s instructions. The cells were then passed through a 45-μm strainer and incubated with the cell-specific beads for 15 minutes. The cells were then loaded on LS columns and separated on a quadroMACS magnet, first with anti-O4 beads (elution was labelled oligodendrocyte lineage), then with anti-ACSA beads to deplete astrocytes, and the remainder (flow-through) was a sample enriched in cells of the neuronal lineage. The target cells were washed once in PBS, then pelleted and snap-frozen. Pellets were subsequently thawed, and lysates were generated for immunoblot analysis.
Light and electron microscopic imaging of myelin in FAM126A KO mice
Male mice at age P15 (three FAM126A KO and two littermate WT controls, on a mixed 129/C57BL/6 background) were perfused transcardially with 2% paraformaldehyde, 2.5% glutaraldehyde in 100 mM cacodylate buffer pH 7.4. Samples were postfixed with 1% osmium tetroxide, 1.5% potassium ferrocyanide in 100 mM cacodylate buffer pH 7.4, en bloc stained with uranyl acetate, dehydrated in increasing concentrations of ethanol and propylene oxide, and finally embedded in Epon. Samples were cured at 60 °C in an oven for 48 h. Epon blocks were sectioned using a Leica EM UC7 ultramicrotome (Leica Microsystems, Vienna). For light microscopy, semithin sections (1 μm) of optic nerve, corpus callosum (sagittal section of the central area), and spinal cord (ventral funiculus cervical region) were generated and stained with toluidine blue. Light microscopy was performed on a Zeiss Axio Imager equipped with a Plan-Apochromat 100x/1.4 oil objective with an AxioCam MRc 5 color CCD camera. For electron microscopy, ultrathin sections (60 nm) were contrasted with 2% uranyl acetate and Sato’s lead solutions and observed with a LEO 912AB Zeiss Transmission Electron Microscope (Carl Zeiss, Oberkochen, Germany). Digital micrographs were taken with a 2kx2k bottom mounted slow-scan Proscan camera (ProScan, Lagerlechfeld, Germany) controlled by the EsivisionPro 3.2 software (Soft Imaging System, Münster, Germany).
Statistics and reproducibility
Statistical analysis was performed using either Microsoft Excel or Prism 6 software. Detailed statistical information (which statistical test was used, number of independent experiments, p values, definition of error bars) is listed in individual figure legends. All immunoblots were repeated at least three times except for some immunoblots shown in Fig. 5, which were repeated twice (see legend for Supplementary Fig. 4a for details). All imaging experiments were repeated three times. No statistical method was used to predetermine samples size for animal experiments, and the experiments were not randomized.
Supplementary Material
Acknowledgments
We thank F. Nakatsu for helpful discussions and J. Chung, H. Czapla, L. Lucast, and F. Wilson for technical assistance. HCC patient and control fibroblasts were kindly provided by the Cell Line and DNA Biobank from Patients Affected by Genetic Diseases (Giannina Gaslini Institute), part of the Telethon Network of Genetic Biobanks (project no. GTB12001). Primate brain samples were generously provided by N. Sestan and A. Sousa (Yale University). This work was supported by the National Institutes of Health (R37NS036251, R01DK082700, and P30DA018343 to P.D.C., R01GM080616 to K.M.R., R01GM095982 to T.C.W., and K99GM110121 to J.M.B.), the Simons Foundation (to P.D.C.), Telethon (GP2007250 to C.M.), the Mariani Foundation (C.M.), the San Paolo Foundation (to C.M.), and the European Leukodystrophy Association Foundation (to C.M.). J.M.B. was recipient of a Jane Coffin Childs Memorial Fund fellowship, and C.M.S. was a recipient of a National Science Foundation Graduate Research Fellowship (DGE-1122492).
Footnotes
Author Contributions
J.M.B., X.W., R.C., T.C.W., K.M.R., and P.D.C. designed the experiments. J.M.B., X.W., R.C., M.S.O., C.M.S., M.M., and A.R. performed the experiments. E.G., S.A., S.B., R.B., F.Z., C.M., and M.S. generated and contributed cells, other tools, and reagents. J.M.B., K.M.R., and P.D.C. wrote the manuscript, with input from all authors.
The authors declare no competing financial interests.
Accession numbers
Structure factors and coordinates for the TTC7/FAM126A-N complex have been deposited in the Protein Data Bank (accession number 5DSE).
References
- 1.Aggarwal S, Yurlova L, Simons M. Central nervous system myelin: structure, synthesis and assembly. Trends in Cell Biology. 2011;21:585–593. doi: 10.1016/j.tcb.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Perlman SJ, Mar S. Leukodystrophies. Adv Exp Med Biol. 2012;724:154–171. doi: 10.1007/978-1-4614-0653-2_13. [DOI] [PubMed] [Google Scholar]
- 3.Zara F, et al. Deficiency of hyccin, a newly identified membrane protein, causes hypomyelination and congenital cataract. Nat Genet. 2006;38:1111–1113. doi: 10.1038/ng1870. [DOI] [PubMed] [Google Scholar]
- 4.Tan J, Brill JA. Cinderella story: PI4P goes from precursor to key signaling molecule. Crit Rev Biochem Mol Biol. 2014;49:33–58. doi: 10.3109/10409238.2013.853024. [DOI] [PubMed] [Google Scholar]
- 5.Nakatsu F, et al. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J Cell Biol. 2012;199:1003–1016. doi: 10.1083/jcb.201206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond GRV, et al. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baird D, Stefan C, Audhya A, Weys S, Emr SD. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nawaz S, et al. Phosphatidylinositol 4,5-bisphosphate-dependent interaction of myelin basic protein with the plasma membrane in oligodendroglial cells and its rapid perturbation by elevated calcium. J Neurosci. 2009;29:4794–4807. doi: 10.1523/JNEUROSCI.3955-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musse AA, Gao W, Homchaudhuri L, Boggs JM, Harauz G. Myelin basic protein as a ‘PI(4,5)P2-modulin’: a new biological function for a major central nervous system protein. Biochemistry. 2008;47:10372–10382. doi: 10.1021/bi801302b. [DOI] [PubMed] [Google Scholar]
- 10.Goebbels S, et al. Elevated phosphatidylinositol 3,4,5-trisphosphate in glia triggers cell-autonomous membrane wrapping and myelination. J Neurosci. 2010;30:8953–8964. doi: 10.1523/JNEUROSCI.0219-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snaidero N, et al. Myelin Membrane Wrapping of CNS Axons by PI(3,4,5)P3-Dependent Polarized Growth at the Inner Tongue. Cell. 2014;156:277–290. doi: 10.1016/j.cell.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin RJM, ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- 13.Crawford AH, Chambers C, Franklin RJM. Remyelination: the true regeneration of the central nervous system. J Comp Pathol. 2013;149:242–254. doi: 10.1016/j.jcpa.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Fancy SPJ, Chan JR, Baranzini SE, Franklin RJM, Rowitch DH. Myelin Regeneration: A Recapitulation of Development? Annu Rev Neurosci. 2011;34:21–43. doi: 10.1146/annurev-neuro-061010-113629. [DOI] [PubMed] [Google Scholar]
- 15.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 16.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiological Reviews. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 18.Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- 19.Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, et al. Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex. Dev Cell. 2014;28:19–29. doi: 10.1016/j.devcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biancheri R, et al. Phenotypic characterization of hypomyelination and congenital cataract. Ann Neurol. 2007;62:121–127. doi: 10.1002/ana.21175. [DOI] [PubMed] [Google Scholar]
- 22.Biancheri R, et al. Hypomyelination and Congenital Cataract Broadening the Clinical Phenotype. Arch Neurol-Chicago. 2011;68:1191–1194. doi: 10.1001/archneurol.2011.201. [DOI] [PubMed] [Google Scholar]
- 23.Traverso M, et al. Novel FAM126A mutations in hypomyelination and congenital cataract disease. Biochem Biophys Res Commun. 2013;439:369–372. doi: 10.1016/j.bbrc.2013.08.077. [DOI] [PubMed] [Google Scholar]
- 24.Trotter PJ, Wu WI, Pedretti J, Yates R, Voelker DR. A Genetic Screen for Aminophospholipid Transport Mutants Identifies the Phosphatidylinositol 4-Kinase, Stt4p, as an Essential Component in Phosphatidylserine Metabolism. J Biol Chem. 1998;273:13189–13196. doi: 10.1074/jbc.273.21.13189. [DOI] [PubMed] [Google Scholar]
- 25.Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan J, Oh K, Burgess J, Hipfner DR, Brill JA. PI4KIIIα is required for cortical integrity and cell polarity during Drosophila oogenesis. J Cell Sci. 2014;127:954–966. doi: 10.1242/jcs.129031. [DOI] [PubMed] [Google Scholar]
- 27.Gazzerro E, et al. Hyccin, the molecule mutated in the leukodystrophy hypomyelination and congenital cataract (HCC), is a neuronal protein. PLoS ONE. 2012;7:e32180. doi: 10.1371/journal.pone.0032180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomasch J. Size, distribution, and number of fibres in the human corpus callosum. Anat Rec. 1954;119:119–135. doi: 10.1002/ar.1091190109. [DOI] [PubMed] [Google Scholar]
- 29.von Keyserlingk DG, Schramm U. Diameter of axons and thickness of myelin sheaths of the pyramidal tract fibres in the adult human medullary pyramid. Anat Anz. 1984;157:97–111. [PubMed] [Google Scholar]
- 30.Kawasoe T, et al. Isolation and characterization of a novel human gene, DRCTNNB1A, the expression of which is down-regulated by beta-catenin. Cancer Res. 2000;60:3354–3358. [PubMed] [Google Scholar]
- 31.Fancy SPJ, et al. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23:1571–1585. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 33.Hubner NC, Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC) Methods. 2011;53:453–459. doi: 10.1016/j.ymeth.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Bojjireddy N, et al. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J Biol Chem. 2014;289:6120–6132. doi: 10.1074/jbc.M113.531426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doublié S. Preparation of selenomethionyl proteins for phase determination. Meth Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Hendrickson WA, Ogata CM. In: Methods in Enzymology: Macromolecular Crystallography. Carter CW, Sweet RM, editors. Vol. 276. Academic Press; 1997. pp. 494–523. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, Grosse-Kunstleve RW, Hung LW. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002 doi: 10.1107/s0907444902016657. [DOI] [PubMed]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Fitzner D, et al. Myelin basic protein-dependent plasma membrane reorganization in the formation of myelin. EMBO J. 2006;25:5037–5048. doi: 10.1038/sj.emboj.7601376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.