Abstract
Viruses have co-evolved with their hosts, acquiring strategies to subvert host cellular pathways for effective viral replication and spread. Human cytomegalovirus (HCMV), a widely-spread β-herpesvirus, is a major cause of birth defects and opportunistic infections in HIV-1/AIDS patients. HCMV displays an intricate system-wide modulation of the human cell proteome. An impressive array of virus–host protein interactions occurs throughout the infection. To investigate the virus life cycle, proteomics has recently become a significant component of virology studies. Here, we review the mass spectrometry-based proteomics approaches used in HCMV studies, as well as their contribution to understanding the HCMV life cycle and the virus-induced changes to host cells. The importance of the biological insights gained from these studies clearly demonstrate the impact that proteomics has had and can continue to have on understanding HCMV biology and identifying new therapeutic targets.
Keywords: virus–host interactions, protein–protein interactions, CMV, proteomics, mass spectrometry, post-translational modifications
Introduction
Human cytomegalovirus (HCMV) is a prominent herpesvirus that infects 40% to nearly 100% of the adult population worldwide1. Similar to other herpesviruses, HCMV establishes a persistent infection, remaining silent in the host and undergoing productive reactivation cycles that contribute to its efficient transmission. HCMV infects and replicates in a wide variety of cells, including epithelial cells of gland and mucosal tissue, smooth muscle cells, fibroblasts, macrophages, dendritic cells, hepatocytes and vascular endothelial cells2. This broad cell tropism facilitates systemic spread in the human body and inter-host spread. In addition, HCMV undergoes latency in myeloid cells of the bone marrow, presumably leading to a life-long infection with sporadic reactivation3. HCMV infection is generally asymptomatic in healthy individuals. However, in immunocompromised individuals, such as organ transplant recipients or human immunodeficiency virus carriers, HCMV poses a life-threatening risk. HCMV is also recognized as the leading infectious cause of congenital neurological disease by transmission through the placenta from the mother to the child4,5. The relevance of this virus and its associated human diseases is highlighted by the decisions of the Institute of Medicine and the National Vaccine Advisory Board to assign HCMV as high priority for vaccine development6.
The social burden caused by HCMV has been a motivation to understand its intricate life cycle (Figure 1). The HCMV genome was estimated to contain ~192 open reading frames (ORFs) with capacity to encode functional proteins, representing the largest genome of the characterized herpesviruses to date7. The HCMV coding potential was recently further expanded, revealing an even higher level of genome complexity8. The virus-coded proteins, either contained in infectious virion particles or expressed in the cell at different stages of infection, interact closely with the cellular machinery. While the majority of these temporal and spatial interactions remain to be defined, these dynamic virus-host interactions form the basis for the exquisite modulation of cellular functions and are required for successful viral replication and spread. During entry, viral envelope glycoproteins, positioned on the outside of infectious virions, interact with host receptors to mediate fusion or endocytosis of the virion into the cell (Figure 1, [A])9. Viral tegument proteins bound to the capsid are believed to interact with the host microtubule machinery to transport viral capsids to the nuclear envelope and into nucleus (Figure 1, [B]), where viral transcription, genome replication and encapsidation occurs10-12. At the same time, other tegument proteins are deposited in infected cells by incoming virions and targeted to different subcellular locations to inhibit the initial steps of immune response and to regulate viral gene expression13-18. Furthermore, many viral-encoded proteins regulate cell-signaling pathways19 and cellular metabolism20 to support viral replication and immune evasion. The expression of these viral proteins occurs as a finely-regulated cascade of events and is divided into several main temporal classes, each regulating different aspects of the infectious cycle (Figure 1; immediate early, IE; delayed early, DE; late, L)21,22. Capsids, assembled in the nucleus, egress through the nuclear double membrane by disruption of the nuclear lamina and formation of a nuclear egress complex (Figure 1, [C])23-26. Once capsids reach the cytoplasm, the assembly and transport of virions occurs via the integration of multiple cellular trafficking pathways27. The cellular secretory machinery, including the endoplasmic reticulum (ER), Golgi apparatus, and endosomal machinery, is hijacked for the formation of a cytoplasmic viral assembly complex (AC; Figure 1)28-30. At the AC, capsids acquire their tegument layer and viral envelope from intracellular vesicles. The generated infectious particles, as well as other non-infectious particles named dense bodies, are next released into the extracellular space (Figure 1, [D]).
Figure 1. Overview of the HCMV life cycle.
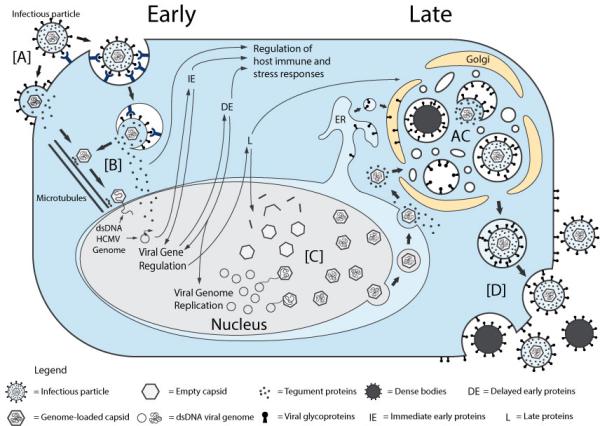
(A) Infectious particles enter the cell through interaction with cellular receptors. Capsid and tegument proteins are delivered to the cytosol. (B) The capsid travels to the nucleus, where the genome is delivered and circularized. Tegument proteins regulate host cell responses and initiate the temporal cascade of the expression of viral I immediate early (IE) genes, followed by delayed early (DE) genes, which initiate viral genome replication, and late (L) genes. (C) Late gene expression initiates capsid assembly in the nucleus, followed by nuclear egress to the cytosol. Capsids associate with tegument proteins in the cytosol and are trafficked to the viral assembly complex (AC) that contains components of the endoplasmic reticulum (ER), Golgi apparatus and endosomal machinery. The capsids further acquire tegument and viral envelope by budding into intracellular vesicles at the AC. (D) Enveloped infectious particles are released along with non-infectious dense bodies.
In recent years, proteomic studies have made a significant contribution to understanding HCMV biology. The integration of proteomic approaches with molecular virology, microscopy, and biochemistry techniques has provided the opportunity to characterize different stages of the HCMV life cycle at a previously unattainable depth. The continuously expanding breadth and sensitivity of mass spectrometry (MS)-based proteomic workflows have been essential for the elucidation of virus-host protein interactions, for defining global changes in cellular protein expression during infection, and for understanding cellular pathways either activated for host defense against infection or modulated for effective viral replication (Figure 2). This review provides an overview of modern MS-based proteomic methodologies used in HCMV research, the biological insight gained from these applications, and the implications these studies have for future therapeutic intervention. We finish with a perspective of current challenges and promising proteomic technologies that can be used to further advance our understanding of HCMV biology and pathogenesis.
Figure 2. Proteomic approaches used for studying HCMV infection.
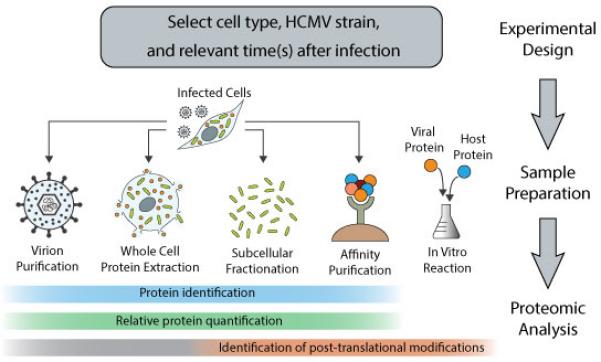
The first step involves the selection of a relevant cell type, the virus strain, and the time(s) of infection for the study, as all these factors will influence the proteome content and the virus-host interactions. These decisions are commonly based on a priori knowledge and/or experimental evidence. To date, mass spectrometry-based proteomics methods have been used for protein identification and quantification in studies of virion composition, whole cell or subcellular proteomes, and virus-host protein interactions during HCMV infection. While posttranslational modifications have been analyzed in subcellular proteomes and protein affinity purification studies, their characterization has the potential to be expanded to virion and whole cell studies.
Looking from the Outside on an Infected Cell: Virion and Cell Surface Proteomes
The infectious HCMV virion contains the necessary material for binding and entry into host cells and for initiation of the virus life cycle. Given the relevance of virus entry into hosts for therapeutic intervention and for understanding the initial steps of infection, determining the content of infectious particles has been of central interest to virologists. The virion glycoproteins must interact with cell surface proteins for the internalization of the infectious virion and, therefore, are considered promising targets for antiviral therapeutics31. Additionally, the host proteins present at the cell surface play critical roles in HCMV entry and replication, as well as in host signaling and immune surveillance9. Modern proteomic technology has provided an effective mean to characterize the virion composition and the regulation of the cell surface proteome during infection.
Methods for Analyzing Infectious Particles and the Cell Surface Proteome
A common workflow for analyzing the protein content of infectious particles involves the collection of viral particles from the culture medium of infected cells, followed by purification by density gradient ultracentrifugation and identification of proteins by mass spectrometry (Figure 2)32-39. For MS analysis, the proteins are usually enzymatically digested and the resulting peptides analyzed by liquid chromatography (LC) and tandem mass spectrometry (MS/MS). The first LC-MS/MS analysis of the HCMV virion was reported ~10 years ago33, at the same time with a similar study on the murine cytomegalovirus (MCMV) virion34. Recently, the proteome of the rhesus cytomegalovirus (RhCMV) virion was also analyzed39. MCMV and RhCMV are commonly used model viruses for in vivo studies of HCMV infection, as HCMV in vivo studies are limited and challenging. These three studies have certainly provided important insights into CMV virion composition (see next section). While at a first glance this MS-based workflow for analyzing virions may seem trivial, it has been and still is challenging. A main challenge is in obtaining a pure and homogeneous population of viral particles. This issue is usually revealed by the assessment of virion preparations by electron microscopy. For example, in the study of HCMV virions, some contamination with dense bodies was observed, and cellular debris was also visible in the RhCMV study33,39. The presence of material other than viral particles limits the accurate determination of virion composition and the interpretation of the findings. For instance, the identification of selected host proteins within infectious particles is of great interest. However, it remains to be determined whether these cellular proteins are present within the mature virion or, instead, associated with the external portion of the virion. The heterogeneity of virion preparations also impacts the ability to accurately quantify and determine the stoichiometry of viral proteins contained within the virion. Another problem is that virion preparations tend to have a high particle to plaque forming unit ratio40 (i.e., defective or damaged viral particles may be present in the preparation), and therefore, distinguishing the protein content of infectious and non-infectious virions remains a major challenge. Recent advances in absolute and relative protein quantification using MS approaches, in conjunction with improvements in virion purification methods that have been applied to other viral systems41,42, have the promise to significantly expand the current understanding of the stoichiometry of proteins within an infectious particle, as well as the direct virus-virus protein interactions.
The investigation of dynamic changes in the cell surface proteome has also benefited from MS-based approaches43,44. In the context of HCMV infection, biotinylated amine-reactive45 and biotinylated sialic acid-reactive groups46,47 have been used to tag cell-surface proteins, allowing for their purification on avidin or streptavidin resins and analysis by LC-MS/MS. These studies quantified a wide range (~500-1,100) of human cell-surface proteins at different stages of viral infection45-47. The differential expression of these proteins was measured using three different quantification approaches: label-free quantification, stable isotope labeling by amino acids in cell culture (SILAC), and isobaric chemical tags (tandem mass tags, TMT) (Figure 3). Label-free quantification approaches provide the ease of comparing a large number of samples, while the protein- (SILAC) and peptide- (TMT) labeling approaches offer increased accuracy of quantification, usually for up to ten samples. Overall, these proteomic-based studies have provided important insights into the virus-induced temporal modulation of cellular surface proteins and activated host defense pathways, as described below. One limitation of these approaches is given by the level of purity of the plasma membrane preparations, as contaminants from other subcellular compartments have been seen in these studies. Future improvements in affinity workflows can help to both improve the specific enrichment of cell surface proteins and to distinguish true plasma membrane constituents from contaminants.
Figure 3. Quantitative proteomic methods used for studying HCMV-infected cells.
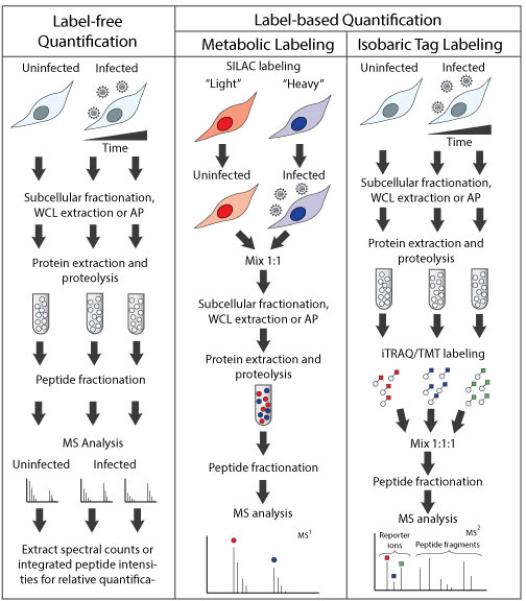
Both label-free and labeling methods have been used in protein quantification during HCMV infection. Label-free quantification can be readily integrated in studies of subcellular fractions, whole cell lysates (WCL), or affinity purified (AP) proteins. The usual workflow involves the digestion of proteins and their analysis by LC-MS/MS using data-dependent acquisition mode. Quantitative values for each protein are obtained by extracting the protein spectral counts or the integrated peptide intensities. Labelling quantification approaches that are commonly used are metabolic labeling and isobaric tag labeling. Stable isotope labeling by amino acids in cell culture (SILAC) incorporates “light” or “heavy” amino acids, allowing the comparison of uninfected and infected cells. Quantification is done at the MS level by comparing the ion intensities of the heavy and light peptides. For labeling with isobaric tag, the samples that are to be compared are processed in parallel, similar to label-free quantification up to the proteolysis steps. The resulting peptides are then labeled using isobaric tags and then mixed. The quantification is done at the MS/MS level, as the spectra contain both peptide fragments for amino acid sequence information and a set of reporter ions from the isobaric tags that illustrate the relative abundances of the peptides originating from the different samples. These compared samples can include cells collected at different times after infection, cells infected with various HCMV strains, or multiple biological replicates.
The HCMV Virion Proteome
In the original MS-based analysis of the HCMV virion proteome, 59 viral proteins and 70 host proteins were identified33. Viral capsid components and glycoproteins necessary for viral entry were confirmed to be virion-associated in this study. The tegument viral protein pUL83 was detected as the most abundant component of virions, accounting for ~15% of the total protein amount. Another abundant tegument protein was ppUL82, estimated to make up ~9% of the virion proteome. These results agree with the critical requirement of these proteins for immune modulation18,48, early viral gene activation17,49, and virion scaffolding50. This MS study also identified several viral proteins that had unknown functions at that time33. Follow-up studies revealed their roles in virion replication (UL7151,52, UL7953, UL9654) and virion extracellular release (UL10355). Host proteins were also detected as possible components of the virion33. These include intracellular transport factors, such as clathrin, later suggested to be used by virion particles for trafficking27. However, it is not clear whether these host proteins are passively integrated into virions, have active roles in virion assembly or initiation of infection, or originate from contamination during the virion preparation with secreted material (e.g., exosomes). One additional aspect to be kept in mind is that the composition of HCMV virions has only been assessed in infected fibroblasts. As HCMV has an exceptionally broad cell tropism, whether the virions assembled in other relevant cell types (e.g., epithelial cells and macrophages) are distinct in composition and infection capacity remains to be elucidated.
To date, the CMV virion proteome studies have identified a similar number of viral proteins, with 59 for HCMV33, 58 for MCMV34, and 70 for RhCMV39. In the MCMV virion proteome analysis, 43 identified proteins were homologous to HCMV proteins, with 32 of these (76%) corresponding to those integrated in the HCMV virion. The RhCMV virion proteome analysis identified 61 proteins homologous to HCMV, with 45 of these (64%) being homologous to HCMV proteins and integrated in the HCMV virion. Therefore, these proteomic analyses revealed conservation between different cytomegalovirus virions, underscoring the utility of the investigation of related viruses and pointing to key viral components of an infectious CMV particle.
HCMV Regulation of the Cell Surface Proteome
The cell surface proteome has been analyzed in the context of both productive and latent HCMV infections. The first analysis of the temporal regulation of cell surface proteins during productive HCMV infection was performed in infected human fibroblasts at 6, 24, and 72 hours post infection (hpi), representing immediate-early, early and late phases of infection, respectively45. HCMV infection was shown to significantly alter the cell surface proteome, with ~24% of the host proteins exhibiting changes in abundance at 72hpi. Regulated proteins included those involved in apoptosis, cell adhesion, immune response, metabolism, transport, and signaling. This study confirmed the upregulation of GLUT4, a glucose transporter important for increased glucose intake in HCMV56, and established the LDL receptor-related protein 1 (LRP1) as a novel modulator of lipid metabolism during HCMV infection45. The second study of the cell surface proteome during productive HCMV infection was performed at seven time points of infection in human fibroblasts47. Many of the previously reported changes, including those of GLUT4, LRP1, and membrane proteins involved in oxidative phosphorylation were confirmed. Furthermore, this study discovered the down regulation of proteins involved in cancer, such as gap junction factors and the majority (11 out of 13) of the wnt receptors.
Recent proteomic-based studies have also investigated latently infected cells. HCMV undergoes latency in cells of the myeloid lineage, causing a lifelong infection in humans57. The impact of UL138, one of the few viral genes expressed during latency, on plasma membrane proteins was investigated in myeloid cells46. This approach identified the multidrug resistance-associated protein (MRP1) as a target of UL138 regulation and a possible marker for latently-infected cells. Cell surface proteins are attractive therapeutic targets due to their accessibility and capacity to regulate several cellular pathways. Therefore, such studies are expected to continue to provide valuable resources for future identification of antiviral targets.
Spatial and Temporal Protein Interactions during HCMV Infection
During the co-evolution with its host, HCMV has acquired a diverse range of mechanisms for subverting cellular pathways either to block host defense or to aid viral replication. At the core of these mechanisms are dynamic, temporally and spatially regulated, virus-host interactions. These interactions encompass protein-protein associations, as well as interactions between proteins and nucleic acids. There is certainly still a long way to go before fully understanding the impressive multitude of virus-host protein interactions at different stages of infection. Nevertheless, the studies reported to date have clearly demonstrated the power of MS-based approaches in defining critical interactions and gaining mechanistic insights. In this section, we present an overview of the proteomic approaches used and the important insights gained with regard to the progression of the virus life cycle and the modulation of immune defense and cell cycle.
Methods for analysis of virus-host and virus-virus protein interactions
The study of protein-protein interactions in the context of HCMV infection has witnessed a substantial growth in recent years, with improvements in both the efficiency of protein complex isolation and the ability to identify co-isolated proteins. These studies have captured virus-host interactions either from the virus perspective, by isolating viral proteins at different time points of infection, or from the host perspective, by focusing on a critical cellular protein and assessing its interactions with viral and other host proteins. Most studies have used the following general workflow: (i) the isolation of a protein of interest during viral infection, (ii) an optional fractionation of co-isolated proteins at either the protein or peptide level, (iii) the identification of protein interacting partners, and (iv) the validation of observed interactions, followed by a variety of molecular biology and biochemistry approaches to define their biological functions.
The isolation and identification of protein complexes is most commonly performed by affinity purification followed by mass spectrometry (AP-MS)58. The viral or host protein of interest is isolated using antibodies directly against the protein59,60 or against a tag, when the protein is engineered to have a single-step or dual-step affinity tag (e.g., FLAG26,60-62, HA26, green fluorescent protein (GFP) tags17,27,63,64, or tandem affinity purification (TAP)65-67). An advantage of using a tag is that researchers can use in-house or comercial high-affinity antibodies that have been already validated for efficient immunoaffinity purifications. However, carefull assessment is required to ensure that the function of the protein is not disrupted by the tag. Small tags, such as FLAG and HA, usually help retain the folding of the protein, sometimes at the expense of the efficiency of isolation. Larger tags, such as GFP and Protein A (within a TAP tag) provide means for effective protein isolation, but have to be placed at locations that allow folding and function of the protein of interest. The use of fluorescent tags in AP-MS studies68 has become increasingly popular in recent years, as they allow the assessment of protein interactions in conjunction with the imaging of protein localization during the progression of an infection in live cells17,27,63,64. An important consideration when using tagged proteins is also the level of overexpression, which can impact protein function and interactions, as well as a range of cellular pathways and overall cell health. Several approaches, including inducible expression systems, have been used to limit overexpression artefacts, as reviewed in69.
Several studies used the ectopic expression of a single viral protein of interest in order to study its interaction. While such studies provide useful information regarding interactions that are independent of the expression of other viral proteins, one important disadvantage is the inability of this system to mimic a natural infection and the loss of temporally regulated interactions. An important advance was the generation of a bacterial artificial chromosome (BACs) containing the entire HCMV genome70,71. This has provided an effective strategy to study protein interactions in a context mirroring a natural infection by introducing the tag at the original viral gene location in the HCMV BAC17,27,61,64,66,67. In this way, the virus can be reconstituted and the gene will be expressed at wild-type levels of expression during the HCMV infection under the control of the original promoter. Another noteworthy addition to the AP-MS workflows has been the use of magnetic beads for immunoaffinity purification 69, providing surface binding for isolating protein complexes of various sizes and minimizing non-specific interactions 17,63,64,66. Non-specific associations frequently occur in AP-MS studies, deriving from binding to the resin, the antibody, the tag, and even to the isolated protein complex of interest. In addition to the choice of resin, different steps of the isolation protocol should be carefully optimized to reduce non-specific associations, including the lysis buffer composition and the selection of appropriate control isolations, as reviewed in58. As protein expression levels can change significantly during the progression of HCMV infection (see sections on Whole Cells and Subcellular Proteomes), it is important to perform the control isolations at the same time point of infection. Quantitative MS approches, using either label-free or metabolic labeling, have been developed to use these parallel control isolations for assessment of specificity58, and promise to be of benefit to HCMV studies. Furthermore, given the demonstration that increased incubation time of the cell lysate with a resin (e.g., beads) results in an accumulation of non-specific associations and a loss of weak interactions68, the current trend of AP-MS workflows is to perform isolations as quickly as possibly (minutes to a maximum of one hour). Magnetic beads, especially those with small diameters, promise to become robust reagents for such rapid one-step isolation experiments that can capture protein complexes close to their original state in the infected cells58. Differential centrifugation has been used to determine the presence of proteins within given fractions and infer their possible association 72. However, controling for contaminants becomes challenging when using this approach.
Following the isolation of a protein with its interacting partners, some studies have used 1D- or 2D-SDS PAGE to fractionate the protein mixture prior to analysis by mass spectrometry. Individual gel bands or spots would then be manually selected and prepared for sequencing by MS59,60,64,73. However, in recent years researchers have opted to analyze the full spectrum of co-isolated proteins by either directly preparing them for MS analysis (without SDS-PAGE separation) or by analyzing the entire 1-D SDS PAGE lane. In this way, large protein interaction networks, also termed protein interactomes, could be generated26,61-63,66,67,74. This approach has benefited from significant recent improvements in the sensitivity and accuracy of the mass spectrometry instrumentation (predominated by the orbitrap technology offered by Thermo Fisher Scientific, Inc.) and in the label-free and labeling approaches for protein quantification (Figure 3). Analyzing the broader interaction network of a protein avoids biases towards just a few proteins of interest and aids the discovery of members of complexes that may be low in abundance and not well visualized on SDS-PAGE separations. Importantly, depending on the stringency of the isolation conditions and on the sensitivity of the MS instrumentation, most AP-MS studies tend to identify tens to hundreds of proteins. Therefore, the use of proper controls, quantification measurements, robust computational analyses, and follow-up validation (e.g., reciprocal AP, microscopy, and functional analyses) are absolutely critical69,75.
Protein Interactions during the HCMV Life Cycle
Although virus-host protein interactions occur throughout the progression of the HCMV life cycle, MS-based studies have so far only been performed on several viral or host proteins and selected stages of infection. While just a few, these studies have provided important insights into the immediate early, early, and late stages of infection (Figure 4). Following its entry into cells, the viral genome is delivered to the nucleus, where it associates with cellular histones 76 (Figure 1). These histones are heavily posttranslationally modified 77 with modifications regulated by various chromatin remodeling enzymes. Among these enzymes are histone deacetylases (HDACs), which have recently been shown to modulate the viral gene expression for several herpesviruses78, including HCMV79. HDAC1, a member of the nucleosome remodeling and deacetylase (NuRD) protein complex, was demonstrated to regulate the expression of IE genes controlled by the viral major immediate-early promoter (MIEP)80 (Figure 4A). Using AP-MS, HDAC1-containing NuRD complex was shown to be targeted by the viral proteins pUL29/28, pUL38, and the viral kinase pUL9763. This study further demonstrated that, via these associations with NuRD and MIEP, pUL29/28 regulates the expression of IE genes. The modification of HDAC1 by pUL97 was later shown to be important for IE gene expression (see posttranslational modification section below)81.
Figure 4. Examples of critical virus-host interactions during the HCMV replication cycle.
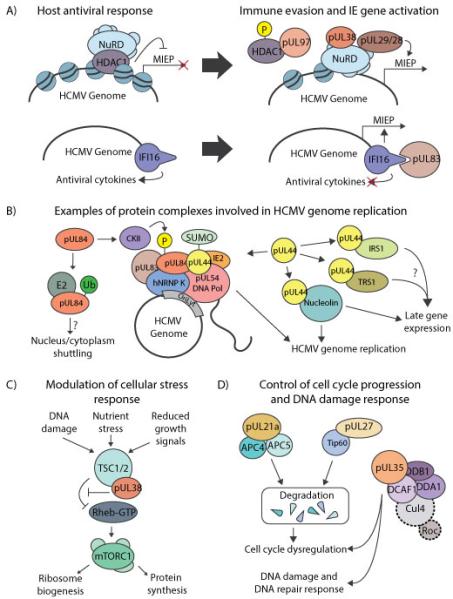
(A) Host and virus control of immediate early (IE) gene expression. pUL38 and pUL29/28 interact with the HDAC1-NuRD complex to help induce expression of IE genes from the MIEP. pUL97 phosphorylates HDAC1 and inhibits its deacetylase activity, promoting viral gene expression by potentially de-stabilizing HDAC1 association with the NuRD complex. The DNA sensor IFI16 binds the HCMV genome to induce antiviral cytokine production. pUL83 interacts with IFI16 to restrict its oligomerization and cytokine production. Additionally, IFI16 is repurposed to activate the MIEP. (B) Protein complexes involved in HCMV genome replication. pUL84 is part of the HCMV DNA replication machinery, interacting with oriLyt and host and viral proteins. To function within the replication complex, pUL84 has to be phosphorylated (P) by CKII. pUL84 also interacts with the ubiquitin-conjugating enzyme E2 and becomes mono ubiquitinated (Ub). The DNA processivity factor, pUL44, is thought to be a part of multiple protein complexes and is important for efficient DNA replication. (C) Viral modulation of cellular stress response through TSC1/2. pUL38 interacts with TSC1/2 to block its activity and maintain an active mTOR pathway following infection. (D) Control of cell cycle progression and induction of the DNA damage response. pUL21a interacts with APC members, leading to their degradation, reduced APC activity, and dysregulation of the cell cycle. pUL27 interacts with Tip60, targeting this protein for degradation and contributing to cell-cycle arrest. pUL35 interacts with subunits of the Cul4 ubiquitin ligase complex, causing cell cycle arrest and induction of DNA damage response.
The progression of the early stages of infection is also influenced by viral proteins that are directly delivered by infectious virions into the cells. Interaction studies of the most abundant viral tegument protein, pUL83 17, revealed that, upon its localization to the nucleus, pUL83 plays roles in both suppressing host immune response18 and regulating viral gene expression17. By directly binding and blocking the pyrin domain of the host interferon inducible protein 16 (IFI16), a nuclear sensor of foreign DNA, pUL83 inhibits the expression of antiviral cytokines that would be induced by IFI16 (Figure 4A)18. Moreover, pUL83 was shown to target IFI16 to the MIEP, helping to activate the MIEP and induce IE gene expression17. The MIEP controls the expression of two immediate early genes—IE1 and IE2. IE2 is a multifunctional viral protein essential for viral replication, modulating host responses to infection and activating the expression of early and late viral genes22. A study of the IE2 interactome at three different time points of infection (8, 24 and 48 hpi) revealed interactions with 9 viral proteins and 75 cellular proteins67. An interaction with C1QBP, a host protein with roles in transcription and splicing, was confirmed and implicated in the regulation of IE genes. This study also highlighted the temporal nature of viral protein interactions, with pUL84 being one of the earliest IE2 interaction partners, followed by the associations with other viral proteins at subsequent time points of infection. Investigation of interactions at different stages of infection can reveal changes in virus-host interactions across the virus life cycle and help to further define the dynamic nature of protein complexes.
The expression of DE genes is next controlled by IE genes and necessary for initiating viral genome replication (Figure 1). A complex is formed at the cis-acting origin of replication, oriLyt, and composed of IE2, the phosphoprotein pUL84, the DNA processivity factor pUL44, the DNA polymerase pUL54, and four other viral factors (pUL70, pUL105, pUL102, and pUL57; reviewed in 82) (Figure 4B). The interaction of pUL84 with pUL44 and IE2 was revealed in an AP-MS study of pUL8459, presumably as part of the DNA replication complex59,60. Other observed binding partners of pUL84 include caseine kinase II, later demonstrated to phosphorylate pUL8483, and C1QBP, potentially in the same complex with IE267. As part of the DNA replication process, pUL44 associates with pUL54 and acts as a processivity factor to allow replication of long DNA strands. Several studies have shown that pUL44 forms numerous interactions and is part of distinct functional complexes60,74,84. While the pUL44 interaction with pUL54 can occur in the absence of DNA replication, its interactions with pUL84, IRS1, and UL25 may require replication74. pUL44 also interacts with TRS1, another viral protein involved in transcriptional activation, and this was suggested to be a separate event from its association to IRS184. Therefore, in addition to its role in genome replication, pUL44 may also stimulate viral gene expression. An unexpected pUL44 interaction was observed with nucleolin, a DNA and RNA binding phosphoprotein that was further shown to be important for HCMV replication and expression of late genes74,85. Finally, using immobilized oriLyt as a bait cellular factors that bind to this region were identified by MS in addition to the previously observed viral proteins86. In particular, the heterogenous ribonuclear protein K (hnRNP K) was observed to interact and shown to be essential for genome replication, similar to what has been reported for other herpesviruses87,88.
Once the capsids are loaded with the viral genome, they undergo nuclear egress by penetrating the nuclear lamina and budding into the nuclear membrane (Figure 1). Milbradt et al. investigated the components of the nuclear egress complex (NEC) that contains the viral proteins pUL50 and pUL53 as its core components26. The NEC was purified by affinity isolation of tagged pUL50 and pUL53 at different times after infection. The list of the main NEC components varied between early and late infection, demonstrating the dynamic nature of this complex as the infection progresses. Emerin was discovered as a component of the NEC and shown to be required for efficient viral replication. Interestingly, knockdown of Emerin caused a disruption of the viral AC, suggesting an intimate connection between nuclear egress and cytoplasmic assembly25,28. An AP-MS approach was also valuable for characterizing intermediate stages of virion assembly during the late stages of infection27. This study determined the localization and interactions of two critical virion components, pUL32—a protein that binds directly to the nucleocapsid and pUL99—a protein required for capsid envelopment. The identified interactions revealed that these viral proteins traffic through distinct pathways, using the host endosomal sorting complex required for transport (ESCRT) and clathrin-associated vesicles. These findings suggested the presence of multiple distinct intermediate virion assemblies that merge later in infection.
Virus Protein Interactions that Modulate Host Responses to Infection and the Cell Cycle
Virus-host protein interactions form the basis of diverse mechanisms through which HCMV modulates and utilizes host cell components to ensure effective viral replication and spread. AP-MS studies have proved valuable in identifying such critical interactions used to suppress cell responses. A striking example is the modulation of stress response and cell growth by the tegument protein pUL38 (Figure 4C). Using AP-MS, pUL38 was shown to interact with the tuberous sclerosis tumor suppressor complex (TSC1/2) 66, a complex that transmits stress signals to the mammalian target of rapamycin complex 1 (mTORC1). By interacting with TSC2, pUL38 block the ability of TSC1/2 complex to negatively regulate mTORC1, thereby ensuring an active mTOR pathway and cell growth. Another characteristic of HCMV is its immunomodulatory capacity. One example is the ability of the viral tegument pUL83 protein to inhibit the DNA sensor IFI16 and immune response18, as mentioned above. HCMV also encodes the RL11 family of largely uncharacterized proteins that contain domains homologous to immunoglobulin. pUL11, a member of the RL11 family, was shown by AP-MS to interact with CD45, thereby limiting CD45 tyrosine kinase activity and suppressing T cell proliferation73.
HCMV infection also triggers changes in cell cycle progression and DNA damage response, with the goal to induce viral gene replication, inhibit cellular DNA replication, and prevent apoptosis 89. Cells infected with HCMV undergo cell cycle arrest in a pseudo-G1 state. Recently, the pUL27 viral protein was identified as an important factor for cell cycle arrest (Figure 4D). AP-MS of pUL27 identified its interaction with Tip6061, an acetyltransferase of histone and non-histone proteins with roles in DNA damage response, apoptosis, and cell-cycle progression. On the basis of this interaction, pUL27 was next shown to be critical for the proteasomal-degradation of Tip60, resulting in cell-cycle arrest. Another mechanism involved in the HCMV-induced cell cycle arrest is the degradation of members of the anaphase promoting complex (APC) and the phosphorylation of an APC co-activator by the viral kinase pUL97. An interactome analysis of pUL21a revealed that this protein is responsible for degradation of the APC subunits APC4 and APC564, with pUL21a and pUL97 acting in synergy to promote cell cycle arrest (Figure 4D). Finally, pUL35 has been shown to interact with components of an E3 ubiquitin ligase complex (DCAF1, DDB, and DDA1) that regulates DNA damage and cell cycle62. pUL35 was found to be necessary for cell cycle arrest and this function required the presence of DCAF1. Overall, these AP-MS studies demonstrate the impressive range of intricate interactions between viral and host proteins that generate an environment permissive for HCMV replication.
Posttranslational Modifications on Virus and Host Proteins during HCMV Infection
HCMV relies on a variety of posttranslational modifications (PTMs) for viral replication and regulation of the host machinery. For example, the two major components of the virion tegument (ppUL83 and p150), as well as other virion components, are phosphorylated90,91, and HCMV encodes its own protein kinase, pUL9722. Host proteins also undergo infection-induced changes in PTMs that can activate or inhibit their functions. Additionally, the epigenetic regulation of viral gene expression occurs through numerous PTMs on histones that are under the direct or indirect control of host and viral proteins77,79. MS-based approaches have been applied to identify a range of PTMs occurring on viral and host proteins during infection, providing the baseline for the characterization of their roles in the HCMV life cycle.
Methods for PTM Analysis of Host and Viral Proteins in the Context of HCMV Infection
MS-based analysis of PTMs on HCMV and host proteins ranged from studies using bacterial expression systems92 and in vitro reactions93 to analyses in human cells94 during HCMV replication18,24,95. Several workflows used in HCMV research involved the enrichment of the protein (or proteins) of interest by affinity purification followed by LC-MS/MS analysis to identify PTMs and the sites at which the modifications occur. Analysis of PTMs requires a good coverage of the amino acid sequence of the protein(s) of interest to avoid missing modified peptides. This coverage can be optimized by using different proteases for protein digestion, and can be affected by the sensitivity and speed of the MS instrument. These analyses have allowed the detection of multiple PTMs on viral proteins, as well as of changes in PTMs on cellular proteins during infection by using different quantitative MS methods (Figure 3). These methods include labeling approaches with isobaric tags (iTRAQ)96 to quantify the relative abundance of PTMs in uninfected (mock-infected) and infected cells24, as well as label-free quantitative approach by extraction of MS peak areas95.
Regulation of Posttranslational Modifications during the HCMV Life Cycle
Mass spectrometry has provided the means to accurately identify PTMs on both viral and host proteins, offering insights into processes crucial for the HCMV life cycle. While numerous PTMs on viral proteins were observed to aid viral replication, PTMs were also found to function in host defense. Upon entry into cells, the most abundant virion component pUL83 was shown by AP-MS to be phosphorylated at numerous sites, likely by host kinases18. While this protein has been known for many years to be phosphorylated97, it was only recently that the sites were identified and functionally characterized18. The phosphorylation within its PAD (pyrin association domain) was suggested to obstruct the ability of this viral protein to inhibit the host defense factor IFI16, thereby allowing IFI16 to oligomerize and trigger the expression of antiviral cytokines.
PTMs were also shown to play important roles in viral gene expression, viral genome replication, nuclear egress, and cell-cycle control. Histone modifications of the chomatinized viral genome are important for viral gene expression and regulation of the viral kinetic classes77,98. A recent study has investigated changes in histone PTMs throughout the infection cycle, revealing numerous dynamic PTMs in both histones H3 and H4. Of interest was the H3K79 methylation, observed to increase during infection, and DOT1L, the methyltransferase regulating this site, was shown to be required for efficient viral replication. Another regulator of histone modifications, the deacetylase HDAC1 was also shown by other studies to be important during infection63,81. Interestingly, the viral kinase pUL97 was reported to phosphorylate HDAC1, a modification suggested to inhibit its association with the NuRD complex (Figure 4A)81. PTMs on viral proteins can also impact viral gene expression. Shen et al. investigated the phosphorylation of the viral transactivator, ppUL8294. This tegument protein activates the MIEP through degradation of Daxx15. Site-specific phosphorylation of ppUL82 modulates its nuclear localization and ability to regulate gene expression early in infection and its localization to the assembly compartment later in infection.
In the context of viral genome replication, the viral protein pUL44 was shown to be modified with Small Ubiquitin-related modifiers (SUMO; Figure 4B)92. The SUMOylation of pUL44 enhances viral DNA replication and production of infection particles. As described above, another PTM involved in viral DNA replication is the phosphorylation of pUL84 by CKII83 (Figure 4B). Following genome replication, capsids assemble in the nucleus and undergo nuclear egress by disruption of the nuclear lamina (Figure 1, [C]). By using an iTRAQ quantitative approach (Figure 3), the nuclear lamina component lamin A/C was shown to be phosphorylated by the viral kinase pUL97, implicating pUL97 in nuclear lamina disruption24. Interestingly, pUL97 has also been implicated in modulating the cell cycle. pUL97 was also shown to phosphorylate the APC co-activator Cdh193. In conjunction with the APC interaction with pUL21, this pUL97-dependent modification is required for dysregulation of the cell cycle through degradation of the APC64.
HCMV Remodeling of Whole Cells and Subcellular Proteomes
In addition to the targeted studies on protein interactions and PTMs during HCMV infection, an area of interest is the characterization of the system-level modulation of the cell following infection. Several research groups have investigated these cellular changes using transcriptomics8,99,100, proteomics47,101-103, metabolomics104,105, and lipidomics106. Advances in MS technology have been at the core of the proteomics, metabolomics, and lipidomics studies, providing increased sensitivity and accuracy of analysis in a high-throughput manner. Analyses were performed in whole cells, as well as on selected subcellular compartments during infection. At the proteome level, the abundances of thousands of proteins have been quantified to reveal functional pathways modulated by HCMV and potential antiviral targets. It is important to note that protein identification by MS requires a reference database to match MS spectra with known virus and host protein sequences. The recent study8 that integrated ribosomal profiling with validation by MS has provided an important step towards generating a complete database of virus-expressed proteins. The ORFs identified in this study have already been included in database searches and peptides originating from these ORFs have been observed47.
MS Analyses of Whole-cell and Subcellular Proteome during Infection
The main goal of proteome studies is to identify proteins differentially expressed in the whole cell or a fraction of the cell proteome. Therefore, protein identification is not sufficient and quantitative approaches have been integrated in all these studies. For analyzing the proteome of a subcellular compartment, several studies have opted for quantification using metabolic labeling101,102. The advantage of this approach is that labeled infected and uninfected cells can be mixed prior to the fractionation steps (Figure 3), ensuring the labeling of all proteins of interest and greatly reducing technical variability. A more recent study took advantage of the multiplexing ability offered by TMT labeling (Figure 3) to quantify changes in the plasma membrane (see cell surface section above)47. In addition to these cell surface analyses, subcellular proteome studies during HCMV infection have focused on fractionating mitochondria and ER mitochondria-associated membranes (MAMs) by density gradient centrifugation101. The secretome of HCMV infected cells has also been analyzed by differential centrifugation in conjunction with label-free LC-MS/MS107. Additionally, a study focused on the analysis of an entire family of proteins by enriching kinases using a column conjugated with kinase inhibitors102. As it is the case with all fractionation studies, the potential for cross-contamination need to be carefully examined. For instance, cytoplasmic proteins were reported in the MAM study101, and it remains to be determined which of these are actual MAM-targeted proteins instead of contaminants found in abundance throughout the density gradient. For analyzing the proteome of whole cell lysates during HCMV infection, both label-free and, more recently, TMT quantifications have been employed47,103. A 10-plex TMT labeling approach was used for whole-cell proteome quantification of cells at seven time points of infection. Although there is a substantial gap in the number of identified proteins between these two studies47,103, this can most likely be attributed to the sample preparation method (i.e., off-line peptide fractionation) and the used instrumentation rather than the quantification approach.
Understanding the HCMV Modulation of Host Cells at Proteome and Subproteome Levels
HCMV targets the mitochondria to control multiple cellular functions, including apoptosis, glucose metabolism and respiration22,104,105,108. The proteome of MAMs, a suborganellar compartment that controls mitochondria activity by interaction with the ER, was investigated during infection101. An overall induction of MAM associated proteins was observed at 72hpi, including ER chaperones, stress modulators, and metabolic enzymes involved in glycolysis and cell respiration. Interestingly, some of the proteins observed as highly enriched in the MAM were only mildly enriched in whole cell lysates. This highlights the value of subcellular proteome analysis during infection, since recruitment of proteins to specific subcellular compartments may cause modulation of their cellular functions. The complement of proteins secreted from HCMV infected cells, termed the secretome, was also investigated107. A remarkable increase in the number of secreted proteins was observed following infection, with >1000 proteins being unique or enriched during infection. Many of these proteins were involved in angiogenesis and wound healing. It was proposed that these secreted factors may explain the association between transplant rejection and HCMV as a result from accelerated transplant vascular sclerosis109,110. In addition to fractionation by purifying specific subcellular compartments, an enrichment of proteins with a particular function can also be performed, as shown for the study of changes in the cell kinome following HCMV infection102. Several kinases important for HCMV life cycle progression were identified with increased abundance and activity. In particular, Aurora A was found to be activated during infection and to have an antiviral function, in agreement with a previous siRNA-based kinome screen during HCMV infection111. Recently, a whole cell proteome analysis of HCMV infected cells has provided insight into pathways modulated by the infection. Over 8000 proteins were quantified at seven time points during HCMV infection47. Bioinformatics analyses of the differentially expressed proteins showed regulation of proteins involved fatty acid metabolism, oxidative phosphorylation, interferon signaling, and mRNA transcription, processes previously shown to be impacted during HCMV infection. The authors also proposed five classes of temporal viral protein expressions based on the observed protein expression patterns. These large proteomic studies provide valuable resources that can be mined to formulate additional hypotheses and to further characterize mechanisms involved in the HCMV-mediated host modulation.
Expert commentary
In recent years, proteomics has become a critical component of discoveries in virology. Viruses have coevolved with their hosts, acquiring mechanisms to capture and manipulate host cellular processes for their replication and spread. Similarly, host cells respond by deploying defense mechanisms or by adapting to the infection environment. The continuous development of diverse proteomics approaches has provided powerful tools for characterizing this dynamic interaction between viruses and hosts. As discussed in this review, MS-based analyses have been used to characterize the composition of virions, to define virus-host protein interactions, to determine the function of posttranslational modifications during infection, and to map global proteome or secretome changes following infection.
While our review has emphasized the power of integrating these two fields of research, proteomics and virology, it has also highlighted the need for future studies that make use of these multidisciplinary technologies. For example, the majority of HCMV-host protein-protein interactions during the progression of HCMV infection still remain to be defined. Similarly, the knowledge regarding the temporal changes in virus and host protein expression levels and their diverse posttranslational modifications remains incomplete and limited to only few cell types and virus strains. When comparing to the broad range of MS-based approaches, it becomes evident that the potential of proteomics has only been exploited at a small extent in such virology studies. Proteomic-based studies applied to different biology area of research, including virology, are many times criticized as being open-ended in scope. However, numerous studies described in this review have beautifully demonstrated the ability of MS-based analyses to provide new questions and concrete hypotheses, to offer answers and proofs for previously established hypotheses, and to address long-standing questions to provide novel mechanistic insights. To take advantage of the continuously developing field of proteomics, there is a need for more scientists at this interface between virology and proteomics, who can understand the promise and value of these emerging technologies, as well as design the appropriate experiments to address important questions with regard to the virus life cycle.
Five-year view
As AP-MS workflows for studying virus-host protein interactions during HCMV infection have become fairly robust, we expect that these approaches will continue to substantially contribute to gaining mechanistic insight into various stages of the infection. The further development of computational and statistical tools for assessment of protein interaction networks and specificity of interactions will be of significant benefit. These interaction studies may help to identify and characterize host factors that can be targeted for therapeutic intervention. Furthermore, as a given protein can be a part of multiple distinct complexes that have different functions, these studies can point to the critical complex that is modulated during HCMV infection. In addition to providing mechanistic insight, this knowledge can help in designing antiviral treatments that are more specific (e.g., targeting a specific complex) and have reduced impact on cellular pathways and reduced toxicity. Another important aspect of HCMV research to which MS-based approaches can contribute is the study of cell-type specific response to infection. The MS technology, in particular the sensitivity of the MS instrumentation and the developments in targeted MS/MS analyses, has reached a stage when it can be applied to analyzing small amounts of sample. HCMV studies will likely be expanded in the coming years to analyze diverse cells that are permissive to infection, and to investigate both productive and latent HCMV infections. Lastly, as proteomic, transcriptomic, metabolomic, and lipidomic resources are continuously expanded, the development of bioinformatic approaches that can integrate the information from these large datasets will be a necessity. The merger of these ‘omic’ studies will provide a systems-view of infection and a global understanding of cellular pathways modulated during infection.
Key issues.
Recent studies have clearly demonstrated the significant contribution of proteomics to understanding important concepts in virology, including studies focused on the HCMV life cycle.
The recent improvements in the sensitivity and accuracy offered by mass spectrometry instrumentation have provided the means to characterize virus and host proteins at a depth and breadth not previously achievable by traditional molecular biology approaches.
Proteomics is readily integrated with molecular virology, biochemistry, and bioinformatics, and has contributed to both discovery/hypothesis-generating and hypothesis-driven studies in virology.
Viral proteomics studies have ranged from the characterization of broad changes in cellular proteomes and posttranslational modifications to the targeted, functional analysis of a given virus-host protein interaction.
In HCMV research, proteomic approaches have helped to functionally characterize the virus-induced modulation of host defense responses and cell cycle progression, as well as processes required for virus gene expression, viral replication, capsid nuclear egress, and virion assembly.
Proteomic approaches still remain to be used at their full capacity for characterizing diverse aspects of the productive and latent HCMV infection and its pathogenicity. We expect that future studies will take advantage of the array of qualitative and quantitative mass spectrometry-based proteomics methods to further expand the understanding of HCMV biology.
Acknowledgments
Financial & competing interests disclosure
This work was supported by National Institutes of Health (NIDA DP1DA026192 and NIAID R21AI102187) to IMC and an HFSPO award RGY0079/2009-C to IMC. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Reviews in medical virology. 2010;20:202–213. doi: 10.1002/rmv.655. doi:10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 2.Sinzger C, Digel M, Jahn G. Cytomegalovirus cell tropism. Current topics in microbiology and immunology. 2008;325:63–83. doi: 10.1007/978-3-540-77349-8_4. [DOI] [PubMed] [Google Scholar]
- 3.Reeves M, Sinclair J. Aspects of human cytomegalovirus latency and reactivation. Current topics in microbiology and immunology. 2008;325:297–313. doi: 10.1007/978-3-540-77349-8_17. [DOI] [PubMed] [Google Scholar]
- 4.Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Reviews in medical virology. 2007;17:253–276. doi: 10.1002/rmv.535. doi:10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- 5.Lanzieri TM, Dollard SC, Bialek SR, Grosse SD. Systematic review of the birth prevalence of congenital cytomegalovirus infection in developing countries. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases. 2014;22:44–48. doi: 10.1016/j.ijid.2013.12.010. doi:10.1016/j.ijid.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arvin AM, et al. Vaccine development to prevent cytomegalovirus disease: report from the National Vaccine Advisory Committee. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2004;39:233–239. doi: 10.1086/421999. doi:10.1086/421999. [DOI] [PubMed] [Google Scholar]
- 7.Murphy E, Rigoutsos I, Shibuya T, Shenk TE. Reevaluation of human cytomegalovirus coding potential. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13585–13590. doi: 10.1073/pnas.1735466100. doi:10.1073/pnas.1735466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stern-Ginossar N, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. doi:10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isaacson MK, Juckem LK, Compton T. Virus entry and innate immune activation. Current topics in microbiology and immunology. 2008;325:85–100. doi: 10.1007/978-3-540-77349-8_5. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa-Goto K, et al. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. Journal of virology. 2003;77:8541–8547. doi: 10.1128/JVI.77.15.8541-8547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalejta RF. Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression. Current topics in microbiology and immunology. 2008;325:101–115. doi: 10.1007/978-3-540-77349-8_6. [DOI] [PubMed] [Google Scholar]
- 12.Gibson W. Structure and formation of the cytomegalovirus virion. Current topics in microbiology and immunology. 2008;325:187–204. doi: 10.1007/978-3-540-77349-8_11. [DOI] [PubMed] [Google Scholar]
- 13.Stamminger T, et al. Open reading frame UL26 of human cytomegalovirus encodes a novel tegument protein that contains a strong transcriptional activation domain. Journal of virology. 2002;76:4836–4847. doi: 10.1128/JVI.76.10.4836-4847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng X, Schroer J, Yu D, Shenk T. Human cytomegalovirus pUS24 is a virion protein that functions very early in the replication cycle. Journal of virology. 2006;80:8371–8378. doi: 10.1128/JVI.00399-06. doi:10.1128/JVI.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang J, Kalejta RF. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology. 2007;367:334–338. doi: 10.1016/j.virol.2007.05.037. doi:10.1016/j.virol.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell DP, Savaryn JP, Moorman NJ, Shenk T, Terhune SS. Human cytomegalovirus UL28 and UL29 open reading frames encode a spliced mRNA and stimulate accumulation of immediate-early RNAs. Journal of virology. 2009;83:10187–10197. doi: 10.1128/JVI.00396-09. doi:10.1128/JVI.00396-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cristea IM, et al. Human cytomegalovirus pUL83 stimulates activity of the viral immediate- early promoter through its interaction with the cellular IFI16 protein. Journal of virology. 2010;84:7803–7814. doi: 10.1128/JVI.00139-10. doi:10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16- mediated DNA sensing for immune evasion. Cell host & microbe. 2013;14:591–599. doi: 10.1016/j.chom.2013.10.007. doi:10.1016/j.chom.2013.10.007. [Integration of virology, biochemistry, and proteomics led to important mechanistic insights into host innate immune response and HCMV immune evasion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yurochko AD. Human cytomegalovirus modulation of signal transduction. Current topics in microbiology and immunology. 2008;325:205–220. doi: 10.1007/978-3-540-77349-8_12. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y, Clippinger AJ, Alwine JC. Viral effects on metabolism: changes in glucose and glutamine utilization during human cytomegalovirus infection. Trends in microbiology. 2011;19:360–367. doi: 10.1016/j.tim.2011.04.002. doi:10.1016/j.tim.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambers J, et al. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. Journal of virology. 1999;73:5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mocarski E, Shenk T, Griffiths P, R.F., P. Knipe DM, editor. Fields Virology. 2013 [Google Scholar]
- 23.Milbradt J, Auerochs S, Marschall M. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. The Journal of general virology. 2007;88:2642–2650. doi: 10.1099/vir.0.82924-0. doi:10.1099/vir.0.82924-0. [DOI] [PubMed] [Google Scholar]
- 24.Hamirally S, et al. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS pathogens. 2009;5:e1000275. doi: 10.1371/journal.ppat.1000275. doi:10.1371/journal.ppat.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchkovich NJ, Maguire TG, Alwine JC. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. Journal of virology. 2010;84:7005–7017. doi: 10.1128/JVI.00719-10. doi:10.1128/JVI.00719-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milbradt J, et al. Proteomic analysis of the multimeric nuclear egress complex of human cytomegalovirus. Molecular & cellular proteomics : MCP. 2014 doi: 10.1074/mcp.M113.035782. doi:10.1074/mcp.M113.035782. [Characterization and functional validation of a complex critical for HCMV life cycle, the nuclear egress complex.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Molecular & cellular proteomics : MCP. 2010;9:851–860. doi: 10.1074/mcp.M900485-MCP200. doi:10.1074/mcp.M900485-MCP200. [This work shows the value of GFP tags for integrating protein interactions with localization during infection. One of the few studies that focuses on late stages of virion assembly.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alwine JC. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS pathogens. 2012;8:e1002878. doi: 10.1371/journal.ppat.1002878. doi:10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S, Vasanji A, Pellett PE. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. Journal of virology. 2007;81:11861–11869. doi: 10.1128/JVI.01077-07. doi:10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das S, Pellett PE. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. Journal of virology. 2011;85:5864–5879. doi: 10.1128/JVI.00155-11. doi:10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freed DC, et al. Pentameric complex of viral glycoprotein H is the primary target for potent neutralization by a human cytomegalovirus vaccine. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4997–5005. doi: 10.1073/pnas.1316517110. doi:10.1073/pnas.1316517110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bortz E, et al. Identification of proteins associated with murine gammaherpesvirus 68 virions. Journal of virology. 2003;77:13425–13432. doi: 10.1128/JVI.77.24.13425-13432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varnum SM, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. Journal of virology. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. doi:10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kattenhorn LM, et al. Identification of proteins associated with murine cytomegalovirus virions. Journal of virology. 2004;78:11187–11197. doi: 10.1128/JVI.78.20.11187-11197.2004. doi:10.1128/JVI.78.20.11187-11197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bechtel JT, Winant RC, Ganem D. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. Journal of virology. 2005;79:4952–4964. doi: 10.1128/JVI.79.8.4952-4964.2005. doi:10.1128/JVI.79.8.4952-4964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu FX, Chong JM, Wu L, Yuan Y. Virion proteins of Kaposi's sarcoma-associated herpesvirus. Journal of virology. 2005;79:800–811. doi: 10.1128/JVI.79.2.800-811.2005. doi:10.1128/JVI.79.2.800-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chertova E, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. Journal of virology. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. doi:10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saphire AC, Gallay PA, Bark SJ. Proteomic analysis of human immunodeficiency virus using liquid chromatography/tandem mass spectrometry effectively distinguishes specific incorporated host proteins. Journal of proteome research. 2006;5:530–538. doi: 10.1021/pr050276b. doi:10.1021/pr050276b. [DOI] [PubMed] [Google Scholar]
- 39.Malouli D, et al. Reevaluation of the coding potential and proteomic analysis of the BAC-derived rhesus cytomegalovirus strain 68-1. Journal of virology. 2012;86:8959–8973. doi: 10.1128/JVI.01132-12. doi:10.1128/JVI.01132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benyesh-Melnick M, Probstmeyer F, McCombs R, Brunschwig JP, Vonka V. Correlation between infectivity and physical virus particles in human cytomegalovirus. Journal of bacteriology. 1966;92:1555–1561. doi: 10.1128/jb.92.5.1555-1561.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammarstedt M, Ahlqvist J, Jacobson S, Garoff H, Fogdell-Hahn A. Purification of infectious human herpesvirus 6A virions and association of host cell proteins. Virology journal. 2007;4:101. doi: 10.1186/1743-422X-4-101. doi:10.1186/1743-422X-4-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ott DE. Purification of HIV-1 virions by subtilisin digestion or CD45 immunoaffinity depletion for biochemical studies. Methods in molecular biology. 2009;485:15–25. doi: 10.1007/978-1-59745-170-3_2. doi:10.1007/978-1-59745-170-3_2. [DOI] [PubMed] [Google Scholar]
- 43.Schiess R, et al. Analysis of cell surface proteome changes via label-free, quantitative mass spectrometry. Molecular & cellular proteomics : MCP. 2009;8:624–638. doi: 10.1074/mcp.M800172-MCP200. doi:10.1074/mcp.M800172- MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elschenbroich S, Kim Y, Medin JA, Kislinger T. Isolation of cell surface proteins for mass spectrometry-based proteomics. Expert review of proteomics. 2010;7:141–154. doi: 10.1586/epr.09.97. doi:10.1586/epr.09.97. [DOI] [PubMed] [Google Scholar]
- 45.Gudleski-O'Regan N, Greco TM, Cristea IM, Shenk T. Increased expression of LDL receptor-related protein 1 during human cytomegalovirus infection reduces virion cholesterol and infectivity. Cell host & microbe. 2012;12:86–96. doi: 10.1016/j.chom.2012.05.012. doi:10.1016/j.chom.2012.05.012. [First study of the cell surface proteome during HCMV infection. Excellent example of HCMV biological insight gained following a large proteomics study.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weekes MP, et al. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science. 2013;340:199–202. doi: 10.1126/science.1235047. doi:10.1126/science.1235047. [A proteomic approach used in studying aspects of latent HCMV infection. Discovery of a potential antiviral target and marker for HCMV latency.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weekes MP, et al. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell. 2014;157:1460–1472. doi: 10.1016/j.cell.2014.04.028. doi:10.1016/j.cell.2014.04.028. [Useful resource for temporal relative abundances of host and viral proteins in whole cells and at the plasma membrane during HCMV infection.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Browne EP, Shenk T. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11439–11444. doi: 10.1073/pnas.1534570100. doi:10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantrell SR, Bresnahan WA. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. Journal of virology. 2005;79:7792–7802. doi: 10.1128/JVI.79.12.7792-7802.2005. doi:10.1128/JVI.79.12.7792-7802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reyda S, et al. The tegument protein pp65 of human cytomegalovirus acts as an optional scaffold protein that optimizes protein uploading into viral particles. Journal of virology. 2014 doi: 10.1128/JVI.01415-14. doi:10.1128/JVI.01415-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Womack A, Shenk T. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. mBio. 2010;1 doi: 10.1128/mBio.00282-10. doi:10.1128/mBio.00282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schauflinger M, et al. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. Journal of virology. 2011;85:3821–3832. doi: 10.1128/JVI.01540-10. doi:10.1128/JVI.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perng YC, Qian Z, Fehr AR, Xuan B, Yu D. The human cytomegalovirus gene UL79 is required for the accumulation of late viral transcripts. Journal of virology. 2011;85:4841–4852. doi: 10.1128/JVI.02344-10. doi:10.1128/JVI.02344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tandon R, Mocarski ES. Cytomegalovirus pUL96 is critical for the stability of pp150- associated nucleocapsids. Journal of virology. 2011;85:7129–7141. doi: 10.1128/JVI.02549-10. doi:10.1128/JVI.02549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahlqvist J, Mocarski E. Cytomegalovirus UL103 controls virion and dense body egress. Journal of virology. 2011;85:5125–5135. doi: 10.1128/JVI.01682-10. doi:10.1128/JVI.01682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. Journal of virology. 2011;85:1573–1580. doi: 10.1128/JVI.01967-10. doi:10.1128/JVI.01967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sinclair J. Human cytomegalovirus: Latency and reactivation in the myeloid lineage. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2008;41:180–185. doi: 10.1016/j.jcv.2007.11.014. doi:10.1016/j.jcv.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 58.Budayeva HG, Cristea IM. A mass spectrometry view of stable and transient protein interactions. Advances in experimental medicine and biology. 2014;806:263–282. doi: 10.1007/978-3-319-06068-2_11. doi:10.1007/978-3-319-06068-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao Y, Colletti K, Pari GS. Identification of human cytomegalovirus UL84 virus- and cell- encoded binding partners by using proteomics analysis. Journal of virology. 2008;82:96–104. doi: 10.1128/JVI.01559-07. doi:10.1128/JVI.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strang BL, Sinigalia E, Silva LA, Coen DM, Loregian A. Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84. Journal of virology. 2009;83:7581–7589. doi: 10.1128/JVI.00663-09. doi:10.1128/JVI.00663-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reitsma JM, et al. Antiviral inhibition targeting the HCMV kinase pUL97 requires pUL27- dependent degradation of Tip60 acetyltransferase and cell-cycle arrest. Cell host & microbe. 2011;9:103–114. doi: 10.1016/j.chom.2011.01.006. doi:10.1016/j.chom.2011.01.006. [Application of AP-MS for characterizing a mechanism of virus-induced cell cycle dysregulation.] [DOI] [PubMed] [Google Scholar]
- 62.Salsman J, et al. Proteomic profiling of the human cytomegalovirus UL35 gene products reveals a role for UL35 in the DNA repair response. Journal of virology. 2012;86:806–820. doi: 10.1128/JVI.05442-11. doi:10.1128/JVI.05442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Terhune SS, et al. Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA. PLoS pathogens. 2010;6:e1000965. doi: 10.1371/journal.ppat.1000965. doi:10.1371/journal.ppat.1000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fehr AR, Gualberto NC, Savaryn JP, Terhune SS, Yu D. Proteasome-dependent disruption of the E3 ubiquitin ligase anaphase-promoting complex by HCMV protein pUL21a. PLoS pathogens. 2012;8:e1002789. doi: 10.1371/journal.ppat.1002789. doi:10.1371/journal.ppat.1002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamil JP, Coen DM. Human cytomegalovirus protein kinase UL97 forms a complex with the tegument phosphoprotein pp65. Journal of virology. 2007;81:10659–10668. doi: 10.1128/JVI.00497-07. doi:10.1128/JVI.00497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moorman NJ, et al. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell host & microbe. 2008;3:253–262. doi: 10.1016/j.chom.2008.03.002. doi:10.1016/j.chom.2008.03.002. [Remarkable example of the application of AP-MS technology to understand HCMV regulation of host stress responses.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du G, Stinski MF. Interaction network of proteins associated with human cytomegalovirus IE2-p86 protein during infection: a proteomic analysis. PloS one. 2013;8:e81583. doi: 10.1371/journal.pone.0081583. doi:10.1371/journal.pone.0081583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Molecular & cellular proteomics : MCP. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. doi:10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 69.Miteva YV, Budayeva HG, Cristea IM. Proteomics-based methods for discovery, quantification, and validation of protein-protein interactions. Analytical chemistry. 2013;85:749–768. doi: 10.1021/ac3033257. doi:10.1021/ac3033257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borst EM, Hahn G, Koszinowski UH, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. Journal of virology. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu D, Smith GA, Enquist LW, Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. Journal of virology. 2002;76:2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prichard MN, Britt WJ, Daily SL, Hartline CB, Kern ER. Human cytomegalovirus UL97 Kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. Journal of virology. 2005;79:15494–15502. doi: 10.1128/JVI.79.24.15494-15502.2005. doi:10.1128/JVI.79.24.15494-15502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gabaev I, et al. The human cytomegalovirus UL11 protein interacts with the receptor tyrosine phosphatase CD45, resulting in functional paralysis of T cells. PLoS pathogens. 2011;7:e1002432. doi: 10.1371/journal.ppat.1002432. doi:10.1371/journal.ppat.1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strang BL, Boulant S, Coen DM. Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication. Journal of virology. 2010;84:1771–1784. doi: 10.1128/JVI.01510-09. doi:10.1128/JVI.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nesvizhskii AI. Computational and informatics strategies for identification of specific protein interaction partners in affinity purification mass spectrometry experiments. Proteomics. 2012;12:1639–1655. doi: 10.1002/pmic.201100537. doi:10.1002/pmic.201100537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nitzsche A, Paulus C, Nevels M. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. Journal of virology. 2008;82:11167–11180. doi: 10.1128/JVI.01218-08. doi:10.1128/JVI.01218-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cuevas-Bennett C, Shenk T. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. Journal of virology. 2008;82:9525–9536. doi: 10.1128/JVI.00946-08. doi:10.1128/JVI.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guise AJ, Budayeva HG, Diner BA, Cristea IM. Histone deacetylases in herpesvirus replication and virus-stimulated host defense. Viruses. 2013;5:1607–1632. doi: 10.3390/v5071607. doi:10.3390/v5071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reeves MB. Chromatin-mediated regulation of cytomegalovirus gene expression. Virus research. 2011;157:134–143. doi: 10.1016/j.virusres.2010.09.019. doi:10.1016/j.virusres.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reeves M, et al. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. Journal of virology. 2006;80:9998–10009. doi: 10.1128/JVI.01297-06. doi:10.1128/JVI.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bigley TM, Reitsma JM, Mirza SP, Terhune SS. Human cytomegalovirus pUL97 regulates the viral major immediate early promoter by phosphorylation-mediated disruption of histone deacetylase 1 binding. Journal of virology. 2013;87:7393–7408. doi: 10.1128/JVI.02825-12. doi:10.1128/JVI.02825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pari GS. Nuts and bolts of human cytomegalovirus lytic DNA replication. Current topics in microbiology and immunology. 2008;325:153–166. doi: 10.1007/978-3-540-77349-8_9. [DOI] [PubMed] [Google Scholar]
- 83.Gao Y, Pari GS. Interaction of human cytomegalovirus pUL84 with casein kinase 2 is required for oriLyt-dependent DNA replication. Journal of virology. 2009;83:2393–2396. doi: 10.1128/JVI.02339-08. doi:10.1128/JVI.02339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strang BL, Geballe AP, Coen DM. Association of human cytomegalovirus proteins IRS1 and TRS1 with the viral DNA polymerase accessory subunit UL44. The Journal of general virology. 2010;91:2167–2175. doi: 10.1099/vir.0.022640-0. doi:10.1099/vir.0.022640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Strang BL, Boulant S, Kirchhausen T, Coen DM. Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments. mBio. 2012;3 doi: 10.1128/mBio.00301-11. doi:10.1128/mBio.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kagele D, Rossetto CC, Tarrant MT, Pari GS. Analysis of the interactions of viral and cellular factors with human cytomegalovirus lytic origin of replication, oriLyt. Virology. 2012;424:106–114. doi: 10.1016/j.virol.2011.12.010. doi:10.1016/j.virol.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmidt T, Striebinger H, Haas J, Bailer SM. The heterogeneous nuclear ribonucleoprotein K is important for Herpes simplex virus-1 propagation. FEBS letters. 2010;584:4361–4365. doi: 10.1016/j.febslet.2010.09.038. doi:10.1016/j.febslet.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 88.Si H, Verma SC, Robertson ES. Proteomic analysis of the Kaposi's sarcoma-associated herpesvirus terminal repeat element binding proteins. Journal of virology. 2006;80:9017–9030. doi: 10.1128/JVI.00297-06. doi:10.1128/JVI.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanchez V, Spector DH. Subversion of cell cycle regulatory pathways. Current topics in microbiology and immunology. 2008;325:243–262. doi: 10.1007/978-3-540-77349-8_14. [DOI] [PubMed] [Google Scholar]
- 90.Meyer H, et al. Identification and procaryotic expression of the gene coding for the highly immunogenic 28-kilodalton structural phosphoprotein (pp28) of human cytomegalovirus. Journal of virology. 1988;62:2243–2250. doi: 10.1128/jvi.62.7.2243-2250.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roby C, Gibson W. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. Journal of virology. 1986;59:714–727. doi: 10.1128/jvi.59.3.714-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sinigalia E, et al. The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner. PloS one. 2012;7:e49630. doi: 10.1371/journal.pone.0049630. doi:10.1371/journal.pone.0049630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tran K, Kamil JP, Coen DM, Spector DH. Inactivation and disassembly of the anaphase- promoting complex during human cytomegalovirus infection is associated with degradation of the APC5 and APC4 subunits and does not require UL97-mediated phosphorylation of Cdh1. Journal of virology. 2010;84:10832–10843. doi: 10.1128/JVI.01260-10. doi:10.1128/JVI.01260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shen W, et al. Nuclear trafficking of the human cytomegalovirus pp71 (ppUL82) tegument protein. Virology. 2008;376:42–52. doi: 10.1016/j.virol.2008.03.007. doi:10.1016/j.virol.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O'Connor C, DiMaggio PA, Jr., Shenk T, Garcia BA. Quantitative Proteomic Discovery of Dynamic Epigenome Changes that Control Human Cytomegalovirus Infection. Molecular & cellular proteomics : MCP. 2014 doi: 10.1074/mcp.M114.039792. doi:10.1074/mcp.M114.039792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine- reactive isobaric tagging reagents. Molecular & cellular proteomics : MCP. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. doi:10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 97.Irmiere A, Gibson W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology. 1983;130:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- 98.Nitzsche A, Steinhausser C, Mucke K, Paulus C, Nevels M. Histone H3 lysine 4 methylation marks postreplicative human cytomegalovirus chromatin. Journal of virology. 2012;86:9817–9827. doi: 10.1128/JVI.00581-12. doi:10.1128/JVI.00581-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hertel L, Mocarski ES. Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of Pseudomitosis independent of US28 function. Journal of virology. 2004;78:11988–12011. doi: 10.1128/JVI.78.21.11988-12011.2004. doi:10.1128/JVI.78.21.11988-12011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fu M, et al. Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA. Gene. 2014;536:272–278. doi: 10.1016/j.gene.2013.12.012. doi:10.1016/j.gene.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 101.Zhang A, et al. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Molecular & cellular proteomics : MCP. 2011;10:M111 009936. doi: 10.1074/mcp.M111.009936. doi:10.1074/mcp.M111.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hutterer C, et al. Profiling of the kinome of cytomegalovirus-infected cells reveals the functional importance of host kinases Aurora A, ABL and AMPK. Antiviral research. 2013;99:139–148. doi: 10.1016/j.antiviral.2013.04.017. doi:10.1016/j.antiviral.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 103.Reyda S, Buscher N, Tenzer S, Plachter B. Proteomic analyses of human cytomegalovirus strain AD169 derivatives reveal highly conserved patterns of viral and cellular proteins in infected fibroblasts. Viruses. 2014;6:172–188. doi: 10.3390/v6010172. doi:10.3390/v6010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS pathogens. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. doi:10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nature biotechnology. 2008;26:1179–1186. doi: 10.1038/nbt.1500. doi:10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu ST, et al. Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12869–12874. doi: 10.1073/pnas.1109796108. doi:10.1073/pnas.1109796108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dumortier J, et al. Human cytomegalovirus secretome contains factors that induce angiogenesis and wound healing. Journal of virology. 2008;82:6524–6535. doi: 10.1128/JVI.00502-08. doi:10.1128/JVI.00502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kaarbo M, et al. Human cytomegalovirus infection increases mitochondrial biogenesis. Mitochondrion. 2011;11:935–945. doi: 10.1016/j.mito.2011.08.008. doi:10.1016/j.mito.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 109.Humar A, et al. Association between cytomegalovirus disease and chronic rejection in kidney transplant recipients. Transplantation. 1999;68:1879–1883. doi: 10.1097/00007890-199912270-00011. [DOI] [PubMed] [Google Scholar]
- 110.Evans PC, et al. An association between cytomegalovirus infection and chronic rejection after liver transplantation. Transplantation. 2000;69:30–35. doi: 10.1097/00007890-200001150-00007. [DOI] [PubMed] [Google Scholar]
- 111.Terry LJ, Vastag L, Rabinowitz JD, Shenk T. Human kinome profiling identifies a requirement for AMP-activated protein kinase during human cytomegalovirus infection. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3071–3076. doi: 10.1073/pnas.1200494109. doi:10.1073/pnas.1200494109. [DOI] [PMC free article] [PubMed] [Google Scholar]


