Abstract
TCF3-HLF-fusion positive acute lymphoblastic leukemia (ALL) is currently incurable. Employing an integrated approach, we uncovered distinct mutation, gene expression, and drug response profiles in TCF3-HLF-positive and treatment-responsive TCF3-PBX1-positive ALL. Recurrent intragenic deletions of PAX5 or VPREB1 were identified in constellation with TCF3-HLF. Moreover somatic mutations in the non-translocated allele of TCF3 and a reduction of PAX5 gene dosage in TCF3-HLF ALL suggest cooperation within a restricted genetic context. The enrichment for stem cell and myeloid features in the TCF3-HLF signature may reflect reprogramming by TCF3-HLF of a lymphoid-committed cell of origin towards a hybrid, drug-resistant hematopoietic state. Drug response profiling of matched patient-derived xenografts revealed a distinct profile for TCF3-HLF ALL with resistance to conventional chemotherapeutics, but sensitivity towards glucocorticoids, anthracyclines and agents in clinical development. Striking on-target sensitivity was achieved with the BCL2-specific inhibitor venetoclax (ABT-199). This integrated approach thus provides alternative treatment options for this deadly disease.
Keywords: acute lymphoblastic leukemia, leukemia xenograft model, next generation sequencing, drug resistance, drug activity profiling, TCF3-PBX1, TCF3-HLF, leukemic stem cell, integrative, genomics, BCL2, BH3-mimetics, venetoclax, ABT-199, cytarabine, vincristine, glucocorticoids, prednisone, dexamethasone, mTOR inhibitor, anthracycline, bortezomib, HSP90, AUY922, panobinostat
One of the hallmarks of pediatric ALL is the presence of subtype-defining chromosomal translocations that cause gene fusions involving master regulators of hematopoietic development. These initiating lesions often cooperate with specific somatic aberrations including monoallelic deletions of B-cell developmental genes, such as PAX5, IKZF1 and EBF11. Other cooperative liaisons are represented by trisomy 21q22 with CRLF2 activation2-4 or near-haploid ALL with activation of receptor tyrosine kinase or RAS signaling5. RAS pathway mutations appear in high-risk ALL, but are often lost with disease progression, suggesting the involvement of additional tumorigenic factors6,7. The patterns of recurrent genomic alterations need to be better understood, because apart from tyrosine kinase inhibitor-supplemented treatment of BCR-ABL1-positive ALL, the only proven successful first-line treatment strategies for high-risk ALL are chemotherapy intensification and early allogeneic hematopoietic stem cell transplantation8.
The translocation t(1;19)9 that fuses the transcriptional activation domain of the B cell developmental transcription factor TCF3 to the DNA-binding domain of PBX1 occurs in about 5-10% of precursor B cell (pre-B) ALL patients and is associated with a median five-year event-free survival probability of 78-85%10. In contrast, the translocation t(17;19)(q22;p13), resulting in the fusion gene TCF3-HLF, defines a rare subtype of ALL (<1% of pediatric ALL) that is typically associated with relapse and death within two years from diagnosis11,12. Both translocations disrupt one allele of TCF3, which drives the B-cell differentiation program upstream of the transcription factor PAX513. As an initiating event, expression of TCF3-HLF leads to transcriptional reprogramming in pre-leukemic cells. Possible direct targets of TCF3-HLF include the transcription factor gene LMO2, which is implicated in initiation of T ALL14,15, and the transcriptional repressor SNAI1 (SLUG), which regulates embryonic development and apoptosis16,17. Further targets were proposed, including BCL214. The TCF3-HLF fusion likely requires additional events to cause leukemia, because TCF3-HLF transgenic and knock-in mice did not recapitulate the human phenotype18,19.
Here, we report that the genomic and transcriptomic landscape of TCF3-HLF-positive ALL differs markedly from TCF3-PBX1-positive ALL. The TCF3-HLF fusion likely occurs in B lymphoid progenitors in the context of PAX5 haploinsufficiency and is associated with transcriptional reprogramming towards an immature, hybrid hematopoietic state. Drug response profiling in patient-derived xenografts, which maintained the genomic and global transcriptome landscapes of the corresponding primary leukemic samples, identified resistance patterns to drugs commonly used for the treatment of TCF3-HLF-positive patients. A general trait of TCF3-HLF-positive ALL in our study is extreme sensitivity towards the BCL2-specific inhibitor ABT-199 (venetoclax), indicating new therapeutic options for this fatal ALL subtype.
RESULTS
Next generation sequencing (NGS) analysis integrating short and large insert size paired-end whole genome (WGS), whole exome (WES) and transcriptome (RNAseq) sequencing was applied to a discovery cohort consisting of five diagnostic pre-treatment samples of TCF3-PBX1-positive (samples 1a-5a) and TCF3-HLF-positive (samples 6a-9a and 11a) ALL, respectively. Matched bone marrow samples collected after induction treatment for minimal residual disease evaluation (MRD, maximum leukemic cell load ≤10−3; samples 1b-9b and 11b) were used as non-tumor controls (Supplementary Table 1). Additional DNA samples from seven TCF3-HLF-positive (diagnostic samples 10a, 12a, 13a, 14a, 15a, 16a, 17a, remission samples 10b, 12b, 13b) and 24 TCF3-PBX1-positive cases were used for validation (Supplementary Tables 2 and 3). In most cases TCF3-HLF-positive ALL responded to induction chemotherapy, but remained MRD positive. Nine children included in this study died due to disease progression and treatment-related toxicities within 2 years on average and only one patient is in remission after a short follow-up time, reflecting the dismal prognosis of TCF3-HLF-positive ALL.
TCF3 breakpoints suggest a committed lymphoid cell of origin
Consistent with previous reports20,21, all TCF3 translocation breakpoints were restricted to three hotspot regions (Fig. 1a and 1b and Supplementary Fig. 1). Those were associated with small non-template nucleotide insertions indicative of terminal deoxynucleotide transferase (TdT) activity characteristic of an early B cell stage (Supplementary Table 4). In TCF3, the breakpoints clustered in close proximity to CpG elements in the absence of classical RAG consensus sequence sites (Supplementary Fig. 1), which is a characteristic feature of translocations that occur in lymphoid progenitors at the pro-/pre-B stage. This may represent illegitimate RAG-mediated recombination at cryptic sites, possibly in the context of deaminated CpG nucleotides as proposed for TCF3-PBX1 translocations22. Consistent with the idea that TCF3-HLF may occur at a lymphoid-committed rather than a pluripotent progenitor stage, we detected this translocation only in sorted pre-B cell populations containing leukemic cells but neither in stem cells nor in myeloid progenitor cells (Supplementary Fig. 2).
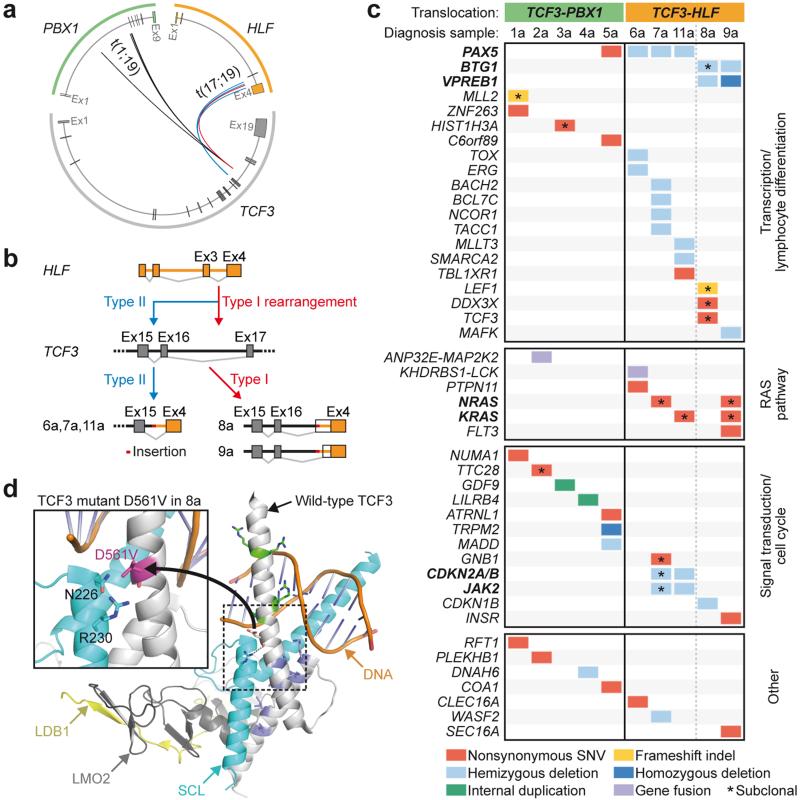
Figure 1. Genetic lesions identified in pediatric TCF3-HLF- and TCF3-PBX1-positive ALL.
(a) Breakpoints in TCF3, PBX1 and HLF cluster in genomic hotspot regions. Boxes correspond to exonic regions; arcs represent fusions in patient samples. (b) TCF3 breakpoints cluster in two TCF3 intronic regions: between exons 16 and 17 (type 1) and between exons 15 and 16 (type 2). On the transcript level, type 1 translocations join TCF3 exon 16 to HLF exon 4, including inserted non-template and intronic sequences and new splice acceptor sites (patients 8 and 9). Type 2 translocations occur downstream of exon 15 and exclude TCF3 exon 16 from the fusion transcript (patients 6, 7 and 11). (c) Absence of overlapping somatic structural and nucleotide variations between the cohorts. TCF3-HLF-positive ALL is characterized by mutually exclusive PAX5, BTG1 and VPREB1 deletions and nonsynonymous nucleotide variations in TCF3 (p.Asp561Val, “D561V” in patient 8 and p.Ser467Gly in patient 13 of the validation cohort, the latter not included here). Subclonal RAS pathway mutations are exclusively detected in HLF-, but not in PBX1-fused cases. (d) Models of wild-type and mutant TCF3 based on the crystal structure of TCF3 in complex with the transcription factors SCL, LMO2 and LDB1 bound to DNA.58 Upon LMO2 binding, bonds are formed between TCF3 and SCL, including a hydrogen bond (dashed line) between D561 and R230 reducing the DNA binding capacity of the complex. Inset: D561V introduces a hydrophobic valine residue close to polar residues that may interfere with hydrogen bonding, thus altering the DNA binding properties of the complex. (For details see Source file for Figure 1.)
TCF3-HLF ALL and impaired pro- to pre-B cell transition
Pre-B cell ALL is frequently associated with somatic copy number alterations affecting B cell developmental genes. PAX5 deletions are generally observed in 13% of ALL cases and in up to 28% of high-risk ALL23. We observed enrichment for monoallelic PAX5 deletions in TCF3-HLF-positive ALL reaching 67% of the cases (Fig. 1c, Supplementary Table 2). Illegitimate RAG-mediated recombination appears to be implicated in the generation of such events in TCF3-HLF-positive ALL given the close proximity to RSS motifs (Supplementary Table 5). In most samples without PAX5 deletion, we identified hemi- and homozygous deletions of VPREB1, a component of the surrogate light chain of the pre-B cell receptor (Fig. 1c, Supplementary Table 6) independent of the lambda light chain locus. VPREB1 deletions in pediatric ALL result in failure to form a viable surrogate light chain in the pre-B cell receptor, an event associated with lower overall survival24. In addition, we detected BTG1 gene deletions in three out of eight TCF3-HLF-positive cases without PAX5 deletions (Supplementary Fig. 3a and 3b). BTG1 deletions occur frequently in ALL positive for ETV6-RUNX1 (19%) or BCR-ABL1 (26%) and may confer a proliferative advantage25. In contrast, no deletion, but only a single PAX5 nonsense mutation could be detected in 29 TCF3-PBX1-positive cases (Fig. 1 and Supplementary Table 3). Our results indicate that cooperative genetic events affecting genes regulating the pro- to pre-B cell transition, in particular PAX5, BTG1 and VPREB1, but not IKZF1, are selected in TCF3-HLF-translocated cells. Other deleted genes associated with pre-B cell ALL26 were JAK2 and CDKN2A/B (patient 7a and 11a) and transcriptional regulators such as ERG, NCOR1, TOX, BACH2, BCL7C, MLLT3, SMARCA2, and MAFK (Fig. 1c).
Recurrent RAS pathway mutations in TCF3-HLF ALL
We identified only a few additional somatic alterations affecting protein-coding sequences in both TCF3-PBX1- and TCF3-HLF-positive ALL (Fig. 1c, Supplementary Tables 6 and 7), involving among others, genes associated with pre-B cell ALL26 (TCF3, PAX5, LEF1) and transcriptional and chromatin regulation (ZNF263, MLL2, HIST1H3A, C6orf89). We observed a prominent association of TCF3-HLF-positive ALL with activating mutations in RAS signaling pathway genes (NRAS, KRAS, and PTPN11), detectable in four out of five discovery cases (Fig. 1c) and in three out of five additional TCF3-HLF-positive validation samples (PTPN11 and SPHK1) (Supplementary Table 2). No RAS pathway mutations were identified in the TCF3-PBX1-positive discovery cohort and only one oncogenic NRAS mutation was detected in the 24 TCF3-PBX1-positive validation cases (Supplementary Table 3). NRAS and KRAS mutations were generally detected in subclones (Supplementary Table 7). Interestingly, we discovered a novel fusion gene, KHDRBS1-LCK, due to an interstitial chromosomal deletion in one TCF3-HLF-positive sample (6a), triggering the overexpression of the LCK tyrosine kinase (Supplementary Fig. 4). This was also present in three out of 74 randomly selected ALL samples, demonstrating that KHDRBS1-LCK fusion is recurrent in ALL (Supplementary Fig. 5). LCK is a drug target in RAS-dependent cancer cells which have higher LCK expression27, suggesting a possible interplay with RAS-related signaling networks in TCF3-HLF-positive ALL. Oncogenic activation of LCK associated with t(1;7)(p34;q34) translocation was reported in the T cell leukemia cell line HSB228. Our data indicate a frequent association of proliferation-driving mutations in TCF3-HLF-positive ALL in the context of stalled B cell differentiation.
Mutations affecting the second TCF3 allele in TCF3-HLF ALL
We identified a mutation in the basic helix-loop-helix region of TCF3 (p.Asp561Val, D561V, Fig. 1c and 1d) affecting the non-translocated chromosome in one TCF3-HLF-positive case (8a). Mutations at this position were reported in sporadic Burkitt lymphoma29 and may reduce binding to its negative regulator ID329. Based on available crystal structure data, p.Asp561Glu may affect the interaction of TCF3 with the transcription factor SCL (also known as TAL1; Fig. 1d), possibly altering TCF3 protein complexes. A second TCF3 mutation (p.Ser467Gly) was detected in another TCF3-HLF-positive case (13a, Supplementary Table 2). The functional consequences of this mutation are currently unclear. Interestingly, we could not detected any somatic mutations in TCF3 by targeted sequencing of 1,033 unselected ALL patients from the European multicenter trial AIEOP-BFM ALL 2000, suggesting a specific association with TCF3-HLF-positive ALL (Supplementary Table 8). Thus, deregulation of normal TCF3 function may also contribute to TCF3-HLF-positive ALL. Corroborating our findings, a recent study included a single TCF3-HLF case, as part of a cohort comparing diagnostic and relapse ALL samples, which showed a PAX5 deletion and two mutations in TCF3 (p.His460Tyr, p.Gly470fs), all of which were conserved at relapse30. Besides, the relapse sample featured a VPREB1 deletion as well as a shift in subclonal mutations in NRAS (p.Gly12Asp and p.Gly12Val), reinforcing the idea of cooperative effects between TCF3-HLF and alteration of PAX5 and VPREB1 gene dosage. Taken together, seven out of eleven TCF3-HLF cases were hemizygous for PAX5, whereas five samples featured VPREB1 deletions (summarized in Supplementary Fig. 6).
Reprogramming towards a more immature state in TCF3-HLF ALL
Consistent with the occurrence of TCF3-HLF and TCF3-PBX1 translocations in lymphoid precursors, both leukemia subtypes shared a gene expression signature of B-lymphoid cells (including PAX5, BLK, CD19, CD22, CD79B, TCF3, EBF1, VPREB1, RAG1, ROR1, BLNK, and DNTT) (Supplementary Tables 9 and 10), but differential expression of 401 genes (false discovery rate ≤ 0.001) strongly distinguished the two TCF3-translocated subtypes (Fig. 2a, Supplementary Tables 11 and 12). In silico prediction of transcription factor binding sites (TFBS) in the corresponding promoter regions revealed significant enrichment for PBX and HLF binding motifs associated with TCF3-PBX1- and TCF3-HLF gene signatures, respectively (Supplementary Tables 13 and 14). Further, PBX1 and HLF were the only transcription factors among those with enriched binding motifs which were significantly differentially expressed between the two ALL subtypes and between leukemia and remissions. The chimeric HLF transcript was strongly induced in TCF3-HLF, but no wild-type HLF expression could be detected. We predicted 39 potential HLF targets, including the known target SNAI2 (SLUG)16, GPC4 and BMP3 involved in stem cell proliferation, which showed induced expression in TCF3-HLF samples (Supplementary Table 15). Other potential TCF3-HLF targets that regulate developmental programs and cell survival, such as LMO214 and BCL214, were not predicted. However, their expression was increased in TCF3-HLF-positive ALL.
Figure 2. TCF3-HLF programs leukemia to a hybrid hematopoietic transcriptional state.
(a) Heatmap of the 401 differentially expressed genes between the two ALL subtypes (edgeR, |log2(fold change)| ≥ 1, FDR ≤ 0.001). (b) Enriched hematopoietic stages in TCF3-HLF-positive (orange) and TCF3-PBX1-positive (green) ALL. Stages shown include hematopoietic stem cells (HSC), common myeloid progenitors (CMP), lymphoid-specified progenitors (GMP and MEP), neutrophils (NEUTRO), monocytes (MONO), multilymphoid progenitor (MLP), early T cell precursors (ETP), pro-B cells (PROB), T cells (TCELL), and B cells (BCELL). Gene set enrichment analysis was carried out using Genomatix genome analyzer and Gene Set Enrichment Analysis (GSEA)59 (GSEA: FDR ≤ 0.02; genomatix genome analyzer: adjusted p-value ≤ 0.02). The source of the significant enriched gene sets is noted by the superscript: 1) curated gene sets of hematopoietic precursors31; 2) human immunologic gene signatures (MSigDB v4.0)32; 3) text mining-based tissue specific gene sets60. (c) Enrichment plot for the HSC signature31. (d) Components of the TCF3-HLF-positive ALL signature reveal functional annotation related to stem cells and their cellular location (Genomatix genome analyzer: p-value = 4.65 × 10−4, adjusted p-value < 0.001). (For details see Source file for Figure 2.)
Gene set enrichment analysis (GSEA) using gene sets from sorted human hematopoietic stem cells (HSC) and early progenitor populations31 as well as curated oncogenic (C6) and human immunologic (C7) signatures from MsigDB32 revealed an enrichment for stem cell and myeloid signatures in TCF3-HLF-positive ALL. In contrast, lymphoid features were more prominent in TCF3-PBX1-positive ALL (Fig. 2b, Supplementary Table 16). The HSC signature l31 ranked among the top gene sets enriched in TCF3-HLF-positive ALL (Fig. 2c, Supplementary Table 17). We obtained similar results using an independent method based on text mining annotations (Fig. 2d, Supplementary Tables 18 and 19). High expression of the stem cell marker LGR533 was consistently detected in TCF3-HLF-positive ALL suggesting a reactivation of immature features shared with other stem cell populations. Consistent with previous reports, the myeloid marker CD33 was expressed in TCF3-HLF-positive blasts, which provides a target for antibody-directed therapy12,34. Other differentially expressed genes, such as BMP235, could present additional therapeutic targets.
Our results are consistent with a model in which TCF3-HLF arises in lymphoid cells and promotes transcriptional reprogramming towards a hybrid hematopoietic state. Features of mesenchyme-derived tissues were also detected in TCF3-HLF-positive ALL, which may indicate a profound cellular reprogramming towards a drug-resistant state.
Mutation profiles of TCF3-HLF ALL are conserved in xenografts
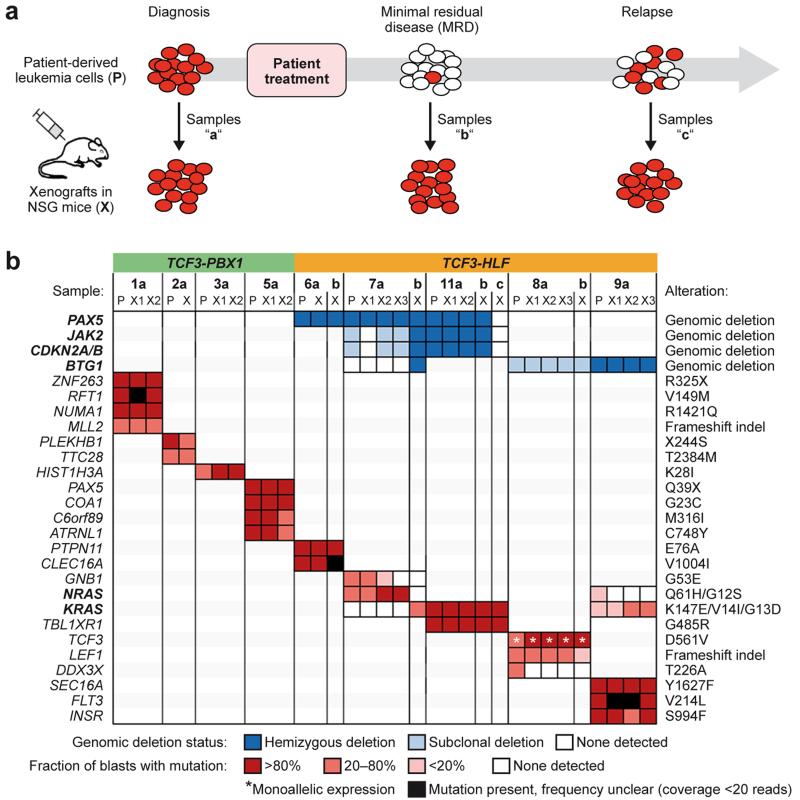
We generated leukemia xenografts in immunodeficient NOD/SCID/IL2rγnull (NSG) mice for all cases included in this study (Supplementary Table 20)36,37. We also established for the first time to our knowledge leukemia xenografts from follow-up samples with MRD, some with less than 0.1% ALL cells after induction chemotherapy (Fig. 3a and Supplementary Tables 1 and 20). Leukemia and MRD engraftment was rapid with conserved and predictable kinetics for xenografts derived from the same patient (Supplementary Fig. 7), suggesting that no major adaptation to the mouse microenvironment was needed for proliferation. Most SNVs and intrachromosomal deletions that were present at diagnosis were conserved in the corresponding xenografts (Fig. 3b, Supplementary Table 7). Only deletions detected in the relapse sample 11c were not conserved in the corresponding xenografts and a deletion in BTG1 emerged in one MRD-derived sample (patient 7b, Supplementary Fig. 3c and 3d). A few mutations were lost in MRD or relapse xenograft samples, including GNB1 and DDX3X indicating that these are probably dispensable or may cause drug sensitivity. Mutations in the RAS pathway were largely maintained in xenografts. However, the NRAS mutation p.Gln61His identified in the primary MRD sample 7b was not detected in the corresponding xenograft. Interestingly, a heterozygous damaging mutation in KRAS (p.Lys147Glu) associated with Noonan syndrome38 emerged instead. In patient 9a we identified two subclones displaying either a KRAS (p.Gly13Asp) or an NRAS (p.Gly12Ser) mutation. The corresponding xenograft retained only the KRAS mutated sub-clone. Thus, maintenance and acquisition of RAS pathway mutations in xenografts support the notion that they occur later during selection at a multiclonal level and confer a selective advantage in TCF3-HLF-positive ALL. No other SNVs emerged de novo in the xenografts. In summary, the molecular characteristics of both leukemia subtypes were largely conserved in the xenografts, confirming the validity of this model.
Figure 3. The genomic landscape of TCF3-HLF- and TCF3-PBX1-positive ALL is preserved in patient-derived leukemia xenografts.
(a) Xenografts were established from cryopreserved patient samples at diagnosis (samples “a”), at follow-up with minimal residual disease (MRD, <1 leukemic cell in 10,000 cells, samples “b”) or from disease progression (samples “c”) and subjected to whole exome and transcriptome sequencing as well as multiplex ligation-dependent probe amplification (MLPA). All available MRD samples from TCF3-HLF-positive cases were successfully engrafted. (b) Comparison of all transcriptionally expressed nucleotide variations and of selected recurrent deletions frequently found in pediatric ALL in corresponding patient (P) and xenograft (X) samples. Deletions and nucleotide variations are colored according to their frequency in the analyzed leukemic cell population. Deletion frequencies were calculated by integrating whole genome and whole exome sequencing data with MLPA data. Nucleotide variation frequencies were calculated by integrating whole genome, whole exome and transcriptome sequencing data. (For details see Source file for Figure 3.)
TCF3-HLF-associated gene expression is maintained in xenografts
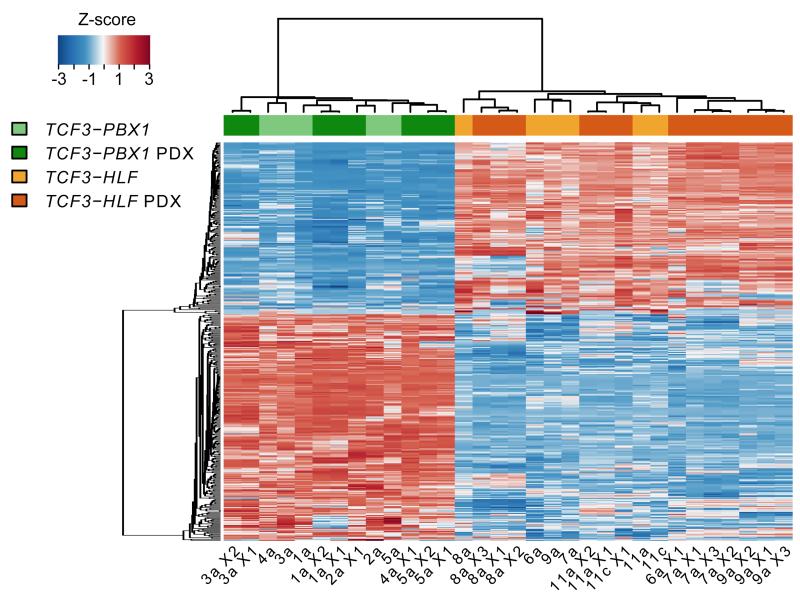
Hierarchical clustering based on the gene signature specifying the two leukemia subtypes showed that the expression profile and the subtype-specificity of the primary leukemia were maintained in the xenografts (Fig. 4). The genes most significantly upregulated in matched patient and xenograft samples from TCF3-HLF-positive leukemia specified stem cell features (Supplementary Table 21 and 22). Similar to patient samples, features of mesenchyme-derived tissues were also detected in xenografts derived from TCF3-HLF-positive ALL. Strikingly, TCF3-HLF-positive leukemias and xenografts displayed systematic down-regulation of PAX5 expression to halved levels. Though mono-allelic deletions of PAX5 were a prominent feature of TCF3-HLF-positive ALL, reduced expression was also seen in diploid cases, hinting at alternative molecular mechanisms. The recapitulation of this pattern in the xenograft samples enforces the notion that TCF3-HLF-positive leukemia emerges in a specific cellular context with reduced PAX5 expression (Supplementary Fig. 8). The essential molecular features of TCF3-HLF-positive samples were maintained in xenografts, providing a useful model of this disease.
Figure 4. Major components of the gene expression signature of TCF3-HLF- and TCF3-PBX1-positive ALL are conserved in patient-derived xenografts.
Hierarchical clustering of primary and patient derived xenograft (PDX) ALL samples based on the expression of the 401 genes of the signature defined with primary samples (Fig. 2) shows that xenografts clearly group with their corresponding primary samples. (For details see Source file for Figure 4.)
Drug activity profiling of TCF3-HLF and TCF3-PBX1 ALL
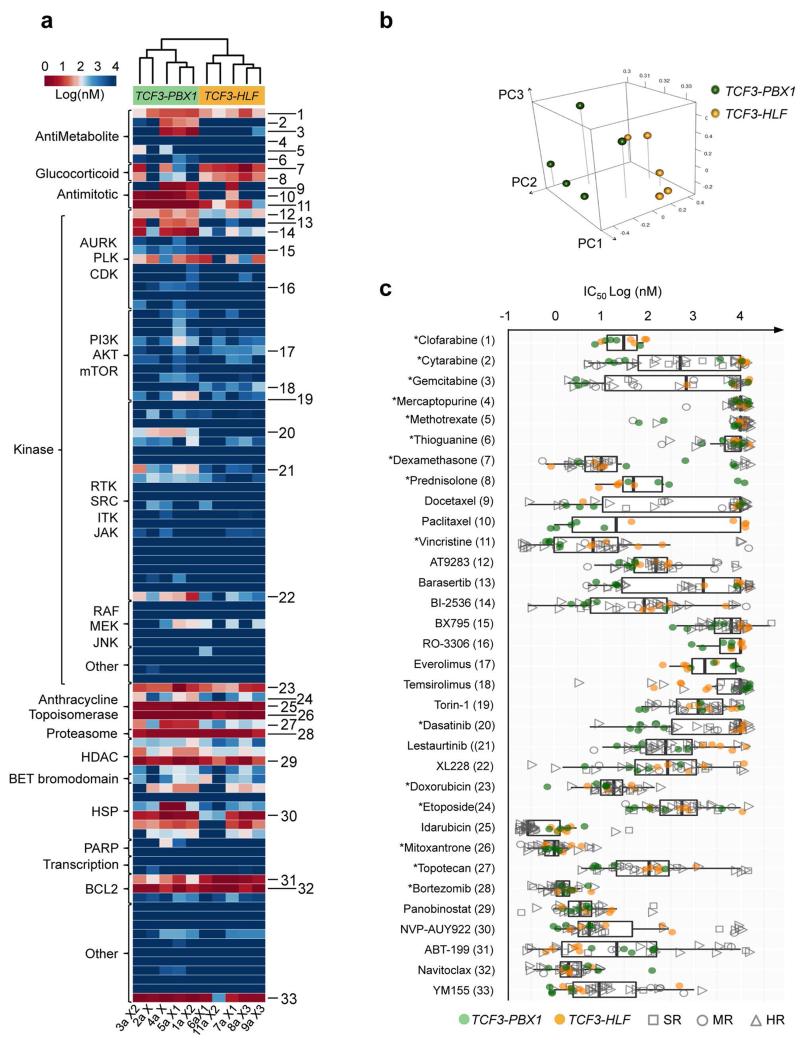
To determine drug sensitivity and resistance profiles, we established ALL co-cultures on human mesenchymal stromal cells (MSC) under serum-free conditions.39 Both subtypes depend on stroma for survival (Supplementary Fig. 9). TCF3-PBX1-positive ALL had a higher proportion of cells in S-phase than TCF3-HLF-positive ALL on such cultures, reflecting consistent biological differences. By screening 98 bioactive agents, including many agents in clinical development (Supplementary Table 23), on an automated-microscopy-based platform, we unambiguously discriminated the two translocations based on their drug sensitivity profiles, using either single (logIC50, Fig. 5a, Supplementary Fig. 10) or multiple response parameters (logIC50, logEC90, logEC50 and AUC, Fig. 5b and Supplementary Table 24). To capture informative differences, we compared the responses of xenografts derived from TCF3-HLF-positive ALL to xenografts derived from other high-risk pre-B and T ALL patients on the same platform (Fig. 5c and Supplementary Table 25). This provided information about the activity range of each drug on the respective ALL subtype. TCF3-HLF-positive cases were consistently more resistant to various drugs from the same class, including nucleotide analogs (e.g. cytarabine), mitotic spindle inhibitors (e.g. vincristine), polo-like and aurora kinase inhibitors. Given the importance of cytarabine and vincristine in standard ALL therapy, the implications of these observations need to be further explored. TCF3-HLF-positive ALL was very resistant to dasatinib in this assay, while TCF3-PBX1-positive ALL responded well. This partly challenges a recent report,40 which proposed dasatinib as an alternative for the treatment of these leukemias based on strong in vitro activity in one TCF3-HLF- and ten TCF3-PBX1-positive primary ALL samples. However, in vivo studies will be required to verify these differences in drug response, as differences in cell cycle activity may influence the pattern of response in vitro.
Figure 5. Drug activity profiling of TCF3-translocated leukemia reveals relevant differences in drug sensitivity.
(a) Unsupervised clustering based on the drug activity profile of 98 compounds (logIC50) separates the two subtypes. Fitted values are provided in the supplementary section (absolute IC50). Numbers identify the compounds shown in (c). (b) Principal component analysis of the response variables IC50, EC50, EC90 and AUC (Supplementary Table 24) show TCF3-PBX1-positive and TCF3-HLF-positive ALL in two distinct clusters. The separation of TCF3-PBX1-positive and TCF3-HLF-positive ALL is determined by responses to topoisomerases, BCL2 inhibitors, glucocorticoids and antimitotic agents, which correlate with the first three principal components. (c) Selection of drugs based on differences in sensitivity or resistance in TCF3-PBX1-positives and TCF3-HLF-positives. For comparison, the corresponding drug activity is indicated for 25 additional ALL samples tested on the same platform, including standard risk (SR, n=5), medium risk (MR, n=4), and high risk (HR, n=16) cases (Supplementary Table 25). Boxplots extend from the first to the third quartiles (hinges) of the response range for each compound. Whiskers correspond to values from the hinge to the lowest or highest values within 1.5x of the distance between the first and third quartiles, respectively. Drugs with differential activity include docetaxel, paclitaxel, vincristine, AT9283, barasertib, BI2536, torin-1, dasatinib, lestaurtinib and XL228 (p≤0.05). Drugs which are active across the patients include doxorubicin, idarubicin, mitoxantrone, bortezomib, panobinostat, NVP-AUY922, ABT-199 (venetoclax) and navitoclax. (For details see Source file for Figure 5.)
TCF3-HLF-positive ALL were sensitive to glucocorticoids (prednisone, dexamethasone) and to other drugs that could be relevant for the treatment of resistant ALL, including mTOR inhibitors, anthracyclines, bortezomib, the HSP90 inhibitor AUY922, and panobinostat. However, in spite of the good response of patients with TCF3-HLF-positive leukemia to prednisone therapy and the observed responsiveness of TCF3-HLF-positive ALL cells to glucocorticoids and anthracyclines that are commonly used in ALL treatment, patients eventually will relapse. Our transcriptome data suggested that resistance to apoptosis due to high expression of the anti-apoptotic oncoprotein BCL2 might promote cancer cell survival and constitute a druggable target (Supplementary Fig. 11). BCL2 is a putative transcriptional target of TCF3-HLF14. Of note, PAX5, commonly deleted in our cohort, normally represses BCL2 transcription41.
TCF3-HLF ALL is extremely sensitive to the BCL2 antagonist venetoclax
To assess the role of BCL2 overexpression in TCF3-HLF-positive ALL and to provide preclinical evidence for therapeutic activity, we tested the BCL2-targeting drug venetoclax (ABT-199) in our xenograft model (Fig. 5c). This BH3-mimetic compound is a highly specific small molecule inhibitor that competes with pro-apoptotic BCL2 family proteins for binding to BCL2 and shifts the balance of pro-death and pro-survival signals inside the cell in favor of cell death42. Venetoclax is in clinical development (phase II and III trials) for chronic lymphocytic leukemia and lymphoma, and holds promises for ALL and acute myeloid leukemia (AML).
TCF3-HLF-positive ALL samples were more sensitive to venetoclax than TCF3-PBX1-positives (Fig. 6a), which correlated with higher BCL2 transcript and protein expression levels (Fig. 6b). A two-week treatment course of daily venetoclax administration delayed leukemia progression significantly in ALL xenografts from three different TCF3-HLF-positive cases (Fig. 6c and 6d). Treatment of mice in the control arm that reached maximal leukemia burden resulted in very rapid reduction of the leukemic load (Fig. 6e). Xenografts from MRD or relapse remained sensitive to venetoclax (Supplementary Fig. 11). Profiling of primary cells from two additional cases with refractory ALL confirmed exquisite sensitivity to venetoclax (Supplementary Fig. 12). Combined treatment of patient-derived xenografts from patients 6 to 11 with venetoclax and either vincristine or dexamethasone indicated a potentially synergistic effect in some of those patients (Supplementary Fig. 13, Supplementary Table 26). Our data identify BCL2-dependency in TCF3-HLF ALL as a druggable target and illustrate how integration of drug response profiling and molecular genetic analyses can inform the development of innovative treatment strategies in patients with unmet therapeutic needs.
Figure 6. The BCL2 antagonist ABT-199 (venetoclax) shows promising anti-leukemic activity in TCF3-HLF-positive xenografts.
(a) In vitro dose response curves normalized against DMSO treated controls. (b) Merged absolute RPKM values of xenografts derived from the same primary leukemia sample (upper panel) and western blot for BCL2 (lower panel) in patient-derived xenografts as indicated. (c,d) In vivo response to ABT-199 on TCF3-HLF-positive xenografts. Treatment (grey bar) with 100 mg kg-1 qd ABT-199 (magenta) or with vehicle control (turquoise) were administered orally for 14 days (6-8 mice per treatment arm). Two treatment courses were administrated to xenograft 7a. For survival analysis an event was defined when at least 25% of leukemic cells were detected by FACS (mCD45−hCD19+hCD45+) in the peripheral blood. Differences in the survival of mice receiving ABT-199 or vehicle control were determined by the Mantel-Cox test and verified by the Gehan-Breslow-Wilcoxon test.
(e) Mice from the control arm of (c,d) were treated with ABT-199 when more than 50% of ALL cells were detected in the blood. Mean and SD are shown (n=4). (For details see Source file for Figure 6.)
DISCUSSION
To our knowledge, long-term cure has never been achieved for patients with TCF3-HLF-positive ALL. Our study revealed a recurrent pattern of TCF3-HLF accompanied by abnormalities that affect transcriptional regulation of lymphoid development. We found frequent deletions of PAX5 and VPREB1 in association with TCF3-HLF, but deletions of Ikaros family members, which are commonly affected in ALL1,23, were not detected. We also uncovered recurrent mutations of the transcription factor TCF3, which acts upstream of PAX5 in lymphoid development, potentially impairing structural interactions with other transcription cofactors29. PAX5 expression was reduced by two-fold in all TCF3-HLF-positive cases, underscoring the possibility of an interaction between TCF3-HLF, TCF3 and PAX5. PAX5 is required for B-lymphoid lineage commitment and maturation43 and frequently deleted in high-risk ALL with complex patterns of copy number abnormalities23. Similarly, deletions in IKFZ1, which is required for the development of B- and T-lymphoid lineages and has additional stem cell-like functions44, are detected both in high-risk BCR-ABL1-positive and -negative ALL and in the more favorable ERG-altered ALL subtype45, but never in TCF3-HLF-positive ALL. Focal deletions of VPREB1 were also detected in TCF3-HLF ALL which may lead to a developmental arrest associated with lack of pre-B cell receptor formation and the resulting loss of negative feedback on RAG-mediated recombination46. VPREB1 deletions were present at a similar frequency compared to other high-risk ALLs, such as BCR-ABL1-like and BCR-ABL1-positive ALL (approximately 30-40% of cases)47 or hypodiploid ALL (≈30%)24, associated with poorer overall survival in high-risk pre-B cell ALL patients24. However, specific ALL subtypes associated with good prognosis (e.g. ETV6-RUNX1-positive ALL) also present high frequencies of VPREB1 deletions24, suggesting an important impact of the genomic context24. Thus, distinct patterns of association emerge that are likely to reflect important underlying biological mechanisms. Based on our results, we propose that a reduction of PAX5 gene dosage constitutes a favorable context for the oncogenic activity of TCF3-HLF.
As observed for hypodiploid ALL5 and in subsets of MLL-rearranged ALL48, we identified mutations in NRAS, KRAS and PTPN11 in TCF3-HLF-positive ALL. In our xenograft models we detected variable persistence of NRAS and a switch to KRAS mutations, indicating that RAS mutations are multiclonal and might not be strictly required for disease progression in TCF3-HLF-positive ALL. Indeed, mutations in the RAS pathway are enriched at relapse in ALL7,30,48, but mostly in a subclonal pattern with losses or switches in NRAS and KRAS from diagnosis to relapse. These represent secondary events, possibly compensating functional effects of the initiating events. Mutations in the RAS pathway might not represent optimal therapeutic targets, given their volatility and the potential to select for slower proliferating, more resistant subclones. The TCF3-HLF gene expression signature, enriched for components of stem cell and myelomonocytic stages, was very similar among leukemias and maintained in xenografts, specifying additional, novel markers associated with stem cell function, such as LGR5, which marks epithelial stem cells49 and embryonic and fetal hematopoietic progenitor cells in mice50. Thus, in analogy to experimental induction of pluripotent stem cells51,52, TCF3-HLF likely induces a whole set of factors that carry out reprogramming and leukemic transformation in the context of low PAX5 expression. Deletion of PAX5 in early B cell progenitors induced dedifferentiation to a state with myeloid and T cell potential43,53. Moreover, rescue with low expression levels of PAX5 in knock-out mice generates a stalled biphenotypic B-lymphoid/myeloid state54. Together with an activating mutation in STAT5, PAX5 haploinsufficiency initiates ALL in mice41. Based on these data, we propose that the initiating TCF3-HLF fusion results in severe transcriptional reprogramming with dedifferentiation. The favorable context for transformation is secured in second line through additional lesions in early B cell differentiation genes including TCF3 and PAX5.
A central question remains pertaining to the cell of origin in different ALL subtypes. Our study provides significant clues that should be further addressed using disease models. The molecular analysis of the TCF3-HLF and TCF3-PBX1 fusion gene breakpoints indicated that the TCF3-HLF, like the TCF-PBX1 translocation, originates in cells already committed to lymphoid differentiation. Furthermore, the associated somatic structural variants were found to be RAG-mediated, which is comparable to patterns identified recently in ETV6-RUNX1-positive ALL, the most frequent pre-B cell ALL subtype, which is consistent with expression of RAG in TCF3-HLF-positive ALL55. We favor the hypothesis that the TCF3-HLF translocation occurs in a B cell progenitor and that the specific lineage context is constrained further in a restricted developmental stage by additional mutations. The detection of TCF3-HLF being restricted to leukemic cells supports this idea, although initiation in a more immature compartment cannot be formally excluded.
The molecular landscapes of TCF3-HLF-positive ALL were largely conserved in xenografts, providing a valuable, well characterized, model for preclinical testing. Drug activity profiling revealed that TCF3-HLF-positive cases were more resistant to several standard ALL drugs, such as nucleotide analogues (e.g. cytarabin) and mitotic spindle inhibitors (e.g. vincristine). Activity was detected for other relevant drug classes, such as mTOR inhibitors, the proteasome inhibitor bortezomib, the HSP90 inhibitor AUY922, and the HDAC inhibitor panobinostat. The BCL2 inhibitor venetoclax (ABT-199)42 was highly active in all TCF3-HLF-positive cases analyzed, which we confirmed using primary ALL cells from two additional cases with refractory disease. These results refine data obtained using the broader spectrum BH3-mimetic ABT-737 in TCF3-HLF-positive cell lines14. Given the activity of venetoclax also in other ALL subsets including immature T ALL (56,57 and own unpublished data) and the lack of on-target thrombocytopenia caused by ABT-737, venetoclax should be explored for experimental therapy in refractory ALL in selected cases based on such functional data. Thus integrated genomic and functional analyses of TCF3-HLF-positive ALL provide novel insight into the molecular context and associated components and offer unprecedented possibilities to investigate new agents for the treatment of these children who currently lack effective therapeutic options.
ONLINE METHODS
Study individuals and sample selection
Samples and associated clinical information from patients included in sequencing and validation analyses were collected from different countries within the International BFM-Study Group (IBFM-SG). All patients were enrolled in multicenter trials on treatment of pediatric ALL conducted by individual member groups of the I-BFM-SG: the AIEOP-BFM study group (Austria, Germany, Italy, Switzerland), the FRALLE study group (France), and the United Kingdom (UK) National Cancer Research Institute (NCRI) Childhood Cancer and Leukaemia Group61,62. All treatment trials were approved by the respective national institutional review boards and informed consent for the use of spare specimens for research was obtained from study individuals, parents or legal guardians. The specific research project reported here was approved by the Ethics Committee of the Medical Faculty of the Christian-Albrechts-University, Kiel, Germany (vote D508/13). Depending on consent and availability of samples, all enrolled patients positive for the rare TCF3-HLF gene fusion were included. These patients were matched with TCF3-PBX1-positive patients. Download of sequencing data is available from the public POPGEN repository (Christian-Albrechts-University, Kiel) upon written request accompanied by a positive internal review board vote for research addressing leukemia-related questions. Data transfer can proceed upon positive review and signing of a material transfer agreement.
Cell isolation and nucleic acid purification
Mononuclear cells (MNCs) were isolated by Ficoll-Paque gradient centrifugation (Pharmacia) from bone marrow or peripheral blood samples followed by extraction of nucleic acids according to standardized protocols using Qiagen DNA Blood Kits (Qiagen) for DNA and Qiagen RNeasy columns (Qiagen) for RNA. The quantity of nucleic acids was determined by spectrophotometry. DNA quality was assessed visually by inspection of agarose gel electrophoresis while RNA integrity was evaluated by using the Bioanalyzer 2100 (Agilent). Nucleic acids isolated from bone marrow aspirates collected in morphological remission served as individual germline surrogates/references.
Sequencing
Whole genome sequencing
For structural variants (SVs), Illumina v2 mate-pair libraries with 5 kbp insert size and 2 × 101 bp reads were prepared from 10 μg of DNA and sequenced on the Illumina HiSeq 2000 platform (Illumina) to obtain a physical coverage of 30x. For copy number alterations, breakpoints and short variants (SNVs, short indels), Illumina TruSeq paired-end libraries with 2 × 101 bp reads were prepared from 1 μg of DNA and sequenced on HiSeq 2000/2500 instruments to a coverage of 40x for reference samples and 80x for tumor samples.
Whole exome sequencing
For increasing the sensitivity of detecting short variants in coding regions, 1 μg of DNA each from the diagnostic leukemic and a corresponding remission sample of patients was used for whole exome sequencing. Whole exome capture employed a TruSeq enrichment kit (Illumina) and paired-end libraries with 2 × 101 bp reads on a HiSeq 2500 according to the manufacturer’s protocol.
Whole transcriptome sequencing
Illumina TruSeq custom stranded paired-end libraries with 2 × 51 bp reads were prepared from 1 μg RNA using the Ribo-Zero Gold Kit (Epicentre) and sequenced on a HiSeq 2000 with a loading of one library per lane.
Sanger sequencing validation
Structural variant breakpoints from mate-pair genome sequencing and SNVs from exome sequencing were validated by Sanger sequencing.
Targeted sequencing of TCF3 and RAS pathway candidate genes
TCF3 binding domain (E47 isoform, exon 18) mutations were screened for in 1033 ALL patients using Sanger sequencing. Primer sequences are listed in Supplementary Table 27. Sanger sequencing was also applied for validation of relative absence of RAS pathway mutations in 24 TCF3-PBX1-positive ALL samples. The latter analysis included KRAS exon 1, NRAS exons 1 and 2, FLT3 exons 14 and 20, PTPN11 exons 3 and 13 and was conducted as previously described63.
Multiplex Ligation-dependent Probe Amplification
Detection of genomic aberrations in B cell differentiation-associated and other genes frequently deleted in ALL (PAX5, IKZF1, ETV6, RB1, BTG1, EBF1, CDKN2A, CDKN2B, and P2RY8-CRLF2) were investigated by the Multiplex Ligation-dependent Probe Amplification (MLPA) assay SALSA p335 kit (MRC-Holland) using 125 ng of genomic DNA. The assays were performed according to the manufacturer’s protocol as previously described64. An intensity ratio between 0.75 and 1.3 was considered to represent a normal copy number, a ratio between 0.25 and 0.75 a monoallelic deletion, and a ratio <0.25 a biallelic deletion.
Bioinformatic analysis
DNA data processing
DNA reads were aligned to the human reference genome hg19 (downloaded from the UCSC Genome browser) using Elandv265 (mate-pairs) and BWA66 (paired-ends). For xenograft samples, the human DNA reads were deconvoluted after mapping to a combined reference consisting of human hg19 and mouse mm9.
Structural variant detection
Structural variants (SVs) were detected using DELLY67 and BIC-seq68 (DNA data) and TopHat269/deFuse70 (RNA data).
SNV detection
Somatic protein-changing SNVs were detected using established pipelines incorporating GATK71, MuTect72, pibase73, Picard, SAMtools74 and VarScan275.
Indel detection
Somatic indels in coding regions were detected using SAMtools followed by Dindel76.
Transcriptome data analysis
RNA reads were aligned to hg19 using BWA and SAMtools and used for integrated data analysis. For xenograft samples, the human RNA reads were deconvoluted after mapping to a combined reference consisting of human hg19 and mouse mm9. Mapped reads were annotated using Ensembl v.70. Gene expression levels were quantified in reads per kilobase of exon model per million mapped reads (RPKM)77. RPKM calculation and differential gene expression (DGE) analysis was performed using the R package edgeR78. To identify DGE between ALL subtypes, and between leukemia and remission the following set-up was performed: TCF3-PBX1 vs. TCF3-HLF (comparison 1), TCF3-PBX1 vs. remission (comparison 2), TCF3-HLF vs. remission (comparison 3). The results were filtered by fold change (FC, |log2(FC)| >= 1) and false discovery rate (FDR, FDR <= 0.001). The final list of 401 genes was created by combining the intersection between comparison 1 and comparison 2 as well as between comparison 1 and comparison 3. The functional analyses of gene lists were done using Gene Set Enrichment Analysis (GSEA)59 and the Genomatix genome analyzer (v. 3.00801; Genomatix Software GmbH). The GeneRanker tool in Genomatix was used to test for enriched gene sets, which were based on gene-tissue annotations obtained by text mining60. For GSEA, protein-coding genes were filtered by a minimum expression of 1 RPKM in at least four samples among the primary pre-B cell ALLs. The remaining 11315 genes were tested for DGE between the ALL subtypes using edgeR. The provided FDR and fold-change values were used to obtain a ranking score to measure the degree of differential expression between the ALL subtype. A pre-ranked classic GSEA was performed using the ranking score, a gene set permutation and a FDR ≤ 0.02. The analysis included gene sets for hematopoietic stages31 and signatures from MsigDB59 pathways (C2): KEGG, BIOCARTA, REACTOME; curated oncogenic signatures (C6); human immunologic signatures (C7).
In silico transcription factor binding site (TFBS) analysis
TFBS in promoter regions of genes (2 kbp upstream region) corresponding to the specific transcriptome signatures of TCF3-PBX1- and TCF3-HLF-positive ALL, respectively, were analyzed using the Genomatix Genome Analyzer (v3.10124). Based on a matrix of known TFBS motifs, the software tool predicted TFBSs in the investigated promoters and compared their frequency against 1) the background of TFBSs in the promoter regions of all known protein-coding genes in the Ensembl database (v.70, 22864 genes) and 2) the background of TFBSs in the whole genome. A Z-score was calculated based on the TFBS frequency in the investigated promoters and the expected frequency and standard deviation were estimated from the background79. The resulting lists were filtered by the Z-scores based on the two backgrounds ( |genomic Z-score| ≥ 2 , promoter Z-score ≥ 2 ). TFBSs overrepresented in genes up-regulated in both TCF3-PBX1- and TCF3-HLF-positive ALL were filered out, to retain only TFBS specifically enriched in the respective subtypes.
Integrated data analysis
SNVs and indels were orthogonally validated by integrating genome, exome, and transcriptome data of patients and xenografts and further confirmed by Sanger sequencing. SVs were validated by integrating whole genome paired-end and mate-pair data and whole transcriptome data and finally by Sanger sequencing. Ensembl v.70 and ANNOVAR80 were used to annotate the variants. Silent variants and known germline variants in the 1000 Genomes Project81 population data, in 136 North German healthy controls (publicly available through GrabBlur82), or in the International Cancer Genome Consortium’s internal healthy controls were eliminated. All final somatic non synonymous variants were inspected using IGV83.
Preclinical characterization
Xenograft model
Animal experiments were approved by the veterinary office of the Canton of Zurich, Switzerland. Approval for experiments with human samples in the mouse xenograft model was obtained from the ethics commission of the Canton Zurich (approval number 2014-0383). In brief, primary ALL cells were recovered from cryopreserved samples and transplanted intrafemorally to NSG mice as previously described36. Mice were between 5 to 10 weeks old, both males and females were randomly used. Leukemia progression was monitored by flow cytometry with rat anti-mouse CD45 (eFluor®450, Clone 30-F11, REF 48-451-82, eBioscience), mouse anti-human CD45 (AlexaFlour647, Clone HI30, REF 304018, BioLegend), and mouse anti-human CD19 (PE, Clone HIB19, REF 302208, BioLegend). ALL cells recovered from spleens of NSG mice were used for molecular characterization in in vitro and in vivo experiments.
Immunophenotyping
Immunophenotyping of patient and xenograft-amplified human ALL cells after recovery from the spleen was performed as described before84. All included xenograft samples consisted of at least 95% human leukemic cells.
Cell culture
Human hTERT immortalized primary bone marrow mesenchymal stromal cells (MSC; provided by D. Campana, St. Jude Children’s Research Hospital, Memphis, TN) were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS h.i.); L-glutamine (2 mM), penicilline/streptomycin (P/S; 100 IU/ml) and hydrocortisone (1 μM). Xenograft-amplified human ALL cells were co-cultured on MSC in AIM V medium (Gibco by Life Technologies) at a ratio of 10:1. All cultured cells were kept in the incubator at 37°C, 5% CO2. For cryopreservation, cells were frozen in FBS h.i. with 10% dimethylsulfoxide and subsequently stored in liquid nitrogen.
Cell viability assay
MSCs were seeded in 24-well plates at a number of 50,000 cells per well in RPMI 1640 medium (10% FBS h.i.). After 24 hours primary ALL cells were thawed and seeded as suspension culture alone or in co-culture with MSCs at a number of 400,000 cells per well in AIM-V medium. Three days later, ALL cells were collected from monoculture or co-culture by scraping and stained with 7-AAD (BD Pharmingen). Cell viability (7-AAD negative population) was measured by FACS using counting beads (SPHERO Accu Count Blanc Particles, Spherotech Inc.) for cell counts normalization. Viabilities shown are average viabilities of duplicate wells (normalized to input) and standard deviations.
Cell cycle assay
MSCs were seeded in 96-well tissue culture plates at a concentration of 10,000 cells per well in 100 μL AIM-V medium. After 24 hours ALL cells were added at a concentration of 100,000 cells per well in 90 μL AIM-V. The Click-iTEdU Alexa Fluor 488 Flow Cytometry Assay Kit (Life Technologies) in combination with propidium iodide was used to measure proliferation and to identify the different phases of the cell cycle on days 1 and 3. Co-cultured cells were incubated with EdU (10 μM) for 20 hours before cell cycle read-out with flow cytometry. The cell cycle assay was performed in triplicates and at least two independent experiments were performed for each sample. Similar variances were obtained between the groups that were statistically compared.
In vitro drug screening and automated microscopy
MSCs were seeded in 384-well plates at a concentration of 2,500 cells per well in 30 μL AIM-V medium. After 24 hours, ALL cells were added at a concentration of 25,000 to 30,000 cells per well in 27.5 μL AIM-V. Drugs were added as single agents after additional 24 hours using the pipetting robot epMotion 5070 (Eppendorf). Drug response was normalized to ALL cells treated with the drug vehicle alone. Experiments were performed in duplicates in five different dilutions (1, 10, 100, 1,000 and 10,000 nM). For two samples comparable results were obtained in two independent drug screening experiments. After 72 or 96 hours of drug incubation, cells were stained using the CyQUANT direct cell proliferation assay (Life technologies). 20 μL staining mix (AIM V medium, CyQUANT (1:300), repressor (1:20)) was added into each well followed by an incubation time of 1 hour at 37°C, 5 % CO2. Subsequently, automated imaging was performed using the ImageXpress Micro microscope (Molecular Devices) equipped with a CoolSNAP HQ camera (Photometrics) and a 10x plan fluor objective with 0.3 NA (Nikon). Nine images were taken per well, covering 50% of each well and captured employing the MetaXpress software (Molecular Devices). Images were processed using CellProfiler software (Broad Institute, Cambridge, MA). Cells were classified and counted using the Advanced Cell Classifier software. This software uses random forest classification to assign ALL cells properly.
Western blot
Whole cell extracts were prepared from 1 × 106 cells using radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-Cl pH 7.5, 150 mMNaCl, 1% NP-40, 1 mM EDTA pH 8.0, 0.1% SDS) supplemented with Complete mini protease inhibitor cocktail (Roche Life Science) for 20 minutes on ice, sonicated as necessary, and diluted with SDS loading buffer (250 mM Tris pH 6.8, 4% SDS, 0.02% bromophenol blue, 40% glycerol, 4% (v/v) β-mercaptoethanol). After SDS-PAGE, proteins were blotted onto nitrocellulose membranes. Membranes were blocked in 5% non-fat dry milk and incubated with primary Bcl-2 (Clone 124; Dako) and tubulin antibodies diluted 1:1,000 in milk. Horseradish peroxidase-labeled anti-mouse antibodies were used for signal detection with chemiluminescence substrate and direct scanning.
In vivo experiments
ALL cells were recovered from cryopreserved xenograft samples, and per thawed sample 12 to 16 mice were transplanted with 1,000,000 cells per mouse. After three days, randomized cohorts were treated with 100 mg/kg of ABT-199 (ABBVIE) or vehicle control with 6 to 8 mice per treatment arm85. ABT-199 or vehicle control were administered orally daily for two weeks. Mice of the ABT-199 group transplanted with sample 7a were additionally treated with a second block (100 mg/kg of ABT-199 for 14 days) starting at day 66, when the frequency of circulating leukemia cells started to increase again. Follow-up of circulating leukemia cells was performed every seven days by flow cytometry with rat anti-mouse CD45, mouse anti-human CD45, and mouse anti-human CD19; frequency of leukemia cells as ratio of mCD45-hCD45+ hCD19+ count to total lymphocytes. The investigator was blinded to the group allocation during the assessment of outcome. To evaluate the ability of ABT-199 to decrease tumor burden, four mice in the control group were treated when the frequency of leukemia cells in the peripheral blood was equal or higher than 50%. Follow-up of circulating leukemia cells was performed every four to seven days. In vivo experiments were terminated when the frequency of circulating leukemia cells reached 50% or earlier if the mice showed abnormal behavior. One in vivo experiment was performed per each sample.
Statistical analysis
Differences in the distribution of categorical variables among patient subsets were analyzed using Fisher’s exact or chi-squared test. Comparisons of continuous variables between groups were performed by t-test or Mann-Whitney U test.
Drug responses were evaluated by fitting DMSO-normalized response data with the four-parameter log-logistic function of the form:
as implemented in the drc package of R (version 2.3-96). Outliers were detected and removed prior to curve-fitting using Bayesian change point analysis25 (R package bcp, version 3.0.1). Non-convergent cases (e.g., drugs with no activity) were identified based on linear fit parameters. Hierarchical clustering was performed to group patients according to their drug-response profiles (R package gplots version 2.14.2). Drugs with differential activity in patients with TCF3-PBX1- compared to TCF3-HLF-positive ALL were identified using a t-test (p-value <= 0.05). In in vivo experiments, 25% of circulating leukemia cells or termination of the experiment if 25% of leukemia was not reached were considered as an event in the Kaplan-Meier analysis. For sample 9a, 50% was used because of the rapid engraftment. Differences in the survival of mice receiving ABT-199 or vehicle control were determined by the Mantel-Cox test and verified by the Gehan-Breslow-Wilcoxon test.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all participants and personnel involved in the involved clinical trials in Austria, France, Germany, United Kingdom and Switzerland. We thank Thomas Radimerski and Novartis for providing essential compounds. We thank the Leukaemia & Lymphoma Research (LLR) Childhood Leukaemia Cell Bank in the UK for providing primary patient samples. This work was supported by the German Federal Office for Radiation Protection (Grant no. St.Sch. 3611S70014), by the Swiss National Research Foundation SNF 310030-133108, the foundation “Kind und Krebs”, the “Krebsliga Zurich”, the Sassella Foundation, the Fondation Panacée, the clinical research focus program “Human Hemato-Lymphatic Diseases“ of the University of Zurich, the “Deutsche Forschungsgemeinschaft” (DFG), Clusters of Excellence “Inflammation at Interfaces”, the EU Seventh Framework Program [FP7/2007-2013, Grant no. 262055, ESGI; FP7-HEALTH-F2-2011 Grant no. 261474, ENCCA; ERA-Net Transcan, Validation of biomarkers for personalized cancer medicine, TRANSCALL; Health-F2-2010 Grant no. 260791, EUROCANPLATFORM], the “Katharina Hardt Stiftung”, the “Deutsche José Carreras Leukämie-Stiftung”, the “Madeleine Schickedanz-Kinderkrebs-Stiftung“, the “Deutsche Krebshilfe – Dr. Mildred Scheel Stiftung” (Grant no. 108613, 102588, 108588), the Foundation of Experimental Biomedicine in Zurich, the Max Planck Society, and the “Verein für krebskranke Kinder Hannover e.V.”. We thank Drs. Anne Dehos, Bernd Grosche, Thomas Jung, Wolfgang Weiss, and Gunde Ziegelberger, German Federal Office for Radiation Protection, as well as Drs. Birger Heinzow, State Office for Social Services of Schleswig-Holstein, and Axel Böttger, German Federal Ministry for the Environment, Nature Conservation, Building and Nuclear Safety for their support and critical discussions. We are grateful for the excellent technical assistance offered by the sequencing team of the Department of Vertebrate Genomics of the Max Planck Institute for Molecular Genetics (Berlin) and by the team of the Genomics Core facility of the European Molecular Biology Laboratory. We further thank Katayoun Alemazkour for excellent technical assistance regarding whole exome sequencing at the Department of Pediatric Oncology, Hematology and Clinical Immunology (Düsseldorf). We cordially thank Prof. Nikolaus Forgo, Institute for Legal Informatics, Leibniz University Hannover, and Prof. Hans-Dieter Tröger, Hannover Medical School, for legal and ethical counselling.
AUTHOR CONTRIBUTIONS
A.B., A.F., J.O.K., J.-P.B., M.-L.Y., and M.S. jointly designed the project. A. Baruchel, A.V., A.V.M., C.C., C.P.K., F.N., G.B., G.C., G.t.K., H.C., M.Sch., M.S., N.v.d.W., and R.P.-G. provided samples or clinical data. A.B., A.C.M., A.F., A.V., C.E., G.H.-S., H.L., J.P.B., J.O.K., J.T., M.D., M.F., M.-L.Y., M.P.D., M.Sch., M.S., M.Z., O.A.H., P.R., S.S., and T.Z. contributed reagents, materials or analysis tools. A.M.S., B.B., B.M., C.E., C.L.W., H.-J.W., J.I.H., J.-P.B., M.F., M.G., S. Sungalee, and U.F. designed experiments. A.M.S., A.R., B.B., B.R., B.S., B.S.P., C.E., C.K., C.L.W., D.D., D.S., H.-J.W., J.T., K.H., L.Z., M.G., M.R., M.Z., M. Sultan, P.K., S.E., S. Sungalee, T.B., U.F., and V.F. performed experiments. A.R., B.S., B.S.P., C.E., C.K., C.L.W., D.S., E.E., J.I.H., H.-J.W., M.G., M.P.D., M.F., N.B., S.G., G.H., P.H., P.K., M.-L.Y., M.R., M.S., M. Schütte, M.Z., S. Sungalee, T.R., U.F., and V.A. analyzed data. A.B., A.C.M., A.F., B.B., H.L., J.-P.B., J.O.K., M.D., M.F., M.-L.Y., M.S., O.H., R.T., and U.F., supervised research. A.R., C.L.W., D.S., H.-J.W., M.F., M.P.D., M.S., P.K., S. Sungalee, T.R., and U.F., prepared tables and figures. J.-P.B., M.S., and M.-L.Y. wrote the manuscript. A.B., A.F., A.R., A.V.M., H.-J.W., J.O.K., M.F., M.P.D., O.H., S.H., S. Sungalee, T.R., and U.F. contributed to the writing of the manuscript. All authors critically reviewed the manuscript for its content.
Footnotes
URLs
Information on the two image processing programs used for in vitro drug screening and automated microscopy can be found at http://acc.ethz.ch/.
DISCLOSURE OF CONFLICTS OF INTEREST
There are no competing financial interests.
REFERENCES
- 1.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 2.Russell LJ, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114:2688–2698. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- 3.Mullighan CG, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hertzberg L, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115:1006–1017. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- 5.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2011;118:3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irving J, et al. Ras pathway mutations are highly prevalent in relapsed childhood acute lymphoblastic leukaemia, may act as relapse-drivers and confer sensitivity to MEK inhibition. Blood. 2014;124:3420–3430. doi: 10.1182/blood-2014-04-531871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381:1943–1955. doi: 10.1016/S0140-6736(12)62187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon K, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Felice MS, et al. Prognostic impact of t(1;19)/ TCF3-PBX1 in childhood acute lymphoblastic leukemia in the context of Berlin-Frankfurt-Munster-based protocols. Leuk Lymphoma. 2011;52:1215–1221. doi: 10.3109/10428194.2011.565436. [DOI] [PubMed] [Google Scholar]
- 11.Hunger SP, Ohyashiki K, Toyama K, Cleary ML. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 1992;6:1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- 12.Inukai T, et al. Hypercalcemia in childhood acute lymphoblastic leukemia: frequent implication of parathyroid hormone-related peptide and E2A-HLF from translocation 17;19. Leukemia. 2007;21:288–296. doi: 10.1038/sj.leu.2404496. [DOI] [PubMed] [Google Scholar]
- 13.Boller S, Grosschedl R. The regulatory network of B-cell differentiation: a focused view of early B-cell factor 1 function. Immunol Rev. 2014;261:102–115. doi: 10.1111/imr.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Boer J, et al. The E2A-HLF oncogenic fusion protein acts through Lmo2 and Bcl-2 to immortalize hematopoietic progenitors. Leukemia. 2011;25:321–330. doi: 10.1038/leu.2010.253. [DOI] [PubMed] [Google Scholar]
- 15.Hirose K, et al. Aberrant induction of LMO2 by the E2A-HLF chimeric transcription factor and its implication in leukemogenesis of B-precursor ALL with t(17;19) Blood. 2010;116:962–970. doi: 10.1182/blood-2009-09-244673. [DOI] [PubMed] [Google Scholar]
- 16.Inukai T, et al. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol Cell. 1999;4:343–352. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- 17.Inoue A, et al. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer Cell. 2002;2:279–288. doi: 10.1016/s1535-6108(02)00155-1. [DOI] [PubMed] [Google Scholar]
- 18.Honda H, et al. Expression of E2A-HLF chimeric protein induced T-cell apoptosis, B-cell maturation arrest, and development of acute lymphoblastic leukemia. Blood. 1999;93:2780–2790. [PubMed] [Google Scholar]
- 19.Smith KS, Rhee JW, Naumovski L, Cleary ML. Disrupted differentiation and oncogenic transformation of lymphoid progenitors in E2A-HLF transgenic mice. Mol Cell Biol. 1999;19:4443–4451. doi: 10.1128/mcb.19.6.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunger SP. Chromosomal translocations involving the E2A gene in acute lymphoblastic leukemia: clinical features and molecular pathogenesis. Blood. 1996;87:1211–1224. [PubMed] [Google Scholar]
- 21.Wiemels JL, et al. Site-specific translocation and evidence of postnatal origin of the t(1;19) E2A-PBX1 fusion in childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2002;99:15101–15106. doi: 10.1073/pnas.222481199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai AG, et al. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moorman AV, et al. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood. 2014;124:1434–1444. doi: 10.1182/blood-2014-03-562918. [DOI] [PubMed] [Google Scholar]
- 24.Mangum DS, et al. VPREB1 deletions occur independent of lambda light chain rearrangement in childhood acute lymphoblastic leukemia. Leukemia. 2014;28:216–220. doi: 10.1038/leu.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waanders E, et al. The origin and nature of tightly clustered BTG1 deletions in precursor B-cell acute lymphoblastic leukemia support a model of multiclonal evolution. PLoS Genet. 2012;8:e1002533. doi: 10.1371/journal.pgen.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tijchon E, Havinga J, van Leeuwen FN, Scheijen B. B-lineage transcription factors and cooperating gene lesions required for leukemia development. Leukemia. 2013;27:541–552. doi: 10.1038/leu.2012.293. [DOI] [PubMed] [Google Scholar]
- 27.Balbin OA, et al. Reconstructing targetable pathways in lung cancer by integrating diverse omics data. Nat Commun. 2013;4:2617. doi: 10.1038/ncomms3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright DD, Sefton BM, Kamps MP. Oncogenic activation of the Lck protein accompanies translocation of the LCK gene in the human HSB2 T-cell leukemia. Mol Cell Biol. 1994;14:2429–2437. doi: 10.1128/mcb.14.4.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz R, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma X, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604. doi: 10.1038/ncomms7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurenti E, et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol. 2013;14:756–763. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 33.Schepers AG, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 34.Akahane K, et al. Specific induction of CD33 expression by E2A-HLF: the first evidence for aberrant myeloid antigen expression in ALL by a fusion transcription factor. Leukemia. 2010;24:865–869. doi: 10.1038/leu.2010.8. [DOI] [PubMed] [Google Scholar]
- 35.Nissim S, et al. Prostaglandin E2 regulates liver versus pancreas cell-fate decisions and endodermal outgrowth. Dev Cell. 2014;28:423–437. doi: 10.1016/j.devcel.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz M, et al. Xenografts of highly resistant leukemia recapitulate the clonal composition of the leukemogenic compartment. Blood. 2011;118:1854–1864. doi: 10.1182/blood-2010-11-320309. [DOI] [PubMed] [Google Scholar]
- 37.Bonapace L, et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest. 2010;120:1310–1323. doi: 10.1172/JCI39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stark Z, et al. Two novel germline KRAS mutations: expanding the molecular and clinical phenotype. Clin Genet. 2012;81:590–594. doi: 10.1111/j.1399-0004.2011.01754.x. [DOI] [PubMed] [Google Scholar]
- 39.Boutter J, et al. Image-based RNA interference screening reveals an individual dependence of acute lymphoblastic leukemia on stromal cysteine support. Oncotarget. 2014;5:11501–11512. doi: 10.18632/oncotarget.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bicocca VT, et al. Crosstalk between ROR1 and the Pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell. 2012;22:656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heltemes-Harris LM, et al. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J Exp Med. 2011;208:1135–1149. doi: 10.1084/jem.20101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 43.Rolink AG, Nutt SL, Melchers F, Busslinger M. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 1999;401:603–606. doi: 10.1038/44164. [DOI] [PubMed] [Google Scholar]
- 44.Joshi I, et al. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol. 2014;15:294–304. doi: 10.1038/ni.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clappier E, et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia. 2014;28:70–77. doi: 10.1038/leu.2013.277. [DOI] [PubMed] [Google Scholar]
- 46.Grawunder U, et al. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 47.Den Boer ML, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersson AK, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–337. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 50.Liu D, et al. Leucine-rich repeat-containing G-protein-coupled Receptor 5 marks short-term hematopoietic stem and progenitor cells during mouse embryonic development. J Biol Chem. 2014;289:23809–23816. doi: 10.1074/jbc.M114.568170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riddell J, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandler VM, et al. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 2014;511:312–318. doi: 10.1038/nature13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 54.Simmons S, et al. Biphenotypic B-lymphoid/myeloid cells expressing low levels of Pax5: potential targets of BAL development. Blood. 2012;120:3688–3698. doi: 10.1182/blood-2012-03-414821. [DOI] [PubMed] [Google Scholar]
- 55.Papaemmanuil E, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peirs S, et al. ABT-199 mediated inhibition of BCL-2 as a novel therapeutic strategy in T-cell acute lymphoblastic leukemia. Blood. 2014;124:3738–3747. doi: 10.1182/blood-2014-05-574566. [DOI] [PubMed] [Google Scholar]
- 57.Chonghaile TN, et al. Maturation stage of T-cell acute lymphoblastic leukemia determines BCL-2 versus BCL-XL dependence and sensitivity to ABT-199. Cancer Discov. 2014;4:1074–1087. doi: 10.1158/2159-8290.CD-14-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El Omari K, et al. Structural basis for LMO2-driven recruitment of the SCL:E47bHLH heterodimer to hematopoietic-specific transcriptional targets. Cell Rep. 2013;4:135–147. doi: 10.1016/j.celrep.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frisch M, Klocke B, Haltmeier M, Frech K. LitInspector: literature and signal transduction pathway mining in PubMed abstracts. Nucleic Acids Res. 2009;37:W135–140. doi: 10.1093/nar/gkp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conter V, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010;115:3206–3214. doi: 10.1182/blood-2009-10-248146. [DOI] [PubMed] [Google Scholar]
- 62.Harrison CJ, et al. Detection of prognostically relevant genetic abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: recommendations from the Biology and Diagnosis Committee of the International Berlin-Frankfurt-Munster study group. Br J Haematol. 2010;151:132–142. doi: 10.1111/j.1365-2141.2010.08314.x. [DOI] [PubMed] [Google Scholar]
- 63.Case M, et al. Mutation of genes affecting the RAS pathway is common in childhood acute lymphoblastic leukemia. Cancer Res. 2008;68:6803–6809. doi: 10.1158/0008-5472.CAN-08-0101. [DOI] [PubMed] [Google Scholar]
- 64.Dorge P, et al. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica. 2012;98:428–432. doi: 10.3324/haematol.2011.056135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bauer MJ, Cox AJ, Evers DJ. Fast gapped read mapping for Illumina reads. In ISMB, ISBC. 2010 [Google Scholar]
- 66.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xi R, et al. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc Natl Acad Sci U S A. 2011;108:E1128–1136. doi: 10.1073/pnas.1110574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McPherson A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forster M, et al. From next-generation sequencing alignments to accurate comparison and validation of single-nucleotide variants: the pibase software. Nucleic Acids Res. 2013;41:e16. doi: 10.1093/nar/gks836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Albers CA, et al. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 78.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ho Sui SJ, et al. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stade B, Seelow D, Thomsen I, Krawczak M, Franke A. GrabBlur--a framework to facilitate the secure exchange of whole-exome and -genome SNV data using VCF files. BMC Genomics. 2014;15(Suppl 4):S8. doi: 10.1186/1471-2164-15-S4-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ratei R, et al. Lineage classification of childhood acute lymphoblastic leukemia according to the EGIL recommendations: results of the ALL-BFM 2000 trial. Klin Padiatr. 2013;225(Suppl 1):S34–39. doi: 10.1055/s-0033-1337961. [DOI] [PubMed] [Google Scholar]
- 85.Festing MF, Altman DG. Guidelines for the design and statistical analysis of experiments using laboratory animals. Ilar J. 2002;43:244–258. doi: 10.1093/ilar.43.4.244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.